Abstract
Genetic discoveries underlie the majority of the current thinking in neurodegenerative disease. This work has been driven by the significant gains made in identifying causal mutations; however, the translation of genetic causes of disease into pathobiological understanding remains a challenge.
The application of a second generation of genetics methods allow the dissection of moderate and mild genetic risk factors for disease. This requires new thinking in two key areas: what constitutes proof of pathogenicity, and how do we translate these findings to biological understanding.
Here we describe the progress and ongoing evolution in genetics. We describe a view that rejects the tradition that genetic proof has to be absolute before functional characterization, and centers on a multi dimensional approach integrating genetics, reference data, and functional work. We also argue that these challenges cannot be efficiently met by traditional hypothesis driven methods, but that high content system wide efforts are required.
Keywords: Alzheimer’s disease, Parkinson’s disease, Genetics, Genome wide association, Exome sequencing
Genetics as the basis for therapeutic development
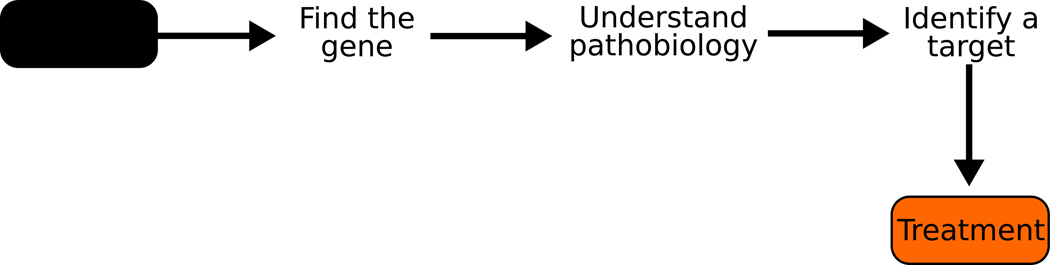
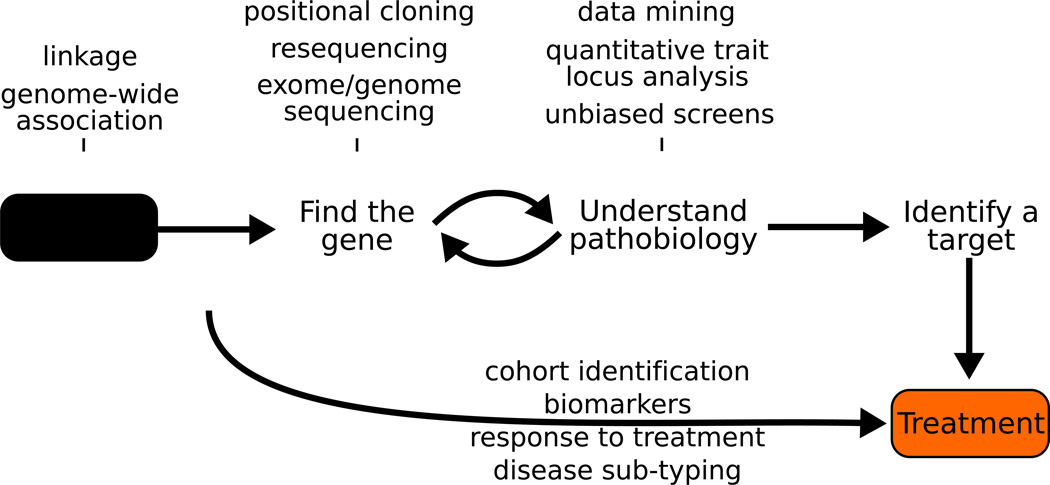
A core aim of human disease genetics is to facilitate the development of etiologic based treatments. The field’s approach has centered on the notion that identifying gene mutations will ultimately allow us to understand the molecular processes that initiate and sustain the disease pathogenesis. This knowledge in turn will allow the development of mechanism-based therapies (Figure 1).
Figure 1.
The simple schema underlying the development of mechanism based therapeutics
There has been a great deal of success in the early parts of this schema. In a little over 20 years human disease genetics has moved from a backwater of biology, to the foundation of much of our understanding of complex biological questions. While this success has been hard won, the tools and skills needed to identify disease linked genes have improved so much that the route to disease gene identification is becoming clearer, more straightforward, and for certain types of risk variation, routine.
While our success in genetics has been both tangible and substantial, translating this understanding to the development of mechanism-based therapies has been a much more difficult, and less productive endeavor. A common theme in the success of genetics is that the rate of progress has been dependent on the use of unbiased and system wide assays; when we have not relied on these tools and instead used our perceived knowledge of the disease to predict genetic targets, our success has been less impressive. We argue here that until now the understanding of pathobiology has been limited by the absence of unbiased and system wide approaches for understanding molecular processes, and that the traditional approach to teasing apart pathogenic function is inefficient and often misleading. We suggest that in order to take advantage of the growing fund of genetic discovery in understanding the molecular basis of disease we will need to develop and use high content system wide approaches.
Monogenic Forms of Disease
The initial major successes in disease genetics were dependent on genetic linkage screens, a genome-wide and unbiased method of identifying segregating chromosomal regions. Linkage, followed by positional cloning was the primary tool of genetic discovery from the late 1980s and is still in use today. While these efforts involved a great deal of time and resources, they were reliable, and served as the mainstay of genetics for a number of years, identifying large numbers of genetic causes of monogenic disease (Bonifati et al., 2003; Goate et al., 1991; Kitada et al., 1998; Paisan-Ruiz et al., 2004; Polymeropoulos et al., 1997; Rogaev et al., 1995; Sherrington et al., 1995; Valente et al., 2004; Zimprich et al., 2004). The prime method in the search for causes of monogenic disease is now second generation sequencing, usually whole exome sequencing (WES) but also whole genome sequencing (WGS). In the context of PD this has lead to the identification of the p.D620N mutation in VPS35 as a cause of disease and the nomination of DNAJC13 and CHCHD2 mutations as disease causing (Chartier-Harlin et al., 2011, p. 1; Funayama et al., 2015, p. 2; Vilariño-Güell et al., 2014, 2011, p. 35; Zimprich et al., 2011). Both WGS and WES will continue to provide insights into the causes of monogenic PD. In each of these approaches it was the development of methods to interrogate an entire system that was key to the success, whether the system comprised chunks of inherited DNA broken down by meiosis, as with genetic linkage studies, or the protein coding regions of the genome as with exome sequencing.
While a detailed discussion of the monogenic forms of these diseases is outside the remit of this review, a catalog of the dominant and recessive mutations for AD and PD can be found online (http://www.molgen.ua.ac.be/ADmutations/; http://www.molgen.vib-ua.be/PDMutDB/) (Cruts et al., 2012).
Common Genetic Risk Factors for Disease
While the success in genetic linkage and positional cloning is illustrative, so too are our failures during the same time. In part because of the broad availability and ease of PCR amplification, candidate gene association studies were widely applied during the mid/late 1990s and early 2000s, with the aim of identifying risk variability for complex disease. These methods were easy to apply, relatively cheap, and extensively used; yet they were also almost universally unsuccessful. With hindsight, the likelihood of nominating the correct gene to be tested and testing the right variants within it was vanishingly small; particularly as we later appreciated that the typical risk effect sizes associated with variants were too small to be seen by the majority of studies. Candidate gene association studies were in essence the antithesis of genome wide unbiased approaches. Our ability to guess at the molecules involved in the rather enigmatic cascade of events that constitutes disease was proven to be poor.
The challenge of identifying common risk variability was eventually met, again by a system wide method: genotyping using whole genome single nucleotide polymorphism (SNP) arrays. The extremely high information content of SNP arrays, and our ability to predict the genotype of many millions of variants based on these data provided a revolutionary tool in risk variant identification: Genome Wide Association (GWA). In this arena, GWA tests broadly and comprehensively, common genetic variation for association with a trait, and once again the system-wide nature of this method is a critical component of its success. GWA studies using these assays were initially controversial, but have proven remarkably successful in identifying the genetic basis of complex disease and traits.
In the fields of AD and PD this work has been driven by large consortia. In Alzheimer’s disease there now exist a large number of GWA efforts that have identified more than 18 loci that contain risk alleles (Harold et al., 2009; Hollingworth et al., 2011; Lambert et al., 2009; Moebus et al., with Lambert et al., European Alzheimer’s Disease Initiative (EADI) et al., 2013; Naj et al., 2011; Seshadri et al., 2010). Likewise there are now a large number of published GWA studies in PD, and since 2009 these have reported reliable and replicable risk loci (Do et al., 2011; Edwards et al., 2010; Elbaz et al., Genetic Epidemiology of Parkinson’s Disease (GEO-PD) Consortium, 2011; Hamza et al., 2010; Hernandez et al., 2012; International Parkinson Disease Genomics Consortium et al., 2011; International Parkinson’s Disease Genomics Consortium and Wellcome Trust Case Control Consortium, 2011; Lill et al., 2012; Liu et al., 2011; Pankratz et al., 2012, p. 2; Pankratz et al., PSG-PROGENI and GenePD Investigators, Coordinators and Molecular Genetic Laboratories, 2009; Pihlstrøm et al., 2013; Saad et al., 2011; Satake et al., 2009; Sharma et al., 2012; Simón-Sánchez et al., 2011). In the latest meta-analysis of PD GWA studies 28 independent risk loci have been identified and confirmed (Nalls, 2014). Like most GWAS hits, individually these PD loci confer only modest risk for disease. If we examine these loci collectively however, the 20% of individuals with the highest burden of genetic risk are about 3.5 times as likely to get disease than those 20% of individuals with the lowest burden of risk.
What Lies Beneath
Following the considerable success of GWAS, one question that remains is how much genetic influence is left to find? An analysis of the heritability of PD using genetic sharing estimates suggests ~30% of the risk for PD can be attributed to genetic factors. This is likely a significant underestimate, because very rare variants are difficult to capture using this method, and this approach, which estimates narrow sense heritability (or the proportion of trait variance that is due to additive genetic factors), does not take into account other genetic factors such as those that are dominant or multiplicative in effect. Notably, however, these analyses show that to date GWAS have identified only one tenth of this narrow sense heritable component: thus we can be sure that there is much left to find (Keller et al., 2012). The next question then, is where to look? Undoubtedly there are common risk loci for AD and PD that remain undiscovered and extending the size of current GWA studies is one approach that will yield results, as will performing GWA in diverse populations, and using alternate data mining methods to prioritize loci. A limiting factor in identifying additional common risk loci through GWA centers both on sample availability and cost. Most of the previous efforts have centered on marginal increases in sample sizes and meta-analysis of existing datasets, however, in large part this pool of extant data has been exhausted. The continued search for such loci will therefore likely require large increases in sample size, and it is unlikely in the current funding climate there will be much appetite for such a substantial resource investment. Thus, gains in this area are likely to be less dramatic than in previous years. However, it is also likely that some of this missing heritability exists in risk alleles that are too rare to detect using traditional GWAS methods, and this is an area of much interest and investigation.
Some success has already occurred in identifying rare risk alleles in AD. In 2012 Guerreiro and colleagues identified homozygous mutation of TREM2 as a cause of frontotemporal dementia (R. J. Guerreiro et al., 2012). Mutations in TREM2 were previously shown to cause Nasu-Hakola disease, a rare disease characterized by bone cysts and early onset progressive dementia (Paloneva et al., 2002). We had previously predicted that variability at genes causing young onset autosomal recessive neurological diseases would contribute to late onset neurological diseases (Singleton and Hardy, 2011). Because of the involvement of TREM2 in recessive diseases with a neurological component, this gene was a natural candidate for investigation in AD. Our work, and that of others revealed that indeed rare variants at TREM2 confer moderate risk for AD (R. Guerreiro et al., 2012, p. 2; Jonsson et al., 2013). In both instances, the foundation for this discovery was next generation sequencing data (both WGS and WES), and it is likely that this method will be the primary means for discovery of additional rare risk loci. The majority of variants that confer moderate risk for disease (OR>2) discovered thus far have altered the amino acid sequence of a protein and this, coupled with the lower price of WES compared to WGS, means that it is often argued that WES will remain the dominant methodology for some time to come. This argument is quite circular, because thus far attempts to look at non-coding sequence for rare risk variants have been quite limited, and it is difficult to interpret the consequences of such variants. Inevitably, as the true cost of whole genome sequencing approaches that of WES, whole genome sequencing will become the dominant approach. Interpretation of non-coding variability will remain a challenge, however as our understanding of the functional relevance of non-coding motifs and regions increases, this will improve. We predict that WGS will become a standard, and that advances in technology will center on more efficient generation of WGS, and the use of single molecule, low error, and long read sequences.
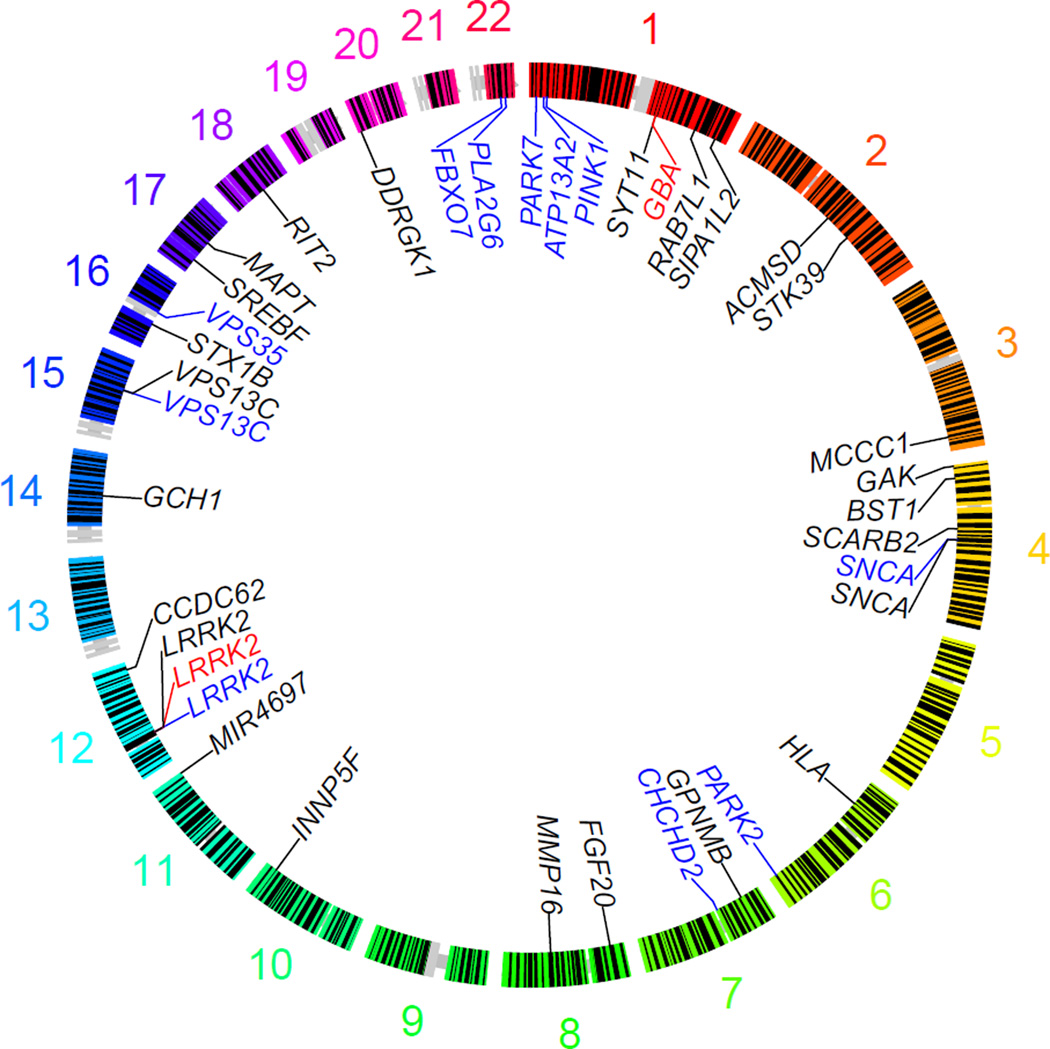
What is notable in the genetic architecture of PD is that the same loci show up under different risk categories i.e. some loci contain an allelic series across a range of frequencies and functional impact. A notable example exists at the gene encoding alpha-synuclein. SNCA point mutations are a rare and highly penetrant cause of young onset PD, SNCA dosage mutations are also rare and penetrance is linked to copy number (Singleton and Gwinn-Hardy, 2004), and common variability at SNCA increases risk for PD by only a modest amount (Odds Ratio~1.5) (figure 2). Likewise, the p.G2019S mutation in LRRK2 occurs in about 2% of North American Caucasian PD patients and is moderately penetrant by late age. There also exist common protein coding variants in LRRK2 that increase risk for disease ~2 fold, and, lastly, there are common non-coding variants at the LRRK2 locus that increase risk for disease ~1.2 fold. This phenomenon of pleomorphic risk does not yet appear generalizeable to every disease or locus, but it is reasonable to suggest that the known risk loci should be investigated for other types of disease risk. Again, second generation sequencing provides an opportunity to address this question with the use of targeted resequencing. In such an approach an entire genomic locus can be sequenced in a very large number of samples. This method has the potential to answer several questions; first, it can be used to fine map association signals identified by GWAS; second, it can be used to identify rare coding, non-coding, and copy number variants associated with disease independent from the GWA signal that nominated the locus; and third, in the identification of a coding variant associated with disease, this effort reveals the gene that is the biologic effector at a particular locus (Figure 2).
Figure 2.
Circular representation of the genome with genes and loci associated with PD indicated. Genes that contain causal mutations are shown in blue; genes that contain moderate effect protein coding risk alleles are shown in red; GWA identified loci that contain modest effect risk alleles are represented by proximal gene symbols in black.
Thus we can see that the likely advances in PD and AD genetics will center on a combination of WGS, WES, deep resequencing of known loci, and GWA studies.
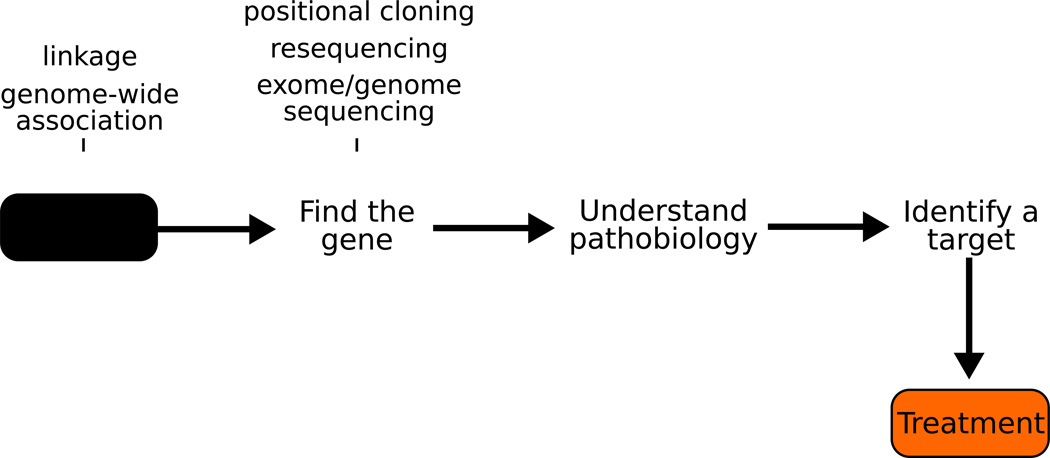
There are many challenges left in understanding the genetic basis of AD and PD and much work to do; however there are other challenges that we must face in parallel. First, how can we efficiently move from a locus to a gene; second, when we have genes, how do we build an accurate picture of pathogenesis; and last, as we move toward potential therapies how do we test these in an efficient manner? See figure 3.
Figure 3.
The application of genetic methods to discovery of novel genetic causes and contributors to disease. These methods, which center on unbiased and largely genome wide approaches, have enabled the majority of our understanding of the etiology of disease
Translating Gene to Pathobiology: Inefficient Success
Insight into the mechanisms underlying disease has been achieved using traditional hypothesis driven functional approaches. In the context of Alzheimer’s disease the most prominent example of this effort has come from translating amyloid precursor and presenilin mutations into an understanding of the amyloid cascade hypothesis. This represents more than two decades of hard won functional effort (Scheuner et al., 1996). While this progress has been absolutely critical in formulating novel therapeutic approaches for Alzheimer’s disease, it has come at a substantial cost. The majority of the pathogenic hypotheses tested have either been shown to be wrong, or ultimately revealed to be true biologically but unimportant pathologically. The approaches used to test these ideas were, out of necessity, performed almost exclusively using traditional hypothesis testing reductionist experiments. In the context of these approaches, we have become used to a scientific culture of failure, punctuated by rare significant success. We argue here that much of the initial work characterizing the biology of disease implicated proteins, could be more efficiently performed using the burgeoning hypothesis generating whole –ome screening methods. Such an approach does not remove the likelihood of chasing false leads, but it may minimize it, and notably it would do so at the earliest stages of investigation. Importantly, this effort does not seek to remove the need for traditional functional work, but simply to refocus it based on largely unbiased hypotheses (or more accurately those without the perception of understanding).
Comprehensive screening and big data integration
Because the early success in human disease genetics was in the identification of gene mutations that underlie monogenic forms of disease, our approaches to understanding the pathobiologic role of disease genes and their products has centered on manipulating systems using protein coding mutations. These approaches are difficult to apply to alleles that confer only low to moderate risk. For most disease-associated loci identified by GWA the effector gene is unknown, so any attempt to understand pathobiology must initially include an assessment that will reveal the gene and protein of interest. For many loci the underlying risk allele (which is often unknown) confers only minor risk for disease, and one might predict that the pathobiological effect is also either minor, or is not evident under basal conditions. Lastly, the majority of low to moderate risk alleles are not associated with protein coding variants, and thus they must confer an effect through altering expression of a transcript. These constraints represent considerable challenges, and they certainly require a rethinking of our approach to understanding pathobiology and a retooling of functional research groups. However, we believe that the return on meeting these challenges is likely to be so meaningful that we must prepare to meet and best them.
There already exists a large number of datasets that can help in this regard, and these have the potential to reveal pathobiology and identify the causal gene at risk loci. A seminal example of this approach was recently published in the field of hereditary spastic paraplegia (HSP) genetics (Novarino et al., 2014). In this work the authors used the large number of known HSP genes and existing protein interaction databases to build an HSPome - a protein interaction network centered on HSP proteins. The construction of this network helped nominate proteins/genes for involvement in the molecular pathogenesis of HSP. Taking this information they were then able to reexamine exome-sequencing data generated in HSP patients, and were able to identify novel genetic causes of this disease. This work illustrates the power of integrating large-scale genetic and functional data, and shows that the combination of these has the power to inform at both the genetic and pathobiological level. Implicitly, this work also answers a common criticism of continued genetic work: Why continue to find low risk genes, when we don’t know how the genes we have are involved in the disease process? In short, the answer is: we believe the larger the number of genes and loci that we have, the better chance we have of connecting their protein products in a pathologic network; in turn allowing us to build a complete picture of the pathogenic process. Work along these lines has been attempted in both AD and PD, most prominently featuring efforts to perform pathway based analysis of GWA implicated genes. This work centers on mining interaction or literature based datasets in an attempt to reveal if the GWA genes collectively highlight pathways of pathobiological relevance. Within both AD and PD the immune system has been highlighted using this approach, although notably there does not appear to be a strong shared genetic component between these two diseases (Dickson et al., with Guerreiro et al., International Parkinson’s Disease Genomics Consortium (IPDGC), 2015; Holmans et al., 2012; International Genomics of Alzheimer’s Disease Consortium (IGAP), 2015; Jones et al., 2010; Moskvina et al., IPDGC and GERAD Investigators, 2013). Additionally it has been argued that cholesterol metabolism may also be a pathway of significance in AD (Jones et al., 2010).
A more commonly used data integration method centers on using quantitative trait locus data, where the quantitative trait is a biologic measure. Most typically this aims to provide broad maps of the genetic control of effects proximal to genetic variability such as gene expression, protein levels, and DNA methylation. These maps can then be used to map the immediate biologic effects of variants linked to disease. It is now fairly typical to combine expression quantitative trait locus (QTL) work with GWAS. There are some limitations to this work; primary of which is that association between a biologic trait and a risk variant does not necessarily imply this effect is disease related. For example, an early identified risk locus for Parkinson’s disease was nominated on the short arm of chromosome 1, and denoted as PARK16 (Satake et al., 2009; Simon-Sanchez et al., 2009). Initial work suggested the risk alleles were also highly significant QTL’s for expression of NUCKS1 and DNA methylation at PM20D1, with a less significant expression QTL for RAB7L1 (now RAB29) (International Parkinson’s Disease Genomics Consortium and Wellcome Trust Case Control Consortium, 2011).; Nevertheless, subsequent functional evidence strongly suggests that RAB29 is the disease related gene at this locus.
There are also limits that depend on the biological source used for the QTL map; for example a typical expression QTL map would examine the relationship between genetic variability and gene expression from a tissue, such as human brain. While quite general genetic effects on constitutive expression are likely to be detectable in such a tissue, expression changes that only occur in a particular cell type, or those that are only evident after induction of expression (for example in response to cell stress) will not be detected. It is notable that for many loci identified by GWA, no QTL effect has been identified.
To date much of the work in the field of expression has relied on array based assays, which have considerable limitations, such as being relatively insensitive to splice changes, unable to detect unknown/ unassayed transcripts, and having a narrow dynamic range for detection. While the resolution of this approach is likely to improve with the application of transcriptome sequencing, the problem of tissue specific and induced expression will remain, and may need to be addressed with reference experiments aimed at recapitulating these effects, such as QTL mapping in differentiated iPSc.
It is notable that analysis of expression as an effector of GWAS signals is being taken a step beyond correlative studies and into mechanistic investigation; this work has shown clearly the complexity of gene regulation and serves as a warning that the most obvious association may not be disease relevant. A prime example comes from the investigation of the FTO locus, where genetic variability is strongly associated with risk for obesity and diabetes; notably FTO expression is also linked with obesity, and it was believed that the genetic risk at this locus was mediated through FTO. Recent work has suggested however that this may not be the case (Claussnitzer et al., 2015; Smemo et al., 2014). A critical resource in this type of mechanistic approach is data derived from the ENCODE project (Encyclopedia of DNA Elements), which aims to build a comprehensive list of functional elements within the human genome. The data underlying ENCODE comes from a wide variety of experimental approaches and tissues, and provides data regarding RNA transcripts, chromatin states, and transcriptional regulation. Such data allow investigators to rapidly take the first steps toward understanding the role of genetic variability linked to disease in the context of altered gene regulation. Likewise public sources of gene expression and genetic data such as that in the GTEx, Braineac, UKBEC, and NABEC data allow the integration of genetics, transriptomic, and regulatory data in order to better understand the genetic control of gene expression in the context of disease (GTEx Consortium, 2013; Nalls et al., International Parkinson’s Disease Genomics Consortium (IPDGC), The Wellcome Trust Case Control Consortium 2 (WTCCC2), North American Brain Expression Consortium (NABEC) and the United Kingdom Brain Expression Consortium (UKBEC), 2014). This work is being extended in creative ways not only to understand the function of disease linked variants, but also to identify new disease linked genes previously undiscovered in traditional GWA studies (Nicolae et al., with Gamazon et al., GTEx Consortium, 2015).
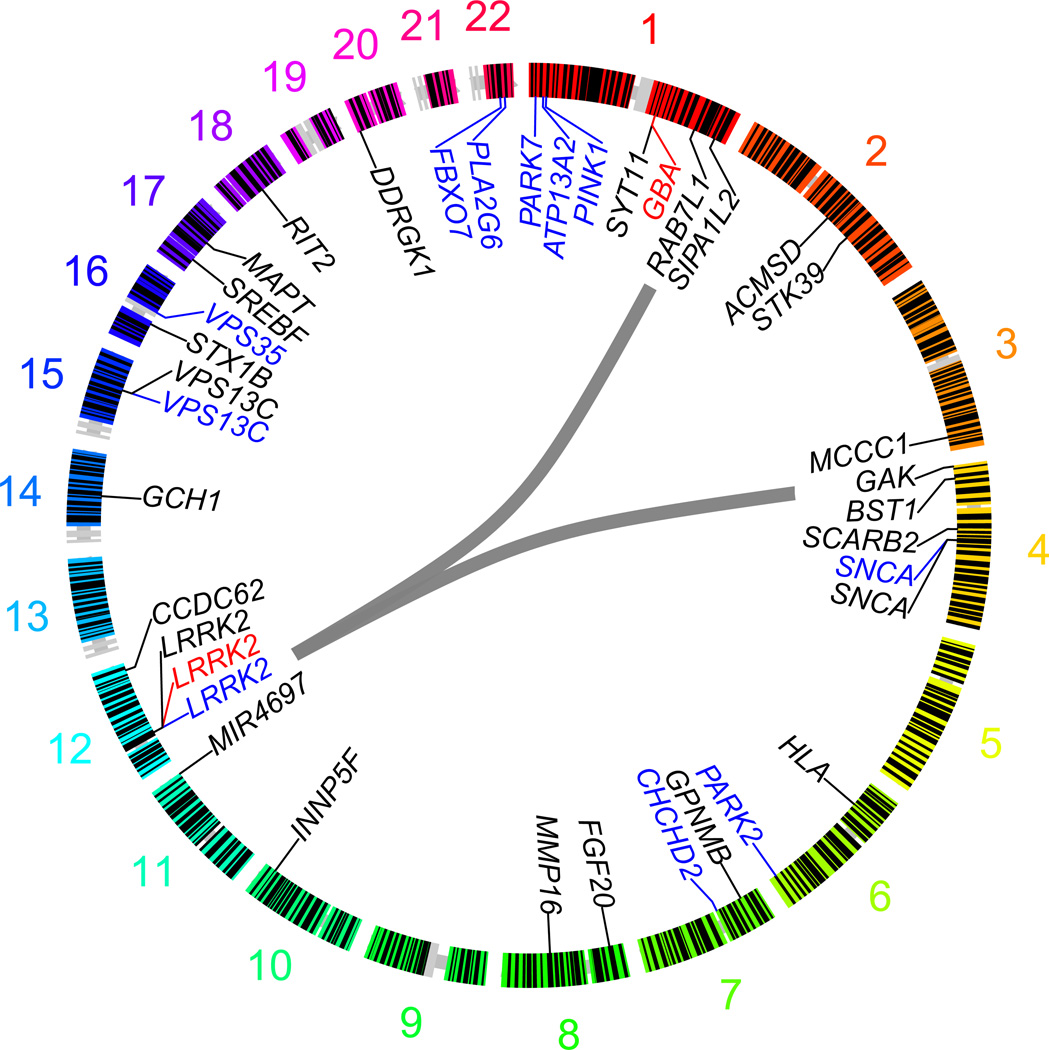
Another broad scale screening approach, in this instance combining genetic and protein interaction data, was recently performed in PD, and we believe this illustrates the power of integrating big data, and a likely path forward for complex disease research (Olszewski et al. with Beilina, Rudenko, et al., International Parkinson’s Disease Genomics Consortium and North American Brain Expression Consortium, 2014). In this work the authors performed unbiased high content screens for interactors of the known PD protein, Lrrk2. As with most high content screens, a large number of potential interactors were identified, and the prioritization of proteins for follow up would typically have been centered around factors such as putative function, and cellular expression. However, the authors used a different approach: combining GWA results with the hits from the Lrrk2 interactor screen showed that two of the hits were encoded by genes under GWA peaks (Pankratz et al., PSG-PROGENI and GenePD Investigators, Coordinators and Molecular Genetic Laboratories, 2009; Satake et al., 2009; Simon-Sanchez et al., 2009). These two proteins, GAK and Rab-7L1 (RAB29) were subsequently shown to form a complex with Lrrk2 that promotes clearance of Golgi-derived vesicles through the autophagy-lysosome system (figure 4).
Figure 4.
The application of both unbiased genetics methods and high content protein-protein interaction assays revealed the interaction between Lrrk2, GAK, and Rab7L1. Not only did this work establish GAK and RAB7L1 as the pathologically relevant genes at these GWA identified risk loci, but it also considerably expanded our understanding of Lrrk2 function, a critical aim in PD research.
What we hope to have illustrated is that these high content data have the ability to be informative about the molecular etiology of disease. This is particularly relevant when integrated with a large list of genes/loci. Notably such experiments have the benefit that they both provide information regarding the molecular etiology of disease, and support the nomination of particular genes within known risk loci, creating a self propagating knowledge generator (figure 5).
Figure 5.
The primary challenge following gene identification remains understanding the pathobiological process; typically this problem has been tackled using limited scale and reductionist methods, which require some a priori hypotheses regarding potential mechanism of action. We suggest here that this understanding can be greatly facilitated using burgeoning high content screening technologies and large-scale data mining. Not only does this have the ability to implicate specific processes in disease, but it can also aid in further mapping of the genetic basis of disease.
Understanding Genetic Uncertainty
As outlined above, we have rapidly moved into a space where the majority of genetic loci associated with disease are not represented by coding mutations that cause disease, but rather by common genetic risk variability with unknown consequences. As second generation sequencing progresses, there will be an additional class of genetic variability, rare protein coding and non-protein coding risk alleles. Notably, because these changes are rare and confer risk rather than cause disease, proving pathogenicity is a considerable challenge. In the context of data heavy genetic approaches the topic of proving pathogenicity is of considerable focus (MacArthur et al., 2014). Several schema for evaluating disease association have been proposed, in general including integration of existing published data, a statistical likelihood based assessment (association/linkage) and, notably the generation of functional evidence (MacArthur et al., 2014). Indeed it is unlikely that the burden of proof for disease involvement can be fully met by genetics alone. Thus, we are rapidly finding ourselves in a situation where alleles of uncertain pathogenic relevance will be commonplace in the literature. In order to move such alleles into an unequivocal category of risk or benign variant, it is absolutely critical that those involved in functional characterization of genetic variants understand the uncertainty surrounding these findings. Further it is extremely important to understand the power and potential bias of the approaches they employ in order to avoid lending false support of these candidates as disease linked genes. The literature in a field can build momentum, which is sometimes inappropriate; for example a very large body of work has been performed on ubiquitin C terminal hydrolase 1, despite the fact that the initial genetic evidence implicating this gene in PD was extremely weak, and remains unsubstantiated (Leroy et al., 1998). The naming of UCHL1 as a genetic locus for PD persists despite this lack of evidence, and a simple pubmed search of “UCHL1” and “Parkinson’s disease” reveals 121 manuscripts on this topic.
We view that, largely, the testing of the involvement of genes/variants for their involvement in pathways implicated in disease as a dangerous approach toward establishing pathogenicity. The majority of such efforts use rather blunt overexpression or knockout models, and the low n associated with most functional experiments, means that it is too easy to ascertain a positive effect on any pathway, regardless of whether this is a true biological effect or not. In this regard, once again the power of unbiased system wide methods is critical. Such work has the ability to test for an effect of a variant/gene on the background of effects seen across the whole genome, and thus to understand what is noise versus what is true signal.
Next Steps for Genetics
Cataloging more risk
As discussed above, there is clearly more genetic risk to be identified for AD and PD, and there exists a compelling rationale to invest time and resources to find it. For both AD and PD GWAS have been successful, however, also for both, tens of thousands of samples have been genotyped, and it is unclear that there will be significant support available to extend this genotyping to ever-increasing cohorts. There will likely be more that can be extracted from the extant data, through simply digging deeper into the sub-significant hits, and through alternative data mining approaches, such as pathways based analysis. Further, it is likely that as we build a greater understanding of the disease network, we will be able to specifically test genes in that network using these existing GWA data.
Second generation sequencing is increasingly accessible to the research community, not only because of reducing costs per base pair, but also with the development of standardized analytical tools. WES continues to be the prime method for gene discovery, and while most of the success has been in identifying disease causing mutations, the accumulation of WES data in large cohorts facilitates the identification of risk alleles using population based efforts. Because the majority of common variants that confer risk should have been identified by GWAS, the remaining risk variants are likely to be rare, and/or to impart very minor alterations in risk. What this means for sequence based risk discovery approaches is that the sample size will need to be high. While GWAS started to identify loci in PD and AD when applied to ~2000 cases, it is likely that exome, genome, and targeted sequencing approaches will need to be applied to tens of thousands of samples in order to reliably find risk alleles without a priori understanding. To date there are no studies that have reached this type of sample size for AD/PD; however, these are now underway for AD and being planned for PD.
One approach that has been used in several diseases, although not yet in AD/PD at a large scale, is resequencing, where candidate genes/loci are sequenced in very large numbers of cases. The genes and regions selected typically include known disease genes and GWA linked loci. This design allows the systematic investigation of nominated genes, including non-protein coding regulatory sequence, quickly and efficiently. The advantage of this approach is not only the speed with which it can be applied, but that it provides genetic information at multiple layers: it is a critical step in fine mapping of a GWA signal, and the identification of the functional risk allele(s); it allows the identification of additional risk at the same locus (for example rare risk alleles at a GWA locus); and, because the sequence is usually very deep, copy number variants are more easily detected.
While WES and resequencing are available now, and offer many benefits such as speed, low costs, and standardized analysis, it is inevitable that these methods will be supplanted by WGS; it is clearly a question of when not whether we will perform extremely large scale whole genome analysis in these diseases. Notably as sequencing reads become longer and contain less error, the alignment and analysis of sequence data will be more routine and less burdensome; such an advance will also lead to a greater resolution of the genetic basis of risk and the consequences of risk in the context of gene expression.
The role of genetics in overcoming other barriers to therapeutic development
Both AD and PD are late onset, progressive, and genetically complex diseases, and the nature of these diseases present significant challenges to therapeutic design and testing.
It is clear that both AD and PD occur for a number of years before the patient presents with clinical signs and symptoms, and that by the time the patient comes to clinic, the disease has progressed considerably, affecting multiple systems. It is most likely that effective therapies will focus on slowing or halting, rather than reversing, the disease process. It is also reasonable to assume that the earlier stages of disease are likely to be more responsive to etiologic based therapeutic intervention, than later, more widespread stages. One challenge therefore lies in identifying patients that are likely to get disease well before the signs and symptoms appear, with the ultimate aim of being able to apply preclinical therapeutic intervention, but with a more immediate aim of identifying cohorts of patients for clinical trials. This is already being pursued in AD, with a current clinical trials of the anti beta-amyloid therapy crenezumab in asymptomatic subjects who carry the PSEN1 p.E280A mutation. There exist a number of causal or highly penetrant gene mutations for both AD and PD, and carriers of these mutations may likewise be recruited into preventative clinical trials. The establishment of multinational networks such as DIAN (Dominantly Inherited Alzheimer Network; http://www.dian-info.org/) aims to study overtly asymptomatic carriers of dominant causal gene mutations in an effort to understand the disease process and identify early indicators of disease onset and progression. Such studies are ideally positioned for the execution of early clinical trials, and indeed the DIAN study has been extended to include such a component, with the testing of gantenerumab and solanezumab in this cohort (https://www.clinicaltrials.gov/ct/show/NCT01760005).
A greater challenge however, lies in the identification of individuals who will go on to become affected with disease, but who do not have a simple genetic cause. It is hoped that genetic risk profiling will help in this regard by estimating risk based on an individual person’s burden of known risk alleles. Progress in refining these pure genetic risk models is modest but steady. The use of genetic risk modeling alone is unlikely to identify soon to be affected patients within the near future. This does promise however to improve, and it will surely be part of a battery of tests aimed at predicting disease likelihood, onset, and course. A recent example of the success of integrating diverse accessible data types to predict disease comes from our efforts in PD [Nalls et al in press 2015]. In this work age, sex, family history of disease, anosmia status, and cumulative genetic risk score were combined to assign individuals an overall risk for PD. Using this model we were able to show an area under the receiver operator curve of ~0.92 across several studies, a remarkable predictive power. Perhaps even more interestingly, this predictive model was able to show that the PD patients who failed to show dopaminergic deficit at presentation represented two distinct risk groups, and that those with the higher risk profile were more likely to progress to show dopaminergic deficit.
Likewise, any efficient clinical trial will require quantitative markers of progression, which perform better than current clinical instruments. Markers of progression will be particularly important in preclinical trials, where changes over a short period are likely to be very subtle; without these, trials would stretch over many years with an associated cost that would be a disincentive to risk averse pharmaceutical companies. As with risk profiling, genetics is likely to form a part of biomarker work, not only in defining high risk individuals for cohorts being used in the development of biomarkers, but also in subsetting of groups of patients who have different biomarker profiles over the course of disease. This latter point touches on a more general theme in disease, subtyping. Our current pathognomonic classification of disease may be too coarse, and it is plausible that there are subtypes with different, or at least not entirely overlapping pathogeneses. This would implicitly suggest that discernable subtypes may require different therapeutic approaches, may respond differently to treatment, and need to be defined and identifiable as a part of clinical trials and biomarker studies (figure 6).
Figure 6.
Genetics can be applied as a tool to facilitate therapeutic trials and ultimately treatment. This includes genetic identification of at risk individuals, use in defining biomarkers, and defining sub-types of both disease and response to treatment. It can also be used as part of the process of identifying people in the prodrome of the disease before clinical symptoms become apparent, which is precisely when one would want to start mechanistic treatment.
A large barrier to the identification of disease subtypes, developing tools to aid in prognosis, and the discovery of biomarkers is cohort availability. Much of the genetic work to date has been appropriately carried out in cross sectional patient collections, where numbers are very high, but clinical details are low. Detailed longitudinal clinical studies are required; these are time consuming, and extremely expensive, however there are several attempts to collect such cohorts in PD and AD including two well know studies, the Parkinson’s Progression Markers Initiative (PPMI) and the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Mueller et al., 2005; Parkinson Progression Marker Initiative, 2011).
Summary
The continuation of an interrogation of the genetic basis of disease is a key step toward effective therapeutic strategies. Genetics will continue to inform researchers and to drive etiologic and clinical research. Critically we argue that in order to take full advantage of these data, a sea change is required in the way in which the field performs functional characterization of disease linked genetic variability. The generation of large unbiased functional datasets that can be integrated with the growing fund of genetic knowledge is a necessary step, and one that should occur before typical reductionist cell biology. In adopting this approach we can more efficiently move toward true disease altering therapeutics.
In Brief.
The piece by Singleton and Hardy describes the evolution and future of the identification and interpretation of genetic influences in neurodegenerative disease. It argues that maximizing the benefits of genetic progress requires a systematic and system wide approach to pathobiology.
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services; project ZO1 AG000949.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MCJ, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin M-C, Dachsel JC, Vilariño-Güell C, Lincoln SJ, Leprêtre F, Hulihan MM, Kachergus J, Milnerwood AJ, Tapia L, Song M-S, Le Rhun E, Mutez E, Larvor L, Duflot A, Vanbesien-Mailliot C, Kreisler A, Ross OA, Nishioka K, Soto-Ortolaza AI, Cobb SA, Melrose HL, Behrouz B, Keeling BH, Bacon JA, Hentati E, Williams L, Yanagiya A, Sonenberg N, Lockhart PJ, Zubair AC, Uitti RJ, Aasly JO, Krygowska-Wajs A, Opala G, Wszolek ZK, Frigerio R, Maraganore DM, Gosal D, Lynch T, Hutchinson M, Bentivoglio AR, Valente EM, Nichols WC, Pankratz N, Foroud T, Gibson RA, Hentati F, Dickson DW, Destée A, Farrer MJ. Translation initiator EIF4G1 mutations in familial Parkinson disease. Am. J. Hum. Genet. 2011;89:398–406. doi: 10.1016/j.ajhg.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, Abdennur NA, Liu J, Svensson P-A, Hsu Y-H, Drucker DJ, Mellgren G, Hui C-C, Hauner H, Kellis M. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum. Mutat. 2012;33:1340–1344. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson D, Hardy J, Singleton A, Bras J, Guerreiro R, Escott-Price V, Darwent L, Parkkinen L, Ansorge O, Hernandez DG, Nalls MA, Clark L, Honig L, Marder K, van der Flier W, Holstege H, Louwersheimer E, Lemstra A, Scheltens P, Rogaeva E, St George-Hyslop P, Londos E, Zetterberg H, Ortega-Cubero S, Pastor P, Ferman TJ, Graff-Radford NR, Ross OA, Barber I, Braae A, Brown K, Morgan K, Maetzler W, Berg D, Troakes C, Al-Sarraj S, Lashley T, Compta Y, Revesz T, Lees A, Cairns NJ, Halliday GM, Mann D, Pickering-Brown S, Powell J, Lunnon K, Lupton MK International Parkinson’s Disease Genomics Consortium (IPDGC) Genome-wide analysis of genetic correlation in dementia with Lewy bodies, Parkinson’s and Alzheimer’s diseases. Neurobiol. Aging. 2015 doi: 10.1016/j.neurobiolaging.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CB, Tung JY, Dorfman E, Kiefer AK, Drabant EM, Francke U, Mountain JL, Goldman SM, Tanner CM, Langston JW, Wojcicki A, Eriksson N. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS Genet. 2011;7:e1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TL, Scott WK, Almonte C, Burt A, Powell EH, Beecham GW, Wang L, Züchner S, Konidari I, Wang G, Singer C, Nahab F, Scott B, Stajich JM, Pericak-Vance M, Haines J, Vance JM, Martin ER. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann. Hum. Genet. 2010;74:97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz A, Ross OA, Ioannidis JPA, Soto-Ortolaza AI, Moisan F, Aasly J, Annesi G, Bozi M, Brighina L, Chartier-Harlin M-C, Destée A, Ferrarese C, Ferraris A, Gibson JM, Gispert S, Hadjigeorgiou GM, Jasinska-Myga B, Klein C, Krüger R, Lambert J-C, Lohmann K, van de Loo S, Loriot M-A, Lynch T, Mellick GD, Mutez E, Nilsson C, Opala G, Puschmann A, Quattrone A, Sharma M, Silburn PA, Stefanis L, Uitti RJ, Valente EM, Vilariño-Güell C, Wirdefeldt K, Wszolek ZK, Xiromerisiou G, Maraganore DM, Farrer MJ Genetic Epidemiology of Parkinson’s Disease (GEO-PD) Consortium. Independent and joint effects of the MAPT and SNCA genes in Parkinson disease. Ann. Neurol. 2011;69:778–792. doi: 10.1002/ana.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama M, Ohe K, Amo T, Furuya N, Yamaguchi J, Saiki S, Li Y, Ogaki K, Ando M, Yoshino H, Tomiyama H, Nishioka K, Hasegawa K, Saiki H, Satake W, Mogushi K, Sasaki R, Kokubo Y, Kuzuhara S, Toda T, Mizuno Y, Uchiyama Y, Ohno K, Hattori N. CHCHD2 mutations in autosomal dominant late-onset Parkinson’s disease: a genome-wide linkage and sequencing study. Lancet Neurol. 2015;14:274–282. doi: 10.1016/S1474-4422(14)70266-2. [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Lohmann E, Brás JM, Gibbs JR, Rohrer JD, Gurunlian N, Dursun B, Bilgic B, Hanagasi H, Gurvit H, Emre M, Singleton A, Hardy J. Using Exome Sequencing to Reveal Mutations in TREM2 Presenting as a Frontotemporal Dementia-like Syndrome Without Bone Involvement. Arch. Neurol. 2012:1–7. doi: 10.1001/jamaneurol.2013.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JSK, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert J-C, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J. TREM2 Variants in Alzheimer’s Disease. N. Engl. J. Med. 2012 doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, Yearout D, Kay DM, Doheny KF, Paschall J, Pugh E, Kusel VI, Collura R, Roberts J, Griffith A, Samii A, Scott WK, Nutt J, Factor SA, Payami H. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat. Genet. 2010;42:781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schürmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Hüll M, Rujescu D, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel K-H, Klopp N, Wichmann H-E, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Ylikotila P, Keller M, Hardy JA, Majamaa K, Singleton AB. Genome wide assessment of young onset Parkinson’s disease from Finland. PloS One. 2012;7:e41859. doi: 10.1371/journal.pone.0041859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ERLC, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Rüther E, Schürmann B, Heun R, Kölsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Gallacher J, Hüll M, Rujescu D, Giegling I, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel K-H, Klopp N, Wichmann H-E, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MMB, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues J-F, Tzourio C, Alpérovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Björnsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossù P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Moskvina V, Jones L, Sharma M, Vedernikov A, Buchel F, Sadd M, Bras JM, Bettella F, Nicolaou N, Simón-Sánchez J, Mittag F, Gibbs JR, Schulte C, Durr A, Guerreiro R, Hernandez D, Brice A, Stefánsson H, Majamaa K, Gasser T, Heutink P, Wood NW, Martinez M, Singleton AB, Nalls MA, Hardy J, Morris HR, Williams NM. A pathway-based analysis provides additional support for an immune-related genetic susceptibility to Parkinson’s disease. Hum. Mol. Genet. 2012 doi: 10.1093/hmg/dds492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Genomics of Alzheimer’s Disease Consortium (IGAP) Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimers Dement. J. Alzheimers Assoc. 2015;11:658–671. doi: 10.1016/j.jalz.2014.05.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Parkinson Disease Genomics Consortium. Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, Saad M, Simon-Sanchez J, Schulte C, Lesage S, Sveinbjornsdottir S, Stefansson K, Martinez M, Hardy J, Heutink P, Brice A, Gasser T, Singleton AB, Wood NW. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. doi:S0140-6736(10)62345-8 [pii] 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Parkinson’s Disease Genomics Consortium, Wellcome Trust Case Control Consortium. A two-stage meta-analysis identifies several new loci for Parkinson’s disease. PLoS Genet. 2011;7:e1002142. doi: 10.1371/journal.pgen.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Holmans PA, Hamshere ML, Harold D, Moskvina V, Ivanov D, Pocklington A, Abraham R, Hollingworth P, Sims R, Gerrish A, Pahwa JS, Jones N, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schürmann B, Heun R, Kölsch H, van den Bussche H, Heuser I, Peters O, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Hüll M, Rujescu D, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel K-H, Klopp N, Wichmann H-E, Rüther E, Carrasquillo MM, Pankratz VS, Younkin SG, Hardy J, O’Donovan MC, Owen MJ, Williams J. Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer’s disease. PloS One. 2010;5:e13950. doi: 10.1371/journal.pone.0013950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MF, Saad M, Bras J, Bettella F, Nicolaou N, Simón-Sánchez J, Mittag F, Büchel F, Sharma M, Gibbs JR, Schulte C, Moskvina V, Durr A, Holmans P, Kilarski LL, Guerreiro R, Hernandez DG, Brice A, Ylikotila P, Stefánsson H, Majamaa K, Morris HR, Williams N, Gasser T, Heutink P, Wood NW, Hardy J, Martinez M, Singleton AB, Nalls MA. Using genome-wide complex trait analysis to quantify “missing heritability” in Parkinson’s disease. Hum. Mol. Genet. 2012;21:4996–5009. doi: 10.1093/hmg/dds335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Lambert J-C, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fiévet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossù P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanché H, Dartigues J-F, Tzourio C, Gut I, Van Broeckhoven C, Alpérovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD, Polymeropoulos MH. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, Schjeide B-MM, Schjeide LM, Meissner E, Zauft U, Allen NC, Liu T, Schilling M, Anderson KJ, Beecham G, Berg D, Biernacka JM, Brice A, DeStefano AL, Do CB, Eriksson N, Factor SA, Farrer MJ, Foroud T, Gasser T, Hamza T, Hardy JA, Heutink P, Hill-Burns EM, Klein C, Latourelle JC, Maraganore DM, Martin ER, Martinez M, Myers RH, Nalls MA, Pankratz N, Payami H, Satake W, Scott WK, Sharma M, Singleton AB, Stefansson K, Toda T, Tung JY, Vance J, Wood NW, Zabetian CP, Young P, Tanzi RE, Khoury MJ, Zipp F, Lehrach H, Ioannidis JPA, Bertram L. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: The PDGene database. PLoS Genet. 2012;8:e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Cheng R, Verbitsky M, Kisselev S, Browne A, Mejia-Sanatana H, Louis ED, Cote LJ, Andrews H, Waters C, Ford B, Frucht S, Fahn S, Marder K, Clark LN, Lee JH. Genome-wide association study identifies candidate genes for Parkinson’s disease in an Ashkenazi Jewish population. BMC Med. Genet. 2011;12:104. doi: 10.1186/1471-2350-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannfelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RFAG, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JSK, Boerwinkle E, Riemenschneider M, Boada M, Hiltunen M, Martin ER, Schmidt R, Rujescu D, Wang L-S, Dartigues J-F, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P, Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, Destefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin C-F, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau M-T, Choi S-H, Reitz C, Pasquier F, Hollingworth P, Ramirez A, Hanon O, Fitzpatrick AL, Buxbaum JD, Campion D, Crane PK, Baldwin C, Becker T, Gudnason V, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MJ, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuinness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Naranjo MCD, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F European Alzheimer’s Disease Initiative (EADI), Genetic and Environmental Risk in Alzheimer’s Disease, Alzheimer’s Disease Genetic Consortium, Cohorts for Heart and Aging Research in Genomic Epidemiology. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvina V, Harold D, Russo G, Vedernikov A, Sharma M, Saad M, Holmans P, Bras JM, Bettella F, Keller MF, Nicolaou N, Simón-Sánchez J, Gibbs JR, Schulte C, Durr A, Guerreiro R, Hernandez D, Brice A, Stefánsson H, Majamaa K, Gasser T, Heutink P, Wood N, Martinez M, Singleton AB, Nalls MA, Hardy J, Owen MJ, O’Donovan MC, Williams J, Morris HR, Williams NM IPDGC and GERAD Investigators. Analysis of genome-wide association studies of Alzheimer disease and of Parkinson disease to determine if these 2 diseases share a common genetic risk. JAMA Neurol. 2013;70:1268–1276. doi: 10.1001/jamaneurol.2013.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. J. Alzheimers Assoc. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JSK, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin L-W, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls M. META analysis GWA placeholder. 2014 [Google Scholar]
- Nalls MA, Saad M, Noyce AJ, Keller MF, Schrag A, Bestwick JP, Traynor BJ, Gibbs JR, Hernandez DG, Cookson MR, Morris HR, Williams N, Gasser T, Heutink P, Wood N, Hardy J, Martinez M, Singleton AB International Parkinson’s Disease Genomics Consortium (IPDGC), The Wellcome Trust Case Control Consortium 2 (WTCCC2), North American Brain Expression Consortium (NABEC) and the United Kingdom Brain Expression Consortium (UKBEC) Genetic comorbidities in Parkinson’s disease. Hum. Mol. Genet. 2014;23:831–841. doi: 10.1093/hmg/ddt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Cox NJ, Im HK, Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC GTEx Consortium. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mansour L, Masri A, Kayserili H, Al-Aama JY, Abdel-Salam GMH, Karminejad A, Kara M, Kara B, Bozorgmehri B, Ben-Omran T, Mojahedi F, Mahmoud IGED, Bouslam N, Bouhouche A, Benomar A, Hanein S, Raymond L, Forlani S, Mascaro M, Selim L, Shehata N, Al-Allawi N, Bindu PS, Azam M, Gunel M, Caglayan A, Bilguvar K, Tolun A, Issa MY, Schroth J, Spencer EG, Rosti RO, Akizu N, Vaux KK, Johansen A, Koh AA, Megahed H, Durr A, Brice A, Stevanin G, Gabriel SB, Ideker T, Gleeson JG. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343:506–511. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski M, Hauser DN, Kumaran R, Lozano AM, Baekelandt V, Greene LE, Taymans J-M, Greggio E, Cookson MR, Beilina A, Rudenko IN, Kaganovich A, Civiero L, Chau H, Kalia SK, Kalia LV, Lobbestael E, Chia R, Ndukwe K, Ding J, Nalls MA International Parkinson’s Disease Genomics Consortium, North American Brain Expression Consortium. Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2626–2631. doi: 10.1073/pnas.1318306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, Tranebjaerg L, Konttinen Y, Peltonen L. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz N, Wilk JB, Latourelle JC, DeStefano AL, Halter C, Pugh EW, Doheny KF, Gusella JF, Nichols WC, Foroud T, Myers RH PSGPROGENI and GenePD Investigators, Coordinators and Molecular Genetic Laboratories. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum. Genet. 2009;124:593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz N, Beecham GW, Destefano AL, Dawson TM, Doheny KF, Factor SA, Hamza TH, Hung AY, Hyman BT, Ivinson AJ, Krainc D, Latourelle JC, Clark LN, Marder K, Martin ER, Mayeux R, Ross OA, Scherzer CR, Simon DK, Tanner C, Vance JM, Wszolek ZK, Zabetian CP, Myers RH, Payami H, Scott WK, Foroud T. Meta-analysis of Parkinson’s Disease: Identification of a novel locus, RIT2. Ann. Neurol. 2012;71:370– 384. doi: 10.1002/ana.22687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson Progression Marker Initiative. The Parkinson Progression Marker Initiative (PPMI) Prog. Neurobiol. 2011;95:629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlstrøm L, Axelsson G, Bjørnarå KA, Dizdar N, Fardell C, Forsgren L, Holmberg B, Larsen JP, Linder J, Nissbrandt H, Tysnes O-B, Ohman E, Dietrichs E, Toft M. Supportive evidence for 11 loci from genome-wide association studies in Parkinson’s disease. Neurobiol. Aging. 2013;34:1708.e7–1708.e13. doi: 10.1016/j.neurobiolaging.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- Saad M, Lesage S, Saint-Pierre A, Corvol J-C, Zelenika D, Lambert J-C, Vidailhet M, Mellick GD, Lohmann E, Durif F, Pollak P, Damier P, Tison F, Silburn PA, Tzourio C, Forlani S, Loriot M-A, Giroud M, Helmer C, Portet F, Amouyel P, Lathrop M, Elbaz A, Durr A, Martinez M, Brice A. Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson’s disease in the European population. Hum. Mol. Genet. 2011;20:615–627. doi: 10.1093/hmg/ddq497. [DOI] [PubMed] [Google Scholar]
- Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, Nakamura Y, Toda T. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. doi:ng.485 [pii] 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D, Younkin S. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat. Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EMC, Ramirez-Lorca R, Debette S, Longstreth WT, Jr, Janssens ACJW, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MMB. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA J. Am. Med. Assoc. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Ioannidis JPA, Aasly JO, Annesi G, Brice A, Van Broeckhoven C, Bertram L, Bozi M, Crosiers D, Clarke C, Facheris M, Farrer M, Garraux G, Gispert S, Auburger G, Vilariño-Güell C, Hadjigeorgiou GM, Hicks AA, Hattori N, Jeon B, Lesage S, Lill CM, Lin J-J, Lynch T, Lichtner P, Lang AE, Mok V, Jasinska-Myga B, Mellick GD, Morrison KE, Opala G, Pramstaller PP, Pichler I, Park SS, Quattrone A, Rogaeva E, Ross OA, Stefanis L, Stockton JD, Satake W, Silburn PA, Theuns J, Tan E-K, Toda T, Tomiyama H, Uitti RJ, Wirdefeldt K, Wszolek Z, Xiromerisiou G, Yueh K-C, Zhao Y, Gasser T, Maraganore D, Krüger R. Large-scale replication and heterogeneity in Parkinson disease genetic loci. Neurology. 2012;79:659–667. doi: 10.1212/WNL.0b013e318264e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin JF, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, Da Silva HA, Haines JL, Perkicak-Vance MA, Tanzi RE, Roses AD, Fraser PE, Rommens JM, St George-Hyslop PH. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Kruger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, van der Brug M, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao J, Huang X, Wood NW, Lorenz D, Deuschl G, Chen H, Riess O, Hardy JA, Singleton AB, Gasser T. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–12. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Sánchez J, van Hilten JJ, van de Warrenburg B, Post B, Berendse HW, Arepalli S, Hernandez DG, de Bie RMA, Velseboer D, Scheffer H, Bloem B, van Dijk KD, Rivadeneira F, Hofman A, Uitterlinden AG, Rizzu P, Bochdanovits Z, Singleton AB, Heutink P. Genome-wide association study confirms extant PD risk loci among the Dutch. Eur. J. Hum. Genet. EJHG. 2011;19:655–661. doi: 10.1038/ejhg.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton A, Gwinn-Hardy K. Parkinson’s disease and dementia with Lewy bodies: a difference in dose? Lancet. 2004;364:1105–1107. doi: 10.1016/S0140-6736(04)17117-1. [DOI] [PubMed] [Google Scholar]
- Singleton A, Hardy J. A generalizable hypothesis for the genetic architecture of disease: pleomorphic risk loci. Hum. Mol. Genet. 2011;20:R158–R162. doi: 10.1093/hmg/ddr358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung H-K, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui C-C, Gomez-Skarmeta JL, Nóbrega MA. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MMK, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, González-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Vilariño-Güell C, Rajput A, Milnerwood AJ, Shah B, Szu-Tu C, Trinh J, Yu I, Encarnacion M, Munsie LN, Tapia L, Gustavsson EK, Chou P, Tatarnikov I, Evans DM, Pishotta FT, Volta M, Beccano-Kelly D, Thompson C, Lin MK, Sherman HE, Han HJ, Guenther BL, Wasserman WW, Bernard V, Ross CJ, Appel-Cresswell S, Stoessl AJ, Robinson CA, Dickson DW, Ross OA, Wszolek ZK, Aasly JO, Wu R-M, Hentati F, Gibson RA, McPherson PS, Girard M, Rajput M, Rajput AH, Farrer MJ. DNAJC13 mutations in Parkinson disease. Hum. Mol. Genet. 2014;23:1794–1801. doi: 10.1093/hmg/ddt570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño-Güell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, Behrouz B, Melrose HL, Hentati E, Puschmann A, Evans DM, Conibear E, Wasserman WW, Aasly JO, Burkhard PR, Djaldetti R, Ghika J, Hentati F, Krygowska-Wajs A, Lynch T, Melamed E, Rajput A, Rajput AH, Solida A, Wu R-M, Uitti RJ, Wszolek ZK, Vingerhoets F, Farrer MJ. VPS35 mutations in Parkinson disease. Am. J. Hum. Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Benet-Pagès A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, Rossle SC, Klopp N, Wolf E, Seppi K, Pirker W, Presslauer S, Mollenhauer B, Katzenschlager R, Foki T, Hotzy C, Reinthaler E, Harutyunyan A, Kralovics R, Peters A, Zimprich F, Brücke T, Poewe W, Auff E, Trenkwalder C, Rost B, Ransmayr G, Winkelmann J, Meitinger T, Strom TM. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Müller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]