Foxtail Mosaic Virus-induced gene silencing works in multiple monocot plants including barley, wheat, and foxtail millet.
Abstract
Virus-induced gene silencing (VIGS) is a powerful technique to study gene function in plants. However, very few VIGS vectors are available for monocot plants. Here we report that Foxtail mosaic virus (FoMV) can be engineered as an effective VIGS system to induce efficient silencing of endogenous genes in monocot plants including barley (Hordeum vulgare L.), wheat (Triticum aestivum) and foxtail millet (Setaria italica). This is evidenced by FoMV-based silencing of phytoene desaturase (PDS) and magnesium chelatase in barley, of PDS and Cloroplastos alterados1 in foxtail millet and wheat, and of an additional gene IspH in foxtail millet. Silencing of these genes resulted in photobleached or chlorosis phenotypes in barley, wheat, and foxtail millet. Furthermore, our FoMV-based gene silencing is the first VIGS system reported for foxtail millet, an important C4 model plant. It may provide an efficient toolbox for high-throughput functional genomics in economically important monocot crops.
Virus-induced gene silencing (VIGS) provides a powerful approach for genetic and functional characterization of genes in plants. VIGS is related to post-transcriptional gene silencing, a plant antiviral defense mechanism that targets and leads to degradation of viral RNAs (Vance and Vaucheret, 2001; Baulcombe, 2004). Post-transcriptional gene silencing in plants is dependent on a relatively high degree of nucleotide homology between RNA transcripts and target endogenous gene sequences (van den Boogaart et al., 1998; Ding, 2000). The recombinant virus-carrying sequence of a host gene generates dsRNAs, which are processed by Dicer-like proteins to produce siRNAs, and the latter triggers silencing of the endogenous gene itself. This forms the basis of plant VIGS technology.
Many plant RNA and DNA viruses have been modified as VIGS vectors and they are used to dissect gene function in diverse dicot plant species (Burch-Smith et al., 2004; Senthil-Kumar and Mysore, 2011; Huang et al., 2012). For instance, Tobacco rattle virus-derived vector is one of the most widely used systems in dicots because of its broad host range and ability to infect meristematic tissue (Ratcliff et al., 2001; Liu et al., 2002a, Liu et al., 2002b; Padmanabhan and Dinesh-Kumar, 2009). Bean pot mottle virus-derived vectors have been developed to induce VIGS in soybean (Glycine max) and common bean (Phaseolus vulgaris; Zhang and Ghabrial, 2006; Zhang et al., 2010). Cabbage leaf curl virus-derived vector has already been developed to trigger VIGS for siRNA-mediated silencing in Arabidopsis (Turnage et al., 2002) and miRNA-mediated silencing in tobacco (Nicotiana benthamiana; Tang et al., 2010).
By contrast, to date only four RNA viruses and one DNA virus have been modified as vectors for VIGS in monocot species, of which, Barley stripe mosaic virus (BSMV)-based VIGS has been applied for functional genomics in barley (Hordeum vulgare L.) and wheat (Triticum aestivum; Holzberg et al., 2002; Tai et al., 2005; Meng et al., 2009; Pacak et al., 2010; Yuan et al., 2011); Brome mosaic virus (BMV) in rice (Oryza sativa L.), barley, and maize (Zea mays L.; Ding et al., 2006); Bamboo mosaic virus and its satellite RNA in N. benthamiana and Brachypodium distachyon (Liou et al., 2014); and Rice tungro bacilliform virus in rice (Purkayastha et al., 2010). Very recently, Cucumber mosaic virus-based VIGS in maize is reported (Wang et al., 2016) BMV and BMV-based vectors are frequently used for VIGS in some monocot plants. However, both virus vectors do not work in many other monocot species and not all cultivars within a particular host species (Ding et al., 2006; Pacak et al., 2010).
Foxtail mosaic virus (FoMV) is a species of the genus Potexvirus and possesses a broad host range including 56 Poaceae species and at least 35 dicot species (Paulsen and Niblett, 1977; Short and Davies, 1987). A VIGS system based on potato virus X (PVX), the type member of Potexvirus, has been developed to silence target genes successfully in plants (Ruiz et al., 1998; Chen et al., 2015; Zhao et al., 2016). FoMV is similar to PVX and other potexviruses in their genomic organization. It consists of a positive-sense single-stranded RNA genome with a 5′-methylguanosine cap, a 3′-polyadenylated tail, and five major open reading frames (ORFs) and a unique 5A gene (Robertson et al., 2000; Bruun-Rasmussen et al., 2008). Each of the five major ORFs encodes a functional protein as in PVX (Bruun-Rasmussen et al., 2008). FoMV has been modified as an RNAi suppressor-dependent expression vector by deleting its coat protein (CP) and triple gene block genetic segments (Liu and Kearney, 2010). However, FoMV has not been successfully engineered as a VIGS vector, although it has been thought to be a potentially useful virus for VIGS in monocot hosts (Scofield and Nelson, 2009).
In this study, we report that a FoMV-based vector can be used to perform VIGS in monocots including barley, wheat, and foxtail millet (Setaria italica), and may provide a useful tool to study gene function in a broad monocot plant species.
RESULTS
Development of FoMV VIGS Vector
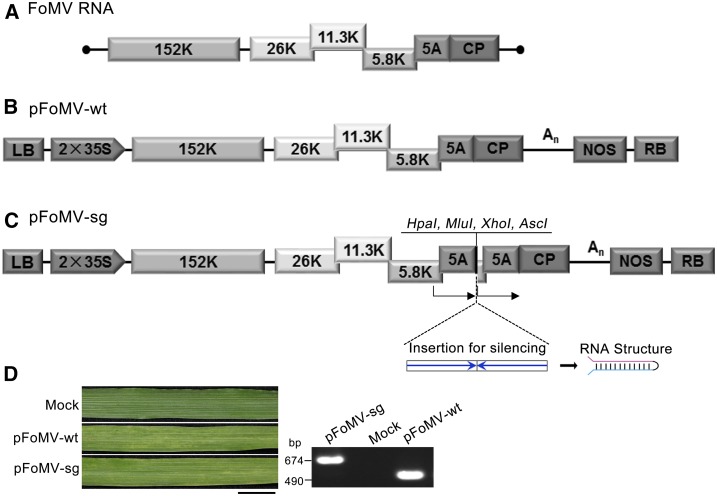
To develop an FoMV VIGS vector, the full-length FoMV cDNA (Fig. 1A, a kind gift from Nancy Robertson) was recloned between the duplicated 35S CaMV promoter and the nitric oxide synthase (NOS) terminator of pBin19-based T-DNA vector pYL44 (Liu et al., 2002a) to generate pFoMV-wt (Fig. 1B). To verify the infectivity of pFoMV-wt, N. benthamiana plants were infiltrated with Agrobacterium-carrying pFoMV-wt. RT-PCR showed the presence of FoMV in systemic leaves at 10 d post-infiltration (dpi). Barley plants inoculated with saps of FoMV-infected N. benthamiana leaves, but not those with saps of mock-inoculated healthy plants, developed a mild mosaic symptom at 10 dpi. These results demonstrate that the cloned FoMV is infectious and FoMV-infected N. benthamiana leaves are an excellent source of FoMV for secondary viral infections in cereals such as barley. Subsequently, all plants of different monocot species were infected through mechanic inoculation with saps of FoMV or the derivatives-infected N. benthamiana leaves.
Figure 1.
Schematic organization of FoMV genome and infectious FoMV vector. A, Genomic organization of FoMV RNA. The ORFs are indicated by boxes. The 152-kD protein is an RNA-dependent RNA polymerase. The three proteins at 26 kD, 11.3 kD, and 5.8 kD, collectively known as a triple gene block, are presumed to mediate viral cell-to-cell moment. The 5A protein was produced in vivo; the 24-kD CP is located at the 3′-terminus. B, Schematic of infectious FoMV T-DNA clone pFoMV-wt. FoMV was cloned between CaMV 35S promoter with duplicated enhancers and a nopaline synthase (NOS) terminator of a T-DNA binary vector. pFoMV-wt has an additional 70 adenosine residues after the viral 3′-terminus. C, Schematic of the FoMV VIGS vector, pFoMV-sg. pFoMV-sg is a pFoMV-wt derivative that contains a direct repeat of the 170-bp putative FoMV CP subgenomic promoter (GenBank accession: EF630359, nt 5202–5371), shown by arrows, and multiple cloning sites (HpaI, MluI, XhoI, and AscI) between CP subgenomic promoters. Right- and left-pointing blue arrows indicate sense and anti-sense orientation of foreign fragments, and the predicted structure of RNA transcript is presented on the right side. D, Symptom development and RT-PCR detection of viral RNA showed that pFoMV-wt and pFoMV-sg are infectious in barley. Bar = 1 cm.
FoMV, similar to PVX, belongs to Potexvirus, and PVX has successfully been used for VIGS. We thus employed a similar strategy for construction of the PVX VIGS vector (Ruiz et al., 1998) to generate our FoMV vector. The resulting vector pFoMV-sg is a pFoMV-wt derivative that contains a direct repeat of the 170-bp putative FoMV CP subgenomic promoter (SGP, which covers the entire 5A sequence) and multiple cloning sites, including HpaI, MluI, XhoI, and AscI between duplicated CP SGP. The modified full-length FoMV cDNA in pFoMV-sg is under control of a CaMV 35S promoter with duplicated enhancers and an NOS terminator (Fig. 1C). When barley plants were inoculated with saps of pFoMV-sg–infected N. benthamiana leaves, the uninoculated systemic leaves of barley plants developed very mild mosaic symptoms at 15 dpi (Fig. 1D). Recombinant FoMV-sg genomic RNA was readily detected by RT-PCR (Fig. 1D).
FoMV-Based VIGS in Barley
To test whether pFoMV-sg can be used to induce gene silencing in barley, a 200-bp barley phytoene desaturase (HvPDS) DNA fragment was cloned into pFoMV-sg to generate FoMV-HvPDS200. It has been reported that VIGS of PDS resulted in a photobleached phenotype (Kumagai et al., 1995; Liu et al., 2002a). Two-leaf-stage barley plants were inoculated with saps of N. benthamiana leaves agroinfiltrated with FoMV-HvPDS200 or empty vector pFoMV-sg. We detected the presence of FoMV in systemic leaves of all infected barley plants. However, we only observed very mild PDS-silencing bleached phenotypes in FoMV-HvPDS200-barley plants, even at 30 dpi.
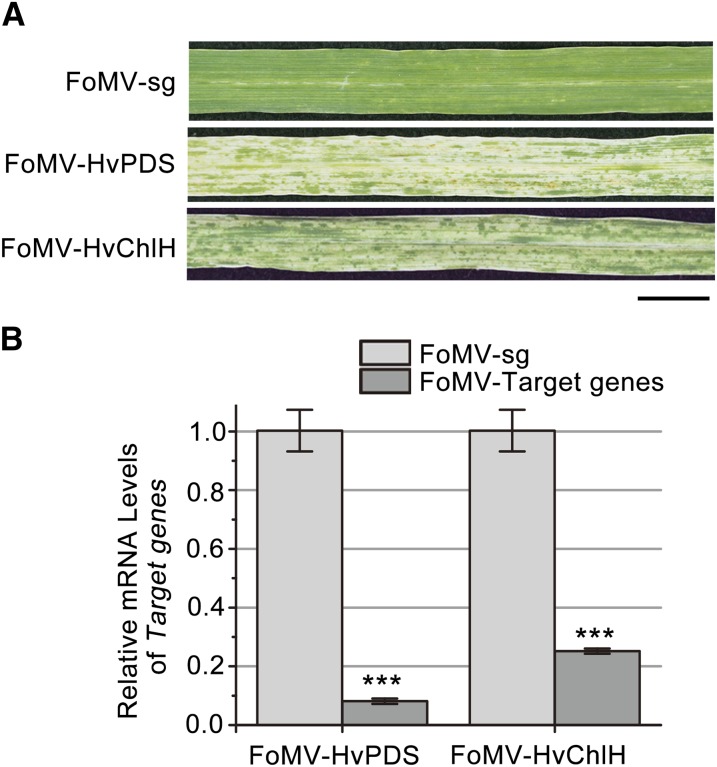
To improve VIGS efficiency, we inserted inverted-repeats into pFoMV-sg, because VIGS can be enhanced through viral-based production of inverted-repeats (Lacomme et al., 2003). We first silenced two barley genes, HvPDS and magnesium chelatase (HvChlH), using this approach. ChlH encodes the H subunit of magnesium chelatase that is required for chlorophyll production (Shen et al., 2006). For this purpose, we cloned 60-bp HvPDS and 55-bp HvChlH of inverted-repeat fragments into pFoMV-sg to generate FoMV-HvPDS and FoMV-HvChlH. Two-leaf-stage barley plants were inoculated with saps of N. benthamiana leaves agroinfiltrated with FoMV-HvPDS, FoMV-HvChlH, or pFoMV-sg. As expected, pFoMV-sg–infected barley plants showed very mild chlorotic symptoms, usually occurring in the first and second leaves above the inoculated leaves. However, all barley plants infected with either FoMV-HvPDS or FoMV-HvChlH exhibited longitudinal white patches and streaks, and developed the strongest photobleached or chlorosis VIGS phenotype in leaf 3, numbered from the bottom of the plant at 21 dpi (Fig. 2A). Real-time RT-PCR showed that the mRNA level of PDS and ChlH in phenotypic tissues was reduced by approximately 90% and 75% in barley plants infected with FoMV-HvPDS and FoMV-HvChlH compared to plants infected with pFoMV-sg, respectively (Fig. 2B). These results clearly suggest that pFoMV-sg containing inverted-repeats of host genes can induce effective silencing in barley.
Figure 2.
FoMV-based VIGS of PDS and ChlH in barley. A, Barley plants were infected with a pFoMV-sg vector carrying an inverted-repeat fragment of HvPDS (FoMV-HvPDS) and HvChlH (FoMV-HvChlH). The third uninoculated leaves were photographed at 21 dpi. Bar = 1 cm. B, Quantification of the mRNA levels of HvPDS and HvChlH of barley plants was analyzed by real-time RT-PCR, normalized relative to the level of 18S rRNA. ***P < 0.01. Measurements were repeated three times, respectively. The error bar indicates se.
FoMV-Based VIGS in Foxtail Millet
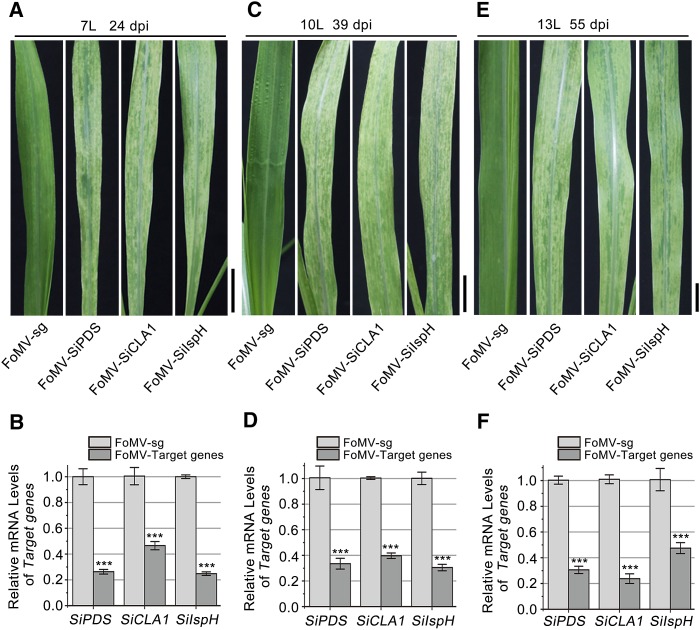
Foxtail millet is an important grain crop widely grown in arid regions and it is a model C4 grass plant. To test whether FoMV VIGS vector is able to induce gene silencing in foxtail millet, we silenced three foxtail millet genes SiPDS, Cloroplastos alterados1 (SiCLA1), and SiIspH. CLA1 encodes 1-deoxy-d-xylulose-5-P synthase (Mandel et al., 1996; Estévez et al., 2001; Tang et al., 2010) and IspH is for an isopentenyl/dimethylallyl diphosphate synthase involved in isoprenoid biosynthesis. Their null mutations or VIGS resulted in albino phenotypes (Rohdich et al., 2002; Hsieh and Goodman, 2005; Yin et al., 2015). 60-bp inverted-repeat fragments targeting SiPDS, SiCLA1, and SiIspH were inserted into pFoMV-sg to generate FoMV-SiPDS, FoMV-SiCLA1, and FoMV-SiIspH, respectively. Three-leaf stage foxtail millet plants were inoculated with saps of N. benthamiana leaves agroinfiltrated with pFoMV-sg or its derivatives. Foxtail millet plants infected with pFoMV-sg exhibited mild chlorotic symptoms. However, these plants infected with FoMV-SiPDS, FoMV-SiCLA1, or FoMV-SiIspH began to develop a mottled photobleached or albino phenotype in new emerging leaves at 10 dpi to 12 dpi. All plants developed larger areas of photobleached phenotype in the newly emerging leaves (the seventh leaf, numbered from the bottom of the plant) at 24 dpi (Fig. 3); in all the cases, the silencing phenotype was maintained and appeared in upper new emerging leaves for up to 60 d (55 dpi in Fig. 3E). Moreover, we performed real-time RT-PCR to determine mRNA levels of SiPDS, SiCLA1, and SiIspH in the silenced leaves. The mRNA levels of SiPDS, SiCLA1, and SiIspH were, respectively, decreased by approximately 66 to 74%, 55 to 76%, and 56 to 73% (Fig. 3) compared with that in FoMV-sg-infected control plants. These results suggest that our FoMV-based VIGS vector can be used to silence genes in foxtail millet.
Figure 3.
FoMV-based VIGS of marker genes in foxtail millet. Foxtail millet plants were infected with a pFoMV-sg or pFoMV-sg vector carrying inverted-repeat fragments of SiPDS (FoMV-SiPDS), SiCLA1 (FoMV-SiCLA1), and SiIspH (FoMV-SiIspH), respectively. Photographs were taken for the seventh leaf (7L) at 24 dpi (A), for the 10th leaf (10L) at 39 dpi (C), and for the 13th leaf (13L) at 55 dpi (E). Bars = 1 cm. Quantification of mRNA levels of three marker genes of foxtail millet plants at 24 dpi (B), 39 dpi (D), and 55 dpi (F) were analyzed by real-time RT-PCR, normalized relative to the value of 18S rRNA. ***P < 0.01. Measurements were repeated three times, respectively. The error bar indicates se.
FoMV-Based VIGS in Wheat
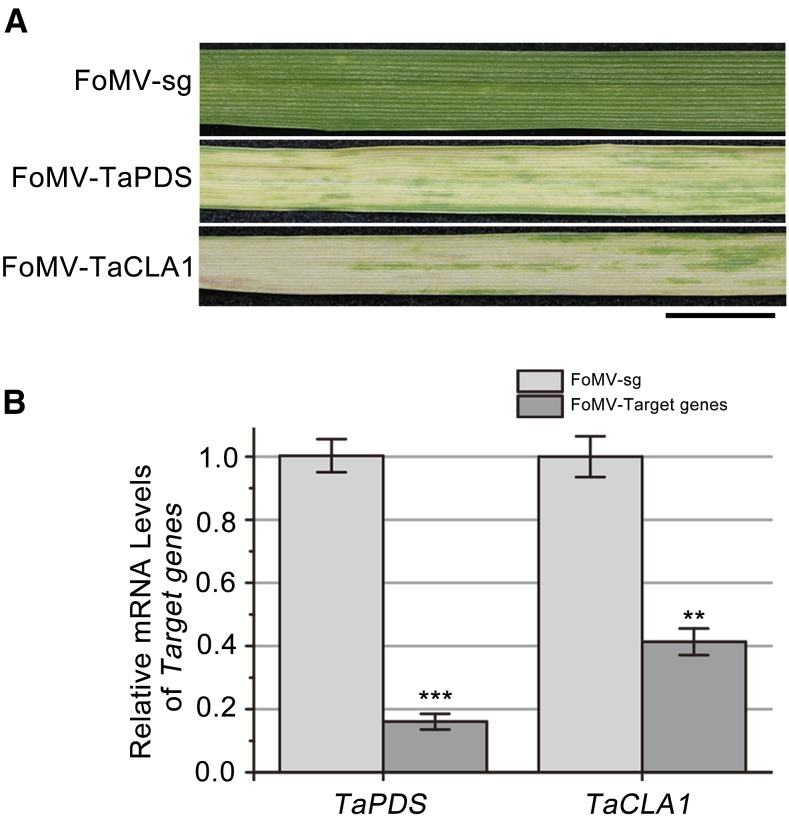
We also tested whether FoMV is able to induce gene silencing in wheat. To achieve this, we first identified two wheat marker genes, TaPDS and TaCLA1. Then we cloned 60-bp TaPDS and TaCLA1 of inverted-repeat fragments into pFoMV-sg to generate FoMV-TaPDS and FoMV-TaCLA1, respectively. Two-leaf-stage wheat plants were inoculated with saps of N. benthamiana leaves agroinfiltrated with pFoMV-sg or its derivatives. All wheat plants infected with FoMV-TaPDS or FoMV-TaCLA1 usually appeared as small white streaks in leaf 3, numbered from the bottom of the plant at 20 dpi; the white patches and streaks or albinism fully expanded in leaf 4 at 35 dpi (Fig. 4A). As plants grew, the bleached patches or albinism spread to the newly emerging leaves. However, no FoMV-sg control plants showed any silencing phenotype. To confirm the silencing of TaPDS and TaCLA1 genes at the molecular level, we performed real-time RT-PCR. As shown in Figure 4B, mRNA levels of TaPDS and TaCLA1 in the silenced leaves were strikingly reduced compared to FoMV-sg–infected plants. Taken together, our results clearly suggest that FoMV can induce efficient gene silencing in wheat.
Figure 4.
FoMV-based VIGS of PDS and CLA1 in wheat. A, Wheat plants were infected with a pFoMV-sg vector carrying an inverted-repeat fragment of TaPDS (FoMV-TaPDS) or TaCLA1 (FoMV-TaCLA1). The fourth leaves were photographed at 35 dpi. Bar = 1 cm. B, Quantification of the mRNA levels of TaPDS and TaCLA1 of wheat plants was analyzed by real-time RT-PCR, normalized relative to the level of 18S rRNA. **P < 0.05; ***P < 0.01. Measurements were repeated three times, respectively. The error bar indicates se.
DISCUSSION
In this study, we investigated whether FoMVs could act as effective inducers for silencing of endogenous genes in monocot plants. Using a FoMV-based viral system, we successfully silenced PDS and ChlH in barley, and PDS and CLA1 genes in wheat. Moreover, we also silenced PDS, CLA1, and IspH in foxtail millet. These results suggest that FoMV vectors carrying inverted-repeat sequences of host endogenous genes effectively induced VIGS in monocot plants. To the best of our knowledge, this is the first report on VIGS in foxtail millet.
VIGS allows silencing of individual gene or families of genes in plant development, disease resistance, and abiotic stress pathways and it is a rapid method to assess gene function (Burch-Smith et al., 2004; Robertson, 2004; Becker and Lange, 2010; Cakir et al., 2010). Among all available VIGS vectors, our (to our knowledge) newly developed pFoMV-sg vector has several characteristics for VIGS. First, FoMV is a flexuous, rod-shaped virus, and has a single, positive-sense single-stranded RNA genome (Bancroft et al., 1991). A VIGS vector-based single viral genome is easy to handle and it usually possesses high efficiency to cause infection of plants, a prerequisite for effective VIGS. Second, FoMV produces only mild symptoms or is latent in many native and adaptive hosts (Robertson et al., 2000). Indeed, our pFoMV-sg VIGS vector generally induces very mild mosaic symptoms in emerging leaves of barley, wheat, and foxtail millet plants. This makes the pFoMV-sg vector extremely useful for functional genomics because severe viral disease symptoms may complicate VIGS phenotypes in testing plants (Ramanna et al., 2013). Third, FoMV can infect a broad range of hosts among the grasses as well as dicot species (Short and Davies, 1987). Thus, our pFoMV-sg VIGS vector should have the potential for VIGS in many monocot and dicot plants although we only tested its usefulness in barley, wheat, and foxtail millet. Fourth, FoMV-sg is highly infectious, and almost 90% of tested plants induced obvious silencing phenotypes. VIGS phenotypes caused by FoMV-sg vectors were stable and persisted for at least two months. In addition, FoMV can penetrate growing points in monocot plants. It may be applied to silence genes and analyze their biological relevance in plant reproductive tissues.
PVX is the type member of the genus Potexvirus and has successfully been developed as both expression and VIGS vectors by including an additional duplicated copy of CP subgenomic promoter in the viral genome (Ruiz et al., 1998). Because FoMV is also a member of Potexvirus, we adopted a similar strategy to generate vector pFoMV-sg, which contains a duplicated copy of the 170-bp putative FoMV CP SGP and a multiple cloning site. Surprisingly, delivery of an anti-sense RNA fragment of HvPDS by pFoMV-sg did not cause any obvious silencing phenotype in barley. To trigger effective VIGS, viral replication should be high enough to produce a threshold level of silencing-trigger RNA molecules (Lacomme et al., 2003). Expression of a hairpin-loop dsRNA structure generated a stronger silencing than expression of the corresponding sense or anti-sense cDNA (Waterhouse et al., 1998; Smith et al., 2000; De Buck et al., 2001). It has been reported that insertion of direct inverted-repeat sequences of target genes that could fold as dsRNA hairpin structures strongly enhance VIGS from Tobacco mosaic virus, BSMV (Lacomme et al., 2003), and Turnip yellow mosaic virus (Pflieger et al., 2008). This is true for pFoMV-sg–mediated VIGS. Expression of inverted-repeats of PDS but not an anti-sense fragment of PDS from FoMV-sg triggered a clear photobleached phenotype in barley (Fig. 2). Similarly, FoMV-based expression of inverted-repeats of ChlH in barley (Fig. 2), of PDS and CLA1 in foxtail millet and wheat plants, of IspH in foxtail millet plants, induced efficient silencing. Moreover, silencing phenotype persisted up to 60 d (Fig. 3). These results support that an insertion of inverted-repeat sequences into the viral genome can enhance the efficiency of VIGS.
Foxtail millet is a member of the Poaceae grasses, and is a model system for understanding functional genomics and photosynthesis in grain crops and C4 plants (Wang et al., 2010). Foxtail millet is also a close relative of switchgrass (Panicum virgatum), an important biofuel crop. Recently, the whole genome sequence of foxtail millet has been published (Bennetzen et al., 2012; Zhang et al., 2012). Therefore this first report of an efficient VIGS system based on pFoMV-sg provides a timely toolbox for high-throughput assays of gene functions in foxtail millet and many other economically important monocot crops.
CONCLUSIONS
Our FoMV-sg vector is able to trigger efficient gene silencing in barley, wheat, and foxtail millet. FoMV-sg–mediated VIGS could possess a great potential for functional genomics in a variety of monocot crops.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Tobacco (Nicotiana benthamiana), barley (Hordeum vulgare L.; Ingrid), wheat (Triticum aestivum.; Zhoumai30), and foxtail millet (Setaria italica; Yugu1) plants were grown in pots at 22–24°C, in a growth chamber under a 16-h light/8-h dark cycle.
Construction of Foxtail mosaic virus-Derived Vectors
The full-length Foxtail mosaic virus (FoMV) cDNA was PCR-amplified using FoMV cDNA clone P9 (Robertson et al., 2000) as a template with primer pairs P1-F/P1-R, and cloned into StuI/XbaI-digested pYL44, a pBIN19-based T-vector containing a duplicated CaMV 35S promoter and nitric oxide synthase (NOS) terminator (Liu et al., 2002b), to generate pFoMV-wt. Further, DNA fragment 1 containing FoMV nt 4258–5371 (GenBank accession: EF630359) and DNA fragment 2 containing FoMV nt 5202–6008 were PCR-amplified using primer pairs P2-F/P2-R and P3-F/P3-R, respectively. SalI/AscI-digested DNA fragment 1 and AscI/AvrII-digested DNA fragment 2 were inserted into SalI/AvrII-digested pFoMV-wt to generate pFoMV-sg. The resulting vector pFoMV-sg is a pFoMV-wt derivative that contains a duplicated copy of 170-bp putative FoMV coat-protein subgenomic promoter and multiple cloning sites between the duplicated coat-protein subgenomic promoters.
Barley PDS fragment (corresponding to nt 345–546 of HvPDS; GenBank accession: AY062039) was PCR-amplified using barley cDNA as template and primer pairs P4-F/P4-R, and cloned into pFoMV-sg to generate FoMV-HvPDS200 for virus-induced gene silencing.
60-bp DNA fragments of barley PDS gene (corresponding to nt 1089–1148 of HvPDS) was RT-PCR-amplified using the primers P5-F/P5-R; 55-bp DNA fragments of barley ChlH gene (corresponding to nt 24–78 of HvChlH; GenBank accession: U26545) was RT-PCR-amplified using the primers P6-F/P6-R, and then PCR products were each digested with SpeI. The inverted-repeats of HvChlH or HvPDS were obtained by self-ligation of SpeI-cut PCR products, and further cloned into the AscI site of pFoMV-sg to generate FoMV-HvPDS or FoMV-HvChlH.
The fragments of wheat PDS and CLA1 genes were RT-PCR–amplified using a primer pair P7-F/P7-R for a PDS gene (corresponding to nt 423–482 of TaPDS; GenBank accession: FJ517553), and P8-F/P8-R for a CLA1 gene (corresponding to nt 1428–1487 of TaCLA; GenBank accession: AK335035). PCR products were digested with SpeI and inverted-repeats of TaPDS or TaCLA1 were obtained by self-ligation of SpeI-cut PCR products, and then cloned into the AscI site of pFoMV-sg to generate FoMV-TaPDS and FoMV-TaCLA1, respectively.
The fragments of foxtail millet PDS, CLA1, and IspH genes were RT-PCR-amplified using primer pairs P7-F/P7-R for PDS (corresponding to nt 558–617 of SiPDS; GenBank accession: XM004985406), and P9-F/P8-R for CLA1 (corresponding to nt 1511–1570 of SiCLA; GenBank accession: XM004962054), and P10-F/P10-R for IspH (corresponding to nt 364–423 of SiIspH; GenBank accession: XM004981826), and then PCR products were digested with SpeI. Inverted-repeats of SiPDS, SiCLA1, and SiIspH were obtained by self-ligation of SpeI-cut PCR products, and then cloned into the AscI site of pFoMV-sg to generate FoMV-SiPDS, FoMV-SiCLA1, and FoMV-SiIspH, respectively.
Agroinfiltration of N. benthamiana and Viral Inoculation of Cereals
FoMV-based VIGS was performed by using N. benthamiana as an intermediate host as described (Yu et al., 2003; Yuan et al., 2011). FoMV-based constructs were introduced into Agrobacterium tumefaciens strain GV3101. Agrobacterium was grown in Luria-Bertani medium containing rifampicin (30 mg l−1) and kanamycin (50 mg l−1) at 28°C for 2 d. Bacterial cells were harvested at 2500 g for 5 min, resuspended in infiltration medium (10 mm MgCl2, 10 mm MES [2-(n-morpholino) ethanesulfonic acid], and 0.2 mm acetosyringone) and adjusted to OD600 = 1.0, and kept at room temperature for 4 h. Agrobacterium were infiltrated into the fully expanded leaves of N. benthamiana using a 1-ml needleless syringe. The infiltrated leaves were harvested for 7–10 d post-infiltration, ground in 20-mm Na-P buffer (pH 7.2), and saps were mechanically inoculated onto leaves of the two-leaf stages of barley and wheat, and three-leaf stages of foxtail millet plants. Each experiment was repeated at least three times, with 20 plants for each viral construct in each experiment.
RNA Extraction and Reverse Transcription-PCR
Total RNA was extracted from plant leaf tissues using the Trizol reagent (Tiangen Biotech, Beijing, China), and then treated with RNase-free DNase I (Sigma-Aldrich, St. Louis, MO) to eliminate genomic DNA (Sigma-Aldrich). First-strand cDNA for RT-PCR was synthesized from 2 mg of RNA by RevertAid Reverse Transcriptase, with Oligo(dT), dNTP, and RiboLock RNase Inhibitors following the instruction of the manufacturer (Tiangen).
Real-time RT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, Cheshire, UK) following the manufacturer’s instructions. Gene-specific primers were designed to prime outside the region targeted for silencing. For barley PDS or ChlH, primer pairs P11-F/P11-R or P12-F/P12-R were used. Barley 18S rRNA gene (GenBank accession: AK251731) was used as an internal control using primer pairs P18-F/P18-R. For wheat PDS or CLA1, primer pairs P13-F/P11-R or P14-F/P14-R were used, respectively. Wheat 18S rRNA gene (GenBank accession: AY049040) was used as an internal control using primer pairs P18-F/P19-R. For foxtail millet PDS, CLA1, or IspH, primer pairs P15-F/P15-R, P16-F/P16-R, or P17-F/P17-R were used, respectively. Foxtail millet 18S rRNA gene (GenBank accession: KC996746) was used as an internal control using primer pairs P20-F/P20-R.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: FoMV (EF630359); HvPDS (AY062039); HvChlH (U26545); TaPDS (FJ517553); TaCLA (AK335035); SiPDS (XM004985406); SiCLA (XM004962054); SiIspH (XM004981826); Barley 18S rRNA (AK251731); Wheat 18S rRNA (AY049040); Foxtail millet 18S rRNA (KC996746).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. List of primers used in this work.
Supplementary Material
Acknowledgments
We thank Dr. Nancy Robertson (USDA, Palmer, Alaska) for kindly providing FoMV cDNA clone P9, and Professors Qianhua Shen and Zhiyong Liu for kindly providing plant seeds.
Glossary
- CP
coat protein
- ORFs
open reading frames
- VIGS
virus-induced gene silencing
Footnotes
Articles can be viewed without a subscription.
References
- Bancroft JB, Rouleau M, Johnston R, Prins L, Mackie GA (1991) The entire nucleotide sequence of foxtail mosaic virus RNA. J Gen Virol 72: 2173–2181 [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Becker A, Lange M (2010) VIGS—genomics goes functional. Trends Plant Sci 15: 1–4 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, Estep M, Feng L, Vaughn JN, Grimwood J, Jenkins J, Barry K, et al. (2012) Reference genome sequence of the model plant Setaria. Nat Biotechnol 30: 555–561 [DOI] [PubMed] [Google Scholar]
- Bruun-Rasmussen M, Madsen CT, Johansen E, Albrechtsen M (2008) Revised sequence of foxtail mosaic virus reveals a triple gene block structure similar to potato virus X. Arch Virol 153: 223–226 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39: 734–746 [DOI] [PubMed] [Google Scholar]
- Cakir C, Gillespie ME, Scofield SR (2010) Rapid determination of gene function by virus-induced gene silencing in wheat and barley. Crop Sci 50: S77–S84 [Google Scholar]
- Chen W, Kong J, Lai T, Manning K, Wu C, Wang Y, Qin C, Li B, Yu Z, Zhang X, He M, Zhang P, et al. (2015) Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci Rep 5: 7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck S, Van Montagu M, Depicker A (2001) Transgene silencing of invertedly repeated transgenes is released upon deletion of one of the transgenes involved. Plant Mol Biol 46: 433–445 [DOI] [PubMed] [Google Scholar]
- Ding SW. (2000) RNA silencing. Curr Opin Biotechnol 11: 152–156 [DOI] [PubMed] [Google Scholar]
- Ding XS, Schneider WL, Chaluvadi SR, Mian MA, Nelson RS (2006) Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol Plant Microbe Interact 19: 1229–1239 [DOI] [PubMed] [Google Scholar]
- Estévez JM, Cantero A, Reindl A, Reichler S, Le NP. 2001. 1-Deoxy-d-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem 276: 22901–22909. [DOI] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30: 315–327 [DOI] [PubMed] [Google Scholar]
- Huang C, Qian Y, Li Z, Zhou X (2012) Virus-induced gene silencing and its application in plant functional genomics. Sci China Life Sci 55: 99–108 [DOI] [PubMed] [Google Scholar]
- Kumagai MH, Donson J, della-Cioppa G, Harvey D, Hanley K, Grill LK (1995) Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci USA 92: 1679–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme C, Hrubikova K, Hein I (2003) Enhancement of virus-induced gene silencing through viral-based production of inverted-repeats. Plant J 34: 543–553 [DOI] [PubMed] [Google Scholar]
- Liou MR, Huang YW, Hu CC, Lin NS, Hsu YH (2014) A dual gene-silencing vector system for monocot and dicot plants. Plant Biotechnol J 12: 330–343 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002a) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002b) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for n-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Liu Z, Kearney CM (2010) An efficient Foxtail mosaic virus vector system with reduced environmental risk. BMC Biotechnol 10: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, León P (1996) CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J 9: 649–658 [DOI] [PubMed] [Google Scholar]
- Meng Y, Moscou MJ, Wise RP (2009) Blufensin1 negatively impacts basal defense in response to barley powdery mildew. Plant Physiol 149: 271–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming-Hsiun H, Goodman HM (2005) The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol 138: 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacak A, Geisler K, Jørgensen B, Barciszewska-Pacak M, Nilsson L, Nielsen TH, Johansen E, Grønlund M, Jakobsen I, Albrechtsen M (2010) Investigations of Barley stripe mosaic virus as a gene silencing vector in barley roots and in Brachypodium distachyon and oat. Plant Methods 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan M, Dinesh-Kumar SP (2009) Virus-induced gene silencing as a tool for delivery of dsRNA into plants. Cold Spring Harb Protoc 2009: pdb.prot5139. doi: 10.1101/pdb.prot5139 [DOI] [PubMed] [Google Scholar]
- Paulsen A, Niblett C (1977) Purification and properties of Foxtail mosaic virus. Phytopathology 67: 1346–1351 [Google Scholar]
- Pflieger S, Blanchet S, Camborde L, Drugeon G, Rousseau A, Noizet M, Planchais S, Jupin I (2008) Efficient virus-induced gene silencing in Arabidopsis using a ‘one-step’ TYMV-derived vector. Plant J 56: 678–690 [DOI] [PubMed] [Google Scholar]
- Purkayastha A, Mathur S, Verma V, Sharma S, Dasgupta I (2010) Virus-induced gene silencing in rice using a vector derived from a DNA virus. Planta 232: 1531–1540 [DOI] [PubMed] [Google Scholar]
- Ramanna H, Ding XS, Nelson RS (2013) Rationale for developing new virus vectors to analyze gene function in grasses through virus-induced gene silencing. Methods Mol Biol 975: 15–32 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Technical Advance: Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Robertson D. (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55: 495–519 [DOI] [PubMed] [Google Scholar]
- Robertson NL, French R, Morris TJ (2000) The open reading frame 5A of Foxtail mosaic virus is expressed in vivo and is dispensable for systemic infection. Arch Virol 145: 1685–1698 [DOI] [PubMed] [Google Scholar]
- Rohdich F, Hecht S, Gärtner K, Adam P, Krieger C, Amslinger S, Arigoni D, Bacher A, Eisenreich W (2002) Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc Natl Acad Sci USA 99: 1158–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Nelson RS (2009) Resources for virus-induced gene silencing in the grasses. Plant Physiol 149: 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS (2011) New dimensions for VIGS in plant functional genomics. Trends Plant Sci 16: 656–665 [DOI] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, Wang XL, Peng CC, Yu XC, Zhu SY, Fan RC, Xu YH, et al. (2006) The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826 [DOI] [PubMed] [Google Scholar]
- Short MN, Davies JW (1987) Host ranges, symptoms and amino acid compositions of eight potexviruses. Ann Appl Biol 110: 213–219 [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407: 319–320 [DOI] [PubMed] [Google Scholar]
- Tai YS, Bragg J, Edwards MC (2005) Virus vector for gene silencing in wheat. Biotechniques 39: 310–314 [DOI] [PubMed] [Google Scholar]
- Tang Y, Wang F, Zhao J, Xie K, Hong Y, Liu Y (2010) Virus-based microRNA expression for gene functional analysis in plants. Plant Physiol 153: 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnage MA, Muangsan N, Peele CG, Robertson D (2002) Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J 30: 107–114 [DOI] [PubMed] [Google Scholar]
- Vance V, Vaucheret H (2001) RNA silencing in plants—defense and counterdefense. Science 292: 2277–2280 [DOI] [PubMed] [Google Scholar]
- van den Boogaart T, Lomonossoff GP, Davies JW (1998) Can we explain RNA-mediated virus resistance by homology-dependent gene silencing? Mol Plant Microbe Interact 11: 717–723 [Google Scholar]
- Wang C, Chen J, Zhi H, Yang L, Li W, Wang Y, Li H, Zhao B, Chen M, Diao X (2010) Population genetics of foxtail millet and its wild ancestor. BMC Genet 11: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Yang X, Wang N, Liu X, Nelson RS, Li W, Fan Z, Zhou T (2016) An efficient virus-induced gene silencing vector for maize functional genomics research. Plant J 86: 102–115 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95: 13959–13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K, Han T, Liu G, Chen T, Wang Y, Yu AY, Liu Y (2015) A Geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep 5: 14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Fan B, Macfarlane SA, Chen Z (2003) Analysis of the Involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol Plant Microbe Interact 16: 206–216 [DOI] [PubMed] [Google Scholar]
- Yuan C, Li C, Yan L, Jackson AO, Liu Z, Han C, Yu J, Li D (2011) A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6: e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Bradshaw JD, Whitham SA, Hill JH (2010) The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol 153: 52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Ghabrial SA (2006) Development of bean pod mottle virus-based vectors for stable protein expression and sequence-specific virus-induced gene silencing in soybean. Virology 344: 401–411 [DOI] [PubMed] [Google Scholar]
- Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, Xie M, Zeng P, Yue Z, Wang W, Tao Y, Bian C, et al. (2012) Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol 30: 549–554 [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu Q, Hu P, Jia Q, Liu N, Yin K, Cheng Y, Yan F, Chen J, Liu Y (2016) An efficient Potato virus X -based microRNA silencing in Nicotiana benthamiana. Sci Rep 6: 20573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.