Evolutionary plasticity of oleosin-lipid droplets in forming novel structures with unique functions in specific tissues/species is exemplified by those in vanilla leaf epidermis and avocado mesocarp.
Abstract
Subcellular lipid droplets (LDs) in diverse plant cells and species are coated with stabilizing oleosins of at least five phylogenic lineages and perform different functions. We examined two types of inadequately studied LDs for coated oleosins and their characteristics. The epidermis but not mesophyll of leaves of vanilla (Vanilla planifolia) and most other Asparagales species contained solitary and clustered LDs (<0.5 μm), some previously studied by electron microscopy and speculated to be for cuticle formation. In vanilla leaves, transcripts of oleosins of the U lineage were present in both epidermis and mesophyll, but oleosin occurred only in epidermis. Immuno-confocal laser scanning microscopy revealed that the LDs were coated with oleosins. LDs in isolated fractions did not coalesce, and the fractions contained heterogeneous proteins including oleosins and diverse lipids. These findings reflect the in situ structure and possible functions of the LDs. Fruit mesocarp of avocado (Persea americana) and other Lauraceae species possessed large LDs, which likely function in attracting animals for seed dispersal. They contained transcripts of oleosin of a novel M phylogenic lineage. Each avocado mesocarp fatty cell possessed one to several large LDs (5 to 20 μm) and at their periphery, numerous small LDs (<0.5 μm). Immuno-confocal laser scanning microscopy revealed that oleosin was present mostly on the small LDs. LDs in isolated fractions coalesced rapidly, and the fraction contained oleosin and several other proteins and triacylglycerols as the main lipids. These two new types of oleosin-LDs exemplify the evolutionary plasticity of oleosins-LDs in generating novel functions in diverse cell types and species.
Subcellular lipid droplets (LDs) containing neutral lipids for food reserves and other purposes are present in eukaryotes and prokaryotes (Hsieh and Huang, 2004; Rajakumari et al., 2008; Thiam et al., 2013; Pol et al., 2014). They exist in seeds, pollen, flowers, fruits, and some vegetative organs of advanced plants; the vegetative and reproductive organs of algae, primitive plants, fungi, and nematodes; mammalian organs/tissues such as mammalian glands and adipose tissues; and bacteria. Of all these LDs, those in seeds, whose constituent oils are extensively used for food and nonfood purposes, are the most prominent and were extensively studied early on (Hsieh and Huang, 2004; Chapman et al., 2012; Murphy, 2012).
Seeds of most plant species store oils (triacylglycerols [TAGs]) as food reserves for germination and post-germination growth. TAGs are present in subcellular spherical LDs (also called “oil bodies” or “lipid bodies”) of approximately 0.5 μm to 2 μm in diameter. Each LD has a matrix of TAGs surrounded by a layer of phospholipids and structural proteins termed “oleosins” (Hsieh and Huang, 2004; Shimada et al., 2008; Chapman et al., 2012; Murphy, 2012). The small size of LDs provides a large surface area per unit TAG, which would facilitate lipase binding and lipolysis during germination. LDs inside the cells of mature seeds or in isolated preparations are stable and do not cluster or coalesce. They are stable because their surface is covered completely with a layer of oleosins. An oleosin molecule has a long hydrophobic hairpin stretch of approximately 72 uninterrupted and conserved non/less-polar residues, with two 30-residue arms joined by a most-conserved 12-residue loop (-PX5SPX3P-, X being a nonpolar residue). The hairpin penetrates the LD matrix and is flanked by less-conserved short or long amphipathic N- and C-terminal peptides residing on the LD surface. These associations result in firm anchorage of the protein and stability of the LD. In maturing seeds, the oleosins, TAGs, and phospholipids are synthesized in endoplasmic reticulum (ER), from which budding LDs are released. Minor proteins termed “caleosin” (Frandsen et al., 2001), lipid droplet-associated protein (Horn et al., 2013), and small rubber particle protein (Oh et al., 1999) have been reported to be associated with LDs in seeds or other organs. These proteins do not have apparent LD-associated molecular structures, and their physical means for association with LDs and their functions have not been examined.
Other than those in seeds, LDs in high or low abundance are present in diverse cell types of all or restricted phylogenic plant groups (Lersten et al., 2006; Kim et al., 2002; Hsieh and Huang, 2004; Chapman et al., 2012; Kwiatkowska et al., 2015). They are present in moderate amounts, along with coating oleosin, in pollen of all examined species. They occur sparsely if at all in vegetative organs/tissues of leaves, roots, and meristems of Arabidopsis and many other studied plant species. The functions of the LDs in pollen and the above-mentioned vegetative organs are unknown and are assumed to be for lipid storage. LD clusters termed “tapetosomes” are present in the tapeta of all Brassicaceae family members (Hsieh and Huang, 2007; Huang et al., 2013). Tapetosomes assemble lipids in oleosin-coated LDs and flavonoids in vesicles, which, upon programmed cell death of the tapetum cells, are discharged to the pollen surface as the pollen coat. Two types of much-less studied LDs are present in special cells or tissues of restricted plant groups. The first type is the LD clusters in the epidermis of ovaries of Ornithogalum and leaves of Haemanthus (Kwiatkowska et al., 2010, 2015) in the Order Asparagales. These LD clusters could include cytoskeleton and other organelles such as mitochondria, Golgi, and small vesicles and vacuoles. They have been examined only by TEM and are speculated to be subcellular lipid sinks for internal metabolic uses and/or for export to become cuticle materials. The second type is the large LDs in fleshy mesocarp of fruits of avocado (Persea americana; Platt-Aloia and Thomson, 1981), oil palm (Elaeis guineensis; Bourgis et al., 2011), olive (Olea europaea; Rangel et al., 1997), and other species of diverse phylogenic origins; they are considered attractants to animals for seed dispersal.
A comprehensive bioinformatics analysis of oleosins based on available information in Huang and Huang (2015) revealed five oleosin phylogenic lineages. Primitive (P) oleosins are present on LDs in primitive organisms including green algae, mosses, and ferns. Genes encoding universal (U) oleosins of unknown function exist in all ferns and seed plants. Seed low (SL) and seed high (SH) molecular-weight oleosins occur on seed storage LDs in seed plants. Tapetum (T) oleosins are on LDs in clusters in tapetosomes in the tapeta of Brassicaceae species.
T oleosins are restricted to Brassicaceae and are absent in the closest family of Cleomaceae (Huang et al., 2013; Huang and Huang, 2015). They are associated with LDs in tapetosomes for the above-mentioned specific metabolic and physiological function. These findings inspired us to examine the LD clusters in Asparagales and the LDs in avocado fruit mesocarp for their possible association with oleosins and the characteristics of these putative oleosins. Avocado mesocarp was not known to possess oleosins, but a recent report found oleosin transcripts in the tissue (Huang and Huang, 2015); whether the LDs in mesocarp are coated with oleosin has not been explored.
We examined comprehensively the morphology and occurrence of LDs and their possible association with oleosins in green organs of diverse species of Asparagales, especially leaves of vanilla (vanilla planifolia) and blood lily (Scadoxus multiflorus, an ornamental plant). Asparagales LDs were coated with oleosins of enhanced quantities of the U oleosin phylogenic lineage. We explored the morphology and occurrence of large LDs (>5 μm) and the neglected numerous small LDs (<0.5 μm) in mesocarp of avocado and related species in the Family Lauraceae and found oleosins associated mainly with small LDs. These Lauraceae oleosins represent a new (6th) oleosin lineage restricted to the fruit mesocarp of Lauraceae. Here, we report our findings.
RESULTS
In Most Asparagales Families, the Epidermis Cells in Green Organs Contained LDs of Moderate Abundance
Earlier, TEM studies showed clustered LDs associated with cytoskeleton and other subcellular structures in the epidermis of the ovary of Ornithogalum and, with lesser detail, leaves of Haemanthus; both species belong to the Order Asparagales (Kwiatkowska et al., 2010, 2015). We used confocal laser scanning microscopy (CLSM) to examine comprehensively LDs in epidermis of leaves and other green organs of species of the Asparagales families obtained locally. The samples were stained for lipids with Nile Red or BODIPY for easy observation.
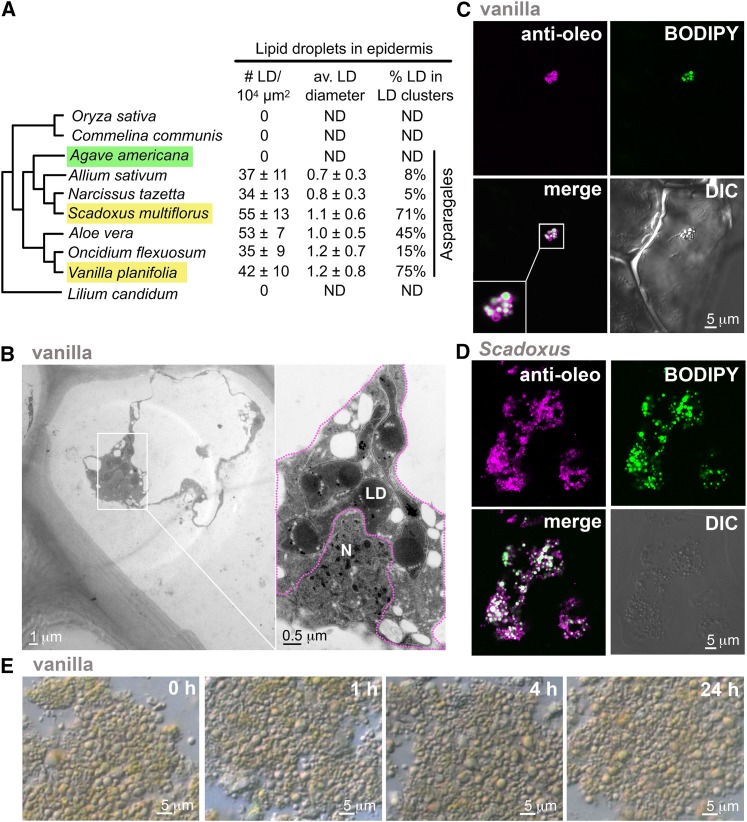
Solitary and clustered LDs were present in the leaf epidermis in species of many Asparagales families (Fig. 1A), with the exception of several agave species of Asparagaceae, which had no detectable LDs in leaf epidermis and mesophyll (Supplemental Fig. S1). In the former species, LDs were present in moderate abundance (approximately 0.3% of total cell volume, calculated from Fig. 1A). They were in solitary and clustered forms of varying proportions in different cells of the epidermis of a mature leaf. LD clusters, each of 10-μm to 20-μm diameter containing about a dozen or more LDs of <0.5 μm in diameter, were present as tight or loose structures. The tight and loose LD clusters could occur in adjacent cells in a leaf or even within the same cell (Supplemental Fig. S1). Figure 1B is a TEM image of a portion of a vanilla epidermis cell that was highly vacuolated and contained a tight LD cluster adjacent to a nucleus. The LD cluster commonly occurred adjacent to the nucleus in vanilla and other Asparagales species. Whether this positioning had subcellular metabolic or regulatory significance remains to be elucidated. Epidermis cells in general were highly vacuolated, and large subcellular organelles, such as the nucleus, LD clusters, non-green plastids, and mitochondria, were present together in large patches of cytoplasm. Solitary and clustered LDs in a low or high proportion in a cell of a mature leaf could be related to biogenesis and function during leaf development. Nevertheless, our general survey findings cannot substantiate this relationship. Within a leaf, LDs were present in both upper and lower epidermis but absent in the mesophyll (Supplemental Fig. S1). In addition to being present in the epidermis of leaves, LDs of solitary and clustered forms were present in the epidermis of green organs (stem, aerial root, and flower parts) of the species.
Figure 1.
Microscopy studies of LDs in epidermis of Asparagales species. A, Solitary and clustered LDs in leaf epidermis of Asparagales and reference species. Epidermis was stained with Nile Red or BODIPY for LDs and observed by light microscopy. The table shows the number of individual LDs (solitary and in clusters) per area, the average diameters of LDs (solitary and individual LDs within clusters), and proportion of LDs in clusters (individual LDs in clusters versus LDs in solitary entities). Agave (highlighted in green) had no LDs, whereas Scadoxus and vanilla (highlighted in yellow) had LDs and were studied further. B, TEM image of a portion of vanilla leaf epidermis. Magnification of the dense cytoplasm patch is shown on the right. The cell was highly vacuolated. The major patch of cytoplasm occurred between large vacuoles and contained a LD cluster, a nucleus (N), and other cytoplasmic materials. The margin of the LD cluster cannot be defined concisely because of its uncertain association with other subcellular structures; nevertheless, it is tentatively indicated with a dotted magenta line. C, Immuno-CLSM image of a vanilla epidermis cell. Magnification of the tight LD cluster is shown on the lower left. The images show a tight LD cluster containing many individual LDs. BODIPY stained (in green) individual LDs in the cluster. Antibodies against vanilla U1 oleosin reacted (in magenta) with the LDs. In the merged image, oleosin appears more on the periphery of individual LDs, resulting in a magenta coat enclosing a white matrix. D, Immuno-CLSM image of a Scadoxus epidermis cell. The images show several loose LD clusters, each containing many individual LDs. BODIPY stained (in green) individual LDs in the clusters. Antibodies against Scadoxus U1 oleosin reacted (in magenta) with the LDs. In the merged image, oleosin appears more on the periphery of individual LDs, resulting in a magenta coat enclosing a white matrix. E, Images of vanilla LDs in an isolated fraction observed by light microscopy at time intervals. The LDs did not coalesce during the 24-h incubation.
We focused our studies on vanilla, because its transcriptomes of different organs were available, and we sought confirmatory or repelling evidence from Scadoxus multiflorus, another Asparagales species. In these two species, solitary and clustered LDs were present in both upper and lower epidermis, and absent in mesophyll (Supplemental Fig. S1), of different green parts of the leaf, including (studied in Scadoxus) the blade, sheath, and mid-vein areas. In general, LDs were present in the epidermis of diverse green (i.e. aerial) parts of the plant, with no apparent pattern correlating the occurrence of clustered versus solitary LDs by age or development.
Leaves of Asparagales Species Contained Transcripts Encoding U Oleosins
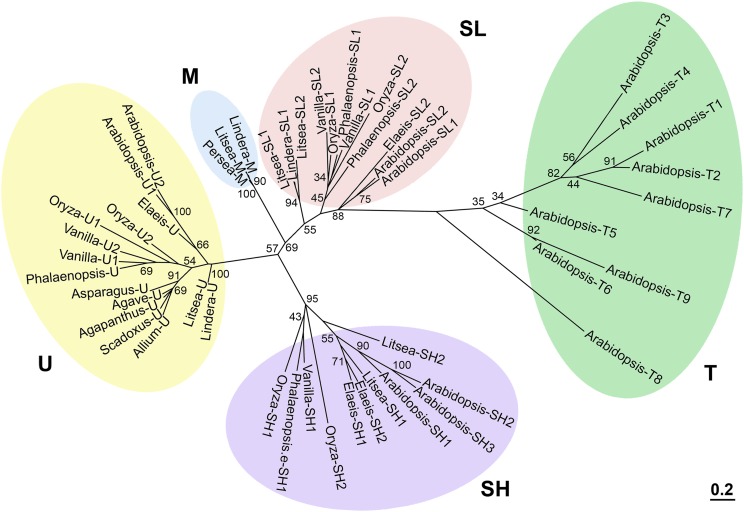
We explored whether oleosin transcripts were present in the leaves of Asparagales species. The dicot Arabidopsis and the monocot rice (Oryza sativa) were used as reference species; they have no oleosin transcripts in their leaf transcriptomes (Supplemental Table S1). With Arabidopsis oleosin amino-acid sequences of the conserved hairpin of approximately 72 uninterrupted non-polar residues as queries, we searched for in silico translated oleosins in available transcriptomes and genome sequences (Supplemental Table S1). In addition, we used RNA-seq to construct transcriptomes of leaves of Scadoxus (yielding one transcript encoding a full-length U oleosin [Supplemental Table S1] and another encoding an incomplete-length oleosin of unknown oleosin lineage) and Ornithogalum caudatum (yielding a transcript encoding an incomplete-length oleosin). Oleosins obtained from these sequence databases are categorized into the U, SL, and SH oleosin phylogenic lineages (Supplemental Table S1) in accordance with the established sequence characteristics of each lineage (Huang and Huang 2015) and their placement in a phylogenic tree of oleosins (Fig. 2). An alignment of residue sequences (Supplemental Fig. S2) shows that all Asparagales leaves contain transcripts encoding U oleosins (Supplemental Table S1). These U oleosins possess (1) the oleosin hallmark hairpin and its loop of PX5SPX3P (X representing a nonpolar residue); (2) a C-terminal sequence highly conserved in residues and length; and (3) the characteristic C-terminal end residues of AAPGA. In a phylogenetic tree (Fig. 2), the U-oleosin lineage includes those from vanilla and other Asparagales species, as well as Arabidopsis and rice. In species with more than one U oleosin, the assignment of U1, U2, etc. was made arbitrarily.
Figure 2.
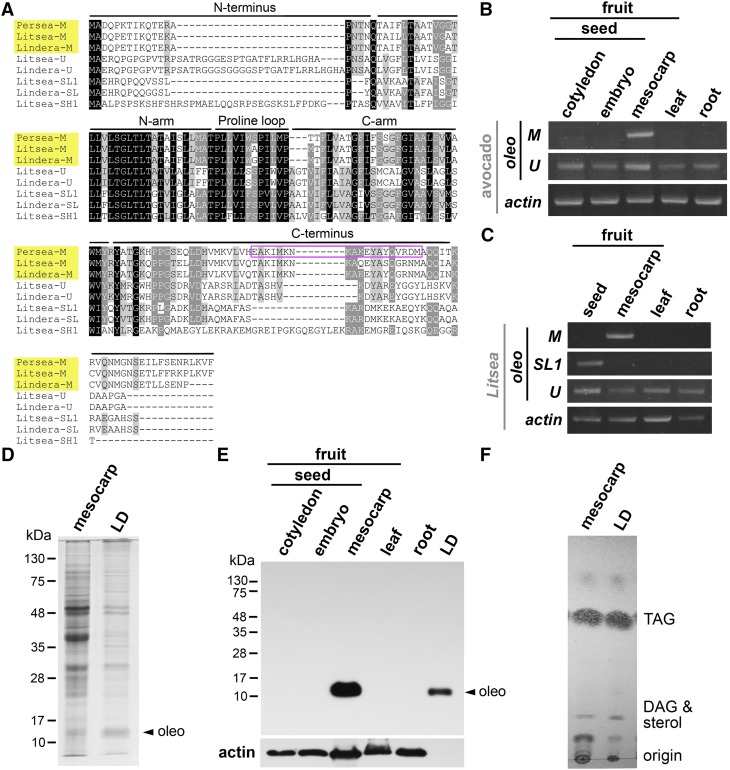
Unrooted phylogenetic tree of oleosins of Asparagales and Lauraceae species and of Arabidopsis and rice. The four previously recognized lineages of U, SL, SH, and T oleosins as well as the new lineage of M oleosins are highlighted with colors. Oleosins of green algae and primitive plants of the recognized P (primitive) lineage, for simplicity, are not included. For presentation clarity, oleosins of plant species are indicated only with the genus names, which include Arabidopsis (Arabidopsis), Rice (O. sativa), Agapanthus (A. praecox), Agave (A. tequilana), Allium (A. sativum), Scadoxus (S. multiflorus), Lindera (L. glauca), Litsea (L. cubeba), Persea (P. Americana), Phalaenopsis (P. equestris), and Vanilla (V. planifolia). Oleosins of the monocot Elaeis (E. oleifera), with LDs in mesocarp but distant from the Family Lauraceae in phylogeny, are included; Elaeis has no M oleosin. GenBank Accession numbers for the sequences are shown in “Materials and Methods.” The tree was constructed by using the Maximum-Likelihood method with a Jones-Taylor-Thornton substitution matrix supported by a bootstrap test of 1000 resamplings from the aligned oleosin sequences with the PHYLIP package (http://evolution.genetics.washington.edu/phylip/doc/main.html). The scale bar indicates the number of amino-acid substitutions per site. The numbers at the nodes represent the percent bootstrap support.
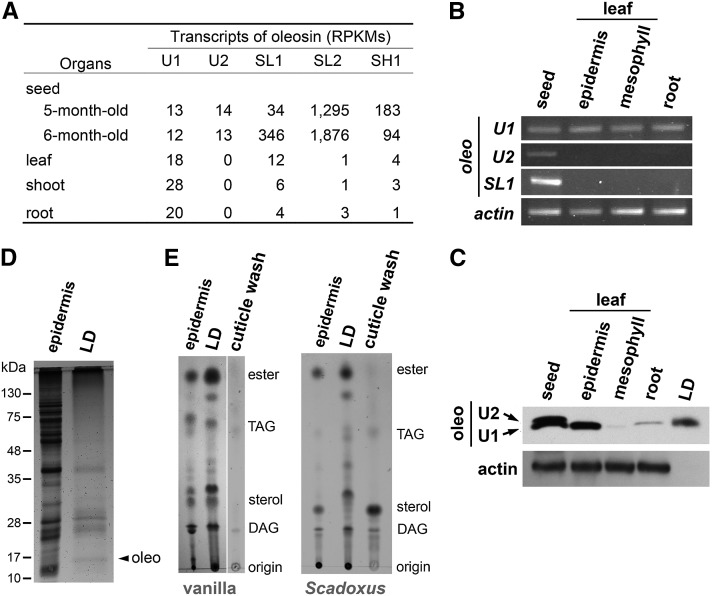
Transcript data for vanilla have the most details among those of Asparagales species (Fig. 3A). Vanilla transcripts of 2 U (U1 and U2), 2 SL (SL1 and SL2), and 1 SH oleosins are present in seed, leaves, shoots, and/or roots. The SL1-2 and SH1 oleosin transcripts are abundant in read per kilobase per million mapped reads (RPKMs) and largely restricted to seed, as expected for SL and SH oleosins in various plant species (Huang and Huang, 2015). U2 oleosin transcript at a low level is also restricted to seed. U1 oleosin transcript, albeit at low levels, is present in all organs but with slightly higher levels in leaves, shoots, and roots. These distributions suggest that U1 oleosin transcript could encode oleosin in leaf epidermis. Our RT-PCR data (Fig. 3B) are consistent with the transcriptome data. SL1 and U2 oleosin transcripts were present mostly if not only in seed, whereas U1 oleosin transcript was present at approximately equal levels in all organs (seed, leaf epidermis, leaf mesophyll, and root). In other Asparagales species, U oleosin transcript is always present in leaf transcriptomes (Supplemental Table S1). Thus, U oleosin transcript is present in leaves and likely other green organs of all the Asparagales species and at elevated levels.
Figure 3.
Oleosin transcripts, oleosins, and lipids in various organs and tissues of vanilla and Scadoxus. A, Transcripts of five oleosins in various organs of vanilla. The five oleosins are categorized into oleosin lineages on the basis of their sequences. The levels of the various oleosin transcripts are in RPKMs. Raw data are from NCBI. B, RT-PCR of transcripts of various oleosins and actin in different organs/tissues of vanilla. Actin transcript was used as a quantity reference. C, Immuno-SDS-PAGE of oleosins in extracts of different organs/tissues and LD fraction of vanilla. Actin protein was used as a quantity reference parallel to actin transcript in RT-PCR (B). The antibodies against U1 oleosin also recognized U2 oleosin in the seed sample; U1 (16.97 kD if no post-translational modification) and U2 (17.00 kD) oleosins were barely separated in the gel. D, SDS-PAGE of total extract and LD fraction of vanilla epidermis. The gel was subjected to silver staining. Oleosins identified with immunoblotting are indicated. Positions of molecular-weight markers are shown on the left. E, TLC of lipids in vanilla and Scadoxus leaf epidermis. Samples included total epidermis extract, LD fraction and cuticle wash (wash of leaf surface with chloroform). Positions of lipid markers are shown on the right. Oleo, Oleosin.
The leaf transcriptomes of several agave species in Asparagales also possess U oleosin transcripts (Supplemental Table S1). Yet, both epidermis and mesophyll cells of Agave americana (Fig. 1A; Supplemental Fig. S1) and two other species (A. victoriae-reginae and A. potatorum) we had examined did not have LDs.
In Vanilla Leaves, Both Epidermis and Mesophyll Contained U1 Oleosin Transcript, But Only the Epidermis had U1 Oleosin
In vanilla leaves, both epidermis and mesophyll contained U1 oleosin transcript, as revealed by RT-PCR (Fig. 3B). Yet, the epidermis but not the mesophyll had LDs (Fig. 1 and Supplemental Fig. S1). We tested whether U1 oleosin protein was present in both tissues or only in the epidermis. The peptide of a sequence unique to vanilla U1 oleosin (also highly similar to that in U2 oleosin; Supplemental Fig. S2) was synthesized and used to produce rabbit antibodies for oleosin detection by immuno-SDS-PAGE (Fig. 3, C and D). Two oleosins, presumably representing U1 and U2 oleosins (of 16.97 and 17.00 kD, respectively, assuming no post-transcriptional modification) and barely separated in the gel, were present in the seed sample. U1 oleosin was present in the leaf epidermis fraction but almost absent in the leaf mesophyll fraction. U1 oleosin was present in the root sample and the leaf epidermis LD fraction (Fig. 3C). These findings for oleosins (Fig. 3C) are consistent with the transcriptome (Fig. 3A) and RT-PCR (Fig. 3B) data indicating that U1 oleosin transcript and U1 oleosin co-existed in all tested organs/tissues, with the clear exception of U1 oleosin present in leaf epidermis but absent in mesophyll.
In the above studies of U1 oleosin transcript by RT-PCR (Fig. 3B) and U1 oleosin protein by immuno-SDS-PAGE (Fig. 3C) in vanilla leaf epidermis and mesophyll, we used similar references of actin gene transcript and protein for sample loading on the gels, and so the findings on transcript and protein are directly comparable. Overall, both epidermis and mesophyll contained U1 oleosin transcript but only the LD-possessing epidermis had U1 oleosin protein.
Immuno-CLSM Shows that U1 Oleosin Coated the Surface of LDs in Vanilla and Scadoxus Epidermis
Immuno-CLSM was performed with vanilla epidermis (Fig. 1C). BODIPY stained (shown in green) individual LDs in a LD cluster, whereas anti-U1-oleosin reacted (magenta) with the oleosin mostly on the LD surface. The merged image shows that each LD has a white matrix and a magenta coating. Thus, individual LDs in a vanilla LD cluster are similar to the well-studied seed LDs in having oleosin enclosing the LDs.
We extended the study from vanilla to Scadoxus (Fig. 1D). Unfortunately, the antibodies against U1 oleosin of vanilla did not react strongly with that in Scadoxus. We synthesized a peptide of a sequence of the Scadoxus U oleosin (Supplemental Fig. S2) and used it to produce rabbit antibodies. In Scadoxus epidermis, individual LDs in loose clusters had the matrix stained (green) with BODIPY and the surface oleosin reacted (magenta) with anti-U-oleosin, and the merged image shows individual LDs with a white matrix and a magenta coating (Fig. 1D). Thus, individual LDs of solitary and clustered entities in the epidermis of Asparagales species are similar to LDs in seeds in having a matrix of lipids enclosed with oleosins.
Solitary and Clustered LDs of Vanilla Did Not Coalesce Upon Isolation and Further Incubation
We prepared an isolated LD fraction from vanilla by gentle grinding of the peeled epidermis and flotation centrifugation. In the fraction, solitary and clustered LDs were in a concentrated suspension and did not coalesce after 24-h incubation (Fig. 1E). Thus, the epidermis LDs are similar to seed LDs in being stable without coalescence in isolated and concentrated preparations. The stability of seed LDs is attributed to oleosins totally covering and shielding the lipid matrix (Hsieh and Huang, 2004). The stability of Asparagales epidermis LDs in solitary or clustered entities should be due to a similar attribution.
Isolated Vanilla LD Preparations Contained Heterogeneous Proteins Including Oleosin and Diverse Lipids
Isolated vanilla LD preparation contained numerous proteins resolved by SDS-PAGE (Fig. 3D). A minor protein of 17 kD was identified as “U1 oleosin” by immunoblotting. The heterogeneity of the proteins in the isolated LD fraction should reflect that in vivo, the LD clusters are associated with cytoskeleton and other subcellular constituents (Fig. 1B; Kwiatkowska et al., 2015).
Isolated epidermis LD fractions of vanilla and Scadoxus contained diverse lipids, as resolved by TLC (Fig. 3E). TAGs were present but were not the dominant lipids. There were substantially more wax esters, fatty esters, and/or steroyl esters, as well as free sterols, tentatively identified via their co-migrations with marker lipids. The lipid diversity of the epidermis LD fraction contrasts with the domination of TAGs in LDs of seeds (Hsieh and Huang, 2004). The epidermis LD lipids differed slightly (in vanilla) or greatly (in Scadoxus) from those of the total epidermis lipids and the cuticle surface-washed lipids (washed with chloroform); all these lipid samples had diverse constituents.
The Fat Cell in the Mesocarp of Mature Avocado Fruit Contained One to Several Large LDs (5–20 μm) and Numerous Small LDs (0.1–0.5 μm)
A mature avocado fruit mesocarp contains approximately half of its dry weight as fats, which are highly saturated TAGs and are solid at room temperature (Kilaru et al., 2015). The mesocarp tissue includes approximately 95% fat-rich cells (herein called fat cells), approximately 2% thick-walled, terpenoid-rich idioblasts (herein called oil cells) scattering among the fat cells, and vascular-bundle cells (Platt-Aloia and Thomson, 1981). Both the fat cells and oil cells have irregular but rather spherical-to-cubical shapes of approximately 0 μm in length at each dimension. A fat cell has one to several large LDs of 5-μm to 20-μm diameter, whereas an oil cell has one large oil drop occupying most of the cell volume. Litsea and Lindera species and other members of Lauraceae also have fruit morphology similar to that of avocado (Little et al., 2009).
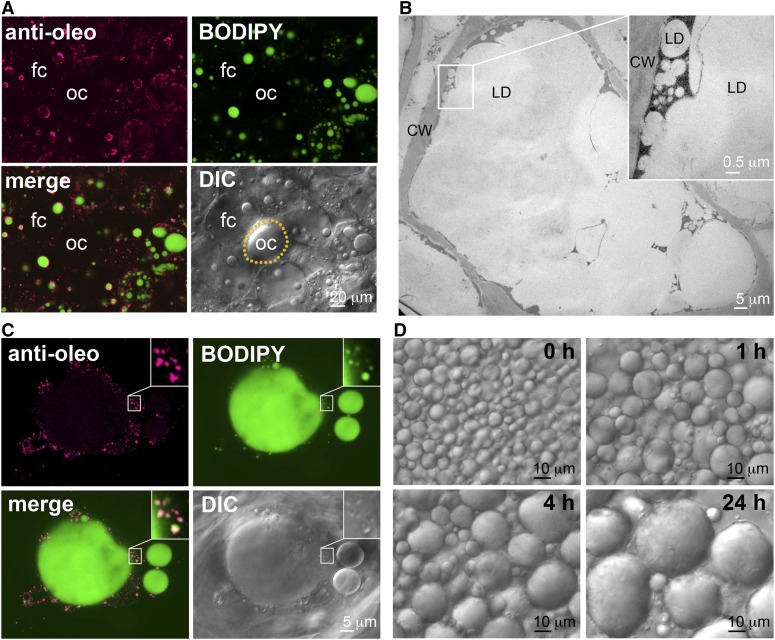
In our studies, the lipid dye BODIPY stained (shown in green) the LDs in the fat cells but not the oil drops in oil cells (Fig. 4A). The BODIPY stain identified the large LDs (5–10 μm) and numerous small LDs (approximately 0.1–0.2 μm) near or on the surface of the large LDs in fat cells (Fig. 4, A and C). These small LDs can be seen by TEM (Platt-Aloia and Thomson, 1981; Fig. 4B) and have not been previously examined.
Figure 4.
Microscopy studies of LDs in avocado mesocarp. A, Immuno-CLSM image of avocado mesocarp cells at low magnification. The images show an oil cell (oc, circled with yellow dots for recognition) at the center adjacent to several fat cells (fc). BODIPY stained (in green) both the large and small LDs in the fat cells but not the large oil drop in the oil cell. Antibodies against avocado M oleosin reacted (in magenta) with the small but not the large LDs in the fat cells and did not react with any constituent in the oil cell. B, TEM image of a portion of an avocado fat cell, showing several large LDs with numerous small LDs on the periphery. Magnification of a junction between a large LD and numerous small LDs is shown on the top right. The LDs and cell wall are labeled. C, Immuno-CLSM image of a portion of an avocado mesocarp fat cell. The images show a large LD and numerous adjacent small LDs. Magnification of a junction between a large LD and numerous small LDs is shown on the top right. BODIPY stained (in green) large and small LDs. Antibodies against avocado M oleosin reacted (in magenta) mostly with the small LDs. In the merged image, oleosin appears more on the periphery of individual small LDs, resulting in a magenta coat enclosing a white matrix. D, Images of avocado LDs in an isolated fraction observed by light microscopy at time intervals. The LDs coalesced during the 24-h incubation. CW, cell wall.
Mesocarp of Avocado and Other Species of Lauraceae Possessed a Mesocarp-Specific Oleosin Transcript Encoding M Oleosin of a New Oleosin Lineage
We reported recently (Huang and Huang, 2015) that an oleosin transcript (sequence shown in Fig. 5A) at a moderate level (several hundred RPKMs; Supplemental Table S1) is present in the transcriptomes of avocado mesocarp of different developmental stages. We explored whether the oleosin transcript was indeed present in and restricted to mesocarp, whether it was present in other species of closely or distantly related phylogeny, whether it produced oleosin, and whether the oleosin was present on the surface of LDs.
Figure 5.
Oleosin transcripts, oleosins, and lipids in various organs and tissues of avocado and other Lauraceae species. A, Alignment of the amino-acid sequences of representative oleosins of Lauraceae. The sequences of M, U, SL, and SH oleosins were derived from transcriptomes of mesocarp of avocado (Persea) and whole fruits of Litsea and Lindera. Names of M oleosins of the three Lauraceae species are highlighted in yellow. The variable N-terminal sequence, the conserved hairpin (divided into N-arm, Pro loop, and C-arm), and the variable C-terminal sequence are indicated. Black and gray colors highlight the highly and moderately conserved residues, respectively. The sequence of Persea M oleosin used for construction of a synthetic peptide for rabbit antibody production is boxed. GenBank Accession numbers for the sequences are shown in “Materials and Methods.” B, RT-PCR of transcripts of M and U oleosins in different avocado organs/tissues. Actin transcript was used as a quantity reference. C, RT-PCR of transcripts of M, SL, and U oleosins in different Litsea organs/tissues. Actin transcript was used as a quantity reference. D, SDS-PAGE of total mesocarp extract and LD fraction of avocado mesocarp. The gel was stained with Coomassie Blue. Oleosin identified with immunoblotting (shown in E) is indicated. Positions of molecular-weight markers are shown on the left. E, Immuno-SDS-PAGE of M oleosin in various organs/tissues and isolated LD fraction of avocado. Actin protein was used as a quantity reference and for a direct comparison with the RT-PCR data (with actin transcript as a quantity reference) in (B). Oleosin identified with immunoblotting is indicated. Positions of molecular-weight markers are shown on the left. F, Thin-layer chromatography of lipids in avocado mesocarp and isolated LD fraction. Positions of lipid markers are shown on the right. Oleo, Oleosin.
RT-PCR with avocado samples showed that the mesocarp oleosin transcript in the above-mentioned transcriptomes was present and restricted to the mesocarp (Fig. 5B), whereas an U-oleosin transcript was present in all samples tested including mesocarp, seed cotyledons, seed embryos, leaves, and roots. We attempted to explore the relationship between the mesocarp oleosin and presumed seed-specific oleosins. The avocado mesocarp transcriptomes contain transcripts of the mesocarp-specific and U oleosins (Supplemental Table S1) and no transcript of seed-specific SL or SH oleosin. No avocado genome sequence is available for retrieval of seed-oleosin genes. We used highly degenerated PCR primers related to the DNA sequences encoding the conserved hairpin peptides of the mesocarp-specific and U oleosins of avocado and seed oleosins of other species and were unsuccessful with RT-PCR or PCR in obtaining a SL or SH oleosin transcript. So, we worked with Litsea cubeba, also a member of Lauraceae, whose total fruit (i.e. including mesocarp and seed) transcriptome is available (Supplemental Table S1). The Litsea fruit transcriptome has one mesocarp-specific, one U, and several SL and SH oleosin transcripts. RT-PCR of Litsea samples (Fig. 5C) showed the mesocarp oleosin transcript restricted to the mesocarp, U oleosin transcript present in all organs tested, and SL1 oleosin transcript confined to seed. An alignment of residues (Fig. 5A) and a phylogenetic tree (Fig. 2) of the oleosins of avocado, Litsea and Lindera, showed that mesocarp-specific oleosins are more similar in sequences among themselves than to those of SL and U oleosins of the same or different Lauraceae species. We did not find oleosins with characteristics of the Lauraceae mesocarp oleosins in available transcriptomes or genome-sequence databases (after in silico translation) of other species in angiosperms we had studied (Huang and Huang, 2015), including the Order Magnoliales (Annona squqmosa and Liriodendron tulipifera fruit transcriptomes) closest to Laurales and the distant Order Piperales (Piper nigrum fruit transcriptome). Thus, current information indicates that mesocarp-specific oleosin is present only in mesocarp of species of Lauraceae.
The Lauraceae mesocarp oleosins, herein termed “M” oleosins, represent a new oleosin lineage, with distinct sequence, tissue location (mesocarp), phylogeny (Lauraceae, which have drupe fruit), and function of its associated LDs (attracting animals for seed dispersion). These distinctions of M oleosins parallel those of T oleosins, which are unique in sequence, tissue location (tapetum), phylogenic occurrence (Brassicaceae), and function of their associated LD clusters (tapetosomes, which assemble, store, and discharge lipids and flavonoids to the surface of adjacent pollen; Hsieh and Huang, 2007; Huang et al., 2013).
M Oleosin Was Present Abundantly on Small LDs But Sparsely or Not At All on Large LDs in Avocado Fat Cells
Rabbit antibodies were prepared against a synthetic peptide of a short sequence of avocado M oleosin (Fig. 5A) and used for immuno-CLSM and immuno-SDS-PAGE. In avocado mesocarp, M oleosin was absent in oil cells but present in fat cells, as shown by immuno-CLSM (Fig. 4A). Within the fat cells, it was present scarcely on large LDs but abundantly on small LDs adjacent to the large LDs (Fig. 4, A and C). BODIPY stained (shown in green) the matrix of the small and large LDs, whereas anti-M-oleosin reacted (in magenta) with the oleosin present only on the small LDs. The merged image shows individual small LDs with a white matrix and a magenta coating (Fig. 4C, inset). This image of oleosin covering the small LDs (Fig. 4C) is less clear than those for the vanilla (Fig. 1C) and Scadoxus LDs (Fig. 1D) because many of the avocado small LDs were less than 0.5 μm (Fig. 4B) and on the borderline of CLSM resolution. The images (Fig. 4, A and C) also show some small LDs with oleosin coating only portions of the sphere or without oleosin coating, and occasional droplets of oleosin apparently without adjacent LDs (which could have resulted from the angle of CLSM observation of LDs with partial oleosin coating).
Avocado LDs Coalesced Rapidly Upon Isolation and Subsequent Incubation
We prepared an isolated LD fraction from avocado mesocarp by gently chopping the tissue in a dilute buffer with a razor blade, letting the extract stand for 5 min, and scooping the fat on the extract surface. In this fraction, the LDs were in a concentrated suspension, and the observable large LDs coalesced in less than 1 h and continuously within 24-h incubation (Fig. 4D). This in vitro instability of LDs from avocado contrasts the in vitro stability of LDs from vanilla leaf epidermis (Fig. 1E) and seeds (Hsieh and Huang, 2004), likely because of the absence of oleosin and other amphipathic proteins and non-proteins on the surface of the avocado large LDs.
Isolated Avocado LD Fractions Contained Several Proteins Including Oleosin and Lipids of Mostly TAGs
As resolved by SDS-PAGE (Fig. 5D), the isolated avocado LD fraction contained several proteins, one of which was identified via immunoblotting as the 17-kD M oleosin (Fig. 5E). The total mesocarp extract possessed numerous proteins including a minor M oleosin. M oleosin was absent in seed cotyledon, seed embryo, leaves, and roots (Fig. 5E). TLC showed that the lipids in the total mesocarp extract and the isolated LD fraction were predominantly TAGs (Fig. 5F).
DISCUSSION
In addition to the oleosin-coated LDs present ubiquitously in seed and pollen, three types of specialized subcellular oleosin-coated solitary or clustered LDs are known to occur in restricted tissues and phylogenic plant groups. The first type is the well-studied tapetosomes of defined morphological and biochemical structures, biogenesis, and function in the tapeta of Brassicaceae species (Wu et al., 1997; Hsieh and Huang, 2007; Huang et al., 2013). Individual LDs within a tapetosome are coated with T oleosin of an independent oleosin lineage. The second type is the much less-studied solitary and clustered LDs in epidermis of green organs of most Asparagales species, whose biochemical structure, biogenesis, and function are mostly unknown (this report; Kwiatkowska et al., 2010, 2015). Pursuit of this knowledge will require different technical approaches in addition to electron microscopy and guidance from the Brassicaceae tapetosome studies. The third type is the large and small LDs aiding seed dispersal in the mesocarp of avocado and other Lauraceae species. Only the small LDs are coated with M oleosin of a novel oleosin lineage (this report). Occurrence of the three types of solitary and clustered LDs in highly restricted cells and phylogenic plant groups, as well as their coating oleosin belonging to unique or existing lineages, suggest that these three types of oleosin-LDs evolved recently at the order or family levels. Other unstudied solitary or clustered LDs of unique structure and functions, with or without oleosin coating, are likely present in specific cells of restricted plant groups. The plasticity of LDs and oleosins to undergo evolutionary changes to exert diversified functions in specific cells is obvious.
The LD fractions of vanilla and Scadoxus contained heterogeneous lipids (TAGs not being the dominating constituents; Fig. 3E), which differed from that of the cuticle lipids obtained with a chloroform wash. The finding neither contradicts nor supports the earlier speculation that the epidermis LDs were involved in cuticle lipid synthesis (Kwiatkowska et al., 2015). If the speculation turns out to be valid, we could offer the following explanations of our findings: (1) Lipids in the cuticle chloroform wash should greatly differ from total cuticle lipids, which would have been modified and covalently linked from putative precursors in the epidermis LD clusters. (2) LD clusters in the isolated LD fraction contained lipids of other subcellular components (Kwiatkowska et al., 2015) unrelated to the lipids of individual LDs. Further studies are required to substantiate or negate the speculation. Green organs such as leaves in Asparagales species contain thick cuticles, and thus their solitary and clustered LDs present in epidermis and absent in mesophyll could be related to cuticle lipid formation. Alternatively, the epidermis LDs may be warehouses of food or lipids for starvation and rapid repair or regeneration of new cells upon damage. These and other alternative possibilities, by themselves or in addition to being cuticle precursors, have not been explored.
In vanilla leaves, the epidermis but not the mesophyll has LDs (Fig. 1A and Supplemental Fig. S1). Both the epidermis and mesophyll have moderate levels of U1 oleosin transcript, but only the epidermis has complementary levels of U1 oleosin (Fig. 3). The very low level of oleosin in the mesophyll (Fig. 3C) may (1) represent oleosins in contaminating epidermis in the mesophyll preparation, or (2) reflect that in the absence of LDs, U1 oleosin was not synthesized or that the synthesized U1 oleosin was not retained. The finding is significant in the exploration of heterologous expression of oleosin genes in different organisms/cells, in that oleosin would be present in significant levels only together with endogenous LDs, whose synthesis may or may not be induced by oleosin. In the various agave species, the leaves possess U oleosin transcripts (Supplemental Table S1) but no LDs (Fig. 1A and Supplemental Fig. S1); whether oleosin is present is unknown. Overall, the U oleosin gene(s) might have evolved to express in the leaves (both epidermis and mesophyll) of all Asparagales species, but only the epidermis of most families (Agavaceae being an exception) also evolved to have LDs, resulting in conferring an advantage in cuticle synthesis or other functions.
The fruit mesocarp of avocado, oil palm, and olive containing abundant large LDs has been used extensively for human consumption. Yet, the structure and biogenesis of these LDs are unknown. In avocado and related species of Lauraceae, the novel M oleosin covers only or mainly the small LDs and not the large LDs, which are highly unstable in vitro. These small LDs could represent various forms of nascent oleosin-coated LDs, still attached to or freshly budded-off from ER, delivering the TAG matrix cargo to the expanding large LDs. After the delivery, the leftover coat oleosin would not be retained on the large LDs and would be degraded. It is unlikely that the oleosin-coated small LDs were those separated from the large LDs during maturation or storage. Future examination of the biogenesis relationship between the small and large LDs throughout the 6-month-long fruit maturation and then storage is warranted. In oil palm and olive, as well as mango (Mangifera indica) and other less-used fruits, the structure and biogenesis of the mesocarp LDs are completely unknown. These and all other fruits of the drupe type (a thin exocarp, fleshy mesocarp, and hard endocarp enclosing a seed) with the mesocarp containing largely fats or carbohydrates are scattered throughout the seed-plant phylogeny; they likely arose via convergent evolution. It is intriguing to comprehend whether mesocarp LDs and oleosins (if present) in these diverse drupes are different or quite similar because of the need for analogous biosynthesis and functioning. TEM shows that large LDs with numerous adjacent small LDs similar to those in avocado are present in the mesocarp of oil palm (Bourgis et al., 2011) and olive (Rangel et al., 1997). Nevertheless, available transcriptomes of oil palm mesocarp (0 RPKM of any oleosin transcript) and olive mesocarp plus other tissues (<10 RPKMs of any oleosin transcript) in NCBI (http://www.ncbi.nlm.nih.gov) have no or negligible oleosin transcripts.
MATERIALS AND METHODS
Plant Materials
Vanilla (Vanilla planifolia) was obtained from Touyuan District Agricultural Research and Extension Station, Taiwan. Vanilla seed was collected from 6-month-old maturing fruit. Scadoxus multiflorus plants were purchased from a local flower market in Taipei, Taiwan and maintained in greenhouses. Vanilla and Scadoxus epidermis and mesophyll were separated by peeling off the upper and lower epidermis with use of forceps via visual observation. The upper and lower epidermis were combined and then washed with distilled water. No cross contamination (<1%) of the epidermis and mesophyll fractions was observed.
Hard and ready for bench-top softening fruit of avocado (Persea Americana, cv Hass) was purchased from a local supermarket or obtained from the Chiayi Agricultural Experiment Branch, Taiwan. Mature Litsea cubeba fruit was collected from a mountain area in Taipei, Taiwan. Avocado and Litsea leaves and roots were obtained from young plants grown in greenhouses.
Retrieval of Oleosin Transcripts from Public and Our Generated RNA-seq Databases
All oleosin gene sequences were retrieved from public transcriptome databases except for S. multiflorus, which we prepared ourselves. Total RNA of Scadoxus leaves was sequenced with use of a Genome Analyzer IIx (Illumina, San Diego, CA) as described previously in Huang et al. (2013). For public databases, the raw data were downloaded from NCBI, and the sequences were assembled with the use of CLC Genomics Workbench (CLC Bio, Cambridge, MA). All analysis methods were previously described in Huang and Huang (2015).
GenBank accession numbers of oleosin genes from RNA-seq databases are as follows: V. planifolia 5-month-old seed, SRX648572; V. planifolia 6-month-old seed, SRX648573; V. planifolia leaf, SRX648194; V. planifolia root, SRX648209; Agapanthus praecox mixed organs, SRX277981; Allium sativum dormant vegetative bud, SRX131656; A. sativum sprouting vegetative bud, SRX131657; A. cepa seedling, SRX540974, A. cepa leaf, SRX393057, Agave americana leaf, SRX585650; A. americana root, SRX585654; A. deserti leaf, SRX252456; A. deserti root, SRX252452; A. tequilana leaf, SRX253018; A. tequilana root, SRX253022; P. americana stage I-V mesocarp: SRX627354, SRX627413, SRX627415, SRX627418, and SRX627420, respectively; L. cubeba fruit, SRX283712; Lindera glauca sarcocarp and kernel, SRX591256; Arabidopsis leaf, SRX463620; and Oryza sativa leaf, SRX017637. Oleosin genes of Phalaenopsis equestris and P. aphrodite were from Orchidbase (http://orchidbase.itps.ncku.edu.tw/).
LD Isolation and In Vitro LD Coalescence
All steps were performed at 4°C. Avocado mesocarp was minced in a grinding medium (0.6 m Suc and 100 mm Tris-HCl, pH 7.5) with use of a razor blade, and the floated LDs were removed with a spatula and used directly for coalescence observation. Leaf epidermis of vanilla and Scadoxus were gently ground in the above grinding medium and sea sand with use of a mortar and pestle. The homogenate was filtered through a 80 × 80 μm nylon mesh and then placed at the bottom of a centrifuge tube. A less-dense medium (0.4 m Suc and 100 mM, Tris-HCl, pH 7.5) was loaded above the homogenate, and the tube was centrifuged at 70,000g for 1 h. Floated LDs were collected with use of a pipette tip for coalescence observation and constituent analysis by SDS-PAGE and TLC. For similar constituent analysis, avocado LDs in the minced mesocarp homogenate were collected after the above-mentioned flotation centrifugation.
For testing in vitro coalescence, immediately after isolation the LDs were placed on a slide with a small cavity and observed by light microscopy at time intervals.
Immunofluorescence Confocal Laser Scanning Microscopy
Plant materials were fixed in a fixation medium (4% paraformaldehyde, 0.15 m Suc, 1× PBS [phosphate-buffered saline; 138 mm NaCl, 2.7 mm KCl and 10 mm K P, pH 7.4]) for 2 h at 24°C, washed with PBS three times, and stored in PBS at 4°C. Before incubation with antibodies, samples were treated with 0.5% Macerozyme R10 (Yakult Honsha, Tokyo, Japan) for 30 min at 24°C for digestion of the cell wall, incubated in a blocking buffer (1% BSA, 1× PBS) for 16 h at 4°C, and washed 3× with PBST (0.5% Tween 20 [w/v] and 1× PBS). Then, samples were incubated sequentially with antibodies against oleosin (1:100 dilution of the serum) for 1 h, cyanine 3-conjugated donkey antibodies against rabbit IgG (1:200) for 1 h, and 1 μm BODIPY 493/503 for 30 min; between two incubation steps, samples were washed 3× with PBST. Samples were placed on a slide and observed with a LSM 510 META confocal microscope (Carl Zeiss, Jena, Germany).
Total RNA Extraction and RT-PCR Analysis
Plant materials were frozen in liquid nitrogen and grounded into fine powder with use of a mortar and pestle. Total RNA was extracted by a procedure used for pine trees (Chang et al., 1993) or with the TRIzol reagent as described by the manufacturer (Invitrogen, Carlsbad, CA), and purified by using the RNAspin Mini RNA Isolation Kit (GE Healthcare, Washington, NY).
For RT-PCR analysis, the first-strand cDNA was synthesized from the purified total RNA with use of SuperScript III Reverse Transcriptase (Invitrogen) and used as templates for RT-PCR with gene-specific primers (Supplemental Table S2). RT-PCR products were analyzed on 1% agarose gel.
SDS-PAGE, Immunoblot Analysis, and Antibody Preparation
Plant materials were placed in tubes (Eppendorf, Hamburg, Germany), frozen with liquid nitrogen, and ground into powder with a pestle. Ground samples or LD fractions were mixed with protein-sample buffer (2% SDS [w/v], 7% glycerol, 5% 2-mercaptoethanol [v/v], and 60 mm Tris-HCl, at pH 6.8) and incubated at 100°C for 5 min. After centrifugation, proteins were separated on 12% SDS-PAGE gel and subjected to immunoblot analysis.
For production of rabbit polyclonal antibodies, the oleosin sequence was analyzed by use of antibody epitope prediction software (http://tools.immuneepitope.org/tools/bcell/iedb_input). The selected sequences in vanilla U1 oleosin and Scadoxus U oleosin (Supplemental Fig. S2) as well as avocado M oleosin (Fig. 5) along the whole sequences are indicated in the respective figures. Synthetic peptides with the selected sequences were used to produce rabbit polyclonal antibodies following a procedure described earlier (Kim et al., 2002). Specificities of the antibodies toward oleosins were confirmed by SDS-PAGE immunoblotting. For immunoblotting analysis, proteins on SDS-PAGE gel were transferred to a nylon membrane by a semidry transfer cell (Bio-Rad, Hercules, CA). The membrane was incubated sequentially with a blocking medium (5% milk in TBST [Tris buffer saline and Tween 20; 50 mm Tris, 150 mm NaCl, 0.05% Tween 20, at pH 7.5]), antibodies for oleosin (1:5000), and HRP-conjugated antibodies for rabbit IgG (1:10,000) at 24°C for 1 h; between two steps, the samples were washed 3× with TBST. After reaction with ECL substrate, the signal was captured by x-ray film.
TEM
Mesocarp of avocado fruit and epidermis of vanilla leaves were fixed with 2.5% glutaraldehyde, 4% paraformaldehyde, and 0.1 m K P (pH 7.0) at 4°C for 24 h. Samples were washed with 0.1 m K P buffer (pH 7.0) for 10 min twice and then treated with 1% OsO4 in 0.1 m K P (pH 7.0) at 24°C for 4 h. Fixed samples were rinsed with 0.1 m K P buffer (pH 7.0), dehydrated through an acetone series, and embedded in Spurr resin. Ultrathin sections (70–90 nm) were stained with uranyl acetate and lead citrate and examined with a model no. CM 100 transmission electron microscope (Philips, Amsterdam, the Netherlands) at 80 kV.
TLC
Total lipid extraction was performed as described in Hara and Radin (1978). A sample of 100-mg plant material or isolated LD fraction from avocado, vanilla, or Scadoxus was mixed with 800-μl isopropanol (v/w) and incubated at 80°C for 5 min. After the addition of 1.2 ml (v/w) of hexane, the sample was homogenized with use of a mortar and pestle and centrifuged for 10 min at 2100g. The supernatant was retained. Volumes of 200-μl isopropanol and 700-μl hexane were added to the pellet to re-extract the lipid. The supernatants from the two extraction steps were combined and dried with nitrogen gas. Cuticle lipid wash involved a chloroform rinse of the leaves for 30 s.
Lipid samples were applied to a TLC plate (Silica gel 60 F254 plate; Merck Minipore, Billerica, MA). The running solvent was hexane/diethyl-ether/acetic acid (70:30:2, v/v/v). Lipids were visualized by charring the plate with sulfuric acid. Lipid standards including triacylglycerol (triolein), diacylglycerol (1,2- and 1,3-diolein), monoacylglycerol (1- and 2-monoolein), free fatty acid (oleoic acid), sterol ester (β-sitosterols from soybean [Glycine max]), and wax ester (cholesteryl palmitate) were from Sigma-Aldrich (St. Louis, MO).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SRX648572, SRX648573, SRX648194, SRX648209, SRX277981, SRX131656, SRX131657, SRX540974, SRX393057, SRX585650, SRX585654, SRX252456, SRX252452, SRX253018, SRX253022, SRX627354, SRX627413, SRX627415, SRX627418, SRX627420, SRX283712, SRX591256, SRX463620, SRX017637.
Supplemental Data
The following materials are available in the on-line version of this article.
Supplemental Figure S1. Light microscopy of leaf epidermis and mesophyll cells of Asparagales and other monocot species.
Supplemental Figure S2. Alignment of residues of U oleosins of Asparagales species.
Supplemental Table S1. Levels of different oleosin transcripts in transcriptomes of Asparagales and Lauraceae species.
Supplemental Table S2. Primers used for RT-PCR.
Supplementary Material
Acknowledgments
We sincerely thank Mr. Chin-Hsin Yeh (Touyuan District Agricultural Research and Extension Station, Taiwan) for the growing vanilla plants and fruits; Miss Hui-Wen Tsai (Chiayi Agricultural Experiment Branch, Taiwan) for unharvested fruit (mature but still hanging on trees) of avocado (cv. Hass); Mr. Guo-Min Lin (Wulai, Taiwan) for samples of Litsea cubeba; and the following persons of the Institute of Plant and Microbial Biology, Academia Sinica: Dr. Yue-Ie Hsing for extensive use of the bioinformatics server, Dr. Wann-Neng Jane for electron microscopy, Dr. Ya-Fang Hong for avocado mesocarp sectioning, and Dr. Tungling Chen and Ms. Pei-Ying Chen for initial microscopy observations of LDs in Asparagales species.
Footnotes
Articles can be viewed without a subscription.
References
- Bourgis F, Kilaru A, Cao X, Ngando-Ebongue GF, Drira N, Ohlrogge JB, Arondel V (2011) Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc Natl Acad Sci USA 108: 12527–12532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report 11: 113–116 [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT (2012) Biogenesis and functions of lipid droplets in plants. Thematic review series: lipid droplet synthesis and metabolism: from yeast to man. J Lipid Res 53: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen GI, Mundy J, Tzen JT (2001) Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant 112: 301–307 [DOI] [PubMed] [Google Scholar]
- Hara A, Radin NS (1978) Lipid extraction of tissues with a low-toxicity solvent. Anal Biochem 90: 420–426 [DOI] [PubMed] [Google Scholar]
- Horn PJ, James CN, Gidda SK, Kilaru A, Dyer JM, Mullen RT, Ohlrogge JB, Chapman KD (2013) Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol 162: 1926–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC (2004) Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol 136: 3427–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC (2007) Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19: 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Chen PY, Huang MD, Tsou CH, Jane WN, Huang AHC (2013) Tandem oleosin genes in a cluster acquired in Brassicaceae created tapetosomes and conferred additive benefit of pollen vigor. Proc Natl Acad Sci USA 110: 14480–14485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MD, Huang AHC (2015) Bioinformatics reveal five lineages of oleosins and the mechanism of lineage evolution related to structure/function from green algae to seed plants. Plant Physiol 169: 453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilaru A, Cao X, Dabbs PB, Sung HJ, Rahman MM, Thrower N, Zynda G, Podicheti R, Ibarra-Laclette E, Herrera-Estrella L, Mockaitis K, Ohlrogge JB (2015) Oil biosynthesis in a basal angiosperm: transcriptome analysis of Persea americana mesocarp. BMC Plant Biol 15: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Hsieh K, Ratnayake C, Huang AHC (2002) A novel group of oleosins is present inside the pollen of Arabidopsis. J Biol Chem 277: 22677–22684 [DOI] [PubMed] [Google Scholar]
- Kwiatkowska M, Polit JT, Stępiński D, Popłońska K, Wojtczak A, Domίnguez E, Heredia A (2015) Lipotubuloids in ovary epidermis of Ornithogalum umbellatum act as metabolons: suggestion of the name ‘lipotubuloid metabolon’. J Exp Bot 66: 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska M, Stepinski D, Poplonska K, Wojtczak A, Polit JT (2010) “Elaioplasts” of Haemanthus albiflos are true lipotubuloids: cytoplasmic domains rich in lipid bodies entwined by microtubules. Acta Physiol Plant 32: 1189–1196 [Google Scholar]
- Lersten NR, Czlapinski AR, Curtis JD, Freckmann R, Horner HT (2006) Oil bodies in leaf mesophyll cells of angiosperms: overview and a selected survey. Am J Bot 93: 1731–1739 [DOI] [PubMed] [Google Scholar]
- Little SA, Stockey RA, Penner B (2009) Anatomy and development of fruits of Lauraceae from the Middle Eocene Princeton Chert. Am J Bot 96: 637–651 [DOI] [PubMed] [Google Scholar]
- Murphy DJ. (2012) The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249: 541–585 [DOI] [PubMed] [Google Scholar]
- Oh SK, Kang H, Shin DH, Yang J, Chow KS, Yeang HY, Wagner B, Breiteneder H, Han KH (1999) Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J Biol Chem 274: 17132–17138 [DOI] [PubMed] [Google Scholar]
- Platt-Aloia KA, Thomson WW (1981) Ultrastructure of the mesocarp of mature avocado fruit and changes associated with ripening. Ann Bot (Lond) 48: 451–465 [Google Scholar]
- Pol A, Gross SP, Parton RG (2014) Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol 204: 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Grillitsch K, Daum G (2008) Synthesis and turnover of non-polar lipids in yeast. Prog Lipid Res 47: 157–171 [DOI] [PubMed] [Google Scholar]
- Rangel B, Platt KA, Thomson WW (1997) Ultrastructural aspects of the cytoplasmic origin and accumulation of oil in olive fruit (Olea europaea). Physiol Plant 101: 109–114 [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Thiam AR, Farese RV Jr., Walther TC (2013) The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 14: 775–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SSH, Platt KA, Ratnayake C, Wang TW, Ting JTL, Huang AHC (1997) Isolation and characterization of neutral-lipid-containing organelles and globuli-filled plastids from Brassica napus tapetum. Proc Natl Acad Sci USA 94: 12711–12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.