HKT1-type transporters with an Asp (D) in the second pore-loop domain display inward-rectification and reduced affinity for Na+, in contrast to those with Asn (N) in this position that transport Na+.
Abstract
A crucial prerequisite for plant growth and survival is the maintenance of potassium uptake, especially when high sodium surrounds the root zone. The Arabidopsis HIGH-AFFINITY K+ TRANSPORTER1 (HKT1), and its homologs in other salt-sensitive dicots, contributes to salinity tolerance by removing Na+ from the transpiration stream. However, TsHKT1;2, one of three HKT1 copies in Thellungiella salsuginea, a halophytic Arabidopsis relative, acts as a K+ transporter in the presence of Na+ in yeast (Saccharomyces cerevisiae). Amino-acid sequence comparisons indicated differences between TsHKT1;2 and most other published HKT1 sequences with respect to an Asp residue (D207) in the second pore-loop domain. Two additional T. salsuginea and most other HKT1 sequences contain Asn (n) in this position. Wild-type TsHKT1;2 and altered AtHKT1 (AtHKT1N-D) complemented K+-uptake deficiency of yeast cells. Mutant hkt1-1 plants complemented with both AtHKT1N-D and TsHKT1;2 showed higher tolerance to salt stress than lines complemented by the wild-type AtHKT1. Electrophysiological analysis in Xenopus laevis oocytes confirmed the functional properties of these transporters and the differential selectivity for Na+ and K+ based on the n/d variance in the pore region. This change also dictated inward-rectification for Na+ transport. Thus, the introduction of Asp, replacing Asn, in HKT1-type transporters established altered cation selectivity and uptake dynamics. We describe one way, based on a single change in a crucial protein that enabled some crucifer species to acquire improved salt tolerance, which over evolutionary time may have resulted in further changes that ultimately facilitated colonization of saline habitats.
As far as current knowledge allows such generalization, plant genomes appear to include most or all functions that are necessary for a halophytic lifestyle, but only a small percentage of all plants are halophytes (Batelli et al., 2014). The requisite functional traits have become modified by diverse influences over approximately 500 million years of evolution on land, generating salt-tolerant species (halophytes) from salt-sensitive plants (mesophytes). Halophytes may have evolved from freshwater algae and, hence, mesophytic traits would have been acquired, while the evolution from saltwater charophytes would have demanded different adaptations (Flowers et al., 2010; Cheeseman, 2015). Multiple evolutionary patterns might be invoked for preadaptive functions in some mesophytes surviving episodes of salt stress (Flowers et al., 2010). A particular gene or a set of starter genes could be imagined necessary for the preadaptation of a mesophyte to such stress (Cheeseman, 2015). Whereas there is some understanding of which genes might have served such a function, there is still limited certainty about the complexity and sequence of genetic modifications required for the generation of plants to successfully colonize unfavorable areas such as saline land (Di Michele et al., 1987).
Increased root-zone salinity leads to cytosolic osmotic stress and sodium ion specific toxicity (Munns and Tester, 2008). In dealing with potential detrimental effects of Na+, plants use several Na+ transporters to achieve protection. These include Na+/H+ antiporters that extrude Na+ from root cells and/or distribute Na+ throughout tissues (Oh et al., 2010b; Quintero et al., 2011) and HKT1-type transporters (Rubio et al., 1995), which retrieve Na+ from the xylem stream to reduce its transport/accumulation to the shoots (Mäser et al., 2002a; Sunarpi et al., 2005; Ren et al., 2005; Davenport et al., 2007; Munns et al., 2012). HKT1-type transporters, in addition to their control of shoot Na+ levels, help by maintaining some balance between Na+ and K+ ions under salt stress by a process that is not fully understood (Berthomieu et al., 2003; Platten et al., 2006; Yao et al., 2010). HKT proteins belong to the HKT/Trk/Ktr-type superfamily of K+ transporters that consists of four repeats of transmembrane/pore-loop/transmembrane motifs, similar to the ion-conducting pore-forming units of K+ channels. However, plant HKTs are Na+ transporters that have been divided into two subclasses based on protein sequence and ion selectivity (Mäser et al., 2002b). Members of class-1 (HKT1) contain a Ser residue at the first pore-loop domain and are highly selective for Na+ over K+, whereas members of class-2 (HKT2) contain a Gly residue at this position and are permeable to Na+ and K+ (Horie et al., 2001). HKT2-type transporters are found in monocots and they are thought to sustain of K+ acquisition under salinity stress and nutritional Na+ uptake into K+-starved plants (Horie et al., 2007; Oomen et al., 2012).
Considering the presence and activity of class-1 HKT1 genes in salt-sensitive species, investigating HKT1 functions in salt-tolerant plants could provide information on the mechanistic nature of plant protection against Na+ excess, which to date is still elusive (Gong et al., 2005; Wu et al., 2012; Vera-Estrella et al., 2014). Arabidopsis contains a single copy AtHKT1;1 gene that codes for a member of class-1 protein that shows highly specific Na+ influx when expressed in Xenopus laevis oocytes and yeast (Saccharomyces cerevisiae; Uozumi et al., 2000; Xue et al., 2011). The genome of Thellungiella salsuginea (Ts), a halophytic relative of Arabidopsis, includes three copies of HKT1-type genes in a tandem array (Wu et al., 2012). Of the three TsHKT1 homologs, only TsHKT1;2 is greatly induced at the transcript level following salt stress (Ali et al., 2012), while the TsHKT1;1 transcript is down-regulated the same way as in AtHKT1 under salt stress (Oh et al., 2010b; Wu et al., 2012). When expressed in yeast cells, TsHKT1;2 shows competence for K+ transport whereas TsHKT1;1 is Na+-selective (Ali et al., 2012). Yet another Arabidopsis halophytic close relative is T. parvula (Tp; now Schrenkiella parvula), whose genome sequence has been determined; it contains two HKT1 genes, TpHKT1;1 and TpHKT1;2 (Dassanayake et al., 2011). The three HKT1 genes in T. salsuginea and two in T. parvula, as they contain a Ser residue at the selectivity filter in the first pore-loop domain, have been defined as class-1 transporters (Platten et al., 2006), despite the K+ uptake capacity of TsHKT1;2.
Alignment of amino-acid sequences of known HKTs with the yeast K+ transporter ScTRK1 provided, to our knowledge, new clues about structural and functional differences between Na+- and K+-selective HKT variants (Ali et al., 2012). Both TsHKT1;2 and TpHKT1;2 contain conserved Asp residues in their second pore-loop domains (Asp-207 and Asp-205, respectively) as well as in the adjacent transmembrane domain (Asp-238 and Asp-236, respectively). Yeast ScTRK1, a known high-affinity K+ transporter, also carries Asp in the second pore-loop position (Ali et al., 2012). However, in published dicot HKT1 protein sequences, either Asn (e.g. Asn-211 in AtHKT1) or Ser residues (Ser-264 and Ser-277, respectively) are found (Wang et al., 2014). These considerations led us to hypothesize that Asp-207, and possibly Asp-238, could impart altered Na+-K+ selectivity to subclass-1 HKT1 (Ali et al., 2012).
Here we present results for Arabidopsis hkt1-1 lines in which AtHKT1N211D and AtHKT1N242D variants have been generated. Moreover, we analyzed functional properties of AtHKT1;1, TsHKT1;2, and the reciprocal mutants AtHKT1N211D and TsHKT1;2D207N in Xenopus oocytes. Changing the Asn residue in the second pore-loop domain of AtHKT1;1 to Asp resulted in a transporter that resembled the TsHKT1;2 with lower affinity for Na+ transport, implying that at physiologically relevant Na+ concentrations TaHKT1;2 and AtHKT1N211D would not carry significant Na+ currents while K+ transport was preserved. Moreover, TaHKT1;2 showed inward-rectifying capacity at physiological membrane potentials and Na+ concentrations whereas AtHKT1;1 did not. Notably, the mutant AtHKT1N211D gained inward-rectification. In addition, select AtHKT1;1 mutant proteins and TsHKT1;2 were tested by expression in yeast, followed by expression in the Arabidopsis hkt1-1 knock-out line. Only TsHKT1;2 and AtHKT1N211D showed improved K+ transport relative to Na+. Modeling Asp or Asn into the second pore-loop region based on the yeast TRK1 structure provided further indication for the crucial role of these amino acids (Corratgé et al., 2007; Corratgé-Faillie et al., 2010; Cao et al., 2011). Transgenic Arabidopsis expressing AtHKT1N211D tolerated salt stress to a higher degree than wild-type AtHKT1;1. Our results indicate monovalent cation selectivity in dicot HKT1 to be determined by the presence of either Asp or Asn residues in the second pore-loop domain.
RESULTS
Plants Expressing TsHKT1;2 Are Tolerant to Salt Stress Compared to AtHKT1;1
We expressed both AtHKT1;1 and TsHKT1;2 under the cauliflower mosaic virus 35S promoter as well as under control of the native AtHKT1;1 promoter (using 1.2 kb upstream of the AtHKT1;1 start codon) in an hkt1-1 knock-out background. When controlled by the 35S promoter, compared to wild type, both AtHKT1;1- and TsHKT1;2-expressing plants were more sensitive to salt stress, as previously reported in Møller et al. (2009). However, plants expressing 35S::TsHKT1;2 showed lower sensitivity than those transformed with 35S::AtHKT1 (Fig. 1A). Under control of the Arabidopsis AtHKT1;1 native promoter, both AtHKT1;1 and TsHKT1;2 lines showed identical phenotypes on plates (Fig. 1A), conditions in which plants are not transpiring. When tested in soil, AtHKT1-expressing plants showed the same phenotypes as wild type. However, compared to wild type and the AtHKT1;1 complementation line, TsHKT1;2-expressing plants were more tolerant to salt stress (Fig. 1, B and C).
Figure 1.
Salt-stress responses of AtHKT1;1- and TsHKT1;2-expressing plants. A, Seeds were surface-sterilized and germinated on 0.5× MS medium with or without 75 mm NaCl. Photographs were taken after one week. B, 10-d-old seedlings grown in MS medium were transferred to soil and further grown for two weeks followed by 300-mm NaCl treatment for two weeks. Photographs were taken at the end of salt treatment. D, Seeds of wild type, hkt1-1, and the transgenic lines (ProAtHKT1::AtHKT1 and ProAtHKT1::TsHKT1;2 in the hkt1-1 background) were grown in hydroponic solution for one week, followed by 20- or 30-mm NaCl treatment as indicated. Photographs were taken two weeks after the addition of salt. C and E, After salt treatment, fresh weights from plants used in (C) and (E) (30 mm NaCl) were measured. Error bars represent sds from three independent repeats (n = 30 in each repeat). Significant difference was determined by a Student’s t -test; single or double stars indicate a P-value of <0.05 or <0.01, respectively. F, Sequence comparison of HKT homologs from Arabidopsis, T. salsuginea, and T. parvula. Amino-acid sequences in the second pore-loop region (PB) and the adjacent transmembrane domain (M2B; red boxes) are aligned with the use of Clustal-W (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The conserved Gly residues in the PB region (Mäser et al., 2002a, 2002b) are indicated by asterisks. The Asp residues specific for TsHKT1;2 (D207) and TpHKT1;2 (D205) are indicated by arrows.
To check the role of AtHKT1;1 and TsHKT1;2 in detail, seeds of ProAtHKT1::AtHKT1 and ProAtHKT1::TsHKT1;2, together with wild type (Col-gl) and the hkt1-1 knock-out line, were grown in hydroponic culture with a modified Long-Ashton mineral solution with 1 mm K+, 2 mm Ca2+, and which was nominally free of Na+ and NH4+ (LAK medium; Barragán et al., 2012). This medium was designed to maximize the toxicity of added Na+ ions, while minimizing the osmotic effects of supplemental NaCl. Under salt stress, ProAtHKT1::AtHKT1-expressing plants showed the same phenotype as the wild type, whereas hkt1-1 plants were hypersensitive to salt stress (Fig. 1, D and E). By contrast, ProAtHKT1::TsHKT1;2-expressing plants were more tolerant to salt stress, accumulated more fresh biomass, and maintained a lower shoots/roots Na+ ratio than wild type or ProAtHKT1::AtHKT1-expressing plants (Fig. 1, D and E; Supplemental Fig. S1). These results are consistent with the previous finding that TsHKT1;2 mediates a better K+/Na+ balance under high salinity, which in turn reduces Na+ toxicity (Ali et al., 2013).
We had previously identified a sequence polymorphism between AtHKT1;1 and TsHKT1;2 as a basis for differential Na+ tolerance in yeast transformants (Ali et al., 2012). This difference was dependent on the presence of the Asp/Asn dichotomy at the second pore-loop domain (Fig. 1F; Supplemental Fig. S2). Whereas Asn is found in all available HKT1 sequences from mesophytic plants, TsHKT1;2 and TpHKT1;2 from the salt-tolerant Arabidopsis relatives T. salsuginea and T. parvula contain Asp in this position. Asp is also found in the yeast TRK1 potassium transporter (Supplemental Fig. S2). We hypothesized that the tolerance to high salinity imparted by TsHKT1;2 could be due to the presence of this Asp residue.
Molecular Modeling of AtHKT1;1 and TsHKT1;2
One argument in favor of such a view is provided by computational analysis of the modeled three-dimensional structure of ion-transport domains of AtHKT1;1 and TsHKT1;2 proteins. HKT proteins consist of four transmembrane/pore-loop/transmembrane domains similar to the ion conducting pore-forming units of K+ channels. Hence, the transmembrane and pore-forming regions for AtHKT1;1 and TsHKT1;2 were modeled using the Membrane-Pore-Membrane motif of K+ channels (Durell et al., 1999; Kato et al., 2001) and refined by 2-ns molecular dynamics simulation. To analyze the possible importance of the Asp and Asn residues at the second pore-loop and the fourth transmembrane domains of the HKT proteins, we carried out an in silico mutation study using the refined structures (Supplemental Fig. S3). For AtHKT1;1, the Asn (n) residues (N211 in second pore-loop, and N242 in the fourth transmembrane) were mutated to Asp (d; Supplemental Fig. S3A), whereas Asp (D207 and D238) residues of TsHKT1;2, were substituted by Asn (n; Supplemental Fig. S3B). The wild-type and mutated models are shown in Figure 2, as wild type-AtHKT1, Mut-AtHKT1 (N211D/N242D), wild type-TsHKT1;2, and Mut-TsHKT1;2 (D207N/D238N). Each system underwent 5-ns molecular dynamics simulation in an explicit membrane environment. The Na+ and K+ ions were placed in the pore-forming regions to investigate interactions with the respective wild-type and mutated residues. The closest frame to the average structure during the last 2 ns was selected as a representative structure. The electrostatic potential distribution was different between the wild-type and the mutant protein models, for both AtHKT1;1 and TsHKT1;2 (Fig. 2, A–D). In the AtHKT1;1 model, a strong negatively charged potential was detected in the mutant (N211D/N242D) system but not in the wild type (Fig. 2, E and F). Wild type-TsHKT1;2 with two Asp residues shows a strong negatively charged surface in the pore-forming region, which is very similar to that shown for Mut-AtHKT1 (Fig. 2, F and G). By contrast, the mutated TsHKT1;2 model (D207N/D238N) resembled the surface in wild type-AtHKT1 (Fig. 2H). In both models with Asp residues, the K+ ion was confined to the pore region of the proteins throughout the simulation as the result of a strong salt-bridge interaction between the K+ ion and oxygen atoms in Asp residues, thereby allowing K+ permeation through the HKT transporter. Stronger K+ retention was dependent on the presence of Asp residues. The weak binding of K+ in the Asn residue models apparently enabled binding of (excess) Na+ ions to successfully replace K+ ions. The modeling studies provide support for the role of Asp residues in the pore region as a critical residue for K+ selectivity in HKT1-type transporters.
Figure 2.
Electrostatic potential distribution of the pore-forming regions of AtHKT1;1, TsHKT1;2, and their mutants. Representative structures of wild type-AtHKT1 (A), Mut-AtHKT1 (B), wild type-TsHKT1;2 (C), and Mut-TsHKT1;2 (D). Pore-forming regions are represented by stick models. Electrostatic potential surfaces of wild-type and Mut-AtHKT1 (E and F) and wild-type and Mut-TsHKT1;2 (G and H) are shown, indicating each pore-forming region. The protein surface structures (E, F, G, and H) are rotated 180°. Key residues in the second pore-forming and fourth transmembrane domain regions are colored yellow and gray, respectively. Na+ and K+ ions are represented by violet and green spheres, respectively. ESP: electrostatic potential distribution.
Ion Selectivity and Pore-Forming Regions of AtHKT1;1 and TsHKT1;2
An earlier study, using yeast cells, had indicated the importance of the Asp (d) residue for cation selectivity in the pore region of HKT1 transporters (Ali et al., 2012). To further explore the importance of altered amino acids in the pore region of different HKT1 proteins, we substituted Asn (n) in AtHKT1;1 with Asp (d) and generated the following variants: AtHKT1N211D, AtHKT1N242D, and AtHKT12N2D, where both N211 and N242 were replaced by d (Fig. 3A).
Figure 3.
Ion selectivity and accumulation of potassium content in AtHKT1 wild-type and mutated proteins. A, Positions of conserved amino acids (Asn) in AtHKT1 that were mutated. AtHKT1 contains Asn residues in the second pore-loop region and in the adjacent transmembrane domain, whereas TsHKT1;2, at the same positions contains Asp residues (red circles and boxes indicate mutation sites, whereas N and C denote N terminus and C terminus of HKT1 protein, respectively). B, The K+-uptake deficient yeast strain CY162 (trk1, trk2) expressing AtHKT1, AtHKT1N211D, AtHKT1N211D, and AtHKT12N2D were grown on SD medium lacking uracil and supplemented with the indicated concentration of KCl. The SD medium has an intrinsic content of 7.5 mm K+. TsHKT1;2 and AtKAT1 were used as negative and positive controls, respectively. C, Potassium content in the CY162 yeast strain expressing AtHKT1, AtHKT1N211D, AtHKT1N211D, AtHKT12N2D, grown in SD medium adjusted to 10-mm KCl final concentration, with error bars representing sds from three independent repeats (n = 3). EV: pYES2 empty vector.
To test the potential effect of these mutations on AtHKT1;1 cation selectivity, we expressed wild-type and mutant versions of AtHKT1;1 and TsHKT1;2 in the K+ uptake-deficient yeast strain CY162 lacking the fungal TRK proteins that are homologous to plant HKTs. In addition, we also expressed the Arabidopsis K+ channel AtKAT1 as a positive control for K+ uptake (Fig. 3B). AtHKT1;1-expressing cells did not grow in low K+ (the SD medium contains 8 mm K+ and 1.7 mm Na+; note that the complete lack of growth of cells expressing AtHKT1;1 compared to the residual growth of the negative control with the empty vector is likely due to the Na+ permeation by AtHKT1;1). On the other hand, TsHKT1;2 complemented the K+ transport deficiency of yeast cells, supporting growth rates that were similar to cells expressing the K+ channel AKT1 in SD medium supplemented with 25 mm KCl. Cells expressing AtHKT1N211D, the mutant version of AtHKT1;1, grew well in basal SD and, as in the case of TsHKT1;2, K+ added to the medium further enhanced growth (Fig. 3B). By contrast, the AtHKT1N242D mutant failed to rescue cell growth in low K+. The double mutant, AtHKT12N2D, also complemented the K+ deficient phenotype and no additional effect was seen based on the second mutation. We further examined the accumulation of K+ contents in cells expressing wild-type and mutant AtHKT1;1 and compared them with TsHKT1;2. Cells transformed with TsHKT1;2 and the mutant forms of AtHKT1;1, AtHKT1N211D, and AtHKT12N2D accumulated more K+ than wild-type AtHKT1;1 (Fig. 3C). Cells with AtHKT1N242D accumulated a lower amount of K+ similar to wild-type AtHKT1;1 (Fig. 3C). These results strongly suggested that the differences in AtHKT1;1 and TsHKT1;2 are based on the presence of either Asn or Asp in the second pore-loop domain of these transporters, which then enables them to take up K+ (D211/207) or not (N211/207).
Expression in X. laevis Oocytes
To investigate the transport characteristics of AtHKT1;1 and TsHKT1;2, we expressed these transporters in X. laevis oocytes. In the presence of varying external Na+ concentrations, the HKT transporters generated Na+-dependent currents that were much higher than those recorded in control oocytes (Supplemental Fig. S4). However, the electrical responses of the Na+-dependent currents were remarkably different between AtHKT1;1 and TsHKT1;2. AtHKT1;1 mediated both inward and outward currents, although the reversal potentials did not correspond to a Nernstian response (Fig. 4A; Supplemental Fig. S4). By contrast, TsHKT1;2 mediated very low outward currents at positive membrane potentials, which revealed a strong inwardly rectifying behavior (Fig. 4A). For both transporters, the conductance shows saturation kinetics. Fitting the inward conductances versus external Na+ to hyperbolic functions revealed half-saturation constants of 8 and 94 mm and maximal conductances of 68 and 90 μS for AtHKT1;1 and TsHKT1;2, respectively (Supplemental Fig. S5). The characteristics of AtHKT1;1 are consistent with those previously reported for this transporter (Uozumi et al., 2000; Xue et al., 2011) and other members of the same family, including OsHKT1;3 (Jabnoune et al., 2009), TdHKT1;4 (Ben Amar et al., 2014), and TmHKT1;5 (Munns et al., 2012). To our knowledge, the remarkable rectifying characteristics of TsHKT1;2 have been previously described only for the rice (Oryza sativa) transporter OsHKT1;1 (Jabnoune et al., 2009).
Figure 4.
Transport characteristics of AtHKT1, TsHKT1;2, and their respective mutants. Representative current-voltage curves from X. laevis oocytes expressing AtHKT1 and TSHKT1;2 (A), and their mutants AtHKT1N211D and TsHKT1;2D207N (B) at increasing Na+-gluconate concentrations (in mm). Dotted line indicates zero current level. The voltage-clamp protocol is given in “Materials and Methods”.
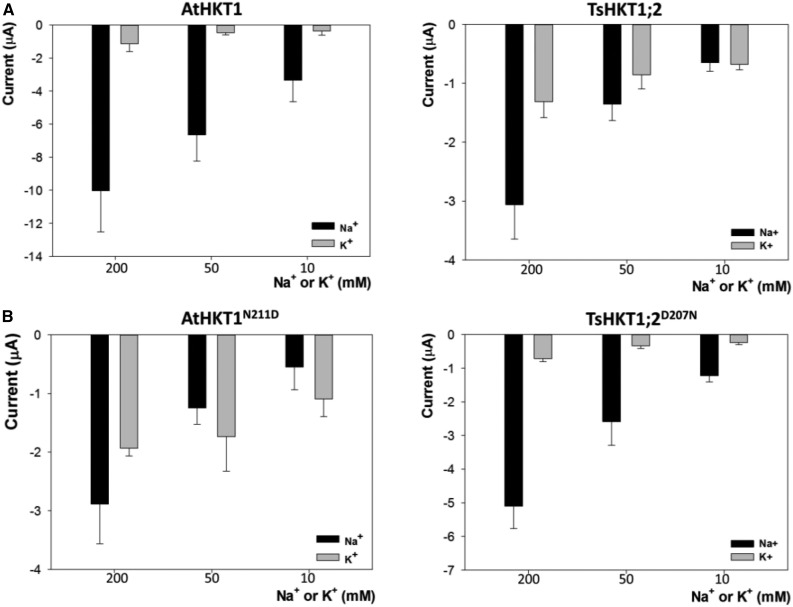
As a question that has long been debated (Xue et al., 2011), it was important for our study to establish whether AtHKT1;1 and TsHKT1;2 transport K+. In our experiments, both transporters showed a very low K+ transport capacity while in the presence of Na+. This was clearly shown by the insignificant effect of increasing the concentrations of K+ over the HKT-dependent currents at a constant Na+ concentration. At 10-mm Na+, increasing K+ from 10 to 100 mm did not affect the magnitude of currents mediated by AtHKT1;1 and TsHKT1;2 (Supplemental Fig. S6). However, currents increased when external Na+ concentration was raised to 100 mm while K+ was kept at 10 mM. Nevertheless, at negative membrane potentials typically found in plant plasma membranes, e.g. −150 mV, weak K+-dependent currents were measurable in both transporters in the absence of Na+ (Fig. 5A). Importantly, in TsHKT1;2, the K+-dependent currents were only 2.4 times lower than the Na+-dependent currents whereas the corresponding ratio for AtHKT1;1 was approximately 10-fold (Fig. 5A). It should be noted that the magnitude of the currents mediated by these two transporters might not be compared directly because the amount of each transporter expressed in the oocyte could be different. However, the ratio between Na+ and K+ currents for each transporter clearly reveals their differential capacity to transport both cations.
Figure 5.
Current responses of AtHKT1, TsHKT1;2, and their mutants at increasing Na+ and K+ concentrations. Currents from X. laevis oocytes expressing AtHKT1 and TSHKT1;2 (A) or their mutants AtHKT1N211D and TsHKT1;2D207N (B) at −150 mV and variable concentrations of either Na+ or K+ (mM). Mean and sd of 15 different oocytes for the AtHKT1, TsHKT1;2, and TsHKT1;2D207N, and nine different oocytes for the AtHKT1N211D.
Next, we studied the effect of the N211D mutation in AtHKT1;1 to find a mechanistic explanation for its effect on the growth of yeast at limiting K+ availability (Fig. 3B). Moreover, the AtHKT1N211D transporter showed inwardly rectifying characteristics very similar to those described for TsHKT1;2 (Fig. 4B; Supplemental Fig. S4). Conductance values plotted against external Na+ concentration were fitted with a single hyperbolic function resulting in a half-saturation constant of 107.2 mm for AtHKT1N211D, a value similar to that obtained for TsHKT1;2 (93.6 mM) but 13-fold higher than that of the wild-type AtHKT1;1 (8.3 mM; Supplemental Fig. S5). Moreover, AtHKT1N211D gained a significant capacity to transport K+ in the absence of Na+ (Fig. 5B). In view of these effects, we tested the D207N mutation in TsHKT1;2. This mutation increased the affinity for Na+ uptake relative to the wild type, reducing the half-saturation constant from 93.6 mm to 52.5 mm (Supplemental Fig. S5) while the capacity to transport K+ decreased (Fig. 5B) without changing the inwardly rectifying properties of the TsHKT1;2 transporter (Fig. 4B; Supplemental Fig. S4). Finally, the currents in both mutants were constant when the concentration of Na+ was fixed and K+ was variable, while the currents increased when the concentration of K+ was fixed and Na+ was raised (Supplemental Fig. S6). However, while the AtHKT1N211D mutation reduced net ion transport compared to the wild type, mutation TsHKT1;2D207N had the opposite effect. In summary, these results signify that the n/d dichotomy at the second pore-loop region of AtHKT1;1 and TsHKT1;2 determines the capacity for Na+ transport and the ratio at which Na+ and K+ are taken up by these proteins (Fig. 5).
The AtHKT1N211D Mutation Enhances Plant Growth under Salt Stress
To extend our data from yeast and oocytes to plants, transgenic lines were generated expressing AtHKT1;1, AtHKT1N211D, AtHKT1N242D, and AtHKT12N2D, where either one or both Asn residues in AtHKT1;1 were replaced by Asp. All sequence variations were expressed in the hkt1-1 mutant background under the control of the Arabidopsis AtHKT1;1 native promoter, ProAtHKT1. Homozygous T3 lines with similar HKT1 transcript levels were selected for further analysis (Supplemental Fig. S7). Plants expressing either AtHKT1N211D or AtHKT12N2D showed significantly improved growth under salt stress, whereas salt-stress responses of lines harboring AtHKT1N242D were similar to wild-type AtHKT1-expressing plants (Fig. 6A). Lines used in Figure 6A were also grown hydroponically in LAK medium for two weeks and treated with 20 mm and 30 mm NaCl for an additional two-week period (Fig. 6B). Plants expressing either AtHKT1N211D or AtHKT12N2D accumulated more fresh weight after salt treatment (Fig. 6, C and D). In contrast, wild-type AtHKT1- and AtHKT1N242D-expressing plants showed similar salt-stress responses, and the hkt1-1 knock-out plants accumulated significantly less fresh weight, compared to wild-type Col-gl plants (Fig. 6D).
Figure 6.
Salt stress-dependent phenotypes of transgenic plants expressing ProAtHKT1:: AtHKT1 and its mutant forms in the hkt1-1 background. A, 10-d-old MS media grown seedlings of Col-gl, hkt1-1, and transgenic plants expressing ProAtHKT1::AtHKT1, ProAtHKT1::AtHKT1N211D, ProAtHKT1::AtHKT1N242D, and ProAtHKT1::AtHKT12N2D in the hkt1-1 background were transferred to soil and further grown for two weeks followed by 300-mm NaCl treatment for another two weeks as described in “Materials and Methods”. Photographs were taken at the end of salt treatment. B, Seeds of wild type, hkt1-1, and all the transgenic lines were grown in hydroponic solution for one week followed by 20- or 30-mm NaCl treatment. Photographs were taken two weeks after the addition of salt. C and D, After salt treatment, fresh weights of plants used in (A) and (B) (30 mm NaCl) were measured. Error bars represent sds from three independent repeats (n = 30). Significant difference was determined by a Student’s t-test; single or double stars indicate a P-value of <0.05 or <0.01, respectively.
We further examined the accumulation of Na+ and K+ contents in plants expressing wild-type or mutant AtHKT1N211D protein in a short-term salinity treatment to monitor long-distance transport of Na+ (Fig. 7). As expected, hkt1-1 plants accumulated more Na+ in shoots and less in roots than wild-type plants, which is consistent with the role of AtHKT1;1 in xylematic Na+ unloading (Fig. 7A). Na+ accumulation in the shoots of ProAtHKT1::AtHKT1N211D plants was similar to wild-type Col-gl plants; however, accumulation of Na+ in the roots of ProAtHKT1::AtHKT1N211D was more than all other lines (Fig. 7B). Compared to wild-type Col-gl, hkt1-1 and ProAtHKT1::AtHKT1, plants with the ProAtHKT1::AtHKT1N211D construct accumulated slightly more K+ in both roots and shoots (Fig. 7, C and D). These results strongly suggest that the differences in AtHKT1;1 and AtHKT1N211D are based on the presence of either Asn or Asp in the second pore-loop domain of these transporters, which enables them to take up preferentially either Na+ (N211) or K+ (D211).
Figure 7.
Na+ and K+ content in plants. Ten-days old grown seedlings of Col-gl, hkt1-1, and transgenic plants expressing ProAtHKT1::AtHKT1 or ProAtHKT1::AtHKT1N211D in the hkt1-1 background were transferred to hydroponic cultures and further grown for two weeks followed by 100 mm NaCl for 24 h in MS medium. Na+ and K+ contents were measured by inductively coupled plasma optical emission spectroscopy. A and B, Na+ contents of shoots or roots. C and D, K+ contents of shoots or roots.
DISCUSSION
Based on protein structure and ion selectivity, HKT-type transporters have been classified into subclasses, 1 and 2 (Horie et al., 2001; Mäser et al., 2002b; Platten et al., 2006). HKT transporters in subclass-1 (HKT1) transport Na+ while the transport capacity of those in subclass-2 (HKT2), either Na+ or K+, is still controversial because their expression in heterologous systems may not reflect their ion selectivity properties in plants (Bañuelos et al., 2008). All HKT1 proteins known from dicots include a Ser at the predicted filter position in the pore-loop A of the protein. Reciprocal amino-acid substitutions in AtHKT1;1 and the wheat (Triticum spp.) TaHKT2;1 transporting Na+ and K+ identified this Ser as an important selectivity filter for the preferential Na+ transport of subclass-1 HKTs (Mäser et al., 2002b). Like AtHKT1;1, TsHKT1;2 also belongs to subclass-1 as it contains the conserved Ser residue in the selectivity filter position in the pore-loop domain (Ali et al., 2012). When aligned with TaHKT1;5, HvHKT1;5, or AtHKT1;1, the TsHKT1;2 protein shows much higher similarity with AtHKT1;1 (Ali et al., 2012). However, while AtHKT1;1 functions as a selective Na+ transporter in yeast and X. laevis oocytes (Fig. 4; Mäser et al., 2002b; Xue et al., 2011), TsHKT1;2 supports K+ uptake in yeast (Fig. 3) and reveals reduced affinity for Na+ transport in oocytes (Figs. 4 and 5). Although categorized as a subclass-1 protein, TsHKT1;2 does not follow the rule established for this class. The conundrum represented by the fundamentally different behavior of the TsHKT1;2 protein (as well as for TpHKT1;2) seems to challenge this dogma that the Ser-Gly dichotomy in the first membrane-pore-membrane domain dictates the Na+-K+ selectivity of HKT proteins (Mäser et al., 2002b).
Our results indicate that cation selectivity of HKT1 transporters is convertible by exchanging a single amino acid in the second pore-loop domain. Ser in the first pore-loop domain appears to be an essential amino acid favoring Na+ uptake, but possibly its function needs to be complemented with specific amino acids in other parts of the protein. We show that the presence of an Asp (d) replacing an Asn residue (n) in the second pore-loop domain greatly reduces Na+ uptake while increasing by 2 to 3-fold the K+ transport capacity of AtHKT1;1 (Fig. 5). Vice versa, replacing d in the TsHKT1;2 protein by n increases Na+ permeation and slightly decreases the K+ uptake of the transporter (Fig. 5). We have shown previously that TsHKT1;2 as well as TpHKT1;2 contain two conserved Asp (d), one in the second pore-loop domain (D207/205) and a second conserved d in the adjacent transmembrane domain (D238/236). Clearly, D207 plays an important role in determining the Na+/K+ transport in TsHKT1;2. In contrast with TsHKT1;2 and TpHKT1;2, most HKTs include either Asn (n), or less frequently Ser (s), instead of the D207/205 (Fig. 1F; Supplemental Fig. S2). Mutation of the corresponding Asn in AtHKT1;1 to Asp (N211D) provided proof for the importance of this residue not only by expression in yeast cells (Fig. 3) but also by expressing the mutated protein in plants (Fig. 6). Unlike AtHKT1;1, AtHKT1N211D (but not AtHKT1N242D) expressed in yeast cells complemented the K+-uptake deficiency of the CY162 mutant (Fig. 3B). The yeast K+ transporter, ScTRK1, also includes an Asp residue in the pore-loop domain but lacks this change in the transmembrane domain (Fig. 1F; Supplemental Fig. S2). This identifies the Asp residue in the pore-loop domain as important for Na+/K+ selectivity. Our conclusions are also supported by molecular modeling studies (Fig. 2; Supplemental Fig. S3). Structures of both AtHKT1;1 and TsHKT1;2 and their mutated versions suggest that the wild type AtHKT1;1 and the mutated TsHKT1;2D207N contain similar charge distributions at the pore. Conversely, the altered AtHKT1N211D showed a charge distribution similar to that of the TsHKT1;2 wild type. Molecular dynamics simulations show Asp residues participating in strong interaction with K+ ions based on their contribution to negatively charged surfaces in the pore. These modeling studies underscore the singular importance of the Asp residue (Fig. 2; Supplemental Fig. S3).
High Na+ content in the cytosol imparts K+ deficiency unless plants can engage countermeasures. One such measure is to activate transporters with high K+ selectivity to maintain ionic balance at the cell level (Maathuis and Amtmann, 1999). Excess of Na+ in the cytosol counteracts the optimal cytosolic Na+/K+ ratio, and is recognized as K+ deficiency. Inducing HKT1 under such K+ deficiency conditions seems detrimental if the HKT1 protein were to be Na+-specific (Kader et al., 2006). Under salt stress, when cytosolic Na+ concentration reaches a toxic level, plants activate high-affinity K+ transporters for K+ uptake that adjust cellular Na+/K+ balance (Yao et al., 2010; Wang et al., 2014). TsHKT1;2 could perform a complementary function in T. salsuginea based on its activation in response to high salinity and its lower Na+/K+ transport (Fig. 5).
Electrophysiological analysis confirmed that TsHKT1;2 can transport Na+ and K+ with similar efficacy, whereas AtHKT1;1 is much more specific for Na+ (Figs. 4 and 5). Therefore, the functional properties of these proteins in X. laevis oocytes are in line with those observed in yeast (Fig. 3) and plants (Figs. 1 and 6). Another key difference between AtHKT1;1 and TsHKT1;2 was inward rectification. Whereas AtHKT1;1 carried both inward and outward currents of Na+, TsHKT1;2 showed strong inward rectification that negated Na+ efflux currents (Fig. 4). Rectification was first described in the rice protein OsHKT1;1 (Jabnoune et al., 2009) and, to our knowledge, other cases have not been described. The rice HKT1 family comprises members displaying high affinity (Km in the low mm range) for Na+ uptake and no rectification, such as OsHKT1;3 and OsHKT1;5, whereas OsHKT1;1 has low affinity (76 mm Na+) and strong inward rectification (Jabnoune et al., 2009; Ren et al.; 2005). These two sets of kinetic properties resemble those of AtHKT1;1 (Km 8.3 mm Na+, no rectification) and TsHKT1;2 (Km 93.6 mm Na+, strong inward rectification), respectively. Remarkably, the N211D mutation converted AtHKT1;1 into an inwardly rectifying transporter, mimicking TsHKT1;2. The physiological implication of inward rectification in HKT transporters is unknown but it might be important, considering the role rectification plays in other ion channels (Xue et al., 2011). Moreover, this is the first finding, to our knowledge, that a point mutation in a pore-loop region gives rise to the rectifying property. Notably, the corresponding opposite mutation in TsHKT1;2, D207N, did not abolish rectification in this transporter. This suggests that the rectification property depends also on other amino acids that are present in AtHKT1;1.
The subfamily-1 of HKT transporters is preferentially expressed in the plant vasculature. Expression patterns of the subfamily-2 transporters also include root periphery cells and have been involved in nutritional K+ and Na+ uptake. HKT1-type transporters play crucial roles in plant adaptation to salinity by mechanisms that are not entirely clear. In one function that is widely acknowledged, they are involved in mediating the distribution of Na+ within the plant by a repeated pattern of removing Na+ from the xylem, particularly in the roots, such that the amount of Na+ arriving in the shoot becomes more easily manageable (Sunarpi et al., 2005; Ren et al., 2005; Munns et al., 2012; Chen et al., 2007; Hauser and Horie, 2010). However, genetic and ionic profiling of Arabidopsis accessions with enhanced salt tolerance identified an hypomorphic allele of AtHKT1;1 with reduced expression in roots as the causal locus driving elevated shoot Na+ content and salt tolerance, which runs opposite to the prevailing model of the shoot avoidance strategy of Arabidopsis (Rus et al., 2006). Moreover, AtHKT1;1 has also been implicated in Na+ uptake from the soil solution and recirculation from leaves to roots via the phloem (Rus et al., 2001; Berthomieu et al., 2003). Of note is that AtHKT1;1 gene expression is induced by mild salinity (30 mm NaCl or KCl) but strongly down-regulated by acute salinity stress by a mechanism that likely involves the ABA- and cytokinin-response regulators ABI4, ARR1, and ARR12 (Sunarpi et al., 2005; Mason et al., 2010; Shkolnik-Inbar et al., 2013). Reduced gene expression at high salinity also runs counterintuitive to the prevailing role of Na+ removal from the transpiration stream as a mechanism for salt adaptation and suggest that curtailing Na+ reabsorption from the xylem in a time- and stress-dependent manner may optimize xylematic Na+ flux and redistribution in whole plants. Together, these data signify that the contribution of AtHKT1;1 to the salt tolerance of Arabidopsis in not univocally linked to the beneficial effect of Na+ retrieval from the transpiration stream, and that alternative scenarios are possible. We show here that HKT1 proteins (TsHKT1;2 and AtHKT1N211D) with reduced Na+ affinity and the capacity for inward-rectification convey salt tolerance that is not necessarily linked to efficacious Na+ retrieval from the xylem sap.
The Na+-permeable HKT1 transporters AtHKT1;1 and OsHKT1;5 preferentially expressed in xylem parenchyma cells inversely control the amount of Na+ and K+ in the xylem sap (Sunarpi et al., 2005; Ren et al., 2005). It has been suggested that Na+ absorption into xylem parenchyma cells via HKT1 transporters depolarizes the plasma membrane, which in turn could activate K+-release channels to achieve K+ loading into the xylem sap (Horie et al., 2009). Our data (Fig. 4) and previous reports (Xue et al., 2011 and references therein) demonstrate the lack of rectification in AtHKT1;1, an attribute that may, under physiologically meaningful conditions, reverse fluxes and mediate the release of Na+ from within the cell. For instance, when expressed in oocytes, AtHKT1-mediated currents shifted from Na+ uptake to Na+ release at 10 mm external Na+ and a membrane potential of −50 mV (Fig. 4), conditions that are well within the physiological range encountered by xylem parenchyma cells (Sunarpi et al., 2005). Note, however, that although the cytosolic concentration of Na+ in oocytes was not controlled in our experiments and are likely to change as soon as the expressed transporters reach the oocyte membrane, outward currents mediated by AtHKT1;1 have indeed been recorded in root stellar cells with clamped cytosolic ion concentrations (Xue et al., 2011); i.e. AtHKT1;1 could effectively transport Na+ efflux from cells upon membrane depolarization. Contrary to AtHKT1;1, reversal of Na+ fluxes were not observed in oocytes expressing TsHKT1;2 or the mutant AtHKT1N211D owing to their rectification capacity (Fig. 4). This critical difference might explain that Arabidopsis plants expressing AtHKT1N211D retained more Na+ in the roots than wild-type AtHKT1;1 despite the reduced Na+ affinity (Fig. 7). We suggest that under acute salinity stress, the amount of Na+ loaded into the xylem sap is determined by active loading driven by the Na+/H+ antiporter SOS1 (Shi et al., 2002; Oh et al., 2010a) and the balance between inward and outward Na+ fluxes through AtHKT1;1 (Fig. 4). The net amount of Na+ loaded or unloaded into the xylem by AtHKT1;1 would be contingent to specific electrophysiological parameters given in xylem parenchyma cells and sections along the root axis. Past a certain threshold of intracellular Na+ concentration and/or plasma membrane depolarization in parenchyma cells, AtHKT1;1 could also contribute to Na+ loading onto the xylem sap and Na+ export to shoots. By negating outward Na+ fluxes, the inward-rectifying AtHKT1N211D mutant protein would impose a greater Na+ retention in roots (Figs. 4 and 7).
Balancing Na+ and K+ ions under salt stress is crucial for plant survival (Qi and Spalding, 2004), yet all the molecular components involved in this process are still unknown. Upon occasional increase of NaCl in the root-zone (a condition that may apply to glycophytes), AtHKT1;1 may contribute to restrict and/or delay Na+ accumulation into the shoot by mediating its retrieval from the xylem and transfer into downward fluxes of phloem (Horie et al., 2009). However, for plants that are continuously exposed to high NaCl, such as halophytes, some HKT1 isoforms might confer the ability to maintain a low cytosolic Na+/K+ ratio in the presence of high salinity stress (Orsini et al., 2010). This K+ transport function would complement the best-documented function of HKT1 in retarding Na+ flux throughout the plant in Thellungiella species. The inwardly rectifying transport properties of TsHKT1;2 and AtHKT1N211D would facilitate shoot K+ over Na+ accumulation and accomplish at least two critical functions for the halophytic habit: (1) compensating the osmotic imbalance due to reduced Na+ arriving to the shoot (Rodríguez-Navarro, 2000); and (2) contrasting the onset of the species-specific Na+ toxicity threshold that would affect normal shoot functions (Maggio et al., 2001; Chen et al., 2007). These findings also support earlier observations demonstrating that plant growth and yield correlate with a measure of the dynamic process of toxic ion accumulation in the shoot and not with root zone salinity per se (Dalton et al., 2000). Suppression of HKT1 expression in T. salsuginea by RNAi leads to hyper-accumulation of Na+ in the shoots, while lowering the Na+ content in the roots compared to wild type, indicating that HKT1 in T. salsuginea regulates Na+ ion uptake under salt stress. The Na+/K+ ratio was likewise disturbed in TsHKT1 RNAi lines (Ali et al., 2012).
The evolution of halophytism is grounds for serious debate. A better understanding of how species may have evolved complex traits such as salt tolerance is of ecological importance as well as helping in attempts to replicate adaptation mechanisms to extreme environments in crop plants. An origin of plants from organisms adapted to the sea is not questioned. However, evolution over time led to fresh water algae that, most likely, gave rise to land plants that carried no salt tolerance (glycophytes; Raven and Edwards, 2001). This view is confirmed by at least two lines of evidence. First, the ability to acquire nutrients from an oligotrophic environment, in which freshwater organisms must have developed, would have preadapted and/or facilitated land colonization where nutrient availability is also limited (Stevens, 2008; Rodríguez-Navarro and Rubio, 2006; Becker and Marin, 2009). Second, phylogenetic analysis shows that species with a halophytic habit represent only a small percentage of plants in all known orders, strongly suggesting that land halophytes evolved from multiple glycophytic ancestors. Although there are examples supporting this view (Flowers et al., 2010), a genetic basis explaining how the transition from a glycophytic to a halophytic habit may have occurred has remained elusive. Here we present the evidence for an evolutionary event that appears to contribute to such a transition. We show that, in the dicot lineage, monovalent cation selectivity—one of the critical mechanisms for the halophytic lifestyle—can be modified by mutating a single Asn to an Asp in the second pore-loop domain of HKT1-type transporters. This substitution altered cation specificity and the rectification of the channel-like Na+ transporter HKT1, which should have helped in the conversion of a salt-sensitive into a somewhat salt-tolerant organism. The change in the nature of HKT1 thus appears as a critical evolutionary change.
Expressing in Arabidopsis a version of the ubiquitous Na+ transporter HKT1 that carries the single amino-acid change found in TsHKT1;2 in cells and tissues that normally express the AtHKT1;1 protein leads to an increase in K+ ions. The significant outcome is that the deleterious effect of higher concentrations of Na+ is counteracted to some degree. While the so-altered Arabidopsis plants are not tolerant to a considerable excess of Na+, tolerance is of such a magnitude that survival of the plants is possible. Arabidopsis and Thellungiella species are separated by some 12 to 14 million years (Oh et al., 2010b; Cheng et al., 2013). Such a time frame allowed for further evolutionary changes to work on genomes, generating additional alterations supporting a truly halophytic phenotype.
MATERIALS AND METHODS
Plant Material
Arabidopsis seeds of Col-gl (wild type), hkt1-1 in Col-gl, as shown by Ali et al. (2012), were used in this study.
Generation of Transgenic Arabidopsis Plants
The cDNAs of TsHKT1;2 and AtHKT1;1 were amplified with primers listed in Supplemental Table S1. The Arabidopsis hkt1-1 mutant line expressing 35S-AtHKT1;1 or 35S-TsHKT1;2 have been previously described by Ali et al. (2012). AtHKT1;1 and TsHKT1;2 cDNAs were fused with the 1.2-Kb fragment of the promoter region of AtHKT1;1 by PCR and inserted between PstI and SpeI sites, in the pCAMBIA 1302 binary vector. hkt1-1 knock-out plants were transformed by floral dip. Transgenic plants were selected based on hygromycin resistance and confirmed by PCR with primers listed in Supplemental Table S1. Lines showing 3:1 segregation with resistance to hygromycin (43 mg/l) were selected and homozygous T3 plants showing similar HKT1 transcript level were used.
Growth Responses of Transgenic Plants to Salt Stress
To test growth responses of the mature plants to salt stress, seeds of transgenic plants expressing ProAtHKT1::AtHKT1 and their mutant version (n-d) together with Col-gl and hkt1-1 were surface-sterilized and grown on 0.5× MS medium in a long day growth chamber with 16-h day, 8-h night, 130 μmol m−2 s−1 light intensity and 22°C to approximately 24°C temperature. Ten-days-old seedlings were transferred to soil, and grown for two weeks. After two weeks, plants were treated with 300 mm NaCl in water every other day. Photographs were taken two weeks after salt treatment. Fresh weights of the plants were measured immediately at the end of the salt treatment.
For salt treatment in hydroponic culture, seeds of the indicated lines were directly grown on hydroponics as described by Barragán et al. (2012). A modified Long-Ashton mineral solution with 1 mm K+ and nominally free of Na+ and NH4+ (LAK medium) was used as base solution for hydroponic cultures. The final composition of the LAK base solution was as follows: 1 mm KH2PO4, 2 mm Ca(NO3)2, 1 mm MgSO4, 30 mm H3BO3, 10 mm MnSO4, 1 mm ZnSO4, 1 mm CuSO4, 0.03 mm (NH4)6Mo7O24, and 100 mm Fe2+ as Sequestrene 138-Fe, at pH 5.3. Seedlings were grown in LAK medium for one week followed by salt treatment at the indicated NaCl concentrations. Photographs were taken two weeks later. Fresh weights of the plants were measured immediately at the end of the salt treatment.
RNA Extraction and RT-PCR
RNA from two-week-old Col-gl1, hkt1-1, and all transgenic plants was extracted with the Qiagen RNeasy plant mini kit (Qiagen, Hilden, Germantown, MD). RT-PCR (Reverse transcriptase PCR) was carried out with 3 μg of total RNA using ThermoScript RT-PCR System (Invitrogen, Carlsbad, CA) with the primers listed in Supplemental Table S1.
Yeast Expression and Growth
The yeast strain (Saccharomyces cerevisiae) CY162 (Mat a, ura3-52, his3∆200, his44-15, trk1∆, trk2::pcK64; Ko and Gaber, 1991) was used for K+ uptake tests. The cDNAs, amplified with primers as listed in Supplemental Table S1, were cloned into the BamHI and NotI sites of the pYES2 vector (Invitrogen) between the GAL1 promoter and the CYC1 terminator sequences. Yeast cells were transformed by the LiAc procedure, selected on SD medium without uracil, and subjected to growth on SD medium supplemented with the indicated concentration of KCl.
Molecular Modeling Studies
The transmembrane regions for two HKT proteins were predicted by TMHMM v2.0 (Center for Biological Sequence Analysis, http://www.cbs.dtu.dk/services/TMHMM). The three-dimensional structures of AtHKT1;1 and TsHKT1;2 were generated using a homology modeling method (the Build Homology Model Protocol), which was implemented in Discovery Studio 3.5 (DS3.5, Studio D; Accelrys, San Diego, CA). The crystal structure of the NaK channel (PDB: 2AHZ), a Na+- and K+-conducting channel, was used as the template. The sequence alignments were performed between our target proteins and the template using the alignment tool in DS3.5. The transmembrane regions of the HKT proteins were separated into four different subunits such as M1A-PA-M2A, M1B-PB-M2B, M1C-PC-M2C, and M1D-PD-M2D, and then aligned with one subunit of the template. To form the pore region responsible for ion permeation in K+ channels, each of the four subunits of the transmembrane regions for AtHKT1;1 and TsHKT1;2 were coordinated as tetramers. Homology-modeled structures were subjected to molecular dynamics (MD) simulation with a DPPC (dipalmitoylphosphatidylcholine) lipid bilayer using the GROMACS program (v. 4.5.3) with a GROMACS 53a6 force field including a Berger lipid parameter (Berger et al., 1997; Oostenbrink et al., 2004; Hess et al., 2008). The 2-ns MD simulations in an explicit membrane environment were applied to refine initial homology-modeled structures. Based on the refined structures, mutant systems (AtHKT12N2D and TsHKT1;22D2N) were constructed by using the Build Mutants protocol in the software DS3.5. Subsequently, 5-ns MD simulations were performed to investigate the mutation effect of experimentally suggested key residues. To construct an explicit membrane environment, GROMACS topologies for the DPPC model, available from the Dr. Peter Tieleman’s Web site (http://moose.bio.ucalgary.ca), were used. Two different ions, Na+ and K+, were placed within the selectivity filter of the structure. The protein structures were incorporated into the DPPC membrane. The membrane systems solvated by SPC (simple point charge) water molecules (Berendsen et al., 1981) and was ionized with 100 mm NaCl. The steepest-descent energy minimization was calculated and each equilibrium simulation of NVT and NPT ensembles was simulated during 100 ps and 1 ns, respectively. The temperature of the system was set to 323 K. The production run was performed until 5 ns, and the GROMACS package was used for analysis of MD simulations. The representative structures of MD simulation were obtained after the last 2 ns, as the frames of 4750 ps for wild type-AtHKT1 and 4752 ps for Mut-AtHKT1 system. For TsHKT systems, representative structures of wild type- and Mut-TsHKT1;2 were selected at 4590 ps and 4708 ps, respectively. The VMD and DS3.5 programs (Humphrey et al., 1996) were utilized for visualization and for structural analysis (Studio D, Accelrys).
Analysis of Ion Content in Plants
Ionic content analyses in plants were carried out as described (Rus et al., 2001) except that plants were grown hydroponically for four weeks. Samples were dried at 65°C for 2 d and 100-mg ground tissue was extracted with 10 ml of 0.1 n HNO3 for 30 min. Samples were filtered and ion content analysis was carried out with inductively coupled plasma optical emission spectroscopy using an OPTIMA 4300DV/5300DV (Perkin-Elmer, Waltham, MA).
Analysis of Ion Content in Yeast and Yeast Cells Growth in Liquid Culture
Analyses of K+ content in yeast followed a protocol by Takahashi et al. (2007) with minor modifications. Briefly, yeast cells of the K+-uptake deficient Saccharomyces cerevisiae strain CY162 (trk1, trk2) transformed with pYES2 (empty vector), AtHKT1;1, AtHKT1N211D, AtHKT1N242D, AtHKT12N2D, and TsHKT1;2 were grown in SD medium without uracil, to OD600 0.7 (2 × 107 cells/ml). Cells were collected and resuspended in the same amount of fresh SD medium with 2.0% Glc, 10 mm MES, pH 6.0 adjusted with Tris base, and supplemented with the indicated concentration of KCl. Cells were incubated for 2 h, collected at 1250 rpm for 5 min, and acid-extracted overnight in 10 ml of 0.1 m HCl. Samples were centrifuged at 5000 g for 5 min to remove debris and ion contents of the supernatant were determined with an inductively coupled plasma optical emission spectroscopy OPTIMA 4300DV/5300DV (Perkin-Elmer). For the growth curves of S. cerevisiae CY162, cells expressing AtHKT1;1, AtHKT1N211D, AtHKT1N242D, and AtHKT12N2D were grown overnight in SD dropout liquid medium without uracil, and the next day new cultures were prepared starting at an OD of 0.04. Then, 200 mm NaCl with 1 mm KCl were added to the cultures. Samples were collected at the indicated time and the rate of cells growth was checked by spectrophotometer.
Site-Directed Mutagenesis in AtHKT1;1
In Arabidopsis HKT1;1 two Asn (n) residues present in the second pore loop domain and in the adjacent transmembrane domain were replaced by conserved aspartic acids (d) as they exist in TsHKT1;2 (Fig. 1G; Supplemental Fig. S2). Individual amino-acid mutations (AtHKT1N211D, AtHKT1N242D) were made to the template of pYES2-AtHKT1 cDNA with their respective primers listed in Supplemental Table S1. To make a double mutation of both N211 and N242 (AtHKT12N2D) the mutated pYES2-AtHKT1N211D was used as a template with the primers designed for AtHKT1N242D.
Electrophysiology in X. laevis Oocytes
cDNAs of AtHKT1;1, TsHKT1;2 and its respective mutants AtHKT1N211D and TsHKT1;2D207N were subcloned into the pNB1U vector downstream from the T7 promoter. One μg of each cDNA linearized with Not1 was placed in a standard in vitro transcription reaction by using a T7 mMessage mMachine Kit (Ambion Europe, Huntingdon, UK). The quantity and purity of the extracted RNA was determined by measuring the A260. The integrity and size range of total purified RNA was checked by denaturing agarose gel electrophoresis (0.7%) and ethidium bromide staining. X. laevis oocytes were defolliculated by collagenase treatment (2 mg ml−1; type IA; Sigma-Aldrich, St. Louis, MO). Defolliculated oocytes were injected with 50 nl of water (control) or 50 ng of mRNA using a Nanoliter 2010 microinjector (World Precision Instruments, Sarasota, FL) and then maintained at 18°C in ND96 medium (mM: 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, 2.5 Na-pyruvate, and 5 HEPES-NaOH, at pH 7.4), which ensured 90% survival. At 2 or 3 d after injection, whole-cell current recordings by two-electrode voltage clamp were performed at room temperature with a GeneClamp500 amplifier (Axon Instruments/Molecular Devices, Sunnyvale, CA). Data were acquired using software Pulse/PulseFit (HEKA, Lambrecht/Pfalz, Germany) after filtering at 10 kHz for analysis. Electrodes filled with 3 m KCl showed resistances of 0.5–1.0 MΩ. The oocyte was continuously perfused during the voltage-clamp experiment. All bath solutions contained a background of 6 mm MgCl2, 1.8 mm CaCl2, and 10 mm MES-1,3 bis[tris(hydroxymethyl)methylamino]propane, at pH 5.5. Monovalent cations were added as Glu salts. d-Mannitol was added to adjust the osmolarity.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Na+ content in ProAtHKT1::AtHKT1- and ProAtHKT1::TsHKT1;2-expressing plants in hkt1-1 background.
Supplemental Figure S2. Comparison of HKT1 homologs from different plant species.
Supplemental Figure S3. Construction of mutation systems for AtHKT1 and TsHKT1;2.
Supplemental Figure S4. Expression of AtHKT1, TSHKT1;2 and mutated proteins in X. laevis oocytes.
Supplemental Figure S5. Inward conductance from oocytes expressing AtHKT1, TsHKT1;2, AtHKT1N211D, and TsHKT1;2D207N in the presence of varying external Na+ concentrations.
Supplemental Figure S6. Permeability in AtHKT1, TsHKT1;2, and their respective mutants.
Supplemental Figure S7. Semiquantitative RT-PCR used to select transgenic lines with comparable levels of HKT1 transcript.
Supplemental Table S1. PCR primer sequences used.
Supplemental Table S2. HKT1-like protein sequences used for amino-acid sequence comparison.
Supplementary Material
Acknowledgments
We are thankful to Professor Alonso Rodriguez-Navarro and Dr. Zahir Ali for their critical discussions and suggestions on the manuscript.
Footnotes
This work was supported by the Next-Generation BioGreen 21 Program (SSAC grant no. PJ01105101); Rural Development Administration, Republic of Korea; and the National Research Foundation of Korea (grant no. 2016R1A2A1A05004931).
Articles can be viewed without a subscription.
References
- Ali Z, Park HC, Ali A, Oh DH, Aman R, Kropornicka A, Hong H, Choi W, Chung WS, Kim WY, Bressan RA, Bohnert HJ, et al. (2012) TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol 158: 1463–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Park HC, Aman R, Ali Z, Yun DJ (2013) Role of HKT1 in Thellungiella salsuginea, a model extremophile plant. Plant Signal Behav 8: e25196.1–e25196.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañuelos MA, Haro R, Fraile-Escanciano A, Rodríguez-Navarro A (2008) Effects of polylinker uATGs on the function of grass HKT1 transporters expressed in yeast cells. Plant Cell Physiol 49: 1128–1132 [DOI] [PubMed] [Google Scholar]
- Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batelli G, Oh DH, D’Urzo MP, Orsini F, Dassanayake M, Zhu JK, Bohnert HJ, Bressan RA, Maggio A (2014) Using Arabidopsis-related model species (ARMS): growth, genetic transformation, and comparative genomics. Methods Mol Biol 1062: 27–51 [DOI] [PubMed] [Google Scholar]
- Becker B, Marin B (2009) Streptophyte algae and the origin of embryophytes. Ann Bot (Lond) 103: 999–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amar S, Brini F, Sentenac H, Masmoudi K, Véry AA (2014) Functional characterization in Xenopus oocytes of Na+ transport systems from durum wheat reveals diversity among two HKT1;4 transporters. J Exp Bot 65: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J (1981) In Pullman B, editor, Interaction models for water in relation to protein hydration. Intermolecular Forces, The Jerusalem Symposia on Quantum Chemistry and Biochemistry, Vol. 14 Springer, New York, pp 331–342 [Google Scholar]
- Berger O, Edholm O, Jähnig F (1997) Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys J 72: 2002–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, Gosti F, Simonneau T, et al. (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Jin X, Huang H, Derebe MG, Levin EJ, Kabaleeswaran V, Pan Y, Punta M, Love J, Weng J, Quick M, Ye S, et al. (2011) Crystal structure of a potassium ion transporter, TrkH. Nature 471: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman JM. (2015) The evolution of halophytes, glycophytes and crops, and its implications for food security under saline conditions. New Phytol 206: 557–570 [DOI] [PubMed] [Google Scholar]
- Chen ZH, Zhou MX, Newman IA, Mendham NJ, Zhang GP, Shabala S (2007) Potassium and sodium relations in salinised barley tissues as a basis of differential salt tolerance. Funct Plant Biol 34: 150–162 [DOI] [PubMed] [Google Scholar]
- Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, Wang X (2013) Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25: 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corratgé C, Zimmermann S, Lambilliotte R, Plassard C, Marmeisse R, Thibaud JB, Lacombe B, Sentenac H (2007) Molecular and functional characterization of a Na+-K+ transporter from the Trk family in the ectomycorrhizal fungus Hebeloma cylindrosporum. J Biol Chem 282: 26057–26066 [DOI] [PubMed] [Google Scholar]
- Corratgé-Faillie C, Jabnoune M, Zimmermann S, Véry AA, Fizames C, Sentenac H (2010) Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family. Cell Mol Life Sci 67: 2511–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton FN, Maggio A, Piccinni G (2000) Simulation of shoot chloride accumulation: separation of physical and biochemical processes governing plant salt tolerance. Plant Soil 219: 1–11 [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, Cheeseman JM (2011) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus A, Tester M (2007) The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 30: 497–507 [DOI] [PubMed] [Google Scholar]
- Di Michele WA, Phillips TL, Olmstead RG (1987) Opportunistic evolution: abiotic environmental stress and the fossil record of plants. Rev Palaeobot Palynol 50: 151–178 [Google Scholar]
- Durell SR, Hao Y, Nakamura T, Bakker EP, Guy HR (1999) Evolutionary relationship between K+ channels and symporters. Biophys J 77: 775–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ, Galal HK, Bromham L (2010) Evolution of halophytes: multiple origins of salt tolerance in land plants. Funct Plant Biol 37: 604–612 [Google Scholar]
- Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44: 826–839 [DOI] [PubMed] [Google Scholar]
- Hauser F, Horie T (2010) A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ 33: 552–565 [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4: 435–447 [DOI] [PubMed] [Google Scholar]
- Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI (2007) Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J 26: 3003–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Hauser F, Schroeder JI (2009) HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci 14: 660–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A (2001) Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J 27: 129–138 [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14: 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Espeout S, Mieulet D, Fizames C, Verdeil JL, Conéjéro G, Rodríguez-Navarro A, Sentenac H, Guiderdoni E, Abdelly C, Véry AA (2009) Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol 150: 1955–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader MA, Seidel T, Golldack D, Lindberg S (2006) Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. J Exp Bot 57: 4257–4268 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakaguchi M, Mori Y, Saito K, Nakamura T, Bakker EP, Sato Y, Goshima S, Uozumi N (2001) Evidence in support of a four transmembrane-pore-transmembrane topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proc Natl Acad Sci USA 98: 6488–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Gaber RF (1991) TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol Cell Biol 11: 4266–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM, Amtmann A (1999) K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann Bot (Lond) 84: 123–133 [Google Scholar]
- Maggio A, Hasegawa PM, Bressan RA, Consiglio MF, Joly RJ (2001) Unraveling the functional relationship between root anatomy and stress tolerance. Aust J Plant Physiol 28: 999–1004 [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, Robertson W, Sussman MR, et al. (2002a) Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Mäser P, Hosoo Y, Goshima S, Horie T, Eckelman B, Yamada K, Yoshida K, Bakker EP, Shinmyo A, Oiki S, Schroeder JI, Uozumi N (2002b) Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc Natl Acad Sci USA 99: 6428–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Jha D, Salt DE, Tester M, Hill K, Kieber JJ, Schaller GE (2010) Type-B response regulators ARR1 and ARR12 regulate expression of AtKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J 64: 753–763 [DOI] [PubMed] [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D, Gilliham M (2012) Wheat grain yield on saline soils is improved by an ancestral Na⁺ transporter gene. Nat Biotechnol 30: 360–364 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Oh DH, Dassanayake M, Haas JS, Kropornika A, Wright C, d’Urzo MP, Hong H, Ali S, Hernandez A, Lambert GM, Inan G, Galbraith DW, et al. (2010a) Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiol 154: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Lee SY, Bressan RA, Yun DJ, Bohnert HJ (2010b) Intracellular consequences of SOS1 deficiency during salt stress. J Exp Bot 61: 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomen RJ, Benito B, Sentenac H, Rodríguez-Navarro A, Talón M, Véry AA, Domingo C (2012) HKT2;2/1, a K⁺-permeable transporter identified in a salt-tolerant rice cultivar through surveys of natural genetic polymorphism. Plant J 71: 750–762 [DOI] [PubMed] [Google Scholar]
- Oostenbrink C, Villa A, Mark AE, van Gunsteren WF (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 25: 1656–1676 [DOI] [PubMed] [Google Scholar]
- Orsini F, D’Urzo MP, Inan G, Serra S, Oh DH, Mickelbart MV, Consiglio F, Li X, Jeong JC, Yun DJ, Bohnert HJ, Bressan RA, et al. (2010) A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana. J Exp Bot 61: 3787–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten JD, Cotsaftis O, Berthomieu P, Bohnert H, Davenport RJ, Fairbairn DJ, Horie T, Leigh RA, Lin HX, Luan S, Mäser P, Pantoja O, et al. (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11: 372–374 [DOI] [PubMed] [Google Scholar]
- Qi Z, Spalding EP (2004) Protection of plasma membrane K+ transport by the Salt Overly Sensitive1 Na+-H+ antiporter during salinity stress. Plant Physiol 136: 2548–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Martinez-Atienza J, Villalta I, Jiang X, Kim WY, Ali Z, Fujii H, Mendoza I, Yun DJ, Zhu JK, Pardo JM (2011) Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Natl Acad Sci USA 108: 2611–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Edwards D (2001) Roots: evolutionary origins and biogeochemical significance. J Exp Bot 52: 381–401 [DOI] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A. (2000) Potassium transport in fungi and plants. Biochim Biophys Acta 1469: 1–30 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A, Rubio F (2006) High-affinity potassium and sodium transport systems in plants. J Exp Bot 57: 1149–1160 [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI (1995) Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270: 1660–1663 [DOI] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM (2001) AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HZ, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Adler G, Bar-Zvi D (2013) ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J. 73: 993–1005 [DOI] [PubMed] [Google Scholar]
- Stevens PF. (2008) Angiosperm Phylogeny. Vol. 9, June 2008. Missouri Botanical Garden, http://www.mobot.org/MOBOT/research/APweb. [Google Scholar]
- Sunarpi HT, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, Konomi M, Osumi M, et al. (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Takahashi R, Liu S, Takano T (2007) Cloning and functional comparison of a high-affinity K+ transporter gene PhaHKT1 of salt-tolerant and saltsensitive reed plants. J Exp Bot 58: 4387–4395 [DOI] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI (2000) The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 122: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, Pantoja O (2014) Comparative 2D-DIGE analysis of salinity responsive microsomal proteins from leaves of salt-sensitive Arabidopsis thaliana and salt-tolerant Thellungiella salsuginea. J Proteomics 111: 113–127 [DOI] [PubMed] [Google Scholar]
- Wang TT, Ren ZJ, Liu ZQ, Feng X, Guo RQ, Li BG, Li LG, Jing H-C (2014) SbHKT1;4, a member of the high-affinity potassium transporter gene family from Sorghum bicolor, functions to maintain optimal Na⁺/K⁺ balance under Na⁺ stress. J Integr Plant Biol 56: 315–332 [DOI] [PubMed] [Google Scholar]
- Wu HJ, Zhang Z, Wang JY, Oh DH, Dassanayake M, Liu B, Huang Q, Sun HX, Xia R, Wu Y, Wang YN, Yang Z, et al. (2012) Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc Natl Acad Sci USA 109: 12219–12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Yao X, Luo W, Jha D, Tester M, Horie T, Schroeder JI (2011) AtHKT1;1 mediates Nernstian sodium channel transport properties in Arabidopsis root stelar cells. PLoS One 6: e24725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Horie T, Xue S, Leung HY, Katsuhara M, Brodsky DE, Wu Y, Schroeder JI (2010) Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiol 152: 341–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.