Abstract
Background
Exposure to chronic ethanol results in changes in expression of proteins that regulate neuronal excitability. The present study examined whether chronic ethanol alters the hippocampal expression and function of Fragile-X mental retardation protein (FMRP), and the role of FMRP in the modulation of chronic ethanol-induced changes in expression of NMDA receptors and Kv4.2 channels.
Methods
For in-vivo studies, C57Bl6/J mice underwent a chronic intermittent ethanol (CIE) vapor exposure procedure. After CIE, hippocampal tissue was collected and subjected to immunoblot blot analysis of NMDA receptor subunits (GluN1, GluN2B), Kv4.2 and its accessory protein KChIP3. For in-vitro studies, hippocampal slice cultures were exposed to 75 mM ethanol for 8 days. Following ethanol exposure, mRNAs bound to FMRP was measured. In a separate set of studies, cultures were exposed to an inhibitor of S6K1 (PF-4708671, 6 μM) in order to assess whether ethanol-induced homeostatic changes in protein expression depend upon changes in FMRP activity.
Results
Immunoblot blot analysis revealed increases in GluN1 and GluN2B but reductions in Kv4.2 and KChIP3. Analysis of mRNAs bound to FMRP revealed a similar bidirectional change observed as reduction of GluN2B and increase in Kv4.2 and KChiP3 mRNA transcripts. Analysis of FMRP further revealed that while chronic ethanol did not alter the expression of FMRP, it significantly increased phosphorylation of FMRP at the S499 residue that is known to critically regulate its activity. Inhibition of S6K1 prevented the chronic ethanol-induced increase in phospho-FMRP and changes in NMDA subunits, Kv4.2 and KChiP3. In contrast, PF-4708671 had no effect in the absence of alcohol, indicating it was specific for the chronic ethanol-induce changes.
Conclusions
These findings demonstrate that chronic ethanol exposure enhances translational control of plasticity related proteins by FMRP, and that S6K1 and FMRP activity are required for expression of chronic ethanol-induced homeostatic plasticity at glutamatergic synapses in the hippocampus.
Keywords: alcohol, plasticity, FMRP, NMDA, Kv4.2
Introduction
Exposure to chronic ethanol results in adaptations at glutamatergic synapses that are thought to reflect, at least in part, homeostatic control of neuronal excitability and synaptic signaling. A well known example of these activity-dependent homeostatic changes induced by chronic ethanol include increases in expression of GluN2B-subunit containing NMDA receptors and a shift in their localization from the extrasynaptic to synaptic region of the membrane (Carpenter-Hyland et al., 2004, Chandler et al., 2006, Hendricson et al., 2007). In addition, we recently demonstrated that changes in the expression of proteins that modulate glutamatergic neurotransmission following chronic ethanol exposure involves bidirectional alterations of the expression of proteins that positively and negatively modulate neuronal signaling and plasticity. Specifically, CIE-induced increases in GluN2B subunit-containing receptors in the hippocampus occur in association with decreases in expression of the A-type K+-channel, Kv4.2 (Mulholland et al., 2015). These K+-channels critically regulate back-propagating action potentials (bAPs) and synaptic integration, and are the major ion channels influencing neuronal excitability of hippocampal pyramidal neurons (Cai et al., 2004, Kim et al., 2007, Carrasquillo et al., 2012). Fully functional Kv4.2 channels require auxiliary proteins that maintain surface expression and determine channel kinetics (Rhodes et al., 2004, An et al., 2000). In particular, K+-channel interacting protein 3 (KChIP3) is a primary binding partner of Kv4.2 in dendrites and dendritic spines in the hippocampus and regulates their membrane trafficking and expression (Shibata et al., 2003, Xiong et al., 2004). Interestingly, KChIP3 has also been shown to negatively regulate the expression and function of NMDA receptors, and thus may play a role in CIE-induced bidirectional modulation of NMDA and Kv4.2 expression (Zhang et al., 2010)
The mRNA-binding protein Fragile-X mental retardation protein (FMRP) is a translational repressor that binds to mRNAs to regulate interaction with the active polyribosome and decrease protein expression (Ascano et al., 2012, Darnell et al., 2011). As one of the main regulators of local activity-dependent translation, FMRP interacts with 3-4% of all mRNAs in dendrites and dendritic spines, including Kv4.2, KChIP3, and NMDA receptor subunits (Darnell et al., 2011, Eadie et al., 2012, Lee et al., 2011). FMRP expression and activity is necessary for proper neuronal function, maintenance of basal protein expression, and mediating mRNA translation (Wang et al., 2014, Zhang et al., 2009, Dictenberg et al., 2008, Mercaldo et al., 2009). Disruption of FMRP expression and function leads to dysregulation of activity-dependent local translation, and affects neuronal mechanisms of plasticity (Wang et al., 2014, Yun and Trommer, 2011). While the importance of FMRP during development has been well established, a potential role for FMRP in mediating alterations in translation in response to chronic ethanol exposure is not known (Saffary and Xie, 2011, Fernandez et al., 2013, Bonaccorso et al., 2015). The goal of the current study was to gain a better understand the role of FMRP in regulating ethanol-induced changes in protein expression at glutamatergic synapses.
Materials and Methods
All procedures were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals, and were approved by the Institutional Animal Care and Use Committees of the Medical University of South Carolina.
In-vivo chronic intermittent ethanol exposure
Male adult mice of C57BL/6 (Charles River Laboratories, Kingston, NY) were single-housed under a 12 hr light/dark cycle with continuous access to food and water. Beginning at approximately 9 weeks of age, mice were subjected to either air or ethanol vapor using chronic intermittent exposure (CIE) procedure as previously described (Becker and Hale, 1993). In brief, this procedure involved exposure to either ethanol or air vapor for 14 hours beginning ~3 hours into the dark cycle followed by an 8-hour withdrawal period. This cycle was repeated for four consecutive days and was followed by a 72 hr withdrawal period. On the days that animals went into the vapor chambers, ethanol intoxication was initiated by administration of 1.6 g/kg ethanol (8% w/v; i.p.) together with the alcohol dehydrogenase inhibitor pyrazole (1 mmol/kg). Mice in the control group received equivalent injections of pyrazole in saline. This weekly CIE exposure and withdrawal cycle was repeated for four consecutive weeks. Chamber ethanol concentrations were measured daily and blood samples were collected from all mice for blood ethanol analysis to ensure stable BEC (target levels of 175-225 mg/dl) throughout the exposure period. At the end of the last week of CIE, mice were removed from the vapor chambers, immediately sacrificed, and the brain removed and dissected on ice. After obtaining bilateral punches of the hippocampus, the tissue was prepared for immunoblot analysis by sonication in 2% LDS and then stored at −80 °C for subsequent western blot analysis.
Hippocampal slice cultures and in-vitro chronic ethanol exposure
Hippocampal slice cultures were prepared from Sprague-Dawley rat pups as previously described (Mulholland et al, 2011). In brief, 6-8 day old rat pups were rapidly decapitated and whole brains were aseptically removed and placed in ice-cold dissection medium consisting of Basal Medium Eagle (with Earle’s salts) plus 25 mM HEPES, 2 mM GlutaMAX, and 100 μg/ml streptomycin (pH 7.2). Bilateral hippocampi were hand dissected and placed in chilled culture medium consisting of dissection medium plus 36 mM glucose, 25% (v/v) Earle’s balanced salt solution, and 25% platelet-poor horse serum (PPHS). Each hippocampus was then coronally sectioned at 400 μm using a McIllwain tissue chopper (Mickle Laboratory Engineering Co. Ltd., Gomshall, UK), and the slices then transferred onto Millicell-CM biopore membrane (0.4-μm pore size) inserts (4 slices/insert) and placed in 35-mm six-well culture plates in pre-incubated culture medium. Cultures were then placed in a humidified 37°C incubator with an atmosphere of 7.5% CO2 and 92.5% air. After 4 days of incubation, the media was changed to one containing a reduced amount of PPHS (5%). This media was replaced every 4 days for the remaining period of culturing. Each set of cultured slices was prepared from 6-8 rats pups and each treatment group within an experiment contained slices from at least 2 separate rat pups. For chronic exposure to alcohol, 8-day old slice cultures were exposed to 75 mM ethanol in sealed vapor chambers also as previously described. In studies using an SK1 inhibitor, PF-4708671 was added to the media every other day. After exposure, slices were scraped from the culture membrane and sonicated in 2% LDS, and prepared for western blot analysis or for co-immunoprecipitation and RT-qPCR.
Western blot analysis
Western blot procedures were carried out as previously described (Trantham-Davidson et al., 2014). In brief, proteins samples were separated by weight via gel electrophoresis and transferred onto a PVDF membrane using Bio-Rad semi-dry transfer apparatus. A Swift Total Protein Stain (G-Biosciences) was used to evaluate protein loading across the different lanes and for normalization of the blot data. Membranes were then blocked in 4% non-fat dry milk and incubated overnight at 4 °C with the primary antibody. Primary antibodies and dilutions included phospho-S499 FMRP (1:1000, Abcam, ab48127), FMRP (1:2000, Milllipore, MAB2160), KChIP3 (1:500, Millipore, 05-756), Kv4.2 (1:1000, NeuroMab, 75-016), GluN1 (1:3000, NeuroMab, 75-272), and GluN2B (1:3000, NeuroMab, 75-101). After incubation with the primary antibody, membranes were then incubated with secondary antibody for 1 hr at room temperature. Membranes were exposed to enhanced chemilluminescence (Bio-Rad,Hercules, CA) and band light intensity measured using a ChemicDoc MP Imaging System (Bio-Rad, Hercules, CA)
Immunoprecipitation of FMRP and real-time quantitative PCR
A co-immunoprecipitation (co-IP) kit from Pierce was used to pull down mRNA bound to FMRP from hippocampal lysates according to the manufacturers instructions. For sample preparation, hippocampal slice cultures were homogenized using a needle and syringe with RNase and protease inhibitors in homogenization buffer consisting of 1.28 M sucrose, 40 mM Tris-HCl pH 7.5, 20 mM MgCl2, and 1% NP-40, with ~ 2 u/mL of RNasin Plus inhibitors (Promega), and 1X cOmplete Mini EDTA-free protease inhibitor (Roche). To pre-clear the lysate, 80 μl of slurry of Control Agarose Resin was cleared on a spin column followed by a wash with 100 μl of Coupling Buffer. One mg of lysate was added to the pre-cleared resin and incubated for 30 min at 4 °C with rotation. Following centrifugation, the eluent was saved to subsequent co-IP. For antibody immobilization, 50 μl of a slurry of AminoLink Coupling Resin was added to a spin column. After brief centrifugation to clear the column, the resin was washed twice with 200 μl of Coupling Buffer consisting of 10 mM NaH2PO4, 150 mM NaCl in nuclease free water. To each pre-cleared spin column, FMRP antibody (6 μg), 200 μl of Coupling Buffer and 3 μl of sodium cyanoborohydride were added. This solution was incubated at room temperature with rotation for 2 hrs. The column was then cleared by centrifugation and the resin washed twice with 200 μl of Coupling Buffer followed by the addition of 200 μl of Quenching Buffer consisting of 1 M Tris-HCl and 3 μl of sodium cyanoborohydride. After 15 min incubation with rotation, the column was cleared by centrifugation, washed twice with 200 μl Coupling Buffer, and then 6 times with Wash Buffer consisting of 0.1M PBS, 200 mM NaCl, and 2 μl/ml of RNase Plus and 1X protease inhibitor. For subsequent co-IP of FMRP-mRNA, the pre-cleared lysate in 500 μl of PBS was added to an antibody immobilized spin column and incubated overnight at 4 °C with rotation, followed by rinsing the column with Wash Buffer. To elute binding of FMRP-mRNA, 50 μl of Elution Buffer was added to the column and incubated at room temperature for 5 min. The eluted FMRP-mRNA was then recovered by centrifugation.
For measurement of mRNAs that were co-IP with FMRP, the mRNA was extracted using a standard chloroform-Trizol (Life Technologies) extraction procedure. The extracted mRNA was then purified using the RNeasy Mini Kit (Qiagen) and nonspecific cDNA was transcribed using a RNA to cDNA Kit (Applied Biosystems). The mRNA reverse transcription protocol involved incubation at 37 °C for 60 min, 95 °C for 5 min, and a 4 °C hold using a Bio-Rad C1000 thermocycler. The primer sequences used for all transcripts examined is provided in Table 1. For qPCR, a Sybr Green qPCR kit (Life Technologies) was used on a Bio-Rad CFX 96 thermocycler with an initial denaturation of 2 min at 94 °C followed by 40 cycles with a 15 sec denaturation at 94 °C, 1 min of annealing and extension at 60 °C, and a 4 °C hold. The comparative ΔCt method was used to calculate fold change. ΔCt = Target cycle # - Ref cycle #; ΔΔCt = ΔCt Ctrl – ΔCt Exp. Fold change was calculated using the ΔΔCt. Fold change = 2−ΔΔC.
Table 1.
Primer sequences for RT-qPCR.
| mRNA | Primer Sequence |
|---|---|
| GAPDH | 5’- AAGGCTCATGACCA 3’- CAGGGATGATGTTCT |
| HPRT | 5’ TTGGATACAGGCCAGACTTTGTT 3’ CTGAAGTACTCATTATAGTCAAGGGCATA |
| FAAH | 5’- ATGAACCCGTGGAAGCCCTC 3’- CGCCGATGTCAGTGCCTAAAC |
| GluN1 | 5’- CTCTAGCCAGGTCTACGCTATCC 3’- GACGGGGATTCTGTAGAAGCCA |
| GluN2B | 5’- CTGGAGTTCTGGTTCCTTACTG 3’- ATTCTCCTATCTTGCCCGGA |
| KChlP3 | 5’ CACCTATGCACACTTCCTCTTCA 3’ ACCACAAAGTCCTCAAAGTGGAT |
| Kv4.2 | 3’-GCCTTCGTTAGCAAATCTGG 3’ GTGACATAAGGACACTGGG |
Statistical Analysis
Experiments conducted with organotypic hippocampal slice cultures were divided into paired treatment groups. Cultures from the same animals were used in both the control and treated groups, such that an n of 1 for both the control and treatment groups came from the same animal, n of 2 from another animal, etc. Each n for every experimental paradigm represents hippocampal tissue taken from the same animal, and this tissue was used only once in the same experiment. Unless otherwise noted, experiments with control and ethanol only groups were analyzed with a paired two-tailed Students t-test with significance of p < 0.05. For experiments with S6K1 inhibitor and concurrent ethanol treatment, a 1-way ANOVA was used to determine significance. For western blots with hippocampal tissue taken from CIE-treated mice, an unpaired Students t-test was used to determined statistical significance between control and ethanol-treated.
Results
Chronic ethanol induces bidirectional changes in NMDA and Kv4.2 expression in the hippocampus
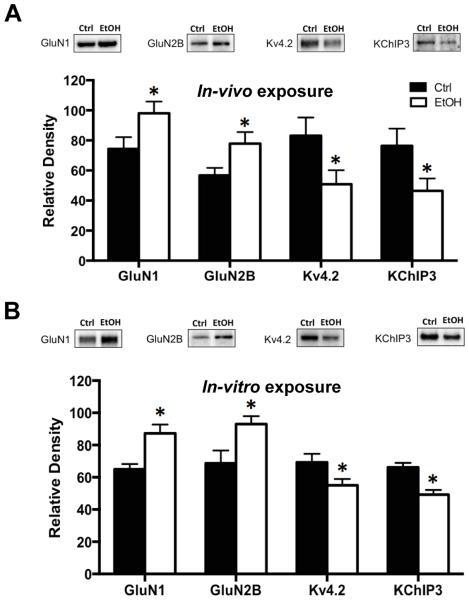
The first set of experiments examined the effects of chronic ethanol exposure on expression of NMDA receptor subunits, Kv4.2 channels and the Kv4.2 auxiliary protein KChIP3. As shown in Figure 1A, immunoblot analysis revealed a significant increase in expression of both GluN1 [t(20) = 2.146, p < 0.05, n = 11] and GluN2B [t(20) = 2.132, p < 0.05, n=11] subunits in hippocampal tissue isolated immediately after the last cycle of CIE exposure. In contrast, there was a significant reduction in expression of both Kv4.2 [t(16) = 2.121, p < 0.05, n = 9] and KChIP3 [t(18) = 2.123, p < 0.05, n = 10]. This confirms a previous observation of bidirectional alterations in the expression of NMDA receptors and Kv4.2 channels, presumably as a homeostatic response to maintain normal levels of neuronal activity (Lee et al., 2011, Andrasfalvy et al., 2008, Jung et al., 2008, Kaufmann et al., 2013, Lei et al., 2010).
Figure 1.
Chronic ethanol exposure induces bidirectional changes in the expression of NMDA and Kv4.2 receptors in the hippocampus. (A) In an in-vivo model of chronic intermittent ethanol exposure (CIE), immunoblot analysis of the expression of GluN1 and GluN2B in the hippocampus was significantly increased when measured immediately following the last episode of ethanol exposure. In contrast, expression of Kv4.2 and KChIP3 was significantly reduced (n = 9-11). (B) Nearly identical bidirectional changes in response to chronic ethanol (75 mM) exposure were obtained in organotypic hippocampal slice cultures (n= 7). Values represent mean ± SEM, * p < 0.05.
The next set of studies examined the expression of these proteins in organotypic hippocampal slice cultures following chronic ethanol exposure. The purpose of these studies using organotypic cultures is to replicate the in-vivo observations so that follow-up mechanistic studies could be performed in this in-vitro model of chronic alcohol exposure. As shown in Figure 1B, chronic ethanol exposure resulted in nearly identical bidirectional changes observed as increases in GluN1 [t(10) = 2.997, p < 0.05, n = 6] and GluN2B [t(12) = 2.612, p < 0.05, n= 7], and reduced expression of Kv4.2 [t(12) = 2.174, p < 0.05, n = 7] and KChIP3 [t(10) = 3.016, p < 0.05, n = 6]. These observations using hippocampal slice cultures provides validation for use of this chronic ethanol exposure model for mechanistic studies described below that examine the role of FMRP in chronic ethanol-induced bidirectional changes in NMDA receptors and Kv4.2 channels.
Chronic ethanol enhances the phosphorylation status of FMRP
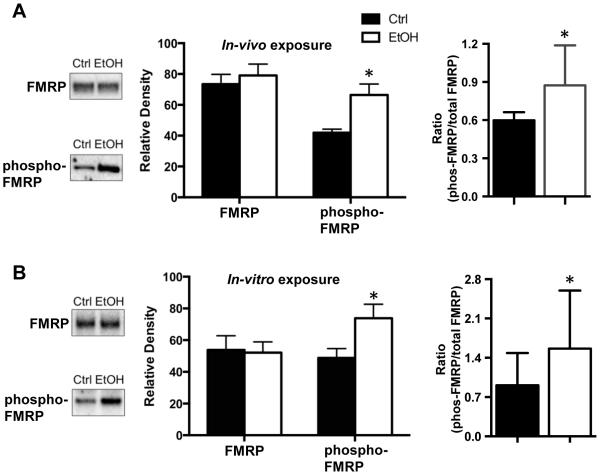
FMRP has been shown to regulate the expression of NMDA receptors, Kv4.2, and KChIP3 in the hippocampus, and may therefore play a critical role in the chronic ethanol-induced bidirectional changes in the expression of these proteins (Darnell et al., 2011, Lee et al., 2011, Shang et al., 2009). In the first set of studies, we examined the effect of both in-vivo (Figure 2A) and in-vitro (Figure 2B) chronic ethanol exposure on expression of FMRP in the hippocampus. This analysis revealed no changes in total FMRP expression following either in-vivo [t(35)=0.0997, p = 0.9211, n = 18,19] or in-vitro [t(5) = 0.7408, p = 0.4921, n = 6] ethanol exposure.
Figure 2.
Chronic ethanol exposure enhances phosphorylation of FMRP. (A) In the in-vivo mouse model of CIE exposure, the basal level of phospho-S499-FMRP was increased in the hippocampus of the ethanol exposed mice (n=21). In contrast, there was no difference between control and CIE in the level of total FMRP (n = 18-19). Expression of the data as a ratio of phospho-FMRP to total FMRP also revealed a significant increase (n = 14-16). (B) Similar chronic ethanol-induced (75 mM) increases in the level of phospho-FMRP and no change total FMRP were obtained in the in-vitro organotypic hippocampal slice culture model (n= 8). Expression of the data as a ratio of phospho-FMRP to total FMRP also revealed a significant increase (n = 6). Data represent mean ± SEM, * p < 0.05.
It has previously been shown that the translational activity of FMRP is critically dependent upon phosphorylation at the S449 residue that appears to promote inhibitory binding of the mRNA transcripts to FMRP and/or stall translation (Ceman et al., 2003, Darnell et al., 2011). Therefore, we used a phospho-specific antibody to determine whether chronic alcohol exposure alters the S499 phosphorylation status of FMRP independent of any changes in total FMRP expression. As shown in Figure 2A and 2B, this analysis revealed an increase in the level of phospho-FMRP following both in-vivo [t(40) = 2.382, p < 0.05, n = 21] and in-vitro [t(5) = 3.149, p < 0.05, n = 6] ethanol exposure. Expression of these results as a ratio of the levels of phospho-FMRP to total FMRP also reveal a significant increase with both in-vivo and in-vitro ethanol exposure compared to controls (one-tailed t-test, p < 0.05).
Chronic ethanol exposure bidirectionally changes mRNA binding to FMRP
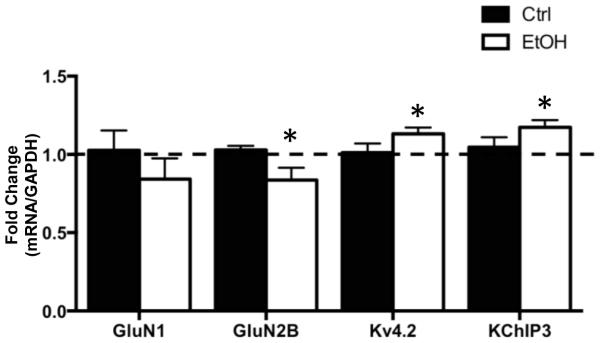
The next set of studies examined the effect of CIE on binding of FMRP to mRNA transcripts for GluN1, GluN2B, Kv4.2, and KChIP3 (Darnell et al., 2011, Gross et al., 2011, Lee et al., 2011). For these studies, a co-IP procedure was used to isolate FMRP, and then RT-qPCR to quantify mRNA transcripts that were pulled down with FMRP. Western blot analysis confirmed successful immmunoprecipitation of FMRP in both control and ethanol-exposed samples (not shown), and RT-qPCR confirmed that mRNA transcripts that code for GluN1, GluN2B, Kv4.2, and KChIP3 co-IP with FMRP (Figure 3). All experimental transcripts were normalized to GAPDH. As a negative control, examination of mRNA for HPRT (which is not a target of FMRP) revealed no measurable levels of this mRNA in the precipitate (data not shown), confirming the specificity of co-IP for mRNAs bound to FMRP (Darnell et al., 2011). Examination of the effect of chronic ethanol exposure on the mRNAs of interest revealed no change in GluN1 [t(7) = 0.9546, p = 0.3716, n = 8] but a significant reduction in the amount of GluN2B [t(8) = 2.402, p < 0.05, n = 9] mRNA. In contrast, chronic ethanol exposure was associated with a significant increase in FMRP binding of mRNA for Kv4.2 [t(8) = 2.340, p < 0.05, n = 9] and KChIP3 [t(7) = 2.561, p < 0.05, n = 6]. It is important to keep in mind that FMRP functions as an inhibitory regulator of protein translation via the sequestration of mRNA transcripts. Thus, all things being equal, an increase or decrease in mRNA bound to FMRP would result in a decrease or increase, respectively, in translation of those transcripts. Therefore, the observations of opposing changes in mRNA associated with FMRP are consistent with a bidirectional change in expression of NMDA receptors and Kv4.2 channels in response to chronic ethanol exposure.
Figure 3.
Bidirectional changes in NMDA receptors and Kv4.2 channels are associated with similar changes in mRNA binding to FMRP. FMRP was immunoprecipitated from homogenates of hippocampal tissue obtained from control and chronic ethanol (75 mM) exposed hippocampal slice cultures. RT-qPCR analysis of the mRNA transcripts in the immunoprecipitate revealed a significant reduction in GluN2B mRNA in CIE exposed slice cultures. In contrast, there was a significant increase in the amount of Kv4.2 and KChIP3 mRNA (n = 7-8). Data represent mean ± SEM, * p < 0.05.
S6K1 and phosphorylation of FMRP are required for chronic ethanol-induced bidirectional changes NMDA and Kv4.2
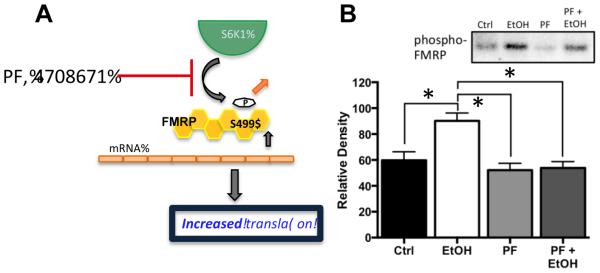
Previous studies have shown that S6K1 is the kinase that phosphorylates FMRP at the S499 site and promotes inhibition of protein translation by FMRP through the mTORC1 pathway (Bartley et al., 2014, Hoeffer et al., 2012). Therefore, the next set of studies examined the effect of inhibition of S6K1 on chronic ethanol-induced changes in FMRP phosphorylation in hippocampal slice cultures using the S6K1 inhibitor PF-4708671 (PF). As shown in Figure 4, while the inclusion of PF (6 μM) in the slice culture media under control conditions had no effect upon the baseline level of phospho-FMRP, co-incubation of PF and ethanol completely prevented chronic ethanol-induced increases in phospho-FMRP [F(3,12) = 6.174 p = 0.0062, Tukey post hoc, p < 0.05, n = 5]. This observation is consistent with a critical requirement for S6K1 in chronic ethanol-induced increases in FMRP phosphorylation.
Figure 4.
Inhibition of S6K1 prevents chronic ethanol-induced increases in phospho-FMRP. (A) Schematic representation of the effect of the S6K1 inhibitor PF-470861 (PF) on phosphorylation of FMRP at S499. Reduced phosphorylation of FMRP results in reduced inhibitory binding of mRNA and an increase in the pool of mRNA transcripts available for translation. (B) Immunoblot analysis revealed that while treatment of organotypic hippocampal cultures with PF (6 μM) had no effect on the phosphorylation status of FMRP at S499 under control conditions, it completely blocked the chronic ethanol-induced increase in phospho-FMRP (n = 5). Data represent mean ± SEM, ** p < 0.01.
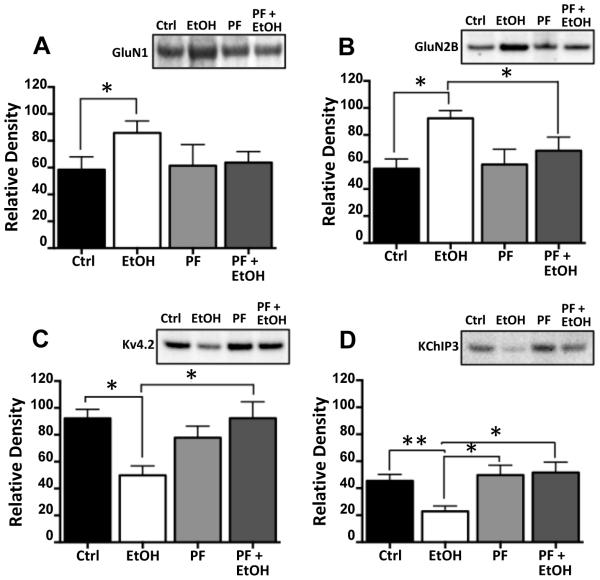
In the next set of studies, we examined the effect of PF on the bidirectional change in NMDA receptors and Kv4.2 in response to chronic ethanol exposure. Consistent with the observations in Figure 1, CIE exposure (Figure 5A-D) was again observed to increase expression of GluN1 [F(3,12) = 6.174 p = 0.0062, Tukey post hoc, p < 0.05, n = 5] and GluN2B [F(3,12) = 5.665 p = 0.0.0272, Tukey post hoc, p < 0.05, n = 5] and reduce expression of Kv4.2 [F(3,12) = 5.509 p = 0.0148, Tukey post hoc, p < 0.05, n = 5] and KChIP3 [F(3,12) = 6.174 p = 0.0.146, Tukey post hoc, p < 0.05, n = 5]. Importantly, co-incubation of PF with ethanol completely blocked the bidirectional changes in NMDA subunits, Kv4.2 and KChIP3 proteins. However, as was the case with phospho-FMRP, chronic exposure to PF had no effect on expression of these proteins in the absence of ethanol.
Figure 5.
Inhibition of S6K1 prevents ethanol-induced changes in protein expression. Hippocampal slice cultures were chronically exposed to ethanol in the presence and absence of the S6K1 inhibitor PF-470861 (PF, 6 μM). Immunoblot analysis revealed that while PF had no effect on the expression of any of the proteins examined (A-D), it completely prevented the increase in expression of GluN1 (A) and GluN2B (B), and the reduction in expression of Kv4.2 (C) and KChiP3 (D) induced by chronic ethanol exposure (n = 7-8). Data represent mean ± SEM, * p < 0.05, ** p < 0.01.
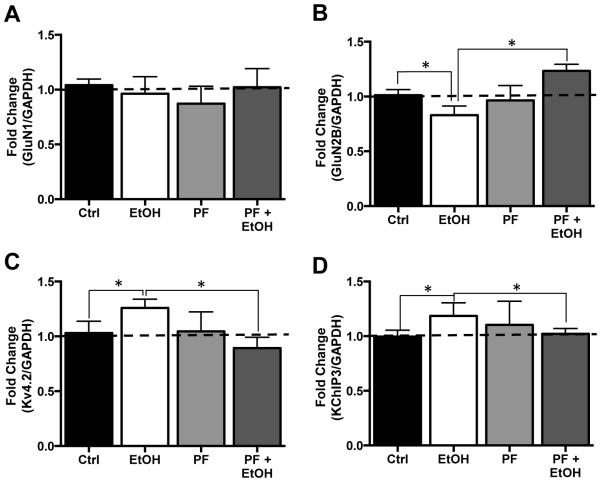
The final next set of studies examined the effect of inhibition of S6K1 on FMRP-mRNA binding. Analysis by RT-qPCR of the mRNA transcripts pulled down by FMRP again showed no change in GluN1 [F(3,12) = 0.3996 p = 0.7558, n = 5] but reduced GluN2B [F(3,15) = 4.232 p = 0.0235, Tukey post hoc, p < 0.05, n = 6] mRNA, and enhanced levels of Kv4.2 [F(3,12) = 3.630 p = 0.0351, Tukey post hoc, p < 0.05, n = 5] and KChIP3 [F(3,12) = 4.465 p = 0.0252, Tukey post hoc, p < 0.05, n = 5] mRNA bound to FMRP (Figure 6A-D). Consistent with the results of PF on FMRP phosphorylation (Figure 4) and protein expression (Figure 5), while chronic exposure of slice cultures to PF and no effect of FMRP-mRNA association in control cultures, it completely blocked the chronic ethanol-induced changes in GluN2, Kv4.2, and KChIP3 mRNA association with FMRP (Figure 6A-D). Taken together, these observations are consistent with the hypothesis that translational control by FMRP plays a critical role in chronic ethanol-induced bidirectional changes in expression of NMDA and Kv4.2 receptors in the hippocampus.
Figure 6.
Inhibition of S6K1 prevents ethanol-induced changes in binding of NMDA and Kv4.2 mRNA transcripts to FMRP. (A) In organotypic hippocampal slice cultures, neither ethanol, (75 mM), the S6K1 inhibitor PF-470861 (PF, 6 μM) alone or ethanol plus PF altered the binding of GluN1 mRNA to FMRP (n=6). (B) GluN2B mRNA binding to FMRP was reduced in response to chronic ethanol exposure alone while exposure to PK had no effect of GluN2B binding of FMRP. The effect of ethanol was blocked by the addition of PK, which resulted in a significant increase in GluN2B mRNA association with FMRP compared to control (Ctrl). In contrast, binding of mRNA coding for Kv4.2 (C) and KChIP3 (D) were significantly increased in cultures chronically exposed the ethanol. This increase was prevented by co-incubation with PK. Data represent mean ± SEM, * p < 0.05.
Discussion
Previous studies have reported alterations in plasticity of glutamatergic synapses in response to both in-vivo and in-vitro chronic ethanol exposure (Harris et al., 2003, Carpenter-Hyland et al., 2004). Many of these changes are thought to reflect homeostatic mechanisms that attempt to reestablish normal levels of synaptic activity and signaling in response to prolonged synaptic depression during chronic ethanol exposure (Chandler et al., 2006). However, in spite of demonstrations of chronic ethanol-induced glutamatergic plasticity, surprising little is known about the mechanisms that lead to changes in protein translation in dendrites and dendritic spines during chronic ethanol exposure. The results of the present study reveal a requirement for S6K1 signaling and FMRP in mediating the expression of homeostatic changes in response to chronic alcohol exposure.
In addition to homeostatic modulation of neuronal activity by changes in expression and function of excitatory and inhibitory neurotransmitter receptors (e.g. glutamate and GABA, respectively), excitability is also regulated by additional receptors and proteins that function to modulate local excitatory inputs and balance neuronal signaling. One prominent change in glutamatergic neurotransmission that has been observed in various brain regions and preparations in response to chronic ethanol exposure is an increase in the synaptic expression of GluN2B-containing NMDA receptors (Carpenter-Hyland et al., 2004, Hendricson et al., 2007). In the hippocampus, we recently demonstrated that chronic ethanol induced changes in GluN2B expression occur in parallel with reduction in Kv4.2 channel expression and function, and it was hypothesized these changes occur through a coupled bidirectional process (Mulholland et al., 2015). Additional studies have reported a reduction in Kv4.2 receptor expression in models of temporal lobe epilepsy, and have observed that increasing Kv4.2 function in the hippocampus can also inhibit NMDA-mediated hyperexcitability (Monaghan et al., 2008, Zhang et al., 2010). The auxiliary protein KChIP3 promotes Kv4.2 channel expression and is necessary for optimal Kv4.2 channel function, and has also been shown to directly interact with NMDA receptors to decrease their surface-expression and reduce NMDA currents (Shibata et al., 2003, Zhang et al., 2010). These observations are consistent with our previous findings that both Kv4.2 and KChIP3 are critical mediators of chronic ethanol-induced plasticity.
The mRNA-binding protein FMRP is a translational repressor that regulates activity-dependent dendritic translation and has been shown to play a critical role in basal-state homeostatic maintenance of proteins in dendrites and spines (Schutt et al., 2009, Dictenberg et al., 2008). FMRP has also been implicated in disorders such as temporal lobe epilepsy and Fragile X syndrome that are characterized by hyperexcitability and dysregulated protein homeostasis (Lee et al., 2011, Hagerman and Stafstrom, 2009). A deletion of FMRP or its dysfunction leads to increases in expression of downstream targets that include Kv4.2 and KChIP3, and loss of the ability of neurons to adapt and adjust protein synthesis in response to changes in neuronal activation (Lee et al., 2011, Gross et al., 2011). FMRP is a component of the mTORC1 pathway, and is regulated by the upstream kinase S6K1 (Gong et al., 2006, Hoeffer et al., 2012). In the nucleus accumbens and central amygdala, it has been reported that rats exposed to chronic ethanol exhibit increases in mTORC1 and S6K1 phosphorylation and activity (Barak et al., 2013, Wang et al., 2010). This increase in S6K1 activity is accompanied by an increase in proteins that promote glutamatergic signaling and whose mRNAs are frequent binding partners of FMRP, including CaMKII, Arc, and PSD-95 (Barak et al., 2013, Schutt et al., 2009, Bhattacharya et al., 2012). Observations in the present study are in agreement with reports that chronic ethanol alters FMRP dependent translation, and further suggest that FMRP dependent translation is, at least in part, a critical mediator of the homeostatic adaptive response to chronic ethanol exposure. Previous studies have shown that inhibition of S6K1 by the mTORC1 inhibitor rapamycin abolished expression of proteins that promote local excitatory signaling and also reduced ethanol-seeking behaviors (Barak et al., 2013, Neasta et al., 2014). The results of the present study further reveal that blockade of S499-FMRP phosphorylation through inhibition of S6K1 activity prevents chronic ethanol-induced changes in expression of key modulators of synaptic plasticity. These experiments highlight the significance of the S6K1 signaling and FMRP in ethanol-induced changes in protein expression in the hippocampus.
Although phosphorylation of FMRP can serve as a useful indicator of its functional activity, it is the binding of specific mRNAs to phosphorylated FMRP that dictates which proteins will be translationally regulated by FMRP. In the present study, we demonstrate that the chronic-ethanol induced increases in phospho-FMRP were indeed associated with selective increases in the binding of mRNA transcripts to FMRP. Specifically, we observed increased binding of FMRP to mRNAs coding for Kv4.2 and KChIP3. In studies of Fragile X syndrome, viral-mediated increases in FMRP protein in fmr1-/- mice prevent hyperexcitability in the hippocampus and return Kv4.2 protein expression to control levels (Lee et al., 2011, Gross et al., 2011). Of note, this decrease in protein expression is due to direct FMRP–Kv4.2 mRNA binding, and mutation of S499 residue of FMRP reduces the amount of Kv4.2 mRNA bound to FMRP (Lee et al., 2011). We further demonstrate that blocking the chronic ethanol-induced increase in S499-FMRP phosphorylation is associated with a reduction in the binding KChIP3 and Kv4.2 mRNA to FMRP. Taken together, these data indicate that increases in FMRP phosphorylation and changes in FMRP-mRNA interactions are an important regulatory component of ethanol-induce plasticity.
Previous studies examining the effect of FMRP on GluN1 expression have yielded mixed results. Developmentally, fmr1-/- show decreased NMDA receptor-mediated current in the CA1 during LTP along with a decrease in expression of GluN1, GluN2B, and GluN2A (Shang et al., 2009, Zhang et al., 2009). Adult mice, however, show no changes in basal GluN1 levels, but still display impaired LTP (Wang et al., 2014, Shang et al., 2009). Other studies in knockout animals suggest different signaling pathways (e.g. ERK/MNK) may have a greater influence over GluN1 expression (Panja and Bramham, 2014, De Rubeis and Bagni, 2011). Clinical studies on post-mortem brains of individuals with co-morbid Fragile X syndrome and temporal lobe epilepsy indicate that although there is dysregulation of NMDA receptor signaling, other downstream targets such as K+ channels may contribute more to FMRP-mediated hyperexcitability than NMDA receptors (Hagerman and Stafstrom, 2009, Nicholas et al., 2010). These findings suggest that FMRP may ‘fine tune’ NMDA-mediated signaling by regulating translation of receptor subunits that affect channel function, and by altering expression of other proteins regulating local excitability in spines, such as Kv4.2 and KChIP3, that suppress excitation in the hippocampus.
Results of the present study also confirm and extend previous observations that chronic ethanol exposure induces homeostatic changes in plasticity related proteins observed as an increase in expression of GluN2B-containing NMDA receptors and reduction in expression of Kv4.2 and KChIP3 (Carpenter-Hyland et al., 2004, Chandler et al., 2006, Mulholland et al., 2015). These changes were found to correlate with a decrease and increase, respectively, in the binding of their mRNA transcripts to FMRP. However, while these observations support the suggestion that FMRP plays an important role in mediating chronic ethanol-induce changes at glutamatergic synapses, they also suggest that other mechanisms likely play an important role and impart specificity. For example, all things being equal, an increase in phosphorylation of FMRP at S499 would be expected to increase the inhibitory binding of GluN2B mRNA, yet the opposite was observed. FMRP contains distinct binding regions that interact with specific RNAs or proteins (Ascano et al., 2012). Along with alterations in phosphorylation, other regulatory components, such as cytoplasmic FMRP interacting proteins and micro-RNAs, bind to FMRP to alter its confirmation and modulate which FMRP binding regions are available for interaction with proteins or mRNAs (Ascano et al., 2012, Chen and Joseph, 2015, Napoli et al., 2008, Edbauer et al., 2010). This suggests that an additional factor(s) are involved that differentially regulate FMRP binding of mRNA transcripts. Regardless, the observation that the ethanol-induced changes in mRNA-FMRP binding and protein expression were prevented by inhibition of S6K1 mediated phosphorylation of FMRP strongly implicates a critical role for FMRP in chronic ethanol-induced homeostatic plasticity. These findings also add to the current literature that local translational control by FMRP in dendrites and dendritic spines induce homeostatic mechanisms to balance neuronal signaling that affect not only glutamatergic function, but have larger behavioral implications in ethanol-induced neuronal plasticity.
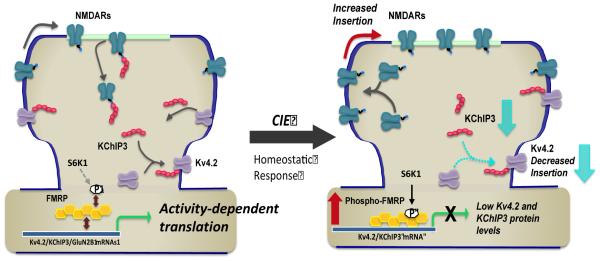
Figure 7.
Model of chronic ethanol induced homeostatic bidirectional changes in expression of NMDA and Kv4.2 channels, and the role of KChIP3 and FMRP in this activity-dependent process. Left panel depicts regulation of the surface expression of NMDA receptors and Kv4.2 via a coupled process involving the multifunctional protein KChiP3. As an accessory protein to Kv4.2, KChiP3 association with Kv4.2 promotes it’s trafficking and enhances its surface expression. In contrast, it functions as a negative modulator of NMDA receptor function and surface expression. The activity dependent expression of all these proteins is controlled by FMRP, which functions as a negative regulator of translation. Right panel depicts homeostatic adaptations in response to chronic attenuation of synaptic activity associated with alcohol exposure. These changes include a increase in the surface expression of GluN2B-containing NMDA receptors and reduction in the surface expression of Kv4.2 channels. This bidirectional change may be mediated, at least in part, by reduction in expression of KChIP3. In addition, these homeostatic changes are dependent on enhanced phosphorylation of S499 of FMRP (via S6K1) that in turn promotes inhibitory binding of mRNA transcripts including Kv4.2 and KChIP3 mRNA. While ethanol exposure is also associated with a reduction in GluN2B binding to FMRP, the mechanism responsible for this reduction is not clear and likely involves additional regulatory processes.
Acknowledgements
This work was funded by NIH grants P50AA010761 (LJC), and R01AA022701 (LJC), R01AA020930 (PJM) and F31AA022032 (KBS)
References
- An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, Rhodes KJ. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- Andrasfalvy BK, Makara JK, Johnston D, Magee JC. Altered synaptic and non-synaptic properties of CA1 pyramidal neurons in Kv4.2 knockout mice. J Physiol. 2008;586:3881–3892. doi: 10.1113/jphysiol.2008.154336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, Tuschl T. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak S, Liu F, Ben Hamida S, Yowell QV, Neasta J, Kharazia V, Janak PH, Ron D. Disruption of alcohol-related memories by mTORC1 inhibition prevents relapse. Nat Neurosci. 2013;16:1111–1117. doi: 10.1038/nn.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley CM, O’Keefe RA, Bordey A. FMRP S499 Is Phosphorylated Independent of mTORC1-S6K1 Activity. PLoS ONE. 2014;9:e96956. doi: 10.1371/journal.pone.0096956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HC, Hale RL. Repeated Episodes of Ethanol Withdrawal Potentiate the Severity of Subsequent Withdrawal Seizures: An Animal Model of Alcohol Withdrawal “Kindling”. Alcoholism: Clinical and Experimental Research. 1993;17:94–98. doi: 10.1111/j.1530-0277.1993.tb00731.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Kaphzan H, Alvarez-Dieppa AC, Murphy JP, Pierre P, Klann E. Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron. 2012;76:325–337. doi: 10.1016/j.neuron.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorso CM, Spatuzza M, Di Marco B, Gloria A, Barrancotto G, Cupo A, Musumeci SA, D'Antoni S, Bardoni B, Catania MV. Fragile X mental retardation protein (FMRP) interacting proteins exhibit different expression patterns during development. Int J Dev Neurosci. 2015;42:15–23. doi: 10.1016/j.ijdevneu.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Cai X, Liang CW, Muralidharan S, Kao JP, Tang CM, Thompson SM. Unique roles of SK and Kv4.2 potassium channels in dendritic integration. Neuron. 2004;44:351–364. doi: 10.1016/j.neuron.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Carpenter-Hyland EP, Woodward JJ, Chandler LJ. Chronic ethanol induces synaptic but not extrasynaptic targeting of NMDA receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:7859–7868. doi: 10.1523/JNEUROSCI.1902-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo Y, Burkhalter A, Nerbonne JM. A-type K+ channels encoded by Kv4.2, Kv4.3 and Kv1.4 differentially regulate intrinsic excitability of cortical pyramidal neurons. J Physiol. 2012;590:3877–3890. doi: 10.1113/jphysiol.2012.229013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Chandler LJ, Carpenter-Hyland E, Hendricson AW, Maldve RE, Morrisett RA, Zhou FC, Sari Y, Bell R, Szumlinski KK. Structural and functional modifications in glutamateric synapses following prolonged ethanol exposure. Alcohol Clin Exp Res. 2006;30:368–376. doi: 10.1097/01.ALC.0000167959.84516.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Joseph S. Fragile X mental retardation protein: A paradigm for translational control by RNA-binding proteins. Biochimie. 2015;114:147–154. doi: 10.1016/j.biochi.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, Bagni C. Regulation of molecular pathways in the Fragile X Syndrome: insights into Autism Spectrum Disorders. J Neurodev Disord. 2011;3:257–269. doi: 10.1007/s11689-011-9087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eadie BD, Cushman J, Kannangara TS, Fanselow MS, Christie BR. NMDA receptor hypofunction in the dentate gyrus and impaired context discrimination in adult Fmr1 knockout mice. Hippocampus. 2012;22:241–254. doi: 10.1002/hipo.20890. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez E, Rajan N, Bagni C. The FMRP regulon: from targets to disease convergence. Front Neurosci. 2013;7:191. doi: 10.3389/fnins.2013.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong R, Park CS, Abbassi NR, Tang SJ. Roles of glutamate receptors and the mammalian target of rapamycin (mTOR) signaling pathway in activity-dependent dendritic protein synthesis in hippocampal neurons. J Biol Chem. 2006;281:18802–18815. doi: 10.1074/jbc.M512524200. [DOI] [PubMed] [Google Scholar]
- Gross C, Yao X, Pong DL, Jeromin A, Bassell GJ. Fragile X mental retardation protein regulates protein expression and mRNA translation of the potassium channel Kv4.2. J Neurosci. 2011;31:5693–5698. doi: 10.1523/JNEUROSCI.6661-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman PJ, Stafstrom CE. Origins of Epilepsy in Fragile X Syndrome. Epilepsy Currents. 2009;9:108–112. doi: 10.1111/j.1535-7511.2009.01309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris BR, Gibson DA, Prendergast MA, Blanchard JA, Holley RC, Hart SR, Scotland RL, Foster TC, Pedigo NW, Littleton JM. The neurotoxicity induced by ethanol withdrawal in mature organotypic hippocampal slices might involve cross-talk between metabotropic glutamate type 5 receptors and N-methyl-D-aspartate receptors. Alcohol Clin Exp Res. 2003;27:1724–1735. doi: 10.1097/01.ALC.0000093601.33119.E3. [DOI] [PubMed] [Google Scholar]
- Hendricson AW, Maldve RE, Salinas AG, Theile JW, Zhang TA, Diaz LM, Morrisett RA. Aberrant synaptic activation of N-methyl-D-aspartate receptors underlies ethanol withdrawal hyperexcitability. J Pharmacol Exp Ther. 2007;321:60–72. doi: 10.1124/jpet.106.111419. [DOI] [PubMed] [Google Scholar]
- Hoeffer CA, Sanchez E, Hagerman RJ, Mu Y, Nguyen DV, Wong H, Whelan AM, Zukin RS, Klann E, Tassone F. Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile X syndrome. Genes Brain Behav. 2012;11:332–341. doi: 10.1111/j.1601-183X.2012.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SC, Kim J, Hoffman DA. Rapid, bidirectional remodeling of synaptic NMDA receptor subunit composition by A-type K+ channel activity in hippocampal CA1 pyramidal neurons. Neuron. 2008;60:657–671. doi: 10.1016/j.neuron.2008.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WA, Matsui K, Jeromin A, Nerbonne JM, Ferraguti F. Kv4.2 potassium channels segregate to extrasynaptic domains and influence intrasynaptic NMDA receptor NR2B subunit expression. Brain structure & function. 2013;218:1115–1132. doi: 10.1007/s00429-012-0450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jung SC, Clemens AM, Petralia RS, Hoffman DA. Regulation of dendritic excitability by activity-dependent trafficking of the A-type K+ channel subunit Kv4.2 in hippocampal neurons. Neuron. 2007;54:933–947. doi: 10.1016/j.neuron.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Ge WP, Huang W, He Y, Wang GX, Rowson-Baldwin A, Smith SJ, Jan YN, Jan LY. Bidirectional regulation of dendritic voltage-gated potassium channels by the fragile X mental retardation protein. Neuron. 2011;72:630–642. doi: 10.1016/j.neuron.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Deng P, Li Y, Xu ZC. Downregulation of Kv4.2 channels mediated by NR2B-containing NMDA receptors in cultured hippocampal neurons. Neuroscience. 2010;165:350–362. doi: 10.1016/j.neuroscience.2009.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercaldo V, Descalzi G, Zhuo M. Fragile X mental retardation protein in learning-related synaptic plasticity. Mol Cells. 2009;28:501–507. doi: 10.1007/s10059-009-0193-x. [DOI] [PubMed] [Google Scholar]
- Monaghan MM, Menegola M, Vacher H, Rhodes KJ, Trimmer JS. Altered expression and localization of hippocampal A-type potassium channel subunits in the pilocarpine-induced model of temporal lobe epilepsy. Neuroscience. 2008;156:550–562. doi: 10.1016/j.neuroscience.2008.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland P, Spencer K, Hu W, Kroener S, Chandler LJ. Neuroplasticity of A-type potassium channel complexes induced by chronic alcohol exposure enhances dendritic calcium transients in hippocampus. Psychopharmacology. 2015;232:1995–2006. doi: 10.1007/s00213-014-3835-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Neasta J, Barak S, Hamida SB, Ron D. mTOR complex 1: a key player in neuroadaptations induced by drugs of abuse. J Neurochem. 2014;130:172–184. doi: 10.1111/jnc.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas A, de Magalhaes JP, Kraytsberg Y, Richfield EK, Levanon EY, Khrapko K. Age-related gene-specific changes of A-to-I mRNA editing in the human brain. Mech Ageing Dev. 2010;131:445–447. doi: 10.1016/j.mad.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja D, Bramham CR. BDNF mechanisms in late LTP formation: A synthesis and breakdown. Neuropharmacology. 2014;76:664–676. doi: 10.1016/j.neuropharm.2013.06.024. Pt C. [DOI] [PubMed] [Google Scholar]
- Rhodes KJ, Carroll KI, Sung MA, Doliveira LC, Monaghan MM, Burke SL, Strassle BW, Buchwalder L, Menegola M, Cao J, An WF, Trimmer JS. KChIPs and Kv4 alpha subunits as integral components of A-type potassium channels in mammalian brain. J Neurosci. 2004;24:7903–7915. doi: 10.1523/JNEUROSCI.0776-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffary R, Xie Z. FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J Neurosci. 2011;31:1427–1439. doi: 10.1523/JNEUROSCI.4854-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt J, Falley K, Richter D, Kreienkamp HJ, Kindler S. Fragile X mental retardation protein regulates the levels of scaffold proteins and glutamate receptors in postsynaptic densities. J Biol Chem. 2009;284:25479–25487. doi: 10.1074/jbc.M109.042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Wang H, Mercaldo V, Li X, Chen T, Zhuo M. Fragile X mental retardation protein is required for chemically-induced long-term potentiation of the hippocampus in adult mice. J Neurochem. 2009;111:635–646. doi: 10.1111/j.1471-4159.2009.06314.x. [DOI] [PubMed] [Google Scholar]
- Shibata R, Misonou H, Campomanes CR, Anderson AE, Schrader LA, Doliveira LC, Carroll KI, Sweatt JD, Rhodes KJ, Trimmer JS. A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J Biol Chem. 2003;278:36445–36454. doi: 10.1074/jbc.M306142200. [DOI] [PubMed] [Google Scholar]
- Trantham-Davidson H, Burnett EJ, Gass JT, Lopez MF, Mulholland PJ, Centanni SW, Floresco SB, Chandler LJ. Chronic Alcohol Disrupts Dopamine Receptor Activity and the Cognitive Function of the Medial Prefrontal Cortex. The Journal of Neuroscience. 2014;34:3706–3718. doi: 10.1523/JNEUROSCI.0623-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Luo Y-x, He Y-y, Li F-q, Shi H-s, Xue L-f, Xue Y-x, Lu L. Nucleus Accumbens Core Mammalian Target of Rapamycin Signaling Pathway Is Critical for Cue-Induced Reinstatement of Cocaine Seeking in Rats. The Journal of Neuroscience. 2010;30:12632–12641. doi: 10.1523/JNEUROSCI.1264-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Peng CZ, Cai WJ, Xia J, Jin D, Dai Y, Luo XG, Klyachko VA, Deng PY. Activity-dependent regulation of release probability at excitatory hippocampal synapses: a crucial role of fragile X mental retardation protein in neurotransmission. Eur J Neurosci. 2014;39:1602–1612. doi: 10.1111/ejn.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Kovacs I, Zhang Z. Differential distribution of KChIPs mRNAs in adult mouse brain. Brain Res Mol Brain Res. 2004;128:103–111. doi: 10.1016/j.molbrainres.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Yun SH, Trommer BL. Fragile X mice: reduced long-term potentiation and N-Methyl-D-Aspartate receptor-mediated neurotransmission in dentate gyrus. J Neurosci Res. 2011;89:176–182. doi: 10.1002/jnr.22546. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hou L, Klann E, Nelson DL. Altered hippocampal synaptic plasticity in the FMR1 gene family knockout mouse models. J Neurophysiol. 2009;101:2572–2580. doi: 10.1152/jn.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su P, Liang P, Liu T, Liu X, Liu XY, Zhang B, Han T, Zhu YB, Yin DM, Li J, Zhou Z, Wang KW, Wang Y. The DREAM protein negatively regulates the NMDA receptor through interaction with the NR1 subunit. J Neurosci. 2010;30:7575–7586. doi: 10.1523/JNEUROSCI.1312-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]