Cytoplasmic acidification is a specific response to freezing; it contributes to activating freezing-tolerance responses including a lipid remodeling enzyme necessary for freezing tolerance.
Abstract
Low temperature is a seasonal abiotic stress that restricts native plant ranges and crop distributions. Two types of low-temperature stress can be distinguished: chilling and freezing. Much work has been done on the mechanisms by which chilling is sensed, but relatively little is known about how plants sense freezing. Recently, Arabidopsis (Arabidopsis thaliana) SENSITIVE TO FREEZING2 (SFR2) was identified as a protein that responds in a nontranscriptional manner to freezing. Here, we investigate the cellular conditions that allow SFR2 activation. Using a combination of isolated organelle, whole-tissue, and whole-plant assays, we provide evidence that SFR2 is activated by changes in cytosolic pH and Mg2+. Manipulation of pH and Mg2+ in cold-acclimated plants is shown to cause changes similar to those of freezing. We conclude that pH and Mg2+ are perceived as intracellular cues as part of the sensing mechanism for freezing conditions. This evidence provides a specific molecular mechanism to combat freezing.
Freezing is a distinct abiotic stress that adds to the stress experienced during chilling (low temperatures above 0°C). There are at least two, possibly related, types of damage during freezing: formation of ice crystals accompanied by cellular dehydration, and membrane leakage (Thomashow, 1999). In Arabidopsis (Arabidopsis thaliana), ice first nucleates outside the cell. The resulting change in the osmotic gradient across the plasma membrane swiftly and severely dehydrates the cell (Steponkus, 1980, 1984). Membrane damage occurs both as a direct response to temperature and as a secondary effect of cellular dehydration. The temperature directly affects membrane fluidity and, therefore, leakage (Xin and Browse, 2000; Hays et al., 2001). During dehydration, membrane damage is heightened because membranes become appressed as the cell shrinks (Steponkus, 1984). This enhanced proximity of membranes, lack of fluidity, and low hydration allow nonlamellar structures to form between membranes, fusing subcellular compartments and ultimately resulting in cell death after rehydration (Webb et al., 1994; Uemura et al., 1995). Multiple mechanisms have evolved in plants to avoid both dehydration and membrane fusion, including solute accumulation, cell wall modification, lipid desaturation, and lipid composition changes (Lineberger and Steponkus, 1980; Browse and Xin, 2001; Degenkolbe et al., 2012; Chen and Thelen, 2013; Ji et al., 2015). These changes typically occur during a period of cold acclimation or cold hardening in which plants are exposed to low, nonfreezing temperatures prior to freezing and transcriptional changes accompany increased freezing tolerance.
An exception to this rule is the gene SENSITIVE TO FREEZING2 (SFR2). It was discovered in an Arabidopsis screen for freezing intolerance (Warren et al., 1996). Mutant plants (sfr2) lacking SFR2 are severely damaged by freezing, but they have no phenotypes under normal growth or a variety of other stress conditions (Fourrier et al., 2008), implying that SFR2’s activity represents a specific adaptation to freezing tolerance. The sfr2 mutation is unusual among freezing-sensitive mutants because the cells of sfr2 remain intact during freezing, as evinced by their lack of ion leakage (Warren et al., 1996). This is likely due to the role of SFR2 in maintaining organellar rather than cellular integrity.
During freezing, SFR2 removes the Gal head group from monogalactosyldiacylglycerol (MGDG) and adds it to a second MGDG. This activity is processive, generating oligogalactolipids (digalactosyldiacylglycerol, trigalactosyldiacylglycerol, and up to hexagalactosyldiacylglycerol) and leaving diacylglycerol as a by-product (Moellering et al., 2010; Roston et al., 2014). SFR2 activity was initially discovered in isolated chloroplasts, where it was referred to as galactolipid:galactolipid galactosyl transferase (Heemskerk et al., 1983, 1986). During freezing conditions, the diacylglycerol is converted into triacylglycerol (TAG), and TAG and oligogalactolipids derived from MGDG specifically increase in response to freezing (Moellering et al., 2010; Vu et al., 2014a). SFR2 is associated with the chloroplast outer envelope membrane (Heemskerk et al., 1986; Roston et al., 2014), where it is anchored by a single transmembrane domain facing the cytoplasm (Roston et al., 2014). The soluble portion of SFR2 is composed primarily of a single glycosyl hydrolase domain. The hydrolase domain was shown to be responsible for the MGDG-specific transferase activity, without measurable hydrolysis activity (Roston et al., 2014). Notably, in original reports of SFR2, its protein and mRNA levels did not change in response to cold (Thorlby et al., 2004), indicating that it may be posttranscriptionally regulated.
Currently, the mechanism by which freezing is sensed at the cellular level is unknown. This lack prevents further understanding of membrane-freezing responses, which are separate from those of cold acclimation and critical for freezing tolerance (Li et al., 2008). Here, we take advantage of SFR2 activation to probe how an enzyme is specifically activated by freezing. We show that SFR2 is posttranslationally activated and probe which cellular responses to freezing activate it in isolated chloroplasts and whole tissues, determining that cytosolic pH and Mg2+ both are involved. We demonstrate that cytosolic acidification occurs in intact plants in response to freezing. It is further shown that SFR2 is not substrate limited under normal conditions and that it has a consistently sized protein complex, implying that acidification may directly activate SFR2. Finally, we investigate whether cytosolic acidification can mimic freezing-like membrane changes in intact tissues.
RESULTS
SFR2 Is Activated Posttranslationally in Response to Freezing
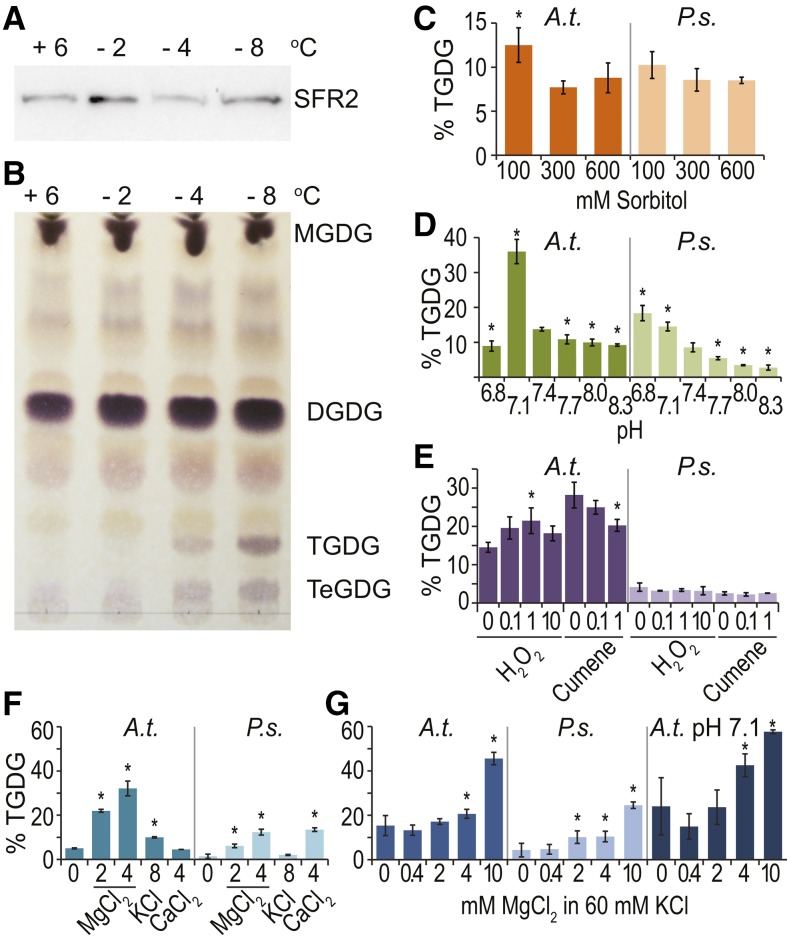
The SFR2 protein is present at all temperatures measured (Fig. 1A; Thorlby et al., 2004). However, SFR2’s specific oligogalactolipid reaction products trigalactolipid (TGDG) and tetragalactolipid were detectable only after plants were incubated at or below −4°C overnight. No detectable accumulation occurred after 1 week of cold acclimation at +6°C, and freezing at −2°C showed little or no oligogalactolipid accumulation (Fig. 1B).
Figure 1.
SFR2 is posttranslationally activated by pH and magnesium ions. Wild-type Arabidopsis plants were cold acclimated at 6°C for 1 week, incubated overnight at the temperatures indicated at top, and then sampled for lipids and proteins. A, Immunoblot detecting SFR2 protein levels. B, Thin-layer chromatogram separating lipids identified at right visualized with a sugar-specific stain. Images shown are representative of three separate plant growth trials. C to G, Isolated chloroplasts were incubated with radiolabeled UDP-Gal in 300 mm sorbitol, 50 mm HEPES, pH 7.5, or modified buffers as indicated below the graph axis. Radiolabel in the oligogalactolipid product TGDG is quantified as a percentage of total radiolabeled lipids. Error bars represent sd of at least three separately grown trials. Asterisks represents significance (P ≤ 0.05) between the treatment and the condition most closely mimicking normal cytoplasm (300 mm sorbitol, pH 7.4, 0 mm hydrogen peroxide [H2O2], 0 mm cumene hydroperoxide, no divalent cations [F], or 0.4 mm MgCl2 [G]).
SFR2 Is Activated by pH and Mg2+
Because SFR2 is present inside the cell, we hypothesized that it may be activated by physical changes in the cell associated with freezing. To test this hypothesis, chloroplasts isolated from Arabidopsis (freezing tolerant) or pea (Pisum sativum; freezing sensitive) were mixed with a radiolabeled precursor of galactolipid synthesis (UDP-Gal) and then incubated under conditions in which a single variable mimicked a possible cellular change in response to freezing. SFR2 activity was measured as production of radiolabeled TGDG. Reactive oxygen species can accumulate during many stresses, including cold (Suzuki and Mittler, 2006), but the addition of water-soluble hydrogen peroxide or lipid-soluble cumene peroxide had little effect on SFR2 activity (Fig. 1E). As lipids approach their transition temperatures, membrane leakage increases (Hays et al., 2001). The vacuole and extracellular space serve as reservoirs of protons, and cytoplasmic pH has been reported to change during cold conditions (Dietz et al., 2001). Low pH values had an activation effect on SFR2 in chloroplast preparations from either Arabidopsis or pea (Fig. 1D). Notably, the effects were not identical, consistent with the two species’ different responses to freezing. The chloroplast stroma is a reservoir of Mg2+ ions, previously shown to activate SFR2 in vitro (Shaul, 2002; Roston et al., 2014). Increases in the Mg2+ concentration specifically affected SFR2 activity (Fig. 1F), and these effects were synergistic with low pH (Fig. 1G). Other cellular cations did not have strong effects when their biologically relevant levels were considered (Fig. 1F). Calcium levels are believed to be nanomolar or picomolar (Monshausen et al., 2008), and potassium levels are near 60 mm (Halperin and Lynch, 2003). When 60 mm potassium was included, it did not prevent further activation by Mg2+ ions (Fig. 1G).
SFR2 Activation by pH and Mg2+ Is Reproducible in Whole Tissues
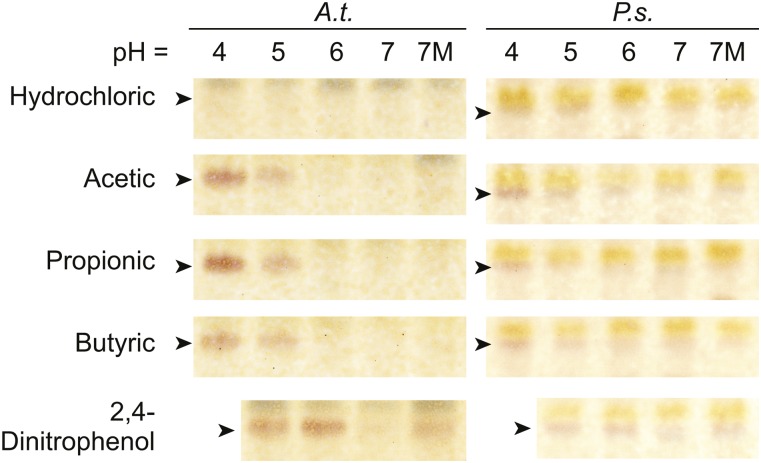
To test if the pH- and Mg2+-based activation of SFR2 was generalizable to whole tissues, 3-week-old Arabidopsis rosettes or 2-week-old pea leaves grown at normal temperatures were excised and floated for 1 h on top of various acids, and oligogalactolipid production was measured (Fig. 2). SFR2 activity occurred only when organic acids were used, presumably because in their protonated form organic acids can carry protons across membranes to affect the cytosolic pH (Plieth et al., 1997). Consistent with this possibility, the membrane-permeable proton carrier 2,4-dinitrophenol had stronger effects at more neutral pH levels, while hydrochloric acid had no measurable effect. SFR2 activity was further enhanced in the 2,4-dinitrophenol sample when 10 mm MgCl2 was added (Fig. 2, lane 7M). Together, these data indicate that SFR2 can be activated in isolated organelles and in whole tissue by lowered pH and increased Mg2+ concentration.
Figure 2.
pH and magnesium changes activate SFR in whole tissues. Thin-layer chromatograms show the separation of lipids from extracts of Arabidopsis (A.t.) shoots or pea (P.s.) leaves floated on 20 mm of the acid indicted at left adjusted to the pH indicated at top with dipotassium phosphate for 1 h. 7M indicates pH 7 with an additional 20 mm MgCl2. TGDG is indicated by arrowheads. Images shown are representative of three separate plant growth trials.
Cytosolic pH Changes in Response to Freezing and Acetic Acid Treatment
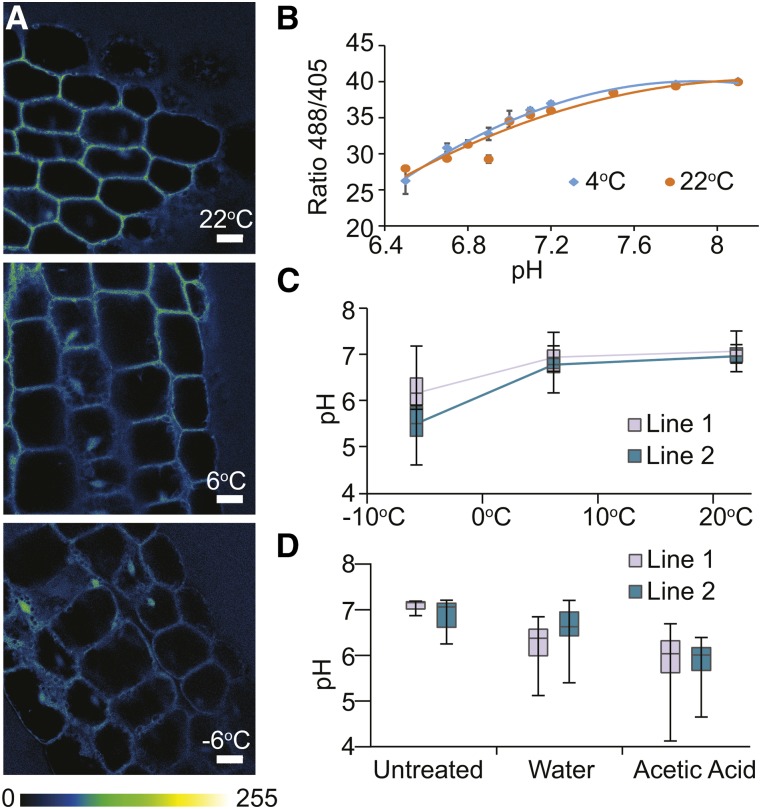
To corroborate the hypothesis of SFR2 activation by pH and Mg2+, physiologically relevant changes in cellular pH were measured during SFR2-activating conditions. This was done using two independent Arabidopsis lines stably expressing a pH-reporting fluorescent protein from sea pen (Ptilosarcus gurneyi; PtGFP) shown to be located in the cytosol (Schulte et al., 2006; Geilfus et al., 2014). These plants were grown at room temperature, cold acclimated for 1 week or cold acclimated and frozen at −6°C overnight, after which ratiometric fluorescence was measured by confocal microscopy (Fig. 3A). Low temperature-treated plants were measured at 4°C immediately after removal from their incubation temperature. A pH decrease was detected between normal and cold-acclimated plants, with a further decrease observable between cold-acclimated and frozen plants (Fig. 3C). The temperature of measurement did not appear to have a large effect on quantification, as ratiometric responses of purified PtGFP buffered at multiple pH values and measured at 4°C and 22°C were nearly identical (Fig. 3B). To compare the level of cytosolic acidification during freezing with that during acid treatment as described in Figure 2, the PtGFP-transformed Arabidopsis lines also were treated with 20 mm acetic acid at pH 5. Plants were grown at room temperature, floated in acetic acid or water for 1 h, and then measured precisely as above (Fig. 3D). After 1 h of flotation on acetic acid, pH decreased significantly in both lines. It should be noted that, using identical microscope parameters, we were able to measure a slight ratiometric response of wild-type plants not transformed with PtGFP. This parameter was used to mathematically correct estimated pH values for all data.
Figure 3.
Cytosolic pH changes during freezing and acetic acid treatment. A, Arabidopsis plants stably transformed with PtGFP were grown under control conditions (22°C), or grown and cold acclimated at 6°C for 1 week, or cold acclimated and frozen overnight at −6°C. A cold stage (4°C) was used to measure chilled plants. Ratiometric fluorescence was measured in hypocotyls, with excitation at 488 nm divided by excitation at 405 nm with detection constantly between 505 and 530 nm. Bars = 22 µm. B, Pure PtGFP protein was measured identically to A in microcapillaries at 22°C or on the cold stage (4°C) to provide a pH scale. C, Ratiometric fluorescence images of two independent lines of PtGFP, including those shown in A, were transformed into pH as described in “Materials and Methods” and are graphed according to most recently exposed temperature. Statistical significance values are as follows: 22°C versus 6°C (all samples), P = 0.0325; 6°C versus −6°C (all samples), P = 5 × 10−8; for line 1 individually: 22°C versus 6°C, P = 0.215; 6°C versus −6°C, P = 0.0006; for line 2 individually: 22°C versus 6°C, P = 0.0661; 6°C versus −6°C, P = 9 × 10−8. D, The same two independent lines of PtGFP used in C and A were untreated or floated on water or 20 mm acetic acid at pH 5 for 1 h, mimicking treatments in Figure 2. Statistical significance values are as follows: acetic acid versus water (all samples), P = 1.21 × 10−16; acetic acid versus untreated (all samples), P = 1.3 × 10−24; water versus untreated (all samples), P = 1.09 × 10−9; for line 1 individually: acetic acid versus water, P = 0.0052; acetic acid versus untreated, P = 2.5 × 10−10; water versus untreated, P = 4.44 × 10−10; for line 2 individually: acetic acid versus water, P = 5.22 × 10−19; acetic acid versus untreated, P = 2.54 × 10−16; water versus untreated, P = 0.0023.
SFR2 Is Not Substrate Limited
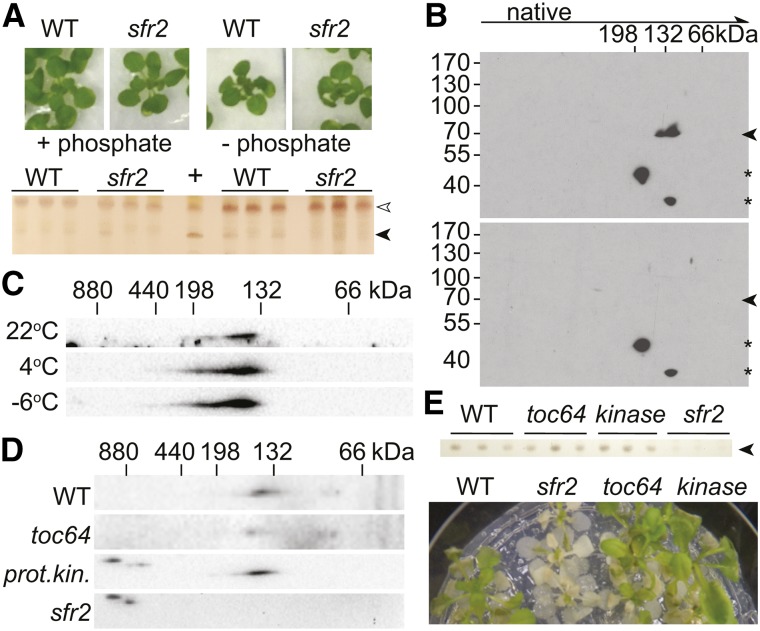
In addition to changes in the aqueous boundary layer that may occur in response to freezing and affect SFR2 activity, changes to the membrane itself may cause SFR2 activation. Specifically, it seemed possible that the substrate MGDG is not accessible to SFR2 in the outer chloroplast envelope membrane under normal conditions but would become accessible following freezing-induced membrane disruption. To test this possibility, oligogalactolipids were quantified under phosphate-limited growth conditions known to induce additional MGDG synthases in the outer envelope membrane, the same suborganellar location as SFR2 (Kobayashi et al., 2009). Plants were grown for 2 weeks and then transferred to medium lacking phosphate for 10 d. If MGDG levels limit SFR2 activity, then oligogalactolipid levels would be expected to increase during phosphate deprivation, because of increased MGDG availability. However, increases in oligogalactolipids were not observed (Fig. 4A). In comparison, a positive control showed production of TGDG after 1 h of flotation on 20 mm acetic acid. Thus, substrate availability is unlikely to play a major role in increasing SFR2 activity following freezing.
Figure 4.
SFR2 is not substrate limited and does not stably interact with other proteins. A, Ten-day-old wild-type (WT) or sfr2 Arabidopsis plants were transferred to regular medium or medium lacking phosphate for 10 d, and then lipids were extracted. Resulting lipids were analyzed by thin-layer chromatography for the presence of TGDG (black arrowhead). The location of digalactosyldiacylglycerol (DGDG) is indicated by the white arrowhead. B, Immunoblot of 40 µg of chlorophyll equivalent wild-type (top) or sfr2 (bottom) chloroplasts solubilized with 2% digitonin separated in two dimensions, 4% to 14% blue-native PAGE in the first dimension and 7.5% denaturing PAGE in the second dimension, detected with the SFR2 antiserum. Arrowheads indicate SFR2-specific signal, while asterisks identify nonspecific signal. C, Comparisons of SFR2 leaf protein two-dimensional immunoblots of plants grown at 22°C, cold acclimated for 1 week (6°C) or cold acclimated and frozen overnight at −6°C. D, Comparisons of SFR2 two-dimensional immunoblots as in B for mutants and controls identified at left. E, Wild-type or mutant Arabidopsis as indicated at top were tested for the ability to produce TGDG (arrowhead) in response to a 1-h incubation in 20 mm acetic acid, pH 5, or to withstand freezing at −6°C (bottom). All portions of the figure are representative of at least three separately grown biological replicates.
SFR2 Does Not Have Stable Protein Partners
In planta, SFR2 appears to form a complex of approximately 140 kD, as determined by native gel electrophoresis (Fig. 4B). This complex does not appear to change with the activity level of SFR2, because the size of the complex did not shift in response to SFR2 activation at −6°C (Fig. 4C). If the complex represents a stable association between SFR2 and other proteins, the other proteins could provide additional information about the mechanism of temperature sensing. To identify SFR2-interacting proteins, SFR2 antibodies were used to precipitate SFR2 in chloroplasts isolated from the wild type or sfr2 transfer DNA (T-DNA) insertion lines that lack SFR2 protein. A cross-linker was used to enhance complex stability during the process. Resulting eluates were analyzed by liquid chromatography-tandem mass spectrometry. Proteins identified in the wild-type samples but not the sfr2 samples in each of three replicates were few (Table I). A full list of identified peptides and proteins is provided in Supplemental Table S1. The most abundant, as judged by numbers of identified spectra, were investigated further. These included the translocon at the outer membrane of chloroplasts, 64 kD, and a protein kinase family protein of unknown function. Arabidopsis insertion lines lacking all paralogs of these genes, toc64 (Aronsson et al., 2007) and protein kinase (Alonso et al., 2003), were obtained and confirmed to obtain genomic insertions by PCR (Supplemental Fig. S1A). These lines were tested for aberrant SFR2 complex formation and activity. The size of the SFR2-containing complex appeared normal (Fig. 4D), and the plants did not have reduced freezing tolerance (Fig. 4E). SFR2 activation also appeared normal, as it could be activated in response to 20 mm acetic acid (Fig. 4E), and was not otherwise active during normal growth (Supplemental Fig. S1B). We concluded that SFR2 does not have stably interacting protein partners and that the higher Mr complex visualized by native gel electrophoresis likely represents a homooligomer. Its size is consistent with a dimer.
Table I. Proteins identified as potential interactors of SFR2.
Numbers of spectra associated with each protein in each sample from the first (1), second (2), and third (3) biological replicates are given. Peptide identifications were accepted to achieve a false discovery rate (FDR) of less than 0.1%. Protein identifications were accepted to achieve an FDR of less than 1%
| Protein Name | Accession No. | sfr2 (1) | Wild Type (1) | sfr2 (2) | Wild Type (2) | sfr2 (3) | Wild Type (3) |
|---|---|---|---|---|---|---|---|
| SFR2, 71 kD | AT3G06510 | 0 | 49 | 0 | 55 | 0 | 55 |
| Translocon at the outer membrane of chloroplasts, 64 kD | AT3G17970 | 0 | 11 | 0 | 10 | 0 | 9 |
| Protein kinase superfamily protein, 68 kD | AT4G32250 | 0 | 5 | 0 | 7 | 0 | 5 |
| Outer membrane protein of 24 kD | AT3G52230 | 0 | 4 | 0 | 3 | 0 | 2 |
| Translocon at the inner envelope membrane of chloroplasts, 214 kD | ATCG01130 | 0 | 8 | 0 | 8 | 0 | 1 |
| Translocon at the outer membrane of chloroplasts, 132 kD | AT2G16640 | 0 | 5 | 0 | 4 | 0 | 1 |
| ATP-binding cassette2-type transporter family protein, 79 kD | AT2G01320 | 0 | 6 | 0 | 2 | 0 | 1 |
| Plastid division2, 34 kD | AT2G16070 | 0 | 2 | 0 | 3 | 0 | 1 |
| Dephospho-CoA kinase family protein, 26 kD | AT2G27490 | 0 | 3 | 0 | 2 | 0 | 1 |
Treatment with Acetic Acid Mimics a Freezing Response
To determine whether treatment of tissues with acetic acid (to lower cytosolic pH) or Mg2+ mimicked freezing responses, lipid changes were measured in cold-acclimated wild-type and sfr2 plants that were frozen or treated with 20 mm acetic acid, pH 5, or acetic acid with 10 mm Mg2+ (Fig. 5). To better mimic the cellular condition of cold-acclimated plants prior to freezing, all plants were cold acclimated whether treated or frozen. Total fatty acid pools, MGDG, TAG, and phosphatidylglycerol (PG), were quantified. PG was included as a representative of a prominent chloroplast lipid without known SFR2-dependent effects. SFR2 dependence was determined by comparing wild-type Arabidopsis changes with those in sfr2. Note that acetic acid and acetic acid with Mg2+ treatments are expected to replicate the direction rather than the precise magnitude of changes due to freezing.
Figure 5.
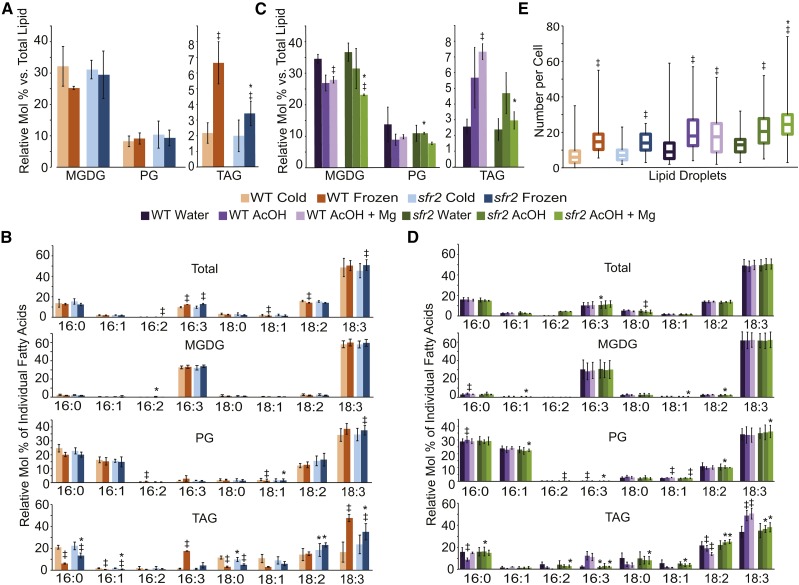
pH and Mg2+ treatments mimic lipid changes due to freezing. A to D, Plants were grown at 22°C for 3 weeks and cold acclimated at 6°C for 1 week for all treatments (Cold). They were subsequently frozen at −6°C overnight (Frozen) or floated on 20 mm acetic acid, pH 5 (AcOH), 20 mm acetic acid, pH 5, with 10 mm magnesium chloride (AcOH + Mg), or water for 3 h. All plants were sampled as rosettes with roots removed. Molar percentage (A and C) of MGDG, PG, and TAG relative to total lipid amount and fatty acid profiles (B and D) of each lipid species relative to total fatty acids for each individual fatty acid were quantified. Values are biological replicate means ± sd. Each biological replicate consists of an average of three or four technical replicates. E, Lipid droplets were visualized with confocal microscopy after Nile Red staining and quantified as the number of lipid droplets per cell. Boxes encompass the interquartile range, with the central line representing the median. Whiskers represent maximum and minimum counts. For all data, significance (P ≤ 0.05) between control and treatment is represented by double daggers, and asterisks represent significance (P ≤ 0.05) between the wild type (WT) and sfr2.
During freezing, a reduction in the amount of MGDG and a corresponding increase in the amount of TAG were observed in wild-type plants, as described previously (Moellering et al., 2010). The profile of TAG fatty acids changed in frozen wild-type plants to contain 16:3, a fatty acid contained primarily in MGDG. However, TAG from frozen sfr2 did not contain significantly more 16:3 after freezing, making this change SFR2 dependent (Fig. 5, A and B). In contrast, TAG levels increased in both wild-type and sfr2 plants, in an SFR2-independent change (Fig. 5, A and B). Total fatty acid compositions were only slightly changed, MGDG fatty acid composition was unchanged, and changes to PG levels and composition were small (Fig. 5B). This is consistent with previous evidence that the species of fatty acids within MGDG do not change during freezing (Li et al., 2008). Treatment with acetic acid or acetic acid with Mg2+ caused increases in TAG levels and decreases in MGDG levels, with relatively small changes to PG levels, in the wild type and sfr2 (Fig. 5C). Few significant changes were observed in the total fatty acid profile or that of MGDG or PG (Fig. 5D). These patterns mimicked the direction and type of change seen during freezing. Notably, the increases in TAG levels in response to acidification were again independent of the sfr2 genotype (i.e. they occurred in the wild type and sfr2), indicating that not only is SFR2 activated similarly in response to acetic acid and freezing, but at least one other lipid-remodeling enzyme is similarly activated by cytosol acidification. Fatty acid changes in TAG of wild-type plants included decreases in 16:0 and 18:0 and increases in 16:3 and 18:3, mimicking TAG fatty acid changes due to freezing (Fig. 5D).
To confirm that the increased levels of TAG depend on cytosolic acidification through an independent method, lipid droplets were observed by Nile Red staining of TAG droplets and subsequent confocal microscopy. Quantification of lipid droplets per cell showed trends consistent with the total lipid changes observed for TAG (Fig. 5, A, C, and E). This again demonstrates that TAG is accumulated and stored in similar ways during freezing and acetic acid or acetic acid with Mg2+ treatments.
DISCUSSION
SFR2 catalyzes a lipid head group transfer reaction that is critical to plant survival of freezing. Because the protein is present in all chloroplasts under all conditions (Fig. 1), it must be activated in a nontranscriptional manner. How plants sense temperature or freezing conditions is unknown. Here, we explored a molecular freezing sensing mechanism at the level of SFR2 activation. To understand the regulation of SFR2 activity through a posttranslational mechanism requires understanding physical changes under freezing conditions inside the cell. We have shown that acidification causes SFR2 activation, and this activation is heightened by the addition of Mg2+ in either isolated chloroplasts or whole shoot tissues (Figs. 1 and 2). In fact, a decrease in cytosolic pH is apparent during both cold and freezing, to an extent consistent with activating SFR2 (Fig. 3). It is likely that this activation occurs through a direct mechanism, as stable interactions of SFR2 with other proteins were not detected, and SFR2 is not substrate limited (Fig. 4). Using whole-tissue assays, changes to the levels and fatty acid profiles of MGDG, TAG, and lipid droplet formation seen during freezing could be mimicked by pH changes. Together, these data provide evidence that pH changes provide a critical link to the activation of SFR2, and this finding can be taken as a paradigm for a molecular mechanism by which plants sense freezing within cells.
Interestingly, the SFR2 response to pH is not due to direct pH manipulation of its glycosyl transferase activity. Yeast-produced SFR2 has a pH optimum of approximately 7.5, although it responds similarly to magnesium ions (Roston et al., 2014). Thus, the need for Mg2+ can be attributed directly to a requirement for catalysis, while the pH change required for activation in situ cannot. SFR2 activation by decreased pH does not coincide with the previously observed pH optimum of 7.5 in vitro for SFR2 (Roston et al., 2014). Hence, proper sensing of freezing by SFR2 must require that it be in its natural local environment, within the outer envelope membrane. It is possible that pH changes affect the properties of the membrane or its constituents and, thus, affect SFR2.
We did not identify stable protein partners that interact with SFR2 by immunoprecipitation using SFR2-specific antisera in wild-type plants (Fig. 4; Supplemental Table S1). Because SFR2 produced heterologously in yeast is always active (Roston et al., 2014), we consider it likely that transient protein interactions or posttranslational modifications play a role in SFR2 activation. Recently, an association of SFR2 with OPEN STOMATA1 (OST1) was reported using tagged OST1 overproduced under the control of the ubiquitin promoter (Waadt et al., 2015). We did not detect OST1 as even a minor component in any of our immunoprecipitations, and it should be noted that SFR2 interaction with OST1 was only reported after abscisic acid treatment (Waadt et al., 2015). Abscisic acid levels are known to increase in response to chilling (Mantyla et al., 1995), and OST1 is active during cold acclimation in Arabidopsis (Ding et al., 2015). Thus, it is unlikely that OST1 is involved directly in the response of SFR2 to below-freezing conditions. However, we cannot rule out the activation of SFR2 by other mechanisms in addition to those described here.
Specifically, the production of oligogalactolipids independent of freezing conditions has been observed in distinct genetic backgrounds or conditions. The TRIGALACTOLIPID (TGD) genes were named for the constitutive production of TGDG in their Arabidopsis mutants (Hurlock et al., 2014). The TGD proteins have been shown to enhance the transport of lipids from the endoplasmic reticulum to the chloroplast, and the respective tgd mutants have altered outer envelope membrane compositions that could contribute to SFR2 activation. Additionally, oligogalactolipid production appears to increase in response to oxidative stress, including ozone fumigation (Sakaki et al., 1990). It is clear that SFR2 does not respond directly to oxidative stress (Fig. 1), but it is unclear if ozone fumigation affects SFR2 activity through changes in pH, Mg2+, or additional factors.
Activation of SFR2 by pH and Mg2+ is relevant to freezing because they likely represent the sensing of membrane damage. Membrane leakage increases when membranes approach phase-transition temperatures of their lipid constituents (Hays et al., 2001), which have been measured in pea chloroplasts to begin at 10°C and continue until −10°C (Leheny and Theg, 1994). Furthermore, membrane damage increases after freezing, as cellular dehydration contracts the cell and osmotic potential increases (Steponkus, 1984). The vacuole and extracellular spaces of plant cells are highly acidic, while the chloroplast stroma has a high Mg2+ concentration, which increases during the day up to 10 mm. As the cells chill and the membranes become partially damaged, leakage of small ions including protons and Mg2+ could provide a convenient mechanism for rapidly activating the membrane protective machinery, beginning with the activation of SFR2 at the outer chloroplast envelope membrane. Consistent with this hypothesis, wounding by a crushing force was observed to cause SFR2 activity (Vu et al., 2014b, 2015). Wounded tissue allows cytoplasmic mixing with acidic apoplastic fluid and, possibly through this simple mechanism, SFR2 activation. Similarly, SFR2 is activated during the isolation of intact chloroplasts (Heemskerk et al., 1983). Both wounding and chloroplast isolation provide stresses that are independent of cold acclimation. During either situation, tissues are broken and multiple forces act on the isolated chloroplasts in ways that may mimic membrane environments during freezing.
The majority of cold- and freezing-tolerance studies have identified transcriptionally controlled genes (Fowler and Thomashow, 2002). SFR2 mRNA levels show little or no response to low temperature (Thorlby et al., 2004), although enzymatic activity increases dramatically below freezing (Fig. 1). The pH changes that activate SFR2 also appear to activate SFR2-independent changes to TAG and MGDG (Fig. 5), which mimic those that occur during freezing (Fig. 5). Changes to cytosolic pH are unlikely to be the only changes that act as signals during freezing, but they appear to play an important role in chloroplast membrane lipid remodeling.
CONCLUSION
Freezing tolerance is a necessary resilience mechanism for plants native to temperate climates. Unlike many proteins required for cold or freezing tolerance, ubiquitous SFR2 is not increased in abundance but activated to combat freezing stress. This provides plants with a rapid-response mechanism during fluctuating weather conditions, which are more frequently encountered as global weather patterns become more unstable and extreme. Here, we showed that SFR2 is activated by cytosolic pH and ionic changes and that these changes can mimic other plant responses to freezing. Specifically, SFR2 activation by relatively moderate pH and ionic changes was supported at the organelle and whole-tissue levels in two species, while pH changes were observed to occur by pH-sensitive GFP responses during freezing of whole Arabidopsis plants. Tissue-level activation of cold-adapted Arabidopsis by pH or pH and Mg2+ was observed to promote freezing-like lipid changes. We conclude that cytoplasmic acidification is a molecular mechanism through which freezing conditions are communicated throughout the plant cell.
MATERIALS AND METHODS
Plant Material
Wild-type Arabidopsis (Arabidopsis thaliana) was of the Columbia ecotype. The Arabidopsis Biological Resource Center supplied a T-DNA insertion in At3g06510, herein referred to as the sfr2 mutant, also published as sfr2-3 (SALK_106253; Moellering et al., 2010), and the protein kinase At4g32250 with a T-DNA inserted in the last exon of the gene (SALK_051823; Alonso et al., 2003). Toc64 has three homologs in Arabidopsis with possible functional redundancy. Arabidopsis with insertions causing the loss of all three Toc64 full-length transcripts was kindly donated by Dr. Paul Jarvis and Sean Maguire. The presence of transgenes was confirmed using primers given by Aronsson et al. (2007) and Moellering et al. (2010) or, for protein kinase, 5′-AGAACATGGATGTGCCAGAAG-3′, 5′-CGCTGCATATACCATGTGATG-3′, and T-DNA-specific primer LB3.1 (Salk Institute).
Plant Growth
Seeds were sterilely planted on Murashige and Skoog medium (Caisson Laboratories) containing 1% Suc and 0.5% MES, pH 5.7, solidified with 6% AgarGel (Sigma). Seeds were exposed to 4°C for 2 d in the dark and then grown in 16-h-day/8-h-night conditions at a constant 22°C. Plants used to test phosphate stress activation of SFR2 were transferred to another plate of the medium described above or similarly prepared medium lacking phosphate 10 d after germination (Caisson Laboratories). Whole shoot tissues were sampled after 11 d of growth on the new medium.
Freeze Testing
All freeze-tested plants were cold acclimated for 1 week at 6°C prior to freezing. During cold acclimation, they were on a 12-h-day/12-h-night cycle. Freeze tolerance tests were performed as described previously (Moellering et al., 2010) with the following exceptions: all freeze testing was performed at the end of the day/beginning of the night cycle; after ice nucleation at −2°C, temperatures were lowered within 1 h to the reported freezing temperature; and postfreezing recovery was performed at 22°C under bench light for 3 d before return to normal growth conditions.
SFR2 Assays in Isolated Chloroplasts
Arabidopsis wild-type plants were grown for 3 to 4 weeks on medium as described above, and garden pea (Pisum sativum “Little Marvel”) was grown for approximately 2 weeks. The plants were not cold acclimated. All shoot tissue was harvested, and chloroplasts were isolated essentially as described previously (Bruce et al., 1994). A total of 100 µg of chlorophyll equivalent chloroplasts was pelleted and resuspended in 98 µL of buffer. The buffer content varied by experiment but included 44 mm HEPES at pH 7.5 unless specified, 300 mm sorbitol or as specified, 0.5 mm glycerol-3-phosphate, 0.3 mm monobasic potassium phosphate, 0.2 mm CoA, and 4 mm magnesium chloride unless specified otherwise. As indicated in the text, specific experiments included one or more of the following: 0.1 to 10 mm hydrogen peroxide, 0.1 to 1 mm cumene hydroperoxide, 0 to 10 mm total magnesium chloride, 8 to 60 mm potassium chloride, or 4 mm calcium chloride, pH 6.8 to 8.3. Immediately after resuspension, 2 µL of 0.1 mCi mL−1 [14C]UDP-Gal (American Radiolabeled Chemicals) was added and mixed by gentle agitation. The chloroplasts were allowed to react for 30 min at room temperature in low bench-top lighting. Following incubation, intact chloroplasts were reisolated on top of a 35% Percoll (Sigma), 330 mm sorbitol, and 50 mm HEPES, pH 7.5, cushion, washed once in buffered sorbitol without Percoll, and then extracted with 200 µL of methanol:chloroform (2:1, v/v). Because of the variance in the recovery of intact chloroplasts from many of the experimental conditions, levels were equalized using chlorophyll fluorescence prior to loading onto a Silica Gel 60 plate (Merck) and separating as described above. MGDG, DGDG, and TGDG bands were identified by comparison with standards purchased or generated using SFR2 expressed in yeast (Roston et al., 2014). Radioactivity in the bands was quantified by scintillation counting. The data presented express the level of radioactivity in TGDG as a percentage of all radioactivity in the sum of MGDG, DGDG, and TGDG as a method to rule out the control of MGDG synthesis, a prerequisite to TGDG radioactivity caused by SFR2.
SFR2 Assay in Whole Tissue
Twenty millimolar hydrochloric acid, acetic acid, propionic acid, butyric acid, or 2,4-dinitrophenol was adjusted to pH 4, 5, 6, or 7 ± 0.01 with dibasic potassium phosphate. 2,4-Dinitrophenol was not adjusted to pH 4 because when dissolved it was already too basic (pKa = 4.09). As indicated, 10 mm magnesium chloride was added. Five milliliters of each solution was used to float either whole, plate-grown Arabidopsis rosettes or two fully expanded pea leaves. The plants were grown under normal conditions (see above) and were not cold acclimated. The thick waxy cuticle of pea leaves was bypassed by cutting five slits across the epidermis of each pea leaf with a fine razor blade. Plants were incubated at room temperature for 1 h, then gently patted dry and analyzed for lipid content.
Lipid Analysis
Plant tissue as described in the text was extracted with a modified Bligh and Dyer protocol to isolate lipids, as described (Wang and Benning, 2011). Sampling of frozen plants was done carefully, to minimize thawing. Frozen plants were protected from thawing during sampling by harvesting with chilled forceps into prechilled tubes and immediately immersing in liquid nitrogen. Comparisons between direct extraction of whole-leaf samples by vigorous shaking and extraction of tissues crushed in liquid N2 did not show noticeable changes in oligogalactolipid levels; therefore, whole-leaf extraction was primarily used. Thin-layer chromatography analysis of oligogalactolipids was performed on Silica Gel 60 thin-layer chromatography plates (Millipore) in resolving solvent composed of chloroform:methanol:acetic acid:water (85:20:10:4, v/v/v/v). Thin-layer chromatography isolation of lipids prior to quantification by gas chromatography was performed on Silica G plates with a preadsorbent zone (SiliCycle) in resolving solvent composed of acetone:toluene:water:acetic acid (91:30:7:2, v/v/v/v), dried, and additionally separated in petroleum ether:diethyl ether:acetic acid (80:20:1, v/v/v). Otherwise, thin-layer chromatography was performed as described (Wang and Benning, 2011). Silica retaining separated lipids was scraped from the plates, pentadecanoic acid was added as a standard, and all lipids were derivatized to fatty acid methyl esters and quantified by gas chromatography coupled to a flame ionization detector. Derivatization and gas chromatography were essentially as described; however, hydrogen was used as the carrier gas for a 30-m capillary HP-Innowax column (Agilent) set at 90°C for 1 min, ramped at 30°C min−1 to 235°C, and held for 5 min. Statistical analysis was by Student’s t test.
Protein Analysis
Blue-native PAGE was done essentially as described previously (Kikuchi et al., 2006). Samples were prepared by extracting leaf tissue by homogenization on ice in ice-cold native sample buffer containing 2% (w/v) digitonin. Particulates were removed by centrifugation at 21,000g for 10 min at 4°C. One percent dodecylmaltoside, decylmaltoside, and Triton X-100 also were screened but did not resolve a single complex species. Immunoblotting using the SFR2 antiserum was as described (Roston et al., 2014). For immunoprecipitation experiments, 400 µL of freshly desalted SFR2N and SFR2C antisera mixed in a 1:1 ratio were coupled to AminoLinkPlus coupling resin (ThermoScientific, Pierce). The resulting resin was split into two microcolumns. Identical amounts of chloroplasts freshly prepared from wild-type or sfr2 plants as described above were cross-linked by incubation with 10 mm dithiobis(succinimidyl propionate) for 2 min at room temperature and 20 min on ice. Fifty millimolar (final concentration) Tris-HCl at pH 7.5 was used to quench the cross-linker reaction by incubation at room temperature for 15 min. Cross-linked chloroplasts were precipitated and resuspended to 2 mg chlorophyll mL−1 in 50 mm HEPES, pH 7.4, 150 mm sodium chloride, 1% dodecylmaltoside, and complete protease inhibitor without EDTA (Roche). After 30 min, insoluble material was precipitated by ultracentrifugation at 100,000g for 10 min at 4°C. This was used as the starting material for immunoprecipitation, which was performed essentially as per AminoLinkPlus instructions. Binding to resin occurred overnight at 4°C in the dark; the column was washed with 80 column volumes of chloroplast solubilization buffer containing 0.1% dodecylmaltoside and then eluted with 90°C nonreducing, SDS-PAGE loading buffer. Mass spectrometry was essentially as described (Roston et al., 2012), except peptides were resuspended in 2% acetonitrile/0.1% trifluoroacetic acid to 25 μL. From this, 5 μL was automatically injected by a Thermo EASYnLC 1000 device onto a Thermo Acclaim PepMap RSLC 0.075-mm × 150-mm C18 column and eluted over 60 min with a gradient of 2% B to 30% B in 49 min, ramping to 100% B at 50 min, and held at 100% B for the duration of the run (buffer A = 99.9% water and 0.1% formic acid, buffer B = 99.9% acetonitrile and 0.1% formic acid) at a constant flow rate of 0.3 nL min−1. Eluted peptides were sprayed into a ThermoFisher Q-Exactive mass spectrometer using a FlexSpray spray ion source. Survey scans were taken in the Orbitrap (35,000 resolution, determined at mass-to-charge ratio 200), and the top 10 ions in each survey scan were then subjected to automatic higher energy collision-induced dissociation with fragment spectra acquired at 17,500 resolution.
Peptide and Protein Identification
Tandem mass spectra without charge state deconvolution or deisotoping were extracted by Mascot Distiller version 2.4 and analyzed with Mascot version 2.5.0 and X! Tandem version CYCLONE (2010.12.01.1). Both Mascot and X! Tandem were set up to search the Arabidopsis genome, annotation version 10 from The Arabidopsis Information Resource, supplemented with common contaminants of the cRAP3 database assuming trypsin digestion. Searches had a fragment ion mass tolerance of 0.30 D and a parent ion tolerance of 10 ppm. Allowed fixed modifications included only carbamidomethyl of Cys. Allowed variable modifications were deamidated Asn and Gln, oxidized Met, and thioacylated Lys. X! Tandem variable modifications additionally included N-terminal pyro-Glu and N-terminal ammonia loss. Scaffold version 4.4.8 was used to validate peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 7% probability to achieve a FDR of less than 0.1%. Peptide probabilities from X! Tandem were assigned by the Peptide Prophet algorithm (Keller et al., 2002) with Scaffold Delta mass correction. Peptide probabilities from Mascot were assigned by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 97% probability to achieve an FDR less than 1% and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins were grouped to satisfy the principles of parsimony if they contained similar peptides and could not be differentiated. If significant peptide evidence was shared, proteins were grouped into clusters.
PtGFP Measurement of Cytosolic pH
Use of cytosolic sea pen (Ptilosarcus gurneyi) PtGFP to measure cytosolic pH was essentially as described (Schulte et al., 2006; Geilfus et al., 2014), with the following exceptions: excitation was with a blue diode laser at 405 nm or an argon gas laser at 488 nm, with emission recorded from 505 to 530 nm. Measurement of tissue was done in unbuffered water for all untreated samples on standard microscope slides and in 20 mm acetic acid, pH 5, for the acetic acid-treated samples. Measurement of purified PtGFP donated by Christoph Plieth was done in 0.5 m buffers used within their pH range on microslides, precision rectangular capillaries (Vitro Dynamics). This study was duplicated at Michigan State University and the University of Nebraska-Lincoln. A Zeiss 10 meta ConfoCor 3 confocal microscope fitted with a PE100-ZAL cooling stage (Linkham Scientific Instruments) at the Michigan State University Center for Advanced Microscopy was used for PtGFP imaging. A similar process to that described was used at the University of Nebraska-Lincoln Morrison Microscopy Core (see microscope description in the following section). ImageJ software with the FIJI plugin package were used to process raw data (Schindelin et al., 2012). For testing pH changes during cold treatments, cold-acclimated and frozen plants were processed as follows. A temporary incubator made of a thick-walled Styrofoam container filled with ice was kept at the same temperature as the plants during the overnight temperature treatment. The plants were kept in these temporary incubators en route to the microscope. Individual plants were removed from the container as quickly as possible, placed on a prechilled slide, and then onto the cooling stage at 4°C. A maximum of two images were taken within 1 min of placing the plant onto the slide. All images were collected within 1 h. Plants used to measure cytosolic pH during cold treatment were 4 weeks old and grown at 22°C; 22°C for 3 weeks and 6°C for 1 week; or 22°C for 3 weeks, 6°C for 1 week, and −6°C overnight. Plants used to measure cytosolic pH during acetic acid treatment were 3 weeks old and were not cold acclimated.
Lipid Droplet Quantification
Wild-type and sfr2 plants were grown for 3 weeks at 22°C and then cold acclimated precisely as described for freeze treatments. Plants were then subsequently frozen at −6°C overnight or floated on 20 mm acetic acid, pH 5, or 20 mm acetic acid, pH 5, with 10 mm magnesium chloride or water for 3 h. After treating, leaves were removed from the rosette and cut into slices for all treatments except freezing, for which leaves were left whole. The leaf sections were soaked in 0.1 mg mL−1 Nile Red stain with 8% dimethyl sulfoxide for 1 h on ice. Leaf sections were then rinsed with deionized water three times before transport to the University of Nebraska-Lincoln Morrison Microscopy Core in deionized water. Measurement of tissue was done in deionized water for all samples on standard microscope slides. Images were taken on a Nikon Eclipse 90i upright fluorescence microscope with excitation at 561.4 nm and emission from 570 to 620 nm for Nile Red stain and with excitation at 640.6 nm and emission from 663 to 738 nm for chloroplast autofluorescence. Images were acquired sequentially and with a Z step of 1 μm. ImageJ software with the FIJI plugin package were used to process raw data. Cells were cropped manually by their dimensions and converted into two-dimensional images using Z projections of maximum intensity. Droplets were then hand counted on a per cell basis. Hand counts were statistically analyzed by ANOVA PROC GLIMMIX using SAS version 9.4 (SAS Institute). Assumptions were satisfied using a Gaussian response distribution, with the response variable recorded as the per cell number of lipid droplets. A completely randomized experimental design was implemented, with treatments considered as fixed effects.
Accession Numbers
The following genes referred to in the text are listed with their accession numbers: SFR2, At3g06510; PROTEIN KINASE, At4g32250; TOC64 has three homologs: TOC64-I, At1g08980; TOC64-III, At3g17970; and TOC64-V, At5g09420.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Confirmation of toc64 and protein kinase disruption lines and test of SFR2 activity during normal growth.
Supplemental Table S1. Proteins and peptides identified by SFR2 immunoprecipitation.
Supplementary Material
Acknowledgments
We thank Christoph Plieth for providing purified PtGFP and microcapillaries; Christian Elowsky and Melinda Frame for assistance with confocal microscopy; Doug Whitten of the Michigan State University Proteomics Facility for assistance with mass spectrometry; Paul Jarvis and Sean Maguire for toc64 mutant plants; and Kun Wang, Anna Hurlock, and Jaruswan Warakanont for helpful comments.
Glossary
- MGDG
monogalactosyldiacylglycerol
- TAG
triacylglycerol
- TGDG
trigalactolipid
- T-DNA
transfer DNA
- PG
phosphatidylglycerol
- FDR
false discovery rate
- DGDG
digalactosyldiacylglycerol
Footnotes
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–98ER20305 to C.B.), the University of Nebraska, and the National Science Foundation (Nebraska EPSCoR First Award to R.L.R.).
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aronsson H, Boij P, Patel R, Wardle A, Töpel M, Jarvis P (2007) Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J 52: 53–68 [DOI] [PubMed] [Google Scholar]
- Browse J, Xin Z (2001) Temperature sensing and cold acclimation. Curr Opin Plant Biol 4: 241–246 [DOI] [PubMed] [Google Scholar]
- Bruce BD, Perry S, Froehlich J, Keegstra K (1994) In vitro import of protein into chloroplasts. In Gelvin SB, Schilperoort RA, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Boston, pp 1–15 [Google Scholar]
- Chen M, Thelen JJ (2013) ACYL-LIPID DESATURASE2 is required for chilling and freezing tolerance in Arabidopsis. Plant Cell 25: 1430–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenkolbe T, Giavalisco P, Zuther E, Seiwert B, Hincha DK, Willmitzer L (2012) Differential remodeling of the lipidome during cold acclimation in natural accessions of Arabidopsis thaliana. Plant J 72: 972–982 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Tavakoli N, Kluge C, Mimura T, Sharma SS, Harris GC, Chardonnens AN, Golldack D (2001) Significance of the V-type ATPase for the adaptation to stressful growth conditions and its regulation on the molecular and biochemical level. J Exp Bot 52: 1969–1980 [DOI] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32: 278–289 [DOI] [PubMed] [Google Scholar]
- Fourrier N, Bédard J, Lopez-Juez E, Barbrook A, Bowyer J, Jarvis P, Warren G, Thorlby G (2008) A role for SENSITIVE TO FREEZING2 in protecting chloroplasts against freeze-induced damage in Arabidopsis. Plant J 55: 734–745 [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geilfus CM, Mühling KH, Kaiser H, Plieth C (2014) Bacterially produced Pt-GFP as ratiometric dual-excitation sensor for in planta mapping of leaf apoplastic pH in intact Avena sativa and Vicia faba. Plant Methods 10: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin SJ, Lynch JP (2003) Effects of salinity on cytosolic Na+ and K+ in root hairs of Arabidopsis thaliana: in vivo measurements using the fluorescent dyes SBFI and PBFI. J Exp Bot 54: 2035–2043 [DOI] [PubMed] [Google Scholar]
- Hays LM, Crowe JH, Wolkers W, Rudenko S (2001) Factors affecting leakage of trapped solutes from phospholipid vesicles during thermotropic phase transitions. Cryobiology 42: 88–102 [DOI] [PubMed] [Google Scholar]
- Heemskerk JW, Bogemann G, Wintermans JFGM (1983) Turnover of galactolipids incorporated into chloroplast envelopes: an assay for galactolipid-galactolipid galactosyltransferase. Biochim Biophys Acta 754: 181–189 [Google Scholar]
- Heemskerk JWM, Wintermans JFGM, Joyard J, Block MA, Dorne AJ, Douce R (1986) Localization of galactolipid-galactolipid galactosyltransferase and acyltransferase in outer envelope membrane of spinach chloroplasts. Biochim Biophys Acta 877: 281–289 [Google Scholar]
- Hurlock AK, Roston RL, Wang K, Benning C (2014) Lipid trafficking in plant cells. Traffic 15: 915–932 [DOI] [PubMed] [Google Scholar]
- Ji H, Wang Y, Cloix C, Li K, Jenkins GI, Wang S, Shang Z, Shi Y, Yang S, Li X (2015) The Arabidopsis RCC1 family protein TCF1 regulates freezing tolerance and cold acclimation through modulating lignin biosynthesis. PLoS Genet 11: e1005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Hirohashi T, Nakai M (2006) Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant Cell Physiol 47: 363–371 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H (2009) Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Leheny EA, Theg SM (1994) Apparent inhibition of chloroplast protein import by cold temperatures is due to energetic considerations not membrane fluidity. Plant Cell 6: 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang R, Li M, Li L, Wang C, Welti R, Wang X (2008) Differential degradation of extraplastidic and plastidic lipids during freezing and post-freezing recovery in Arabidopsis thaliana. J Biol Chem 283: 461–468 [DOI] [PubMed] [Google Scholar]
- Lineberger RD, Steponkus PL (1980) Cryoprotection by glucose, sucrose, and raffinose to chloroplast thylakoids. Plant Physiol 65: 298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantyla E, Lang V, Palva ET (1995) Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol 107: 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Muthan B, Benning C (2010) Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330: 226–228 [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Messerli MA, Gilroy S (2008) Imaging of the Yellow Cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol 147: 1690–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Plieth C, Sattelmacher B, Hansen UP (1997) Cytoplasmic Ca2+-H+-exchange buffers in green algae. Protoplasma 198: 107–124 [Google Scholar]
- Roston RL, Gao J, Murcha MW, Whelan J, Benning C (2012) TGD1, -2, and -3 proteins involved in lipid trafficking form ATP-binding cassette (ABC) transporter with multiple substrate-binding proteins. J Biol Chem 287: 21406–21415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roston RL, Wang K, Kuhn LA, Benning C (2014) Structural determinants allowing transferase activity in SENSITIVE TO FREEZING 2, classified as a family I glycosyl hydrolase. J Biol Chem 289: 26089–26106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki T, Saito K, Kawaguchi A, Kondo N, Yamada M (1990) Conversion of monogalactosyldiacylglycerols to triacylglycerols in ozone-fumigated spinach leaves. Plant Physiol 94: 766–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte A, Lorenzen I, Böttcher M, Plieth C (2006) A novel fluorescent pH probe for expression in plants. Plant Methods 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul O. (2002) Magnesium transport and function in plants: the tip of the iceberg. Biometals 15: 309–323 [DOI] [PubMed] [Google Scholar]
- Steponkus PL. (1980) Cellular and subcellular aspects of freezing injury and cold acclimation in higher plants. Cryobiology 17: 620–621 [Google Scholar]
- Steponkus PL. (1984) Role of the plasma membrane in freezing injury and cold acclimation. Annu Rev Plant Physiol Plant Mol Biol 35: 543–584 [Google Scholar]
- Suzuki N, Mittler R (2006) Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol Plant 126: 45–51 [Google Scholar]
- Thomashow MF. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Thorlby G, Fourrier N, Warren G (2004) The SENSITIVE TO FREEZING2 gene, required for freezing tolerance in Arabidopsis thaliana, encodes a β-glucosidase. Plant Cell 16: 2192–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Joseph RA, Steponkus PL (1995) Cold-acclimation of Arabidopsis thaliana: effect on plasma membrane lipid composition and freeze-induced lesions. Plant Physiol 109: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu HS, Roston R, Shiva S, Hur M, Wurtele ES, Wang X, Shah J, Welti R (2015) Modifications of membrane lipids in response to wounding of Arabidopsis thaliana leaves. Plant Signal Behav 10: e1056422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu HS, Shiva S, Hall AS, Welti R (2014a) A lipidomic approach to identify cold-induced changes in Arabidopsis membrane lipid composition. Methods Mol Biol 1166: 199–215 [DOI] [PubMed] [Google Scholar]
- Vu HS, Shiva S, Roth MR, Tamura P, Zheng L, Li M, Sarowar S, Honey S, McEllhiney D, Hinkes P, et al. (2014b) Lipid changes after leaf wounding in Arabidopsis thaliana: expanded lipidomic data form the basis for lipid co-occurrence analysis. Plant J 80: 728–743 [DOI] [PubMed] [Google Scholar]
- Waadt R, Manalansan B, Rauniyar N, Munemasa S, Booker MA, Brandt B, Waadt C, Nusinow DA, Kay SA, Kunz HH, et al. (2015) Identification of Open Stomata1-interacting proteins reveals interactions with Sucrose Non-fermenting1-Related Protein Kinases2 and with type 2A protein phosphatases that function in abscisic acid responses. Plant Physiol 169: 760–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Benning C (2011) Arabidopsis thaliana polar glycerolipid profiling by thin layer chromatography (TLC) coupled with gas-liquid chromatography (GLC). J Vis Exp 49: 2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, McKown R, Marin AL, Teutonico R (1996) Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MS, Uemura M, Steponkus PL (1994) A comparison of freezing injury in oat and rye: two cereals at the extremes of freezing tolerance. Plant Physiol 104: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Browse J (2000) Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ 23: 893–902 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.