Floral symmetry regulators play antagonistic roles in establishing two distinct floral symmetries within a capitulum in Asteraceae.
Abstract
All members of Asteraceae, the largest flowering family, have a unique compressed inflorescence known as a capitulum, which resembles a solitary flower. The capitulum often consists of bilateral (zygomorphic) ray florets and radial (actinomorphic) disc florets. In Antirrhinum majus, floral zygomorphy is established by the interplay between dorsal petal identity genes, CYCLOIDEA (CYC) and RADIALIS (RAD), and a ventral gene DIVARICATA (DIV). To investigate the role of CYC, RAD, and DIV in the development of ray and disc florets within a capitulum, we isolated homologs of these genes from an Asteraceae species, Senecio vulgaris (common groundsel). After initial uniform expression of RAY3 (CYC), SvRAD, and SvDIV1B in ray florets only, RAY3 and SvRAD were exclusively expressed in the ventral petals of the ray florets. Our functional analysis further showed that RAY3 promotes and SvDIV1B represses petal growth, confirming their roles in floral zygomorphy. Our results highlight that while floral symmetry genes such as RAY3 and SvDIV1B appear to have a conserved role in petal growth in both Senecio and Antirrhinum, the regulatory relationships and expression domains are divergent, allowing ventral petal elongation in Senecio versus dorsal petal elongation in Antirrhinum. In S. vulgaris, diversification of CYC genes has led to novel interactions; SvDIV1B inhibits RAY3 and SvRAD, and may activate RAY2. This highlights how recruitment of floral symmetry regulators into dynamic networks was crucial for creating a complex and elaborate structure such as the capitulum.
Floral symmetry plays an essential role in pollination efficiency and, thus, in flower evolution. Based on floral symmetry, flowers can be classified as zygomorphic or actinomorphic. Zygomorphic flowers have a single plane of symmetry (bilateral), whereas actinomorphic flowers have multiple planes of symmetry (radial; Endress, 1999). Floral zygomorphy has independently evolved multiple times within the angiosperm lineage (Citerne et al., 2010). It has been proposed that zygomorphic flowers first arose from actinomorphic flowers around 70 million years ago, which coincided with the species radiation of pollinators. This suggests that flower-pollinator interactions may be a major driver in the evolution of floral zygomorphy (Crepet, 1996).
In Antirrhinum majus (Lamiales), the key genetic regulators of floral zygomorphy have been identified. The TCP transcription factors CYCLOIDEA (CYC) and DICHOTOMA (DICH) have partially redundant functions and determine the dorsal region of the flower by differentially regulating the rate of cell growth in the developing floral organs (Luo et al., 1999, 1996; Zhang et al., 2010; Preston and Hileman, 2009). These genes belong to the CYC2 clade (Howarth and Donoghue, 2006) and are expressed in the dorsal petals of flowers (Almeida et al., 1997; Luo et al., 1999, 1996). The MYB-domain transcription factors RADIALIS (RAD; Corley et al., 2005) and DIVARICATA (Almeida and Galego, 2005; Almeida et al., 1997) also participate in regulating floral symmetry. RAD is expressed dorsally and is positively regulated by CYC (Costa et al., 2005), and rad mutants, in similarity to cyc mutants, form partially ventralized flowers (Luo et al., 1996). The DIV gene promotes ventral petal identity and loss of DIV function leads to lateralized ventral petals (Almeida et al., 1997). RAD acts antagonistically to DIV (Corley et al., 2005), repressing its activity in the dorsal petals and causing DIV activity to be restricted to the ventral and lateral petals (Hileman, 2014; Raimundo et al., 2013).
In the evolution of floral zygomorphy, CYC activity has been independently recruited in many species. While CYC orthologs maintained a conserved role in controlling petal growth, changes in their expression domains were crucial for the shift between zygomorphic and actinomorphic flowers (Busch and Zachgo, 2007; Kim et al., 2008; Zhang et al., 2010; Howarth et al., 2011; Hileman, 2014; Feng et al., 2006; Wang et al., 2008; Zhong and Kellogg, 2015a, 2015b). For example, a gain of dorsal-specific CYC expression caused floral zygomorphy in the Malpighiaceae family (Zhang et al., 2010), whereas a uniform CYC expression in all petals resulted in a regain of floral actinomorphy in a legume species (Citerne et al., 2006). While the importance of CYC genes in the evolution of floral zygomorphy has been studied extensively in many species, evidence of RAD and DIV involvement is very limited. Recently, it has been proposed that the reversion to actinomorphic flowers in Plantago lanceolata is due to a loss of RAD from the genome (Reardon et al., 2014). In Bournea leiophylla (Gesneriaceae), loss of dorsal expression of BlCYC and BlRAD during the later stages of flower development led to up-regulation of BlDIV in the entire flower, which in turn generated a final actinomorphic structure (Zhou et al., 2008). In Lamiales, the RAD expression domain plays an important role in flower zygomorphy: The basal actinomorphic flowers in Lamiales had uniform RAD expression, whereas zygomorphic flowers in the clade showed dorsal/lateral RAD expression (Zhong and Kellogg, 2015b). In Dipsacales, RAD genes are duplicated (RAD1, RAD2, and RAD3 groups) and only a subset of RAD genes appears to be expressed in the dorsal regions (Boyden et al., 2012). Furthermore, it has been shown that in the Dipsacales species, Heptacodium miconioides, DIV-like genes have also been duplicated and are differentially expressed among petals within a flower, suggesting HmDIV genes are vital for floral zygomorphy in this species (Howarth and Donoghue, 2009). Although these studies imply that RAD and DIV are involved in zygomorphy evolution, their suggested roles require further validation by functional analyses.
In the sunflower family (Asteraceae), the evolution of floral zygomorphy is more complex. Asteraceae species are characterized by having a capitulum, which is a compressed inflorescence consisting of two types of flowers: ray and disc florets. In the capitulum, disc florets are positioned in the center, surrounded by ray florets at the margin (Fig. 1, A–D). A disc floret has five evenly sized petals with radial symmetry (actinomorphic; Fig. 1G), while a ray floret has bilateral symmetry (zygomorphic; Fig. 1F) with three fused and elongated ventral petals and two reduced dorsal petals (Trow, 1912). In addition to controlling zygomorphy in solitary flowers such as Antirrhinum, the floral symmetry regulator CYC has acquired a novel role in determining floret identity (disc versus ray) within the capitulum (Kim et al., 2008; Broholm et al., 2008; Chapman et al., 2008). Much of this work has focused on the three model Asteraceae species: Senecio vulgaris (common groundsel), Helianthus annuus (sunflower), and Gerbera hybrida (gerbera; Kim et al., 2008; Broholm et al., 2008; Chapman et al., 2008).
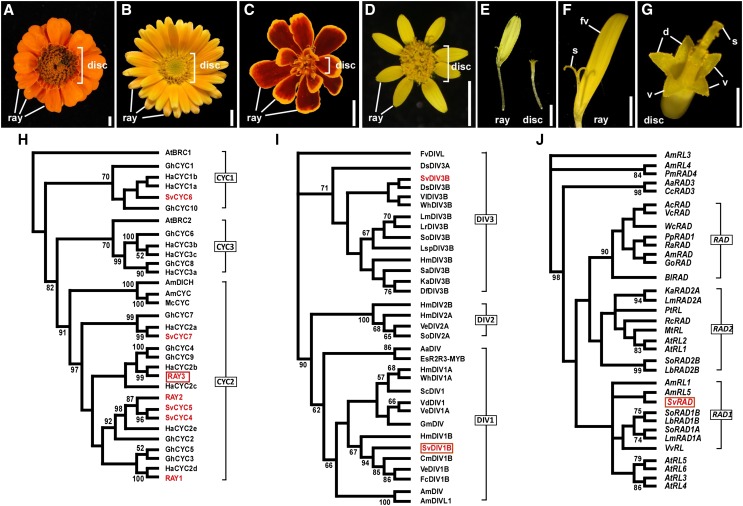
Figure 1.
Capitulum and floret morphology in Asteraceae. A to D, Capitulum of Zinnia elegans (A), Calendula officinalis (B), Tagetes patula (C), and S. vulgaris (D). E to G, zygomorphic ray (E and F) and actinomorphic disc (E and G) floret of S. vulgaris. d, Dorsal petal; fv, fused three ventral petals; S, stigma; V, ventral petal. Bars = 5 mm (A–D), 2 mm (E), 1 mm (F), and 0.5 mm (G). H to J, ML phylogenetic tree analyses showing floral symmetry gene relationships among several species. ML trees were generated based on 99 amino acids including the conserved TCP and R domains for CYC, 95 amino acids that included the conserved SANT domain for DIV, and 83-bp DNA sequences including the conserved SANT domain for RAD. S. vulgaris (Sv) genes are colored in red. H, RAY3 clusters together with HaCYC2b (red rectangle). I, The ML three major clades are labeled as DIV1, DIV2, and DIV3. SvDIV1B protein falls into the DIV1 clade (red rectangle). J, SvRAD grouped with AmRL5 (red rectangle). See “Materials and Methods” for the species abbreviations. Bootstrap values (500 replicates) greater than 50% are shown.
Of the three CYC clades of the CYCLOIDEA/TEOSINTE BRANCHED1-LIKE (CYC/TB1) subfamily, the CYC2 clade has highly diversified and duplicated within the Asteraceae lineage; six CYC2 clade genes have been identified in both G. hybrida and S. vulgaris, and five in H. annuus (Tähtiharju et al., 2012; Broholm et al., 2008; Kim et al., 2008). Of these CYC2 clade genes, some members are expressed exclusively in ray florets and also cause altered ray floret morphology when disrupted. For example, in H. annuus, HaCYC2c and HaCYC2d showed ray floret-specific expression (Chapman et al., 2008; Tähtiharju et al., 2012). Furthermore, overexpression of HaCYC2c due to an insertion in the promoter generated capitula with only ray florets, providing functional evidence for its role in ray formation (Chapman et al., 2012). In addition to ray and disc florets, the G. hybrida capitulum has zygomorphic trans florets that have intermediate features of both ray and disc florets. Among the G. hybrida CYC2 clade members, GhCYC2, GhCYC3, GhCYC4, GhCYC5, and GhCYC9 also showed ray/trans floret-specific expression (Broholm et al., 2008; Juntheikki-Palovaara et al., 2014) and overexpression of GhCYC2 generated disc florets with ray floret features (Broholm et al., 2008; Preston and Hileman, 2009; Juntheikki-Palovaara et al., 2014). In agreement with the other species, two CYC2 genes in S. vulgaris, RAY1 and RAY2, were expressed only in the ray florets and were also shown to control ray versus disc floret identity (Kim et al., 2008).
In addition to determining floret identity, CYC control of ray floral zygomorphy also appears to be conserved in some cases. Functional analyses in S. vulgaris revealed that overexpression of RAY1 reduced ventral petal growth in ray florets, while overexpression of RAY2 promoted dorsal ray petal outgrowth to produce tubular ray florets (Kim et al., 2008). Further work in gerbera suggests that localized expression of GhCYC2 to the ventral petals explains this zygomorphic phenotype (Broholm et al., 2008; Juntheikki-Palovaara et al., 2014); however, differential expression of CYC2 clade genes in ventral and dorsal petals in other Asteraceae species has not yet been seen.
In order to better understand how floral symmetry is established in two distinct ray and disc florets within a capitulum in the Asteraceae family, we have investigated the role of a CYC2 clade gene (RAY3) as well as two floral symmetry MYB domain regulators (SvRAD and SvDIV1B) in S. vulgaris. Our study revealed that during the early capitulum development, RAY3, SvRAD, and SvDIV1B showed uniform ray floret specific expression, suggesting that the CYC-RAD-DIV gene network was recruited into S. vulgaris capitula for zygomorphic ray floret formation. During ray floret petal development, RAY3 and SvRAD expression was confined to emerging ray ventral petals, suggesting that ventral petal specific expression could play a role in establishing ray floret bilateral symmetry. Our functional analysis further showed that RAY3 promotes and SvDIV1B represses petal growth. At the later capitula stages, prolonged RAY3 and decreasing SvDIV1B expression may be crucial for the rapid ventral petal growth in the ray florets. Furthermore, during capitulum development, RAY3 promotes RAY1 and RAY2, while SvDIV1B inhibits RAY3 and SvRAD, and activates RAY2. As well as reaffirming the recruitment of CYC2 genes for ray identity in the Asteraceae lineage, our results highlight that floral symmetry genes such as RAD and DIV appear to have a conserved role in petal growth in both Antirrhinum and Senecio; however, their expression domains and regulatory relationships may differ to account for the varying flower symmetries in these two distant species. It appears that the CYC-RAD-DIV gene network has been reinvented in Asteraceae to recapitulate a functional flower by the arrangement of florets.
RESULTS
Gene Isolation and Phylogenetic Relationships
To determine the mechanism that controls floral symmetry within the capitulum, we have isolated a CYC homolog gene fragment (297 bp) from S. vulgaris. To be consistent with previously reported RAY1 and RAY2 CYC-like proteins in S. vulgaris (Kim et al., 2008), we have named our sequence RAY3. BLAST analysis showed that its protein sequence shared 100% similarity with Senecio squalidus CYC-like (accession no. JF299257) and 60% with H. annuus CYC2b proteins. The maximum likelihood (ML) phylogenetic relationship analysis based on partial TCP and R domain protein sequences (Broholm et al., 2008; Cubas et al., 1999) placed RAY3 in the CYC2 subclade as a HaCYC2b homolog (Fig. 1H; RAY3 in red rectangle), unlike other Senecio CYC2 proteins (SvCYC4, SvCYC5, and svCYC7).
Two DIV genes were also isolated from S. vulgaris. ML phylogenetic analysis based on MYB protein sequences (which included the conserved SANT domain and the DIV-like typical conserved Trp residues; Cubas et al., 1999; Corley et al., 2005) placed these two DIV genes distinctly in the clade of class DIV1B and DIV3B genes, respectively (Fig. 1I). We have therefore designated them as SvDIV1B and SvDIV3B. However, in this article, we focused on SvDIV1B (Fig. 1I, red rectangle), which we found to be involved in capitulum development. The DIV1B protein sequence (274 amino acids) shared 67% identity with A. majus DIV.
Moreover, we have also isolated a 299-bp RAD-related gene fragment and showed that its protein (SvRAD) sequence was 72% identical to RAD from Gratiola officinalis (GoRAD). ML phylogenetic analysis revealed that our RAD-related protein was highly similar to RAD and RAD-LIKE proteins. Further ML phylogenetic analysis based on RAD and RAD-like DNA sequences that included the conserved SANT domain, placed SvRAD with AmRAD-like5 in the RAD1 clade (Fig. 1J, red rectangle).
Expression of RAY3, SvDIV1B, and SvRAD during S. vulgaris Capitulum Development
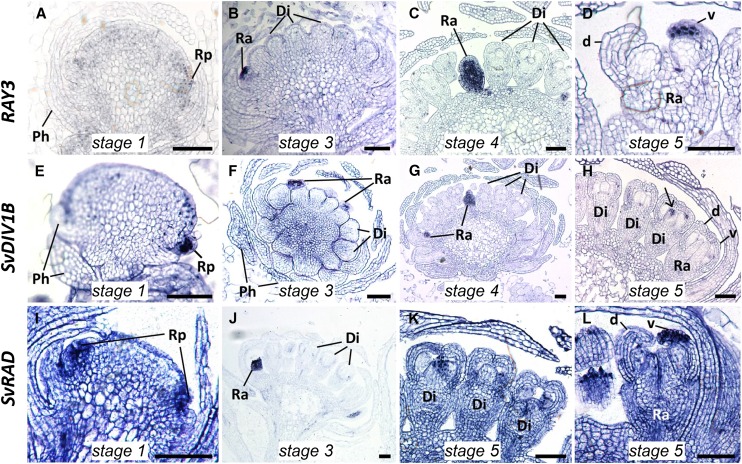
To reveal the roles of RAY3, SvDIV1B, and SvRAD during capitulum development, we investigated their expression patterns in stage 1 to 5 S. vulgaris capitula (for the details of stages, see “Materials and Methods”) by RNA in situ hybridization. Results showed that all RAY3, SvDIV1B, and SvRAD mRNA transcripts accumulated in the emerging ray floret primordia of the stage 1 capitulum (Fig. 2, A, E, and I). Once disc and ray florets were formed (stage 3-4), high and uniform expression was detected for all three genes exclusively in ray florets but absent in disc florets (Fig. 2, B, C, F, G, and J). In stage 5 of capitulum development, RAY3 and SvRAD were still highly expressed in ray florets but only in the ventral petals (Fig. 2, D and L). In contrast, SvDIV1B transcripts were not detectable in either dorsal or ventral petals of ray and disc florets (Fig. 2H), although very faint expression was detected in carpels (Fig. 2H, arrow).
Figure 2.
RAY3, SvDIV1B, and SvRAD RNA in situ expression profiles during S. vulgaris capitulum development. RAY3 (A–D), SvDIV1B (E–H), and SvRAD (I–L) transcripts were restricted to ray primordia (Rp) in stage 1 (A, E, and I) and developing ray florets (Ra) in stage 3 (B, F, and J), stage 4 (C and G) and stage 5 (D, H, K, and L) capitula. Unlike SvDIV1B (G and H), transcripts of RAY3 and SvRAD accumulated in the developing ventral (v) petals of ray florets (D and L) but were absent in the disc florets (C and K). Di, Disc florets; Ra, ray florets; Rp, ray primodium; Ph, phyllaries; d, dorsal petal; v, ventral petal. Bars = 100 µm.
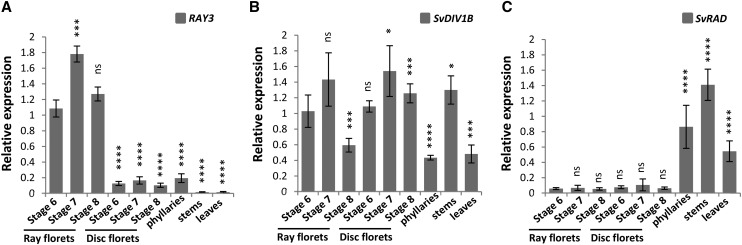
In order to quantify RAY3, SvDIV1B, and SvRAD gene expression during later stages of S. vulgaris capitulum development, we performed quantitative RT-PCR (qRT-PCR) using ray florets, disc florets, and phyllaries from capitula at stages 6 to 8, as well as stems and leaves. The RAY3 transcripts showed capitulum-specific expression (Fig. 3A). For all three stages, RAY3 transcripts were significantly higher in ray florets, lower in disc florets and phyllaries, and almost absent from stems and leaves (Fig. 3A). RAY3 is therefore constantly expressed in ray florets in all the developmental stages analyzed. Conversely, similar levels of SvDIV1B expression were initially detected for both ray and disc florets (Fig. 3B) as lateral organ expanded (stage 6) and ray ventral petals started to elongate (stage 7). However, a significant decrease of SvDIV1B transcript levels was detected only in ray florets (stage 8), which coincided with rapid ventral petal elongation (Fig. 4E). In capitulum stage 6-7, SvDIV1B in situ hybridization showed that SvDIV1B was ubiquitously expressed in florets, including petals of ray florets (Supplemental Fig. S2, A and B). In contrast, very low expression levels were detected by qRT-PCR for SvRAD in both ray and disc florets of the six to eight developmental stages analyzed but significantly higher expression levels were observed in all vegetative tissues (Fig. 3C). This is supported by SvRAD in situ data, which showed that at late developmental stages, SvRAD is almost nondetectable in florets but highly expressed in the vegetative tissues (Supplemental Fig. S2, C and D).
Figure 3.
qRT-PCR analysis of RAY3, SvDIV1B, and SvRAD transcripts in developing wild-type S. vulgaris capitulum tissues. A, RAY3 transcription levels were significantly higher in ray floret developmental stages than all the other plant tissues. B, SvDIV1B transcription levels were higher in stage 6 and 7, compared to stage 8 ray florets, while disc florets showed similar expression levels in all developmental stages. SvDIV1B transcripts were also detected in vegetative tissues. C, SvRAD expression levels were very low in all ray and disc floret stages but high in vegetative tissues. Gene expression levels were normalized to 5.8s rRNA. Asterisks indicate significance using two tailed Student's t test when compared to stage 6 ray florets: *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001, and nsP > 0.05 (ns, not significant). Error bars are ± sd values of three biological and four technical replicates.
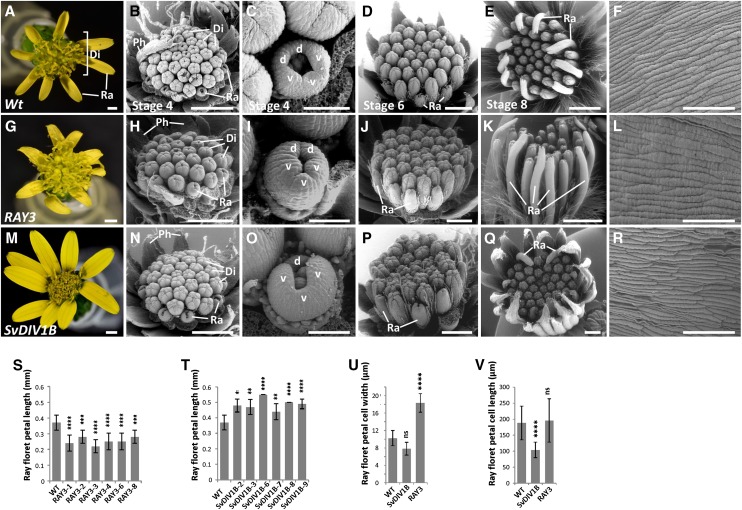
Figure 4.
Flower phenotypes of wild-type, RAY3, and SvDIV1B transgenic plants. A to F, Wild-type (nontransformed) control. G to R, RAY3 (G–L) and SvDIV1B (M–R) down-regulation capitulum and ray floret phenotypes. Di, Disc florets; Ra, ray florets; Ph, phyllaries; d, dorsal petal; v, ventral petal. Bars = 100 µm (C, F, I, L, O, and R), 500 µm (B, D, H, J, N, and P), and 1 mm (A, E, G, K, M, and Q). S to V, Quantitative ray floret phenotypic analysis of wild-type control, SvDIV1B, and RAY3 down-regulation plants. S and T, Ray floret length of wild-type control and several RAY3 and SvDIV1B transgenic lines, respectively. Down-regulation of RAY3 and SvDIV1B had antagonistic effects on ray floret petal length. Down-regulation of SvDIV1B significantly increased ray petal length, while RAY3 down-regulation decreased petal length in all transgenic lines. U and V, Average mature cell width and length of ray floret petal epidermal cells in wild-type control and in several SvDIV1B and RAY3 transgenic lines. U, Cell width was significantly increased in RAY3 down-regulated lines, while it was not changed in SvDIV1B transgenic lines. V, Down-regulation of SvDIV1B expression levels led to a significant ray floret petal cell length reduction, while RAY3 down-regulation had no effect. Error bars indicate sd values for n = 10 plants (S and T) and n = 20 plants (U and V) relative to wild-type controls. Asterisks indicate significance in two tailed Student’s t test when compared to the wild type: **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, and nsP > 0.05 (ns, not significant).
Down-Regulation of a CYC2 Homolog, RAY3, Led to Shorter Ray Florets
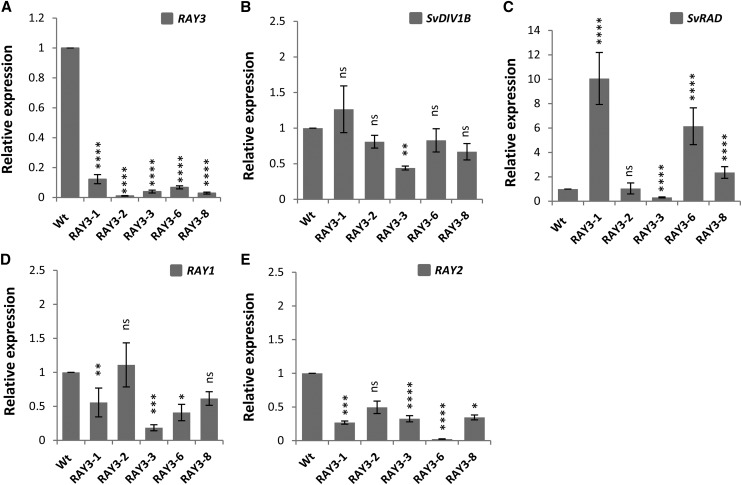
To determine the role of ray floret specific RAY3 expression, we generated S. vulgaris transgenic plants with RAY3 down-regulation. We obtained five independent transgenic lines (Supplemental Fig. S1A), all of which were confirmed to have decreased RAY3 expression by qRT-PCR (Fig. 5A). In these transgenic lines, mature capitula consistently had significantly shorter ray floret ventral petals while no difference was observed in disc florets, compared to nontransformed controls (Fig. 4, A, G, and S).
Figure 5.
qRT-PCR analysis of flower symmetry gene regulators in S. vulgaris wild-type and RAY3 transgenic capitula (stage 6-8). A, RAY3 transcription levels were significantly down-regulated in all transgenic lines. B, SvDIV1B transcription levels were only reduced in one RAY3 down-regulated transgenic line. C, SvRAD expression levels were significantly increased in three out of five transgenic lines in response to RAY3 down-regulation. D, RAY1 transcription levels were significantly reduced in most lines in response to RAY3 down-regulation. E, RAY2 transcription expression levels were significantly reduced in most lines in response to RAY3 down-regulation. Gene expression levels were normalized with 5.8S rRNA and are shown relative to the wild-type control expression levels. Asterisks indicate significance using two tailed Student's t test when compared to the wild type: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, and nsP > 0.05 (ns, not significant). Error bars are ± sd values of four technical replicates on biological triplicates.
To further investigate the morphological changes in petal length at earlier stages, transgenic and wild-type control plants were analyzed by scanning electron microscopy (SEM). In wild-type S. vulgaris, ray florets initially consist of five similarly sized petals (two dorsal and three ventral; Fig. 4C, stage 4). As the capitulum develops (Fig. 4E, stage 8), the three fused ventral petals elongate rapidly to form a tongue-shaped ray floret (Fig. 4A, Ra). SEM images showed that the shorter ventral petal phenotype of RAY3 antisense plants (Fig. 4, A and G) was not evident in stage 4 (Fig. 4, C and I), suggesting that RAY3 is involved in ventral petal elongation during later developmental stages.
In order to determine whether changes in cell shape or cell number caused the final shorter ray petal phenotype of RAY3 transgenic plants, we measured width and length of adaxial epidermal cells in ray ventral petals (Fig. 4, F and L). Results showed that ray ventral petal cell width increased in RAY3 transgenic lines but not cell length, compared to wild-type controls (Fig. 4, U and V). This suggests that down-regulation of RAY3 caused fewer cells along the proximal-distal axis of the ray ventral petal, indicating that decreased cell division caused shorter ray florets.
SvDIV1B Down-Regulation Led to Longer Ray Petals
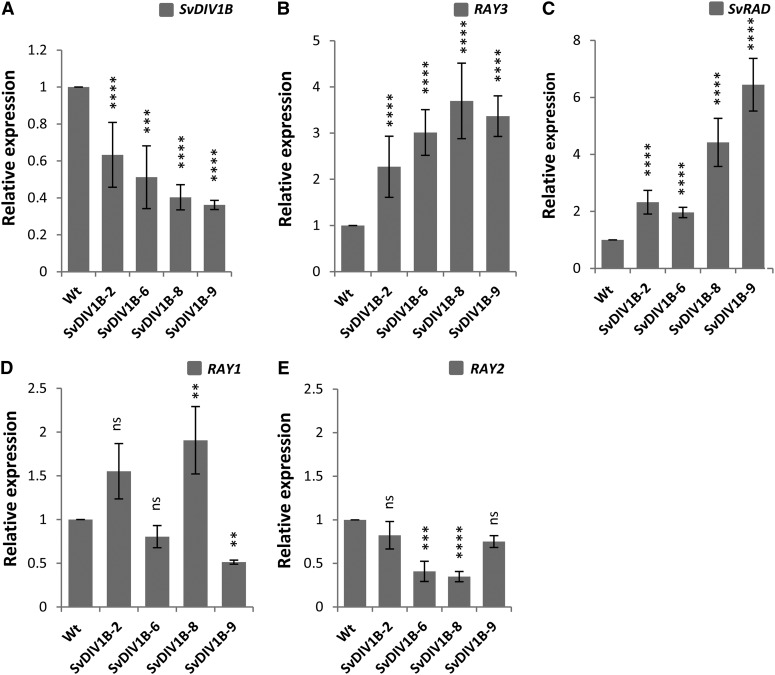
qRT-PCR expression data suggested that SvDIV1B might play a role in ray and disc floret development. To further determine the role of SvDIV1B in floret development, we generated transgenic plants that down-regulated SvDIV1B in S. vulgaris. We obtained six independent transgenic lines (Supplemental Fig. S1B) and decreased expression levels were confirmed in four independent transgenic lines by qRT-PCR (Fig. 6A). In mature capitula of these SvDIV1B transgenic plants, ray ventral petals were significantly longer than in wild-type controls, while no effect was observed in disc florets (Fig. 4, A, M, and T).
Figure 6.
qRT-PCR analysis of flower symmetry gene regulators in S. vulgaris wild-type and SvDIV1B transgenic capitula (stage 6-8). A, SvDIV1B transcription levels were significantly down-regulated in all transgenic lines. B and C, RAY3 (B) and SvRAD (C) transcription levels were significantly increased in all SvDIV1B down-regulated transgenic lines. D, RAY1 transcription levels were variable for all the SvDIV1B down-regulated transgenic lines. E, RAY2 transcription expression levels were significantly reduced in half of the SvDIV1B down-regulated transgenic lines. Gene expression levels were normalized with 5.8S rRNA and are shown relative to the expression levels of wild-type control. Asterisks indicate significance using two tailed Student's t test when compared to the wild type: **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, and nsP > 0.05 (ns, not significant). Error bars are ± sd values of three biological and four technical replicates.
To study how this long petal phenotype developed in earlier stages, SEM images were taken of SvDIV1B transgenic capitula. Longer ventral petals were seen as early as stage 4 (Fig. 4, N and O, stage 4), suggesting that the initiation of ray ventral petals in SvDIV1B transgenic plants occurs earlier than in the wild type. After stage 4, longer ray ventral petals were maintained through all the later stages of capitulum development (Fig. 4, M, P, and Q). However, disc floret morphology was not affected by SvDIV1B down-regulation at any of the stages studied.
As in the RAY3 transgenic analysis, we measured individual cell width and length of ray ventral petal epidermal cells of SvDIV1B transgenic and wild-type plants (Fig. 4, F and R). Results showed that cell width was not significantly different (Fig. 4U); however, cell length was reduced in transgenic lines compared to wild-type controls (Fig. 4V). Both longer final petal length and shorter individual cell length suggests that the number of cells along the proximal-distal petal axis increased, which implies that reduction of SvDIV1B RNA levels led to increased cell division.
Interaction among Floral Symmetry Regulators
In S. vulgaris, down-regulation of RAY3 and SvDIV1B had an opposite effect on ray petal length, suggesting their antagonistic roles during ray floret development. To investigate their genetic relationships and interactions with other known floral symmetry regulators, we have analyzed SvDIV1B, RAY1, RAY2, RAY3, and SvRAD expression levels by qRT-PCR in both RAY3 and SvDIV1B transgenic plants. Results showed that down-regulation of RAY3 led to reduction of SvDIV1B levels in one transgenic line (Fig. 5B) and up-regulation of SvRAD in three out of five transgenic lines (Fig. 5C). Furthermore, RAY1 and RAY2 transcription levels were significantly reduced in most lines in response to RAY3 down-regulation (Fig. 5, D and E).
In contrast, down-regulation of SvDIV1B expression levels led to up-regulation of both RAY3 and SvRAD in all lines (Fig. 6, B and C) and to down-regulation of RAY2 in half of the transgenic lines (Fig. 6E). However, RAY1 transcription levels were variable in all the SvDIV1B transgenic lines (Fig. 6D). Together, our gene expression data suggest that RAY3 appears to promote RAY1 and RAY2, while SvDIV1B inhibits RAY3 and SvRAD and activates RAY2 (directly or indirectly) during capitulum development.
DISCUSSION
Expression of RAY3, SvDIV1B, and SvRAD Is Restricted to Ray Florets within the Young Capitulum
Here, our phylogenetic analyses showed that RAY3 was clustered within the CYC2 subfamily of TCP genes known to control flower asymmetry (Fig. 1H). In particular, the RAY3 gene fragment was shown to be a homolog of the sunflower HaCYC2b gene and gerbera GhCYC4 and GhCYC9. While RAY3 expression was consistent with GhCYC4 and GhCYC9 (Tähtiharju et al., 2012), its expression differed from that of HaCYC2b (Tähtiharju et al., 2012; Chapman et al., 2008). Our in situ hybridization and qRT-PCR data showed that RAY3 was expressed mainly in ray florets throughout all stages (Figs. 2 and 3A). The RAY3 sunflower homolog HaCYC2b was initially expressed in ray florets as seen here in Senecio; however, later, HaCYC2b was expressed across multiple floral and vegetative tissues including disc florets (Tähtiharju et al., 2012; Chapman et al., 2008). Instead, RAY3 expression was more similar to the sunflower paralogs, HaCYC2c and HaCYC2d, which were expressed mainly in ray florets throughout all stages. Moreover, two S. vulgaris CYC genes, RAY1 and RAY2, were also shown to be specifically expressed in ray floret primordia (Kim et al., 2008). Therefore, although capitulum patterning relies on CYC2 clade ray floret specific expression, it appears that different Asteraceae lineages have independently recruited distinct CYC paralogs for ray floret differentiation (Chapman et al., 2012; Hileman, 2014). Perhaps this is not surprising since ray florets are considered to have evolved multiple times in this family (Panero and Funk, 2008; Hileman, 2014).
Notably, in the early stages of capitulum development, RAY3 was ubiquitously expressed across the ray floret, which contrasts with the dorsal-specific expression patterns of their homologs in other species with zygomorphic solitary flowers (Corley et al., 2005; Luo et al., 1996; Hileman et al., 2003). The uniform RAY3 expression in the early ray floret primordium suggests that RAY3 may be involved in the initiation of all five petals. In the Malpighiaceae-Elatinaceae families, basal species have actinomorphic flowers with uniform CYC2 expression, whereas derived species have zygomorphic flowers with dorsal-specific CYC2 expression (Zhang et al., 2010). Therefore, this early uniform RAY3 expression in the ray floret may resemble the ancestral expression pattern of CYC2 homologs in these families. However, the fact that their expression was uniform in the young ray floret cannot explain how CYC genes establish bilateral symmetry in the ray floret. Unlike RAY1 and RAY2, which are expressed in both dorsal and ventral petals (Supplemental Fig. S2, E–H), in the later stages of capitulum development (stage 5), RAY3 was exclusively expressed in ray ventral petals (Fig. 2D), which led to their elongation and, therefore, floral zygomorphy in the ray floret. This is in agreement with studies in G. hybrida, which show ventral expression of GhCYC2 in ray florets (Broholm et al., 2008; Juntheikki-Palovaara et al., 2014). In the zygomorphic Antirrhinum flower, however, CYC is expressed in the dorsal petals (Luo et al., 1996). Although these CYC genes have evolved to express differently in dorsal (CYC in A. majus) versus ventral (RAY3 in S. vulgaris and GhCYC2 in G. hybrida) domains, it is likely that both genes have maintained similar functions in promoting petal elongation. The fact that loss of RAY3 expression only partly affected ray petal elongation and did not lead to fully actinomorphic flowers indicates that additional factors such as RAY2 (Kim et al., 2008) may contribute to the ray petal elongation.
As with RAY3, we have found that SvRAD expression during early capitulum development (stage 1-4) was restricted to ray florets and absent from disc florets (Fig. 2, I and J). After initial uniform SvRAD expression in the ray florets, expression is later restricted to the ventral petals (Fig. 2L), suggesting a role in controlling zygomorphy. This contrasts with dorsal RAD expression in A. majus but matches the expression pattern of RAY3. Our qPCR data showing the low SvRAD expression during the later capitulum stages suggests that SvRAD may only have an active role in the initial establishment of zygomorphy but not in the elongation of petals. Moreover, this is supported by the fact that variable SvRAD expression in RAY3 transgenics (Fig. 5C) did not correspond to the expected petal phenotypes based on RAD function in A. majus. To verify this, further functional analyses are required. It is also possible that other Senecio RAD genes (yet to be identified) are involved in later petal elongation. Notably, our phylogenetic analysis showed that SvRAD is not an AmRAD ortholog, either suggesting that an AmRAD paralog was independently recruited in Senecio or that multiple paralogs control floral symmetry in this species.
The initial SvDIV1B expression pattern on the other hand is conserved with the A. majus DIV ortholog. In A. majus, DIV expresses throughout the floral meristem and is excluded from dorsal petals at later stages of flower development, which corresponds with dorsal petal elongation (Almeida et al., 1997). In S. vulgaris, after the expression of SvDIV1B in ray and disc florets, phyllaries, and vegetative tissues (Fig. 3B), a drastic decrease of SvDIV1B transcripts was detected in stage 8 ray florets (Fig. 3B), which coincided with the rapid ventral petal elongation (Fig. 4E). Together, these data suggest that SvDIV1B needs to be down-regulated in order to achieve ray petal elongation and thus bilateral symmetry. This reduction in SvDIV1B expression across the whole ray floret differs from the ventral localization seen in A. majus. Together, these SvDIV1B and SvRAD data provide the first explanation for how RAD and DIV genes in Asteraceae contribute to bilateral symmetry establishment in the developing ray floret.
RAY3 and SvDIV1B Play Antagonistic Roles during Ray Petal Development
Consistent with the role suggested by our RAY3 and SvDIV1B expression data, transgenic plants down-regulating RAY3 and SvDIV1B showed altered petal elongation in ray florets but not in disc florets. In mature capitula, down-regulation of RAY3 and SvDIV1B RNA levels led to significantly shorter or longer ray ventral petals, respectively, while no effect was observed in disc florets (Fig. 4). These changes in ray petal length in both RAY3 and SvDIV1B transgenic plants were caused by alterations in cell division. Ray ventral petal cell width increased (Fig. 4U) in RAY3 transgenic lines but not cell length (Fig. 4V). Contrarily, in SvDIV1B, cell width of ray ventral petals was similar (Fig. 4U), while cell length was reduced (Fig. 4V) in transgenic plants. Therefore, these results indicate that down-regulation of RAY3 causes a decrease in cell division leading to shorter ray florets, whereas reducing SvDIV1B RNA levels increases cell division, which in turn leads to longer ray floret phenotypes. Taken together, these results suggest that both genes have antagonist effects on ray petal length via cell division and that RAY3 is likely to promote while SvDIV1B is likely to repress cell division during wild-type ray petal development. These results are consistent with several other studies, which connect TCP genes to cell proliferation in various plant developmental processes (Efroni et al., 2008; Nath et al., 2003; Palatnik et al., 2003; Koyama et al., 2007; Hervé et al., 2009; Costa et al., 2005).
These results also suggest that the recruitment of RAY3 and SvDIV1B activity to ray florets is likely to be a critical evolutionary step to differentiate bilateral ray florets from radial disc florets. However, the fact that down-regulation of RAY3 alone could not convert ray florets into disc florets suggests that several CYC genes, e.g. RAY1 and RAY2, were recruited for ray floret identity and zygomorphy. Given that SvDIV1B transgenic phenotypes were confined to ray ventral petals but SvDIV1B RNA expression was uniform in the ray floret, other dorsoventral genetic components should interact with SvDIV1B for bilateral symmetry to be established in these florets. SvRAD could be a gene candidate to determine the dorsoventral symmetry of ray florets by interacting with SvDIV1B as in other species (Raimundo et al., 2013). However, to confirm this hypothesis, the function of SvRAD needs to be further investigated.
Regulatory Relationships of Floral Symmetry Genes Are Altered to Promote Ray Florets in S. vulgaris
We have found that novel gene regulatory interactions among floral symmetry genes exist in S. vulgaris. Our results showed that during capitulum development, down-regulation of RAY3 led to reduction of RAY1 and RAY2 RNA levels (Fig. 5, D and E). As the DNA sequence identity in the cloned RAY3 region and corresponding RAY1 and RAY2 fragments are 66% and 59%, respectively, cross-suppression based on sequence identity is unlikely. For this reason, the qRT-PCR data are likely to reflect a positive regulatory relationship between CYC paralogs. In A. majus, the backpetals mutant, caused by ectopic expression of CYC, did not alter the other CYC2 gene, DICH expression in the dorsal petals (Luo et al., 1999). Furthermore, CYC activity was shown to be unaltered in dich mutants, indicating that the regulation of these CYC genes is independent during dorsal petal development (Luo et al., 1999). However, in Primulina heterotricha, CYC1C regulates CYC2D and vice versa, showing the presence of regulatory relationships between CYC genes as seen here (Yang et al., 2012).
In addition, in A. majus, CYC activates RAD, which is a key interaction in determining floral dorsal and lateral identity (Corley et al., 2005). However, our results showed that in RAY3 down-regulation transgenic lines, SvDIV1B and SvRAD expression levels were variable, suggesting that in S. vulgaris capitula RAY3 does not interact (directly) with these genes to regulate ray floret dorsoventral identity. One possibility is that in S. vulgaris, the CYC-RAD interaction may involve CYC paralogs other than RAY3 alone.
While RAY3 promoted RAY2 expression during ray floret development, SvDIV1B repressed RAY3 and SvRAD but activated RAY2. Floral bilateral symmetry in A. majus involves the action of CYC and DICH that activate RAD, which in turn down-regulates DIV activity in dorsal petals (Raimundo et al., 2013; Corley et al., 2005; Costa et al., 2005; Almeida et al., 1997). However, there has been no evidence as to whether DIV can feedback to regulate these genes during flower development. To the best of our knowledge, this is the first report that shows a regulatory role of SvDIV1B on dorsal identity genes (RAY2, RAY3, and SvRAD), suggesting that a more complex gene network may exist in capitulum development. Interestingly, in SvDIV1B antisense transgenic lines, up-regulation of RAY3 (by DIV1B down-regulation) did not lead to RAY2 up-regulation; RAY2 was instead down-regulated. This suggests that regulation of RAY2 by RAY3 involves a more complex network and may rely on the presence/activity of other genetic components such as SvDIV1B. However, further studies are required to identify such interactions in Senecio.
Inference for Ray Floret Dorsoventral Symmetry
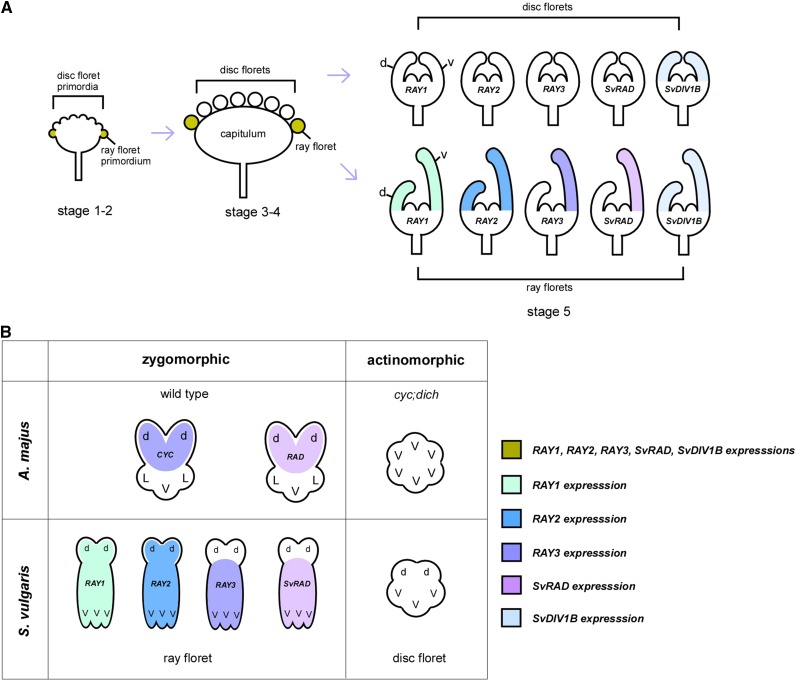
Based on our expression and functional analyses of RAY3 and SvDIV1B, we propose the following model (Fig. 7) to explain the establishment of floral symmetry in a capitulum. During early capitulum development (Fig. 7A, stage 1-4), key players of floret symmetry, RAY1, RAY2, RAY3, SvRAD, and SvDIV1B are expressed in the ray florets to give the competency to develop bilateral symmetry in these florets, which sets them apart from radial disc florets. As petals emerge (stage 5), RAY3 accumulates in the ventral ray petals, which leads to rapid ventral petal elongation, thus to the bilateral symmetry in the ray floret (Fig. 7A). Given that RAY3 promotes and SvDIV1B represses petal growth, persistent RAY3 expression (accompanied by decreasing SvDIV1B levels) may explain rapid ventral petal elongation in the ray florets at the later stages (stage 8). This also highlights that in S. vulgaris, the roles of CYC (RAY3) and DIV (SvDIV1B) in petal elongation are conserved. However, to create ray floret bilateral symmetry via ventral petal outgrowth, similar to GhCYC2 in gerbera (Broholm et al., 2008), the RAY3 activity was shifted to the ventral region of the ray floret in S. vulgaris, contrasting to the CYC-RAD activity in the dorsal region in (non-Asteraceae) species with solitary flowers (Luo et al., 1996; Corley et al., 2005; Zhang et al., 2010). In this model (Fig. 7), all RAY genes are exclusively recruited to the zygomorphic ray florets but not to actinomorphic disc florets. Moreover, a subset of RAY genes, such as RAY3, acquired ray ventral petal specific expression, which in turn promotes petal growth in this domain. This highlights how the diversification of CYC genes played an important role in creating a complex structure such as the capitulum in the Asteraceae family and how different regulatory interactions between floral symmetry genes have been recruited in angiosperms to control flower shape.
Figure 7.
A schematic model showing flower symmetry gene expression during capitulum and floret development in S. vulgaris. A, Initially, RAY (CYC-like) genes, SvRAD and SvDIV1B are expressed in the emerging ray floret primordia to give the competency to develop bilateral symmetry in these florets, which sets them apart from radial disc florets. Later, as petals emerge in a ray floret, RAY3 and SvRAD are expressed in the ventral petals to set up the dorsoventral symmetry by promoting the ventral petal elongation. During ray ventral petal elongation, RAY1 and RAY2 are also present in the ventral domain and involved in petal growth (Kim et al., 2008). SvDIV1B appears to play a role in counterbalancing the RAY gene activities although its role in dorsal ray petal and disc floret development remains to be investigated. B, The expression patterns of CYC genes are diversified in S. vulgaris. To generate two different florets, RAY genes are exclusively recruited only to the zygomorphic ray florets. However, a subset of RAY genes such as RAY3 acquired ventral petal specific expression and promotes ventral petal growth. This is comparable to the CYC activity in the dorsal petal domain where it promotes dorsal petal growth to establish zygomorphy in A. majus. No RAY gene expression in actinomorphic disc florets is analogous to actinomorphic cyc;dich flowers. d, Dorsal petal; L, lateral petal; V, ventral petal.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Senecio vulgaris RR genotype plants were grown from seed for in vitro tissue culture. Seeds were vapor-gas sterilized (Clough and Bent, 1998) prior to transferring to germination media (half-strength Murashige and Skoog [MS] [w/v] media and 0.8% [w/v] plant agar [Melford] at pH 5.8). In order to increase S. vulgaris seed germination rate, filter sterilized 0.1% (w/v) Gibberellic Acid A3 (Melford) was added to seeds. Plates were transferred to a Percival tissue culture cabinet (22°C, 16 h light, 100 μmol m−2 s−1) for 2 weeks. Seedlings were transferred to magenta boxes containing full-strength (w/v) MS medium (3% Suc [w/v] and 0.8% [w/v] plant agar [Melford] at pH 5.8). After 1 month, plants were ready to be used for tissue culture transformation. Plants for seed harvesting and phenotypic analysis were grown in pots with Sinclair compost in a growth chamber under 16-h-light photoperiod (150 µmol m−2 s−1) at 24°C until flowering stage. Images of flower phenotypes were recorded with a dissecting scope (Leica S8AP0) using a Nikon D3100 camera. Several stages of S. vulgaris capitula were used for in situ hybridizations (stage 1-5), qRT-PCR (stage 6-8), and SEM (stages 4, 6, and 8). For wild-type qRT-PCR tissue specificity analyses, ray and disc florets were dissected separately under a dissecting microscope (Leica S8AP0), and phyllaries, developing leaves (1-cm-long leaves), and stems were also harvested. For RAY3 and SvDIV1B down-regulation transgenic qRT-PCR analyses, inflorescences consisting of capitula (stages 6–8) were harvested. We defined S. vulgaris capitulum stages 1 to 8 of development as follows: stage 1, capitula with emerging ray floret primordia; stage 2, capitula with emerging disc floret primordia; stage 3, capitula with ray and disc florets already formed but without floret lateral organs such as sepals and petals; stage 4, capitula with lateral organ primordia emerging within both florets; stage 5, lateral organs expanding within both florets; stage 6, ventral petal start to elongate within the ray florets; stage 7, ray and disc reach similar florets length; stage 8, further elongation of ventral petals within the ray florets.
Gene Isolation and Vector Constructs
A CYC homolog genomic DNA (gDNA) fragment (RAY3) of 297 bp was cloned from S. vulgaris RR genotype (DNA from the inflorescence consisting of stage 1-8 capitula) using degenerative primers (DeCYC-1/DeCYC-2; Supplemental Table S1). These primers were designed based on several Senecio and sunflower (Helianthus annuus) CYC2 sequences from two plants species obtained from the GenBank database (FJ356699.1, FJ356698.1, FJ356702.1, FJ356701.1, EU088371, and EU088372), and the following gradient PCR conditions were used for amplification: 98°C for 3 min, 98°C 15 s, temperature of 45 to 51°C for 30 s, and 72°C for 1 min, repeated for 36 cycles and followed by an extension of 5 min at 72°C. Partial sequences for SvCYC4, SvCYC5, SvCYC6, and SvCYC7 genes were obtained by RACE-PCR from total RNAs of Senecio R/R young inflorescence (Edinburgh accession), with degenerate primers designed according to a conserved region of TCP genes: G775 and G873 (Supplemental Table S1). A 824-bp DIV homolog was also cloned from S. vulgaris RR genotype (DNA from the inflorescence consisting of stage 1-8 capitula) using previously reported degenerative primers (Howarth and Donoghue, 2009). The DeDIVForw-1/DeDIVRev-1 pair (Supplemental Table S1) was used to amplify DIV1B fragment using the above PCR conditions with a few modifications (temperature of 41 to 48°C). Moreover, we isolated a 299-bp RAD homolog fragment from S. vulgaris cDNA of capitulum inflorescences. We also used this template to clone a 179-bp RAD gene fragment using degenerative primers (DeRADForw/DeRADRev; Supplemental Table S1). These primers were designed on RAD-like and MYB domain gene sequences from five plant species: NM_001050197.1 (Oryza sativa), NM_106181.1 (Arabidopsis thaliana), NM_120086.2 (Arabidopsis), NM_001246920.1 (Solanum lycopersicum), NM_001249151.1 (Glycine max), and AY954971.1 (Antirrhinum majus). PCR conditions were the same as described above with a few modifications (temperature of 42°C). Amplified PCR products were cloned into the pGEM-T easy vector (Promega) for sequencing. To make the antisense constructs, specific primers were then designed for both SvDIV1B and RAY3 gene fragments using restriction enzyme sites for directional cloning into the binary vector pBI121 (Clontech). All PCR reactions were performed using Phusion DNA Polymerase (Thermo Fisher Scientific), 2.5 mm dNTPs (Bioline), and 10 µm primers (Eurofins). See primer sequences in Supplemental Table S1.
Phylogenetic Analysis
ML phylogenetic trees for RAY3, SvDIV1B, and SvRAD were obtained using amino acid and DNA sequences. Protein alignments were performed with the ClustalX2 program (Thompson et al., 2002), and ML trees were generated using the RAXML website (http://phylobench.vital-it.ch/raxml-bb/index.php; Stamatakis et al., 2008) with 500 bootstrap replications. For the CYC proteins, 99 amino acids were used, which included the conserved TCP and R domains. For DIV proteins, we generated the ML tree using 95 amino acids, which included the conserved SANT domains. The ML RAD tree was generated using 83-bp DNA sequences, which also included the conserved SANT domain. The species abbreviations are shown in as follows: Aragoa abietina (Aa), Aragoa cundinamarcensis (Ac), Antirrhinum majus (Am), Arabidopsis thaliana (At), Bournea leiophylla (Bl), Cryptothladia chinensis (Cc), Centranthus macrosiphon (Cm), Diervilla sessilifolia (Ds), Digitalis purpurea (Dp), Dipelta floribunda (Df), Epimedium sagittatum (Es), Fedia cornucopiae (Fc), Fragaria vesca (Fv), Gerbera hybrida (Gh), Glycine max (Gm), Gratiola officinalis (Go), Helianthus annuus (Ha), Heptacodium miconioides (Hm), Kolkwitzia amabilis (Ka), Lonicera x bella (Lb), Lonicera morrowii (Lm), Lonicera reticulata (Lr), Leycesteria sp. (Lsp), Medicago truncatula (Mt), Mohavea confertiflora (Mc), Plantago major (Pm), Polypremum procumbens (Pp), Populus trichocarpa (Pt), Rhytidophyllum auriculatum (Ra), Ricinus communis (Rc), Sixalix atropurpurea (Sa), Sambucus cerulea (Sc), Senecio vulgaris (Sv), Symphoricarpos orbiculatus (So), Valerianella eriocarpa (Ve), Veronica chamaedrys (Vc), Viburnum davidii (Vd), Vitis vinifera (Vv), Weigela hortensis (Wh), and Wulfenia carinthiaca (Wc).
In Situ Hybridizations
Riboprobe synthesis and in situ hybridizations of RAY3, SvDIV1B, and SvRAD genes were performed using the protocol described previously (Garcês and Sinha, 2009b). In situ hybridization for RAY1 and RAY2 were performed as previously described (Kim et al., 2008). To clone the SvDIV1B 191-bp exon from 824-bp gDNA fragment in the pGEM easy-T (Promega), we used the specific primer set SvDIV1BF/SvDIV1BR (see Supplemental Table S1 for primer sequences). To synthesize the in situ hybridization RNA probes, the 270-bp RAY3, 179-bp SvRAD, and 191-bp SvDIV1B gene fragments, which were cloned in the pGEM easy-T vector, were PCR amplified using the universal M13 forward and reverse primers present in the plasmid backbone and treated as described previously (Garcês and Sinha, 2009b). All fragments were amplified using Phusion DNA polymerase (Thermo Fisher Scientific) and the following PCR conditions: 98°C for 3 min, 98°C 15 s, temperature of 55°C for 30 s, and 72°C for 1 min, repeated for 36 cycles and followed by an extension of 5 min at 72°C. One microgram of PCR product was used to transcribe both sense (−) and antisense (+) single-stranded RNA probes using T7 and SP6 promoters, digoxigenin-UTP, and the T7 and SP6 RNA polymerases contained in DIG labeling kit (Roche Applied Science). In situ hybridizations were performed according to the previous protocol (Garcês and Sinha, 2009b) with the following modifications: Inflorescences of S. vulgaris capitula (1 to 5 capitula stages) were fixed in paraformaldehyde (Alfa Aesar) solution (4% paraformaldehyde [v/v], 100 mm PBS, and 1% DMSO) for 16 h at 4°C, embedded in Surgipath Paraplast Plus paraffin (Leica Biosystems), and sectioned into 9-µm sections. Sections were deparaffinized with Histo-clear II (Agar Scientific) and rehydrated via an ethanol series. Slides were then treated for 25 min at 37°C with 0.065 mg/mL proteinase K (Sigma-Aldrich). Probe hybridization was carried out overnight at 50°C. After hybridization, slides were treated as described previously (Garcês and Sinha, 2009b). Signal detection was performed using antidigoxigenin conjugated to alkaline phosphatase fab fragments (Roche) as the secondary antibody. The alkaline phosphatase substrate 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium (BCIP/NBT; Promega), was prepared according to the manufacturer’s specifications prior to application to the sections. Tissues were incubated for 3 to 6 h on average with BCIP/NBT or until a purple/blue color developed (gene expression). Samples were viewed using a Leica DMR microscope, and electronic images were acquired using SPOT advanced software (SPOT Imaging Solutions). Image color balancing and cropping was performed using Adobe Photoshop CS6, and figures were executed using Canvas X software (ADC Systems).
qRT-PCR Expression Analysis
For qRT-PCR analysis of wild-type S. vulgaris tissue-specific gene expression, ray and disc florets were dissected and harvested from capitula at several developmental stages (stage 6-8), as well as phyllaries, developing leaves (1-cm-long leaves), and stems (Fig. 3). For gene regulatory relationship studies, inflorescences of capitula (stage 6-8) were used for qRT-PCR analyses (Figs. 5 and 6). All tissues were immediately frozen after harvesting into liquid nitrogen and saved at −80°C until further analyses. Total RNA was extracted using RNeasy Mini Kit (Qiagen). Five micrograms of total RNA was DNase I treated according to the manufacture’s specifications (Promega) and used for cDNA synthesis using the Untergasser protocol (Untergasser et al., 2007). qRT-PCR primers were designed using Primer3 software (Untergasser et al., 2007), and qRT-PCR was performed on a ABI Prism 7000 PCR machine (Applied Biosystems). Reactions were performed using SensiFAST SYBR Hi-ROX Kit (Bioline) according to manufacturer’s specifications with primers at a final concentration of 500 nm and RNA at 10 ng per reaction. Annealing temperatures were kept to ±1°C of 60°C with target GC content of 50 to 60%. The PCR efficiency of each target gene was calculated using LinReg (Hårdstedt et al., 2005), and the gene expression levels were determined using the comparative threshold cycle method (Schmittgen and Livak, 2008). All qRT-PCR was performed on biological triplicates in technical quadruplicates, as described previously (Etchells et al., 2012). Statistical analysis and graphs were performed in Excel (Microsoft). Samples were normalized to 5.8s rRNA and 18s rRNA control genes. Both control genes gave similar expression levels for all the genes tested. We chose 5.8s rRNA control gene for gene analysis, and ± sd error bars were calculated on three biological and four technical replicates per sample.
Plant Transformation
For the plant transformation, RAY3 and DIV1B, 270- and 824-bp gDNA fragments, respectively, were cloned in antisense orientation into pBI121 carrying the 35S promoter and the neomycin phosphotransferase II (NPTII) gene, which confers kanamycin resistance to positive transformants. One-month-old S. vulgaris RR genotype plants grown in sterile conditions in magenta boxes were used for the transformation experiments. Leaves were harvest and sectioned in 1-cm2 explants for transformation. Agrobacterium tumefaciens (GV3101) carrying the antisense constructs was inoculated into liquid LB medium containing 100 mg/L kanamycin (Melford) and 50 mg/L Rifampicin (Melford) and shaken overnight at 30°C. Cells were collected by centrifugation and resuspended to an OD600 of 0.5 to 0.8 nm in liquid full-strength (w/v) MS medium with no antibiotics (MS0) for plant infection. Acetosyringone (100 µm; Sigma-Aldrich) was added to this suspension to enhance DNA transfer. The bacterial suspension was shaken in the dark and at room temperature until needed. S. vulgaris leaf explants were cocultivated with the A. tumefaciens suspension for 20 min, blotted dry, and transferred to petri dishes containing coculture (without selection) callus/shoot inducing media (full-strength MS salts, 3% Suc [w/v], 1 mg/L thidiazuron [Sigma-Aldrich], 0.1 mg/L 1-naphthaleneacetic acid [Sigma-Aldrich] at pH 5.8, and 7.5g/L Phytoagar [Sigma-Aldrich]). Plates were transferred to a Percival tissue culture cabinet at 22°C for 3 d in the dark. Explants were then transferred to calli/shoot inducing media containing 40 mg/L kanamycin (Melford) and 250 mg/L cefotaxime (Melford) antibiotics to select for positive transformants (MS1). These plates were incubated in the Percival tissue culture cabinet at 22°C with a photoperiod of 16 h light (100 μmol m−2 s−1). Explants were subcultured to fresh MS1 medium every 2 weeks until the appearance of calli with shoots. Once shoots were formed, they were removed from calli and transferred to magenta boxes containing root induction media (MS2) (half-strength MS salts, 3% [w/v] Suc, pH 5.8, and 7.5g/L Phytoagar) with antibiotics (100 mg/L kanamycin and 250 mg/L cefotaxime). Rooted shoots were transferred to new fresh MS2 media in Magentas every month or earlier if needed. Once transgenic plants had a well-developed root system, plants were transferred to soil. Plant trays were covered with a humidity lid, and this lid was slowly removed over the period of a week to adjust plants to their new environment. Transgenic plants were ready to be analyzed 2 months later. In this species, the whole process to obtain transgenic plants ready to be analyzed can take a total of 6 to 8 months. The presence of RAY3 and SvDIV1B antisense transgenes was confirmed by PCR using gDNA extracted from five and six independent transgenic lines with 35S Forw and RAY3-5′SacI reverse primer or SvDIV1B-5′SacI, respectively (Supplemental Table S1).
SEM Analysis
Several flower head developmental stages (very young, 1–2 mm; medium, 2–3 mm; and mature, 3–4 mm flowers) from wild-type nontransformed plants and from several SvDIV1B and RAY3 down-regulation transgenic lines were fixed for SEM and viewed as described previously (Garcês and Sinha, 2009a). Dehydrated samples were taken through critical point drying using a Polaron critical point dryer (Quorum Technologies) and were mounted onto SEM stubs (Agar Scientific) using carbon tape (Agar Scientific). Samples were then sputter coated with carbon for 90 s using a Polaron E5100 sputter coater (Quorum Technologies). SEM images were obtained directly from a FEI Quanta 250 FEG electron microscope (FEI) at 5 to 10 kV accelerating voltage. Image color balancing and cropping were performed using Adobe Photoshop CS6, and figures were executed using Canvas X software (ADC Systems).
Capitulum Phenotypic Analysis
SEM images were used to measure mature ray floret petal adaxial epidermis cell length and width in wild-type controls, RAY3 and SvDIV1B down-regulation transgenic lines. Statistical analysis and graphs were performed in Excel. The sd values were calculated on n = 10 plants for ray floret length an on n = 20 plants for ray floret width relative to wild-type controls. Stars indicate significance in two-sample tailed Student's t test assuming unequal variances with **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, and nsP > 0.05 (ns, not significant).
Accession Numbers
Gene sequence information can be found on the NCBI website under the following accession numbers: KT722935 (RAY3), KT722933 (SvDIV1B), KT722936 (SvRAD), SvCYC4 (KU663021), SvCYC5 (KU663022), SvCYC6 (KU663023), SvCYC7 (KU663024), and DIV3B (KU666938).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PCR amplification of RAY3 and SvDIV1B transgenes.
Supplemental Figure S2. In situ hybridization showing the expression pattern of SvDIV1B, SvRAD, RAY1, and RAY2 in S. vulgaris.
Supplemental Table S1. Primer sequences used for cloning genes, making constructs, and expression analysis.
Supplementary Material
Acknowledgments
We thank Dr. Tobias Starborg in the Electron Microscopy Facility of the Faculty of Life Sciences for his assistance and the Wellcome Trust for equipment grant support to the Electron Microscopy Facility.
Glossary
- ML
maximum likelihood
- SEM
scanning electron microscopy
- gDNA
genomic DNA
Footnotes
Articles can be viewed without a subscription.
References
- Almeida J, Galego L (2005) Flower symmetry and shape in Antirrhinum. Int J Dev Biol 49: 527–537 [DOI] [PubMed] [Google Scholar]
- Almeida J, Rocheta M, Galego L (1997) Genetic control of flower shape in Antirrhinum majus. Development 124: 1387–1392 [DOI] [PubMed] [Google Scholar]
- Boyden GS, Donoghue MJ, Howarth DG (2012) Duplications and expression of radialis-like genes in Dipsacales. Int J Plant Sci 173: 971–983 [Google Scholar]
- Broholm SK, Tähtiharju S, Laitinen RAE, Albert VA, Teeri TH, Elomaa P (2008) A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci USA 105: 9117–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Zachgo S (2007) Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc Natl Acad Sci USA 104: 16714–16719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Leebens-Mack JH, Burke JM (2008) Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol Biol Evol 25: 1260–1273 [DOI] [PubMed] [Google Scholar]
- Chapman MA, Tang S, Draeger D, Nambeesan S, Shaffer H, Barb JG, Knapp SJ, Burke JM (2012) Genetic analysis of floral symmetry in Van Gogh’s sunflowers reveals independent recruitment of CYCLOIDEA genes in the Asteraceae. PLoS Genet 8: e1002628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citerne H, Jabbour F, Nadot S, Damerval C. 2010. The evolution of floral symmetry. In J. C. Kader, M. Delseny, eds, Advances in Botanical Research, Vol 54, pp. 85–137, Elsevier [Google Scholar]
- Citerne HL, Pennington RT, Cronk QCB (2006) An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc Natl Acad Sci USA 103: 12017–12020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corley SB, Carpenter R, Copsey L, Coen E (2005) Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc Natl Acad Sci USA 102: 5068–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MMR, Fox S, Hanna AI, Baxter C, Coen E (2005) Evolution of regulatory interactions controlling floral asymmetry. Development 132: 5093–5101 [DOI] [PubMed] [Google Scholar]
- Crepet WL. (1996) Timing in the evolution of derived floral characters: Upper Cretaceous (Turonian) taxa with tricolpate and tricolpate-derived pollen. Rev Palaeobot Palynol 90: 339–359 [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. (1999) Symmetry in flowers: Diversity and evolution. Int J Plant Sci (Suppl) 160: S3–S23 [DOI] [PubMed] [Google Scholar]
- Etchells JP, Provost CM, Turner SR (2012) Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling. PLoS Genet 8: e1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Zhao Z, Tian Z, Xu S, Luo Y, Cai Z, Wang Y, Yang J, Wang Z, Weng L, et al. (2006) Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA 103: 4970–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcês H, Sinha N (2009a) Fixing and sectioning tissue from the plant Kalanchoë daigremontiana. Cold Spring Harb Protoc 2009: t5301. [DOI] [PubMed] [Google Scholar]
- Garcês H, Sinha N (2009b) In situ hybridization in the plant Kalanchoë daigremontiana. Cold Spring Harb Protoc 2009: t5302. [DOI] [PubMed] [Google Scholar]
- Hårdstedt M, Finnegan CP, Kirchhof N, Hyland KA, Wijkstrom M, Murtaugh MP, Hering BJ (2005) Post-transplant upregulation of chemokine messenger RNA in non-human primate recipients of intraportal pig islet xenografts. Xenotransplantation 12: 293–302 [DOI] [PubMed] [Google Scholar]
- Hervé C, Dabos P, Bardet C, Jauneau A, Auriac MC, Ramboer A, Lacout F, Tremousaygue D (2009) In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol 149: 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC. (2014) Bilateral flower symmetry--how, when and why? Curr Opin Plant Biol 17: 146–152 [DOI] [PubMed] [Google Scholar]
- Hileman LC, Kramer EM, Baum DA (2003) Differential regulation of symmetry genes and the evolution of floral morphologies. Proc Natl Acad Sci USA 100: 12814–12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth DG, Donoghue MJ (2006) Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc Natl Acad Sci USA 103: 9101–9106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth DG, Donoghue MJ (2009) Duplications and expression of DIVARICATA-like genes in dipsacales. Mol Biol Evol 26: 1245–1258 [DOI] [PubMed] [Google Scholar]
- Howarth DG, Martins T, Chimney E, Donoghue MJ (2011) Diversification of CYCLOIDEA expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Ann Bot (Lond) 107: 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntheikki-Palovaara I, Tähtiharju S, Lan T, Broholm SK, Rijpkema AS, Ruonala R, Kale L, Albert VA, Teeri TH, Elomaa P (2014) Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J 79: 783–796 [DOI] [PubMed] [Google Scholar]
- Kim M, Cui M-L, Cubas P, Gillies A, Lee K, Chapman MA, Abbott RJ, Coen E (2008) Regulatory genes control a key morphological and ecological trait transferred between species. Science 322: 1116–1119 [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E (1999) Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376 [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Nath U, Crawford BCW, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Panero JL, Funk VA (2008) The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol Phylogenet Evol 47: 757–782 [DOI] [PubMed] [Google Scholar]
- Preston JC, Hileman LC (2009) Developmental genetics of floral symmetry evolution. Trends Plant Sci 14: 147–154 [DOI] [PubMed] [Google Scholar]
- Raimundo J, Sobral R, Bailey P, Azevedo H, Galego L, Almeida J, Coen E, Costa MMR (2013) A subcellular tug of war involving three MYB-like proteins underlies a molecular antagonism in Antirrhinum flower asymmetry. Plant J 75: 527–538 [DOI] [PubMed] [Google Scholar]
- Reardon W, Gallagher P, Nolan KM, Wright H, Cardeñosa-Rubio MC, Bragalini C, Lee C-S, Fitzpatrick DA, Corcoran K, Wolff K, Nugent JM (2014) Different outcomes for the MYB floral symmetry genes DIVARICATA and RADIALIS during the evolution of derived actinomorphy in Plantago. New Phytol 202: 716–725 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57: 758–771 [DOI] [PubMed] [Google Scholar]
- Tähtiharju S, Rijpkema AS, Vetterli A, Albert VA, Teeri TH, Elomaa P (2012) Evolution and diversification of the CYC/TB1 gene family in Asteraceae--a comparative study in Gerbera (Mutisieae) and sunflower (Heliantheae). Mol Biol Evol 29: 1155–1166 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG 2002. Multiple sequence alignment using ClustalW and ClustalX. In A.D. Baxevanis, ed, Current Protocols in Bioinformatics, 00:2.3:2.3.1–2.3.22 [DOI] [PubMed] [Google Scholar]
- Trow AH. (1912) On the inheritance of certain characters in the common groundsel, Senecio vulgaris L., and its segregates. J Genet 2: 239–276 [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35: W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Luo Y, Li X, Wang L, Xu S, Yang J, Weng L, Sato S, Tabata S, Ambrose M, et al. (2008) Genetic control of floral zygomorphy in pea (Pisum sativum L.). Proc Natl Acad Sci USA 105: 10414–10419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Pang H-B, Liu B-L, Qiu Z-J, Gao Q, Wei L, Dong Y, Wang Y-Z (2012) Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 24: 1834–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Kramer EM, Davis CC (2010) Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc Natl Acad Sci USA 107: 6388–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Kellogg EA (2015a) Duplication and expression of CYC2-like genes in the origin and maintenance of corolla zygomorphy in Lamiales. New Phytol 205: 852–868 [DOI] [PubMed] [Google Scholar]
- Zhong J, Kellogg EA (2015b) Stepwise evolution of corolla symmetry in CYCLOIDEA2-like and RADIALIS-like gene expression patterns in Lamiales. Am J Bot 102: 1260–1267 [DOI] [PubMed] [Google Scholar]
- Zhou X-R, Wang Y-Z, Smith JF, Chen R (2008) Altered expression patterns of TCP and MYB genes relating to the floral developmental transition from initial zygomorphy to actinomorphy in Bournea (Gesneriaceae). New Phytol 178: 532–543 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.