The methyl-transferases OsDDM1 and OsDRM2 display functional specificities that define distinct DNA methylation pathways in the bulk of DNA methylation of the rice genome.
Abstract
Plant DNA methylation that occurs at CG, CHG, and CHH sites (H = A, C, or T) is a hallmark of the repression of repetitive sequences and transposable elements (TEs). The rice (Oryza sativa) genome contains about 40% repetitive sequence and TEs and displays specific patterns of genome-wide DNA methylation. The mechanism responsible for the specific methylation patterns is unclear. Here, we analyzed the function of OsDDM1 (Deficient in DNA Methylation 1) and OsDRM2 (Deficient in DNA Methylation 1) in genome-wide DNA methylation, TE repression, small RNA accumulation, and gene expression. We show that OsDDM1 is essential for high levels of methylation at CHG and, to a lesser extent, CG sites in heterochromatic regions and also is required for CHH methylation that mainly locates in the genic regions of the genome. In addition to a large member of TEs, loss of OsDDM1 leads to hypomethylation and up-regulation of many protein-coding genes, producing very severe growth phenotypes at the initial generation. Importantly, we show that OsDRM2 mutation results in a nearly complete loss of CHH methylation and derepression of mainly small TE-associated genes and that OsDDM1 is involved in facilitating OsDRM2-mediated CHH methylation. Thus, the function of OsDDM1 and OsDRM2 defines distinct DNA methylation pathways in the bulk of DNA methylation of the genome, which is possibly related to the dispersed heterochromatin across chromosomes in rice and suggests that DNA methylation mechanisms may vary among different plant species.
DNA methylation in flowering plants occurs in three sequence contexts: the so-called symmetric CG and CHG and the asymmetric CHH, where H is any nucleotide except G. Methylation in each context is believed to be catalyzed primarily by a specific family of DNA methyltransferases: MET1 (homologous to animal Dnmt1) for CG, plant-specific chromomethylases (CMT3 and CMT2) for CHG, and DRM2 (homologous to animal Dnmt3) for CHH (Law and Jacobsen, 2010). Recently, it was shown that CMT2 is a major CHH methyltransferase in Arabidopsis (Arabidopsis thaliana; Zemach et al., 2013; Stroud et al., 2014). The majority of plant methylation is found in transposable elements (TEs) and transcribed TE genes (known as transposable element-related genes [TEGs]) and is crucial for the repression of TE expression and transposition (Law and Jacobsen, 2010). Substantial methylation also is found in the bodies of active genes, where methylation is generally restricted to the CG context (Law and Jacobsen, 2010; Zemach et al., 2010). Methylation in all sequence contexts also is found in the promoter regions of many genes, which represses gene expression.
In Arabidopsis, the maintenance of CHH methylation is mediated by RNA-directed DNA methylation (RdDM) involving the methyltransferase activity of DRM2 (Law and Jacobsen, 2010). DNA methylation also is influenced by chromatin factors. For instance, the Arabidopsis Snf2 family nucleosome remodeler DDM1, which can shift nucleosomes in vitro (Brzeski and Jerzmanowski, 2003), is essential for normal DNA methylation (Jeddeloh et al., 1999; Lippman et al., 2004). The loss of DDM1 leads to a profound decrease of methylation from some TEs and repeats and strong transcriptional activation of TEs (Jeddeloh et al., 1999; Lippman et al., 2004), and inbred ddm1 mutant lines have increased rates of transposition and produce severe developmental abnormalities (Tsukahara et al., 2009). Mutation of Lsh, the mouse homolog of DDM1, causes similar methylation phenotypes (Tao et al., 2010). Recent data indicate that Arabidopsis DDM1 is important for DNA methylation in TEs, and the strength of the DDM1 requirement is positively correlated with heterochromatin (Zemach et al., 2013). It is believed that DDM1 can facilitate CMT2-mediated CHH methylation independently of RdDM and that DDM1 and RdDM synergistically mediate nearly all Arabidopsis TE methylation, prevent transposition, and maintain proper patterns of gene expression (Zemach et al., 2013).
Rice (Oryza sativa) is one of the most important food crops in the world and has been established as a model plant for genome research. The rice genome, one of the most accurately sequenced eukaryotic genomes, contains about 40% repetitive sequence and TEs (Paterson et al., 2009). In contrast to the more localized pattern of repeats close to centromeres in Arabidopsis, repeats in rice are widely spread on the chromosomes. In addition, the rice genome contains a large number of miniature inverted-repeat transposable elements (MITEs), a majority of which are associated with genes (Feschotte and Wessler, 2002; Lu et al., 2012). Extensive genome-wide DNA methylation and histone modification data sets have been generated in rice (Feng et al., 2010; He et al., 2010; Zemach et al., 2010; Li et al., 2012; Liu et al., 2015). Compared with Arabidopsis, the rice genome displays a different global DNA methylation profile and has a much higher level of genome-wide cytosine methylation (Feng et al., 2010; Li et al., 2012). Rice genes involved in DNA methylation are generally conserved (Chen and Zhou, 2013), and some of them have been studied. For instance, knockdown of OsDRM2 by homologous recombination-mediated gene targeting produces very severe defects at the seedling stage (Moritoh et al., 2012). Except for OsMET1 (Hu et al., 2014), the function of rice methylation genes in genome-wide DNA methylation is unknown. In particular, it is unclear whether a similar mechanism is conserved for the bulk of methylation of the rice genome that displays significant structural differences from that of Arabidopsis.
Here, we report a genome-wide analysis of DNA methylation, gene expression, and small RNA abundance in rice plants lacking OsDDM1 and OsDRM2. We find that loss of OsDDM1 and OsDRM2 severely affects rice plant growth in the initial generation and causes the deregulation of a large number of genes, indicating an essential role of the proteins in maintaining proper patterns of gene expression in rice. We find that OsDDM1 is required for heterochromatic CHG and CG methylation and euchromatic CHH methylation. We find that, in contrast to Arabidopsis DRM2, which is required for a minor part of CHH methylation of the genome, OsDRM2 is required for most of the CHH methylation in rice, which is essentially located in small TEs such as MITEs that are scattered in the genic regions. Importantly, we find that OsDDM1-mediated CHH methylation sites totally overlapped with that by OsDRM2, suggesting that OsDDM1 may be involved in OsDRM2-mediated CHH methylation. These data revealed specific pathways that mediate the bulk of genomic DNA methylation in rice, suggesting that specific epigenetic mechanisms may be elaborated to adapt to genome structure variation that is strikingly high in plants.
RESULTS
OsDDM1 Genes Are Required Mainly for Heterochromatic CHG and CG Methylation and Euchromatic CHH Methylation
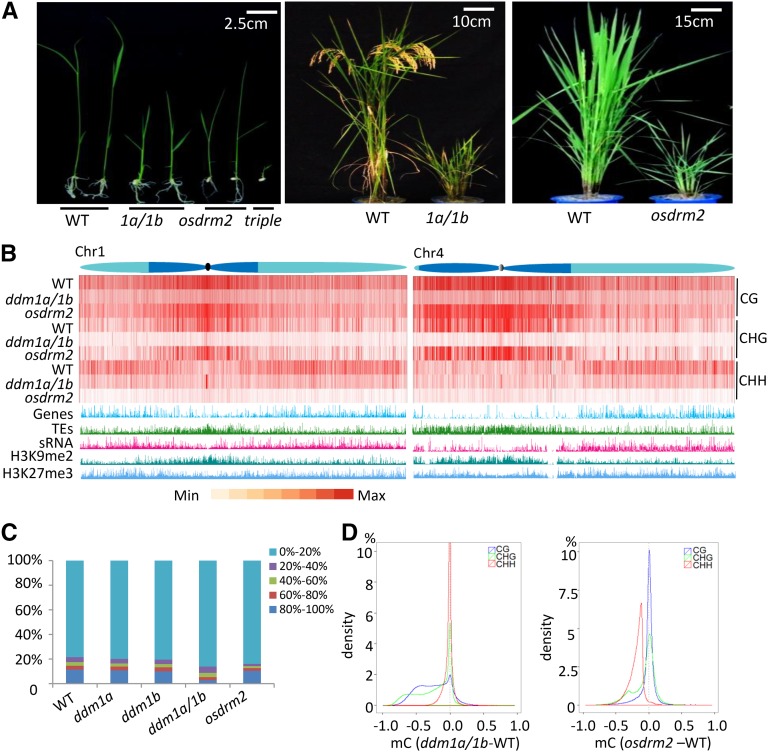
Rice has two DDM1 homologous genes, OsDDM1a (Os09g27060) and OsDDM1b (Os03g51230), which share 93% amino acid identity (Higo et al., 2012). Both genes displayed a similar expression pattern, with the transcript levels of OsDDM1b higher than that of OsDDM1a in the tested organs (Supplemental Fig. S1A). A previous study showed that down-regulation of both genes by antisense produced a dwarf phenotype in the transgenic plants, some of which progressively lost fertility and became completely infertile (Higo et al., 2012). Here, we isolated homozygous loss-of-function transfer DNA (T-DNA) insertion mutants of the two genes (Supplemental Fig. S1, B and C). However, the single osddm1a or osddm1b mutation did not produce any clear phenotype during either the vegetative or reproductive stage, even after several generations (Supplemental Fig. S1D). By contrast, the osddm1a/1b double mutation obtained by genetic crosses produced many severe developmental abnormalities, including an extremely reduced plant height at the mature stage and complete sterility (Fig. 1A; Supplemental Fig. S1E), suggesting that the two rice DDM1 orthologs acted redundantly to ensure normal plant growth and development. To study the effects of the mutations on DNA methylation, we first estimated overall methylated cytosine levels by mass spectrometry (MS). The analysis revealed that the osddm1a and osddm1b single mutations reduced cytosine methylation by 3% and 11.5%, respectively, whereas the osddm1a/1b double mutation reduced the methylation by 53.6% (Supplemental Table S1). The synergistic effects on phenotype production and cytosine methylation suggested that the two genes had a redundant function in cytosine methylation and gene regulation. To study the precise effects on genome-wide DNA methylation, we generated single-nucleotide resolution maps of cytosine methylation in seedling leaf tissues of osddm1a, osddm1b, and the double mutants by bisulfite sequencing (BS-seq; Supplemental Table S2). Because the rice T-DNA mutants were obtained after callus culture, in this work we used callus culture-regenerated wild-type plants as a control. Compared with the wild type, methylation at CG, CHG, and CHH sites in osddm1a was reduced by 5.5%, 14%, and 18.9%, respectively, while that in osddm1b was reduced by 8.3%, 26%, and 32%, respectively. The double mutation caused 44.9%, 73.5%, and 49% reductions of CG, CHG, and CHH methylation, respectively (Table I). More dramatic losses of CHG and CG than CHH methylation were found in the double mutant, suggesting that the two rice DDM1 orthologs functioned redundantly to mostly regulate CHG and CG methylation. However, it is possible that deregulation of methylation-related genes or disrupted methylation homeostasis might account for the divergence in CHG/CG and CHH methylation changes between single and double mutants.
Figure 1.
Effects of the osddm1a/1b and osdrm2 mutations on plant growth and genome-wide cytosine methylation. A, The osddm1a/1b and osdrm2 mutations severely reduced plant height. Plants at seedling and mature stages are shown. The osdrm2 osddm1a/1b triple mutants were seedling lethal. B, Heat map of methylation levels at CG, CHG, and CHH sites in chromosomes 1 and 4 of the wild type (WT), osddm1a/1b, and osdrm2. Average methylation levels per 1-kb bin were calculated and are shown as red bars. The distribution of histones H3K9me2 and H3K27me3 and small interfering RNA (siRNAs) identified in this work are shown along the chromosomes. Genes and TEs are indicated. The heterochromatin (dark blue) and euchromatin (light blue) regions of the chromosomes are drawn according to Cheng et al. (2001). C, Effects of osdrm2 and osddm1a/1b mutations on cytosine methylation at different levels. Genome-wide cytosine methylation is divided into five intervals (from lowest [0%–20%] to highest [80% or greater], represented by the indicated colors). The percentage of each interval (detected in the wild type and the mutants) is shown on the y axis. D, Density plots of differential methylation levels at CG (blue), CHG (green), and CHH (red) sites in osddm1 (left) and osdrm2 (right) compared with the wild type. The y axis shows the percentage of differential methylation levels, and the x axis shows differential methylation levels (from −1 to +1) in the mutants relative to the wild type (0), with the area of each sequence context set as 100%. Only methylation differences for 1-kb windows containing at least five informative sequenced cytosines were considered.
Table I. Cytosine methylation levels in wild-type and mutant plants.
| Genotype | Methylation Levels | Percentage of Decrease | ||||||
|---|---|---|---|---|---|---|---|---|
| C | CG | CHG | CHH | C | CG | CHG | CHH | |
| Wild type | 17.60% | 63.90% | 30.60% | 5.30% | ||||
| ddm1a | 15.90% | 60.40% | 26.30% | 4.30% | 9.66% | 5.48% | 14.05% | 18.87% |
| ddm1b | 14.70% | 58.60% | 22.60% | 3.60% | 16.48% | 8.29% | 26.14% | 32.08% |
| osdrm2 | 12.60% | 57.00% | 23.60% | 0.80% | 28.41% | 10.80% | 22.88% | 84.91% |
| ddm1a/1b | 8.10% | 35.20% | 8.10% | 2.70% | 53.98% | 44.91% | 73.53% | 49.06% |
In rice chromosomes, the majority of heterochromatin is distributed in the pericentromeric regions, with chromosome 4 having a distinct pattern in which the entire left (short) arm is highly heterochromatinized (Cheng et al., 2001; Fig. 1B). We observed that methylation at CG and CHG sites was enriched in heterochromatic regions, whereas CHH methylation was located essentially in euchromatic regions (Fig. 1B). For comparison, we analyzed genome-wide H3K9me2 (a heterochromatic gene mark), H3K27me3 (a euchromatic gene mark; Supplemental Table S3), and small RNAs in the same tissues as for BS-seq of wild-type plants. H3K9me2 was relatively more enriched in the heterochromatic and pericentromeric regions, while more peaks of H3K27me3 were found in euchromatic regions (Fig. 1B). siRNA also was enriched in the euchromatic region in rice, consistent with previous observations in maize (Zea mays; Gent et al., 2014). The analysis revealed that the osddm1a/1b double mutation resulted in predominant losses of CHG and, to a lesser extent, CG methylation, mostly in heterochromatin regions, and clear CHH methylation losses in euchromatic regions of the chromosomes (Fig. 1B), suggesting that OsDDM1 is involved in the methylation of both heterochromatic and euchromatic regions of the genome.
OsDRM2 Is Required for Most of the CHH Methylation in Rice
The rice genome contains three DRM1/2 homologous genes. OsDRM2 (Os03g0110800) is the major DRM1/2-type methyltransferase gene in rice, as OsDRM1a (Os11g0109200) and OsDRM1b (Os12g01018900) are not expressed and/or lack methyltransferase motifs (Moritoh et al., 2012). To study the function of OsDRM2 in DNA methylation, we characterized a homozygous T-DNA insertion mutant of the gene (Supplemental Fig. S1, B and C). This mutant displayed severe pleiotropic growth phenotypes at both the vegetative and reproductive stages, including a dwarfed stature, reduction in tiller number, delayed heading or no heading, abnormal panicle and spikelet morphology, and complete sterility, which are similar to the previously characterized osdrm2 knockout plants (Fig. 1A; Supplemental Fig. S1E; Moritoh et al., 2012). Analysis of the single-nucleotide resolution maps of cytosine methylation of seedling leaves by BS-seq revealed that the osdrm2 mutation greatly reduced CHH methylation (84.9%), but with much weaker effects on CG (10.8%) and CHG (22.9%) methylation (Table I). As in the osddm1a/1b double mutant, losses of CHH methylation in osdrm2 plants were located primarily in euchromatin domains (Fig. 1B), suggesting that OsDRM2 may play an important role in gene regulation.
To study the characteristics of methylated cytosines targeted by OsDDM1 and OsDRM2, we divided the cytosine methylation levels into five intervals (20% each from 0% to 100%) and calculated the percentages of each interval in the wild type and mutants. The analysis revealed that the osddm1a/1b double mutation preferentially reduced the percentage of the highest interval (greater than 80% methylation; Fig. 1C), indicating that OsDDM1 mostly targeted to heavily methylated regions of the genome. By contrast, the osdrm2 mutation mainly reduced the percentages of moderately methylated intervals (20%–40% and 40%–60% methylation; Fig. 1C). To further refine the methylation losses at each sequence context in the mutants, we plotted the percentage (density) of the methylation changes (from −1 to +1) in the mutants relative to the wild type (set at 0) at CG, CHG, and CHH sites. The analysis indicated that the areas of heavy CHG and CG methylation losses (greater than 50%; from −0.5 to −1) were much larger than that of CHH methylation in osddm1a/1b (Fig. 1D). In contrast, in osdrm2, CHH methylation losses were most prevalent (e.g. about 75% at −0.2), while CG methylation loss was not clearly observed (Fig. 1D). This analysis confirmed the above observations that OsDDM1 was mainly required for CHG and CG methylation, whereas OsDRM2 was essentially required for CHH methylation.
Function of OsDDM1 and OsDRM2 in DNA Methylation of Protein-Coding Genes
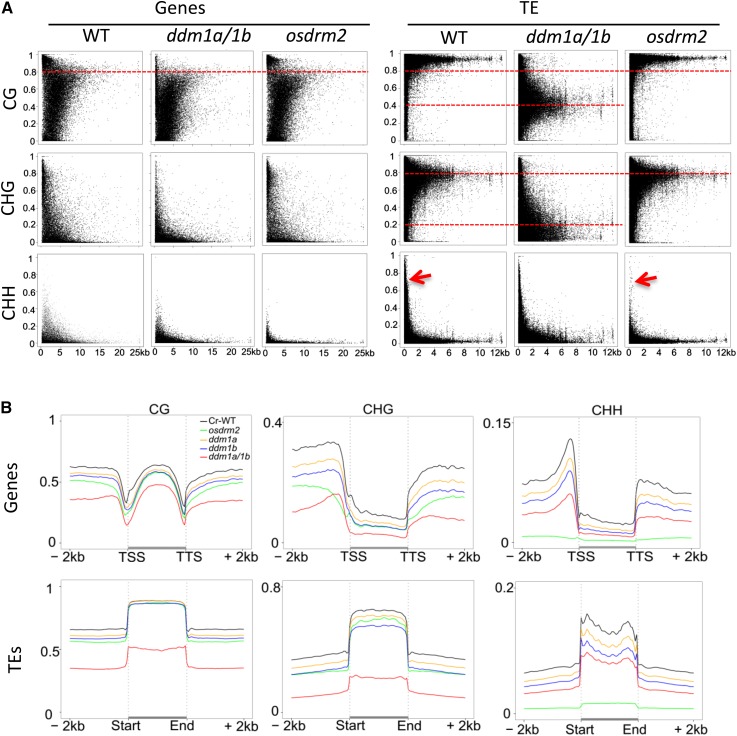
To study the effects of the mutations on methylation levels at different genomic elements, we calculated the percentages of methylated cytosines of each of the rice protein-coding genes, TEGs, and TEs and plotted them according to the length of each element. This analysis indicated that the rice protein-coding genes had lower methylation levels than TEs and TEGs (Fig. 2; Supplemental Fig. S2). Like TEs or TEGs, a small group of protein-coding genes displayed high levels of CG methylation (greater than 80%; Fig. 2A; Supplemental Fig. S2). The osddm1a/1b double mutation eliminated the high level of CG methylation and reduced CHG and CHH methylation in protein-coding genes (Fig. 2A). To analyze the mutation effects on methylation levels at different regions of rice protein-coding genes, we calculated the average methylation levels for every 100-bp interval of each gene and its 2-kb upstream and downstream flank regions in wild-type and mutant plants. The analysis revealed that, in the wild type, CG methylation was at about similar levels between the 5′ and 3′ regions and gene body (except the ends of the body), while non-CG methylation was much higher in the 5′ and 3′ regions than the body (Fig. 2B), confirming previous data (Zemach et al., 2010). In osddm1a/1b, CHG and CHH methylation was reduced mainly from the 5′ and 3′ regions of protein-coding genes (Fig. 2B). By contrast, the osdrm2 mutation largely reduced CHH methylation (Fig. 2A), which was located mostly at the 5′ and 3′ ends of protein-coding genes (Fig. 2B). McrBC digestion and PCR analysis of methylation levels of several genes confirmed the BS-seq data (Supplemental Fig. S3). This analysis indicated that both OsDDM1 and OsDRM2 were mainly involved in methylation of the 5′ and 3′ ends of protein-coding genes.
Figure 2.
Average cytosine methylation levels in all sequence contexts in genes and TEs. A, Average cytosine methylation levels in all sequence contexts plotted against gene (left) and TE (right) sizes. Note that high levels of wild-type (WT) CG methylation (greater than 80%) in genes and TEs were reduced to about 40% on average in osddm1a/1b, and wild-type CHG levels (80% on average) in TEs were reduced to lower levels (about 20% on average) in osddm1a/1b but remained unchanged in osdrm2, which mostly reduced CHH methylation in genes and small TEs (less than 1 kb; arrows). The percentage of methylated cytosines out of total cytosines of each protein-coding gene or TE was plotted in terms of the lengths of the elements. Each dot represents a gene or TE. B, Patterns and genome-wide average levels of cytosine methylation (CG, CHG, and CHH) of genes (top row) and TEs (bottom row) in wild-type and mutant rice plants. Protein-coding genes and TEs with their contiguous 2-kb upstream and downstream flanks were aligned at the 5′ and 3′ ends, and average cytosine methylation levels within each 100-bp interval are plotted.
Function of OsDDM1 in TE and TEG Methylation
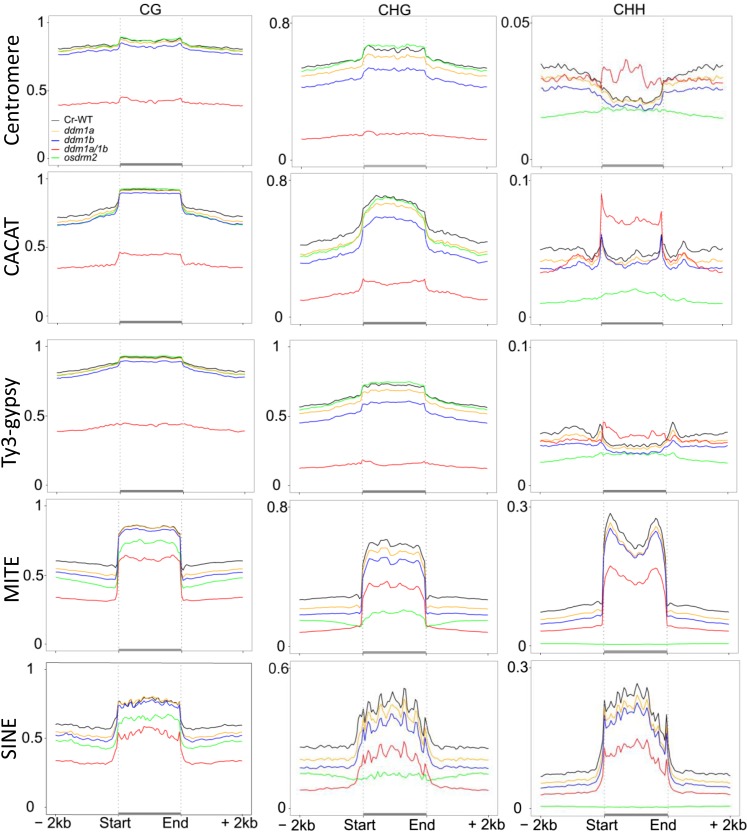
TE and repetitive sequences represent about 40% of the rice genome. In addition, the rice genome contains a large number of TEGs (16,924, compared with 38,622 protein-coding genes; http://rice.plantbiology.msu.edu/). Similar analysis to that with the protein-coding genes revealed that rice TEs and TEGs were highly methylated at CG and CHG sites, but only small TEs (less than 1 kb) showed relatively high levels of CHH methylation (Fig. 2A; Supplemental Fig. S2). In osddm1a/1b plants, CHG and CG methylation in TEs and TEGs were greatly reduced (Fig. 2; Supplemental Fig. S2). However, CG methylation reduction (from an average of more than 80% to about 40%) appeared less important than that of CHG methylation (from about 80% to about or below 20%) of long TEs and TEGs (Fig. 2A; Supplemental Fig. S2). Consistent with the observation in Figure 1B, these data indicated that OsDDM1 was required predominantly for CHG methylation. McrBC digestion and PCR analysis of four TEGs confirmed the BS-seq data (Supplemental Fig. S3). Analysis of average cytosine methylation levels within each 100-bp interval of the different classes of TEs and repeats revealed hyper-CHH methylation of centromere repeats, CACAT DNA elements, and long terminal repeat retrotransposons (Ty3-gypsy) in osddm1a/1b plants (Fig. 3; Supplemental Fig. S4). Hyper-CHH methylated TEs were located mostly in the pericentromeric regions (Supplemental Fig. S5). The ectopic CHH methylation may be caused by an internal balancing mechanism to compensate for the extensive loss of CHG and CG methylation found in heterochromatic TEs due to the loss of OsDDM1 function. Considering that DRM2 or RdDM seems not to be required for CHH methylation of heterochromatic or long TEs in Arabidopsis and maize (Zemach et al., 2013; Li et al., 2014), the ectopic CHH methylation in heterochromatic TEs may be mediated by a CMT2-like activity that was shown to methylate CHH in heterochromatic regions in Arabidopsis (Zemach et al., 2013). However, the osddm1a/1b mutation clearly reduced CHH methylation of small TEs (i.e. MITEs and SINEs; Fig. 3). Thus, OsDDM1 was required mainly for CHG and, to a lesser extent, CG methylation of TEs and TEGs. OsDDM1 also was required for CHH methylation of small TEs, which are located mainly in the euchromatic region (see below).
Figure 3.
Genome-wide average levels of methylation in each sequence context of the indicated TE families and repeats in culture-regenerated wild-type (Cr WT) and mutant plants. TEs with their contiguous 2-kb upstream and downstream flanks were aligned at the 5′ and 3′ ends, and average methylation for all cytosines within each 100-bp interval is plotted. Note the hyper-CHH methylation of centromere repeats, CACAT, and Ty3-gypsy retroelements in osddm1a/1b plants and the nearly complete losses of CHH and CHG methylation of MITEs and short interspersed nuclear elements (SINEs) in ordrm2 plants.
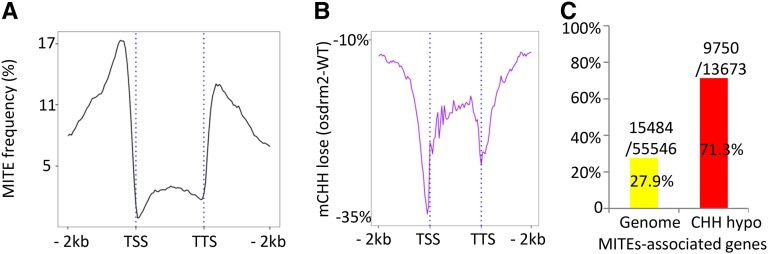
OsDRM2 Is Required for CHH Methylation of Gene-Associated Small TEs
The osdrm2 mutation clearly reduced CHH methylation of TEs, especially small TEs (less than 1 kb), but had little effect on CG or CHG methylation of TEs or TEGs (Fig. 2; Supplemental Fig. S2). When we examined the methylation levels of different classes of TEs, we found that the osdrm2 mutation generally reduced CHH methylation of TEs, but it almost completely eliminated CHH methylation from small TEs such as MITEs and SINEs (Fig. 3; Supplemental Fig. S4). This is consistent with the function of Arabidopsis DRM2 (Tran et al., 2005). Small TEs, especially MITEs, are the most abundant short DNA elements in rice and are dispersed in genic regions (Bureau and Wessler, 1994; Zemach et al., 2010). MITEs are located at high frequencies at the 5′ and 3′ ends of protein-coding genes (Fig. 4A). MITEs are highly methylated at CHH sites in different rice tissues, and MITE distribution closely parallels that of CHH methylation (Zemach et al., 2010). In osdrm2 plants, CHH methylation losses peaked at the 5′ and 3′ ends of protein-coding genes (Fig. 4B). We found that about 28% (15,484 of 55,546) of the total rice genes had MITE sequences detected within the range from 200 bp upstream to 200 bp downstream to the coding regions (Fig. 4C). About 71% (9,750 of 13,673) of the hypo-CHH methylated genes (see below) in osdrm2 were MITE-associated genes (Fig. 4C). Analysis by McrBC and HaeIII digestion coupled with PCR of MITE-associated genes confirmed the BS-seq data (Supplemental Fig. S3). Thus, OsDRM2-dependent CHH methylation mainly targets gene-associated small TEs, thereby affecting the methylation of protein-coding genes.
Figure 4.
The osdrm2 mutation reduces CHH methylation of MITEs that are located close to genes. A, Frequency of the occurrence of MITEs in rice protein-coding genes. The rice protein-coding genes (including the body and the 2-kb upstream and downstream regions) were divided into 120 intervals (x axis) and searched for the presence of MITE sequences at each interval. The y axis shows the percentage of rice genes with the presence of MITEs. B, Distribution of genome-wide average loses of CHH methylation in the rice protein-coding genes in ordrm2. All rice protein-coding genes (including the body and the 2-kb upstream and downstream regions) were divided into 120 intervals (x axis), and CHH methylation losses at each interval in osdrm2 compared with the wild type (WT) are shown as minus percentages (y axis). C, High enrichment of MITE-associated genes in hypo-CHH methylation (71.4%) in osdrm2, compared with 27.9% of MITE-associated genes genome wide. TSS, Transcription start site; TTS: transcription terminal site.
OsDDM1-Dependent CHH Methylation Sites Overlap with That Methylated by OsDRM2
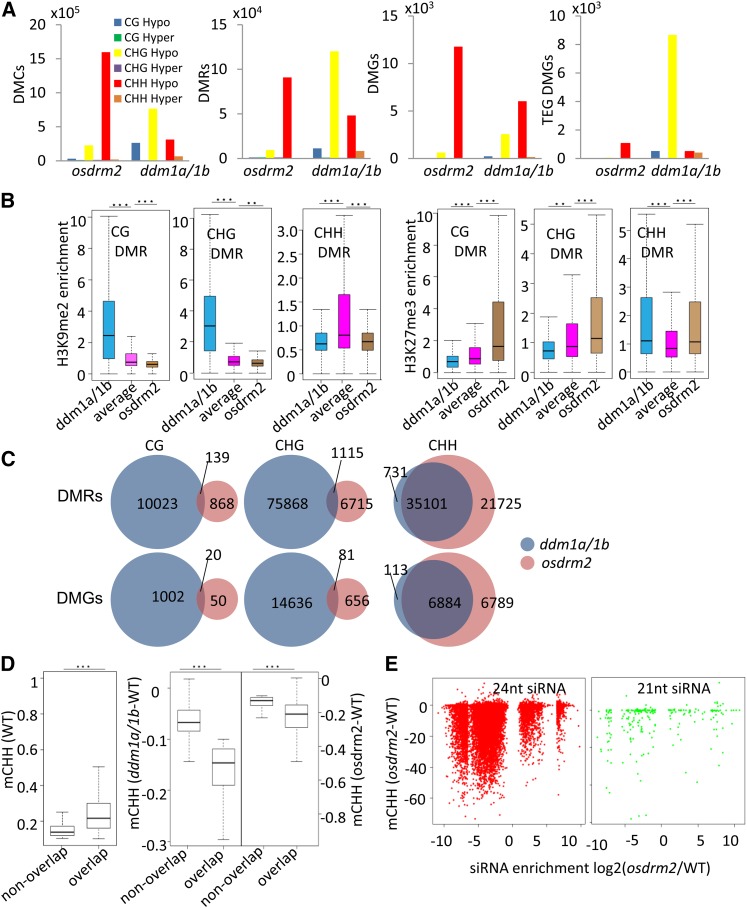
Next, we analyzed differentially methylated cytosines (DMCs), differentially methylated regions (DMRs), and differentially methylated genes (DMGs) in all sequence contexts (see “Materials and Methods”) and found that the ddm1a/1b double mutation mostly produced hypomethylated DMCs or DMRs at CHG and, to a lesser extent, at CHH and CG sites, while the osdrm2 mutation produced hypomethylated DMCs/DMRs essentially at CHH sites (Fig. 5A; Supplemental Table S5). Relatively smaller numbers of hypermethylated CHH DMCs/DMRs also were produced in osddm1a/1b plants (Fig. 5A). In terms of DMGs, the osddm1a/1b mutation produced not only a large number of hypo-CHG methylated TEGs (about 8,600) but also many hypo-CHG (about 2,500) and hypo-CHH (about 6,000) methylated protein-coding genes (Fig. 5A; Supplemental Table S5). The osdrm2 mutation produced about 11,700 hypo-CHH methylated protein-coding genes (Fig. 5A; Supplemental Table S5). These data indicated that OsDDM1 and OsDRM2 controlled the methylation of many functional genes in rice, suggesting that they might play an important role in gene regulation.
Figure 5.
Analysis of DMCs, DMRs, and DMGs detected in osddm1a/1b and osdrm2 mutants. A, Numbers of hypo- and hyper-DMCs/DMRs/DMGs detected in osddm1a/1b and osdrm2. DMGs of protein-coding genes and TEGs are shown. B, Enrichment of H3K9me2 or H3K27me3 of hypo-CG, -CHG, and -CHH DMRs in osddm1a/1b and osdrm2 plants relative to genome-wide averages. Significant enrichments are indicated by double asterisks (P < 0.01) or triple asterisks (P < 0.001, Student’s t test). C, Venn diagrams of hypo-CG, -CHG, and -CHH DMRs and DMGs in osddm1a/1b and osdrm2. D, Box plots of CHH methylation levels of overlapped and nonoverlapped hypo-CHH DMRs in wild-type (WT) plants (left) and CHH methylation losses of overlapped hypo-CHH DMRs and nonoverlapped DMRs in osdrm2 and osddm1a/1b plants (right). Significant differences are indicated by triple asterisks (P < 0.001, Student’s t test). E, CHH methylation level changes (y axis; in minus percentage) of each bin (1 kb) with differential 24- and 21-nucleotide siRNA levels (x axis; false discovery rate < 0.01, Student’s t test) in osdrm2 compared with the wild type.
Hypo-DMRs at CG and CHG sites in osddm1a/1b had lower H3K27me3 but higher H3K9me2 levels than the genome-wide average, while hypo-CHH DMRs in both osddm1a/1b and osdrm2 plants displayed higher H3K27me3 and lower H3K9me2 than the genome-wide average (Fig. 5B). These data were consistent with the above observations that CHH methylation losses in both osddm1a/1a and osdrm2 plants were located essentially in euchromatic regions and that CHG methylation losses in osddm1a/1b were mainly in heterochromatic regions (Fig. 1B). Interestingly, nearly all of the hypo-CHH DMRs in osddm1a/1b were overlapped by those in osdrm2 (Fig. 5C), while there was little overlap of hypo-CG or hypo-CHG DMRs between the two mutants (Fig. 5C). We observed that the wild-type methylation levels of the overlapped regions were higher than the regions affected only in osdrm2 (Fig. 5D). The overlapped regions displayed more significant losses of CHH methylation in both osdrm2 and osddm1a/1b compared with nonoverlapped regions (Fig. 5D). We noted that the decreases of CHH methylation were more important in osdrm2 than in osddm1a/1b (Fig. 5D). Slight decreases of CHH methylation of nonoverlapped regions also could be detected in osddm1a/1b plants (Fig. 5D). These data suggest that OsDDM1 and OsDRM2 may cooperate for high levels of CHH methylation in the rice genome.
Given the intrinsic relationship between DRM2-mediated DNA methylation and small RNAs, we conducted small RNA sequencing (RNA-seq) of osdrm2 plants using the same tissues as for the BS-seq. About 30 million reads for the wild type and 44 million reads for osdrm2 were obtained. In total, 17,932 (15,189 down-regulated and 2,743 up-regulated) differential 24-nucleotide siRNA regions were identified between the wild type and osdrm2, indicating that OsDRM2 was required for the production of a large number of siRNAs. To explore a possible relationship between the regional abundance of siRNAs and hypo-CHH DMRs found in osdrm2, we binned the abundance of 24- and 21-nucleotide siRNAs into 1-kb windows in the genome (as for DMRs) and searched for differences between the wild type and osdrm2. We plotted the differential 24- and 21-nucleotide siRNA regions (false discovery rate < 0.01) that were at the same time ordrm2 hypo-CHH DMRs (P ≤ 0.01) in terms of relative siRNA abundance (x axis) and DNA methylation levels in ordrm2 versus the wild type (y axis; Fig. 5E). We found that a majority of hypo-CHH DMRs showed reduced 24-nucleotide siRNA levels in ordrm2, indicating a positive correlation between the 24-nucleotide siRNA abundance and OsDRM2-mediated CHH methylation.
Effects of the osddm1a/1b and osdrm2 Mutations on Gene Expression
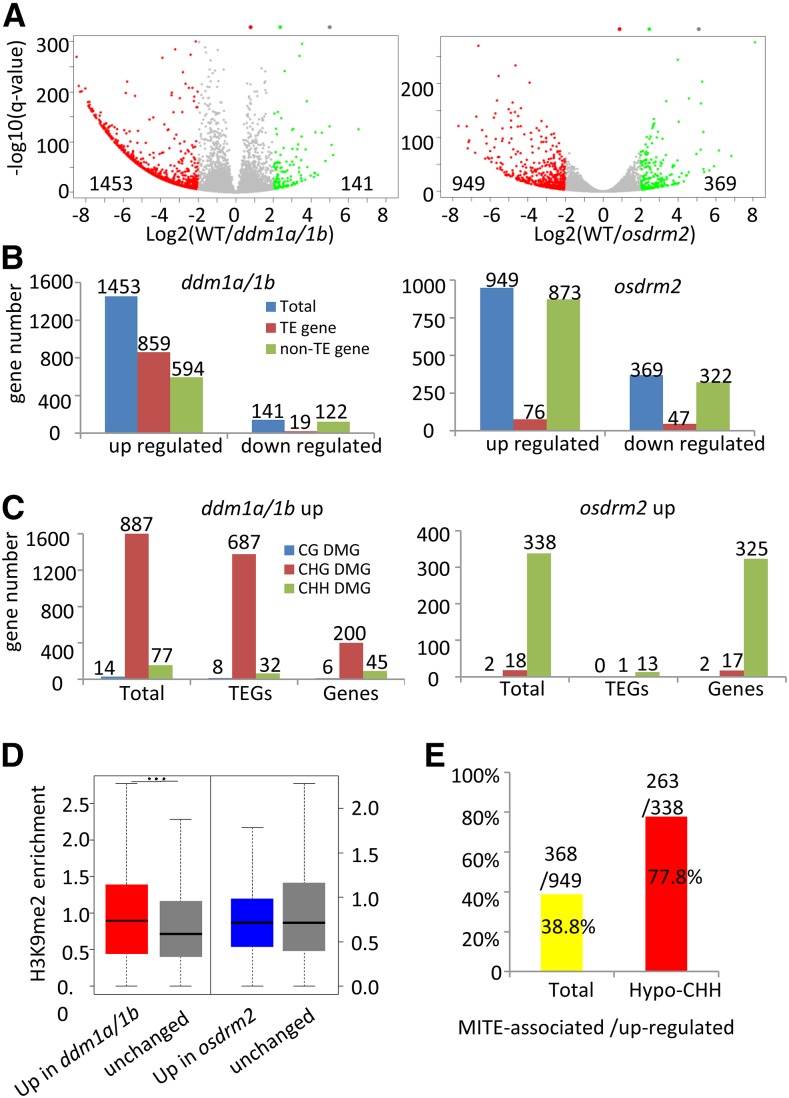
To study the mutation effects of osddm1a/1b and osdrm2 on gene expression, we analyzed the transcriptomes of the same tissues by RNA-seq (Supplemental Table S4). Two biological repeats were analyzed for both the wild type and the mutants. The sequence data displayed a high reproducibility (r2 > 0.99; Supplemental Fig. S6A). Analysis of the data revealed 1,453 up-regulated and 141 down-regulated genes in osddm1a/1b and 949 up-regulated and 369 down-regulated genes in osdrm2 compared with the wild type (greater than 4-fold; P < 0.001; Fig. 6, A and B). The many more up-regulated than down-regulated genes found in the mutants highlighted the repressive function of OsDDM1 and OsDRM2 in gene repression. In osddm1a/1b, about 60% (859 of 1,453) of the up-regulated genes were TEGs (Fig. 6C), among which 80.3% (687 of 859) were hypo-CHG DMGs, compared with about 1% (eight of 859) and 3.7% (32 of 859) of the up-regulated hypo-CG or hypo-CHH methylated TEGs, respectively (Fig. 6C). Quantitative reverse transcription (qRT)-PCR analysis of five TEGs that lost methylation confirmed the data (Supplemental Figs. S3 and S7A). These data suggested that OsDDM1-mediated methylation was required for the repression of TEGs. Moreover, the up-regulated protein-coding genes were more enriched for H3K9me2 than unaffected genes (Fig. 6D). About 34% (200 of 594) of the up-regulated protein-coding genes were hypo-CHG DMGs, compared with only 1% (six of 594) hypo-CG and 8% (45 of 594) hypo-CHH DMGs (Fig. 6C; Supplemental Data Set S1). qRT-PCR analysis of the five protein-coding genes that lost methylation confirmed the data (Supplemental Figs. S3 and S7B). These results indicated that OsDDM1-mediated CHG methylation was required for the repression of TEGs as well as a subset of protein-coding genes that display heterochromatic features.
Figure 6.
Differentially expressed protein-coding genes and TEGs in osddm1a/1b and osdrm2 mutants. A, Total numbers of differentially expressed genes (greater than 4-fold; P < 0.01) in osddm1a/1b and osdrm2 compared with the wild type (WT). B, Numbers of up- and down-regulated protein-coding genes and TEGs in the mutants. C, Numbers of up-regulated protein-coding genes and TEGs that are at the same time hypo-CG, -CHG, and -CHH DMGs in osddm1a/1b and osdrm2. D, Relative enrichment of H3K9me2 of up-regulated genes compared with unaffected genes in osddm1a/1b and osdrm2. Significant enrichments are indicated by triple asterisks (P < 0.001, Student’s t test). E, Most of the up-regulated and hypo-CHH DMGs are associated with MITEs.
By contrast, in osdrm2 plants, most (873 of 949) up-regulated genes were protein-coding genes (Fig. 6B). About 36% (338 of 949) of up-regulated genes were hypo-CHH DMGs (Fig. 6C; Supplemental Data Set S1). There were very few or no up-regulated hypo-CHG or hypo-CG DMGs. qRT-PCR analysis of the four genes that lost CHH methylation in the mutant confirmed the data (Supplemental Figs. S3 and S7C). Most (263 of 338) of the hypo-CHH methylated and up-regulated genes were associated with MITEs (Fig. 6E), supporting the assumption that OsDRM2 targeted MITEs for genic CHH methylation and gene repression.
DISCUSSION
Our data indicate that the two OsDDM1 genes have redundant functions in rice genome-wide DNA methylation, which differs in several respects from that of Arabidopsis DDM1. First, our results show that OsDDM1 plays an essential role in gene regulation and plant growth, as the loss of OsDDM1 function produced very severe growth defects in the first generation, while Arabidopsis ddm1 mutants grew relatively normally in the initial generations and produced developmental abnormalities only after multiple rounds of self-pollination (Kakutani et al., 1996). These gradually developed abnormalities are mainly due to the insertion of derepressed TEs or the rearrangement of repeats during generational propagation (Miura et al., 2001; Singer et al., 2001; Tsukahara et al., 2009; Yi and Richards, 2009), although others are due to DNA methylation changes that affect gene expression (Soppe et al., 2000; Saze and Kakutani, 2007). Maize also has two DDM1 orthologs, Chr101 and Chr106. While single gene loss-of-function alleles are viable, double mutation is likely lethal, as no double mutant could be obtained (Li et al., 2014). Those results together with our data suggest that the loss of DDM1 genes in grasses may have stronger deleterious phenotypic effects than in Arabidopsis.
The developmental defects of osddm1a/1b plants are likely due to the alteration of epigenetic changes of many protein-coding genes. OsDDM1-dependent DNA methylation is likely to be involved directly in the epigenetic repression of functional genes in rice, as many derepressed genes in osddm1a/1b have been shown to be involved in rice development and growth (Supplemental Data Set S1). Among these genes, we checked DNA methylation and transcript levels of OsFIE1, OsMPS, Kala4, and several others (Supplemental Figs. S3 and S7B). OsFIE1 encodes a key component of POLYCOMB REPRESSIVE COMPLEX2 and is repressed in rice vegetative tissues. Loss of DNA methylation from OsFIE1 during tissue culture leads to stable derepression of the gene, which severely affects rice plant growth, leading to a dwarf phenotype (Zhang et al., 2012). Overexpression of OsMPS also resulted in decreased plant stature (Schmidt et al., 2013). Kala4 encodes a basic helix-loop-helix protein whose ectopic expression produces black grains (Oikawa et al., 2015), which also were observed in osddm1a/1b (Supplemental Fig. S1E). The function of OsDDM1 in maintaining the proper expression pattern of a large number of genes may be related at least partly to DNA methylation in genic regions. However, it is not ruled out that some of the phenotypes also may be caused by the derepression of TEs during the production of the double mutants. A relatively high cooccurrence between hypo-CHG DMG and up-regulated genes suggested that OsDDM1-mediated CHG methylation played an important role in developmental gene repression (Fig. 6C). However, only 8% of the up-regulated genes were hypo-CHH DMGs in osddm1a/1b, and only about 20% of the up-regulated genes (greater than 2-fold; P < 0.01) in osddm1a/1b overlapped with those in osdrm2 (Supplemental Fig. S6B). This could be explained by relatively low levels of decreases of CHH methylation in osddm1a/1b (Figs. 1, B and D, and 2).
The Arabidopsis ddm1 mutation mostly reduces methylation in all sequence contexts of heterochromatic repeats, TEs, and TEGs, but it produces ectopic CHG methylation in gene bodies (Lippman et al., 2004; Zemach et al., 2013). However, our data indicate that, among the three sequence contexts, CHG methylation was affected mostly by the osddm1a/1b mutation. Similarly, loss-of-function alleles of one maize DDM1 gene, Chr106, also results in more reduction of non-CG than CG methylation (Li et al., 2014). It is established that CHG methylation and H3K9me2 form a positive feedback loop in Arabidopsis: CHG methylation by CMT3 requires H3K9me2, to which CMT3 binds via its chromo- and bromo-adjacent homology domains (Law and Jacobsen, 2010; Du et al., 2012). H3K9 dimethylation by SUVH proteins requires CHG methylation, to which SUVH proteins bind via the SRA domain (Johnson et al., 2007; Rajakumara et al., 2011). CHG methylation is kept out of genes by a histone demethylase, IBM1, which removes H3K9me2 from gene bodies (Saze and Kakutani, 2011). It seemed that CHG methylation loss in osddm1a/1b plants was not due to up-regulation of the rice homolog of IBM1 (JMJ719) or repression of OsCMT3 or OsCMT2, as their transcript levels were not clearly altered in the mutant (Supplemental Fig. S6D). The more important role of OsDDM1 in heterochromatic CHG methylation suggests that OsDDM1-mediated chromatin remodeling is essential to facilitate the OsCMT3-mediated CHG methylation of heterochromatic regions of the rice genome (Cheng et al., 2015).
In Arabidopsis, RdDM operates primarily through DRM2 and is responsible for a relatively minor fraction of genomic CHH methylation (Wierzbicki et al., 2012; Zemach et al., 2013), and CMT2 mediates most CHH methylation, separately from the RdDM pathway (Zemach et al., 2013). It has been shown that Arabidopsis DDM1 facilitates CMT2-mediated CHH methylation in body regions of long (or heterochromatic) TEs, while RdDM-dependent CHH methylation mostly targets short TEs and TE edges (Zemach et al., 2013). Our data here suggest that OsDDM1 may be involved in OsDRM2-mediated CHH methylation at genic regions (Fig. 5C). OsDDM1 may facilitate OsDRM2 or RdDM-mediated CHH methylation in the rice genome by the remodeling of dispersed small heterochromatin in genic regions by displacing histone H1 (Zemach et al., 2013). However, it is possible that the loss of CHH methylation is coordinated with that of CHG in osddm1a/1b, as observed in Arabidopsis and maize (Li et al., 2014; Stroud et al., 2014).
Our data indicate that, in rice, CHH methylation was located essentially in the euchromatic regions of the genome (Fig. 1B), in agreement with the previous observations that CHH peaks are absent in the rice pericentromeric regions, in contrast to the more localized pattern of repeats close to centromeres in Arabidopsis (Feng et al., 2010). It is believed that this might reflect the fact that repeats are widely spread across the rice genome (Feng et al., 2010). The massive loss of CHH methylation (about 85%; Table I) in ordrm2 plants suggests that the OsDRM2-mediated RdDM pathway in rice may play a major role for CHH methylation. Our results indicate that, in fact, rice CHH methylation essentially targets small TEs such as MITEs that are largely scattered in euchromatic regions, most at the 5′ and 3′ ends of genes. MITE-associated genes that have significantly lower level expression than genes away from MITEs (Lu et al., 2012) are likely to be repressed by OsDRM2-mediated CHH methylation, as a large portion of up-regulated genes were MITE-associated hypo-CHH DMGs in osdrm2 (Fig. 6). This suggests an important role of OsDRM2 and RdDM in gene regulation in rice. Many up-regulated DMGs were important rice developmental genes, such as OsGA2ox5 and Os08g19420 (Supplemental Data Set S1). The overexpression of OsGA2ox5 produces a severe dwarf phenotype (Shan et al., 2014), and plants with increased expression of Os08g19420 showed exaggerated flag leaf angle at the heading stage (Wei et al., 2014). These phenotypes were observed in osdrm2 (Fig. 1A; Supplemental Fig. S1F). Interestingly, OsMPS, hypo-CHH methylated and up-regulated in osddm1a/1b, also was hypo-CHH methylated and up-regulated in osdrm2 (Supplemental Figs. S3 and S6, B and C). RdDM is a DNA methylation process triggered by double-stranded RNA from which siRNAs are generated and used by the RNA interference machinery to guide DRM2-mediated DNA methylation. Conversely, the production of siRNA also depends on DRM2 (Zilberman et al., 2004; Henderson et al., 2010). The correlation between the decrease of 24-nucleotide siRNA abundance and the loss of CHH methylation in osdrm2 suggests that OsDRM2 is required for siRNA production in the RdDM pathway that regulates many protein-coding genes in rice. This hypothesis is supported by hundreds of up-regulated genes and the severe phenotypes of ordrm2 mutants and those of other rice RdDM genes, such OsDCL3a, OsDCL4, and OsRDR6 (Wu et al., 2010; Song et al., 2012a, 2012b; Arikit et al., 2013). By comparison, mutants of the Arabidopsis homologous genes display no or only subtle phenotypes (Cao and Jacobsen, 2002), whereas no double mutants of the maize DRM2 orthologs Zmet3 and Zmet7 could be recovered (Li et al., 2014). Likely, DNA methylation mediated by DRM2-like proteins in grasses is crucial for normal plant growth and viability.
These specific features of OsDDM1 and OsDRM2 in DNA methylation and epigenetic regulation of gene expression, together with both the conserved and specific functions of OsMET1 in DNA methylation (Hu et al., 2014), indicate that the precise functions of DNA methylation regulators and the identities of rice pathways that mediate the bulk of methylation of the rice genome differ from those in Arabidopsis. The distinct feature of the rice DNA methylation pattern and the unique roles of OsDDM1 and OsDRM2 may be related to the differences in genome and chromatin organization of rice and Arabidopsis. The majority of CHH methylation locates in euchromatic regions in rice (Fig. 1B), whereas most CHH methylation is in the pericentromeric heterochromatin in Arabidopsis (Lister et al., 2008; Stroud et al., 2014). This is likely due to the fact that heterochromatic regions are relatively dispersed across chromosomes in rice but are much more focused in pericentromeric regions in Arabidopsis.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa L. ssp. japonica ‘DongJin’) plants were used in this study. T-DNA insertion lines of osddm1a (PFG_3A-51065), osddm1b (PFG_2B-60109), and osdrm2 (PFG_3A-04110) were obtained from the Postech rice mutant database (http://www.postech.ac.kr/life/pfg/risd/). The homozygous mutants of osddm1a, osddm1b, osdrm2, osddm1a/1b double mutant, osddm1a/1b/osdrm2 triple mutant, and callus culture-regenerated wild type were germinated and grown on hormone-free, one-half-strength Murashige and Skoog medium under 16/8 h of light/dark at 30°C/25°C or in the field. Twelve-day-old seedling leaves from each genotype were harvested for chromatin, genomic DNA, or total RNA extraction.
Genotyping of osddm1a, osddm1b, and osdrm2 Plants
The insertion of osddm1a was confirmed by PCR using the primers 3A-51065-P1 and 3A-51065-P2 and the T-DNA-specific primer P3, the osddm1b mutant by the primers 2B-60109-P1 and 2B-60109-P2 and the T-DNA-specific primer P3, and the osdrm2 mutant by the primers 3A-04110-P1 and 3A-04110-P2 and the T-DNA-specific primer P4. The primers used for genotyping and reverse transcription-PCR analysis are listed in Supplemental Data Set S2.
Semiquantitative Reverse Transcription-PCR and qRT-PCR Analysis
Total RNA was isolated from 12-d-old seedling leaves using TRIzol reagent (Invitrogen/Life Technologies). Four micrograms of treated total RNA was used to synthesize first-strand complementary DNA with a reverse transcription kit (Promega). The rice Actin gene was used as an internal control.
Methylation-Sensitive Endonuclease Digestion
Genomic DNA (1 μg) isolated from 12-d-old seedlings was digested with 40 units of the methylation-sensitive restriction enzyme McrBC (New England Biolabs; M0272L) for 6 h, which cuts methylated DNA, followed by PCR with specific primer pairs (Supplemental Data Set S2).
Enzymatic Hydrolysis of DNA and Liquid Chromatography-Mass Spectrometry Analysis
Enzymatic hydrolysis of DNA and liquid chromatography-mass spectrometry analysis were conducted according to protocols described previously, with slight modifications (Friso et al., 2002). DNA was extracted from mature leaves using the DNeasy plant mini kit (Qiagen). About 0.5 µg of DNA was denatured by heating at 100°C for 3 min and subsequently chilled in ice slush. One-tenth volume of 0.1 m ammonium acetate (pH 5.3) and 2 units of benzonase P1 (Sigma) were added and incubated at 37°C for 2 h. To the solution were subsequently added one-tenth volume of 1 m ammonium bicarbonate (Sigma) and 0.002 units of venom phosphodiesterase I (Sigma). The incubation was continued for an additional 2 h at 37°C. Thereafter, the mixture was incubated for 1 h at 37°C with 0.5 units of alkaline phosphatase (New England Biolabs). After digestion, 0.4 mL of ultrapure water filtered through a 0.22-μm filter for liquid chromatography-mass spectrometry analysis was added. Nucleoside standards including deoxycytidine and 5-methyldeoxycytidine were used to determine the peak position of each nucleoside.
Chromatin Immunoprecipitation
Chromatin was isolated from 2 g of rice seedling leaves. After sonication, chromation fragments were incubated with antibody (anti-H3K9me2 or anti-H3K27me3)-coated beads (Invitrogen/Life Technologies; 10001D) overnight. After wash and elution, products were reverse cross-linked. Then, the products were treated with protease K (Takara; 9034). Anti-H3K9me2 (ab1220; Abcam) and anti-H3K27me3 (A2363; ABclonal) antibodies were used.
Whole-Genome BS-Seq Library Construction and Sequencing
For genomic BS-seq, the total genomic DNA of callus culture-regenerated wild-type and mutant plants was extracted from 12-d-old seedling leaves using the DNeasy plant mini kit (Qiagen). The library was prepared and sequenced at the Novogene Bioinformatics Institute on an Illumina HiSeq 2500 platform.
BS-Seq Data Processing
Quality control, mapping, and processing of BS-seq reads were performed as follows. Briefly, low-quality reads were removed from raw data by Trimmomatic (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic/Trimmomatic-Src-0.35.zip), and then the clean data were mapped to the MSU7.0 rice reference genome (ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_7.0/all.dir/) by BatMeth (Lim et al., 2012). Only uniquely mapped reads were retained for further analysis. To calculate the methylation density, we first counted the total number of nucleotides C and T that overlap with each genomic cytosine site across the whole genome. The methylation level for each cytosine site was calculated by the sequencing depth of this site divided by the number of unconverted cytosines.
Identification of DMCs, DMRs, and DMGs
The DMCs between any two genotypes were defined by a binomial test (methylSig; Park et al., 2014). Only cytosine sites whose adjusted q values were 0.01 or less and had DNA methylation differences of 0.7, 0.5, or 0.1 between any two genotypes were designated for DMCs (Stroud et al., 2013). The DMRs were discriminated by comparison of the methylation levels of 1-kb windows/regions throughout the genome between any two genotypes. For the identification of DMGs, either the methylation level of the gene body or 2-kb upstream flanks (promoter) of the gene has a difference (calculated the same as DMCs).
mRNA Sequencing and Small RNA-Seq and Analysis
Total RNA was extracted from 12-d-old seedlings using TRIzol reagent (Invitrogen). RNA-seq libraries were constructed from total RNA isolated using TRIzol reagent (Invitrogen/Life Technologies) from leaf tissues. Total RNA (10 μg) for each sample was used to purify poly(A) mRNA; this mRNA was used for the synthesis and amplification of complementary DNA. The RNA-seq libraries were prepared using the TruSeq RNA Sample Preparation Kit from Illumina. Libraries were sequenced on an Illumina HiSeq 2000.
Small RNA-seq libraries were constructed from total RNAs isolated from the same tissues as for the mRNA libraries, using the TruSeq Small RNA Sample Preparation Kit from Illumina. The libraries were sequenced on the same Illumina HiSeq 2000 as the mRNA sequencing libraries.
RNA-seq data were first cleaned by removing contaminations and low-quality reads by Trimmomatic (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic/Trimmomatic-Src-0.35.zip). The clean data were mapped against the MSU7.0 rice genome with corresponding annotation by Tophat2.0 (Kim et al., 2013; http://tophat.cbcb.umd.edu/) using default parameters. Then, the gene counts were calculated with htseq-count, and the differentially expressed genes (log [fold change] ≥ 2, q < 0.05) were calculated by DESeq2.
Small RNA sequence raw data were cleaned by removing adaptor contamination and low-quality reads by Cutadapt (Martin, 2011). MicroRNA (http://www.mirbase.org/ftp.shtml), transfer RNA (http://rice.plantbiology.msu.edu/analyses_search_tRNA.shtml), ribosomal RNA (Rice Genome Annotation MSU7.0), small nucleolar RNA, and small nuclear RNA (http://www.arb-silva.de/no_cache/download/archive/release_111/Exports/) also were removed. Clean reads were aligned to the rice MSU7.0 reference genome by Bowtie (Langmead et al., 2009), allowing zero mismatches and unique mapping. Two types of small RNA abundance were calculated by 21- or 24-nucleotide read numbers falling into 1-kb windows/regions throughout the whole genomes, and windows containing more than three reads were tested by edgeR (Robinson et al., 2010) between samples. Windows with corrected P ≤ 0.01 were defined as differential small RNA regions.
Chromatin Immunoprecipitation Sequencing and Analysis
DNA from chromatin immunoprecipitation was used to construct sequencing libraries following the protocol provided by the Illumina TruSeq ChIP Sample Prep Set A and sequenced by HiSeq 2000.
Trimmomatic (version 0.32) was used to filter out low-quality reads and crop all reads to a uniform length (45 bp). Clean reads were mapped to the rice genome (Rice Genome Annotation Project version 7) by default, allowing up to two mismatches using Bowtie2 (version 2.1.0).
The sequence data and analyses generated in this work have been submitted to the National Center for Biotechnology Information databases under the accession number GSE81436.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of expression and T-DNA insertion mutants of OsDDM1a, OsDDM1b, and OsDRM2.
Supplemental Figure S2. Cytosine methylation average levels in all sequence contexts in TEGs.
Supplemental Figure S3. Genome browser and validation of DNA methylation loss in the mutants.
Supplemental Figure S4. Patterns of cytosine methylation (CG, CHG, and CHH) of additional TEs and repeats in the wild type and osddm1a, osddm1b, osddm1a/1b, and osdrm2 mutants.
Supplemental Figure S5. Heterochromatin localization of hyper-CHH methylated TEs in osddm1a/1b plants.
Supplemental Figure S6. Transcriptomic analysis.
Supplemental Figure S7. Transcript levels (relative to Actin1) tested by qRT-PCR.
Supplemental Table S1. Cytosine methylation levels in wild-type and mutant rice leaves measured by mass spectrometry.
Supplemental Table S2. BS‐seq data of the wild type and mutants (two repeats per sample).
Supplemental Table S3. H3K27me3 and H3K9me2 chromatin immunoprecipitation sequencing data.
Supplemental Table S4. RNA‐seq data.
Supplemental Table S5. Numbers of hypo- or hyper-DMCs, DMRs, and DMGs in CG, CHG, and CHH contexts in the mutants.
Supplemental Data Set S1. Differentially methylated and differentially expressed genes in osddm1 and ordrm2 mutants.
Supplemental Data Set S2. Sequences of primers used in this study.
Supplementary Material
Acknowledgments
We thank Dengnian Liu for the initial work in OsDDM1 and Dr. Qinghua Zhang for sequencing using Illumina HiSeq 2000.
Glossary
- TE
transposable element
- TEG
transposable element-related gene
- RdDM
RNA-directed DNA methylation
- MITE
inverted-repeat transposable element
- T-DNA
transfer DNA
- BS-seq
bisulfite sequencing
- siRNA
small interfering RNA
- SINEs
short interspersed nuclear element
- DMCs
differentially methylated cytosine
- DMRs
differentially methylated region
- DMGs
differentially methylated gene
- RNA-seq
RNA sequencing
- qRT
quantitative reverse transcription
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31371241 and 31571256), the National High Technology Research and Development Program of China (grant no. 2014AA10A600), the French Agence Nationale de la Recherche (Nerdpath), and the Huazhong Agricultural University Scientific and Technological Self-Innovation Foundation (grant no. 2016RC003).
References
- Arikit S, Zhai J, Meyers BC (2013) Biogenesis and function of rice small RNAs from non-coding RNA precursors. Curr Opin Plant Biol 16: 170–179 [DOI] [PubMed] [Google Scholar]
- Brzeski J, Jerzmanowski A (2003) Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem 278: 823–828 [DOI] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR (1994) Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc Natl Acad Sci USA 91: 1411–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002) Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12: 1138–1144 [DOI] [PubMed] [Google Scholar]
- Chen X, Zhou DX (2013) Rice epigenomics and epigenetics: challenges and opportunities. Curr Opin Plant Biol 16: 164–169 [DOI] [PubMed] [Google Scholar]
- Cheng C, Tarutani Y, Miyao A, Ito T, Yamazaki M, Sakai H, Fukai E, Hirochika H (2015) Loss of function mutations in the rice chromomethylase OsCMT3a cause a burst of transposition. Plant J 83: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Buell CR, Wing RA, Gu M, Jiang J (2001) Toward a cytological characterization of the rice genome. Genome Res 11: 2133–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. (2012) Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Wessler SR (2002) Mariner-like transposases are widespread and diverse in flowering plants. Proc Natl Acad Sci USA 99: 280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso S, Choi SW, Dolnikowski GG, Selhub J (2002) A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem 74: 4526–4531 [DOI] [PubMed] [Google Scholar]
- Gent JI, Madzima TF, Bader R, Kent MR, Zhang X, Stam M, McGinnis KM, Dawe RK (2014) Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26: 4903–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F, et al. (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Deleris A, Wong W, Zhong X, Chin HG, Horwitz GA, Kelly KA, Pradhan S, Jacobsen SE (2010) The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet 6: e1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo H, Tahir M, Takashima K, Miura A, Watanabe K, Tagiri A, Ugaki M, Ishikawa R, Eiguchi M, Kurata N, et al. (2012) DDM1 (decrease in DNA methylation) genes in rice (Oryza sativa). Mol Genet Genomics 287: 785–792 [DOI] [PubMed] [Google Scholar]
- Hu L, Li N, Xu C, Zhong S, Lin X, Yang J, Zhou T, Yuliang A, Wu Y, Chen YR, et al. (2014) Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc Natl Acad Sci USA 111: 10642–10647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 22: 94–97 [DOI] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE (2007) The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ (1996) Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA 93: 12406–12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Eichten SR, Hermanson PJ, Zaunbrecher VM, Song J, Wendt J, Rosenbaum H, Madzima TF, Sloan AE, Huang J, et al. (2014) Genetic perturbation of the maize methylome. Plant Cell 26: 4602–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu J, Hu F, Ge S, Ye M, Xiang H, Zhang G, Zheng X, Zhang H, Zhang S, et al. (2012) Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics 13: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JQ, Tennakoon C, Li G, Wong E, Ruan Y, Wei CL, Sung WK (2012) BatMeth: improved mapper for bisulfite sequencing reads on DNA methylation. Genome Biol 13: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476 [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhou S, Wang W, Ye Y, Zhao Y, Xu Q, Zhou C, Tan F, Cheng S, Zhou DX (2015) Regulation of histone methylation and reprogramming of gene expression in the rice inflorescence meristem. Plant Cell 27: 1428–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Chen J, Zhang Y, Hu Q, Su W, Kuang H (2012) Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol 29: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12 [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T (2001) Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214 [DOI] [PubMed] [Google Scholar]
- Moritoh S, Eun CH, Ono A, Asao H, Okano Y, Yamaguchi K, Shimatani Z, Koizumi A, Terada R (2012) Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J 71: 85–98 [DOI] [PubMed] [Google Scholar]
- Oikawa T, Maeda H, Oguchi T, Yamaguchi T, Tanabe N, Ebana K, Yano M, Ebitani T, Izawa T (2015) The birth of a black rice gene and its local spread by introgression. Plant Cell 27: 2401–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Figueroa ME, Rozek LS, Sartor MA (2014) MethylSig: a whole genome DNA methylation analysis pipeline. Bioinformatics 30: 2414–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE (2011) A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev 25: 137–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Kakutani T (2007) Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J 26: 3641–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Kakutani T (2011) Differentiation of epigenetic modifications between transposons and genes. Curr Opin Plant Biol 14: 81–87 [DOI] [PubMed] [Google Scholar]
- Schmidt R, Schippers JH, Mieulet D, Obata T, Fernie AR, Guiderdoni E, Mueller-Roeber B (2013) MULTIPASS, a rice R2R3-type MYB transcription factor, regulates adaptive growth by integrating multiple hormonal pathways. Plant J 76: 258–273 [DOI] [PubMed] [Google Scholar]
- Shan C, Mei Z, Duan J, Chen H, Feng H, Cai W (2014) OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS ONE 9: e87110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, Yordan C, Martienssen RA (2001) Robertson’s Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev 15: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Li P, Zhai J, Zhou M, Ma L, Liu B, Jeong DH, Nakano M, Cao S, Liu C, et al. (2012a) Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J 69: 462–474 [DOI] [PubMed] [Google Scholar]
- Song X, Wang D, Ma L, Chen Z, Li P, Cui X, Liu C, Cao S, Chu C, Tao Y, et al. (2012b) Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J 71: 378–389 [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802 [DOI] [PubMed] [Google Scholar]
- Stroud H, Ding B, Simon SA, Feng S, Bellizzi M, Pellegrini M, Wang GL, Meyers BC, Jacobsen SE (2013) Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife 2: e00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xi S, Briones V, Muegge K (2010) Lsh mediated RNA polymerase II stalling at HoxC6 and HoxC8 involves DNA methylation. PLoS ONE 5: e9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran RK, Zilberman D, de Bustos C, Ditt RF, Henikoff JG, Lindroth AM, Delrow J, Boyle T, Kwong S, Bryson TD, et al. (2005) Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol 6: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T (2009) Bursts of retrotransposition reproduced in Arabidopsis. Nature 461: 423–426 [DOI] [PubMed] [Google Scholar]
- Wei L, Gu L, Song X, Cui X, Lu Z, Zhou M, Wang L, Hu F, Zhai J, Meyers BC, et al. (2014) Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc Natl Acad Sci USA 111: 3877–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS (2012) Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev 26: 1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y (2010) DNA methylation mediated by a microRNA pathway. Mol Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Yi H, Richards EJ (2009) Gene duplication and hypermutation of the pathogen resistance gene SNC1 in the Arabidopsis bal variant. Genetics 183: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D (2010) Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA 107: 18729–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cheng Z, Qin R, Qiu Y, Wang JL, Cui X, Gu L, Zhang X, Guo X, Wang D, et al. (2012) Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24: 4407–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE (2004) Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol 14: 1214–1220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.