Abstract
Cell death and inflammation are ancient processes of fundamental biological importance in both normal physiology and human disease pathologies. The recent observation that apoptosis regulatory components have dual roles in cell death and inflammation suggests that these proteins function, not primarily to kill, but to coordinate tissue repair and remodeling. This perspective unifies cell death components as positive regulators of tissue repair that replaces malfunctioning or damaged tissues and enhances the resilience of epithelia to insult. It is now recognized that cells that die by apoptosis do not do so silently, but release a variety of paracrine signals to communicate with their cellular environment to ensure tissue regeneration, and wound healing. Moreover, inflammatory signalling pathways, such as those emanating from the TNF-receptor or Toll-related receptors, take part in cell competition to eliminate developmentally aberrant clones. Ubiquitylation has emerged as crucial mediator of signal transduction in cell death and inflammation. Here we focus on recent advances on ubiquitin-mediated regulation of cell death and inflammation, and how this is used to regulate the defense of homeostasis.
Keywords: caspases, Ubiquitin, IAPs, RIPK1, homeostasis
Introduction
The remarkable capacity to correct for tissue stress and malfunction is one of the most fascinating hallmarks of multicellular organisms. While we marvel at the high level of tissue plasticity during animal development, we struggle with the incredible ability of cancer cells to escape growth inhibitory signals or develop resistance to treatments. For the maintenance of tissue homeostasis and tissue fitness it is essential that damaged or malfunctioning cells are detected, eliminated and replaced (Jacobson, Weil, & Raff, 1997). The decision as to whether a cell lives or dies relies not just on the regulation of the intrinsic apoptotic programme, but critically depends on paracrine interactions between damaged cells and cells of the surrounding tissue (Neves, Demaria, Campisi, & Jasper, 2015). It is now clear that cellular malfunction, and this includes activation of oncogenes or loss of tumour suppressor proteins, results in the production of cytokines, chemokines and mitogens, which stimulate an adaptive response for the defense of homeostasis (Medzhitov, 2008). Activation of the cell death programme is, thereby, critically important to eliminate and replace malfunctioning or damaged cells (Thompson, 1995).
Studies in flies show that tissue homeostasis in epithelia is governed by ‘collective’ decision mechanisms that determine cell death and proliferation across tissues (Vincent, Fletcher, & Baena-Lopez, 2013). These mechanisms include cell competition and apoptosis-induced compensatory proliferation (Fig. 1). Cell competition is a process in which fast-growing fitter cells (winners) kill neighboring slow growing, ‘less fit’ (losers) cells, even when the weaker cells are fully viable in a non-mosaic tissue. While cell competition and compensatory proliferation have been studied extensively in flies, recent studies reveal the existence and importance of similar processes in mammals (Bondar & Medzhitov, 2010; Oertel, Menthena, Dabeva, & Shafritz, 2006; Oliver, Saunders, Tarle, & Glaser, 2004). For example, cell competition selects the fittest stem cell in the epiblast that gives rise to the entire organism (Claveria, Giovinazzo, Sierra, & Torres, 2013), while in young individuals, cell competition is used to ensure tissue fitness (Amoyel & Bach, 2014). Due to chronic engagement of the tissue repair programme in older individuals, age-related decline of tissue homeostasis can occur, resulting in degeneration, metabolic dysfunction, and cancer (Martins et al., 2014; Neves et al., 2015). Consistently, loss of homeostasis is one of the hallmarks of aging (Chovatiya & Medzhitov, 2014).
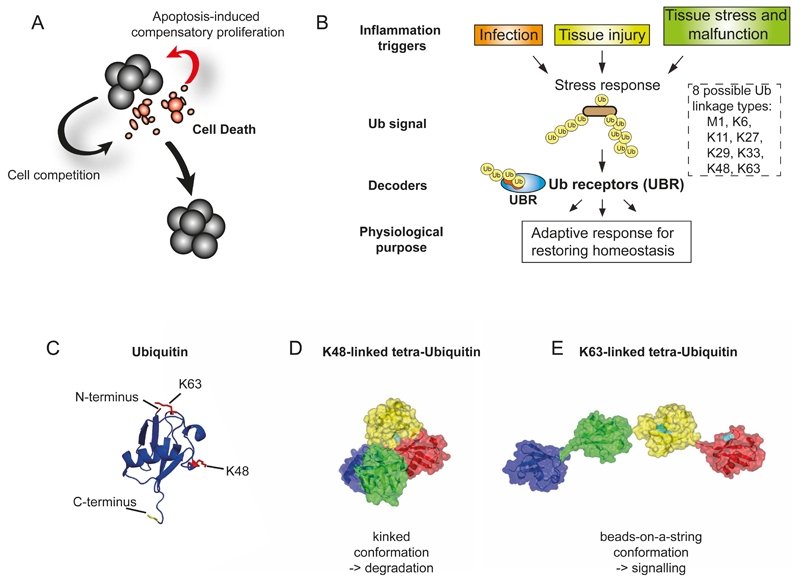
Figure 1. Ubiquitylation as mediator and regulator of signal transduction in cell death, inflammation, and defense of homeostasis.
(A) Tissue malfunction results in a secretery programme and inflammatory response whose purpose it is to restore homeostasis. Tissue homeostasis is regulated by a collective decision mechanism that influences cell death and proliferation across tissues. These include cell competition and apoptosis-induced compensatory proliferation. (B) Tissue stress response and inflammation underlie a common principle in which the conjugation of typical Ub-chains produces robust networks that are decoded by Ub-receptors whose actions serve to coordinate adaptation to tissue stress. (C) Ribbon Structure of Ub. Lysines at position 48 and 63 of Ub are highlighted. (D, E)Topology of K48- (C) and K63-linked (D) tetra Ub chains.
Ubiquitin as mediator of signalling events
While superficially tissue stress, inflammation and cell death appear separate phenomena, recent advances in our understanding of these stress responses strongly indicate that the underlying signalling events are in fact linked. Ubiquitylation, thereby, has emerged as crucial mediator and regulator of signal transduction in cell death and inflammation (Dikic, Wakatsuki, & Walters, 2009) (Fig. 1).
The covalent attachment of ubiquitin (Ub) to target proteins can alter the protein’s conformation or binding properties and thus influences protein activities, localization or stability. Ub is a small protein modifier that is covalently attached to proteins in a step-wise process that involves Ub activating enzymes (E1), Ub conjugating enzymes (E2) and Ub protein ligases (E3) (Hershko, Ciechanover, & Varshavsky, 2000). E3s confer substrate specificity by bringing Ub-loaded E2 to targets substrates and promoting the formation of an isopeptide linkage between the carboxyl-terminus of Ub (glycine (G)76) and the amino group of a lysine (K) residue of the substrate.
Ub can be conjugated either as a single moiety or as chains of variable length (Komander, 2009) (Fig. 1). Different linkage types provide further complexity, as Ub moieties can be conjugated to one another via each of the seven K residues within Ub, or via ubiquitin’s N terminal methionine. This allows the formation of homotypic chains linked via ubiquitin’s K6, K11, K27, K29, K33, K48, K63 or M1 (Kirisako et al., 2006). In addition to homotypic chains that are sequentially linked through the same successive linkage type, mixed-linkage chain types also exist in which several distinct K residues are used to connect consecutive Ub moieties (Meyer & Rape, 2014). The complexity and versatility of Ub-dependent modifications are further increased through the generation of heterologous chain types where Ub is connected with other Ub-like modifiers, such as SUMO (Tatham et al., 2008).
The eight different types of homotypic Ub chains exert distinct effects on cellular processes (Bhoj & Chen, 2009). This is because the differently linked poly-Ub chains adopt distinct structures. For instance, K48-linked poly-Ub chains take up a kinked topology while K63- and M1-linked chains adopt an open configuration that resembles ‘beads-on-a-string’ (Komander, 2009). While it is well established that K48-linked modifications can promote degradation through recognition by the 26S proteasome, recent evidence indicate that other linkage types, such as M1, K29, K33 and K63, can regulate biological processes in a degradation-independent manner (Komander & Rape, 2012). Whether ubiquitylation targets proteins for degradation or mediates non-degradative signalling depends on protein-interactions between the ubiquitylated protein and Ub-binding proteins - often termed Ub "receptors" (Hoeller, Hecker, & Dikic, 2006). Ub-receptors (UBRs) carry small Ub-binding domains (UBDs) that bind to the Ub modification via low-affinity, non-covalent interactions. Currently more than 20 different types of UBDs are known that detect overlapping as well as distinct Ub modifications. UBRs that selectively recognize K48-linked poly-Ub chains, such as the proteasome subunit Rpn13 (Lundgren, Masson, Realini, & Young, 2003), recruit modified proteins to the proteasome for degradation. In contrast, UBRs that bind to mono-Ub, K63-linkages or linear (M1) Ub allow Ub-dependent association with signalling molecules (Hoeller et al., 2006). In particular, K63-linked Ub chains, and their respective UBRs, are critical for tumour necrosis factor (TNF)-mediated NFkB activation and cell survival (Stickle et al., 2004).
Ub as arbiter of life and death – TNF signalling as paradigm
The role of Ub as crucial mediator and regulator of signal transduction in the defense of homeostasis is best illustrated by the signalling pathways emanating from the TNF receptor (Fig. 2). TNF is a major inflammatory cytokine that was first identified for its ability to induce rapid hemorrhagic necrosis of experimental cancers (Carswell et al., 1975). Now it is clear that TNF functions as a master regulator of the cytokine network that coordinates defense of homeostasis via controlling inflammation, cell proliferation, differentiation, survival and death (Balkwill, 2009).
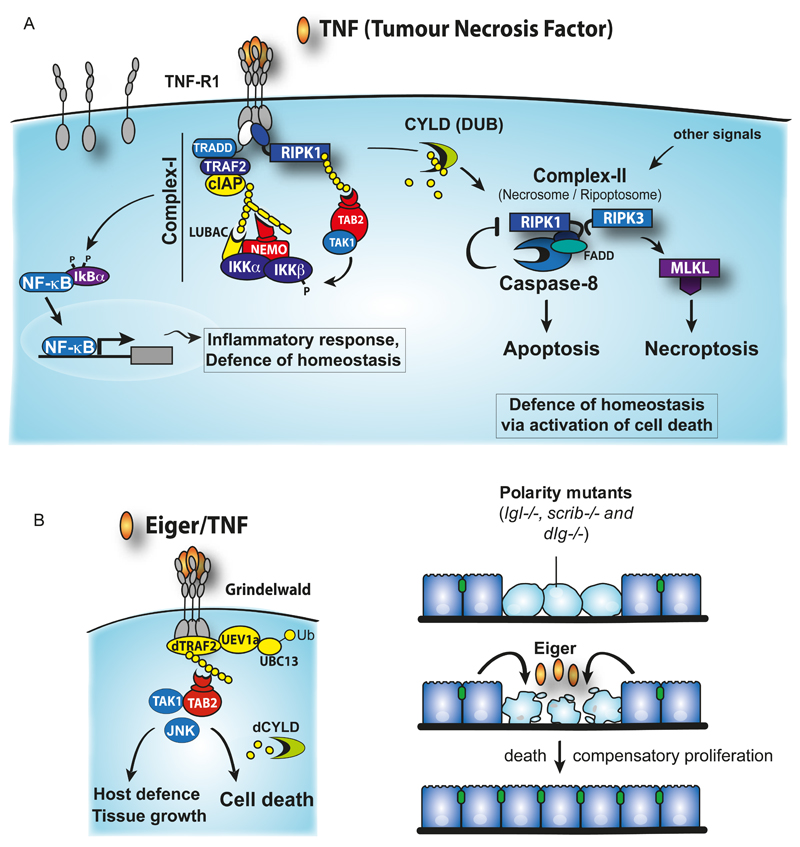
Figure 2. Evolutionary conservation of TNF-induced cell death and the defense of homeostasis.
(A) TNF signaling and the transition of complex-I to complex-II. Upon TNF-binding, cIAPs are recruited to the TNF-R1 signaling complex (Complex-I) via TRADD/TRAF2. cIAPs ubiquitylate several molecules within the complex. RIPK1 ubiquitylation is the most readily observed. Ubiquitylation of components of complex-I, such as RIPK1, drives the recruitment of HOIL-1/HOIP/Sharpin that together form the Linear Ubiquitin Assembly Complex (LUBAC). LUBAC generates linear Ub chains on NEMO and RIPK1 that in turn recruits more NEMO molecules via its linear Ub binding UBAN domain. NEMO is probably constitutively associated with IKKa/IKKb and IKKb is phosphorylated and activated by TAK1 that is independently recruited to ubiquitylated complex-I via its Ub receptors TAB2 and TAB3 that bind only to K63-linked Ub chains. Phosphorylated and activated IKKb in turn phosphorylates IκBα, which leads to recruitment of a HECT E3 ligase. This E3 ligase promotes K48-linked ubiquitylation and proteasomal degradation of IκBα, allowing translocation of NFkB subunits p50/p65 to drive production of cytokines. p50/p65 also promote expression of IκBα, to cause feedback inhibition, as well as genes such as cFLIP that are required to protect cells from complex-II-induced cell death. The numbered arrows provide a tentative indication of temporal sequence. Complex-II is most likely generated from complex-I, in an as yet undefined manner, and comprises RIPK1, FADD and caspase-8. Deubiquitylation by CYLD is thereby a decisive step in the transition of complex-I to complex-II. Caspase-8 limits Complex-II formation by cleaving and inactivating RIPK1. Consequently, loss of IAPs, LUBAC or caspase-8 activity results in formation of Complex-II that is able to drive necroptosis. Formation of complex-II, Necrosome or ripoptosome can also occur following stimulation of Pattern Recognition receptors or genotoxic stress. (B) Eiger-mediated signalling that regulates a variety of cellular and tissue processes, including the elimination of polarity mutant cells. Eiger mediates its effect through binding to its cognate receptor Grindelwald. This results in activation of JNK in a DTRAF2/Bendless/dUev1A dependent manner. The Drosophila homologue of TAB2/3 (dTAB2) links TAK1 to the presumptive Ub chains conjugated by DTRAF2. dCYLD influences the decision as to whether JNK drives cell death or non-cell death processes (see text for further details).
In mammals, binding of TNF to its extracellular receptor TNFR1 triggers either prosurvival/inflammatory or pro-death signaling pathways in a strictly Ub-dependent manner (Walczak, 2013) (Fig. 2). TNF can regulate tissue homeostasis in at least three different ways: through 1) activation of NF-kB-dependent and MAPK/JNK-dependent transcriptional programmes, 2) induction of caspase-8-dependent apoptosis or 3) stimulation of Receptor interacting protein kinase (RIPK)-mediated necrosis (necroptosis) (Declercq, Vanden Berghe, & Vandenabeele, 2009).
Binding of TNF to TNFR1 results in the assembly of a protein complex at the receptor’s cytoplasmic tail. This complex, which is frequently referred to as complex-I (Micheau & Tschopp, 2003), consists of TNFR1, the adaptors TRADD, TRAF2, the kinase RIPK1 and the E3 Ub-ligases cellular Inhibitor of Apoptosis cIAP1 and cIAP2. Within this complex, RIPK1 is rapidly conjugated with poly-Ub chains by cIAPs. cIAP-mediated conjugation of Ub to RIPK1 allows subsequent recruitment of the Linear Ubiquitin chain Assembly Complex (LUBAC, composed of HOIL/HOIP/Sharpin), the kinase complexes TAK1/TAB2/TAB3, and IKK (composed of NEMO/IKKa/IKKb) (Silke, 2011). Ub-dependent recruitment of LUBAC, TAK1/TAB2/TAB3, and IKKs is mediated by UBDs present in TAB2, NEMO and HOIP. Once recruited, LUBAC then modifies NEMO and RIPK1 with M1-linked Ub chains, resulting in increased stability of the TNF signalling complex. Additionally, the binding of NEMO to M1-linked Ub chains causes a conformational change of the IKK complex that is thought to facilitate its activation (Rahighi et al., 2009). Complex-I then signals inflammation and cell survival through TAK1 and IkB kinase (IKK)-dependent activation of NFkB. cIAPs are also required for JNK signaling (Gardam et al., 2011; Matsuzawa et al., 2008). This has been most clearly demonstrated for signaling that emanates from CD40, a TNF-super family receptor, but similar concepts likely hold true for TNFR1 signaling too. Ultimately this drives expression of a transcriptional programme that restores homeostasis and lowers the apoptotic threshold through the induction of anti-apoptotic molecules (Fig. 2).
As its name suggest, TNF can also potently induce cell death. This is mediated by a RIPK1-based secondary complex that is frequently referred to as complex-II or necrosome (Pasparakis & Vandenabeele, 2015; L. Wang, Du, & Wang, 2008). This RIPK1-based complex can either kill via caspase-8 leading to apoptosis, or through RIPK3 and MLKL, which results in necroptosis. Under normal conditions, a small fraction of RIPK1 dissociates from complex-I within 30min to three hours, and together with TRADD associates with the adaptor protein FADD and procaspase-8 to form complex-II (Micheau & Tschopp, 2003) or necrosome (Pasparakis & Vandenabeele, 2015). Whether lethal levels of complex-II forms critically depends on the ubiquitylation status of RIPK1 in complex-I, RIPK1 deubiquitylation by deubiquitylating enzymes such as CYLD, or NF-kB-dependent expression of anti-apoptotic genes, such as cFLIP or cIAP2. The formation and activity of complex-II is tightly regulated by cIAPs, LUBAC, NEMO and TAK1. cIAPs, NEMO and TAK1 not only suppress formation complex-II via activation of NF-kB but also thwart transition of complex-I to complex-II through mechanisms that are NF-kB-independent. It is believed that the Ub chains conjugated to RIPK1 by cIAP1/2 and LUBAC in complex-I constitute the decisive factor preventing RIPK1 from forming complex-II, and limiting its killing potential. The ability of IAPs to ubiquitylate RIPK1 and suppress its killing potential is antagonist by the deubiquitylating enzyme CYLD, which can cleave M1, and K63-linked Ub chains on RIPK1 (Komander et al., 2008). It is currently thought that deubiquitylation of RIPK1 drives complex-II formation and favors RIPK1-dependent apoptosis. However, when the levels of RIPK3 and MLKL are sufficiently high and caspase-8 activity is reduced, blocked or absent, complex-II can recruit and activate RIPK3, which in turn can drive MLKL-dependent necroptosis (Pasparakis & Vandenabeele, 2015). Caspase-8 together with cFLIPL reportedly inhibits necroptosis through cleaving RIPK1 and RIPK3 (Feng et al., 2007; Lin, Devin, Rodriguez, & Liu, 1999). In addition, caspase-8 can also cleave CYLD (O'Donnell et al., 2011), which removes Ub chains from RIPK1 and contributes to necroptosis in vitro and in vivo.
Ub-dependent regulation of TNF-induced cell death is evolutionary conserved, and is critically important for the maintenance of tissue fitness and the elimination of developmentally aberrant cells in Drosophila (Igaki & Miura, 2014). The Drosophila genome encodes a single member of the TNF family, named Eiger (Igaki et al., 2002; Moreno, Yan, & Basler, 2002) (Fig. 2). Eiger is required for inducing the death of cells mutant for apico-basal polarity genes, such as scribbled (scrib), discs large (dlg), and lethal giant larvae (lgl). In the absence of Eiger, such mutant clones grow aggressively and develop into neoplastic tumours. This suggests that in Drosophila Eiger functions as part of a surveillance programme that actively eliminates oncogenic polarity-deficient cells from the tissue (Igaki & Miura, 2014). Intriguingly, the Eiger-dependent elimination system is the result of cell competition that only operates when mutant cells are confronted with wild-type cells in a mosaic tissue. While Drosophila imaginal epithelium entirely mutant for scribbled or discs large results in tumorous overgrowth, such polarity-deficient oncogenic mutant cells, in an otherwise wild-type tissue, do not overgrow but are instead eliminated from the tissue by cell death.
Eiger-dependent cell elimination critically depends on Ub-dependent activation of TAK1 and JNK signalling, which in turn drives a cell death programme that relies on inputs from Drosophila initiator caspase DRONC and the metabolic state of a cell (Igaki & Miura, 2014). Eiger-mediated cell death requires the E3 ligase TRAF2 and the K63-selective E2-heteromeric complex Bendless (Ubc13)/dUev1a (Ma et al., 2014; Ma et al., 2013). In addition, the E3 ligase NOPO (no poles), which is the Drosophila orthologue of mammalian TRAF-interacting protein TRIP, also contributes to Eiger-induced cell death (Ma et al., 2012). Most likely, TRAF2 in conjunction with Bendless/dUev1a, promotes the conjugation of K63-linked Ub chains that in turn allows recruitment and activation of TAK1 via its Ub-receptor TAB2. Activation of TAK1 subsequently results in activation of Hemipterous (also known as JNKK or MKK7) and the Drosophila JNK orthologue Basket (Igaki & Miura, 2014). Through a process that is molecularly ill defined, Eiger-mediated activation of Basket results in the induction of cell death signalling.
Intriguingly, Eiger-mediated cell death is strictly dependent on deubiquitylation. In the absence of the deubiquitylating enzyme CYLD, Eiger-mediated cell death, and elimination of neoplastic tumours is blocked (Xue et al., 2007). Given that mammalian CYLD is an M1 and K63-selective deubiquitylating enzyme (Komander et al., 2008), and that the catalytic domains of Drosophila and mammalian CYLD are 53% identical, it is highly likely that the removal of non-degradative, K63-linked Ub chains is critical for the execution of cell death. The current literature suggests that CYLD promotes Eiger-induced cell death via the removal of degradative K48-poly-Ub chains on dTRAF2, thereby stabilizing TRAF2 and allowing efficient TRAF2-dependent activation of TAK1, Hep and JNK (Xue et al., 2007). Such a model is, however, inconsistent with the notion that TRAF2 and Bendless/Uev1a exclusively promotes the formation of K63-linked non-degradative Ub chains, and that CYLD, at least in mammals, lacks affinity for K48 di-Ub chains and instead preferentially cleaves K63- and M1-Ub linkages (Komander, Clague, & Urbe, 2009).
Taken together, in both Drosophila and mammals, ligation of TNF and Eiger to their cognate receptors results in the transient formation of Ub-dependent signalling hubs that allow recruitment and activation of kinases via specialized adaptor molecules with Ub-binding domains. While assembly of these Ub-dependent signalling centers can mediate various cellular phenotypes ranging from cytokine production, proliferation, canalization, pain sensitization to host defense, TNF/Eiger-dependent cell death appears to be achieved through specific deubiquitylation events that tip the balance of TNF/Eiger-signalling in favor of death.
Ub-dependent regulation of RIPK1 and the ripoptosome
In mammals, RIPK1-dependent cell death not only occurs in response to TNF, but also operates downstream of many other cytokine receptors, damage associated molecular pattern receptors, pathogen associated molecular pattern receptors or in response to genotoxic stress (Pasparakis & Vandenabeele, 2015) (Fig. 2). This suggests that RIPK1 functions as a more generic stress sentinel. Under these conditions, RIPK1 assembles a protein complex containing the core components RIPK1, FADD, and caspase-8 (Feoktistova et al., 2011; Tenev et al., 2011a). While these are the same components as the ones of complex-II (Micheau & Tschopp, 2003; L. Wang et al., 2008), the fact that this complex forms independently of TNFR1 indicates that it cannot constitute complex-II, which, per definition, originates from complex-I (Micheau & Tschopp, 2003; L. Wang et al., 2008). To uncouple it from the TNF-dependent complex-II, TNFR1-independent assembly of the RIPK1/FADD/caspase-8 complex is, therefore, better referred to as ‘ripoptosome’ (Feoktistova et al., 2011; Tenev et al., 2011a). Although the core of this complex consists of RIPK1, FADD and caspase-8, the ripoptosome can also include additional proteins such as FLIP and RIPK3, depending on cell type and stimulus (Green, Oberst, Dillon, Weinlich, & Salvesen, 2011).
Proper regulation of RIPK1, and the ripoptosome, is critically important for normal development and physiology (Bonnet et al., 2011; Dannappel et al., 2014; Declercq et al., 2009; Dondelinger et al., 2013; Duprez et al., 2011; Ermolaeva et al., 2008; Green et al., 2011; O'Donnell et al., 2011; Oberst et al., 2011; Pasparakis & Vandenabeele, 2015; Polykratis et al., 2014; Vandenabeele, Galluzzi, Vanden Berghe, & Kroemer, 2010; Welz et al., 2011). Consistently, genetic deletion of RIPK1 in mouse models results in lethality due to systemic inflammation. Although cell death is frequently considered to be the result of inflammation, recent evidence suggest that cell death may in fact precede, trigger or amplify the inflammatory response. Indeed, neither apoptosis nor necroptosis are ‘silent’ processes. Although caspase-mediated apoptosis is widely thought to be immunologically inert or even tolerogenic, it is now clear that activation of caspases not only contributes to apoptosis but also leads to the generation of paracrine signals that ensure tissue homeostasis and coordinate tissue repair (Fuchs & Steller, 2011; Martin, Henry, & Cullen, 2012). Further, a complex relationship exists between caspase activation, apoptosis and necroptosis (Green et al., 2011). Accordingly, necroptosis typically occurs under circumstances in which caspase activation is absent or blocked. Because necroptotic death results in the spillage of cytosolic components and alarmins, this form of death triggers secondary inflammation. The important physiological role of necroptosis was highlighted by a number of genetic studies showing that caspase-8 or FADD deficiency cause embryonic lethality and trigger inflammation in vivo by sensitizing cells to RIPK3-mediated necroptosis (Bonnet et al., 2011; Dannappel et al., 2014; Declercq et al., 2009; Dondelinger et al., 2013; Duprez et al., 2011; Ermolaeva et al., 2008; Green et al., 2011; O'Donnell et al., 2011; Oberst et al., 2011; Pasparakis & Vandenabeele, 2015; Polykratis et al., 2014; Vandenabeele et al., 2010; Welz et al., 2011). The activity of RIPK3 is subject to modulation by RIPK1. This is achieved through the RIP homotypic interaction motif (RHIM) present in both RIPK1 and RIPK3 that allows the formation of RIPK1:RIPK3 protein complexes. The recruitment of RIPK1 to RIPK3 not only allows activation of RIPK3, but also permits RIPK1-dependent negative regulation of RIPK3, most likely via recruitment of caspase-8/FLIP heterodimers, which cleave and inactivate RIPK3 (Feng et al., 2007).
Formation and activity of the ripoptosome is also subject to tight regulation by multiple members of the IAP protein family. Of the IAPs, cIAP1 and cIAP2 are the most critical regulators of ripoptosome assembly (Feoktistova et al., 2011; Geserick et al., 2009; Tenev et al., 2011a). Nevertheless, XIAP also contributes to the regulation of this RIPK1-based platform because in the absence of XIAP, depletion of cIAPs results in increased assembly of this complex (Tenev et al., 2011a). The extent to which individual IAPs contribute to the inhibition of ripoptosome assembly most likely depends upon cell type and stimulus. IAP-mediated inactivation of RIPK1 and/or ripoptosome occurs in an Ub-dependent manner, most likely by targeting RIPK1, and other components of the ripoptosome, for proteasomal degradation. Caspase-8-mediated cleavage of cFLIP generates cFLIP(p43), permitting its binding to TRAF2 and the formation of cFLIP(p43)-caspase-8-TRAF2 tertiary complex (Kataoka & Tschopp, 2004; Micheau et al., 2002). TRAF2 then recruits cIAPs which target cFLIP(p43) and caspase-8 for ubiquitylation (Tenev et al., 2011b). This indicates that cIAP1 and cIAP2 target ‘active’ cFLIP-caspase-8 complexes for ubiquitylation and inactivation. The importance of IAP-mediated regulation of RIPK1-based death complexes is illustrated by the notion that Xiap-/-Ciap1-/- and Ciap1-/- Ciap2-/- animals are embryonic lethal, and that this lethality is rescued by crossing the Xiap-/-Ciap1-/- and Ciap1-/-Ciap2-/- mice to Ripk1-/- and Ripk3-/- mice (Moulin et al., 2012). This demonstrates that multiple IAPs function together to regulate an embryonic decision point involving RIP kinase activity.
IAP-mediated regulation of caspases
In both Drosophila and mammals, members of the IAP protein family are the most prominent E3 ligases that modulate caspases and apoptosis. Apoptosis regulatory IAPs carry either two or three NH2-terminal BIR domains and a C-terminal RING finger that provides them with Ub E3-ligase activity (Silke & Meier, 2013). The BIR domain mediates protein interactions, and in most cases, binds to IAP-binding motifs (IBM) present in active caspases and IAP-antagonists such as mammalian Smac/DIABlO and Omi/HtrA2 or Drosophila REAPER, GRIM and Head Involution Defective (HID) (Shi, 2002a). The main feature of an IBM is the presence of an NH2-terminal alanine (Fig. 3). However, in some cases IBMs can also harbor a serine at the first position (Verhagen et al., 2007). The NH2-terminal alanine or serine, which must be exposed and unblocked (devoid of NH2-terminal acetylation), inserts into the extensive hydrophobic cleft on the surface of BIRs and forms hydrogen bonds with neighboring residues, thereby anchoring the IBM-carrying protein to IAPs (Wu et al., 2000). Subtle changes in the peptide-binding groove of BIR domains alter their preference for particular client proteins with IBMs. Therefore, proteins with IBMs display differential and selective binding to specific BIR domains. Apoptosis regulatory IAPs such as XIAP, cIAP1, cIAP2 and Drosophila IAP1 (DIAP1) and DIAP2 carry two BIR domains capable of binding IBMs. The tandem arrangement (i) increases the repertoire of proteins with which they can interact, and (ii) potentially enhances the binding-affinity to particular IBM-containing target proteins, particularly when they are dimeric or oligomeric in nature.
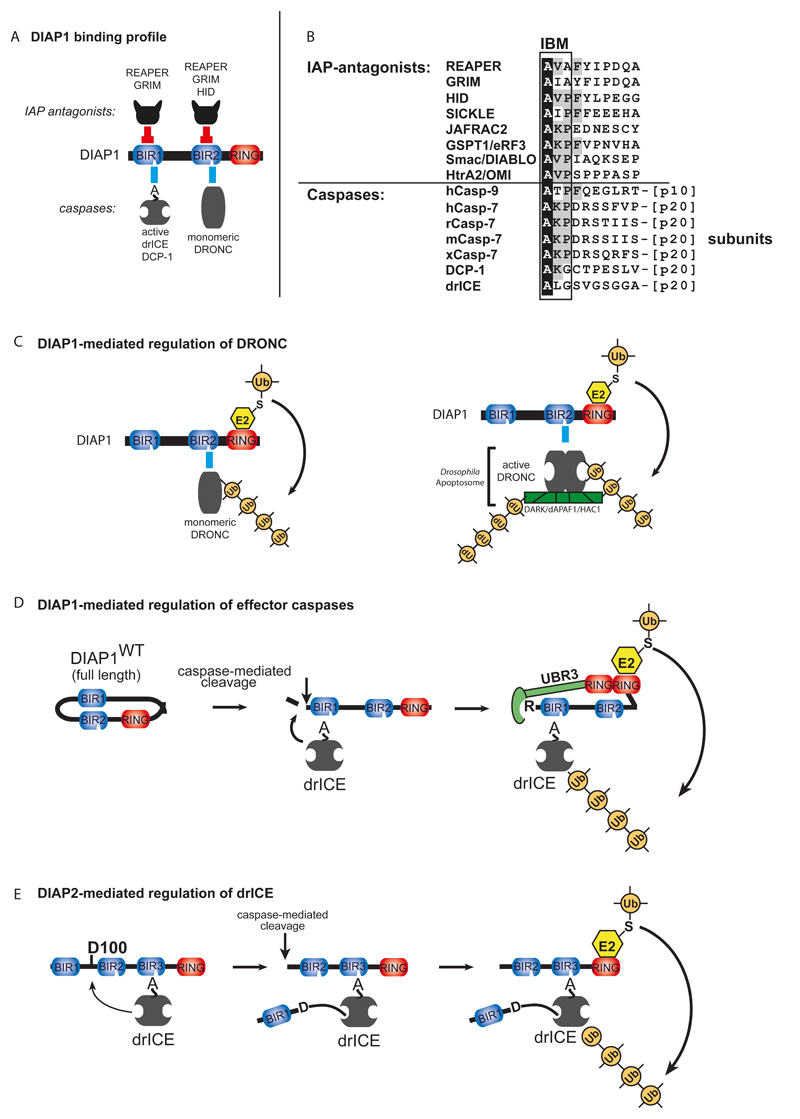
Figure 3. IAP-mediated regulation of caspases in Drosophila.
(A) Binding profile of DIAP1 with caspases and IAP antagonists. Direct physical interaction with the effector caspases drICE or DCP-1 and the initiator caspase DRONC is mediated through DIAP1’s BIR1 and BIR2 domains, respectively. Following their activation, drICE and DCP-1 expose an NH2-terminal IBM (depicted as A), which allows their binding to BIR1. (B) Sequence alignment of IBM-bearing proteins. Identical residues are highlighted in black. Residues conserved in four or more IBM-proteins are indicated in grey. (C) DIAP1’s BIR2-DRONC association is essential for DIAP1 to neutralise DRONC. Following binding, DIAP1’s RING-finger promotes Ub conjugation of DRONC, leading to its inactivation through non-degradative ubiquitylation of monomeric DRONC (left panel), and by targeting apoptosome-associated active DRONC for degradation (right panel). (D-E) Mechanism of effector caspase (drICE) inactivation by DIAP1 (D) and DIAP2 (E). (D) Full-length wild-type DIAP1 is held in an inactive conformation, and requires caspase-mediated proteolytic cleavage at residue 20 for its activation. After cleavage, BIR-mediated caspase-binding occurs more efficiently. Cleavage also facilitates recruitment of N-end rule UBR E3 ligases, which together with DIAP1’s RING domain, promote ubiquitylation and inactivation of drICE and DCP-1. (E) drICE is also subject to regulation by DIAP2. drICE binds to the BIR3 of DIAP2 in an IBM-dependent manner, and following binding cleaves DIAP2 at D100. DIAP2 cleavage results in a covalent adduct between D100 and the catalytic machinery of drICE, trapping the caspase. Full inactivation of drICE is achieved through RING-mediated ubiquitylation.
In Drosophila, DIAP1, encoded by the thread (th) gene, was the first BIR-containing IAP identified. DIAP1-mediated inhibition of caspases is essential for cell survival as loss of DIAP1 function instigates spontaneous caspase-mediated cell death (Goyal, McCall, Agapite, Hartwieg, & Steller, 2000; Lisi, Mazzon, & White, 2000; S. L. Wang, Hawkins, Yoo, Muller, & Hay, 1999). Conversely, gain-of-function mutations significantly suppress cell death, and lead to over-growth phenotypes due to supernumerary cells (Goyal et al., 2000; Lisi et al., 2000; S. L. Wang et al., 1999).
The BIR domain, in combination with its flanking regions, functions as a protein interaction module that, for DIAP1, mediates binding to both initiator (DRONC) and effector caspases (drICE and DCP-1) (Fig. 3). Importantly, different caspases bind to distinct BIRs: while the BIR1 region of DIAP1 is essential for binding to the effector caspases drICE and DCP-1 (Hawkins, Wang, & Hay, 1999; Kaiser, Vucic, & Miller, 1998; Zachariou et al., 2003), the BIR2 region directly associates with the initiator caspase DRONC (Meier, Silke, Leevers, & Evan, 2000). As a consequence of this differential binding, one molecule of DIAP1 can bind simultaneously to DRONC and drICE or DCP-1 (Zachariou et al., 2003). Physical association between DIAP1 and effector caspases is essential for cell survival. Embryos homozygous for diap1 loss-of-function mutations that completely abrogate binding to effector or initiator caspases, die during embryogenesis due to inappropriate cell death (Goyal et al., 2000; Lisi et al., 2000; Rodriguez, Chen, Oliver, & Abrams, 2002; S. L. Wang et al., 1999; Zachariou et al., 2003). On the other hand, mutations that enhance DIAP1’s ability to associate with activated effector caspases result in a gain-of-function phenotype (Goyal et al., 2000; Zachariou et al., 2003). Therefore, DIAP1:caspase association represents a pivotal step in the regulation of the apoptotic caspase cascade.
The mechanism of caspase-binding differs greatly depending on the caspase involved (Fig. 3). While zymogenic DRONC readily binds to the BIR2 region of DIAP1, DIAP1 only associates with proteolytically cleavage forms of drICE/DCP-1. As proteolytic cleavage and removal of the pro-domain of drICE or DCP-1 results in activation of thee effector caspases, this indicates that DIAP1 only regulates active versions of drICE and DCP-1. The mechanism behind this selectivity resides in the exposure of an IBM at the neo-NH2 terminus of drICE and DCP-1, which is uncovered following cleavage and activation of these effector caspases (Tenev, Zachariou, Wilson, Ditzel, & Meier, 2005). Although IAP:caspase association is the decisive step in the regulation of apoptosis in Drosophila, physical interaction between DIAP1 and caspases alone is insufficient to regulate caspases. This is evident because DIAP1-bound effector caspases remain catalytically active under in vitro conditions (Tenev et al., 2005). Moreover, DIAP1 mutants with a dysfunctional RING finger completely fail to suppress caspase-mediated cell death, even though these proteins bind to caspases with the same affinity as their wild-type counterparts. Ultimately, suppression of caspases and apoptosis results from DIAP1-mediated ubiquitylation of the zymogenic form of DRONC and active drICE or DCP-1 (Chai et al., 2003; Ditzel et al., 2008; Lisi et al., 2000; Wilson et al., 2002).
The mechanism by which ubiquitylation of DRONC causes its inactivation appears to be context-dependent, involving degradative as well as non-degradative ubiquitylation. Outside of the apoptosome, DIAP1-mediated ubiquitylation of DRONC neutralizes it through an unknown mechanism that operates independent of the proteasome (Lee et al., 2011) (Fig. 3). However, when part of the apoptosome, DIAP1 conjugates K48-linked poly-Ub chains to DRONC, targeting it for proteasomal destruction (Shapiro, Hsu, Jung, Robbins, & Ryoo, 2008). Hence, only apoptosome-associated DRONC (as well as DARK itself), but not free monomeric DRONC, is targeted for proteasomal degradation. Interestingly, DRONC-mediated cleavage of DARK is required for proteasomal degradation of the DRONC/DARK complex. This suggests that the cleavage event recruits the E3 ligase (Shapiro et al., 2008).
DIAP1-mediated regulation of effector caspases is also dependent on the conjugation of Ub. Attachment of non-degradative (K63-linked) poly-Ub chains to the effector caspase drICE (homologue of caspase-3/-7) directly reduces its proteolytic potency, affecting kinetic parameters of the enzyme (Ditzel et al., 2008). Computational modelling of an ubiquitylated effector caspase suggests that the Ub chains sterically occlude the catalytic pocket of the caspase and would interfere with substrate entry.
In addition to Ub, DIAP1 can also inactivate effector caspases via the covalent attachment of the Ub-like modifier NEDD8. NEDD8-mediated suppression of drICE occurs via a mechanism that relies on non-competitive inhibition, most likely through a NEDD8-induced conformational change of the caspase. Disruption of drICE ubiquitylation or NEDDylation, either by loss of DIAP1's E3 activity or generation of a non-modifyable form of drICE, renders this effector caspase resistant to DIAP1-mediated inactivation (Broemer et al., 2010; Ditzel et al., 2008).
Surprisingly, DIAP1 in its full-length form is incapable of binding and regulating caspases, or acting as an E3 ligase. To activate its anti-apoptotic potential, it requires proteolytic cleavage. Removal of the first 20 amino acid residues of DIAP1 radically changes its properties: the cleaved form interacts with caspases far better than full-length, non-cleaved DIAP1. It seems that full-length DIAP1 resides in an inactive, ‘closed’ configuration that precludes caspase binding (Yan, Wu, Chai, Li, & Shi, 2004). Only when it is cleaved can it bind tightly to DRONC or effector caspases, and function as an E3 (Ditzel et al., 2008). In this respect, caspases activate their own inhibition in a regulatory feedback loop.
Cleavage of DIAP1 not only removes the presumptive inhibitory NH2-terminal portion of the protein, it also exposes a new docking site for UBR-containing E3 ligases of the NH2-end rule pathway (Ditzel et al., 2003; Herman-Bachinsky, Ryoo, Ciechanover, & Gonen, 2007) (Fig. 3). DIAP1’s two ubiquitylation-associated activities - UBR-E3 recruitment and DIAP1’s own RING finger – are both required for its full anti-apoptotic activity. The presence of a functional RING alone seems not to be sufficient, since a DIAP1 mutant that retains a functional RING but fails to bind UBRs also fails to protect from cell death induced by ectopic expression of proapoptotic proteins. Likewise, DIAP1 that has a defective RING but is otherwise fully competent in recruiting UBR-E3s also fails to regulate apoptosis properly. Among the different UBRs, UBR3 appears to be particularly important for the regulation of caspases (Q. Huang et al., 2014), as RNAi-mediated knockdown of ubr3 leads to caspase activation and cell death. Intriguingly, in the absence of ubr3, over-expression of DIAP1 fails to suppress caspases; corroborating the notion that DIAP1 relies on recruitment of the NH2-end rule system to suppress caspase activity. Therefore, UBR-recruitment and DIAP1’s RING are both required for DIAP1’s full anti-apoptotic activity (Ditzel et al., 2008; Ditzel et al., 2003; Muro, Means, & Clem, 2005).
drICE, but not DRONC or DCP-1, is also regulated by the second Drosophila IAP, DIAP2. Intriguingly, DIAP2 functions as a mechanism-based regulator of drICE, acting as a pseudo-substrate, which, following cleavage, traps the active caspase via a covalent linkage between DIAP2 and the catalytic machinery of drICE (Ribeiro et al., 2007) (Fig. 3). Despite direct inhibition of drICE’s catalytic cysteine, DIAP2’s E3 ligase activity also contributes to proper drICE inhibition (Ribeiro et al., 2007). Despite the ability of DIAP2 to regulate drICE, this function seems not to be essential for cell viability as diap2 mutant animals are fertile and fully viable. A cell death phenotype in diap2-mutant animals is only revealed when flies are subjected to ionizing radiation (Ribeiro et al., 2007).
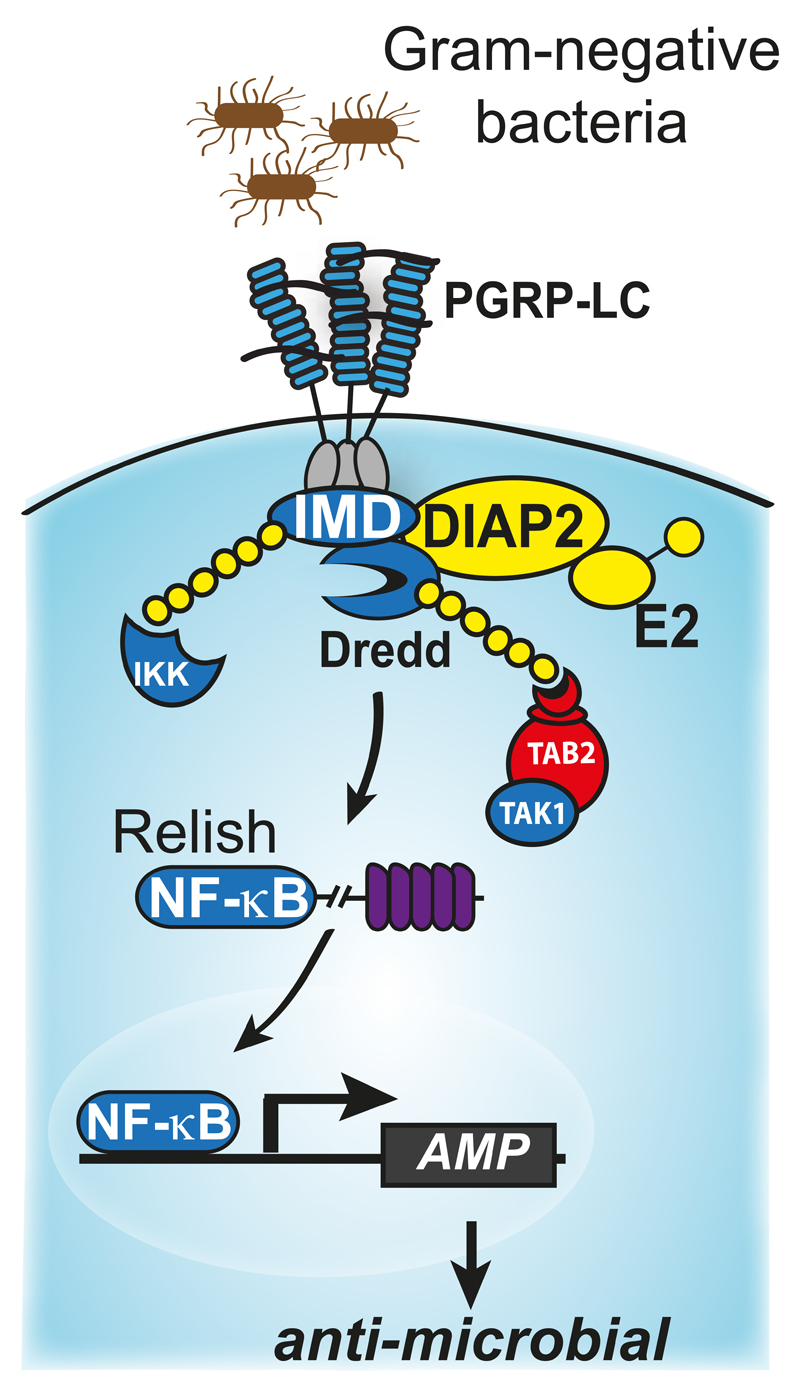
While DIAP2-mediated regulation of caspases is less important for the regulation of cell death, recent evidence indicate that the ability of DIAP2 to regulate caspases is essential for proper regulation of the Drosophila innate immune response to infection with Gram-negative bacteria. DIAP2-mediated conjugation of K63-linked Ub chains on DREDD, the Drosophila orthologue of caspase-8, thereby allows Ub-mediated aggregation and activation of DREDD (Meinander et al., 2012) (Fig. 4). Active DREDD subsequently cleaves IMD (immune deficiency) (Paquette et al., 2010). Upon cleavage, IMD exposes an IBM at its neo-NH2 terminus, which binds to the BIR2/3 of DIAP2. This provides DIAP2 with an additional docking site, reinforcing complex stability and allowing DIAP2-mediated ubiquitylation of IMD, and quite possibly other components of the signaling complex (Paquette et al., 2010). The Ub chains on IMD and DREDD appear to serve as scaffolds for the recruitment of dTAK1, IKK and the precursor form of the NF-kB transcription factor RELISH (Ferrandon, Imler, Hetru, & Hoffmann, 2007; Kanayama et al., 2004; Kleino et al., 2005; Lu, Wu, & Anderson, 2001; Rutschmann et al., 2000; Rutschmann, Kilinc, & Ferrandon, 2002; Silverman et al., 2003; Silverman et al., 2000; Vidal et al., 2001; Zhuang et al., 2006). This brings RELISH into close proximity of ubiquitylated and active DREDD, allowing DREDD-mediated proteolysis of Relish. The proximity to the signaling complex also allows phospho-mediated activation of RELISH (Erturk-Hasdemir et al., 2009). Subsequently, cleaved and phosphoryated RELISH translocates to the nucleus where it drives expression of anti-microbial peptide genes.
Figure 4. Model depicting Ub-dependent activation of NF-κB during Imd signalling.
Ligand binding induces recruitment of Imd and dFADD to PGRP-LC. This allows binding of DREDD to dFADD, which leads to dimerisation-induced activation of DREDD. Active DREDD subsequently cleaves IMD, which leads to the exposure of an IBM at the neo-amino terminus of cleaved IMD. This in turn allows recruitment of DIAP2 into the signalling complex, followed by DIAP2-mediated ubiquitylation of various components of this complex, such as IMD and DREDD. Ubiquitylation of DREDD is key for DREDD-mediated cleavage of RELISH. The Ub chains of DREDD may serve as a scaffold for the recruitment of IKK. Since IKK binds to RELISH, as evidenced by its ability to phosphorylate RELISH, IKK might bring RELISH into close proximity of DREDD for proteolysis. Ubiquitylation of components of the IMD pathway also leads to activation of TAK1 and IKK, which in turn phosphorylates RELISH. (6.) Phosphorylated and cleaved RELISH subsequently translocates to the nucleus where it drives expression of RELISH target genes.
While ubiquitylation of DREDD and IMD are essential for activation of RELISH, several deubiquitylating enzymes have been identified that negatively regulate the ability of IMD and DREDD to drive induction of RELISH-dependent target genes (Engel et al., 2014; Thevenon et al., 2009). Among these are USP36, USP2 and USP34, which prevent inappropriate activation of IMD-dependent immune signalling in unchallenged conditions.
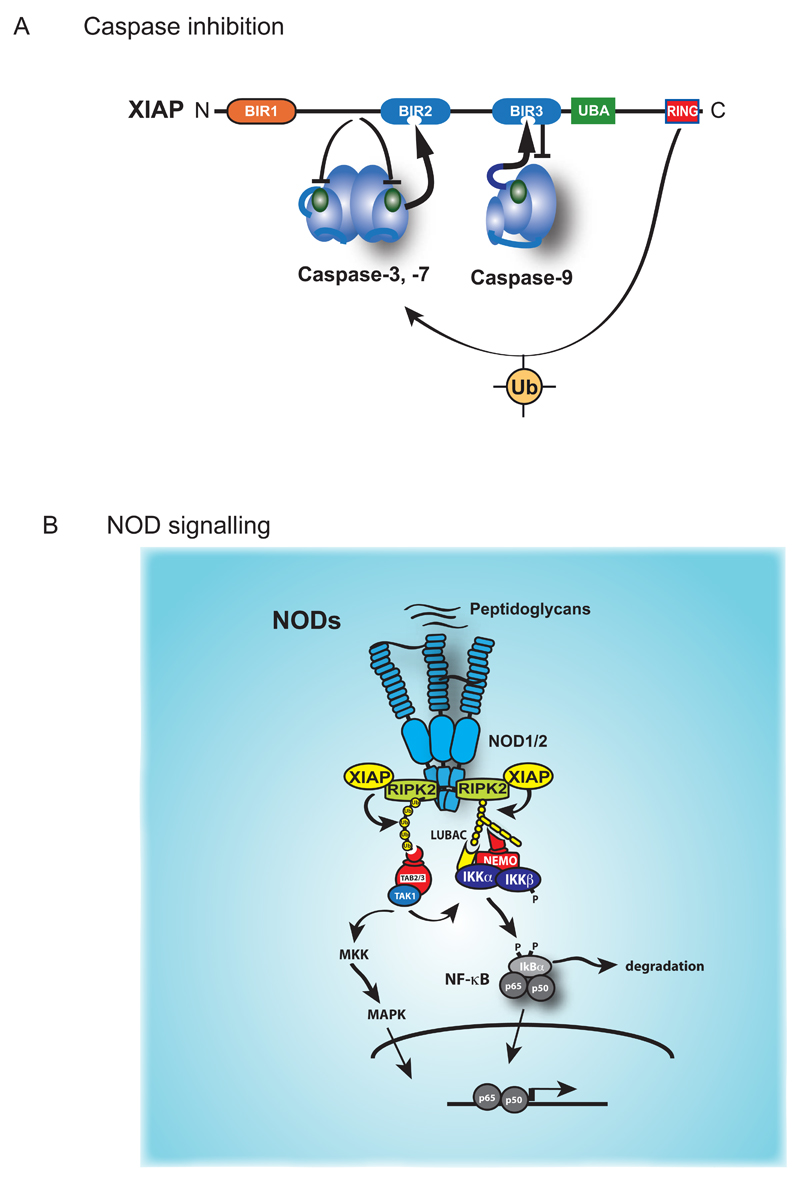
Like DIAP2, mammalian XIAP similarly functions both as mechanism-based caspase inhibitor and potent regulator of Ub-dependent activation of NF-kB (Fig. 5). XIAP-mediated inactivation of caspase-3, -7 and -9 does not require a functional RING finger under in vitro conditions or when over-expressed (Shi, 2002b). Residues located immediately upstream of XIAP’s BIR2 domain directly bind to the active site pocket of caspase-3 and –7 and obstruct substrate entry (Chai et al., 2001; Y. Huang et al., 2001; Riedl et al., 2001; Silke et al., 2001; Suzuki, Nakabayashi, Nakata, Reed, & Takahashi, 2001). XIAP-mediated inactivation of caspase-9 occurs differently, namely by keeping caspase-9 in a monomeric, inactive state (Shiozaki et al., 2003).
Figure 5. XIAP-mediated regulation of caspases and NF-κB.
(A) XIAP directly inhibits the effector caspase-3 and caspase-7, and the initiator caspase-9. The sequence preceding the BIR2 domain of XIAP occupies the catalytic pocket of caspase-3 or caspase-7, thereby blocking substrate entry. In addition, the BIR2 domain interacts with the IBM of caspase-3 or caspase-7 that is exposed following their proteolytic activation (shown as an arrow). XIAP-mediated inhibition of caspase-9 requires proteolytic cleavage of caspase-9, which exposes an IBM that binds to the BIR3 of XIAP. Caspase-9 activity is blocked because XIAP prevents caspase 9 dimerization, a prerequisite for initiator caspasenactivity. The RING domain of XIAP also contributes to caspase inhibition. (B) NOD-mediated activation of NF-kB and MAPK signalling. Detection of bacterial peptidoglycans by NOD1 and NOD2 results in the formation of an oligomeric signalling complex that recruits RIPK2, and XIAP. XIAP mediated ubiquitylation of RIPK2 allows the recruitment of TAB2/TAB3/TAK1 and IKKs, thereby triggering NF-kB and MAPK signalling.
Even though the RING domain of XIAP is not required for caspase inhibition in vitro, it contributes to XIAP’s function in vivo. A recent report now finds that XIAP’s RING indeed is critical for its anti-apoptotic function (Schile, Garcia-Fernandez, & Steller, 2008). Using gene targeting of endogenous XIAP, the authors show that deletion of the RING finger sensitized fibroblasts to TNFα-induced cell death and led to increased rates of apoptosis in an Eµ-Myc mouse lymphoma model. In both these systems, XIAPΔRING mutant cells responded in the same way as XIAP null cells, even though XIAPΔRING protein was abundantly expressed. Moreover, following apoptosis induction caspase activity was significantly higher in XIAPΔRING cells compared to wild-type controls. This indicates that the BIR domains are not sufficient on their own to block caspase activity in vivo. Consistent with a requirement of the RING finger, caspase-3 poly-Ubiquitylation was reduced in mutant cells. Remarkably, no increased levels of caspase-3 were observed, suggesting a non-degradative mode of caspase ubiquitylation, a finding that contradicts an earlier study (Suzuki, Nakabayashi, & Takahashi, 2001). In addition, XIAP reportedly also ubiquitylates caspase-9 (Morizane, Honda, Fukami, & Yasuda, 2005), though the functional outcome of this yet to be determined.
As indicated above, XIAP not only regulates cellular processes by controlling caspases, but also by mediating activation of NF-kB. In particular, XIAP is indispensable for innate immune signalling triggered by NOD1 and NOD2 (Bauler, Duckett, & O'Riordan, 2008; Bertrand et al., 2009; Krieg et al., 2009). NODs belong to the nucleotide-binding oligomerisation domain family that contain an NH2-terminal caspase recruitment domain (CARD), a centrally located nucleotide-binding and oligomerisation domain (NBD), and 10 tandem leucine-rich repeats (LRRs) in their C-terminus through which they detect bacterial peptidoglycans shed during bacterial growth (Chen, Shaw, Kim, & Nunez, 2009).
The pathways activated by NODs are highly reminiscent to the Ub-dependent signal transduction cascade downstream of TNF-R1 (Fig. 5). Like TNF-R1, NODs stimulate Ub-mediated activation of NF-kB and the MAP kinases p38 and JNK. Stimulation of NOD1 and NOD2 receptors triggers the formation of a NOD-dependent multi-protein complex that recruits RIPK2, a close homologue of RIPK1 (Inohara et al., 2000). XIAP in conjunction with LUBAC then conjugate RIPK2 with M1- and K63-linked Ub chains (Bertrand et al., 2009; Hasegawa et al., 2008; Yang et al., 2007), which subsequently result in TAK1/TAB2/3-mediated activation of MAPK and NF-kB pathways leading to production of cytokines, chemokines and antimicrobial peptides that help to defend against invading microbes and ensures the defense of homeostasis (Bertrand et al., 2009; Hasegawa et al., 2008; Park et al., 2007; Yang et al., 2007).
Although XIAP is involved in regulating NOD signalling, XIAP deficiency manifests in a wide range of clinical immune phenotypes, including EBV-associated hemophagocytic lymphohistiocytosis, Crohn-like bowel disease, severe infectious mononucleosis, splenomegaly, uveitis, periodic fever, fistulating skin abscesses, and severe Giardia enteritis (Speckmann et al., 2013). These clinical phenotypes are not shared by mutations in other components of the NOD-dependent multi-protein complex, and thus suggest other immune-related roles of XIAP.
IAPs other than cIAPs and XIAP also play important roles in regulating apoptosis as well as non-apoptotic signalling events in an Ub-dependent fashion. BRUCE/Apollon is a membrane-associated IAP that carries only one BIR domain. Additionally, it also contains an Ub-conjugating (UBC) motif that can function as an Ub-E2, transferring Ub to substrates. In addition to contributing to cytokinesis, BRUCE/Apollon also safe guards cell viability by targeting caspase-9, and the IAP-antagonist protein SMAC/Diablo, for Ub-mediated proteasomal degradation (Bartke, Pohl, Pyrowolakis, & Jentsch, 2004; Hao et al., 2004). In Drosophila, the activity of dBRUCE is indispensable for controlled activation of caspases required for spermatide individualization (Arama, Agapite, & Steller, 2003). Further, dBRUCE also targets the IAP-antogonist Reaper and Grim for proteasomal degradation, thereby contributing to the apoptotic threshold (Domingues & Ryoo, 2012; Vernooy et al., 2002).
Although IAPs are the most prominent class of E3 ligases that target caspases for ubiquitylation, IAPs are not the only E3s that control caspases. Components of a testis specific Cullin-3 based E3 Ub-ligase complex is particularly important for caspase activation during sperm differentiation (Arama, Bader, Rieckhof, & Steller, 2007; Bader, Arama, & Steller, 2010; Kaplan, Gibbs-Bar, Kalifa, Feinstein-Rotkopf, & Arama, 2010). Defects in any of the Cullin-3 components decreases caspase activity and causes male sterility due to individualization defects. Similarly caspases are activated during mammalian spermatogenesis and disrupting the cell death machinery results in infertile males due to defects during late spermatid maturation. In Drosophila, spermatid individualization is a process in which 64 interconnected spermatids separate from one another and eliminate the majority of their cytoplasmic contents (Feinstein-Rotkopf & Arama, 2009). This process requires the activity of Cullin-3, the substrate binding BTB protein KLHL10 and the pseudosubstrate inhibitor Soti (Arama et al., 2007; Bader et al., 2010; Kaplan et al., 2010). Cullin-3/KLHL10 allows sub lethal activation of caspases by targeting the caspase inhibitor dBRUCE for ubiquitylation and proteasomal degradation. Interestingly, it was recently reported that mutations in the human form of KLHL10 are associated with male infertility and low sperm count (Yatsenko et al., 2006), indicating that ubiquitylation may be used in a similar fashion for cell sculpting in mammals.
Concluding remarks
The maintenance of homeostasis in multicellular organisms depends on a continuous, coordinated response to external and internal insults that challenge cellular and tissue integrity throughout life. Loss of homeostasis is a hallmark of aging, resulting in pathologies often caused by defective or deregulated tissue damage responses. Inflammation and cell death are essential defense responses that are induced by infection or injury. However, inflammation and cell death are also induced by tissue stress and malfunction to maintain tissue homeostasis under a variety of noxious conditions. A number of dedicated sensors have evolved to detect different stressors and induce appropriate adaptive responses. Common to these pathways is the conjugation of non-degradative Ub chains that produce robust signalling networks, which coordinate tissue remodeling and adaptation to tissue stress via transcriptional programmes, the induction of apoptosis or necroptosis. Most, if not all, cellular stress responses, in addition to cell–autonomous adaptive changes, produce secreted factors that affect other cells in the tissue. This coordinates tissue remodeling that replaces malfunctioning or damaged tissues (Grivennikov, Greten, & Karin, 2010).
Whatever the cause of the inflammatory response, its ‘purpose’ is to remove the source of the disturbance, to allow the host to adapt to the abnormal conditions, and, ultimately, to restore functionality and homeostasis to the tissue. Much more needs to be learnt about how the Ub-signalling system impacts on inflammation and adaptation to tissue stress. Further, unraveling how the Ub-signal is conjugated, recognized and disassembled will be critically important to gain a better understanding of the regulatory processes that control cell death, inflammation and stress responses, and might contribute to disease pathologies such as cancer.
Acknowledgments
We regret not being able to discuss many studies relevant to this topic. PM and OM are supported by funds from Breakthrough Breast Cancer and The Institute of Cancer Research, and MB is supported by the Deutsche Forschungsgemeinschaft (DFG) (BR3442/2-1). We acknowledge NHS funding to the NIHR Biomedical Research Centre.
References
- Amoyel M, Bach EA. Cell competition: how to eliminate your neighbours. Development. 2014;141(5):988–1000. doi: 10.1242/dev.079129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4(5):687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 2007;5(10):e251. doi: 10.1371/journal.pbio.0050251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader M, Arama E, Steller H. A novel F-box protein is required for caspase activation during cellular remodeling in Drosophila. Development. 2010;137(10):1679–1688. doi: 10.1242/dev.050088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9(5):361–371. doi: 10.1038/nrc2628. doi: nrc2628 [pii] [DOI] [PubMed] [Google Scholar]
- Bartke T, Pohl C, Pyrowolakis G, Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14(6):801–811. doi: 10.1016/j.molcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Bauler LD, Duckett CS, O'Riordan MX. XIAP regulates cytosol-specific innate immunity to Listeria infection. PLoS Pathog. 2008;4(8):e1000142. doi: 10.1371/journal.ppat.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M. Cellular Inhibitors of Apoptosis cIAP1 and cIAP2 Are Required for Innate Immunity Signaling by the Pattern Recognition Receptors NOD1 and NOD2. Immunity. 2009 doi: 10.1016/j.immuni.2009.04.011. doi: S1074-7613(09)00203-9 [pii] [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458(7237):430–437. doi: 10.1038/nature07959. doi: nature07959 [pii] [DOI] [PubMed] [Google Scholar]
- Bondar T, Medzhitov R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell. 2010;6(4):309–322. doi: 10.1016/j.stem.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet MC, Preukschat D, Welz PS, van Loo G, Ermolaeva MA, Bloch W, Pasparakis M. The adaptor protein FADD protects epidermal keratinocytes from necroptosis in vivo and prevents skin inflammation. Immunity. 2011;35(4):572–582. doi: 10.1016/j.immuni.2011.08.014. doi: S1074-7613(11)00401-8 [pii] [DOI] [PubMed] [Google Scholar]
- Broemer M, Tenev T, Rigbolt KT, Hempel S, Blagoev B, Silke J, Meier P. Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Mol Cell. 2010;40(5):810–822. doi: 10.1016/j.molcel.2010.11.011. doi: S1097-2765(10)00849-X [pii] [DOI] [PubMed] [Google Scholar]
- Carswell EA, Old LJ, Kassel RL, Green S, Fiore N, Williamson B. An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci U S A. 1975;72(9):3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J, Shiozaki E, Srinivasula SM, Wu Q, Datta P, Alnemri ES, Dataa P. Structural basis of caspase-7 inhibition by XIAP. Cell. 2001;104(5):769–780. doi: 10.1016/s0092-8674(01)00272-0. [DOI] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol. 2003;10(11):892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Chen G, Shaw MH, Kim YG, Nunez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol. 2009;4:365–398. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- Chovatiya R, Medzhitov R. Stress, inflammation, and defense of homeostasis. Mol Cell. 2014;54(2):281–288. doi: 10.1016/j.molcel.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claveria C, Giovinazzo G, Sierra R, Torres M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature. 2013;500(7460):39–44. doi: 10.1038/nature12389. [DOI] [PubMed] [Google Scholar]
- Dannappel M, Vlantis K, Kumari S, Polykratis A, Kim C, Wachsmuth L, Pasparakis M. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature. 2014 doi: 10.1038/nature13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declercq W, Vanden Berghe T, Vandenabeele P. RIP kinases at the crossroads of cell death and survival. Cell. 2009;138(2):229–232. doi: 10.1016/j.cell.2009.07.006. doi: S0092-8674(09)00845-9 [pii] [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10(10):659–671. doi: 10.1038/nrm2767. doi: nrm2767 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KT, Meier P. Inactivation of effector caspases through nondegradative polyubiquitylation. Mol Cell. 2008;32(4):540–553. doi: 10.1016/j.molcel.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, Meier P. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5(5):467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- Domingues C, Ryoo HD. Drosophila BRUCE inhibits apoptosis through non-lysine ubiquitination of the IAP-antagonist REAPER. Cell Death Differ. 2012;19(3):470–477. doi: 10.1038/cdd.2011.116. doi: cdd2011116 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondelinger Y, Aguileta MA, Goossens V, Dubuisson C, Grootjans S, Dejardin E, Bertrand MJ. RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death Differ. 2013;20(10):1381–1392. doi: 10.1038/cdd.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T, Vandenabeele P. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity. 2011;35(6):908–918. doi: 10.1016/j.immuni.2011.09.020. doi: S1074-7613(11)00508-5 [pii] [DOI] [PubMed] [Google Scholar]
- Engel E, Viargues P, Mortier M, Taillebourg E, Coute Y, Thevenon D, Fauvarque MO. Identifying USPs regulating immune signals in Drosophila: USP2 deubiquitinates Imd and promotes its degradation by interacting with the proteasome. Cell Commun Signal. 2014;12:41. doi: 10.1186/PREACCEPT-1588328929121802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Michallet MC, Papadopoulou N, Utermohlen O, Kranidioti K, Kollias G, Pasparakis M. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat Immunol. 2008;9(9):1037–1046. doi: 10.1038/ni.1638. 1046. doi: ni.1638 [pii] [DOI] [PubMed] [Google Scholar]
- Erturk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Silverman N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci U S A. 2009;106(24):9779–9784. doi: 10.1073/pnas.0812022106. doi: 0812022106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein-Rotkopf Y, Arama E. Can't live without them, can live with them: roles of caspases during vital cellular processes. Apoptosis. 2009;14(8):980–995. doi: 10.1007/s10495-009-0346-6. [DOI] [PubMed] [Google Scholar]
- Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N, Wu M. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal. 2007;19(10):2056–2067. doi: 10.1016/j.cellsig.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Leverkus M. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell. 2011;43(3):449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7(11):862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147(4):742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardam S, Turner VM, Anderton H, Limaye S, Basten A, Koentgen F, Brink R. Deletion of cIAP1 and cIAP2 in murine B lymphocytes constitutively activates cell survival pathways and inactivates the germinal center response. Blood. 2011;117(15):4041–4051. doi: 10.1182/blood-2010-10-312793. doi: blood-2010-10-312793 [pii] [DOI] [PubMed] [Google Scholar]
- Geserick P, Hupe M, Moulin M, Wong WW, Feoktistova M, Kellert B, Leverkus M. Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting RIP1 kinase recruitment. J Cell Biol. 2009;187(7):1037–1054. doi: 10.1083/jcb.200904158. doi: jcb.200904158 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. Embo J. 2000;19(4):589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Oberst A, Dillon CP, Weinlich R, Salvesen GS. RIPK-dependent necrosis and its regulation by caspases: a mystery in five acts. Mol Cell. 2011;44(1):9–16. doi: 10.1016/j.molcel.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Sekine K, Kawabata A, Nakamura H, Ishioka T, Ohata H, Naito M. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol. 2004;6(9):849–860. doi: 10.1038/ncb1159. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G, Inohara N. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. Embo J. 2008;27(2):373–383. doi: 10.1038/sj.emboj.7601962. doi: 7601962 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins CJ, Wang SL, Hay BA. A cloning method to identify caspases and their regulators in yeast: Identification of Drosophila IAP1 as an inhibitor of the Drosophila caspase DCP-1. Proc Natl Acad Sci U S A. 1999;96(6):2885–2890. doi: 10.1073/pnas.96.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman-Bachinsky Y, Ryoo HD, Ciechanover A, Gonen H. Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ. 2007;14(4):861–871. doi: 10.1038/sj.cdd.4402079. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat Med. 2000;6(10):1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6(10):776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- Huang Q, Tang X, Wang G, Fan Y, Ray L, Bergmann A, Lin X. Ubr3 E3 ligase regulates apoptosis by controlling the activity of DIAP1 in Drosophila. Cell Death Differ. 2014;21(12):1961–1970. doi: 10.1038/cdd.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Park YC, Rich RL, Segal D, Myszka DG, Wu H. Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain. Cell. 2001;104(5):781–790. [PubMed] [Google Scholar]
- Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, Miura M. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. Embo J. 2002;21(12):3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T, Miura M. The Drosophila TNF ortholog Eiger: emerging physiological roles and evolution of the TNF system. Semin Immunol. 2014;26(3):267–274. doi: 10.1016/j.smim.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Inohara N, Koseki T, Lin J, del Peso L, Lucas PC, Chen FF, Nunez G. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem. 2000;275(36):27823. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88(3):347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Vucic D, Miller LK. The Drosophila inhibitor of apoptosis D-IAP1 suppresses cell death induced by the caspase drICE. FEBS Lett. 1998;440(1-2):243–248. doi: 10.1016/s0014-5793(98)01465-3. [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chen ZJ. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15(4):535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Kaplan Y, Gibbs-Bar L, Kalifa Y, Feinstein-Rotkopf Y, Arama E. Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Dev Cell. 2010;19(1):160–173. doi: 10.1016/j.devcel.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Tschopp J. N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol Cell Biol. 2004;24(7):2627–2636. doi: 10.1128/MCB.24.7.2627-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. Embo J. 2006;25(20):4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleino A, Valanne S, Ulvila J, Kallio J, Myllymaki H, Enwald H, Ramet M. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. Embo J. 2005;24(19):3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37(Pt 5):937–953. doi: 10.1042/BST0370937. doi: BST0370937 [pii] [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. doi: nrm2731 [pii] [DOI] [PubMed] [Google Scholar]
- Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29(4):451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Krieg A, Correa RG, Garrison JB, Le Negrate G, Welsh K, Huang Z, Reed JC. XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci U S A. 2009;106(34):14524–14529. doi: 10.1073/pnas.0907131106. doi: 0907131106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TV, Fan Y, Wang S, Srivastava M, Broemer M, Meier P, Bergmann A. Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation. PLoS Genet. 2011;7(9):e1002261. doi: 10.1371/journal.pgen.1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13(19):2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154(2):669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wu LP, Anderson KV. The antibacterial arm of the drosophila innate immune response requires an IkappaB kinase. Genes Dev. 2001;15(1):104–110. doi: 10.1101/gad.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren J, Masson P, Realini CA, Young P. Use of RNA interference and complementation to study the function of the Drosophila and human 26S proteasome subunit S13. Mol Cell Biol. 2003;23(15):5320–5330. doi: 10.1128/MCB.23.15.5320-5330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Huang J, Yang L, Yang Y, Li W, Xue L. NOPO modulates Egr-induced JNK-independent cell death in Drosophila. Cell Res. 2012;22(2):425–431. doi: 10.1038/cr.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Li W, Yu H, Yang Y, Li M, Xue L, Xu T. Bendless modulates JNK-mediated cell death and migration in Drosophila. Cell Death Differ. 2014;21(3):407–415. doi: 10.1038/cdd.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Yang L, Yang Y, Li M, Li W, Xue L. dUev1a modulates TNF-JNK mediated tumor progression and cell death in Drosophila. Dev Biol. 2013;380(2):211–221. doi: 10.1016/j.ydbio.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Henry CM, Cullen SP. A perspective on Mammalian caspases as positive and negative regulators of inflammation. Mol Cell. 2012;46(4):387–397. doi: 10.1016/j.molcel.2012.04.026. doi: S1097-2765(12)00348-6 [pii] [DOI] [PubMed] [Google Scholar]
- Martins VC, Busch K, Juraeva D, Blum C, Ludwig C, Rasche V, Rodewald HR. Cell competition is a tumour suppressor mechanism in the thymus. Nature. 2014;509(7501):465–470. doi: 10.1038/nature13317. [DOI] [PubMed] [Google Scholar]
- Matsuzawa A, Tseng PH, Vallabhapurapu S, Luo JL, Zhang W, Wang H, Karin M. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science. 2008;321(5889):663–668. doi: 10.1126/science.1157340. doi: 1157340 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454(7203):428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. Embo J. 2000;19(4):598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinander A, Runchel C, Tenev T, Chen L, Kim CH, Ribeiro PS, Meier P. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. Embo J. 2012;31(12):2770–2783. doi: 10.1038/emboj.2012.121. doi: emboj2012121 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157(4):910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW, Grutter MG. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J Biol Chem. 2002;277(47):45162–45171. doi: 10.1074/jbc.M206882200. [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114(2):181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Moreno E, Yan M, Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol. 2002;12(14):1263–1268. doi: 10.1016/s0960-9822(02)00954-5. [DOI] [PubMed] [Google Scholar]
- Morizane Y, Honda R, Fukami K, Yasuda H. X-linked inhibitor of apoptosis functions as ubiquitin ligase toward mature caspase-9 and cytosolic Smac/DIABLO. J Biochem (Tokyo) 2005;137(2):125–132. doi: 10.1093/jb/mvi029. [DOI] [PubMed] [Google Scholar]
- Moulin M, Anderton H, Voss AK, Thomas T, Wong WW, Bankovacki A, Vaux DL. IAPs limit activation of RIP kinases by TNF receptor 1 during development. Embo J. 2012;31(7):1679–1691. doi: 10.1038/emboj.2012.18. doi: emboj201218 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro I, Means JC, Clem RJ. Cleavage of the apoptosis inhibitor DIAP1 by the apical caspase DRONC in both normal and apoptotic Drosophila cells. J Biol Chem. 2005 doi: 10.1074/jbc.M501206200. [DOI] [PubMed] [Google Scholar]
- Neves J, Demaria M, Campisi J, Jasper H. Of flies, mice, and men: evolutionarily conserved tissue damage responses and aging. Dev Cell. 2015;32(1):9–18. doi: 10.1016/j.devcel.2014.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R, Ting AT. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol. 2011;13(12):1437–1442. doi: 10.1038/ncb2362. doi: ncb2362 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Green DR. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3- dependent necrosis. Nature. 2011;471(7338):363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel M, Menthena A, Dabeva MD, Shafritz DA. Cell competition leads to a high level of normal liver reconstitution by transplanted fetal liver stem/progenitor cells. Gastroenterology. 2006;130(2):507–520. doi: 10.1053/j.gastro.2005.10.049. quiz 590. [DOI] [PubMed] [Google Scholar]
- Oliver ER, Saunders TL, Tarle SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131(16):3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette N, Broemer M, Aggarwal K, Chen L, Husson M, Erturk-Hasdemir D, Silverman N. Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol Cell. 2010;37(2):172–182. doi: 10.1016/j.molcel.2009.12.036. doi: S1097-2765(10)00039-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, Nunez G. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178(4):2380–2386. doi: 10.4049/jimmunol.178.4.2380. doi: 178/4/2380 [pii] [DOI] [PubMed] [Google Scholar]
- Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517(7534):311–320. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- Polykratis A, Hermance N, Zelic M, Roderick J, Kim C, Van TM, Kelliher MA. Cutting Edge: RIPK1 Kinase Inactive Mice Are Viable and Protected from TNF-Induced Necroptosis In Vivo. J Immunol. 2014;193(4):1539–1543. doi: 10.4049/jimmunol.1400590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136(6):1098–1109. doi: 10.1016/j.cell.2009.03.007. doi: S0092-8674(09)00264-5 [pii] [DOI] [PubMed] [Google Scholar]
- Ribeiro PS, Kuranaga E, Tenev T, Leulier F, Miura M, Meier P. DIAP2 functions as a mechanism-based regulator of drICE that contributes to the caspase activity threshold in living cells. J Cell Biol, 179. 2007;179(7):1467–1480. doi: 10.1083/jcb.200706027. doi: jcb.200706027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Salvesen GS. Structural basis for the inhibition of caspase-3 by XIAP. Cell. 2001;104(5):791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Chen P, Oliver H, Abrams JM. Unrestrained caspase-dependent cell death caused by loss of Diap1 function requires the Drosophila Apaf-1 homolog, Dark. Embo J. 2002;21(9):2189–2197. doi: 10.1093/emboj/21.9.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann S, Jung AC, Zhou R, Silverman N, Hoffmann JA, Ferrandon D. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat Immunol. 2000;1(4):342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- Rutschmann S, Kilinc A, Ferrandon D. Cutting edge: the toll pathway is required for resistance to gram-positive bacterial infections in Drosophila. J Immunol. 2002;168(4):1542–1546. doi: 10.4049/jimmunol.168.4.1542. [DOI] [PubMed] [Google Scholar]
- Schile AJ, Garcia-Fernandez M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22(16):2256–2266. doi: 10.1101/gad.1663108. 22/16/2256 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro PJ, Hsu HH, Jung H, Robbins ES, Ryoo HD. Regulation of the Drosophila apoptosome through feedback inhibition. Nat Cell Biol. 2008;10(12):1440–1446. doi: 10.1038/ncb1803. ncb1803 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. A conserved tetrapeptide motif: potentiating apoptosis through IAP-binding. Cell Death Differ. 2002a;9(2):93–95. doi: 10.1038/sj.cdd.4400957. [DOI] [PubMed] [Google Scholar]
- Shi Y. Mechanisms of Caspase Activation and Inhibition during Apoptosis. Mol Cell. 2002b;9(3):459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. MolCell. 2003;11(2):519. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Silke J. The regulation of TNF signalling: what a tangled web we weave. Curr Opin Immunol. 2011;23(5):620–626. doi: 10.1016/j.coi.2011.08.002. doi: S0952-7915(11)00105-1 [pii] [DOI] [PubMed] [Google Scholar]
- Silke J, Ekert PG, Day CL, Hawkins CJ, Baca M, Chew J, Vaux DL. Direct inhibition of caspase 3 is dispensable for the anti-apoptotic activity of XIAP. Embo J. 2001;20(12):3114–3123. doi: 10.1093/emboj/20.12.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silke J, Meier P. Inhibitor of apoptosis (IAP) proteins-modulators of cell death and inflammation. Cold Spring Harb Perspect Biol. 2013;5(2) doi: 10.1101/cshperspect.a008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Erlich RL, Hunter M, Bernstein E, Schneider D, Maniatis T. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J Biol Chem. 2003;278(49):48928–48934. doi: 10.1074/jbc.M304802200. M304802200 [pii] [DOI] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14(19):2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckmann C, Lehmberg K, Albert MH, Damgaard RB, Fritsch M, Gyrd-Hansen M, Ehl S. X-linked inhibitor of apoptosis (XIAP) deficiency: the spectrum of presenting manifestations beyond hemophagocytic lymphohistiocytosis. Clin Immunol. 2013;149(1):133–141. doi: 10.1016/j.clim.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG, Jr, Ohh M. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol. 2004;24(8):3251–3261. doi: 10.1128/MCB.24.8.3251-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi Y, Nakata K, Reed JC, Takahashi R. X-linked inhibitor of apoptosis protein (XIAP) inhibits caspase-3 and - 7 in distinct modes. J Biol Chem. 2001;276(29):27058–27063. doi: 10.1074/jbc.M102415200. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci U S A. 2001;98(15):8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10(5):538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Meier P. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell. 2011a;43(3):432–448. doi: 10.1016/j.molcel.2011.06.006. doi: S1097-2765(11)00420-5 [pii] [DOI] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Meier P. The Ripoptosome, a Signaling Platform that Assembles in Response to Genotoxic Stress and Loss of IAPs. Mol Cell. 2011b doi: 10.1016/j.molcel.2011.06.006. doi: S1097-2765(11)00420-5 [pii] [DOI] [PubMed] [Google Scholar]
- Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol. 2005;7(1):70–77. doi: 10.1038/ncb1204. [DOI] [PubMed] [Google Scholar]
- Thevenon D, Engel E, Avet-Rochex A, Gottar M, Bergeret E, Tricoire H, Fauvarque MO. The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe. 2009;6(4):309–320. doi: 10.1016/j.chom.2009.09.007. doi: S1931-3128(09)00316-3 [pii] [DOI] [PubMed] [Google Scholar]
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267(5203):456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11(10):700–714. doi: 10.1038/nrm2970. doi: nrm2970 [pii] [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Kratina TK, Hawkins CJ, Silke J, Ekert PG, Vaux DL. Identification of mammalian mitochondrial proteins that interact with IAPs via N-terminal IAP binding motifs. Cell Death Differ. 2007;14(2):348–357. doi: 10.1038/sj.cdd.4402001. doi: 4402001 [pii] [DOI] [PubMed] [Google Scholar]
- Vernooy SY, Chow V, Su J, Verbrugghe K, Yang J, Cole S, Hay BA. Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not Hid-dependent cell death. Curr Biol. 2002;12(13):1164–1168. doi: 10.1016/s0960-9822(02)00935-1. [DOI] [PubMed] [Google Scholar]
- Vidal S, Khush RS, Leulier F, Tzou P, Nakamura M, Lemaitre B. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 2001;15(15):1900–1912. doi: 10.1101/gad.203301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JP, Fletcher AG, Baena-Lopez LA. Mechanisms and mechanics of cell competition in epithelia. Nat Rev Mol Cell Biol. 2013;14(9):581–591. doi: 10.1038/nrm3639. [DOI] [PubMed] [Google Scholar]
- Walczak H. Death receptor-ligand systems in cancer, cell death, and inflammation. Cold Spring Harb Perspect Biol. 2013;5(5) doi: 10.1101/cshperspect.a008698. a008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133(4):693–703. doi: 10.1016/j.cell.2008.03.036. doi: S0092-8674(08)00501-1 [pii] [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98(4):453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernandez-Majada V, Ermolaeva M, Pasparakis M. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477(7364):330–334. doi: 10.1038/nature10273. doi: nature10273 [pii] [DOI] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408(6815):1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- Xue L, Igaki T, Kuranaga E, Kanda H, Miura M, Xu T. Tumor suppressor CYLD regulates JNK-induced cell death in Drosophila. Dev Cell. 2007;13(3):446–454. doi: 10.1016/j.devcel.2007.07.012. doi: S1534-5807(07)00273-0 [pii] [DOI] [PubMed] [Google Scholar]
- Yan N, Wu JW, Chai J, Li W, Shi Y. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat Struct Mol Biol. 2004;11(5):420–428. doi: 10.1038/nsmb764. [DOI] [PubMed] [Google Scholar]
- Yang Y, Yin C, Pandey A, Abbott D, Sassetti C, Kelliher MA. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J Biol Chem. 2007;282(50):36223–36229. doi: 10.1074/jbc.M703079200. doi: M703079200 [pii] [DOI] [PubMed] [Google Scholar]
- Yatsenko AN, Roy A, Chen R, Ma L, Murthy LJ, Yan W, Matzuk MM. Non-invasive genetic diagnosis of male infertility using spermatozoal RNA: KLHL10 mutations in oligozoospermic patients impair homodimerization. Hum Mol Genet. 2006;15(23):3411–3419. doi: 10.1093/hmg/ddl417. [DOI] [PubMed] [Google Scholar]
- Zachariou A, Tenev T, Goyal L, Agapite J, Steller H, Meier P. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. Embo J. 2003;22(24):6642–6652. doi: 10.1093/emboj/cdg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang ZH, Sun L, Kong L, Hu JH, Yu MC, Reinach P, Ge BX. Drosophila TAB2 is required for the immune activation of JNK and NF-kappaB. Cell Signal. 2006;18(7):964–970. doi: 10.1016/j.cellsig.2005.08.020. doi: S0898-6568(05)00220-2 [pii] [DOI] [PubMed] [Google Scholar]