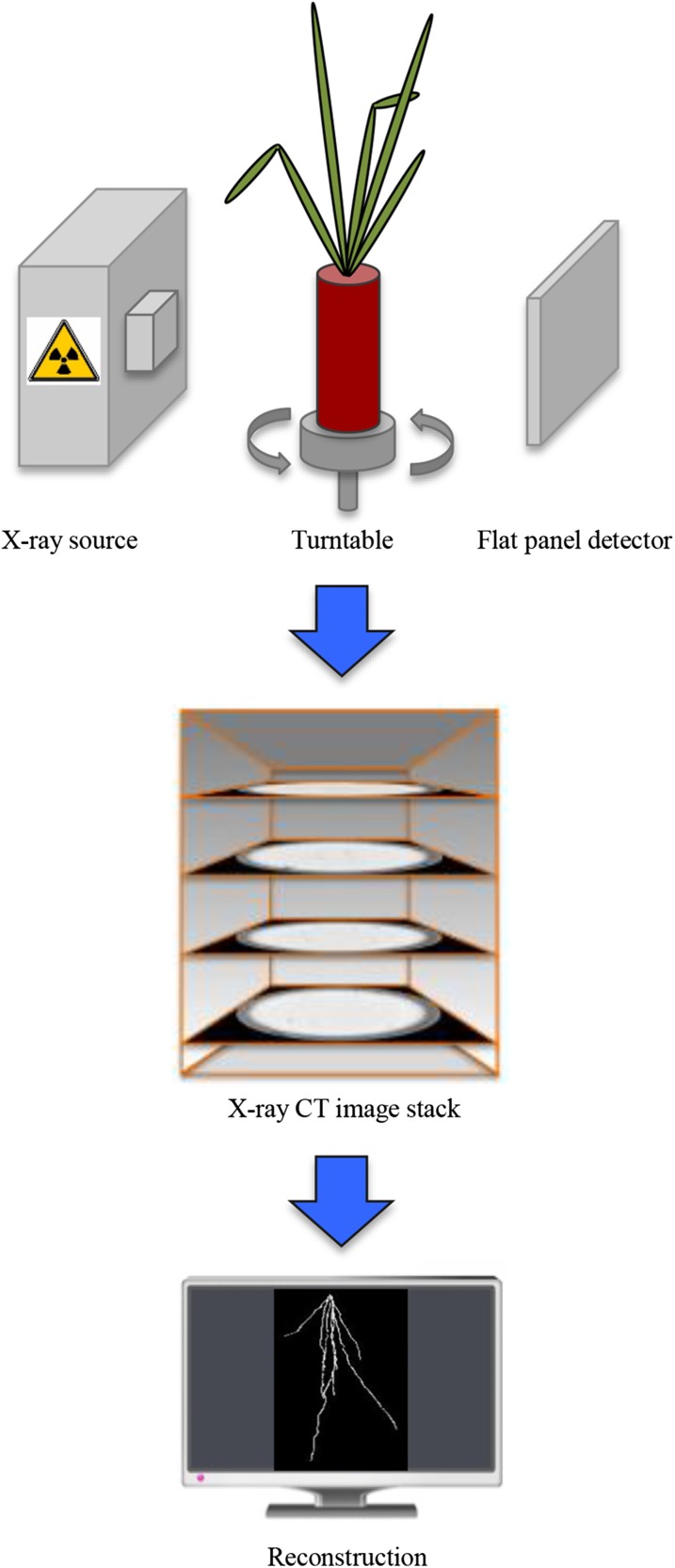
X-ray computed tomography of rice root system architecture in different substrate textures reveals genotype by environment interactions.
Abstract
Root system architecture (RSA) impacts plant fitness and crop yield by facilitating efficient nutrient and water uptake from the soil. A better understanding of the effects of soil on RSA could improve crop productivity by matching roots to their soil environment. We used x-ray computed tomography to perform a detailed three-dimensional quantification of changes in rice (Oryza sativa) RSA in response to the physical properties of a granular substrate. We characterized the RSA of eight rice cultivars in five different growth substrates and determined that RSA is the result of interactions between genotype and growth environment. We identified cultivar-specific changes in RSA in response to changing growth substrate texture. The cultivar Azucena exhibited low RSA plasticity in all growth substrates, whereas cultivar Bala root depth was a function of soil hardness. Our imaging techniques provide a framework to study RSA in different growth environments, the results of which can be used to improve root traits with agronomic potential.
Root system architecture (RSA) describes the spatial arrangement of roots within the soil and plays a major role in crop performance (Lynch, 1995; Beebe et al., 2006; de Dorlodot et al., 2007; Magalhaes et al., 2007; Hodge et al., 2009; Tester and Langridge, 2010; Gamuyao et al., 2012; Lynch and Brown, 2012; Munns et al., 2012). RSA impacts fitness, performance, and grain yield by facilitating efficient nutrient and water uptake from the soil (Lynch, 1995; Beebe et al., 2006; Hodge et al., 2009; Gamuyao et al., 2012; Rich and Watt, 2013; Uga et al., 2013). Many reports have shown that RSA is plastic and shaped by interactions between genotype and components of the local soil environment, which include nutrient and water localization, the soil microbiome, and the physical properties of soil (Zobel, 1996; Kano-Nakata et al., 2011; Band et al., 2012; Gouda et al., 2012; Gowda et al., 2012). Poor soil fertility and environmental stresses suppress crop yields in many parts of the world. Thus, identifying RSA traits that could mitigate these effects would have far-reaching implications (Morita and Nemoto, 1995; Eshel and Waisel, 1996; Zobel, 1996; McCully, 1999; Shi et al., 2013; Intergovernmental Panel on Climate Change, 2014). To this end, many studies have found that deeper rooting may contribute to drought tolerance in the field (Lynch and Ho, 2005; Trachsel et al., 2011; Uga et al., 2011; Venuprasad et al., 2011; Zhu et al., 2011; Fang et al., 2013), whereas shallow roots are more beneficial where there is limited phosphorous (Clark et al., 2011; Iyer-Pascuzzi et al., 2011; Galkovskyi et al., 2012). Thus, it has been proposed that different RSA ideotypes can maximize soil exploration and plant fitness (Feldman, 1994; McCully, 1995). Despite the importance of RSA, how roots respond to differences in soil composition remains to be elucidated and could be an untapped resource for agricultural improvement.
Soil consists of organic matter, inorganic compounds, microbes, water, air, and a combination of solid particles of different sizes: sand (0.05–2 mm), silt (0.002–0.05 mm), and clay (less than 0.002 mm; Kalita, 2011). Soil texture, defined as the relative proportion of these solid particles, significantly impacts soil porosity, compaction, and mechanical impedance (Kalita, 2011). Collectively, these soil properties control the water, oxygen, and nutrients available to plants and greatly impact RSA and crop yield (Rich and Watt, 2013). It also has been suggested that the granular nature of soil affects plant root growth (Wendell et al., 2012). Studies with wheat (Triticum aestivum) and tomato (Solanum lycopersicum) reported that compacted soil consisting of smaller particles resulted in roots that were shorter, thicker, and explored less of the soil environment (Tracy et al., 2012a, 2012b). In addition, root elongation in cereal crops is more influenced by mechanical and physical properties compared with chemical properties of soil (Whitmore and Whalley, 2009; White and Kirkegaard, 2010; Bengough et al., 2011; Valentine et al., 2012; Fang et al., 2013).
Root penetration of soil is a complex trait influenced by many factors. In many cereal crops, root elongation is reduced proportional to the mechanical impedance, and significant changes in root growth are observed within 2 weeks of growth in compacted soils (Ehlers et al., 1983; Goodman and Ennos, 1999; Merotto and Mundstock, 1999). Despite its importance, the ability to penetrate different types of soil is largely unexplored; however, evidence in rice (Oryza sativa) and maize (Zea mays) indicates a strong genetic component (Yu et al., 1995; Ray et al., 1996; Bushamuka and Zobel, 1998; Zheng et al., 2000). Previous studies in rice have identified quantitative trait loci that are associated with differential soil penetration that shows a tradeoff between root thickness and length (Price et al., 2000).
A major limitation to understanding how soil properties influence RSA is that soil is heterogenous, opaque, and constantly changing within a field and across a growing season. As a result, techniques have been developed to characterize RSA that involve transparent soils (Clark et al., 2011; Iyer-Pascuzzi et al., 2011; Downie et al., 2012; Galkovskyi et al., 2012; Topp et al., 2013; Le Marié et al., 2014). These have the advantage of RSA visibility but do not model the heterogenous and mechanical properties of soil. An alternative approach is to use x-ray computed tomography (CT), which allows for nondestructive visualization of three-dimensional (3-D) RSA and its surrounding soil environment including pores and obstructions (Moran et al., 2000; Gregory et al., 2003; Jenneson et al., 2003; Tracy et al., 2010; Mairhofer et al., 2012, 2013; Mooney et al., 2012; Schulz et al., 2013). X-ray CT has been demonstrated to be an effective tool to visualize plant roots growing in soil, but limited resources are available for data analysis (Mairhofer et al., 2012).
In this study, we used x-ray CT to quantify changes in rice RSA in response to varied growth substrates. We conducted a systematic analysis of root responses to different growth substrates using eight different rice cultivars. We defined genotype-by-environment (GxE) interactions and identified cultivar-specific RSA responses. We discovered that one drought-resistant rice cultivar displayed low RSA plasticity. By comparing the RSA of plants grown in different particle sizes with that of growth in a transparent gel substrate, we identified a genotype that exhibited decreased root growth in response to growth substrates with low penetration resistance.
RESULTS
Modification of an X-Ray Imaging Platform to Analyze Rice RSA
Soil characteristics are known to affect RSA, but studies have been limited by the opacity, complexity, and heterogeneity of soil (Rich and Watt, 2013). To perform a systematic analysis of the effect of soil parameters on rice RSA, we used simplified growth substrates (Supplemental Fig. S1; Supplemental Table S1). Rice cultivars were grown in three granular substrates of different particle size (d): sieved sand (d < 0.71 mm), fine (d = 0.24 mm), and coarse silica beads (d = 0.70 mm) saturated with nutrient solution. To image roots in these granular substrates, we modified an x-ray imaging platform used previously to investigate fire ant nest excavation (Monaenkova et al., 2015) consisting of an x-ray source, a flat panel detector, and a turntable (Fig. 1). Plants were grown in aluminum cans enabling the acquisition of high-contrasting x-ray CT slices. The turntable was positioned to maximize the can width in the field of view. A scissor lift was used to capture two sets of images (top and bottom) that encompass the entire RSA. To quantitate RSA, we developed a pipeline using Avizo software to identify and reconstruct 3-D models of roots, providing input into GiA Roots to quantify RSA (Galkovskyi et al., 2012).
Figure 1.
Visualization of roots in different granular substrates. Data acquisition consists of an x-ray source, turntable, and flat panel detector to acquire CT slices of plants growing in aluminum soda cans with dimensions of 6.6 × 10.8 cm (width × height). CT slices are aligned with Cobra software and exported with MATLAB, and the root systems were reconstructed using Avizo. Root reconstructions are input into the GiA Roots pipeline to quantify root traits.
X-ray reconstructions captured the primary root and crown roots larger than 0.4 mm in diameter. As a result of the flat panel detector resolution, roots were not visible prior to day 7 and thin crown roots and lateral roots could not be discerned from voids in the granular substrate. We focused on five traits calculated by GiA Roots that were not affected by the absence of fine root structure and captured the overall shape, size, and extent of RSA exploration: maximum root depth (depth), maximum network width (width), total root length (total length), network convex area (convex area), and median root number. Two additional traits were obtained after roots were removed from the growth substrates, which were independent of the x-ray reconstructions: crown root number (manual counting) and root diameter (semiautomated using GiA Roots). Both crown root number and median root number were used because they capture different aspects of root number (for details, see “Materials and Methods”).
To assess the accuracy of x-ray reconstructions, we utilized a ground truth model based upon a simple root system for which the RSA trait values are known (Supplemental Fig. S2). RSA traits calculated from the x-ray reconstruction were compared with known values and exhibited an error of 5% to 12% (Supplemental Table S2).
Varying the Growth Substrate Identifies Unique Cultivar Responses
We analyzed eight rice cultivars (cv Azucena, Basmati 217, Bala, Caiapo, CO39, IR64, Moroberekan, and Swarna) based on their agronomic importance and/or the availability of genetic tools that could facilitate future analysis. We used a gel-based imaging platform (Topp et al., 2013) to characterize the RSA of these cultivars at day 7 post planting. When grown in 1× gel, these cultivars showed diverse RSA characteristics ranging from deep and wide (cv Azucena and Swarna) to shallow and narrow (cv Bala; Fig. 2; Table I).
Figure 2.
Rice root architecture is dependent upon particle size and growth substrate. Eight cultivars of rice (rows) were grown in two concentrations of Gelzan and three different granular substrates (columns). Columns 1 and 2 are representative camera images after thresholding using GiA Roots of plants grown in 1× and 6× gel. Columns 3 to 5 are representative x-ray reconstructions of plants grown in granular substrates. Plants were chosen to have a depth and width that are most similar to the mean of the group and specific rotation selected that showed the majority of the roots. Bar = 25 mm for all images.
Table I. RSA traits for cultivars grown in Gelzan.
Values shown are means and sd. n, Replicates.
| Growth Substrate | Cultivar | n | Depth | Width | Total Length | Convex Area | Diameter | Median Root No. |
|---|---|---|---|---|---|---|---|---|
| mm | mm | mm | mm2 | mm | ||||
| 1× gel | Azucena | 25 | 102.9 ± 23.7 | 29.0 ± 7.6 | 538.4 ± 136.9 | 2,404.6 ± 1,046.5 | 0.3 ± 0.0 | 4.1 ± 0.7 |
| Basmati 217 | 20 | 70.4 ± 7.8 | 21.8 ± 5.3 | 292.2 ± 43.8 | 1,128.7 ± 268.6 | 0.2 ± 0.0 | 3.4 ± 0.7 | |
| Bala | 28 | 44.5 ± 9.0 | 21.0 ± 4.1 | 382.0 ± 83.2 | 682.3 ± 187.3 | 0.3 ± 0.0 | 7.2 ± 1.7 | |
| Caiapo | 24 | 89.5 ± 14.1 | 29.5 ± 7.6 | 429.4 ± 64.7 | 1,931.8 ± 557.4 | 0.3 ± 0.0 | 4.2 ± 1.0 | |
| CO39 | 38 | 60.0 ± 8.7 | 26.2 ± 8.9 | 439.6 ± 140.1 | 1,108.9 ± 348.4 | 0.3 ± 0.0 | 5.9 ± 2.6 | |
| IR64 | 24 | 54.6 ± 10.9 | 30.3 ± 8.1 | 296.2 ± 77.6 | 1,180.0 ± 387.9 | 0.2 ± 0.0 | 4.0 ± 1.4 | |
| Moroberekan | 38 | 80.6 ± 12.2 | 35.0 ± 10.7 | 496.0 ± 158.8 | 2,110.6 ± 863.2 | 0.3 ± 0.0 | 4.7 ± 1.6 | |
| Swarna | 21 | 99.5 ± 19.5 | 27.1 ± 7.4 | 486.5 ± 90.2 | 1,898.0 ± 342.4 | 0.3 ± 0.0 | 4.3 ± 1.4 | |
| 6× gel | Azucena | 10 | 89.1 ± 9.9 | 41.5 ± 7.1 | 625.2 ± 131.8 | 2,766.7 ± 446.2 | 0.3 ± 0.0 | 4.9 ± 1.2 |
| Basmati 217 | 10 | 65.4 ± 6.9 | 24.9 ± 5.1 | 272.0 ± 66.4 | 1,227.3 ± 310.2 | 0.2 ± 0.0 | 3.1 ± 0.7 | |
| Bala | 10 | 72.3 ± 3.4 | 30.1 ± 5.1 | 701.7 ± 69.5 | 1,664.5 ± 211.8 | 0.3 ± 0.0 | 7.2 ± 1.0 | |
| Caiapo | 10 | 89.5 ± 8.1 | 28.7 ± 5.7 | 358.7 ± 48.0 | 1,911.0 ± 309.5 | 0.3 ± 0.0 | 3.2 ± 0.5 | |
| CO39 | 10 | 113.7 ± 18.6 | 36.3 ± 9.6 | 692.7 ± 131.8 | 2,829.7 ± 619.2 | 0.3 ± 0.0 | 3.8 ± 3.0 | |
| IR64 | 10 | 69.7 ± 7.3 | 34.3 ± 9.9 | 310.9 ± 110.6 | 1,626.9 ± 518.9 | 0.2 ± 0.0 | 2.9 ± 1.1 | |
| Moroberekan | 9 | 73.7 ± 7.6 | 36.2 ± 7.9 | 502.6 ± 147.3 | 2,097.1 ± 498.5 | 0.3 ± 0.0 | 4.7 ± 1.2 | |
| Swarna | 8 | 112.4 ± 14.6 | 22.5 ± 6.0 | 407.7 ± 60.8 | 1,818.4 ± 484.9 | 0.3 ± 0.0 | 3.2 ± 0.7 |
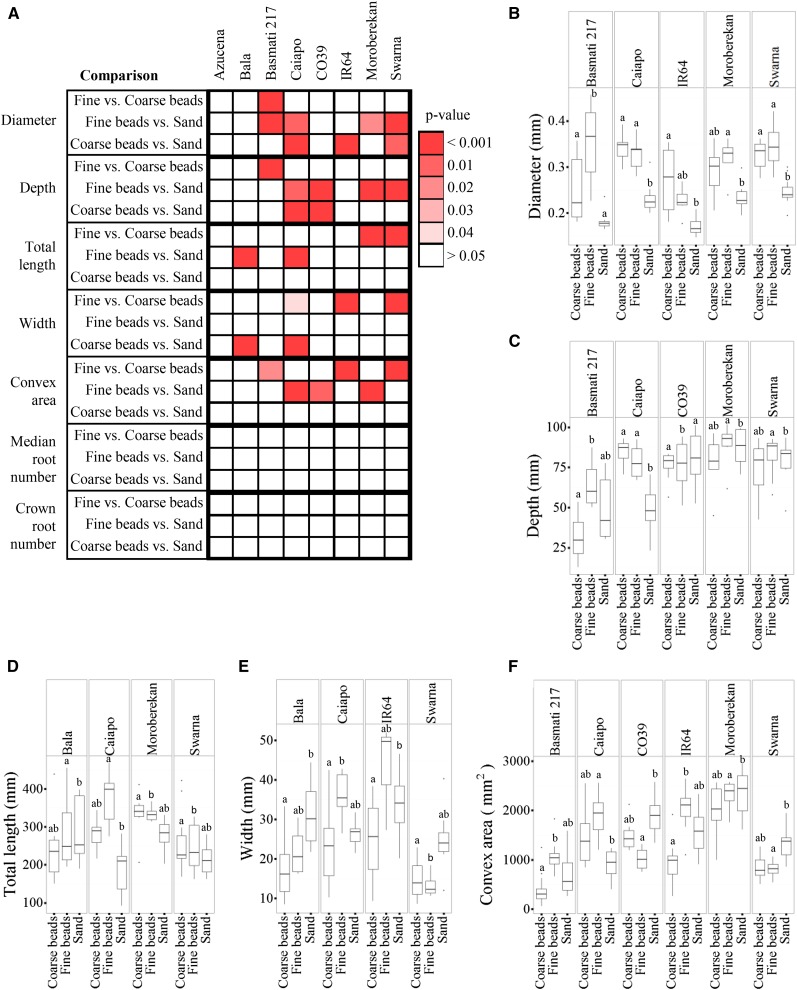
We next compared the RSA response of each cultivar with the three different granular substrates (Fig. 2; Table II; Supplemental Tables S3 and S4). RSA depth was similar across all cultivars grown in fine beads but varied widely in other granular substrates (Table II). For at least half of the cultivars, their diameter, depth, width, total length, and convex area were influenced by particle size (Tukey’s honestly significant difference [HSD] P < 0.05; Fig. 3). By contrast, the crown root number and median root number were not influenced by particle size in any of the cultivars (Tukey’s HSD P < 0.05; Fig. 3A). Similar to the 1× gel-grown plants, each cultivar had a unique RSA, and striking differences were seen among cultivars. Most of the traits were influenced by the substrate particle size (Table II), although cv Azucena and Moroberekan generally had the largest RSA in terms of depth, width, and convex area, whereas, cv Basmati 217 and Caiapo tended to have the smallest RSA and were most impacted by substrate particle size (Figs. 2 and 3A). The cv Swarna maintained a deep and narrow RSA in all growth substrates. Interestingly, the RSA of cv Azucena had almost no response to the different particle sizes. These data indicate that the RSA response to particle size is cultivar dependent.
Table II. RSA traits for cultivars grown in granular substrates.
Values shown are means and sd. n, Replicates.
| Granular Substrate | n | Cultivar | Depth | Width | Total Length | Convex Area | Diameter | Median Root No. | Crown Root No. |
|---|---|---|---|---|---|---|---|---|---|
| mm | mm | mm | mm2 | mm | |||||
| Fine beads | 6 | Azucena | 79.6 ± 9.1 | 35.9 ± 3.6 | 322.9 ± 48.4 | 2,132.0 ± 324.5 | 0.3 ± 0.1 | 3.2 ± 0.4 | 7.7 ± 1.5 |
| 8 | Basmati 217 | 64.8 ± 15.0 | 23.4 ± 6.1 | 182.9 ± 47.8 | 1,084.3 ± 355.2 | 0.4 ± 0.1 | 2.2 ± 0.6 | 6.3 ± 1.8 | |
| 8 | Bala | 72.5 ± 14.8 | 21.9 ± 5.7 | 283.6 ± 93.7 | 1,211.8 ± 499.0 | 0.3 ± 0.0 | 3.0 ± 0.9 | 10.9 ± 2.6 | |
| 9 | Caiapo | 78.7 ± 9.1 | 36.3 ± 5.6 | 379.0 ± 69.1 | 1,928.6 ± 412.8 | 0.3 ± 0.0 | 3.3 ± 0.6 | 6.8 ± 1.7 | |
| 8 | CO39 | 76.3 ± 15.9 | 19.9 ± 6.7 | 236.6 ± 49.1 | 1,023.1 ± 212.6 | 0.3 ± 0.1 | 2.1 ± 1.3 | 10.0 ± 1.5 | |
| 7 | IR64 | 69.1 ± 15.1 | 44.0 ± 10.3 | 301.0 ± 69.3 | 2,015.9 ± 496.9 | 0.2 ± 0.0 | 2.8 ± 0.9 | 8.0 ± 1.8 | |
| 6 | Moroberekan | 88.9 ± 14.1 | 36.5 ± 5.8 | 333.9 ± 19.7 | 2,307.4 ± 311.0 | 0.3 ± 0.0 | 2.9 ± 0.9 | 7.9 ± 2.3 | |
| 8 | Swarna | 82.2 ± 13.2 | 13.1 ± 2.6 | 245.4 ± 63.0 | 825.1 ± 169.8 | 0.3 ± 0.0 | 2.0 ± 0.9 | 5.2 ± 2.8 | |
| Coarse beads | 6 | Azucena | 71.5 ± 17.3 | 32.3 ± 5.3 | 290.7 ± 45.5 | 1,596.2 ± 227.6 | 0.3 ± 0.1 | 3.0 ± 0.9 | 8.0 ± 2.6 |
| 15 | Basmati 217 | 31.5 ± 13.4 | 13.7 ± 6.5 | 124.7 ± 89.7 | 378.7 ± 298.9 | 0.3 ± 0.1 | 2.6 ± 0.9 | 7.1 ± 1.7 | |
| 13 | Bala | 59.1 ± 16.1 | 17.5 ± 7.5 | 229.7 ± 77.7 | 827.9 ± 471.2 | 0.2 ± 0.0 | 2.8 ± 1.0 | 10.7 ± 4.6 | |
| 10 | Caiapo | 84.5 ± 7.9 | 24.3 ± 10.9 | 281.1 ± 35.0 | 1,453.7 ± 562.3 | 0.3 ± 0.0 | 2.7 ± 0.7 | 6.6 ± 1.5 | |
| 8 | CO39 | 76.1 ± 9.2 | 25.6 ± 5.1 | 353.3 ± 71.8 | 1,492.6 ± 310.0 | 0.2 ± 0.0 | 3.6 ± 0.7 | 10.8 ± 2.6 | |
| 10 | IR64 | 48.3 ± 15.6 | 25.0 ± 9.7 | 200.5 ± 81.7 | 940.5 ± 459.8 | 0.3 ± 0.1 | 3.2 ± 1.3 | 8.6 ± 3.1 | |
| 6 | Moroberekan | 77.3 ± 18.2 | 33.3 ± 6.3 | 330.7 ± 68.2 | 1,987.3 ± 579.8 | 0.3 ± 0.0 | 3.5 ± 0.4 | 6.8 ± 2.6 | |
| 15 | Swarna | 73.9 ± 15.4 | 14.5 ± 4.2 | 253.5 ± 71.8 | 834.8 ± 236.1 | 0.3 ± 0.0 | 2.9 ± 1.0 | 6.4 ± 2.2 | |
| Sand | 6 | Azucena | 61.5 ± 12.5 | 41.1 ± 7.3 | 249.6 ± 66.0 | 1,860.0 ± 687.0 | 0.3 ± 0.0 | 2.4 ± 1.1 | 9.8 ± 2.1 |
| 13 | Basmati 217 | 48.2 ± 18.0 | 19.3 ± 8.1 | 150.7 ± 49.5 | 699.8 ± 393.2 | 0.2 ± 0.0 | 1.8 ± 0.9 | 6.9 ± 2.8 | |
| 10 | Bala | 66.3 ± 8.6 | 31.3 ± 8.3 | 291.5 ± 86.4 | 1,437.3 ± 523.7 | 0.2 ± 0.0 | 2.5 ± 1.4 | 13.2 ± 4.3 | |
| 8 | Caiapo | 48.9 ± 15.4 | 26.3 ± 3.2 | 190.2 ± 64.3 | 910.2 ± 305.3 | 0.2 ± 0.0 | 2.5 ± 0.6 | 7.0 ± 2.2 | |
| 6 | CO39 | 80.4 ± 18.6 | 30.8 ± 5.5 | 281.8 ± 62.3 | 1,905.4 ± 435.1 | 0.2 ± 0.0 | 2.4 ± 0.6 | 9.8 ± 2.0 | |
| 11 | IR64 | 70 ± 20.0 | 33.5 ± 8.0 | 259.6 ± 69.3 | 1,556.4 ± 438.1 | 0.2 ± 0.0 | 2.5 ± 1.4 | 8.9 ± 3.3 | |
| 8 | Moroberekan | 87.6 ± 11.5 | 39.6 ± 4.8 | 280.0 ± 42.0 | 2,352.8 ± 472.2 | 0.2 ± 0.0 | 2.2 ± 0.4 | 8.2 ± 2.0 | |
| 8 | Swarna | 78.9 ± 14.1 | 24.9 ± 8.0 | 214.2 ± 41.7 | 1,320.0 ± 346.7 | 0.3 ± 0.0 | 1.7 ± 0.9 | 5.3 ± 2.9 |
Figure 3.
Cultivars have unique responses to each granular substrate texture. A, Depiction of significant differences between granular substrates for each cultivar and root trait. Shades of red indicate that a trait value is significantly different between indicated granular substrates; white indicates no significant difference. Pairwise significance for each granular substrate and cultivar was tested by post hoc one-to-one comparisons of the GxE term using the Tukey-Kramer HSD test. B to F, Quantification of diameter (B), depth (C), total length (D), width (E), and convex area (F) for each cultivar with significant differences based on granular substrate. Error bars represent values that are within 1.5 multiplied by the interquartile range of the first and third quartiles. Lowercase letters represent significance at P < 0.05 between each granular substrate within a cultivar.
Genetic and Environmental Interactions Influence Rice RSA
When grown in the three granular substrates, there was significant variation of five traits (depth, total length, width, convex area, and diameter) in response to both granular substrate and cultivar (GxE; ANOVA P < 0.004; Table III). The GxE interaction explained 11.7% to 15.5% of the variance in RSA traits. The genetic component explained 19.5% to 42.8% of the variance and contributed more than the environmental component. The one exception was root diameter, which had a greater environmental component (27%) than genetic component (19.5%). This was also the largest environmental contribution detected. Excluding diameter, the granular substrate environment explained 3.6% to 9.2% of the variance in RSA traits. Of the seven traits analyzed, root diameter was the only one influenced primarily by granular substrate, while the others were influenced predominantly by genotype.
Table III. ANOVA P values for the GxE interaction on RSA traits.
| Sample | Depth | Width | Total Length | Convex Area | Diameter | Median Root No. | Crown Root No. |
|---|---|---|---|---|---|---|---|
| Cultivar | 0.0021 | 0.0021 | 0.0021 | 0.0021 | 0.0021 | 0.0600 | 0.0021 |
| Granular substrate | 0.0021 | 0.0021 | 0.0040 | 0.0021 | 0.0021 | 0.0021 | 0.1356 |
| GxE | 0.0021 | 0.0021 | 0.0021 | 0.0021 | 0.0021 | 0.2230 | 0.1910 |
Two traits did not have a significant GxE interaction: crown root number and median root number. Median root number lacked a genetic component (ANOVA P = 0.06) and was statistically the same in all cultivars but was influenced by granular substrate (ANOVA P = 0.0021). Crown root number had a strong genetic component (ANOVA P = 0.0021) but no influence from the environment (ANOVA P = 0.1356). These differences are likely due to how each trait is calculated. Crown root number encompasses both the number of older/thicker roots and younger/thinner roots, whereas median root number counts only the older/thicker roots. Median root number is also highly dependent upon root diameter and depth, both influenced by granular substrate, which likely contributes to the environmental component observed. Taken together, these data suggest that crown root number is influenced predominantly by genotype and support the conclusion that root diameter and depth are influenced by the substrate texture (Fig. 3; Table II).
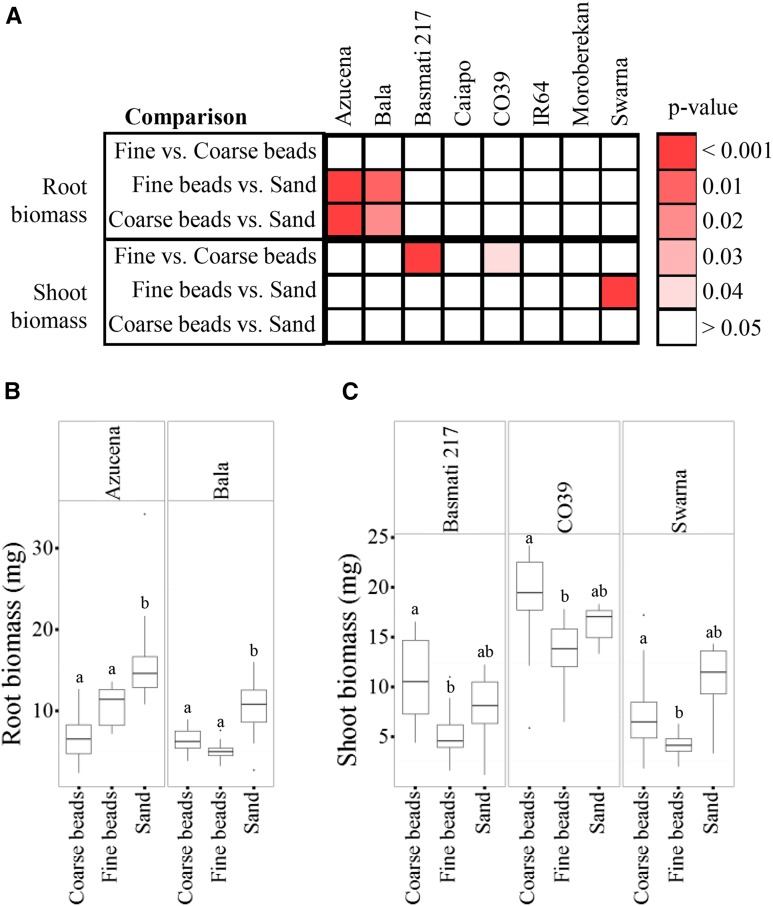
Granular Substrate Texture Impacts Plant Biomass
To determine if granular substrate texture impacts root biomass or shoot growth, we measured the dry weight for all cultivars (Table IV; Supplemental Table S5). We saw cultivar-specific changes dependent upon granular substrate in both root and shoot biomass (Fig. 4). However, none of the cultivars had a significant change in both root and shoot biomass. The shoot biomass in cv Basmati 217, CO39, and Swarna was affected by granular substrate, suggesting that shoot growth was related to the RSA response in some cultivars.
Table IV. Cultivar dry weight in granular substrates.
Values shown are means and sd of at least 10 plants.
| Granular Substrate | Cultivar | Root | Shoot | Root:Shoot Ratio |
|---|---|---|---|---|
| mg | ||||
| Fine beads | Azucena | 10.6 ± 2.5 | 13.9 ± 3.7 | 0.8 ± 0.3 |
| Basmati 217 | 9.1 ± 2.1 | 5.2 ± 2.3 | 2.3 ± 1.7 | |
| Bala | 5.1 ± 1.2 | 10.4 ± 2.9 | 0.5 ± 0.1 | |
| Caiapo | 15.6 ± 4.7 | 8.3 ± 2.7 | 2.0 ± 0.8 | |
| CO39 | 6.0 ± 1.4 | 13.5 ± 3.3 | 0.5 ± 0.2 | |
| IR64 | 11.5 ± 2.6 | 4.8 ± 2.0 | 2.8 ± 1.6 | |
| Moroberekan | 14.9 ± 2.6 | 5.8 ± 1.9 | 3.2 ± 2.5 | |
| Swarna | 6.8 ± 2.3 | 4.1 ± 1.2 | 1.8 ± 0.6 | |
| Coarse beads | Azucena | 6.7 ± 3.1 | 16.9 ± 4.2 | 0.5 ± 0.3 |
| Basmati 217 | 8.5 ± 2.2 | 10.6 ± 4.3 | 0.9 ± 0.4 | |
| Bala | 6.3 ± 1.4 | 14.2 ± 2.6 | 0.5 ± 0.2 | |
| Caiapo | 15.4 ± 3.0 | 12.5 ± 4.2 | 1.4 ± 0.7 | |
| CO39 | 6.3 ± 4.7 | 18.8 ± 5.3 | 0.5 ± 0.9 | |
| IR64 | 12.6 ± 1.4 | 8.6 ± 4.2 | 1.9 ± 1.0 | |
| Moroberekan | 10.4 ± 1.8 | 10.0 ± 4.0 | 1.2 ± 0.5 | |
| Swarna | 6.0 ± 2.5 | 7.3 ± 4.3 | 1.2 ± 0.9 | |
| Sand | Azucena | 16.3 ± 5.7 | 15.8 ± 3.8 | 1.0 ± 0.3 |
| Basmati 217 | 9.9 ± 1.8 | 7.8 ± 3.2 | 1.9 ± 1.9 | |
| Bala | 10.5 ± 3.4 | 15.3 ± 2.9 | 0.7 ± 0.2 | |
| Caiapo | 14.8 ± 4.1 | 12.9 ± 4.6 | 1.3 ± 0.9 | |
| CO39 | 7.3 ± 2.4 | 16.4 ± 1.8 | 0.4 ± 0.1 | |
| IR64 | 13.6 ± 4.7 | 8.7 ± 3.2 | 1.7 ± 0.9 | |
| Moroberekan | 14.2 ± 2.7 | 10.7 ± 1.1 | 1.3 ± 0.2 | |
| Swarna | 7.3 ± 3.5 | 10.9 ± 3.4 | 0.7 ± 0.3 | |
Figure 4.
Granular substrate texture influences root and shoot biomass. A, Depiction of significant differences between granular substrates for each cultivar and root trait. Shades of red indicate that a trait value is significantly different between indicated granular substrates; white indicates no significant difference. Pairwise significance for each granular substrate and cultivar was tested by post hoc one-to-one comparisons of the GxE term using the Tukey-Kramer HSD test. B and C, Quantification of root (B) and shoot (C) biomass for each cultivar with significant differences based on granular substrate. Error bars represent values that are within 1.5 multiplied by the interquartile range of the first and third quartiles. Lowercase letters represent significance at P < 0.05 between each granular substrate within a cultivar.
Comparison across all growth substrates revealed that cv Azucena RSA was largely unaltered, suggesting that RSA has a strong genetic influence in this cultivar. However, cv Azucena is one of two cultivars that had a slight increase in root biomass when grown in sand. These data suggest that genetics largely influence the RSA traits measured in this study but that additional traits exist that are influenced by the growth environment.
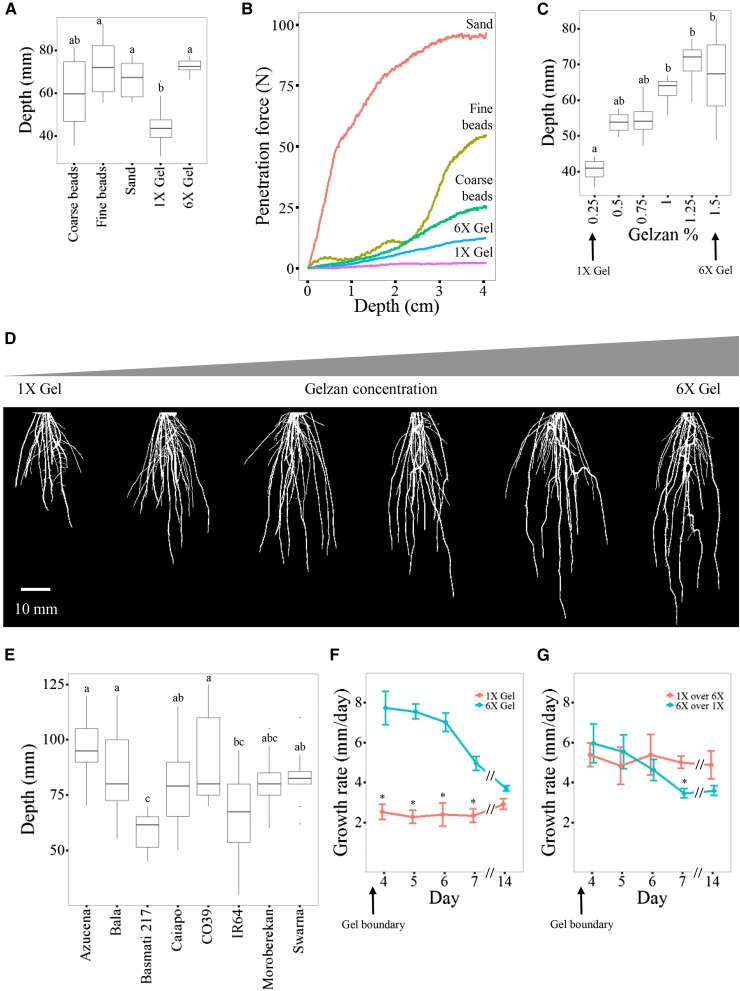
RSA Depth in cv Bala Is Decreased in Growth Substrates with Low Penetration Forces
We found that the roots of cv Bala, a drought-tolerant rice cultivar, were shallow when grown in 1× gel (Tukey’s HSD P ≤ 0.016, 1× gel compared with granular substrates) but deep when grown in granular substrate (Tukey’s HSD P > 0.9, between granular substrates; Fig. 5A). The maximum force (see “Materials and Methods”) required to penetrate the granular substrates was much greater than for the 1× gel, raising the possibility that cv Bala roots may grow less deep in soft substrates (Fig. 5B). To separate the effects of substrate texture from substrate hardness, we increased the concentration of Gelzan in the gel (6× gel; Fig. 2) and found that this caused an increase in RSA depth equal to what was seen when cv Bala was grown in granular substrates (Tukey’s HSD P > 0.945; Fig. 5A; Table I). We did not detect any relationship between gel hardness and depth in the other cultivars (Table I; Fig. 2). The cv Bala also was grown in increasing concentrations of Gelzan, and a strong correlation between depth and higher Gelzan concentrations was observed (Fig. 5, C and D). We also measured the maximum root depth of all eight cultivars when grown in coarse potting soil used for rice cultivation in the greenhouse. Under these conditions, maximum depth was not significantly different from that observed in granular substrates for all the cultivars (Fig. 5E). We also tested the pH, dissolved oxygen, and water percentage in the gel and in granular substrates. Similar amounts of dissolved oxygen were present in all granular substrates, and sand was the only growth substrate with a significant decrease in oxygen levels over time (Tukey’s HSD P = 0.0495; Table V). We found that the pH remained constant in both gel substrates but was considerably higher than expected in the coarse and fine beads (Table V). Comparison between trait values in plants grown in 1× gel and granular substrates suggests that the RSA response to soil texture is independent of the pH at this stage of development (Tables I and II). The fine beads had a small but significantly greater amount of water content than the coarse beads or sand (20% versus 19%; Table V). The 1× and 6× gels had equal amounts of water.
Figure 5.
The cv Bala RSA depth responds to the penetration force in the growth substrate. A, The cv Bala RSA depth in all growth substrates. B, Penetration force in all growth substrates. C, The cv Bala RSA depth in increasing concentrations of Gelzan. D, Representative camera images for plants in C after thresholding using GiA Roots. Bar = 10 mm. E, Mean RSA depth of all cultivars when grown in potting soil. F, The cv Bala growth rate in 1× and 6× gel. G, The cv Bala growth rate in layers of 1× and 6× gel. All roots have entered the lower gel layer by day 4. Values plotted represent the change in depth for each consecutive day. Day 14 represents the average depth change from day 7 to day 14. Error bars represent values that are within 1.5 multiplied by the interquartile range of the first and third quartiles (A, C, and E) or the se (F and G). Lowercase letters and asterisks represent significance at P < 0.05 between each growth substrate using Tukey’s HSD test.
Table V. Chemical properties of the growth substrates.
| Growth Substrate | Day | Dissolved Oxygen | pH | Water Content |
|---|---|---|---|---|
| mg L−1 | % | |||
| Deionized water | – | 3.3 ± 0.0 | 5.8 ± 0.0 | – |
| Yoshida’s solution | 0 | 4.6 ± 0.0 | 5.8 ± 0.0 | – |
| 7 | 4.2 ± 0.0 | 6.4 ± 0.0 | – | |
| 1× gel | 0 | 2.3 ± 0.2 | 5.8 ± 0.0 | 99.7 ± 0.1 |
| 7 | 2.0 ± 0.3 | 5.7 ± 0.1 | 99.7 ± 0.1 | |
| 6× gel | 0 | 2.2 ± 0.4 | 5.6 ± 0.0 | 98.3 ± 0.0 |
| 7 | 2.4 ± 0.2 | 5.5 ± 0.0 | 98.3 ± 0.0 | |
| Fine beads | 0 | 2.4 ± 0.1 | 10.2 ± 0.3 | – |
| 7 | 2.02 ± 0.5 | 9.8 ± 0.2 | 20.1 ± 0.1 | |
| Coarse beads | 0 | 2.4 ± 0.3 | 9.8 ± 0.6 | – |
| 7 | 1.82 ± 0.4 | 8.5 ± 0.3 | 19.3 ± 0.1 | |
| Sand | 0 | 2.3 ± 0.4 | 6.31 ± 0.0 | – |
| 7 | 1.44 ± 0.3 | 6.6 ± 0.1 | 19.5 ± 0.3 |
To further investigate the relationship between substrate hardness and cv Bala root growth, we quantified RSA depth over time and found that Bala has a much higher growth rate in the 6× gel compared with the 1× gel for at least 7 d (Fig. 5F). We also tested the dynamics of cv Bala root growth when encountering a sudden change in penetration force using containers that consisted of two layers of gel, either 1× gel on top of 6× gel or vice versa (Fig. 5G). Using this two-layer system, the oldest roots began to reach the bottom gel layer at approximately 3 d post planting. When roots transitioned from 6× gel to 1× gel, the growth rate decreased, and by day 6 it was equal to that of 1× gel alone. When roots transitioned from 1× gel to 6× gel, the roots grew parallel to the 6× gel prior to entering it. Upon penetration of the 6× gel layer, the growth rate increased rapidly to levels equal to that of the 6× gel alone. This increase in growth rate was maintained for several days but was equal to that of the 1× gel alone on day 14. We conclude that cv Bala root growth responds to a decrease in substrate penetration force by decreasing the rate of root growth and ultimately the RSA depth.
DISCUSSION
We used nondestructive x-ray CT imaging of rice roots to study the response of RSA to changes in the physical properties of a growth substrate. A major limitation to the use of x-ray CT is the inability to distinguish smaller root structures from the organic matter of soil, making it primarily suited to plants with larger root structures such as maize, wheat, and tomato. By taking a systematic approach, we were able to reliably distinguish rice roots from a granular substrate and capture the spatial arrangement of the majority of crown roots. This was made possible through the use of granular substrates with defined particle sizes. The bulk density was similar in the granular substrates, which allowed us to compare the RSA response to particle size and should facilitate future studies using mixtures of particle sizes that more closely resemble natural soil.
Our experiments revealed that RSA was shaped through interactions of genotype by growth substrate texture in 7-d-old plants and that genetic factors were more influential than environment at this time. The impact of soil texture on RSA may have been underestimated in this study based on previous reports using tomato, for which the greatest impact of soil texture on root growth was at 10 d after transplanting (Tracy et al., 2013). Nevertheless, we found that traits varied widely among the cultivars, suggesting that many cultivar-specific root traits have evolved independently, likely due to cultivation in different environments such as wet lowland paddies versus drier upland fields. A paddy-grown plant is likely to have adapted to the presence of adequate water and would benefit from increased root exploration near the soil surface, whereas upland rice survival is likely to be more dependent upon having deeper root systems that can reach water reserves. This could explain why we see cultivar-specific patterns of growth in our experiments and why the GxE response of root traits is largely controlled by genetic factors. We predict that each response has a biological underpinning and determines overall plant fitness, either through enhanced nutrient and water acquisition as a seedling or through root establishment that benefits the adult plant. For example, cv Azucena, an upland variety with known drought resistance (Price and Tomos, 1997), showed little response to varying growth substrate texture and had a consistent depth and spatial arrangement of roots. We hypothesize that one of the factors that makes cv Azucena drought resistant is its ability to maintain a consistent spatial growth pattern in the presence of hard soil and that this growth pattern allows the roots to quickly penetrate a layer of compacted soil in an attempt to reach deeper soil with higher water availability. Therefore, the RSA established during the seedling stage has major implications for survival of the adult plant and ultimately the next generation.
Our results show that both the growth substrate texture and hardness can have direct impacts on rice RSA (Table II; Fig. 5). Unlike in the homogenous gel substrate, which is formed via chemical cross-links, the stress in granular soils is localized in force chains, and the level of contact forces at the granular level depends on the size of the soil particles and compaction (Santamarina, 2003; Wendell et al., 2012). We observed that coarse beads were much easier to penetrate than fine beads and that sand was even more difficult to penetrate. The heterogeneity of stress on the granular level could be an important factor defining the RSA response to soil texture. Future studies will be required to tease apart the effects of hardness and texture on RSA. It has been reported in many cereal crops that root elongation decreases and root thickness increases as soil compaction increases, limiting soil exploration. Interestingly, we did not always see a correlation between harder growth substrate and reduced root length. Root traits of cv Bala, CO39, Moroberekan, and Swarna were unchanged in coarse beads and sand. We also observed that roots were thinner, not thicker, in the hardest granular substrate (Fig. 3B). One possible explanation for these differences are the unique cultivation practices of rice. Rice is one of the few cereal crops grown in a wide range of ecosystems that include lowland paddy fields and upland fields. An alternative explanation for differences in root thickness could be due to the roots being grown in a saturated substrate. Previous studies in rice have tested penetration ability as it pertains to drought avoidance or tolerance (Champoux et al., 1995; Ray et al., 1996; Price et al., 2000, 2002a, 2002b; Cairns et al., 2004, 2009; Bengough et al., 2006, 2011). These experiments involved withholding water or rain from fields or pots or the use of paraffin wax layers to simulate soil hardpan. These methods likely generate much harder soil than we used, but the phenotypes detected also may be caused by drought stress rather than increased penetration resistance.
In our experiments, we used equal concentrations of nutrient solution in all growth conditions to minimize the influence of nutrient availability on RSA. It is still possible that plants grown in gel substrate experience lower levels of calcium and magnesium because these elements are used to cross-link with Gelzan during solidification. However, we do not think nutrient levels influenced these experiments, because all of the cultivars had comparable RSA between at least one of the gel substrates and one of the granular substrates. Additionally, it is highly unlikely that any nutrient is limiting in our growth conditions during the first 7 d, because of the high levels of these molecules being present in Yoshida’s solution and because many of the nutrients for early root growth come from seed stores.
We identified a condition in which cv Bala RSA depth was greatly reduced in 1× gel, a substrate with low penetration force. The cv Bala RSA depth is considerably shorter than that of cv Azucena when grown in the field; as a result, these cultivars have been used for genetic studies to identify quantitative trait loci for root traits (Price et al., 1997, 2000, 2002b; Topp et al., 2013). Previous studies showed that the maximum root length of cv Azucena and Bala was equivalent when grown in soil cores until day 14, after which the cv Bala growth rate decreased (Price et al., 1997). Hydroponic conditions caused a reduction in cv Bala growth rate at day 7, while cv Azucena growth rate remained constant. These data support our findings that cv Bala root length is equal to that of cv Azucena when plants are grown in potting soil, granular substrates, and 6× gel but considerably shorter when grown in 1× gel. One would predict that our 1× gel would mirror what is seen using hydroponics, because the least amount of Gelzan required for solidification is used, thus creating a solidified hydroponic environment. Data from Price et al. (1997) support our findings that the cv Bala growth rate in 6× gel does not persist after 14 d, which we hypothesize is an energy-saving mechanism. The presence of shorter roots in hydroponics combined with our observation of cv Bala root length in different concentrations of Gelzan (Fig. 5, C and D) leads us to hypothesize that there is a minimum soil penetration force, between 2 and 12 n, required to elicit rapid root elongation in cv Bala. This allows the establishment of a framework for soil exploration, and below this level of force, root elongation is reduced drastically in order to conserve resources. This hypothesis is consistent with previous reports that indicated that soil structure and moderate compaction influence root establishment and growth (Hamza and Anderson, 2005; Atkinson et al., 2009; Tracy et al., 2012a, 2012b).
CONCLUSION
RSA traits have an impact on plant fitness and can provide a growth advantage during times of environmental stress, which ultimately impacts yield. Despite its importance, the response of RSA to soil is largely unexplored due to the difficulty of studying belowground structures. Our studies provide the foundation to understand how local soil forces influence RSA development. We have shown that x-ray CT analyses can uncover the RSA response to changes in the soil environment and can define novel areas for potential RSA improvement.
MATERIALS AND METHODS
Plant Growth
Seeds of rice (Oryza sativa) were dehulled and germinated in the dark at 30°C for 2 to 4 d in water (x-ray imaging) or on agar plates (gel imaging). Gel-based imaging was conducted at Duke University and performed as described (Topp et al., 2013), except that either 1× (0.25%) or 6× (1.5%) Gelzan was used in the glass cylinders. Glass cylinders containing layers of both 1× and 6× Gelzan were prepared by first allowing the bottom layer to solidify prior to pouring the top layer (3 cm thick). Plants of equal size were transferred to either 2-L glass cylinders containing Gelzan or aluminum soda cans (6.6 cm × 10.8 cm) containing either 0.24-mm-diameter (fine beads) or 0.7-mm-diameter (coarse beads) silica beads (Surface Preparation), sieved play sand (less than 0.71 mm; Quikrete), or potting soil composed of equal parts Fafard 52 (Wyatt Quarles) and Turface mound clay (Turface; Supplemental Table S1). Soda cans were prepared by removing the top and punching drainage holes in the bottom. The cans were filled with sand, silica beads (2.5 g cm−3), and potting soil that were saturated with Yoshida’s nutrient solution, pH 5.8. The soil bulk density in sand, coarse beads, fine beads, and potting soil was 1.384, 1.438, 1.431, and 0.044 g cm−3, respectively. The cans were submerged in Yoshida’s nutrient solution, and the level was kept constant at 1 cm below the top of the can with daily addition of water. All plants were grown for 7 d with a 12-h day/night cycle (550–600 µmol m−2 s−1), 28°C day, 25°C night in climate-controlled growth chambers at both universities. Eight rice cultivars were used: cv Azucena (tropical japonica), Basmati 217 (basmati), Bala (indica), Caiapo (tropical japonica), CO39 (indica), IR64 (indica), Moroberekan (tropical japonica), and Swarna (indica). X-ray reconstructions were generated for six to 15 individuals of each cultivar; 21 to 38 individuals were used for 1× gel analysis, and eight to 10 individuals were used for 6× gel analysis.
Root Traits
A detailed description of all root traits can be found in Topp et al. (2013), with the exception of crown root number and diameter, which were measured as described below. Crown root number represents the number of crown roots emerged that could be seen with the naked eye. Diameter is the root diameter calculated by GiA Roots, from a scanned flattened image of the root system. Maximum root depth represents the deepest extent of the root system. Maximum network width represents the widest extent of the root system. Network convex area, also known as convex hull area, is equal to the overall extent of space the network explores. Total root length represents the sum total of all root lengths in the network. Median root number represents the median number of roots that a horizontal line crosses from top to bottom of the network.
Root Diameter, Crown Root Number, and Dry Weight
Plants were grown at Duke University in soda cans containing granular substrate as described above alongside plants for gel imaging. The plants were removed 7 d post planting and washed. Crown roots were counted by hand, and the root system was imaged on a flatbed scanner. The scanned root images were input into GiA Roots, and the root diameter was calculated. After scanning, the plants were dried in a 60°C oven for 1 week prior to being weighed.
Gelzan Concentration Gradient
Two-liter glass cylinders were prepared as described above using 0.25% (1× gel), 0.5%, 0.75%, 1%, 1.25%, or 1.5% (6× gel) Gelzan. RSA depth was calculated after 7 d of growth using GiA Roots. Four replicates for each concentration were performed.
X-Ray Imaging and 3-D Reconstructions
All x-ray imaging was performed at the Georgia Institute of Technology. A custom x-ray CT system was used to capture 3-D root structure. A Spellman Monoblock 502 x-ray source paired with a Varian flat panel detector were used to capture CT sections. Soda cans were revolved on a home-built turntable fixed to a scissor lift at a rate of 10 steps per second to capture 200 images per rotation; top and bottom rotations were captured separately. The turntable and flat panel detector were positioned 52.1 ± 3 cm and 158.8 ± 3 cm from the x-ray source, respectively. After x-ray imaging, the plants were removed and the roots were photographed with a digital camera to verify the recovery of all roots after 3-D reconstruction.
Top and bottom CT sections were reconstructed separately using Cobra software (Exxim Computing) based on the filtered back-projection algorithm (Feldkamp). The preprocessing step included standard Exxim Computing algorithms and filters. The size of the reconstructed voxel was 0.2 × 0.2 × 0.2 mm and was exported as an image stack using MATLAB. Individually, top and bottom image stacks were input into Avizo 8.1 software (FEI), and intensity threshold-based semiautomatic segmentation was performed to generate a 3-D reconstruction. The top and bottom reconstructions were merged in Avizo using two reference points (metal bead or wire) that were attached to the outside of each can prior to imaging. 3-D reconstructions were exported from Avizo, reformatted, and input into GiA Roots. Reformatting involved generating two-dimensional image stacks (MATLAB) that were reassembled into a 3-D voxel model, and 40 rotational snapshots were captured (SciPy) and input into GiA Roots. All of the image manipulation techniques were validated by a ground truth model that was created as described (Topp et al., 2013). Three identical ground truth models were 3-D printed (Proto Labs) and imaged in a soda can containing fine beads. The average of all three models for each trait was used to calculate percentage error.
Penetration Force Measurements
Substrate hardness was measured by the insertion of a stainless steel cylindrical probe (diameter, 1.6 cm; length, 3.81 cm) into the substrate 4 cm (to approximate half of the maximum root depth). The cylinder was attached via a support rod (diameter, 0.63 cm) to a force-torque sensor (ATI Industrial) mounted on a robotic arm (CRS Robotics). We defined soil penetration force as the peak penetration force achieved during substrate penetration. All measurements were performed in 2-gallon paint buckets (Leaktite) with a diameter of 23 cm and a height of 24 cm, to minimize the effect on force from the container walls. The buckets were filled to a height of 16 cm with sand, fine beads, coarse beads, or gel. The bottom of the buckets containing sand or silica beads was perforated and immersed in a larger bucket (28 cm diameter) containing water. The water level was allowed to equilibrate prior to measuring penetration force to mimic the moisture content and growing conditions inside a soda can.
Soil Chemical Properties
Levels of dissolved oxygen (mg L−1) were measured at a depth of 4 cm following equilibration using a hand-held dissolved oxygen meter (Extech) with a polarographic type oxygen probe with automatic temperature compensation. A standard bench-top pH meter was used to measure the pH of the growth substrate. Water content was measured by comparing the dry and wet weights of each growth substrate and calculated as a percentage of water in the wet substrate.
Statistical Analysis
All statistical calculations were performed using JMP Pro version 11. To analyze the GxE interaction, a biological model that includes genotype, environment, and GxE was tested for each trait using linear regression calculated as the standard least squares (JMP). ANOVA P values were collected for all models and adjusted using the p.adjust function in R according to the Holm’s sequential Bonferroni method. The percentage of variance explained by each model was calculated as the R2 value. To calculate the percentage explained by each fixed variable, first the full model (genotype, environment, and GxE) was analyzed, followed by analysis with each variable removed. The R2 value for GxE was calculated by subtracting the R2 from a model containing genotype and environment from the full model. The R2 value for genotype was calculated by subtracting the R2 from a model containing only environment from the model containing genotype and environment. The R2 value for environment was calculated by subtracting the R2 from a model containing only genotype from the model containing genotype and environment. Pairwise significance for each combination of granular substrate and cultivar was tested by post hoc one-to-one comparisons of the GxE term using the Tukey-Kramer HSD test. Student’s t test was used to compare penetration forces in each growth substrate, because the variance was significantly lower in the two gel substrates compared with the granular substrates (P < 0.0001) using Brown-Forsythe, O’Brien, Levene, and Bartlett tests.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Images of particles used in this study.
Supplemental Figure S2. X-ray reconstruction of a ground truth model.
Supplemental Table S1. Particle size distribution in granular substrates.
Supplemental Table S2. Accuracy of RSA traits using a ground truth model.
Supplemental Table S3. Significant difference between granular substrates for each cultivar.
Supplemental Table S4. Least square means and connecting letters reports between simulated soils for each trait.
Supplemental Table S5. Cultivar dry weight in gel.
Supplementary Material
Acknowledgments
We thank Paul R. Zurek for providing Python scripts used for image manipulation, Maggie R. Wagner for help with statistical analyses, members of the Benfey laboratory for helpful discussion and feedback, and Nick Gravish for initial help with x-ray experiments.
Glossary
- RSA
root system architecture
- CT
computed tomography
- 3-D
three-dimensional
- GxE
genotype-by-environment
- HSD
honestly significant difference
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–14–11750 to P.N.B. and D.I.G. and Physics of Living Systems grant no. PHY–0957659 to D.I.G., D.M., and A.N.) and by the Gordon and Betty Moore Foundation (grant no. GBMF3405 to P.N.B.).
Articles can be viewed without a subscription.
References
- Atkinson BS, Sparkes DL, Mooney SJ (2009) Effect of seedbed cultivation and soil macrostructure on the establishment of winter wheat (Triticum aestivum). Soil Tillage Res 103: 291–301 [Google Scholar]
- Band LR, Fozard JA, Godin C, Jensen OE, Pridmore T, Bennett MJ, King JR (2012) Multiscale systems analysis of root growth and development: modeling beyond the network and cellular scales. Plant Cell 24: 3892–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe SE, Rojas-Pierce M, Yan XL, Blair MW, Pedraza F, Muñoz F, Tohme J, Lynch JP (2006) Quantitative trait loci for root architecture traits correlated with phosphorus acquisition in common bean. Crop Sci 46: 413–423 [Google Scholar]
- Bengough AG, Bransby MF, Hans J, McKenna SJ, Roberts TJ, Valentine TA (2006) Root responses to soil physical conditions: growth dynamics from field to cell. J Exp Bot 57: 437–447 [DOI] [PubMed] [Google Scholar]
- Bengough AG, McKenzie BM, Hallett PD, Valentine TA (2011) Root elongation, water stress, and mechanical impedance: a review of limiting stresses and beneficial root tip traits. J Exp Bot 62: 59–68 [DOI] [PubMed] [Google Scholar]
- Bushamuka VN, Zobel RW (1998) Differential genotypic and root type penetration of compacted soil layers. Crop Sci 38: 776–781 [Google Scholar]
- Cairns JE, Audebert A, Mullins CE, Price AH (2009) Mapping quantitative trait loci associated with root growth in upland rice (Oryza sativa L.) exposed to soil water-deficit in fields with contrasting soil properties. Field Crops Res 114: 108–118 [Google Scholar]
- Cairns JE, Audebert A, Townend J, Price AH, Mullins CE (2004) Effect of soil mechanical impedance on root growth of two rice varieties under field drought stress. Plant Soil 267: 309–318 [Google Scholar]
- Champoux MC, Wang G, Sarkarung S, Mackill DJ, O’Toole JC, Huang N, McCouch SR (1995) Locating genes associated with root morphology and drought avoidance in rice via linkage to molecular markers. Theor Appl Genet 90: 969–981 [DOI] [PubMed] [Google Scholar]
- Clark RT, MacCurdy RB, Jung JK, Shaff JE, McCouch SR, Aneshansley DJ, Kochian LV (2011) Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol 156: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dorlodot S, Forster B, Pagès L, Price A, Tuberosa R, Draye X (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 12: 474–481 [DOI] [PubMed] [Google Scholar]
- Downie H, Holden N, Otten W, Spiers AJ, Valentine TA, Dupuy LX (2012) Transparent soil for imaging the rhizosphere. PLoS ONE 7: e44276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers W, Kopke U, Hesse F, Bohm W (1983) Penetration resistance and root-growth of oats in tilled and untilled loess soil. Soil Tillage Res 3: 261–275 [Google Scholar]
- Eshel A, Waisel Y. 1996. Multiform and multifunction of various constituents of one root system In Waisel Y, Eshel A, Kafkafi U, eds, Plant Roots: The Hidden Half. Marcel Dekker, New York, pp 175–191 [Google Scholar]
- Fang S, Clark RT, Zheng Y, Iyer-Pascuzzi AS, Weitz JS, Kochian LV, Edelsbrunner H, Liao H, Benfey PN (2013) Genotypic recognition and spatial responses by rice roots. Proc Natl Acad Sci USA 110: 2670–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman L. (1994) The maize root. In M Freeling, V Walbot, eds, The Maize Handbook. Springer-Verlag, New York, pp 29–37 [Google Scholar]
- Galkovskyi T, Mileyko Y, Bucksch A, Moore B, Symonova O, Price CA, Topp CN, Iyer-Pascuzzi AS, Zurek PR, Fang S, et al. (2012) GiA Roots: software for the high throughput analysis of plant root system architecture. BMC Plant Biol 12: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488: 535–539 [DOI] [PubMed] [Google Scholar]
- Goodman AM, Ennos AR (1999) The effects of soil bulk density on the morphology and anchorage mechanics of the root systems of sunflower and maize. Ann Bot (Lond) 83: 293–302 [Google Scholar]
- Gouda PK, Kumar Varma CM, Saikumar S, Kiran B, Shenoy V, Shashidhar HE (2012) Direct selection for grain yield under moisture stress in Oryza sativa cv. IR58025B × Oryza meridionalis population. Crop Sci 52: 644–653 [Google Scholar]
- Gowda VRP, Henry A, Vadez V, Shashidhar HE, Serraj R (2012) Water uptake dynamics under progressive drought stress in diverse accessions of the OryzaSNP panel of rice (Oryza sativa). Funct Plant Biol 39: 402–411 [DOI] [PubMed] [Google Scholar]
- Gregory PJ, Hutchison DJ, Read DB, Jenneson PM, Gilboy WB, Morton EJ (2003) Non-invasive imaging of roots with high resolution X-ray micro-tomography. Plant Soil 255: 351–359 [Google Scholar]
- Hamza MA, Anderson WK (2005) Soil compaction in cropping systems: a review of the nature, causes and possible solutions. Soil Tillage Res 82: 121–145 [Google Scholar]
- Hodge A, Berta G, Doussan C, Merchan F, Crespi M (2009) Plant root growth, architecture and function. Plant Soil 321: 153–187 [Google Scholar]
- Intergovernmental Panel on Climate Change (2014) Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, UK
- Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN (2011) Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell 21: 770–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenneson PM, Gilboy WB, Morton EJ, Gregory PJ (2003) An x-ray micro-tomography system optimised for the low-dose study of living organisms. Appl Radiat Isot 58: 177–181 [DOI] [PubMed] [Google Scholar]
- Kalita UC. (2011) Soil Mechanics and Foundation Engineering. PHI Learning, New Delhi, India [Google Scholar]
- Kano-Nakata M, Inukai Y, Wade LJ, Siopongco JDLC, Yamauchi A (2011) Root development, water uptake, and shoot dry matter production under water deficit conditions in two CSSLs of rice: functional roles of root plasticity. Plant Prod Sci 14: 307–317 [Google Scholar]
- Le Marié C, Kirchgessner N, Marschall D, Walter A, Hund A (2014) Rhizoslides: paper-based growth system for non-destructive, high throughput phenotyping of root development by means of image analysis. Plant Methods 10: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J, Ho M (2005) Rhizoeconomics: carbon costs of phosphorus acquisition. Plant Soil 269: 45–56 [Google Scholar]
- Lynch JP, Brown KM (2012) New roots for agriculture: exploiting the root phenome. Philos Trans R Soc Lond B Biol Sci 367: 1598–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes JV, Liu J, Guimarães CT, Lana UG, Alves VM, Wang YH, Schaffert RE, Hoekenga OA, Piñeros MA, Shaff JE, et al. (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39: 1156–1161 [DOI] [PubMed] [Google Scholar]
- Mairhofer S, Zappala S, Tracy S, Sturrock C, Bennett MJ, Mooney SJ, Pridmore TP (2013) Recovering complete plant root system architectures from soil via x-ray μ-computed tomography. Plant Methods 9: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairhofer S, Zappala S, Tracy SR, Sturrock C, Bennett M, Mooney SJ, Pridmore T (2012) RooTrak: automated recovery of three-dimensional plant root architecture in soil from x-ray microcomputed tomography images using visual tracking. Plant Physiol 158: 561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCully M. (1995) Water efflux from the surface of field-grown grass roots: observations by cryo-scanning electron microscopy. Physiol Plant 95: 217–224 [Google Scholar]
- McCully ME. (1999) Roots in soil: unearthing the complexities of roots and their rhizospheres. Annu Rev Plant Physiol Plant Mol Biol 50: 695–718 [DOI] [PubMed] [Google Scholar]
- Merotto AJ, Mundstock C (1999) Wheat root growth as affected by soil strength. Rev Bras Cienc Solo 23: 197–202 [Google Scholar]
- Monaenkova D, Gravish N, Rodriguez G, Kutner R, Goodisman MA, Goldman DI (2015) Behavioral and mechanical determinants of collective subsurface nest excavation. J Exp Biol 218: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Mooney SJ, Pridmore TP, Helliwell J, Bennett MJ (2012) Developing x-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil 352: 1–22 [Google Scholar]
- Moran C, Pierret A, Stevenson A (2000) X-ray absorption and phase contrast imaging to study the interplay between plant roots and soil structure. Plant Soil 223: 101–117 [Google Scholar]
- Morita S, Nemoto K (1995) Morphology and anatomy of rice roots with special reference to coordination in organo- and histogenesis. In F Baluska, M Ciamporova, O Gasparikova, PW Barlow, eds, Structure and Function of Roots. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 75–86 [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, et al. (2012) Wheat grain yield on saline soils is improved by an ancestral Na⁺ transporter gene. Nat Biotechnol 30: 360–364 [DOI] [PubMed] [Google Scholar]
- Price AH, Steele KA, Gorham J, Bridges JM, Moore BJ, Evans JL, Richardson P, Jones RGW (2002a) Upland rice grown in soil-filled chambers and exposed to contrasting water-deficit regimes I. Root distribution, water use and plant water status. Field Crops Res 76: 11–24 [Google Scholar]
- Price AH, Steele KA, Moore BJ, Barraclough PB, Clark LJ (2000) A combined RFLP and AFLP linkage map of upland rice (Oryza sativa L.) used to identify QTLs for root-penetration ability. Theor Appl Genet 100: 49–56 [Google Scholar]
- Price AH, Steele KA, Moore BJ, Jones RGW (2002b) Upland rice grown in soil-filled chambers and exposed to contrasting water-deficit regimes II. Mapping quantitative trait loci for root morphology and distribution. Field Crops Res 76: 25–43 [Google Scholar]
- Price AH, Tomos AD (1997) Genetic dissection of root growth in rice (Oryza sativa L). 2. Mapping quantitative trait loci using molecular markers. Theor Appl Genet 95: 143–152 [Google Scholar]
- Price AH, Tomos AD, Virk DS (1997) Genetic dissection of root growth in rice (Oryza sativa L). 1. A hydrophonic screen. Theor Appl Genet 95: 132–142 [Google Scholar]
- Ray JD, Yu L, McCouch SR, Champoux MC, Wang G, Nguyen HT (1996) Mapping quantitative trait loci associated with root penetration ability in rice (Oryza sativa L.). Theor Appl Genet 92: 627–636 [DOI] [PubMed] [Google Scholar]
- Rich SM, Watt M (2013) Soil conditions and cereal root system architecture: review and considerations for linking Darwin and Weaver. J Exp Bot 64: 1193–1208 [DOI] [PubMed] [Google Scholar]
- Santamarina JC. (2003) Soil behavior at the microscale: particle forces. In J Germaine, T Sheahan, R Whitman, eds, Proceedings: Soil Behavior and Soft Ground Construction. American Society of Civil Engineers, Cambridge, MA, pp 25–56 [Google Scholar]
- Schulz H, Postma J, van Dusschoten D, Scharr H, Behnke S (2013) Plant root system analysis from MRI images. In A Ranchordas, J Madeiras Pereira, HJ Araujo, J Tavares, eds, Computer Vision, Imaging and Computer Graphics: Theory and Application. Springer, Berlin, pp 411–425 [Google Scholar]
- Shi L, Shi T, Broadley MR, White PJ, Long Y, Meng J, Xu F, Hammond JP (2013) High-throughput root phenotyping screens identify genetic loci associated with root architectural traits in Brassica napus under contrasting phosphate availabilities. Ann Bot (Lond) 112: 381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327: 818–822 [DOI] [PubMed] [Google Scholar]
- Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee CR, Zurek PR, Symonova O, Zheng Y, Bucksch A, Mileyko Y, Galkovskyi T, et al. (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci USA 110: E1695–E1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler S, Brown K, Lynch J (2011) Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341: 75–87 [Google Scholar]
- Tracy SR, Black CR, Roberts JA, McNeill A, Davidson R, Tester M, Samec M, Korošak D, Sturrock C, Mooney SJ (2012a) Quantifying the effect of soil compaction on three varieties of wheat (Triticum aestivum L.) using x-ray micro computed tomography (CT). Plant Soil 353: 195–208 [Google Scholar]
- Tracy SR, Black CR, Roberts JA, Mooney SJ (2013) Exploring the interacting effect of soil texture and bulk density on root system development in tomato (Solanum lycopersicum L.). Environ Exp Bot 91: 38–47 [Google Scholar]
- Tracy SR, Black CR, Roberts JA, Sturrock C, Mairhofer S, Craigon J, Mooney SJ (2012b) Quantifying the impact of soil compaction on root system architecture in tomato (Solanum lycopersicum) by X-ray micro-computed tomography. Ann Bot (Lond) 110: 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy SR, Roberts JA, Black CR, McNeill A, Davidson R, Mooney SJ (2010) The X-factor: visualizing undisturbed root architecture in soils using x-ray computed tomography. J Exp Bot 61: 311–313 [DOI] [PubMed] [Google Scholar]
- Uga Y, Okuno K, Yano M (2011) Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J Exp Bot 62: 2485–2494 [DOI] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45: 1097–1102 [DOI] [PubMed] [Google Scholar]
- Valentine TA, Hallett PD, Binnie K, Young MW, Squire GR, Hawes C, Bengough AG (2012) Soil strength and macropore volume limit root elongation rates in many UK agricultural soils. Ann Bot (Lond) 110: 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuprasad R, Impa S, Gowda R, Atlin G, Serraj R (2011) Rice near-isogenic-lines (NILs) contrasting for grain yield under lowland drought stress. Field Crops Res 123: 38–46 [Google Scholar]
- Wendell DM, Luginbuhl K, Guerrero J, Hosoi AE (2012) Experimental investigation of plant root growth through granular substrates. Exp Mech 52: 945–949 [Google Scholar]
- White RG, Kirkegaard JA (2010) The distribution and abundance of wheat roots in a dense, structured subsoil: implications for water uptake. Plant Cell Environ 33: 133–148 [DOI] [PubMed] [Google Scholar]
- Whitmore AP, Whalley WR (2009) Physical effects of soil drying on roots and crop growth. J Exp Bot 60: 2845–2857 [DOI] [PubMed] [Google Scholar]
- Yu LX, Ray JD, Otoole JC, Nguyen HT (1995) Use of wax-petrolatum layers for screening rice root penetration. Crop Sci 35: 684–687 [Google Scholar]
- Zheng HG, Babu RC, Pathan MS, Ali L, Huang N, Courtois B, Nguyen HT (2000) Quantitative trait loci for root-penetration ability and root thickness in rice: comparison of genetic backgrounds. Genome 43: 53–61 [PubMed] [Google Scholar]
- Zhu J, Ingram PA, Benfey PN, Elich T (2011) From lab to field, new approaches to phenotyping root system architecture. Curr Opin Plant Biol 14: 310–317 [DOI] [PubMed] [Google Scholar]
- Zobel M. (1996) Report of the Root Nomenclature Workshop ISRR V. 7th Symposium of the International Society of Root Research (ISRR) on “Root Research and Application.” Austrian Society for Root Research, Vienna [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.