The peroxisomal NAD carrier from Arabidopsis contributes to the NAD homeostasis by supplying peroxisomes with cytosolic NAD in exchange with peroxisomal AMP.
Abstract
Cofactors such as NAD, AMP, and Coenzyme A (CoA) are essential for a diverse set of reactions and pathways in the cell. Specific carrier proteins are required to distribute these cofactors to different cell compartments, including peroxisomes. We previously identified a peroxisomal transport protein in Arabidopsis (Arabidopsis thaliana) called the peroxisomal NAD carrier (PXN). When assayed in vitro, this carrier exhibits versatile transport functions, e.g. catalyzing the import of NAD or CoA, the exchange of NAD/NADH, and the export of CoA. These observations raise the question about the physiological function of PXN in plants. Here, we used Saccharomyces cerevisiae to address this question. First, we confirmed that PXN, when expressed in yeast, is active and targeted to yeast peroxisomes. Secondl, detailed uptake analyses revealed that the CoA transport function of PXN can be excluded under physiological conditions due to its low affinity for this substrate. Third, we expressed PXN in diverse mutant yeast strains and investigated the suppression of the mutant phenotypes. These studies provided strong evidences that PXN was not able to function as a CoA transporter or a redox shuttle by mediating a NAD/NADH exchange, but instead catalyzed the import of NAD into peroxisomes against AMP in intact yeast cells.
NAD is a ubiquitous biological molecule that participates in many fundamental processes within the living cell (Pollak et al., 2007; Houtkooper et al., 2010). NAD and its phosphorylated form act as electron acceptors and donors in numerous redox reactions, and it is also involved in the generation and scavenging of reactive oxygen species that serve as signaling molecules (Dröge, 2002; Mittler, 2002). Furthermore, NAD is also a precursor for cADP-ribose, which modulates the release of calcium as a second messenger (Pollak et al., 2007; Houtkooper et al., 2010). Moreover, it also plays a role in transcription and post-translational modification through histone deacetylation and ADP ribosylation of proteins (Chang and Guarente, 2014; Bai et al., 2015).
Since NAD has multiple essential functions, its cellular levels need to be maintained either through de novo synthesis or salvage pathways, which involves recycling of NAD degradation products. In eukaryotic cells, both pathways take place in the cytosol, and thus NAD has to be always distributed to diverse cell compartments. As a consequence, transport proteins are required to shuttle NAD across intracellular membranes. In plants, humans, and fungi, members of the mitochondrial carrier family mediate the import of NAD into mitochondria, plastids, and peroxisomes (Todisco et al., 2006; Palmieri et al., 2009; Agrimi et al., 2012a, 2012b; Bernhardt et al., 2012). These transporters have been characterized by in vitro uptake assays using recombinant protein reconstituted into lipid vesicles (liposomes). Based on these biochemical data, the NAD carriers function as antiporters, importing NAD in a strict counter-exchange with another molecule (Todisco et al., 2006; Palmieri et al., 2009; Agrimi et al., 2012a, 2012b; Bernhardt et al., 2012).
In the case of plastids and mitochondria, the carriers catalyze the uptake of NAD in counter-exchange with AMP or ADP (Todisco et al., 2006; Palmieri et al., 2009). The efflux of adenine nucleotides is compensated by their de novo synthesis in the stroma or unidirectional uptake into the matrix (Palmieri et al., 2008, 2009). The importance of such a NAD import system has been established in the yeast Saccharomyces cerevisiae (Todisco et al., 2006; Agrimi et al., 2011). Yeast cells lacking the mitochondrial NAD carrier proteins Ndt1p and Ndt2p were unable to grow on nonfermentable carbon sources, such as ethanol (Todisco et al., 2006). The utilization of ethanol requires the operation of the NAD-dependent tricarboxylic acid (Krebs) cycle and the mitochondrial respiratory chain. The ndt1p/ndt2p double mutant lacks mitochondrial NAD, which renders the two pathways inactive, thus explaining the mutant growth phenotype (Todisco et al., 2006; Agrimi et al., 2011). This finding indicates that mitochondrial NAD transport proteins are essential for providing NAD to mitochondria.
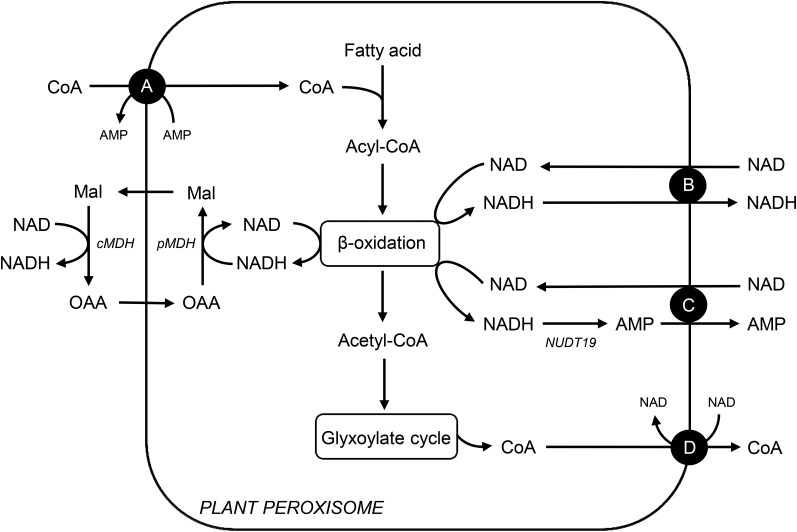
A peroxisomal NAD transporter from Arabidopsis (Arabidopsis thaliana), called peroxisomal NAD carrier (PXN), has been previously identified (Agrimi et al., 2012a, 2012b; Bernhardt et al., 2012). In comparison to the plastidial and mitochondrial NAD carriers, PXN exhibits unique transport properties. The recombinant PXN protein accepts NADH and Coenzyme A (CoA) as suitable substrates in addition to NAD, AMP, and ADP (Agrimi et al., 2012a, 2012b; Bernhardt et al., 2012). Based on its broad substrate specificity, four transport functions can be postulated for PXN in plants (Fig. 1). In plants, NAD- and CoA-dependent processes play fundamental roles in peroxisomal metabolism. For example, fatty acid degradation via β-oxidation requires NAD and CoA as co-factor (Graham, 2008). Before fatty acids can enter β-oxidation, they need to be activated to the corresponding acyl-CoA esters, which occurs in the intraperoxisomal matrix (Fulda et al., 2004). Therefore, a specific transport protein is required to import CoA into plant peroxisomes. PXN might be a good candidate for catalyzing this transport step, since it accepts CoA as a substrate in vitro (Fig. 1A). During β-oxidation, NAD is reduced to NADH. To maintain the flux through this pathway, NAD has to be regenerated via the reversible reduction of oxaloacetate (Mettler and Beevers, 1980). The resulting malate is exported to the cytosol, where it is reoxidized to oxaloacetate, which in return is imported into peroxisomes (Fig. 1). Such a malate/oxaloacetate shuttle allows the indirect exchange of the oxidized and reduced forms of NAD with the cytosol (Mettler and Beevers, 1980). As a redundant system, PXN might catalyze the import of NAD in exchange with NADH. This leads to the transfer of reducing equivalents across the peroxisomal membrane (Fig. 1B). Alternatively, PXN imports NAD in exchange with AMP to supply β-oxidation system with NAD, as known for the plastidial and mitochondrial NAD transporters (Fig. 1C). The counter-exchange substrate for the NAD import might be generated via the hydrolysis of NADH to AMP catalyzed by the NADH pyrophophatase NUDT19 (At5g20070) in Arabidopsis. This member of the NUDIX hydrolase family exhibits activity toward NADH and was shown to be targeted to peroxisomes (Ogawa et al., 2008; Lingner et al., 2011). An additional transport scenario for PXN is the export of CoA (Fig. 1D). High levels of CoA are released when acetyl-CoA, the end product of the β-oxidation, is further metabolized via the glyoxylate cycle (Graham, 2008). To maintain CoA homeostasis in peroxisomes and cytosol, PXN might mediate the CoA export. These predicted transport modes raise the question: what is the physiological function of PXN in living cells?
Figure 1.
Possible transport functions in vivo for the peroxisomal NAD(H) or CoA carrier (PXN) in plants. A, PXN imports CoA against AMP to fuel fatty acid activation required for β-oxidation. B, PXN functions as redox shuttle by transferring NAD versus NADH across the peroxisomal membrane to regenerate NAD for β-oxidation, redundant to the malate/oxaloacetate shuttle via the peroxisomal and cytosolic malate dehydrogenases. C, PXN mediates the NAD uptake against AMP to provide β-oxidation with its cofactor. In Arabidopsis, the peroxisomal NADH pyrophosphatase NUDT19 might generate the counter-exchange substrate AMP for the PXN carrier via NADH hydrolysis. D, PXN exports CoA, which is released during the glyoxylate cycle to prevent an accumulation of CoA in the peroxisomal matrix. Mal, Malate; OAA, oxaloacetate; pMDH, peroxisomal malate dehydrogenase; cMDH, cytosolic malate dehydrogenase; NUDT19, a putative NADH pyrophosphatase in peroxisomes.
In this work, we used different deletion mutants from S. cerevisiae to explore the transport role of PXN. To this end, we functionally expressed the Arabidopsis carrier protein in yeast mutant strains and investigated the suppression of the mutant yeast phenotype in the presence of recombinant PXN. Our study demonstrated that PXN is required to supply peroxisomes with NAD by importing NAD in exchange with AMP.
RESULTS
PXN Protein Is Functional When Expressed in Yeast
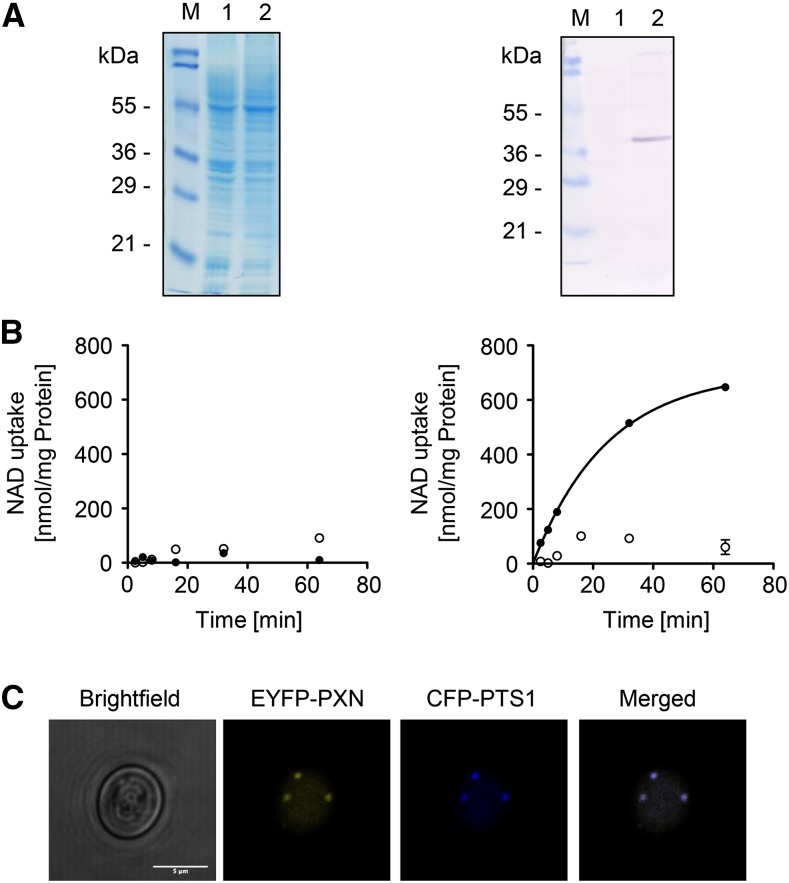
In this study we used several deletion mutants from S. cerevisiae to dissect the physiological transport function of PXN. Our approach was to express PXN in the corresponding yeast mutant and analyze the suppression of the respective yeast mutant phenotype. To establish the feasibility of this approach, we first expressed recombinant PXN and assessed its functionality by analyzing the NAD uptake rates in yeast. Therefore, we expressed PXN with a C-terminal His affinity tag (His) in the S. cerevisiae wild-type strain FGY217 (Kota et al., 2007). The expression of PXN was driven by a Gal-inducible promoter 1 from S. cerevisiae (pGAL1) that led to sufficient amounts of recombinant protein to analyze its transport activity. We extracted membranes from yeast cells expressing PXN-His (pMSU219) or transformed with the empty vector (pNL14) and separated the membrane proteins by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (Fig. 2A, left panel). Proper expression of PXN-His, which has a calculated molecular mass of 37.1 kD, was checked by immunoblot analysis with a His-tag antibody (Fig. 2A, right panel). Total yeast membranes containing recombinant PXN-His protein were reconstituted into lipid vesicles. The uptake of radioactively labeled [α-32P]-NAD into these liposomes was measured in the presence or absence of 10 mm NAD as counter-exchange substrate. We used total membrane fractions from yeast cells transformed with the empty vector as controls, to estimate the background activity of yeast endogenous NAD carriers. NAD import into liposomes reconstituted with yeast membranes from the control cells was below the detection limit of the employed assay system (Fig. 2B, left panel). In contrast, reconstituted PXN-His protein mediated rapid uptake of NAD into liposomes, but only when they were preloaded with NAD (Fig. 2B, right panel). The NAD/NAD exchange mediated by PXN-His followed first-order kinetics and reached a maximum uptake rate of 703 nmol/mg protein per minute. This finding confirmed that PXN functioned as an antiporter. Taken together, our in vitro activity experiments showed that PXN was functionally expressed in yeast cells.
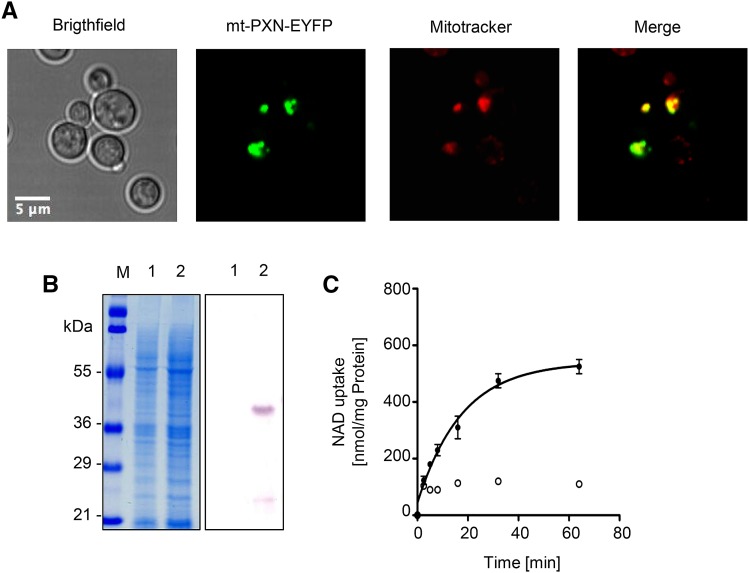
Figure 2.
Functional expression of PXN in yeast. A, Gal-inducible expression of PXN-His (37.1 kD) in FGY217 yeast cells. Left, Coomassie-stained sodium dodecyl-sulfate polyacrylamide gel electrophoresis gel; right, immunoblot analysis treated with His-tag antibody. Lane 1, Yeast membrane proteins isolated from yeast cells containing pNL14 vector; lane 2, yeast membrane proteins from yeast cells expressing PXN-His (pMSU219); M, protein marker. B, Time-dependent uptake of radioactively labeled [α-32P]-NAD (0.2 mm) was measured into liposomes reconstituted with total yeast membranes in the absence (left) or in the presence of PXN-His (right). The proteoliposomes were preloaded internally with 10 mm NAD (black symbols) or in the absence of NAD as counter-exchange substrate (white symbols). All graphs represent the arithmetic mean ± se of three technical replicates. C, PXN when expressed in yeast is targeted to peroxisomes. Confocal microscopic images were taken from yeast cells coexpressing EYFP-PXN and the peroxisomal marker CFP-PTS1. Scale bar = 5 µm.
In addition, we demonstrated that PXN fused at the N terminus to the enhanced yellow fluorescent marker (EYFP) was targeted to yeast peroxisomes. The expression of the EYFP-PXN protein (pMSU70) in the FGY217 yeast strain was driven by the constitutive promoter of the plasma membrane H+-ATPase from S. cerevisiae (pPMA1). These yeast cells were cotransformed with the pNL6 marker construct expressing the cyan fluorescent protein (CFP) tagged at the C terminus with the peroxisomal targeting signal 1 (PTS1) under the control of the alcohol dehydrogenase promoter from S. cerevisiae (pADH). This constitutive promoter was chosen to ensure the synthesis of PXN-EYFP in the presence of glycerol and fatty acids as carbon sources for the complementation studies. Furthermore, the proliferation of peroxisomes was found to be induced in yeast under those growth conditions (Gurvitz and Rottensteiner, 2006). Co-localization of EYFP-PXN and the peroxisomal fluorescent chimera (CFP-PTS1) was observed by fluorescence microscopy when both were expressed in yeast (Fig. 2C), indicating that Arabidopsis PXN was targeted to yeast peroxisomes.
Analysis of the PXN-Mediated CoA Transport Function
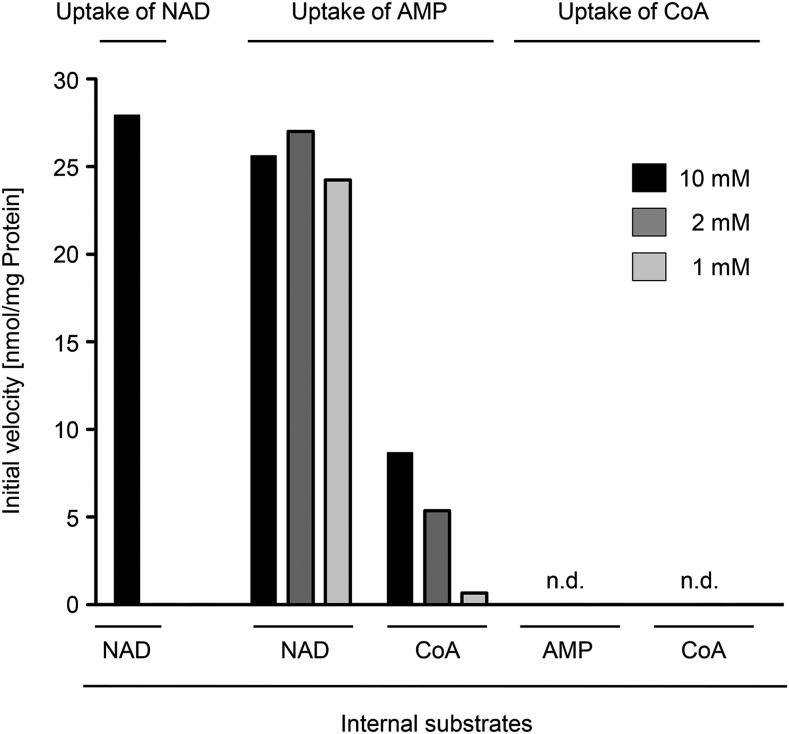
To investigate the physiological relevance of a CoA transport activity mediated by the PXN protein, we conducted concentration-dependent in vitro uptake assays into liposomes with yeast-expressed PXN-His protein (pMSU219), as described above. Therefore, we decreased stepwise the concentration of the internal exchange substrate from 10 mm to 2 mm, and finally 1 mm (NAD, CoA, or AMP) and kept the concentration of the radiolabeled uptake substrate constant at 0.2 mm: [α-32P]-NAD, [3H]CoA, or [α-32P]-AMP (Fig. 3). The initial velocity of the NAD uptake against 10 mm NAD for PXN-His shown in Figure 2B was determined as 27.3 nmol/mg protein per min. We observed significant and similar initial rates of AMP uptake for PXN-His against the counter-exchange substrate NAD at all tested concentrations. In contrast, the velocity of AMP import into CoA-preloaded liposomes at time point zero had only a residual rate of 30% at the highest internal CoA concentration of 10 mm compared to 10 mm NAD. Furthermore, initial rates of AMP uptake drastically decreased to almost zero with 1 mm of internal CoA. In addition, neither 10 mm CoA nor 10 mm AMP as counter-exchange partner facilitated any uptake of labeled CoA (0.2 mm) into PXN-containing liposomes. Based on our findings, we concluded that PXN preferred AMP and NAD as substrates, leading to high AMP/NAD exchange activities even at low internal NAD levels. PXN had a lower affinity to CoA and showed a marginal CoA import even under very high nonphysiological CoA and AMP concentrations in the liposomes, thereby calling into question its in vivo relevance.
Figure 3.
Initial velocities of PXN for the import of NAD, AMP, and CoA against various exchange partners in respect to internal concentrations. Time-dependent uptake of [α-32P]-NAD, [α-32P]-AMP, or [3H]CoA (0.2 mm) into liposomes reconstituted with total yeast membranes containing recombinant PXN-His was measured. The proteoliposomes were preloaded internally with 10 mm, 2 mm, or 1 mm NAD, CoA, or AMP. Initial velocities of technical triplicates were determined using global fitting parameters extracted from nonlinear regression analyses. n.d., Not detectable.
PXN Partially Restores the mdh3Δ Mutant Phenotype
Based on biochemical data PXN was able to catalyze NAD/NADH exchange in our liposome system (Agrimi et al., 2012a, 2012b; Bernhardt et al., 2012). We hypothesized that PXN transfers reducing equivalents across the peroxisomal membrane in addition to the peroxisomal malate/oxaloacetate shuttle (Pracharoenwattana et al., 2007; Fig. 1B). To test this hypothesis, we aimed for the complementation of a yeast mutant deficient in Mdh3p (peroxisomal malate dehydrogenase 3; van Roermund et al., 1995). This enzyme is part of the peroxisomal malate/oxaloacetate redox shuttle, which regenerates NAD in the peroxisomal matrix. Since NAD was required for fatty acid breakdown via peroxisomal β-oxidation, the loss of Mdh3p led to a yeast mutant that was unable to grow on oleate as sole carbon source (van Roermund et al., 1995).
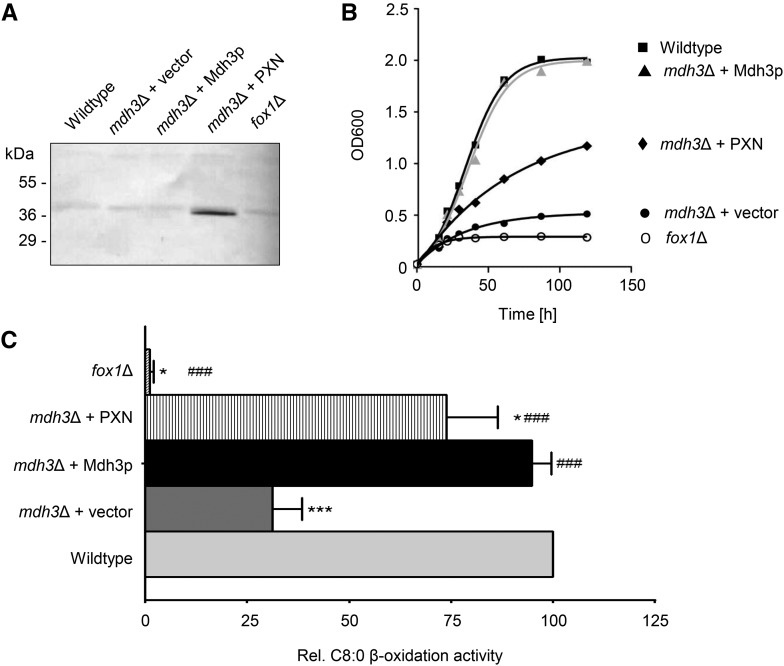
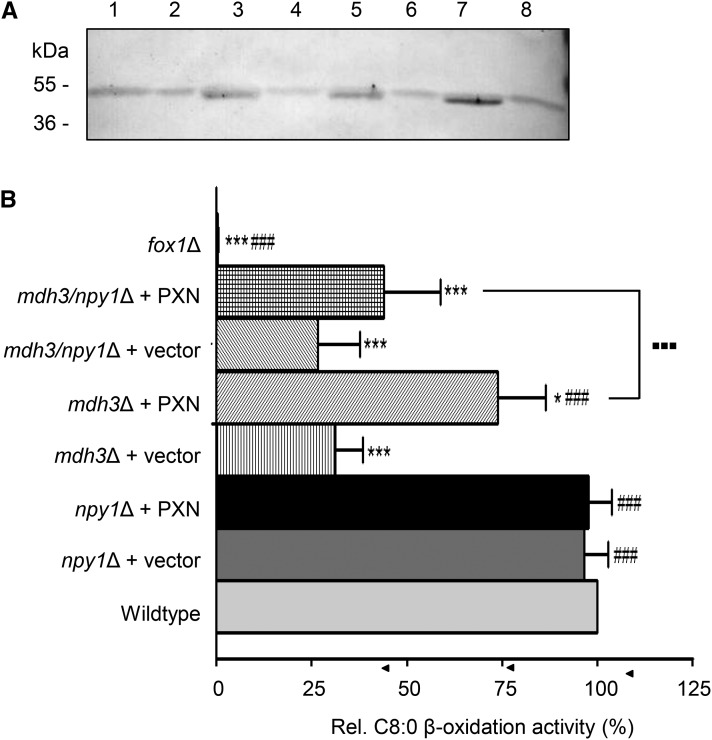
We investigated whether PXN represented an alternative route for supplying peroxisomal β-oxidation with NAD in the mdh3Δ mutant via a direct NAD/NADH exchange. To do so, we expressed the PXN protein (36.1 kD) in this yeast mutant under the control of the oleate-inducible catalase promoter from S. cerevisiae (pCAT1). The presence of oleate as a sole carbon source results in the up-regulation of genes encoding β-oxidation enzymes (Gurvitz and Rottensteiner, 2006). Since the yeast mutant successfully expressed PXN (Fig. 4A), we analyzed the growth of mdh3Δ cells transformed with MDH3 (van Roermund et al., 1995), PXN (pHHU274), or an empty vector (pEL30) on oleate (C18:1) as sole carbon source. In addition, we used as a control the yeast mutant fox1Δ, which lacks the acyl-CoA oxidase, the first enzyme in peroxisomal β-oxidation (Hiltunen et al., 1992). As expected, the mdh3Δ cells transformed with the empty vector did not grow on oleate, whereas the complementation of mdh3Δ mutant with Mdh3p led to complete growth recovery on oleate (van Roermund et al., 1995). Expression of PXN in the mdh3Δ mutant partially suppressed the mutant phenotype (Fig. 4B).
Figure 4.
Partial rescue of FAO in mdh3Δ mutant by PXN. A, Oleate-inducible expression of PXN (pHHU274; 36.1 kD) in the mdh3Δ mutant background was confirmed by immunoblot using PXN-specific antibody. Total yeast membrane proteins isolated from the wild type and different yeast mutants were analyzed. B, Growth curves of wild-type and mutant strains on oleate (C18:1) medium as sole carbon source. The strains shown are: wild-type cells (▪), mdh3Δ cells transformed with empty vector (●), mdh3Δ cells expressing Mdh3p (▴ in gray), and mdh3Δ cells expressing PXN (♦). As negative control fox1Δ cells (○) were used. C, Wild-type and mdh3Δ cells transformed with the empty vector or expressing PXN or Mdh3p, grown on oleate medium, were incubated with [1-14C]-octanoate and β-oxidation activity was measured. The β-oxidation activity of wild-type cells was taken as reference (100%). Data are represented as arithmetic means ± sd of two to five technical replicates. Asterisks indicate statistical differences to wild type with ***P < 0.001 as extremely significant and *P = 0.01 to 0.05 as significant. Hash tags indicate statistical differences to mdh3Δ cells with ###P < 0.001 as extremely significant.
To examine whether the partial suppression of the mdh3Δ mutant by PXN was associated with an increased rate of β-oxidation, we measured the rate of fatty acid oxidation (FAO) using octanoate (C8:0) as substrate in different mutant yeasts. Figure 4C shows the relative rates for C8:0 β-oxidation of mutant cells compared to the wild-type strain, which was set to 100%. In fox1Δ cells lacking the acyl-CoA oxidase, the β-oxidation of octanoate was blocked, whereas deletion of the MDH3 gene resulted in a decreased FAO rate (31%). Complementation with the endogenous MDH3 led to a full recovery of β-oxidation function (94%). When PXN was expressed in the mdh3Δ cells, we determined 73% of the wild-type FAO rates, and in comparison with β-oxidation activities in mdh3Δ we measured a significant increase of 2.4-fold. These data strongly indicated that PXN was able to exchange NAD for NADH in vivo, which was required to suppress the mdh3Δ growth defect and restore FAO function.
Further Suppressor Analysis to Confirm PXN as Redox Shuttle Candidate
We performed an additional phenotype suppression analysis in yeast, to investigate the NAD/NADH exchange function of PXN. S. cerevisiae contains a peroxisomal NADH pyrophosphatase, called Npy1p (AbdelRaheim et al., 2001). This enzyme catalyzes the hydrolysis of NADH to AMP, as postulated for NUDT19 in Arabidopsis (see Fig. 1). Npy1p regulates NADH homeostasis, in particular when reducing equivalents accumulate inside the peroxisomal matrix (AbdelRaheim et al., 2001). Thus, the possibility exists that the loss of Mdh3p in the presence of oleate as a sole carbon source leads to high levels of NADH inside peroxisomes. As a consequence, Npy1p might reduce the peroxisomal NADH levels by converting it to AMP in the mdh3Δ mutant. As a second consequence, mainly AMP (and not NADH) might be available as transport substrate for PXN (see Fig. 1C).
To exclude a role of Npy1p in the conversion of NADH to AMP in the mdh3Δ mutant, we deleted the NPY1 gene in this mutant background by PCR-mediated replacement of the NPY1 gene with the antibiotic resistance gene cassette BLE. For the FAO analyses we compared wild type with the mdh3Δ, npy1Δ, and mdh3/npy1Δ mutants, which were transformed either with the empty vector (pEL30) or with the PXN expression construct (pHHU274), as described above. We confirmed the expression of PXN (36.1 kD) driven by the pCAT1 promoter in the yeast mutants under oleate growth conditions by immunoblot analysis using a PXN-specific antibody (Fig. 5A). Next we grew the corresponding yeast cells on oleate and measured octanoate FAO rates (Fig. 5B). The loss of Npy1p did not affect β-oxidation activities (96%). We measured comparable wild-type octanoate oxidation activities in npy1Δ, and also in npy1Δ cells with an overexpressed PXN protein (Fig. 5B; 97%). In the mdh3Δ and mdh3/npy1Δ double mutant, however, we observed reduced rates of octanoate oxidation (30% of the wild-type control; Fig. 5B). To test the PXN function as an NAD/NADH antiporter, we transformed mdh3/npy1Δ cells with PXN and measured octanoate β-oxidation. We observed that the expression of PXN did not restore, to the same extent, as the FAO phenotype when Npy1p is absent (Fig. 5B). The octanoate oxidation activities of the double mutant mdh3/npy1Δ with PXN were significantly decreased compared to mdh3Δ expressing PXN.
Figure 5.
NPY1p regulates the peroxisomal NADH homeostasis in the mdh3Δ mutant. A, Constitutive expression of PXN (pHHU274; 36.1 kD) in the npy1Δ, mdh3Δ, and mdh3/npy1Δ mutant was confirmed by immunoblot analysis using PXN-specific antibody. Arrowhead indicates the detected protein band for the recombinant PXN. Lane 1, Wild-type cells; lane 2, npy1Δ cells; lane 3, npy1Δ cells expressing PXN; lane 4, mdh3Δ cells; lane 5, mdh3Δ cells expressing PXN; lane 6, mdh3/npy1Δ cells; lane 7, npy1Δ cells expressing PXN; lane 8, fox1Δ cells. B, Octanoic acid β-oxidation activity in oleate-induced yeast wild-type and mutant cells. The strains shown are as follows: wild type, mdh3Δ, npy1Δ, and mdh3/npy1Δ transformed with empty vector or expressing PXN. Relative FAO rates were calculated, in which the activities in wild-type cells were taken as reference (100%). Data are represented as arithmetic means ± sd of technical triplicates. Asterisks indicate statistical differences to the wild type with *** P < 0.001 as very significant and *P < 0.05 as significant. Hash tags indicate statistical differences to mdh3Δ cells with ###P < 0.001 as very significant. Large dots indicate statistical differences between mdh3Δ and mdh3/npy1Δ expressing PXN with ▪▪▪P < 0.001 as very significant.
In summary, our results suggest that—due to the loss of Npy1p—the increased NADH levels in the peroxisomal matrix of mdh3/npy1Δ cells could not stimulate the NAD import required for higher FAO under these conditions. We assume that peroxisomes of the mdh3Δ single mutant contained less NADH and more AMP produced by the active Npy1p when compared to mdh3Δ/npy1Δ cells. Thus we hypothesize that PXN did not function as a redox shuttle by exchanging NAD for NADH across the peroxisomal membrane in intact yeast cells, but was instead catalyzing the exchange of NAD with AMP.
PXN Catalyzes an NAD/AMP Exchange in Vivo
We used a yeast mutant lacking both mitochondrial NAD carriers Ndt1p and Ndt2p (Todisco et al., 2006) to verify the NAD/AMP exchange function of PXN in vivo. This double mutant displays a growth delay when ethanol is used as nonfermentable sole carbon source (Todisco et al., 2006). In this ndt1/ndt2Δ double mutant, the TCA cycle and mitochondrial respiration are inhibited due to the lack of mitochondrial NAD, which normally is imported by Ndt proteins in exchange for AMP (Todisco et al., 2006).
To be able to restore the impaired NAD uptake into the mitochondria of the ndt1/ndt2Δ double mutant, the peroxisomal PXN had to be targeted to mitochondria. To this end, the mitochondrial target peptide of the mitochondrial succinate/fumarate translocator from Arabidopsis was fused to the N terminus of PXN (Catoni et al., 2003). In addition, mt-PXN was tagged at the C terminus with the EYFP. This reporter allowed visualization of the subcellular localization of the fusion protein. We expressed mt-PXN-EYFP driven by the constitutive pPMA1 promoter (pMSU388) in FGY217 yeast cells, which were stained with the mitochondria-specific dye MitoTracker Orange (see “Materials and Methods” for vendor information). Confocal microscopy revealed that the fluorescence pattern of EYFP matched with the distribution of the MitoTracker signal (Fig. 6A), confirming that PXN fused with a mitochondrial target signal was localized to mitochondria in yeast.
Figure 6.
Mitochondrial targeted PXN protein is functional. A, Fusion of the mitochondrial target signal (mt) allows the localization of PXN to yeast mitochondria. The mt-PXN-EYFP fusion protein was constitutively expressed in BY4741 yeast cells (pMSU388). Localization study was analyzed by fluorescence microscopy. To visualize mitochondria, yeast cells were stained with MitoTracker Orange. Scale bar = 5 μm. B, Gal-inducible expression of mt-PXN-His (39.1 kD) in the FGY217 yeast strain. Left, Coomassie-stained SDS-PAGE gel. Right, Immunoblot blot treated with His-tag antibody. Lane 1, Yeast membrane proteins isolated from yeast cells containing empty vector pNL33; lane 2, yeast membrane proteins isolated from yeast cells expressing mt-PXN-His (pMSU237); M, protein marker. C, Time-dependent uptake of radioactively labeled [α-32P]-NAD (0.2 mm) was measured in liposomes reconstituted with total yeast membranes containing mt-PXN-His. The proteoliposomes were preloaded internally with 10 mm NAD (black symbols) or lack NAD as counter-exchange substrate (white symbols). Graphs represent the arithmetic mean ± se of three technical replicates.
To show that the fusion with the mitochondrial target peptide did not affect the transport function of PXN, we conducted a Gal-inducible expression of mt-PXN-His (pMSU237) in FGY217 yeast cells. The mt-PXN-His protein was expressed in yeast with the expected calculated mass of 39.1 kD visualized by immunoblot analysis using an His-tag antibody (Fig. 6B). We reconstituted lipid vesicles with yeast membranes containing mt-PXN-His fusion protein and measured the uptake of radioactively labeled [α-32P]-NAD (0.2 mm) in the absence or presence of internal NAD (10 mm). Reconstitution of the mt-PXN-His protein led to increased NAD transport activities at least when a counter-exchange substrate was present (Fig. 6C), indicating that the recombinant fusion protein was active.
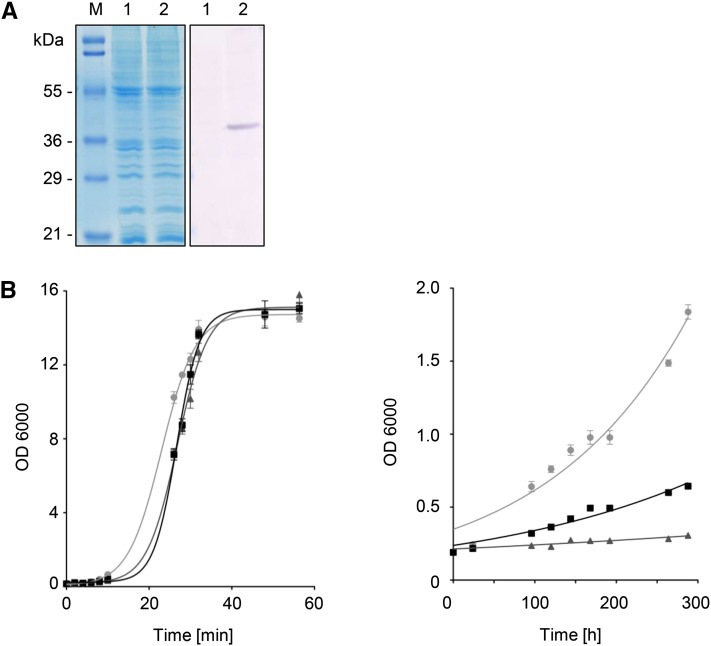
The ndt1/ndt2Δ double mutant was used to test the hypothesis that mt-PXN could complement NAD import into mitochondria in counter-exchange with AMP, as previously described for the yeast Ndt carriers (Todisco et al., 2006). Therefore, we expressed the mt-PXN-His fusion protein (39.1 kD) under the control of the constitutive pPMA1 promoter (pMSU377) in the double mutant (Fig. 7A). We grew the ndt1/ndt2Δ mutant carrying either the empty vector pNL24 or the mt-PXN-His expression plasmid on Glc or ethanol as sole carbon source at 28°C. Over time we monitored the growth of liquid yeast cultures by measuring the optical density at 600 nm (OD600). In the presence of Glc no obvious growth difference of wild type, ndt1/ndt2Δ, or ndt1/ndt2Δ expressing mt-PXN was observed (Fig. 7B, left panel). In contrast, the growth rate on the nonfermentable carbon source ethanol was significantly reduced in the ndt1/ndt2Δ double mutant compared to wild type (Fig. 7B, right panel), in line with earlier results (Todisco et al., 2006). Expression of mt-PXN-His in this mutant background partially suppressed the growth defect on ethanol. From the resulting growth curves shown in Figure 7B (right panel), we calculated the growth rates. Compared to the exponential growth phase of wild-type cells with a doubling time of 121 h, ndt1/ndt2Δ cells were unable to grow on ethanol, indicated by the doubling time of 566 h. This severe phenotype was suppressed by the expression of mt-PXN-His as reflected in a doubling time of 182 h. These results suggested that mt-PXN-His partially compensated for the loss of both Ndt1p and Ndt2p isoforms and supplied mitochondrial enzymes with NAD, which was essential for growth on ethanol. Since Ndt1p and Ndt2p catalyzed the import of NAD against AMP, we suggested that PXN mediated the same exchange.
Figure 7.
Rescue of ethanol growth by the mitochondrial-targeted Arabidopsis PXN in ndt1/ndt2Δ. A, Constitutive expression of mt-PXN-His (39.1 kD) in the ndt1/ndt2Δ double mutant. Left, Coomassie-stained SDS-PAGE gel; right, immunoblot blot treated with His-tag antibody. Lane 1, yeast membrane proteins isolated from yeast cells containing pNL24 vector; lane 2, yeast membrane proteins isolated from yeast cells expressing mt-PXN-His (pHHU377); M, protein marker. B, Suppression of the ndt1/ndt2Δ growth phenotype in the presence of mt-PXN-His: wild type transformed with empty vector (● in gray), ndt1/ndt2Δ transformed with the empty vector pDR195 (▴), and ndt1/ndt2Δ expressing mt-PXN (▪) were inoculated in yeast nitrogen base-uracil medium supplemented with 2% (w/v) Glc (left panel) and 2% (v/v) ethanol (right panel). OD600 of liquid cultures were measured using a spectrophotometer. Data from a representative experiment are shown. Graphs represent the arithmetic mean ± sd of three technical replicates. Similar results were obtained in three biological replicates.
DISCUSSION
Our knowledge of how peroxisomes are supplied with essential cofactors is still incomplete. In this study we elucidated the potential routes by which peroxisomes acquire NAD(H) and/or CoA. Like plastids and mitochondria, peroxisomes are absolutely dependent on supply with these cofactors from the cytoplasm (Linka and Esser, 2012). Since the last steps of NAD and CoA biosynthesis take place in the cytosol, these molecules must enter peroxisomes by specific transport proteins. The loss of the peroxisomal NAD carrier (PXN) compromises seedling growth in Arabidopsis (Bernhardt et al., 2012). This phenotype was explained by a delayed of storage lipid mobilization due to inefficient degradation of fatty acids via peroxisomal β-oxidation (Bernhardt et al., 2012). This previous work raised the question: How does PXN support FAO? Previous in vitro studies using artificial liposomes provided evidence that PXN is able to catalyze the following exchange modes: NAD/AMP, NAD/NADH, and AMP/CoA (Agrimi et al., 2012a, 2012b; Bernhardt et al., 2012). Consequently, several transport scenarios for a role of PXN in β-oxidation can be postulated as shown in Figure 1. Here, we performed analyses using baker’s yeast to elucidate PXN function in a living system.
First, we confirmed that PXN can be properly expressed in S. cerevisiae and that the protein is both functional and targeted to the correct organelle, the peroxisome (Fig. 2). The transport activity of PXN was analyzed in a reconstituted system. Measurement of the uptake of labeled substrate into lipid vesicles against a preloaded counter-exchange substrate led to high NAD exchange activities against NAD (Fig. 2B, right panel).
In the same experimental setup, we tested CoA as a putative substrate for PXN and compared the initial velocities of different CoA exchange combinations to the AMP/NAD exchange (Fig. 3). In contrast to the high uptake rates and high affinity for the AMP import against NAD, we detected only marginal CoA transport activities of PXN when CoA was offered at higher concentrations, indicating a lower affinity of PXN to this substrate. In context to the physiological situation, the free CoA levels are rather low. In animal tissue, the cytosolic and peroxisomal CoA concentrations are estimated between 0.02 and 0.14 mm and approximately 0.7 mm, respectively (Leonardi et al., 2005). Under our experimental conditions, we do not exhibit a significant CoA exchange with 10 mm of CoA or AMP. Therefore, a physiological role of PXN in supplying peroxisomes with CoA in plants is unlikely.
To address the in vivo transport function of PXN, we used different S. cerevisiae mutant strains. First, we analyzed a yeast strain deficient in the redox shuttle due to the deletion of the peroxisomal malate dehydrogenase. This mutant is unable to grow on fatty acids, because the NAD required for β-oxidation cannot be regenerated via the malate/oxaloacetate shuttle (van Roermund et al., 1995). Our complementation assay revealed that the growth on oleate and the activity of octanoate oxidation were partially restored in the presence of PXN (Fig. 4), indicating that PXN might catalyze a NAD/NADH exchange. When the peroxisomal enzyme Npy1p, which degrades NADH to AMP (thereby preventing NADH accumulation; AbdelRaheim et al., 2001, was deleted in the mdh3Δ mutant, PXN was unable to suppress the β-oxidation phenotype of the corresponding double mutant (Fig. 5). From this result, we conclude that PXN substitutes the loss of MDH3 via the import of cytosolic NAD in exchange with peroxisomal AMP, the latter being generated by Npy1-linked NADH degradation. This PXN-catalyzed transport mode allows β-oxidation with NAD even when the redox shuttle is compromised. We confirmed this hypothesis with a second independent yeast mutant. We artificially targeted PXN to yeast mitochondria (Fig. 5) and demonstrated that PXN was able to replace the endogenous NAD carriers Ndt1p and Ndt2p (Fig. 6). The successful suppression of ndt1/ndt2Δ mutant phenotype with PXN indicated that this carrier catalyzes the NAD/AMP exchange under in vivo conditions.
Our work provides strong evidence that the main function of PXN is to supply peroxisomes with NAD. In other eukaryotic organisms, an analogous peroxisomal NAD import system has not yet been identified (Visser et al., 2007; Linka and Theodoulou, 2013). However, the loss of the MDH3 gene in S. cerevisiae did not fully abolish FAO activity. Indeed, in the mdh3Δ mutant we detected significant residual FAO activity that amounts to approximately 30% of wild type (Fig. 4). Thus, we assume that the impaired redox shuttle across the peroxisomal membrane is partially compensated by an alternative redox shuttle or/and endogenous NAD carriers of yeast peroxisomes. The abundance of this last protein is lower than the constitutively expressed PXN in the yeast mutant background. The higher import activity of the recombinant PXN protein led to a significant restoration of the mutant phenotype (Fig. 4). One prime candidate for mediating the NAD import into yeast peroxisomes is the peroxisomal ATP carrier Ant1p (Palmieri et al., 2001; van Roermund et al., 2001), which is the closest relative of PXN in yeast. Uptake studies using recombinant protein confined the substrate specificity of Ant1p to ATP, ADP, and AMP (Tumaney et al., 2004). However, only (desoxy) nucleotides were tested in this study (Palmieri et al., 2001). Thus, we investigated whether cell-free expressed Ant1p has a larger substrate spectrum (Supplemental Fig. S1). Based on our analysis, Ant1p is unable to transport NAD across the liposomal membrane, implying that other putative carrier proteins are involved in the uptake of NAD into yeast peroxisomes. Further yeast candidates for the peroxisomal NAD transport are the two mitochondrial Ndt proteins (Todisco et al., 2006). These carriers are able to specifically import NAD and might be dual-targeted to yeast peroxisomes, since several mitochondrial proteins have been also found in peroxisomes (Theodoulou et al., 2013; Ast et al., 2013; Carrie et al., 2009; Yogev and Pines, 2011). Ndt1p has been demonstrated by GFP fusion to be located to mitochondria (Todisco et al., 2006), whereas the mitochondrial localization of Ndt2p has not been experimentally validated. Alternative routes for supplying yeast peroxisomes with NAD are the glycerol 3-P shuttle or via vesicular traffic from the endoplasmic reticulum, but up to now there is no evidence for this route (Hoepfner et al., 2005).
In this work we showed that PXN did not mediate a CoA transport or NAD/NADH exchange under physiologically relevant conditions. Consequently, additional transport systems in the peroxisomal membrane must exist to mediate CoA transport or the redox shuttle in plants. In humans, a peroxisomal CoA carrier has been identified that belongs to the mitochondrial carrier family (MCF; Agrimi et al., 2012a), as is the case for PXN. Recently, two MCF transport proteins have been identified as mitochondrial CoA carriers in Arabidopsis (Zallot et al., 2013). Thus, one of the 58 MCF members encoded by the Arabidopsis genome might provide the peroxisomal β-oxidation with CoA (Haferkamp and Schmitz-Esser, 2012). An independent uptake route for this cofactor might be also the peroxisomal fatty acid transporter, called Comatose (also known as PXA1, PED3, ACN2, and AtABCD1 (Hu et al., 2012)). It has been suggested that this membrane protein has an additional intrinsic enzymatic function. During the import of activated fatty acids into plant peroxisomes, the thioesterase activity of Comatose releases the CoA moiety into the cytosol or in the peroxisomal lumen. In the latter case, the CoA release results in a net CoA uptake into peroxisomes (De Marcos Lousa et al., 2013).
A peroxisomal pore-forming channel (porin) has been proposed to mediate the exchange of malate and oxaloacetate, which acts as a redox shuttle that connects the cytosolic and peroxisomal NAD/NADH pools (Linka and Theodoulou, 2013). Such a porin has been characterized by electrophysiological experiments using spinach leaf peroxisomes and castor bean glyoxysomes (Reumann et al., 1995, 1996, 1997), but the identity of the corresponding proteins has remained elusive. The Arabidopsis ortholog of peroxisomal Pxmp2, which has been characterized as a channel-forming protein in mammals (Rokka et al., 2009), might represent a candidate protein for linking the peroxisomal and cytosolic pools of NAD(H) in plants (Linka and Theodoulou, 2013). A major challenge for the future will be to identify the genes responsible for the transport processes mentioned above, which are required to execute the great diversity of peroxisomal functions in plants.
CONCLUSION
We showed that PXN acts as a NAD/AMP transporter in vivo while other postulated transport functions for PXN could be excluded in physiologically relevant conditions. Thus, PXN imports cytosolic NAD into plant peroxisomes. The uptake of NAD by PXN occurs in a strict counter-exchange with peroxisomal AMP. Our yeast complementation studies revealed that the NAD transport into yeast peroxisomes is linked to a peroxisomal NADH pyrophosphatase. This enzyme maintains the NADH homeostasis by degrading NADH and thus preventing the accumulation of NADH, for example, during high flux of FAO. In Arabidopsis, NUDT19 might be responsible for hydrolysis of NADH to AMP. The resulting AMP might be used by PXN as a counter-exchange substrate for the NAD import. The exported AMP can be fed into the salvage pathway to synthesize NAD in the cytosol. Future studies are required to investigate whether the interplay between PXN and NUDT19 in Arabidopsis plays a role in regulating the NAD/NADH redox state as an ancillary system to the malate/oxaloacetate shuttle.
MATERIALS AND METHODS
Materials
Chemicals were purchased from Sigma-Aldrich (www.sigmaaldrich.com). Reagents and enzymes for recombinant DNA techniques were obtained from New England Biolabs (www.neb.com), Qiagen (www.qiagen.com), ThermoFischer Scientific (www.thermofisher.com), and Promega (www.promega.com). Anion-exchange resin was purchased from Bio-Rad Laboratories (www.bio-rad.com). Radiochemical [α-32P]-AMP, [α-32P]-NAD, and [3H]CoA were obtained from Hartmann Analytic (www.hartmann-analytic.de) and Perkin Elmer (www.perkinelmer.de).
Cloning Procedures
In silico DNA sequences for cloning were retrieved from the Aramemnon database (aramemnon.uni-koeln.de) and the Saccharomyces Genome database (www.yeastgenome.org). Cloning was performed according to standard molecular techniques (Sambrook and Russell, 2001). Sequences were verified by DNA sequencing (Macrogen; dna.macrogen.com). Primers were synthesized by Sigma-Aldrich. The oligonucleotide sequences used in this study are listed in Supplemental Table S1. The coding DNA sequence (CDS) of PXN (At2g39970) was amplified via PCR using the cDNA clone pda.00682 provided by the Riken BioResource Center (www.en.brc.riken.jp) as a template.
For the in vitro uptake studies, the peroxisomal NAD carrier, PXN, was fused at the C terminus with a His-tag under the control of the Gal-inducible promoter pGAL1. The pYES2 vector (ThermoFischer Scientific) was chosen, in which a linker sequence (NL230/NL231) encoding for 6 His residues was introduced via BamHI and XbaI, yielding pNL14. The gene-specific primers NL33 (HindIII) and NL34 (BamHI) were used to clone the PXN CDS into the pNL14 expression vector, resulting in pMSU219.
To generate the constructs for the peroxisomal localization studies in yeast, PXN CDS was amplified using the primers NL7 (XhoI) and NL8 (BamHI). The resulting PCR product was cloned into pDR195 (Rentsch et al., 1995). The primers NL9 and NL10 (XhoI) were used to clone EYFP CDS via XhoI into pDR195. The insertion of both DNA fragments led to pMSU70 expressing EYFP-PXN via the constitutive pMA1 promoter. The clone (a peroxisomal fluorescence marker protein, CFP-PTS1) and the sequence of the Ser-Lys-Leu motif were fused to the CFP sequence via the primers NL254/NL255. The CFP-PTS1 sequence was then inserted via HindIII into the expression vector pACTII (Clontech Laboratories; www.clontech.com) carrying the constitutive pADH promoter, yielding the pNL6 construct.
For the complementation of the mdh3Δ and mdh3/nyp1Δ mutants, the cells were transformed with the yeast expression vector pEL30 containing the PXN coding sequence. The primers NL631 and NL632 were used to clone PXN CDS into pEL30 via BamHI and HindIII. The expression of PXN was under the control of the oleate-inducible catalase A promoter (pCTA1; Elgersma et al., 1993).
To complement the ndt1/ndt2Δ mutant with PXN, the transporter had to be targeted to yeast mitochondria. Therefore, the target sequence from the mitochondrial succinate/fumarate carrier from Arabidopsis (Arabidopsis thaliana; At5g01340) was fused to the N terminus of PXN. To do so, the mitochondrial target peptide (mt) was amplified from Arabidopsis cDNA using the primer (P75/P76) and introduced via HindIII and BamHI into pYES2 (ThermoFischer Scientific). The inducible pGAL1 promoter was exchanged with the constitutive pPMA1 promoter from pDR195 (Rentsch et al., 1995). The EYFP CDS (amplified by NL350/NL351) was introduced via BamHI and XbaI into pNL24. Finally, the PXN CDS (amplified by NL335/NL34) was cloned in frame with the mt and EYFP sequence via BamHI, resulting pMSU388.
To investigate whether the mt-PXN fusion protein is functionally expressed in yeast, the mitochondrial target peptide was amplified as described above and introduced via HindIII and BamHI into pNL14, leading to pNL33. The PXN CDS was amplified by NL335/NL34 and inserted into pNL33 via BamHI. The resulted pMSU237 construct was used for the Gal-inducible expression of mt-PXN-His fusion protein in yeast, which was functionally analyzed by in vitro uptake studies.
The mt-PXN-His expressing construct was cloned for rescuing the ndt1/ndt2Δ yeast mutant phenotype. Therefore, the PXN CDS was amplified by NL335/NL34 and inserted into the frame with a mitochondrial target sequence of pNL24 via BamHI, leading to the construct pMSU377. The synthesis of mt-PXN-His was driven by the constitutive pPMA1 promoter.
Yeast Strains and Culture Conditions
The Saccharomyces cerevisiae strain FGY217 (MATα, ura3-52, lys2Δ201, pep4Δ; Kota et al., 2007) was used for PXN uptake studies and subcellular localization analyses of EYFP-PXN and mt-PXN-EYFP.
For complementing the mdh3Δ and mdh3/npy1Δ mutant phenotype, the BJ1991 yeast cells (MATα, leu2, trp1, ura3-251, prb1-1122, pep4-3, gal2) were used as a wild-type strain. The following derivatives of this strain, all impaired in β-oxidation, were used: fox1Δ and mdh3Δ (carrying a deletion of the acyl-CoA oxidase or peroxisomal malate dehydrogenase, respectively). These mutants were constructed from BJ1991 as described by van Roermund et al. (1995). The mdh3/npy1Δ double mutant was made by replacing the whole NPY1 CDS from the mdh3Δ:LEU mutant by the BLE gene conferring resistance to Zeocin. The NPY1 deletion construct was made by PCR using the pUG66 plasmid (Gueldener et al., 2002) as a template and the CVR1 and CVR2 primers. BLE+ transformants were selected for integration in the NPY1 gene by PCR analysis. Yeast transformants were selected and grown in minimal medium containing 6.7 g/l yeast nitrogen base without amino acids, supplemented with 5 g/l Glc and amino acids (20 mg/l) if required. For the induction of peroxisome proliferation, cells were shifted to YPO medium containing 5 g/l potassium P buffer pH 6.0, 3 g/l yeast extract, 5 g/l peptone, 1.2 g/l oleate, and 2 g/l Tween-80. Prior to shifting to these media, the cells were grown in minimal 5 g/l Glc medium for at least 24 h.
For the generation of the ndt1/ndt2Δ double mutant, the ndt1Δ single mutant (BY4741; MATα, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0; YIL006w::kanMX4) was obtained from the EUROSCARF collection (web.uni-frankfurt.de/fb15/mikro/euroscarf). The NDT2 gene in ndt1Δ single mutant was disrupted by PCR-mediated gene replacement using the LEU2 cassette, amplified from pUG73 (Gueldener et al., 2002) with the primer set NL352/353. Transformed yeast cells that were able to grow in the absence of Leu were selected and the presence of the LEU2 cassette was verified by PCR analysis.
Yeast cells were transformed according to standard protocols for lithium acetate/PEG transformation (Gietz and Schiestl, 2007). Yeast cells were selected on synthetic complete medium (SC; 0.67% [w/v] yeast nitrogen base supplemented with appropriate amino acids and bases for auxotrophy and a carbon source).
Heterologous Protein Expression in Yeast for Uptake Studies
For the heterologous expression, the FGY217 strain was transformed with the empty vectors pNL14 and pNL33 or the expression constructs pMSU219 (PXN-His) and pMSU237 (mt-PXN-His). Fifty milliliters of SC-Ura liquid medium supplemented with 2% (w/v) Glc were inoculated with an overnight culture and cultured to an OD600 of 0.4. The yeast cells were grown for 6 h aerobically at 30°C. Control cultures with the empty vectors were processed in parallel. Harvest and enrichment of total yeast membranes without and with heterologously expressed PXN proteins were achieved according to Linka et al. (2008).
Protein Biochemistry
Sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblot analyses were conducted as described in Sambrook and Russell (2001). For immunodetection, either α-poly-His HPR-conjugated mouse IgG1 antibody (MACS Molecular; www.miltenyibiotec.com) or the PXN-specific antibody (Bernhardt et al., 2012) was used. No tagged PXN variants were visualized using anti-PXN-specific serum (Bernhardt et al., 2012) and AP-conjugated anti-rabbit IgG (Promega). PageRuler Prestained Protein Ladder (New England Biolabs) was used to estimate molecular masses. Protein concentrations were determined using a bicinchoninic acid assay (ThermoFischer Scientific).
Reconstitution of Transport Activities into Liposomes
Yeast membranes were reconstituted into 3% (w/v) l-α-phosphatidylcholine by a freeze-thaw-sonication procedure for in vitro uptake studies, as described in Linka et al. (2008). Proteoliposomes were either preloaded with 10 mm NAD; 10 mm, 2 mm, or 1 mm AMP; 10 mm, 2 mm, or 1 mm CoA; or produced without preloading (negative control). Counter-exchange substrate, which was not incorporated into proteoliposomes, was removed by gel filtration on Sephadex G-25M columns (GE Healthcare; www.gehealthcare.com).
Transport assays were started via adding 0.2 mm [α-32P]-AMP (6000 Ci/mmol), [α-32P]-NAD (800 Ci/mmol), or [3H]CoA (5 Ci/mmol). The uptake reaction was terminated via passing proteoliposomes over AG1-X8 Dowex anion-exchange columns (Merck Millipore; www.merckmillipore.com). The incorporated radiolabeled compounds were analyzed by liquid scintillation counting. Time-dependent uptake data were fitted using nonlinear regression analysis based on one-phase exponential association using GraphPad Prism 5.0 software (GraphPad; www.graphpad.com). The background activities in the absence of PXN-His were subtracted. The initial velocity of uptakes were calculated using the equation 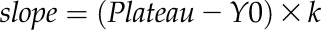 , whereas Y0 was set to 0. The values for the Plateau and k were extracted from the nonlinear regression analyses using a global fit from three technical replicates.
, whereas Y0 was set to 0. The values for the Plateau and k were extracted from the nonlinear regression analyses using a global fit from three technical replicates.
Suppression Analysis of the mdh3Δ and mdh3/npy1Δ Mutant
PXN expression construct pHHU274 and empty pEL30 vector were transformed into the following strains: BJ1991 wild-type cells, mdh3Δ single mutant, npy1Δ single mutant, and mdh3/npy1Δ double mutant. Transformed yeast cells were selected accordingly to the yeast strain genotype and pEL30 autotrophy.
β-Oxidation Activity Measurements
β-oxidation assays in intact yeast cells were performed as described previously by van Roermund et al. (1995) and optimized for the pH and the amount of protein. Oleate-grown cells were washed in water and resuspended in 0.9% NaCl (OD = 0.5). Aliquots of 2 μl of cell suspension were used for β-oxidation measurements in 200 μl of 50 mm MES (pH 6.0) and 0.9% (w/v) NaCl supplemented with 10 μm [1-14C]-octanoate. Subsequently, [1-14C]-CO2 was trapped with 2 m NaOH and used to quantify the rate of FAO. Results are presented as percentage relative activity to the rate of oxidation of wild-type cells. In wild-type cells, the rates of octanoate oxidation is 7.84 ± 1.09 nmol−1 min−1 OD cells−1.
Suppression Analysis of the ndt1/ndt2Δ Double Mutant
Wild-type strain BY4741 and ndt1/ndt2Δ double mutant were transformed with the empty vector pNL24 and pMSU377 expressing mt-PXN-His. Yeast cells were grown on SC-Ura supplemented with either 2% (w/v) Glc or 2% (v/v) ethanol, as described in Todisco et al. (2006).
Fluorescence Microscopy
For peroxisomal localization studies in yeast, FGY217 cells were cotransformed with pMSU70 and pNL6 expressing EYFP-PXN or peroxisomal fluorescent chimera (CFP-PTS1), respectively. Both fluorescent proteins were synthetized under the control of the constitutive pPMA1 promoter. Yeast cells harboring both constructs were selected on SC agar medium with 2% (w/v) Glc in the absence of uracil (pMSU70) and Leu (pNL6). The selected yeast cells were grown overnight at 30°C in SC-Leu-Ura with 3% (w/v) glycerol, 0.1% (w/v) oleate, and 0.2% (w/v) Tween-80 to induce peroxisome proliferation. The expression of the EYFP-PXN and CFP-PTS1 was analyzed with a confocal laser scanning microscope model no. 510 Meta (Carl Zeiss www.zeiss.de). CFP fluorescence was excited at 405 nm; fluorescence emission was detected with a 470- to 500-nm band-pass filter. EYFP fluorescence was excited at 514 nm, and the emission was recorded with a 530- to 600-nm band-pass filter.
Targeting of mt-PXN to yeast mitochondria was validated by transforming FGY217 yeast cells with pMSU388 ensuring constitutive expression of the mt-PXN-EYFP fusion protein via the pPMA1 promoter. Yeast cells carrying pMSU388 were grown in liquid culture overnight in SC-Ura with 2% (w/v) Glc. Yeast cells were harvested by centrifugation for 5 min at 3000g, washed with 25 mm HEPES-KOH (pH 7.3) and 10 mm MgCl2 and then stained with 50 μm MitoTracker ORANGE CMTMRos (Life Technologies/Thermo Scientific) for 10 min at room temperature. Residual MitoTracker dye was removed by washing with wash buffer twice. Yeast cells were immobilized on poly-l-Lys coated microscope slides for confocal microscopy. Analysis of yeast cells was performed with a confocal laser scanning microscope model no. 510 Meta (Carl Zeiss). MitoTracker fluorescence was excited at 561 nm, fluorescence emission was detected with a 575- to 615-nm band-pass filter. EYFP fluorescence was excited at 514 nm and the emission was recorded with a 530- to 600-nm band-pass filter.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_181526.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Primer used in this study; restriction sites (RS) are underlined.
Supplemental Figure S1. Substrate specificity of the peroxisomal ATP carrier Ant1p from S. cerevisiae.
Acknowledgments
Authors thank Jeanine Schlebusch and Daniel Wrobel for assistance with obtaining yeast growth curves. For technical assistance, the authors acknowledge Kirsten Abel. We thank Dr. R. van der Bend from Saxion Hogeschool (Life Science, Engineering, and Design) in Deventer for assistance in generating the npy1Δ and mdh3/npy1Δ double mutants and grateful to Lodewijk IJlst for stimulating discussion.
Glossary
- FAO
fatty acid oxidation
Footnotes
This work was supported by Deutsche Forschungsgemeinschaft (grant nos. 1781/2-1 to N.L., GRK1525 to N.L., and EXC1028 to A.P.M.W.).
Articles can be viewed without a subscription.
References
- AbdelRaheim SR, Cartwright JL, Gasmi L, McLennan AG (2001) The NADH diphosphatase encoded by the Saccharomyces cerevisiae NPY1 nudix hydrolase gene is located in peroxisomes. Arch Biochem Biophys 388: 18–24 [DOI] [PubMed] [Google Scholar]
- Agrimi G, Brambilla L, Frascotti G, Pisano I, Porro D, Vai M, Palmieri L (2011) Deletion or overexpression of mitochondrial NAD+ carriers in Saccharomyces cerevisiae alters cellular NAD and ATP contents and affects mitochondrial metabolism and the rate of glycolysis. Appl Environ Microbiol 77: 2239–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrimi G, Russo A, Scarcia P, Palmieri F (2012a) The human gene SLC25A17 encodes a peroxisomal transporter of coenzyme A, FAD and NAD+. Biochem J 443: 241–247 [DOI] [PubMed] [Google Scholar]
- Agrimi G, Russo A, Pierri CL, Palmieri F (2012b) The peroxisomal NAD+ carrier of Arabidopsis thaliana transports coenzyme A and its derivatives. J Bioenerg Biomembr 44: 333–340 [DOI] [PubMed] [Google Scholar]
- Ast J, Stiebler AC, Freitag J, Bölker M (2013) Dual targeting of peroxisomal proteins. Front Physiol 4: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Nagy L, Fodor T, Liaudet L, Pacher P (2015) Poly(ADP-ribose) polymerases as modulators of mitochondrial activity. Trends Endocrinol Metab 26: 75–83 [DOI] [PubMed] [Google Scholar]
- Bernhardt K, Wilkinson S, Weber APM, Linka N (2012) A peroxisomal carrier delivers NAD⁺ and contributes to optimal fatty acid degradation during storage oil mobilization. Plant J 69: 1–13 [DOI] [PubMed] [Google Scholar]
- Carrie C, Kühn K, Murcha MW, Duncan O, Small ID, O’Toole N, Whelan J (2009) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Catoni E, Schwab R, Hilpert M, Desimone M, Schwacke R, Flügge UI, Schumacher K, Frommer WB (2003) Identification of an Arabidopsis mitochondrial succinate-fumarate translocator. FEBS Lett 534: 87–92 [DOI] [PubMed] [Google Scholar]
- Chang HC, Guarente L (2014) SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab 25: 138–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marcos Lousa C, van Roermund CWT, Postis VLG, Dietrich D, Kerr ID, Wanders RJA, Baldwin SA, Baker A, Theodoulou FL (2013) Intrinsic acyl-CoA thioesterase activity of a peroxisomal ATP binding cassette transporter is required for transport and metabolism of fatty acids. Proc Natl Acad Sci USA 110: 1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge W. (2002) Free radicals in the physiological control of cell function. Physiol Rev 82: 47–95 [DOI] [PubMed] [Google Scholar]
- Elgersma Y, van den Berg M, Tabak HF, Distel B (1993) An efficient positive selection procedure for the isolation of peroxisomal import and peroxisome assembly mutants of Saccharomyces cerevisiae. Genetics 135: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda M, Schnurr J, Abbadi A, Heinz E, Browse J (2004) Peroxisomal Acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2: 31–34 [DOI] [PubMed] [Google Scholar]
- Graham IA. (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvitz A, Rottensteiner H (2006) The biochemistry of oleate induction: transcriptional upregulation and peroxisome proliferation. Biochim Biophys Acta 1763: 1392–1402 [DOI] [PubMed] [Google Scholar]
- Haferkamp I, Schmitz-Esser S (2012) The plant mitochondrial carrier family: functional and evolutionary aspects. Front Plant Sci 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen JK, Wenzel B, Beyer A, Erdmann R, Fosså A, Kunau WH (1992) Peroxisomal multifunctional β-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the fox2 gene and gene product. J Biol Chem 267: 6646–6653 [PubMed] [Google Scholar]
- Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF (2005) Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122: 85–95 [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Cantó C, Wanders RJ, Auwerx J (2010) The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 31: 194–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Gilstring CF, Ljungdahl PO (2007) Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD. J Cell Biol 176: 617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi R, Zhang YM, Rock CO, Jackowski S (2005) Coenzyme A: back in action. Prog Lipid Res 44: 125–153 [DOI] [PubMed] [Google Scholar]
- Lingner T, Kataya AR, Antonicelli GE, Benichou A, Nilssen K, Chen X-Y, Siemsen T, Morgenstern B, Meinicke P, Reumann S (2011) Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell 23: 1556–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka N, Esser C (2012) Transport proteins regulate the flux of metabolites and cofactors across the membrane of plant peroxisomes. Front Plant Sci 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka N, Theodoulou FL (2013) Metabolite transporters of the plant peroxisomal membrane: known and unknown. Subcell Biochem 69: 169–194 [DOI] [PubMed] [Google Scholar]
- Linka N, Theodoulou FL, Haslam RP, Linka M, Napier JA, Neuhaus HE, Weber APM (2008) Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20: 3241–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler IJ, Beevers H (1980) Oxidation of NADH in glyoxysomes by a malate-aspartate shuttle. Plant Physiol 66: 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yoshimura K, Miyake H, Ishikawa K, Ito D, Tanabe N, Shigeoka S (2008) Molecular characterization of organelle-type Nudix hydrolases in Arabidopsis. Plant Physiol 148: 1412–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F, Rieder B, Ventrella A, Blanco E, Do PT, Nunes-Nesi A, Trauth AU, Fiermonte G, Tjaden J, Agrimi G, et al. (2009) Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J Biol Chem 284: 31249–31259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Rottensteiner H, Girzalsky W, Scarcia P, Palmieri F, Erdmann R (2001) Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J 20: 5049–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Santoro A, Carrari F, Blanco E, Nunes-Nesi A, Arrigoni R, Genchi F, Fernie AR, Palmieri F (2008) Identification and characterization of ADNT1, a novel mitochondrial adenine nucleotide transporter from Arabidopsis. Plant Physiol 148: 1797–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak N, Dölle C, Ziegler M (2007) The power to reduce: pyridine nucleotides—small molecules with a multitude of functions. Biochem J 402: 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM (2007) Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J 50: 381–390 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370: 264–268 [DOI] [PubMed] [Google Scholar]
- Reumann S, Bettermann M, Benz R, Heldt HW (1997) Evidence for the presence of a porin in the membrane of glyoxysomes of castor bean. Plant Physiol 115: 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Maier E, Benz R, Heldt HW (1996) A specific porin is involved in the malate shuttle of leaf peroxisomes. Biochem Soc Trans 24: 754–757 [DOI] [PubMed] [Google Scholar]
- Reumann S, Maier E, Benz R, Heldt HW (1995) The membrane of leaf peroxisomes contains a porin-like channel. J Biol Chem 270: 17559–17565 [DOI] [PubMed] [Google Scholar]
- Rokka A, Antonenkov VD, Soininen R, Immonen HL, Pirilä PL, Bergmann U, Sormunen RT, Weckström M, Benz R, Hiltunen JK (2009) Pxmp2 is a channel-forming protein in Mammalian peroxisomal membrane. PLoS One 4: e5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Theodoulou FL, Bernhardt K, Linka N, Baker A (2013) Peroxisome membrane proteins: multiple trafficking routes and multiple functions? Biochem J 451: 345–352 [DOI] [PubMed] [Google Scholar]
- Todisco S, Agrimi G, Castegna A, Palmieri F (2006) Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J Biol Chem 281: 1524–1531 [DOI] [PubMed] [Google Scholar]
- Tumaney AW, Ohlrogge JB, Pollard M (2004) Acetyl coenzyme A concentrations in plant tissues. J Plant Physiol 161: 485–488 [DOI] [PubMed] [Google Scholar]
- van Roermund CWT, Drissen R, van Den Berg M, Ijlst L, Hettema EH, Tabak HF, Waterham HR, Wanders RJA (2001) Identification of a peroxisomal ATP carrier required for medium-chain fatty acid β-oxidation and normal peroxisome proliferation in Saccharomyces cerevisiae. Mol Cell Biol 21: 4321–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund CWT, Elgersma Y, Singh N, Wanders RJA, Tabak HF (1995) The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J 14: 3480–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser WF, van Roermund CWT, Ijlst L, Waterham HR, Wanders RJA (2007) Metabolite transport across the peroxisomal membrane. Biochem J 401: 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O, Pines O (2011) Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim Biophys Acta 1808: 1012–1020 [DOI] [PubMed] [Google Scholar]
- Zallot R, Agrimi G, Lerma-Ortiz C, Teresinski HJ, Frelin O, Ellens KW, Castegna A, Russo A, de Crécy-Lagard V, Mullen RT, et al. (2013) Identification of mitochondrial coenzyme A transporters from maize and Arabidopsis. Plant Physiol 162: 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]