The plasma membrane aquaporin AtPIP1;4 facilitates the diffusion of pathogen-induced apoplastic H2O2 to activate plant immunity.
Abstract
Hydrogen peroxide (H2O2) is a stable component of reactive oxygen species, and its production in plants represents the successful recognition of pathogen infection and pathogen-associated molecular patterns (PAMPs). This production of H2O2 is typically apoplastic but is subsequently associated with intracellular immunity pathways that regulate disease resistance, such as systemic acquired resistance and PAMP-triggered immunity. Here, we elucidate that an Arabidopsis (Arabidopsis thaliana) aquaporin (i.e. the plasma membrane intrinsic protein AtPIP1;4) acts to close the cytological distance between H2O2 production and functional performance. Expression of the AtPIP1;4 gene in plant leaves is inducible by a bacterial pathogen, and the expression accompanies H2O2 accumulation in the cytoplasm. Under de novo expression conditions, AtPIP1;4 is able to mediate the translocation of externally applied H2O2 into the cytoplasm of yeast (Saccharomyces cerevisiae) cells. In plant cells treated with H2O2, AtPIP1;4 functions as an effective facilitator of H2O2 transport across plasma membranes and mediates the translocation of externally applied H2O2 from the apoplast to the cytoplasm. The H2O2-transport role of AtPIP1;4 is essentially required for the cytoplasmic import of apoplastic H2O2 induced by the bacterial pathogen and two typical PAMPs in the absence of induced production of intracellular H2O2. As a consequence, cytoplasmic H2O2 quantities increase substantially while systemic acquired resistance and PAMP-triggered immunity are activated to repress the bacterial pathogenicity. By contrast, loss-of-function mutation at the AtPIP1;4 gene locus not only nullifies the cytoplasmic import of pathogen- and PAMP-induced apoplastic H2O2 but also cancels the subsequent immune responses, suggesting a pivotal role of AtPIP1;4 in apocytoplastic signal transduction in immunity pathways.
Hydrogen peroxide (H2O2) is a stable component of reactive oxygen species (ROS) compared with other ROS molecules, such as the superoxide anion and hydroxyl radical. In plants, the rapid production of ROS, especially H2O2, represents the successful recognition of pathogen infection and pathogen-associated molecular patterns (PAMPs; Torres, 2010). Well-known examples of PAMPs are invariant microbial epitopes like fungal chitin (Kaku et al., 2006) and bacterial flagellin (Zipfel et al., 2004) and harpin (Sang et al., 2012; Choi et al., 2013) proteins. These PAMPs can be recognized by plasma membrane (PM) integral pattern receptors to induce immune responses (Ausubel, 2005), including H2O2 production in plants (Felix et al., 1992; Levine et al., 1994 Newman et al., 2013; Galletti et al., 2011). The production of H2O2 is typically apoplastic, resulting mainly from the enzymatic activity of NADPH oxidase (NOX) located on the PM (Sagi and Fluhr, 2006). Then, H2O2 experiences cross talk with immunity pathways, such as systemic acquired resistance (SAR) and pathogen-associated molecular pattern-triggered immunity (PTI), to regulate plant disease resistance (Torres, 2010). SAR is characteristic of the induced expression of PATHOGENESIS-RELATED (PR) genes, typically PR-1 and PR-2, under the regulation of the NONINDUCER OF PR GENES1 (NPR1) protein (Cao et al., 1997). NPR1 functions by conformational changes under cytoplasmic redox conditions (Tada et al., 2008) and proteasome-mediated turnover in the nucleus (Spoel et al., 2009). The PTI pathway deploys a cytoplasmic mitogen-activated protein kinase (MAPK) cascade (Asai et al., 2002; Pitzschke et al., 2009) with a branch in which MPK3 and MPK6 phosphorylate different substrates (Bigeard et al., 2015; Pitzschke, 2015) to activate a set of immune responses, including H2O2 and callose production (Bethke et al., 2012; Daudi et al., 2012). Callose is a β-1,3-glucan synthesized by glucan synthase-like (GSL) enzymes, with GSL5 playing a critical role in cellular immune responses (Lü et al., 2011). Therefore, both the SAR and PTI pathways constitute pivotal tiers of intracellular responses in cross talk with the H2O2 signal following its production in the apoplast (Sagi and Fluhr, 2006). Obviously, a cytological gap exists between H2O2 generation and functional performance. In fact, how the apoplastic H2O2 penetrates plant PMs to enter the cytoplasm and regulate immunity remains a long-unanswered question.
It has been proposed that H2O2 transport across a biomembrane is mediated by particular aquaporin (AQP) isoforms in addition to certain roles of membrane lipids (Bienert et al., 2006, 2007; Bienert and Chaumont, 2014; Aguayo et al., 2015). AQPs are biomembrane channels essential for the transport of water, H2O2, and other small substrates in all living cells (Maurel, 2007; Gomes et al., 2009). In this role, AQPs can modulate many physiological and/or pathological processes (Maurel, 2007; Ji and Dong, 2015a, 2015b). Plant AQPs fall into five major phylogenic families, and in most plant species, the PM intrinsic protein (PIP) family comprises 13 members assigned to two highly conserved subfamilies, PIP1 (PIP1;1–PIP1;5) and PIP2 (PIP2;1–PIP2;8; Abascal et al., 2014). They are believed to mediate the transport of different substrates across plant PMs in an overlapping or redundant manner for substrate selectivity (Maurel, 2007; Péret et al., 2012; Prado et al., 2013). To date, five AtPIP2 isoforms (AtPIP2;1, AtPIP2;2, AtPIP2;4, AtPIP2;5, and AtPIP2;7) have been assumed to mediate H2O2 transport in engineered yeast cells (Bienert and Chaumont, 2014). Under de novo expression, these PIPs are able to increase H2O2 sensitivity and decrease the viability of yeast (Dynowski et al., 2008; Hooijmaijers et al., 2012). However, only AtPIP2;1 has been elucidated to increase H2O2 uptake by yeast cells (Dynowski et al., 2008; Bienert and Chaumont, 2014). In fact, as yet there is no study to show a definite role of any PIP isoform in H2O2 transport across plant PMs.
As PMs directly face the environment, PIPs are implicated in cellular responses to extracellular signals (Gomes et al., 2009; Ji and Dong, 2015a, 2015b). For example, the harpin Hpa1 from rice (Oryza sativa) bacterial blight pathogen (Zhu et al., 2000) recognizes a rice PIP1 isoform to regulate virulence (Ji and Dong, 2015a). In Arabidopsis (Arabidopsis thaliana), externally applied or de novo-expressed Hpa1 is located at the PMs (Li et al., 2015) and acts as a PAMP to induce apoplastic H2O2 production through the NOX activity (Sang et al., 2012). Then, a large proportion of apoplastic H2O2 moves into the cytoplasm to enhance plant resistance to the virulent strain DC3000 of Pseudomonas syringae pv tomato (DC3000). This pathogen causes plant bacterial speck by secreting virulence effectors, which repress plant innate immunity (Oh and Collmer, 2005; Zhang et al., 2007; Guo et al., 2009), after translocation with the aid of four harpins, including the PAMP HrpZ1 (Lee et al., 2001; Kvitko et al., 2007). While an effector executes its virulence role by inhibiting H2O2 production and the MAPK cascade (Zhang et al., 2007), this PTI inhibitory effect may be impaired by a PAMP. For example, when HrpZ1 is directed to the apoplast in transgenic plants (Pavli et al., 2011) or infiltrated into the apoplastic spaces of normal plants (Kvitko et al., 2007), it recognizes a PM lipid sensor (Lee et al., 2001; Haapalainen et al., 2011) and induces immune responses, including H2O2 production (Dayakar et al., 2003). Under different conditions, harpin-induced immunity is attributable to SAR (Strobel et al., 1996; Dong et al., 1999 Peng et al., 2004) or PTI (Lee et al., 2001; Fu et al., 2014) activated following the cytoplasmic import of apoplastic H2O2 (Sang et al., 2012).
Therefore, the translocation of apoplastic H2O2 may be an integral component of the immunity systems of plants. Based on the potential role of PIP isoforms in H2O2 transport (Bienert and Chaumont, 2014; Aguayo et al., 2015), the translocation of apoplastic H2O2 is likely to recruit particular PIPs, which may function to connect the induction of H2O2 with the activation of the immunity pathways. In this study, we elucidate that AtPIP1;4 is a significant facilitator of H2O2 transport across PMs. We present evidence that this role of AtPIP1;4 mechanistically links the induction of apoplastic H2O2 to the activation of the SAR and PTI pathways in response to DC3000 and two typical PAMPs (flagellin and chitin), respectively.
RESULTS
AtPIP1;4 Affects Plant Immunity and Cytoplasmic H2O2 Accumulation
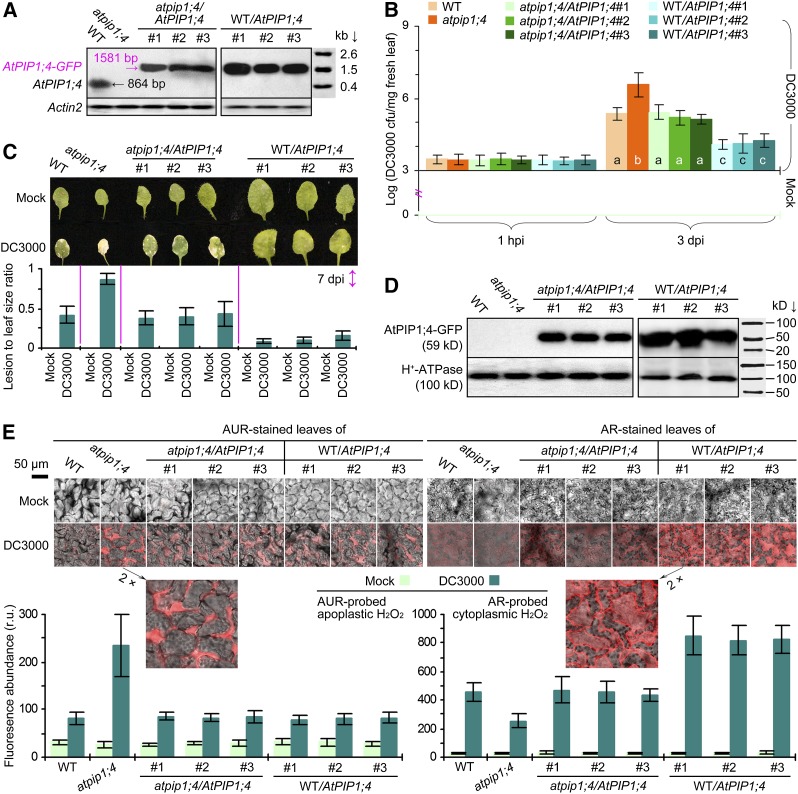
In order to identify the AtPIP isoforms that might affect the infection of Arabidopsis by DC3000, we analyzed the expression of 13 AtPIP genes in plants inoculated with the bacterial suspension containing 10 mm MgCl2 or mock inoculated with 10 mm MgCl2. At 24 h post inoculation (hpi), the AtPIP1;2 expression level changed little, the expression levels of AtPIP1;1, AtPIP1;4, AtPIP2;1, AtPIP2;3, AtPIP2;4, and AtPIP2;5 increased, and those of six additional AtPIPs decreased in plants inoculated with DC3000 in contrast to the mock agent (Supplemental Fig. S1). In comparison, AtPIP1;4, AtPIP2;3, and AtPIP2;4 were highly induced by DC3000 and exhibited 2.3-, 3.5-, and 3.6-fold increased expression levels in the inoculated plants. Thus, we deduced that these AtPIPs might be closely related to Arabidopsis immunity against the pathogen. Here, we tested this hypothesis by focusing on the immune role of AtPIP1;4, since the Arabidopsis atpip1;4 mutant (The Arabidopsis Information Resource; www.arabidopsis.org) has been well characterized (Supplemental Fig. S2).
We assessed immunity in AtPIP1;4 functional plants and the loss-of-function atpip1;4 mutant. This mutant was created previously by transfer DNA insertion at site 1,434 in the AtPIP1;4 coding region (Supplemental Fig. S2A), carrying a transfer DNA-indexed AtPIP1;4 sequence (Supplemental Fig. S2B) that was unable to express (Fig. 1A). Compared with the wild-type plant, atpip1;4 was more susceptible to DC3000 infection (Fig. 1, B and C). DC3000 was present in similar quantities in all plants 1 hpi, indicating uniform inoculation, and multiplied to a higher population in the mutant than in the wild type within 3 d post inoculation (dpi), in contrast to mock inoculation (Fig. 1B). By 7 dpi, chlorosis and/or necrosis symptoms became evident on the leaves of DC3000-inoculated plants compared with the apparently healthy leaves of mock-inoculated plants (Fig. 1C, photograph). Compared with the wild-type plant, the atpip1;4 mutant exhibited more severe necrosis and had a higher lesion area-to-leaf size ratio (Fig. 1C, bar graph). We complemented atpip1;4 with the wild-type AtPIP1;4 gene fused to the coding sequence of GFP and generated transgenic atpip1;4/AtPIP1;4 lines (Li et al., 2015). Three lines were characterized to resemble the wild type in AtPIP1;4 expression (Fig. 1A) and in response to DC3000 infection (Fig. 1, B and C). These atpip1;4/AtPIP1;4 plants exclusively produced the AtPIP1;4-GFP fusion protein (Fig. 1D). We also transformed the wild-type plant with the AtPIP1;4-GFP fusion gene and created AtPIP1;4-overexpressing WT/AtPIP1;4 lines (Li et al., 2015). Three WT/AtPIP1;4 lines were characterized based on the overexpression of AtPIP1;4 (Fig. 1A) and the production of the AtPIP1;4-GFP fusion protein (Fig. 1D). The WT/AtPIP1;4 lines acquired a high level of immunity, evidenced by the marked reduction of the bacterial population in the leaves (Fig. 1B) and the substantial alleviation of leaf necrosis severity (Fig. 1C). These genetic data suggest that AtPIP1;4 is a necessary regulator of Arabidopsis immunity against the bacterial pathogen. In addition, AtPIP1;4 overexpression caused a promoting effect on plant growth (Li et al., 2015; i.e. larger leaves; Fig. 1C), complying with the antagonism between growth (e.g. leaf size) and immunity (such as SAR) reported previously (Wang et al., 2007). Since AtPIP1;4 affects the growth of every leaf on a single plant (Li et al., 2015), it is improper to test the immune responses of the different genotypes by using leaves with equivalent size; instead, we used leaves with different sizes but the same position on plants.
Figure 1.
AtPIP1;4 affects plant immunity and cytoplasmic H2O2 accumulation. A, Northern-blot hybridization with probes specific for AtPIP1;4 and for the reference gene ACTIN2. B, Logarithmic colony formation units (cfu) of bacteria recovered from plant leaves. Six replicates were used; error bars indicate se. Different letters indicate significant differences in a multiple comparison (P < 0.01). C, Leaves at 7 dpi and necrosis severity (mean ± se; n = 6). D, Western blotting of the leaf PM fraction hybridized with antibodies against GFP and the PM marker protein H+-ATPase. E, Imaging of AUR- or AR-stained leaves and H2O2 content in leaf cells (mean ± se; n = 3). r.u., Relative units; WT, wild type.
To correlate immunity with H2O2 production, we employed H2O2-specific probes (i.e. Amplex Red [AR] and Amplex Ultra Red [AUR]) to detect H2O2 signals in the leaves of inoculated and mock-inoculated plants; both probes can be oxidized in reaction with H2O2 to produce strong crimson fluorescence (Ashtamker et al., 2007). While AR is able to penetrate PMs and thus probe cytoplasmic H2O2, AUR is impermeable to PMs and only detects H2O2 present in the apoplast (Ashtamker et al., 2007; Rhee et al., 2010; Deng et al., 2011; Sang et al., 2012). By laser confocal microscopy performed on leaves 1 hpi, we found that AUR and AR probing well visualized the H2O2 present in the apoplast and cytoplasm, respectively (Fig. 1E, photographs). We employed the scanning tool in the microscope to quantify the crimson fluorescence density and used this parameter to estimate the relative levels of AUR-probed apoplastic H2O2 or AR-probed cytoplasmic H2O2 (Fig. 1E, bar graphs). Relative levels of apoplastic H2O2 were highly increased in all plants following inoculation and reached the greatest value in atpip1;4 compared with basal levels in mock-inoculated plants. Meanwhile, relative levels of cytoplasmic H2O2 varied greatly in different plants. The cytoplasmic H2O2 quantities were highest (820‒850 relative units) in the WT/AtPIP1;4 lines, moderate (436‒462) in wild-type and atpip1;4/AtPIP1;4 plants, and lowest (253) in the atpip1;4 mutant. Thus, AtPIP1;4 overexpression caused an approximately 46% increase while the loss-of-function mutation caused an approximately 44% decrease in cytoplasmic H2O2 quantities. Clearly, AtPIP1;4 is unrelated to the DC3000-induced production of apoplastic H2O2 but is responsible for H2O2 accumulation in the cytoplasm. On the basis of our previous demonstration that apoplastic H2O2 mandatorily underwent cytoplasmic import (Sang et al., 2012), we surmised that reduced cytoplasmic H2O2 levels in the atpip1;4 mutant might be caused by impaired translocation of apoplastic H2O2 and that AtPIP1;4 might play a role in H2O2 transport across plant PMs.
De Novo-Expressed AtPIP1;4 Mediates H2O2 Translocation in Yeast
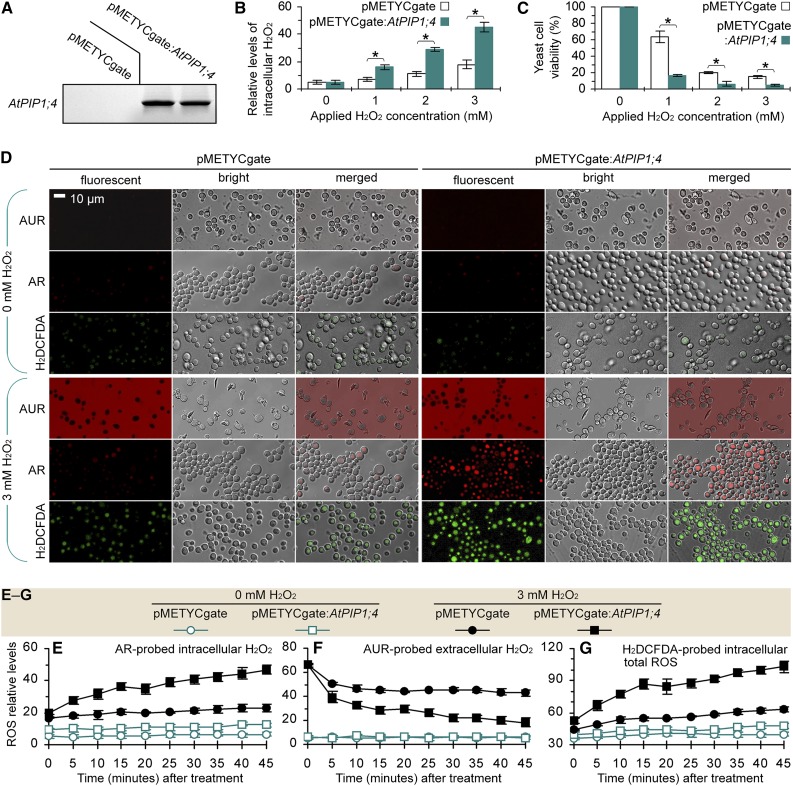
To infer the role of AtPIP1;4 in H2O2 transport, we analyzed the toxicity and translocation of externally applied H2O2 in yeast (Saccharomyces cerevisiae) cells transformed with the recombinant vector pMETYCgate:AtPIP1;4 for de novo expression of AtPIP1;4 or with the empty vector in a transgenic control (Fig. 2A). We used a luminometer to quantify AR-probed H2O2 inside yeast cells 45 min after treatment with a range of H2O2 concentrations. As shown in Figure 2B, H2O2 treatment significantly (P < 0.01) increased the intracellular H2O2 levels, and this effect was further enhanced by AtPIP1;4 expression compared with the transgenic control. Increased intracellular H2O2 caused toxicity, which became evident 3 d after H2O2 application (Supplemental Fig. S3). Toxicity was shown to significantly (P < 0.01) reduce yeast viability (Fig. 2C), and the viability was further reduced by AtPIP1;4 expression (Fig. 2C; Supplemental Fig. S3). The increase of intracellular H2O2 and the toxicity were H2O2 dosage dependent, with 3 mm H2O2 being highly effective (Fig. 2, B and C; Supplemental Fig. S3). Confocal microscopy performed 45 min after yeast treatment with 3 mm H2O2 revealed that AUR and AR probing well visualized the H2O2 signals outside and inside the yeast cells (Fig. 2D). In particular, AR detected ample H2O2 distributed exclusively in the cytoplasm of H2O2-treated AtPIP1;4-expressing cells but small amounts in the H2O2-null treatment control or the transgenic control cells (Fig. 2D).
Figure 2.
De novo AtPIP1;4 expression mediates H2O2 translocation in yeast. A, AtPIP1;4 probe hybridization to the blot of total RNA from yeast cells transformed with the empty vector and the AtPIP1;4-containing vector. B and C, Yeast H2O2 content and viability after 45 min of H2O2 treatment (mean ± se). Asterisks indicate significant differences in paired comparisons (n = 6; P < 0.01). D, AUR and AR probing of yeast cells 45 min after treatment with 0 or 3 mm H2O2. E to G, Chronological changes in the H2O2 content in yeast cells after treatment with 0 or 3 mm H2O2 (mean ± se; n = 3).
To elucidate H2O2 translocation, chronological variations of relative levels of H2O2 in yeast were monitored at 5-min intervals for 45 min after treatment with 0 or 3 mm H2O2 (Fig. 2, E and F). While the H2O2 quantities remained low in the H2O2-null treatment control, AtPIP1;4-dependent alterations of H2O2 levels were found in the H2O2-treated yeast cells. With AtPIP1;4 expression, the intracellular H2O2 quantities started to increase within 5 min and were increased by approximately 2-fold compared with the transgenic control at each time point after 10 min. By contrast, the extracellular H2O2 levels declined consistently, and the extent of the decrease was approximately 2-fold greater because of AtPIP1;4 expression (Fig. 2F). In addition, the green fluorescent dye 2,7-dichlorofluorescein diacetate (H2DCFDA) was used in cell imaging to visualize total ROS inside the living cells (Wang et al., 2009). Based on confocal microscopy (Fig. 2D) and fluorescence luminometry (Fig. 2G), H2DCFDA-probed ROS levels fluctuated (Fig. 2G) similarly to AR-probed H2O2 (Fig. 2F) during the chronological course, indicating that H2O2 translocation from the extracellular supply might be a major source of intracellular ROS.
To validate this hypothesis, we measured the enzymatic activities that control intracellular H2O2 generation. In living cells, H2O2 can be produced by numerous processes (Giorgio et al., 2007; Li et al., 2014) but is linked exclusively to superoxide dismutase (SOD) enzymes, which catalyze the dismutation reaction of 2O2∙− and 2H+ to produce H2O2 and oxygen (Gralla and Kosman, 1992). We determined that externally applied H2O2 did not induce SOD activities in yeast cells; by contrast, the SOD activities changed little in 50 min after yeast treatment with 0 or 3 mm H2O2 (Supplemental Fig. S4). This finding excluded the possibility of intracellular H2O2 production induced by applied H2O2. In fact, the priority for living cells under oxidative stress is to scavenge rather than to produce ROS or H2O2 (Martins and English, 2014). Therefore, the increase in intracellular H2O2 levels is attributed to translocation of the extracellular supply because of de novo expression of AtPIP1;4.
AtPIP1;4 Contributes to H2O2 Translocation in Plants
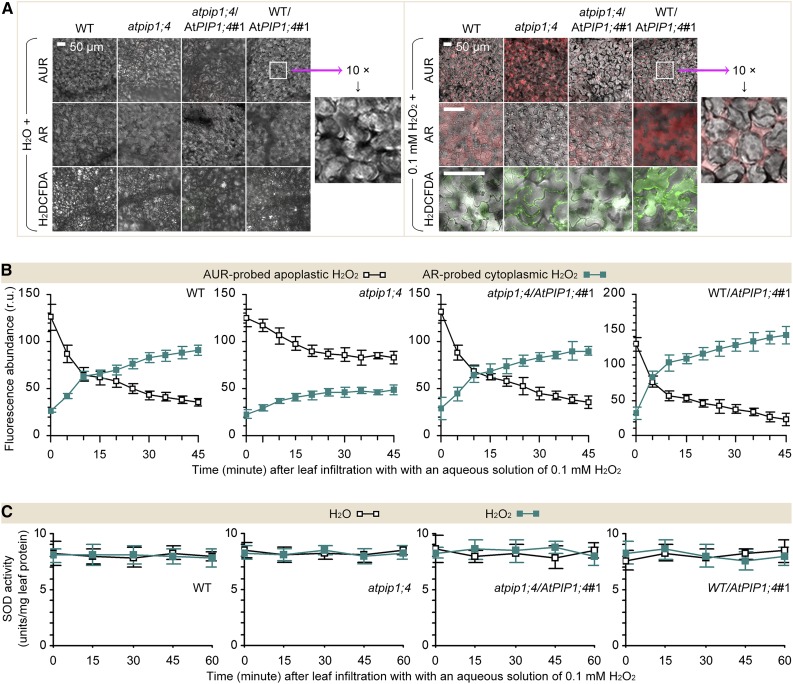
To test the possibility of H2O2 translocation and the effect of AtPIP1;4 in Arabidopsis, we compared AtPIP1;4-functional plants and the atpip1;4 mutant in terms of apoplastic and cytoplasmic H2O2 levels following leaf infiltration with water in the control or with H2O2 at 0.1 mm, a known effective dosage in plants (Sang et al., 2012). Within 45 min, there was considerable H2O2 or ROS in the leaves treated with H2O2 compared with low quantities in the water-treated leaves (Fig. 3A; Supplemental Fig. S5). Using confocal microscopy, we monitored apoplastic and cytoplasmic H2O2 at 5-min intervals for 45 min. After 10 min, a large proportion of the applied H2O2 moved into the cytoplasm in the AtPIP1;4-functional plants, while quantities of translocated H2O2 were highly decreased in the atpip1;4 mutant (Fig. 3B). Meanwhile, the possibility of induced H2O2 production in the cytoplasm was excluded, as the SOD activities were not induced under all circumstances (Fig. 3C). In this case, the relative levels of cytoplasmic H2O2 were increased by 2.4-fold in the wild type, 2.1-fold in atpip1;4/AtPIP1;4#1, and 3.6-fold in WT/AtPIP1;4#1 compared with initially measured values (Fig. 3B). By contrast, the apoplastic H2O2 quantities were decreased by 2.6-fold in the wild type and atpip1;4/AtPIP1;4#1 and 3.7-fold in WT/AtPIP1;4#1. In atpip1;4, H2O2 was largely retained in the apoplast, while the quantity of translocated H2O2 was 35% to 48% smaller than that of the wild type or atpip1;4/AtPIP1;4 (Fig. 3B). These data suggest that the increase in the cytoplasmic H2O2 content is a result of translocation of the externally applied H2O2 and that AtPIP1;4 is, indeed, a facilitator of H2O2 transport across plant PMs.
Figure 3.
AtPIP1;4 governs H2O2 translocation in plants. A, Imaging of leaves 45 min after infiltration with water or H2O2. B and C, Chronological changes of H2O2-probing fluorescence densities and SOD activities in leaf cells (mean ± se; n = 3). r.u., Relative units; WT, wild type.
AtPIP1;4 Links Pathogen-Induced Apoplastic H2O2 to the SAR Pathway
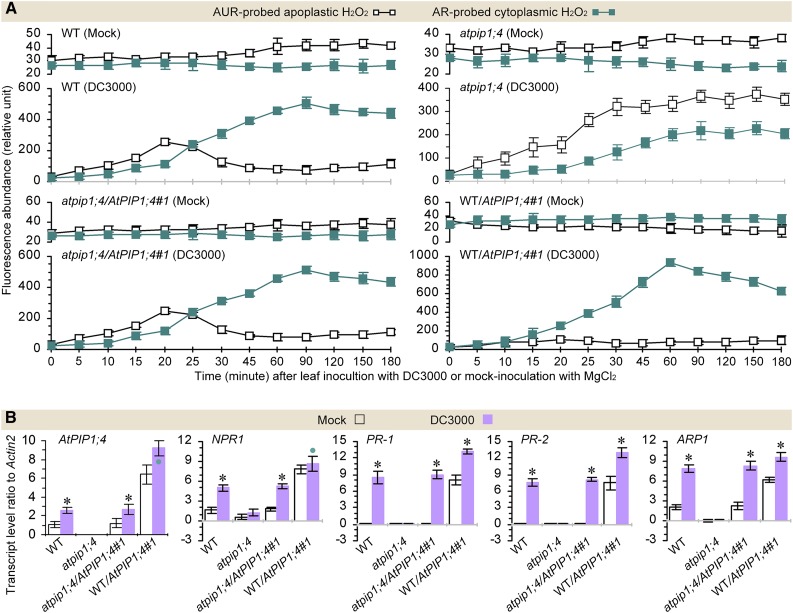
H2O2 production was induced in Arabidopsis leaves inoculated by leaf infiltration with DC3000 in contrast to the mock agent (Fig. 4A; Supplemental Fig. S6A). DC3000-induced H2O2 accumulated not only in the apoplast but also in the cytoplasm of the leaf cells based on the AUR and AR fluorescence densities quantified at various intervals for 3 hpi (Supplemental Fig. S6A). In this period, cytoplasmic H2O2 was not likely to be produced, since the SOD activities were not induced in all cases (Supplemental Fig. S6B), as compared with the steady-state levels in mock-inoculated plants (8.5‒11.3 ± 0.5‒2.8 units mg−1 fresh weight). Thus, H2O2 accumulation in the cytoplasm indeed resulted from translocation of the apoplastic H2O2 originally induced by DC3000 (Fig. 4A; Supplemental Fig. S6A). Under this condition, relative levels of apoplastic and cytoplasmic H2O2 altered in an AtPIP1;4-dependent manner (Fig. 4A). The H2O2 levels were highly elevated in the cytoplasm and synchronously reduced in the apoplast of the wild type, atpip1;4/AtPIP1;4#1, and WT/AtPIP1;4#1. Meanwhile, WT/AtPIP1;4#1 more vigorously supported H2O2 translocation. In WT/AtPIP1;4#1, the H2O2 content in the cytoplasm increased to a level higher than that measured in the apoplast; this occurred 15 min earlier (30 min was reduced to 15 min), and the translocated H2O2 amounts were approximately 1.5 times those of the wild type and atpip1;4/AtPIP1;4#1 from 20 min onward (Fig. 4A). In atpip1;4, the apoplastic and cytoplasmic H2O2 levels experienced a constant increase; H2O2 translocation also took place, but the extent was decreased by 37% to 45% compared with that in the wild type or atpip1;4/AtPIP1;4#1 (Fig. 4A). Evidently, AtPIP1;4 acts as an H2O2 transport facilitator to dominate the cytoplasmic import of H2O2 from the apoplastic origin induced by the pathogen.
Figure 4.
AtPIP1;4 links pathogen-induced apoplastic H2O2 to the SAR pathway. A, Chronological changes in the H2O2-probing fluorescence densities in DC3000-inoculated and mock-inoculated leaves (mean ± se; n = 3). B, SAR gene expression levels in leaves 45 min after inoculation or mock inoculation (n = 6; *, P < 0.01 and •, P < 0.05 in paired comparisons between DC3000-inoculated and mock-inoculated plants). WT, Wild type.
The role of AtPIP1;4 in H2O2 translocation was related to the SAR pathway, which involves the transcriptional regulation of NPR1 and PR genes. NPR1 is constitutively expressed at a steady-state level, allowing the production of a basal amount of NPR1 protein to maintain the immune threshold that prevents supersusceptibility once infection has occurred (Fu et al., 2012). In plants under infection, NPR1 expression is enhanced to produce sufficient NPR1 protein required for PR gene activation (Cao et al., 1997; Spoel et al., 2009). Since the auxin-repressed protein ARP1 regulates NPR1 and PR expression following induction by pathogens and PAMPs in Nicotiana benthamiana (Zhao et al., 2014), expression of the Arabidopsis ARP1 homolog (dormancy/auxin-associated protein mRNA; called ARP1 hereafter) also was tested as an SAR response. Based on real-time quantitative reverse transcription RT-qPCR analyses, the expression of ARP1, NPR1, PR-1, and PR-2 was induced by DC3000 concurrently with AtPIP1;4 in AtPIP1;4-functional plants, but not in the atpip1;4 mutant following inoculation, in contrast to mock inoculation (Fig. 4B). In order to determine AtPIP1;4 expression in the different plant genotypes, primers used in qRT-PCR were designed to amplify the 1,056 to 1,234 region of the 2,592-bp AtPIP1;4 coding sequence. Thus, DC3000-enhanced AtPIP1;4 expression was detected not only in wild-type and atpip1;4/AtPIP1;4 plants but also in the AtPIP1;4-overexpressing plant (Fig. 4B). However, compared with the wild type and atpip1;4/AtPIP1;4#1, gene expression was induced more vigorously in WT/AtPIP1;4#1. In particular, expression of the NPR1 and PR genes was enhanced by DC3000 infection in wild-type, atpip1;4/AtPIP1;4, and WT/AtPIP1;4 plants but not in the atpip1;4 mutant (Fig. 4B), suggesting the important role of AtPIP1;4 in SAR activation. This immune difference was coincident with that of H2O2 translocation. Therefore, activation of the SAR pathway relies on the role of AtPIP1;4 in the cytoplasmic import of apoplastic H2O2 induced by DC3000, and this mechanism is effective to repress the pathogenicity of the pathogen itself.
AtPIP1;4 Links PAMP-Induced Apoplastic H2O2 to the PTI Pathway
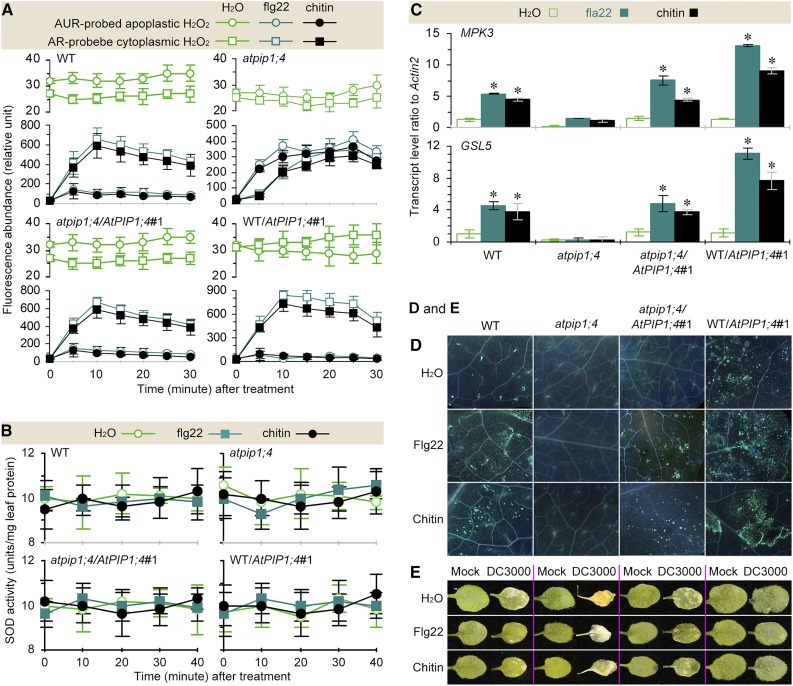
To study the effect of AtPIP1;4 on PTI, we treated atpip1;4 and AtPIP1;4-functional plants by leaf infiltration with water (in the control), or with 1 μm aqueous solution of flg22, the active module of flagellin containing the first 22 residues (Asai et al., 2002), or with an aqueous suspension of 0.1 mg mL−1 chitin based on known effective dosages (Zhao et al., 2014). We found that apoplastic H2O2 was induced in all plants within 5 min after treatment with flg22 or chitin compared with the control, and cytoplasmic H2O2 accumulated in an AtPIP1;4-dependent manner within 30 min (Fig. 5A). In this period, PAMPs did not affect the SOD activities, suggesting that intracellular H2O2 generation was not induced (Fig. 5B). Under this condition, cytoplasmic H2O2 existed at the minimal level in atpip1;4 but accumulated strongly in AtPIP1;4-functional plants and reached a maximum in WT/AtPIP1;4#1 at each time point during 30 min (Fig. 5A). In the chronological course, cytoplasmic H2O2 appeared to peak at 25 min in atpip1;4 and at 10 min in the other plants after treatment with flg22 or chitin. However, the quantity of translocated H2O2 was approximately 45% more in WT/AtPIP1;4#1 and approximately 42% less in atpip1;4 than in wild-type and atpip1;4/AtPIP1;4#1 plants. Clearly, a functional AtPIP1;4 is required for the cytoplasmic import of PAMP-induced apoplastic H2O2.
Figure 5.
AtPIP1;4 links PAMP-induced apoplastic H2O2 to the PTI pathway. A and B, Chronological changes in the H2O2-probing fluorescence densities and SOD activities in leaves infiltrated with water or an aqueous solution of PAMPs (mean ± se; n = 3). C, PTI gene expression levels in leaves 45 min after different treatments (mean ± se; n = 3; *, P < 0.01 in paired comparisons between treatments with water and flg22 or chitin). D, Callose visualization in leaves 45 min after treatment with the agents shown at left. E, Leaves at 7 dpi inoculated or mock inoculated and treated in advance with the agents shown at left. WT, Wild type.
AtPIP1;4 was further required for flg22 and chitin to activate MPK3 and GSL5, which were expressed to higher degrees in WT/AtPIP1;4#1 than in wild-type and atpip1;4/AtPIP1;4#1 plants but to a lesser extent in atpip1;4 (Fig. 5C). However, both flg22 and chitin were unable to induce MPK6 expression (Supplemental Table S1). Instead, both PAMPs were effective in inducing callose deposition in an AtPIP1;4-dependent manner (Fig. 5D). While atpip1;4 produced little callose, callose deposition was robust in the wild-type, atpip1;4/AtPIP1;4#1, and WT/AtPIP1;4#1, with higher densities in the latter plant following treatment with flg22 or chitin compared with the control. Coincidently, the bacterial population in the leaves was reduced (Supplemental Fig. S7) and leaf necrosis severities were alleviated (Fig. 5E) following flg22 or chitin treatment in the AtPIP1;4-functional plants but not in the atpip1;4 mutant. Thus, the activation of the PTI pathway relies on functional AtPIP1;4. These analyses, together with the H2O2 data, suggest that AtPIP1;4-mediated cytoplasmic transport of PAMP-induced apoplastic H2O2 is an integral component of the PTI pathway.
NOX and AtPIP1;4 Play Independent Roles in H2O2 Production and Translocation
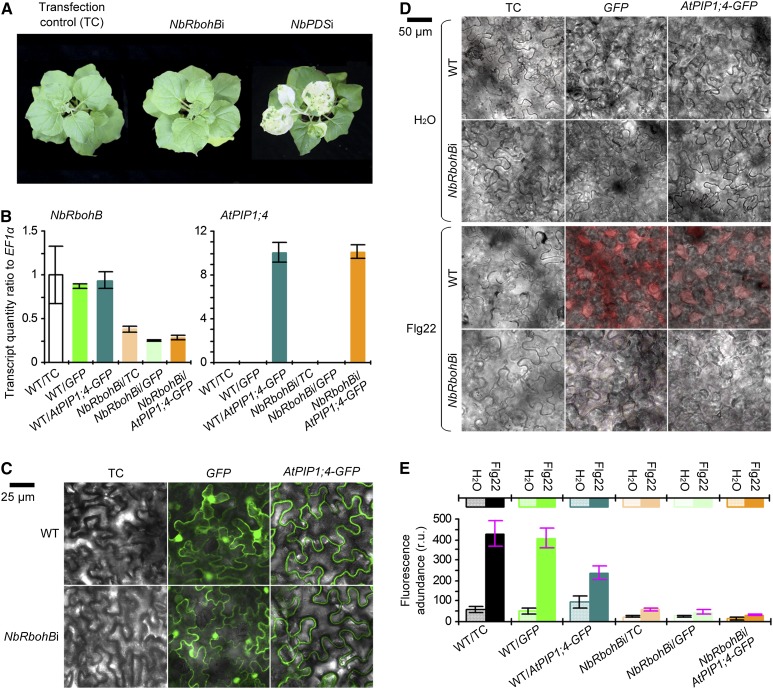
In DC3000-inoculated or PAMP-treated plants, the time to produce H2O2 and the levels of total H2O2 in the apoplast and cytoplasm at a given time point were equivalent in the AtPIP1;4-functional plants and the atpip1;4 mutant (Figs. 4A and 5A). Thus, AtPIP1;4 might only be responsible for the cytoplasmic import of apoplastic H2O2 without affecting H2O2 generation. The latter role is presumably attributable to the PM-integrated NOX. This notion was confirmed by genetic modulation of a NOX-encoding gene, namely RESPIRATORY BURST OXIDASE HOMOLOG B (RbohB), in N. benthamiana (Yoshioka et al., 2003). NbRbohB is a PM NOX with a prominent role in apoplastic H2O2 generation (Yoshioka et al., 2003; Zhang et al., 2009). We applied a virus-induced gene silencing (VIGS) protocol (Zhang et al., 2009) to NbRbohB while using the PHYTOENE DESATURASE (PDS) gene as a reference, since PDS silencing caused a mottled photobleaching phenotype (Travella et al., 2006). Here, the phenotype was observed, indicating the desired performance of the protocol (Fig. 6A). A high efficiency of NbRbohB silencing (NbRbohBi) was achieved; relative levels of NbRbohB expression were decreased by approximately 80% in the NbRbohBi background compared with the wild type (Fig. 6B). With NbRbohBi or the wild type, GFP and AtPIP1;4-GFP proteins were produced following gene transient expression (Fig. 6, B and C) and were used to evaluate the subsequent effect on flag22-induced apoplastic H2O2 generation. In fluorescence imaging, GFP was found in the PM, cytoplasm, and nucleus, while AtPIP1;4-GFP was localized only to the PM (Fig. 6C). Based on AUR probing, flg22 treatment effectively induced apoplastic H2O2 production in the wild-type and transfection control plants, but the extent of H2O2 induction was considerably lower in NbRbohBi (Fig. 6, D and E). Transient expression of AtPIP1;4-GFP markedly reduced the amounts of apoplastic H2O2 in the wild-type background and further reduced apoplastic H2O2 quantities in NbRbohBi cells, but GFP had no effect (Fig. 6E). Clearly, NOX, rather than AtPIP1;4, is responsible for the generation of apoplastic H2O2 in N. benthamiana, confirming the results from Arabidopsis.
Figure 6.
NbRbohB gene silencing impairs apoplastic H2O2 generation. A, Plants 14 d after gene silencing. B, qRT-PCR analyses (n = 3). C, Imaging of leaves 36 h after transformation with GFP or AtPIP1;4-GFP. D, Cell imaging of AUR-stained leaves 30 min after treatment with water or aqueous flg22 solution. E, Quantification of fluorescence densities in leaves from D (n = 3). r.u., Relative units; WT, wild type.
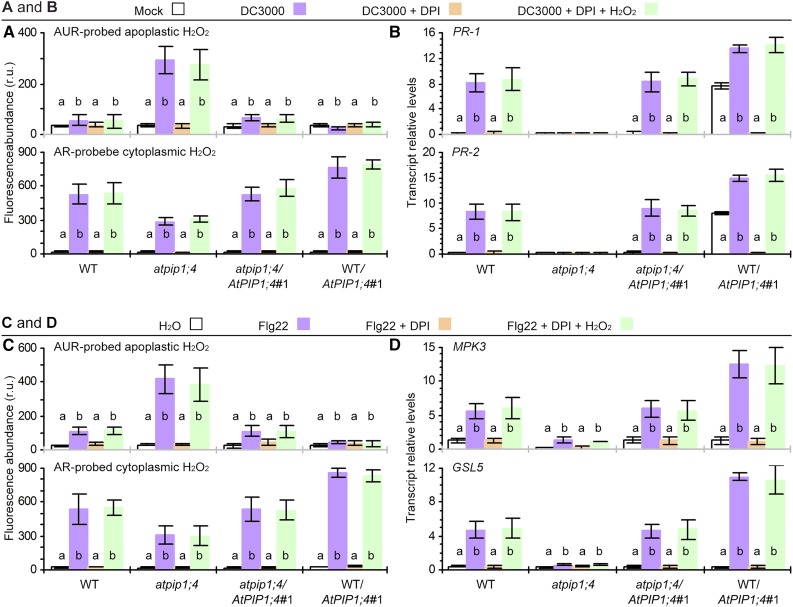
In Arabidopsis, NOX is largely responsible for apoplastic H2O2 production (Sagi and Fluhr, 2006) and has been associated with the activation of the SAR pathway (Mammarella et al., 2015). We found that the application of diphenyleneiodonium (DPI), a specific inhibitor that represses NOX activity (Wang et al., 2009) but does not affect plant infection at least by DC3000 (Sang et al., 2012), eliminated some of the induced apoplastic H2O2 in all plants inoculated with DC3000. In this case, the apoplastic H2O2 quantities in inoculated plants were equivalent to the basal levels found in mock-inoculated plants (Fig. 7A). The inhibitory effect of DPI was extended to the SAR responses, abrogating pathogen-induced expression of PR-1 and PR-2 (Fig. 7B). The inhibitory effect of DPI also was observed in flg22-treated plants. With DPI treatment, cytoplasmic H2O2 in flg22-treated plants no longer accumulated to detectably higher quantities than the basal level scored in the water treatment control (Fig. 7C). DPI further cancelled the flg22-induced expression of MPK3 and GSL5 (Fig. 7D). If plants were treated with DPI and H2O2, H2O2 translocation was resumed (Fig. 7, A and C), while both the SAR and PTI pathways were reactivated (Fig. 7, B and D) in AtPIP1;4-functional plants. Moreover, the pharmacological effects on H2O2 production were observed in all plants, including atpip1;4, but the mutant remained less active in supporting H2O2 translocation (Fig. 7, A and C). The mutant also was inactive in PR-1, PR-2, MPK3, and GSL5 expression (Fig. 7, B and D). Based on these data, we propose that the functions of the PM-localized NOX and AtPIP1;4 are independent of each other (i.e. to generate apoplastic H2O2 and govern its translocation into the cytoplasm, respectively).
Figure 7.
NOX and AtPIP1;4 play independent roles in H2O2 production and translocation. A and B, H2O2 content and SAR gene expression in leaves 45 min after treatment with the indicated agents. C and D, H2O2 content and PTI gene expression in leaves 45 min after treatment with the indicated agents. Data are means ± se (n = 6); different letters on bar graphs indicate significant differences (P < 0.01) in multiple comparisons for every plant genotype. r.u., Relative units; WT, wild type.
DISCUSSION
We have demonstrated, to our knowledge, the first case that an Arabidopsis AQP, AtPIP1;4, can regulate the SAR and PTI pathways to confer plant immunity against the bacterial pathogen P. syringae pv tomato. This newly appreciated function of AtPIP1;4 is an extension of the primary roles in substrate transport assigned to different AQPs initially in mammals (Preston and Agre, 1991; Preston et al., 1992) and subsequently in plants (Maurel et al., 1993). On the one hand, AtPIP1;4-dependent SAR responses that are induced by the bacterial pathogen effectively repress the pathogenicity of the pathogen itself. In this case, plant immunity repressors of the pathogen (Oh and Collmer, 2005; Zhang et al., 2007; Guo et al., 2009) may be repressed, or their immunity-repressive functions may be counteracted by the role of AtPIP1;4 in H2O2 translocation linked to the immunity pathway. At present, however, we do not have any evidence to support this postulation. On the other hand, AtPIP1;4 is an integral component of PTI in response to typical PAMPs, which represent conserved microbial cell surface composition, such as flagellin (Zipfel et al., 2004) and chitin (Kaku et al., 2006; Shimizu et al., 2010). Despite different biochemical natures, both PAMPs commonly require AtPIP1;4 to induce PTI responses, except for the absence of induced MPK6 expression. This result is in line with previous findings that the MAPK cascade diverges at MPK3 and MPK6 (Asai et al., 2002; Bigeard et al., 2015) to regulate distinct substrates in response to different PAMPs (Galletti et al., 2011; Pitzschke, 2015) and that induced expression of MPK3 represents a circuit of the MAPK cascade in response to H2O2 (Gudesblat et al., 2007).
The H2O2 signal has multifaceted functions, regulating many processes in living organisms, and is particularly associated with the PTI and SAR pathways (Torres, 2010) following apoplastic generation (Sagi and Fluhr, 2006; Mammarella et al., 2015) and subcellular trafficking in plants (Ashtamker et al., 2007; Wang et al., 2009). In fact, the conventionally undersized yet life-essential H2O2 molecule is not as unhindered as was thought previously to penetrate membranes. Instead, H2O2 transport across biomembranes is subject to refined control by a specific gateway assumed to involve AQPs (Bienert and Chaumont, 2014). Since induced production of H2O2 is apoplastic, it needs to overcome the PM seclusion to participate in intracellular immune responses (Sang et al., 2012; Bienert and Chaumont, 2014). PM-traversing cytoplasmic import has been proposed as a sensible solution to close the cytological distance for H2O2 generation and functional performance (Bienert et al., 2006; Sang et al., 2012). However, this hypothesis has not been validated until now.
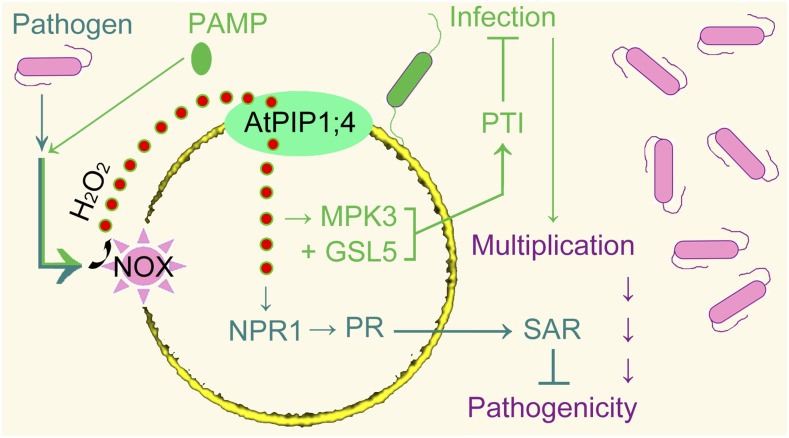
Our data demonstrate the pivotal role of AtPIP1;4 in connecting the induction of apoplastic H2O2 with the activation of immunity pathways under different conditions. In plants infected by the bacterial pathogen, apoplastic H2O2 is generated through the PM-located NOX enzyme and is rapidly translocated into the cytoplasm. There, the H2O2 signal contributes to the activation of the SAR pathway, which represses the bacterial pathogenicity (Fig. 8, watery green pathway). In response to a PAMP, the cytoplasmic import of apoplastic H2O2 takes place to activate the PTI pathway, which confers resistance to the pathogen (Fig. 8, green pathway). Both immunity pathways rely on functional AtPIP1;4; indeed, it is an H2O2 translocator and enables extracellular H2O2 to traverse PMs. As a result, cytoplasmic H2O2 levels are increased in the absence of the induced production of intracellular H2O2. The role of AtPIP1;4 in H2O2 translocation establishes a mechanistic connection between the production of apoplastic H2O2 and its cross talk with the SAR and PTI pathways.
Figure 8.
Model of AtPIP1;4-mediated linkage of apoplastic H2O2 to SAR (watery green) and PTI (green) pathways. Upon induction by the bacterial pathogen or PAMPs, apoplastic H2O2 is generated through the NOX activity and moves rapidly into cytoplasm under the regulation by AtPIP1;4. Translocated H2O2 cooperates with SAR or PTI to repress the pathogenicity.
However, AtPIP1;4 may not be an exclusive facilitator of H2O2 transport across PMs, because H2O2 translocation decreases but is not eliminated in the atpip1;4 mutant. In addition to AtPIP1;4, five AtPIP2 isoforms (AtPIP2;1, AtPIP2;2, AtPIP2;4, AtPIP2;5, and AtPIP2;7) also were shown to increase H2O2 sensitivity and decrease the viability of yeast under the de novo expression condition (Dynowski et al., 2008; Hooijmaijers et al., 2012). Whether these isoforms mediate H2O2 transport in plants is unknown. The expression levels of 13 AtPIPs in H2O2-treated Arabidopsis plants did not necessarily indicate a role of the gene expression in yeast sensitivity to H2O2 (Hooijmaijers et al., 2012). It was proposed that H2O2 treatment of Arabidopsis reduced the expression of AtPIP2s in roots but not in leaves, while the expression of AtPIP1s was not affected. However, H2O2 treatment also highly induced AtPIP1;3, AtPIP1;4, and AtPIP2;8 expression in leaves and also induced AtPIP2;1, AtPIP2;5, AtPIP2;6, and AtPIP2;8 expression in roots. It was further proposed that de novo expression of AtPIP2;2, AtPIP2;4, AtPIP2;5, or AtPIP2;7, but not AtPIP1s, impaired yeast growth and survival. However, de novo expression of AtPIP1;4 also was able to reduce yeast viability in response to 1 mm H2O2 (Hooijmaijers et al., 2012). To date, the most convincing candidate in the PIP2 channel for H2O2 transport is AtPIP2;1, the only PIP2 ortholog that has been demonstrated to increase H2O2 uptake by yeast cells (Dynowski et al., 2008; Bienert and Chaumont, 2014). Therefore, a definite conclusion about whether those AtPIP2s truly play a role in H2O2 transport must await direct evidence like translocation data.
Once the factual roles that the five AtPIP2s (Bienert and Chaumont, 2014) play in H2O2 transport are verified, it will be worthwhile to study how these redundant H2O2 transporters coordinate their functions. An interesting subject could be to characterize whether those AtPIP2s, and AtPIP1;4 as well, form a consortium or work alone in plants under certain circumstances, such as in response to H2O2 stress (Hooijmaijers et al., 2012), PAMP stimulation, or pathogen infection (Ji and Dong, 2015a). These circumstances represent apocytoplastic signal transduction not only immunity but also to developmental regulation (Gomes et al., 2009; Ji and Dong, 2015b). Our demonstration of the function of AtPIP1;4 in H2O2 translocation provides a paradigm for studies in the future to characterize apocytoplastic communication, with a broad significance for both immunity and development (Maurel, 2007; Gomes et al., 2009; Ji and Dong, 2015a, 2015b). At present, although the exact mechanism that underpins H2O2 signaling cross talk with immunity pathways is an open question, our findings coherently bridge the prolonged cytological gap between H2O2 generation and its function in plants following pathogen infection or pattern recognition (Levine et al., 1994; Mammarella et al., 2015). In addition, it was suggested previously that plant cell wall peroxidases were the initial origin of apoplastic H2O2 production, while NOX might amplify this H2O2 signal (Bindschedler et al., 2006). In fact, amplification of a signaling decibel is nothing but the increased production and activities of signaling compounds in a regulatory cascade. Our data suggest that the independent roles of NOX and AtPIP1;4 in H2O2 generation and translocation are sufficient to activate the PTI and SAR pathways.
The role of AtPIP1;4 in cytoplasmic import of the apoplastic H2O2 signal may have more biological importance than the substrate transport and immune role, since the signal frequently experiences extensive cross talk with phytohormones, such as abscisic acid (Grondin et al., 2015) and salicylic acid (Tada et al., 2008; Torres, 2010). Moreover, as PMs directly face the environment, PIPs are also implicated in cellular responses to a variety of extracellular signals in addition to substrate transport (Gomes et al., 2009; Ji and Dong, 2015a, 2015b; Li et al., 2015). Thus, a particular PIP must be subject to multiple mechanisms for functional regulation, at the transcriptional and posttranscriptional levels, for example (Maurel, 2007; Gomes et al., 2009). The latter is basically related to the topological structure of AQPs. AQPs possess six α-helical TM (TM1–TM6) domains that are tilted along the plane of the PM and are linked one to the other by five connecting loops (LA–LE). LB, LD, and both the N-terminal and C-terminal regions are located inside the cell and potentially bind to cytosolic substrates. Inversely, LA, LC, and LE face the apoplasm and have the opportunity to contact the apoplastic substrates. Presumably, LA, LC, and LE enable PIPs to sense biotic signals and, therefore, extend their functional scopes beyond substrate transport (Bienert et al., 2006, 2014, 2005; Ji and Dong, 2015a, 2015b). This structural feature and functional flexibility of AQPs provide the molecular basis for PIP sensing of H2O2 and hormone signals that may enable cross talk to regulate immunity or other processes, such as stomatal closure and lateral root emergence (Péret et al., 2012; Grondin et al., 2015). Studies in the future that characterize whether the topological distribution of a PIP on the PM changes upon interacting with a particular environmental signal will be critical to elucidate the mechanisms that underpin the functional overlapping of the protein.
MATERIALS AND METHODS
Plant Growth Conditions
Nicotiana benthamiana plants were grown in a greenhouse. All Arabidopsis (Arabidopsis thaliana) genotypes were produced and their seeds were maintained in the laboratory (Li et al., 2015). The seeds were germinated in flat plastic trays filled with a substrate containing peat, sand, and vermiculite (1:1:1, v/v). Three days later, germinal seedlings were moved to 60-mL pots (three plants per pot) filled with the same substrate and grown in plant growth chambers at 24°C ± 1°C under 12 h of light at 250 ± 50 μmol quanta m−2 s−1. The plants were grown for 35 d before use in all experiments.
Gene Expression Analysis
Gene expression analysis followed standard methods. Total RNA was isolated from 3-d-old yeast (Saccharomyces cerevisiae) cultures, and the top third to fifth unfolded leaves of Arabidopsis, or transfected leaves of N. benthamiana, were analyzed by northern-blot hybridization with specific probes. Uniformly loaded 20 μg of total RNA was resolved on a 0.8% (w/v) agarose gel and transferred to a nylon membrane, followed by hybridization to a digoxigenin-labeled AtPIP1;4 full-length probe prepared using the DIG Nucleic Acid Detection Kit (Roche Diagnostics; Liu et al., 2011). Similar hybridization was performed with the ACTIN2 probe to verify uniform loading of the RNA samples on the gel. To verify the specificity of probe hybridization with respect to the conservation of the AtPIP sequences, the probe was hybridized to the blot of sequenced reverse transcription-PCR products of all 13 AtPIPs. The hybridization signal with AtPIP1;4 was detected at a high density, but weak hybridization occurred with the other 12 genes, especially AtPIP1;2, AtPIP1;3, AtPIP1;5, and AtPIP2;8 (Supplemental Fig. S8). Thus, AtPIP1;4 expression levels were quantified by RT-qPCR using specific primers (Supplemental Table S2), which were designed to amplify a less conserved region (1,056‒1,234) of the AtPIP1;4 sequence. After the PCR product was confirmed as an exclusive AtPIP1;4 transcript, the primers were used in RT-qPCR to assess relative levels of AtPIP1;4 expression in different plants. In addition to AtPIP1;4, immunity-related genes also were analyzed by qRT-PCR. All RT-qPCR analyses were carried out using the SuperScript II RNase Hˉ Reverse Transcriptase Kit (Invitrogen) according to the manufacturer’s guide book, and the constitutively expressed ACTIN2 or EF1α gene was used as a reference (Chen et al., 2008). All reactions were performed in triplicate with null-template (complementary DNA-absent) controls. The expression level of a gene was normalized to that of the null-template control. The relative level of gene expression was quantified as the transcript ratio to ACTIN2.
Plant Inoculation and Immunity Evaluation
Inoculation was performed on 35-d-old plants in the absence of any other treatments or 37-d-old plants that had been treated 2 d previously with other agents. DC3000 inoculum was made as a suspension (optical density at 600 nm = 0.05) containing 10 mm MgCl2 (Kvitko et al., 2007). This inoculum and 10 mm MgCl2 used as a mock agent were amended with the surfactant Silwet77 (0.02%, v/v) and applied by dipping over the plant tops. The plants were immediately placed into a bell jar and subjected to vacuum and air exchange three times within 30 min by the aid of a vacuum pump. Using this method, the inoculum or mock agent was infiltrated into the leaf intercellular spaces based on microscope observations of leaves in the primary experiments. Inoculation was thought to be accomplished after the pump-aided leaf infiltration, and inoculated plants were grown under the conditions stated above to induce disease. The bacterial number in the leaves was determined at 1 hpi to verify uniform inoculation. The bacterial population in the leaves was further determined at 3 dpi to assess the degree of infection. Leaf chlorosis and/or necrosis symptoms were observed at 7 dpi; leaves were photographed, and symptom severities were scored as the ratio of lesion area to leaf size. Variations in the bacterial population and symptom severities in different plants were used as criteria to assess the immunity levels affected by AtPIP1;4.
Plant Treatment
Aqueous solutions of H2O2 in a range of concentrations, an aqueous solution of flg22 (Absin Biosci) at 1 µm, and an aqueous suspension of chitin (Sigma-Aldrich) at 0.1 mg mL−1 were mixed with 0.02% Silwet77. Each solution was employed alone or in combination with 5 μm DPI (Sigma-Aldrich) and applied by spraying over the tops of 35-d-old plants. The plants were subjected to pump-aided leaf infiltration similar to the inoculated plants described above. The leaves were treated similarly with pure water in the control. The top third fully unfolded leaves were used at designated intervals for analyses of immune responses, including ROS or H2O2 accumulation.
ROS Detection
ROS detection was performed on liquid yeast cultures or the top third fully unfolded leaves of plants by staining with the ROS-probing dye AR, AUR, or H2DCFDA (Sigma-Aldrich) used at a final concentration of 10 μm (Wang et al., 2009; Sang et al., 2012). To ensure sufficient diffusion into living cells, the three dyes were applied 30 min earlier to the yeast suspension that was to be treated with H2O2 and 30 min earlier to the leaves of plants that were to be inoculated, mock inoculated, or treated with different agents.
Engineered yeast cells were cultured in liquid Yeast Extract-Peptone-Dextrose medium (Macierzyńska et al., 2007) for 16 to 18 h in a shaker at 160 rpm and 30°C, centrifuged at 1,000 rpm, and washed twice with phosphate buffer solution (PBS; 0.2 mm, pH 7.4). Precipitated cells were suspended with PBS containing an ROS-probing dye, incubated for 30 min, and then supplied with H2O2. Yeast cells were observed at 5-min intervals for 45 min to monitor AUR-probed extracellular H2O2 and AR-probed intracellular H2O2 or H2DCFDA-probed intracellular ROS signals.
To detect H2O2 or ROS in plants, a solution of AR, AUR, or H2DCFDA was infiltrated into the intercellular spaces of the top third of the leaves of plants; infiltration was performed near the central sites of leaf moieties using a needleless syringe. Infiltrated leaves were excised and used to monitor apoplastic H2O2 and cytoplasmic H2O2 or ROS from 45 min to 3 h depending on the study purposes.
Previously described protocols were used in the ROS-probing analyses (Wang et al., 2009; Sang et al., 2012). Stained yeast cells or plant leaves were observed with a Zeiss LSM700 laser scanning confocal microscope. The fluorescence emission of oxidized AR, AUR, or H2DCFDA in the yeast and plant cells was observed between 585 and 610 nm using 543-nm argon laser excitation. The AUR, AR, or H2DCFDA florescence densities in leaves were quantified with the equipped scanner to estimate relative levels of extracellular and intracellular H2O2 or intracellular ROS. Quantification of the AUR probing of fluorescence signals was restricted to apoplastic spaces, while that of AR or H2DCFDA was directed to cytoplasmic areas. Fluorescence densities in yeast cells were quantified with a SpectramaMax M5 96 microplate luminometer (Molecular Devices) to estimate relative levels of intracellular and extracellular H2O2 or ROS. The relative levels of ROS or H2O2 were scored in contrast to 1,000 yeast cells and single leaf cells or intercellular spaces.
Yeast Growth and Viability Scoring
The yeast strain NMY51 was incubated in liquid Yeast Extract-Peptone-Dextrose medium (Macierzyńska et al., 2007) containing 0 (control), 1, 2, or 3 mm H2O2 in a shaker at 160 rpm and 30°C. One hour later, the yeast culture suspensions were centrifuged for 10 min at 700 rpm and 4°C. The precipitate was washed three times with 0.2 mm PBS (pH 7.4) by centrifugation and resuspension. The last precipitate was suspended in the buffer, and the cell number in 5 µL of the suspension (placed on a slide) was counted using an optical microscope; on this basis, the total number of yeast cells in the suspension was calculated. Alternatively, yeast cells from the last precipitate were incubated on agar YPD medium after the cell number was determined. Three days later, colonies were collected and diluted with water to count the number of yeast cells by microscopy. The viability of the yeast cells under each H2O2 dose was scored in contrast to the control.
SOD Activity Measurements
The Total Superoxide Dismutase Assay Kit with WST-8 (Beyotime Biotech) was used to determine total intracellular SOD activities in yeast cultures and plant leaves according to the manufacturer’s instructions. The yeast culture suspension was centrifuged at 1,000 rpm for 5 min at 4°C, and the precipitate was washed with PBS twice and used to prepare the yeast extract. Plant leaf moieties were dissected by cutting the midribs and were infiltrated with pure water with the aid of a vacuum pump. Then, infiltrated leaf moieties were placed cut side down into a centrifuge tube, separated by nylon mesh from the glass bead padding on the bottom of the centrifuge tube, and centrifuged at 1,000 rpm to remove intracellular fluids (Sang et al., 2012). Centrifuged leaf samples were used in SOD extraction (Macierzyńska et al., 2007). All operations were carried out at –4°C, and the yeast or plant material was amended with phenylmethylsulfonyl fluoride at 100 μg mL−1 to inhibit enzyme decomposition. Enzymatic activities were quantified relative to the amount of total proteins from yeast cells or plant leaves. Protein concentrations were quantified with the BCA Protein Assay Regent Kit (Pierce).
Callose Visualization
Callose deposition in leaves was detected as described previously (Lü et al., 2011). The top third fully unfolded leaves were infiltrated with 5 mL of a solution made of phenol, glycerol, lactic acid, water, and 95% ethanol (1:1:1:1:2, v/v). Leaves in solution were incubated in a 65°C bath until they were judged clear and then stained with Aniline Blue. The staining reaction was performed in the dark for 4 h. The leaf samples were observed by microscopy under an ultraviolet light field, and callose deposition in the vascular bundles of the middle veins of the leaves was visualized as a blue color.
N. benthamiana Gene Silencing and Transient Expression
The pTRV VIGS system was used to construct the NbRbohBi or NbPDSi unit based on specific primers (Supplemental Table S2) and previous protocols (Zhang et al., 2009). VIGS-directed transfection was performed on the first and second leaves of 35-d-old plants. The same leaves of equivalent plants were transfected with the empty vector, which did not contain any gene-silencing construct, in the transfection control. After 14 d, a mottled photobleaching phenotype of the NbPDSi plants was observed, and NbRbohBi efficiency in cognate plants was analyzed by qRT-PCR. The top two leaves were transformed with a plant binary vector in the transformation control or with recombinant vectors containing GFP and AtPIP1;4-GFP fused to a constitutive promoter (Li et al., 2015). Two days later, the transient expression of both genes was analyzed by qRT-PCR, and proteins in transformed leaves were observed by laser confocal microscopy. Independent plants were treated with flg22, and H2O2 was detected as stated above.
Statistical Analysis
Quantitative data were analyzed using the commercial IBM SPSS19.0 software package (Shi, 2012). The homogeneity of variance was determined using Levene’s test, and the formal distribution pattern of the data was confirmed by the Kolmogorov-Smirnov test and P-P plots, an SPSS tool that yields a graph to assess whether the data are normal or not. Then, data were subjected to ANOVA along with Fisher’s lsd test and the Tukey-Kramer test. Differences between AtPIP1;4-transformed and control yeast cells, H2O2-treated and control yeast cultures, inoculated and mock-inoculated plants, and PAMP-treated and control plants were tested for significance. Differences among multiple treatments in a single plant genotype or among the several genotypes under a single treatment or inoculation condition also were tested for significance.
Sequence data from this article can be found in Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Differential expression of AtPIP genes in DC3000-inoculated plants.
Supplemental Figure S2. An insertional mutation of AtPIP1;4.
Supplemental Figure S3. Yeast colonies grown with H2O2 supplied at the indicated concentrations.
Supplemental Figure S4. Yeast SOD activity measurements.
Supplemental Figure S5. Imaging of apoplastic water and cytoplasmic H2O2 or ROS in leaves of plants treated with H2O2 or water.
Supplemental Figure S6. H2O2 or ROS visualization and SOD activities in leaves of inoculated and mock-inoculated plants.
Supplemental Figure S7. Effects of PAMPs on DC3000 population growth in plant leaves.
Supplemental Figure S8. Northern-blot analysis with AtPIP1;4 probe hybridized to complementary DNAs of AtPIPs.
Supplemental Table S1. Relative levels of MPK6 expression in leaves.
Supplemental Table S2. Information on genes tested and primers used in this study.
Supplementary Material
Acknowledgments
We thank Steven Beer and Alan Collmer for gifts of the bacterial strain and prokaryotic vectors, respectively.
Glossary
- H2O2
hydrogen peroxide
- ROS
reactive oxygen species
- PAMP
pathogen-associated molecular pattern
- PM
plasma membrane
- SAR
systemic acquired resistance
- PTI
pathogen-associated molecular pattern-triggered immunity
- MAPK
mitogen-activated protein kinase
- AQP
aquaporin
- DC3000
virulent strain DC3000 of Pseudomonas syringae pv tomato
- hpi
hours post inoculation
- dpi
days post inoculation
- AR
Amplex Red
- AUR
Amplex Ultra Red
- H2DCFDA
2,7-dichlorofluorescein diacetate
- SOD
superoxide dismutase
- qRT
quantitative reverse transcription
- VIGS
virus-induced gene silencing
- DPI
diphenyleneiodonium
- PBS
phosphate buffer solution
Footnotes
This work was supported by the Natural Science Foundation of China (grant no. 31272027), the China National Basic Research and Scientific Development Program (973 Plan grant no. 2012CB114000), the Novel Transgenic Organisms Breeding Project (grant no. 2014ZX0800910B), the Special Public Welfare Industry Program (grant no. 201303015), and the Specialized Research Fund for University Doctoral Program (grant no. YXFZ2012).
Articles can be viewed without a subscription.
References
- Abascal F, Irisarri I, Zardoya R (2014) Diversity and evolution of membrane intrinsic proteins. Biochim Biophys Acta 1840: 1468–1481 [DOI] [PubMed] [Google Scholar]
- Aguayo D, Pacheco N, Morales EH, Collao B, Luraschi R, Cabezas C, Calderón P, González-Nilo F, Gil F, Calderón IL, et al. (2015) Hydrogen peroxide and hypochlorous acid influx through the major S. typhimurium porin OmpD is affected by substitution of key residues of the channel. Arch Biochem Biophys 568: 38–45 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Ashtamker C, Kiss V, Sagi M, Davydov O, Fluhr R (2007) Diverse subcellular locations of cryptogein-induced reactive oxygen species production in tobacco Bright Yellow-2 cells. Plant Physiol 143: 1817–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6: 973–979 [DOI] [PubMed] [Google Scholar]
- Bethke G, Pecher P, Eschen-Lippold L, Tsuda K, Katagiri F, Glazebrook J, Scheel D, Lee J (2012) Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol Plant Microbe Interact 25: 471–480 [DOI] [PubMed] [Google Scholar]
- Bienert GP, Chaumont F (2014) Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim Biophys Acta 1840: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Bienert GP, Møller AL, Kristiansen KA, Schulz A, Møller IM, Schjoerring JK, Jahn TP (2007) Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol Chem 282: 1183–1192 [DOI] [PubMed] [Google Scholar]
- Bienert GP, Schjoerring JK, Jahn TP (2006) Membrane transport of hydrogen peroxide. Biochim Biophys Acta 1758: 994–1003 [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8: 521–539 [DOI] [PubMed] [Google Scholar]
- Bindschedler LV, Dewdney J, Blee KA, Stone JM, Asai T, Plotnikov J, Denoux C, Hayes T, Gerrish C, Davies DR, et al. (2006) Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J 47: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Chen L, Qian J, Qu S, Long J, Yin Q, Zhang C, Wu X, Sun F, Wu T, Hayes M, et al. (2008) Identification of specific fragments of HpaG Xooc, a harpin from Xanthomonas oryzae pv. oryzicola, that induce disease resistance and enhance growth in plants. Phytopathology 98: 781–791 [DOI] [PubMed] [Google Scholar]
- Choi MS, Kim W, Lee C, Oh CS (2013) Harpins, multifunctional proteins secreted by gram-negative plant-pathogenic bacteria. Mol Plant Microbe Interact 26: 1115–1122 [DOI] [PubMed] [Google Scholar]
- Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, Bolwell GP (2012) The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24: 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayakar BV, Lin HJ, Chen CH, Ger MJ, Lee BH, Pai CH, Chow D, Huang HE, Hwang SY, Chung MC, et al. (2003) Ferredoxin from sweet pepper (Capsicum annuum L.) intensifying harpinpss-mediated hypersensitive response shows an enhanced production of active oxygen species (AOS). Plant Mol Biol 51: 913–924 [DOI] [PubMed] [Google Scholar]
- Deng B, Deng S, Sun F, Zhang S, Dong H (2011) Down-regulation of free riboflavin content induces hydrogen peroxide and a pathogen defense in Arabidopsis. Plant Mol Biol 77: 185–201 [DOI] [PubMed] [Google Scholar]
- Dong H, Delaney TP, Bauer DW, Beer SV (1999) Harpin induces disease resistance in Arabidopsis through the systemic acquired resistance pathway mediated by salicylic acid and the NIM1 gene. Plant J 20: 207–215 [DOI] [PubMed] [Google Scholar]
- Dynowski M, Schaaf G, Loque D, Moran O, Ludewig U (2008) Plant plasma membrane water channels conduct the signalling molecule H2O2. Biochem J 414: 53–61 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fu M, Xu M, Zhou T, Wang D, Tian S, Han L, Dong H, Zhang C (2014) Transgenic expression of a functional fragment of harpin protein Hpa1 in wheat induces the phloem-based defence against English grain aphid. J Exp Bot 65: 1439–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R, Ferrari S, De Lorenzo G (2011) Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol 157: 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio M, Trinei M, Migliaccio E, Pelicci PG (2007) Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat Rev Mol Cell Biol 8: 722–728 [DOI] [PubMed] [Google Scholar]
- Gomes D, Agasse A, Thiébaud P, Delrot S, Gerós H, Chaumont F (2009) Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim Biophys Acta 1788: 1213–1228 [DOI] [PubMed] [Google Scholar]
- Gralla EB, Kosman DJ (1992) Molecular genetics of superoxide dismutases in yeasts and related fungi. Adv Genet 30: 251–319 [DOI] [PubMed] [Google Scholar]
- Grondin A, Rodrigues O, Verdoucq L, Merlot S, Leonhardt N, Maurel C (2015) Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 27: 1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat GE, Iusem ND, Morris PC (2007) Guard cell-specific inhibition of Arabidopsis MPK3 expression causes abnormal stomatal responses to abscisic acid and hydrogen peroxide. New Phytol 173: 713–721 [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR (2009) The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe Interact 22: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapalainen M, Engelhardt S, Küfner I, Li CM, Nürnberger T, Lee J, Romantschuk M, Taira S (2011) Functional mapping of harpin HrpZ of Pseudomonas syringae reveals the sites responsible for protein oligomerization, lipid interactions and plant defence induction. Mol Plant Pathol 12: 151–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooijmaijers C, Rhee JY, Kwak KJ, Chung GC, Horie T, Katsuhara M, Kang H (2012) Hydrogen peroxide permeability of plasma membrane aquaporins of Arabidopsis thaliana. J Plant Res 125: 147–153 [DOI] [PubMed] [Google Scholar]
- Ji H, Dong H (2015a) Key steps in type III secretion system (T3SS) towards translocon assembly with potential sensor at plant plasma membrane. Mol Plant Pathol 16: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji HT, Dong HS (2015b) Biological significance and topological basis of aquaporin-partnering protein-protein interactions. Plant Signal Behav 10: e1011947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko BH, Ramos AR, Morello JE, Oh HS, Collmer A (2007) Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J Bacteriol 189: 8059–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Klessig DF, Nürnberger T (2001) A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell 13: 1079–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Li L, Hu L, Han LP, Ji H, Zhu Y, Wang X, Ge J, Xu M, Shen D, Dong H (2014) Expression of turtle riboflavin-binding protein represses mitochondrial electron transport gene expression and promotes flowering in Arabidopsis. BMC Plant Biol 14: 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang H, Gago J, Cui H, Qian Z, Kodama N, Ji H, Tian S, Shen D, Chen Y, et al. (2015) Harpin Hpa1 interacts with aquaporin PIP1;4 to promote the substrate transport and photosynthesis in Arabidopsis. Sci Rep 5: 17207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Chen L, Jia Z, Lü B, Shi H, Shao W, Dong H (2011) Transcription factor AtMYB44 regulates induced expression of the ETHYLENE INSENSITIVE2 gene in Arabidopsis responding to a harpin protein. Mol Plant Microbe Interact 24: 377–389 [DOI] [PubMed] [Google Scholar]
- Lü B, Sun W, Zhang S, Zhang C, Qian J, Wang X, Gao R, Dong H (2011) HrpNEa-induced deterrent effect on phloem feeding of the green peach aphid Myzus persicae requires AtGSL5 and AtMYB44 genes in Arabidopsis thaliana. J Biosci 36: 123–137 [DOI] [PubMed] [Google Scholar]
- Macierzyńska E, Grzelak A, Bartosz G (2007) The effect of growth medium on the antioxidant defense of Saccharomyces cerevisiae. Cell Mol Biol Lett 12: 448–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammarella ND, Cheng Z, Fu ZQ, Daudi A, Bolwell GP, Dong X, Ausubel FM (2015) Apoplastic peroxidases are required for salicylic acid-mediated defense against Pseudomonas syringae. Phytochemistry 112: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins D, English AM (2014) Catalase activity is stimulated by H2O2 in rich culture medium and is required for H2O2 resistance and adaptation in yeast. Redox Biol 2: 308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C. (2007) Plant aquaporins: novel functions and regulation properties. FEBS Lett 581: 2227–2236 [DOI] [PubMed] [Google Scholar]
- Maurel C, Reizer J, Schroeder JI, Chrispeels MJ (1993) The vacuolar membrane protein γ-TIP creates water specific channels in Xenopus oocytes. EMBO J 12: 2241–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Sundelin T, Nielsen JT, Erbs G (2013) MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci 4: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HS, Collmer A (2005) Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins. Plant J 44: 348–359 [DOI] [PubMed] [Google Scholar]
- Pavli OI, Kelaidi GI, Tampakaki AP, Skaracis GN (2011) The hrpZ gene of Pseudomonas syringae pv. phaseolicola enhances resistance to rhizomania disease in transgenic Nicotiana benthamiana and sugar beet. PLoS ONE 6: e17306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JL, Bao ZL, Ren HY, Wang JS, Dong HS (2004) Expression of harpinxoo in transgenic tobacco induces pathogen defense in the absence of hypersensitive cell death. Phytopathology 94: 1048–1055 [DOI] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, Band LR, Voß U, Postaire O, Luu DT, Da Ines O, Casimiro I, Lucas M, et al. (2012) Auxin regulates aquaporin function to facilitate lateral root emergence. Nat Cell Biol 14: 991–998 [DOI] [PubMed] [Google Scholar]
- Pitzschke A. (2015) Modes of MAPK substrate recognition and control. Trends Plant Sci 20: 49–55 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Prado K, Boursiac Y, Tournaire-Roux C, Monneuse JM, Postaire O, Da Ines O, Schäffner AR, Hem S, Santoni V, Maurel C (2013) Regulation of Arabidopsis leaf hydraulics involves light-dependent phosphorylation of aquaporins in veins. Plant Cell 25: 1029–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston GM, Agre P (1991) Isolation of the cDNA for erythrocyte integral membrane protein of 28 kilodaltons: member of an ancient channel family. Proc Natl Acad Sci USA 88: 11110–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston GM, Carroll TP, Guggino WB, Agre P (1992) Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science 256: 385–387 [DOI] [PubMed] [Google Scholar]
- Rhee SG, Chang TS, Jeong W, Kang D (2010) Methods for detection and measurement of hydrogen peroxide inside and outside of cells. Mol Cells 29: 539–549 [DOI] [PubMed] [Google Scholar]
- Sagi M, Fluhr R (2006) Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol 141: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang S, Li X, Gao R, You Z, Lü B, Liu P, Ma Q, Dong H (2012) Apoplastic and cytoplasmic location of harpin protein Hpa1Xoo plays different roles in H2O2 generation and pathogen resistance in Arabidopsis. Plant Mol Biol 79: 375–391 [DOI] [PubMed] [Google Scholar]
- Shi LW. (2012) SPSS19.0: Statistical Analysis from Accidence to Conversance. [In Chinese.] Tsinghua University Press, Beijing, pp 109–143 [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel RN, Gopalan JS, Kuc JA, He SY (1996) Induction of systemic acquired resistance in cucumber by Pseudomonas syringae pv. syringae HrpZPss protein. Plant J 9: 431–439 [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA. (2010) ROS in biotic interactions. Physiol Plant 138: 414–429 [DOI] [PubMed] [Google Scholar]
- Travella S, Klimm TE, Keller B (2006) RNA interference-based gene silencing as an efficient tool for functional genomics in hexaploid bread wheat. Plant Physiol 142: 6–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17: 1784–1790 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu R, Chen L, Wang Y, Liang Y, Wu X, Li B, Wu J, Liang Y, Wang X, et al. (2009) Nicotiana tabacum TTG1 contributes to ParA1-induced signalling and cell death in leaf trichomes. J Cell Sci 122: 2673–2685 [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Numata N, Nakajima K, Katou S, Kawakita K, Rowland O, Jones JD, Doke N (2003) Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15: 706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Fang Q, Zhang Z, Wang Y, Zheng X (2009) The role of respiratory burst oxidase homologues in elicitor-induced stomatal closure and hypersensitive response in Nicotiana benthamiana. J Exp Bot 60: 3109–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li C, Ge J, Xu M, Zhu Q, Wu T, Guo A, Xie J, Dong H (2014) Recessive mutation identifies auxin-repressed protein ARP1, which regulates growth and disease resistance in tobacco. Mol Plant Microbe Interact 27: 638–654 [DOI] [PubMed] [Google Scholar]
- Zhu W, MaGbanua MM, White FF (2000) Identification of two novel hrp-associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae. J Bacteriol 182: 1844–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.