Abstract
The second messenger bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) has emerged as a broadly conserved intracellular signaling molecule. This soluble molecule is important for controlling biofilm formation, adhesion, motility, virulence, and cell morphogenesis in diverse bacterial species. But how is the typical bacterial cell able to coordinate the actions of upward of 50 proteins involved in synthesizing, degrading, and binding c-di-GMP? Understanding the specificity of c-di-GMP signaling in the context of so many enzymes involved in making, breaking, and binding the second messenger will be possible only through mechanistic studies of its output systems. Here we discuss three newly characterized c-di-GMP effector systems that are best understood in terms of molecular and structural detail. As they are conserved across many bacterial species, they likely will serve as central paradigms for c-di-GMP output systems and contribute to our understanding of how bacteria control critical aspects of their biology.
Keywords: motility, adhesion, exopolysaccharide, diguanylate cyclase, phosphodiesterase
INTRODUCTION
Finely tuned sensory systems enable bacteria to sense and respond to fluctuating environments and coordinate adaptive changes in metabolic pathways and physiological outputs. By integrating environmental cues, bacteria are able to make important decisions regarding how to respond to their constantly changing environs. Formation of a community of bacterial cells, or a biofilm, is one possible bacterial adaptation. These communities are often more advantageous than life as a planktonic cell, as biofilms have increased tolerance to antibiotics and the ability to survive periods of environmental stresses (Costerton et al. 1995, Hogan & Kolter 2002). Biofilms are found in natural, industrial, and clinical settings. Bacterial biofilms in nature play key roles in the production and degradation of organic matter, the degradation of many pollutants, and the cycling of nitrogen and sulfur. Industrial biofilms are utilized to process sewage and treat contaminated groundwater. Biofilms within the medical setting are quite problematic; they are much more antibiotic tolerant than their planktonic counterparts, and contamination of medical implants can occur owing to the bacteria's ability to form biofilms on abiotic surfaces (Davey & O'Toole 2000).
Bacteria transition from a planktonic to a sessile lifestyle in response to environmental cues such as osmolarity, pH, carbon availability, iron availability, oxygen tension, and temperature (Jackson et al. 2002a, 2002b; Lugtenberg et al. 1999; O'Toole & Kolter 1998b; O'Toole et al. 2000; Prigent-Combaret et al. 1999, 2001; Sauer et al. 2004; Singh et al. 2002; Thormann et al. 2006). Extensive research has revealed an array of cellular factors and diverse molecular mechanisms that are required for the transition from a planktonic to a sessile mode of life and the subsequent development of a biofilm. A unifying theme across bacterial species, however, is that biofilm formation coincides with the synthesis of the cellular signaling molecule bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP; Figure 1).
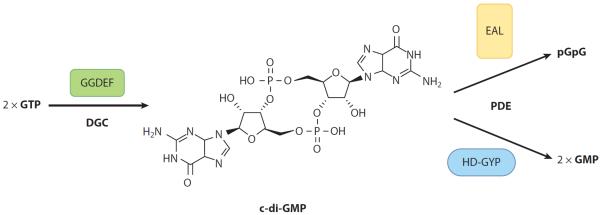
Figure 1.
c-di-GMP synthesis and degradation. Diguanylate cyclases (DGCs) containing the GGDEF domain synthesize c-di-GMP (shown center) from two molecules of GTP. Phosphodiesterases (PDEs) containing the EAL domain or HD-GYP domain degrade c-di-GMP to pGpG or GMP, respectively. Illustration courtesy of William Scavone, MA, CMI, Kestrel Illustration Studio, LLC. Abbreviations: GMP, guanosine monophosphate; pGpG, 5′-phosphoguanylyl-(3′-5′)-guanosine.
c-di-GMP has emerged as a broadly conserved bacterial intracellular second messenger. c-di-GMP was initially described as an activator of cellulose synthase in Gluconacetobacter xylinus, formerly Acetobacter xylinum (Ross et al. 1987). More recently this soluble molecule has been shown to also be important for controlling adhesion, motility, virulence, and cell morphogenesis in diverse bacterial species by exerting control at the level of transcription, translation, and posttranslation (Hengge 2009, Romling et al. 2005, Wolfe & Visick 2008). Over the past five to seven years, we have learned a great deal about how c-di-GMP is synthesized and degraded. Diguanylate cyclases (DGCs) containing the GGDEF domain, defined by this highly conserved set of amino acids, synthesize c-di-GMP from two molecules of GTP (Paul et al. 2004), whereas phosphodiesterases (PDEs) containing the EAL domain (Christen et al. 2005, Tischler & Camilli 2004) or HD-GYP domain (Ryan et al. 2006a), defined by these respective amino acids motifs, degrade c-di-GMP to 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG) or guanosine monophosphate (GMP), respectively (Figure 1). Many proteins contain both GGDEF and EAL domains but typically exhibit only DGC or PDE activity. GGDEF and EAL domains often occur in multidomain proteins in combination with diverse regulatory domains common to bacterial signaling proteins (Galperin et al. 2001). These regulatory domains, in response to the environment and environmental cues, can induce the synthesis or degradation of c-di-GMP in response to signals such as light (Barends et al. 2009, Tarutina et al. 2006) and oxygen (Chang et al. 2001, Qi et al. 2009, Sawai et al. 2010, Tuckerman et al. 2009, Wan et al. 2009), or through posttranslational modifications such as phosphorylation (Paul et al. 2007, Rao et al. 2009) or proteolysis (Tarutina et al. 2006).
c-di-GMP Signaling Pathways
Whole-genome sequencing has revealed that GGDEF and EAL domains are ubiquitous in bacteria; often dozens of these domain-containing proteins are found within a single bacterium (Hengge 2009). Escherichia coli contains 29 predicted c-di-GMP DGC or PDE proteins, whereas Salmonella enterica has 19 such proteins, Pseudomonas aeruginosa 41 proteins, Vibrio cholerae 72 proteins (Povolotsky & Hengge 2012), and Pseudomonas fluorescens 43 proteins (Newell et al. 2011b). Early studies focused on functionally analyzing GGDEF and EAL domain-containing proteins to biochemically demonstrate their ability to synthesize and degrade c-di-GMP, respectively (Christen et al. 2005, Hickman et al. 2005, Paul et al. 2004, Schmidt et al. 2005, Tamayo et al. 2005).
Although the transition from a motile to a sessile lifestyle seems to generally require an increase in the concentration of c-di-GMP, until recently, the specific outputs impacted by c-di-GMP have been understudied. The surprisingly large number of predicted c-di-GMP signaling proteins in many bacterial species suggests that c-di-GMP signaling networks are extraordinarily complex. How microbes integrate multiple input signals to regulate critical biological processes and how the action of multiple enzymes involved in c-di-GMP metabolism can be integrated to control bacterial behaviors are critical questions to address if we hope to understand the mechanisms underlying how this second messenger exerts its affects.
Systematic analyses of DGCs and PDEs in E. coli, Salmonella, V. cholerae, and P. fluorescens have revealed that motility, virulence, and biofilm formation are regulated by c-di-GMP synthesized by a subset of DGCs. Boehm et al. (2010) identified a c-di-GMP network consisting of the DGCs YfiN, YegE, YedQ, and YddV as well as the PDE YhjH that synthesizes and degrades c-di-GMP to regulate motility in E. coli. In Salmonella, Ahmad et al. (2011) have revealed a complex c-di-GMP network, whereby distinct groups of GGDEF and EAL domain-containing proteins affect virulence phenotypes, including invasion of epithelial cells, induction of the proinflammatory cytokine interleukin-8, and biofilm formation. Systematic mutagenesis studies of c-di-GMP signaling proteins in V. cholerae conducted by the Yildiz laboratory (Beyhan & Yildiz 2007; Beyhan et al. 2006, 2007, 2008; Lim et al. 2006) identified several specific DGCs and PDEs that impact colony rugosity at transcriptional and posttranscriptional levels. Epistasis experiments suggest c-di-GMP signaling proteins may act in parallel to control rugosity. In P. fluorescens, Newell et al. (2011b) identified a c-di-GMP network consisting of four DGCs that synthesize c-di-GMP for biofilm formation. Newell and colleagues suggest that a particular subset of DGCs specifically control c-di-GMP-regulated outputs such as adhesin localization and motility. Thus, understanding how multiple c-di-GMP signaling pathways are isolated from each other, or integrated to produce coherent outputs, is a question of fundamental importance for understanding an array of c-di-GMP-mediated biological processes in bacteria.
c-di-GMP Effectors in a World of Complex Input Pathways
c-di-GMP networks must have mechanisms to both generate this signal and respond to its changing levels. Thus, to complement those enzymes involved in the synthesis and degradation of c-di-GMP, effector systems allow microbes to respond to changing levels of this second messenger. c-di-GMP receptors or effectors are c-di-GMP binding proteins or RNA; c-di-GMP binds to and exerts its control by altering the structure and output function of the effector (Hengge 2009).
Amikam & Galperin (2006) were the first to implicate the PilZ domain as a specific c-di-GMP binding motif. PilZ domains are defined by an amino acid motif of extensively conserved residues (Amikam & Galperin 2006). Their bioinformatic study suggested that the PilZ domain is present in numerous bacterial cellulose synthases, including Alg44, an alginate biosynthesis protein found in P. aeruginosa, and the YcgR protein of E. coli. Functional studies later confirmed that the PilZ domain of Alg44 and YcgR binds c-di-GMP (Merighi et al. 2007, Ryjenkov et al. 2006). Importantly, the bioinformatic study of Amikam & Galperin (2006) revealed that for some bacterial genomes that encode GGDEF-containing proteins, PilZ domains are not always present, which suggests that protein domains other than PilZ may be able to act as c-di-GMP effectors.
Additional protein domains and motifs shown to bind c-di-GMP (summarized in Table 1) include the EAL domain of FimX from P. aeruginosa and of LapD from P. fluorescens (Navarro et al. 2009, Newell et al. 2009), the RxxD motif of PopA from Caulobacter crescentus and of PelD from P. aeruginosa (Duerig et al. 2009, Lee et al. 2007), the W[F/L/M][T/S]R motif of CpsQ from Vibrio parahaemolyticus and of VpsT from V. cholerae (Ferreira et al. 2011, Krasteva et al. 2010), the interface between a cyclic nucleotide-binding and DNA-binding domain of Clp from Xanthomonas campestris (Chin et al. 2010, Tao et al. 2010), the GEMM motif of c-di-GMP riboswitch class I (Sudarsan et al. 2008), and the pseudoknot motif of c-di-GMP riboswitch class II (Smith et al. 2011). Although Bcam1349 from Burkholderia cenocepacia, FleQ from P. aeruginosa, Clp from Xanthomonas axonopodis, and polynucleotide phosphorylase (PNPase) from E. coli (Fazli et al. 2011, Hickman & Harwood 2008, Leduc & Roberts 2009, Tuckerman et al. 2011) bind c-di-GMP, which domain or motif is necessary for binding is unclear.
Table 1.
c-di-GMP effector systems
| Effector | Motif/domain | Organism | Mechanism | Regulated output | Reference (s) |
|---|---|---|---|---|---|
| Alg44 | PilZ | Pseudomonas aeruginosa | Protein-protein interaction | Production of alginate | Merighi et al. (2007), Oglesby et al. (2008) |
| MrkH | PilZ | Klebsiella pneumoniae | Transcriptional activation, DNA binding | Expression of type 3 flmbriae and biofllm formation | Wilksch et al. (2011) |
| PlzA | PilZ | Borrelia burgdorferi | Unknown | Motility, infectivity | Freedman et al. (2010), Pitzer et al. (2011) |
| DgrA | PilZ | Caulobacter crescentus | Protein-protein interaction | Motility | Christen et al. (2007) |
| BcsA | PilZ | Gluconacetobacter xylinus | Unknown | Cellulose synthase | Ryjenkov et al. (2006), Weinhouse et al. (1997) |
| PlzD | PilZ | Vibrio cholerae | Unknown | Motility, biofllm formation, virulence | Pratt et al. (2007) |
| FleQ | Unknown | Pseudomonas aeruginosa | Derepression of pel operon | Biofllm formation | Hickman & Harwood (2008) |
| Clp | Unknown | Xanthomonas axonopodis | Inhibition of Clp-DNA binding | Likely virulence | (Leduc & Roberts 2009) |
| Clp | Interface between cyclic nucleotide binding and DNA binding domains | Xanthomonas campestris | Inhibition of Clp-DNA binding | Virulence | Chin et al. (2010), Tao et al. (2010) |
| Bcaml349 | Unknown | Burkholderia cenocepacia | Transcriptional regulation, c-di-GMP enhances DNA binding | Biofllm formation, virulence | Fazli et al. (2011) |
| FimX | EAL | Pseudomonas aeruginosa | Conformational change | Twitching motility | Huang et al. (2003), Kazmierczak et al. (2006), Navarro et al. (2009), Qi et al. (2011) |
| PelD | RxxD | Pseudomonas aeruginosa | Unknown | Exopolysaccharide production | Lee et al. (2007) |
| PopA | RxxD | Caulobacter crescentus | Localization to the pole | Cell cycle progression | Abel et al. (2011), Duerig et al. (2009) |
| PNPase | Unknown | Escherichia coli | Enhances PNPase activity | RNA processing | Tuckerman et al. (2011) |
| Vc2 Riboswitch Class I | GEMM | Vibrio cholerae | Conformational change | Gene expression | Kulshina et al. (2009), Smith et al. (2009), Sudarsan et al. (2008) |
| 84Cd Riboswitch Class II | Pseudoknot | Clostridium difficile | Triggers alternative splicing | Altered transcripts | Lee et al. (2010), Smith et al. (2011) |
Effectors bound to c-di-GMP utilize a range of mechanisms to relay signals to cellular processes and impact exopolysaccharide synthesis (Lee et al. 2007, Merighi et al. 2007, Weinhouse et al. 1997), motility (Boehm et al. 2010, Christen et al. 2007, Fang & Gomelsky 2010, Guzzo et al. 2009, Paul et al. 2010, Pratt et al. 2007, Ryjenkov et al. 2006), transcription (Hickman & Harwood 2008, Krasteva et al. 2010, Leduc & Roberts 2009), and subcellular (Duerig et al. 2009) or cell-surface protein localization (Monds et al. 2007, Newell et al. 2011a). Clearly, c-di-GMP modulates multiple outputs through discrete mechanisms. Even though many c-di-GMP metabolizing enzymes and effectors are expressed simultaneously, subsets of enzymes impact discrete phenotypic outputs, and bacterial cells are able to coordinate the actions of complex c-di-GMP networks. This coordination is likely achieved through both temporal and spatial regulation (Hengge 2009). Some DGCs, PDEs, and effectors may be expressed only under particular growth conditions or in response to environmental changes (Hengge 2009). For example, rapA, which encodes a PDE in P. fluorescens, is transcribed in response to low levels of inorganic phosphate (Pi) (Monds et al. 2007), whereas YcgR, an effector protein in E. coli, is coregulated with the genes required for the synthesis of the flagellum (Girgis et al. 2007). c-di-GMP signaling components may associate with one another in multiprotein complexes or microcompartments within the cell to influence the local concentration of c-di-GMP (Hengge 2009). In support of this idea, fluorescent c-di-GMP sensors have demonstrated both temporal and spatial separation of c-di-GMP pools (Christen et al. 2010), whereas work in P. aeruginosa has shown a lack of correlation between total c-di-GMP pools and particular phenotypic outputs (Merritt et al. 2010). Additionally, DGCs and PDEs in the same pathway can interact with one another (Andrade et al. 2006, Bobrov et al. 2008), and direct communication between an effector protein and a downstream target has been suggested (Oglesby et al. 2008). The diverse ways in which effector proteins bind to c-di-GMP and interact with DGCs and PDEs allow bacteria to confer the specificity needed for the regulation of various outputs, a notion first suggested by early work characterizing this signal (Ross et al. 1987).
In this review we first focus our discussion on characterized c-di-GMP effector systems for which the mechanism has been explored in some detail. Specifically, we provide an overview of three systems that illustrate, in detail, distinct mechanisms that serve to sense and respond to c-di-GMP. These three systems highlight the diversity of c-di-GMP effector mechanisms, which likely only scratch the surface in regard to how cells sense and respond to this dinucleotide. We focus our discussion on the LapD protein of P. fluorescens, the YcgR protein of E. coli, and the VpsT protein of V. cholerae, which function in the transition from a motile to a sessile mode of life. LapD, YcgR, and VpsT bind c-di-GMP via distinct domains and impact discrete outputs through cell-surface protein localization, protein-protein interaction, and transcription, respectively. We discuss the role of these effector systems in the complexity of c-di-GMP signaling in the context of where the field needs to go next. The review concludes by highlighting some newly discovered or poorly understood effectors that warrant additional studies, as these effectors play a variety of roles in bacterial biology.
c-di-GMP EFFECTOR SYSTEMS
LapD Binds c-di-GMP to Regulate Cell Surface Protein Localization
Studies of P. fluorescens have identified LapD as an inner membrane effector protein that binds c-di-GMP via a degenerate EAL domain and, through an inside-out signaling mechanism, impacts cell-surface levels of the biofilm adhesin LapA (Newell et al. 2009, 2011a). To initiate the transition from a planktonic to a sessile mode of life, and thus formation of a biofilm, the large adhesin protein LapA must be present at the cell surface (Hinsa et al. 2003). Presumably, such radical changes in lifestyle occur only in response to defined signals. For example, in P. fluorescens, restriction of Pi availability to low micromolar levels results in loss of biofilm formation with little or no impact on planktonic growth, which indicates a biofilm-specific regulatory signal. Growth in low-Pi medium resulted in loss of the critical LapA adhesin from the cell surface (Monds et al. 2007).
What mechanism was employed to regulate cell-surface localization of LapA, and thus biofilm formation, in response to changing Pi availability? Initial genetic and biochemical analyses indicated that when cells are grown in a low-Pi medium, the cytoplasmic PDE RapA is expressed and degrades c-di-GMP to pGpG. Genetic studies supported the conclusion that reduced cellular levels of c-di-GMP resulted in the loss of LapA from the cell surface and thus eliminated biofilm formation (Monds et al. 2007). These results demonstrated that the modulation of intracellular levels of c-di-GMP regulates cell-surface protein localization and prompted us to investigate the mechanisms through which c-di-GMP levels were being sensed and monitored within the cell. Importantly, these findings suggested that changing levels of c-di-GMP in the cytoplasm impacted the localization of an adhesin on the cell surface, that is, two membranes away. These data suggested the possibility of a novel mechanism to monitor and regulate this c-di-GMP-dependent output.
A transposon mutagenesis screen identified the lapA gene as required for biofilm formation (O'Toole & Kolter 1998b), and subsequent work demonstrated that maintenance of the LapA adhesin at the cell surface required LapD (Hinsa & O'Toole 2006). That is, growth in low-Pi medium yielded a phenotype quite similar to that of loss of LapD function, which indicates that Pi may exert its effects through this protein. LapD is an inner membrane protein (Hinsa & O'Toole 2006) that contains degenerate and inactive GGDEF and EAL domains (Newell et al. 2009), which have been proposed to function as regulatory domains that bind c-di-GMP (Christen et al. 2006, Ryan et al. 2006b). Therefore, given that c-di-GMP and LapD are both required for biofilm formation and maintenance of LapA at the cell surface, we hypothesized that LapD may bind c-di-GMP through its GGDEF or EAL domain. Binding assays and mutagenesis revealed that the EAL domain of LapD is necessary and sufficient to bind c-di-GMP (Newell et al. 2009).
LapD contains a HAMP domain, a motif often found in transmembrane signaling proteins such as the sensor histidine kinases of two-component regulatory systems and chemotaxis receptors (Taylor 2007, Williams & Stewart 1999). Therefore, we hypothesized that upon sensing cytoplasmic levels of c-di-GMP, the HAMP domain relays this signal to the cell surface through activation of the periplasmic domain. In support of this model, deletion of a portion of the HAMP domain known to be required for signaling resulted in a hyperbiofilm phenotype, which was interpreted to result from the constitutive activation of the LapD output. The HAMP deletion mutant bound c-di-GMP at levels comparable with the wild type, whereas a HAMP deletion mutation combined with the c-di-GMP binding mutation LapDE617A could not bind c-di-GMP but still formed a hyperbiofilm. This intramolecular epistasis analysis suggested that a HAMP domain deletion locks LapD in an active state regardless of whether or not c-di-GMP is bound (Newell et al. 2009).
Predictions of LapD membrane topology, confirmed experimentally, indicated that this protein has two transmembrane domains separated by a periplasmic loop near its N terminus (Newell et al. 2009). To assess the role of the LapD periplasmic domain in mediating biofilm formation, a mutation was made within the periplasmic loop, LapDL152P. This mutation reduced the extent of biofilm formation (i.e., the phenotype was similar to the lapD null mutation and the opposite of that observed for the HAMP locked-on mutation), thus suggesting the necessity of the periplasmic domain for the LapD output. Combination of the LapDL152P mutation and the HAMP deletion resulted in loss of the HAMP deletion-mediated hyperbiofilm phenotype, which suggests that the c-di-GMP signal is propagated from the inside out.
These mutational analyses are consistent with a model wherein LapD senses the cytoplasmic levels of c-di-GMP, and through an inside-out signaling mechanism, the periplasmic domain mediates the LapD output (Newell et al. 2009). However, the mechanism by which an inner membrane effector protein bound to c-di-GMP impacts a cell-surface output remained unclear. We hypothesized that a gene encoding a periplasmic protein may regulate LapA at the cell surface. The lapG gene located directly upstream of lapD presented itself as a good candidate. Genetic analyses indicated that lapG is epistatic to lapD, whereas lapA is epistatic to lapG, thus placing lapG in the LapA-dependent biofilm pathway. Biochemical and enzyme activity assays showed that LapG removes LapA from the cell surface through proteolytic cleavage of the N terminus of LapA (Newell et al. 2011a). Furthermore, LapD and LapG interact in a c-di-GMP-dependent manner (Newell et al. 2011a). We proposed a model whereby LapD, when bound to c-di-GMP, interacts with LapG such that it prevents LapG from cleaving and releasing LapA, thus promoting biofilm formation. In conditions unfavorable for biofilm formation and in which c-di-GMP is low, LapD is no longer able to interact with LapG, and LapG through its cysteine protease activity cleaves LapA from the cell surface (Newell et al. 2009, 2011a).
X-ray crystallographic studies have elucidated the mechanistic basis for c-di-GMP-dependent regulation of LapA (Navarro et al. 2011). Three crystal structures of LapD were solved, including the c-di-GMP-unbound cytoplasmic portion of the protein (excluding the HAMP domain), the c-di-GMP-bound EAL domain dimer, and the periplasmic domain. The overall GGDEF and EAL domain folds of LapD are very similar to those of both catalytically active and inactive DGCs and PDEs, but nonconservative amino acid substitutions in the LapD GGDEF and EAL domains affect catalytic activity, thus rendering the domains nonfunctional (Navarro et al. 2011).
Studies of the c-di-GMP-unbound cytoplasmic LapD domain suggested that the protein attains an autoinhibited state. That is, the GGDEF domain likely restricts access of c-di-GMP to the EAL domain, whereas the signaling helix motif (S-helix), a helical extension of the HAMP domain, buttresses the EAL domain (Navarro et al. 2011). X-ray crystallographic studies of the c-di-GMP-bound EAL domain revealed that the EAL domain dimerizes at the surface occupied by the S-helix. Thus, upon c-di-GMP binding and EAL domain dimerization, the S-helix and GGDEF domain undergo a conformational change and displacement (Navarro et al. 2011). On the basis of these results, a model was proposed in which intramolecular interactions between the EAL domain, the GGDEF domain, and the S-helix prevent c-di-GMP from binding to the EAL domain. Autoinhibited LapD would then undergo conformational changes upon c-di-GMP binding (Figure 2). Structure-guided mutagenesis studies to test c-di-GMP binding and biofilm formation in both sufficient and insufficient Pi conditions confirmed that c-di-GMP binding and dimerization of the EAL domain are likely interdependent events and suggested that the S-helix stabilizes the autoinhibited state of c-di-GMP, whereas the positioning of the GGDEF domain blocks c-di-GMP from accessing the binding pocket within the EAL domain (Navarro et al. 2011).
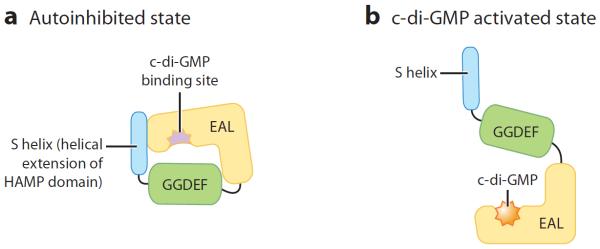
Figure 2.
LapD binds c-di-GMP. c-di-GMP binding to LapD induces a conformational change in the protein. (a) Autoinhibited state. Intramolecular interactions among the EAL domain, the GGDEF domain, and the S-helix prevent c-di-GMP from binding to the EAL domain. Structure-guided mutagenesis studies suggest that the S-helix stabilizes the autoinhibited state of LapD, whereas the positioning of the GGDEF domain blocks c-di-GMP from accessing the c-di-GMP binding pocket with the EAL domain (Navarro et al. 2011). (b) c-di-GMP activated state. Upon c-di-GMP binding to the EAL domain, LapD undergoes a conformational change as the S-helix and GGDEF domain are displaced (Navarro et al. 2011; Illustration courtesy of William Scavone, MA, CMI, Kestrel Illustration Studio, LLC). Abbreviations: GMP, guanosine monophosphate.
The cytoplasmic conformation of LapD, which is indicative of c-di-GMP levels, must be relayed to the cell surface to regulate cell-surface adhesion through the modulation of LapG activity. To better understand how the cytoplasmic level of c-di-GMP is sensed in the periplasm, the structure of the periplasmic output domain of LapD was determined. It forms a domain-swapped dimer and is connected to the HAMP domain (Navarro et al. 2011). A sequence alignment of the LapD periplasmic domain with LapD homologs revealed a conserved tryptophan residue located at the most distal point of the periplasmic domain. Mutagenesis of this residue prevented biofilm formation and disrupted the interaction between LapG and LapD, which suggests its requirement for interaction with and modulation of LapG activity (Navarro et al. 2011).
From these structure-based mechanistic studies the following model (Figure 3) was proposed: in the absence of c-di-GMP, LapD exists in an autoinhibited state, as the S-helix and the GGDEF domain block c-di-GMP from accessing the EAL domain (Navarro et al. 2011). In this autoinhibited state, the periplasmic domain of LapD attains a LapG-binding-incompetent state. LapG through its cysteine protease function cleaves LapA from the cell surface and thereby prevents biofilm formation. It is proposed that the HAMP domain relays the cytoplasmic state of LapD to the periplasmic domain of LapD. Upon an increase in cytoplasmic c-di-GMP, a large conformational change within the GGDEF and S-helix occurs that allows for EAL domain dimerization. This signal is relayed to the periplasm, which allows this domain to attain a LapG-binding competent state. LapG is bound by LapD and no longer able to cleave LapA from the cell surface, which results in biofilm formation (Navarro et al. 2011).
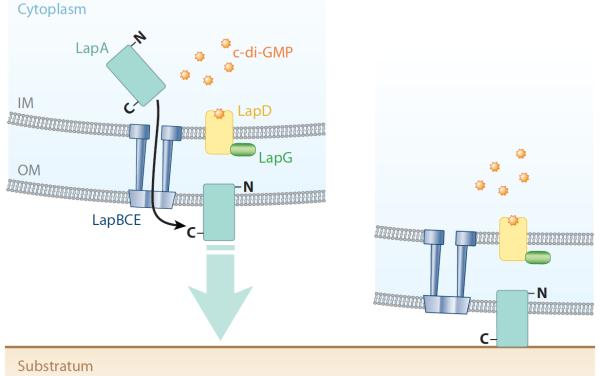
Figure 3.
c-di-GMP effector system in Pseudomonas fluorescens. This diagram depicts a summary of the current model for the c-di-GMP effector system in P. fluorescens, which impacts the motile-to-sessile transition. The LapA protein, a predicted cell-surface adhesion, is transported to the cell surface through the ABC transporter, comprised of the LapBCE proteins. LapD binds c-di-GMP, and through an inside-out signaling mechanism, the periplasmic domain of LapD binds LapG. Thus, LapG is prevented from cleaving and releasing LapA from the cell surface, thereby promoting biofilm formation (Newell et al. 2011a; Illustration courtesy of William Scavone, MA, CMI, Kestrel Illustration Studio, LLC). Abbreviations: GMP, guanosine monophosphate; IM, inner membrane; OM, outer membrane.
Navarro et al. (2011) reported that the LapD-LapG c-di-GMP signaling system is conserved in many diverse environmental and pathogenic bacterial species including those in the Pseudomonas, Legionella, and Vibrio genera. Although studies of the effector protein LapD detail the mechanisms of c-di-GMP binding and regulation of biofilm formation in P. fluorescens, the types of behaviors and outputs these conserved LapD-LapG signaling systems regulate in other bacteria are largely unknown. As these LapD-LapG systems are studied in more detail, we expect similar mechanistic details will be elucidated, potentially unraveling the regulation of novel outputs.
Motile-to-Sessile Transition in E. coli Is Induced Via YcgR
Flagellar motility is critical to biofilm formation in many microbes (Deflaun et al. 1990; Genevaux et al. 1996; O'Toole & Kolter 1998a, 1998b; Pratt & Kolter 1998; Watnick et al. 1999). Thus, understanding how the cell controls flagellar motility is integral to understanding the planktonic-to-biofilm transition. Studies in E. coli have demonstrated that c-di-GMP binding to the PilZ domain of the effector protein YcgR stimulates its interaction with the flagellar motor and/or switch complex to positively affect the motile-to-sessile transition (Boehm et al. 2010, Fang & Gomelsky 2010, Paul et al. 2010). However, as outlined in detail below, these three studies identified different components of the flagellar motor as the downstream target.
The motor is comprised of the stator proteins MotA and MotB and the rotor proteins FliG, FliM, and FliN, which together function to drive and regulate flagellar rotation. The current model for motor function predicts that protons flow through an ion channel within the stator and bind to an Asp residue in MotB. A subsequent conformational change in MotA stimulates electrostatic interactions between MotA and FliG to generate torque and turning of the rotor.
Although ion flux and torque generation power flagellar rotation, in the case of swimming motility, a phosphorelay signaling cascade and the switch complex composed of FliG, FliM, and FliN control the direction of rotation. Chemotaxis proteins sense and respond to environmental stimuli and transmit signals to the motor. The phosphorylated state of the response regulator, CheY, impacts its ability to interact with the switch complex. The current model of flagellum switching suggests that phosphorylated CheY-P interacts with FliM, which leads to an alteration in the interaction between FliM and the C terminus of FliG, which induces a modification of the rotor-stator interface between FliG-MotA to result in a switch in rotational direction (Terashima et al. 2008).
Studies by Ko & Park (2000) first identified the ycgR gene in a suppressor mutagenesis screen. The nucleoid protein H-NS is a positive regulator of the flagellar regulon (Bertin et al. 1994) and interacts with FliG to modulate the rotational speed of the flagellum (Donato & Kawula 1998). To examine H-NS involvement in flagellum function, a suppressor mutagenesis screen was performed in hns-deficient cells constitutively expressing flhDC in an H-NS-independent manner (to allow for flagellum production). Cells lacking H-NS but constitutively expressing flhDC contain paralyzed flagella and are nonmotile (Ko & Park 2000). Mutations in the ycgR gene were able to suppress the motility defect, whereas the yhjH gene was identified in a search for multicopy suppressors of the motility defect (Ko & Park 2000).
An analysis of the ycgR and yhjH promoters suggested that both are part of the flagellar regulon (Ko & Park 2000). Phenotypic assays showed that deletion of ycgR or overexpression of yhjH in the flhDC-constitutive hns mutant background resulted in an increase in swimming speeds (Ko & Park 2000). The percentage of rotating cells in flhDC-constitutive hns mutants was drastically less than that of the ycgR mutant or the yhjH overexpression strain, but the rotational speed was similar in all strains (Ko & Park 2000). Overexpression of genes encoding the motor components, namely, motB, motAB, and fliG, in the flhDC-constitutive hns mutant did not restore motility; however, overexpression of motA increased motility slightly (Ko & Park 2000). The study suggested that H-NS, YcgR, and YhjH participate in motor function, but how was unclear. Ko & Park (2000) hypothesized that the stator proteins were not able to associate properly with the components of the rotor in the hns mutant, which resulted in a defective motor complex.
Later studies identified YcgR as a PilZ domain-containing protein (Amikam & Galperin 2006) and YhjH as a PDE (Schmidt et al. 2005). Ryjenkov et al. (2006) tested the hypothesis that YcgR binds c-di-GMP through its PilZ domain. Using equilibrium dialysis and size exclusion chromatography, YcgR was shown to bind c-di-GMP, and results suggested that YcgR binds one c-di-GMP dimer at a single binding site (Ryjenkov et al. 2006). The YcgRR118D mutant was unable to bind c-di-GMP, whereas the PilZ domain alone binds c-di-GMP but at a decreased efficiency when compared with the full-length protein (Ryjenkov et al. 2006). These data suggested that other regions within the protein are important in c-di-GMP binding or that c-di-GMP binding induces conformational changes within YcgR (Ryjenkov et al. 2006). Ryjenkov et al. also tested whether c-di-GMP binding induced oligomerization. Size exclusion chromatography results suggested that YcgR, the PilZ domain of YcgR, and the PilZ domain of BcsA from G. xylinus exist as monomers in the presence and absence of c-di-GMP (Ryjenkov et al. 2006). This biochemical study established YcgR as a c-di-GMP-binding protein in E. coli.
Structures of three PilZ domain-containing proteins bound to c-di-GMP have revealed differences in binding stoichiometries and oligomeric states. PlzD from V. cholerae exists as a dimer in the unbound state and when bound to c-di-GMP. One molecule of c-di-GMP binds to the C-terminal PilZ domain, which brings this domain in closer proximity to the N terminus (Benach et al. 2007). PP4397 from Pseudomonas putida binds two molecules of c-di-GMP and undergoes a dimer-to-monomer transition (Ko et al. 2010). PA4608 from P. aeruginosa is a single-domain-containing protein that binds to one molecule of c-di-GMP and undergoes rearrangement to expose a negative surface of the protein that is predicted to function in downstream processes (Habazettl et al. 2011, Shin et al. 2011). These differences in stoichiometry and oligomeric states are predicted to allow for diverse forms of c-di-GMP-dependent regulation (Habazettl et al. 2011, Ko et al. 2010, Shin et al. 2011).
In E. coli and the Salmonella rdar morphotype (rough, dry, and rugose colony), adherence to an abiotic surface, biofilm formation, and swimming and swarming motility are dependent on c-di-GMP level (Ryjenkov et al. 2006). To assess the effects of YcgR on outputs regulated by c-di-GMP, ycgR was deleted in the yhjH mutant background (in which c-di-GMP levels were elevated, as the strain lacks the YhjH PDE). The yhjH ycgR double mutant showed an increase in both swimming and swarming motility when compared with the yhjH deletion mutant. The YcgRR118D point mutant could not complement the swimming and swarming motilities of the yhjH ycgR double mutant, which suggests that YcgR regulates motility in a c-di-GMP-dependent manner (Ryjenkov et al. 2006). Phenotypes for rdar morphotype, adherence to an abiotic surface, and biofilm formation were similar in the ycgR mutant and yhjH ycgR double mutant. Taken together, these results suggested that YcgR specifically regulates flagellum-based motility in a c-di-GMP-dependent manner (Ryjenkov et al. 2006).
Ko & Park (2000) established that YcgR participates in motor function, whereas Ryjenkov et al. (2006) demonstrated that YcgR regulates flagellum-based motility in a c-di-GMP-dependent manner. In reviewing c-di-GMP regulation of flagellum-based motility, Wolfe & Visick (2008, p. 469) put forth the following model: “YhjH and an unknown DGC set the levels of c-di-GMP, which binds to YcgR. In a mechanism yet to be determined, this complex interferes with the proper association of the Mot proteins with FliG and the rest of the switching device. The result is a paralyzed flagellum.” They suggested that H-NS could enhance the interaction of the Mot proteins with FliG, whereas the YcgR-c-di-GMP complex could inhibit this interaction.
Within the same month three groups published studies exploring the posttranslational mechanism whereby c-di-GMP-bound YcgR interacts with components of the flagellar motor to facilitate the transition from a motile to a sessile lifestyle, but each identified different components of the flagellar motor as the downstream target (Boehm et al. 2010, Fang & Gomelsky 2010, Paul et al. 2010). Fang & Gomelsky (2010) hypothesized that YcgR binding to FliG in a c-di-GMP-dependent manner disrupts FliG-FliM interactions to result in a counterclockwise flagellum rotation. Cells unable to switch from counterclockwise to clockwise flagellum rotation migrate poorly in semisolid agar, thus facilitating the motile-to-sessile transition. In liquid, these cells are unable to change the direction of their movement and are set on a crash course with a surface. To test the hypothesis that YcgR interacts with components of the switch complex, pull-down assays and a bacterial two-hybrid system were used to determine binding partners of YcgR. YcgR pulled down the soluble FliGMN switch complex expressed in the nonflagellated E. coli strain. FliG interacted with YcgR in the bacterial two-hybrid system. Furthermore, when c-di-GMP levels were manipulated in the bacterial two-hybrid system, YcgR still interacted with FliG in the absence of c-di-GMP, but the interaction was enhanced in the presence of c-di-GMP. Interestingly, YcgRR118D, which has impaired c-di-GMP binding, interacted weakly with FliM in the bacterial two-hybrid system (Fang & Gomelsky 2010). Interaction between YcgR and FliG was predicted to disrupt the energy transfer to the flagellum rotor, impair flagellum assembly or stability, or disrupt the FliG-FliM interaction to impede flagellum rotational behavior and the switch from counterclockwise to clockwise rotation (Fang & Gomelsky 2010). When analyzed under the microscope, the wild type, yhjH mutant, and yhjH ycgR double mutant were similarly motile, which suggests that energy transfer is not hindered. Levels of the flagellin subunit, FliC, were also similar in the three strains, which suggests that the stability and assembly of the flagellum is not impaired. Tethering assays to measure flagellum rotation revealed that the yhjH mutant rotated counterclockwise and reversed to clockwise much less frequently than did the wild type. The strong counterclockwise rotational bias of the yhjH mutant was first reported by Girgis et al. (2007). The flagellum was effectively locked in the counterclockwise direction. Deletion of ycgR in the yhjH mutant background reversed the counterclockwise rotation bias, restoring a bias similar to that of the wild type (Fang & Gomelsky 2010). On the basis of these results, Fang & Gomelsky (2010) proposed the following model (Figure 4a): In response to c-di-GMP levels, YcgR binds to FliG and increases flagellum counterclockwise rotational bias by disrupting the FliG-FliM interaction, which leads to poor migration, and thus facilitates the motile-to-sessile transition.
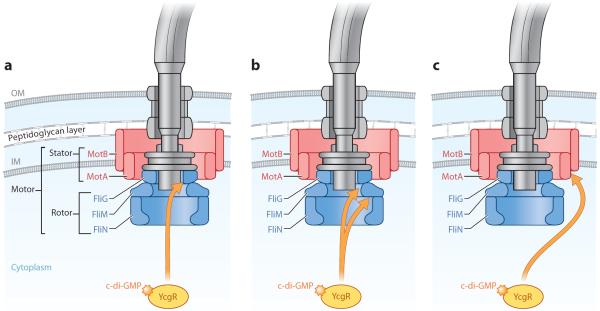
Figure 4.
c-di-GMP effector system in Escherichia coli. This diagram depicts a summary of the current models of the c-di-GMP effector system in E. coli, which impacts the motile-to-sessile transition. YcgR bound to c-di-GMP stimulates YcgR to interact with one or more components of the flagellar motor, reducing or altering motor function and thus promoting the motile-to-sessile transition. (a) Fang & Gomelsky (2010) suggest that c-di-GMP-bound YcgR interacts with FliG and disrupts the FliG-FliM interaction. An increased bias in counterclockwise flagellar rotation ensues, which leads to poor migration and thus facilitates the motile-to-sessile transition. (b) Paul et al. (2010) suggest that c-di-GMP-bound YcgR binds to FliM, disrupting the FliG-FliM interaction and thereby promoting a counterclockwise rotational bias. They also propose that YcgR also interacts with FliG, which leads to a disruption in the FliG-MotA interactions and a reduction in torque generation. (c) Boehm et al. (2010) suggest that c-di-GMP-bound YcgR interacts with MotA to reduce flagellar motor function. Illustration courtesy of William Scavone, MA, CMI, Kestrel Illustration Studio, LLC. Abbreviations: GMP, guanosine monophosphate; IM, inner membrane; OM, outer membrane.
Paul et al. (2010) similarly hypothesized that YcgR interacts with FliG and FliM in a c-di-GMP-dependent manner to reduce the efficiency of torque generation and induce a counterclockwise flagellum rotation. To more closely examine the effects of the yhjH deletion and the overexpression of ycgR on motor performance, tethered cell assays were used to assess motor speed as an indicator of motor torque and rotation bias. Although Fang & Gomelsky (2010) did not visually observe differences in the rotational speeds of tethered cells, Paul et al. (2010) noted that motor torque is reduced by 30%, but only in the presence of nonphysiologically increased c-di-GMP levels and overexpression of ycgR. As in the Fang & Gomelsky study, Paul et al. observed wild type cells rotating counterclockwise and clockwise, whereas yhjH mutants overexpressing ycgR predominately rotated counterclockwise. Chemotaxis mutants, wherein the level of CheY-P is elevated, are biased for clockwise rotation. Deletion of yhjH or overexpression of ycgR in such a chemotaxis mutant background resulted in many cells switching from a clockwise bias to a counterclockwise bias (Paul et al. 2010). As in the Fang & Gomelsky study, Paul et al. hypothesized that YcgR targeted components of the switch complex. Pull-down assays showed that FliG and FliM, but not FliN or MotA, interact with YcgR in both the presence and absence of c-di-GMP (Paul et al. 2010). As observed in Salmonella by fluorescence microscopy, YcgR-GFP (green fluorescent protein) displayed punctate fluorescence in cells, which became more apparent in the yhjH deletion background. Deletion of fliM or fliG, but not motA, eliminated the punctate fluorescence, which suggests that YcgR localizes to the rotor (Paul et al. 2010).
To identify regions in FliM and FliG that interact with YcgR, mutations were made in fliM and fliG and tested for their ability to interact with YcgR using pull-down assays, their effects on YcgR-GFP localization by fluorescence microscopy, and their ability to disrupt the in vivo interaction with YcgR and improve motility. Mutations in fliM were constructed at residues encoding surface regions. FliMN155E and FliML160E mutants weakened and eliminated binding to YcgR, respectively, and YcgR-GFP puncta were eliminated in the FliMN155E and FliML160E mutants (Paul et al. 2010). Mutations in fliG were also constructed at residues encoding various surface regions (Brown et al. 2007) near the charged ridge that interacts with the stator (Lloyd & Blair 1997, Zhou et al. 1998). FliGQ252W, FliGN292W, and FliGP295W caused a significant reduction in YcgR binding, whereas FliGD248W resulted in a slight reduction. YcgR-GFP puncta were not eliminated in the FliGP295W mutant. Because YcgR interacts with FliG and FliM, it was predicted that motility may improve if this interaction was disrupted by mutating residues in FliG and FliM shown to be involved in interacting with YcgR. Indeed, in Salmonella, overexpression of fliM or fliG mutants improved motility when compared with strains expressing wild type copies of fliM or fliG (Paul et al. 2010).
To identify residues in YcgR important for interacting with FliM and FliG, a series of amino acid substitutions in the C-terminal α-helix of YcgR were constructed on the basis of the crystal structure of PlzD, the YcgR-like protein in V. cholerae (Benach et al. 2007), and residues of DgrA of C. crescentus predicted to interact with downstream targets (Christen et al. 2007). YcgRQ223W and YcgRI227W could not bind to FliM, even in the presence of c-di-GMP, and the YcgRQ223W-GFP mutant displayed no punctate localization. When overexpressed in the yhjH mutant, YcgRQ223W improved the motility defect and motor speed as well as corrected the rotational bias of the yhjH mutant. YcgR α-helix mutants were able to bind FliG, which suggests that the C-terminal α-helical region of YcgR is important for binding FliM. The N terminus of YcgR was hypothesized to be important for binding to FliG. When YcgRK42D, YcgRN62W, or YcgRK81D was overexpressed in the yhjH mutant, it showed improved motility in comparison with the yhjH mutant, whereas the interactions between FliG and YcgRN62W or YcgRK81D were weakened. Binding of N-terminal YcgR point mutants to FliM was unchanged. These results suggested that the N terminus of YcgR is important for binding to FliG.
To establish whether the motility defect in the yhjH deletion mutant is only due to a counterclockwise motor bias, the gene encoding the clockwise-signaling protein CheY was overexpressed in the yhjH mutant background. Motility was not improved, which suggested that another factor was contributing to the motor defect. Therefore, Paul et al. (2010) overexpressed components of the stator, as Ko & Park (2000) had done previously. Overexpression of motA or motAB in the yhjH mutant background improved the motility defect. Motility was further enhanced upon overexpression of motA and cheY in the yhjH mutant background. These results suggested that not only does YcgR induce a rotational bias, but it may also impact torque generation (Paul et al. 2010).
Paul et al. (2010) hypothesized that YcgR may disrupt the organization of the C-terminal portion of FliG within the motor. To test this hypothesis, cross-linking experiments to detect FliG dimers and multimers in the yhjH mutant and the yhjH mutant overexpressing ycgR were performed. A FliG Cys replacement at residue 297 cross-linked into dimers in the wild type background and in the yhjH mutant overexpressing the gene encoding YcgRR118D, but in the yhjH mutant strain and the yhjH mutant strain overexpressing ycgR, cross-linking was reduced. This result suggested that YcgR bound to c-di-GMP may disrupt the ability of FliG to properly organize within the motor (Paul et al. 2010).
On the basis of the above experiments, Paul et al. (2010) suggested a backstop break model whereby c-di-GMP-bound YcgR reduces torque generation and biases counterclockwise flagellum rotation to promote a motile-to-sessile transition. c-di-GMP-bound YcgR binds to FliM, which disrupts the FliG-FliM interaction and promotes a counterclockwise rotational bias (Figure 4b). For YcgR to interact with FliG, FliG must reorient its charged ridge within proximity to YcgR. This reorientation disrupts the FliG-MotA interactions and reduces torque generation. Paul et al. (2010) also suggest that YcgR interaction with one FliG may disrupt orientation of neighboring FliG. Therefore, “YcgR could thus function as a `backstop brake,' both slowing the motor and inhibiting preferentially its rotation in one direction” (Paul et al. 2010, p. 136). Interestingly, using the bacterial two-hybrid system, Fang & Gomelsky (2010) showed that YcgRR118D, which is unable to bind c-di-GMP, binds to FliM, whereas Paul et al. (2010) found that YcgRR118D interacts with FliM and this interaction is stimulated in the presence of c-di-GMP. YcgRR118D retains the ability to bind to FliG, but binding is reduced when compared with wild type YcgR (Paul et al. 2010). Fang & Gomelsky (2010) did not detect an interaction between FliG and YcgRR118D. Overexpression of YcgRR118D in the yhjH mutant background improved the motility defect. The experiments with YcgRR118D indicate that interaction with FliM occurs in both the presence and absence of c-di-GMP. As stated above, YcgRR118D is unable to disrupt FliG organization within the motor. Therefore, YcgR may be positioned to associate with the flagellar switch complex to allow for rapid c-di-GMP-dependent regulation of flagellar motility. In the absence of c-di-GMP, YcgR associates with FliM, but once levels of c-di-GMP increase within the cell, YcgR binds c-di-GMP and impacts torque generation and rotational bias through interactions with FliG and FliM (Paul et al. 2010).
In a third study, Boehm et al. (2010) suggest that YcgR, in a c-di-GMP-dependent manner, interacts with the stator protein MotA to reduce flagellar motor function by inactivating individual stator units in a brake-like fashion. To establish that the yhjH mutant motility defect is due to a chemotaxis or velocity defect, the trajectories of individual cells were measured in conditions that eliminated chemical gradients. Decreased swimming speeds for yhjH mutant cells were observed when compared with wild type cells or cells harboring a yhjH ycgR double mutant (Boehm et al. 2010). Deletion of four DGCs shown to regulate cell motility in the yhjH mutant background restored swimming velocity to wild type levels (Boehm et al. 2010). To establish that reduced swimming speeds are not due to a chemotaxis defect, in vivo fluorescence resonance energy transfer (FRET) was used to assess the interaction between CheY and FliM in the wild type and yhjH mutant background. FliM and CheY interacted similarly in both backgrounds, which suggests that c-di-GMP regulates motility through adjustment of swimming speeds (Boehm et al. 2010). The analysis of this result implied that the interaction of c-di-GMP and YcgR at the motor does not induce a rotational defect, although Fang & Gomelsky (2010) and Paul et al. (2010) observed such rotational defects. Rather, Boehm et al. (2010) suggested that YcgR and c-di-GMP decrease swimming speed.
As YcgR binds c-di-GMP (Ryjenkov et al. 2006), a screen to isolate spontaneous motile suppressors of the yhjH mutant was performed. It was predicted that mutations would be identified in YcgR-binding partners, as a mutation would render the binding partner insensitive to elevated levels of c-di-GMP. Four point mutants were identified in motA, which led Boehm et al. (2010) to hypothesize that YcgR and MotA interact. YcgR colocalizes with FliM to the cell envelope, as observed by fluorescence microscopy. YcgR localization was disrupted upon decreases in c-di-GMP through overexpression of yhjH, upon deletion of motA, or when a motA suppressor allele was overexpressed in a motA deletion background. Acceptor photobleaching FRET experiments were used to measure an interaction between MotA and YcgR in vivo. The FRET signal increased in the presence of elevated levels of c-di-GMP, whereas the FRET signal decreased in the absence of c-di-GMP or when a motA suppressor allele was overexpressed in a motA deletion background (Boehm et al. 2010). The identification of motA suppressor alleles and the results of the colocalization and acceptor photobleaching FRET experiments suggested that MotA and YcgR interact, and this interaction is predicted to interfere with the stator-rotor interaction (Boehm et al. 2010).
Boehm et al. (2010) predicted that YcgR may regulate flagellar motility by disengaging rotor and stator units or by decreasing and preventing flagellar rotation. Tethering assays were employed to evaluate whether the wild type, the yhjH mutant, and the yhjH ycgR double mutant displayed actively rotating behavior, indicative of a functional flagellar motor, passively rotating behavior, indicative of disengaged rotors and stators, or static cell behavior, indicative of a locked motor. Populations of the wild type, yhjH mutant, and yhjH ycgR double mutant contained similar proportions of passively rotating cells, whereas yhjH mutants displayed more static cells and fewer actively rotating cells than did the wild type or yhjH ycgR double mutants. From this analysis, Boehm et al. (2010) concluded that YcgR regulates flagellar motors by decreasing torque generation. The yhjH mutant cells exhibit some rotating behavior; therefore, a subset of flagellar motors apparently were functional. To test whether YcgR inactivation of some stator units is sufficient to completely inhibit flagellar motility or if flagellar motility gradually decreases as more stator units are inactivated, combinations of chromosomal and plasmid wild type motA and motAG93E (a mutant motA gene that encodes MotAG93E) suppressor alleles were expressed in the yhjH deletion background under varying inducing conditions and motility was quantified. In suboptimal inducing conditions, yhjH mutants expressing only the wild type motA gene had the slowest swimming velocity, followed by the yhjH mutant expressing chromosomal motAG93E and the wild type motA gene on a plasmid, followed by yhjH mutants expressing chromosomal wild type motA and motAG93E on a plasmid; yhjH mutants expressing only the protein encoded by the motAG93E allele had the greatest swimming velocity. This analysis suggested that both the wild type and mutant MotA comprise the flagellum motor and that inactivation of one stator unit by YcgR does not inhibit all stator units but rather leads to a gradual decrease in the motility output (Boehm et al. 2010). Boehm et al. propose the following model to explain the mechanism of c-di-GMP-dependent YcgR regulation of motility: c-di-GMP-bound YcgR interacts with MotA to reduce flagellar motor function by inactivating individual stator units in a brake-like fashion to facilitate the transition from a motile to a sessile lifestyle (Figure 4c).
Clearly, YcgR and c-di-GMP function posttranslationally to regulate flagellum motility by interacting with motor components. The YcgR- and c-di-GMP-regulated motor components identified by Paul et al. and Fang & Gomelsky are more consistent with one another than with those described by Boehm et al. It is difficult to compare the studies directly, as different assays were used to measure outputs or similar assays conferred different results. Additionally, some assays were performed in E. coli or Salmonella, and the strains used in each study were not consistent. Reanalysis of mutants in the same strains and with the same assays would be useful. Through extensive experimentation, amazingly detailed models of flagellum torque generation and switching have been developed, but without more detailed structural studies of the motor and stator, it is not yet possible to understand the complete molecular mechanism (Sowa & Berry 2008). As these mechanistic details are unknown, it is rather challenging to assert confidently how and where YcgR affects the flagellar motor. Thus, further structure-function analyses of the flagellum motor will provide much insight into the specific mechanisms of the YcgR-c-di-GMP-regulated motile-to-sessile transition.
Transcriptional Regulation of the Vibrio Polysaccharide Genes Proceeds Via VpsT Bound to c-di-GMP
Unlike YcgR in E. coli, the PilZ domain-containing proteins in the V. cholerae biotype El Tor are not essential for biofilm formation, which indicates that non-PilZ domain-containing effector proteins function to bind c-di-GMP and regulate biofilm formation (Beyhan et al. 2008). Studies of V. cholerae have identified that c-di-GMP binding to the effector protein VpsT, a transcriptional regulator, induces oligomerization, which is required for DNA recognition and transcriptional regulation of the Vibrio polysaccharide (vps) genes (Krasteva et al. 2010).
Biofilm formation in V. cholerae depends upon the production of VPS, a major component of the extracellular matrix (Yildiz & Schoolnik 1999). V. cholerae undergoes phase variation to produce either smooth or rugose variants (White 1938); rugose variants produce more VPS than do smooth variants (Yildiz & Schoolnik 1999). Through whole-genome expression profiling it became apparent that the expression of VpsT is elevated in the V. cholerae rugose variant when compared with the smooth variant (Casper-Lindley & Yildiz 2004). VpsT is homologous to the transcriptional regulators found in E. coli and Salmonella that are required for the production of biofilm-associated products (Casper-Lindley & Yildiz 2004). Therefore, mutagenesis studies of vpsT were carried out to assess its role in regulating VPS production and biofilm formation. VpsT is required for maintaining the rugose variant colony phenotype, contributes to rugose variant biofilm formation, and induces vps gene expression (Casper-Lindley & Yildiz 2004). These results suggested that VpsT regulates biofilm formation through positively regulating the expression of vps genes (Casper-Lindley & Yildiz 2004).
Rugose variants contain higher levels of c-di-GMP than do smooth variants (Lim et al. 2006). When compared with a wild type rugose variant, deletion of the gene vpvC, which is predicted to encode a DGC, resulted in a decrease in intracellular concentrations of c-di-GMP, a decrease in the expression of vps genes and vpsT, and alterations in the thickness and heterogeneity of the biofilm (Beyhan & Yildiz 2007). These studies suggested that levels of c-di-GMP may impact phenotypes associated with the rugose variant (Beyhan & Yildiz 2007, Lim et al. 2006). Given that VpsT may regulate biofilm formation through the positive regulation of the vps genes (Casper-Lindley & Yildiz 2004), it was suggested that VpsT integrates c-di-GMP signals to induce transcription of the vps genes (Krasteva et al. 2010).
To elucidate the mechanism by which VpsT integrates c-di-GMP levels to positively regulate vps expression and biofilm formation, X-ray crystallographic analyses of VpsT were undertaken. The structure of VpsT was solved in both the presence and the absence of c-di-GMP. VpsT contains an N-terminal receiver (REC) domain and a C-terminal helix-turn-helix domain that mediates DNA binding. An additional helix (helix α6) extends the C-terminal portion of the REC domain. Two nonoverlapping dimerization interfaces are present in VpsT. The first mediates c-di-GMP-independent dimerization of VpsT through a methionine residue at amino acid position 17. The second mediates c-di-GMP-dependent dimerization of VpsT, as c-di-GMP binds to helix α6 and stabilizes dimerization. The c-di-GMP binding motif in VpsT was determined to be W[F/L/M][T/S]R (Krasteva et al. 2010). Isothermal titration calorimetry (ITC) analyses suggested that a dimer of c-di-GMP is bound to a dimer of VpsT. Mutations within the c-di-GMP binding motif, namely, VpsTW131F, VpsTT133V, and VpsTR134A, abolished c-di-GMP binding as assessed by ITC. Additionally, a mutation in helix α6, VpsTI141E, also disrupted c-di-GMP binding, which suggests that c-di-GMP binding is dependent upon dimerization of VpsT. A VpsTM17D mutation at the c-di-GMP-independent dimerization interface did not disrupt c-di-GMP binding (Krasteva et al. 2010). Dimerization of VpsT at both c-di-GMP-dependent and c-di-GMP-independent interfaces was further assessed by static multiangle light scattering of various VpsT mutants. VpsTM17D remained monomeric in the absence of c-di-GMP, whereas VpsTR134A and VpsTI141E formed both monomeric and dimeric species. Addition of c-di-GMP to VpsTM17D resulted in the formation of dimeric species (Krasteva et al. 2010).
The c-di-GMP-binding mutant VpsTR134A and dimerization interface mutants VpsTI141E and VpsTM17D were used in electrophoretic mobility shift assays to assess the effects of c-di-GMP binding and VpsT dimerization on VpsT regulation of vps genes. VpsTR134A and VpsTI141E were unable to bind to the promoter of vpsL, a gene positively regulated by VpsT (Krasteva et al. 2010). The three mutants were then used to assess the requirements of c-di-GMP binding and VpsT dimerization for transcription of vps genes. Results from vpsLp-lacZ transcriptional fusion assays demonstrated that VpsTR134A and VpsTI141E were unable to increase the expression of vpsL to the same extent observed for the wild type VpsT and the VpsTM17D mutant protein. These results suggested that both c-di-GMP binding and c-di-GMP-dependent dimerization of VpsT are critical for VpsT transcriptional regulation of the vps genes (Krasteva et al. 2010).
Whole-genome expression profiling of the vpsT deletion mutant overexpressing VpsTI141E, VpsTM17D, or VpsTR134A revealed that c-di-GMP binding of VpsT and c-di-GMP-dependent dimerization of VpsT are required for induction of vps genes. vps genes were induced at significantly lower levels when VpsTI141E or VpsTR134A was overexpressed as compared with the overexpression of VpsTM17D or wild type VpsT. Whole-genome expression profiling also revealed that overexpression of VpsTI141E or VpsTR134A did not reduce flagellar gene expression, as did overexpression of VpsTM17D or wild type VpsT. Motility assays confirmed that c-di-GMP binding to VpsT and c-di-GMP-dependent dimerization of VpsT are required to reduce motility (Krasteva et al. 2010). Lastly, the conversion from smooth to rugose colony morphology requires c-di-GMP binding and VpsT dimerization (Krasteva et al. 2010). The above studies demonstrate that c-di-GMP binding to and c-di-GMP-dependent oligomerization of VpsT is necessary and sufficient for DNA regulation and transcription of vps genes. Thus, VpsT integrates c-di-GMP to inversely regulate biofilm formation and motility in V. cholerae (Krasteva et al. 2010) (Figure 5).
Figure 5.
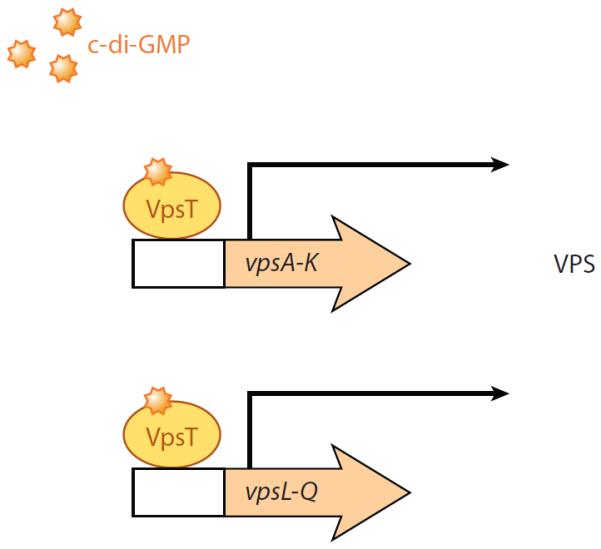
c-di-GMP effector system in Vibrio cholerae. This diagram depicts a summary of the current model of the c-di-GMP effector system in V. cholerae, which impacts the motile-to-sessile transition. The transcriptional regulator VpsT binds c-di-GMP to positively regulate the transcription of the vps genes encoding the proteins needed for VPS production. The VPS polysaccharide is required for biofilm formation (Krasteva et al. 2010; Illustration courtesy of William Scavone, MA, CMI, Kestrel Illustration Studio, LLC).
c-di-GMP levels affect the biofilm-associated curli fimbriae through the transcriptional regulator CsgD found in Salmonella and E. coli (Kader et al. 2006, Weber et al. 2006). CsgD is a VpsT homolog (Casper-Lindley & Yildiz 2004), but the binding pocket of CsgD, YF[T/S]Q, differs from that of the VpsT c-di-GMP binding pocket and is unlikely to be able to bind c-di-GMP in this pocket (Krasteva et al. 2010). Furthermore, whereas VpsT does not seem to be regulated by phosphorylation (Krasteva et al. 2010), CsgD DNA binding is (Zakikhany et al. 2010). Krasteva et al. suggest that whereas dimerization to induce DNA binding may be a general mechanism in these homologous transcriptional regulators, the mechanisms of c-di-GMP binding and dimerization in VpsT may be conserved only in similar proteins.
In V. parahaemolyticus the cps genes encode proteins required for the production of a sticky capsular polysaccharide required for biofilm formation (Boles & McCarter 2002) in response to c-di-GMP levels (Ferreira et al. 2008). Ferreira et al. (2011) suggest that CpsQ, a homolog of VpsT that contains a strongly conserved c-di-GMP binding pocket in comparison to that of VpsT, binds c-di-GMP and is a direct transcriptional regulator of the cps genes. Whether the mechanism of c-di-GMP binding to CpsQ is similar to that of VpsT and whether binding of c-di-GMP induces oligomerization of CpsQ remain to be determined.
Other Potential Effector Mechanisms
In recent years several other c-di-GMP effector systems have been identified, but the mechanisms by which these effectors function are less well understood. However, the study of such systems is an exciting future avenue of research that will likely unveil new secrets regarding mechanisms by which c-di-GMP regulates bacterial biology. An overview of other identified c-di-GMP effector systems is summarized in Table 1.
CONCLUSION
The diversity of c-di-GMP control mechanisms and their varied targets highlights the scope and intricacy of signaling by this second messenger. c-di-GMP binding induces effectors to undergo conformational changes, transcriptional activation, DNA binding, protein-protein interactions, derepression of genes, localization, and enhanced enzymatic activity. The diversity of mechanisms through which c-di-GMP impacts effector function suggests that effectors are able to impact a variety of outputs, such as motility, biofilm formation, virulence, cell cycle regulation, RNA processing, gene expression, and likely others that are yet to be uncovered.
In the context of c-di-GMP signaling, bacteria possess a complex network of enzymes and effectors that sense and respond to environmental signals, adjust cellular levels of c-di-GMP, and regulate phenotypic outputs. Understanding how multiple c-di-GMP signaling pathways are isolated from each other, or integrated to produce coherent outputs, is a question of fundamental importance for understanding an array of biological outputs in bacteria. Assigning molecular functions to DGCs and PDEs as well as determining which specific outputs are regulated by the coordinated action of these enzymes will aid in that understanding. Additionally, studies must focus on identifying new effectors and deciphering the mechanisms by which c-di-GMP effectors appropriately coordinate inputs. Going forward, much progress needs to be made in understanding interactions between specific DGCs, PDEs, and effectors. Such studies will provide a better understanding of c-di-GMP signaling mechanisms that are critical for bacterial biology.
ACKNOWLEDGMENTS
This work was supported by a National Institutes of Health T32 AI007519 predoctoral fellowship to C.D.B. and by National Science Foundation grant MCB-9984521 to G.A.O. We also thank M. Gomelsky and H. Sondermann for critical reading of the manuscript.
DISCLOSURE STATEMENT The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Glossary
- Biofilm
a community of bacterial cells attached to a surface
- Bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP)
bacterial intracellular second messenger that stimulates biofilm formation and inhibits motility
- Diguanylate cyclase (DGC)
enzyme that synthesizes c-di-GMP
- GGDEF
core amino acid motif conserved in DGCs for synthesizing c-di-GMP
- Phosphodiesterase (PDE)
enzyme that degrades c-di-GMP
- EAL
core amino acid motif conserved in PDEs for degrading c-di-GMP to pGpG
- HD-GYP
core amino acid motif conserved in PDEs for degrading c-di-GMP to GMP
- Effector
c-di-GMP receptor; protein or RNA that binds c-di-GMP
- PilZ
c-di-GMP binding domain found in some effector proteins
- HAMP
domain of approximately 50 amino acids found in transmembrane signaling proteins; functions to mediate signal transduction
LITERATURE CITED
- Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, et al. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol. Cell. 2011;43:550–60. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad I, Lamprokostopoulou A, Le Guyon S, Streck E, Barthel M, et al. Complex c-di-GMP signaling networks mediate transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLoS ONE. 2011;6:e28351. doi: 10.1371/journal.pone.0028351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- Andrade MO, Alegria MC, Guzzo CR, Docena C, Rosa MC, et al. The HD-GYP domain of RpfG mediates a direct linkage between the Rpf quorum-sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv citri. Mol. Microbiol. 2006;62:537–51. doi: 10.1111/j.1365-2958.2006.05386.x. [DOI] [PubMed] [Google Scholar]
- Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–18. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–66. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin P, Terao E, Lee EH, Lejeune P, Colson C, et al. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 1994;176:5537–40. doi: 10.1128/jb.176.17.5537-5540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 2007;189:388–402. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Odell LS, Yildiz FH. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J. Bacteriol. 2008;190:7392–405. doi: 10.1128/JB.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 2006;188:3600–13. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Yildiz FH. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 2007;63:995–1007. doi: 10.1111/j.1365-2958.2006.05568.x. [DOI] [PubMed] [Google Scholar]
- Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ. Microbiol. 2008;10:1419–32. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, et al. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–16. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Boles BR, McCarter LL. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 2002;184:5946–54. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PN, Terrazas M, Paul K, Blair DF. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 2007;189:305–12. doi: 10.1128/JB.01281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper-Lindley C, Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 2004;186:1574–8. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AL, Tuckerman JR, Gonzalez G, Mayer R, Weinhouse H, et al. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry. 2001;40:3420–26. doi: 10.1021/bi0100236. [DOI] [PubMed] [Google Scholar]
- Chin KH, Lee YC, Tu ZL, Chen CH, Tseng YH, et al. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J. Mol. Biol. 2010;396:646–62. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- Christen B, Christen M, Paul R, Schmid F, Folcher M, et al. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 2006;281:32015–24. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- Christen M, Christen B, Allan MG, Folcher M, Jeno P, et al. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA. 2007;104:4112–17. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 2005;280:30829–37. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science. 2010;328:1295–97. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu. Rev. Microbiol. 1995;49:711–45. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- Davey ME, O'Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000;64:847–67. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deflaun MF, Tanzer AS, McAteer AL, Marshall B, Levy SB. Development of an adhesion assay and characterization of an adhesion-deficient mutant of Pseudomonas fluorescens. Appl. Environ. Microbiol. 1990;56:112–19. doi: 10.1128/aem.56.1.112-119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato GM, Kawula TH. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli. J. Biol. Chem. 1998;273:24030–36. doi: 10.1074/jbc.273.37.24030. [DOI] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, et al. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Gomelsky M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 2010;76:1295–305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, et al. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 2011;82:327–41. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- Ferreira RB, Antunes LC, Greenberg EP, McCarter LL. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 2008;190:851–60. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RBR, Chodur DM, Antunes LCM, Trimble MJ, McCarter LL. Output targets and transcriptional regulation by a cyclic dimeric GMP-responisve circuit in Vibrio parahaemolyticus Scr network. J. Bacteriol. 2011;194:914–24. doi: 10.1128/JB.05807-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman JC, Rogers EA, Kostick JL, Zhang H, Iyer R, et al. Identification and molecular characterization of a cyclic-di-GMP effector protein, PlzA (BB0733): additional evidence for the existence of a functional cyclic-di-GMP regulatory network in the Lyme disease spirochete, Borrelia burgdorferi. FEMS Immunol. Med. Microbiol. 2010;58:285–94. doi: 10.1111/j.1574-695X.2009.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- Genevaux P, Muller S, Bauda P. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol. Lett. 1996;142:27–30. doi: 10.1111/j.1574-6968.1996.tb08402.x. [DOI] [PubMed] [Google Scholar]
- Girgis HS, Liu Y, Ryu WS, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:1644–60. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo CR, Salinas RK, Andrade MO, Farah CS. PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis. J. Mol. Biol. 2009;393:848–66. doi: 10.1016/j.jmb.2009.07.065. [DOI] [PubMed] [Google Scholar]
- Habazettl J, Allan MG, Jenal U, Grzesiek S. Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J. Biol. Chem. 2011;286:14304–14. doi: 10.1074/jbc.M110.209007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009;7:263–73. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008;69:376–89. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA. 2005;102:14422–27. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003;49:905–18. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- Hinsa SM, O'Toole GA. Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology. 2006;152:1375–83. doi: 10.1099/mic.0.28696-0. [DOI] [PubMed] [Google Scholar]
- Hogan D, Kolter R. Why are bacteria refractory to antimicrobials? Curr. Opin. Microbiol. 2002;5:472–77. doi: 10.1016/s1369-5274(02)00357-0. [DOI] [PubMed] [Google Scholar]
- Huang B, Whitchurch CB, Mattick JS. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 2003;185:7068–76. doi: 10.1128/JB.185.24.7068-7076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DW, Simecka JW, Romeo T. Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 2002a;184:3406–10. doi: 10.1128/JB.184.12.3406-3410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 2002b;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader A, Simm R, Gerstel U, Morr M, Romling U. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2006;60:602–16. doi: 10.1111/j.1365-2958.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- Kazmierczak BI, Lebron MB, Murray TS. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 2006;60:1026–43. doi: 10.1111/j.1365-2958.2006.05156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Ryu KS, Kim H, Shin JS, Lee JO, et al. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by PilZ domain proteins. J. Mol. Biol. 2010;398:97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Ko M, Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 2000;303:371–82. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–68. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshina N, Baird NJ, Ferre-D'Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 2009;16:1212–17. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J. Bacteriol. 2009;191:7121–22. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–48. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 2007;65:1474–84. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Beyhan S, Meir J, Yildiz FH. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 2006;60:331–48. doi: 10.1111/j.1365-2958.2006.05106.x. [DOI] [PubMed] [Google Scholar]
- Lloyd SA, Blair DF. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 1997;266:733–44. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- Lugtenberg BJ, Kravchenko LV, Simons M. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ. Microbiol. 1999;1:439–46. doi: 10.1046/j.1462-2920.1999.00054.x. [DOI] [PubMed] [Google Scholar]
- Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 2007;65:876–95. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- Merritt JH, Ha DG, Cowles KN, Lu W, Morales DK, et al. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio. 2010;1:e00183–10. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds RD, Newell PD, Gross RH, O'Toole GA. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 2007;63:656–79. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- Navarro MV, De N, Bae N, Wang Q, Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure. 2009;17:1104–16. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro MV, Newell PD, Krasteva PV, Chatterjee D, Madden DR, et al. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol. 2011;9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PD, Boyd CD, Sondermann H, O'Toole GA. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 2011a;9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PD, Monds RD, O'Toole GA. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. USA. 2009;106:3461–66. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O'Toole GA. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J. Bacteriol. 2011b;193:4685–98. doi: 10.1128/JB.05483-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby LL, Jain S, Ohman DE. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology. 2008;154:1605–15. doi: 10.1099/mic.0.2007/015305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Gibbs KA, Hager PW, Phibbs PV, Jr, Kolter R. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 2000;182:425–31. doi: 10.1128/jb.182.2.425-431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998a;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 1998b;28:449–61. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell. 2010;38:128–39. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J. Biol. Chem. 2007;282:29170–77. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, et al. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–27. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzer JE, Sultan SZ, Hayakawa Y, Hobbs G, Miller MR, Motaleb MA. Analysis of the Borrelia burgdorferi cyclic-di-GMP-binding protein PlzA reveals a role in motility and virulence. Infect. Immun. 2011;79:1815–25. doi: 10.1128/IAI.00075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povolotsky TL, Hengge R. “Life-style” control networks in Escherichia coli: signaling by the second messenger c-di-GMP. J. Biotechnol. 2012;160:10–16. doi: 10.1016/j.jbiotec.2011.12.024. [DOI] [PubMed] [Google Scholar]
- Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 2007;282:12860–70. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998;30:285–93. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Brombacher E, Vidal O, Ambert A, Lejeune P, et al. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 2001;183:7213–23. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Chuah ML, Dong X, Xie K, Luo Z, et al. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J. Biol. Chem. 2011;286:2910–17. doi: 10.1074/jbc.M110.196220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Rao F, Luo Z, Liang ZX. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry. 2009;48:10275–85. doi: 10.1021/bi901121w. [DOI] [PubMed] [Google Scholar]
- Rao F, Pasunooti S, Ng Y, Zhuo W, Lim L, et al. Enzymatic synthesis of c-di-GMP using a thermophilic diguanylate cyclase. Anal. Biochem. 2009;389:138–42. doi: 10.1016/j.ab.2009.03.031. [DOI] [PubMed] [Google Scholar]
- Romling U, Gomelsky M, Galperin MY. C-di-GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol. 2005;57:629–39. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–81. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA. 2006a;103:6712–17. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ryan RP, Fouhy Y, Lucey JF, Dow JM. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 2006b;188:8327–34. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: The PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 2006;281:30310–14. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- Sauer K, Cullen MC, Rickard AH, Zeef LA, Davies DG, Gilbert P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 2004;186:7312–26. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H, Yoshioka S, Uchida T, Hyodo M, Hayakawa Y, et al. Molecular oxygen regulates the enzymatic activity of a heme-containing diguanylate cyclase (HemDGC) for the synthesis of cyclic di-GMP. Biochim. Biophys. Acta. 2010;1804:166–72. doi: 10.1016/j.bbapap.2009.09.028. [DOI] [PubMed] [Google Scholar]
- Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 2005;187:4774–81. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JS, Ryu KS, Ko J, Lee A, Choi BS. Structural characterization reveals that a PilZ domain protein undergoes substantial conformational change upon binding to cyclic dimeric guanosine monophosphate. Protein Sci. 2011;20:270–77. doi: 10.1002/pro.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–55. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 2009;16:1218–23. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Shanahan CA, Moore EL, Simon AC, Strobel SA. Structural basis of differential ligand recognition by two classes of bis-(3′-5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc. Natl. Acad. Sci. USA. 2011;108:7757–62. doi: 10.1073/pnas.1018857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa Y, Berry RM. Bacterial flagellar motor. Q. Rev. Biophys. 2008;41:103–32. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–13. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Tischler AD, Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 2005;280:33324–30. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao F, He YW, Wu DH, Swarup S, Zhang LH. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J. Bacteriol. 2010;192:1020–29. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarutina M, Ryjenkov DA, Gomelsky M. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 2006;281:34751–58. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- Taylor BL. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol. Microbiol. 2007;65:1415–24. doi: 10.1111/j.1365-2958.2007.05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Kojima S, Homma M. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 2008;270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, et al. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 2006;188:2681–91. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 2004;53:857–69. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J. Mol. Biol. 2011;407:633–39. doi: 10.1016/j.jmb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Tuckerman JR, Gonzalez G, Sousa EH, Wan X, Saito JA, et al. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry. 2009;48:9764–74. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- Wan X, Tuckerman JR, Saito JA, Freitas TA, Newhouse JS, et al. Globins synthesize the second messenger bis-(3′-5′)-cyclic diguanosine monophosphate in bacteria. J. Mol. Biol. 2009;388:262–70. doi: 10.1016/j.jmb.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Fullner KJ, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 1999;181:3606–9. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. Cyclic-di-GMP-mediated signaling within the sigma network of Escherichia coli. Mol. Microbiol. 2006;62:1014–34. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- Weinhouse H, Sapir S, Amikam D, Shilo Y, Volman G, et al. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1997;416:207–11. doi: 10.1016/s0014-5793(97)01202-7. [DOI] [PubMed] [Google Scholar]
- White BP. The rugose variant of Vibrios. J. Pathol. 1938;46:1–6. [Google Scholar]
- Wilksch JJ, Yang J, Clements A, Gabbe JL, Short KR, et al. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011;7:e1002204. doi: 10.1371/journal.ppat.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SB, Stewart V. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 1999;33:1093–102. doi: 10.1046/j.1365-2958.1999.01562.x. [DOI] [PubMed] [Google Scholar]
- Wolfe AJ, Visick KL. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol. 2008;190:463–75. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA. 1999;96:4028–33. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakikhany K, Harrington CR, Nimtz M, Hinton JC, Romling U. Unphosphorylated CsgD controls biofilm formation in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2010;77:771–86. doi: 10.1111/j.1365-2958.2010.07247.x. [DOI] [PubMed] [Google Scholar]
- Zhou J, Lloyd SA, Blair DF. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA. 1998;95:6436–41. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]