The tomato SlMYB12 transcription factor regulates flavonol glycoside biosynthesis and primary-secondary metabolism homeostasis.
Abstract
The identification and characterization of new tomato (Solanum lycopersicum) mutants affected in fruit pigmentation and nutritional content can provide valuable insights into the underlying biology, as well as a source of new alleles for breeding programs. To date, all characterized pink-pigmented tomato fruit mutants appear to result from low SlMYB12 transcript levels in the fruit skin. Two new mutant lines displaying a pink fruit phenotype (pf1 and pf2) were characterized in this study. In the pf mutants, SlMYB12 transcripts accumulated to wild-type levels but exhibited the same truncation, which resulted in the absence of the essential MYB activation domain coding region. Allelism and complementation tests revealed that both pf mutants were allelic to the y locus and showed the same recessive null allele in homozygosis: Δy. A set of molecular and metabolic effects, reminiscent of those observed in the Arabidopsis (Arabidopsis thaliana) myb11 myb12 myb111 triple mutant, were found in the tomato Δy mutants. To our knowledge, these have not been described previously, and our data support the idea of their being null mutants, in contrast to previously described transcriptional hypomorphic pink fruit lines. We detected a reduction in the expression of several flavonol glycosides and some associated glycosyl transferases. Transcriptome analysis further revealed that the effects of the pf mutations extended beyond the flavonoid pathway into the interface between primary and secondary metabolism. Finally, screening for Myb-binding sites in the candidate gene promoter sequences revealed that 141 of the 152 co-down-regulated genes may be direct targets of SlMYB12 regulation.
In Asia, consumption of pink-colored tomato (Solanum lycopersicum) varieties is high (Ballester et al., 2010; Lin et al., 2014) and their popularity is increasing worldwide. The pink fruit trait was described by Lindstrom (1925) in fruit with a transparent epidermis that lacks its natural yellow flavonoid compounds. Fruits from homozygous plants that contain the recessive “y” allele in the y locus in chromosome 1 failed to accumulate the yellow flavonoid precursor, naringenin chalcone, in the fruit peel, resulting in a pink-colored fruit phenotype (Lindstrom, 1925; Rick and Butler, 1956; Adato et al., 2009; Ballester et al., 2010). Flavonoids are class of phenolic compounds that have a broad range of ecological and physiological functions (Winkel-Shirley, 2001; Grotewold, 2006; Domínguez et al., 2009a, 2009b, 2011) and are widely distributed in the plant kingdom (Domínguez et al., 2011). There is interest in flavonoids due to their effect on fruit appearance and their nutritional and health-related properties (Rein et al., 2006; Butelli et al., 2008; Luo et al., 2008; Martin et al., 2011). In tomato fruit, they have also been reported to contribute to correct cuticle function as they affect its rigidity and susceptibility to cracking and, consequently, postharvest behavior (Domínguez et al., 2009a, 2011; Lara et al., 2014). Increasing the accumulation, or compositional diversity, of polyphenol species, including flavonoids, to improve fruit nutritional value and tolerance to environmental stress, is an important goal of biotechnology and breeding (Luo et al., 2008; Zhang et al., 2015). The reduced content of flavonoids in pink-fruited tomatoes may not be desirable in the context of human health, but it represents an interesting trait in terms of organoleptic fruit quality (e.g. color and texture). Moreover, pink-fruited tomatoes provide excellent material to study the molecular basis of the phenotype, investigate other processes that are affected by flavonoids, such as cuticle biosynthesis (Adato et al., 2009), and to identify additional candidate genes for breeding programs.
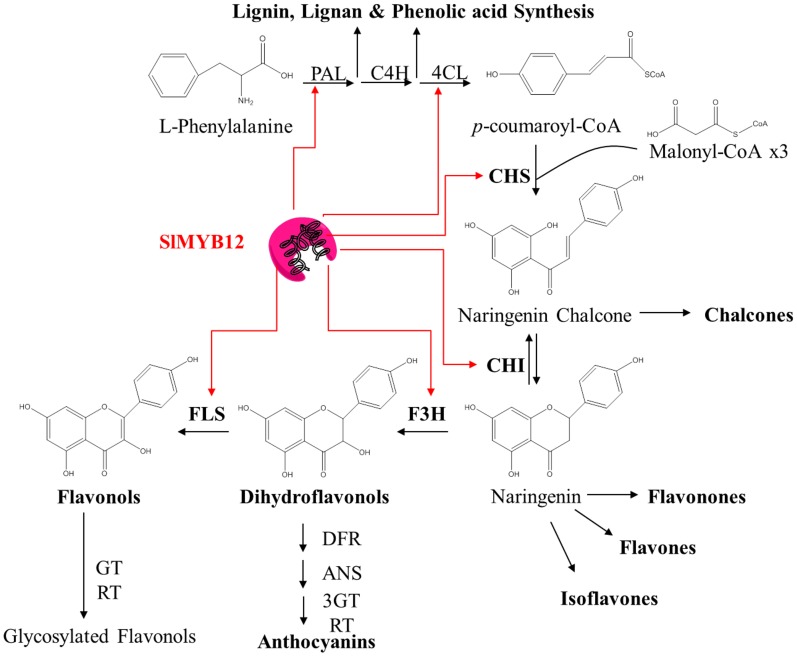
The biosynthesis of flavonoids has been studied thoroughly (Grotewold, 2006). Flavonoids comprise a group of polyphenols that are generated through the phenylpropanoid biosynthetic pathway. Phenylpropanoids (PPs) are derived from l-Phe, an amino acid synthesized by the shikimic acid and chorismic acid biosynthetic pathways. At the start of the PP pathway, Phe is converted to 4-coumaroyl-CoA, the precursor of flavonoid biosynthesis, by the successive activities of PHE AMMONIA-LYASE (PAL), CINNAMATE 4-HYDROXYLASE (C4H), and 4-COUMARATE:CoA LIGASE (4CL) enzymes. Next, CHALCONE SYNTHASE (CHS) produces naringenin chalcone by incorporating three molecules of malonyl-CoA into one molecule of 4-coumaroyl-CoA. A series of enzymatic steps then converts naringenin chalcone into different flavonoids (i.e. flavones, flavonones, isoflavones, dihydroflavonols, and flavonols), proanthocyanidins, and anthocyanins through distinct branches of the main pathway (Fig. 1). More than 70 different flavonoids have been identified in tomato fruit, including glycosylated and other conjugated forms (Moço et al., 2006; Mintz-Oron et al., 2008; Iijima et al., 2008; Adato et al., 2009; Ballester et al., 2010). Studies of different plants (Arabidopsis [Arabidopsis thaliana], maize [Zea mays], snapdragon [Antirrhinum majus], rose [Rosa spp.], gentian [Gentiana lutea], tomato, or grape [Vitis vinifera], among others) have revealed that R2R3-MYB-type transcription factors can regulate both CHS and CHALCONE ISOMERASE (CHI) gene expression, as well as that of many other genes in the PP, flavonoid, and anthocyanin biosynthetic pathways (Grotewold et al., 1994; Chopra et al., 1996; Kranz et al., 1998; Stracke et al., 2007; Adato et al., 2009; Czemmel et al., 2009; Ballester et al., 2010; Lin-Wang et al., 2010; Hichri et al., 2011; Du et al., 2012; Nakatsuka et al., 2012). Specific R2R3-MYB transcription factors (TFs) capable of regulating the expression of genes in the flavonoid branch are characterized by the presence of the SG7 motif (Kranz et al., 1998; Stracke et al., 2007; Czemmel et al., 2009), which is positioned downstream of the R2R3 DNA-binding domain. This group includes a set of flavonol-specific TFs, termed the SG7-2 subgroup, which also have a small motif at the C terminus that is able to recognize and bind specific domains in those genes involved in the flavonol biosynthetic branch (Mehrtens et al., 2005; Stracke et al., 2007; Czemmel et al., 2009). The most widely studied flavonol-specific regulator in the SG7-2 subgroup is MYB12. In Arabidopsis, MYB12 activates flavonol glycoside biosynthesis, and this function is shared by two paralogs, MYB111 and MYB11. These three TFs act in an additive manner and exhibit differential spatial activity (Stracke et al., 2007, 2010). They regulate flavonol moiety production, but characterization of the Arabidopsis myb11 myb12 myb111 triple mutant showed that their function includes controlling flavonol glycosylation (Stracke et al., 2007, 2010). Overexpression of AtMYB12 in tomato fruit resulted in an increase in levels of flavonol glycosides and caffeoylquinic acid (Luo et al., 2008; Pandey et al., 2015; Zhang et al., 2015). Moreover, overexpression of AtMYB11 in tomato fruit, driven by the E8 promoter, produced a specific increase in caffeoylquinic acid content (Li et al., 2015). In tomato, SlMYB12 has been described as the ortholog of AtMYB12 (Adato et al., 2009; Ballester et al., 2010), and the tomato AtMYB111 homolog, SlMYB111, may have a different function in tomato fruit (Adato et al., 2009). Thus, SlMYB12 appears to be the single MYB type regulator of flavonol biosynthesis in tomato, in contrast to the coordinated action of AtMYB11, AtMYB12, and AtMYB111 in Arabidopsis. To date, all studies have reported a reduction in flavonol content, but not in caffeoylquinic acids (Adato et al., 2009), associated with a decrease in SlMYB12 expression, suggesting that AtMYB12 overexpression in tomato might reveal effects on flavonoid biosynthesis regulation in the tomato fruit other than those occurring during the regulation of endogenous SlMYB12.
Figure 1.
Schematic overview of the flavonoid biosynthetic pathway and regulation in tomato fruit. The amino acid l-Phe is transformed into naringenin chalcone by the action of PAL, C4H, 4CL, and CHS enzymes. Naringenin chalcone is the precursor of different flavonoid classes: chalcones, flavonones, flavones, isoflavones, dihydroflavonols, and flavonols. SlMYB12 TF is the main activator of the flavonol biosynthetic branch to act on PAL, 4CL, CHS, CHI, F3H, and FLS genes (Adato et al., 2009; Ballester et al., 2010). 3GT, 3-O-GLYCOSYL TRANSFERASE; RT, RHAMNOSYL TRANSFERASE.
Although overexpression of CHI and silencing of CHS were reported to result in a reduction in naringenin chalcone levels in tomato fruit skin and in a pink fruit phenotype (Muir et al., 2001; Schijlen et al., 2007; Bovy et al., 2007), all pink fruit/y mutants characterized to date have been associated with low expression levels of SlMYB12 (described as the “y” locus in Ballester et al., 2010). The low number of SlMYB12 transcripts has been associated with the down-regulation of CHS, CHI, FLAVONOID 3-HYDROXYLASE (F3H), and FLAVONOL SYNTHASE (FLS) gene expression and in a partial block of flavonol biosynthesis (Fig. 1; Adato et al., 2009; Ballester et al., 2010). This is the underlying molecular basis of the pink phenotype of modern cultivars carrying the “y” mutation and of some introgression lines derived from Solanum chmielewskii in the cv Moneyberg background (Ballester et al., 2010), as well as the LA3189 line in the cv Ailsa Craig background (Adato et al., 2009). This is probably also the case for a large number of characterized pink accessions (Lin et al., 2014).
As a result of the economic and nutritional importance of flavonoids in tomato fruit, identifying new mutants, studying the genetic variability associated with the pink-fruit trait, and deciphering their biosynthetic regulatory network are important research targets. In this article, we characterized two pink-fruit tomato mutants by performing a metabolomic study using liquid chromatography coupled with mass spectrometry (LC-MS) and a gene expression analysis by next-generation sequencing (i.e. RNA-Seq). Combined with this, we also conducted an in silico search to identify potential target genes affected by this new pink fruit phenotype that may be of value in breeding programs.
RESULTS AND DISCUSSION
Morphological and Genetic Characterization of the New pink fruit Phenotype
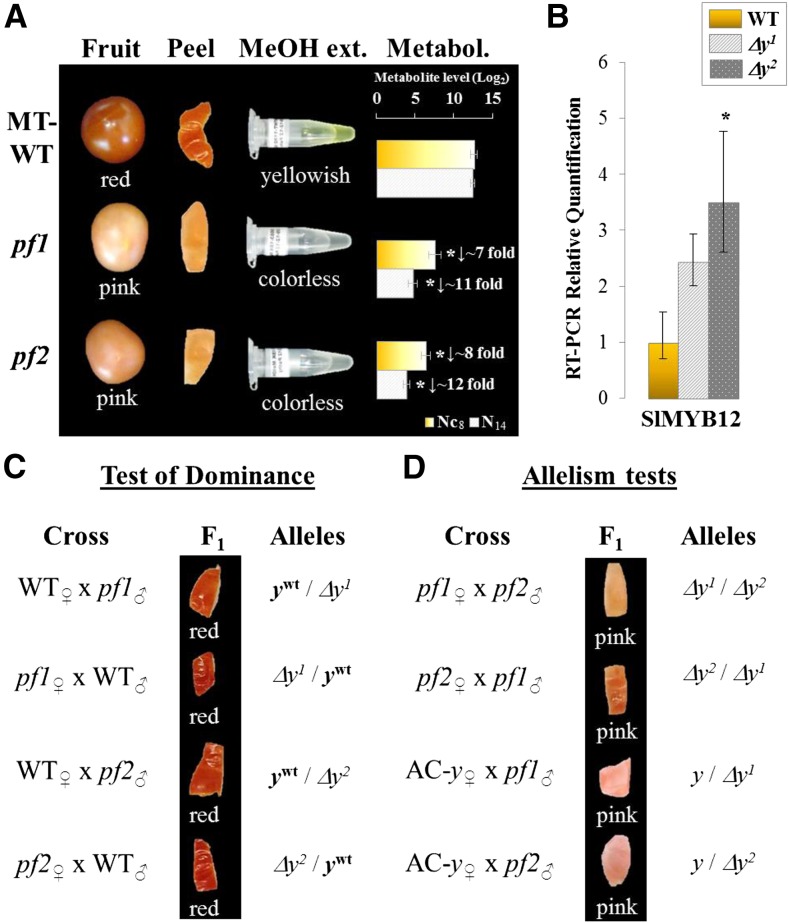
A phenotypic description of tomato mutants (cultivar Micro-Tom) available in the TOMATOMA database (Kazusa, Japan; http://tomatoma.nbrp.jp/), revealed two mutant lines showing pink-colored fruits (database numbers TOMJPG-4498 and TOMJPG-5663), which were named pink fruit1 (pf1) and pf2, respectively. We observed that the skin of both pf mutants showed no yellowish coloration, which was confirmed by methanolic extracts; the control extract was shiny yellow, but the pf mutant extracts were colorless and showed a reduced accumulation of naringenin chalcone (Fig. 2A). The visual pink fruit phenotype of the pf mutants prompted us to check for any alteration of SlMYB12 function. To determine SlMYB12 gene expression in the pf mutants, RT-PCR analysis was conducted of mRNA from the skin of breaker fruits. In contrast to previous studies of y fruits (Adato et al., 2009; Ballester et al., 2010; Lin et al., 2014), SlMYB12 transcripts showed equal, or even higher, expression levels in the pf mutants than in those of the wild-type fruit (Fig. 2B).
Figure 2.
The pink fruit phenotype in both the pf1 and pf2 mutants. A, The phenotype for tomato fruit, fruit skin, and methanolic extracts in both pf mutants and the wild type. Relative quantifications for both naringenin chalcone (Nc) and naringenin (N) metabolites (Metabol.) are also shown (see metabolites #8 and #14, respectively, in Supplemental Table S1). B, Gene expression level of SlMYB12 detected by RT-PCR analysis in both pf mutants and wild-type fruit skin. C, Allelism test of both pf mutants and the wild type. D, Complementation tests comparing pf1 and pf2 mutants, and both pf mutants and the y mutant (cv Ailsa Craig; AC-y). Alleles in C and D are based on molecular results (see about Δy further in the text). MeOH Extr., methanol extracts; ywt, wild-type allele for the y locus.
To define the pink fruit mutation in the new mutants, we performed both allelism and complementation diagnostic tests. The allelism test comparing the pf mutants and the wild type resulted in F1 individuals with only red fruit. This was consistent with the pink fruit mutation being caused by a recessive allele of a single gene in both new mutants (Fig. 2C). In addition, we performed complementation tests to compare the pf1 and pf2 mutants, and both pf mutants and the y mutant (Adato et al., 2009). These resulted in F1 individuals with only pink fruit that had a colorless skin (Fig. 2D). Hence, both allelism and complementation tests revealed that both new pink fruit mutants were carrying a recessive allele in the SlMYB12/y locus that did not affect SlMYB12 gene expression levels, in contrast with previously characterized y mutants (Adato et al., 2009; Ballester et al., 2010; Lin et al., 2014).
Molecular Characterization of New pink fruit Mutants Revealed a New Allele for SlMYB12: Δy
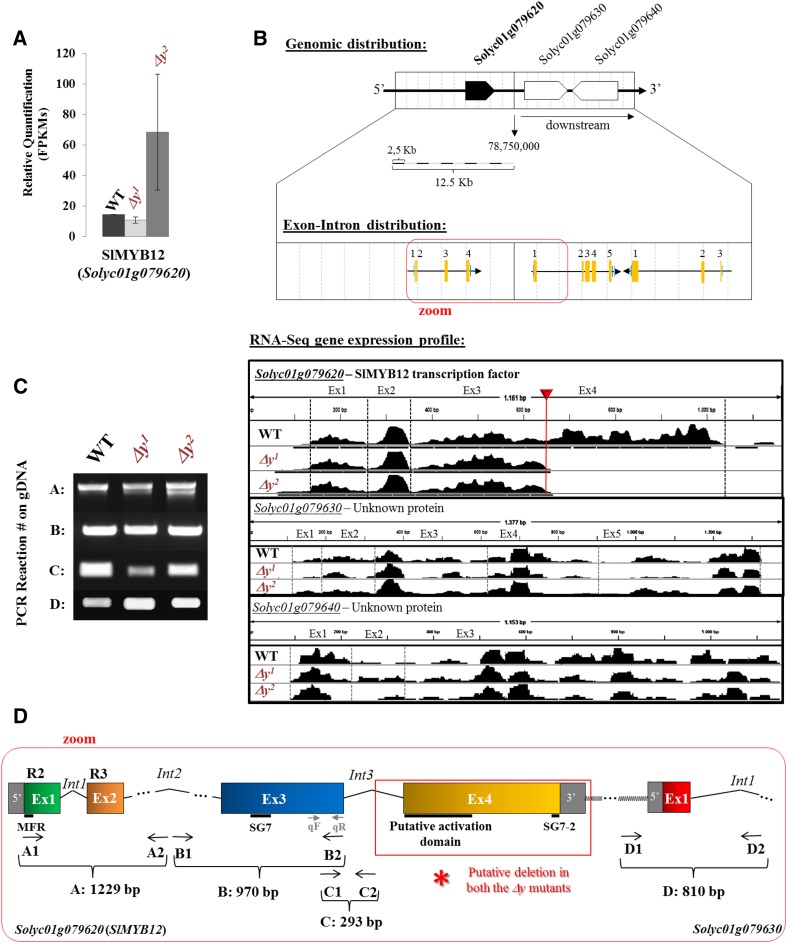
To determine the underlying molecular basis of the new pink fruit phenotypes, we performed RNA-Seq analysis of breaker stage fruit skins from both pf mutants. This confirmed that SlMYB12 was expressed at equal or slightly higher levels in the pf mutant fruits compared with the wild type (Fig. 3A). RNA-Seq read mapping revealed that SlMYB12 transcripts were truncated at the same position in both pf mutants (Fig. 3B). The missing region corresponded to exon 4, which starts after nucleotide 661 (see the red headed arrow in Fig. 3B). In that position, we detected a nucleotide change (A:T > G:C). To define this change at the DNA level in the two pf mutants, we amplified the SlMYB12 genomic region from exon 1 to the 3′-untranslated region (see zoom in Fig. 3, B and D). While we amplified exons 1 to 3 and the beginning of intron 3 (nucleotides 1 to 2,405; Fig. 3, C and D), all attempts to amplify the 3′ end of the gene from the pf mutants, or to cover the gap, were unsuccessful (Fig. 3D). These results suggested that the same deletion, between intron 3 and the end of the gene, was present in the two pf mutants, in contrast to other y mutants described to date (i.e. alterations in the promoter region [Lin et al., 2014] or in exon 3 [Adato et al., 2009; Ballester et al., 2010]). Neighbor genes located between the 5 kb upstream (i.e. Solyc01g079610, Solyc01g079600, and Solyc01g079590) and 3 kb downstream (i.e. Solyc01g079630, Solcy01g079640, Solyc01g079650, and Solcy01g079660) genomic regions of SlMYB12 showed almost unaltered RNA-Seq mapping profiles (Supplemental Fig. S1A), indicating that they were not affected by the deletion in MYB12. PCR analysis of the 5′-untranslated region and the first exon and intron of the Solyc01g079630 gene, located immediately downstream of SlMYB12, showed that Solyc01g079630 expression was similar in both pf mutants and the wild type (Fig. 3C). These results suggested that both pf mutants were probably derived from a single initial line and were affected by the same mutagenic event. Thus, this initial pink fruit mutant line containing the truncated SlMYB12 may have been inadvertently segregated in two independent lines during generation of the M3 lines. Therefore, this new Slmyb12 allele was named Δy, and both pf1 and pf2 mutants were renamed Δy1 and Δy2 (see “alleles” in Fig. 2, C and D), although we believe that Δy1 and Δy2 are probably the same mutation.
Figure 3.
The Slmyb12-Δy allele in the pf mutants. A, SlMYB12 gene expression levels (FPKMs) in RNA-Seq analysis for both Δy mutants and the wild type. B, SlMYB12 genomic region and RNA-Seq read mapped gene expression profile. The two genes placed downstream of SlMYB12 in the genome are also shown. C, SlMYB12 PCR amplicons using genomic DNA (gDNA) templates from the wild type, Δy1, and Δy2 mutants. See both size and location of the PCR reactions in D. D, SlMYB12 genomic sequence (available in tomato database: http://www.solgenomics.net/) and the PCR/RT-PCR oligonucleotide distribution in the gene sequence. The location of the putative deletion in SlMYB12 is indicated by a red asterisk. MFR, Myb-like flavonol regulation motif; SG7, subgroup 7 with the flavonol regulation motif; SG7-2, subgroup 7-2 with the specific flavonol regulation motif.
To further investigate the effects of the Δy allele, we considered the different domains and motifs in the SlMYB12 protein. Exons 1 and 2 corresponded to the R2 and R3 DNA-binding domains described for a typical MYB R2R3-type TF protein (Fig. 3D; Supplemental Fig. S2). At the N terminus, immediately upstream of the R2 motif, we found a well-conserved motif for the flavonol and proanthocyanidin regulator group (Czemmel et al., 2009). Exon 3 contained a flavonol regulation motif, which was shared by the R2R3-type TFs belonging to subgroup 7 (SG7), as described in Arabidopsis (Kranz et al., 1998; Stracke et al., 2007; Czemmel et al., 2009). Finally, exon 4 of the wild-type version of SlMYB12 encoded a putative activation domain (defined by a negatively charged amino acid-rich sequence; Chopra et al., 1996) and a very well-conserved flavonol-specific regulation motif (SG7-2; Czemmel et al., 2009) located at the C terminus of the protein. Thus, the Δy mutants were predicted to lack the activation domain and SG7-2 motif coding motifs. This suggested that the Δy mutant could be considered a Slmyb12 null mutant as the absence of the SG7-2 motif in the truncated protein renders it unable to recognize and activate the target genes.
Accumulation of Glycosylated Flavonols Is Diminished in the Δy Mutants
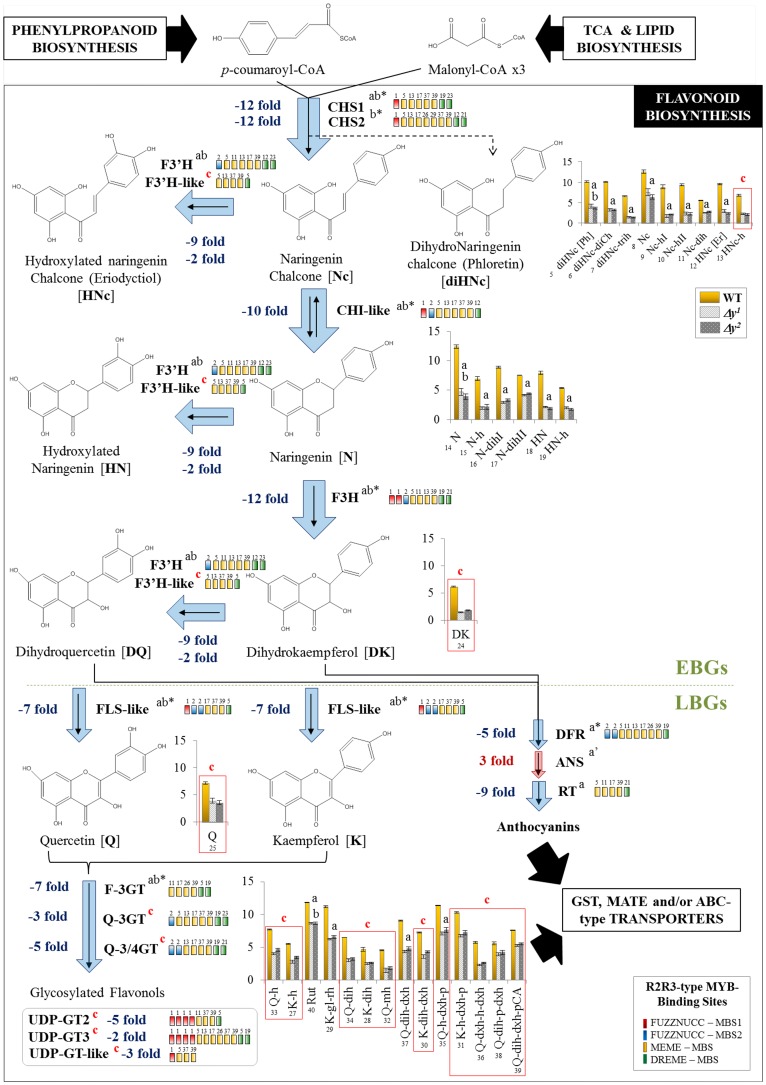
Although skin color was similar in the new and previously described pink fruit mutants, the novel nature of the Δy allele led us to investigate possible consequences on metabolite profiles. LC-MS analysis revealed 40 PP compounds that were differentially accumulated in the fruit skin between the Δy mutants and the wild type (Supplemental Table S1). As was reported to be the case with other y fruits (Adato et al., 2009; Ballester et al., 2010), levels of naringenin, naringenin chalcone, and their hydroxylated (i.e. phloretin and eriodyctiol) and glycosylated derivatives were reduced in abundance in both Δy mutants (Fig. 4; Supplemental Table S1). However, we also observed reduced levels of dihydrokaempferol, quercetin, and 13 glycosylated flavonol derivatives that contained one to four glycosyl groups in the Δy mutants. Of these 15 compounds, only four were previously reported as showing altered abundance in y fruits compared with the wild type: rutin, quercetin-dihexose-deoxyhexose, quercetin-hexose-deoxyhexose-pentose, and kaempferol-Glc-rhamnose (see 40, 37, 35, and 29, respectively, in Supplemental Table S1). Furthermore, we found a greater than 7-fold reduction in the levels of two phenolic acids derived from the l-Tyr pathway (hydroxycinnamic acid-hexose and hydroxybenzoic acid-hexose); these were also described as having altered levels in another y mutant, but not to such a great extent (Adato et al., 2009). Finally, we found increased levels of l-Phe in the pf mutants compared with the wild type (Fig. 4; Supplemental Table S1), which again differed from what was previously reported for other y mutants.
Figure 4.
Flavonol biosynthesis is blocked in the Δy mutants. Genes differentially expressed in the Δy mutants are indicated by blue (down-regulated) or red (up-regulated) arrows (thicker arrows represent a higher-fold change). The MBS motifs found in the promoter region of the down-regulated genes are also included (in colored boxes). Bar graphs show the metabolite levels for flavonols. The wild type is represented in yellow (left), the Δy1 mutant in dashed gray (middle), and the Δy2 mutant in dotted gray (right). Error bars are standard deviations (n = 3). Genes and metabolites differentially expressed also in the y mutant and in the other y lines are marked “a” and “b,” respectively, and those differentially expressed only in the Δy mutants are marked “c” (in red). Numbers in bar graphs correspond to metabolites in Supplemental Table S1. EBGs, early biosynthetic genes; N, naringenin; Nc, naringenin chalcone; HNc, hydroxylated naringenin chalcone (eriodictyol-chalcone); diHNc, dihydronaringenin chalcone (phloretin); HN, hydroxylated naringenin (eriodictyol); DK, dihydrokaempferol; DQ, dihydroquercetin; K, kaempferol; Q, quercetin; H, hexose; dxh, deoxyhexose; P, pentose; gl, Glc; rh, rhamnose; mh, malonyl-hexose; pCA, p-coumaric acid; Rut, rutin; a’, the ANS gene was down-regulated in the y mutant.
A Complex Alteration in the Gene Expression Program Associated with the new pink fruit Phenotype Is Identified in the Δy Mutants
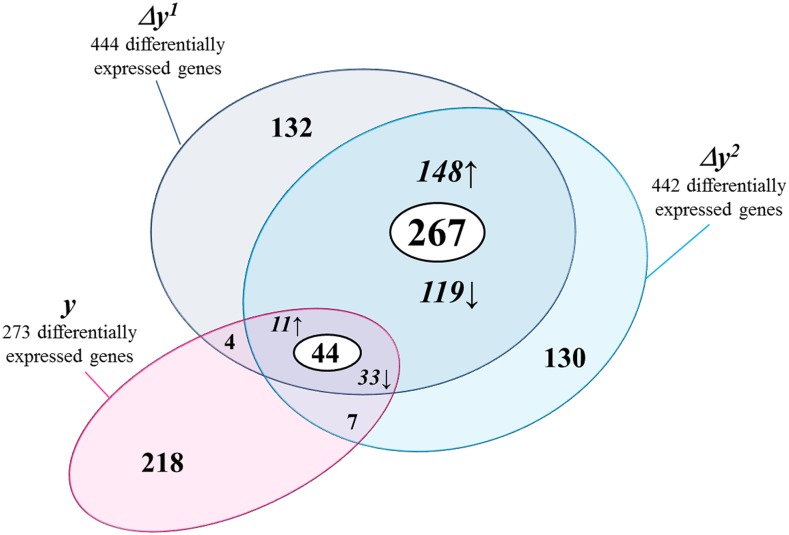
Since the PP metabolite profiles of the Δy mutants and the previously described y mutants clearly differed, we used RNA-Seq transcriptome analysis to elucidate the underlying molecular basis of the Δy mutant metabolic phenotype. Gene expression analysis revealed that 444 genes were differentially expressed in the skin of the Δy1 mutant and 442 in the Δy2 mutant. Of these, 311 showed similar patterns of altered expression in both Δy mutants when compared with the wild type (Fig. 5), and, of these, 159 were up-regulated and 152 down-regulated in the mutants. These differences could be explained by the putative segregation of the initial line containing the truncated Slmyb12, which may have affected different cosegregating mutations in each Δy mutant line. Our RNA-Seq analysis detected an ∼ 2-fold increase in the number of differentially expressed genes in the Δy mutants compared to the 273 differentially expressed genes in the y mutants using Affymetrix microarray technology (Adato et al., 2009). Only 14% (44) of the 311 genes were shared by both the Δy and y mutants (Fig. 5; Supplemental Table S2). Among the remaining 267 genes, we found some had a functional annotation similar to that shown to be differentially expressed in the y mutant but with a different tomato gene ID (http://www.solgenomics.net/). This could be explained by differences in the profiling technologies and by the genetic backgrounds used for the y and Δy mutant experiments (cultivars Ailsa Craig and MT, respectively). For detailed analysis, we focused on the 311 differentially expressed genes shared by both Δy1 and Δy2 mutants, including the 44 genes reported in previous y mutant studies.
Figure 5.
Genes differentially expressed in the Δy mutants detected by the RNA-Seq approach. Venn diagram including all genes differentially expressed in mutant Δy1 and Δy2 (dark-blue and light-blue circles, respectively). Those genes also detected as being differentially expressed in the y mutant (pink circle) by the Affymetrix microarray approach were also included. The genes shared between mutant Δy1 and Δy2, and between both the Δy mutants and the y mutant, were represented in a white circle. Shared up-regulated genes (↑) and shared down-regulated genes (↓) are also represented.
The Flavonoid Biosynthetic Pathway Was Severely Blocked
Expression of early flavonoid biosynthetic genes, namely, CHS1 (Solyc09g091510), CHS2 (Solyc05g053550), CHI-like (Solyc05g052240), and F3H (Solyc02g083860), was considerably reduced (i.e. 10- to 12-fold) in both Δy mutants (Fig. 4; Supplemental Table S3A). F3′H (Solyc03g115220) and F3′H-like (Solyc12g088460), encoding FLAVONOID 3′ HYDROXYLASE, were also down-regulated in both mutants (9- and ∼2-fold reductions, respectively). However, we also found that some late flavonoid biosynthetic genes (LBGs) were differentially expressed in the Δy mutants. These belong to both the flavonol biosynthetic branch (a highly reduced FLS-like gene, Solyc11g013110) and the proanthocyanidin biosynthetic branch (a highly reduced DIHYDROFLAVONOL REDUCTASE [DFR; Solyc02g089770] and a slightly increased ANTHOCYANIDIN SYNTHASE [ANS; Solyc10g076660]). The above-described early flavonoid biosynthetic genes and the LBGs were also down-regulated in the previously described y lines (Adato et al., 2009; Ballester et al., 2010). Interestingly, and similarly to what was observed in Δy, the proanthocyanidin LBGs, DFR and ANS, were also co-down-regulated in the y mutants. DFR (Solyc02g089770) was annotated as CCR-like by Adato et al. (2009) given its similarity with an Arabidopsis CCR-related gene, but has recently been annotated as DFR in tomato databases (http://www.solgenomics.net/; Supplemental Fig. S3). Finally, four flavonoid GLYCOSYL TRANSFERASES (GTs), probably involved in the decoration of the flavonoid aglycones, showed highly reduced expression levels in the Δy mutants (Fig. 4; Supplemental Table S3A), including three FLAVONOL 3-GTs, i.e. F-3GT (Solyc10g083440), QUERCETIN 3-GT (Q-3GT; Solyc01g107780), and Q-3/4GT (Solyc02g081690), and an ANTHOCYANIN 3-GT (RT; Solyc09g059170). We also found three additional GTs that were down-regulated in the Δy mutants, UDP-GT2 (Solyc11g010760), UDP-GT3 (Solyc03g078770), and UDP-GT-like (Solyc05g053890), which are probably related to glycosylation of flavonoids prior to their transport to the vacuole. Of these six GTs, only F-3GT has previously been reported as being down-regulated in y mutants (Adato et al., 2009).
In Arabidopsis, PP pathway regulation (including flavonol biosynthesis) is controlled by three genes: AtMYB12, AtMYB111, and AtMYB11 (Stracke et al., 2007). Each of these TFs has redundant activity in different organs (Stracke et al., 2007, 2010; Franco-Zorrilla et al., 2014; Li et al., 2015). Thus, the Arabidopsis myb11 myb12 myb111 triple mutant is described as a null mutant for the regulation of PP biosynthesis (Stracke et al., 2007). In this triple mutant, the levels of glycosyl flavonols are reduced, as is the expression of several GTs (Stracke et al., 2007, 2010). Similar effects were observed here for both Δy mutants, which included additional glycosyl flavonol species and a larger number of GTs differentially accumulated in our Δy mutants compared to y mutants. In fact, the example that is closest to a null mutant for SlMYB12 in tomato fruit was reported by Adato et al. (2009), who developed an artificial microRNA to target SlMYB12 (amiR-Slmyb12) in the cv MT background. The amiR-Slmyb12 line showed lower levels of three additional flavonol glycosides compared to the original y mutant. However, neither the y mutant nor the amiR-Slmyb12 transgenic line displayed the substantial changes observed in the flavonol glycoside content detected in the Δy mutants. Moreover, F3′H and UDP-GT2 were down-regulated in the Δy mutants in the same way as was described previously in the Arabidopsis myb11 myb12 myb111 triple mutant (Stracke et al., 2007), but not in any of the previous y mutants (Adato et al., 2009). Additionally, CHS2, CHI-like, F3H, FLS-like, and F3GT homologs were co-down-regulated in the Arabidopsis triple mutant, as well as in Δy and y mutants. Therefore, a biosynthetic gene set seems to be affected in the tomato Δy mutants similar to that described in the Arabidopsis triple null mutant. This result supports the proposition that the Δy mutants are tomato null mutants for flavonol glycoside biosynthesis. Nevertheless, we also observed some differences between the Arabidopsis triple null mutant and tomato Δy mutants. For instance, we found no significant changes in dihydroquercetin accumulation levels but, rather, in dihydrokaempferol levels. Production of dihydroquercetin in Arabidopsis is attributed to F3′H enzyme activity. Hence, the gene function of F3′H in tomato fruit could differ from that in Arabidopsis, or it could be subject to different posttranscriptional control. Furthermore, we stress the fact that no proanthocyanidin LBG was co-down-regulated in the Arabidopsis myb11 myb12 myb111 triple mutant. However, we found that DFR and RT genes were differentially down-regulated in both the Δy and y mutants. Interestingly, the overexpression of AtMYB12 in tomato fruit revealed that the known target genes CHS1, CHS2, CHI-like, F3H, and FLS were up-regulated, as were DFR and ANS (Zhang et al., 2015). These combined results could advance research into the role of SlMYB12 in the regulation of the proanthocyanidin biosynthetic branch in tomato fruit.
Genes Up- and Downstream of the Flavonoid Biosynthetic Branch Are Also Affected in the Δy Mutants
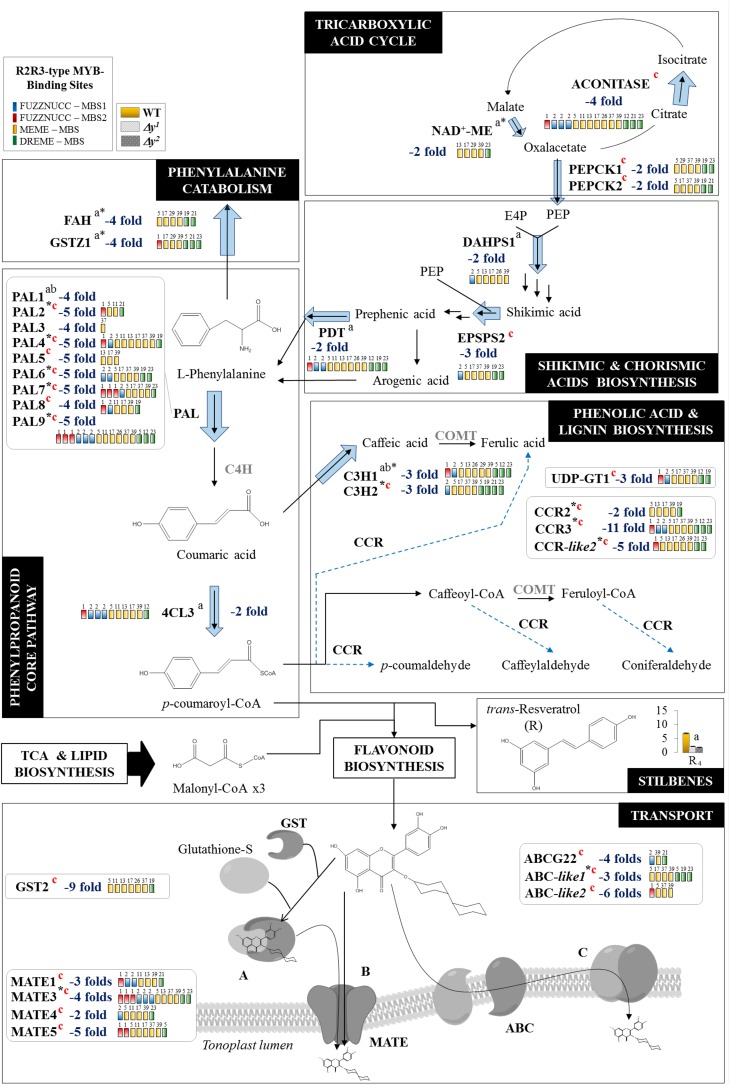
Recently, Zhang et al. (2015) reported that overexpression of the Arabidopsis MYB12 ortholog, AtMYB12, driven by the E8 promoter in tomato fruit can result in fruit with large amounts of flavonols in their flesh. This is due in part to the activation of various genes involved in primary metabolism, the functions of which are to increase the supply of precursors for the flavonol biosynthetic pathway. Moreover, Adato et al. (2009) also described genes upstream of the flavonol biosynthetic pathway affected by the y mutation. Here, we detected genes differentially expressed upstream of the PP biosynthetic pathway, including those in the shikimate and chorismate pathway and even those in the tricarboxylic acid (TCA) cycle and gluconeogenesis (Fig. 6; Supplemental Table S3, A and B). Specifically, we found down-regulated expression for ACONITASE (Solyc12g005860) and NAD+-MALIC ENZYME (ME; Solyc08g066360) in the TCA cycle and both PHOSPHOENOLPYRUVATE CARBOXYKINASE1 (PEPCK1; Solyc12g088160) and PEPCK2 (Solyc04g076880) in the gluconeogenesis pathway. PEPCK catalyzes the synthesis of phosphoenolpyruvate (PEP) from the oxaloacetate produced by ME in the TCA cycle and, together with erythrose-4-phosphate (E4P), PEP is the precursor of the shikimic acid pathway (Fig. 6). Both E4P and PEP are used by 3-DEOXY-d-ARABINOHEPTULOSONATE 7-PHOSPHATE SYNTHASE (DAHPS; Solyc04g074480) to produce 3-deoxy-d-arabinoheptulosonic acid 7-phospate, the first precursor of shikimic acid synthesis. PEP is also used by the 5-ENOLPYRUVYLSHIKIMATE 3-PHOSPHATE SYNTHASE (EPSPS; Solyc05g050980) enzyme, acting downstream of DAHPS, to produce 5-enoylpyruvilshikimate-3-phosphate from shikimate-3-phosphate, the precursor of chorismic acid. Expression of both the DAHPS and EPSPS genes was significantly reduced in both the Δy mutants (Fig. 6; Supplemental Table S3A). In the E8:AtMYB12 tomato lines ME, DAHPS, and EPSPS genes were also differentially expressed (Zhang et al., 2015). However, in these lines, ENOLASE (Solyc03g114500), an enzyme involved in the synthesis of PEP from glycerate-2-phosphate, was affected and not PEPCK. Downstream in the pathway, CHORISMATE MUTASE1 (Solyc02g088460), the enzyme involved in the biosynthesis of prephenic acid, was down-regulated in the y mutant (Adato et al., 2009) and up-regulated in tomato E8:AtMYB12 lines (Zhang et al., 2015). However, in the Δy mutants a <2-fold gene expression reduction was detected and so it was not considered as differentially regulated in our analyses. Additionally the PREPHENATE DEHYDRATASE (Solyc06g074530), which catalyzes the synthesis of l-Phe from arogenic acid, was also down-regulated in the Δy and y mutants and E8:AtMYB12 (Zhang et al., 2015). These results indicate that the alternative synthesis of l-Phe by the phenylpyruvate pathway (Tzin et al., 2009) may be partially blocked when SlMYB12 is mutated, whereas the main biosynthetic pathway through arogenate (Maeda et al., 2011) is active and produces l-Phe (Fig. 6; Supplemental Table S3A) in the Δy mutants. We propose that in tomato, SlMYB12 could be activating the alternative biosynthetic pathway of l-Phe to feed in the PP synthesis.
Figure 6.
Genes up- and downstream of flavonol biosynthesis are also affected in the Δy mutants. The figure represents the next biosynthetic processes upstream of flavonoid biosynthesis: TCA cycle; shikimate, chorismate, and arogenate pathways; core phenylpropanoid pathway; phenolic acid and lignin pathways; resveratrol biosynthesis; and l-Phe catabolism. In addition, the flavonoid transport process is shown downstream of flavonoid biosynthesis. Those genes differentially down-regulated (blue) are shown by arrows (the thicker the arrow, the higher the fold change). MBS motifs found in their promoters were also included (in colored boxes). Genes in gray were not differentially expressed in the Δy mutants. Genes and metabolites differentially expressed both in the y mutant and in other y-lines are marked “a” and “b,” respectively; those differentially expressed only in the Δy mutants are marked “c” (in red). NAD+-ME, NAD+-MALIC ENZYME; PDT, PREPHRENATE DEHYDRATASE; COMT, CAFFEIC ACID O-METHYLTRANSFERASE; HCT, CINNAMOYL COA TRANSFERASE; UDP-GT, GLUCURONOSYL TRANSFERASE.
Subsequently, l-Phe is converted to cinnamic acid by the enzyme PAL, the first step in the PP biosynthetic pathway. Our RNA-Seq analysis detected up to nine homologous PAL genes with expression levels that were reduced by between 3- and 6-fold with respect to the wild type (Fig. 6; Supplemental Table S3A). Interestingly, we detected two genes involved in the catabolism of l-Phe showing down-regulation: MALEYLACETOACETATE ISOMERASE (GSTZ1; Solyc01g091330) and FUMARYLACETOACETATE HYDROLASE (FAH; Solyc11g012160). Thus, the higher levels of l-Phe in the Δy tomato skin could be due to the activation of the main l-Phe biosynthetic pathway in combination with the down-regulation of nine PAL genes (representing more than half of the 16 annotated PAL genes in tomato; http://www.solgenomics.net/), together with the down-regulation of two genes involved in l-Phe catabolism (GSTZ1 and FAH).
Downstream from PAL into the PP core biosynthetic pathway, expression of a 4CL gene (4CL3, Solyc03g097030) was also significantly reduced in the Δy mutants, although no C4H homologous gene was differentially expressed. These results are consistent with those reported for y fruits (Adato et al., 2009; Ballester et al., 2010). Moreover, similar effects were observed in E8:AtMYB12 tomato lines: C4H was not differentially expressed and two 4CL genes other than 4CL3 were up-regulated (Zhang et al., 2015). To conclude, gene expression of C4H was not altered in any of the aforementioned tomato MYB12 mutants or line backgrounds, but it was altered in Arabidopsis myb11 myb12 myb111 triple mutants (Stracke et al., 2007). These results indicate that MYB12 does not regulate C4H and, consequently, appears to have no specific role in the biosynthesis of synapoyl or lignin derivatives in tomato fruit.
On the other hand, we found some evidence that SlMYB12 could be involved in the regulation of other phenolic compounds derived from the PP core pathway. Notably, some genes in the phenolic acid biosynthetic branch (Liu et al., 2015) were also differentially expressed in the Δy mutants (Fig. 6; Supplemental Table S3A). These included two P-COUMAROYL 3-HYDROXYLASE (C3H), several CYNNAMOYL-CoA REDUCTASES (CCRs), and a UDP-GT. Most of these genes were down-regulated, but we found that CCR-like4 (Solyc06g061280) was up-regulated. Despite this, we observed no C3H or CCR metabolite product to be altered in the Δy mutants. Of this last set of genes, only C3H1 (Solyc03g097030) had been reported as differentially down-regulated in the y mutant, whereas both C3H and three CCR genes were differentially expressed in the E8:AtMYB12 tomato line. However, the overexpression of AtMYB12 in tomato fruit (Pandey et al., 2015; Zhang et al., 2015) also produced an increase in caffeoylquinic acid (CQA) levels by up-regulating several genes in the CQA biosynthetic branch, although not C3H or CCR. We found that neither the CQA compounds nor their related biosynthetic genes differentially accumulated in the Δy mutants and concluded that SlMYB12 does not regulate the synthesis of CQAs in tomato fruit skin during ripening. These differences between our results and those previously reported could be due to the different approaches used. The overexpression of AtMYB12 in tomato under the control of a strong promoter may lead to the binding of the overexpressed protein to promoters that are not normally the target of endogenous SlMYB12.
Another differentially accumulated phenolic compound was transresveratrol. In contrast to the scenario described above, we found reduced levels of transresveratrol that were not accompanied by differentially expressed STILBENE SYNTHASE (STS). Similar effects were reported earlier for the y mutants. Even the E8:AtMYB12 tomato lines showed high levels of resveratrol in tomato fruits without changes in STS levels. The STS and CHS enzymes are members of the CHS superfamily of the type III polyketide synthases that catalyze similar condensation reactions, although different cyclization reactions, to produce resveratrol or naringenin chalcone, respectively (Supplemental Fig. S4B; Yamaguchi et al., 1999; Yu et al., 2012; Stewart et al., 2013). They appear to be promiscuous to the extent that STS can produce relatively small amounts of naringenin chalcone, while CHS can generate small amounts of resveratrol (Yamaguchi et al., 1999). Resveratrol production in tomato fruit may perhaps be catalyzed by a protein encoded by one of the down-regulated CHS homologous genes (Supplemental Fig. S4C).
Finally, downstream in the flavonol biosynthetic pathway, eight genes probably involved in flavonoid glycosides transport to the vacuole were differentially expressed in both Δy mutants (Fig. 6; Supplemental Table S3A). These included a GLUTATHIONE S-TRANSFERASES (GSTs) and several MATE-type (MULTI-ANTIMICROBIAL EXTRUSION) and ABC-type (ATP-BINDING CASSETTE) transporter genes, which were mostly down-regulated (Fig. 6; Supplemental Table S3A). Only the E8:AtMYB12 tomato line showed two of these eight genes differentially expressed (Supplemental Table S6) with no effect reported in the previous y mutants. The GSTs are a family of transporters mainly involved in the conjugation and transport of secondary metabolites to the vacuole (Dixon et al., 2002; Petrussa et al., 2013). Flavonols could be found in the cuticle and also inside the vacuoles as flavonol glycosides. However, the mechanism by which this occurs remains unclear. The eight transporter transcripts differentially accumulated in the Δy mutants represent candidate genes to be investigated for their involvement in this mechanism.
Additional Processes Likely Affected in the Δy Mutants
Of the 311 genes differentially expressed in the Δy mutants, only 44 could be related to PP biosynthesis, as described above. The other 267 genes could be associated with a range of processes (Supplemental Fig. S5), including primary metabolism (i.e. photosynthesis or carbohydrate or energy metabolism; Supplemental Fig. S6 and Supplemental Table S3B), secondary metabolism other than PP (Supplemental Table S3B), biotic and abiotic stress responses (Supplemental Table S3C), transcriptional and hormonal regulation (Supplemental Table S3D), and additional cellular processes (i.e. signal transduction or DNA or protein metabolism; Supplemental Table S3E).
Since flavonols are important for fruit cuticle biomechanics and postharvest behavior (Domínguez et al., 2009a, 2011; Lara et al., 2014), we investigated whether some of the genes differentially expressed in the Δy mutants might be involved in cuticle development. Of these 267 genes, we identified a set of 18 in the biosynthesis of cutin monomers, cuticular waxes (i.e. alkanes and esters) and triterpenoids (i.e. amyrins) (Supplemental Table S3B). Interestingly, most were up-regulated in the Δy mutants, which is consistent with the increased levels of alkanes and amyrin and quantitative changes in cutin composition observed earlier in y mutant breaker fruits. We also observed that genes associated with sugar and amino acid production, as well as with cell wall biosynthesis and developmental processes, were altered in the Δy mutants (Supplemental Table S3B). These processes are probably part of the fruit ripening process (Giovannoni, 2004; Pandey et al., 2015). Thus, the Δy mutants might provide a new opportunity to study the role of SlMYB12 TF during fruit ripening, as well as in the homeostasis between primary and secondary metabolisms. Furthermore, a key role for MYB12 in biotic (i.e. fungi and bacterial infections) and abiotic (i.e. phosphate, nitrogen, light, temperature, UV, or drought) stress responses has recently been described for Arabidopsis (Nakabayashi et al., 2014; Schenke and Cai, 2014; Zoratti et al., 2014). We identified a set of 19 genes that were differentially expressed in the Δy mutants that might be involved in these processes (Supplemental Table S3C). Interestingly, most of the down-regulated genes were annotated as likely being involved in biotic stress responses (i.e. those encoding defensin, α-dioxygenase, or peroxidases), while the up-regulated genes were annotated as contributing to abiotic stress responses (i.e. those encoding heat shock proteins, dehydrins, or cold-induced proteins). In addition, we found flavonoid glycosides whose levels were substantially reduced in the Δy mutants (Fig. 4; Supplemental Table S1), These compounds have been described as being involved in several stress response mechanisms (Nakabayashi et al., 2014), and even transporters like GST, ABC, and MATE have also been reported as being involved in these responses (Petrussa et al., 2013).
Finally, we investigated differentially accumulated transcripts annotated as regulatory genes. We detected 33 regulators: 12 TFs and 21 genes involved in hormone biosynthetic processes (Supplemental Table S3D). Two of these were MYB-TFs putatively involved in the regulation of PP biosynthesis: SlTHM27 (Solyc10g055410) and SlMYB11 (Solyc12g049350). In Arabidopsis, C4H gene expression is repressed by AtMYB4 TF (Hemm et al., 2001). Adato et al. (2009) described SlTHM27 (Solyc10g055410) as the tomato ortholog of Arabidopsis AtMYB4 TF. In the y mutant, SlMYB12 and SlTHM27 were co-down-regulated, suggesting that SlTHM27 acts downstream of SlMYB12. In support of this hypothesis, we also found that SlTHM27 was down-regulated in the Δy mutants (Supplemental Table S3D). However, the C4H gene was not differentially expressed in the Δy or y mutants, suggesting a different role for SlTHM27 in tomato fruit. The second down-regulated MYB-related TF was the SlMYB12 homolog SlMYB11 (Solyc12g04930). This newly identified TF was not described in the earlier literature. Adato et al. (2009) described the homolog SlMYB111 (Solyc06g009710), the expression of which was not perturbed in the y mutant. Both SlMYB111 and SlMYB11 have three, rather than four, exons (Supplemental Fig. S6), as is also the case with Arabidopsis AtMYB12, AtMYB111, and AtMYB11 TFs (http://www.arabidopsis.org/) and in contrast with SlMYB12 TF. The overexpression of AtMYB11 in tomato fruit has been described as affecting the synthesis of CQAs, rather than flavonoids (Li et al., 2015). However, as mentioned above, no CQA was found to be differentially accumulated in the Δy mutants. Hence, although SlMYB11 is down-regulated in the Δy mutants and therefore acts downstream of SlMYB12, its function in tomato fruit remains unknown. It appears that the flavonoid pathway in the tomato fruit peel is mainly regulated by SlMYB12 and not SlMYB111 or SlMYB11. However, other regulators are likely to be involved in the biosynthesis of flavonoids in the Δy mutants, as we detected basal levels of flavonoids in all cases. The remaining differentially expressed regulatory genes appear to be involved in the biosynthesis and transport of different plant hormones, including those affecting abscisic acid, auxins, gibberellins, brassinosteroids, ethylene, jasmonic acid, and salicylic acid (Supplemental Table S3D). Of the 21 differentially expressed hormone-related genes, six were down-regulated (belonging to abscisic acid, auxins, gibberellins, and brassinosteroid biosynthesis) and only three showed equivalent patterns in the y mutant (Supplemental Table S3D), while only 3 of the 21 were also affected in the E8:AtMYB12 tomato line.
An Extended Target Gene Set for SlMYB12 Regulation Was Identified in the Δy Mutants
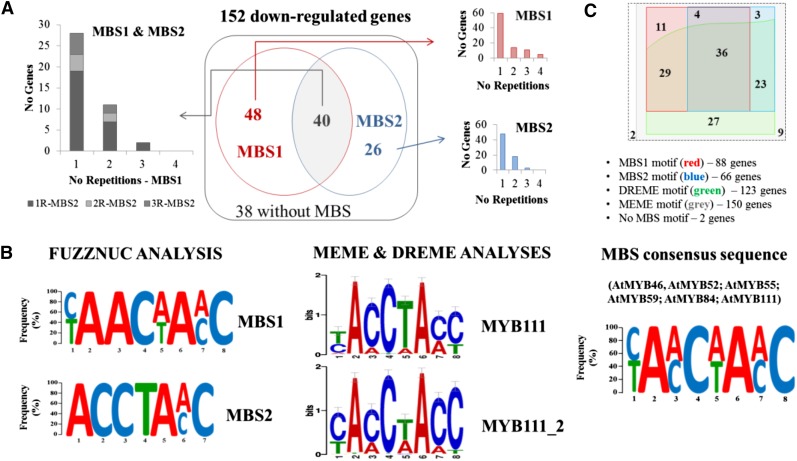
To identify potential target genes affected by the loss of function of SlMYB12 in the null Δy mutants, we investigated the presence of Myb-binding sites (MBSs) in the promoter sequences of the 152 down-regulated genes in these Δy mutants. Myb-recognition elements have been described as essential to activate the flavonol biosynthetic pathway by MYB12 TF in Arabidopsis (Hartmann et al., 2005; Mehrtens et al., 2005). We performed three independent in silico MBS searches by using three online tools: FUZZNUC (http://www.emboss.bioinformatics.nl/cgi-bin/emboss/fuzznuc/), MEME (Multiple Em for Motif Elicitation; Bailey and Elkan, 1994), and DREME (Discriminative Regular Expression Motif Elicitation; Bailey, 2011). The FUZZNUC tool allowed us to identify known MBS motifs including some mismatches, MEME found ungapped wider motifs corresponding to complexes, and DREME identified short ungapped and monomeric motifs and cofactors. The presence of MBS motifs detected by two or three of these approaches in the promoter region of the down-regulated genes in the Δy mutants was considered to be sufficient to define a Myb-recognition element recognizable to SlMYB12 and, therefore, a potential target gene of SlMYB12 activity. First, we used FUZZNUC to scan the promoter sequences for two different MBS motifs that have been previously reported for Arabidopsis AtMYB12 and its functionally redundant homologous TFs AtMYB11 and AtMYB111 (Hartmann et al., 2005; Mehrtens et al., 2005; Franco-Zorrilla et al., 2014). These were (1) MBS1, [T/C]AAC[T/A]A[A/C]C (MBSII) and (2) MBS2, ACCTA[A/C] (MBSIIG) (Franco-Zorrilla et al., 2014). We observed nucleotide frequencies for the MBS1 and MBS2 consensus sequences in our data sets similar to those described for the Arabidopsis myb11 myb12 myb111 triple mutant (Franco-Zorrilla et al., 2014). Of the 152 down-regulated genes, 114 exhibited MBS motifs in their promoters (Fig. 7A; Supplemental Table S4); 88 contained the MBS1 motif (77.2%), showing the following consensus sequence [C(73%)/T(67%)]AAC[A(81%)/T(59%)]A[A(84%)/C(56%)]C (Fig. 7B); 66 contained the MBS2 motif (57.9%), showing the consensus sequence ACCTA[C(47%)/A(46%)] (Fig. 7B); and 40 exhibited both types of MBS motifs in their promoters. Furthermore, most of these 114 genes had one or two MBS1 and/or MBS2 motifs in their promoters, and very few genes (<20%) had more than two MBS1 and/or MBS2 motifs (Fig. 7A). Most of the promoters of the co-down-regulated genes in the Arabidopsis myb11 myb 12 myb111 triple mutant have been described as containing an MBS2 motif in their promoter, whereas only a small set of genes contain the MBS1 motif. Moreover, it has been suggested that the presence of several MBS motifs in promoters indicates primary target genes of the MYB11, MYB12, and MYB111 TFs in Arabidopsis, whereas genes with a single MBS motif could represent secondary targets of these TFs (Franco-Zorrilla et al., 2014). Yet, unlike Arabidopsis, the number of down-regulated genes with promoters containing the MBS1 motif was higher than that of those containing the MBS2 motif in tomato fruit, and most of these genes contained one or two, rather than several, MBS motifs (Fig. 7A).
Figure 7.
In silico MBS motifs found in potential target genes of SlMYB12. A, Venn diagram representing the MBSs (MBS1 and MBS2) recognized by AtMYB12 TF and found using FUZZNUC in 114 of the 152 genes in the down-regulated gene set of Δy mutants. Bar graphs represent the number of genes with promoters containing one or more than one MBS1 (in red), MBS2 (in blue), or both MBS (in gray) motifs. B, Consensus sequences for MBS1 and MBS2 FUZZNUC motifs detected in the 114 genes. MYB12-related MBS motifs detected by the Tomtom tool in both MEME and DREME motifs are also shown. These were AtMYB111 (primary motif) and AtMYB111_2 (secondary motif) MBSs. Finally, the consensus sequence for the six R2-R3-type MBSs detected in the analyses was also provided (see Supplemental Tables S5, A–D). C, Venn diagram showing the distribution of those down-regulated genes containing MBS motifs based on the different MBS search tools used.
Next, we investigated other possible MBS motifs that could be recognized by SlMYB12. We performed a nontargeted search for DNA-binding domains (DBDs) using two search tools: MEME and DREME. In combination with these tools, we also used the online tool Tomtom (motif comparison tool; Gupta et al., 2007) to detect MYB R2R3-type DBDs in the DNA motifs obtained by MEME and DREME. Two DBD databases were used by Tomtom: Arabidopsis PBM motifs (Franco-Zorrilla et al., 2014) and the updated JASPAR CORE PLANT (2016) (http://www.meme-suite.org/db/motifs/). Finally, we used FIMO (Find Individual Motif Occurrences; Grant et al., 2011) to determine the occurrence of different MYB-DBDs in the gene set under study. These combined analyses of the 2-kb promoter sequences of the 152 down-regulated genes in the Δy mutants resulted in the detection of 39 DNA motifs by MEME and 24 by DREME tools. Of these, only eight MEME motifs (Supplemental Table S5A) and five DREME motifs (Supplemental Table S5C) corresponded to possible MYB R2R3-type DBDs (Supplemental Table S5, B and D). Tomtom identified the AtMYB111 DBD among these 13 motifs, using both PBM and JASPAR databases, as well as other MYB R2R3 DBDs (i.e. AtMYB46, AtMYB52, AtMYB55, AtMYB59, and AtMYB84), which were very much like the DBD recognized by AtMYB111 (Supplemental Table S5, A–D). A total of 150 genes contained some of the eight MEME motifs in their promoters and 123 of those also included some of the five DREME motifs (Supplemental Table S4), as predicted by FIMO.
In total, 141 of the 152 down-regulated genes contained MBS motives detected by at least two of the online tools and, we propose, represent targets for SlMYB12. The MBS consensus sequence obtained by combining the three search tools for the 141 down-regulated genes was [C/T]A[A/C]C[T/A]A[A/C]C. Interestingly, the MBS consensus sequence obtained recently (Zhang et al., 2015) by the ChIP-Seq methodology in the promoters of the target genes for both AtMYB12 and SlMYB12 TFs in tomato fruit was almost identical, other than the third nucleotide position, which was [C/G] instead of [A/C]. These results supported our 141 candidate genes as potential direct targets of SlMYB12 in tomato fruit.
CONCLUSION
In this study, we characterized a null allele (Δy) associated with the pink fruit phenotype in two new tomato mutants. In contrast to the previously characterized y lines, the Δy mutants accumulated normal levels of a truncated version of the Slmyb12 transcript and did not accumulate flavonol glycosides in their skin. Transcriptome analysis allowed us to identify a larger set of genes that were differentially expressed in the Δy mutants than those reported in the previously characterized y lines. Moreover, it allowed us to examine the molecular program that underlies the Δy mutation with higher resolution. Furthermore, we gained new insights into the role of SlMYB12 in primary and secondary metabolism homeostasis. Finally, we identified a large set of potential candidate target genes of SlMYB12 activity, which extend beyond the flavonol biosynthetic branch and that will be the subject of future in-depth characterization.
MATERIALS AND METHODS
Plant Material
Mutant seeds were obtained from mutagenized tomato population (Solanum lycopersicum cv ‘MicroTom’) developed by Tsukuba University in Japan (TOMATOMA, http://www.tomatoma.nbrp.jp/; Saito et al., 2011). M3 seeds from two mutant lines with a pink fruit phenotype, TOMJPG-4497 and TOMJPG-5663, were used. Wild-type, pf1, and pf2 mutant plants were grown under natural light and controlled temperature conditions (24°C during the day, 18°C at night) in a greenhouse. Once fruit reached the mature green stage, plants were checked daily to harvest fruits at breaker stage. Skins from five to eight breaker fruits per genotype were manually separated, gently cleaned of excess mesocarp, as described by Adato et al. (2009), and frozen in liquid nitrogen until further use. Three biological replicates, which represented the skin of 5 to 10 fruits, were collected from three independent plants and were used to represent each experimental data point.
UPLC-QTOF-MS Profiling of Semipolar Compounds and Data Analysis
An analysis of semipolar compounds, including flavonoids and other phenylpropanoids, was conducted using an UPLC-ESI-QTOF instrument (HDMS Synapt; Waters) with the UPLC column connected online to a photodiode array detector and then to the MS detector, following the methodology described by Rogachev and Aharoni (2012). Methanolic extracts prepared from frozen tissue were analyzed. A 100 × 2.1-mm i.d., 1.7-μm UPLCBEH C18 column (Waters) was used to separate metabolites. The photodiode array detector was set to 210 to 500 nm. Masses of the eluted compounds were detected by a QTOF mass spectrometer, equipped with an electrospray ionization (ESI) source, in positive and negative ionization modes, within the 50 to 1500Da m/z range. XCMS open-source LC-MS-based data analysis software (Smith et al., 2006) was used for peak picking and peak alignment, as described elsewhere (Rogachev and Aharoni, 2012). Compounds were identified using standards, injected in the same LC-MS conditions, or putatively identified, as previously described (Adato et al., 2009). Statistical analysis of the XCMS outputs was performed using a custom-made computer program (Adato et al., 2009; Mintz-Oron et al., 2008). Only metabolites exhibiting changes (log2) greater than 2-fold between mutants and control samples were considered differentially accumulating.
RNA-Seq Gene Expression Analysis
For whole-transcriptome shotgun sequencing (WTSS or RNA-Seq), total RNA was extracted, and short cDNA libraries of 50 bp length were produced (Zhong et al., 2011), quality tested in an Agilent 2100 bioanalyzer (Faculties of Life Sciences, Weizmann Institute of Science, Israel), and sequenced by Illumina technology (Crown Institute of Genomics, Weizmann Institute of Science, Israel). High-quality reads were filtered by CASAVA 1.8 software. Clean reads were mapped to the tomato genome using TopHat (Trapnell et al., 2012). The resulting alignment files were used to generate a transcriptome assembly for both pf mutants and the MicroTom wild-type (MT-WT) with the help of CUFFLINKS (Trapnell et al., 2012). These assemblies were merged to calculate gene and transcript expressions with the CUFFMERGE utility of the CUFFLINKS package. We used CUFFDIFF to calculate expression levels and test the statistical significance of the CUFFMERGE results. The differentially expressed (P value < 0.05) transcripts between each mutant and control were aligned using BWA software (Li and Durbin, 2009) to create consensus sequences with iAssembler software (Zheng et al., 2011). Integrative Genomics Viewer freeware was used to view the consensus sequences (Robinson et al., 2011).
PCR and RT-PCR Gene Expression Analyses
For PCR analysis, genomic DNA was extracted by the CTAB-chloroform procedure (15 g CTAB, 40.9 g NaCl, 0.5 m EDTA, pH 8, 1 m Tris-HCl, pH 8, and 0.2% β-mercaptoethanol). The aqueous phase was treated with RNase (50 mg/mL), and DNA was precipitated with 3 m sodium acetate and isopropanol. The pellet was washed with 70% and 80% of ethanol and resuspended in DDW. The PCR reaction was set as follows: 1 μL of diluted (1:100) genomic DNA, 12.5 μL of GOTaq Green Master Mix (Promega), and 0.5 μL of each SlMYB12-specific forward (F) and reverse (R) oligonucleotide primers in a 25-μL reaction. PCR conditions were as follows: 94°C, 4 min (94°C, 30 s; 50–60°C [depending on the primer sequence], 30 s; 72°C, 1–4 min [depending on the primer length]) × 35 cycles; 72°C, 5 min; 8°C, overnight. The gene-specific oligonucleotide primers were designed using Vector NTI 10.3.0 (2006; Invitrogen; Supplemental Table S7).
For RT-PCR analysis, total RNA was extracted using Tri reagent 100 mg/mL (38% water-saturated phenol, 0.8 m guanidine thiocyanate, 0.4 m ammonium thiocyanate, 0.1 m sodium acetate, pH 5, and 5% glycerol). The aqueous phase was extracted twice with chloroform and precipitated with isopropanol. The RNA pellet was washed twice with 70% ethanol and resuspended in DDW. Isolated RNA was treated with DNase I to be reverse-transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Real-time PCR analysis was performed as previously described (Adato et al., 2009; Mintz-Oron et al., 2008). The SlMYB12-specific oligonucleotide primers were designed using Primer Express Software (Applied Biosystems; Supplemental Table S7). The TIP41-like gene (Solyc10g049850) was used as an endogenous control (Expósito-Rodríguez et al., 2008).
Protein Sequence Alignments
Protein sequences were obtained from the Arabidopsis (Arabidopsis thaliana; http://www.arabidopsis.org/), tomato (http://www.solgenomics.net/), and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) databases. Alignments were performed by using EMBL-EBI ClustalW 2.1 online freeware (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Identification of TF DNA Recognition Elements
The gene promoter region for a selection of differentially down-regulated genes in the Δy mutants was extracted in FASTA format (http://www.solgenomics.net/). The promoter region was defined as that of the 2,000 bp upstream and 100 bp downstream of the start codon (Veerla and Höglund, 2006). Then, the primary (MBSII) and secondary (MBSIIG) MBS motifs for the target genes of AtMYB111 in the Arabidopsis myb111 myb11 myb12 triple mutant, as described by Franco-Zorrilla et al. (2014), were used as a string search query of the gene promoter regions: MBS1, [T/C]AAC[T/A]A[A/C]C (based on the MBSII motif); and MBS2, ACCTA[A/C]C (based on the MBSIIG motif from Franco-Zorrilla et al., [2014] and also on the MBS motif proposed by Hartmann et al. [2005] and Mehrtens et al. [2005]). FUZZNUC tool (http://www.emboss.bioinformatics.nl/cgi-bin/emboss/fuzznuc/) was used to search the MBS patterns in the promoter sequences of interest.
In addition, using the 2,100-bp promoter sequences, DNA motifs were also found with the MEME 4.9.0 (Bailey and Elkan, 1994; Bailey et al., 2009). MEME analysis was run to search for the 100 motifs of between 6 and 50 bp in width using the default distribution model (Zero Or One Per Sequence [ZOOPS]) and stopping if the motif E-value was above 1e−03. The analysis included those motifs that were repeated between 2 and 600 times and considered the maximum data set size as 1,000,000 characters. DREME 4.11.0 (Bailey, 2011) was used to search the same 2,100-bp promoter sequences for DNA motifs no longer than 8 bp, using the default settings. Then, Tomtom 4.11.0 (Gupta et al., 2007) was used to identify the DBDs in the significant DNA motifs obtained by MEME and DREME analysis. To identify different DBDs, we ran the Tomtom freeware against two independent motif databases: the Arabidopsis PBM motif database (Franco-Zorrilla et al., 2014), containing 6- to 8-bp motifs, and the 2016 updated JASPAR CORE PLANT (Portales-Casamar et al., 2010), containing 6- to 21-bp motifs. Only the MEME and DREME motifs containing MBSs detected in both databases were considered in our study. Next, FIMO 4.11.0 (Grant et al., 2011) was used to determine the occurrence of MBS in the gene set under study. Tomtom and FIMO analysis were conducted with the default settings (http://www.meme-suite.org/).
Gene Ontology Analysis for Primary Metabolism
The differentially expressed gene set shared by pf1 and pf2 mutants was analyzed with MapMan 3.6.0 RC1 software (http://mapman.gabipd.org/eb/guest/mapman/) to identify those genes involved in primary metabolism processes in the differentially expressed gene set. To map the genes into the primary metabolism processes, we used an updated tomato mapping file available on the GoMapMan open Web-accessible resource for gene functional annotations (http://www.gomapman.org/).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RNA-Seq gene expression profiles for genes up- and downstream of the SlMYB12 genomic sequence.
Supplemental Figure S2. SlMYB12 protein alignment for the Δy mutant and y lines.
Supplemental Figure S3. FLS and DFR phylogenetic comparison for tomato (Solanum lycopersicum) and Arabidopsis thaliana.
Supplemental Figure S4. Similarities between both STILBENE SYNTHASE and CHALCONE SYNTHASE enzymes.
Supplemental Figure S5. General overview of the metabolic processes affected in the Δy mutants.
Supplemental Figure S6. Protein alignment for tomato SlMYB12, Slmyb12-Δy, SlMYB111, and SlMYB11.
Supplemental Table S1. Metabolites differentially accumulated in the breaker fruit skin of both Δy mutants.
Supplemental Table S2. Genes differentially expressed in breaker fruit skin shared by both the Δy mutants and y mutant.
Supplemental Table S3. Genes belonging to the phenylpropanoid biosynthetic pathway differentially expressed in breaker fruit skins and shared by both Δy1 and Δy2 mutants.
Supplemental Table S4. List of potential candidate target genes of SlMYB12 transcription factor activity.
Supplemental Table S5. DNA motifs containing R2R3-type Myb-binding sites identified in the Δy mutant.
Supplemental Table S6. Genes differentially expressed in both the Δy mutants and the A8:AtMYB12 overexpression tomato mutant line.
Supplemental Table S7. PCR and RT-PCR oligonucleotides.
Supplementary Material
Acknowledgments
We are grateful to Prof. Daisuke Shibata and Prof. Tohru Ariizumi for providing the pink fruit mutant lines. We thank Justin Lashbrooke for his collaboration in establishing optimal conditions in order to obtain the RNA-Seq cDNA libraries in tomato fruit. We thank Shelly Hen-Avivi for growing the mutant and wild plant types. We are also grateful to Dr. Kristina Gruden for providing us with the tomato mapping file to identify the genes involved in primary metabolism by using the online GoMapMan tool. We thank the Bioinformatics Core Service at the IBMCP for the support provided. Our special thanks to Joss Rose for his comments and careful editing of the final version of the manuscript.
Glossary
- PP
phenylpropanoid
- TF
transcription factor
- LC-MS
liquid chromatography coupled with mass spectrometry
- LBG
late flavonoid biosynthetic gene
- TCA
tricarboxylic acid
- PEP
phosphoenolpyruvate
- CQA
caffeoylquinic acid
- MBS
Myb-binding site
- DBD
DNA-binding domain
References
- Adato A, Mandel T, Mintz-Oron S, Venger I, Levy D, Yativ M, Domínguez E, Wang Z, De Vos RCH, Jetter R, et al. (2009) Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet 5: e1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. (2011) DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, AAAI Press, Menlo Park, CA, pp 28–36 [PubMed] [Google Scholar]
- Bailey TL, Bodén M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester AR, Molthoff J, de Vos R, Hekkert Bt, Orzaez D, Fernández-Moreno JP, Tripodi P, Grandillo S, Martin C, Heldens J, et al. (2010) Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol 152: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy A, Schijlen E, Hall RD (2007) Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): the potential for metabolomics. Metabolomics 3: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock H-P, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, Martin C (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Chopra S, Athma P, Peterson T (1996) Alleles of the maize P gene with distinct tissue specificities encode Myb-homologous proteins with C-terminal replacements. Plant Cell 8: 1149–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czemmel S, Stracke R, Weisshaar B, Cordon N, Harris NN, Walker AR, Robinson SP, Bogs J (2009) The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol 151: 1513–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R (2002) Plant glutathione transferases. Genome Biol. 3: reviews3004.1–reviews3004.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez E, Cuartero J, Heredia A (2011) An overview on plant cuticle biomechanics. Plant Sci 181: 77–84 [DOI] [PubMed] [Google Scholar]
- Domínguez E, España L, López-Casado G, Cuartero J, Heredia A (2009a) Biomechanics of isolated tomato (Solanum lycopersicum) fruit cuticles during ripening: the role of flavonoids. Funct Plant Biol 36: 613–620 [DOI] [PubMed] [Google Scholar]
- Domínguez E, Luque P, Heredia A (2009b) Sorption and interaction of the flavonoid naringenin on tomato fruit cuticles. J Agric Food Chem 57: 7560–7564 [DOI] [PubMed] [Google Scholar]
- Du H, Feng BR, Yang SS, Huang YB, Tang YX (2012) The R2R3-MYB transcription factor gene family in maize. PLoS One 7: e37463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111: 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. (2006) The Science of Flavonoids. Springer, New York [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553 [DOI] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS (2007) Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57: 155–171 [DOI] [PubMed] [Google Scholar]
- Hemm MR, Herrmann KM, Chapple C (2001) AtMYB4: a transcription factor general in the battle against UV. Trends Plant Sci 6: 135–136 [DOI] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62: 2465–2483 [DOI] [PubMed] [Google Scholar]
- Iijima Y, Nakamura Y, Ogata Y, Tanaka K, Sakurai N, Suda K, Suzuki T, Suzuki H, Okazaki K, Kitayama M, et al. (2008) Metabolite annotations based on the integration of mass spectral information. Plant J 54: 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al. (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16: 263–276 [DOI] [PubMed] [Google Scholar]
- Lara I, Belge B, Goulao LF (2014) The fruit cuticle as a modulator of postharvest quality. Postharvest Biol Technol 87: 103–112 [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen M, Shaoli W, Ning J, Ding X, Chu Z (2015) AtMYB11 regulates caffeoylquinic acid and flavonol synthesis in tomato and tobacco. Plant Cell Tissue Organ Cult 122: 309–319 [Google Scholar]
- Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, et al. (2014) Genomic analyses provide insights into the history of tomato breeding. Nat Genet 46: 1220–1226 [DOI] [PubMed] [Google Scholar]
- Lindstrom EW. (1925) Inheritance in tomatoes. Genetics 10: 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10: 50–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Plant Physiol 8: 689–708 [DOI] [PubMed] [Google Scholar]
- Luo J, Butelli E, Hill L, Parr A, Niggeweg R, Bailey P, Weisshaar B, Martin C (2008) AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J 56: 316–326 [DOI] [PubMed] [Google Scholar]
- Maeda H, Yoo H, Dudareva N (2011) Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Brief communication. Nat Chem Biol 7: 19–21 [DOI] [PubMed] [Google Scholar]
- Martin C, Butelli E, Petroni K, Tonelli C (2011) How can research on plants contribute to promoting human health? Plant Cell 23: 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, Aharoni A (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol 147: 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moço S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoort J, de Vos CHR (2006) A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol 141: 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir SR, Collins GJ, Robinson S, Hughes S, Bovy A, Ric De Vos CH, van Tunen AJ, Verhoeyen ME (2001) Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat Biotechnol 19: 470–474 [DOI] [PubMed] [Google Scholar]
- Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, et al. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka T, Saito M, Yamada E, Fujita K, Kakizaki Y, Nishihara M (2012) Isolation and characterization of GtMYBP3 and GtMYBP4, orthologues of R2R3-MYB transcription factors that regulate early flavonoid biosynthesis, in gentian flowers. J Exp Bot 63: 6505–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Misra P, Choudhary D, Yadav R, Goel R, Bhambhani S, Sanyal I, Trivedi R, Trivedi PK (2015) AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues. Sci Rep 5: 12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrussa E, Braidot E, Zancani M, Peresson C, Bertolini A, Patui S, Vianello A (2013) Plant flavonoids--biosynthesis, transport and involvement in stress responses. Int J Mol Sci 14: 14950–14973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A (2010) JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 38: D105–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein D, Schijlen E, Kooistra T, Herbers K, Verschuren L, Hall R, Sonnewald U, Bovy A, Kleemann R (2006) Transgenic flavonoid tomato intake reduces C-reactive protein in human C-reactive protein transgenic mice more than wild-type tomato. J Nutr 136: 2331–2337 [DOI] [PubMed] [Google Scholar]
- Rick CM, Butler L (1956) Cytogenetics of the tomato. Advan Genet 8: 267–382 [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogachev I, Aharoni A (2012) UPLC-MS-based metabolite analysis in tomato. Methods Mol Biol 860: 129–144 [DOI] [PubMed] [Google Scholar]
- Saito T, Ariizumi T, Okabe Y, Asamizu E, Hiwasa-Tanase K, Fukuda N, Mizoguchi T, Yamazaki Y, Aoki K, Ezura H (2011) TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol 52: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenke D, Cai D (2014) The interplay of transcription factors in suppression of UV-B induced flavonol accumulation by flg22. Plant Signal Behav 9: e28745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schijlen EGWM, de Vos CHR, Martens S, Jonker HH, Rosin FM, Molthoff JW, Tikunov YM, Angenent GC, van Tunen AJ, Bovy AG (2007) RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol 144: 1520–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78: 779–787 [DOI] [PubMed] [Google Scholar]
- Stewart C Jr, Vickery CR, Burkart MD, Noel JP (2013) Confluence of structural and chemical biology: plant polyketide synthases as biocatalysts for a bio-based future. Curr Opin Plant Biol 16: 365–372 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B (2010) Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol 188: 985–1000 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzin V, Malitsky S, Aharoni A, Galili G (2009) Expression of a bacterial bi-functional chorismate mutase/prephenate dehydratase modulates primary and secondary metabolism associated with aromatic amino acids in Arabidopsis. Plant J 60: 156–167 [DOI] [PubMed] [Google Scholar]
- Veerla S, Höglund M (2006) Analysis of promoter regions of co-expressed genes identified by microarray analysis. BMC Bioinformatics 7: 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Kurosaki F, Suh D-Y, Sankawa U, Nishioka M, Akiyama T, Shibuya M, Ebizuka Y (1999) Cross-reaction of chalcone synthase and stilbene synthase overexpressed in Escherichia coli. FEBS Lett 460: 457–461 [DOI] [PubMed] [Google Scholar]
- Yu D, Xu F, Zeng J, Zhan J (2012) Type III polyketide synthases in natural product biosynthesis. IUBMB Life 64: 285–295 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Alseekh S, Tohge T, Rallapalli G, Luo J, Kawar PG, Hill L, Santino A, Fernie AR, Martin C (2015) Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat Commun 6: 8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhao L, Gao J, Fei Z (2011) iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics 12: 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ (2011) High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011: 940–949 [DOI] [PubMed] [Google Scholar]
- Zoratti L, Karppinen K, Luengo Escobar A, Häggman H, Jaakola L (2014) Light-controlled flavonoid biosynthesis in fruits. Front Plant Sci 5: 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.