Phenylalanine 165 is important for PIN1 endocytosis and its trafficking through the secretory pathway.
Abstract
In contrast with the wealth of recent reports about the function of μ-adaptins and clathrin adaptor protein (AP) complexes, there is very little information about the motifs that determine the sorting of membrane proteins within clathrin-coated vesicles in plants. Here, we investigated putative sorting signals in the large cytosolic loop of the Arabidopsis (Arabidopsis thaliana) PIN-FORMED1 (PIN1) auxin transporter, which are involved in binding μ-adaptins and thus in PIN1 trafficking and localization. We found that Phe-165 and Tyr-280, Tyr-328, and Tyr-394 are involved in the binding of different μ-adaptins in vitro. However, only Phe-165, which binds μA(μ2)- and μD(μ3)-adaptin, was found to be essential for PIN1 trafficking and localization in vivo. The PIN1:GFP-F165A mutant showed reduced endocytosis but also localized to intracellular structures containing several layers of membranes and endoplasmic reticulum (ER) markers, suggesting that they correspond to ER or ER-derived membranes. While PIN1:GFP localized normally in a μA (μ2)-adaptin mutant, it accumulated in big intracellular structures containing LysoTracker in a μD (μ3)-adaptin mutant, consistent with previous results obtained with mutants of other subunits of the AP-3 complex. Our data suggest that Phe-165, through the binding of μA (μ2)- and μD (μ3)-adaptin, is important for PIN1 endocytosis and for PIN1 trafficking along the secretory pathway, respectively.
PIN-FORMED (PIN) family proteins are auxin efflux carriers that are localized asymmetrically in plant cells, and their polarity determines the directionality of intercellular auxin flow in plants (Petrásek et al., 2006; Wisniewska et al., 2006; Adamowski and Friml, 2015). Eight different PIN proteins have been described in Arabidopsis (Arabidopsis thaliana). All of them have a similar structure, with a central cytoplasmic hydrophilic loop separating two hydrophobic domains with five predicted transmembrane regions each (Křeček et al., 2009). While the hydrophobic domains of these proteins are extremely conserved, the central cytoplasmic domain varies, dividing PIN proteins into two subfamilies: type 1 or long PINs, which include PIN1, PIN2, PIN3, PIN4, and PIN7, all characterized by their long hydrophilic domain and its localization at the plasma membrane; and type 2 or short PINs, which encompass PIN5, PIN6, and PIN8, localized at the endoplasmic reticulum (ER) and with a shorter (PIN6) or strongly reduced (PIN5 and PIN8) hydrophilic loop (Supplemental Fig. S1; Křeček et al., 2009; Mravec et al., 2009; Ding et al., 2012). The predominant localization of long PINs to the plasma membrane suggests that their hydrophilic loop may provide molecular cues for their trafficking along with cell type-specific factors (Wisniewska et al., 2006; Ganguly et al., 2014).
We have focused in this work on PIN1, a type 1 PIN localized to the plasma membrane. PIN1 cycles between the plasma membrane and endosomal compartments. PIN1 endocytosis has been shown to be clathrin dependent (Dhonukshe et al., 2007; Robert et al., 2010), while its recycling from endosomes to the plasma membrane depends on the ADP ribosylation factor-guanine nucleotide exchange factor (ARF-GEF) GNOM and is inhibited by the ARF-GEF inhibitor brefeldin A (BFA; Geldner et al., 2001, 2003). However, there is no information on the sorting signals allowing the sorting of PIN1 within clathrin-coated vesicles, which could contribute to its polar localization.
Sorting within clathrin-coated vesicles is mediated by adaptor protein (AP) complexes, which recognize cytosolic sorting signals in membrane proteins. AP complexes are composed of four adaptin subunits: two large subunits of around 100 kD, one α-type subunit (γ, α, δ, or ε) that mediates membrane binding and one β-subunit that interacts with clathrin; one medium (μ) subunit, of around 50 kD, that recognizes sorting signals in cargo proteins; and a small σ-subunit of unknown function (Boehm and Bonifacino, 2001). Several clathrin adaptors have been shown to mediate basolateral polarity in epithelial cells and neurons, including AP-1A, AP-1B, and AP-4 (Gravotta et al., 2012; Bonifacino, 2014; Guo et al., 2014). The Arabidopsis genome encodes subunits of four types of putative AP complexes (AP-1 to AP-4), including five medium subunits, named μA (μ2), μB1 (μ1-1), μB2 (μ1-2), μC (μ4), and μD (μ3). There might be an additional AP complex, because Arabidopsis appears to have orthologs of several subunits of the recently identified mammalian AP-5 complex; however, no matching σ-subunit has been identified (Boehm and Bonifacino, 2001; Park et al., 2013). Several AP complexes already have been functionally characterized in Arabidopsis. μA-adaptin was initially proposed to be the putative medium subunit of the AP-1 complex, as inferred from its localization at the trans-Golgi in Arabidopsis and from its in vitro interaction with the Tyr-based sorting sequence of VSR-PS1 or TGN38 (Happel et al., 2004). However, there is no functional in vivo evidence supporting this notion. More recently, it has been shown that μB1 and μB2 adaptins are functionally redundant subunits of the Arabidopsis AP-1 complex acting at the trans-Golgi network (TGN) to promote late secretory and vacuolar traffic (Park et al., 2013). In particular, a μB2 adaptin mutant showed inhibition of multiple trafficking pathways from the TGN to the vacuole and to the plasma membrane in interphase and to the plane of cell division in cytokinesis (Park et al., 2013).
It has been described that the AP-1 complex is required for cell plate-targeted trafficking of the cytokinesis-specific syntaxin KNOLLE in dividing plant cells (Park et al., 2013; Teh et al., 2013; Wang et al., 2013). The AP-3 complex also has been studied extensively. Consisting of δ/β3/μD/σ3, AP-3 seems to be involved in vacuolar biogenesis, including the transition between storage and lytic vacuolar types (Feraru et al., 2010; Zwiewka et al., 2011), and may play a role in intracellular pH homeostasis (Niñoles et al., 2013). In addition, it has been shown that the vesicle transport regulator EpsinR2 (see below) interacts with the putative δ-subunit of AP-3 and binds to clathrin and phosphatidylinositol 3-phosphate (Lee et al., 2007). EpsinR2 also interacts with the SNARE VTI12 that has a role in protein trafficking to the protein storage vacuole (Lee et al., 2007; Sanmartín et al., 2007). Furthermore, TERMINAL FLOWER1, a protein that plays a role in trafficking to the protein storage vacuole, colocalized with the AP-3 δ-subunit (Sohn et al., 2007). Interestingly, mutants defective in β3-adaptin (protein affected trafficking2 [pat2]) or δ3-adaptin (pat4), as well as dominant negative AP-3μ transgenic lines, showed strong intracellular accumulation of membrane proteins (including PIN1) in aberrant vacuolar structures (Feraru et al., 2010; Zwiewka et al., 2011). On the other hand, elimination of μ3-adaptin or other subunits of AP-3 suppresses the knockout mutant phenotype of the soluble N-ethylmaleimide-sensitive factor attachment receptor Qb-SNARE VTI11, which is involved in trafficking from the TGN to the prevacuolar compartment (PVC; Niihama et al., 2009). Concerning AP-2, the ANTH domain-bearing monomeric adaptor AP180 (see below) has been shown to interact with Arabidopsis αC-adaptin, one of the putative large subunits of AP-2 (Barth and Holstein, 2004). In the case of μA (μ2)-adaptin, a component of the AP-2 complex (Yamaoka et al., 2013), there are conflicting results concerning its localization. While, in some cases, μ2-adaptin was found to localize exclusively at the plasma membrane and the cytosol (Di Rubbo et al., 2013; Kim et al., 2013), other reports found it to localize also to intracellular compartments (Golgi/TGN; Happel et al., 2004; Bashline et al., 2013). On the other hand, it has been demonstrated that AP-2 is involved in the endocytosis of cellulose synthase (Bashline et al., 2013) or brassinosteroid receptor1 (Di Rubbo et al., 2013) as well as in the polar localization of PIN1 (Fan et al., 2013) and PIN2 (Kim et al., 2013). Recently, it was shown that TPLATE is part of a unique multisubunit protein complex (the TPLATE complex) that acts in concert with the AP-2 complex for clathrin-mediated endocytosis in plants (Gadeyne et al., 2014).
In contrast with the wealth of recent reports about the function of μ-adaptins and clathrin AP complexes, there is very little information about the motifs that determine the sorting of membrane proteins within clathrin-coated vesicles in plants. One of the best characterized sorting signals, which is recognized by the medium (μ) subunit of adaptor complexes, is the YXXΦ motif (where Y is Tyr, X is any amino acid, and Φ is a bulky hydrophobic residue). Other signals for sorting within clathrin-coated vesicles include a di-Leu motif or acidic clusters (Traub, 2009; Canagarajah et al., 2013) as well as other less characterized signals, like a Phe-based motif in the Man-6-P receptor (Denzer et al., 1997; Höning et al., 1997). LeEix2, the tomato (Solanum lycopersicum) receptor-like protein for the fungal elicitor ethylene-inducing xylanase, is a cell surface glycoprotein possessing a Tyr-based endocytosis signal: a point mutation (Tyr-993 to Ala) within its endocytosis signal abolished the ability of LeEix2 to induce a hypersensitive response (Ron and Avni, 2004). The products of the race-specific Ve1 and Ve2 disease resistance genes from tomato contain a typical acidic or Tyr-based sorting motif (Kawchuk et al., 2001). Tyr and di-Leu motifs in KORRIGAN, an endo-1,4-β-glucanase required for cell wall modification, determine its polar localization to the cell plate (Zuo et al., 2000). The polar localization and vacuolar targeting of boron transporters also have been shown to depend on several YXXΦ motifs (Takano et al., 2010). However, there are not many reports showing a direct interaction between sorting signals in membrane proteins and the μ-subunit of adaptor complexes. In this respect, a member of the Leu-rich repeat subfamily of receptor protein kinase contains a YXXΦ motif within its cytoplasmic tail that binds to the receptor-binding domain of the Arabidopsis μA-adaptin (Holstein, 2002). Arabidopsis μA-adaptin has been shown to interact with the Tyr motif of the vacuolar sorting receptor VSR-PS1 (Happel et al., 2004). A model receptor for receptor-mediated endocytosis in animal cells, the human transferrin receptor, also has been shown to interact with a μ-adaptin subunit from Arabidopsis cytosol, a process that was blocked by tyrphostin A23, which inhibits the interaction between the YTRF endocytosis signal in the human transferrin receptor cytosolic tail and the μ2-subunit of the AP-2 complex (Ortiz-Zapater et al., 2006). More recently, a Tyr-based motif (YMPL) in VSR4 was found to interact with μ1-adaptin (Nishimura et al., 2016).
In this study, we searched for putative sorting signals in PIN1 that may be involved in the binding of μ-adaptins and thus in its trafficking and polar localization. We found that Phe-165 and Tyr-260, Tyr-328, and Tyr-394 have the ability to interact with several μ-adaptins. However, only Phe-165, which has the ability to interact with μA- and μD-adaptins in vitro, appeared to be essential for PIN1 trafficking and localization in vivo, since the PIN1:GFP-F165 mutant showed reduced endocytosis but also accumulated in intracellular structures containing double or multiple membranes and ER markers.
RESULTS
The PIN1 Cytosolic Loop Binds Arabidopsis μ-Adaptins and Clathrin
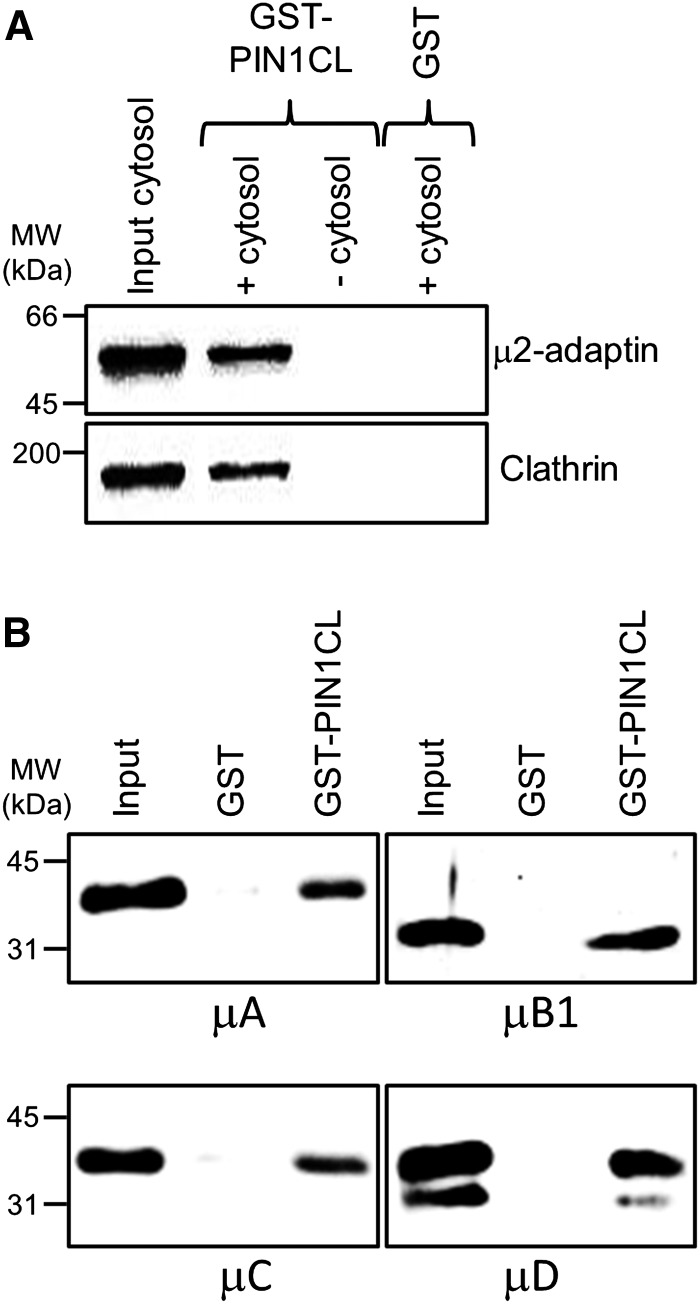
Putative sorting signals in PIN1 are likely to be located within its large cytosolic loop. This cytosolic loop contains several Tyr residues, some of them possibly matching the consensus YXXΦ motif, which has been shown to be recognized by the μ-subunit of several adaptor complexes (Traub, 2009; Canagarajah et al., 2013). This is the case of Tyr-260 (YSMM), Tyr-328 (YSTA), and Tyr-394 (YPAP; Supplemental Fig. S1). Interestingly, the first portion of this hydrophilic loop also contains a motif remarkably similar to one of the endocytosis signals present in the cytosolic tail of the Man-6-P receptor (MEQFP; Supplemental Fig. S1). In this motif it is essential the presence of the Phe residue, and it was shown to interact specifically with AP-2 (the clathrin adaptor specifically involved in endocytosis) but not with AP-1 (Denzer et al., 1997; Höning et al., 1997). This motif is present in all long PINs, which are typically targeted polarly to the plasma membrane. PIN3, PIN4, and PIN7 contain exactly the same MEQFP motif, while PIN1 and PIN2 contain a very similar one (SEQFP; Supplemental Fig. S1); PIN6, which accumulates in internal membranes consistent with the ER (Bender et al., 2013), has the most divergent version (only FP residues conserved). In contrast, this motif is not present in PIN5 and PIN8, which localize to the ER (Mravec et al., 2009; Ding et al., 2012; Ganguly et al., 2014; Supplemental Fig. S1). To test for the ability of the cytosolic loop of PIN1 to interact with plant μ-adaptins, we used an in vitro binding assay, with an Arabidopsis cytosolic extract as a source of plant adaptins and clathrin and a glutathione S-transferase (GST) fusion protein containing the complete cytosolic loop of PIN1 (GST-PIN1CL). As shown in Figure 1, GST-PIN1CL bound specifically cytosolic μ2-adaptin and clathrin. Therefore, the cytosolic loop of PIN1 contains sorting signals to interact with the clathrin machinery.
Figure 1.
The PIN1 cytosolic loop binds μ-adaptins and clathrin. A, Binding of μ2-adaptin and clathrin from Arabidopsis cytosol to the cytosolic loop of PIN1 fused to GST (GST-PIN1CL; Supplemental Fig. S3) detected by western-blot analysis with antibodies against mouse μ2-adaptin or rat clathrin H chain, respectively. As a control, we used GST alone or omitted the cytosol. Input was 2% of the cytosol used in the pull-down assay. B, Binding of the receptor binding domain (RBD) of Arabidopsis μ-adaptins A to D with an N-terminal His tag (Supplemental Fig. S2) to the cytosolic loop of PIN1 (GST-PIN1CL) detected by western blotting with His antibodies. As a control, His-tagged μ-adaptins were incubated with GST alone. Input was 5% of the amount of the μ-adaptin used in the pull-down assay. MW, Molecular mass.
Our next goal was to identify which of the μ-adaptin(s) was able to interact with the cytosolic loop of PIN1 and the residue(s) within this hydrophilic loop involved in the interaction with μ-adaptins. To this end, we expressed in bacteria the receptor-binding domain (RBD) of Arabidopsis μ-adaptins with a C-terminal His tag for purification (His-RBD-μA-D; Supplemental Fig. S2), and GST-PIN1CL was incubated in vitro with each of the purified His-RBD-μ-adaptins. Binding of μ-adaptins to the cytosolic loop of PIN1 was analyzed by western-blot analysis using His antibodies. Figure 1B shows that the cytosolic loop of PIN1 binds specifically to the receptor-binding domain of all μ-adaptins.
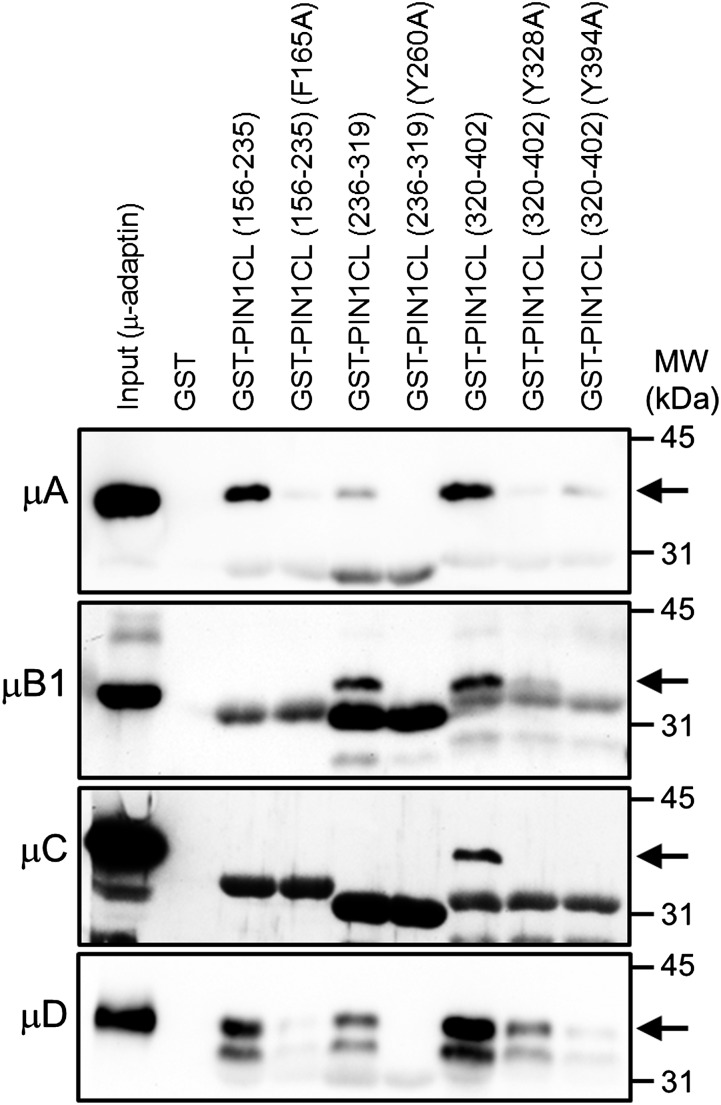
To test whether Phe-165, Tyr-260, Tyr-328, or Tyr-394 was involved in the interaction with μ-adaptins, we prepared GST fusion proteins with different portions of the cytosolic loop: PIN1-CL(156-235), including Phe-165; PIN1CL(236-319), including Tyr-260; and PIN1CL(320-402), including Tyr-328 and Tyr-394 (Supplemental Fig. S3). GST fusion proteins were incubated with purified His-RBD-μ-adaptins, and binding was analyzed. As shown in Figure 2, PIN1CL(156-235) bound specifically μA- and μD-adaptin but not μB1- or μC-adaptin. μ-adaptin binding required Phe-165, since a mutant version where Phe-165 was changed to Ala (F165A) showed strongly reduced μ-adaptin binding. PIN1CL(236-319) bound specifically μA-, μB1-, and μD-adaptin but not μC-adaptin, and μ-adaptin binding required Tyr-260, since almost no binding was detected with a mutant version where Tyr-260 was changed to Ala (Y260A). Finally, PIN1CL(320-402) bound specifically all μ-adaptins, and μ-adaptin binding required Tyr-328 or Tyr-394, since reduced binding was detected with mutant versions where Tyr-328 or Tyr-394 was changed to Ala (Y328A and Y394A). Therefore, Phe-165, Tyr-260, Tyr-328, and Tyr-394 were candidate residues to be involved in PIN1 trafficking based in their ability to bind μ-adaptins.
Figure 2.
Binding of the RBD of Arabidopsis μ-adaptins to different regions of the cytosolic loop of PIN1 and mutant versions. Pull-down experiments used different regions of the cytosolic loop of PIN1 fused with GST (GST-PIN1CL) or mutant versions (Supplemental Fig. S3) and the RBD of Arabidopsis μ-adaptins, with an N-terminal (His)6-tag used for purification (Supplemental Fig. S2). As a control, we used GST. Pull-down results were analyzed by western blotting with His antibodies. The input lane contains 5% of the amount of the μ-adaptin used in the pull-down assay. Arrows point at the expected molecular masses (MW) of the RBD of μ-adaptins A to D. Lower molecular weight bands correspond to the GST-PIN1CL fragments used for the pull-down assays, which cross-reacted with the His antibodies.
Phe-165 Is Essential for PIN1 Localization
We next investigated whether Phe-165 or Tyr-260, Tyr-328, and Tyr-394, which are involved in the binding of μ-adaptins in vitro, were important for PIN1 localization in vivo. To this end, we first generated transgenic lines expressing mutant versions of PIN1-GFP where Phe-165 or Tyr-260, Tyr-328, and Tyr-394 were changed to Ala. Then, these mutant versions also were expressed in a pin1 background (Okada et al., 1991). To this end, PIN1:GFP (Benková et al., 2003), PIN1:GFP-F165A, PIN1:GFP-Y260A, PIN1:GFP-Y328A, and PIN1:GFP-Y394A plants from three independent transgenic lines (lines 1–3) were crossed with a pin1 transfer DNA (T-DNA) insertion mutant (Furutani et al., 2004), and double homozygous plants were selected for analysis. The PIN1:GFP-Y260A and PIN1:GFP-Y328A mutants were able to complement pin1 defects, as does wild-type PIN1:GFP. However, the PIN1:GFP-Y394A mutant could only complement pin1 defects if highly expressed, as was the case for lines 1 and 3 (Supplemental Fig. S4). This suggests that Tyr-394, which is not present in other PINs, may somehow be important for the function of the protein. The most striking phenotypic defects were observed in the PIN1:GFP-F165A mutant (Fig. 3). Two out of three lines of this mutant (lines 1 and 3) showed a partial rescue of the pin1 phenotype, reflected by fewer cotyledon anomalies, and at the flowering stage they produced occasionally defective flowers that gave rise to aberrant siliques with one to 10 seeds inside in addition to pin-like inflorescences (Fig. 3). The third line (line 2) did not recover the pin1 phenotype at all, and plants produced only needle-shaped inflorescences. These phenotypic defects were not related to low expression levels of the mutant, since the three transgenic lines showed expression levels comparable to those of the PIN1:GFP control line (Supplemental Fig. S4).
Figure 3.
Phenotypic analysis of PIN1:GFP-F165A plants in the pin1 background. The table at top shows the phenotypic analysis of 350 F2 plants from the cross of pin1 and three independent homozygous lines of PIN1:GFP-F165A (F165A1–F165A3). The images at bottom show examples of the partial pin1 phenotype found in those lines. Flowers of PIN1:GFP in the pin1 background (PIN1-GFP) were like wild-type Columbia-0 (Col-0) flowers. In contrast, flowers of PIN1:GFP-F165A in the pin1 background from lines 1 and 3 (F165A) showed abnormal shapes and numbers of flower-related structures (for details, see text). The arrow points to a pin-like inflorescence.
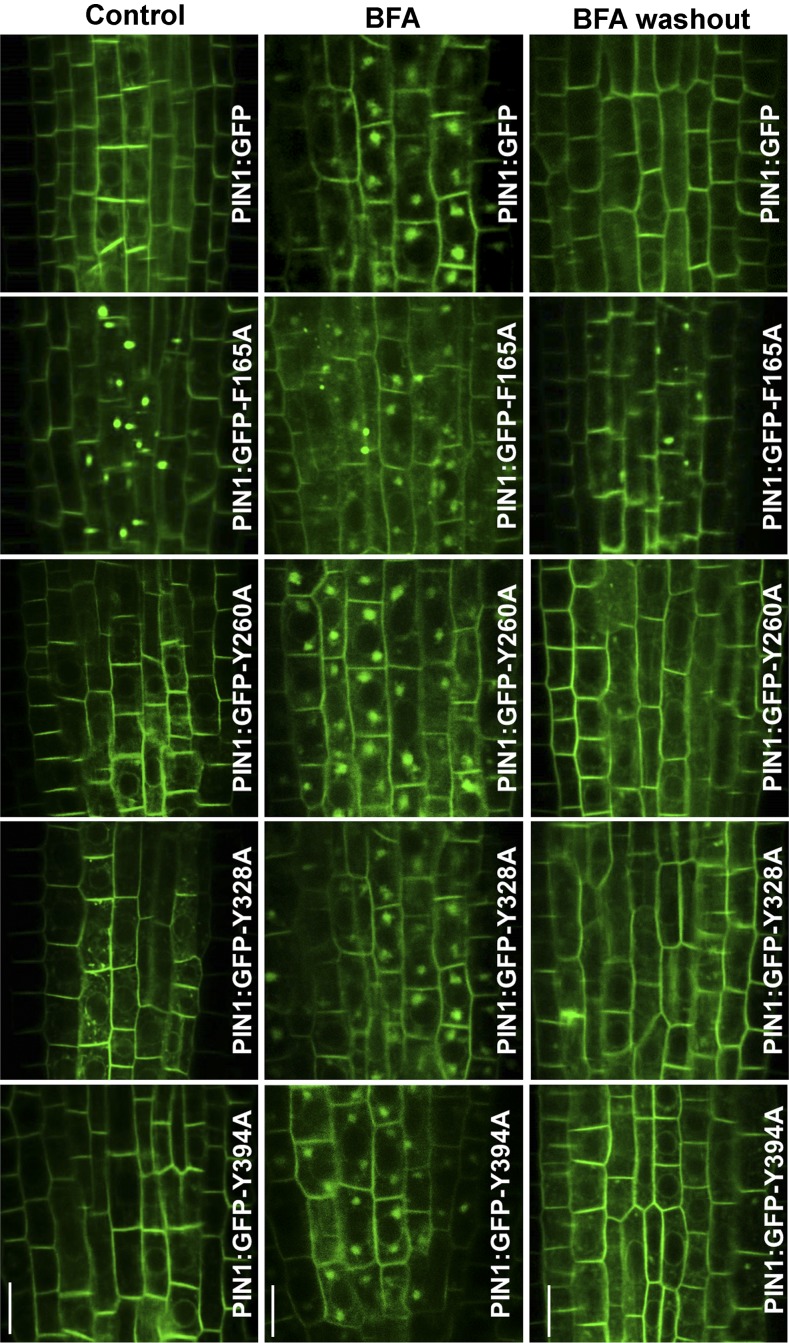
Next, we investigated whether these mutant defects correlated with changes in the subcellular localization of the PIN1-GFP mutants. To this end, roots of 4-d-old seedlings from the different lines of PIN1:GFP and mutant versions, in the pin1 background, were grown in vitro and analyzed by confocal laser scanning microscopy (CLSM). As shown in Figure 4, the PIN1:GFP-Y260A, PIN1:GFP-Y328A, and PIN1:GFP-Y394A mutants localized mainly to the plasma membrane of stele cells, like wild-type PIN1:GFP. This suggests that Tyr-260, Tyr-328, and Tyr-394 are not essential for steady-state PIN1 localization. In clear contrast, the PIN1:GFP-F165A mutant localized only partially to the plasma membrane of stele cells and was present mainly in big intracellular punctae (Figs. 4 and 6A). Interestingly, these structures often were localized in close proximity to the plasma membrane (Figs. 4 and 6A). Therefore, Phe-165 is required for the correct subcellular localization of PIN1. To rule out that the appearance of the mutant in these structures was due to the presence of the GFP tag, we generated a hemagglutinin (HA)-tagged version of the PIN1-F165A mutant. As shown in Supplemental Figure S5, PIN1:HA-F165A also accumulated in big intracellular structures, very similar to those in which the PIN1:GFP-F165A mutant was found, in contrast to PIN1-HA, which was found only at the plasma membrane.
Figure 4.
Localization and trafficking of PIN1:GFP and different mutant versions of PIN1:GFP at stele cells of Arabidopsis roots. CLSM was performed on primary roots of 4-d-old seedlings expressing PIN1:GFP and different mutant versions of PIN1:GFP in the pin1 background. The left row shows PIN1:GFP-Y260A, PIN1:GFP-Y328A, and PIN1:GFP-Y394A mutants localized to the plasma membrane, like wild-type PIN1:GFP. In contrast, PIN1:GFP-F165A (line 1) showed intracellular protein accumulations that do not appear in roots expressing wild-type PIN1:GFP. This intracellular location also was found for the other PIN1:GFP-F165A lines, either in the pin1 or wild-type background. In the middle row, roots were incubated with 50 μm BFA for 60 min. All lines were able to form BFA compartments, including the PIN1:GFP-F165A mutant. In the right row, BFA was washed out after BFA treatment and roots were analyzed after 90 min. In all lines, most BFA compartments disappeared and fluorescence was visible only at the plasma membrane, except in the PIN1:GFP-F165A mutant, which still contained intracellular punctae similar to those found in the absence of BFA (for details, see text). Bars = 10 μm.
Figure 6.
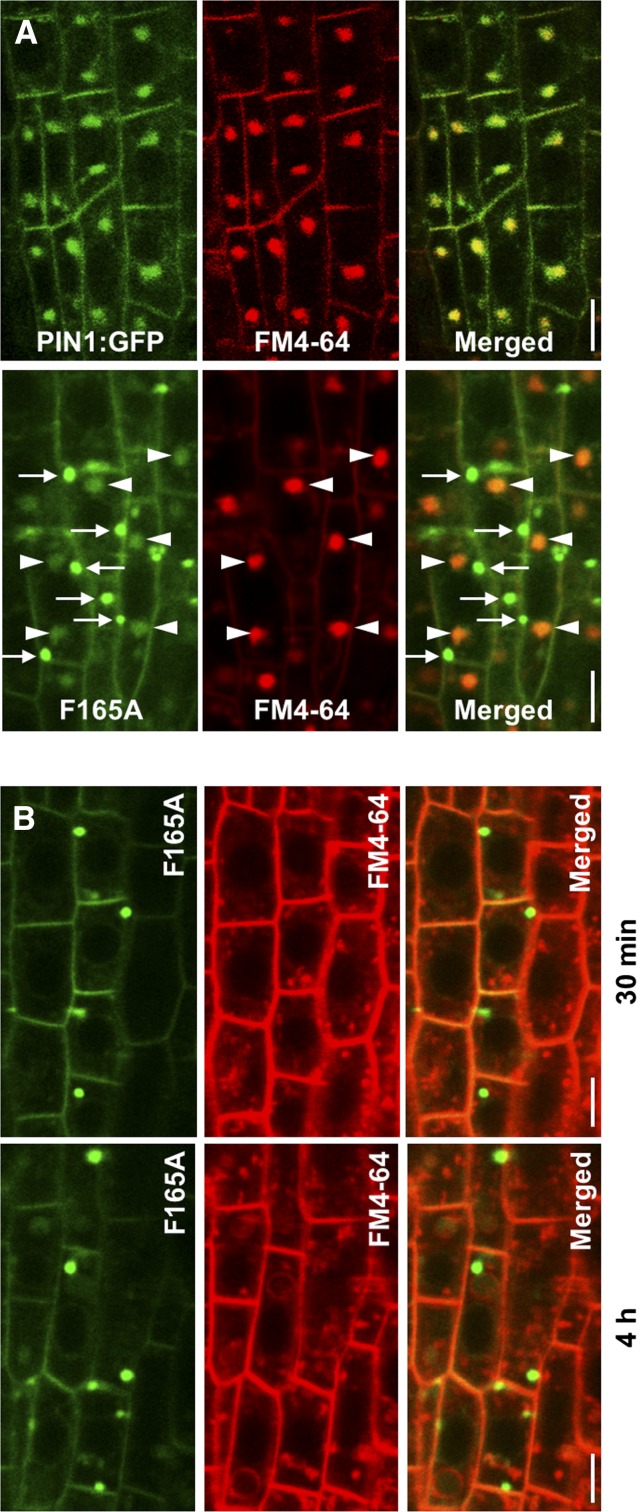
PIN1:GFP-F165A does not localize to BFA compartments or FM4-64-labeled compartments. CLSM was performed on primary roots of 4-d-old seedlings expressing PIN1:GFP and PIN1:GFP-F165A. A, Roots were incubated with 4 μm FM4-64 for 10 min and then in the presence of 50 μm BFA during 60 min. Arrowheads point to putative BFA compartments, labeled by both FM4-64 and PIN1:GFP-F165A, while arrows point to structures containing PIN1:GFP-F165A but not FM4-64. B, PIN1:GFP-F165A mutant roots were incubated with 10 μm FM4-64 for 10 min and analyzed after the indicated periods of time (for details, see text). Bars = 5 μm.
The PIN1:GFP-F165A Mutant Shows Reduced Endocytosis
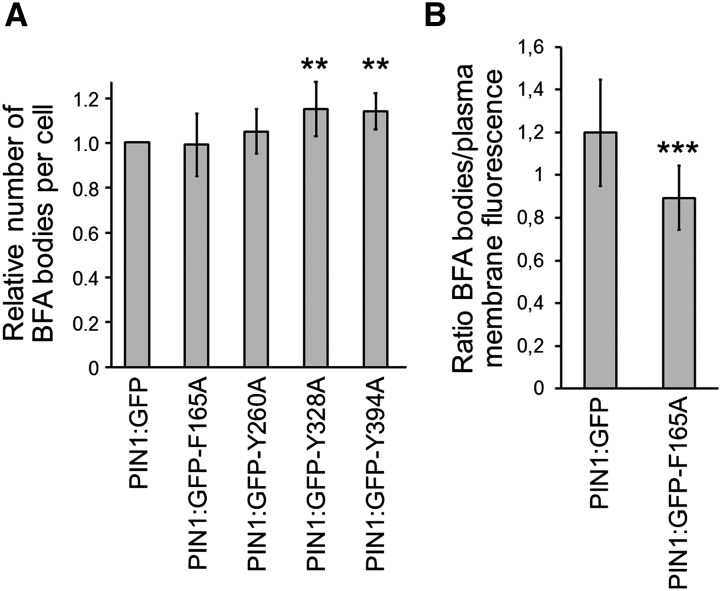
We next investigated whether the mutations in the putative sorting signals affect the dynamics of endocytosis and/or recycling of PIN1:GFP. Transgenic seedlings expressing these mutant versions were treated with BFA to monitor the appearance of the mutant protein in BFA compartments. As shown in Figure 4, PIN1:GFP-Y260A, PIN1:GFP-Y328A, and PIN1:GFP-Y394A mutants were able to form well-defined BFA compartments after 1 h of BFA treatment, very similar to what was found for wild-type PIN1:GFP. Indeed, the relative number of BFA bodies per cell in the PIN1:GFP-Y260A mutant was not statistically different from that of PIN1:GFP, while PIN1:GFP-Y328A and PIN1:GFP-Y394A mutants showed slight increases (Fig. 5A). Therefore, these mutants do not seem to have any defect in endocytosis. We next monitored the rate of recycling. To this end, we did a BFA washout after BFA treatment and monitored the disappearance of BFA compartments and the reappearance of PIN1:GFP-Y260A, PIN1:GFP-Y328A, and PIN1:GFP-Y394A mutants at the plasma membrane. As shown in Figure 4, these three mutants relocated from BFA compartments to the plasma membrane after 90 min, very similar to what was found for wild-type PIN1:GFP. Therefore, PIN1:GFP-Y260A, PIN1:GFP-Y328A, and PIN1:GFP-Y394A mutants do not seem to have any obvious defect in recycling. Altogether, these data suggest that Tyr-260, Tyr-328, and Tyr-394 are not essential for PIN1 endocytosis and recycling. This is consistent with the phenotypic analysis showing that these mutant proteins were able to rescue the pin1 phenotype.
Figure 5.
A, Relative number of BFA bodies per cell. Roots from PIN1:GFP and the different mutants were incubated with 50 μm BFA for 60 min, and the number of BFA bodies per cell was analyzed. To clearly identify BFA bodies in the PIN1:GFP-F165A mutant, they were labeled with FM4-64 before BFA treatment, and only intracellular structures that colocalized with FM4-64 were recorded. B, Ratio of BFA bodies to plasma membrane fluorescence. The fluorescence of BFA bodies (labeled with FM4-64) and the plasma membrane was recorded and expressed as a ratio. n = 3 independent experiments, and at least 100 cells were counted for each assay. Statistical analysis (Mann-Whitney, two-tailed, nonparametric test) was performed using PRISM software (version 5.0a; GraphPad Software). Error bars represent sd. **, P < 0.01; and ***, P < 0.001.
As described above, the PIN1:GFP-F165A mutant was present mainly as big intracellular punctae, with a morphology apparently similar to that of BFA compartments. Therefore, we decided to investigate the dynamics of endocytosis and recycling of this mutant using BFA treatment and washout, as described above. As shown in Figure 4, BFA treatment of the PIN1:GFP-F165A mutant produced internal structures with the typical pattern of BFA compartments that were apparently different from the internal structures already present in this mutant before BFA treatment. In order to distinguish whether these structures were (or were not) the same, BFA treatment was performed after FM4-64 labeling of the plasma membrane. Under these conditions, BFA compartments become labeled by internalized FM4-64. As shown in Figure 6A, the PIN1:GFP-F165A mutant was able to reach BFA compartments, where it colocalized extensively with FM4-64. In addition, PIN1:GFP-F165A also was found in internal structures that were not labeled with FM4-64, probably corresponding to the structures already observed in the absence of BFA. This suggests that the appearance of the PIN1:GFP-F165A mutant protein in BFA compartments derives from the fraction that localizes to the plasma membrane at steady state. A quantification of the number of BFA bodies per cell in this mutant showed that it was nearly identical to that found for wild-type PIN1:GFP (Fig. 5A). However, a quantification of the ratio between the GFP fluorescence associated with BFA compartments (labeled by FM4-64) and plasma membrane fluorescence showed that the PIN1:GFP-F165A mutant has a significantly lower capacity to enter BFA compartments compared with wild-type PIN1:GFP (Fig. 5B). This suggests that Phe-165, which binds μA (μ2)-adaptin, may be important for PIN1 endocytosis, although other residues also may play a role, since a very significant proportion of the mutant still localizes to BFA compartments upon BFA treatment. On the other hand, the fraction of the mutant that was included in BFA compartments following BFA treatment relocated to the plasma membrane after BFA washout with a normal kinetics, suggesting that the F165A mutation did not affect recycling (Fig. 4C). Therefore, the PIN1:GFP-F165A mutant protein is competent for endocytic cycling between the plasma membrane and endosomal compartments, although it shows reduced endocytosis. Since most of the mutant protein localized to an unidentified intracellular compartment (in contrast to other mutants), we decided to focus on further characterization of the PIN1:GFP-F165A mutant.
PIN1:GFP-F165A Localizes to a Compartment Containing ER Markers
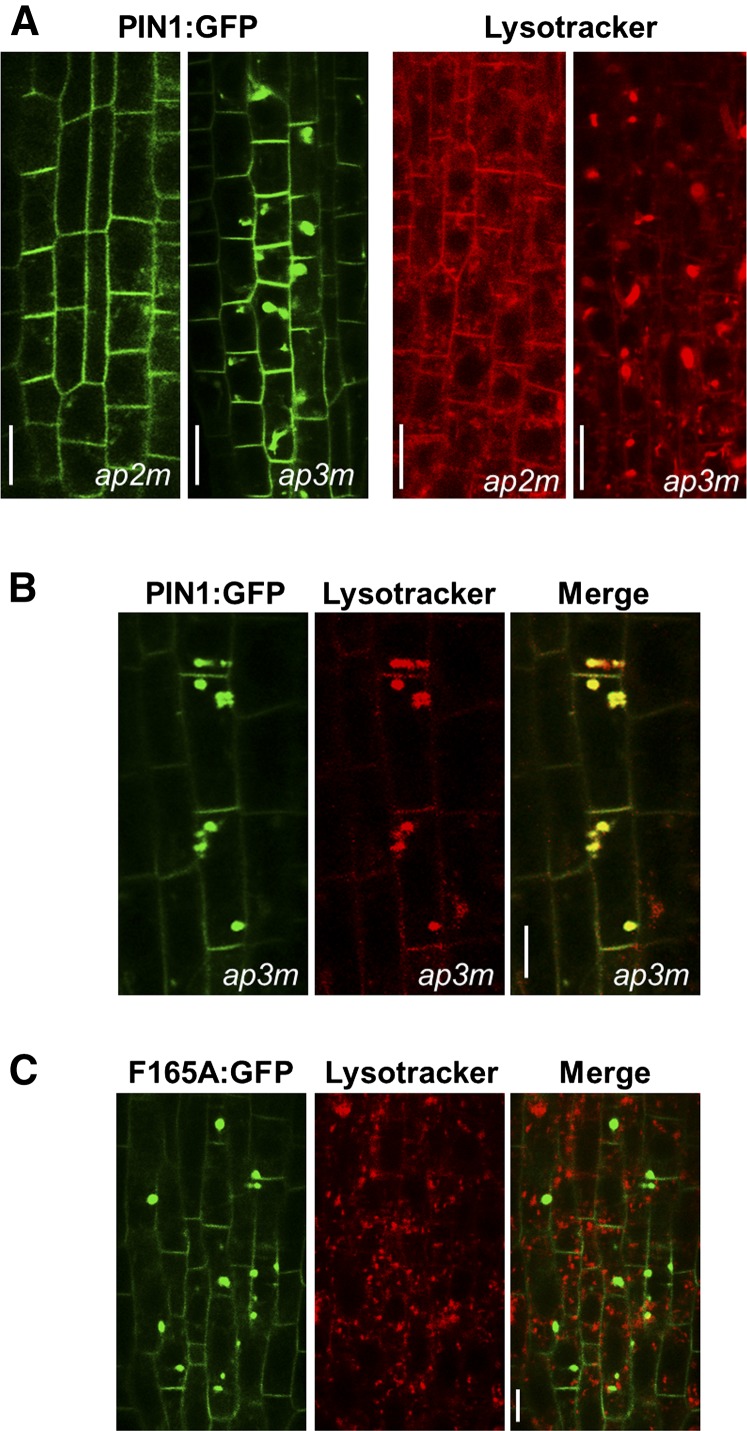
To test whether PIN1:GFP-F165A localized to endosomal compartments, mutant seedlings were incubated for different time periods with FM4-64 in the absence of BFA. As shown in Figure 6B, the compartments containing PIN1:GFP-F165A were not stained significantly with FM4-64, either after 30 min or 4 h of internalization. Consistent with these results, these structures did not colocalize with LysoTracker, a marker of acidic compartments (see below). These observations strongly suggest that the PIN1:GFP-F165A mutant protein does not localize to compartments along the endocytic pathway.
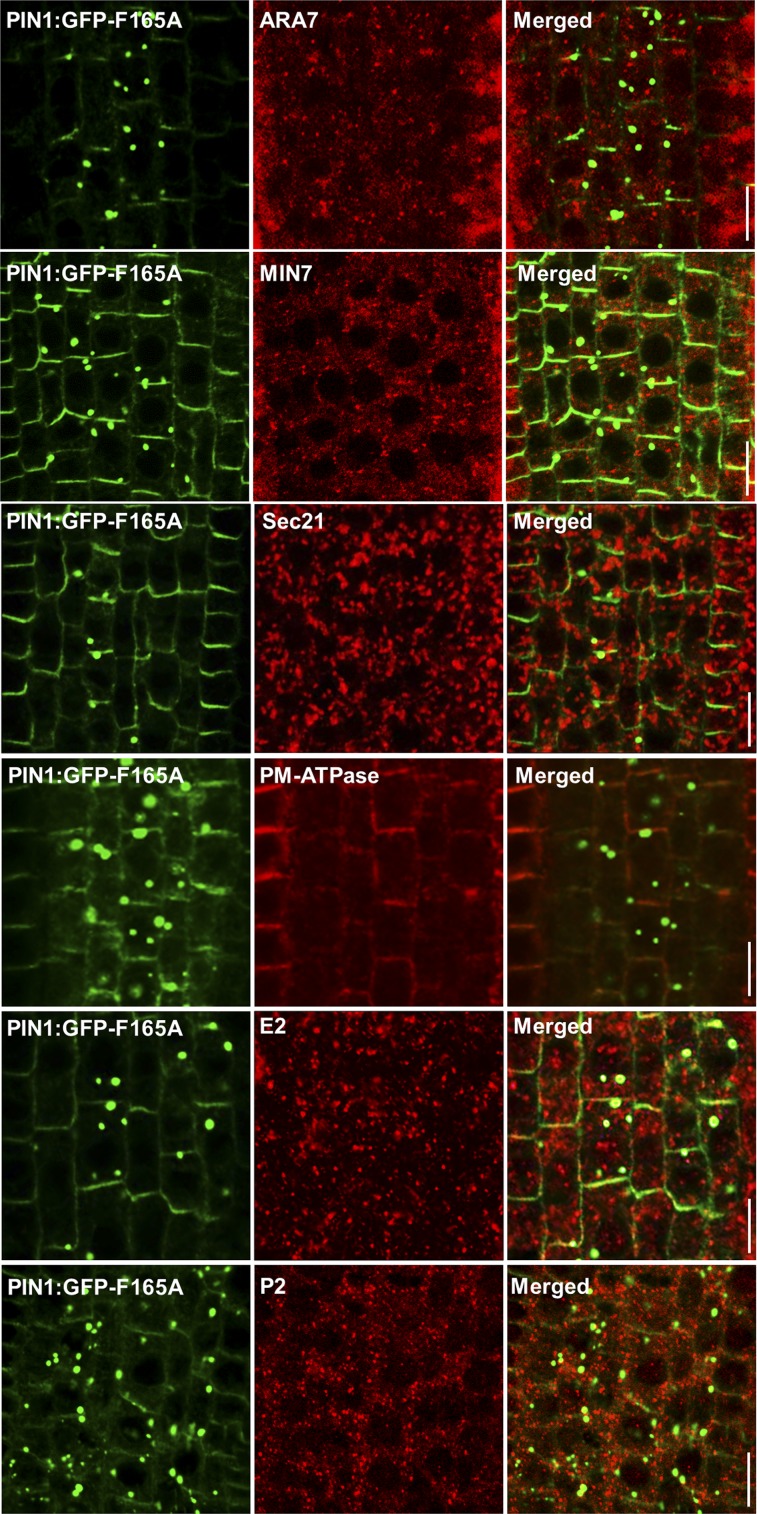
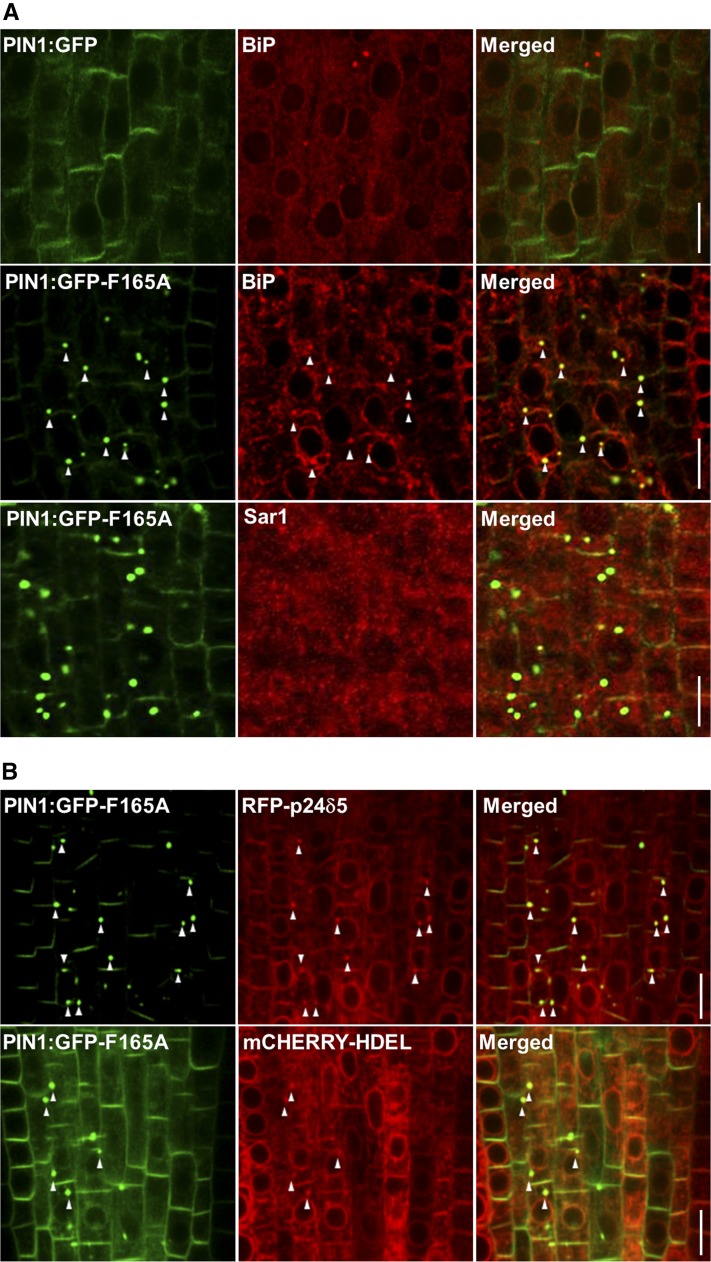
To further investigate the identity of the compartments where PIN1:GFP-F165A localizes, we performed immunolocalization with antibodies against different organelle marker proteins (Sauer et al., 2006; see “Materials and Methods”). These experiments showed that PIN1:GFP-F165A did not colocalize with markers of the Golgi complex (Sec21), TGN/endosomes (MIN7), or the PVC (ARA7), which showed a rather normal localization (Fig. 7). We also analyzed the localization of a plasma membrane marker different from PIN1, the plasma membrane ATPase, which localized normally in the PIN1:GFP-F165A mutant and did not accumulate in intracellular punctae (Fig. 7). We also analyzed whether the structures containing the PIN1:GFP-F165A mutant were part of the unconventional secretion route, which is mediated by a compartment labeled by components of the exocyst complex, the exocyst-positive (EXPO) compartment (Wang et al., 2010). Therefore, we tested whether the compartments that contain the PIN1:GFP-F165A mutant could correspond to EXPO compartments using an antibody against one of the components of the exocyst complex (E2; Wang et al., 2010). As shown in Figure 7, no significant colocalization was observed between both proteins, indicating that PIN1:GFP-F165A does not localize to EXPO compartments. Additionally, we also tested whether these structures could correspond to autophagosomes. To this end, we used an antibody against a component of these structures, the SH3P2 protein (Zhuang and Jiang, 2014), but again no obvious colocalization was found (Fig. 7). Finally, we tested for colocalization with ER markers. As shown in Figure 8A, most PIN1:GFP-F165A mutants colocalized extensively with BiP, which, in addition to their typical ER pattern, also accumulated in the same big punctae, which are not observed in roots expressing wild-type PIN1:GFP (Fig. 8A). These data suggest that the PIN1:GFP-F165A mutant may accumulate at the ER or ER-derived structures. To rule out that these structures could correspond to endoplasmic reticulum export sites (ERES), we also performed a double immunolocalization using Sar-1, an ERES marker (daSilva et al., 2004). As shown in Figure 8A, no significant colocalization was found between PIN1:GFP-F165A and Sar-1, suggesting that this mutant does not accumulate significantly at ERES. In summary, the colocalization studies with markers for different subcellular structures revealed an extensive presence of the PIN1:GFP-F165A mutant protein at compartments characterized by the presence of ER markers, without any discernible effect on other subcellular compartments.
Figure 7.
Immunolocalization of PIN1:GFP-F165A with different organelle markers. Immunolocalization of PIN1:GFP-F165A in primary roots of 4-d-old seedlings was performed as described in “Materials and Methods” with the following organelle markers: ARA7 (PVC), MIN7 (endosomes), Sec21 (Golgi), plasma membrane (PM) ATPase, E2 (EXPO compartments), and P2 (SH3P2). Bars = 10 μm.
Figure 8.
PIN1:GFP-F165A colocalizes with ER markers. A, Coimmunolocalization of PIN1:GFP or PIN1:GFP-F165A with ER markers. Intracellular punctae containing PIN1:GFP-F165A colocalized with BiP but not with Sar-1. In addition, the localization pattern of BiP changed in the PIN1:GFP-F165A mutant but not in PIN1:GFP roots (for details, see text). B, CLSM of primary roots of 4-d-old seedlings expressing PIN1:GFP-F165A and two different ER markers, RFP-p24δ5 or mCherry-HDEL. In addition to their typical ER pattern, RFP-p24δ5 and mCherry-HDEL also localized to intracellular punctae, where they colocalized with PIN1:GFP-F165A. Bars = 10 μm.
PIN1:GFP-F165A Colocalizes with ER Markers in Vivo
To confirm whether these compartments correspond to ER membranes, Arabidopsis PIN1:GFP-F165A plants were transformed with constructs of two different ER markers: a soluble ER marker, mCherry-HDEL (Nelson et al., 2007), which contains the HDEL signal that results in the retention of soluble proteins within the ER in plant cells (Denecke et al., 1992); and red fluorescent protein (RFP)-p24δ5, a membrane protein that has been shown previously to localize to the ER in Arabidopsis as a consequence of highly efficient COPI-dependent Golgi-to-ER transport (Langhans et al., 2008; Montesinos et al., 2012, 2013). T1 seedlings were then observed with the confocal microscope. Both markers showed its typical ER pattern in PIN1:GFP plants, without any obvious accumulation in intracellular punctae (Supplemental Fig. S6). As shown in Figure 8B, RFP-p24δ5 also showed its typical ER pattern in most cells of PIN1:GFP-F165A plants. However, in cells containing the PIN1:GFP-F165A punctae, RFP-p24δ5 also accumulated within the same punctae, as observed with the ER marker BiP. mCherry-HDEL also colocalized partially with these structures. The fact that two ER marker proteins, other than the chaperone BiP, also accumulate at these structures may indicate that the colocalization of the PIN1:GFP-F165A mutant with BiP is not simply a consequence of an improper folding of this mutant. Alternatively, the inability of the mutant to exit the ER may cause an accumulation of the protein at ER membrane structures where other ER membrane proteins (like RFP-p24δ5) also may be trapped. These observations confirm that PIN1-GFP:F165A localizes at the ER or ER-derived structures.
Trafficking of Newly Synthesized PIN1:GFP-F165A
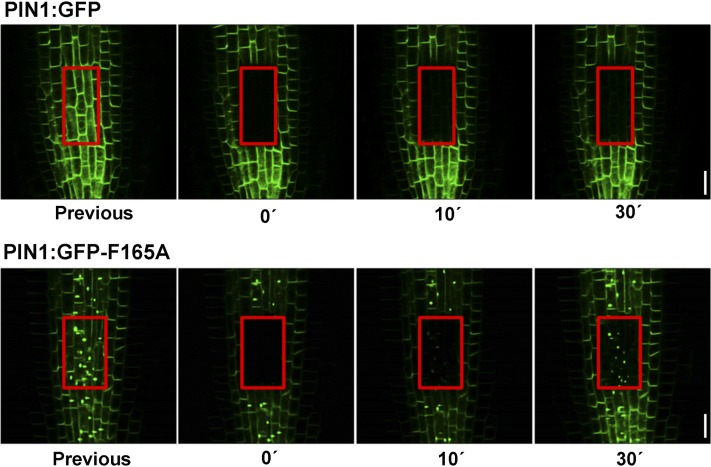
To investigate the kinetics of the appearance of PIN1:GFP-F165A at intracellular punctae, we performed experiments using fluorescence recovery after photobleaching (FRAP). To this end, an area of the stele cells containing these intracellular punctae was photobleached, and fluorescence recovery was analyzed by CLSM. Under these conditions of complete photobleaching, the recovered signal should come mainly from the newly synthesized protein. These experiments showed that PIN1:GFP-F165A accumulated in these structures as early as 10 min after photobleaching (Fig. 9). Actually, these are the first structures that became labeled, which would be consistent with a putative ER localization. As a control, we performed a parallel experiment with wild-type PIN1:GFP, which did not show any accumulation at intracellular punctae during photobleaching; instead, it was seen only at the plasma membrane after 0.5 to 1 h of recovery.
Figure 9.
Trafficking of newly synthesized PIN1:GFP-F165A. FRAP analysis is shown for PIN1:GFP (top) and PIN1:GFP-F165A (bottom) roots. Roots expressing PIN1:GFP or PIN1:GFP-F165A were imaged before and during recovery after bleaching an area containing stele cells. Images were taken at the indicated times after the bleach pulse (red squares). Bars = 10 μm.
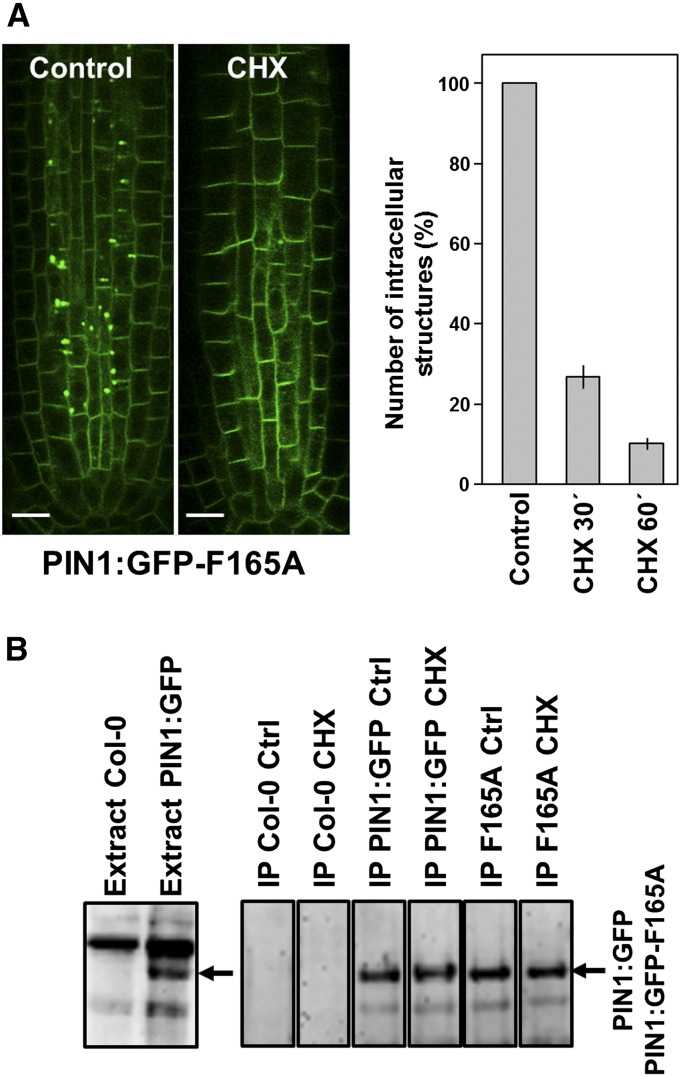
To confirm that the intracellularly accumulated PIN1:GFP-F165A protein corresponds to the de novo-synthesized protein in the secretory pathway, we inhibited protein synthesis by treating the seedlings with 100 μm cycloheximide (CHX). Figure 10A shows that CHX treatment caused a very significant decrease in the number of intracellular punctae, roughly 70% after 30 min and 90% after 1 h of CHX addition. To investigate if the loss of internal fluorescence could be a consequence of protein degradation or due to its relocalization to the plasma membrane, we analyzed the levels of PIN1:GFP-F165A before and after CHX treatment in total protein extracts of Arabidopsis roots by western-blot analysis using GFP antibodies. Since the GFP antibodies cross-reacted with other proteins in total protein extracts, in particular with a protein with a molecular mass similar to that of PIN1:GFP, immunoprecipitation prior to western-blot analysis also was performed to facilitate the comparison between the levels of PIN1:GFP-F165A with or without CHX. As shown in Figure 10B, the levels of PIN1:GFP-F165A were not significantly different from those of wild-type PIN1:GFP, either in the absence of CHX or after a 2-h CHX treatment. These data suggest that the PIN1:GFP-F165A mutant is not degraded during CHX treatment. Therefore, the loss of internal fluorescence following CHX treatment suggests that the PIN1:GFP-F165A mutant accumulates intracellularly during the secretion of de novo-synthesized protein to the plasma membrane.
Figure 10.
CHX treatment of PIN1:GFP and PIN1:GFP-F165A seedlings. A, The left images show CLSM analysis of primary roots of 4-d-old seedlings expressing PIN1:GFP-F165A treated with 100 μm CHX or dimethyl sulfoxide (Control) for 1 h. Bars = 10 μm. The graph at right shows a quantification of the number of intracellular structures in roots from the PIN1:GFP-F165A mutant after CHX treatment for 30 and 60 min expressed as a percentage of the number of intracellular structures in the absence of CHX (Control). n = 3 independent experiments, and at least 100 different seedlings were analyzed per condition. Error bars represent sd. B, Western-blot analysis of the amount of PIN1:GFP or PIN1:GFP-F165A with or without CHX treatment. The left gel shows the western-blot analysis of protein extracts from 5-d-old Col-0 and PIN1:GFP seedlings with GFP antibodies. A total of 30 μg of protein was loaded in each lane. The arrow points to the position of PIN1:GFP. The right gels show the western-blot analysis of immunoprecipitation (IP) experiments performed as described in “Materials and Methods” using mouse anti-GFP antibodies. Ctrl and CHX correspond to seedlings treated for 2 h with dimethyl sulfoxide and CHX, respectively. The arrow points to the position of PIN1:GFP or PIN1:GFP-F165A.
PIN1:GFP-F165A Localizes to Multilayered Membrane Structures
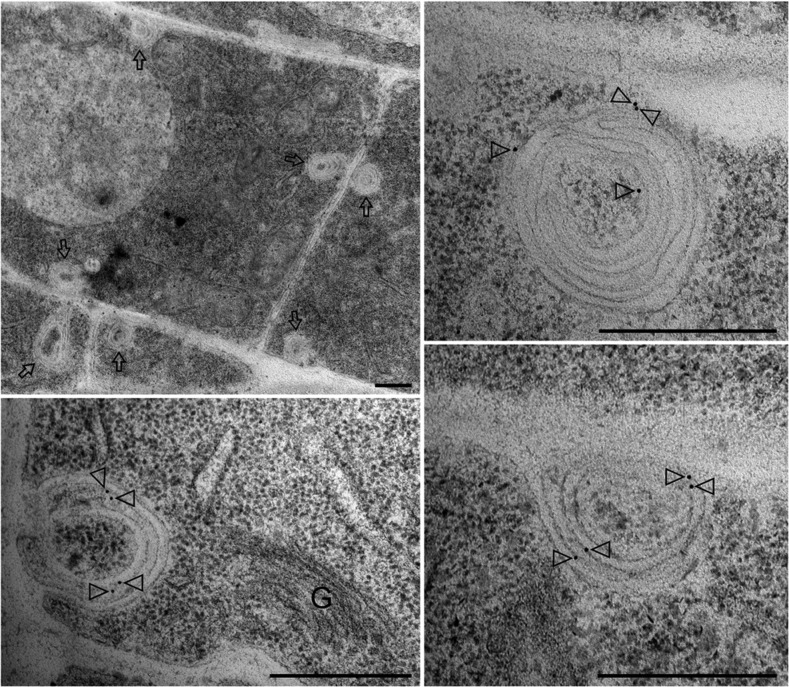
We next investigated the nature of the structures where PIN1:GFP-F165A accumulates by immunogold labeling on ultrathin sections of cryofixed samples using GFP antibodies. As shown in Figure 11 and Supplemental Figure S7A, PIN1:GFP-F165A localized to vesicular structures containing sometimes several layers of membranes, folded over each other, which were abundant in the PIN1:GFP-F165A transgenic lines but rarely seen in the wild type. The size of the structures observed by electron microscopy (200–500 nm) was consistent with that of the structures observed by CLSM (Supplemental Fig. S7). These structures sometimes appeared to be ordered into concentric or spiral-like structures and often were found close to the plasma membrane, in some cases fusing with it (Fig. 11; Supplemental Fig. S7A), which would explain why the mutant protein disappears from intracellular punctae during CHX treatment to be delivered to the plasma membrane. Table I shows that the gold labeling was specific for these multilayered structures. A few gold particles also could be seen at the plasma membrane, in particular in regions in close proximity to the place where these structures were found to fuse with the plasma membrane (Fig. 11; Supplemental Fig. S7). However, ultrastructural analysis of the PIN1:GFP-F165A mutant showed no obvious alterations in other endomembrane compartments, including the ER, Golgi apparatus, multivesicular bodies, or mitochondria (Supplemental Fig. S8).
Figure 11.
Localization of PIN1:GFP-F165A by immunogold labeling in roots of seedlings expressing PIN1:GFP-F165A. Labeling with GFP antibodies showed that the PIN1:GFP-F165A mutant accumulates in big structures, from 200 to 500 nm diameter, in stele cells. Lower magnification (top left) shows that these structures (labeled by arrows) are often localized near the plasma membrane. A higher magnification shows that these structures contain multiple membranes and can be seen fusing with the plasma membrane. Arrowheads point to gold particles. G, Golgi. Bars = 500 nm.
Table I. Distribution of immunogold particles with GFP antibodies in transgenic PIN1:GFP-F165A Arabidopsis root cells.
Significant differences between the two compartments/organelles (multilayered structures and Golgi) were analyzed using a two-tailed paired Student’s t test (**, P < 0.001).
| Compartment/Organelle | No. of Organelles | No. of Immunogold Particles | Immunogold Particles per Organelle |
|---|---|---|---|
| Multilayered structures | 30 | 80 | 2.66** |
| Golgi | 30 | 5 | 0.16** |
| Multivesicular bodies | 30 | 6 | 0.20 |
| Mitochondrion | 30 | 5 | 0.16 |
PIN1 Localization in gnl1 and μ2- and μ3-Adaptin Mutants
To further investigate the mechanisms involved in the trafficking of PIN1 to the plasma membrane, we first analyzed the trafficking of newly synthesized PIN1-RFP in a gnl1 mutant expressing BFA-sensitive GNL1 (Richter et al., 2007). To this end, we used estradiol-inducible PIN1-RFP (Richter et al., 2014) and followed the fate of newly synthesized PIN1-RFP 6 h after estradiol induction in the absence or presence of 10 μm BFA. As shown in Figure 12, BFA treatment of these seedlings led to the intracellular accumulation of estradiol-induced PIN1-RFP at structures with a morphology consistent with that of the ER. This would fit with the transport of PIN1 to the plasma membrane taking place via the Golgi apparatus and with the requirement of GNL1 for the transport of PIN1 along the early secretory pathway, as suggested recently (Doyle et al., 2015).
Figure 12.
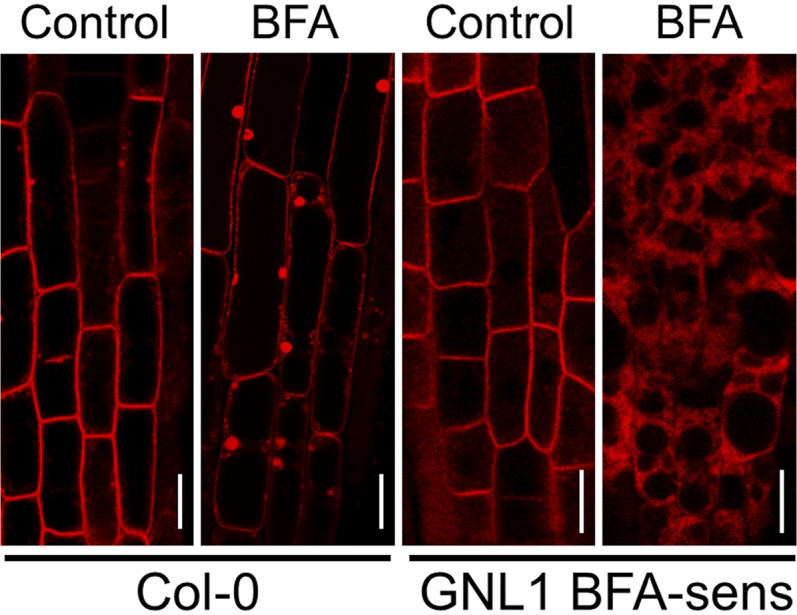
Trafficking of PIN1 in the early secretory pathway requires GNL1 function. Col-0 and gnl1 mutant plants expressing BFA-sensitive GNL1 (GNL1 BFA-sens) were transformed with estradiol-inducible PIN1-RFP. T2 4-d-old-seedlings from these plants were treated with 20 μm estradiol in the absence (Control) or presence (BFA) of 10 μm BFA for 6 h and analyzed by CLSM. Bars = 10 μm.
We next focused on the putative role of μ-adaptins in PIN1 trafficking. Since Phe-165 interacts specifically with μA (μ2)- and μD (μ3)-adaptins (but not with μB- or μC-adaptin), we hypothesized that the function of μ2- and μ3-adaptins may be required for the proper trafficking and localization of PIN1. Therefore, we decided to analyze the localization of wild-type PIN1-GFP in the roots of μA- and μD-adaptin mutant seedlings. In the case of μA (μ2)-adaptin, we used the knockout mutant ap2m-1, which was characterized previously (Bashline et al., 2013; Kim et al., 2013). As shown in Figure 13A, the localization of PIN1:GFP in the ap2m-1 mutant was very similar to that in the wild-type background, without any obvious accumulation in big intracellular structures similar to those found in the PIN1:GFP-F165A mutant. To analyze the role of μ3-adaptin in the trafficking and localization of PIN1-GFP, we used the knockout mutant ap3m-3, which was characterized previously (Niihama et al., 2009). As shown in Figure 13A, PIN1-GFP showed strong intracellular accumulation in root stele cells of this mutant. Based on previous results with mutants of the AP-3 β- and δ-subunits (pat2 and pat4 mutants), which showed a strong intracellular accumulation of PIN1-GFP in aberrant vacuolar structures (Feraru et al., 2010; Zwiewka et al., 2011), we performed a 1-h treatment with LysoTracker, which labels acidic compartments, including vacuoles. As shown in Figure 13A, labeling of the ap3m-3 mutant with LysoTracker revealed an aberrant morphology of acidic compartments with big intracellular structures very similar to those found in pat2 and pat4 mutants. In contrast, labeling of the ap2m-1 mutant with LysoTracker did not show any obvious alteration (Fig. 13A). The PIN1-GFP intracellular structures found in the ap3m-3 mutant showed extensive colocalization with LysoTracker (Fig. 13B), which suggests that, indeed, PIN1-GFP also may accumulate in aberrant vacuolar structures in this mutant. Very similar structures have been found previously upon the expression of a dominant negative version of μ3-adaptin (Zwiewka et al., 2011). These results confirm that the AP-3 complex is required for the correct trafficking and localization of PIN1. In contrast, the PIN1:GFP-F165A mutant did not colocalize with LysoTracker (Fig. 13C).
Figure 13.
Localization of PIN1-GFP in μA (μ2)- and μD (μ3)-adaptin mutants. A, CLSM of primary roots of 4-d-old seedlings of the ap2m-1 and ap3m-3 mutants. Left images show the localization of PIN1:GFP expressed in both mutants, while right images show the labeling of these mutants with 50 μm LysoTracker for 1 h. B, Colocalization of PIN1-GFP and LysoTracker in intracellular aggregates in the ap3m-3 mutant. C, Labeling of the PIN1:GFP-F165A mutant with LysoTracker. Bars = 10 μm (A) and 5 μm (B and C).
DISCUSSION
In this study, we analyzed sorting signals that may be involved in PIN1 trafficking and localization. Since PIN1 endocytosis, which is essential for PIN1 polar localization, has been shown to be clathrin dependent (Dhonukshe et al., 2007), we focused on those signals that could match the consensus for binding AP complexes. Adaptor proteins (in particular AP-1 and AP-4) also have been shown to be involved in sorting proteins to the basolateral plasma membrane in epithelial cells, and the signals involved in basolateral sorting very often overlap with those involved in endocytosis, including Tyr-based or di-Leu motifs but also other noncanonical motifs (Canagarajah et al., 2013; Bonifacino, 2014). Sorting of membrane proteins within clathrin-coated vesicles is thought to be mediated by the μ-adaptin subunit of AP complexes (Boehm and Bonifacino, 2001). The best characterized motif that is recognized by μ-adaptins is the YXXΦ motif (Canagarajah et al., 2013). Previously, the role of a different type of Tyr-based motif (NPXY; Traub, 2009) in PIN1 trafficking was investigated. A triple PIN1:GFP mutant (NPNTY to NSLSL) involving Tyr-480 was found to accumulate at the ER, suggesting the involvement of this motif in PIN1 localization (Mravec et al., 2009). Therefore, in this study, we focused on YXXΦ motifs. Three Tyr-based motifs (involving Tyr-260, Tyr-328, and Tyr-394) were analyzed. It was found that they had the ability to bind different μ-adaptins in vitro. However, when single mutants in these residues were expressed in Arabidopsis, there was no obvious change in their steady-state localization. It cannot be ruled out that these Tyr residues are functionally redundant in μ-adaptin binding, which would explain why steady-state localization is not affected in single Tyr mutant versions. In contrast, a less well-characterized motif based on a Phe residue (Phe-165), which was found to bind μ2- and μ3-adaptin in vitro, also was shown to be essential for PIN1 trafficking and localization in vivo. Consistent with the role of a similar motif as one of the endocytosis signals of the Man-6-P receptor, we found that Phe-165 may be important for PIN1 endocytosis. Strikingly, this mutant accumulated in multilayered intracellular structures containing ER markers. Therefore, we focused our attention on the trafficking of PIN1 along the early secretory pathway. In this respect, it was shown recently that ENDOSIDIN8 interferes with the localization of PIN1 at the basal plasma membrane without affecting the polarity of apical proteins, and it was proposed that transport of PIN1 in the early secretory pathway may require GNOM/GNL1 function (Doyle et al., 2015). Notably, it has been shown that GNOM localizes mainly to the Golgi apparatus (Naramoto et al., 2014). Here, we found that interfering with the function of GNL1 causes an accumulation of newly synthesized PIN1 at internal structures with a morphology consistent with the ER. These data suggest that trafficking of PIN1 along the secretory pathway involves the Golgi apparatus and requires GNL1 function.
In order to investigate whether the defects in the trafficking and localization of the PIN1:GFP-F165A mutant correlated with its inability to bind μ2- or μ3-adaptin, we analyzed the localization of wild-type PIN1:GFP in previously characterized μ2- and μ3-adaptin mutants (ap2m-1 and ap3m-1, respectively). The ap2m-1 mutant was shown previously to have reduced internalization of FM4-64 and cellulose synthase in root epidermal cells, consistent with a role of AP-2 in endocytosis, but normal secretion of cellulose synthase complexes (Bashline et al., 2013). On the other hand, PIN2-GFP showed a punctate pattern in the filaments of this mutant, although these punctae were located along the plasma membrane (Kim et al., 2013). In an ap2 σ-subunit mutant, PIN1-GFP was reported previously to accumulate in large intracellular aggregates at the globular stage. In root cells of this mutant, PIN1-GFP was shown to form BFA compartments of smaller size and lower intensity, consistent with a role of AP-2 in PIN1 endocytosis (Fan et al., 2013). However, no image was shown for the localization of PIN1-GFP in root cells in the absence of BFA. Here, we found that the PIN1-GFP-F165A mutant showed less accumulation in BFA compartments, consistent with a role of Phe-165, which binds μ2-adaptin in vitro, in PIN1 endocytosis in vivo. To our knowledge, this is the first report of a putative endocytosis signal in the cytosolic loop of PIN1. On the other hand, the localization of PIN1-GFP in root stele cells of the ap2m-1 mutant was very similar to that in the wild-type background, without any obvious intracellular accumulation. This suggests that the lower binding of μA (μ2)-adaptin may not be the primary cause of the accumulation of the PIN1:GFP-F165A mutant in intracellular structures in root stele cells. In contrast, PIN1-GFP accumulated in big intracellular structures containing LysoTracker in the ap3m-1 mutant, suggesting that the AP-3 complex is essential for PIN1 trafficking and localization, as described previously using mutants in the AP-3 β- and δ- subunits (pat2 and pat4 mutants). The most striking morphological defect in those mutants was an abnormal vacuole morphology, which led to the suggestion that the AP-3 complex is required for the biogenesis and function of the lytic vacuole (Feraru et al., 2010; Zwiewka et al., 2011; Niñoles et al., 2013). In particular, these aberrant vacuoles were often multilamellar, with various multilayered endomembrane enclosures (Feraru et al., 2010; Viotti et al., 2013). Interestingly, these structures showed enhanced accumulation of PIN1-GFP (Feraru et al., 2010).
Other mutants affecting Golgi- and post-Golgi trafficking, like vps45 and amsh3, also presented alterations in vacuole morphology (Zouhar et al., 2009; Isono et al., 2010). In the case of the vps45 mutant, it also showed multilayered provacuoles, like the pat2 mutant. This suggests that impaired Golgi- and post-Golgi trafficking may interfere with vacuole biogenesis from ER membranes, with the result of a proliferation of membranes that may, at some point, start to curl concentrically to form multilayered compartments (Viotti, 2014). On the other hand, the PIN1:GFP-F165A mutant, which cannot bind μ3-adaptin, accumulated in structures containing ER markers. There are a number of other examples where a defect in post-Golgi trafficking seems to affect the ER exit of post-Golgi cargos. For instance, the expression of mutants of the sorting nexins (which locate to the TGN) inhibits vacuolar protein transport and leads to the accumulation of vacuolar cargo at the ER (Niemes et al., 2010). Vacuolar proteins also have been shown to accumulate in the ER in vsr1 and vsr3 double mutants (Lee et al., 2013). Concerning adaptins, certain vacuolar (Arabidopsis aleurin-like protease) and secretory (invertase) cargos have been shown to accumulate at the ER in a mutant of the μ-subunit of the AP-1 complex, which is involved in late secretory and vacuolar traffic (Park et al., 2013). As indicated by Park et al. (2013), “it is poorly understood how a defect in post-Golgi trafficking might affect the ER exit of post-Golgi cargos,” while Robinson and Pimpl (2014) suggested that there is “some kind of feedback between correct TGN function and ER exit.”
It is striking that the PIN1:GFP-F165A mutant contains multilayered membrane structures similar to those found in the pat2 mutant, although one important difference is that PIN1:GFP-F165A mutant aggregates are not acidic (as judged by LysoTracker labeling). Nevertheless, both structures were not expected to be exactly the same, since the pat2 and μ3-adaptin mutants (in contrast to the PIN1:GFP-F165A mutant) should show affected trafficking of many different proteins (in addition to PIN1-GFP) that use the AP-3 complex to be included in clathrin-coated vesicles, including perhaps ion channels or transporters (Niñoles et al., 2013). Both structures, however, accumulated PIN1-GFP. Therefore, the precise molecular explanation linking Phe-165 with the AP-3 complex and the transport of PIN1 along the secretory pathway still needs to be elucidated. On the other hand, there is increasing evidence that the ER is the main source of membrane for the biogenesis of lytic vacuoles (Viotti et al., 2013; Viotti, 2014). In this respect, direct connections between the ER and vacuoles have been detected at the ultrastructural level, and provacuoles were found to contain the ER chaperone calnexin (Viotti et al., 2013), consistent with the colocalization of the PIN1:GFP-F165A mutant with BiP. Vacuoles have been found previously to fuse directly with the plasma membrane as an unusual form of secretion (Hatsugai et al., 2009), which indicates that tonoplast membranes (probably derived from the ER) are competent for fusion with the plasma membrane. The fact that the structures found in the PIN1:GFP-F165A mutant often are observed close to the plasma membrane may indicate that their fusion with the plasma membrane is not very efficient. However, these structures disappear during CHX treatment, and the PIN1:GFP-F165A mutant is transported to the plasma membrane, perhaps with a delayed kinetics.
Alternative explanations for the formation of the structures found in the PIN1:GFP-F165A mutant have been described previously. In this respect, it has been reported that protein accumulation in discrete subdomains of the ER may favor adjacent ER membranes to become zippered together by low-affinity protein interactions generating onion-like membrane structures (Gong et al., 1996; Snapp et al., 2003). The proliferation of ER-derived membrane structures, either in the cytosol or in the nucleus, has been observed previously upon the overexpression of integral ER membrane proteins, such as 3-hydroxy-3-methyl glutaryl-CoA reductase, cytochrome P450, and IP3 receptor, and different models have been proposed to explain their formation (Chin et al., 1982; Schunck et al., 1991; Takei et al., 1994; Koning et al., 1996). These membranous structures also were found to contain integral membrane proteins, like calnexin, an ER chaperone, but also luminal ER proteins, like BiP or PDI (Isaac et al., 2001; Sørensen et al., 2004). Since the levels of PIN1:GFP-F165A in the mutant lines were not significantly different from those of PIN1:GFP in the control plants (Fig. 10; Supplemental Fig. S4), the appearance of these structures cannot be due to overexpression of the protein per se. However, a defect in the ER export of the mutant may cause an accumulation of the protein at ER membrane domains, mimicking the effect of the overexpression of the protein.
In any case, while interference with the function of adaptins (in particular AP-3 subunits) may cause more pleiotropic effects in trafficking, in particular for tonoplast proteins, which still need to be elucidated in plants, this study aimed to investigate PIN1 trafficking from the point of view of sorting signals within the PIN1 sequence. In this work, we analyzed the contributions of specific residues within the cytosolic loop of PIN1 to its trafficking and localization. From these studies, we propose that Phe-165, through the binding of μ2- and μ3-adaptins, may play a role in PIN1 endocytosis and in PIN1 trafficking along the secretory pathway, respectively.
MATERIALS AND METHODS
Expression and Purification of Fusion Proteins and Pull-Down Assays
The sequences of the complete cytosolic loop of PIN1 (residues156–482) and of different regions of this loop (residues156–235, 236–319, and 320–402) were commercially synthesized de novo (Geneart) and cloned into the pGEX-4T-3 vector, in order to express them in bacteria as GST fusion proteins. In the case of μ-adaptins, we made use of the fact that µ-adaptins have a bipartite structure, with the N-terminal one-third spanning the β-adaptin-binding domain and the C-terminal two-thirds comprising the receptor-binding domain. Separate expression of either region does not result in a loss of functionality (Aguilar et al., 1997). Therefore, the sequences of the RBD of μ-adaptins A (At5g46630; residues 144–441), B1 (At1g10730; residues 145–428), C (At4g24550; residues 144–451), and D (At1g56590; residues 142–415) were commercially synthesized de novo (Geneart) and cloned into the pQE30 vector (Qiagen) in order to express them in bacteria as His-tagged proteins: (His)6x-RBD-µ-adaptins A to D. Cultures of Escherichia coli strain BL21-DE3 (containing the GST constructs) or M15 (pREP4; containing the His-tagged constructs) were grown at 37°C and induced for 2 h with 1 mm isopropylthio-β-galactoside at 28°C, after which cells were harvested by centrifugation and frozen at −80°C. Purification of GST fusion proteins and (His)6x-RBD of µ-adaptins A to D was performed following the instructions from the manufacturer (GE Healthcare). For pull-down assays, both the GST fusion proteins and the (His)6x fusion proteins were changed into binding buffer (100 mm Tris-HCl [pH 7.5], 5 mm EDTA, and 0.1% [v/v] Triton X-100) using PD-10 columns (Amersham Pharmacia Biotech). A total of 100 µg of packed glutathione-Sepharose beads was prepared according to the manufacturer’s instructions (Amersham Pharmacia Biotech) and subsequently incubated with 50 µg of the GST fusion proteins for 30 min at 4°C. The beads were washed three times with 500 µL of binding buffer and centrifuged at 510g. For each binding experiment, 30 µL of the preincubated glutathione-Sepharose beads was incubated with 1 to 16 µg of the (His)6x-µA-RBD construct and bovine serum albumin at a final concentration of 1% (w/v), filled up with binding buffer to 100 µL final volume, and incubated for 1.5 h at 4°C on a rotator. The beads were washed two times with 200 µL of binding buffer, and the final pellet was resuspended in 2-fold sample buffer (Laemmli, 1970). The samples were boiled at 95°C for 1 min and subjected to SDS-PAGE. Each binding assay was performed independently three times.
Plant Material
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used. The loss-of-function pin1 mutant (SALK_047613), the AP2M knockout mutant ap2m-1 (SALK_083693), and the AP3M knockout mutant ap3m-3 (SALK_127431) were from the Salk Institute Genomic Analysis Laboratory and obtained from the Nottingham Arabidopsis Stock Centre. GNL1-LM-myc (GNL1-BFAS) gnl1 and pMDC7-PIN1-RFP seeds were provided by Dr. Gerd Jürgens. Arabidopsis plants were grown in growth chambers as described previously (Ortiz-Masia et al., 2007). For immunogold electron microscopy, seedlings were grown on Murashige and Skoog (MS) medium containing 0.5% agar, and the roots were harvested after 5 d.
Recombinant Plasmid Production, Plant Transformation, and Transformant Selection
The sequence of PIN1::PIN1:GFP:PIN1 (PIN1:GFP) was inserted into the pBINPLUS vector (12,396 bp) through the site SalI, generating a pBINPLUS-PIN1:GFP construct of 18,759 bp (Benková et al., 2003), from which the different mutant versions were obtained. Different DNA fragments were synthesized de novo (Geneart) to generate the different mutant versions of PIN1:GFP (mutations F165A, Y260A, Y328A, and Y394A), and they were introduced into the pBINPLUS-PIN1:GFP construct through the sites AscI (pBINPLUS cloning site) and XhoI (inside PIN1:GFP sequence), except for the F165A mutant. The DNA fragment containing the F165A mutation was introduced into SpeI and XhoI sites, both included in the PIN1:GFP sequence.
In total, four different mutant versions of PIN1:GFP were generated. Each of these mutants had one point mutation in the cytoplasmic loop of PIN1:GFP that results in the substitution of one amino acid by Ala. The names of these mutations refer to the original amino acid name and their position followed by A for Ala: PIN1:GFP-F165A, PIN1:GFP-Y260A, PIN1:GFP-Y328A, and PIN1:GFP-Y394A. Arabidopsis plants were transformed with the pBINPLUS constructs, via Agrobacterium tumefaciens, by the floral dip method according to standard procedures (Clough and Bent 1998). To estimate the number of T-DNA insertions in the transgenic plants, 40 seeds of each T1 line were plated on 0.5× MS basal salts, 1% Suc, 0.6% agar, with 50 mg L−1 kanamycin. Lines where the proportion of kanamycin-resistant to kanamycin-sensitive plants in their progeny fitted to a 3:1 ratio were considered to contain the T-DNA inserted in a single locus. Homozygous and hemizygous T2 plants from those lines were identified by analyzing their progeny by the same method. Alternatively, selection of transformants also was performed by GFP detection in roots of 4-d-old seedlings grown on MS plates through a fluorescence microscope (Olympus szx9).
Homozygous lines of PIN1:GFP and mutant versions were crossed with the heterozygous line of pin1 (SALK_047613). Selection of F1 and F2 progeny was performed by PCR using genomic DNA isolated following the protocol described previously (Edwards et al., 1991; Blakeslee et al., 2007) as a template. Two pairs of primers, GFP3 (specific for GFP)/LPM pin (specific for PIN1) and RPIN (specific for PIN1)/LBb1 (specific for T-DNA; Supplemental Table S1) were used for F1 selection, and LPIN and RPIN were used for F2 selection. In addition, homozygous lines for a GFP-tagged protein were selected by looking at F3 progeny 4-d-old seedlings using the fluorescence microscope (Olympus szx9), choosing the ones in which all the progeny showed green fluorescence from GFP in the roots. In total, 350 F2 plants were analyzed for rescue.
A PIN1 genomic DNA fragment corresponding to the 3′ end of the PIN1 genomic DNA, which contains at the 5′ end the XhoI site and one HA tag DNA before the stop codon, was synthesized de novo (Geneart). The fragment was introduced into pBINPLUS-PIN1:GFP and pBINPLUS-PIN1:GFP-F165 constructs through the sites AscI (pBINPLUS cloning site) and XhoI (inside PIN1:GFP sequence) to generate the pBINPLUS-PIN1-HA and pBINPLUS-PIN1-HA-F165 constructs, respectively. Arabidopsis plants were transformed with the constructs, and the transformants were selected as above. We obtained two independent lines of PIN1:HA-F165A that accumulated the mutant PIN1-F165A in intracellular structures, very similar to its GFP version, over subsequent generations. GNL1-LM-myc (GNL1-BFAS) gnl1 plants were transformed with pMDC7-PIN1-RFP (Richter et al., 2007, 2014), and transformants were selected by looking at 4-d-old seedlings after 6 h of estradiol treatment using the fluorescence microscope, choosing the ones with red fluorescence from RFP in the roots.
Reverse Transcription-PCR
Total RNA was extracted from seedlings using a Qiagen RNeasy plant mini kit, and 1 μg of the RNA solution obtained was reverse transcribed using 0.1 μg of oligo(dT)15 primer and Expand Reverse Transcriptase (Roche) to finally obtain a 40-μL complementary DNA solution. PCR amplifications were performed on 3 μL of complementary DNA template using the kit PCR Master (Roche). The sequences of the primers used for PCR amplifications are included in Supplemental Table S1.
Preparation of Protein Extracts, Coimmunoprecipitation Experiments, and Western Blotting
Immunoprecipitation experiments were performed using total protein extracts of Arabidopsis roots (Col-0, PIN1:GFP, and PIN1:GFP-F165A) treated or not with CHX. To this end, 100 to 150 seeds were sown on MS plates over a nylon mesh (Sefar) and grown at 21°C under a 16-h/8-h photoperiod. Five-day-old seedlings were transferred to another MS plate containing or not 50 μm CHX. After 2 h of incubation in the dark, roots were collected and frozen in liquid nitrogen. Next, they were ground with mortar and pestle, and the powder obtained was transferred to a microcentrifuge tube with 0.2 to 0.3 mL of lysis buffer (0.05 m Tris-HCl, 0.5% Triton X-100, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 0.15 m NaCl, pH 7.5) and 0.1% protease inhibitor cocktail (Sigma) and incubated on ice for 10 min. The resultant supernatant after two centrifugations of 5 and 3 min, at 13,000 rpm and 4°C, was the total protein extract. Protein extracts were used for SDS-PAGE followed by western-blot analysis (using the antibodies listed in Supplemental Table S2) or immunoprecipitation experiments (performed as described by Montesinos et al. [2012]) using a monoclonal anti-GFP antibody (MA1; Thermo Fisher Scientific). Western-blot analysis was performed using peroxidase-labeled secondary antibodies (GE Healthcare) and the SuperSignal West Pico chemiluminiscent substrate (Pierce, Thermo Scientific). The intensity of the bands obtained after western blotting was analyzed using the ChemiDoc XRS+ imaging system and Quantity One software (Bio-Rad Laboratories).
Immunolocalization
For immunolocalization assays, 4-d-old Arabidopsis seedlings were used. Whole-mount samples were analyzed by immunofluorescence microscopy, as described previously (Sauer et al., 2006), using the InsituPro VSi (Intavis) robot. Immunolocalization data were generated from three independent experiments per genotype or treatment using 30 to 40 roots in total. The antibodies and dilutions used for immunolocalization are described in Supplemental Table S2.
FRAP
FRAP experiments were performed with the Olympus FV100 confocal microscope in stele cells of 4-d-old Arabidopsis roots expressing PIN1:GFP or PIN1:GFP-F165A. Seedlings were placed on chambered cover glasses (Nunc Lab-Tek), and they were covered with 0.2-mm-thin square blocks of solid MS medium. Bleaching was performed in a specific zone of the root containing more than six inner stele cells for 2 min at 100% main laser power. Confocal fluorescent images were collected as described above.
Immunogold Labeling
The general procedures for transmission electron microscopy sample preparation, thin sectioning, and immunogold labeling were performed essentially as described previously (Tse et al., 2004; Gao et al., 2012). Root tips of 4-d-old Arabidopsis seedlings expressing PIN1:GFP or PIN1:GFP-F165A were cut and immediately frozen in a high-pressure freezer (EM PACT2; Leica), followed by subsequent freeze substitution in dry acetone containing 0.1% uranyl acetate at −85°C in an AFS freeze substitution unit (Leica). Infiltration with Lowicryl HM20, embedding, and UV light polymerization were performed stepwise at −35°C. Immunogold labeling was performed with rabbit anti-GFP antibodies (40 μg mL−1) and 10-nm gold-coupled secondary antibodies (Supplemental Table S2). Transmission electron microscopy examination was done with a Hitachi H-7650 transmission electron microscope with a CCD camera (Hitachi High-Technologies) operating at 80 kV.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: PIN1 (At1G73590)/p24δ5 (At1g21900)/AP1M1 (At1g10730)/AP2M (At5g46630)/AP3M (At1g56590)/AP4M (At4g24550)/GNL1 (At5g39500)/ACT7 (AT5G09810)
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of the cytosolic loop of PIN proteins.
Supplemental Figure S2..Purification of His-RBD-μ-adaptins.
Supplemental Figure S3. Purification of GST-PIN1CL and GST-PIN1CL portions.
Supplemental Figure S4. Reverse transcription-PCR, western-blot, and phenotypic analyses in the pin1 background of PIN1:GFP-F165A, PIN1:GFP-Y260A, PIN1:GFP-Y328A, and PIN1:GFP-Y394A lines.
Supplemental Figure S5. Localization of PIN1:HA and PIN1:HA-F165A mutants in Arabidopsis roots.
Supplemental Figure S6. Localization of mCherry-HDEL and RFP-p24δ5 in PIN1:GFP plants.
Supplemental Figure S7. Localization of PIN1:GFP-F165A by immunogold labeling and CLSM in roots of seedlings expressing PIN1:GFP-F165A.
Supplemental Figure S8. Ultrastructural analysis of the PIN1:GFP-F165A mutant.
Supplemental Table S1. Primers used for PCR amplification.
Supplemental Table S2. Antibodies used for western blotting and immunolocalization.
Supplementary Material
Acknowledgments
We thank Dr. R. Offringa (Leiden University) for providing the GST-PIN-CL construct; Sandra Richter and Gerd Jurgens (University of Tübingen) for providing the estradiol-inducible PIN1-RFP construct and the gnl1 mutant expressing BFA-sensitive GNL1; F.J. Santonja (University of Valencia) for help with the statistical analysis; Jurgen Kleine-Vehn, Elke Barbez, and Eva Benkova for helpful discussions; the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants; and the greenhouse section and the microscopy section of SCSIE (University of Valencia) and Pilar Selvi for excellent technical assistance.
Glossary
- ER
endoplasmic reticulum
- TGN
trans-Golgi network
- T-DNA
transfer DNA
- CLSM
confocal laser scanning microscopy
- BFA
brefeldin A
- EXPO
exocyst-positive
- ERES
endoplasmic reticulum export sites
- PVC
prevacuolar compartment
- FRAP
fluorescence recovery after photobleaching
- CHX
cycloheximide
- Col-0
Columbia-0
- MS
Murashige and Skoog
Footnotes
This work was supported by the Ministerio de Economía y Competitividad (grant no. BFU2012–33883 to F.A. and M.J.M.), the Generalitat Valenciana (grant nos. ISIC/2013/004 and GVACOMP2014–202 to F.A. and M.J.M.), the European Research Council (grant no. ERC–2011–StG–20101109–PSDP to J.F.), the Research Grants Council (grant no. AoE/M–05/12 to L.J.), and the Universitat de Valencia (V. Segles Program fellowship to G.S.-A.).
References
- Adamowski M, Friml J (2015) PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar RC, Ohno H, Roche KW, Bonifacino JS (1997) Functional domain mapping of the clathrin-associated adaptor medium chains mu1 and mu2. J Biol Chem 272: 27160–27166 [DOI] [PubMed] [Google Scholar]
- Barth M, Holstein SE (2004) Identification and functional characterization of Arabidopsis AP180, a binding partner of plant alphaC-adaptin. J Cell Sci 117: 2051–2062 [DOI] [PubMed] [Google Scholar]
- Bashline L, Li S, Anderson CT, Lei L, Gu Y (2013) The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol 163: 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender RL, Fekete ML, Klinkenberg PM, Hampton M, Bauer B, Malecha M, Lindgren K, A Maki J, Perera MA, Nikolau BJ, et al. (2013) PIN6 is required for nectary auxin response and short stamen development. Plant J 74: 893–904 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al. (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Bonifacino JS (2001) Adaptins: the final recount. Mol Biol Cell 12: 2907–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS. (2014) Adaptor proteins involved in polarized sorting. J Cell Biol 204: 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canagarajah BJ, Ren X, Bonifacino JS, Hurley JH (2013) The clathrin adaptor complexes as a paradigm for membrane-associated allostery. Protein Sci 22: 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin DJ, Luskey KL, Anderson RG, Faust JR, Goldstein JL, Brown MS (1982) Appearance of crystalloid endoplasmic reticulum in compactin-resistant Chinese hamster cells with a 500-fold increase in 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci USA 79: 1185–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- daSilva LL, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F (2004) Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16: 1753–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J, De Rycke R, Botterman J (1992) Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. EMBO J 11: 2345–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzer K, Weber B, Hille-Rehfeld A, Figura KV, Pohlmann R (1997) Identification of three internalization sequences in the cytoplasmic tail of the 46 kDa mannose 6-phosphate receptor. Biochem J 326: 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J (2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Ding Z, Wang B, Moreno I, Dupláková N, Simon S, Carraro N, Reemmer J, Pěnčík A, Chen X, Tejos R, et al. (2012) ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat Commun 3: 941. [DOI] [PubMed] [Google Scholar]
- Di Rubbo S, Irani NG, Kim SY, Xu ZY, Gadeyne A, Dejonghe W, Vanhoutte I, Persiau G, Eeckhout D, Simon S, et al. (2013) The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25: 2986–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Haeger A, Vain T, Rigal A, Viotti C, Łangowska M, Ma Q, Friml J, Raikhel NV, Hicks GR, et al. (2015) An early secretory pathway mediated by GNOM-LIKE 1 and GNOM is essential for basal polarity establishment in Arabidopsis thaliana. Proc Natl Acad Sci USA 112: E806–E815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Hao H, Xue Y, Zhang L, Song K, Ding Z, Botella MA, Wang H, Lin J (2013) Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 140: 3826–3837 [DOI] [PubMed] [Google Scholar]
- Feraru E, Paciorek T, Feraru MI, Zwiewka M, De Groodt R, De Rycke R, Kleine-Vehn J, Friml J (2010) The AP-3 β adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell 22: 2812–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M (2004) PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131: 5021–5030 [DOI] [PubMed] [Google Scholar]
- Gadeyne A, Sánchez-Rodríguez C, Vanneste S, Di Rubbo S, Zauber H, Vanneste K, Van Leene J, De Winne N, Eeckhout D, Persiau G, et al. (2014) The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156: 691–704 [DOI] [PubMed] [Google Scholar]
- Ganguly A, Park M, Kesawat MS, Cho HT (2014) Functional analysis of the hydrophilic loop in intracellular trafficking of Arabidopsis PIN-FORMED proteins. Plant Cell 26: 1570–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Yu CK, Qu S, San MWY, Li KY, Lo SW, Jiang L (2012) The Golgi-localized Arabidopsis endomembrane protein12 contains both endoplasmic reticulum export and Golgi retention signals at its C terminus. Plant Cell 24: 2086–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Gong FC, Giddings TH, Meehl JB, Staehelin LA, Galbraith DW (1996) Z-membranes: artificial organelles for overexpressing recombinant integral membrane proteins. Proc Natl Acad Sci USA 93: 2219–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravotta D, Carvajal-Gonzalez JM, Mattera R, Deborde S, Banfelder JR, Bonifacino JS, Rodriguez-Boulan E (2012) The clathrin adaptor AP-1A mediates basolateral polarity. Dev Cell 22: 811–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Sirkis DW, Schekman R (2014) Protein sorting at the trans-Golgi network. Annu Rev Cell Dev Biol 30: 169–206 [DOI] [PubMed] [Google Scholar]
- Happel N, Höning S, Neuhaus JM, Paris N, Robinson DG, Holstein SE (2004) Arabidopsis μA-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J 37: 678–693 [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I (2009) A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev 23: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein SE. (2002) Clathrin and plant endocytosis. Traffic 3: 614–620 [DOI] [PubMed] [Google Scholar]
- Höning S, Sosa M, Hille-Rehfeld A, von Figura K (1997) The 46-kDa mannose 6-phosphate receptor contains multiple binding sites for clathrin adaptors. J Biol Chem 272: 19884–19890 [DOI] [PubMed] [Google Scholar]
- Isaac C, Pollard JW, Meier UT (2001) Intranuclear endoplasmic reticulum induced by Nopp140 mimics the nucleolar channel system of human endometrium. J Cell Sci 114: 4253–4264 [DOI] [PubMed] [Google Scholar]
- Isono E, Katsiarimpa A, Müller IK, Anzenberger F, Stierhof YD, Geldner N, Chory J, Schwechheimer C (2010) The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell 22: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, et al. (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA 98: 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Xu ZY, Song K, Kim DH, Kang H, Reichardt I, Sohn EJ, Friml J, Juergens G, Hwang I (2013) Adaptor protein complex 2-mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell 25: 2970–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning AJ, Roberts CJ, Wright RL (1996) Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isozymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol Biol Cell 7: 769–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Křeček P, Skůpa P, Libus J, Naramoto S, Tejos R, Friml J, Zazímalová E (2009) The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Langhans M, Marcote MJ, Pimpl P, Virgili-López G, Robinson DG, Aniento F (2008) In vivo trafficking and localization of p24 proteins in plant cells. Traffic 9: 770–785 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Kim H, Kang H, Jang M, Lee DW, Lee S, Hwang I (2007) EpsinR2 interacts with clathrin, adaptor protein-3, AtVTI12, and phosphatidylinositol-3-phosphate: implications for EpsinR2 function in protein trafficking in plant cells. Plant Physiol 143: 1561–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jang M, Song K, Kang H, Lee MH, Lee DW, Zouhar J, Rojo E, Sohn EJ, Hwang I (2013) Functional identification of sorting receptors involved in trafficking of soluble lytic vacuolar proteins in vegetative cells of Arabidopsis. Plant Physiol 161: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos JC, Langhans M, Sturm S, Hillmer S, Aniento F, Robinson DG, Marcote MJ (2013) Putative p24 complexes in Arabidopsis contain members of the delta and beta subfamilies and cycle in the early secretory pathway. J Exp Bot 64: 3147–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos JC, Sturm S, Langhans M, Hillmer S, Marcote MJ, Robinson DG, Aniento F (2012) Coupled transport of Arabidopsis p24 proteins at the ER-Golgi interface. J Exp Bot 63: 4243–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Krecek P, Bielach A, Petrásek J, Zhang J, Gaykova V, Stierhof YD, et al. (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- Naramoto S, Otegui MS, Kutsuna N, de Rycke R, Dainobu T, Karampelias M, Fujimoto M, Feraru E, Miki D, Fukuda H, et al. (2014) Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26: 3062–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Niemes S, Labs M, Scheuring D, Krueger F, Langhans M, Jesenofsky B, Robinson DG, Pimpl P (2010) Sorting of plant vacuolar proteins is initiated in the ER. Plant J 62: 601–614 [DOI] [PubMed] [Google Scholar]
- Niihama M, Takemoto N, Hashiguchi Y, Tasaka M, Morita MT (2009) ZIP genes encode proteins involved in membrane trafficking of the TGN-PVC/vacuoles. Plant Cell Physiol 50: 2057–2068 [DOI] [PubMed] [Google Scholar]
- Niñoles R, Rubio L, García-Sánchez MJ, Fernández JA, Bueso E, Alejandro S, Serrano R (2013) A dominant-negative form of Arabidopsis AP-3 β-adaptin improves intracellular pH homeostasis. Plant J 74: 557–568 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Matsunami E, Yoshida S, Kohata S, Yamauchi J, Jisaka M, Nagaya T, Yokota K, Nakagawa T (2016) The tyrosine-sorting motif of the vacuolar sorting receptor VSR4 from Arabidopsis thaliana, which is involved in the interaction between VSR4 and AP1M2, μ1-adaptin type 2 of clathrin adaptor complex 1 subunits, participates in the post-Golgi sorting of VSR4. Biosci Biotechnol Biochem 80: 694–705 [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Masia D, Perez-Amador MA, Carbonell J, Marcote MJ (2007) Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett 581: 1834–1840 [DOI] [PubMed] [Google Scholar]
- Ortiz-Zapater E, Soriano-Ortega E, Marcote MJ, Ortiz-Masiá D, Aniento F (2006) Trafficking of the human transferrin receptor in plant cells: effects of tyrphostin A23 and brefeldin A. Plant J 48: 757–770 [DOI] [PubMed] [Google Scholar]
- Park M, Song K, Reichardt I, Kim H, Mayer U, Stierhof YD, Hwang I, Jürgens G (2013) Arabidopsis μ-adaptin subunit AP1M of adaptor protein complex 1 mediates late secretory and vacuolar traffic and is required for growth. Proc Natl Acad Sci USA 110: 10318–10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Richter S, Geldner N, Schrader J, Wolters H, Stierhof YD, Rios G, Koncz C, Robinson DG, Jürgens G (2007) Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448: 488–492 [DOI] [PubMed] [Google Scholar]
- Richter S, Kientz M, Brumm S, Nielsen ME, Park M, Gavidia R, Krause C, Voss U, Beckmann H, Mayer U, et al. (2014) Delivery of endocytosed proteins to the cell-division plane requires change of pathway from recycling to secretion. eLife 3: e02131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Čovanová M, et al. (2010) ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Pimpl P (2014) Clathrin and post-Golgi trafficking: a very complicated issue. Trends Plant Sci 19: 134–139 [DOI] [PubMed] [Google Scholar]
- Ron M, Avni A (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín M, Ordóñez A, Sohn EJ, Robert S, Sánchez-Serrano JJ, Surpin MA, Raikhel NV, Rojo E (2007) Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc Natl Acad Sci USA 104: 3645–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Paciorek T, Benková E, Friml J (2006) Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc 1: 98–103 [DOI] [PubMed] [Google Scholar]
- Schunck WH, Vogel F, Gross B, Kärgel E, Mauersberger S, Köpke K, Gengnagel C, Müller HG (1991) Comparison of two cytochromes P-450 from Candida maltosa: primary structures, substrate specificities and effects of their expression in Saccharomyces cerevisiae on the proliferation of the endoplasmic reticulum. Eur J Cell Biol 55: 336–345 [PubMed] [Google Scholar]
- Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J (2003) Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol 163: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn EJ, Rojas-Pierce M, Pan S, Carter C, Serrano-Mislata A, Madueño F, Rojo E, Surpin M, Raikhel NV (2007) The shoot meristem identity gene TFL1 is involved in flower development and trafficking to the protein storage vacuole. Proc Natl Acad Sci USA 104: 18801–18806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen V, Brech A, Khnykin D, Kolpakova E, Citores L, Olsnes S (2004) Deletion mutant of FGFR4 induces onion-like membrane structures in the nucleus. J Cell Sci 117: 1807–1819 [DOI] [PubMed] [Google Scholar]
- Takano J, Tanaka M, Toyoda A, Miwa K, Kasai K, Fuji K, Onouchi H, Naito S, Fujiwara T (2010) Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc Natl Acad Sci USA 107: 5220–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Mignery GA, Mugnaini E, Südhof TC, De Camilli P (1994) Inositol 1,4,5-trisphosphate receptor causes formation of ER cisternal stacks in transfected fibroblasts and in cerebellar Purkinje cells. Neuron 12: 327–342 [DOI] [PubMed] [Google Scholar]
- Teh OK, Shimono Y, Shirakawa M, Fukao Y, Tamura K, Shimada T, Hara-Nishimura I (2013) The AP-1 μ adaptin is required for KNOLLE localization at the cell plate to mediate cytokinesis in Arabidopsis. Plant Cell Physiol 54: 838–847 [DOI] [PubMed] [Google Scholar]
- Traub LM. (2009) Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol 10: 583–596 [DOI] [PubMed] [Google Scholar]
- Tse YC, Mo B, Hillmer S, Zhao M, Lo SW, Robinson DG, Jiang L (2004) Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C. (2014) ER and vacuoles: never been closer. Front Plant Sci 5: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C, Krüger F, Krebs M, Neubert C, Fink F, Lupanga U, Scheuring D, Boutté Y, Frescatada-Rosa M, Wolfenstetter S, et al. (2013) The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25: 3434–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ding Y, Wang J, Hillmer S, Miao Y, Lo SW, Wang X, Robinson DG, Jiang L (2010) EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 22: 4009–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JG, Li S, Zhao XY, Zhou LZ, Huang GQ, Feng C, Zhang Y (2013) HAPLESS13, the Arabidopsis μ1 adaptin, is essential for protein sorting at the trans-Golgi network/early endosome. Plant Physiol 162: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Yamaoka S, Shimono Y, Shirakawa M, Fukao Y, Kawase T, Hatsugai N, Tamura K, Shimada T, Hara-Nishimura I (2013) Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell 25: 2958–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Jiang L (2014) Autophagosome biogenesis in plants: roles of SH3P2. Autophagy 10: 704–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J, Rojo E, Bassham DC (2009) AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol 149: 1668–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Nishizawa N, Wu Y, Kost B, Chua NH (2000) KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12: 1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiewka M, Feraru E, Möller B, Hwang I, Feraru MI, Kleine-Vehn J, Weijers D, Friml J (2011) The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res 21: 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.