Arabidopsis Xyloglucan Xylosyltransferase 5 uses an acceptor substrate similar to that of Xylosyltransferases 1 and 2 but at a lower rate.
Abstract
Xyloglucan, the most abundant hemicellulosic component of the primary cell wall of flowering plants, is composed of a β-(1,4)-glucan backbone decorated with d-xylosyl residues. Three xyloglucan xylosyltransferases (XXTs) participate in xyloglucan biosynthesis in Arabidopsis (Arabidopsis thaliana). Two of these, XXT1 and XXT2, have been shown to be active in vitro, whereas the catalytic activity of XXT5 has yet to be demonstrated. By optimizing XXT2 expression in a prokaryotic system and in vitro activity assay conditions, we demonstrate that nonglycosylated XXT2 lacking its cytosolic amino-terminal and transmembrane domain displays high catalytic activity. Using this optimized procedure for the expression of XXT5, we report, to our knowledge for the first time, that recombinant XXT5 shows enzymatic activity in vitro, although at a significantly slower rate than XXT1 and XXT2. Kinetic analysis showed that XXT5 has a 7-fold higher Km and 9-fold lower kcat compared with XXT1 and XXT2. Activity assays using XXT5 in combination with XXT1 or XXT2 indicate that XXT5 is not specific for their products. In addition, mutagenesis experiments showed that the in vivo function and in vitro catalytic activity of XXT5 require the aspartate-serine-aspartate motif. These results demonstrate that XXT5 is a catalytically active xylosyltransferase involved in xylosylation of the xyloglucan backbone.
Plant cell walls consist of cellulose (Endler and Persson, 2011), hemicellulose (Scheller and Ulvskov, 2010), pectin (Atmodjo et al., 2013), lignin (Vanholme et al., 2010), and glycoproteins (Josè-Estanyol and Puigdomènech, 2000). In plants, the cell wall strengthens the cell to resist turgor pressure and acts in intracellular communication and defense against biotic and abiotic stresses (Somerville et al., 2004; Keegstra, 2010). In addition, the plant cell wall has many potential and current industrial applications in biomaterials or biofuels (Somerville et al., 2004; McCann and Carpita, 2008; Abramson et al., 2010; Carpita, 2012)
Xyloglucan (XyG), the most abundant hemicellulose in the primary cell wall of dicotyledonous plants, has many proposed functions (Hayashi and Kaida, 2011; Park and Cosgrove, 2015), including cross-linking adjacent cellulose microfibrils (Hayashi, 1989; Carpita and Gibeaut, 1993; Somerville et al., 2004) and acting as a spacer to prevent the aggregation of cellulose microfibrils (Thompson, 2005). In Arabidopsis (Arabidopsis thaliana), XyG is composed of a β-(1,4)-glucan backbone with 50% to 75% of the d-Glc residues substituted with α-(1,6)-linked d-Xyl residues. The side chain Xyl residues can be further decorated with α-(1,2)-linked d-Gal or l-Fucp-(1,2)-β-d-Galp. According to the established nomenclature for the description of glucan backbone substitution patterns, G represents the unsubstituted glucosyl residue, X represents the glucosyl residue substituted at O-6 with α-d-Xyl, L represents the Xyl substituted with α-(1,2)-linked d-Galp, and F represents the Xyl substituted with α-(1,2)-linked l-Fucp-(1,2)-β-d-Galp (Fry et al., 1993). Arabidopsis XyG is composed mainly of XXXG, XXFG, and XLFG subunits (Vanzin et al., 2002; Madson et al., 2003).
Enzymes involved in XyG biosynthesis in Arabidopsis have been identified through reverse genetics and biochemical approaches. All of the enzymes that decorate the glucan backbone of XyG are type II transmembrane proteins containing a short N terminus localized in the cytosol, a transmembrane domain, a stem region, and a large catalytic domain localized in the Golgi lumen. Xylosylation of the glucan backbone is catalyzed by at least three glycosyltransferases, Xyloglucan Xylosyltransferase 1 (XXT1), XXT2, and XXT5, which are members of the GT34 family (Faik et al., 2002; Cavalier and Keegstra, 2006; Cavalier et al., 2008; Zabotina et al., 2008, 2012; Vuttipongchaikij et al., 2012) in the Carbohydrate Active Enzyme database (Campbell et al., 1997; Coutinho et al., 2003). Analysis of a phylogenetic tree of GT34 glycosyltransferases revealed three major clades (Mansoori et al., 2015). Clade A included AtXXT1 and AtXXT2, clade B included AtXXT3 to AtXXT5, and clade C included other glycosyltranferases that are involved in mannan biosynthesis (Mansoori et al., 2015). Glycosyltransferases are enzymes that transfer a sugar moiety from an activated donor substrate to an acceptor substrate (Lairson et al., 2008). XXTs are α-1,6-xylosyltranferases that transfer a Xyl residue from UDP-Xyl onto the glucan backbone of XyG, synthesizing XXXG-type oligosaccharide (Supplemental Fig. S1A)
Predictions based on their primary amino acid sequences indicate that XXTs contain a GT-A fold and catalyze the formation of a glycosidic bond in which the product retains the anomeric configuration of UDP-Xyl. XXT1 and XXT2 showed α-xylosyltransferase activity in vitro when cellohexaose was used as an acceptor (Faik et al., 2002; Cavalier and Keegstra, 2006). XXT1 and XXT2 primarily xylosylate the fourth d-glucosyl unit from the reducing end of cellohexaose, producing GGXGGG. Catalysis mainly continues on the third d-glucosyl unit from the reducing end, producing GGXXGG with small amounts of GXXGGG (Cavalier and Keegstra, 2006). The generation of knockout mutants in Arabidopsis showed that these enzymes, along with the putative xylosyltransferase XXT5, function to synthesize XyG (Cavalier et al., 2008; Zabotina et al., 2008, 2012). Another xylosyltransferase, XXT4, was later demonstrated to be catalytically active in vitro, although xxt4 mutants had no detectable decrease in XyG content (Vuttipongchaikij et al., 2012). The xxt1 xxt2 double mutant, xxt1 xxt2 xxt5 triple mutant, and xxt5 single mutant all show severe root hair phenotypes, displaying root hairs that were short and thick, formed bubble-like extrusions near the tips, and were lower in abundance compared with wild-type roots (Cavalier et al., 2008; Zabotina et al., 2008, 2012). By contrast, the xxt1 and xxt2 single mutants had normal root hairs, indicating that these enzymes are partially redundant (Cavalier et al., 2008). Analysis of the XyG content demonstrated that the xxt5 single mutant plants had a 50% reduction in total XyG content, while the xxt1 xxt2 double mutants had no detectable XyG (Cavalier et al., 2008; Zabotina et al., 2008). Although XXT5 clearly plays a significant role in XyG biosynthesis, its enzymatic activity has yet to be demonstrated. XXTs, along with all other GTs involved in XyG biosynthesis, form homodimers and heterodimers and have been proposed to form multiprotein complexes localized in the Golgi; these interactions may be required for the high efficiency of these enzymes in vivo (Chou et al., 2012, 2015; Lund et al., 2015).
In this study, we optimized the conditions for the expression of XXTs in Escherichia coli through the systematic investigation of several of the most commonly used E. coli strains, various solubility tags, and storage conditions. We demonstrate that XXT1, XXT2, and XXT5 can be expressed in a prokaryotic system, producing catalytically active recombinant proteins. Using this protocol, we demonstrate that XXT5 has enzymatic activity and transfers a d-xylosyl residue from UDP-Xyl onto cellohexaose at a similar position to XXT2. However, XXT5 has significantly lower activity compared with XXT1 and XXT2. To our knowledge for the first time, we show that XXT5 is catalytically active in vitro, and we demonstrate that its activity does not depend on the products of XXT1 and XXT2. We also identified an Asp-Ser-Asp motif that is essential for XXT5 function as a xylosyltransferase.
RESULTS
Optimization of XXT2 Expression
Due to the hydrophobic nature of transmembrane domains, to increase protein solubility and expression levels, we produced truncated versions of XXT1, XXT2, and XXT5 that lacked the transmembrane domain and contained only the protein stem region and catalytic domain (Supplemental Fig. S1B). A previous study reported the expression of truncated XXTs with an N-terminal glutathione S-transferase tag in E. coli BL21 cells (Vuttipongchaikij et al., 2012). Here, we tested different E. coli strains and various N-terminal tags to determine the optimal conditions for expressing XXTs (Supplemental Fig. S2)
We investigated E. coli BL21 (DE3)-derived cell lines, which were engineered for different purposes (Francis and Page, 2010). We examined BL21 (DE3), CodonPlus, SoluBL21, Rosetta 2 (DE3), Arctic Express (DE3) codon plus, and BL21 (DE3) pLysS. SoluBL21 was engineered for the expression of insoluble proteins, and BL21 CodonPlus and Rosetta contain extra copies of tRNA genes, allowing for the expression of proteins with codons rarely used in E. coli (Francis and Page, 2010). The pET20b-His-GB1-XXT2 plasmid was transformed into each cell line, and protein expression was carried out in parallel. Cells were lysed, and XXT2 was purified using nickel-nitrilotriacetic acid agarose (Ni-NTA) resin. We estimated XXT2 levels by analyzing the intensity of the corresponding band observed after immunoblotting using antibody against the GB1 tag (Supplemental Fig. S2A). The highest expression level of XXT2 was obtained in SoluBL21, and a slightly lower expression level was obtained in BL21 CodonPlus and Rosetta (Supplemental Fig. S2A).
Glycerol can improve protein stability and increase the specific activity of enzymes (Bradbury and Jakoby, 1972; Leandro et al., 2001). Therefore, we investigated the effect of the addition of glycerol following protein purification. Glycerol was added to the proteins eluted from Ni-NTA columns to a final concentration of 0%, 20%, or 50%, and the samples were frozen overnight at −80°C. The following day, the proteins were analyzed by activity assays using equal protein concentrations in all reactions. XXT2 stored in 20% glycerol had 1.4-fold higher activity compared with XXT2 stored in 0% or 50% glycerol (Supplemental Fig. S2B).
To further improve the expression levels and affinity chromatography purification efficiency, we fused various solubility tags, along with a 6xHis tag, to the N terminus of XXT2 (Supplemental Fig. S1C). We tested the following tags: the immunoglobulin-binding domain of streptococcal protein G (GB1; Huth et al., 1997), maltose-binding protein (MBP; Nallamsetty and Waugh, 2006), GFP (Chalfie, 1995), and the 6xHis tag alone. Constructs containing XXT2 and the N-terminal tags were expressed in SoluBL21 cells and purified using Ni-NTA resin (Supplemental Fig. S2C). Activity assays of these XXT2 constructs showed that the 6xHis-GB1- and 6xHis-MBP-tagged proteins have the highest activity (Supplemental Fig. S2D). Although XXT2 with only the 6xHis tag had a very large band at the expected size, the activity was significantly lower in comparison with XXT2 tagged with 6xHis-GB1 or 6xHis-MBP. The 6xHis-GB1- and 6xHis-MBP-tagged proteins produced similar xylosylated products but, as observed by SDS-PAGE, the 6xHis-GB1 tagged protein had a much higher purity (Supplemental Fig. S2C). Based on these results, all further experiments were performed using 6xHis-GB1 fusion proteins expressed in SoluBL21 cells and stored in 20% glycerol until analysis.
Optimization of Activity Assay Conditions
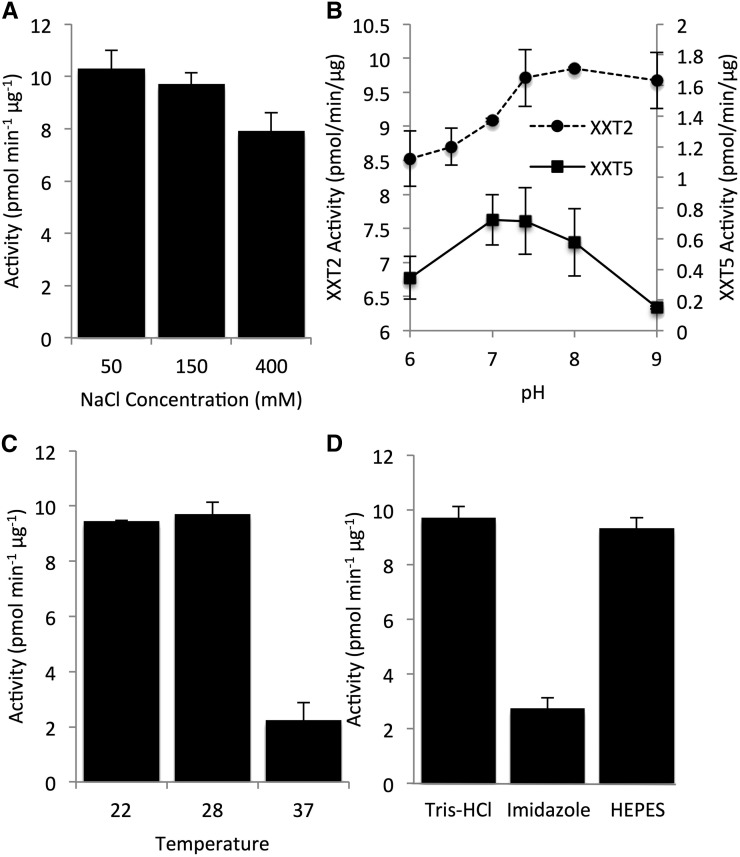
A previous study showed that the maximum activity of XXT1 and XXT2 requires the presence of Mn2+ and an acceptor glucan chain with a degree of polymerization of at least 6 (Cavalier and Keegstra, 2006). Here, we investigated the effect of various salt concentrations, pH, temperature, and two buffers on XXT2 activity (Fig. 1). Increasing the sodium chloride concentration in the assay reaction mixture decreased XXT2 activity (Fig. 1A). Low pH in the assay reaction also decreased XXT2 activity, while pH between 7.4 and 9 did not significantly affect XXT2 activity (Fig. 1B). Activity assay reactions incubated at 22°C or 28°C showed no difference in XXT2 activity, but increasing the incubation temperature to 37°C decreased XXT2 activity 4-fold (Fig. 1C).
Figure 1.
Optimization of assay conditions for XXT2 and XXT5. A to C, Effects of sodium chloride concentration (A), pH (B), and temperature (C) on His-GB1-XXT2 or His-GB1-XXT5 activity. D, Effects of imidazole and HEPES on His-GB1-XXT2 activity. The imidazole sample was in Tris-HCl buffer. All assays were with XXT2 unless indicated. All buffers were adjusted to pH 7.4 unless indicated. All constructs were expressed in SoluBL21 cells and purified with Ni-NTA columns. Assays contained 25 μL with concentrated His-GB1-XXT2 (3 μm, 2.5 μL) or His-GB1-XXT5 (5.5 μm, 8.25 μL), 12.5 μL of 2× buffer, 2 mm UDP-Xyl, 0.5 mm cellohexaose, and 2 mm MnCl2 and were incubated for 4 h; 2× buffer is a buffer that contains a concentration 2 times higher than that of the desired final concentration of all components in the assay reaction. The final concentration of buffers was 50 mm for His-GB1-XXT2 reactions; the buffer concentration of His-GB1-XXT5 was 250 mm to reduce the effect of the larger volume of His-GB1-XXT5 protein that was added to the reaction on the overall reaction conditions. Products of the enzyme assays were analyzed by high-performance anion-exchange chromatography (HPAEC), quantified by peak integration, and are presented as pmol UDP-Xyl min−1 μg−1 XXT protein. All enzyme assays were performed in duplicate.
Next, two types of buffer (Tris-HCl and HEPES) and the presence of imidazole in the reaction mixture were investigated. A previous study reported that Tris-type buffers can inhibit enzymes by forming a complex with the nucleotide sugar (Mukerjea et al., 2012). To investigate the effect of Tris-HCl buffer on XXT2 activity, we performed activity assays in Tris-HCl (pH 7.4) or HEPES (pH 7.4) buffers and found that reactions in Tris-HCl or HEPES buffer showed similar XXT2 activities (Fig. 1D). In addition, XXT2 was eluted from the Ni-NTA column using a buffer containing imidazole, which has been shown to affect the activity of other enzymes (Cotovio et al., 1996; McGee et al., 2004; Zhang et al., 2011). To investigate the effect of imidazole on XXT2, activity assays were carried out in the presence of 250 mm imidazole and without imidazole in the reaction mixture. The activity of XXT2 with 250 mm imidazole was 4-fold lower compared with the XXT2 activity without imidazole (Fig. 1D). Based on these results, all further activity assays were performed in 50 mm Tris-HCl buffer (pH 7.4) and 150 mm NaCl at 28°C without imidazole.
Expression and Catalytic Activities of XXT1, XXT2, and XXT5
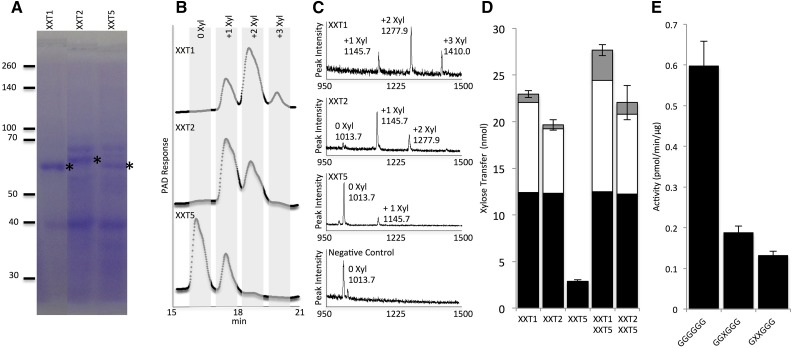
XXT1, XXT2, and XXT5 lacking their cytosolic N termini and transmembrane domains were fused with a 6xHis-GB1 tag and expressed in SoluBL21. When expressed in parallel using the same conditions, XXT1 showed an approximately 2.5-fold higher expression level compared with XXT2; the expression of XXT5 was slightly lower than that of XXT2 (Fig. 2A). SDS-PAGE and quantitative western blots were used to estimate the concentration of XXT proteins obtained after affinity purification (Supplemental Fig. S3). The purified XXT proteins were then assayed for enzymatic activity in a reaction containing 2 mm UDP-Xyl, 0.5 mm cellohexaose, and 2 mm MnCl2. Enzyme assays contained a maximum volume of XXT solution (maximum protein) to determine if XXT1, XXT2, and XXT5 can xylosylate cellohexaose at one, two, or three positions. Cellohexaose from reactions with XXT1 (6.2 µm) and XXT2 (2.5 µm) was completely xylosylated, and XXT1 was capable of xylosylating cellohexaose at three positions (Fig. 2B). XXT5 had very low activity under these conditions, yet when concentrated 6-fold (6 µm), XXT5 was capable of xylosylating cellohexaose (Fig. 2B). All product sizes were verified with matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS; Fig. 2C). Negative controls were assayed in parallel with XXT reactions (Fig. 2C; Supplemental Fig. S4). Localization of XXT5 in the cell or the XyG biosynthetic complex might differ from the localization XXT2, and XXT5 might have a different optimal pH compared with XXT2. Thus, we investigated various pH values in the XXT5 activity assay. XXT5 showed highest activity between pH 7 and 8 (Fig. 1B). XXT5 also showed higher sensitivity to increased pH compared with XXT2. To our knowledge, this is the first demonstration of XXT5 enzymatic activity in vitro.
Figure 2.
Protein expression and enzyme activity of XXT1, XXT2, and XXT5. All His-GB1-XXT proteins were expressed in SoluBL21 cells and purified on Ni-NTA resin. A, SDS-PAGE of XXT1, XXT2, and XXT5 with 10 μL (5.6 μg), 20 μL (12 μg), and 25 μL (12.5 μg) of the elution loaded in each well, respectively. The positions of XXT proteins are shown with asterisks. B, HPAEC of XXT enzyme assay products. XXT1 and XXT2 concentration in assay reactions was 3 μm; XXT5 was concentrated 6-fold, and the concentrated protein was assayed at 11.5 μm. PAD, Pulsed-amperometric detection. C, MALDI-TOF chromatograms of enzyme assay products. Mass in MALDI-TOF chromatograms represents the mass of the oligosaccharide plus a sodium ion; the negative control consists of the activity assay reaction with no enzyme. D, Enzyme assay of XXT1, XXT2, and XXT5 in combination. All XXT concentrations were 8.5 μm, with reaction time of 20 h. Black bars indicate monoxylosylated cellohexaose, white bars indicate dixylosylated cellohexaose, and gray bars indicate trixylosylated cellohexaose. E, XXT5 activity with various acceptors. Acceptor substrates were synthesized as described previously (Fauré et al., 2007). All reactions contained 2 mm UDP-Xyl, 2 mm MnCl2, 0.5 mm acceptor substrate, and 14 μm XXT5 with reaction time of 4 h. Products of the enzyme assay were analyzed by HPAEC, quantified by peak integration, and are presented as pmol UDP-Xyl min−1 μg−1 XXT protein. All enzyme assays were performed in duplicate.
To determine whether XXT5’s preferred substrate is the product of XXT1 or XXT2 (GGXXGG), we studied XXT5 activity in combination with either XXT1 or XXT2. Single assays were performed with only one XXT (8.5 µm), and combination assays were performed with XXT5 (8.5 µm) and XXT1 or XXT2 (8.5 µm). In the single-enzyme reactions, XXT1 showed the highest activity, followed by XXT2. XXT5 showed the lowest catalytic activity, transferring only 2.9 nmol of d-Xyl to cellohexaose (Fig. 2D). The combination XXT1+XXT5 transferred 27.7 nmol of d-Xyl to cellohexaose, slightly higher xylosylation than the sum of the single-enzyme reactions of XXT1 and XXT5 (25.8 nmol). Similarly, the combination XXT2+XXT5 showed a slight decrease in xylosylation (22 nmol) compared with the single-enzyme reactions, transferring 22 nmol of Xyl to cellohexaose, which was lower than the sum of the single-enzyme reactions of XXT2 and XXT5 (22.5 nmol). To further investigate if XXT5 is specific for xylosylated glucan, we ran XXT5 activity assays with different acceptor substrates, including GGXGGG and GXXGGG. Acceptor substrates were chemically synthesized as described previously (Fauré et al., 2007). XXT5 showed a 3-fold higher activity when cellohexaose (GGGGGG) was used as an acceptor, compared with the reactions when monoxylosylated or dixylosylated cellohexaose was used (Fig. 2E).
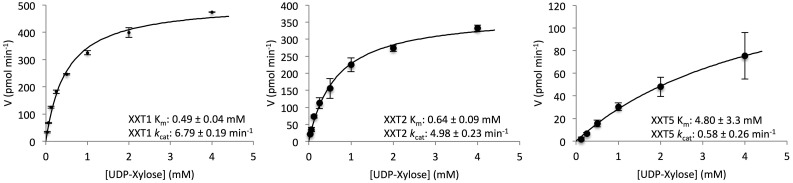
Previous studies of XXTs used activity assays that were run for the maximum time period to determine the ability of the XXTs to xylosylate cellohexaose at one, two, or three glucoses. Here, we performed kinetic studies to investigate the initial velocities of XXTs with various UDP-Xyl concentrations and a constant cellohexaose concentration (1 mm). XXT1 and XXT2 assays required 30 min of reaction time and an enzyme concentration of 3 µm to produce detectable levels of xylosylated product, using HPAEC for detection. XXT5 assays required 60 min of reaction time and an enzyme concentration of 11.5 µm to produce detectable levels of xylosylated product. In these conditions, XXT1 showed the lowest Km and highest kcat and XXT5 showed the highest Km and lowest kcat. XXT5 showed a 7-fold higher Km and 9-fold lower kcat compared with XXT1 and XXT2 (Fig. 3).
Figure 3.
Kinetic analysis of XXT1, XXT2, and XXT5. XXT1 and XXT2 assays contained 3 μm XXT and were performed for 30 min; XXT5 assays contained 11.5 μm XXT5 and were performed for 60 min. All reactions contained 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm cellohexaose, and 2 mm MnCl2 and were incubated at 28°C. The kinetic constants Km and kcat were calculated by fitting initial velocities as a function of UDP-Xyl concentration using nonlinear curve fitting with Dynafit (Kuzmic, 1996). Products of the enzyme assay were analyzed by HPAEC, quantified by peak integration, and are presented as pmol UDP-Xyl min−1. All enzyme assays were performed in duplicate.
XXT5 and XXT2 Produce Similar Xylosylation Patterns
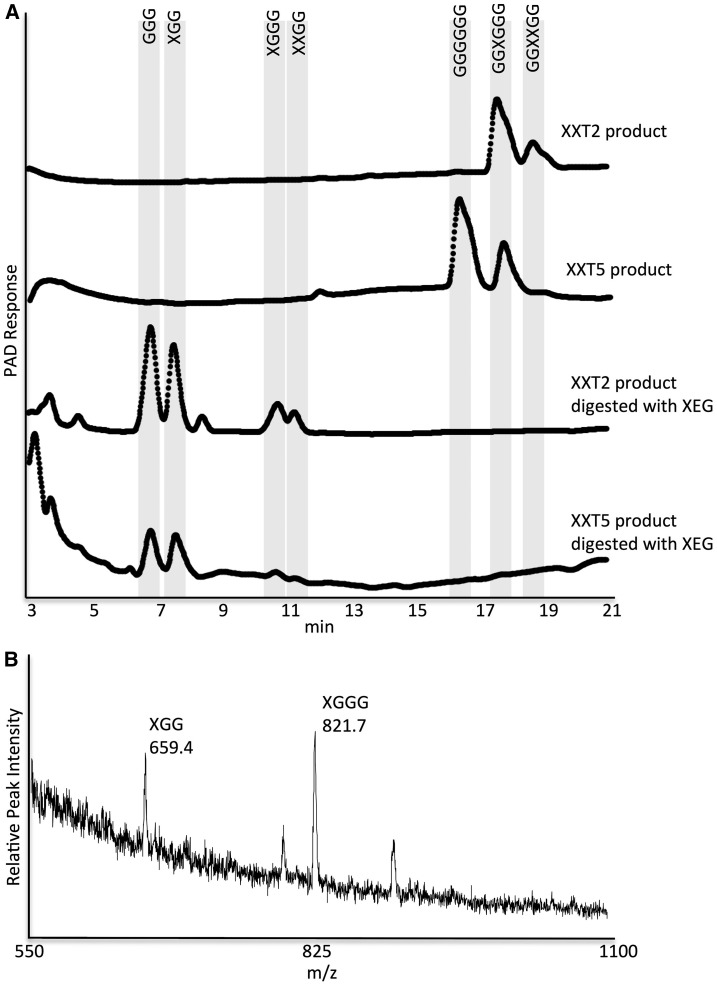
A previous study reported that both XXT1 and XXT2 link the first d-Xyl to the fourth d-Glc from the reducing end of the cellohexaose oligosaccharide (Cavalier and Keegstra, 2006). Next, the enzymes link a second d-Xyl preferentially to the third d-Glc from the reducing end; they also can add the second d-Xyl to the fifth d-Glc, albeit at a lower frequency. Finally, the enzymes link a third Xyl to the fifth or the third Glc, forming a trixylosylated product with three adjacent xyloses (Cavalier and Keegstra, 2006). To further investigate the activity of XXT5 in vitro, we determined the position of the d-Xyl unit in the monoxylosylated product. Xylosylation positions were investigated by digesting the products of XXT2 and XXT5 reactions with xyloglucan-specific endo-β-1,4-glucanase (XEG), which cleaves the glucan backbone at the nonreducing end of a Glc substituted with Xyl (Pauly et al., 1999; Yaoi et al., 2009). Digestion of the products of XXT2 (monoxylosylated and dixylosylated) produced mainly XGG and XGGG. Similarly, digestion of the XXT5 product (only monoxylosylated) produced mainly XGG with minor amounts of XGGG (Fig. 4A). Product sizes were verified with MALDI-TOF MS (Fig. 4B). These results indicated that XXT2 and XXT5 add d-Xyl to cellohexaose at a similar position.
Figure 4.
Analysis of the products of reactions catalyzed by XXT2 and XXT5 with HPAEC and MALDI-TOF. The products were precipitated (see “Materials and Methods”) and treated with XEG for 12 h. A, HPAEC chromatogram of enzyme assay products and products following XEG digestion. PAD, Pulsed-amperometric detection. B, MALDI-TOF chromatogram of the XXT2 reaction to confirm product sizes. m/z, Mass-to-charge ratio.
XXT5 Activity Requires the DxD Motif
In GT-A fold glycosyltransferases, which include XXTs, the DxD motif (Asp-x-Asp) plays an important role in coordinating an essential divalent cation (Busch et al., 1998; Wiggins and Munro, 1998; Zhang et al., 2001; Boeggeman and Qasba, 2002; Götting et al., 2004; Cavalier and Keegstra, 2006; Bobovská et al., 2014). Structural characterization of a putative glycosyltransferase from the Paramecium bursaria chlorella virus-1 (PBCV-1), which is predicted to belong to the same GT34 gene family as the XXTs, showed that the two Asp residues in the DxD motif participate in the coordination of the divalent cation (Zhang et al., 2007). Sequence alignment of the PBCV-1 glycosyltransferase with XXT1, XXT2, and XXT5 demonstrated that both Asp residues within the DxD motif are conserved in each of the XXTs (Supplemental Fig. S1D).
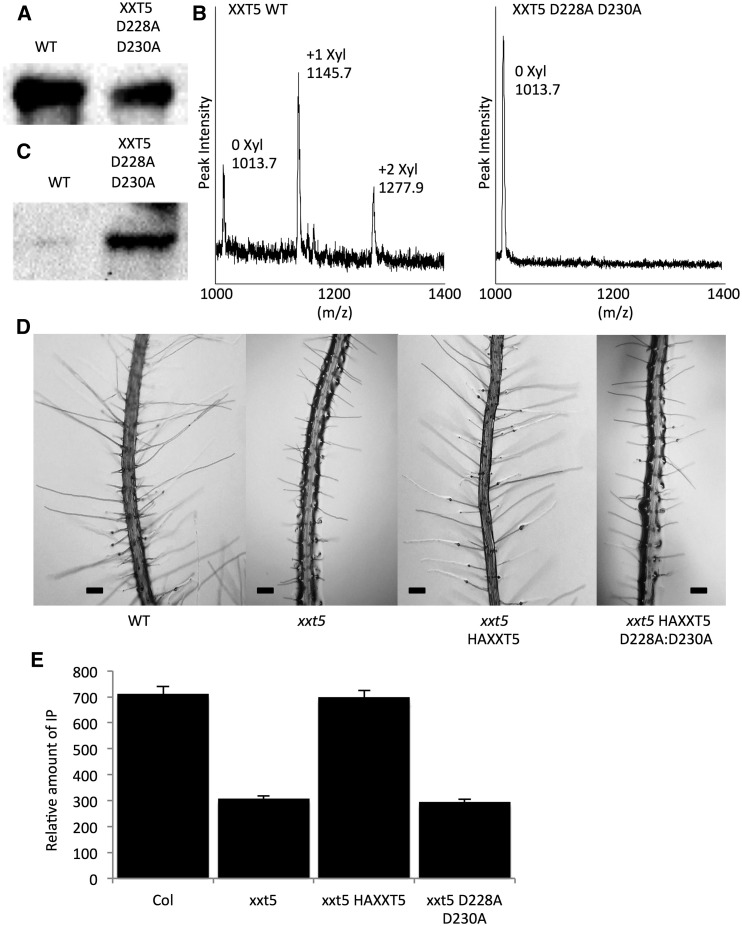
To test whether XXT5 catalytic activity requires the DxD motif, we mutated both Asp residues in the DxD motif of XXT5 (Asp-Ser-Asp) to Ala (Ala-Ser-Ala; D228A:D230A). The resulting protein was expressed, purified (Fig. 5A), and assayed for transferase activity. These activity assays showed no detectable xylosylated product for the XXT5 D228A:D230A mutant when analyzed by MALDI-TOF MS. To ensure that the lack of xylosylated product is not due to products below the sensitivity of MALDI-TOF MS, we concentrated both wild-type XXT5 and the XXT5 D228A:D230A mutant protein 15-fold and performed activity assays with increased concentrations of UDP-Xyl and MnCl2 (4 mm) and a reaction time of 30 h (Fig. 5B). Under these conditions, wild-type XXT5 added two d-xyloses, producing dixylosylated cellohexaose, while the XXT5 D228A:D230A mutant did not produce any xylosylated oligosaccharide (Fig. 5B).
Figure 5.
Characterization of the XXT5 D228A/D230A mutant. A, MALDI-TOF chromatograms of enzyme assay products of XXT5 D228A/D230A. The enzyme assay was modified slightly. Affinity-purified protein was concentrated 15-fold; enzyme assays ran for 30 h and contained 4 mm UDP-Xyl and 4 mm MnCl2. B, Driselase digestion of plant cell wall extracts. C, Western blot of bacterially expressed XXT5 proteins. D, Western blot of proteins extracted from transgenic plants. E, Root hair phenotype of wild-type (WT) and transgenic plants.
To investigate the function of enzymatically inactive XXT5 in vivo, we overexpressed the XXT5 D228A:D230A mutant protein in the xxt5 knockout mutant (Zabotina et al., 2008). The mutant complementary DNA of XXT5 D228A:D230A was fused with a hemagglutinin (HA) tag and introduced into the binary vector pEarleyGate 201. The resulting construct was transformed into xxt5 homozygous mutant plants (Zabotina et al., 2008). In planta expression of XXT5 D228A:D230A was confirmed by immunoblot using HA antibodies (Fig. 5C). Overexpression of XXT5 D228A:D230A did not complement the root hair phenotype of the xxt5 mutants (Fig. 5D). To measure XyG content in the transgenic plants, we used Driselase to digest cell walls extracted from the wild type, xxt5, xxt5 complemented with HA-XXT5, and xxt5 complemented with HA-XXT5 D228A:D230A. Driselase is a mixture of hydrolases that digests most cell wall polysaccharides but lacks α-xylosidase activity. Hence, hydrolysis of XyG will produce the signature disaccharide isoprimeverose [d-Xylp-α-(1-6)-d-Glc]. Driselase digestion of the cell wall from xxt5 plants complemented with XXT5 D228A:D230A resulted in isoprimeverose levels similar to those of the xxt5 knockout mutant, indicating that XXT5 D228A:D230A cannot rescue XyG biosynthesis in the xxt5 mutant (Fig. 5E).
DISCUSSION
Three Arabidopsis xylosyltransferases, XXT1, XXT2, and XXT5, have been shown to be involved in XyG biosynthesis (Faik et al., 2002; Cavalier et al., 2008; Zabotina et al., 2008). The xxt1 and xxt2 single knockout mutant plants had no apparent phenotype, whereas the xxt1 xxt2 double mutants had no detectable XyG and a strong root hair phenotype, indicating that XyG biosynthesis requires the presence of at least one of these enzymes (Zabotina et al., 2012). The xxt5 mutant had a 50% reduction in XyG content as well as a distinct root hair phenotype. In addition, XXT1 and XXT2 have been confirmed to be catalytically active in vitro as α-1,6-xylosyltransferases (Cavalier and Keegstra, 2006), while the catalytic activity of XXT5 has not been shown despite several attempts (Faik et al., 2002; Zabotina et al., 2008; Vuttipongchaikij et al., 2012). Therefore, it was suggested that XXT5 might lack enzymatic activity and, most likely, perform other functions. Here, we demonstrate, to our knowledge for the first time, that XXT5 is enzymatically active in vitro, catalyzing the formation of a xylosylated oligosaccharide product, but at a significantly lower rate compared with XXT1 and XXT2.
Plant glycosyltransferases have been heterologously expressed mainly in eukaryotic cells for in vitro characterization (Faik et al., 2002; Cavalier and Keegstra, 2006; Jensen et al., 2014; Urbanowicz et al., 2014). Here, we expressed truncated versions of XXTs in prokaryotic cells, demonstrating that catalytic activity does not require the N terminus and transmembrane domains or glycosylation; however, glycosylation may improve protein folding and/or solubility. Negative controls of reactions lacking either UDP-Xyl or enzyme were used to confirm that the xylosylation is the product of XXTs (Fig. 2C; Supplemental Fig. S4). In addition, the XXT5 D228A:D230A mutant showed no xylosylation, demonstrating that other proteins copurified from E. coli cells do not xylosylate cellohexaose (Fig. 5A). Vuttipongchaikij et al. (2012) reported the only attempt to produce recombinant XXTs in E. coli cells, expressing XXTs without transmembrane domains in BL21 cells, and demonstrated enzymatic activity for XXT1, XXT2, and XXT4. However, protein expression levels in that study were low, as indicated by faint bands on SDS-PAGE and by reported yields of 40 to 90 μg of total protein per 1 L of cell culture. Vuttipongchaikij et al. (2012) showed no in vitro activity for XXT5 with an enzyme concentration of 0.3 μg of total protein per 25-μL reaction. Here, increasing the XXT5 concentration 26-fold to 6 μm (7.8 μg) showed catalytic activity of XXT5 in vitro (Fig. 2). Thus, the lack of XXT5 activity described previously is most likely due to low protein levels in activity assay reactions. However, the demonstration of XXT5 catalytic activity in vitro does not exclude any of the noncatalytic functions proposed earlier for this protein in vivo.
XXT1 showed a higher degree of xylosylated product (Fig. 2), lower Km, and higher kcat (Fig. 3) compared with XXT2. This confirms previous reports showing higher activity for XXT1 compared with XXT2 when expressed in Pichia pastoris (Faik et al., 2002), Drosophila Schneider 2 cells (Cavalier and Keegstra, 2006), and Spodoptera frugiperda 21 cells (Cavalier and Keegstra, 2006), although the kinetic parameters for XXTs were not determined in those studies. Due to limitations in the product detection by HPAEC, the values reported here are only an estimation of kinetic parameters, yet they allow comparison between the XXTs. The development of more sensitive detection methods for more accurate determination of kinetic parameters remains a topic for future research. In the conditions used in this study, XXT5 showed a 7-fold higher Km and 9-fold lower kcat in comparison with XXT1 or XXT2 (Fig. 3). This slower rate of XXT5 activity might be the reason why XXT5 cannot fully xylosylate the glucan backbone in vivo in the xxt1 xxt2 double mutant.
It was proposed that XXT5 might require prexylosylated product formed by XXT1 or XXT2 to add d-Xyl onto the third d-Glc in the XXGG subunit to form an XXXG oligosaccharide (Cavalier et al., 2008). The results in our study do not support this hypothesis. First, both XXT1 and XXT2 can xylosylate cellohexaose at three positions, as shown here (Fig. 2) and reported earlier (Cavalier and Keegstra, 2006). Second, characterization of the reaction products by XEG digestion shows that both XXT2 and XXT5 add the first d-Xyl at the same position on the cellohexaose acceptor (Fig. 4). XXT5 also was able to add the second d-Xyl when the enzyme was present at high concentration and a higher amount of donor substrate was provided (Fig. 5A). Third, in the conditions used in this study, XXT5 in combination with either XXT1 or XXT2 did not significantly increase the xylosylation of cellohexaose compared with the sum of the single-protein reactions (Fig. 2D). Finally, XXT5 showed reduced activity when xylosylated cellohexaose was used as an acceptor compared with nonxylosylated cellohexaose (Fig. 2E), and this is similar to what was shown before for XXT1 and XXT2 (Fauré et al., 2007). This suggests that XXT5 is not specific for the products of XXT1 or XXT2. However, it is possible that XXT5 might require the acceptor with the higher degree of polymerization or the one containing the XLGG subunit. Further work will be needed to investigate this possibility.
XXT5 can form strong heterocomplexes with XXT2 and CSLC4 (Chou et al., 2012, 2015; Lund et al., 2015). Protein-protein interaction between XXT5 and XXT2 or CSLC4 could produce a conformational change in XXT5 to increase its activity. Combination assays with XXT2+XXT5 did not increase xylosylation compared with the sums of the XXT2 and XXT5 single-enzyme reactions (Fig. 2D). This indicates that the protein-protein interaction with XXT2 does not increase the activity of XXT5. Further work is needed to investigate if the interaction between XXT5 and CSLC4 increases XXT5 activity.
Our mutagenesis studies showed that the XXT5 D228A:D230A mutant has no detectable activity in vitro, demonstrating the importance of the DxD motif for the catalytic activity of XXT5. The DxD motif functions to coordinate the divalent cation that forms ionic interactions with the negatively charged diphosphate moiety in UDP-Xyl and is critical for catalytic activity in numerous other glycosyltransferases (Busch et al., 1998; Wiggins and Munro, 1998; Li et al., 2001; Götting et al., 2004). To determine the functional role of XXT5 in vivo, we overexpressed the XXT5 D228A:D230A mutant in the xxt5 knockout mutant plant. Based on the XyG content and root hair phenotype (Fig. 5), we conclude that the XXT5 D228A:D230A mutant is not functional in vivo, indicating that XXT5 function requires the DxD putative donor substrate-binding motif. It is possible that XXT5 binds UDP-Xyl and functions to channel the substrate to other XXTs, such as XXT1 or XXT2. This also has been suggested for Irregular Xylem 14 (IRX14), which is involved in xylan biosynthesis, since the DxD motif is required for complementation of the irx14 mutant phenotype, while IRX14 lacks in vitro activity (Ren et al., 2014). Our demonstration of the importance of the DxD motif for the function of XXT5 in vivo does not exclude the possibility that XXT5 also has a similar channeling function.
CONCLUSION
We demonstrate that catalytically active truncated XXTs can be expressed in a prokaryotic system with high yield and do not require glycosylation for activity. With this optimized procedure, we show that XXT5 is catalytically active, yet with a significantly slower rate of catalysis than XXT1 or XXT2. XXT5 requires a DxD motif for both in vivo function and in vitro activity. In addition, XXT5 produces a xylosylation pattern similar to XXT2 and, most likely, does not require the XXT1 or XXT2 products for activity. To our knowledge, this is the first time that XXT5 has been shown to be enzymatically active in vitro.
MATERIALS AND METHODS
XXT Cloning
The pET20b expression vectors were modified as described previously (Boyken et al., 2012) to incorporate an N-terminal His-GB1, His-MBP, or His-GFP tag using NdeI and XhoI restriction sites. Overhangs of the forward primers were designed to contain the NdeI restriction site and an N-terminal 6xHis tag, and the reverse primers were designed to contain the BamHI and XhoI restriction sites. Coding sequences of the Arabidopsis (Arabidopsis thaliana) xylosyltransferase genes XXT1 (At3g62720), XXT2 (At4g02500), and XXT5 (At1g74380) were PCR amplified with N-terminal truncations of 135, 126, and 213 bp, respectively, and were cloned into the pET20b plasmid. XXT2 was cloned using BamHI and XhoI restriction sites, whereas the compatible restriction enzymes SalI and BglII were used to clone XXT1 and XXT5, respectively, due to the presence of an XhoI restriction site in XXT1 and a BamHI restriction site in XXT5 (Supplemental Table S1). All forward primers were designed to produce the tobacco etch virus protease site (ENLYFQG) at the N terminus of XXT. The resulting pET20b constructs were then transformed into Escherichia coli DH10b for plasmid amplification, and the sequence was verified by sequencing. Confirmed plasmids were then transformed into the various E. coli transgenic cells that were used for protein expression: BL21 (DE3) (Invitrogen), CodonPlus (Agilent Technologies), SoluBL21 (AMS Biotechnology), Rosetta 2 (DE3) (EMD Millipore), Arctic Express (DE3) CodonPlus (Agilent Technologies), and BL21 (DE3) pLysS (Agilent Technologies). To generate the XXT5 D228A:D230A mutant, two fragments for each mutant were PCR amplified using primers designed to contain the mutated base pairs (Supplemental Table S1). The XXT5 D228A:D230A mutant fragment 1 was generated with XXT5 forward and XXT5 D228A:D230A reverse primers, and fragment 2 was generated using XXT5 D228A:D230A forward and XXT5 reverse primers. The resulting fragments containing the mutated base pairs were then used as the template in a fusion PCR using XXT5 forward and XXT5 reverse primers to generate the final mutated gene fragment.
Protein Expression
E. coli cells harboring the plasmids were grown at 37°C with shaking at 200 rpm in 500 mL of Luria-Bertani broth. When the cell culture reached an optical density at 600 nm of 0.5, the temperature was lowered to 18°C, and protein expression was induced by adding isopropylthio-β-galactoside to a final concentration of 0.5 mm. Cells were incubated for 18 h at 18°C and then harvested by centrifugation. Pelleted cells were resuspended in 12.5 mL of 25 mm Tris-HCl (pH 7.4), 300 mm NaCl, and 0.1 mm EDTA and rapidly frozen in liquid nitrogen. Cells were lysed by thawing and incubating for 30 min with 1 mg mL−1 lysozyme and then sonicated for 15 s a total of five times. Solubilized proteins were collected by centrifugation at 20,000g for 30 min.
Protein Purification
Crude lysate was loaded onto an Ni-NTA column with a lysate:resin ratio of 20:1. Affinity resin was incubated on a shaker for 1 h at 4°C. Unbound proteins were removed as a flow-through fraction, and the resin was washed four times with washing buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 20 mm imidazole. The protein of interest was eluted in three fractions with a total volume of 6 mL using elution buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 300 mm imidazole. Buffer switch into 50 mm Tris-HCl (pH 7.4), and 150 mm NaCl was performed by concentrating the 6-mL elution fraction to 1 mL, then diluting it to 15 mL with buffer, and repeating this a total of four times. Glycerol was added to the purified protein to a final concentration of 20%, and proteins were stored at −80°C.
SDS-PAGE and Western Blots
Proteins were analyzed by SDS-PAGE in reducing conditions and stained with Coomassie Blue G250. Western-blot analysis was performed as described previously (Chou et al., 2015). Proteins were electrophoretically transferred to a nitrocellulose membrane and blocked with nonfat milk. The GB1 tag was probed with horseradish peroxidase-conjugated goat anti-rabbit antibody (Invitrogen) at a 10,000-fold dilution, developed using HyGLO quick spray, and visualized by ChemiDoc XRS+ (Bio-Rad). Prestained size markers were visualized on the same membrane using visible light. To estimate the concentration of XXT1 protein, XXT1 purity was estimated by determining the intensity of bands on Coomassie Blue-stained SDS-PAGE gels using ImageJ, and total protein concentration was measured by Bradford assay (Quick Start Bradford Dye reagent 1X; catalog no. 500-0205) according to the manufacturer’s instructions. Next, quantitative western blotting was performed using several volumes of protein loaded on the gel and quantifying the intensities of the XXT band with ImageJ (Supplemental Fig. S3). Only full-length bands were used for quantitating XXT concentration.
Enzyme Activity Assay and Product Precipitation
The enzymatic activity assay was performed in a total volume of 25 µL, consisting of 2 mm UDP-Xyl (CarboSource Services), 0.5 mm cellohexaose (Megazyme), and 2 mm MnCl2. The reaction mixture was incubated at 28°C for 20 h with shaking at 100 rpm. All reactions were performed in duplicate. The reaction was stopped by adding 1 mL of 100% ethanol and incubating at −22°C for 6 h; the precipitated reaction product was collected by centrifugation at 21,000g for 30 min. Combination assays were all performed as described above with equal protein concentration for each XXT.
XXT Kinetic Parameter Estimations
Kinetic experiments were performed with 3 μm XXT1 or XXT2 with reactions containing various UDP-Xyl concentrations, 1 mm cellohexaose, 2 mm MnCl2, 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, and reaction time of 30 min at 28°C. XXT5 reactions contained 11.5 μm XXT5, and the reaction time was 60 min. Kinetic parameters Km and kcat were calculated by nonlinear curve fitting with the software Dynafit (Kuzmic, 1996). The reaction times were selected based on minimum reaction time to allow for detection by HPAEC.
HPLC and MALDI-TOF MS Characterization of Catalysis Products
Products obtained from the enzymatic activity assays were analyzed by HPAEC with a pulsed-amperometric detector (Dionex), as described (Cavalier and Keegstra, 2006). Briefly, the aqueous solution of the product was injected onto a CarboPac PA-20 column and eluted using the following gradient: 30 to 100 mm sodium acetate for 25 min, with 100 mm NaOH remaining constant through the entire run; after the 25-min run, the column was reequilibrated for 15 min to the initial conditions. Nonxylosylated, monoxylosylated, dixylosylated, and trixylosylated cellohexaose eluted from the column with retention times of approximately 16.4, 17.8, 18.9, and 20.2 min, respectively. Quantitative estimation of Xyl transfer was performed by peak integration of xylosylated cellohexaose to determine the relative percentage of each product (nonxylosylated, monoxylosylated, dixylosylated, or trixylosated cellohexaose). For example, if 40% of total cellohexaose (12.5 nmol) is monoxylosylated, then 5 nmol of Xyl was transferred to cellohexaose. Response factors for nonxylosylated, monoxylosylated, dixylosylated, and trixylosylated glucan might vary when using pulsed-amperometric detection; nevertheless, this enabled estimation of the amount of Xyl transfer in the reaction. MALDI-TOF MS analysis of the products was performed as described earlier (Zabotina et al., 2012). Briefly, 1 μL of the product solution was spotted onto a MALDI-TOF sample plate using a 2,5-dihydroxybenzoic acid matrix at a sample:matrix ratio of 1:1. The sample spectra were analyzed on an Applied Biosystems VOYAGER-DE Pro MALDI-TOF MS instrument in positive reflection mode with an acceleration voltage of 20 kV and extraction delay time of 350 ns.
Characterization of Enzyme Assay Products
Enzyme assays and product precipitations were performed as described above, except that the product was resuspended in 25 mm sodium acetate buffer (pH 5). Three micrograms of XEG (Megazyme) was added to the reaction and incubated at 37°C for 12 h. The sample was heated for 10 min at 100°C and centrifuged at 21,000g for 10 min. The resulting supernatant was used for analysis by HPAEC and MALDI-TOF.
Production of Transgenic Plants
PCR-amplified coding sequences were ligated into the pCR8/GW/TOPO entry vector (Invitrogen) as described in the manufacturer’s instructions. The ligation product was transformed into E. coli DH10b and selected on Luria-Bertani agar plates with 50 µg mL−1 spectinomycin. The mutated gene in the entry construct was transferred into the binary destination vector pEarleyGate 201 (Basta resistant; The Arabidopsis Information Resource). The binary destination vector was transformed into Agrobacterium tumefaciens GV3101 by electroporation. Arabidopsis was transformed using the floral dip method (Clough and Bent, 1998). Plants were grown under long-day conditions (16 h of light/8 h of dark) at 22°C in a growth chamber.
Examination of Protein Expression in Transgenic Plants
Total membrane protein was extracted from 40 to 80 seedlings. The seedlings were ground in 10 mL of protein extraction buffer (40 mm HEPES, 0.45 m Suc, 1 mm EDTA, 1 mm MgCl2, 1 mm KCl, 1 mm dithiothreitol, and protease inhibitor cocktail [Roche], pH 8). The extract was homogenized using a Polytron homogenizer three times, 10 s each, at 10,000 rpm. The extract was filtered through three layers of Miracloth and centrifuged for 30 min at 10,000 rpm at 4°C. The supernatant was transferred to a polycarbonate tube with aluminum cap assembly and ultracentrifuged at 37,000 rpm (100,000g) for 45 min with a 70Ti fixed rotor. After centrifugation, the supernatant was removed, and the pellet was resuspended in 200 μL of suspension buffer (40 mm HEPES and 0.2 m Suc, pH 7.3). The membrane protein fraction was treated with 1% Triton X-100 for 30 min at 4°C to solubilize membrane-bound proteins. After solubilization, proteins were ultracentrifugated at 37,000 rpm for 45 min to precipitate nonsoluble membrane fragments. The protein concentration of the supernatant was measured using the Bradford protein assay. The supernatant was concentrated by precipitating with 10% TCA, and the precipitated protein was resuspended in loading buffer with β-mercaptoethanol. Proteins were separated using SDS-PAGE and transferred to a nitrocellulose membrane (0.2 μm; Bio-Rad) for immunodetection. Polyclonal anti-HA antibodies were used (1:500 dilution) to detect HA-fused proteins.
Root Hair Phenotype Analysis and Measurement of Xyloglucan
Analysis of plant root hairs was done by plating sterilized seeds on one-half-strength Murashige and Skoog medium with 0.3% GelRite. After the seeds germinated and the roots grew into the medium, the plates were placed at a 45° angle. Photographs of 10-d-old roots were taken using a Leica DMIRE2 light microscope with a Retiga 1300 camera.
Alcohol-insoluble residues from wild-type Columbia-0, xxt5, and xxt5 complemented either with HA-XXT5 or HA-XXT5(D228A:D230A) seedlings were prepared and digested with Driselase as described (Zabotina et al., 2012). Five milligrams of each alcohol-insoluble residue was homogenized in 0.5 mL of 20 mm sodium acetate buffer (pH 5), approximately 0.5 units of Driselase was added, and the mixtures were incubated at 37°C for 48 h. Supernatants were separated by centrifugation and analyzed by HPAEC.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers Q9LZJ3 (XXT1), O22775 (XXT2), and Q9CA75 (XXT5).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Predicted topology of XXTs according to the Universal Protein Resource (http://www.uniprot.org).
Supplemental Figure S2. Optimization of expression conditions for XXT2.
Supplemental Figure S3. Quantitative western blots of XXT1, XXT2, and XXT5.
Supplemental Figure S4. Negative controls of the XXT2 activity assay.
Supplemental Table S1. Forward and reverse primers used for the amplification of the XXTs and the truncations of XXT2.
Supplementary Material
Acknowledgments
We thank Dr. Amy Andreotti for the pET20b-GB1 and pET20b-MBP plasmids.
Glossary
- XyG
xyloglucan
- Ni-NTA
nickel-nitrilotriacetic acid agarose
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
- MS
mass spectrometry
- HPAEC
high-performance anion-exchange chromatography
- XEG
xyloglucan-specific endo-β-1,4-glucanase
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–1121163), the ICMG (grant no. FR 2607 to R.F. and S.C.), the PolyNat Carnot Institute (to R.F. and S.C.), and LabEx ARCANE (grant no. ANR–11–LABX–0003–01 to R.F. and S.C.)
Articles can be viewed without a subscription.
References
- Abramson M, Shoseyov O, Shani Z (2010) Plant cell wall reconstruction toward improved lignocellulosic production and processability. Plant Sci 178: 61–72 [Google Scholar]
- Atmodjo MA, Hao Z, Mohnen D (2013) Evolving views of pectin biosynthesis. Annu Rev Plant Biol 64: 747–779 [DOI] [PubMed] [Google Scholar]
- Bobovská A, Tvaroška I, Kóňa J (2014) A theoretical study on the catalytic mechanism of the retaining α-1,2-mannosyltransferase Kre2p/Mnt1p: the impact of different metal ions on catalysis. Org Biomol Chem 12: 4201–4210 [DOI] [PubMed] [Google Scholar]
- Boeggeman E, Qasba PK (2002) Studies on the metal binding sites in the catalytic domain of beta1,4-galactosyltransferase. Glycobiology 12: 395–407 [DOI] [PubMed] [Google Scholar]
- Boyken SE, Fulton DB, Andreotti AH (2012) Rescue of the aggregation prone Itk Pleckstrin Homology domain by two mutations derived from the related kinases, Btk and Tec. Protein Sci 21: 1288–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury SL, Jakoby WB (1972) Glycerol as an enzyme-stabilizing agent: effects on aldehyde dehydrogenase. Proc Natl Acad Sci USA 69: 2373–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch C, Hofmann F, Selzer J, Munro S, Jeckel D, Aktories K (1998) A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J Biol Chem 273: 19566–19572 [DOI] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B (1997) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J 326: 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC. (2012) Progress in the biological synthesis of the plant cell wall: new ideas for improving biomass for bioenergy. Curr Opin Biotechnol 23: 330–337 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Keegstra K (2006) Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J Biol Chem 281: 34197–34207 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M. (1995) Green fluorescent protein. Photochem Photobiol 62: 651–656 [DOI] [PubMed] [Google Scholar]
- Chou YH, Pogorelko G, Young ZT, Zabotina OA (2015) Protein-protein interactions among xyloglucan-synthesizing enzymes and formation of Golgi-localized multiprotein complexes. Plant Cell Physiol 56: 255–267 [DOI] [PubMed] [Google Scholar]
- Chou YH, Pogorelko G, Zabotina OA (2012) Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol 159: 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cotovio J, Roguet R, Pion FX, Rougier A, Leclaire J (1996) Effect of imidazole derivatives on cytochrome P-450 enzyme activities in a reconstructed human epidermis. Skin Pharmacol 9: 242–249 [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317 [DOI] [PubMed] [Google Scholar]
- Endler A, Persson S (2011) Cellulose synthases and synthesis in Arabidopsis. Mol Plant 4: 199–211 [DOI] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K (2002) An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA 99: 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauré R, Cavalier D, Keegstra K, Cottaz S, Driguez H (2007) Glycosynthase-assisted synthesis of xylo-gluco-oligosaccharide probes for α-xylosyltransferases. Eur J Org Chem 2007: 4313–4319 [Google Scholar]
- Francis DM, Page R (2010) Strategies to optimize protein expression in E. coli. Curr Protoc Protein Sci Chapter 5: Unit 5.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, McNeil M (1993) An unambiguous nomenclature for xyloglucan‐derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Götting C, Müller S, Schöttler M, Schön S, Prante C, Brinkmann T, Kuhn J, Kleesiek K (2004) Analysis of the DXD motifs in human xylosyltransferase I required for enzyme activity. J Biol Chem 279: 42566–42573 [DOI] [PubMed] [Google Scholar]
- Hayashi T. (1989) Xyloglucans in the primary cell wall. Annu Rev Plant Biol 40: 139–168 [Google Scholar]
- Hayashi T, Kaida R (2011) Functions of xyloglucan in plant cells. Mol Plant 4: 17–24 [DOI] [PubMed] [Google Scholar]
- Huth JR, Bewley CA, Jackson BM, Hinnebusch AG, Clore GM, Gronenborn AM (1997) Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci 6: 2359–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Johnson NR, Wilkerson CG (2014) Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J 80: 207–215 [DOI] [PubMed] [Google Scholar]
- Josè-Estanyol M, Puigdomènech P (2000) Plant cell wall glycoproteins and their genes. Plant Physiol Biochem 38: 97–108 [Google Scholar]
- Keegstra K. (2010) Plant cell walls. Plant Physiol 154: 483–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmic P. (1996) Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem 237: 260–273 [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77: 521–555 [DOI] [PubMed] [Google Scholar]
- Leandro P, Lechner MC, Tavares de Almeida I, Konecki D (2001) Glycerol increases the yield and activity of human phenylalanine hydroxylase mutant enzymes produced in a prokaryotic expression system. Mol Genet Metab 73: 173–178 [DOI] [PubMed] [Google Scholar]
- Li J, Rancour DM, Allende ML, Worth CA, Darling DS, Gilbert JB, Menon AK, Young WW Jr (2001) The DXD motif is required for GM2 synthase activity but is not critical for nucleotide binding. Glycobiology 11: 217–229 [DOI] [PubMed] [Google Scholar]
- Lund CH, Bromley JR, Stenbæk A, Rasmussen RE, Scheller HV, Sakuragi Y (2015) A reversible Renilla luciferase protein complementation assay for rapid identification of protein-protein interactions reveals the existence of an interaction network involved in xyloglucan biosynthesis in the plant Golgi apparatus. J Exp Bot 66: 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoori N, Schultink A, Schubert J, Pauly M (2015) Expression of heterologous xyloglucan xylosyltransferases in Arabidopsis to investigate their role in determining xyloglucan xylosylation substitution patterns. Planta 241: 1145–1158 [DOI] [PubMed] [Google Scholar]
- McCann MC, Carpita NC (2008) Designing the deconstruction of plant cell walls. Curr Opin Plant Biol 11: 314–320 [DOI] [PubMed] [Google Scholar]
- McGee DJ, Zabaleta J, Viator RJ, Testerman TL, Ochoa AC, Mendz GL (2004) Purification and characterization of Helicobacter pylori arginase, RocF: unique features among the arginase superfamily. Eur J Biochem 271: 1952–1962 [DOI] [PubMed] [Google Scholar]
- Mukerjea R, McIntyre AP, Robyt JF (2012) Potent inhibition of starch-synthase by Tris-type buffers is responsible for the perpetuation of the primer myth for starch biosynthesis. Carbohydr Res 355: 28–34 [DOI] [PubMed] [Google Scholar]
- Nallamsetty S, Waugh DS (2006) Solubility-enhancing proteins MBP and NusA play a passive role in the folding of their fusion partners. Protein Expr Purif 45: 175–182 [DOI] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2015) Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol 56: 180–194 [DOI] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinen S, Kofod LV, York WS, Albersheim P, Darvill A (1999) A xyloglucan-specific endo-beta-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Ren Y, Hansen SF, Ebert B, Lau J, Scheller HV (2014) Site-directed mutagenesis of IRX9, IRX9L and IRX14 proteins involved in xylan biosynthesis: glycosyltransferase activity is not required for IRX9 function in Arabidopsis. PLoS ONE 9: e105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Thompson DS. (2005) How do cell walls regulate plant growth? J Exp Bot 56: 2275–2285 [DOI] [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS (2014) Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J 80: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuttipongchaikij S, Brocklehurst D, Steele-King C, Ashford DA, Gomez LD, McQueen-Mason SJ (2012) Arabidopsis GT34 family contains five xyloglucan α-1,6-xylosyltransferases. New Phytol 195: 585–595 [DOI] [PubMed] [Google Scholar]
- Wiggins CA, Munro S (1998) Activity of the yeast MNN1 α-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc Natl Acad Sci USA 95: 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaoi K, Kondo H, Hiyoshi A, Noro N, Sugimoto H, Tsuda S, Miyazaki K (2009) The crystal structure of a xyloglucan-specific endo-beta-1,4-glucanase from Geotrichum sp. M128 xyloglucanase reveals a key amino acid residue for substrate specificity. FEBS J 276: 5094–5100 [DOI] [PubMed] [Google Scholar]
- Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou YH, Eberhard S, Danhof L, Keegstra K, Hahn MG (2012) Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol 159: 1367–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, van de Ven WT, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV (2008) Arabidopsis XXT5 gene encodes a putative alpha-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J 56: 101–115 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang X, Wu C, Lu D, Guo G, Mao X, Zhang Y, Wang DC, Li D, Zou Q (2011) Expression, purification and characterization of arginase from Helicobacter pylori in its apo form. PLoS ONE 6: e26205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang PG, Brew K (2001) Specificity and mechanism of metal ion activation in UDP-galactose:beta-galactoside-alpha-1,3-galactosyltransferase. J Biol Chem 276: 11567–11574 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiang Y, Van Etten JL, Rossmann MG (2007) Structure and function of a chlorella virus-encoded glycosyltransferase. Structure 15: 1031–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.