A WRKY transcription factor promotes root-to-shoot Fe translocation under Fe deficiency via direct regulation of a vacuolar Fe transporter gene in Arabidopsis.
Abstract
Iron (Fe) deficiency affects plant growth and development, leading to reduction of crop yields and quality. Although the regulation of Fe uptake under Fe deficiency has been well studied in the past decade, the regulatory mechanism of Fe translocation inside the plants remains unknown. Here, we show that a WRKY transcription factor WRKY46 is involved in response to Fe deficiency. Lack of WRKY46 (wrky46-1 and wrky46-2 loss-of-function mutants) significantly affects Fe translocation from root to shoot and thus causes obvious chlorosis on the new leaves under Fe deficiency. Gene expression analysis reveals that expression of a nodulin-like gene (VACUOLAR IRON TRANSPORTER1-LIKE1 [VITL1]) is dramatically increased in wrky46-1 mutant. VITL1 expression is inhibited by Fe deficiency, while the expression of WRKY46 is induced in the root stele. Moreover, down-regulation of VITL1 expression can restore the chlorosis phenotype on wrky46-1 under Fe deficiency. Further yeast one-hybrid and chromatin immunoprecipitation experiments indicate that WRKY46 is capable of binding to the specific W-boxes present in the VITL1 promoter. In summary, our results demonstrate that WRKY46 plays an important role in the control of root-to-shoot Fe translocation under Fe deficiency condition via direct regulation of VITL1 transcript levels.
Iron (Fe) is an essential microelement for both plants and animals. Despite its abundance in the soil, Fe is only slightly soluble under aerobic conditions, especially in high-pH and calcareous soils, which results in Fe deficiency (Kobayashi and Nishizawa, 2012). Fe deficiency affects plant growth and development, leading to reduction of crop yield and quality, and causing health problems to human beings.
To cope with Fe deficiency, plants have developed two main strategies for Fe uptake. Except the gramineae, which use the strategy II mechanism to take up Fe from the soil, all other plants acquire Fe via strategy I mechanism (Schmidt, 2003). In strategy I, two main processes are involved, including the reduction of ferric chelates at the root surface and the absorption of the generated ferrous irons across the root plasma membrane (Kobayashi and Nishizawa, 2012). The dominant genes responsible for these processes were first cloned from Arabidopsis (Arabidopsis thaliana), known as the ferric-chelate reductase oxidase gene FRO2 and the Fe-regulated transporter gene IRT1 (Eide et al., 1996; Robinson et al., 1999). The expression of both genes is induced by Fe deficiency and is tightly regulated at multiple levels (Connolly et al., 2002, 2003; Barberon et al., 2011, 2014; Shin et al., 2013; Ivanov et al., 2014).
After acquisition of Fe from soils, plants transport the Fe from root epidermal cells to other tissues. Fe translocation in plants involves various steps, including radial transport across the root tissues, xylem loading and unloading, xylem-to-phloem transfer, phloem transport, symplastic movement toward the site of demand, and retranslocation from source or senescing tissue (Kobayashi and Nishizawa, 2012). Among these processes, xylem loading plays an essential role in root-to-shoot Fe translocation. A few transporter genes involved in xylem Fe loading have been isolated so far in Arabidopsis. FERRIC REDUCTASE DEFFECTIVE3 (FRD3), encoding a member of MATE (multidrug and toxin efflux) transporter family, facilitates citrate efflux into xylem (Rogers and Guerinot, 2002; Durrett et al., 2007). Mutation of FRD3 results in Fe localization to the central vascular cylinder of the roots and failure to transport it to aerial parts (Green and Rogers, 2004). Arabidopsis ferroportin1/iron regulated 1 (AtFPN1/AtIREG1), a potential novel effluxer, is expected to be responsible for free Fe transport into xylem, although the transport activity for IREG1 has not been reported (Morrissey et al., 2009). Besides, a group of nodulin-like genes, whose expression is dramatically down-regulated by Fe deficiency, show high similarity of protein sequence to AtVIT1 (vacuolar Fe uptake transporter 1; Kim et al., 2006) and are thought to function in Fe homeostasis in root vascular tissue, thus controlling the distribution of Fe between roots and shoots (Gollhofer et al., 2011, 2014). Nevertheless, little is known about the molecular regulatory mechanism of Fe translocation in plants.
Transcriptional regulation of Fe deficiency-responsive genes has been intensively studied in the past decade. The key regulator gene in Arabidopsis is FIT1 (Fe-DEFICIENCY INDUCED TRANSCRIPTION FACTOR1, also known as FIT/FRU/bHLH29), which encodes a basic helix-loop-helix (bHLH) transcription factor (Colangelo and Guerinot, 2004). FIT1 is indeed an ortholog of the tomato (Solanum lycopersicum) gene FER that is the first identified regulator of Fe deficiency responses in nongraminaceous plants (Ling et al., 2002). FIT1 loss-of-function mutants show severe chlorosis and even die without excess Fe supply. While a great many Fe deficiency-inducible genes, including IRT1 and FRO2, have significantly decreased expression in fit1 mutant, it is not sufficient to induce the expression of these genes by only overexpressing FIT1 under Fe supply (Colangelo and Guerinot, 2004). Yuan et al. (2008) further demonstrated that FIT1 needed another two bHLH proteins, bHLH38 or bHLH39, to activate the Fe deficiency responses. bHLH38 and bHLH39 belong to the subgroup Ib bHLH genes, together with bHLH100 and bHLH101 (Wang et al., 2007). These four bHLH genes function redundantly in Fe deficiency responses, and each of them can interact with FIT1 at protein level (Wang et al., 2013). Furthermore, some other bHLH proteins, e.g. POPEYE (PYE) and its homologs, are capable of forming heterodimers and play an important role in the regulation of Fe homeostasis probably independent of FIT1-mediated signaling pathway (Long et al., 2010; Selote et al., 2015). This evidence reveals that the bHLH transcription factors have key roles in regulating Fe uptake and homeostasis in plants. Recently, the EIN3/EIL1 transcription factors in ethylene signaling pathway were shown to maintain FIT1 protein stability and enhance Fe deficiency response via interacting with FIT1 and the Mediator subunit MED16/25 complex (Lingam et al., 2011; Yang et al., 2014, 2016). Besides, two MYB transcription factors (MYB10/72) downstream of FIT1 are also involved in Fe deficiency response by regulating expression of the nicotianamine synthase gene NAS4 in Arabidopsis (Palmer et al., 2013). The question is raised if there is any other transcription factors involved in Fe deficiency responses.
WRKY proteins comprise one of the largest transcription factor families in plants (Eulgem et al., 2000). There are 72 WRKY members in Arabidopsis (Rushton et al., 2010). WRKY proteins contain one or two domains composed of the conserved amino acid sequence WRKYGQK, together with a zinc-finger-like motif (Ulker and Somssich, 2004). Using these domains, WRKYs can activate or repress transcription through directly binding to the W-box [core sequence: (T)(T)TGAC(C/T)] present in the promoters of target genes (Eulgem et al., 2000). WRKY transcription factors play important roles in a series of plant biological processes, including senescence, seed development, seed dormancy and germination, biotic and abiotic stress responses, etc. (Ulker and Somssich, 2004; Rushton et al., 2010). Recently, accumulating evidence has demonstrated that WRKY transcription factors also take part in the responses to nutritional stresses. For example, some WRKYs (WRKY6/42/45/75) regulate phosphate starvation responses in Arabidopsis (Devaiah et al., 2007; Chen et al., 2009; Wang et al., 2014; Su et al., 2015). Meanwhile, WRKY6 is involved in the response to boron deficiency (Kasajima et al., 2010). Another WRKY gene WRKY46 functions negatively in resistance to aluminum stress (Ding et al., 2013).
Interestingly, there are a number of W-boxes present in the promoters of many Fe deficiency-responsive genes (e.g. IRT1 and NAS4; Li et al., 2015), suggesting that WRKY transcription factor may play a role in Fe deficiency response. Here, we identified a WKRY gene, WRKY46, that is involved in the regulation of Fe translocation in Arabidopsis, through screening 72 WRKYs via gene expression and mutant analysis. WRKY46 has been reported to function in various processes, e.g. biotic and abiotic stress responses (Hu et al., 2012; Ding et al., 2013, 2014) and lateral root development (Ding et al., 2015), indicating WRKY46 as an important regulator in Arabidopsis. In this study, we discovered a previously unknown role for WRKY46 and demonstrated that WRKY46 promotes Fe translocation from root to shoot by suppressing the expression of a nodulin-like gene (VIT1-like1) under Fe deficiency.
RESULTS
WRKY46 Is Involved in Fe Deficiency Response
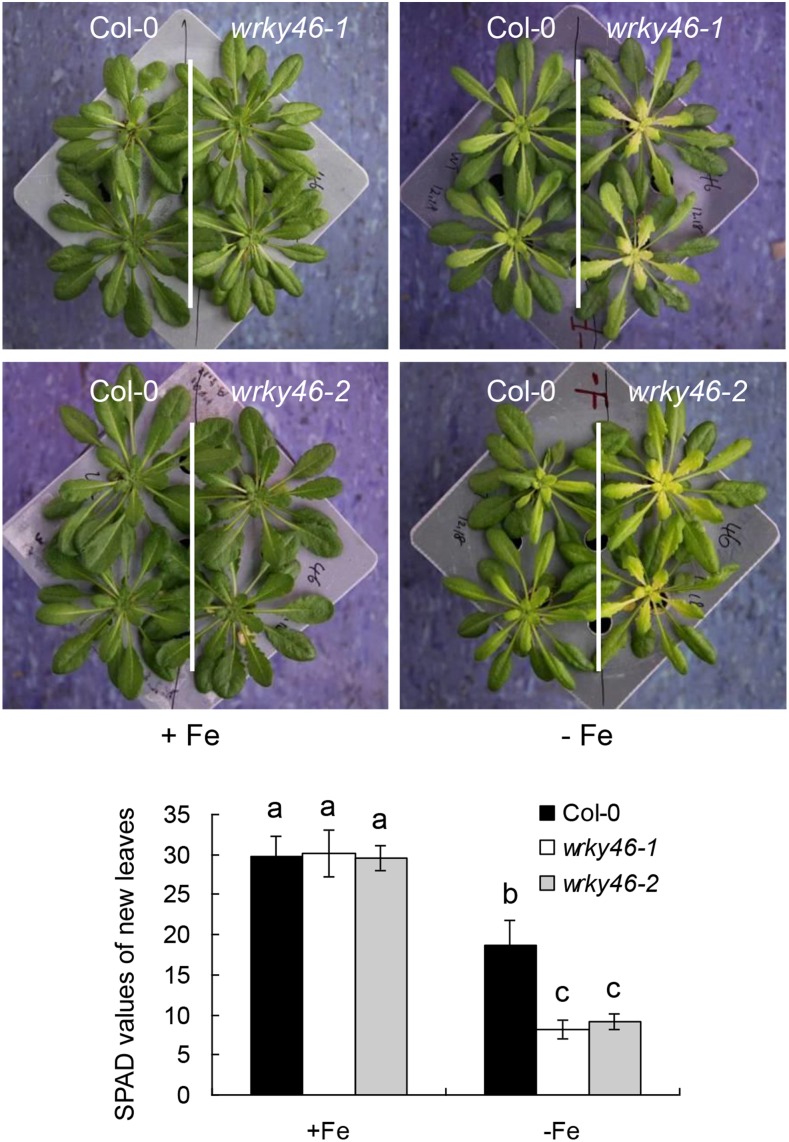
To evaluate the role of WRKY transcription factor in Fe deficiency response, we previously analyzed the expression pattern of WRKY genes that are present in the public microarray data sets under Fe deficiency treatment and found that five WKRYs are obviously induced by Fe deficiency (Supplemental Fig. S1). By further mutant analysis, we found that wrky46 is more responsive to Fe deficiency than other wrky mutants. To confirm the role of WRKY46 in Fe deficiency response, we used two independent WRKY46 T-DNA insertion mutant alleles, wrky46-1 and wrky46-2, as previously described (Ding et al., 2013). Six-week-old wild-type and wrky46 mutant plants grown in half-strength Murashige and Skoog (MS) nutrient solution were transferred into the same fresh solution with or without Fe supply for 6 d. There was no visible difference between the wild type and wrky46 mutants on growth when sufficient Fe was supplied. However, in Fe free condition, the shoots, especially the new leaves of both wrky46 mutants, were more chlorotic than those of the wild type (Fig. 1). This was also revealed by the corresponding SPAD (soil plant analysis development) values (Fig. 1), which indicate the chlorophyll content and leaf greenness. Actually, the leaves of wrky46 mutants became obvious chlorotic after 3 d of Fe deficiency treatment, while the wild-type leaves showed slight chlorosis even after 5 d of treatment. To validate the function of WRKY46, we further performed experiments with a complemented wrky46-1 mutant, which harbors a construct of WRKY46pro:WRKY46-GFP (as previously described; Ding et al., 2013). The complemented wrky46-1 mutant plants displayed similar phenotypes to those of the wild type in both normal and Fe-deficient conditions (Supplemental Fig. S2). Taken together, these results indicate that WRKY46 is involved in Fe deficiency response.
Figure 1.
Lack of WRKY46 shows increased sensitivity to Fe deficiency. Growth of 6-week-old wild-type and wrky46 loss-of-function mutant (wrky46-1 and wrky46-2) plants under normal and Fe deficiency treatments for 6 d. The chlorophyll content or leaf greenness in the new leaves was indicated as SPAD values. Three independent experiments were done, each containing at least 10 plants per genotype for one treatment. Data were analyzed by one-way ANOVA following Duncan’s test. Error bars with different letters represent a statistical difference (P < 0.05, Duncan’s test).
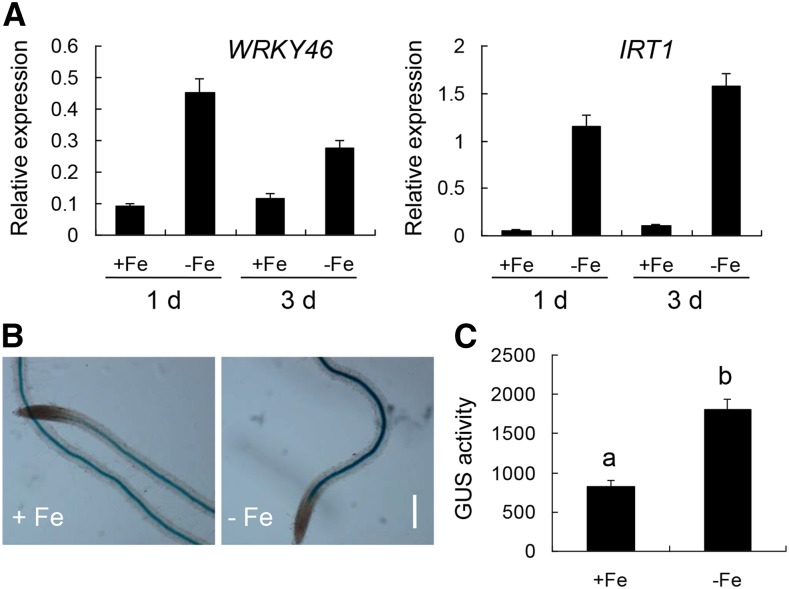
To verify if the expression of WRKY46 is truly responsive to Fe deficiency, we detected the expression by RT-qPCR. It was evident that the transcript level of WRKY46 was induced during different periods of Fe deficiency treatment (IRT1 was used as a positive control; Fig. 2A). In accordance with this, the expression and activity of GUS driven by WRKY46 native promoter were significantly increased in the root stele under Fe deficiency (Fig. 2, B and C). These results suggest that WRKY46 expression is responsive to Fe deficiency.
Figure 2.
WRKY46 expression is induced by Fe deficiency. A, Expression of WRKY46 and IRT1 in the wild type with or without Fe deficiency treatment. RNAs were extracted from roots of 6-week-old wild-type plants with or without Fe deficiency treatment for indicated days. Gene expression was determined by RT-qPCR using ACT mRNA as internal reference. B, Expression of WRKY46 in roots via GUS staining without or with Fe deficiency treatment for 2 d. One-week-old transgenic plants carrying a WRKY46pro:GUS construct were used for GUS staining. Bar = 100 μm. C, GUS activity measured in protein extracts from roots (in B) with or without Fe deficiency treatment. Activity units are given nmol methylumbelliferone (μg protein)−1 min−1. Three independent experiments were done. Data were analyzed by one-way ANOVA following Duncan’s test. Error bars with different letters represent a statistical difference (P < 0.05, Duncan’s test).
WRKY46 Regulates Root-to-Shoot Fe Translocation
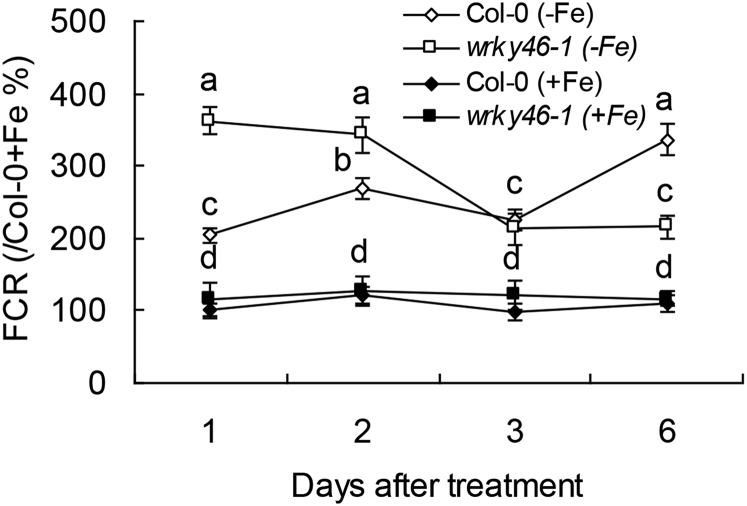
WRKY46 was previously demonstrated to modulate lateral root development under water stress via regulation of auxin levels in root (Ding et al., 2015). To confirm if the increased sensitivity of wrky46 mutants to Fe deficiency results from the lack of lateral roots in the mutants, we compared the development of lateral roots in wrky46 and wild-type plants and found that WRKY46 does not significantly affect lateral root development under Fe-deficient condition (Supplemental Fig. S3). Because auxin plays a potentially positive role in Fe deficiency responses (Chen et al., 2010), we also detected the effect of exogenous naphthylacetic acid (a permeable auxin analog) on Fe deficiency response of the wild type and wrky46-1 and found that naphthylacetic acid could not obviously rescue chlorotic phenotype on wrky46-1 mutant (Supplemental Fig. S4), which indicates that the increased sensitivity of wrky46-1 to Fe deficiency might not be related to auxin homeostasis in roots. To further determine how WRKY46 controls responses to Fe deficiency, we analyzed ferric chelate reductase (FCR) activity, the typical indicator of Fe deficiency. Both the wild type and wrky46-1 mutant increased FCR activity under different periods of Fe deficiency (versus Fe supplied condition; Fig. 3). There was no significant difference between the wild type and wrky46-1 in FCR activity when Fe was sufficient, while under Fe deficiency conditions, wrky46-1 had higher activity than the wild type under the first 2 d of treatment but had lower activity later at sixth day under Fe deficiency (Fig. 3). As wrky46-1 mutant always shows obvious chlorosis after 3 d of Fe deficiency treatment, the effect of wrky46 mutation on FCR activity thus seems to be more indirect.
Figure 3.
Effect of WRKY46 on the induction of FCR activity under Fe deficiency. Relative FCR activity (the data of Col-0 + Fe at 1 d were set as 100%) of wild-type and wrky46-1 mutant plants with or without Fe deficiency treatment for indicated days. Three independent experiments were done. Data were analyzed by one-way ANOVA following Duncan’s test. Error bars with different letters represent a statistical difference (P < 0.05, Duncan’s test).
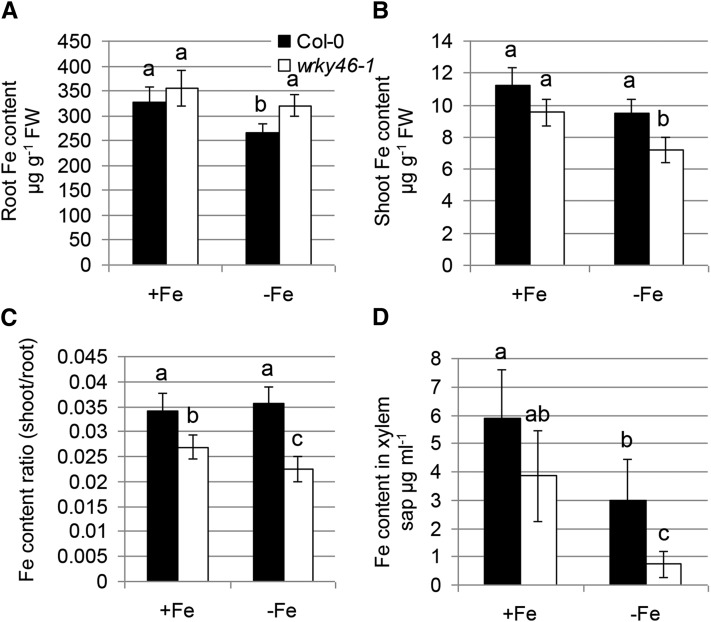
To detect how WRKY46 contributes to Fe deficiency responses, we further measured Fe content in both wild-type and wrky46-1 mutant plants with or without Fe supply. There was no significant difference between the wild type and wrky46-1 mutants in Fe content in both roots and shoots when Fe was sufficient. However, wrky46-1 mutants (versus the wild type) showed substantially higher Fe concentrations in roots but significantly lower in shoots under Fe deficiency (Fig. 4, A–C). Similarly, Perl staining revealed that wrky46-1 mutants contained visibly elevated amounts of ferric Fe in their roots compared with wild-type plants (Supplemental Fig. S5A). This indicates that WRKY46 may play a role in Fe translocation from root to shoot. Consistent with this, the Fe content was dramatically lower in xylem sap of wrky46-1 than in that of the wild type, especially under Fe deficiency (Fig. 4D).
Figure 4.
Lack of WRKY46 affects root-to-shoot Fe translocation. A and B, Fe content measured in roots (A) and shoots (B) of wild-type and wrky46-1 mutant plants with or without Fe deficiency treatment for 6 d. C, The ratio of shoot Fe content (in B) to root Fe content (in A). D, Fe content in xylem sap of wild-type and wrky46-1 mutant plants with or without Fe deficiency treatment. Three independent experiments were done. Data were analyzed by one-way ANOVA following Duncan’s test. Error bars with different letters represent a statistical difference (P < 0.05, Duncan’s test).
Plants commonly contain elevated amounts of Zn and Mn under Fe-deficient conditions (Vert et al., 2002) because some Fe transporters (e.g. IRT1) nonspecifically take up or transport these metals. Interestingly, wrky46-1 plants displayed decreased Zn and Mn levels in roots but increased that in shoots under both Fe-sufficient and -deficient conditions (Supplemental Fig. S5B), suggesting that the homeostasis of Zn and Mn is affected by altered translocation of Fe in wrky46-1 mutant.
Besides, we also measured Fe concentrations in young leaves and old leaves separately and found that lack of WRKY46 does not significantly affect the distribution of Fe between old and young leaves (Supplemental Fig. S6). Taken together, these results demonstrate that WRKY46 contributes to root-to-shoot Fe translocation under Fe-deficient conditions.
WRKY46 Regulates Expression of the Genes Involved in Fe Translocation
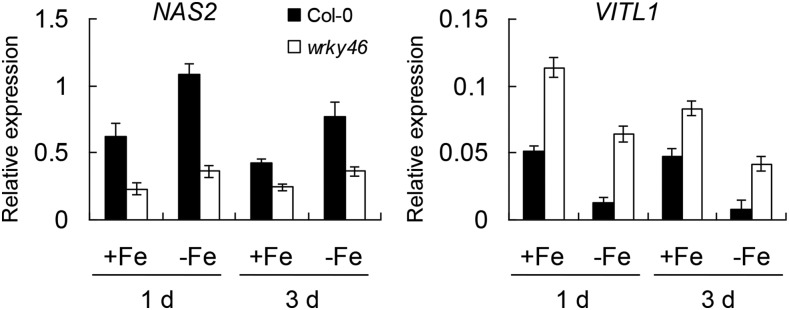
Previously, our microarray data revealed that the expression of two Fe translocation-related genes, NICOTIANAMINE SYNTHASE2 (NAS2) and VACUOLAR IRON TRANSPORTER1-LIKE1 (VITL1), was affected by WRKY46 overexpression (Ding et al., 2014). NAS2 encodes an enzyme that synthesizes nicotianamine and is responsible for Fe transport inside the body of plants (Schuler et al., 2012). VITL1 encodes a potential Fe transporter involved in Fe uptake into vacuole (Gollhofer et al., 2014) and is proposed to function in Fe homeostasis in vascular tissues (Gollhofer et al., 2011). To confirm whether these two genes (NAS2 and VITL1) are indeed regulated by WRKY46, we detected their expression in wild-type and wrky46-1 mutant plants under Fe-sufficient or -deficient conditions. RT-qPCR data revealed that NAS2 had obviously decreased expression in wrky46-1 (versus the wild type) under both Fe-sufficient and -deficient conditions, while VITL1 substantially increased expression in wrky46-1 plants (Fig. 5). Moreover, we determined the expression of other Fe deficiency-responsive genes in wild-type and wrky46-1 plants and found that most of these genes were not affected by loss of WRKY46 function, except IRT1 and FRO2, whose expression was slightly increased in wrky46-1 under Fe deficiency (Supplemental Figs. S7 and S8).
Figure 5.
WRKY46 regulates the expression of Fe translocation genes. Expression of NAS2 and VITL1 in wild-type and wrky46-1 mutant plants with or without Fe deficiency treatment. RNAs were extracted from roots of 6-week-old wild-type and wrky46-1 mutant plants with or without Fe deficiency treatment for indicated days. Gene expression was determined by RT-qPCR using ACT mRNA as internal reference. Three independent repeats were done with similar results. Error bars indicate sd (n = 3).
Lack of VITL1 Rescues Responses of wrky46-1 Mutant to Fe Deficiency
To verify if WRKY46 controls Fe translocation via the regulation of NAS2 or VITL1 expression, we first overexpressed NAS2 in wrky46-1 plants. The NAS2/wrky46 transgenic plants, which harbor high expression levels of NAS2, showed similar phenotypes as those of wrky46-1 mutants under Fe-deficient condition (Supplemental Fig. S9), indicating that NAS2 does not dominantly contribute to the regulation of Fe deficiency response by WRKY46.
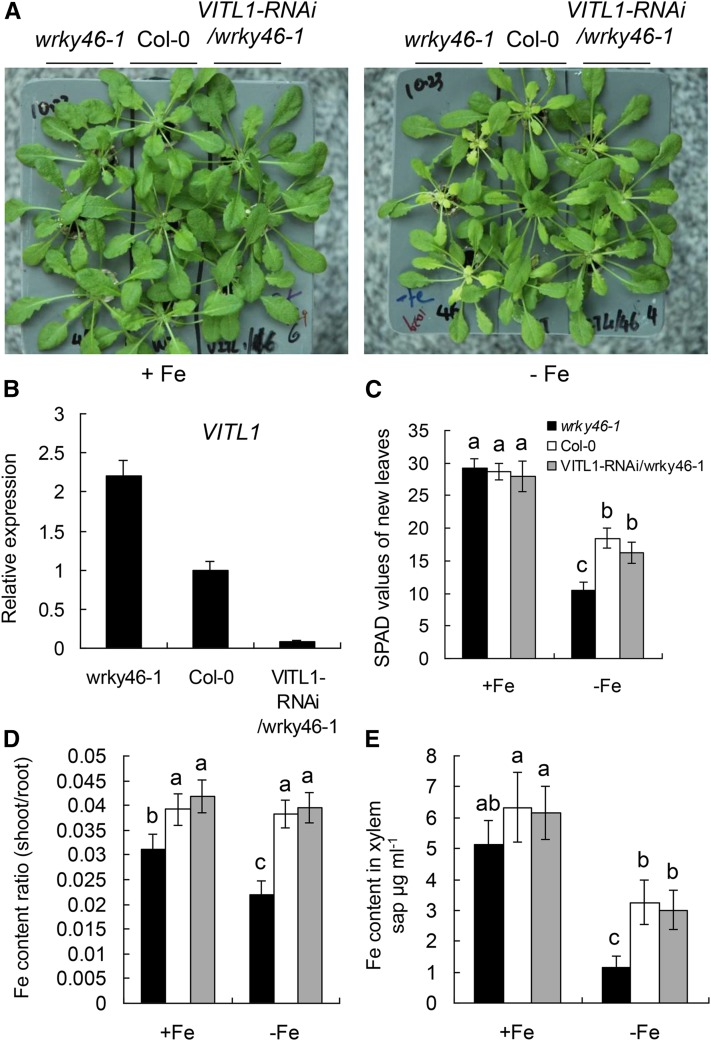
Next, we reduced the expression of VITL1 on the wrky46-1 mutant background using the RNA interference (RNAi) technique. The VITL1-RNAi/wrky46 transgenic lines displayed less chlorotic than wrky46-1 mutant plants under Fe deficiency at varying degrees, corresponding to the expression levels of VITL1 in each line (Supplemental Fig. S10). The lowest expression of VITL1 (in VITL1-RNAi/wrky46-1 plants) restored the Fe-deficient phenotypes of wrky46-1 mutant to that of the wild type (Fig. 6). These results suggest that VITL1 is the major contributor to Fe translocation regulated by WRKY46.
Figure 6.
Loss of VITL1 restores wrky46-1 Fe deficiency phenotype. A, Growth of 6-week-old indicated genotypes with or without Fe deficiency treatment for 6 d. B, Expression of VITL1 in roots of indicated genotypes. C, SPAD values of new leaves from indicated genotypes in A. D, Ratio of shoot-to-root iron content in indicated genotypes with or without Fe deficiency treatment (as in C). E, Fe content in xylem sap of indicated genotypes with or without Fe deficiency treatment. Three independent experiments were done. Data were analyzed by one-way ANOVA following Duncan’s test. Error bars with different letters represent a statistical difference (P < 0.05, Duncan’s test).
WRKY46 Binds to the Promoter Region of VITL1
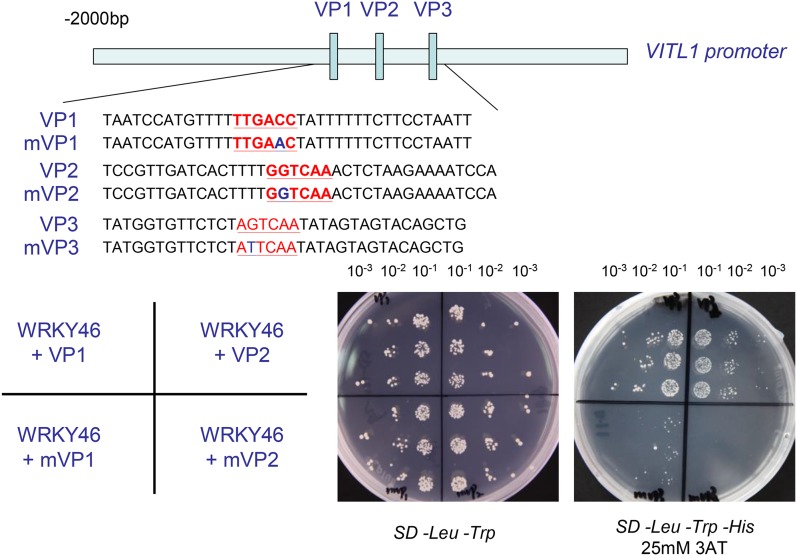
WRKY proteins recognize the W-boxes containing TTGACC/T core sequence present in the promoters of target genes. We found that there are three typical W-boxes in the VITL1 promoter (2-kb region), suggesting that WRKY46 may regulate VITL1 expression in a direct manner. To verify this possibility, three regions about 50 bp length surrounding each W-box, as shown in Figure 7, and the same regions with the mutant W-boxes were used as baits for binding assays in the yeast one-hybrid system (Fig. 7; Vidal and Legrain, 1999). The interactions between WRKY46 and these promoter fragments were tested by growth on media lacking Trp, Leu, and His. The His synthase inhibitor 3-aminotriazole (3AT) was added to the media to suppress background activation and assess the strength of the interaction. It was evident that yeast cotransformed with WRKY46 and the natural promoter regions (VP1 and VP2), but not the corresponding mutant fragments (mVP1 and mVP2), grew well in the selective media (Fig. 7). Besides, yeast cotransformed with WRKY46 and VP3 or mVP3 could not grow in the selective media. This indicates that WRKY46 is able to bind the specific regions (VP1 and VP2) in the VITL1 promoter through the W-boxes.
Figure 7.
WRKY46 binds to VITL1 promoter region in yeast. The 2-kb promoter region of VITL1 was indicated. The small blue boxes in the promoter represent three W-boxes (corresponding to the sequences in red). VP1-3 indicate the promoter fragments (about 50 bp) surrounding each W-box, and mVP1-3 indicate the mutated VP1-3 (with 1 bp mutation in each W-box), respectively. Yeast cells were cotransformed with a bait vector, containing a promoter fragment (VP1-3 and mVP1-3) fused to a HIS2 reporter gene, and a prey vector, containing WRKY46 fused to a GAL4 activation domain. Cells were grown in liquid media to OD600 of 0.1 (10−1) and diluted in a 10× dilution series (10−2 to 10−3). Of each dilution, 5 µL was spotted on media selecting for both plasmids (SD –Trp –Leu) and selecting for interaction (SD –Trp –Leu –His), supplemented with 25 mm 3AT to suppress background growth and test the strength of the interaction.
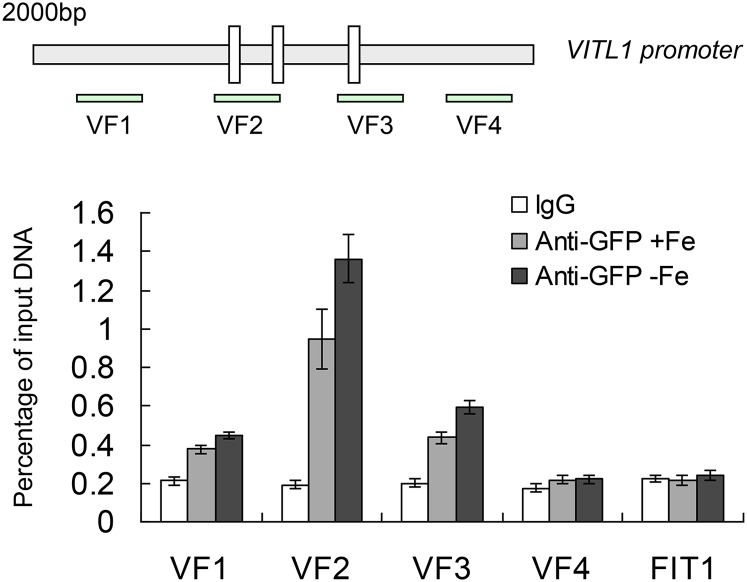
We further performed chromatin immunoprecipitation (ChIP) analysis using the WRKY46pro:WRKY46-GFP transgenic line under Fe-sufficient and -deficient treatments (Ding et al., 2013) to determine whether WRKY46 directly binds to the VITL1 promoter in vivo. After ChIP with anti-GFP antibody, the enrichment of specific VITL1 promoter fragments in the immune precipitant was determined by qPCR using four primer pairs (VF1-4; Fig. 8). The normal mouse lgG and a specific FIT1 promoter fragment containing the W-box were used as negative controls. We noticed that the VF2 region had a substantially stronger enrichment of WRKY46 than the negative controls, especially under Fe deficiency treatment, while VF1 and VF3 had little enrichment of WRKY46 (Fig. 8). These results suggest that WRKY46 regulates the expression of VITL1 by binding to its promoter.
Figure 8.
WRKY46 binds to VITL1 promoter in vivo. Diagram of VITL1 promoter showing three W-boxes. VF1-4 indicate genomic DNA fragments around the VITL1 promoter for ChIP assay. ChIP analysis showing the direct binding of WRKY46 to the VITL1 promoter in vivo. Eight-day-old WRKY46pro-WRKY46-GFP transgenic seedlings with or without Fe deficiency treatment were used for ChIP analysis. The lgG and a fragment of FIT1 promoter were used as negative controls. Three independent repeats were done with similar results. Error bars indicate sd (n = 3).
WRKY46 Acts Probably Independently of FIT1- or PYE-Mediated Signaling Pathway in Response to Fe Deficiency
The bHLH proteins, including FIT1 and PYE and their related bHLH transcription factors (e.g. bHLH38/39), have been considered as the key regulators of Fe uptake or homeostasis in Arabidopsis (Colangelo and Guerinot, 2004; Yuan et al., 2008; Long et al., 2010). Gene expression analysis revealed that the expression of FIT1 (as well as other bHLH genes) was not affected by loss of WRKY46 function (Supplemental Fig. S7). To further investigate the relationship between WRKY46 and FIT1 or PYE signaling, we detected the transcript levels of WRKY46 in the wild type, fit1 loss-of-function mutant, the transgenic plants simultaneously overexpressing FIT1 and bHLH38 (OX29/38) or FIT1 and bHLH39 (OX29/39), and pye-1 mutant plants under both Fe-sufficient and -deficient conditions. The qPCR revealed that WRKY46 had similar expression levels in these genotypes (fit1, OX29/38, OX29/39, and pye-1 versus the wild type; IRT1 was used as a positive control; Supplemental Fig. S11). Taken together, these results suggest that WRKY46 may act independently of FIT1 or PYE signaling in Fe deficiency responses.
DISCUSSION
Fe is an essential micronutrient for plants, and its acquisition is tightly controlled, as both deficient and excess Fe are pernicious for plants to grow and develop. The bHLH protein-dependent transcriptional regulation (FIT1- and PYE-dependent) plays a key role in the control of Fe uptake and homeostasis in plants (Colangelo and Guerinot, 2004; Long et al., 2010; Selote et al., 2015). Here, we show another kind of transcription factor that is from WRKY family participating in Fe deficiency response in Arabidopsis.
WRKY transcription factors have important roles in various plant biological processes, including developmental regulation and stress responses (Rushton et al., 2010). Accumulating evidence reveals that WRKY genes are also involved in plant responses to nutrient deficiency or toxicity. For example, a number of WRKYs regulate phosphate uptake and translocation in Arabidopsis (Devaiah et al., 2007; Chen et al., 2009; Wang et al., 2014; Su et al., 2015). Interestingly, W-boxes, the DNA binding sites of WRKY proteins, are enriched in the promoters of many Fe deficiency-responsive genes (Li et al., 2015). Although some rice (Oryza sativa) WRKY genes may play a role in response to Fe excess stress possibly through responding to disturbed redox homeostasis (Ricachenevsky et al., 2010), little is known about whether WRKY proteins take part in Fe deficiency responses. Among total WRKY genes present in public microarray data, there are five WRKYs that are responsive to Fe deficiency at transcript levels (Supplemental Fig. S1), including WRKY46. Our preliminary mutant analysis revealed that the wrky46 loss-of-function mutants show obvious chlorosis other than other wrky mutants under Fe deficiency conditions, indicating that WRKY46 may function specifically or have less redundancy with other WRKYs in Fe deficiency response. Further using independent wrky46 knockout mutants as well as the complemented wrky46-1 transgenic lines, we confirmed the response of WRKY46 to Fe deficiency (Fig. 1; Supplemental Fig. S2).
FCR activity is a typical indicator of Fe deficiency (Yi and Guerinot, 1996). Because the mutants lacking WRKY46 generally show visible chlorosis on new leaves in 3 d of Fe deficiency treatment, together with the evidence that WRKY46 is expressed in root stele (Fig. 2), the mutants displaying higher FCR activity (versus the wild type) at the early Fe deficiency treatment (for 1 and 2 d; Fig. 3) may suggest that Fe deficiency is more severe in the mutants during these periods and that lack of WRKY46 function has an indirect effect on FCR activity as well as on Fe uptake processes. Accordingly, the expression of Fe uptake related genes (IRT1 and FRO2) is higher in wrky46 mutants (versus the wild type; Supplemental Fig. S7). This can also be found in other Fe deficiency-responsive mutants. For example, frd3 mutants that are defective in Fe translocation from root to shoot resulted from the failure of loading of citrate into xylem (Green and Rogers, 2004) have higher levels of Fe deficiency responses (including FCR activity) under both Fe-sufficient and -deficient conditions (Rogers and Guerinot, 2002; Durrett et al., 2007). However, wrky46-1 mutants show a lower FCR activity than the wild type at the sixth day following onset of Fe deficiency treatment (Fig. 3), which might be due to more seriously secondary effects raised by long-term exposure to Fe deficiency stress in the mutants, although we can’t rule out the possibility that WRKY46 regulates Fe uptake in a non-cell-autonomous way.
Further analysis revealed that wrky46-1 mutants showed lower ratios of shoot-to-root Fe content (versus the wild type; Fig. 4, A–C) and that Fe concentrations in xylem sap were significantly lower in the mutants lacking WRKY46 than in the wild type (Fig. 4D). This suggests that wrky46 mutant plants had decreased capacity of root-to-shoot Fe translocation, which was potentially caused by the reduced loading of iron into the xylem in the mutants, thus indicating a role of WRKY46 in promoting root Fe transport to shoot. Furthermore, the content of other divalent metals (e.g. Zn and Mn) increased in shoots and decreased in roots, which was opposite to the change of Fe content in wrky46-1 mutants (versus the wild type; Supplemental Fig. S5B). Since the concentrations of these metals can efficiently reflect the Fe nutritional status (Baxter et al., 2008), such altered homeostasis of these metals (Zn and Mn) thereby again supported the contribution of WRKY46 to Fe translocation from root to shoot. But unlike in other research (Vert et al., 2002; Long et al., 2010), the total content of Zn and Mn (in whole plant) was not significantly increased in wrky46-1 mutants (versus the wild type) under Fe deficiency (Supplemental Fig. S5B). This might be because Fe deficiency in wrky46-1 was not severe enough to induce higher expression levels of IRT1 that is responsible for nonselective uptake of different kind of divalent metals in addition to Fe (Supplemental Fig. S7; Vert et al., 2002), thus making it unable to accumulate more non-Fe metals in wrky46 plants. Besides, the expression of WRKY46 in root stele also favors our conclusion that WRKY46 is involved in regulation of root-to-shoot Fe translocation under Fe-deficient condition (Fig. 2).
A small family of nodulin-like genes (including VITL1) that are induced by elevated Fe concentrations at transcript levels while inhibited by Fe deficiency encode putative cationic metal transporters and are thought to function in Fe homeostasis to protect plants from excess Fe stress (Gollhofer et al., 2011). Lack of some of these genes (e.g. Nodulin-like3/21) increased the ratios of shoot-to-root Fe content, suggesting that these nodulin-like genes are probably involved in Fe distribution between shoots and roots (Gollhofer et al., 2011). VITL1 (also named Nodulin-like1) localized to the vacuolar membrane and is responsible for Fe uptake into vacuoles (Gollhofer et al., 2011, 2014). We speculate that VITL1 is the major contribution to the root-to-shoot Fe translocation regulated by WRKY46. First, the transcript levels of VITL1 are obviously elevated in wrky46-1 mutants (versus the wild type) under both Fe-sufficient and -deficient conditions, and Fe deficiency inhibits VITL1 expression dramatically in the wild type but less in the mutants (Fig. 5). Consistent with this, WRKY46 is induced by Fe deficiency at transcript level, whereas VITL1 is down-regulated (Figs. 2 and 5; Gollhofer et al., 2011). Second, both WRKY46 and VITL1 are expressed in root stele as well as in vasculature tissues in shoots (Fig. 2; Gollhofer et al., 2011; Ding et al., 2014), suggesting they have similar expression patterns. Third, down-regulation of VITL1 significantly increases Fe concentrations in xylem sap of wrky46 mutants and thus restores Fe homeostasis and Fe deficiency response of wrky46 mutants to that of the wild type (Fig. 6), indicating that VITL1 is epistatic to WRKY46 in response to Fe deficiency. Finally, the yeast one-hybrid experiment and ChIP assay reveal that WRKY46 can bind to the promoter of VITL1 through specific W-boxes (Figs. 7 and 8), suggesting the regulation of VITL1 by WRKY46 is in a direct manner.
In summary, we established a role for WRKY46 in Fe deficiency responses. We present that WRKY46 is induced under Fe-deficient conditions and that it directly inhibits the transcription of VITL1, which is responsible for transport of iron into vacuoles in the root stele cells, thus promoting root-to-shoot Fe translocation by releasing more iron into root xylem. The function of WRKY46 is probably independent of the well-known bHLH transcription factors (FIT and PYE; Supplemental Fig. S11), suggesting that WRKY46 may contribute to another signaling pathway to complement the bHLH protein-mediated Fe deficiency responses. In addition, it’s interesting to find that WRKY46 expression is inhibited by aluminum stress (Ding et al., 2013), but induced by Fe deficiency (as shown in this article). This seems to be evolutionarily reasonable, as both of the stresses are present in the different kind of soil conditions respectively: Aluminum stress exists in acidic soils, while Fe deficiency usually can be seen in basic soils. It is economical and advisable for plants to have such genes (e.g. WRKY46) that simultaneously control multiple biological processes, making them adaptive to variable environments.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in a controlled environment growth room programmed for a 12-h-light/12-h-dark cycle with a constant temperature of 22°C. T-DNA insertion mutants wrky46-1 (SAIL_1230_H01) and wrky46-2 (SALK_134310C) were used as previously described (Ding et al., 2013). The fit1 mutants and OX29/38-5 and OX29/39-12 transgenic lines were obtained from Prof. Hongqing Lin (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences). pye-1 T-DNA insertion mutants (SALK_021217) were obtained from the ABRC. WRKY46pro:WRKY46-GFP was transformed into wrky46-1 plants, and WRKY46pro:GUS were transformed into wild-type (Col-0) plants, respectively (Ding et al., 2013). 35S:NAS2 was constructed in pCambia1301 vector; for VITL1-RNAi construction, the specific DNA fragments from VITL1 were first subcloned into pAGRIKOLA vector and then constructed into pCambia1301 vector driven by 35S promoter. Both of the 35S:NAS2 and VITL1-RNAi constructs were further transformed into wrky46-1 mutant plants through Agrobacterium tumefaciens GV3101, and homozygous lines were selected via screening on media containing hygromycin B and via further PCR-based confirmation. All genotypes used in this study have the Col-0 genetic background.
Plants were grown on modified half-strength MS agar medium or solution (pH 5.6) as described previously (Ding et al., 2014). For hydroponic Fe deficiency treatment, 6-week-old various genotypes grown in half-strength MS nutrient solution were transferred into the same fresh solution with or without Fe supply for different periods (3–6 d). The nutrient solution was changed every 2 d. For Fe deficiency treatment on agar plates, 3- to 8-d-old seedlings grown on half-strength MS medium were transferred onto the same fresh medium containing 300 μm ferrozine (an Fe chelator) for the indicated days of treatment.
Chlorophyll Content Analysis
Chlorophyll content was expressed in terms of SPAD value, which was determined on the fully expanded youngest leaves of seedlings with a portable chlorophyll meter (Konica Minolta SPAD-502).
FCR Activity Measurement
FCR activity was determined according to Lei et al. (2014). Briefly, whole excised roots were placed into a falcon tube filled with 20 mL of assay solution consisting of 0.5 mm CaSO4, 0.1 mm 4-morpho-lineethanesulfonic acid, 0.1 mm bathophenanthroline-disulfonic acid disodium salt hydrate (BPDS), and 100 mm Fe- EDTA at pH 5.5, adjusted with 1 m NaOH. The tubes were placed on a shaker (60 rpm) in a dark room at 25°C for 1 h. The absorbance of the assay solutions was recorded by a spectrophotometer (Shimadzu) at 535 nm, and the concentration of Fe(II)[BPDS]3 was calculated using an extinction coefficient of 22.14 mm−1 cm−1. The data were expressed as the means of relative root FCR activity.
Perl Staining
Perl’s staining for Fe3+ localization was performed according to Long et al. (2010). Briefly, 7-d-old seedlings grown on sufficient media were vacuum infiltrated with Perl’s stain solution (equal volumes of 4% [v/v] HCl and 4% [w/v] K-ferrocyanide) for 30 min. Seedlings were then incubated for another 30 min in Perl staining solution, washed three times with water, observed, and photographed with a stereoscope (Nikon) equipped with a CCD camera.
Fe Content Determination
Roots and shoots from treated plants were harvested separately, washed three times with ddH2O, weighed, and digested with HNO3:HClO4 (4:1, v/v). The total Fe concentrations were determined by inductively coupled plasma-atomic emission spectrometry (ICP optical emission spectrometry). The young leaves were indicated as half the number of total leaves that were counted outward from the center of the rosette. The rest of the rosette leaves were indicated as old leaves.
Xylem Sap Collection and Fe Content Measurement
Xylem sap was collected by excision of the hypocotyls below the rosette with a sharp scalpel as previously described (Schuler et al., 2012). The first droplet was discarded to minimize possible contamination. Then, a pipette tip (1–10 μL size) was carefully mounted over the stem and xylem sap was subsequently soaked into the mounted pipette tip due to capillary forces. The collection was carried out for 30 min, and the total volume was determined. Collected sap samples were later diluted in 5% HNO3 solution, and the Fe content was determined by ICP-OES.
GUS Analysis
GUS staining of 1-week-old transgenic lines with or without Fe deficiency treatment (on plate) for 2 d was performed by immersing seedlings in a staining solution [100 mm sodium-phosphate buffer, pH 7, 2 mm K4Fe(CN)6, 2 mm K3Fe(CN)6, 0.2% Triton X-100, 10 mm EDTA, and 2 mm X-Gluc] in a 10-mL tube for 12 h at 37 in the dark followed by two times washes with 70% ethanol to remove chlorophyll. Samples were photographed using a stereoscope (Nikon) equipped with a CCD camera.
For GUS activity quantification, the seedlings with or without Fe deficiency treatment for 2 d were collected and lysed for assays in a extraction buffer (50 mm NaH2PO4, pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 0.1% sodium lauryl sarcosine, and 10 mm β-mercaptoethanol) by freezing with liquid nitrogen and grinding with mortar and pestle. The GUS activity assay was performed using the substrate 4-methylumbelliferyl glucuronide (Sigma-Aldrich) and the standards of methylumbelliferone (Sigma-Aldrich) as previously described (Jefferson et al., 1987).
Gene Expression Analysis
For real-time qPCR, total RNAs from roots of various genotypes were extracted using the RNAprep pure Plant Kit (from TianGen Biotech Co. of Qiagen) according to the manufacturer’s protocol. Total RNAs treated with DNase I (TianGen Biotech Co. of Qiagen) were converted into cDNAs using the PrimeScript RT reagent kit (Takara). Real-time qPCR analysis was carried out using the SYBR Premix Ex Taq II (Takara) on a Roche LightCycler480 real-time qPCR system, following the manufacturer’s instructions. Transcript levels of each mRNA were determined and normalized with the level of ACT mRNAs using the delta Ct method (Czechowski et al., 2005; Schmittgen and Livak, 2008). Gene-specific primers are listed in Supplemental Table S1.
The expression browser tool provided by BAR (The BioArray Resource for Arabidopsis Functional Genomics; http://bbc.botany.utoronto.ca/; Toufighi et al., 2005) was employed to show the heat map of gene expression patterns in Supplemental Figure S1.
Yeast One-Hybrid Experiments
WRKY46 coding regions were amplified and cloned into the pGADT7-rec2 prey vector (Clontech), creating a translational fusion between the GAL4 activation domain and the transcription factor. For construction of the pHIS2 vector, forward and reverse oligonucleotides (Fig. 7) were annealed, digested by EcoRI/SacI, and subcloned into EcoRI/SacI-linearized pHIS2 vector. Competent yeast cells were prepared according to the Clontech Yeast Protocols Handbook using the Y187 yeast strain. For yeast transformation, 50 µL of competent yeast cells were incubated with 100 ng of pHIS2 bait vector and 100 ng of pGADT7-Rec2 prey vector, 50 µg salmon sperm Carrier DNA (Invitrogen), and 0.5 mL PEG/LiAc solution. Cells were transformed according to the manufacturer’s instructions. Transformations were plated onto SD media –Leu –Trp to select for cotransformed cells and incubated at 28°C for 4 d. Transformed yeast cells were subsequently grown in SD –Trp –Leu liquid media to OD600 of 0.1 and diluted in a 10× dilution series. From each dilution, 5 µL was spotted on SD –Trp –Leu and on SD –Trp –Leu –His media plates supplemented with 25 mm 3AT (Sigma-Aldrich). The plates were then incubated for 3 d at 28°C.
ChIP Assay
The EpiQuik Plant ChIP kit (Epigentek) was used to perform the ChIP assays. In brief, 0.8 to 1.0 g of 8-d-old Pro-46-WRKY46-sGFP seedlings with or without Fe deficiency treatment (on plate) for 2 d were fixed with 20 mL of 1.0% formaldehyde by vacuuming for 10 min. The chromatin DNA was extracted and sheared to 200- to 1000-bp fragments by sonicating. One hundred microliters of the sheared DNA was immunoprecipitated with 3 to 5 μg anti-GFP (Abcam) for 90 min at 50 to 100 rpm at room temperature. In addition, 1 µL of normal mouse IgG (Epigentek) was used as a negative control. DNA fragments that specifically associated with WRKY46 were released, purified, and used as templates for real-time qPCR using specific primers (Supplemental Table S1).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: WRKY46 (At2g46400), VITL1 (At1g21140), IRT1 (At4g19690), FRO2 (At1g01580), FIT1 (At2g28160), bHLH38 (At3g56970), bHLH39 (At3g56980), bHLH100 (At2g41240), bHLH101 (At5g04150), PYE (At3g47640), FRD3 (At3g08040), NAS1 (At5g04950), NAS2 (At5g56080), NAS4 (At1g56430), YSL1 (At4g24120), YSL2 (At5g24380), NRAMP3 (At2g23150), NRAMP4 (At5g67330), IREG1 (At2g38460), Nodulin-like2 (At1g76800), Nodulin-like3 (At3g43630), Nodulin-like4 (At3g43660), and Nodulin-like21 (At3g25190).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression pattern of WRKY genes in response to Fe deficiency revealed by BAR Expression Browser.
Supplemental Figure S2. Lack of WRKY46 affects sensitivity to Fe deficiency.
Supplemental Figure S3. Lack of WRKY46 does not significantly affect lateral root development in Fe deficiency condition.
Supplemental Figure S4. Auxin cannot rescue response of wrky46-1 to Fe deficiency.
Supplemental Figure S5. Microelements content analysis in wild-type and wrky46-1 mutant plants.
Supplemental Figure S6. WRKY46 does not significantly affect old to young leaves in Fe translocation.
Supplemental Figure S7. Expression analysis of Fe deficiency-responsive genes in wild-type and wrky46-1 mutant plants.
Supplemental Figure S8. Expression of nodulin-like genes in wild-type and wrky46-1 mutant plants.
Supplemental Figure S9. Increased expression of NAS2 can’t restore Fe deficiency phenotype on wrky46-1 mutant.
Supplemental Figure S10. Lack of VITL1 rescues wrky46-1 sensitivity to Fe deficiency.
Supplemental Figure S11. WRKY46 expression is not regulated by FIT1 or PYE transcription factors.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Prof. HongQing Lin (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for kindly providing the fit1 mutant and OX29/38-5 and OX29/39-12 transgenic line seeds.
Glossary
- SPAD
soil plant analysis development
- FCR
ferric chelate reductase
- 3AT
3-aminotriazole
- ChIP
chromatin immunoprecipitation
Footnotes
Articles can be viewed without a subscription.
References
- Barberon M, Dubeaux G, Kolb C, Isono E, Zelazny E, Vert G (2014) Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc Natl Acad Sci USA 111: 8293–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G (2011) Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter IR, Vitek O, Lahner B, Muthukumar B, Borghi M, Morrissey J, Guerinot ML, Salt DE (2008) The leaf ionome as a multivariable system to detect a plant’s physiological status. Proc Natl Acad Sci USA 105: 12081–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Yang JL, Qin C, Jin CW, Mo JH, Ye T, Zheng SJ (2010) Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis. Plant Physiol 154: 810–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML (2003) Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol 133: 1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding ZJ, Yan JY, Xu XY, Li GX, Zheng SJ (2013) WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J 76: 825–835 [DOI] [PubMed] [Google Scholar]
- Ding ZJ, Yan JY, Xu XY, Yu DQ, Li GX, Zhang SQ, Zheng SJ (2014) Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J 79: 13–27 [DOI] [PubMed] [Google Scholar]
- Ding ZJ, Yan JY, Li CX, Li GX, Wu YR, Zheng SJ (2015) Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J 84: 56–69 [DOI] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W, Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Gollhofer J, Schläwicke C, Jungnick N, Schmidt W, Buckhout TJ (2011) Members of a small family of nodulin-like genes are regulated under iron deficiency in roots of Arabidopsis thaliana. Plant Physiol Biochem 49: 557–564 [DOI] [PubMed] [Google Scholar]
- Gollhofer J, Timofeev R, Lan P, Schmidt W, Buckhout TJ (2014) Vacuolar-Iron-Transporter1-Like proteins mediate iron homeostasis in Arabidopsis. PLoS One 9: e110468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LS, Rogers EE (2004) FRD3 controls iron localization in Arabidopsis. Plant Physiol 136: 2523–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Dong Q, Yu D (2012) Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci 185-186: 288–297 [DOI] [PubMed] [Google Scholar]
- Ivanov R, Brumbarova T, Blum A, Jantke A-M, Fink-Straube C, Bauer P (2014) SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON-REGULATED TRANSPORTER1. Plant Cell 26: 1294–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasajima I, Ide Y, Yokota Hirai M, Fujiwara T (2010) WRKY6 is involved in the response to boron deficiency in Arabidopsis thaliana. Physiol Plant 139: 80–92 [DOI] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Lei GJ, Zhu XF, Wang ZW, Dong F, Dong NY, Zheng SJ (2014) Abscisic acid alleviates iron deficiency by promoting root iron reutilization and transport from root to shoot in Arabidopsis. Plant Cell Environ 37: 852–863 [DOI] [PubMed] [Google Scholar]
- Li H, Wang L, Yang ZM (2015) Co-expression analysis reveals a group of genes potentially involved in regulation of plant response to iron-deficiency. Gene 554: 16–24 [DOI] [PubMed] [Google Scholar]
- Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23: 1815–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J, Baxter IR, Lee J, Li L, Lahner B, Grotz N, Kaplan J, Salt DE, Guerinot ML (2009) The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell 21: 3326–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML (2013) MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet 9: e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricachenevsky FK, Sperotto RA, Menguer PK, Fett JP (2010) Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol Biol Rep 37: 3735–3745 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rogers EE, Guerinot ML (2002) FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 14: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Schmidt W. (2003) Iron solutions: acquisition strategies and signaling pathways in plants. Trends Plant Sci 8: 188–193 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schuler M, Rellán-Álvarez R, Fink-Straube C, Abadía J, Bauer P (2012) Nicotianamine functions in the Phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell 24: 2380–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA (2015) Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol 167: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin LJ, Lo JC, Chen GH, Callis J, Fu H, Yeh KC (2013) IRT1 degradation factor1, a ring E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 25: 3039–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Xu Q, Zhang FC, Chen Y, Li LQ, Wu WH, Chen YF (2015) WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol 167: 1579–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7: 491–498 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Legrain P (1999) Yeast forward and reverse ‘n’-hybrid systems. Nucleic Acids Res 27: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xu Q, Kong YH, Chen Y, Duan JY, Wu WH, Chen YF (2014) Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol 164: 2020–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908 [DOI] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ (2013) Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant 6: 503–513 [DOI] [PubMed] [Google Scholar]
- Yang Y, Li L, Qu LJ (2016) Plant Mediator complex and its critical functions in transcription regulation. J Integr Plant Biol 58: 106–118 [DOI] [PubMed] [Google Scholar]
- Yang Y, Ou B, Zhang J, Si W, Gu H, Qin G, Qu LJ (2014) The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J 77: 838–851 [DOI] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.