Abstract
Background
Laterality in the vertebrate embryo is determined by left–right asymmetric gene expression driven by the flow of extraembryonic fluid across the embryonic node. Defects in these processes cause heterotaxy, the abnormal formation and arrangement of visceral organs that can range from complete inversion of symmetry to the selective misarrangement of organs. However, our understanding of the genetic causality for laterality defects in human beings remains relatively limited.
Methods
We performed whole exome sequencing in a consanguineous family with heterotaxia. To interrogate the pathogenic potential of the discovered variant, we used an in vivo system in which the potential of the candidate gene to induce L-R asymmetry was tested by transient suppression and CRISPR/Cas9-induced deletions. We also used in vitro assays to test a possible link between our exome-derived candidate and Notch signaling.
Results
We identified a homozygous 2 bp deletion in MMP21, encoding matrix metalloproteinase-21, as the sole coding mutation that segregated with the phenotype. Transient suppression or CRISPR/Cas9-mediated deletion of mmp21 in zebrafish embryos induced cardiac looping defects, with concomitant disruption of laterality markers in the lateral plate mesoderm and disrupted notch signalling in vitro and in vivo.
Conclusions
Our data implicate loss of MMP21 as a cause of heterotaxy in humans with concomitant defects in Notch signaling. In support of this finding, a homozygous missense mutation in MMP21 was identified previously in mice with N-Ethyl-N-Nitrosourea (ENU)-induced heterotaxy. Taken together, these observations suggest a role of matrix metalloproteinases in the establishment of asymmetric organ development, likely through the regulation of morphogenetic signals.
INTRODUCTION
Defects during the establishment of left–right (L–R) asymmetry lead to heterotaxy, occurring in approximately one in 8000–25 000 live births.1 Derived from Greek words (hetero, ‘different’, and taxy, ‘arrangement’), this condition is hallmarked by misplacement of the visceral organs across the L–R axis of the body. In vertebrates, the establishment of L–R asymmetry is a conserved process initiated by breaking symmetry at an organising structure, consisting of an embryonic cavity at the midline that is filled with extraembryonic fluid.2 Although named differently across vertebrate taxa, this structure has been observed in many species such as mouse (node), rabbit (Hensen’s node) and zebrafish (Kupffer’s vesicle, KV). The KV in the zebrafish embryo contains cells with motile cilia that generate an anticlockwise fluid flow. In most vertebrates, nodal cilia are tilted towards the posterior axis.3,4 This posterior tilt is considered to be an important element for generation of the leftward unidirectional flow at the node, which is essential for proper L–R asymmetry.5,6 In concordance with the role of nodal cilia in L–R asymmetry, mutations in the motor proteins Kif3A and Kif3B, and the transcription factor Foxj1 in the mouse have demonstrated that ciliary motility is important for L–R asymmetry.5,7,8
Once a break of symmetry is initiated, leftward flow induces the asymmetric distribution of Nodal (southpaw, in zebrafish) in the lateral plate mesoderm (LPM) where Lefty and Pitx2 are subsequently expressed.9 Nodal, identified originally as a gene for normal mouse gastrulation and embryonic development,10 is a transforming growth factor β superfamily factor. Both Nodal and southpaw (spaw) are initially expressed bilaterally around the node/KV.11 Inhibited by Cerl2 on the right side, Nodal activity is then transmitted exclusively to the left side of the LPM,12 leading to asymmetrical activation of genes such as Lefty and Pitx2, to mediate asymmetric organogenesis of the heart and other visceral organs. Nodal activity is restricted to the left side of the LPM by a negative feedback loop of its target genes, Lefty1 and Lefty2, determining leftness. In zebrafish, engineered mutants or targeted knockdown using morpholinos have broadened our understanding of the mechanisms of laterality establishment. For example, knockdown of spaw leads to defects in heart looping as well as abnormal expression of pitx2, lefty1 and lefty2 in the LPM.13
The proper organ asymmetry across the L–R axis is referred to as situs solitus. Perturbation of this asymmetry causes either situs inversus totalis (SIT), an often asymptomatic condition, or situs ambiguous (heterotaxia), leading to complex cardiovascular malformations such as transposition of the great arteries (TGA), double outlet right ventricle, double inlet left ventricle, atrioventricular septal defects and total anomalous pulmonary venous connection. This disorder is often associated with anomalies of the spleen and the gastrointestinal system, such as asplenia or polysplenia, and bowel malrotation, which may lead to bowel obstruction.14,15
Heterotaxy is typically diagnosed in the newborn period due to cyanotic congenital heart disease.16 Mutations in components of Nodal signaling, including NODAL itself, as well as ACVR2B, LEFTYA, CFC1 and FOXA2 have been reported.17–22 Moreover, mutations in other laterality-related genes, such as CFC1 (a NODAL cofactor) and FOXH1, have been associated with complex cardiovascular malformations.23,24 Nonetheless, despite progress in identifying genetic lesions that contribute to heterotaxy and L–R asymmetry defects, our understanding of the genetic causality for laterality defects in human beings remains relatively limited.
Here, we report a recessive mutation in MMP21 that causes laterality defects in multiple affected members of a consanguineous family; in support of this finding, we show that transient knockdown, CRISPR/Cas9-mediated generation of mosaic F0 mutants or generation of germline F1 mutants targeting the sole zebrafish mmp21 ortholog produces similar phenotypes to that observed in our patients, and we propose excessive Notch signalling as one candidate driver of this pathology.
METHODS
Whole exome sequencing and analysis
Exonic sequences were enriched with the SureSelect Human All Exon 50 Mb Kit (Agilent Technologies, Santa Clara, California, USA). Sequences (100 bp paired-end) were generated on a HiSeq2000 (Illumina, San Diego, California, USA). Read alignment and variant calling were performed with DNAnexus (Palo Alto, California, USA) using default parameters with the human genome assembly hg19 (GRCh37) as reference. Parental consent was given for genetic studies. The study was performed with the approval of the ethical committees of Hadassah Medical Center and the Israeli Ministry of Health.
Morpholino injections and RT-PCR
Zebrafish (Danio rerio) embryos were raised and maintained as described.25 The AB and ZDR strains were used in this study. A splice blocking (SB) morpholino (MO, Gene Tools) was designed to target the splicing donor site (exon 3/intron 3) of mmp21 (5′-AAATGTGCGATTTAAAACCTGTGCA-3′) based on the Zebrafish genome database (Zv9). The mmp21 splice-blocking morpholino (SB-MO) was injected into the yolk of zebrafish embryos at one-cell to four-cell stage as described.26 RT-PCR of cDNA generated from whole embryos followed by Sanger sequencing was carried out to determine SB-MO efficiency. Total RNA was extracted from 2 days post fertilisation (dpf) embryos with TRIzol Reagent (Life Technologies), followed by DNase treatment. First-strand cDNA was synthesised with Superscript III with oligo(dT) primer, and PCR amplification (72°C, 5 min extension) was performed (Forward primer, 5′-TCCACAGCAGAGACCATTCA-3′ and Reverse, 5′-AGCACACTTTTGGAGAAGGC-3′). For real-time qPCR, cDNA was applied with Power SYBR Green PCR Master Mix (Applied Biosystems) onto a 7900HT Fast Real-Time PCR System (Applied Biosystems). Primer sequence is available on request. Real-time data were collected and analysed with the Sequence Detection System software package V.2.3 (Applied Biosystems). Sequence information of primers used for qPCR is available on request.
Generation of mmp21 mutants by CRISPR/CAS9
A guide RNA was generated as described.27 Cloning of a guide RNA template was initiated by annealing two oligonucleotides (Forward, 5′-TAGGCGGAGCTGATCACTGACA-3′ and Reverse, 5′-AAACTGTCAGTGATCAGCTCCG-3′) and ligated into Bsm BI site of the pT7Cas9sgRNA vector (Addgene). To generate the guide RNA, template DNA was linearised with Bam HI, and purified by phenol/chloroform extraction. The guide RNA was transcribed in vitro with MEGAshortscript T7 kit (Invitrogen). Subsequently, 100 pg of guide RNA and 200 ng of Cas9 protein (PNA bio, CP01) were injected directly into one-cell stage embryos. To test the efficiency of the guide RNA, genomic DNA from each embryo (F0) was prepared by proteinase K digestion (Life technologies, AM2548). By using a primer set (Forward, 5′-CCAGACCTTCATCTTCATCACA-3 and Reverse, 5′-AAAACAAAATGTCCAATGCAAA-30) flanking the targeted genomic editing site; the genomic region was amplified, followed by denaturing and slow reannealing to facilitate heteroduplex formation (denaturing step: 5 min, followed by reannealing step: cooling to 85°C at −2°C/s and additional to 25°C at −0.1°C/s). The reannealed PCR amplicon was subject to digestion by T7 endonuclease I (New England Biolabs, M0302L) at 37°C for 1 h, and was visualised on 1.5% of agarose gel. For Sanger sequencing of individual products from the mmp21 locus, PCR fragments were cloned into the pCR4/TOPO TA cloning vector (Life technologies, 450030), and each clone was Sanger sequenced.
RNA in situ hybridisation
cmlc2 and spaw riboprobes (kind gift from Drs Kenneth D Poss and Michel Bagnat, Duke university). For generating a riboprobe against zebrafish mmp21, PCR-amplified product using primer set (Forward 5′-GACACTGCACAGCGTTTTCT-3′, Reverse 5′-TACACACTCCGTACAGCCTC-3′) was synthesised with the digoxigenin (DIG) RNA Labeling Kit, following the manufacturer’s instruction. Whole-mount in situ hybridisation was performed as described.28 Briefly, embryos were fixed overnight at 4°C in 4% paraformaldehyde (PFA), followed by overnight methanol fixation at −20°C. Fixed embryos were washed by serial dilution of methanol and an additional wash with PBST containing 0.1% tween 20, followed by proteinase K for 30 s for 23 somite stage embryo and 10 min for 1 dpf embryo. Followed by postfixation by PFA, samples were prehybridised (50% formamide, 5× SSC, 50 µg/mL heparin, 50 µg/mL tRNA and 10 mM citric acid) at 60°C for 1 h. After overnight hybridisation with a DIG-labelled riboprobe at 60°C, samples were washed several times with 2× SSC and 0.1× SSC at 60°C for 10–20 min each. The sample was then incubated with AP-conjugated anti-DIG antibody (Roche) for 2 h at room temperature. NBT/BCIP (Roche) was used as the chromogenic substrate. Embryos were imaged with Nikon NIS Elements software; statistical analyses were performed by comparing embryo batches with a χ2 test.
Luciferase assay
Luciferase reporter assays were carried out as described.29 HEK-293-FT cells were seeded in 24-well plates at a density of 1×104 cells/well. After 24 h, cells were transfected with shorthairpin plasmids (Sigma, TRCN0000050933 for MMP21), and a CBF-luciferase reporter (a kind gift from Dr Nicholas Gaiano, Johns Hopkins University). A pRL-SV40 plasmid expressing Renilla luciferase was used as an internal control. At 72 h post transfection, cells were lysed with Passive Lysis Buffer (Promega). The luciferase activity of lysates was measured with the Dual Luciferase Reporter Assay System (Promega) on a FLUOstar Omega microplate reader (BMG LABTECH), and was analysed with the MARS Data Analysis Software (BMG LABTECH). Statistical analyses were performed with a student’s t test.
Western blotting
Transfected cells were lysed in modified RIPA buffer (150 mM sodium chloride, 50 mM Tris-HCl, pH7.4, 1% nonidet P-40, 0.1% sodium deoxycholate, 1 mM EDTA) with 1× proteasome inhibitor (Roche) and centrifuged at 4°C for 15 min. Equal amounts of protein in each sample was resolved by SDS-PAGE on 4%–15% Mini-PROTEAN TGX Precast Gel (Bio-Rad) and transferred to an Immuno-Blot PVDF Membrane (Bio-Rad). Then the membrane was blocked with 5% non-fat milk and incubated with anti-GFP (1:1000, Roche, 11814460001) and antiactin (1:1000, sc-1615, Santa Cruz Biotech) antibodies. After washing with PBST three times for 100 each, horseradish peroxidase (HRP)-conjugated antigoat IgG (1:2000, sc-2020, Santa Cruz Biotech) and antimouse IgG (1:2000, GE Healthcare, LNA931V/AG) were applied. Images were taken with Image Lab (Bio-Rad).
RESULTS
Patients
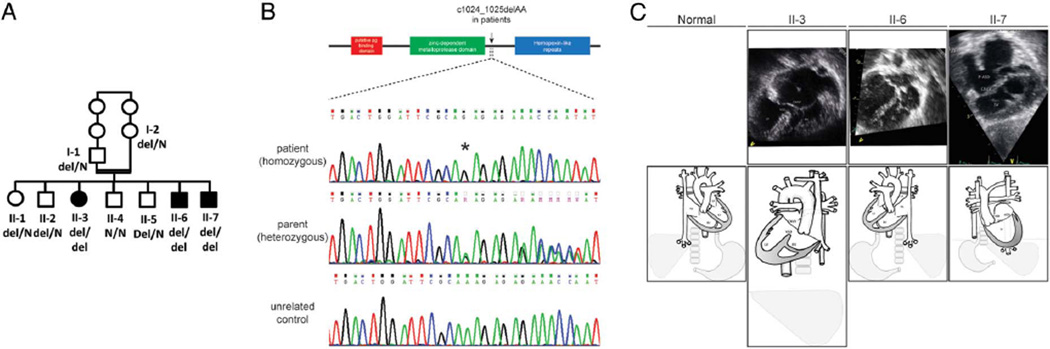
The index patient, patient II-3, was the third of seven children born to a consanguineous Arab couple (figure 1A). At 3 months of age, due to cyanotic events, she was diagnosed with {I,L,L} dextrocardia with atrial situs inversus, complete atrioventricular canal defect, TGA and pulmonary atresia with a duct-like aortopulmonary collateral (figure 1B). At 8 years of age, she underwent a palliative Blalock–Taussig shunt due to worsening cyanosis, with subsequent improvement in exercise tolerance and reduction in frequency and severity of cyanotic episodes. At the time of reporting, age 23, she is treated with penicillin prophylaxis due to her asplenia and with aspirin to keep the shunt patency, but is extremely limited in terms of physical activity.
Figure 1.
Family pedigree and the MMP21 mutation. (A) A family pedigree. Patients are denoted by filled symbols with the genotype of the MMP21 c.1024_1025delAA. (B) Schematic of the MMP21 protein and Sanger sequencing results. MMP21 consists of a putative peptidoglycan (pg) binding domain (red), a zinc-dependent metalloprotease domain (green) and hemopexin-like repeats (blue). The mutation site is indicated by an arrow. Sanger sequencing results showed the mutation site (asterisk) in patients (upper panel), parent (middle panel) and an unrelated control (lower panel). (C) Echocardiogram and schematic representation of the heart malformations. Patient II-3: {I,L,L} dextrocardia with atrial situs inversus, complete atrioventricular canal defect, transposition of the great arteries and pulmonary atresia with a duct-like aortopulmonary collateral midline liver. The echocardiogram from the subcostal view (top) is aligned with the diagram (bottom). Patient II-6: {I,L,I} situs inversus totalis of the heart and visceral organs. The echocardiogram from the subcostal view (top) is aligned with the diagram (bottom). Patient II-7: {A(I),D,D} visceral and atrial situs inversus with bilateral superior vena cavas, complete atrioventricular canal defect and transposition of the great vessels with pulmonary atresia, after performance of bilateral bidirectional cavopulmonary communication. The echocardiogram from the apical four-chamber view (top) is aligned with the diagram (bottom). CAVV, common atrioventricular valve; LA, left atrium; LV, left ventricle; P-ASD, primum atrial septal defect; RA, right atrium; RV, right ventricle; SV, single ventricle; VSD, ventricular septal defect.
Her brother, patient II-7, was born after uneventful pregnancy, birth weight 2880 grams. Severe cyanosis was observed immediately after birth with a 4/6 continuous systolic and diastolic murmur over the left upper sternal border. An echocardiogram revealed {A(I),D,D}, visceral and atrial situs inversus with bilateral superior vena cava, a complete atrioventricular canal defect and transposition of the great vessels with pulmonary atresia. He underwent Blalock–Taussig shunt palliation and, at 2 years of age, a bilateral bidirectional Glenn shunt (figure 1B). At age 4 years, a seizure disorder was noted. The brain CT scan was normal, but electroencephalogram revealed a right temporo-occipital epileptic focus. He was started on carbamazepine with a favourable response. Currently at age 10, his psychomotor development is age appropriate, and he is capable of moderate exercise.
None of the sibs had a history of recurrent lung infections, chronic sinusitis, otitis media or nasal polyps; nasal NO screening performed in the exhalation against resistance technique (velum closure breath hold) was normal for the affected patients II-3 and II-7.
Whole exome sequencing and genetic analysis of variants
Given the suspected genetic basis of heterotaxy in this consanguineous family, we performed whole exome sequencing (WES) under a rare allele, recessively inherited hypothesis. WES of the index patient II-7 yielded 58.63 million mapped reads with a mean coverage of 69×. Following alignment and variant calling, we performed a series of filtering steps. These included removing variants called less than X8, were off-target, heterozygous, synonymous, on the chromosome X, had minor allel frequency (MAF) >1% at ExAC (Exome Aggregation Consortium, Cambridge, Massachusetts, USA (URL: http://exac.broadinstitute.org)) and MAF >4% in the Hadassah in-house dbSNP.30 Thirteen variants survived this process (see online supplementary table S1), and we examined by Sanger sequencing those predicted to affect a residue with a high conservation score (Genomic Evolutionary Rate Profiling). There was a single variant that survived segregation in the parents and the second affected individual: the parents were heterozygous, and patient II-3 was homozygous for a 2 bp deletion in MMP21, which is predicted to induce a premature termination codon (chr10: 127459115 delAA, NM_147191: c.1024_1025delAA, p. K342fs). Subsequent testing of the reportedly healthy children indicated that the variant segregated with the disease in the family (figure 1A) except for individual II-6 who was homozygous for the mutant allele. Physical examination and echocardiography in this individual disclosed {I,L,I} SIT. Nasal NO screening performed on this sibling in the technique described above was normal. Post hoc examination of all other available family members revealed no additional individuals with situs inversus. Given this observation, we performed exome analysis in individual II-6 as well. The yield was 50.3 million mapped reads with a mean coverage of 84×. Within the variant list, filtered purely at the 1% MAF in ExAC and without consideration for evolutionary conservation (see online supplementary table S1), only two variants were shared in common with the affected sib II-7: Ser57Leu in FGFR2 (a known gene for Antley–Bixler syndrome) and c.1024_1025delAA in MMP21. In further support of the pathogenic effect of this allele, the c.1024_1025delAA deletion was absent from the ~63 000 exomes in ExAC (accessed June 2015); importantly, within this cohort, MMP21 appears intolerant to recessive deleterious mutations. In contrast, 600 healthy individuals, eight of whom are homozygous, carry the FGFR2 Ser57Leu variant. RT-PCR from white blood cells from the index patient showed expression of MMP21, suggesting that the c.1024_1025delAA mutation is unlikely to trigger nonsense-mediated decay, thus predicting the translation of a truncated protein that lacks the hemopexin repeat domain (figure 1B; see online supplementary figure S1).
In vivo modelling of MMP21
Given that our patients were diagnosed with L–R axis determination defects, a process conserved across vertebrates,31 and that the discovered mutation is predicted to diminish or abolish protein function, we hypothesised that suppression or deletion of MMP21 should induce asymmetry defects. We, thus, turned to a zebrafish model, in which, suppression or mutation of a number of laterality loci gives rise to either inversion or randomisation of the L–R axis of symmetry.32 Reciprocal BLAST identified a single mmp21 ortholog in the zebrafish genome (48.3% identity; 63.2% similarity to human MMP21 at the protein level), against which, we designed a SB-MO (see online supplementary figure S2A). RT-PCR and sequencing from morphants injected with 6 and 9 ng of SB-MO showed abnormal splicing, leading to the inclusion of intron 3 and the introduction of a premature termination codon (see online supplementary figure S2B).
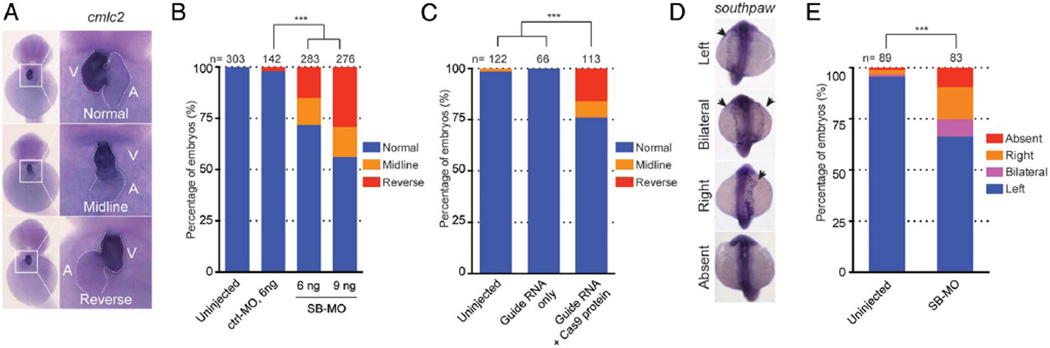
During cardiac development in zebrafish, the future ventricle begins to bend towards the right side at 1.5 dpf.33 By 2 dpf, heart looping (and its direction) is observable unambiguously under light microscopy. We, therefore, injected a control-MO or an SB-MO, and scored the orientation of the heart. Phenotyping results, at 2 dpf by two investigators blind to injection cocktails, showed embryos with no gross morphological defects such as cardiac/yolk oedema and body curvature (data not shown). However, we did observe a dosage-dependent induction of positional heart defects that were consistent with the progressive efficacy of the MO (see online supplementary figure S2B); 28% (n=80/283 with 6 ng of SB-MO) to 44% (n=121/276 with 9 ng of SB-MO) of morphants showing heartlooping defects, with the heart either positioned in the midline or on the left (figure 2A, B).
Figure 2.
Knockdown or genome editing on zebrafish mmp21 leading to heart-looping defects. (A) Representative images of heart looping observed in mmp21 morphants at 2 dpf. Embryos injected with mmp21 splice-blocking morpholino (SB-MO) showed abnormal heart looping such as midline or reversely looped heart whereas uninjected embryos showed normal heart looping. (B) Bar graph indicating the percentage of the embryos with normal, midline or reversely looped heart. The presence of abnormal heart looping was observed from the embryos injected with mmp21 SB-MO in a dose-dependent manner. Statistical significance of morphants versus uninjected embryos are depicted as ***, p<0.001 by χ2 test. (C) Quantification of the embryo batches with a normal, midline or reversely looped heart. Embryos injected with cocktail (guide RNA with Cas9 protein) showed the presence of a heart-looping defect, compared with the embryos injected with guide RNA only. (D) Representative images of expression of southpaw (spaw) in mmp21 morphants at 2 dpf. Embryos injected with mmp21 SB-MO showed bilateral, right or absence of spaw expression. (E) Bar graph indicating the percentage of the embryos with abnormal expression of spaw. Statistical significance of morphants versus uninjected embryos are depicted as ***, p< 0.001 by χ2 test.
Despite the apparent reproducibility of this phenotype, we were challenged to confirm its specificity because of a technical limitation in generating stable mRNA to rescue morphants. As such, we turned to an alternative approach, in which, we asked whether we could replicate the phenotype by inducing deletions by CRISPR/Cas9. For this, we designed a guide RNA targeting exon 3 of mmp21. We injected the guide RNA and Cas9 protein directly at the one-cell stage to induce mosaic mutations. We validated their presence in injected F0 embryos at 2 dpf by a T7 endonuclease I assay; we identified aberrant bands in all 20 embryos tested (see online supplementary figure S2C). Subsequent PCR from the locus in five 2 dpf embryos, followed by cloning and Sanger sequencing of individual clones, revealed exonic insertions/deletions (indel) in ~80% of clones, indicating a high abundance of mosaicism. Almost all indels are predicted to induce premature termination and, thus, loss of function.
Based on these data, we injected embryos with a cocktail of either the validated guide RNA and Cas9 protein or guide RNA alone, and phenotyped them similar to that of the SB-MO paradigm. Similar to our morphant studies, we found that 24% (n=27/113) of embryos injected with guide plus CAS9 cocktail, but none of the guide RNA-alone injected embryos, induced heart-looping defects (figure 2C). If the robust mosaicism we observed was true, we hypothesised that progeny from intercross of F0 founders would also show heart-looping defects. We, thus, intercrossed five pairs of F0s and scored their progeny. Consistent with our expectation and earlier data, 20%–40% of F1 mutants showed defects in heart looping (see online supplementary figure S3).
To study further whether the observed heart-looping defect is associated with L–R patterning, we evaluated the asymmetric distribution of the left-identity marker, southpaw (spaw). The nodal-related gene spaw is one of the earliest markers to be detected asymmetrically in the left LPM.13 We, thus, injected SB-MO and analysed the expression profile of spaw. We observed spaw expression primarily on the left side in most uninjected embryos (95%). In contrast, 34% of embryos injected with SB-MO showed an abnormal expression pattern of spaw such as right-sided, bilateral expression or absence (figure 2D, E).
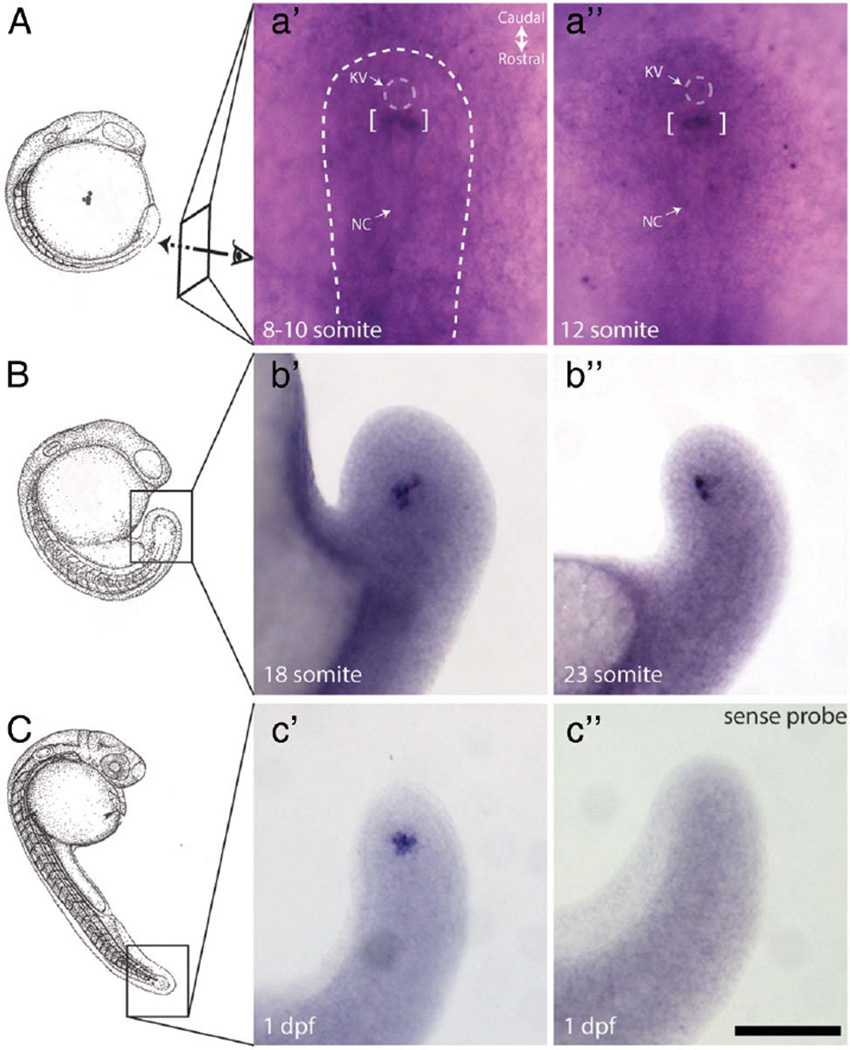
To gain preliminary insight as to the temporal involvement of MMP21 in L–R asymmetry, we examined the developmental profile of mmp21 expression by whole-mount RNA in situ hybridisation, paying particular attention to the developmental stages when L–R asymmetry begins to be established. In contrast to spaw expression during early development, we detected specific mmp21 expression close to and rostral to KV between 8 and 23 somite stages (figure 3A, B). Expression persisted in this region at 1 dpf, while no signal was detectable in embryos hybridised with a sense riboprobe, confirming the specificity of the signal (figure 3C). Taken together, our data suggest that mmp21 in zebrafish is required for heart looping and likely acts prior to the establishment of L–R asymmetry, supporting the dextrocardia phenotype found in the three patients.
Figure 3.
Specific mmp21 expression nearby KV during zebrafish development. (A–C) schematic of embryo (adapted from34-see below) between 8 and 12 somite stage (A), 18–23 somite stage (B) and 1 dpf embryo (C). Specific signal marked by brackets was detected rostrally near the KV at 8–10 somite (a0) and 12 somite (a″) stage of embryo. This signal stayed close to the end of the tail tip in 18 somite (b′) and 23 somite (b″), and remained detectable at 1 dpf (c′). A sense riboprobe was used as negative control (exemplar shown in c″). Dotted line indicating outline of developing tail area. scale bar, 100 µm. KV, Kupffer’s vesicle; NC, Notocord.
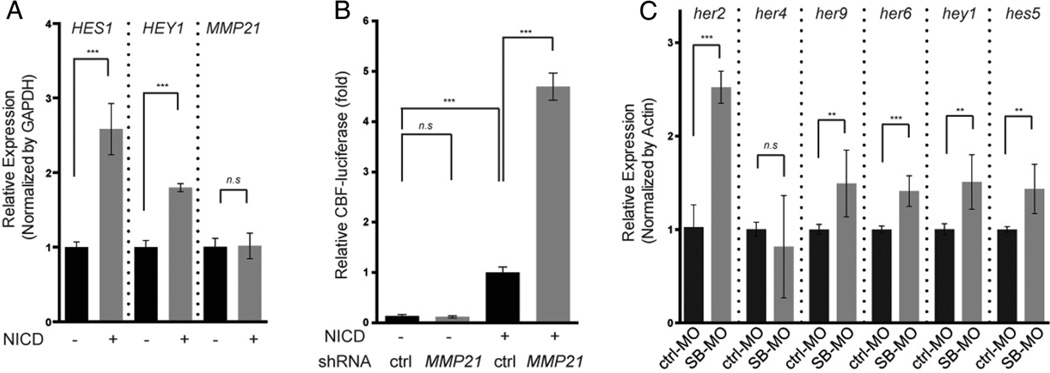
MMP21 encodes a member of the metalloproteinase superfamily that is known to hydrolyse extracellular matrix components,35 but has no prior associations to establishing L–R symmetry. We, thus, wondered what might be a possible link between loss of mmp21 and heart-looping defects. A previous study of the MMP21 locus indicated the presence of the binding sites for several paracrine signalling components in the upstream of the transcription initiation site,36 including an RBP-Jk binding site. This element is the main transcriptional mediator of Notch signalling, a pathway necessary for establishment of L–R asymmetry.37 We first tested the idea that expression of MMP21 is regulated by Notch signalling. We transiently transfected Hela cell with Notch1 intracellular domain (NICD) and measured the expression of mmp21 by quantitative real time PCR. However, we observed no changes in expression on NICD-mediated induction of signalling (figure 4A). Next, we asked whether MMP21 might have an effect on Notch signalling itself. For this purpose, we transiently transfected GFP-tagged human MMP21 with shRNA against MMP21 or control shRNA into HEK293 cells, and we monitored the abundance of GFP-tagged MMP21 protein. We quantified GFP-positive cells as well as the level of GFP-tagged MMP21 by western blotting. Approximately 6% (73/1225) of cells transfected with shRNA were positive for GFP compared with 29% (354/1217) of cells transfected with control shRNA. Western blotting also indicated that transfection of shRNA reduced the level of GFP-tagged MMP21 by ~90% (see online supplementary figure S4), suggesting potency in suppressing MMP21. We, therefore, cotransfected cells with the verified shRNA, a plasmid encoding the NICD and a CBF1-luciferase reporter containing four CBF1-responsive elements and the SV40 promoter for Notch activity. In three independent experiments within which transfection was done in triplicate (for a total of nine measurements), knockdown of MMP21 increased luciferase activity by 4.7 fold (±0.4, p<0.0001); transfection of shRNA alone did not induce the phenotype (figure 4B), suggesting a role of the mmp21 as a negative regulator for Notch signalling. Next, the Notch reporter result led us to ask whether target genes of Notch signalling might be misregulated in mmp21 zebrafish morphants. Quantitative real time PCR on cDNA generated from embryos injected with control-MO or SB-MO showed that most of the target genes we tested (her2, her6, her9, hey1 and hes5) were also upregulated at 2 dpf, in concordance with the luciferase assay result (figure 4C).
Figure 4.
Activation of Notch signalling on knockdown of MMP21. (A) MMP21 expression in Hela cells transfected transiently with Notch1 intracellular domain (NICD). Quantitative real time PCR result showed no statistical difference in the expression of human MMP21. (B) Quantitative real time PCR result from two independent experiments. Most of target genes for Notch signalling (her2, her6, her9, hey1 and hes5) are statistically upregulated in splice-blocking morpholino (SB-MO) injected embryos at 2 dpf. (C) CBF1-luciferase reporter assays to detect the transcriptional activity of NICD in HEK293 cells with shRNA against MMP21. Knockdown of MMP21 leads to increased CBF1-reporter activity. Statistical significance is depicted as ***, p<0.001; **, p<0.01—by student’s t test.
DISCUSSION
Here, we used WES in a consanguineous family with heterotaxia followed by filtering and in vivo modelling to demonstrate a causal role for mutations in MMP21 in disrupting L–R asymmetry. Our data indicate that homozygous mutation in our patients may lead to a spectrum of laterality disorders: either asymptomatic SIT or heterotaxia with complex congenital heart anomalies. There is a growing body of evidence that these two laterality disorders, SIT and heterotaxia, are part of a phenotypic continuum, with the same genetic defects underlying them. This phenotypic variability was described by us in individuals with mutations in CCDC11 and WDR1638 and by others in patients harbouring deleterious changes in ZIC3.39 We also tested mmp21 suppression or mutation in zebrafish. Knockdown or genetic editing of mmp21 recapitulated the heart-looping defects of our patients, and affected the expression of the early laterality marker southpaw. The mutation and suppression experiments in zebrafish do not recapitulate the human genotype, in which, the hemopexin domain is absent. However, the direction of effect is the same (loss of function) and consistent with the recently reported ENU-induced p. W177L mutation in the mouse Mmp21 locus that causes heterotaxy with TGA.40 Together, the model organism mutant data support the causality of the MMP21 mutation in our family, although a larger allelic series in human beings will be required to determine whether different mutations and deletions might affect the penetrance and expressivity of the phenotype.
The human and mouse genomes contain at least 23 MMPs. Testament to their biological importance, MMP defects have been implicated in a host of human genetic disorders, including arthritis, vascular disease, lung injury, cancer and some neurodegenerative disorders.41 For example, mutations in MMP2 cause multicentric osteolysis or ‘vanishing’ bone syndrome,42 while a mouse model lacking Mmp2 has defects in osteblast and osteoclast growth.43 A missense mutation in MMP13 results in an autosomal dominant disorder influencing growth defect and remodelling of long bones.44 Likewise, Mmp13-null mice exhibit several defects in plate cartilage growth with increased hypertrophic domains and delayed endochondral ossification.45,46 In addition, MMPs are thought to play a role in cancer progression, possibly because four hallmarks of cancer (migration, invasion, metastasis and angiogenesis) are influenced by the local microenvironment, including extracellular matrix (ECM) cues.47 Several studies of MMP21 in cancer indicate that the expression level of MMP21 is elevated in progression of colorectal cancer, squamous cell carcinoma, Merkel cell carcinoma and gastric cancer.48–51 In that context, mutations that reduce available MMP21 might have a protective role. However, MMP proteins have not been reported with human structural heart defects, potentially expanding our appreciation of the biological roles of this protein family.
Most MMPs are composed of multidomains, which contain a signal peptide, a propeptide, a catalytic domain, a linker domain and a hemopexin-like domain.35 The catalytic domain containing a zinc-binding motif (HEXXHXXGXXH) contributes to the proteolytic function of MMPs. The hemopexin-like domain, composed of four-bladed β-propeller structure and predicted to be absent from MMP21 in our patients, determines substrate and/or ligand specificity, subcellular localisation and activation/inhibition.52 We, thus, speculate that loss of the function of this domain is a potential driver of pathology in our patients, although it is also possible that the generated protein is more broadly unstable or dysfunctional, since the mouse ENU mutation is on the N-terminal side of this domain; engineering the discovered mutation in the mouse will be required to model the biochemistry of this allele further.
In terms of molecular mechanism, our data suggest that Notch signalling was increased by knockdown MMP21 from the in vitro study, and target genes of Notch signalling was increased by knockdown of zebrafish mmp21. This observation is interesting, given that Notch signalling is known to be crucial to L–R asymmetry by balancing the number of motile and immotile cilia at the L–R organiser.53 Gain or loss of function in Notch signalling is also known to affect L–R asymmetry by controlling ciliary length in KV.37 This link between Notch signalling and MMP21 may imply the possible pathomechanism that abnormal notch signalling might be participating in the dextrocardia phenotype found in our patients. Additionally, it is reported that upstream of MMP21 locus has a TCF4 binding site,35 which might indicate that MMP21 is also subject to Wnt regulation; this pathway is also involved in L–R axis specification, in part by signalling in left-sided perinodal cells.54 The presence of putative PAX2 binding site would intimate further complexity in MMP21 transcriptional regulation by multiple developmental signals. In that context, it will be important to identify MMP21 interacting partners; MMP21 is known to regulate both ECM components, such as gelatin, but mode of action on ECM components is not well understood.35 However, MMP21 could modulate indirectly the signalling by regulation of the ECM through cleaving other factors or their receptors required for L–R asymmetry such as southpaw in the ECM. For examples, the ECM protein microfibril-associated glycoprotein 2 (MAGP2) affects cellular differentiation through modulating the Notch signaling,55,56 and the treatment of pan-MMP inhibitor, GM6001, at 24 h post fertilisation in zebrafish led to defect in gut looping phenotype.57 In summary, our discovery of MMP21 in human patients with laterality defects and supporting data from zebrafish and mouse mutant40 offer an opportunity to explore hitherto unknown roles of matrix metalloproteinases in the establishment of L–R asymmetry during development. Mutations in genes of matrix metalloproteinases and their interacting partners might also contribute to heterotaxia in human beings.
Supplementary Material
Acknowledgments
We thank the family for their participation, Drs Yangfan Liu, I-Chun Tsai and Edwin Oh for assistance and advice on signalling assays and Dr Erica Davis for her critical evaluation of the manuscript.
Funding This work was supported by grants from the National Institutes of Health (R01DK072301 and R01DK075972) to NK.
Footnotes
Contributors ZP, AT-S, BY, SE AJJTR and OE led the patient recruitment, clinical phenotyping, genetic analysis and expression studies in patient cells. SM, LF and NK performed and analysed all functional studies. All authors assisted in the assembly and editing of the manuscript.
Competing interests None declared.
Patient consent Obtained.
Ethics approval Ethical committees of Hadassah Medical Center and the Israeli Ministry of Health.
REFERENCES
- 1.Zhu L, Belmont JW, Ware SM. Genetics of human heterotaxias. Eur J Hum Genet. 2006;14:17–25. doi: 10.1038/sj.ejhg.5201506. [DOI] [PubMed] [Google Scholar]
- 2.Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nat Rev Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- 3.Nonaka S, Shiratori H, Saijoh Y, Hamada H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature. 2002;418:96–99. doi: 10.1038/nature00849. [DOI] [PubMed] [Google Scholar]
- 4.Okada Y, Takeda S, Tanaka Y, Izpisúa Belmonte J-C, Hirokawa N. Mechanism of nodal flow: a conserved symmetry breaking event in left-right axis determination. Cell. 2005;121:633–644. doi: 10.1016/j.cell.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 6.Okada Y, Nonaka S, Tanaka Y, Saijoh Y, Hamada H, Hirokawa N. Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol Cell. 1999;4:459–468. doi: 10.1016/s1097-2765(00)80197-5. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Knowles HJ, Hebert JL, Hackett BP. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J Clin Invest. 1998;102:1077–1082. doi: 10.1172/JCI4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda S, Yonekawa Y, Tanaka Y, Okada Y, Nonaka S, Hirokawa N. Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J Cell Biol. 1999;145:825–836. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshiba S, Hamada H. Roles of cilia, fluid flow, and Ca2+ signaling in breaking of left-right symmetry. Trends Genet. 2014;30:10–17. doi: 10.1016/j.tig.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes Dev. 2002;16:2339–2344. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rankin CT, Bunton T, Lawler AM, Lee SJ. Regulation of left-right patterning in mice by growth/differentiation factor-1. Nat Genet. 2000;24:262–265. doi: 10.1038/73472. [DOI] [PubMed] [Google Scholar]
- 12.Oki S, Kitajima K, Marques S, Belo JA, Yokoyama T, Hamada H, Meno C. Reversal of left-right asymmetry induced by aberrant Nodal signaling in the node of mouse embryos. Development. 2009;136:3917–3925. doi: 10.1242/dev.039305. [DOI] [PubMed] [Google Scholar]
- 13.Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- 14.Kosaki K, Casey B. Genetics of human left-right axis malformations. Semin Cell Dev Biol. 1998;9:89–99. doi: 10.1006/scdb.1997.0187. [DOI] [PubMed] [Google Scholar]
- 15.Kathiriya IS, Srivastava D. Left-right asymmetry and cardiac looping: implications for cardiac development and congenital heart disease. Am J Med Genet. 2000;97:271–279. doi: 10.1002/1096-8628(200024)97:4<271::aid-ajmg1277>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 16.Sutherland MJ, Ware SM. Disorders of left-right asymmetry: heterotaxy and situs inversus. Am J Med Genet C Semin Med Genet. 2009;151C:307–317. doi: 10.1002/ajmg.c.30228. [DOI] [PubMed] [Google Scholar]
- 17.Gebbia M, Ferrero GB, Pilia G, Bassi MT, Aylsworth AS, Penman-Splitt M, Bird LM, Bamforth JS, Burn J, Schlessinger D, Nelson DL, Casey B. X-linked situs abnormalities result from mutations in ZIC3. Nat Genet. 1997;17:305–308. doi: 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- 18.Kosaki K, Kosaki R, Bassi MT, Lewin M, Belmont J, Schauer G, Casey B. Characterization and mutation analysis of human LEFTY A and LEFTY B, homologues of murine genes implicated in left-right axis development. Am J Hum Genet. 1999;64:712–721. doi: 10.1086/302289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosaki R, Gebbia M, Kosaki K, Lewin M, Bowers P, Towbin JA, Casey B. Left-right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am J Med Genet. 1999;82:70–76. doi: 10.1002/(sici)1096-8628(19990101)82:1<70::aid-ajmg14>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 20.Bamford RN, Roessler E, Burdine RD, Saplakoğlu U, dela Cruz J, Splitt M, Goodship JA, Towbin J, Bowers P, Ferrero GB, Marino B, Schier AF, Shen MM, Muenke M, Casey B. Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat Genet. 2000;26:365–369. doi: 10.1038/81695. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami Y, Raya A, Raya RM, Rodríguez-Esteban C, Izpisúa Belmonte JC. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435:165–171. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- 22.Mohapatra B, Casey B, Li H, Ho-Dawson T, Smith L, Fernbach SD, Molinari L, Niesh SR, Jefferies JL, Craigen WJ, Towbin JA, Belmont JW, Ware SM. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet. 2009;18:861–871. doi: 10.1093/hmg/ddn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selamet Tierney ES, Marans Z, Rutkin MB, Chung WK. Variants of the CFC1 gene in patients with laterality defects associated with congenital cardiac disease. Cardiol Young. 2007;17:268–274. doi: 10.1017/S1047951107000455. [DOI] [PubMed] [Google Scholar]
- 24.Roessler E, Ouspenskaia MV, Karkera JD, Vélez JI, Kantipong A, Lacbawan F, Bowers P, Belmont JW, Towbin JA, Goldmuntz E, Feldman B, Muenke M. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am J Hum Genet. 2008;83:18–29. doi: 10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) (3rd) 1995 [Google Scholar]
- 26.Stuart GW, Mcmurray JV, Westerfield M. Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development. 1988;103:403–412. doi: 10.1242/dev.103.2.403. [DOI] [PubMed] [Google Scholar]
- 27.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 29.Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, Robinson JF, Murdoch H, Dunlop AJ, Kubo K, Furukori K, Huang B, Zeledon M, Hayashi-Takagi A, Okano H, Nakajima K, Houslay MD, Katsanis N, Sawa A. DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. 2011;473:92–96. doi: 10.1038/nature09859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damseh N, Danson CM, Al-Ashhab M, Abu-Libdeh B, Gallon M, Sharma K, Yaacov B, Coulthard E, Caldwell MA, Edvardson S, Cullen PJ, Elpeleg O. A defect in the retromer accessory protein, SNX27, manifests by infantile myoclonic epilepsy and neurodegeneration. Neurogenetics. 2015;16:215–221. doi: 10.1007/s10048-015-0446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blum M, Feistel K, Thumberger T, Schweickert A. The evolution and conservation of left-right patterning mechanisms. Development. 2014;141:1603–1613. doi: 10.1242/dev.100560. [DOI] [PubMed] [Google Scholar]
- 32.Matsui T, Bessho Y. Left-right asymmetry in zebrafish. Cell Mol Life Sci. 2012;69:3069–3077. doi: 10.1007/s00018-012-0985-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res. 2011;91:279–288. doi: 10.1093/cvr/cvr098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 35.Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562–573. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Marchenko GN, Marchenko ND, Strongin AY. The structure and regulation of the human and mouse matrix metalloproteinase-21 gene and protein. Biochem J. 2003;372(Pt 2):503–515. doi: 10.1042/BJ20030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopes SS, Lourenco R, Pacheco L, Moreno N, Kreiling J, Saude L. Notch signalling regulates left-right asymmetry through ciliary length control. Development. 2010;137:3625–3632. doi: 10.1242/dev.054452. [DOI] [PubMed] [Google Scholar]
- 38.Perles Z, Cinnamon Y, Ta-Shma A, Shaag A, Einbinder T, Rein AJJT, Elpeleg O. A human laterality disorder associated with recessive CCDC11 mutation. J Med Genet. 2012;49:386–90. doi: 10.1136/jmedgenet-2011-100457. [DOI] [PubMed] [Google Scholar]
- 39.D’Alessandro LC, Casey B, Siu VM. Situs inversus totalis and a novel ZIC3 mutation in a family with X-linked heterotaxy. Congenit Heart Dis. 2013;8:E36–E40. doi: 10.1111/j.1747-0803.2011.00602.x. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, Chang C, Yagi H, San Agustin JT, Thahir M, Anderton S, Lawhead C, Vescovi A, Pratt H, Morgan J, Haynes L, Smith CL, Eppig JT, Reinholdt L, Francis R, Leatherbury L, Ganapathiraju MK, Tobita K, Pazour GJ, Lo CW. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fingleton B. Matrix metalloproteinases as valid clinical targets. Curr Pharm Des. 2007;13:333–346. doi: 10.2174/138161207779313551. [DOI] [PubMed] [Google Scholar]
- 42.Martignetti JA, Aqeel AA, Sewairi WA, Boumah CE, Kambouris M, Mayouf SA, Sheth KV, Eid WA, Dowling O, Harris J, Glucksman MJ, Bahabri S, Meyer BF, Desnick RJ. Mutation of the matrix metalloproteinase 2 gene (MMP2) causes a multicentric osteolysis and arthritis syndrome. Nat Genet. 2001;28:261–265. doi: 10.1038/90100. [DOI] [PubMed] [Google Scholar]
- 43.Mosig RA, Dowling O, Difeo A, Ramirez MCM, Parker IC, Abe E, Diouri J, Aqeel AA, Wylie JD, Oblander SA, Madri J, Bianco P, Apte SS, Zaidi M, Doty SB, Majeska RJ, Schaffler MB, Martignetti JA. Loss of MMP-2 disrupts skeletal and craniofacial development and results in decreased bone mineralization, joint erosion and defects in osteoblast and osteoclast growth. Hum Mol Genet. 2007;16:1113–1123. doi: 10.1093/hmg/ddm060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy AM, Inada M, Krane SM, Christie PT, Harding B, López-Otín C, Sánchez LM, Pannett AAJ, Dearlove A, Hartley C, Byrne MH, Reed AAC, Nesbit MA, Whyte MP, Thakker RV. MMP13 mutation causes spondyloepimetaphyseal dysplasia, Missouri type (SEMD(MO) J Clin Invest. 2005;115:2832–2842. doi: 10.1172/JCI22900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inada M, Wang Y, Byrne MH, Rahman MU, Miyaura C, Lopez-Otin C, Krane SM. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc Natl Acad Sci USA. 2004;101:17192–17197. doi: 10.1073/pnas.0407788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stickens D, Behonick DJ, Ortega N, Heyer B, Hartenstein B, Yu Y, Fosang AJ, Schorpp-Kistner M, Angel P, Werb Z. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131:5883–5895. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 48.Suomela S, Koljonen V, Skoog T, Kukko H, Böhling T, Saarialho-Kere U. Expression of MMP-10, MMP-21, MMP-26, and MMP-28 in Merkel cell carcinoma. Virchows Arch. 2009;455:495–503. doi: 10.1007/s00428-009-0856-1. [DOI] [PubMed] [Google Scholar]
- 49.Huang Y, Li W, Chu D, Zheng J, Ji G, Li M, Zhang H, Wang W, Du J, Li J. Overexpression of matrix metalloproteinase-21 is associated with poor overall survival of patients with colorectal cancer. J Gastrointest Surg. 2011;15:1188–1194. doi: 10.1007/s11605-011-1519-5. [DOI] [PubMed] [Google Scholar]
- 50.Wu T, Li Y, Liu X, Lu J, He X, Wang Q, Li J, Du X. Identification of high-risk stage II and stage III colorectal cancer by analysis of MMP-21 expression. J Surg Oncol. 2011;104:787–791. doi: 10.1002/jso.21970. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Z, Yan L, Li S, Sun H, Zhou Y, Li X. Increased MMP-21 expression in esophageal squamous cell carcinoma is associated with progression and prognosis. Med Oncol. 2014;31:91. doi: 10.1007/s12032-014-0091-8. [DOI] [PubMed] [Google Scholar]
- 52.Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- 53.Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, Brueckner M, Khokha MK. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature. 2013;504:456–459. doi: 10.1038/nature12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitajima K, Oki S, Ohkawa Y, Sumi T, Meno C. Wnt signaling regulates left-right axis formation in the node of mouse embryos. Dev Biol. 2013;380:222–232. doi: 10.1016/j.ydbio.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 55.Nehring LC, Miyamoto A, Hein PW, Weinmaster G, Shipley JM. The extracellular matrix protein MAGP-2 interacts with Jagged1 and induces its shedding from the cell surface. J Biol Chem. 2005;280:20349–20355. doi: 10.1074/jbc.M500273200. [DOI] [PubMed] [Google Scholar]
- 56.Albig A. Extracellular matrix controls Notch signaling via integrin activation and NICD ubiquitination. FASEB J. 2014;28 [Google Scholar]
- 57.Yin C, Kikuchi K, Hochgreb T, Poss KD, Stainier DYR. Hand2 regulates extracellular matrix remodeling essential for gut-looping morphogenesis in zebrafish. Dev Cell. 2010;18:973–984. doi: 10.1016/j.devcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.