Bacterial AvrRpt2-like cysteine proteases specifically suppress flagellin-induced phosphorylation of specific Arabidopsis mitogen-activated protein kinases.
Abstract
To establish infection, pathogens deliver effectors into host cells to target immune signaling components, including elements of mitogen-activated protein kinase (MPK) cascades. The virulence function of AvrRpt2, one of the first identified Pseudomonas syringae effectors, involves cleavage of the plant defense regulator, RPM1-INTERACTING PROTEIN4 (RIN4), and interference with plant auxin signaling. We show now that AvrRpt2 specifically suppresses the flagellin-induced phosphorylation of Arabidopsis (Arabidopsis thaliana) MPK4 and MPK11 but not MPK3 or MPK6. This inhibition requires the proteolytic activity of AvrRpt2, is associated with reduced expression of some plant defense genes, and correlates with enhanced pathogen infection in AvrRpt2-expressing transgenic plants. Diverse AvrRpt2-like homologs can be found in some phytopathogens, plant-associated and soil bacteria. Employing these putative bacterial AvrRpt2 homologs and inactive AvrRpt2 variants, we can uncouple the inhibition of MPK4/MPK11 activation from the cleavage of RIN4 and related members from the so-called nitrate-induced family as well as from auxin signaling. Thus, this selective suppression of specific mitogen-activated protein kinases is independent of the previously known AvrRpt2 targets and potentially represents a novel virulence function of AvrRpt2.
One of the first layers of plant immunity is activated by plasma membrane-resident pattern recognition receptors upon sensing of conserved microbial molecular structures (so-called pathogen-associated molecular patterns [PAMPs]), such as bacterial flagellin or translation elongation factor Tu. The resulting PAMP-triggered immunity (PTI) leads to multiple processes including ion fluxes, an oxidative burst, the activation of mitogen-activated protein kinase (MAPK/MPK) cascades and calcium-dependent protein kinases, defense gene expression, and the synthesis of antimicrobial compounds (Boller and Felix, 2009). To suppress PTI, pathogens evolved effector molecules to interfere with plant defenses. For instance, gram-negative bacteria use a type III secretion system (T3SS) to inject effectors directly into plant cells (Büttner and He, 2009). Based on genomic information, individual Pseudomonas syringae species may translocate some 20 to 30 effectors to target diverse steps of host defense regulation and to promote infection (Hann et al., 2010). Susceptible host plants may eventually evolve resistance (R) proteins to (in)directly detect effector proteins, activating effector-triggered immunity (ETI) that often includes a hypersensitive cell death to restrict the spread of biotrophic pathogens (Jones and Dangl, 2006).
MAPK cascades play key roles in diverse developmental and stress-related adaptation processes (Meng and Zhang, 2013; Lee et al., 2015b). They represent critical modules for mounting an adequate defense response. The hierarchically organized MAPK cascade is initiated by a mitogen-activated protein triple kinase (e.g. MEKK1) phosphorylating the MAPK kinase (MKK) that then phosphorylates a MAPK (MPK). Two main MAPK cascades are activated upon PAMP treatment, involving MKK4/5-MPK3/6 and MEKK1-MKK1/2-MPK4 (Meng and Zhang, 2013). A fourth MAPK, MPK11, was found recently to be activated during PTI signaling (Bethke et al., 2012; Eschen-Lippold et al., 2012). Several pathogen effectors target either MAPK cascade components directly or upstream signaling components to suppress defense signaling. Among the latter, the P. syringae effectors AvrPto, AvrPtoB, and HopF2 target PAMP receptor complex components to block PTI signaling (Göhre et al., 2008; Shan et al., 2008; Xiang et al., 2008; Gimenez-Ibanez et al., 2009; Zhou et al., 2014). In addition, HopF2 ADP ribosylates MKK5 (and possibly other MKKs), thereby blocking phosphorylation activity and interfering with immune signaling (Wang et al., 2010). At the MAPK level, MPK3, MPK4, and MPK6 are inactivated by P. syringae HopAI1, a phospho-Thr lyase, removing the phosphate group within the activation loop motif (Zhang et al., 2007, 2012).
Like MAPKs, RPM1-INTERACTING PROTEIN4 (RIN4) is a vital defense regulator (Kim et al., 2005b). The importance of RIN4 in immunity is underscored by it being targeted by several pathogen effectors, namely AvrB, AvrRpm1, AvrRpt2, HopF2, AvrPto, and AvrPtoB (Deslandes and Rivas, 2012). AvrB and AvrRpm1 induce RIN4 phosphorylation (Mackey et al., 2002; Chung et al., 2011) via kinases such as RIN4-INTERACTING RECEPTOR-LIKE PROTEIN KINASE (Liu et al., 2011), to enhance the RIN4 defense-suppressing activity (Kim et al., 2005b; Chung et al., 2014; Lee et al., 2015a). Plants with RPM1 (for resistance to P. syringae pv maculicola) R protein recognize RIN4 phosphorylation and mount ETI, arresting bacterial colonization (Grant et al., 1995; Chung et al., 2011). AvrRpt2, by contrast, is a Cys-protease that undergoes self-cleavage and activation within plant cells to cleave RIN4 at the plasma membrane (Axtell and Staskawicz, 2003; Mackey et al., 2003; Coaker et al., 2006). The homology of the cleaved sequences in RIN4 and AvrRpt2 suggests a specific recognition/cleavage motif (Chisholm et al., 2005; Takemoto and Jones, 2005). RIN4 processing by AvrRpt2 may thus represent a bacterial strategy to circumvent ETI induced by AvrB or AvrRpm1 in the presence of RPM1 (Kim et al., 2005a). Additionally, RIN4 cleavage products have stronger PTI-suppressing activity than membrane-anchored RIN4 (Afzal et al., 2011). Some plants may subsequently acquire the RPS2 (for resistance to P. syringae) R protein to monitor RIN4 disappearance (Kunkel et al., 1993; Axtell and Staskawicz, 2003; Mackey et al., 2003).
Virulence functions of AvrRpt2 independent of RIN4 also have been demonstrated (Lim and Kunkel, 2004); one of these is the alteration of plant auxin levels and the proteasomal degradation of auxin/indole-3-acetic-acid (Aux/IAA) transcriptional repressors to induce auxin signaling to promote disease (Chen et al., 2007; Cui et al., 2013). Here, we report a potentially novel virulence function of AvrRpt2, where it specifically suppresses MPK4/MPK11 activation induced by the flagellin-derived PAMP, flg22.
RESULTS
AvrRpt2 Suppresses flg22-Induced MPK4 and MPK11 Activation
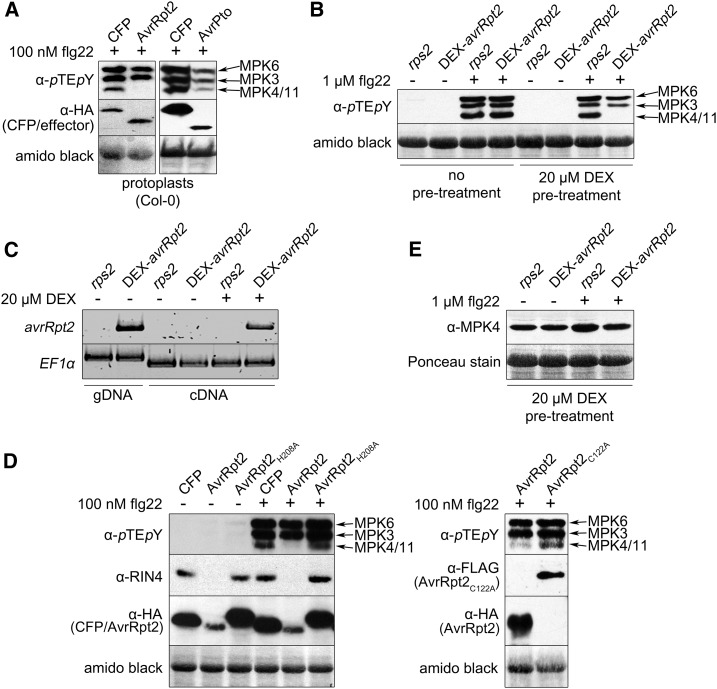
While screening for PAMP signaling interference by bacterial effectors transiently expressed in Arabidopsis (Arabidopsis thaliana) protoplasts, we found that while the bacterial AvrPto effector has a general suppressive effect on all MAPKs, AvrRpt2 expression specifically blocked the flg22-induced activation of MPK4/MPK11 but not of MPK3/MPK6 (Fig. 1A). To determine if this involves the corresponding RPS2 signaling pathway and to exclude any artifacts of the protoplast system, we used transgenic plants expressing avrRpt2 under the control of the dexamethasone (DEX)-inducible promoter (in an rps2 background). As in protoplasts, in planta expression of AvrRpt2 by DEX induction suppressed the flg22-induced MPK4/MPK11 activation (Fig. 1B). AvrRpt2 expression (Fig. 1C) upon DEX treatment correlated with the inhibition of flg22-induced MPK4/MPK11 activation. Since the inhibition did not occur in the rps2 background in the presence of DEX (Fig. 1B), the phenotype is specific for AvrRpt2 expression.
Figure 1.
AvrRpt2 suppresses MPK4 and MPK11 activation. A, MAPK phosphorylation in protoplasts (Columbia-0 [Col-0]; 15 min after flg22 treatment) was monitored with anti-pTEpY and expression of hemagglutinin (HA)-tagged AvrRpt2, cyan fluorescent protein (CFP), or AvrPto with anti-HA antibodies. The latter two acted as a negative control and a positive control for an effector that globally reduced the activation of all MAPKs, respectively. The identities of MAPK bands are according to Bethke et al. (2012). B, DEX-avrRpt2 (rps2) and rps2 plants were treated with DEX (for 5 h), infiltrated with flg22 (15 min), and MAPK phosphorylation was analyzed as in A. C, Plant material from B was used for reverse transcription (RT)-PCR expression analysis with avrRpt2- and elongation factor (EF1α)-specific primers. (Note that genomic DNA [gDNA] contamination can be excluded, since genomic DNA-amplified EF1α bands will be larger due to an intron.) D, Flg22-induced MAPK phosphorylation and expression of HA-CFP or AvrRpt2-HA variants in protoplasts were assayed as in A using the indicated antibodies (Col-0; 10 min; + or − 100 nm flg22). E, MPK4/MPK11 levels were visualized by immunoblot with an anti-MPK4 antibody on DEX-treated plants (as in B). Amido Black staining after immunoblotting (showing the large subunit of Rubisco) was used to demonstrate equal loading in all western-blot experiments. Experiments were performed at least three times with similar results.
RIN4 cleavage and AvrRpt2 autocleavage are blocked when the AvrRpt2 Cys-protease catalytic triad is mutated (Axtell et al., 2003). We transiently expressed two catalytically inactive AvrRpt2 variants (C122A and H208A) with C-terminal HA or FLAG epitope tags, respectively. Neither mutant version suppressed the flg22-induced MPK4/MPK11 activation (Fig. 1D), suggesting that this process requires the AvrRpt2 Cys-protease activity. Since no reduction in the overall MPK4/MPK11 protein levels was detected in the presence of AvrRpt2 (Fig. 1E) when probed with an antibody recognizing both MPK4 and MPK11 (Bethke et al., 2012), the suppression of MPK4/MPK11 activation is not due to direct proteolytic cleavage of the MAPKs by AvrRpt2.
The Suppression of MPK4 and MPK11 Activation Is Independent of RIN4 Presence and Localization
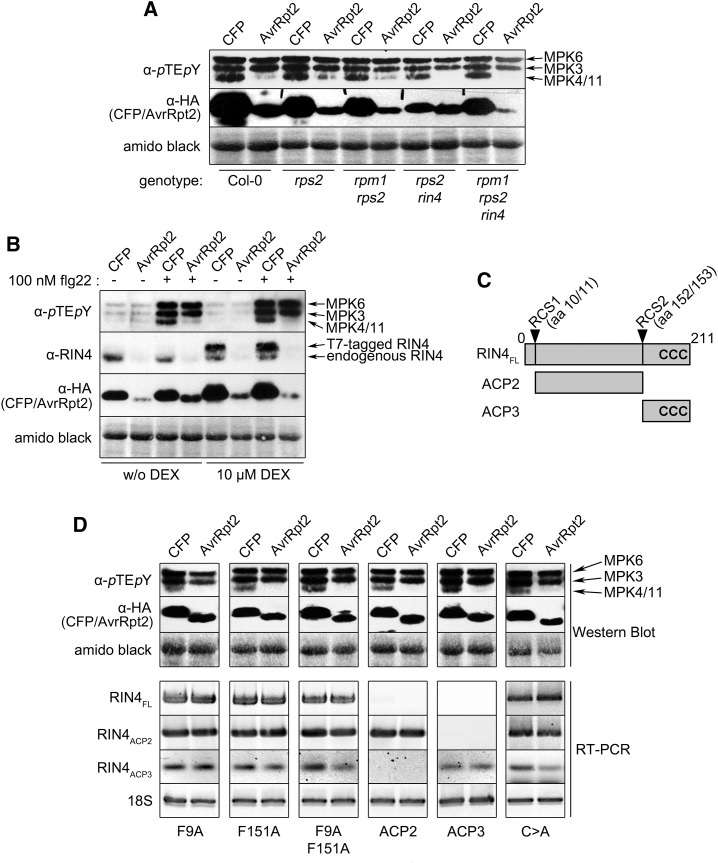
Since the AvrRpt2-mediated suppression of MPK4/MPK11 activation is accompanied by RIN4 disappearance, we investigated the role of RIN4. Due to constitutive RPS2 activation, rin4 mutants can be propagated only as the rps2rin4 double mutant. To avoid the light-dependent RPS2-mediated hypersensitive response upon AvrRpt2 expression in the Col-0 background (Zeier et al., 2004), we also kept the protoplasts in the dark for our assays. As the protoplasts appear intact, AvrRpt2 expression can be detected (Fig. 2A), and they still respond to flg22 elicitation (as evident from MPK3/MPK6 activation), AvrRpt2 expression did not cause substantial hypersensitive response-related cell death when using the Col-0 genotype in the protoplast system. In all tested rps2 or rin4 genotypes, AvrRpt2 expression interfered with flg22-induced MPK4/MPK11 activation (Fig. 2A). Thus, this AvrRpt2 effect is independent of RPS2 and RPM1, the R proteins guarding RIN4. Furthermore, the ability of AvrRpt2 to inhibit MPK4/MPK11 phosphorylation also is independent of RIN4.
Figure 2.
RIN4 is not required for the AvrRpt2-mediated suppression of MPK4/11 activation. A, Flg22 treatment, visualization of MAPK phosphorylation, and expression of HA-CFP or AvrRpt2-HA in protoplasts of the indicated genotypes were performed as in Figure 1D. B, Effect of RIN4 overexpression on MAPK activation was examined after DEX-induced expression using protoplasts prepared from DEX-T7-RIN4 (Col-0) plants (as described above). A RIN4 antibody was used to visualize RIN4 levels. C, Schematic representation of RIN4 and its cleavage products. aa, Amino acids; CCC, membrane-tethering Cys residues at Cys-203 to Cys-205; FL, full length; RCS1 and RCS2, RIN4 cleavage sites. D, The effect of expressing the indicated RIN4 variants on MPK4/MPK11 suppression was determined as in A using Col-0 protoplasts. RT-PCR with specific primers was used to confirm the expression of full-length RIN4 or ACP2 and ACP3 fragments. 18S, 18S ribosomal RNA-specific primers; F9A/F151A, RCS1/2 mutants. Experiments were repeated twice with similar results. Amido Black staining after immunoblotting (showing the large subunit of Rubisco) was used to demonstrate equal loading.
To check the effect of RIN4 overexpression, protoplasts were isolated from plants expressing RIN4 under the control of the DEX-inducible promoter and transfected with avrRpt2 or CFP. The suppression of flg22-induced MPK4/MPK11 activation by AvrRpt2 was not abolished after elevating RIN4 levels (Fig. 2B). However, note that while western blot using a RIN4-specific antibody showed increased overall RIN4 levels upon DEX treatment, both endogenous and DEX-induced T7-tagged RIN4 were cleaved by AvrRpt2 (Fig. 2B). Thus, it is not possible to evaluate if RIN4 overexpression can negate the AvrRpt2 effect.
We next assessed the importance of RIN4 cleavage. RIN4 has two conserved cleavage sites (RCSs; Fig. 2C) that are homologous to the AvrRpt2 autocleavage site (Chisholm et al., 2005). In the rps2rin4 background, overexpression of the individual RCS1 (F9A), RCS2 (F151A), or the F9A/F151A double mutant did not prevent MPK4/MPK11 inactivation by AvrRpt2 (Fig. 2D). Thus, RIN4 cleavage is not required, per se, for the observed interference of MPK4/MPK11 activation. Since the AvrRpt2 cleavage products of RIN4 (ACP2/3; Fig. 2C) are potent suppressors of defense (Afzal et al., 2011), we checked if overexpressing the ACP2 or ACP3 protein fragment is sufficient to block flg22-induced MPK4/MPK11 activation (in the absence of AvrRpt2), but this was not the case. Suppression of MPK4/MPK11 activation was observed only if AvrRpt2 was expressed. Finally, to address the role of proper RIN4 cellular localization, a non-membrane-tethering mutant (RIN4 C203A/C204A/C205A; Kim et al., 2005a; Afzal et al., 2011) was overexpressed. This also did not abrogate the MPK4/MPK11 inactivation phenotype (Fig. 2D). Taken together, the AvrRpt2 suppression of flg22-induced MPK4/MPK11 activation appears to be either independent of RIN4 or there are RIN4 homologs facilitating the effect.
Role of Selected RIN4 Family Members in AvrRpt2 Activity
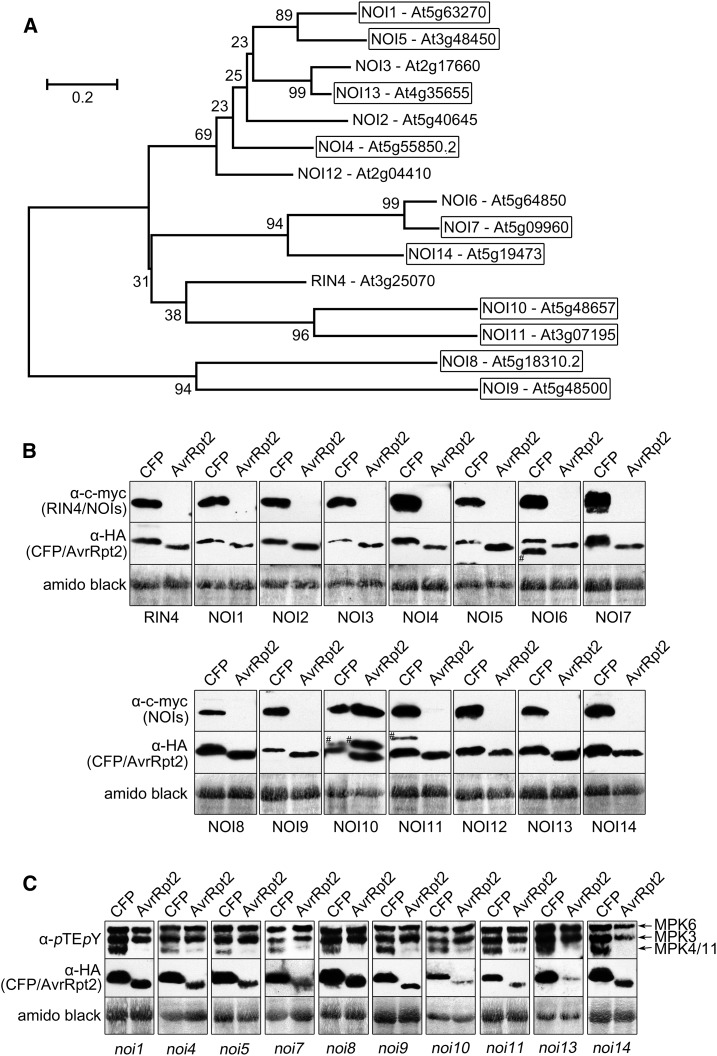
RIN4 belongs to the plant-specific nitrate-induced (NOI) protein family (Fig. 3A; Kim et al., 2005a; Afzal et al., 2013). All NOIs were cleaved by AvrRpt2 when expressed in protoplasts (Fig. 3B), except for NOI10, which lacks the conserved cleavage site in the splice variant deduced from the representative gene model, AT5G48657.1 (for both NOI10 gene models and the predicted AvrRpt2 cleavage sites of all the NOI proteins, see Supplemental Fig. S1). Thus, at least 14 of the 15 NOIs are AvrRpt2 substrates, with the consensus AvrRpt2 cleavage site being [LVI]PxFGxW (where x represents any amino acid).
Figure 3.
Several RIN4 protein family members are not required for the suppression of MPK4/11 activation. A, Phylogenetic analyses of RIN4/NOI protein family members. Bootstrap test values (%) are depicted next to the branches (for details, see Supplemental Methods S1). Boxes mark members where homozygous T-DNA insertion lines were isolated. B, Disappearance of c-myc-RIN4/NOIs in the presence of AvrRpt2 was monitored in protoplasts (rps2) after successive immunoassays with the indicated antibodies (# indicates remnant c-myc-signals after stripping). C, AvrRpt2-mediated suppression of MPK4/MPK11 action was tested in protoplasts prepared from the indicated genotypes as in Figure 1D (15 min after 100 nm flg22 treatment). Note that only noi4, noi7, noi8, noi9, and noi10 are true knockdown/knockout mutants (see quantitative reverse transcription [qRT]-PCR; Supplemental Fig. S3). Experiments were performed three times with similar results. Amido Black staining after immunoblotting (showing the large subunit of Rubisco) was used to demonstrate equal loading.
We next screened for transfer DNA (T-DNA) insertion lines of NOIs, where T-DNA insertions within the coding region were identified for NOI9 and NOI10, while insertions for the others were located either in introns (NOI7, NOI8, and NOI14) or in the 5′ (NOI1, NOI11, and NOI13) and 3′ (NOI4 and NOI5) flanking regions (Supplemental Fig. S2). When these homozygous lines (highlighted by boxes in the phylogenetic tree shown in Fig. 3A) were tested in the transient protoplast assay, the flg22-mediated MPK4/MPK11 activation was suppressed in the presence of AvrRpt2 in all cases (Fig. 3C). However, note that only noi4, noi7, noi8, noi9, and noi10 could be confirmed as knockdown or knockout mutants through qRT-PCR (Supplemental Fig. S3). Hence, at least for the NOI members where mutants or knockdowns are available, lack of individual NOIs (or RIN4) is insufficient to revoke the negative effect of AvrRpt2 on MPK4/MPK11 activation.
AvrRpt2 Suppresses the Expression of a Subset of Defense-Related Genes
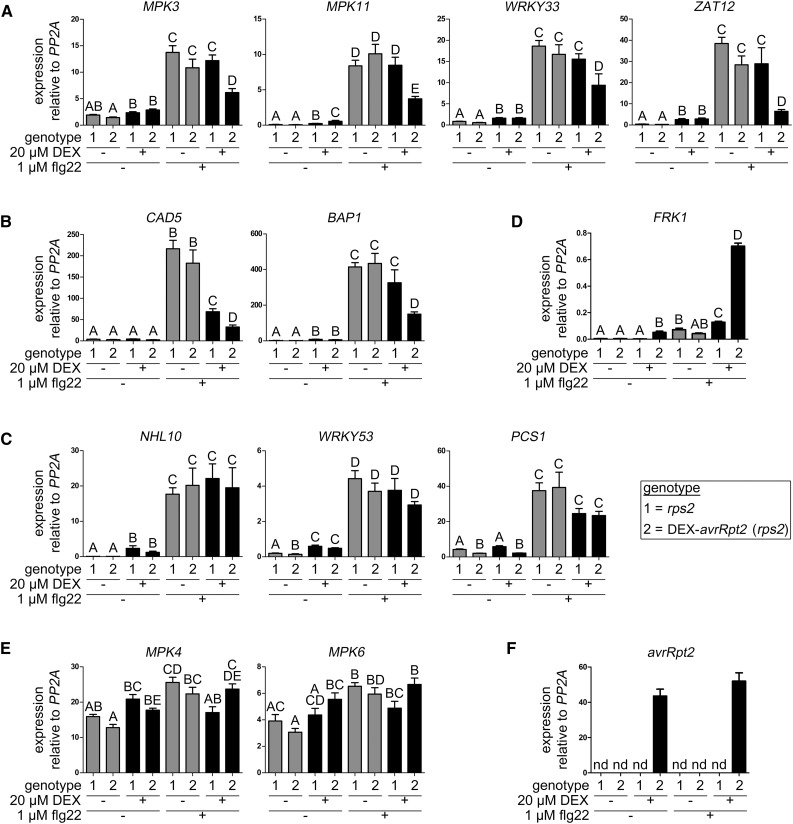
AvrRpt2 suppresses Arabidopsis PATHOGENESIS-RELATED (PR) gene expression upon infection with P. syringae carrying the effector (Chen et al., 2004). To test AvrRpt2 suppression of PAMP-induced defense genes (in the absence of pathogens), we monitored the expression of various known flg22-induced genes in transgenic plants expressing avrRpt2 under the control of the DEX-inducible promoter in the rps2 background.
Since specific MAPK activation appears to be targeted by AvrRpt2, we first addressed if the expression of MPK genes is altered upon PAMP perception or AvrRpt2 expression. Among the four PAMP-responsive MAPKs, MPK3 and MPK11 are transcriptionally up-regulated upon PAMP treatment (Bethke et al., 2012). AvrRpt2 attenuated the flg22-induced expression of MPK3 and MPK11 and also that of WRKY33, ZAT12, CAD5, and BAP1 (Fig. 4, A and B). For CAD5, although DEX treatment already has a negative effect, AvrRpt2 further reduced the flg22-induced expression. Interestingly, flg22-induced expression of CAD5 (Bethke et al., 2012) and BAP1 (Frei dit Frey et al., 2014) is known to be regulated by MPK4 and/or MPK11. Conversely, there are other genes (e.g. NHL10, WRKY53, or PCS1) where the flg22-induced transcript accumulation was not affected by AvrRpt2 (Fig. 4C). Consistent with our observation that PAMP-induced MPK3/MPK6 activation was not suppressed by AvrRpt2, flg22-induced expression of the MPK3/MPK6-regulated gene, FRK1 (Boudsocq et al., 2010), also was not reduced but was even elevated by AvrRpt2 (Fig. 4D). Furthermore, AvrRpt2 weakly increased the basal expression of MPK4 and MPK6 (Fig. 4E). As a control, specific avrRpt2 expression also was confirmed (Fig. 4F). Altogether, AvrRpt2 does not globally suppress all but targets a subset of flg22-activated genes, including some MPK4/MPK11-regulated genes.
Figure 4.
AvrRpt2 specifically suppresses the expression of a subset of defense-related genes. DEX-avrRpt2 (rps2 background) and rps2 control plants were treated with 20 µm DEX (5 h) to induce AvrRpt2 expression and expression of the indicated genes tested by qRT-PCR 1 h after water or flg22 infiltration. A, Flg22-inducible genes that are negatively affected by AvrRpt2. B, Flg22-inducible genes that are negatively affected by AvrRpt2 as in A and reported to be regulated by MPK4 and/or MPK11 (Bethke et al., 2012; Frei dit Frey et al., 2014). C, Flg22-inducible genes that are unaffected by AvrRpt2. D, Flg22-inducible MPK3/MPK6-regulated gene, FRK1, positively affected by AvrRpt2. E, MPK genes without known flg22-inducible expression. Note the slightly increased expression upon flg22 treatment in the presence of AvrRpt2. F, Validation of specific avrRpt2 transcript accumulation upon DEX treatment. Letters indicate statistically significant differences (n = 12; one-way ANOVA with Newman-Keuls multiple comparison test after log2 transformation of the data). nd, Not detectable. Experiments were repeated twice with similar results.
AvrRpt2 Suppresses Other PTI Responses
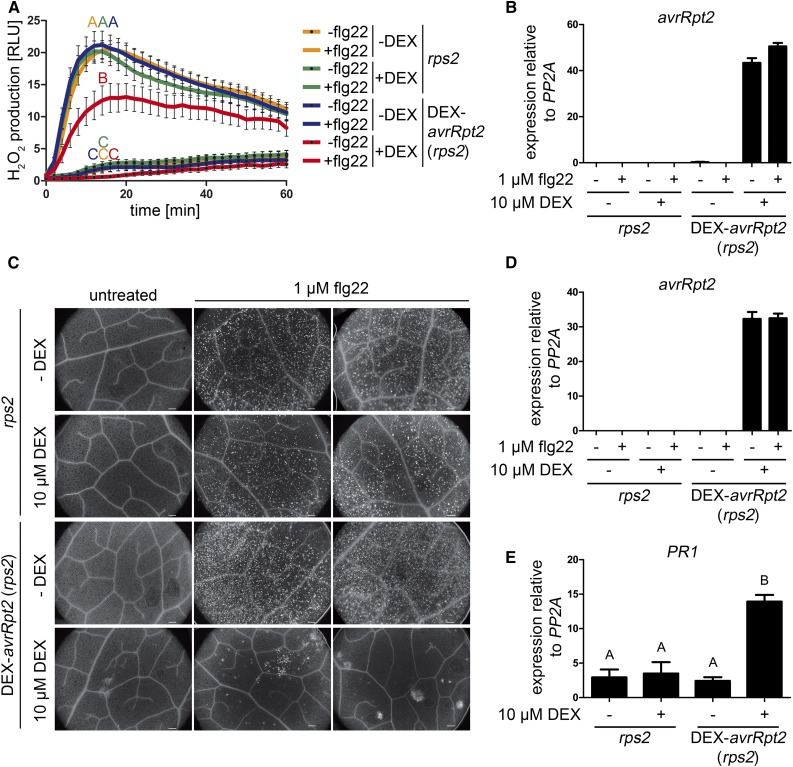
Since the mpk4 mutant shows constitutive salicylic acid (SA) and reactive oxygen species (ROS) accumulation (Petersen et al., 2000), we wondered if the AvrRpt2 suppression of MPK4/MPK11 activation would affect such defense responses. Basal ROS levels in the DEX-treated avrRpt2 transgenic plants were not significantly higher than those in the control lines, but the flg22-induced ROS burst was attenuated (Fig. 5A) by DEX-induced expression of AvrRpt2 (Fig. 5B). Callose deposition, a late PTI response, occurs constitutively in mpk4 (Frei dit Frey et al., 2014) and also in mekk1 (Ichimura et al., 2006) mutants. Consistent with prior work (Kim et al., 2005b), no enhanced callose deposition was observable in the DEX-treated avrRpt2 transgenic lines, but the number of flg22-induced callose deposits was reduced dramatically upon AvrRpt2 expression (Fig. 5, C and D). Thus, AvrRpt2 also suppresses other PTI responses, such as callose deposition and ROS accumulation, but it is difficult to associate this directly to the observed suppressive effect on MPK4/MPK11 activities at this stage. The lack of constitutive callose deposition and ROS accumulation, despite the inhibition of MPK4/MPK11 activity after an overnight (approximately 16-h) DEX-induced expression of AvrRpt2 in these transgenic plants, may imply that additional (stress or developmental) signals are required to trigger these responses typically seen in mpk4 or mekk1 mutants. To exclude that the DEX treatment time may be insufficient, we always monitored the expression of avrRpt2 by qRT-PCR (Fig. 5, B and D). Additionally, PR1 expression (as an SA-responsive gene and a proxy for SA levels) is enhanced in DEX-treated avrRpt2 transgenic plants without flg22 treatment (Fig. 5E). This is in contrast to the suppression of PR1 expression when infected with P. syringae carrying avrRpt2 (Chen et al., 2004), which probably is due to the complex interaction involving multiple PAMPs or other bacterial components. In our system, the enhanced PR1 expression upon expressing avrRpt2 presumably reflects elevated basal SA levels caused by the inhibition of basal MPK4 activity.
Figure 5.
AvrRpt2 affects other PTI responses. A, Leaf discs of the indicated genotypes were incubated for approximately 16 h in water (+ or − 10 µm DEX) in 96-well plates. Hydrogen peroxide (H2O2) accumulation was measured upon treatment with or without 100 nm flg22 and is depicted as relative light units (RLU). Different letters denote statistically significant differences (n = 12; two-way Repeated-Measures-ANOVA with Bonferroni posttests). The experiment was performed four times with similar results. B, qRT-PCR quantification of avrRpt2 expression of the samples in A. C, Callose deposition was analyzed by fluorescence microscopy after Aniline Blue staining. Plants were sprayed with 10 µm DEX, and after 5 h, leaves were infiltrated with 1 µm flg22. Sixteen hours later, staining with Aniline Blue was performed. Bars = 200 µm. The experiment was repeated twice with similar results. D, Corresponding qRT-PCR quantification of avrRpt2 expression of the samples in C. E, As a proxy for SA levels, qRT-PCR of PR1 expression was measured after 16 h of DEX treatment. Letters indicate statistically significant differences (n = 4; one-way ANOVA with Newman-Keuls multiple comparison test after log2 transformation of the data). The experiment was performed four times with similar results.
AvrRpt2 Enhances Bacterial and Fungal Infection
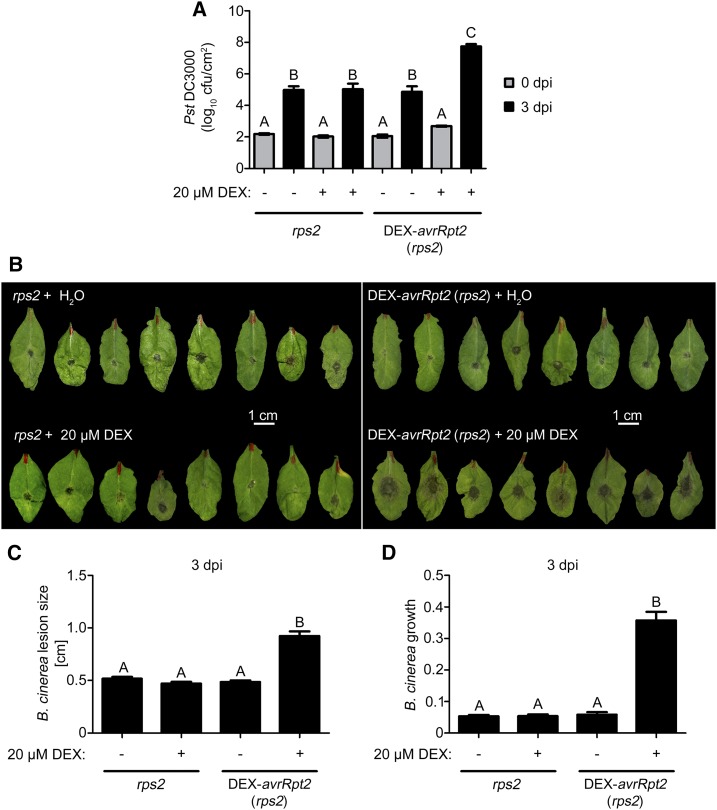
The reduced expression of PAMP-inducible genes and PTI responses may act as a proxy for the plant’s resistance status. Transgenic plants expressing avrRpt2 under the control of the RPS2 promoter support better growth of attenuated (i.e. T3SS-defective) bacteria but do not exhibit enhanced infection by Erysiphe cichoracearum or Hyaloperonospora arabidopsidis (Chen et al., 2004). To compare with these previous studies, where there was basal expression through the RPS2 promoter, here we used transgenic plants with controlled AvrRpt2 expression. Only after DEX induction did the plants show enhanced susceptibility after spray inoculation of (nonattenuated) virulent P. syringae pv tomato DC3000 (Fig. 6A). Similarly, DEX-induced AvrRpt2 expression also led to enhanced Botrytis cinerea infection, as seen by increased lesion sizes on leaves and quantitative PCR (qPCR)-quantified fungal biomass (Fig. 6, B–D). Hence, in the absence of RPS2-mediated resistance, AvrRpt2 confers enhanced susceptibility. This suggests that its virulence targets are conserved immunity components essential for resistance to certain bacterial and fungal pathogens.
Figure 6.
AvrRpt2 expression enhances infection with bacterial and fungal pathogens. A, DEX-avrRpt2 (rps2 background) and rps2 control plants were treated with DEX (5 h) to induce AvrRpt2 expression, spray inoculated with P. syringae pv tomato DC3000 (5 × 108 bacteria mL−1), and bacterial growth was determined (0 or 3 d post infection [dpi]) by counting colony-forming units (cfu) after plating serial dilutions. The experiment was repeated twice with similar results; the diagram shows combined data sets (n ≥ 20). B, Plants were DEX treated as in A. After 6 h, 10-µL droplets of B. cinerea (strain B05.10) spores (2 × 105 spores mL−1) were inoculated on the leaves and photographed 3 d later. The experiment was repeated twice with similar results. C, Lesion size determination of B. cinerea infection sites of samples described in B. The diagram shows combined data sets (n ≥ 26). D, Fungal biomass in leaves at day 3 was measured by qPCR on the basis of fungal genomic DNA (n = 12). Letters denote statistically significant differences (one-way ANOVA with Tukey’s multiple comparison test, P < 0.001).
Several Bacterial Homologs Show AvrRpt2-Like Activities
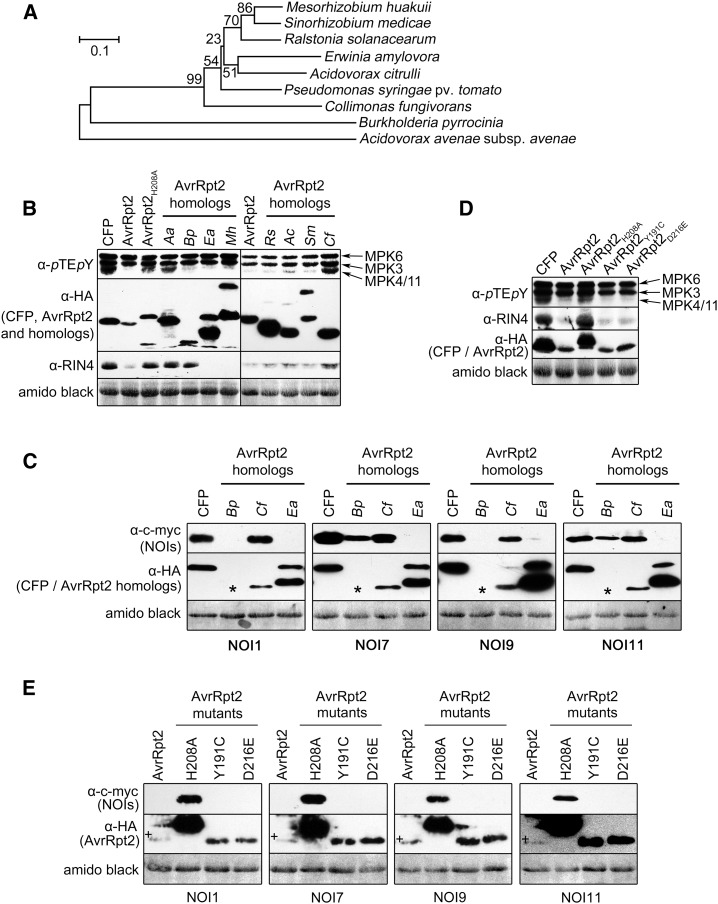
Since AvrRpt2 virulence function may benefit the pathogenic lifestyle of microbes, we wondered if AvrRpt2 homologs exist in other bacteria. We identified several putative avrRpt2 homologs in sequenced bacterial genomes (Supplemental Table S1), including sequences from known phytopathogens (e.g. Erwinia amylovora, Ralstonia solanacearum, Acidovorax citrulli, and Acidovorax avenae), plant-associated bacteria (e.g. Mesorhizobium huakuii and Sinorhizobium medicae), or soil/rhizosphere (e.g. Burkholderia pyrrocinia). Notably, the homolog with one of the highest sequence identities (61%) to the P. syringae AvrRpt2 is from Collimonas fungivorans, a chitinolytic bacterium that infects living soil fungi (de Boer et al., 2004). Five of the deduced homologs cover the full-length P. syringae AvrRpt2, with the N terminus followed by the autocleavage site and the putative Cys-protease domain, while three homologs resemble the cleaved AvrRpt2 (i.e. only the putative Cys-protease domain; for alignment and identity matrix, see Supplemental Fig. S4). Without corresponding mRNA analysis, it is unclear if these are truly truncated or misannotated. Except for the A. avenae homolog, phylogenetic analyses clustered protein sequences derived from phytopathogenic and plant-associated bacteria together. Likewise, the homologs derived from the chitinolytic bacterium C. fungivorans or the soil bacterium B. pyrrocinia form separate branches (Fig. 7A). Note that the hosts of A. avenae are mostly monocots, including crops such as oat (Avena sativa), rice (Oryza sativa), maize (Zea mays), barley (Hordeum vulgare), sorghum (Sorghum bicolor), and various folder grasses. Hence, the phylogenetic relationship between the AvrRpt2 homologs (Fig. 7A) mirrors the host range or ecological niche of the bacteria.
Figure 7.
Functional comparison of AvrRpt2 homologs from different bacteria. A, Phylogenetic analysis of AvrRpt2 homologs was performed as in Figure 3A. B, MAPK activation, expression of AvrRpt2-HA homologs (or HA-CFP control), or RIN4 levels in protoplasts (rps2; 100 nm flg22, 15 min) were visualized with the indicated antibodies. Aa, A. avenae ssp. avenae; Ac, A. citrulli; Bp, B. pyrrocinia; Cf, C. fungivorans; Ea, E. amylovora; Mh, M. huakuii; Rs, R. solanacearum; Sm, S. medicae. C, Effect of the AvrRpt2-HA homologs (or HA-CFP control) on selected c-myc-NOIs in protoplasts (rps2) was assessed after successive immunoblotting with the indicated antibodies. Asterisks indicate weak expression of the B. pyrrocinia homolog (Supplemental Fig. S5). D and E, Effects of the AvrRpt2 mutants on MPK4/MPK11 suppression and RIN4/NOI cleavage analyzed as in B and C (+ indicates AvrRpt2 bands). All experiments were performed three times with similar results. Amido Black staining after immunoblotting (showing the large subunit of Rubisco) was used to demonstrate equal loading.
When transiently expressed in protoplasts, all the tested AvrRpt2 homologs (except the A. avenae and C. fungivorans variants) suppressed flg22-induced MPK4/11 activation, which is typically accompanied by RIN4 cleavage (Fig. 7B). Interestingly, the B. pyrrocinia homolog suppressed MPK4/11 activation but did not cleave RIN4. Thus, in agreement with the data above (Fig. 2), the suppression of MPK4/11 activation can be uncoupled from RIN4 cleavage (Fig. 7B).
To extend this analysis to the NOIs, we selected three AvrRpt2 homologs displaying different activities: (1) the B. pyrrocinia homolog showing MPK4/11 activation suppression but no RIN4 cleavage; (2) the (inactive) C. fungivorans homolog mediating neither MPK4/11 activation suppression nor RIN4 cleavage; and (3) the E. amylovora homolog suppressing MPK4/11 activation and cleaving RIN4. A representative NOI protein family member from each of the four main clades of the NOI/RIN4 phylogenetic tree (Fig. 3A) was selected. As observed for RIN4, the C. fungivorans homolog did not cleave any of the NOI proteins, while the E. amylovora homolog cleaved all four tested NOI proteins (Fig. 7C). The B. pyrrocinia homolog cleaved NOI1 and NOI9 more efficiently than NOI7 and NOI11 (Fig. 7C). Thus, while still speculative, members from the NOI1 and NOI9 clusters may contribute to the MPK4/11 suppression activity of AvrRpt2.
To provide a possible link of this newly discovered MAPK-suppressive activity of AvrRpt2 to its virulence functions, we tested two P. syringae AvrRpt2 mutants, Y191C and D216E, which have compromised virulence-promoting effects on susceptible plants (Lim and Kunkel, 2004). These were selected for the following reasons: (1) Asp-216 of the P. syringae AvrRpt2, while highly conserved in all the AvrRpt2 homologs tested in this study, is replaced by Gln in the B. pyrrocinia homolog (Supplemental Fig. S5A); and (2) Tyr-191, heterogenous among the homologs, is replaced by Asp or Glu in the inactive A. avenae or C. fungivorans homolog, respectively. However, the active E. amylovora homolog has a Cys at this position (i.e. like the Y191C mutant with compromised virulence-promoting function). We compared the ability of these AvrRpt2 variants to suppress MPK4/MPK11 activation, using the H208A mutant as a control. Upon expression in protoplasts, both of the AvrRpt2 mutants displayed wild-type characteristics (i.e. autocleavage, suppression of MPK4/11 activation upon flg22 treatment, and cleavage of RIN4 and the tested NOIs; Fig. 7, D and E). Thus, the reduced virulence-promoting effect of both AvrRpt2 mutant versions (Lim and Kunkel, 2004) is unrelated to impaired protease activity or the loss of MPK4/11 activation suppression.
Auxin Signaling Is Not Involved in the Suppression of MPK4/11 Activation by AvrRpt2
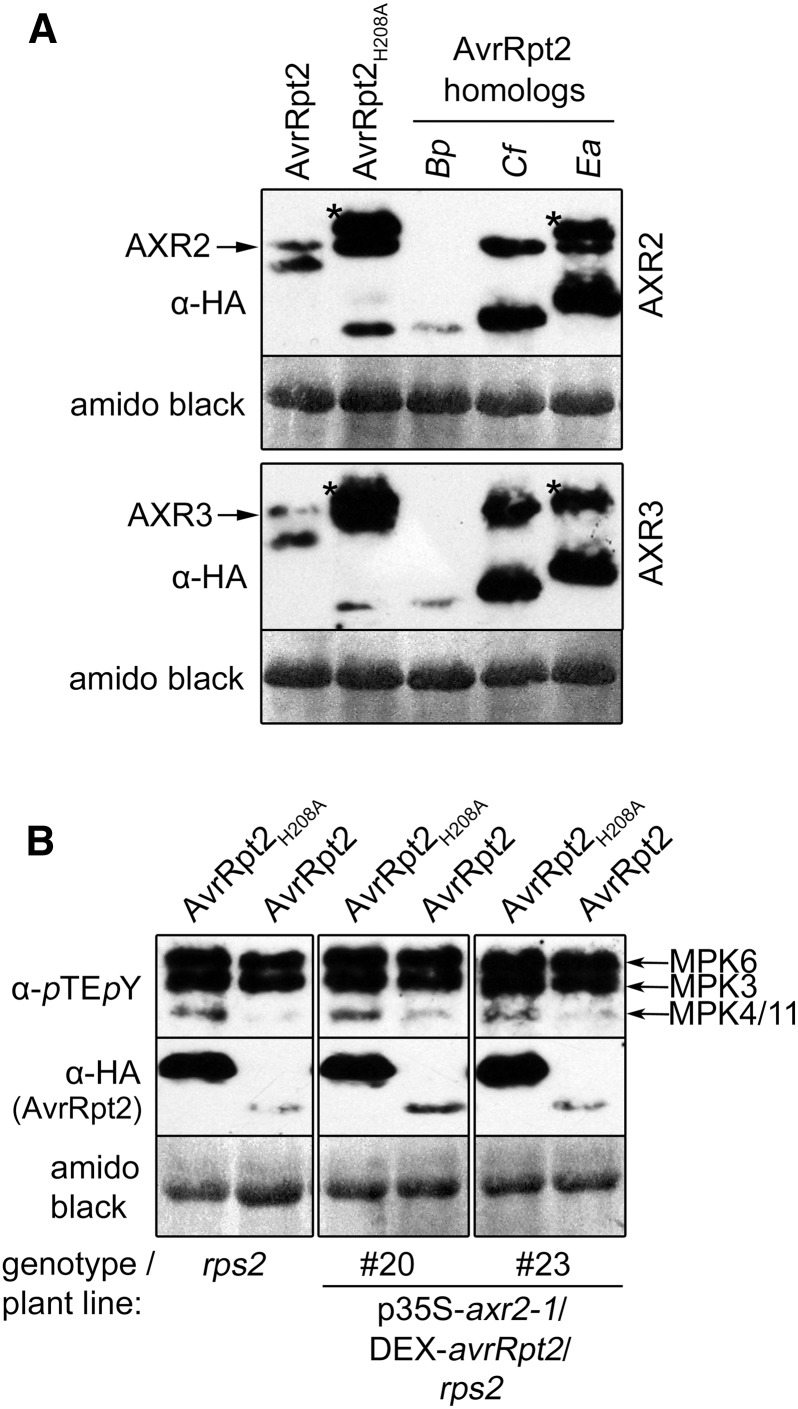
One AvrRpt2 virulence mechanism is the promotion of auxin signaling by inducing the proteasome-dependent degradation of members of the Aux/IAA family (Cui et al., 2013). We tested three selected AvrRpt2 homologs for their ability to induce the destabilization of two Aux/IAA transcriptional repressor members, AUXIN-RESISTANT2 (AXR2) and AXR3. Interestingly, the C. fungivorans homolog that was inactive in suppressing MPK4/MPK11 activation did not cause the disappearance of AXR2 or AXR3 (Fig. 8A). Conversely, the B. pyrrocinia and E. amylovora homologs that suppressed MPK4/MPK11 activation (Fig. 7B) also induced the disappearance of AXR2 or AXR3, although the AXR2 disappearance induced by the E. amylovora homolog was only partial (Fig. 8A). This correlation hinted at a possible link between auxin signaling and the MPK4/MPK11 suppression activity.
Figure 8.
Auxin is not involved in the suppression of MPK4/11 activation by AvrRpt2. A, Destabilization of AXR2-/AXR3-HA by AvrRpt2 homologs in protoplasts (rps2). (AXR2/AXR3 bands are indicated by arrows to distinguish them from the uncleaved or partially cleaved AvrRpt2 precursors, which are indicated by asterisks). Bp, B. pyrrocinia; Cf, C. fungivorans; Ea, E. amylovora. B, Suppression of flg22-activated MPK4/MPK11 was assessed in protoplasts prepared from two independent axr2-1-overexpressing lines. DEX induction and flg22 treatment were performed as in Figure 1B. The experiments were repeated twice with similar results. Amido Black staining after immunoblotting (showing the large subunit of Rubisco) was used to demonstrate equal loading.
To investigate if the AvrRpt2-mediated suppression of MPK4/11 activation is dependent on auxin signaling, we used the gain-of-function mutation of AXR2, axr2-1 (Wilson et al., 1990). This P87S mutation increases its stability by preventing proteasomal turnover, leading to the repression of auxin responses. We used two independent axr2-1-overexpressing transgenic lines (Cui et al., 2013) to express native AvrRpt2 or nonfunctional AvrRpt2-H208A. Similar to the situation in Col-0 wild-type or rps2 mutant plants, native AvrRpt2, but not AvrRpt2-H208A, suppressed the flg22-induced MPK4/11 activation (Fig. 8B). Since the axr2-1 protein is not cleaved by AvrRpt2 (Cui et al., 2013) and the auxin response is blocked in these transgenic lines, the AvrRpt2-mediated suppression of MPK4/11 activation is independent of auxin signaling.
Mode of Action for the AvrRpt2 Suppression of flg22-Induced MPK4/MPK11 Activation
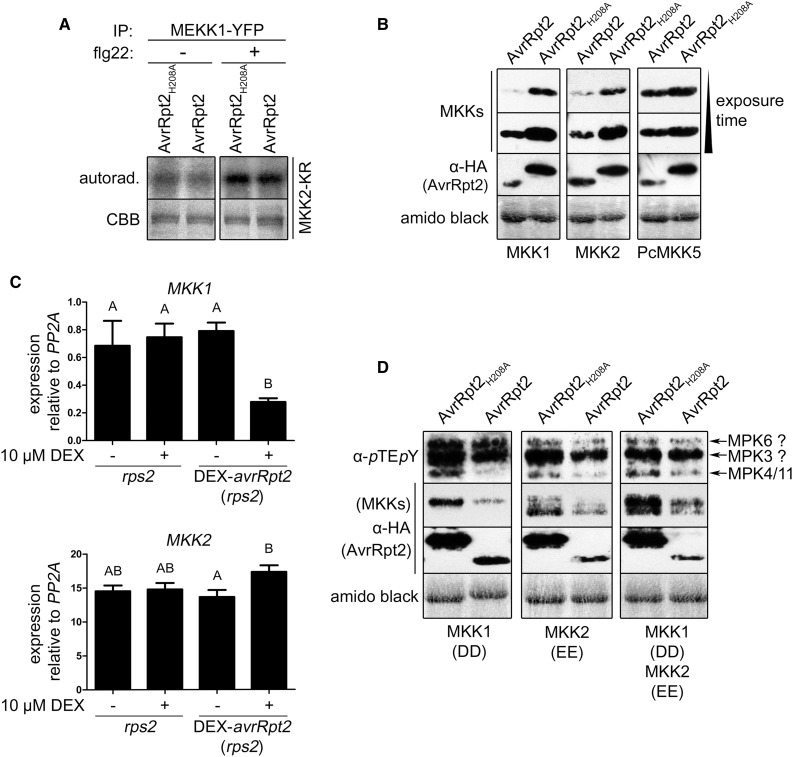
We next sought to understand how AvrRpt2 suppresses MPK4/11 activation. Since MPK4/11 protein levels were not changed by AvrRpt2 (Fig. 1E), we focused on components upstream of MPK4/11. When yellow fluorescent protein (YFP)-tagged MEKK1, the mitogen-activated protein triple kinase upstream of the MPK4 pathway (Ichimura et al., 2006), was expressed in protoplasts and the immunoprecipitated kinase activity was analyzed using recombinant MKK2-KR (a kinase-inactive MKK2 variant) as a substrate, there was no reduction of MEKK1 kinase activity in the presence of AvrRpt2 (Fig. 9A). However, it should be mentioned that MEKK1-YFP expression alone led to a low but clear basal activation of at least three MAPKs prior to PAMP treatment. On the basis of their sizes and the increased intensities after flg22 treatment, these three bands are likely to be MPK3, MPK6, and MPK4/MPK11 (Supplemental Fig. S6). Thus, overexpression of MEKK1-YFP can apparently override the AvrRpt2 suppression of PAMP-induced MPK4/MPK11 activation. Nevertheless, we still conclude from the MEKK1 kinase activity on MKK2-KR (Fig. 9A) that AvrRpt2 does not inactivate MEKK1 directly.
Figure 9.
Possible mechanism for the suppression of MPK4/11 activation by AvrRpt2. A, MEKK1 was immunoprecipitated (IP) with GFP-trap beads from protoplasts (rps2) coexpressing MEKK1-YFP and AvrRpt2-HA variants (+ or − 100 nm flg22 treatment, 1 h) and used to phosphorylate recombinant MKK2-KR as a substrate in radioactive in vitro kinase assays. autorad., Autoradiography; CBB, Coomassie Brilliant Blue stain. B, Effect of (in)active AvrRpt2 on the levels of Arabidopsis MKK1/MKK2 (or the parsley MKK5 [PcMKK5] as a control) in protoplasts (rps2). Western blots were successively probed with anti-c-myc (for PcMKK5) and anti-HA (for MKKs and AvrRpt2 variants) antibodies. C, Effect of AvrRpt2 on MKK1/MKK2 transcript levels. DEX-avrRpt2 (in the rps2 background) and rps2 control plants were treated with 10 µm DEX (16 h) to induce AvrRpt2 expression, and the expression of MKK1/MKK2 was analyzed by real-time PCR. Letters indicate statistically significant differences (n = 4; one-way ANOVA with Newman-Keuls multiple comparison test after log2 transformation of the data). The experiment was repeated four times with similar results. D, Effect of AvrRpt2 on the MPK4/MPK11 phosphorylation induced by constitutively active MKK1T218D,S224D (DD) and/or MKK2T229E,T235E (EE) in protoplasts (rps2). MKK levels were analyzed by immunoblotting as described in B. Experiments A, B, and D were repeated twice with similar results. Amido Black staining (B and D) after immunoblotting (showing the large subunit of Rubisco) was used to demonstrate equal loading.
MKK1 and MKK2 act downstream of MEKK1 to redundantly activate MPK4 (Gao et al., 2008; Qiu et al., 2008b). Both MKK1 and MKK2 accumulated to a lesser extent in the presence of functional AvrRpt2 compared with inactive AvrRpt2-H208A (Fig. 9B). As a control, we used MKK5, the parsley (Petroselinum crispum) functional MKK4/5 ortholog upstream of the second PAMP-induced MAPK pathway leading to MPK3/MPK6 (Lassowskat et al., 2014). Unlike MKK1 or MKK2, MKK5 was not or was only slightly reduced (Fig. 9B). To determine if this is an effect on the MKK1/2 proteins or mRNAs, we also analyzed the MKK1/2 transcripts by qRT-PCR. Interestingly, AvrRpt2 suppressed MKK1 expression but had little to no effect on MKK2 transcript levels (Fig. 9C). Thus, the negative impact of AvrRpt2 on MKK1 and MKK2 proteins may be due to transcriptional and posttranscriptional/(post)translational control, respectively.
To further pinpoint AvrRpt2 action(s) independent of other PAMP-induced pathways and to focus on the levels of the MKKs, we used constitutively active MKK1 or MKK2 to phosphorylate MPK4/11 (Asai et al., 2002). MKK1DD and MKK2EE phosphomimetics, either individually or in combination, caused MPK4/11 activation without PAMP stimulus, although two MAPKs with similar sizes to MPK6 and MPK3 also were activated (Fig. 9D). MKK2 is known to activate MPK4 and MPK6 (Teige et al., 2004). Besides interaction with MPK4/MPK11, yeast two-hybrid assays showed interaction between MKK2 and MPK6, MPK10, or MPK13 (Lee et al., 2008). Therefore, the two additional MKK1/2-induced MAPK bands may be MPK6/MPK10 (both approximately 45 kD) or MPK3/MPK13 (approximately 42–43 kD). AvrRpt2 specifically reduced the MPK4/MPK11 phospho status (but not that of the other two MAPK bands; Fig. 9D). As for wild-type MKK1/2, AvrRpt2 also led to decreased levels of MKK1DD and MKK2EE. Taken together, these data suggest that AvrRpt2 may act at the level of the MKKs by stimulating MKK turnover at the mRNA or protein level but specifically for the MKKs upstream of the MPK4/MPK11 pathway. However, for the protein turnover hypothesis, since MKK1 and MKK2 lack AvrRpt2 cleavage sites in their primary sequences, AvrRpt2 most likely targets unknown component(s) that control the turnover of specific MKK proteins/mRNAs.
DISCUSSION
We report a potential novel virulence function of AvrRpt2, in which it specifically blocks PAMP-induced MPK4/MPK11 activation, which is accompanied by reduced expression of selected defense genes, including MPK4/MPK11-regulated genes. MAPKs have critical regulatory roles in diverse developmental aspects and stress adaptation (Meng and Zhang, 2013; Lee et al., 2015b) and are targeted by P. syringae effectors such as HopAI1 and AvrB (Zhang et al., 2007, 2012). Animal pathogens also employ effectors such as HopAI1 orthologs (Shigella OspF or Salmonella SpvC) or Yersinia YopJ orthologs (Salmonella AvrA or Vibrio VopA) to inactivate MAPK elements (for review, see Shan et al., 2007), highlighting that MAPK-induced defense interference is a common virulence strategy for animal and plant pathogens. While the phospho-Thr lyase activity of HopAI1/OspF/SpvC should act generally on all MAPKs (Li et al., 2007), OspF specifically inactivated ERK and p38, but not JNK MAPKs (Arbibe et al., 2007). The P. syringae HopAI1 inactivated Arabidopsis MPK3, MPK4, and MPK6 (Zhang et al., 2007, 2012). By contrast, AvrRpt2 specifically prevents the activation of only MPK4 and its closest homolog, MPK11. Since MPK4 negatively regulates SA-mediated defense (Petersen et al., 2000), perturbing MPK4 appears counterintuitive, but this implies that MPK4 (and MPK11) must have positive regulatory roles in resistance. One may be the role of MPK4 in the release of WRKY33 transcription factor from preexisting inhibitory complexes for the proper expression of PAD3, a key biosynthetic gene for antimicrobial camalexin production (Qiu et al., 2008a).
In the absence of RPS2, AvrRpt2 promotes the ability of bacteria mutated in the T3SS apparatus to grow in planta (Chen et al., 2000). We extended this finding by showing that, besides attenuated bacteria, AvrRpt2 also boosts the infection of fully virulent P. syringae and additionally the necrotrophic fungus B. cinerea. The improved sensitivity in virulence detection may be due to the transgene expression strength and the defined DEX-inducible expression compared with the basal expression of AvrRpt2 controlled by the RPS2 promoter (Chen et al., 2000). One of the AvrRpt2 virulence functions contributing to immunity attenuation may be the MPK4/MPK11 suppression reported here, but this is difficult to test genetically due to the dwarf phenotype and enhanced SA content of mpk4 mutants (Petersen et al., 2000) and the lethality of mpk4mpk11 double mutants (Bethke et al., 2012). An argument against the inhibition of MPK4/MPK11 as a virulence strategy is the observation that the Y191C and D216E AvrRpt2 mutants, previously described to compromise virulence function, can suppress flg22-induced MPK4/MPK11 activation (Fig. 7D; Table I). However, in this work, we expressed these proteins directly in plant cells, while the previous work was based on AvrRpt2 delivery after P. syringae infection. Notably, the Y191C and D216E AvrRpt2 mutants showed activities of native AvrRpt2 (cleaving RIN4/NOIs and suppressing MPK4/MPK11 activation) when expressed in plant cells (Fig. 7, D and E). Thus, it is possible that the previously described reduced virulence of P. syringae expressing the Y191C or D216E AvrRpt2 mutant is caused by poorer delivery through the T3SS rather than by loss of functionality.
Table I. Summary of activities of AvrRpt2 homologs and variants.
Ps, P. syringae pv tomato JL1065; Rs, R. solanacearum CMR15; Ea, E. amylovora ATCC 49946; Ac, A. citrulli strain tw6; Aa, A. avenae ssp. avenae ATCC 19860; Mh, M. huakuii 7653R; Sm, S. medicae WSM1369; Bp, B. pyrrocinia Lyc2; Cf, C. fungivorans; Y, yes; N, no; n.t., not tested.
| Classification | AvrRpt2 Homolog | Percentage Identitya | Autocleavageb | MPK4/MPK11 Inhibition | RIN4 Cleavage | NOI Cleavagec | AXR2/AXR3 Destabilization |
|---|---|---|---|---|---|---|---|
| Phytopathogens | Ps | 100 | Y | Y | Y | Y | Y |
| Rs | 54 | Y | Y | Y | n.t. | n.t. | |
| Ea | 58 | Partial | Y | Y | (Y) | Y | |
| Ac | 49 | Y | Y | Y | n.t. | n.t. | |
| Aa | 27* | ? | N | N | n.t. | n.t. | |
| Plant associated | Mh | 51 | Partial | Y | Y | n.t. | n.t. |
| Sm | 55 | Partial | Y | Y | n.t. | n.t. | |
| Soil/rhizosphere | Bp | 25* | ? | Y | N | (specificNOIs) | Y |
| Fungus infecting | Cf | 61* | ? | N | N | (N) | N |
| P. syringae AvrRpt2 mutants | H208A | 99.6 | N | N | N | (N) | N |
| C122A | 99.6 | N | N | n.t. | n.t. | n.t. | |
| Y191C | 99.6 | Y | Y | Y | (Y) | n.t. | |
| D216E | 99.6 | Y | Y | Y | (Y) | n.t. |
Sequence identities (as calculated by Clustal2.1) are based on the complete sequence, resulting in lower scores for truncated sequences, which are marked with asterisks. b?, Autocleavage is unclear, since truncated sequences without cleavage sites were used; partial or incomplete cleavage occurred when multiple bands (including presumed precursor bands) were seen. cInterpretation of NOI cleavage based only on NOI1, NOI7, NOI9, and NOI11 is shown in parentheses.
As summarized in Table I, there is no absolute correlation between RIN4/NOI cleavage and the suppression of flg22-induced MPK4/MPK11 activation between the various AvrRpt2 putative homologs. For instance, the B. pyrrocinia homolog interfered with flg22-induced MPK4/MPK11 activation but did not cleave RIN4 and cleaved only selected NOIs (Fig. 7, B and C). Thus, taken together with the analysis of the rin4 mutant, the MPK4/MPK11-suppressive activity of AvrRpt2-like proteins is independent of established AvrRpt2 targets such as RIN4 and auxin signaling or the AvrRpt2-activated RPS2 pathway. In agreement, the AvrRpt2-mediated enhanced virulence in terms of bacterial growth in rps2 plants and the suppression of PR gene expression are independent of RIN4 presence, indicating that AvrRpt2 has additional virulence targets (Lim and Kunkel, 2004). Thus, like other pathogen effectors (Deslandes and Rivas, 2012), AvrRpt2 targets multiple cellular targets in the host to suppress immunity. The mechanistic target mediating the suppression of MPK4/11 activation is currently unknown, but since the AvrRpt2 Cys-protease activity is essential, only AvrRpt2 protease substrates are likely candidates. Our analysis excludes the direct cleavage of MPK4/MPK11 (or MEKK1) but points to the MKKs as a likely level for AvrRpt2 activity. Remarkably, AvrRpt2 regulates MKK1 mRNA levels while MKK2 is regulated posttranscriptionally, probably at the protein level (Fig. 9C). It is also conceivable that MKK1 is regulated at both levels. In view of the proteasome-dependent degradation of Aux/IAA proteins by AvrRpt2 (Cui et al., 2013), it is tempting to postulate a similar mechanism for the MKK proteins. The lack of putative AvrRpt2 cleavage sites within MKK1/MKK2 also indicates an indirect effect. Bioinformatics analyses of Arabidopsis protein sequences containing conserved AvrRpt2 cleavage sites identified the NOI protein family and additional proteins (Chisholm et al., 2005). We can now exclude a role of individual RIN4/NOI members where mutant lines are available. Notably, correlative evidence between MPK4/MPK11 suppression and the cleavage of selected NOIs (Fig. 7C) suggests the importance of members from the NOI1 and NOI9 clades. Thus, future analysis could be focused on these NOI members and should take into consideration functional redundancies. In addition, inspection of autocleavage showed that the E. amylovora, M. huakuii, and S. medicae AvrRpt2 homologs have an Ala at the partially variable [LVI] position of the consensus AvrRpt2 cleavage site ([LVI]PxFGxW). These are less efficiently processed (double bands of precursor and processed forms visible; Fig. 7B). Continued analysis with these AvrRpt2 homologs should allow refining of the prediction algorithm for the canonical AvrRpt2 cleavage site and aid in finding the host targets.
CONCLUSION
In this work, we show that AvrRpt2 specifically blocks the flagellin-induced activation of MPK4/MPK11 (but not MPK3/MPK6) independently of the previously identified AvrRpt2 targets. In view of the roles of MAPKs in immune signaling, such a selective inhibition of a specific branch of MAPK signaling is likely a novel virulence function of AvrRpt2. Furthermore, AvrRpt2-like homologs exist in many bacteria (including pathogens of crop plants), and these may have a more widespread role for bacterial interactions with hosts or the environment than previously anticipated.
MATERIALS AND METHODS
Plant Growth Conditions, Protoplast Assays, and Immunoblot Analysis
Arabidopsis (Arabidopsis thaliana) plants were grown on soil in climate chambers for 5 to 6 weeks (22°C, 8 h of light and 16 h of darkness, and 140 µE). Protoplast isolation and transfection were performed as described (Yoo et al., 2007). Proteins were extracted by directly adding SDS-loading buffer to the pelleted protoplasts and processed for immunoblotting as described (Lee et al., 2004).
General Molecular Cloning
Coding sequences of genes used in transient expression experiments were subcloned in either pDONR201/pDONR221 or pENTR-D-TOPO vectors (Thermo Fisher Scientific). Gateway LR Clonase II Enzyme mix (Thermo Fisher Scientific) was used to generate 35S promoter-driven expression constructs in pUGW14, pUGW15, or pUGW18 (Nakagawa et al., 2007). For details, primers and transient expression constructs used in this study are listed in Supplemental Tables S2 and S3, respectively. Site-directed mutagenesis was performed as described (Palm-Forster et al., 2012; Eschen-Lippold et al., 2014).
Cloning of AvrRpt2 Homologs
The Pseudomonas syringae pv tomato (strain JL1065) AvrRpt2 protein sequence was searched against predicted sequences translated from genome sequence projects (National Center for Biotechnology Information database). Selected homologs (Supplemental Table S1) were created by gene synthesis (Life Technologies) after optimizing for Arabidopsis codon usage and cloned into pDONR221 (Thermo Fisher Scientific). Note that the avrRpt2 sequence from Acidovorax citrulli strain tw6 is interrupted by an ISRso4-like IS element; the predicted intact sequence was recreated by in silico fusion of the genome sequence (gi|802681671|ref|NZ_JXDJ01000021.1) at 176,210 to 176,740 bp and 178,724 to 178,966 bp.
Pathogen Infection
Infection experiments were performed with Botrytis cinerea B05.10 and P. syringae pv tomato DC3000 as described previously (Bethke et al., 2012).
Statistical Analyses
Statistical significance was analyzed with Prism 5 (GraphPad) software.
Accession Numbers
The following Arabidopsis or P. syringae pv tomato genes were used in this work: MPK3 (At3g45640), MPK4 (At4g01370), MPK6 (At2g43790), MPK11 (At1g01560), MKK1 (At4g26070), MKK2 (At4g29810), MEKK1 (At4g08500), RIN4 (At3g25070), NOI1 (At5g63270), NOI2 (At5g40645), NOI3 (At2g17660), NOI4 (At5g55850), NOI5 (At3g48450), NOI6 (At5g64850), NOI7 (At5g09960), NOI8 (At5g18310), NOI9 (At5g48500), NOI10 (At5g48657), NOI11 (At3g07195), NOI12 (At2g04410), NOI13 (At4g35655), NOI14 (At5g19473), and avrRpt2 from P. syringae (Q6LAD6).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignments of both NOI10 gene models and of the predicted AvrRpt2 cleavage sites of all the NOI proteins.
Supplemental Figure S2. Positions of T-DNA insertions in the NOI genes.
Supplemental Figure S3. Verification of knockout/knockdown of transcript accumulation in noi T-DNA insertion lines.
Supplemental Figure S4. Multiple sequence alignment and identity matrix of AvrRpt2 homologs.
Supplemental Figure S5. Detection of low expression of the B. pyrrocinia AvrRpt2 homolog (longer exposure of Fig. 7C).
Supplemental Figure S6. Western blot corresponding to the experiment in Figure 9A showing MAPK activation (α-pTEpY), accumulation of MEKK1-YFP, and HA-tagged CFP or AvrRpt2 variants.
Supplemental Table S1. AvrRpt2 homologs used in this study.
Supplemental Table S2. Primers and qPCR probes used in this study.
Supplemental Table S3. Vectors and DNA constructs used for transient expression in this study.
Supplemental Methods S1. Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center/Nottingham Arabidopsis Stock Centre for mutant seeds, Nico Dissmeyer (IPB) for RCS1/2 mutant constructs, Tino Unthan for the MKK1/MKK2 Gateway-compatible Entry clones, and Nicole Bauer for excellent technical support.
Glossary
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- T3SS
type III secretion system
- ETI
effector-triggered immunity
- DEX
dexamethasone
- Col-0
Columbia-0
- T-DNA
transfer DNA
- qRT
quantitative reverse transcription
- RT
reverse transcription
- SA
salicylic acid
- ROS
reactive oxygen species
- qPCR
quantitative RCR
Footnotes
This work was supported by ProNET-T3 (grant nos. 03ISO2211B and SFB648/TP–B1).
Articles can be viewed without a subscription.
References
- Afzal AJ, da Cunha L, Mackey D (2011) Separable fragments and membrane tethering of Arabidopsis RIN4 regulate its suppression of PAMP-triggered immunity. Plant Cell 23: 3798–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal AJ, Kim JH, Mackey D (2013) The role of NOI-domain containing proteins in plant immune signaling. BMC Genomics 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, Parsot C, Sansonetti PJ (2007) An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat Immunol 8: 47–56 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ (2003) Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol 49: 1537–1546 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Bethke G, Pecher P, Eschen-Lippold L, Tsuda K, Katagiri F, Glazebrook J, Scheel D, Lee J (2012) Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol Plant Microbe Interact 25: 471–480 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D, He SY (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol 150: 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN (2007) Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA 104: 20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Boch J, Katagiri F, Kunkel BN (2000) The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol Plant Microbe Interact 13: 1312–1321 [DOI] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Cuzick A, Moeder W, Tang D, Innes RW, Klessig DF, McDowell JM, Kunkel BN (2004) The Pseudomonas syringae type III effector AvrRpt2 functions downstream or independently of SA to promote virulence on Arabidopsis thaliana. Plant J 37: 494–504 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ (2005) Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc Natl Acad Sci USA 102: 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL (2011) Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EH, El-Kasmi F, He Y, Loehr A, Dangl JL (2014) A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immune receptors. Cell Host Microbe 16: 484–494 [DOI] [PubMed] [Google Scholar]
- Coaker G, Zhu G, Ding Z, Van Doren SR, Staskawicz B (2006) Eukaryotic cyclophilin as a molecular switch for effector activation. Mol Microbiol 61: 1485–1496 [DOI] [PubMed] [Google Scholar]
- Cui F, Wu S, Sun W, Coaker G, Kunkel B, He P, Shan L (2013) The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol 162: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer W, Leveau JH, Kowalchuk GA, Klein Gunnewiek PJ, Abeln EC, Figge MJ, Sjollema K, Janse JD, van Veen JA (2004) Collimonas fungivorans gen. nov., sp. nov., a chitinolytic soil bacterium with the ability to grow on living fungal hyphae. Int J Syst Evol Microbiol 54: 857–864 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Rivas S (2012) Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci 17: 644–655 [DOI] [PubMed] [Google Scholar]
- Eschen-Lippold L, Bauer N, Löhr J, Palm-Forster MA, Lee J (2014) Rapid mutagenesis-based analysis of phosphorylation sites in mitogen-activated protein kinase substrates. Methods Mol Biol 1171: 183–192 [DOI] [PubMed] [Google Scholar]
- Eschen-Lippold L, Bethke G, Palm-Forster MA, Pecher P, Bauer N, Glazebrook J, Scheel D, Lee J (2012) MPK11: a fourth elicitor-responsive mitogen-activated protein kinase in Arabidopsis thaliana. Plant Signal Behav 7: 1203–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei dit Frey N, Garcia AV, Bigeard J, Zaag R, Bueso E, Garmier M, Pateyron S, de Tauzia-Moreau ML, Brunaud V, Balzergue S, et al. (2014) Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol 15: 87R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y (2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S (2008) Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Hann DR, Gimenez-Ibanez S, Rathjen JP (2010) Bacterial virulence effectors and their activities. Curr Opin Plant Biol 13: 388–393 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K (2006) MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem 281: 36969–36976 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL (2005a) The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA 102: 6496–650115845764 [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D (2005b) Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121: 749–759 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ (1993) RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassowskat I, Böttcher C, Eschen-Lippold L, Scheel D, Lee J (2014) Sustained mitogen-activated protein kinase activation reprograms defense metabolism and phosphoprotein profile in Arabidopsis thaliana. Front Plant Sci 5: 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Bourdais G, Yu G, Robatzek S, Coaker G (2015a) Phosphorylation of the plant immune regulator RPM1-INTERACTING PROTEIN4 enhances plant plasma membrane H⁺-ATPase activity and inhibits flagellin-triggered immune responses in Arabidopsis. Plant Cell 27: 2042–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Eschen-Lippold L, Lassowskat I, Böttcher C, Scheel D (2015b) Cellular reprogramming through mitogen-activated protein kinases. Front Plant Sci 6: 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Rudd JJ, Macioszek VK, Scheel D (2004) Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J Biol Chem 279: 22440–22448 [DOI] [PubMed] [Google Scholar]
- Lee JS, Huh KW, Bhargava A, Ellis BE (2008) Comprehensive analysis of protein-protein interactions between Arabidopsis MAPKs and MAPK kinases helps define potential MAPK signalling modules. Plant Signal Behav 3: 1037–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F (2007) The phosphothreonine lyase activity of a bacterial type III effector family. Science 315: 1000–1003 [DOI] [PubMed] [Google Scholar]
- Lim MTS, Kunkel BN (2004) The Pseudomonas syringae type III effector AvrRpt2 promotes virulence independently of RIN4, a predicted virulence target in Arabidopsis thaliana. Plant J 40: 790–798 [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJ, Coaker G (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF III, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway Binary Vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Palm-Forster MA, Eschen-Lippold L, Lee J (2012) A mutagenesis-based screen to rapidly identify phosphorylation sites in mitogen-activated protein kinase substrates. Anal Biochem 427: 127–129 [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al. (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, et al. (2008a) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, Mackinlay J, Loake GJ, Mundy J, Morris PC (2008b) Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Sheen J (2007) Intercepting host MAPK signaling cascades by bacterial type III effectors. Cell Host Microbe 1: 167–174 [DOI] [PubMed] [Google Scholar]
- Takemoto D, Jones DA (2005) Membrane release and destabilization of Arabidopsis RIN4 following cleavage by Pseudomonas syringae AvrRpt2. Mol Plant Microbe Interact 18: 1258–1268 [DOI] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15: 141–152 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, Zhou JM (2010) A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22: 2033–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222: 377–383 [DOI] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeier J, Pink B, Mueller MJ, Berger S (2004) Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683 [DOI] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y (2012) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wu S, Chen X, Liu C, Sheen J, Shan L, He P (2014) The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J 77: 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.