Wild-type shot WUS expression and cytokinin responses can be repressed by a signal produced by bps1 mutant roots.
Abstract
The bypass1 (bps1) mutant of Arabidopsis (Arabidopsis thaliana) produces a root-sourced compound (the bps signal) that moves to the shoot and is sufficient to arrest growth of a wild-type shoot; however, the mechanism of growth arrest is not understood. Here, we show that the earliest shoot defect arises during germination and is a failure of bps1 mutants to maintain their shoot apical meristem (SAM). This finding suggested that the bps signal might affect expression or function of SAM regulatory genes, and we found WUSCHEL (WUS) expression to be repressed in bps1 mutants. Repression appears to arise from the mobile bps signal, as the bps1 root was sufficient to rapidly down-regulate WUS expression in wild-type shoots. Normally, WUS is regulated by a balance between positive regulation by cytokinin (CK) and negative regulation by CLAVATA (CLV). In bps1, repression of WUS was independent of CLV, and, instead, the bps signal down-regulates CK responses. Cytokinin treatment of bps1 mutants restored both WUS expression and activity, but only in the rib meristem. How the bps signal down-regulates CK remains unknown, though the bps signal was sufficient to repress expression of one CK receptor (AHK4) and one response regulator (AHP6). Together, these data suggest that the bps signal pathway has the potential for long-distance regulation through modification of CK signaling and altering gene expression.
Leaves and other lateral organs arise from a stem cell population called the shoot apical meristem (SAM). In Arabidopsis (Arabidopsis thaliana), the SAM initiates during embryogenesis in a rectangular area between the two cotyledons (Medford et al., 1992; Barton and Poethig, 1993). Recruitment of leaf founder cells from the SAM periphery causes a temporary reduction of SAM size, which is rapidly restored by homeostatic mechanisms; thereafter, the SAM is maintained at a stereotypical size. The SAM is divided into domains based on function and gene expression patterns. The central zone (CZ) is a narrow domain at the SAM’s apex and is the expression domain of CLAVATA3 (CLV3), which encodes a signaling peptide (Clark et al., 1995; Fletcher et al., 1999). The CLV3 peptide is perceived by a set of partially redundant receptors, encoded by CLV1, CLV2, CORYNE, and RECEPTOR-LIKE KINASE2 (Clark et al., 1997; Jeong et al., 1999; Miwa et al., 2008; Müller et al., 2008; Kinoshita et al., 2010), and functions to constrain cell proliferation. Another important SAM domain is the organizing center (OC), a small set of cells located just below the CZ that expresses WUSCHEL (WUS), a homeodomain protein that promotes SAM cell proliferation and specifies stem cell identity in adjacent cells (Laux et al., 1996; Mayer et al., 1998). Subjacent to the OC is the rib meristem (RM), whose activity contributes to stem growth. An elegant model for SAM homeostasis features interactions between WUS, which activates CLV3 expression, and the CLV pathway, which is proposed to repress WUS expression (Brand et al., 2000; Schoof et al., 2000). WUSCHEL-LIKE HOMEOBOX (WOX) genes appear to confer stem cell identity in other plant stem cell niches; e.g. WOX5 regulates activity of the root meristem, WOX4 regulates the activity of the vascular cambium, and in Arabidopsis, WOX1 and WOX3 together promote leaf blade outgrowth (Sarkar et al., 2007; Hirakawa et al., 2010; Nakata et al., 2012). What is less understood is how signaling originating from other organs regulates the SAM.
Coordination of SAM homeostasis relies in part on mobile signaling molecules. An important mobile signal for SAM homeostasis is cytokinin (CK). This compound is a potent activator of WUS expression that is normally produced in the upper tiers of the SAM’s CZ (Gordon et al., 2009; Zhao et al., 2010; Chickarmane et al., 2012). This localized production of active CK is important for SAM maintenance, as rice (Oryza sativa) mutants defective in its production have SAM maintenance defects (Kurakawa et al., 2007). Equally important for WUS expression is the localized perception of CK, which is carried out by two partially redundant CK receptors, ARABIDOPSIS HIS KINASE2 (AHK2) and AHK4 (Gordon et al., 2009; Chickarmane et al., 2012). Another mobile signal that is important for SAM homeostasis is the active CLV3 gene product, which is derived from a precursor protein that is processed into a secreted 12-amino acid glycopeptide (Fletcher et al., 1999; Kondo et al., 2006). The WUS transcription factor itself is also mobile, but instead of apoplastic movement, it travels through plasmodesmata (Yadav et al., 2011; Daum et al., 2014). Plasmodesmata are critical for SAM maintenance, presumably because they transport essential signaling molecules, such as WUS, and their aperture is also developmentally regulated to define symplastic domains (Rinne and van der Schoot, 1998; Kim et al., 2005; Benitez-Alfonso et al., 2009). However, how plasmodesmata gating regulation might play into stem cell homeostasis is unknown.
While plant stem cell populations typically function as intrinsically regulated drivers of growth, their activity can also be affected by environmental conditions (Xiong et al., 2002; Wolters and Jürgens, 2009). A large variety of environmental conditions lead to temporary growth reduction, including shoot arrest following drought perception by roots (Gowing et al., 1990; Mulholland et al., 1996; Jackson, 2002; Schachtman and Goodger, 2008). These observations suggest the existence of a mobile compound that is produced at sites where environmental stress is perceived, moves to the shoot, and regulates growth. Recently, translational regulation emerged as a possible mechanism coordinating SAM homeostasis with stress responses; an internal ribosome entry site in the WUS 5′ untranslated region appears to be used to restore normal SAM size as plants transition back to growth following stress perception (Cui et al., 2015). However, how stress perception leads to the initial growth arrest is not understood.
One key for understanding growth arrest is to identify the causal mobile signaling molecules. Both abscisic acid (ABA) and reactive oxygen species are candidates for the mobile stress-induced signaling compound (Munns and Sharp, 1993; Ishitani et al., 1997; Zhu, 2002; Miller et al., 2009); however, it is unclear whether either of these induces growth arrest. Another candidate for the growth-arrest pathway was discovered when analyzing the bypass1 (bps1) mutant (At1g01550) of Arabidopsis. Shortly after germination, bps1 mutants arrest shoot growth due to a mobile root-synthesized compound, which we call the bps signal (Van Norman et al., 2004). Because bps1 mutants are recessive, and BPS1 overexpression rescues bps1 mutants but only to a wild-type phenotype, we proposed that BPS1 is a negative regulator whose normal function is to prevent (or modulate) synthesis of the bps signal. The predicted BPS1 protein has no functionally characterized domains, but it does contain a large uncharacterized plant-specific domain of unknown function. Genetic analysis showed that synthesis of the bps signal occurs across multiple cell types in the root, requires an intact carotenoid biosynthesis pathway, and requires active root growth (Van Norman and Sieburth, 2007; Van Norman et al., 2011). Although the link between a carotenoid biosynthesis requirement and root-to-shoot signaling made it appealing to consider the possibility that the bps signal was ABA, double mutants that combine bps1 with ABA biosynthesis deficiency show the same severe bps1 phenotype, indicating that the bps signal continues to be synthesized, even in mutants not producing ABA. Because the BPS1 protein appears to negatively regulate bps signal synthesis, we suspect that the very severe bps1 phenotype arises because this mobile compound is produced in vast excess. We are pursuing characterization of this pathway to determine whether wild-type plants use the bps signal as part of a normal physiological response. For example, the bps signal might be used for growth arrest of salt-stressed plants, as both salt stress and the bps signal halt cell cycle progression (Burssens et al., 2000; Van Norman et al., 2011; Adhikari et al., 2013).
Here, we undertook a detailed analysis of shoot responses to the bps signal. We identified SAM maintenance as the earliest developmental defect in shoots and showed that the bps signal leads to both repression of WUS expression and diminished CK responses, even in wild-type shoots that were transiently grafted to the bps1 root. These data lead to an attractive model of long-distance growth control by interfering with stem cell homeostasis through diminished CK responses.
RESULTS
bps1 Mutants Show SAM Maintenance Defects
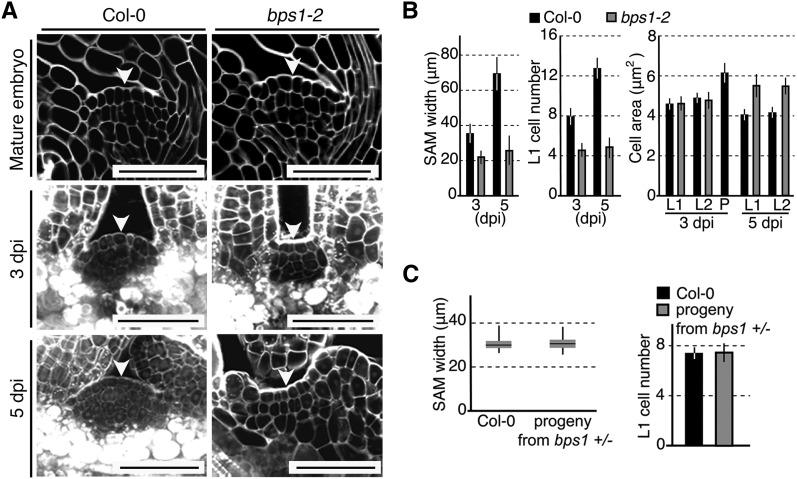
Previously, we showed that the growth arrest phenotype in bps1 shoots arises from a non-cell-autonomous activity derived from the root and that this activity is sufficient to repress growth of wild-type shoots (Van Norman et al., 2004, 2011). Those studies revealed that an early developmental defect was leaf blade outgrowth, which we detected in 5 d postimbibition (dpi) seedlings. Here, we sought to uncover the molecular and genetic basis for shoot arrest, and so reexamined very young bps1 seedlings for possible earlier shoot development phenotypes (Supplemental Fig. S1A). We found that bps1 mutants show defects in SAM maintenance. The normal SAM is a dome-shaped structure with distinct cell layers; by contrast, the 3 dpi bps1 SAM was small and flat, and it remained small at day five (Fig. 1, A and B; Supplemental Fig. S1B). We also examined SAM cell sizes; in the wild-type SAM, cells showed a cross-sectional area of 4 to 4.8 µm2, while the cells in nearby differentiating organs were larger (6 to 6.5 µm2). The bps1 SAM cell sizes were indistinguishable from those of the wild type at 3 dpi, but showed a modest size increase at 5 dpi (Fig. 1, A and B; Supplemental Fig. S1B). These observations suggested that bps1 mutants might have defects in stem cell maintenance. Consistent with this, we were unable to find the SAM in the 7 dpi bps1 seedlings (Supplemental Fig. S1D).
Figure 1.
BPS1 is required for postembryonic shoot apical meristem maintenance. A, Confocal images of the SAM of the wild type (Col-0) and bps1-2 mutants. White arrowheads indicate the center of the SAM, the CZ. For mature embryos and 5 dpi seedlings, the images show frontal longitudinal sections, while for 3 dpi seedlings, the images show sagittal longitudinal sections. Bars = 50 µm. B, Quantitative analysis of SAM sizes in 3 and 5 dpi seedlings. Between 16 and 24 SAMs were analyzed for each developmental stage and for each genotype. A diagram depicting the measurements is provided in Supplemental Figure S1B. Data are shown as the mean ± sd. C, Quantitative analysis of SAM size in mature embryos from the wild type and from the progeny of a self-pollinated bps1-2 heterozygote. The box and whisker plots depict the SAM width, with the box showing the sizes ranging from the 25th to the 75th quartile, and the whiskers showing the data extremes. L1 cell numbers show the mean ± sd.
We examined SAM morphology in mature embryos to determine whether the bps1 SAM defect arose during embryogenesis or during germination. We dissected mature embryos from seeds of a self-pollinated bps1-2 heterozygote and compared their SAM morphology to that of mature wild-type embryos. If the bps1 SAM defect arose during embryogenesis, we expected to find SAM defects in 22 to 23 of the 90 embryos examined. However, we found no embryonic SAM defects (Fig. 1, A and C), indicating that the bps1 SAM is established normally, and because defects arose after germination, that the bps1 SAM size defect is due to a lack of SAM maintenance. This timing of SAM defects is consistent with a previous study that found the bps signal to be largely produced post embryogenesis (Van Norman et al., 2011).
WUS Expression Is Lost in the bps1 SAM
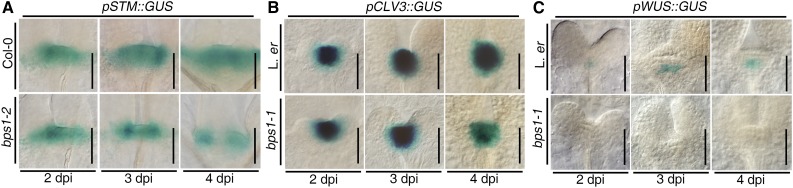
Maintenance of the SAM is largely governed by three well-established activities (Supplemental Fig. S1C). The first is SHOOTMERISTEMLESS (STM), which encodes a knotted-like homeobox gene that is required for establishment of the SAM during embryogenesis and for SAM maintenance after germination (Long et al., 1996; Kirch et al., 2003). We analyzed STM expression using pSTM::GUS, which in the wild type reveals the normal pattern of STM expression across the SAM (Fig. 2A). In bps1 mutants, pSTM::GUS expression looked normal in 2 and 3 dpi seedlings, though the expression area was smaller, consistent with the small and flat SAM in these mutants. However, in 4 dpi seedlings, pSTM::GUS expression was discontinuous. Staining appeared normal around the perimeter of the SAM but was lost from the central region. This change in expression occurred later than the appearance of SAM morphological defects, and so it is unlikely to explain the bps1 SAM maintenance defects.
Figure 2.
The bps1 SAM fails to express WUS. A to C, GUS staining in the SAM of 2, 3, and 4 dpi seedlings. In each, the top row shows the pattern in the wild type (Col-0 or Ler), and the bottom row shows the pattern in bps1 mutants. A, pSTM::GUS; B, pCLV3::GUS; C, pWUS::GUS. Bars = 50 µm.
A second activity that is important in regulation of SAM maintenance is the CLV3 signaling peptide pathway. Perception of the CLV3 peptide by its receptors leads to repression of WUS expression (Brand et al., 2000; Schoof et al., 2000), and we reasoned that SAM arrest might be brought about by CLV3 overexpression. We used pCLV3::GUS to compare expression in wild-type and bps1 seedlings. In both the wild type and bps1 mutants, pCLV3::GUS expression was found in its normal CZ expression domain (Mayer et al., 1998; Fig. 2B). To more carefully visualize these GUS staining patterns, we also compared a GUS incubation time course (Supplemental Fig. S1E); this revealed that while the pCLV3::GUS expression domain was smaller in bps1, the staining intensity in the two genotypes was very similar, indicating that shoot arrest in bps1 is unlikely to be a consequence of the CLV pathway.
A third important activity that governs normal SAM maintenance is WUS, which encodes a transcription factor that is expressed in the OC, a small domain at the center of the SAM whose function is to promote SAM identity in nearby cells (Laux et al., 1996; Mayer et al., 1998). We characterized WUS expression using pWUS::GUS (Bäurle and Laux, 2005). Expression in the wild type matched its normal embryonic expression in 2, 3, and 4 dpi seedlings, namely, in the top central L3 as well as the OC (Fig. 2C; Supplemental Fig. S1G). By 5 dpi, WUS expression matched the adult pattern, in the OC below L3 and at the top central regions of the RM (Supplemental Fig. S1G). By contrast, pWUS::GUS expression was not detectable in bps1 mutants at any of the time points examined (Fig. 2C; Supplemental Fig. S1F). A loss of WUS expression is an attractive explanation for the bps1 shoot phenotype, as it is consistent with the observed loss of SAM maintenance.
The bps Signal Is Sufficient to Reduce WUS Expression
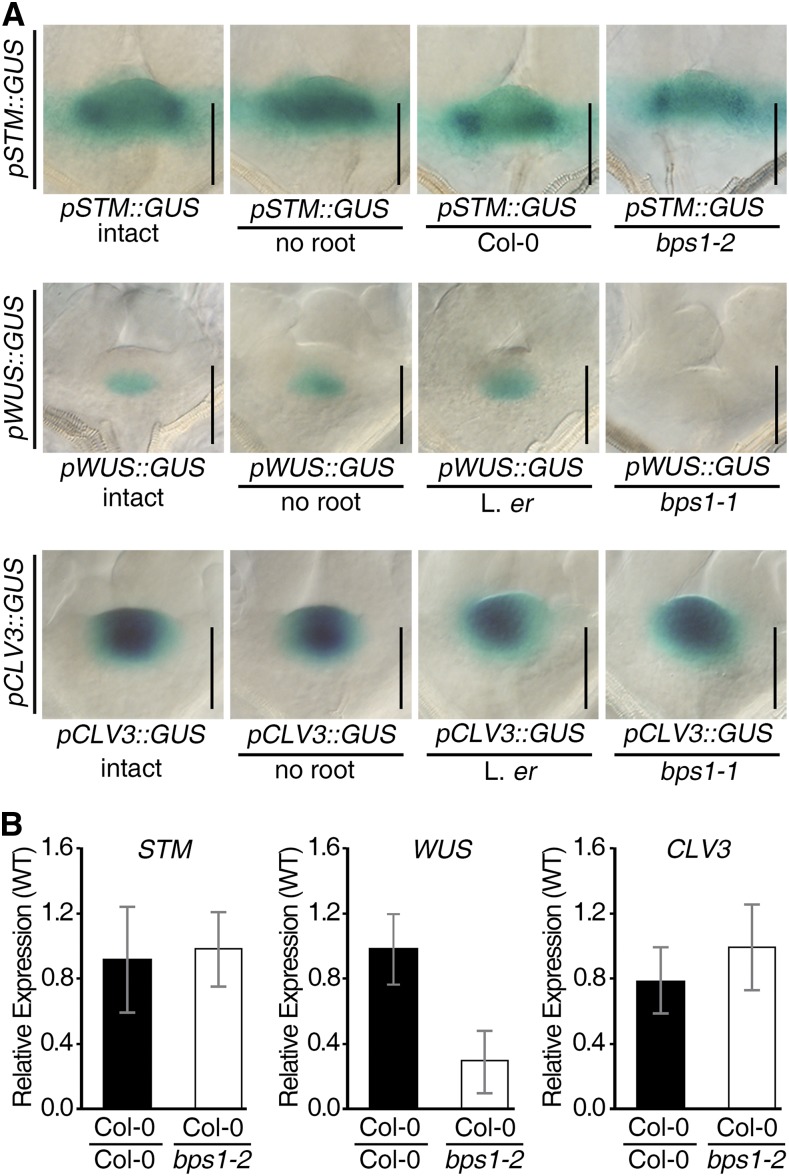
Previously, we used traditional micrografts to show that the bps1 root was sufficient to arrest shoot development in the wild type (Van Norman et al., 2004). Here, we extended the grafting approach to test whether the bps1 root was also sufficient to repress pWUS::GUS expression. We used a transient micrograft assay (Adhikari et al., 2013), which, by providing an aqueous cushion between the scion and rootstock (agarose in water, ∼0.3 mm; Supplemental Fig. S2A), allows the transfer of hydrophilic molecules between scion and rootstock without the delay from tissue repair that occurs with traditional grafting. The 4 dpi wild-type scions carrying SAM markers were coupled to either wild-type or bps1 roots or applied to just agarose (no root) and GUS stained 24 h after coupling. Scions carrying pSTM::GUS and pCLV3::GUS produced GUS staining patterns that were identical, regardless of whether they had been transiently grafted to bps1 (Fig. 3A). However, scions carrying pWUS::GUS showed a root genotype-dependent change in GUS expression. pWUS::GUS expression was undetectable in wild-type shoots coupled to the bps1 root, whereas in transient micrograft controls (no root or wild-type root), we found normal pWUS::GUS expression (Fig. 3A; Supplemental Fig. S2B). This specific response to the bps1 root indicated that the mobile root-sourced bps signal affected SAM maintenance by interfering with WUS expression.
Figure 3.
The bps1 root is sufficient to repress WUS expression in wild-type scions. A, Analysis of shoot expression of pSTM::GUS, pWUS::GUS, and pCLV3::GUS in 5-d wild-type plants that were intact or in scions that were applied to the agarose grafting apparatus but no root or coupled to wild-type or bps1 roots. GUS staining was carried out 24 h after transient graft establishment. Bars = 50 µm. B, Quantitative analysis of STM, WUS, and CLV3 expression in 5-d wild-type shoots transiently grafted to either a wild-type (Col-0) or bps1-2 root. Data are shown as the mean ± sd of three biological and two technical replicates.
Because these analyses relied on pWUS::GUS, a transgenic marker, we addressed whether the bps1 root response was specific for WUS or whether it might be associated with the pWUS::GUS transgene, e.g. because of its insertion site. We tested two independently produced pWUS::GUS markers, FspBst and ApaBst (Bäurle and Laux, 2005). Both of these lines showed identical robust and specific bps1 root-induced repression in transient graft assays (Supplemental Fig. S2C), which ruled out insertion site as a confounding influence. We also tested whether the bps1 root affected expression from the endogenous WUS locus. We generated transient micrografts that coupled wild-type scions to either bps1 or wild-type roots for 24 h, isolated RNA from the wild-type scions, and used qRT-PCR to analyze transcript levels for STM, WUS, and CLV3. Only WUS showed a significant change in expression (−5.3-fold; P < 0.05; Fig. 3B; Supplemental Table S1), and there was no significant impact on CLV3 or STM transcript levels. This finding, together with bps1’s repressive effect on three independently produced pWUS::GUS marker lines, indicates that the bps1 root leads to WUS repression.
CLV-Independent Repression of WUS Expression
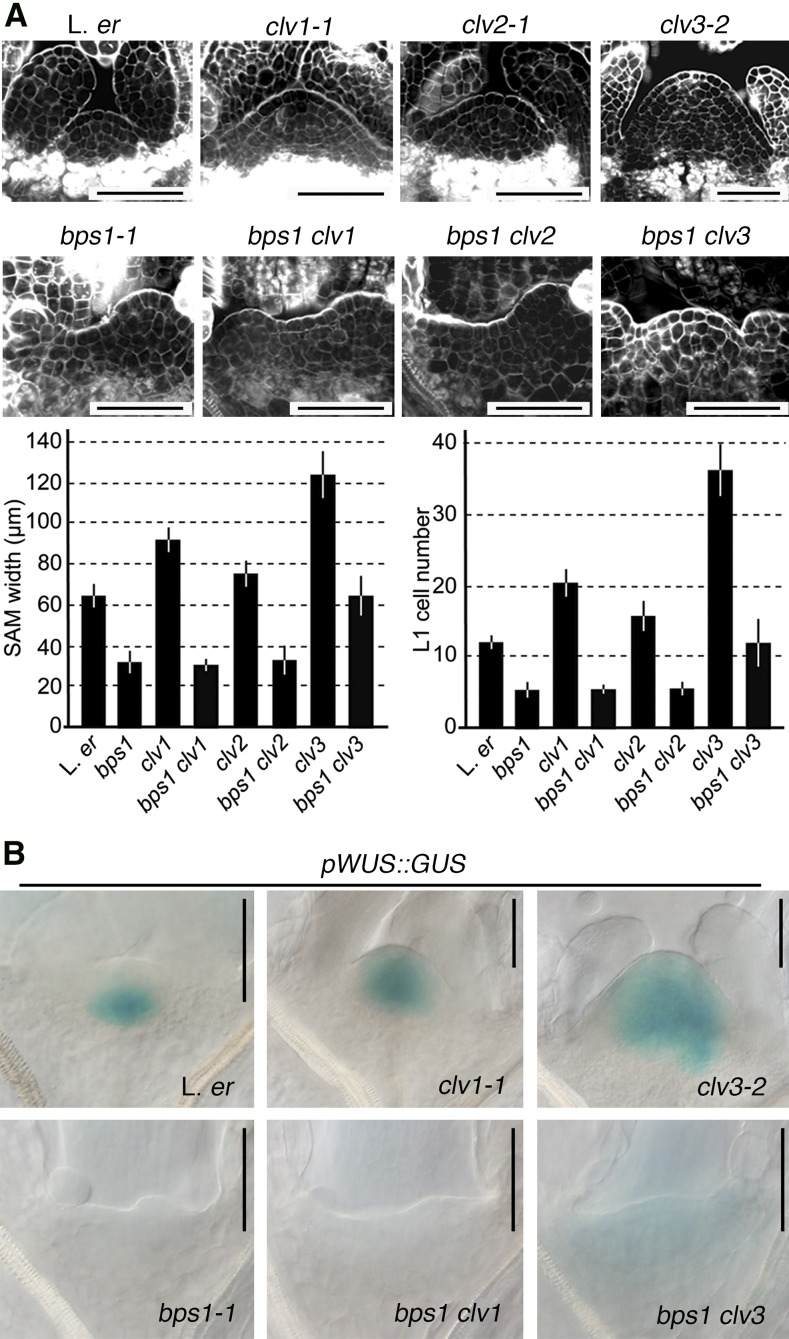
Finding that the bps1 root could repress WUS expression led us to next consider possible mechanisms. Despite not observing CLV3 overexpression (Figs. 2 and 3), we considered it still possible that the bps signal might work through the CLV3 peptide signaling pathway, which functions to specify the apical boundary of WUS expression (Brand et al., 2000; Schoof et al., 2000; Chickarmane et al., 2012). We tested this idea genetically, reasoning that if SAM arrest required CLV, then bps1 clv double mutants would be less effective at repressing WUS, and so SAM maintenance would be at least partially restored. However, the size and organization of bps1 clv1-1 and bps1 clv2-1 SAMs were indistinguishable from those of bps1 single mutants (Fig. 4A), indicating that neither CLV1 nor CLV2 were required for bps signal-mediated SAM arrest.
Figure 4.
CLV3 peptide signaling and the bps signal are independent negative regulators of pWUS::GUS expression. A, Confocal images depict the SAM of single and double mutants (top) and a quantitative analysis of SAM size at 5 dpi. Graphs depict data from 17 to 20 seedlings for each genotype and are shown as the mean ± sd. B, pWUS::GUS expression in single and double mutants at 5 dpi. Bars = 50 µm.
We also analyzed the bps1 clv3 double mutant and found that it produced a SAM that was intermediate in size between the clv3-2 single mutant and the bps1-2 single mutant (Fig. 4A). This intermediate size could mean that the bps signal worked through CLV3 to induce SAM arrest (independently of CLV1 or CLV2). However, another possibility was that the intermediate phenotype resulted from different developmental timing: CLV3 is required both during embryogenesis and after germination, while production of the bps signal starts late in embryogenesis in bps1 mutants (Schoof et al., 2000; Van Norman et al., 2004, 2011). To distinguish between these possibilities, we compared pWUS::GUS expression in bps1 clv1 and bps1 clv3 double mutants. As in bps1 single mutants, pWUS::GUS expression was not detectable in the bps1 clv1 mutants (Fig. 4B). Similarly, pWUS::GUS expression in the bps1 clv3 double mutants ranged from undetectable to very faint. In addition, we examined older bps1 clv3 seedlings; if shoot arrest by the bps signal requires CLV3, then we would expect to observe continued SAM maintenance and expansion in bps1 clv3 double mutants. However, at 12 d, the bps1 clv3 seedlings were indistinguishable from bps1 (Supplemental Fig. S3). Both bps1 and bps1 clv3 had only a single pair of small aberrant leaf primordia, in contrast to the four leaves produced by the wild type and the six leaves produced by clv3. In addition, the SAM of bps1 clv3 seedlings failed to be maintained during continued growth, and by 12 d, its size was indistinguishable from that of the bps1 single mutant. Thus, these data confirmed that the bps signal functions independently of CLV signaling and indicated that the bps signal can repress WUS expression even in the very broad expression domain conditioned by the loss of CLV3.
The Root-Derived bps Signal Is Sufficient to Reduce Cytokinin Responses
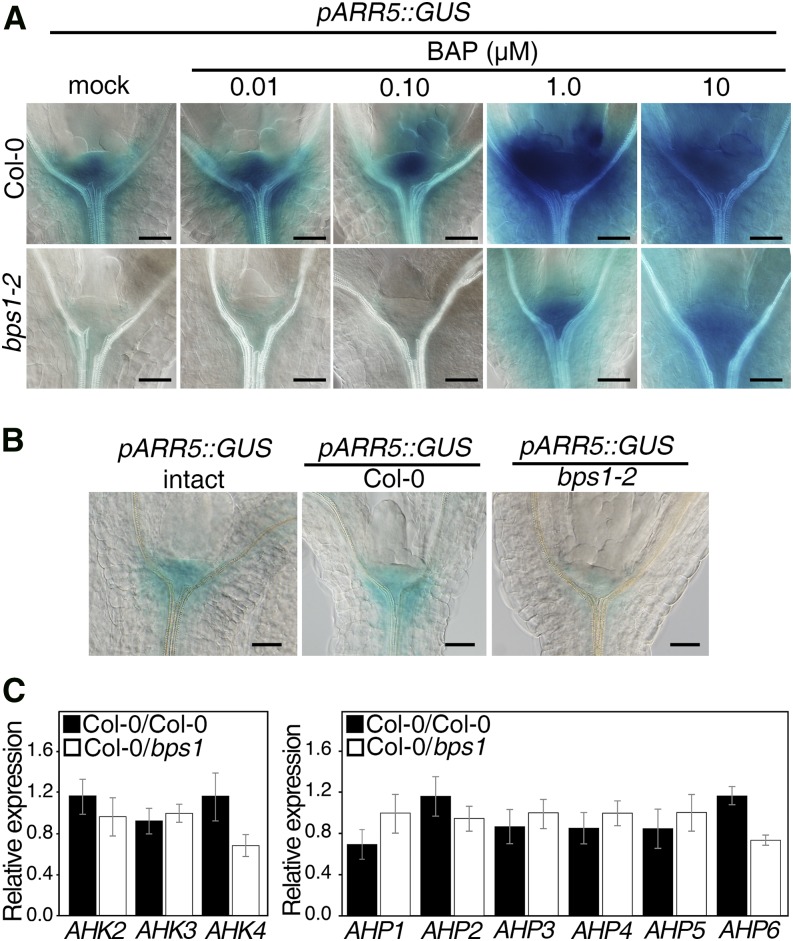
A second potential mechanism by which the bps signal might affect WUS expression is by interfering with its positive regulation by CK (Kurakawa et al., 2007; Gordon et al., 2009; Chickarmane et al., 2012). We tested whether the bps signal affected general CK signaling using the CK response reporter pARR5::GUS (D’Agostino et al., 2000). Under normal growth conditions, the wild type showed broad expression across much of the SAM and especially strong expression in the SAM’s center (Fig. 5A). By contrast, bps1 mutants showed only faint expression that was largely found beneath the SAM, and associated with the vascular tissue, suggesting a defect in CK responses in bps1 mutants. We next analyzed pARR5::GUS expression in seedlings 24 h after being transferred to CK-containing medium (N6-benzylaminopurine [BAP]; 0.01–10 µM). The wild type transferred to 0.01 and 0.10 µM BAP showed expression that was very similar to the mock-treated control and progressively stronger expression following transfer to 1.0 and 10.0 µM BAP media (Fig. 5A). The results from bps1 mutants were similar, except that for all CK concentrations, expression was much lower than that of the wild type. Low expression that was predominantly in the RM was observed for mock-treated, 0.01 µM, and 0.10 µM BAP, while expression was progressively stronger in mutants that had been transferred to 1.0 and 10.0 µM BAP. These observations indicate that bps1 mutants fail to produce a robust CK response.
Figure 5.
The bps signal is sufficient to reduce cytokinin responses. A, Shoot expression of the CK response marker pARR5::GUS in 5-d wild-type (Col-0) and bps1-2, mock-treated or transferred to 0.01 to 10 µM CK (BAP) for 24 h. Bars = 50 µm. B, Transient graft assay of pARR5::GUS in 5-d wild-type plants that were intact or scions that were coupled to wild-type or bps1 roots. GUS staining was carried out 24 h after transient graft establishment. Bars = 50 µm. C, Quantitative analysis of AHK and AHP expression in 5-d wild-type shoots transiently grafted to either a wild-type (Col-0) or bps1-2 root. Data are shown as the mean ± sd of three biological and two technical replicates.
To determine whether the reduced CK response was induced by the root-derived bps signal, we used transient micrografts; wild-type scions carrying pARR5::GUS were coupled to either bps1 or wild-type roots, and GUS expression analyzed 24 h after coupling (Fig. 5B; Supplemental Fig. S4). The intact and wild-type root graft controls showed indistinguishable patterns and intensity of GUS staining, indicating that pARR5::GUS expression is not sensitive to the experimental treatments involved in micrograft establishment. However, wild-type shoots coupled to bps1 roots showed strongly reduced pARR5::GUS expression, and residual expression was largely restricted to the RM. This pattern matched that of pARR5::GUS in bps1 mutants and indicated that the root-derived bps signal was sufficient to reduce pARR5::GUS expression.
To get at how the bps signal might be interfering with CK responsiveness, we tested whether the bps signal repressed expression of cytokinin receptors or response regulators. The Arabidopsis genome encodes three CK receptor genes, ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and CRE1/WOL1/AHK4 (Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001; Yamada et al., 2001). We used qRT-PCR to assess their relative expression in wild-type shoots transiently micrografted to either wild-type or bps1 roots (Fig. 5C) and required a fold change of <−1.5 or >1.5 and P value < 0.05 for significance. The expression of AHK2 and AHK3 was not significantly affected by grafting to the bps1 roots; however, wild-type shoots coupled to bps1 root showed a significant decrease in AHK4 expression (−1.7-fold change, P < 0.05; Supplemental Table S1). Although WUS expression has been linked to CK perception through AHK2 and AHK4 (Gordon et al., 2009), the impact of the bps signal on WUS expression (Fig. 3B) was much stronger than its effect on AHK4 expression.
We also used qRT-PCR to assess the impact of the bps1 root on expression of the ARABIDOPSIS HIS PHOSPHOTRANSFER (AHP) gene family, of which six genes are encoded by the Arabidopsis genome (Suzuki et al., 2000; Fig. 5C). Only AHP6 showed a significant response to the bps1 root (−1.6-fold change, P < 0.05; Supplemental Table S1). AHP6 is an unusual member of this gene family, as it inhibits rather than promotes phosphotransfer, thus inhibiting cytokinin signaling (Mähönen et al., 2006). In the root, AHP6 plays important roles in regulating differentiation of protoxylem, while in the inflorescence meristem (IM), AHP6 expression at the flanks of the IM appears to negatively regulate IM size (Mähönen et al., 2006; Bartrina et al., 2011). However, if APH6 function in the SAM is similar to its functions in the IM, we would expect its reduced expression to lead to enhanced SAM size, the opposite of the bps1 phenotype. Thus, reduced AHP6 is unlikely to explain the diminished bps1 SAM size or the loss of WUS expression in bps1 mutants.
CK Treatment Recovers WUS Expression in bps1, But Only in the RM
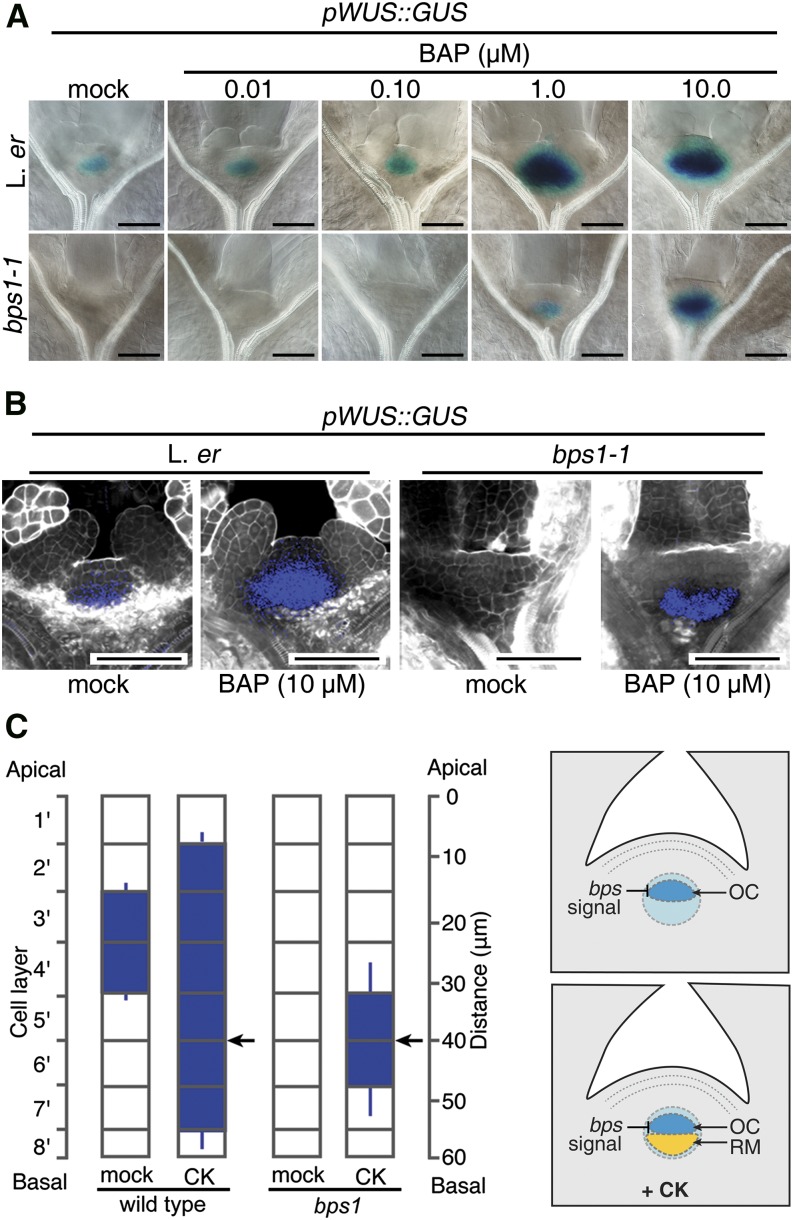
Because CK provided to bps1 mutants led to partially restored pARR5::GUS expression, we tested whether pWUS::GUS expression in bps1 also responded to exogenous CK. As with the stronger and broader WUS expression reported for CK-treated wild-type inflorescences (Lindsay et al., 2006; Gordon et al., 2009; Chickarmane et al., 2012), wild-type seedlings transferred to media supplemented with CK (BAP) also showed broader pWUS::GUS expression, though only at concentrations of 1 µM + (Fig. 6A), in agreement with our earlier results using pARR5::GUS (Fig. 5A). Similarly, bps1 mutants transferred to 0, 0.01, and 0.10 µM BAP showed no pWUS::GUS expression, but expression was partially recovered in seedlings transferred to 1.0 and 10 µM BAP (24 h). This expression, though, was not in its normal domain; instead, it was limited to the RM and did not include the OC (Fig. 6, B and C). These data suggest that these high CK treatments allow WUS expression to occur in two separable domains, an upper OC domain and a lower RM domain, and highlight the importance of the bps signal in specifically inhibiting expression in the OC domain.
Figure 6.
CK treatment restores WUS expression in the bps1 rib meristem. A, Shoot expression of the pWUS::GUS in 5-d wild-type (Ler) and bps1-1, mock-treated or transferred to 0.01 to 10 µM CK (BAP) for 24 h. Bars = 50 µm. B, Confocal images of pWUS::GUS expression (4 dpi) in mock-treated or in seedlings transferred to 10 µM BAP for 24 h. C, Quantitative analysis of CK-treated pWUS::GUS expression domain positions; scale on the left indicates cell layers, and scale on the right indicates distance from the apex (n = 20). Arrows point to strongest GUS staining (error bar, sd). Cartoon to the right depicts the two domain model for WUS expression.
WUS Repression by the bps Signal: Altered CK Signaling or SAM Differentiation?
Our analysis of SAM arrest in bps1 mutants showed that the bps signal repressed WUS expression and reduced CK responsiveness, but whether the reduction in WUS expression was a direct result of altered CK, was unclear. An alternative explanation of these linked responses was that the bps signal induces general SAM differentiation. To distinguish between these possibilities, we assessed SAM differentiation status using several approaches.
First, we tested whether the CK-induced WUS expression in bps1 mutants (in the RM) conferred stem cell maintenance. In wild-type seedlings, CK treatment resulted in a SAM that was wider than the mock-treated control and that had small meristematic cells extending deeper into the RM (Fig. 7A). The SAM from CK-treated bps1-1 mutants was also deeper and had additional layers of small meristem-like cells in the RM. However, the SAM apex remained narrow and flat. These observations revealed that the RM domain of the bps1 SAM remained responsive to WUS expression and was consistent with the pattern of pARR5::GUS induction (Fig. 5). Nevertheless, it was unclear why the SAM L1 or L2 cells failed to respond to this restored WUS expression, but possibilities include their distance from the bps1 WUS expression domain (the RM), because the bps signal had altered the responsiveness of these cells, or because these cells had differentiated.
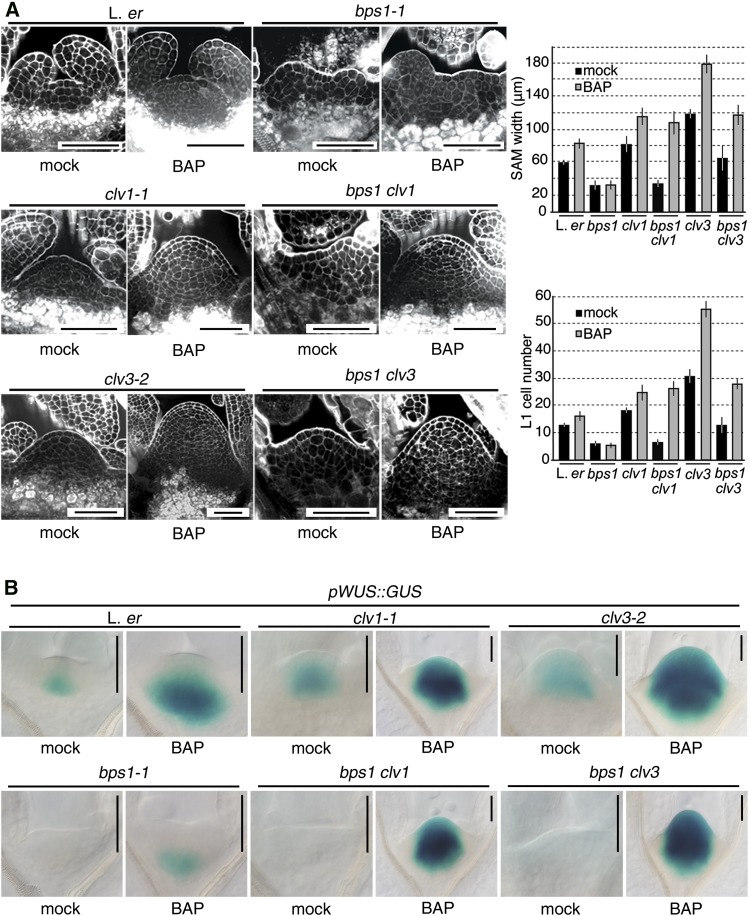
Figure 7.
In clv mutant backgrounds, CK overrides the repression of WUS expression provided by the bps signal. A, Confocal images of the SAM of 5 dpi seedlings, mock-treated and grown on medium containing CK (0.1 µM BAP). Seed germination and plant growth were on GM without or with 0.1 µM BAP until 5 dpi. To the right, histograms depict SAM sizes of mock-treated and CK-treated seedlings. Error bars indicate sd (n = 10, at 5 dpi). B, pWUS::GUS expression in single and double mutants at 5 dpi from the experiment condition in A. Bars = 50 µm.
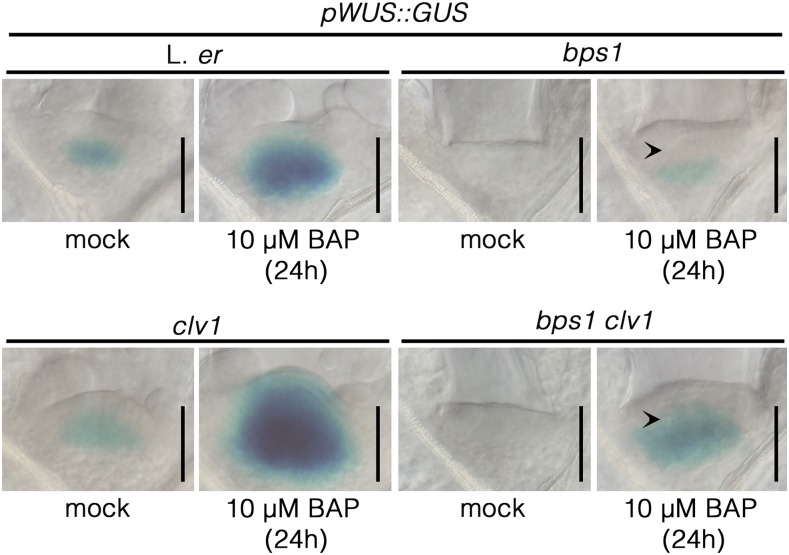
As another strategy, we used a genetic approach and assessed the response of bps1 clv double mutants to exogenous CK. As reported by others, clv1 and clv3 mutants produce a very large SAM in response to exogenous CK (Lindsay et al., 2006; Gordon et al., 2009; Zhao et al., 2010). Although bps1 mutants do not respond to CK by SAM apex expansion and do not require CLV signaling for repression of WUS expression, we found that both bps1 clv1-1 and bps1 clv3-2 double mutants grown on CK-supplemented medium produced large SAMs (Fig. 7A); this revealed that cells of the L1 and L2 were able to proliferate in bps1 backgrounds. Furthermore, this proliferation appeared to be driven by WUS, as pWUS::GUS expression extended to the L1 of CK-supplemented bps1 clv double mutants (Fig. 7B). Notably, these seedlings (12 d) still showed other features of the bps1 phenotype, including short abnormal roots and small cotyledons, which indicated that the bps signal continued to be produced (Supplemental Fig. S5). Because this experiment involved germinating the bps1 clv mutants on CK-containing medium, and we were interested in whether the bps1 SAM had differentiated, we repeated the bps1 clv CK induction experiment but by transferring germinated bps1 clv1 double mutants to CK-supplemented growth medium (GM). Because bps1 and clv1 bps1 are phenotypically indistinguishable when grown on GM, we expected that if the bps1 SAM was partially differentiated, then the same would be true for bps1 clv1 double mutants. Seedlings transferred to CK-supplemented medium were analyzed for pWUS::GUS expression 24 h later (Fig. 8). In bps1 clv1 double mutants, transfer induced pWUS::GUS in a pattern that was very similar to that of the wild type; expression was detected in the OC and also deeper into the RM (Fig. 8). Remarkably, these bps1 clv1 double mutants also had a slightly dome-shaped SAM, suggesting that in the absence of CLV, restored WUS expression also restored normal SAM maintenance. These observations revealed that the CZ cells in bps1 clv1 double mutants remained able to respond to WUS, even when it is expressed several days after germination. Moreover, this restored WUS response required enhancing CK responsiveness, which was conditioned through the loss of CLV signaling.
Figure 8.
CK restores pWUS::GUS expression and rescues SAM maintenance in 4-d seedlings. Three-day seedlings that germinated on GM were transferred to medium containing 10 µM BAP and GUS stained 24 h later. Arrowhead in BAP-treated bps1 and bps1 clv1 indicates the OC position. Bars = 50 µm.
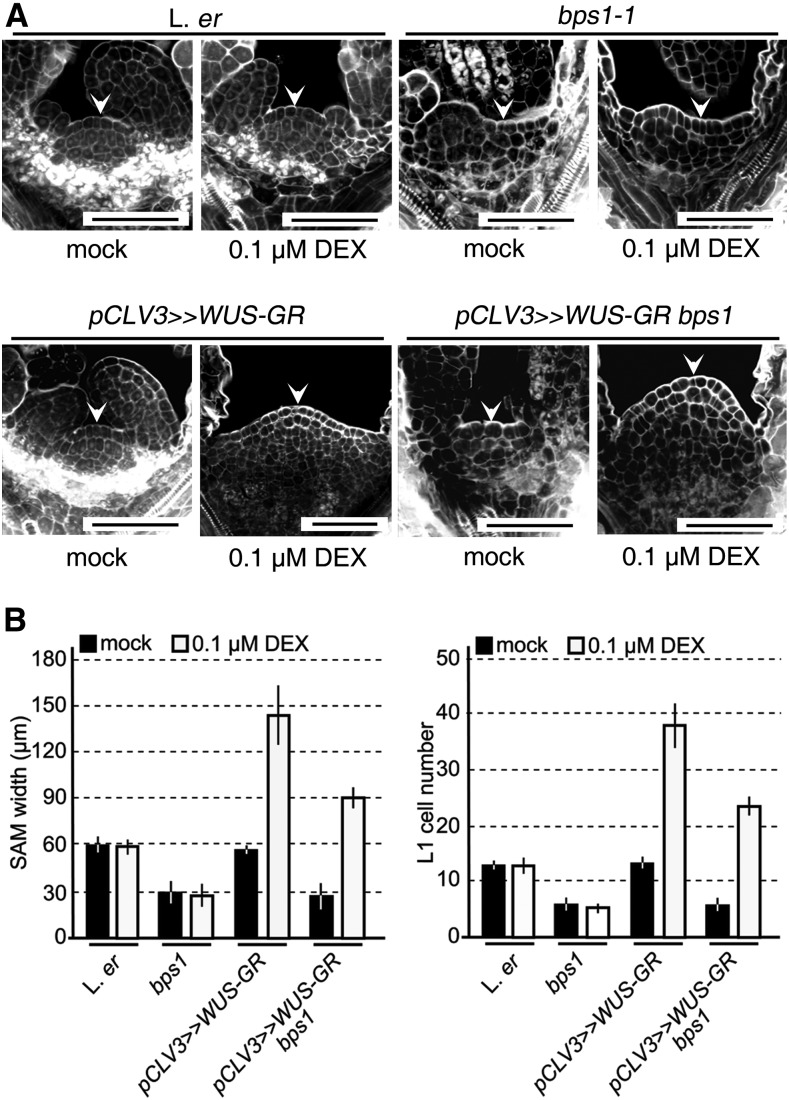
We also used the pCLV3>>WUS-GR system (Yadav et al., 2010) to induce WUS expression in the upper layers of the bps1 SAM. This construct produces an inactive form of WUS (WUS-GR) in the SAM’s central zone, and addition of dexamethasone (DEX) allows WUS-GR to be activated. In wild-type seedlings, this leads to strong SAM expansion (Fig. 9, A and B). Transgenic bps1 mutants supplied with DEX also produced an enlarged and dome-shaped SAM that included normal-sized cells in the upper SAM. Although ectopic WUS expression has been shown to lead to SAM formation, even in differentiated tissues (Gallois et al., 2002, 2004; Xu et al., 2005), these results support the CK-mediated rescue of SAM maintenance in bps1 clv1 double mutants.
Figure 9.
Ectopic WUS expression in the CZ is sufficient to restore stem cell maintenance in bps1. WUS expression was under control of the CLV3 promoter, and activity was regulated by protein fusion with GR. A, Confocal images of SAM of mock-treated and DEX-treated seedlings, including nontransgenic controls, at 5 dpi. Bars = 50 µm. B, Quantitative analysis of the SAM from the experiment depicted in A. Error bars indicate sd (n = 20 for each genotype).
DISCUSSION
Long-distance signaling in plants is necessary for both normal development and to coordinate physiological responses. Signaling between roots and shoots is especially important for coordinating stress perception (e.g. water availability and nutrient deficiencies) with growth. A possible new pathway functioning in root-to-shoot signaling was implicated by analyses of the Arabidopsis bps1 mutant. This mutant overproduces a small metabolite (which we call the bps signal) in its roots that moves to the shoot, reversibly arrests shoot growth, and is sufficient to arrest shoot growth in wild-type plants (Van Norman et al., 2004). The intriguing hormone-like activity of the bps signal prompted us to characterize how it affected shoot growth. The findings presented here revealed that the bps signal leads to repression of WUS expression, perhaps by repression of CK responses (Supplemental Fig. S6).
The first hint that the bps signal might interfere with SAM maintenance came from comparing the SAM morphology of 3 d bps1 and wild type; although the embryonic SAMs were indistinguishable, the 3 d bps1 SAM was small and flat in contrast to the domed SAM of the 3 d wild type. This timing was consistent with previous analysis of bps signal responses (Van Norman et al., 2011), suggesting that the loss of BPS1 allows synthesis of the bps signal during germination. The change in SAM size further suggested that following recruitment of the first pair of leaf primordia (Medford et al., 1992), the SAM failed to restore a normal number of cells.
We linked the bps1 SAM maintenance defect to a loss of WUS expression. pWUS::GUS transgenes were not expressed in bps1 mutants. Moreover, transient micrograft experiments revealed that the bps1 root was sufficient to repress WUS expression in wild-type shoots. Experiments coupling the bps1 root to a wild-type shoot indicated that the bps signal can exit the root, diffuse across a small agarose block, enter the wild-type scion through the hypocotyl, and repress WUS expression, all in 24 h. Thus, the bps signal appears to be highly potent and suggested an attractive model for the long-distance regulation of plant growth: root production of the bps signal might modulate WUS expression to coordinate shoot growth with conditions in the rhizosphere.
Normally the size of the SAM is primarily governed by a WUS/CLV3 feedback loop (Brand et al., 2000; Schoof et al., 2000), and so we considered that WUS repression might arise if the bps signal induced higher or ectopic CLV activity. We showed that neither the CLV1 nor the CLV2 receptors were required for bps signal-induced arrest; however, it is a formal possibility that the other CLV3 receptors, CORYNE and RECEPTOR-LIKE KINASE2, do function in this pathway. We consider this unlikely, though, as we still observed down-regulation of pWUS::GUS in the modestly enlarged 5 d SAM of bps1 clv3 double mutants, and the 12 d bps1 clv3 double mutants showed shoot growth defects very similar to those of bps1 single mutants. Although we cannot rule out that the bps signal itself might be perceived by the other CLV3 receptors, leading to WUS repression, a simpler interpretation is that the bps signal influenced WUS expression through a CLV-independent mechanism.
Instead, our data suggest that the bps signal interferes with activation of WUS expression by disrupting CK signaling. The general CK reporter pARR5::GUS (D’Agostino et al., 2000) showed low expression in bps1 mutants, and in transient micrografts, the bps1 root was sufficient to induce a similar low expression in wild-type scions. An attractive mechanism for bps signal repression of CK responsiveness is down-regulation of CK receptors; indeed, we found AHK4, a receptor previously linked to reduced WUS expression (Gordon et al., 2009), to be down-regulated in wild-type tissue transiently grafted to the bps1 root. However, within these same wild-type scions, the bps1 root led to a much stronger repression of WUS expression (AHK4 transcripts down 1.7-fold and WUS transcripts down 5-fold). Our expression analysis does not exclude the possibility that AHK4 is highly down-regulated in just the SAM nor that the bps signal might also AHK protein activity; however, the differential impact of the bps signal on WUS and AHK4 transcripts is difficult to reconcile with a simple receptor repression model.
Elements of CK signaling remained intact in bps1 mutants, as providing CK to bps1 mutants partially restored WUS expression, albeit limited to the internal RM. This restricted WUS expression domain was supported by the restored stem cell maintenance that was restricted to the RM when bps1 mutants were provided an exogenous supply of CK. The level of CK required to activate WUS, however, was quite high and thus not a physiologically normal level, though it was the same as that required to expand the WUS expression domain in the wild type. Nevertheless, the data do indicate that it is possible to separate WUS expression into two domains. How the bps signal restricts CK responsiveness from the upper tier of the SAM is currently unclear. One possibility is that the bps signal could function in a manner analogous to CLV3 and provide stronger CK buffering (Lindsay et al., 2006; Gordon et al., 2009; Chickarmane et al., 2012). Alternatively, the bps signal might, in addition to reducing AHK4 expression, disrupt production of CK in the SAM epidermis (Kurakawa et al., 2007; Kuroha et al., 2009), enhance CK degradation (Bartrina et al., 2011), or affect mechanical signaling from the epidermis (Savaldi-Goldstein et al., 2007; Hamant et al., 2008; Bozorg et al., 2014; Gruel et al., 2016).
The bps1 mutant provided a unique opportunity to examine the activity of WUS when its expression was limited to the RM domain. This localized activity induced a strictly localized response; small meristem-like cells were restored to the RM, but the SAM apex showed no restored maintenance and instead remained flat, with no increase in SAM width or L1 cell number. The localized response to restored WUS might not be surprising, as it was consistent with the pattern of pWUS::GUS expression; however, an important feature of WUS is its movement, which allows it to confer stem cell identity to the surrounding cells (Daum et al., 2014). Because studies of WUS movement have focused on OC-specific expression, and we find WUS expression lower in the RM, it is possible that WUS mobility might depend on where it is expressed. In bps1, the failure of RM-expressed WUS to rescue the stem cell identity in the tunica (L1 and L2) might be caused by a loss of cell-to-cell movement, e.g. if the bps signal altered plasmodesmata aperture. Indeed, the induced closure of the plasmodesmata in the vicinity of the OC using pWUS::CalS3m also caused SAM termination, albeit more modest than the termination induced by the bps signal (Daum et al., 2014). It is also possible that plasmodesmata connecting cells of the RM and OC are normally closed, regardless of the bps signal. Supporting this idea, a tracer study mapping symplastic domains in Arabidopsis inflorescences showed that at that developmental stage, the RM was symplastically uncoupled from the upper three cell layers (Gisel et al., 1999).
Given the hormone-like activity of the bps signal, an attractive model is that the bps signal can be deployed to alter the balance of WUS activity in the upper and lower regions of the SAM’s OC. WUS activity in the RM leads to stem elongation, while expression in the adjacent OC promotes the activity of the overlying stem cells, which are available to contribute to organogenesis. Separable regulation of these two domains might have important roles in controlling plant architecture, or it could be deployed to arrest organogenesis while still maintaining a residual stem cell population. Indeed, studies of rhizosphere stress implicate long-distance growth coordination by unknown mobile signaling compounds (Blackman and Davies, 1985; Saab and Sharp, 1989; Gowing et al., 1990; Mulholland et al., 1996; Stuurman et al., 2002). Root signaling associated with drought causes production of both fewer and smaller leaves, which is strikingly similar to the bps1 mutant. If the bps signal is the drought-induced compound, then we would predict its activity in the shoot to result from specific targets that cause reversible effects. We previously showed that growth arrest was reversible when we removed the source of the bps signal (the root; Van Norman et al., 2004). Moreover, although wus mutants have severe defects in SAM maintenance, they also frequently produce a new SAM (Laux et al., 1996). Whether the bps signal is the compound produced by a drought-treated root, however, awaits a deeper understanding of the full set of responses to the bps signal and biochemical identification of this enigmatic molecule.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Plants used in this study were Arabidopsis (Arabidopsis thaliana), and specifically the Columbia-0 (Col-0) and Landsberg erecta (Ler) accessions. The bps1-1 (Ler) and bps1-2 (Col-0) alleles have been described previously (Van Norman et al., 2004). For double mutant analyses, the bps1-1 allele was crossed to clv1-1 (Ler), clv2-1 (Ler), and clv3-2 (Ler) (Clark et al., 1993; Laux et al., 1996; Kayes and Clark, 1998). Molecular markers included pSTM::GUS (Col-0), pWUS::GUS (Ler), pCLV3::GUS (Ler), and pARR5::GUS (Col-0) (D’Agostino et al., 2000; Kirch et al., 2003; Bäurle and Laux, 2005; Williams et al., 2005). Homozygous markers in heterozygous bps1 and wild-type background were identified and then the expression was tested in next generation. Seeds were plated on plant GM (0.5× MS salts [Caisson Labs], 1% Suc, 0.5 g/L MES [Fisher Scientific], and 0.8% agar [MP Biomedicals]), incubated for 2 or 3 d in the dark at 4°C, and transferred to the growth chamber at 22°C under continuous light (80–140 µE).
Transient Micrografts
Transient micrografts, a modification of the Arabidopsis hypocotyl graft, were described previously (Turnbull et al., 2002; Adhikari et al., 2013). Briefly, the rootstock-scion junction was stabilized inside a silicone sleeve (0.012-in. internal diameter; Helix Mark Co.) filled with 0.8% agarose (Fisher Scientific, Molecular Biology Grade). Grafts used 4 dpi seedlings, which were incubated on sterile GM (2% agar) for 24 h prior to analysis.
Microscopy
SAM anatomy was analyzed using a Zeiss 510 Meta laser scanning confocal microscope. Embryos (isolated from mature seeds) and seedlings were fixed in 50% methanol and 10% acetic acid, and then cell walls were stained using a modified pseudo-Schiff propidium iodide method (Truernit et al., 2008). Confocal microscopy to analyze the GUS staining patterns was carried out using the same modified pseudo-Schiff propidium iodide staining procedure, with the confocal microscope set to reflection mode. Quantitative analysis used 71 to 90 mature seeds, 16 to 24 individuals for each seedling stage, and 17 to 20 for each genotype of double mutants. SAM serial sections were generated using sectioned wax-embedded tissue (8 µm thick and stained with toluidine blue) as previously described (Long and Barton, 1998).
GUS Analysis
GUS staining followed previously described methods (Sieburth and Meyerowitz, 1997). Staining incubation time was optimized for each marker using 2 mm 5-bromo-4-chloro-3-indoxyl-beta-d-glucuronide (X-gluc). After GUS staining, tissue was incubated in 70% ethanol for 2 d and cleared with saturated chloral hydrate for 6 to 24 h. Observation of the GUS staining pattern was carried out using an Olympus BX-50 microscope and images collected using a digital camera system (Olympus PD71).
Quantitative Analysis of Transient Micrografts
Each transient micrograft experiment included three controls: intact (scion genotype), root cut (placed into the agarose-containing sleeve but with no rootstock), and coupled to a wild-type rootstock. Each experiment used between 16 and 48 micrografted plants and repeated with similar results in at least three independent experiments. Staining intensity was classified as strong if the staining was easily observed, classified as weak if the blue staining product was difficult to observe and/or diffuse, and classified as absent if no trace of the blue staining product was detectable.
Quantitative Real-Time RT-PCR
Total RNA was isolated from 5 dpi wild-type (Col-0) scions 24 h after transient micrografting to bps1-2 rootstocks using the Qiagen Plant RNeasy Kit. The quantity, quality, and purity of the total RNA was estimated using a NanoDrop 2000 (Thermo Scientific) and a Bioanalyzer 2100 (Agilent). RNA was converted to cDNA using the Reverse Transcription System (Promega) and analyzed using the Maxima SYBR Green qPCR Master Mix (Fermentas) and the Mastercycler realplex EP (Eppendorf). ACTIN2 was used as an internal control to normalize gene expression. The qRT-PCR was carried out using three biological and two technical replicates. Gene-specific primers used for qRT-PCR are listed in Supplemental Table S2.
Cytokinin Treatment
The cytokinin BAP (Acros Organics) was used for WUS induction experiments. BAP was provided to the seedlings in two ways. For transient induction, seedlings carrying pARR5::GUS and pWUS::GUS (3 or 4 dpi) were transferred to GM supplemented with BAP and GUS staining 24 h later. Optimum BAP concentrations were established by testing 0.01 to 10 µM, and 10 µM BAP was selected for the remaining experiments; this is a 100× lower concentration than those used for induction in floral and inflorescence meristems (Lindsay et al., 2006; Gordon et al., 2009; Chickarmane et al., 2012). CK induction experiment were also carried out by germinating seedlings on GM supplemented with 0.1 µM BAP, as higher concentrations inhibited germination. For testing of SAM differentiation, 4 dpi seedlings were transferred to GM with 10 µM BAP and analyzed 24 h later.
DEX Induction
WUS expression was induced in bps1-1 plants by introducing pCLV3::LhG4 6XOP::WUS-GR (pCLV3>>WUS-GR) (Yadav et al., 2010), and homozygous pCLV3>>WUS-GR lines that segregated bps1-1 were established. The minimal effective dose of DEX (Sigma-Aldrich) for pCLV3>>WUS-GR was determined by germination on GM supplemented with 0.01 to 10 µM DEX; 0.1 µM DEX was selected because it was the lowest DEX concentration that led to SAM expansion in the wild type. Quantitative analyses were carried out with 20 individuals for each genotype.
Accession Numbers
Genes from this article can be found in the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) under the following accession numbers: BPS1 (At1g01550), STM (At1g62360), CLV1 (At1g75820), CLV2 (At1g65380), CLV3 (At2g27250), WUS (At2g17950), ARR5 (At3g48100), AHK2 (At5g37550), AHK3 (At1g27320), AHK4 (At2g01830), AHP1 (At3g21510), AHP2 (At3g29350), AHP3 (At5g39340), AHP4 (At3g16360), AHP5 (At1g03430), and AHP6 (At1g80100).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The bps signal disrupts shoot apical meristem maintenance.
Supplemental Figure S2. Transient micrograft establishment and analysis.
Supplemental Figure S3. Seedling and SAM phenotypes of 12 d bps1 clv3 seedlings reveal SAM arrest in bps1 clv3 double mutants.
Supplemental Figure S4. Quantitative analysis of pARR5::GUS by transient grafting assay.
Supplemental Figure S5. CK-treated 12 d bps1 clv3 seedlings.
Supplemental Figure S6. Model for mechanism by which the root-derived bps signal arrests SAM maintenance.
Supplemental Table S1. Expression in wild-type shoots transiently grafted to Col-0 and bps1-2 roots.
Supplemental Table S2. qRT-PCR primers.
Supplementary Material
Acknowledgments
We thank J. Fletcher for providing pWUS::GUS and pCLV3::GUS, P. Springer for providing pSTM::GUS, V. Reddy for providing pCLV3>>WUS-GR, and the Arabidopsis Biological Resource Center for providing clv1-1, clv2-1, and clv3-2. We are also grateful to J. Van Norman for critical reading of this manuscript and H. Tian, G. Drews, and M. Deshotel for discussion.
Glossary
- SAM
shoot apical meristem
- CZ
central zone
- OC
organizing center
- RM
rib meristem
- CK
cytokinin
- ABA
abscisic acid
- dpi
days postimbibition
- BAP
N6-benzylaminopurine
- IM
inflorescence meristem
- DEX
dexamethasone
- GM
growth medium
Footnotes
Articles can be viewed without a subscription.
References
- Adhikari E, Lee D-K, Giavalisco P, Sieburth LE (2013) Long-distance signaling in bypass1 mutants: bioassay development reveals the bps signal to be a metabolite. Mol Plant 6: 164–173 [DOI] [PubMed] [Google Scholar]
- Barton M, Poethig R (1993) Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823–831 [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäurle I, Laux T (2005) Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17: 2271–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D (2009) Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman PG, Davies WJ (1985) Root to shoot communication in maize plants of the effects of soil drying. J Exp Bot 36: 39–48 [Google Scholar]
- Bozorg B, Krupinski P, Jönsson H (2014) Stress and strain provide positional and directional cues in development. PLOS Comput Biol 10: e1003410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Burssens S, Himanen K, van de Cotte B, Beeckman T, Van Montagu M, Inzé D, Verbruggen N (2000) Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta 211: 632–640 [DOI] [PubMed] [Google Scholar]
- Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM (2012) Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc Natl Acad Sci USA 109: 4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S, Running M, Meyerowitz E (1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121: 2057–2067 [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM (1993) CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Cui Y, Rao S, Chang B, Wang X, Zhang K, Hou X, Zhu X, Wu H, Tian Z, Zhao Z, Yang C, Huang T (2015) AtLa1 protein initiates IRES-dependent translation of WUSCHEL mRNA and regulates the stem cell homeostasis of Arabidopsis in response to environmental hazards. Plant Cell Environ 38: 2098–2114 [DOI] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Medzihradszky A, Suzaki T, Lohmann JU (2014) A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci USA 111: 14619–14624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Gallois J-L, Nora FR, Mizukami Y, Sablowski R (2004) WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev 18: 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois J-L, Woodward C, Reddy GV, Sablowski R (2002) Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129: 3207–3217 [DOI] [PubMed] [Google Scholar]
- Gisel A, Barella S, Hempel FD, Zambryski PC (1999) Temporal and spatial regulation of symplastic trafficking during development in Arabidopsis thaliana apices. Development 126: 1879–1889 [DOI] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowing D, Davies WJ, Jones HG (1990) A Positive root-sourced signal as an indicator of soil drying in apple, Malus x domestica Borkh. J Exp Bot 41: 1535–1540 [Google Scholar]
- Gruel J, Landrein B, Tarr P, Schuster C, Refahi Y, Sampathkumar A, Hamant O, Meyerowitz EM, Jönsson H (2016) An epidermis-driven mechanism positions and scales stem cell niches in plants. Sci Adv 2: e1500989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jönsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, Couder Y, Traas J (2008) Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H (2010) TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22: 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MB. (2002) Long-distance signalling from roots to shoots assessed: the flooding story. J Exp Bot 53: 175–181 [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes JM, Clark SE (1998) CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125: 3843–3851 [DOI] [PubMed] [Google Scholar]
- Kim I, Cho E, Crawford K, Hempel FD, Zambryski PC (2005) Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc Natl Acad Sci USA 102: 2227–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S (2010) RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kirch T, Simon R, Grünewald M, Werr W (2003) The DORNROSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem ccll fate and lateral organ development. Plant Cell 15: 694–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H (2009) Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21: 3152–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lindsay DL, Sawhney VK, Bonham-Smith PC (2006) Cytokinin-induced changes in CLAVATA1 and WUSCHEL expression temporally coincide with altered floral development in Arabidopsis. Plant Sci 170: 1111–1117 [Google Scholar]
- Long JA, Barton MK (1998) The development of apical embryonic pattern in Arabidopsis. Development 125: 3027–3035 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y (2006) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Medford JI, Behringer FJ, Callos JD, Feldmann KA (1992) Normal and abnormal development in the Arabidopsis vegetative shoot apex. Plant Cell 4: 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal 2: ra45. [DOI] [PubMed] [Google Scholar]
- Miwa H, Betsuyaku S, Iwamoto K, Kinoshita A, Fukuda H, Sawa S (2008) The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol 49: 1752–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland B, Black C, Taylor I, Roberts J, Lenton J (1996) Effect of soil compaction on barley (Hordeum vulgare L.) growth I. Possible role for ABA as a root-sourced chemical signal. J Exp Bot 47: 539–549 [Google Scholar]
- Munns R, Sharp RE (1993) Involvement of abscisic acid in controlling plant growth in soil of low water potential. Funct Plant Biol 20: 425–437 [Google Scholar]
- Müller R, Bleckmann A, Simon R (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Matsumoto N, Tsugeki R, Rikirsch E, Laux T, Okada K (2012) Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24: 519–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne PL, van der Schoot C (1998) Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development 125: 1477–1485 [DOI] [PubMed] [Google Scholar]
- Saab IN, Sharp RE (1989) Non-hydraulic signals from maize roots in drying soil: inhibition of leaf elongation but not stomatal conductance. Planta 179: 466–474 [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J (2007) The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Goodger JQ (2008) Chemical root to shoot signaling under drought. Trends Plant Sci 13: 281–287 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman J, Jäggi F, Kuhlemeier C (2002) Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev 16: 2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T (2001) The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Sakurai K, Imamura A, Nakamura A, Ueguchi C, Mizuno T (2000) Compilation and characterization of histidine-containing phosphotransmitters implicated in His-to-Asp phosphorelay in plants: AHP signal transducers of Arabidopsis thaliana. Biosci Biotechnol Biochem 64: 2486–2489 [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui J-C (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull CGN, Booker JP, Leyser HMO (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Sato S, Kato T, Tabata S (2001) The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol 42: 751–755 [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Frederick RL, Sieburth LE (2004) BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr Biol 14: 1739–1746 [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Murphy C, Sieburth LE (2011) BYPASS1: synthesis of the mobile root-derived signal requires active root growth and arrests early leaf development. BMC Plant Biol 11: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman JM, Sieburth LE (2007) Dissecting the biosynthetic pathway for the bypass1 root-derived signal. Plant J 49: 619–628 [DOI] [PubMed] [Google Scholar]
- Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132: 3657–3668 [DOI] [PubMed] [Google Scholar]
- Wolters H, Jürgens G (2009) Survival of the flexible: hormonal growth control and adaptation in plant development. Nat Rev Genet 10: 305–317 [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu J-K (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-Y, Wang X-M, Li J, Li J-H, Wu J-S, Walker JC, Xu Z-H, Chong K (2005) Activation of the WUS gene induces ectopic initiation of floral meristems on mature stem surface in Arabidopsis thaliana. Plant Mol Biol 57: 773–784 [DOI] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Tavakkoli M, Reddy GV (2010) WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development 137: 3581–3589 [DOI] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42: 1017–1023 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU (2010) Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]
- Zhu J-K. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.