Abstract
Combination antiretroviral therapy (ART) for HIV-1 infection reduces plasma virus levels to below the limit of detection of clinical assays. However, even with prolonged suppression of viral replication with ART, viremia rebounds rapidly after treatment interruption. Thus ART is not curative. The principal barrier to cure is a remarkably stable reservoir of latent HIV-1 in resting memory CD4+ T cells. Here we consider explanations for the remarkable stability of the latent reservoir. Stability does not appear to reflect replenishment from new infection events but rather normal physiologic processes that provide for immunologic memory. Of particular importance are proliferative processes that drive clonal expansion of infected cells. Recent evidence suggests that in some infected cells, proliferation is a consequence of proviral integration into host genes associated with cell growth. Efforts to cure HIV-1 infection by targeting the latent reservoir may need to consider the potential of latently infected cells to proliferate.
Introduction
In 2014, ~37 million people were living with HIV-1 infection (www.unaids.org). Optimal patient outcomes are achieved by initiating combination antiretroviral therapy (ART) as soon infection is diagnosed, regardless of the CD4+ T cell count (1–3). ART reduces plasma virus levels to below the clinical detection limit (20–50 copies of HIV-1 RNA/ml) and halts disease progression (4–6). Recommended initial regimens consist of two nucleoside analog reverse transcriptase inhibitors and a third drug, either an integrase inhibitor or the protease inhibitor darunavir (3). Although ART effectively suppresses viremia, it is not curative, and viremia rebounds upon ART cessation (7, 8). Therefore, lifelong treatment is required. Providing lifelong treatment for all infected individuals poses a major economic and logistical challenge. Only 15 million people currently receive ART. The tolerability of ART regimens has improved dramatically, but long term drug toxicity is also a concern. Other problems include the emergence of resistance with suboptimal treatment and the stigma associated with the infection. For these reasons, there is great current interest in a cure (9, 10).
The principal barrier to cure is a stable reservoir of latent HIV-1 in resting CD4+ T cells (11, 12). The reservoir persists even in patients on long term ART who have no detectable viremia (13–18). The cells comprising this reservoir have a memory phenotype (12, 19–23). Direct measurements of the latent reservoir in patients on ART show a very slow decay rate (t1/2=3.7 years) (16, 17). At this rate, eradication of a reservoir of 106 cells would require 73 years, making cure unlikely even with lifelong ART. Thus, research towards a cure focuses on eliminating this reservoir. Recent reviews have discussed molecular mechanisms of HIV-1 latency (24–27) and approaches for eliminating the reservoir (10, 28–30). Here we consider explanations for its remarkable stability.
Why does HIV-1 establish latent infection?
Viral latency is a reversibly nonproductive state of infection of individual cells (31). Latently infected cells contain a stable form of the viral genome, either as a circular plasmid in the case of herpesviruses or as a linear provirus stably integrated into host cell DNA in the case of HIV-1. During latency, there is highly restricted expression of viral genes (31). For some herpesviruses, latency evolved as an essential mechanism of immune evasion and viral persistence (31, 32). For HIV-1, latency is not necessary for persistence as active viral replication occurs throughout the course of infection in untreated patients (33). Escape from immune responses is through rapid evolution of variants not recognized by cytolytic T lymphocytes (CTL) or neutralizing antibodies (34–41). Nevertheless, a latent reservoir is established rapidly in all HIV-1-infected individuals (42). Latently infected cells can be detected in the rare individuals who spontaneously control HIV-1 infection without ART (43). Early ART restricts the size of the reservoir (22, 44) but does not block its establishment (42). In rhesus macaques infected with simian immunodeficiency virus (SIV), which also establishes a latent reservoir in resting CD4+ T cells (45, 46), initiation of ART on day 3 post infection prevents detectable viremia but not the establishment of a latent reservoir (47). Thus it is difficult to prevent the establishment of the latent reservoir.
A recent theory suggests that HIV-1 evolved a mechanism for rapid establishment of latent infection to facilitate transmission across mucosal barriers (48, 49). Latency is proposed to serve as a “bet-hedging strategy” that allows some infected cells to survive long enough to transit the mucosa. However, as is discussed below, infected cells can remain in a latent state for years, and a long time interval between mucosal exposure and viremia has never been documented.
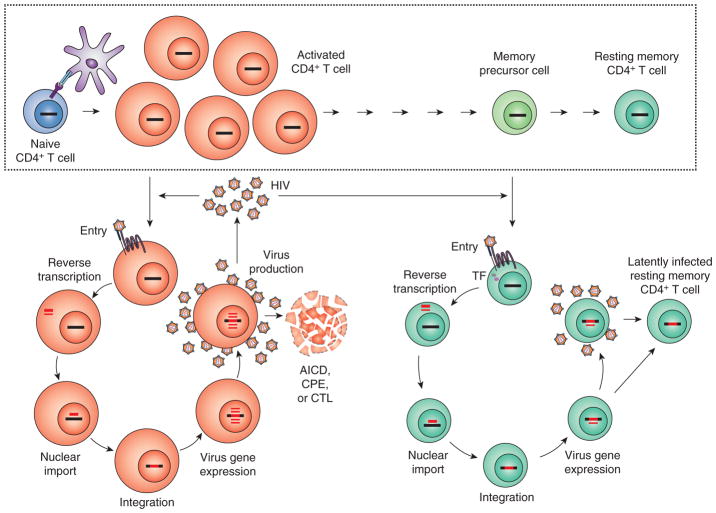
Latency is most simply explained as a consequence of viral tropism for activated CD4+ T cells which can transition to a resting memory state that is non-permissive for replication (Fig. 1). HIV-1 has a strong propensity to infect activated CD4+ T cells (50, 51). CCR5, a critical co-receptor for entry of the commonly transmitted forms of HIV-1 (52–57), is upregulated on CD4+ T cell activation (58). Following entry, reverse transcription of the viral RNA genome into DNA and integration of the resulting provirus into host cell DNA occur within hours (59). Transcription of the integrated provirus then begins because active nuclear forms of key host factors needed for the initiation and elongation of viral transcription, including NFκB, NFAT, and pTEFb, are present in activated cells (60–67). In contrast, resting CD4+ T cells mostly lack CCR5 expression (58), and other factors interfere with HIV-1 replication even when the virus has successfully entered. The cellular protein SAMHD1, a deoxynucleoside triphosphate triphosphohydrolase, depletes dNTP levels, thus impeding reverse transcription (68–70). It is expressed at high levels in myeloid cells and resting CD4+ T cells (52–55). Interestingly, SIV and HIV-2 encodes a protein, Vpx, that promotes SAMHD1 degradation (68, 71). However, HIV-1 lacks Vpx, and thus reverse transcription in resting CD4+ T cells is inefficient, taking as long as 3 days (72–74). The static nature of the actin cytoskeleton in resting cells inhibits delivery of the reverse transcribed viral genome to the nucleus (75). These delays facilitate recognition of DNA intermediates generated during reverse transcription by a host innate DNA sensor, IFI16, leading to caspase-1 activation and a proinflammatory form of cell death known as pyroptosis (76–78). Additional barriers to replication in resting CD4+ T cells include the lack of active forms of NFκB, NFAT, and pTEFb needed for transcription of the provirus (60–63, 65, 66).
Figure 1.
Model for the establishment of latent HIV-1 infection in resting memory CD4+ T cells. The normal process of memory cell generation (boxed) involves the exposure of a resting CD4+ T cells to antigen which leads to blast transformation, proliferation, and differentiation into effector cells. Many effector cells die during the contraction phase of the immune response, but a fraction survive and gradually return to a quiescent state as long-lived resting memory cells. Most resting CD4+ T cells lack expression of CCR5, a critical coreceptor for HIV-1 entry. Activation of resting cells by antigen (Ag) upregulates CCR5 expression and reverses other blocks to HIV-1 replication in resting CD4+ T cells, allowing productive infection of these cells. Most productively infected CD4+ T lymphoblasts die rapidly from activation-induced cells death (AICD), viral cytopathic effects (CPE), or lysis by CTL. As activated cells transition back to a resting state, active forms of key host transcription factors needed for HIV-1 gene expression are sequestered. Infection at this stage may lead to latent infection rather than cell death. Other models posit direct infection of resting cells. Please see text for references.
Although activated CD4+ T cells are the principle target for HIV-1, they die quickly after infection. Classic studies of viral dynamics revealed a rapid decay in viremia when new infection events are blocked with ART (6, 79–81). This decay reflects the short half-life of plasma virions (t1/2 ~ minutes) and of the infected cells that produce most of the plasma virus (t1/2 ~ 1 day). Activated T cells are prone to die in the contraction phase of immune responses due to activation-induced cell death (82). In addition, productively infected cells may die from other cell death pathways triggered by viral proteins or by integration of the provirus into the host cell genome (83, 84). Infected CD4+ T lymphoblasts may also be lysed by CD8+ CTL (34, 85–87). Surprisingly CTL do not appear to shorten the t1/2 of productively infected cells (88, 89). Nevertheless, it appears that most productively infected CD4+ T lymphoblasts are short-lived.
Given that resting CD4+ T cells are resistant to infection and that activated CD4+ T cells die quickly after infection, how is the latent reservoir established? Some infected CD4+ T lymphoblasts may survive long enough to revert to a resting memory state that is non-permissive for viral gene expression (11), particularly if they are infected within a narrow time window when still permissive for steps in the life cycle up through integration, but not for high level gene expression (Fig. 1). Thus establishment of latent infection is a rare event, consistent with the low frequency of latently infected cells in vivo (1/106) (13, 16–18). Latency may be further enforced by silencing epigenetic modifications of the integrated provirus (90–92). In this latent state, the virus persists essentially as genetic information. When antigen or cytokines subsequently activate the cell, the provirus is transcribed, viral proteins are produced, and virus particles are released. Given the long t1/2 of memory T cell responses and the fact that the latent proviruses in these cells are not detected by the immune system or targeted by ART, stable persistence of HIV-1 is not surprising. This simple model views latency in the context of the normal physiology of immunologic memory, thereby explaining all clinical observations regarding HIV-1 persistence without requiring the evolution of special viral mechanisms for latency.
Residual viremia, the latent reservoir, and viral rebound
Trace levels of free virus (~1 copy/ml) are present in the plasma of most patients on ART (93–95). Sequence analysis of the residual viremia (RV) reveals that these viruses resemble viremia present earlier in infection, are sensitive to the patient’s current ART regimen, and generally do not show evidence of ongoing evolution (96–100). These features all suggest RV originates from a stable reservoir (101). In situations where evolution has been detected, suboptimal ART may be the cause (102). Importantly, intensification of standard three drug ART with additional antiretroviral drugs from a different class does not reduce RV (103–105), indicating that it originates from long-lived cells infected prior to ART. Latently infected resting CD4+ T cells are at least one source of RV. The presence of RV suggests that multiple latently infected cells are become activated every day. While patients remain on ART, the viruses released do not infect additional cells. However, if ART is interrupted, viral rebound occurs. Rebound is typically seen within 2 weeks (7, 8), the time required for washout of antiretroviral drugs and growth of the recently released viruses to detectable levels. The rebound virus is archival in character, consistent with the conclusion that it originates from a stable latent reservoir (106). The limited variation in time to rebound, despite a two log variation in reservoir size, also suggests that multiple cells are activated per day (107). This conclusion is consistent with a recent analysis of viral rebound which detected multiple viral lineages emerging in multiple sites (lymph node, ileum, and rectum) (8).
Evidence for latent infection of resting memory CD4+ T cells
This model for latent HIV-1 infection as a barrier to cure is supported by several lines of evidence:
Replication-competent HIV-1 can be readily recovered from highly purified resting CD4+ T cells from essentially all infected individuals, regardless of the duration of ART (12, 13, 15, 16, 18, 108). Recovery requires activating the cells to reverse latency, as predicted by the model. As is discussed below, recovery only fails when the size of the latent reservoir is substantially reduced (44, 109–111). Controversy remains over the question of whether other cell types including macrophages serve as stable HIV-1 reservoirs (112–123). To date, long term persistence of replication-competent HIV-1 in the setting of optimal ART has only been demonstrated for resting CD4+ T cells (124). This may in part reflect the difficulty of sampling tissue macrophages, particularly in sites such as the central nervous system. Persistence in tissue macrophages can in principle be studied in novel humanized mouse models (123) and in the SIV model, but only through the use of animals on fully suppressive, long term ART and with the caveat that restriction by SAMHD1 is counteracted by SIV Vpx (68, 69).
Latent HIV-1 is found in resting memory CD4+ T cells but only to a limited extent in naïve CD4+ T cells (12, 19–23).
The generation of latently infected cells can be reproduced in vitro in primary CD4+ T cells that have been activated in some manner, infected, and then cultured to allow reversion to a resting state (125–129). Restimulation of these cells through the T cell receptor (TCR) leads to HIV-1 gene expression. Together, these results support persistence of latent HIV-1 in resting CD4+ T cells that have been previously infected while in an activated state.
The general concept of HIV-1 latency is also strongly supported by the cure and “near cure” cases of the “Berlin patient” (109), the two “Boston patients” (110, 111), and the “Mississippi baby” (44). The Boston and Berlin patients were HIV-1-infected individuals who developed malignancies requiring hematopoietic stem cell transplantation (HSCT) resulting in immune reconstitution with donor cells. The Berlin patient received HSCT from a donor whose cells were homozygous for a deletion in CCR5, and was cured as the reconstituting T cells were not permissive for entry of R5-tropic HIV-1 (109). Attempts to reproduce this cure have thus far been unsuccessful due in large part to progression of the malignancy. In one case, the appearance of viral variants that utilize the alternative HIV-1 coreceptor, CXCR4, has been noted (130). The Boston patients received HSCT from donors with wild-type CCR5, and ART was continued throughout the transplant period to protect donor cells from infection. When apparently complete reconstitution with donor T cells had occurred, and HIV-1 was no longer detectable by standard assays, ART was interrupted. The patients maintained suppression of viremia for 3 and 8 months before sudden and dramatic rebounds.
The Mississippi baby, born to an infected mother who had no prenatal care, had a plasma HIV-1 RNA level of ~20,000 copies/ml shortly after birth and was immediately started on ART. Plasma HIV-1 RNA declined to below the limit of detection and remained there even after treatment was interrupted against medical advice between 15 and 18 months of age. Treatment was not restarted, and viremia remained undetectable for over 2 years before suddenly rebounding. Importantly, HIV-1-specific T cell responses were absent in all three subjects because of the transplant process or early treatment. Antibodies to HIV-1 were not detected in the Mississippi baby and were markedly decreased in the Boston patients. Because HIV-1 replication is exponential in the absence of immune responses and ART, HIV-1 persistence for months or years in these patients can be best explained by the non-replicating or latent form of the virus. In these cases, HSCT or early ART delayed rebound by reducing the number of latently infected cells to the point where stochastic reactivation was a rare event (107).
Explanations for the long t1/2 of the latent reservoir
The decay rate of the latent reservoir was originally measured using a viral outgrowth assay (VOA), which quantifies viral outgrowth from limiting dilutions of mitogen-stimulated resting CD4+ T cells from patients on ART (13, 131–133). The original VOA-based measurements of the reservoir decay, published in 1999 and 2003, indicated a t1/2 of 3.7 years (16, 17). This value was confirmed in a more recent study (t1/2 =3.6 years), indicating that despite the development of newer, less toxic, and more convenient ART regimens, the fundamental problem of the reservoir as a barrier to cure has not been overcome (18). A critical issue is whether the remarkable stability of the reservoir is the result of normal homeostatic mechanisms that maintain immunologic memory or other factors.
One controversial explanation for stability is that the reservoir is constantly replenished by a low level of de novo infection that continues despite ART (134). This replication may reflect inadequate drug levels in certain anatomical sites including the lymph nodes (135, 136) or cell-to-cell spread which is more difficult to block with ART (137). However, multiple lines of evidence indicate that ART effectively curtails new infection of susceptible cells. Because HIV-1 replication is invariably accompanied by the progressive accumulation of mutations (138) reflecting the error prone nature of reverse transcriptase (139) and possibly hypermutation by the host restriction factor APOBEC3G (40, 140–143), the lack of sequence evolution in the viral reservoir (22, 97, 100, 144, 145) indicates that ART blocks ongoing cycles of viral replication. A recent report claiming viral evolution is confounded by sampling only in the first 6 months of ART, a period during which short-lived populations of infected cells not representative of the stable reservoir are dominant (146). Prior to the development of effective ART, a dominant clinical issue was the evolution of drug resistance (147–151), but the incidence of resistance is now decreasing (152–154). Indeed there are overwhelming clinical data that ART is effective, and treated patients can expect a near normal life expectancy (3, 155–158). As mentioned above, the failure of ART intensification to reduce RV indicates that current ART regimens stop new infection events (103–105). Finally, in the delayed rebound cases mentioned above, HIV-1 persistence during ART cannot be explained by ongoing replication as this would have led to immediate rebound. Therefore, the stability of the latent reservoir is most likely due to the long t1/2 of memory T cells and their renewal through proliferation.
Functional studies have shown that memory CD4+ T cell responses in humans can provide lifelong immunity. In individuals who received the smallpox vaccine or cleared Hepatitis C infection, virus-specific CD4+ T cell responses persist for decades despite the absence of further antigen exposure (159, 160). Early studies of the memory cell lifespan in humans examined radiation-induced chromosomal abnormalities that preclude cell proliferation. This allowed estimates of the intermitotic t1/2 of lymphocytes (161, 162). The measured t1/2 of 22 weeks for memory T cells is roughly consistent with subsequent in vivo measurements using glucose or deuterium labeling which indicate a t1/2 on the order of months for human memory CD4+ T cells (163, 164; also reviewed in 165). This is substantially shorter than the t1/2 of individual naïve T cells (1–8 years). Importantly, it is shorter than the t1/2 of the HIV-1 reservoir (3.7 years) and of functional memory T cell responses (8–12 years). The discrepancy between the half-life of individual memory T cells and the overall memory immune response suggests that proliferation of memory cells must contribute to the stability of the latent reservoir. However, as discussed above, productively infected cells have a very short t1/2, and therefore the concept that infected cells can proliferate is not well appreciated. The HIV-1 Vpr protein induces cell cycle arrest at G2 by interacting with a host E3 ubiquitin ligase (166, 167) and stimulating the degradation of host proteins including the DNA replication factor MCM10 (168). For cells in a latent state of infection, this block to proliferation is not operative, and latently infected cells can, in principle, proliferate if the driving stimulus does not strongly upregulate HIV-1 gene expression (169–172).
Memory CD4+ T cell proliferation can be driven by antigen, cross-reactive recognition of other self or foreign peptides presented with MHC class II, or cytokines. In the murine system, neither antigen nor class II MHC is required for memory T cell persistence (173), although memory CD4+ T cells that persist in the absence of MHC class II are functionally impaired (174). The requirements for maintenance of human memory CD4+ T cell responses is less clear, and could include interactions with cognate signals or cytokines. Little is currently known about the antigen specificity of cells harboring latent HIV-1, although a small subset of them may be HIV-1-specific (175). However, human memory CD4+ T cells exhibit cross-reactivity, and specificities for antigens never encountered can be detected among memory CD4+ T cells in peripheral blood (176). As discussed above, it is expected that stimuli acting through the TCR will upregulate expression of latent HIV-1, but this may not be the case for cytokine-driven proliferation. The cytokines implicated in memory T cell homeostasis and survival are IL-7 and IL-15 (reviewed in 177). IL-7 is required for stimulating homeostatic proliferation of memory CD4+ T cells. Mice deficient in IL-7 (or IL-7R) have severely reduced total T lymphocyte levels, and reduced splenic size and cellularity (178). IL-15 also plays a role in homeostatic proliferation of memory CD4+ T cells (179). Early in vitro studies indicated that IL-7 can actually induce expression of latent HIV-1 (180, 181). However, in patients on ART, infusion of IL-7 leads to the proliferation of memory CD4+ T cells, including latently infected cells, with little or no induction of HIV-1 gene expression (182, 183). In vitro studies in a primary cell model of HIV-1 latency confirm that latently infected cells can proliferate in response to IL-7 (plus IL-2) without upregulation of HIV-1 gene expression (169). These studies suggest that the latent reservoir can be maintained within memory T cells undergoing homeostatic turnover. Analysis of memory T cell subsets has provided additional insight into this issue.
Memory CD4+ T cell subsets
Memory T cells can be divided into two main subsets, central memory (TCM) and effector memory cells (TEM), based on expression of homing and chemokine receptors involved in preferential trafficking to secondary lymphoid organs or peripheral sites, respectively (184). HIV-1 DNA is preferentially harbored in TCM and another subset of memory T cells, transitional memory T cells (TTM) (21). TTM have a phenotype (CD45RA−, CD27+, CCR7−) intermediate between TCM and TEM. The two main subsets of CD4+ memory T cells that harbor latent HIV-1, TCM and TTM, may provide a more stable reservoir for HIV-1 than TEM cells, which have a higher proliferative index and are more susceptible to programmed cell death (21, 185). A recent study using the viral outgrowth assay rather than PCR demonstrated replication-competent HIV-1 persisting in TCM but to a much lesser extent in TTM, indicating that TCM may represent the major source of persistent HIV-1 in most patients (186).
Another recently defined subset of memory CD4+ T cells that may contribute to HIV-1 persistence is the stem cell-like memory T cell subset (TSCM) (187). TSCM are phenotypically similar to naïve T cells (TN) in that they are CD45RO−, CD45RA+, and CCR7+. However, they also express surface markers characteristic of memory cells, such as CD95 and IL-2Rβ (187). TSCM rapidly respond to antigen and secrete IFN-γ, IL-2, and TNF. They are also stimulated to proliferate by IL-7. A stepwise progression from TN to TSCM to TCM to TEM has been proposed, with TSCM potentially able to give rise to other types of memory T cells and self-renew upon stimulation. TSCM can be infected with HIV-1 in vitro, and in patients on ART, HIV-1 DNA is present in TSCM at higher levels than in other memory subsets (188). Although latently infected TSCM represent only a small fraction of the total reservoir, they may be of particular importance because of their stability and capacity for self-renewal (23, 188).
In summary, analysis of memory subsets reveals HIV-1 genomes distributed in multiple memory cell subsets, with higher frequencies in subsets with greater potential to survive. Several issues remain. One concern is that many studies of the distribution of HIV-1 genomes in T cell subsets rely primarily on PCR-based measures of the proviral DNA. This is problematic in that the vast majority of proviruses in resting CD4+ T cells from treated patients are highly defective (189). There is also substantial patient-to-patient variability in the distribution of viral genomes within these subsets. Finally, these subsets are not static and can interconvert in ways that are not yet fully understood, and it is therefore unclear whether latent HIV-1 stably persists in a given subset.
Anatomical distribution of the latent reservoir
Most studies of the latent reservoir sample CD4+ T cells from peripheral blood. Given the continuous recirculation and wide tissue distribution of memory T cells, it is generally presumed that latently infected resting CD4+ T cells will be present in most secondary lymphoid organs and in non–lymphoid tissues (190–192). Early studies demonstrated latently infected cells at roughly equal frequency in blood and lymph nodes (12). In the SIV model, latently infected resting CD4+ T cells were demonstrated in blood, lymph node, and spleen (45, 46). Interestingly, as is discussed below, some recently described memory cell populations that are not present in the blood may also contribute to HIV-1 persistence.
HIV-1 can infect follicular helper T cells (TFH) (193–196), and this population has received considerable attention because CD8+ CTL lack chemokine receptors needed for migrating into B cell follicles (196), thus making the follicles a site of “immune privilege”. In the subset of rhesus macaques that spontaneously control SIV, viral replication is restricted to TFH, presumably because CD8+ CTL lyse infected cells elsewhere in the node (196). The extent to which TFH serve as a long term reservoir for HIV-1 in the setting of optimal ART remains to be determined. If latently infected TFH persist, HIV-1 eradication strategies may need to include not only latency reversing agents and stimuli to enhance the CD8+ CTL response (197), but also interventions to disrupt B cell follicles to permit access by CTL (196).
Another population of memory T cells that could potentially harbor latent HIV-1 is the tissue resident memory T cell (TRM) populations (198, 199). Pioneering studies in the murine system demonstrated wide distribution of memory CD4+ T cells, including in non-lymphoid tissues such as liver and lung (190). Subpopulations of memory cells may be generated in or recruited to particular non- lymphoid tissues where they reside for long periods of time (192, 199). These TRM lack expression of CCR7 and share phenotypic and functional properties with TEM. However, unlike other memory subsets, they express CD69, a cell surface lectin that is upregulated at early times following T cell activation. In humans, the majority of TEM cells in lymphoid and mucosal tissues, including lungs and intestines, express CD69 and therefore may be retained in these sites as TRM (191, 200). Human skin also contains significant TRM populations (198). Thus far, TRM have not been directly examined for the presence of latent HIV-1. However, persistent HIV-1 has been detected in gut associated lymphoid tissue of individuals on ART (201, 202). TRM are prominent in the lamina propria and among intraepithelial lymphocytes, and it is possible that TRM harbor HIV-1. Many CD4+ TRM exhibit activated phenotypes, with reduced surface expression of CD28 (191), and therefore it is unclear whether latent infection can be established in these cells. Further characterization of tissue-specific reservoirs for HIV-1 is an important research priority.
Evidence for clonal expansion of infected cells
Consideration of the mechanism of memory cell homeostasis suggests that the stability of the latent reservoir is at least partially dependent upon the ability of infected cells to proliferate. Several studies have provided direct evidence for clonal expansion of infected cells, beginning with studies of RV (100, 102). Although patients starting ART during chronic infection harbor diverse viral quasispecies (138), the RV is often dominated by identical sequences detected on independent sampling over months to years. The origin of these sequences is unknown, but may reflect infection of cells that then proliferate giving rise to multiple progeny cells carrying identical proviruses (100, 102, 203). The fraction of identical HIV-1 sequences within samples from patients on ART increases with time, consistent with proliferation of infected cells (145). More recent studies have used integration site analysis to provide definitive evidence for the proliferation of infected cells. The sites of integration in different cells are generally different and are distributed widely throughout the human genome. Early studies in cell lines infected in vitro with HIV-1 (204) and in resting CD4+ T cells from patients on ART (205) revealed a strong preference for integration within active transcriptional units. However, integration occurs in either orientation with respect to the host gene, and there is no consensus sequence at the integration site. Therefore, the precise human sequence at the junction between host and HIV-1 DNA uniquely identifies individual infection events and thus all the clonal progeny of a single infected cell. In addition, novel deep sequencing analysis allows enumeration of the clonal progeny of a single infected cell within a sample by detection of differences in the random break points in fragments of sheared DNA containing the same integration site (170, 206). Application of this and related approaches to CD4+ T cells from patients on ART has provided dramatic evidence for clonal expansion (170, 171, 207). Maldarelli et al. showed that 43% of 2410 integration sites in CD4+ T cells from 5 patients were in clonally expanded cells (170). The finding that multiple cells with the same integration site can be captured in a single blood sample reflects dramatic clonal expansion in vivo.
Interestingly, some expanded clones had proviruses integrated in human genes associated with cell growth, and some of these genes have been observed to contain integrated proviruses in multiple independent studies (170, 171, 205, 207, 208). These include myocardin-like protein 2 (MKL2), a transcription factor, and basic leucine zipper transcription factor 2 (BACH2), a transcription regulator affecting lymphocyte growth, activation, senescence, and cytokine homeostasis. For these genes, integration events were found in a specific regions of the gene and in the same transcriptional orientation as the host gene. This skewed pattern reflects a post-integration selection process that favors the in vivo growth and survival of cells with those integration events since these patterns were not seen in in vitro infections (170, 171, 204). These results raise the interesting possibility that integration into certain host genes contributes to HIV-1 persistence by stimulating infected cells to proliferate in a manner distinct from homeostatic proliferation. The molecular mechanisms are currently unclear.
A caveat to these studies is that the methods used do not capture the full sequence of the integrated provirus. Some methods capture only the junction between host and HIV-1 DNA. Given that the vast majority of proviruses are defective, as a result of large internal deletions or APOBEC3G-mediated hypermutation (189, 207), it must be assumed that most expanded clones carry defective proviruses. There is no selective pressure against cells carrying defective proviruses that do not produce viral proteins. Previous studies have described expanded clones carrying defective proviruses, some of which persist for many years (22, 209). However, a recent report has described dramatic in vivo expansion of an infected CD4+ T cell clone in a treated patient who also had squamous cell carcinoma (210). Importantly, this clone was capable of producing replication-competent virus. The integration site could not be precisely localized because it was in a region of repetitive sequence. The clone was found widely distributed in sites of metastatic tumor throughout the body, raising the possibility that the clonal expansion occurred in response to tumor antigen. A current issue of great importance is the extent to which expanded cellular clones harbor replication-competent HIV-1.
Implications
The stability of the latent reservoir is the principal reason that HIV-1 infection cannot be cured. The normal mechanisms that maintain immunologic memory provide a simple explanation for this stability. However, the pool of latently infected cells is not static. While the total pool size decreases only very slowly, cells in the reservoir are continually being activated to produce virus that is evident as residual viremia. These cells may die, but homeostatic proliferation of memory cells helps to balance the loss. In addition, a more cell autonomous process of proliferation driven by integration-site dependent alterations in host gene expression may allow some infected cells to undergo dramatic clonal expansion. Efforts to target the latent reservoir have generally assumed that intervention-dependent reductions in the frequency of latently infected cells will be stable so that repeated interventions will ultimately allow cure. The possibility that subpopulations of infected cells can continue to proliferate may further complicate eradication efforts.
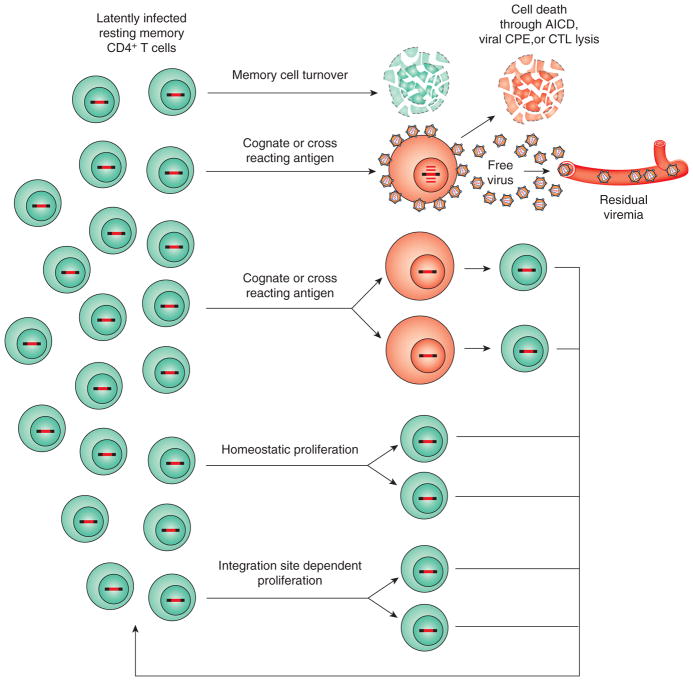
Figure 2.
Dynamics of the latent reservoir. ART largely blocks new infection of susceptible cells. In patients on long term ART, the pool of latently infected cells is extremely stable (t½ = 3.7 years) so that memory cell turnover must be largely balanced by proliferation of previously infected cells. Latently infected resting memory CD4+ T cells occasionally encounter the relevant cognate antigen (or a cross-reacting antigen) and become activated. Activation reverses latency, allowing viral gene expression and virus production. In patients on ART, the released viruses do not successfully infect new cells, but may be detected at very low levels in the plasma where they constitute the residual viremia (RV). Most productively infected cells die quickly from AICD, CPE, or lysis by CTL. It is possible that some degree of antigen-driven proliferation may occur without activation of viral gene expression. Homeostatic proliferation of memory cells may also occur without reactivating viral gene expression. For some infected cells, integration of the provirus into genes associated with cell growth may also stimulate proliferation. See text for references.
Acknowledgments
We thank Dr. Janet Siliciano for critically reading the manuscript.
Grant support
This work was supported by the Martin Delaney CARE and DARE Collaboratories (NIH grants AI096113 and 1U19AI096109), by an ARCHE Collaborative Research Grant from the Foundation for AIDS Research (amFAR 108165-50-RGRL), by the Johns Hopkins Center for AIDS Research (P30AI094189), by NIH grant 43222, and by the Howard Hughes Medical Institute and the Bill and Melinda Gates Foundation. D.L.F. is supported by NIH grants AI100119, AI106697, and HL116136.
References
- 1.INSIGHT START Study Group. Lundgren JD, Babiker AG, Gordin F, Emery S, Grund B, Sharma S, Avihingsanon A, Cooper DA, Fatkenheuer G, Llibre JM, Molina JM, Munderi P, Schechter M, Wood R, Klingman KL, Collins S, Lane HC, Phillips AN, Neaton JD. Initiation of Antiretroviral Therapy in Early Asymptomatic HIV Infection. N Engl J Med. 2015;373:795–807. doi: 10.1056/NEJMoa1506816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danel C, Moh R, Gabillard D, Badje A, Le Carrou J, Ouassa T, Ouattara E, Anzian A, Ntakpe JB, Minga A, Kouame GM, Bouhoussou F, Emieme A, Kouame A, Inwoley A, Toni TD, Ahiboh H, Kabran M, Rabe C, Sidibe B, Nzunetu G, Konan R, Gnokoro J, Gouesse P, Messou E, Dohoun L, Kamagate S, Yao A, Amon S, Kouame AB, Koua A, Kouame E, Ndri Y, Ba-Gomis O, Daligou M, Ackoundze S, Hawerlander D, Ani A, Dembele F, Kone F, Guehi C, Kanga C, Koule S, Seri J, Oyebi M, Mbakop N, Makaila O, Babatunde C, Babatounde N, Bleoue G, Tchoutedjem M, Kouadio AC, Sena G, Yededji SY, Assi R, Bakayoko A, Mahassadi A, Attia A, Oussou A, Mobio M, Bamba D, Koman M, Horo A, Deschamps N, Chenal H, Sassan-Morokro M, Konate S, Aka K, Aoussi E, Journot V, Nchot C, Karcher S, Chaix ML, Rouzioux C, Sow PS, Perronne C, Girard PM, Menan H, Bissagnene E, Kadio A, Ettiegne-Traore V, Moh-Semde C, Kouame A, Massumbuko JM, Chene G, Dosso M, Domoua SK, N’Dri-Yoman T, Salamon R, Eholie SP, Anglaret X TEMPRANO ANRS 12136 Study Group. A Trial of Early Antiretrovirals and Isoniazid Preventive Therapy in Africa. N Engl J Med. 2015;373:808–822. doi: 10.1056/NEJMoa1507198. [DOI] [PubMed] [Google Scholar]
- 3.Gunthard HF, Aberg JA, Eron JJ, Hoy JF, Telenti A, Benson CA, Burger DM, Cahn P, Gallant JE, Glesby MJ, Reiss P, Saag MS, Thomas DL, Jacobsen DM, Volberding PA International Antiviral Society- USA Panel. Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA. 2014;312:410–425. doi: 10.1001/jama.2014.8722. [DOI] [PubMed] [Google Scholar]
- 4.Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, McMahon D, Richman DD, Valentine FT, Jonas L, Meibohm A, Emini EA, Chodakewitz JA. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 5.Hammer SM, Squires KE, Hughes MD, Grimes JM, Demeter LM, Currier JS, Eron JJ, Jr, Feinberg JE, Balfour HH, Jr, Deyton LR, Chodakewitz JA, Fischl MA. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 6.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 7.Davey RT, Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS, Lane HC. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A. 1999;96:15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, Schmidt TE, Thorkelson A, Reilly C, Perkey K, Reimann TG, Utay NS, Nganou Makamdop K, Stevenson M, Douek DC, Haase AT, Schacker TW. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A. 2015;112:E1126–34. doi: 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 10.Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, Chun TW, Churchill M, Mascio MD, Katlama C, Lafeuillade A, Landay A, Lederman M, Lewin SR, Maldarelli F, Margolis D, Markowitz M, Martinez-Picado J, Mullins JI, Mellors J, Moreno S, O’Doherty U, Palmer S, Penicaud MC, Peterlin M, Poli G, Routy JP, Rouzioux C, Silvestri G, Stevenson M, Telenti A, Lint CV, Verdin E, Woolfrey A, Zaia J, Barre-Sinoussi F The International AIDS Society Scientific Working Group on HIV Cure. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol. 2012;12:607–614. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1:1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 12.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 13.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 14.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 15.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 17.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 18.Crooks AM, Bateson R, Cope AB, Dahl NP, Griggs MK, Kuruc JD, Gay CL, Eron JJ, Margolis DM, Bosch RJ, Archin NM. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J Infect Dis. 2015 doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierson T, Hoffman TL, Blankson J, Finzi D, Chadwick K, Margolick JB, Buck C, Siliciano JD, Doms RW, Siliciano RF. Characterization of chemokine receptor utilization of viruses in the latent reservoir for human immunodeficiency virus type 1. J Virol. 2000;74:7824–7833. doi: 10.1128/jvi.74.17.7824-7833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol. 2004;78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, Epling L, Tan A, Ho T, Lemey P, Shao W, Hunt PW, Somsouk M, Wylie W, Douek DC, Loeb L, Custer J, Hoh R, Poole L, Deeks SG, Hecht F, Palmer S. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A. 2013;110:E4987–96. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaafoura S, de Goer de Herve MG, Hernandez-Vargas EA, Hendel-Chavez H, Abdoh M, Mateo MC, Krzysiek R, Merad M, Seng R, Tardieu M, Delfraissy JF, Goujard C, Taoufik Y. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4(+) memory T Cells. Nat Commun. 2014;5:5407. doi: 10.1038/ncomms6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karn J. The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS. 2011;6:4–11. doi: 10.1097/COH.0b013e328340ffbb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taube R, Peterlin M. Lost in transcription: molecular mechanisms that control HIV latency. Viruses. 2013;5:902–927. doi: 10.3390/v5030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruelas DS, Greene WC. An integrated overview of HIV-1 latency. Cell. 2013;155:519–529. doi: 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dahabieh MS, Battivelli E, Verdin E. Understanding HIV latency: the road to an HIV cure. Annu Rev Med. 2015;66:407–421. doi: 10.1146/annurev-med-092112-152941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spivak AM, Planelles V. HIV-1 Eradication: Early Trials (and Tribulations) Trends Mol Med. 2016;22:10–27. doi: 10.1016/j.molmed.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Archin NM, Sung JM, Garrido C, Soriano-Sarabia N, Margolis DM. Eradicating HIV-1 infection: seeking to clear a persistent pathogen. Nat Rev Microbiol. 2014;12:750–764. doi: 10.1038/nrmicro3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katlama C, Deeks SG, Autran B, Martinez-Picado J, van Lunzen J, Rouzioux C, Miller M, Vella S, Schmitz JE, Ahlers J, Richman DD, Sekaly RP. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet. 2013;381:2109–2117. doi: 10.1016/S0140-6736(13)60104-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speck SH, Ganem D. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe. 2010;8:100–115. doi: 10.1016/j.chom.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perng GC, Jones C. Towards an understanding of the herpes simplex virus type 1 latency-reactivation cycle. Interdiscip Perspect Infect Dis. 2010;2010:262415. doi: 10.1155/2010/262415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piatak M, Jr, Saag MS, Yang LC, Clark SJ, Kappes JC, Luk KC, Hahn BH, Shaw GM, Lifson JD. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 34.Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 35.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 36.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 38.Jones NA, Wei X, Flower DR, Wong M, Michor F, Saag MS, Hahn BH, Nowak MA, Shaw GM, Borrow P. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J Exp Med. 2004;200:1243–1256. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frost SD, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, Petropoulos CJ, Little SJ, Richman DD. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A. 2005;102:18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, Daniels M, Ferrari G, Haynes BF, McMichael A, Shaw GM, Hahn BH, Korber B, Seoighe C. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, Lai J, McHugh HL, Hao H, Zhang H, Margolick JB, Gurer C, Murphy AJ, Valenzuela DM, Yancopoulos GD, Deeks SG, Strowig T, Kumar P, Siliciano JD, Salzberg SL, Flavell RA, Shan L, Siliciano RF. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buckheit RW, 3rd, Salgado M, Martins KO, Blankson JN. The implications of viral reservoirs on the elite control of HIV-1 infection. Cell Mol Life Sci. 2013;70:1009–1019. doi: 10.1007/s00018-012-1101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Persaud D, Gay H, Ziemniak C, Chen YH, Piatak M, Jr, Chun TW, Strain M, Richman D, Luzuriaga K. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N Engl J Med. 2013;369:1828–1835. doi: 10.1056/NEJMoa1302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen A, Zink MC, Mankowski JL, Chadwick K, Margolick JB, Carruth LM, Li M, Clements JE, Siliciano RF. Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J Virol. 2003;77:4938–4949. doi: 10.1128/JVI.77.8.4938-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dinoso JB, Rabi SA, Blankson JN, Gama L, Mankowski JL, Siliciano RF, Zink MC, Clements JE. A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J Virol. 2009;83:9247–9257. doi: 10.1128/JVI.00840-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, Smith JY, Brinkman AL, Peter LE, Mathew SI, Smith KM, Borducchi EN, Rosenbloom DI, Lewis MG, Hattersley J, Li B, Hesselgesser J, Geleziunas R, Robb ML, Kim JH, Michael NL, Barouch DH. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014;512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouzine IM, Weinberger AD, Weinberger LS. An evolutionary role for HIV latency in enhancing viral transmission. Cell. 2015;160:1002–1012. doi: 10.1016/j.cell.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razooky BS, Pai A, Aull K, Rouzine IM, Weinberger LS. A hardwired HIV latency program. Cell. 2015;160:990–1001. doi: 10.1016/j.cell.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Margolick JB, Volkman DJ, Folks TM, Fauci AS. Amplification of HTLV-III/LAV infection by antigen-induced activation of T cells and direct suppression by virus of lymphocyte blastogenic responses. J Immunol. 1987;138:1719–1723. [PubMed] [Google Scholar]
- 51.Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, Reimann KA, Reinhart TA, Rogan M, Cavert W, Miller CJ, Veazey RS, Notermans D, Little S, Danner SA, Richman DD, Havlir D, Wong J, Jordan HL, Schacker TW, Racz P, Tenner-Racz K, Letvin NL, Wolinsky S, Haase AT. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science. 1999;286:1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 52.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 53.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 54.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 55.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 56.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 57.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 58.Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci U S A. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohammadi P, Desfarges S, Bartha I, Joos B, Zangger N, Munoz M, Gunthard HF, Beerenwinkel N, Telenti A, Ciuffi A. 24 hours in the life of HIV-1 in a T cell line. PLoS Pathog. 2013;9:e1003161. doi: 10.1371/journal.ppat.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 61.Bohnlein E, Lowenthal JW, Siekevitz M, Ballard DW, Franza BR, Greene WC. The same inducible nuclear proteins regulates mitogen activation of both the interleukin-2 receptor-alpha gene and type 1 HIV. Cell. 1988;53:827–836. doi: 10.1016/0092-8674(88)90099-2. [DOI] [PubMed] [Google Scholar]
- 62.Duh EJ, Maury WJ, Folks TM, Fauci AS, Rabson AB. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci U S A. 1989;86:5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams M, Sharmeen L, Kimpton J, Romeo JM, Garcia JV, Peterlin BM, Groudine M, Emerman M. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc Natl Acad Sci U S A. 1994;91:3862–3866. doi: 10.1073/pnas.91.9.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kinoshita S, Chen BK, Kaneshima H, Nolan GP. Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell. 1998;95:595–604. doi: 10.1016/s0092-8674(00)81630-x. [DOI] [PubMed] [Google Scholar]
- 65.Rice AP, Herrmann CH. Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages. Curr HIV Res. 2003;1:395–404. doi: 10.2174/1570162033485159. [DOI] [PubMed] [Google Scholar]
- 66.Lin X, Irwin D, Kanazawa S, Huang L, Romeo J, Yen TS, Peterlin BM. Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J Virol. 2003;77:8227–8236. doi: 10.1128/JVI.77.15.8227-8236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pessler F, Cron RQ. Reciprocal regulation of the nuclear factor of activated T cells and HIV-1. Genes Immun. 2004;5:158–167. doi: 10.1038/sj.gene.6364047. [DOI] [PubMed] [Google Scholar]
- 68.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, Mulder LC, Fernandez-Sesma A, Rutsch F, Simon V, Konig R, Flory E. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7:e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Romani B, Cohen EA. Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase. Curr Opin Virol. 2012;2:755–763. doi: 10.1016/j.coviro.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 73.Pierson TC, Zhou Y, Kieffer TL, Ruff CT, Buck C, Siliciano RF. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J Virol. 2002;76:8518–8531. doi: 10.1128/JVI.76.17.8518-8531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taylor HE, Simmons GE, Jr, Mathews TP, Khatua AK, Popik W, Lindsley CW, D’Aquila RT, Brown HA. Phospholipase D1 Couples CD4+ T Cell Activation to c-Myc-Dependent Deoxyribonucleotide Pool Expansion and HIV-1 Replication. PLoS Pathog. 2015;11:e1004864. doi: 10.1371/journal.ppat.1004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoder A, Yu D, Dong L, Iyer SR, Xu X, Kelly J, Liu J, Wang W, Vorster PJ, Agulto L, Stephany DA, Cooper JN, Marsh JW, Wu Y. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell. 2008;134:782–792. doi: 10.1016/j.cell.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Munoz-Arias I, Greene WC. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Munoz-Arias I, Doitsh G, Yang Z, Sowinski S, Ruelas D, Greene WC. Blood-Derived CD4 T Cells Naturally Resist Pyroptosis during Abortive HIV-1 Infection. Cell Host Microbe. 2015;18:463–470. doi: 10.1016/j.chom.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 80.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 81.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 82.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 83.Sakai K, Dimas J, Lenardo MJ. The Vif and Vpr accessory proteins independently cause HIV-1-induced T cell cytopathicity and cell cycle arrest. Proc Natl Acad Sci U S A. 2006;103:3369–3374. doi: 10.1073/pnas.0509417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 85.Walker BD, Chakrabarti S, Moss B, Paradis TJ, Flynn T, Durno AG, Blumberg RS, Kaplan JC, Hirsch MS, Schooley RT. HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature. 1987;328:345–348. doi: 10.1038/328345a0. [DOI] [PubMed] [Google Scholar]
- 86.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 88.Wong JK, Strain MC, Porrata R, Reay E, Sankaran-Walters S, Ignacio CC, Russell T, Pillai SK, Looney DJ, Dandekar S. In vivo CD8+ T-cell suppression of siv viremia is not mediated by CTL clearance of productively infected cells. PLoS Pathog. 2010;6:e1000748. doi: 10.1371/journal.ppat.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, Miller MD, Else J, Pandrea I, Estes JD, Apetrei C, Schmitz JE, Ribeiro RM, Perelson AS, Silvestri G. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog. 2010;6:e1000747. doi: 10.1371/journal.ppat.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He G, Ylisastigui L, Margolis DM. The regulation of HIV-1 gene expression: the emerging role of chromatin. DNA Cell Biol. 2002;21:697–705. doi: 10.1089/104454902760599672. [DOI] [PubMed] [Google Scholar]
- 91.Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS. 2004;18:1101–1108. doi: 10.1097/00002030-200405210-00003. [DOI] [PubMed] [Google Scholar]
- 92.West MJ, Lowe AD, Karn J. Activation of human immunodeficiency virus transcription in T cells revisited: NF-kappaB p65 stimulates transcriptional elongation. J Virol. 2001;75:8524–8537. doi: 10.1128/JVI.75.18.8524-8537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L, Jr, Ingerman MJ, Witek J, Kedanis RJ, Natkin J, DeSimone J, Pomerantz RJ. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 1999;282:1627–1632. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 94.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, Mellors JW, Coffin JM. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;41:4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, Kovacs JA, Davey RT, Rock-Kress D, Dewar R, Liu S, Metcalf JA, Rehm C, Brun SC, Hanna GJ, Kempf DJ, Coffin JM, Mellors JW. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 2007;3:e46. doi: 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hermankova M, Ray SC, Ruff C, Powell-Davis M, Ingersoll R, D’Aquila RT, Quinn TC, Siliciano JD, Siliciano RF, Persaud D. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA. 2001;286:196–207. doi: 10.1001/jama.286.2.196. [DOI] [PubMed] [Google Scholar]
- 97.Kieffer TL, Finucane MM, Nettles RE, Quinn TC, Broman KW, Ray SC, Persaud D, Siliciano RF. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J Infect Dis. 2004;189:1452–1465. doi: 10.1086/382488. [DOI] [PubMed] [Google Scholar]
- 98.Persaud D, Siberry GK, Ahonkhai A, Kajdas J, Monie D, Hutton N, Watson DC, Quinn TC, Ray SC, Siliciano RF. Continued production of drug-sensitive human immunodeficiency virus type 1 in children on combination antiretroviral therapy who have undetectable viral loads. J Virol. 2004;78:968–979. doi: 10.1128/JVI.78.2.968-979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nettles RE, Kieffer TL, Kwon P, Monie D, Han Y, Parsons T, Cofrancesco J, Jr, Gallant JE, Quinn TC, Jackson B, Flexner C, Carson K, Ray S, Persaud D, Siliciano RF. Intermittent HIV-1 viremia (Blips) and drug resistance in patients receiving HAART. JAMA. 2005;293:817–829. doi: 10.1001/jama.293.7.817. [DOI] [PubMed] [Google Scholar]
- 100.Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, Haggerty CM, Kamireddi AR, Liu Y, Lee J, Persaud D, Gallant JE, Cofrancesco J, Jr, Quinn TC, Wilke CO, Ray SC, Siliciano JD, Nettles RE, Siliciano RF. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nickle DC, Jensen MA, Shriner D, Brodie SJ, Frenkel LM, Mittler JE, Mullins JI. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol. 2003;77:5540–5546. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tobin NH, Learn GH, Holte SE, Wang Y, Melvin AJ, McKernan JL, Pawluk DM, Mohan KM, Lewis PF, Mullins JI, Frenkel LM. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J Virol. 2005;79:9625–9634. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dinoso JB, Kim SY, Wiegand AM, Palmer SE, Gange SJ, Cranmer L, O’Shea A, Callender M, Spivak A, Brennan T, Kearney MF, Proschan MA, Mican JM, Rehm CA, Coffin JM, Mellors JW, Siliciano RF, Maldarelli F. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 2009;106:9403–9408. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gandhi RT, Zheng L, Bosch RJ, Chan ES, Margolis DM, Read S, Kallungal B, Palmer S, Medvik K, Lederman MM, Alatrakchi N, Jacobson JM, Wiegand A, Kearney M, Coffin JM, Mellors JW, Eron JJ AIDS Clinical Trials Group A5244 team. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med. 2010;7:e1000321. doi: 10.1371/journal.pmed.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McMahon D, Jones J, Wiegand A, Gange SJ, Kearney M, Palmer S, McNulty S, Metcalf JA, Acosta E, Rehm C, Coffin JM, Mellors JW, Maldarelli F. Short-course raltegravir intensification does not reduce persistent low-level viremia in patients with HIV-1 suppression during receipt of combination antiretroviral therapy. Clin Infect Dis. 2010;50:912–919. doi: 10.1086/650749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Boni J, Hirschel B, Weber R, Trkola A, Gunthard HF Swiss HIV Cohort Study. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci U S A. 2008;105:16725–16730. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hill AL, Rosenbloom DI, Fu F, Nowak MA, Siliciano RF. Predicting the outcomes of treatment to eradicate the latent reservoir for HIV-1. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1406663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O’Doherty U, Palmer S, Deeks SG, Siliciano JD. Comparative Analysis of Measures of Viral Reservoirs in HIV-1 Eradication Studies. PLoS Pathog. 2013;9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 110.Henrich TJ, Hu Z, Li JZ, Sciaranghella G, Busch MP, Keating SM, Gallien S, Lin NH, Giguel FF, Lavoie L, Ho VT, Armand P, Soiffer RJ, Sagar M, Lacasce AS, Kuritzkes DR. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis. 2013;207:1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee TH, Robles YP, Davis BT, Li JZ, Heisey A, Hill AL, Busch MP, Armand P, Soiffer RJ, Altfeld M, Kuritzkes DR. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med. 2014;161:319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 113.Koenig S, Gendelman HE, Orenstein JM, Dal Canto MC, Pezeshkpour GH, Yungbluth M, Janotta F, Aksamit A, Martin MA, Fauci AS. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 114.Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc Natl Acad Sci U S A. 2001;98:658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Babas T, Munoz D, Mankowski JL, Tarwater PM, Clements JE, Zink MC. Role of microglial cells in selective replication of simian immunodeficiency virus genotypes in the brain. J Virol. 2003;77:208–216. doi: 10.1128/JVI.77.1.208-216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gonzalez-Scarano F, Martin-Garcia J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5:69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- 117.Peng G, Greenwell-Wild T, Nares S, Jin W, Lei KJ, Rangel ZG, Munson PJ, Wahl SM. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 2007;110:393–400. doi: 10.1182/blood-2006-10-051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arfi V, Riviere L, Jarrosson-Wuilleme L, Goujon C, Rigal D, Darlix JL, Cimarelli A. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J Virol. 2008;82:6557–6565. doi: 10.1128/JVI.02321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schnell G, Spudich S, Harrington P, Price RW, Swanstrom R. Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1-associated dementia. PLoS Pathog. 2009;5:e1000395. doi: 10.1371/journal.ppat.1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Redel L, Le Douce V, Cherrier T, Marban C, Janossy A, Aunis D, Van Lint C, Rohr O, Schwartz C. HIV-1 regulation of latency in the monocyte-macrophage lineage and in CD4+ T lymphocytes. J Leukoc Biol. 2010;87:575–588. doi: 10.1189/jlb.0409264. [DOI] [PubMed] [Google Scholar]
- 121.Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog. 2011;7:e1002286. doi: 10.1371/journal.ppat.1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cribbs SK, Lennox J, Caliendo AM, Brown LA, Guidot DM. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res Hum Retroviruses. 2015;31:64–70. doi: 10.1089/aid.2014.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, Wietgrefe S, Caro-Vegas C, Madden V, Sharpe G, Haase AT, Eron JJ, Garcia JV. Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest. 2016 doi: 10.1172/JCI84456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Eisele E, Siliciano RF. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity. 2012;37:377–388. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sahu GK, Lee K, Ji J, Braciale V, Baron S, Cloyd MW. A novel in vitro system to generate and study latently HIV-infected long-lived normal CD4+ T-lymphocytes. Virology. 2006;355:127–137. doi: 10.1016/j.virol.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 126.Bosque A, Planelles V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood. 2009;113:58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang HC, Xing S, Shan L, O’Connell K, Dinoso J, Shen A, Zhou Y, Shrum CK, Han Y, Liu JO, Zhang H, Margolick JB, Siliciano RF. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest. 2009;119:3473–3486. doi: 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tyagi M, Pearson RJ, Karn J. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J Virol. 2010;84:6425–6437. doi: 10.1128/JVI.01519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saleh S, Solomon A, Wightman F, Xhilaga M, Cameron PU, Lewin SR. CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: a novel model of HIV-1 latency. Blood. 2007;110:4161–4164. doi: 10.1182/blood-2007-06-097907. [DOI] [PubMed] [Google Scholar]
- 130.Kordelas L, Verheyen J, Beelen DW, Horn PA, Heinold A, Kaiser R, Trenschel R, Schadendorf D, Dittmer U, Esser S Essen HIV AlloSCT Group. Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N Engl J Med. 2014;371:880–882. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- 131.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 132.Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, Siliciano JD, Siliciano RF. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Laird GM, Rosenbloom DI, Lai J, Siliciano RF, Siliciano JD. Measuring the Frequency of Latent HIV-1 in Resting CD4(+) T Cells Using a Limiting Dilution Coculture Assay. Methods Mol Biol. 2016;1354:239–253. doi: 10.1007/978-1-4939-3046-3_16. [DOI] [PubMed] [Google Scholar]
- 134.Chun TW, Nickle DC, Justement JS, Large D, Semerjian A, Curlin ME, O’Shea MA, Hallahan CW, Daucher M, Ward DJ, Moir S, Mullins JI, Kovacs C, Fauci AS. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J Clin Invest. 2005;115:3250–3255. doi: 10.1172/JCI26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, Anderson J, Perkey K, Stevenson M, Perelson AS, Douek DC, Haase AT, Schacker TW. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A. 2014;111:2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Kosakovsky Pond SL, Chung YS, Penugonda S, Chipman JG, Fletcher CV, Schacker TW, Malim MH, Rambaut A, Haase AT, McLean AR, Wolinsky SM. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016 doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Martin AR, Siliciano RF. Progress Toward HIV Eradication: Case Reports, Current Efforts, and the Challenges Associated with Cure. Annu Rev Med. 2015 doi: 10.1146/annurev-med-011514-023043. [DOI] [PubMed] [Google Scholar]
- 138.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 141.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 142.Kieffer TL, Kwon P, Nettles RE, Han Y, Ray SC, Siliciano RF. G-->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol. 2005;79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cuevas JM, Geller R, Garijo R, Lopez-Aldeguer J, Sanjuan R. Extremely High Mutation Rate of HIV-1 In Vivo. PLoS Biol. 2015;13:e1002251. doi: 10.1371/journal.pbio.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mens H, Pedersen AG, Jorgensen LB, Hue S, Yang Y, Gerstoft J, Katzenstein TL. Investigating signs of recent evolution in the pool of proviral HIV type 1 DNA during years of successful HAART. AIDS Res Hum Retroviruses. 2007;23:107–115. doi: 10.1089/aid.2006.0089. [DOI] [PubMed] [Google Scholar]
- 145.Wagner TA, McKernan JL, Tobin NH, Tapia KA, Mullins JI, Frenkel LM. An increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J Virol. 2013;87:1770–1778. doi: 10.1128/JVI.01985-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Blankson JN, Finzi D, Pierson TC, Sabundayo BP, Chadwick K, Margolick JB, Quinn TC, Siliciano RF. Biphasic decay of latently infected CD4+ T cells in acute human immunodeficiency virus type 1 infection. J Infect Dis. 2000;182:1636–1642. doi: 10.1086/317615. [DOI] [PubMed] [Google Scholar]
- 147.Larder BA, Darby G, Richman DD. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 148.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 149.Gunthard HF, Wong JK, Ignacio CC, Guatelli JC, Riggs NL, Havlir DV, Richman DD. Human immunodeficiency virus replication and genotypic resistance in blood and lymph nodes after a year of potent antiretroviral therapy. J Virol. 1998;72:2422–2428. doi: 10.1128/jvi.72.3.2422-2428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shafer RW. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin Microbiol Rev. 2002;15:247–277. doi: 10.1128/CMR.15.2.247-277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 152.Theys K, Snoeck J, Vercauteren J, Abecasis AB, Vandamme AM, Camacho RJ Portuguese HIV-1 Resistance Study Group. Decreasing population selection rates of resistance mutation K65R over time in HIV-1 patients receiving combination therapy including tenofovir. J Antimicrob Chemother. 2013;68:419–423. doi: 10.1093/jac/dks380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Charpentier C, Lambert-Niclot S, Visseaux B, Morand-Joubert L, Storto A, Larrouy L, Landman R, Calvez V, Marcelin AG, Descamps D. Evolution of the K65R, K103N and M184V/I reverse transcriptase mutations in HIV-1-infected patients experiencing virological failure between 2005 and 2010. J Antimicrob Chemother. 2013 doi: 10.1093/jac/dkt184. [DOI] [PubMed] [Google Scholar]
- 154.Bontell I, Haggblom A, Bratt G, Albert J, Sonnerborg A. Trends in antiretroviral therapy and prevalence of HIV drug resistance mutations in Sweden 1997–2011. PLoS One. 2013;8:e59337. doi: 10.1371/journal.pone.0059337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nakagawa F, May M, Phillips A. Life expectancy living with HIV: recent estimates and future implications. Curr Opin Infect Dis. 2013;26:17–25. doi: 10.1097/QCO.0b013e32835ba6b1. [DOI] [PubMed] [Google Scholar]
- 156.van Sighem AI, Gras LA, Reiss P, Brinkman K, de Wolf F ATHENA national observational cohort study. Life expectancy of recently diagnosed asymptomatic HIV-infected patients approaches that of uninfected individuals. AIDS. 2010;24:1527–1535. doi: 10.1097/QAD.0b013e32833a3946. [DOI] [PubMed] [Google Scholar]
- 157.Mills EJ, Bakanda C, Birungi J, Chan K, Ford N, Cooper CL, Nachega JB, Dybul M, Hogg RS. Life expectancy of persons receiving combination antiretroviral therapy in low-income countries: a cohort analysis from Uganda. Ann Intern Med. 2011;155:209–216. doi: 10.7326/0003-4819-155-4-201108160-00358. [DOI] [PubMed] [Google Scholar]
- 158.Johnson LF, Mossong J, Dorrington RE, Schomaker M, Hoffmann CJ, Keiser O, Fox MP, Wood R, Prozesky H, Giddy J, Garone DB, Cornell M, Egger M, Boulle A International Epidemiologic Databases to Evaluate AIDS Southern Africa Collaboration. Life expectancies of South African adults starting antiretroviral treatment: collaborative analysis of cohort studies. PLoS Med. 2013;10:e1001418. doi: 10.1371/journal.pmed.1001418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hammarlund E, Lewis MW, Hansen SG, Strelow LI, Nelson JA, Sexton GJ, Hanifin JM, Slifka MK. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 160.Takaki A, Wiese M, Maertens G, Depla E, Seifert U, Liebetrau A, Miller JL, Manns MP, Rehermann B. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat Med. 2000;6:578–582. doi: 10.1038/75063. [DOI] [PubMed] [Google Scholar]
- 161.Michie CA, McLean A, Alcock C, Beverley PC. Lifespan of human lymphocyte subsets defined by CD45 isoforms. Nature. 1992;360:264–265. doi: 10.1038/360264a0. [DOI] [PubMed] [Google Scholar]
- 162.Mclean AR, Michie CA. In vivo estimates of division and death rates of human T lymphocytes. Proc Natl Acad Sci U S A. 1995;92:3707–3711. doi: 10.1073/pnas.92.9.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hellerstein MK, Hoh RA, Hanley MB, Cesar D, Lee D, Neese RA, McCune JM. Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection. J Clin Invest. 2003;112:956–966. doi: 10.1172/JCI17533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vrisekoop N, den Braber I, de Boer AB, Ruiter AF, Ackermans MT, van der Crabben SN, Schrijver EH, Spierenburg G, Sauerwein HP, Hazenberg MD, de Boer RJ, Miedema F, Borghans JA, Tesselaar K. Sparse production but preferential incorporation of recently produced naive T cells in the human peripheral pool. Proc Natl Acad Sci U S A. 2008;105:6115–6120. doi: 10.1073/pnas.0709713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.De Boer RJ, Perelson AS. Quantifying T lymphocyte turnover. J Theor Biol. 2013;327:45–87. doi: 10.1016/j.jtbi.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, Chen IS. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gerard FC, Yang R, Romani B, Poisson A, Belzile JP, Rougeau N, Cohen EA. Defining the interactions and role of DCAF1/VPRBP in the DDB1-cullin4A E3 ubiquitin ligase complex engaged by HIV-1 Vpr to induce a G2 cell cycle arrest. PLoS One. 2014;9:e89195. doi: 10.1371/journal.pone.0089195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Romani B, Shaykh Baygloo N, Aghasadeghi MR, Allahbakhshi E. HIV-1 Vpr Protein Enhances Proteasomal Degradation of MCM10 DNA Replication Factor through the Cul4-DDB1[VprBP] E3 Ubiquitin Ligase to Induce G2/M Cell Cycle Arrest. J Biol Chem. 2015;290:17380–17389. doi: 10.1074/jbc.M115.641522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V. Homeostatic Proliferation Fails to Efficiently Reactivate HIV-1 Latently Infected Central Memory CD4+ T Cells. PLoS Pathog. 2011;7:e1002288. doi: 10.1371/journal.ppat.1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, Wells D, Su L, Luke BT, Halvas EK, Besson G, Penrose KJ, Yang Z, Kwan RW, Van Waes C, Uldrick T, Citrin DE, Kovacs J, Polis MA, Rehm CA, Gorelick R, Piatak M, Keele BF, Kearney MF, Coffin JM, Hughes SH, Mellors JW, Maldarelli F. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A. 2016;113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Swain SL, Hu H, Huston G. Class II-independent generation of CD4 memory T cells from effectors. Science. 1999;286:1381–1383. doi: 10.1126/science.286.5443.1381. [DOI] [PubMed] [Google Scholar]
- 174.Kassiotis G, Garcia S, Simpson E, Stockinger B. Impairment of immunological memory in the absence of MHC despite survival of memory T cells. Nat Immunol. 2002;3:244–250. doi: 10.1038/ni766. [DOI] [PubMed] [Google Scholar]
- 175.Brenchley JM, Ruff LE, Casazza JP, Koup RA, Price DA, Douek DC. Preferential infection shortens the life span of human immunodeficiency virus-specific CD4+ T cells in vivo. J Virol. 2006;80:6801–6809. doi: 10.1128/JVI.00070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 178.Maeurer MJ, Lotze MT. Interleukin-7 (IL-7) knockout mice. Implications for lymphopoiesis and organ-specific immunity. Int Rev Immunol. 1998;16:309–322. doi: 10.3109/08830189809042999. [DOI] [PubMed] [Google Scholar]
- 179.Purton JF, Tan JT, Rubinstein MP, Kim DM, Sprent J, Surh CD. Antiviral CD4+ memory T cells are IL-15 dependent. J Exp Med. 2007;204:951–961. doi: 10.1084/jem.20061805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Scripture-Adams DD, Brooks DG, Korin YD, Zack JA. Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J Virol. 2002;76:13077–13082. doi: 10.1128/JVI.76.24.13077-13082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang FX, Xu Y, Sullivan J, Souder E, Argyris EG, Acheampong EA, Fisher J, Sierra M, Thomson MM, Najera R, Frank I, Kulkosky J, Pomerantz RJ, Nunnari G. IL-7 is a potent and proviral strain-specific inducer of latent HIV-1 cellular reservoirs of infected individuals on virally suppressive HAART. J Clin Invest. 2005;115:128–137. doi: 10.1172/JCI22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Vandergeeten C, Fromentin R, DaFonseca S, Lawani MB, Sereti I, Lederman MM, Ramgopal M, Routy JP, Sekaly RP, Chomont N. Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood. 2013;121:4321–4329. doi: 10.1182/blood-2012-11-465625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Katlama C, Lambert-Niclot S, Assoumou L, Papagno L, Lecardonnel F, Zoorob R, Tambussi G, Clotet B, Youle M, Achenbach CJ, Murphy RL, Calvez V, Costagliola D, Autran B EraMune-01 study team. Treatment intensification followed by interleukin-7 reactivates HIV without reducing total HIV DNA: a randomized trial. AIDS. 2016;30:221–230. doi: 10.1097/QAD.0000000000000894. [DOI] [PubMed] [Google Scholar]
- 184.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 185.Riou C, Yassine-Diab B, Van grevenynghe J, Somogyi R, Greller LD, Gagnon D, Gimmig S, Wilkinson P, Shi Y, Cameron MJ, Campos-Gonzalez R, Balderas RS, Kelvin D, Sekaly RP, Haddad EK. Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells. J Exp Med. 2007;204:79–91. doi: 10.1084/jem.20061681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol. 2014;88:14070–14077. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, Wang E, Douek DC, Price DA, June CH, Marincola FM, Roederer M, Restifo NP. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. HIV-1 persistence in CD4(+) T cells with stem cell-like properties. Nat Med. 2014;20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Reinhardt RL, Khoruts A, Merica R, Zell T, Jenkins MK. Visualizing the generation of memory CD4 T cells in the whole body. Nature. 2001;410:101–105. doi: 10.1038/35065111. [DOI] [PubMed] [Google Scholar]
- 191.Thome JJ, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliyawala T, Kato T, Lerner H, Shen Y, Farber DL. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Farber DL, Yudanin NA, Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014;14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hong JJ, Amancha PK, Rogers K, Ansari AA, Villinger F. Spatial alterations between CD4(+) T follicular helper, B, and CD8(+) T cells during simian immunodeficiency virus infection: T/B cell homeostasis, activation, and potential mechanism for viral escape. J Immunol. 2012;188:3247–3256. doi: 10.4049/jimmunol.1103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Connick E, Folkvord JM, Lind KT, Rakasz EG, Miles B, Wilson NA, Santiago ML, Schmitt K, Stephens EB, Kim HO, Wagstaff R, Li S, Abdelaal HM, Kemp N, Watkins DI, MaWhinney S, Skinner PJ. Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL. J Immunol. 2014;193:5613–5625. doi: 10.4049/jimmunol.1401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, Morcock D, Swanson T, Legasse AW, Axthelm MK, Hesselgesser J, Geleziunas R, Hirsch VM, Edlefsen PT, Piatak M, Jr, Estes JD, Lifson JD, Picker LJ. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, Zhang H, Margolick JB, Blankson JN, Siliciano RF. Stimulation of HIV-1-Specific Cytolytic T Lymphocytes Facilitates Elimination of Latent Viral Reservoir after Virus Reactivation. Immunity. 2012;36:491–501. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Clark RA. Resident memory T cells in human health and disease. Sci Transl Med. 2015;7:269rv1. doi: 10.1126/scitranslmed.3010641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Thome JJ, Farber DL. Emerging concepts in tissue-resident T cells: lessons from humans. Trends Immunol. 2015;36:428–435. doi: 10.1016/j.it.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Purwar R, Campbell J, Murphy G, Richards WG, Clark RA, Kupper TS. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One. 2011;6:e16245. doi: 10.1371/journal.pone.0016245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chun TW, Nickle DC, Justement JS, Meyers JH, Roby G, Hallahan CW, Kottilil S, Moir S, Mican JM, Mullins JI, Ward DJ, Kovacs JA, Mannon PJ, Fauci AS. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis. 2008;197:714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 202.Lerner P, Guadalupe M, Donovan R, Hung J, Flamm J, Prindiville T, Sankaran-Walters S, Syvanen M, Wong JK, George MD, Dandekar S. The gut mucosal viral reservoir in HIV-infected patients is not the major source of rebound plasma viremia following interruption of highly active antiretroviral therapy. J Virol. 2011;85:4772–4782. doi: 10.1128/JVI.02409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Anderson JA, Archin NM, Ince W, Parker D, Wiegand A, Coffin JM, Kuruc J, Eron J, Swanstrom R, Margolis DM. Clonal sequences recovered from plasma from patients with residual HIV-1 viremia and on intensified antiretroviral therapy are identical to replicating viral RNAs recovered from circulating resting CD4+ T cells. J Virol. 2011;85:5220–5223. doi: 10.1128/JVI.00284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 205.Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu X, Pierson TC, Margolick JB, Siliciano RF, Siliciano JD. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol. 2004;78:6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Berry CC, Gillet NA, Melamed A, Gormley N, Bangham CR, Bushman FD. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28:755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, McElrath MJ, Lehmann C, Klein F, Caskey M, Walker BD, Siliciano JD, Siliciano RF, Jankovic M, Nussenzweig MC. HIV-1 integration landscape during latent and active infection. Cell. 2015;160:420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ikeda T, Shibata J, Yoshimura K, Koito A, Matsushita S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J Infect Dis. 2007;195:716–725. doi: 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- 209.Imamichi H, Natarajan V, Adelsberger JW, Rehm CA, Lempicki RA, Das B, Hazen A, Imamichi T, Lane HC. Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS. 2014;28:1091–1099. doi: 10.1097/QAD.0000000000000223. [DOI] [PubMed] [Google Scholar]
- 210.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, Wells D. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proceedings of the National Academy of Sciences. 2016;113(7):1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]