Sensor-based proteomics allows the identification of proteins carrying specific types of polyubiquitin chains.
Abstract
The polybiquitination of proteins can take on different topologies depending on the residue from ubiquitin involved in the chain formation. Although the role of lysine-48 (K48) polyubiquitination in proteasome-mediated degradation is fairly well characterized, much less is understood about the other types of ubiquitin chains and proteasome-independent functions. To overcome this, we developed a K63 polyubiquitin-specific sensor-based approach to track and isolate K63 polyubiquitinated proteins in plants. Proteins carrying K63 polyubiquitin chains were found to be enriched in diverse membrane compartments as well as in nuclear foci. Using liquid chromatography-tandem mass spectrometry, we identified over 100 proteins from Arabidopsis (Arabidopsis thaliana) that are modified with K63 polyubiquitin chains. The K63 ubiquitinome contains critical factors involved in a wide variety of biological processes, including transport, metabolism, protein trafficking, and protein translation. Comparison of the proteins found in this study with previously published nonresolutive ubiquitinomes identified about 70 proteins as ubiquitinated and specifically modified with K63-linked chains. To extend our knowledge about K63 polyubiquitination, we compared the K63 ubiquitinome with K63 ubiquitination networks based on the Arabidopsis interactome. Altogether, this work increases our resolution of the cellular and biological roles associated with this poorly characterized posttranslational modification and provides a unique insight into the networks of K63 polyubiquitination in plants.
The posttranslational modification of proteins by the 76-amino acid polypeptide ubiquitin, a process known as ubiquitination, plays a pivotal role in biology. Ubiquitination affects target proteins by altering their stability, interaction with other proteins, enzymatic activity, and subcellular localization (Mukhopadhyay and Riezman, 2007). Ubiquitination occurs through the sequential activation of a cascade of reactions catalyzed by three classes of enzymes (E1, E2, and E3), resulting ultimately in the covalent attachment of ubiquitin to a Lys (K) residue in the target protein. Often, several ubiquitin moieties are added to the target protein in the form of polyubiquitin chains, and these can take on distinct forms: nonlinear topologies, depending on which of the seven Lys residues present in a ubiquitin molecule is used to form the chain (K6, K11, K27, K29, K33, K48, and K63), or head-tail linear repeats (Peng et al., 2003; Pickart and Fushman, 2004; Kirisako et al., 2006). Different ubiquitin-linkage topologies are associated with diverse biological functions (Woelk et al., 2007). The best characterized polyubiquitin chain is the one involving K48 linkage, where at least four ubiquitin subunits trigger degradation of the target protein by the 26S proteasome (Pickart and Fushman, 2004). Much less is known about the other polyubiquitin chain linkages (Pickart and Fushman, 2004; Kirisako et al., 2006; Woelk et al., 2007). K63-linked ubiquitination does not induce proteasome-dependent degradation but serves as a molecular platform for protein-protein interaction. The roles of polyubiquitination involving residue K63 from ubiquitin, hereafter called K63 polyubiquitination, have been widely studied for the past decade in the context of ubiquitin-mediated endocytosis in yeast and mammals. Other roles for K63 polyubiquitin emerged in DNA damage responses and, more marginally, in autophagy and signaling (Woelk et al., 2007; Adhikari and Chen, 2009; Komander and Rape, 2012).
In plants, only a handful of proteins were shown to be K63 polyubiquitinated, despite K63 polyubiquitination being reported to be the second most abundant ubiquitination type after K48-linked chains (Kim et al., 2013). These include the PIN2 auxin efflux carrier, the BOR1 boron transporter, and the BRI1 steroid hormone receptor (Kasai et al., 2011; Leitner et al., 2012; Martins et al., 2015). In all three cases, K63 polyubiquitination of these membrane proteins has been associated with their endocytosis and degradation in the vacuole. Covalent linkage of K63 polyubiquitin chains to a target protein, however, is not likely restricted only to endocytosis and is expected to control other cellular functions in plants (Tomanov et al., 2014). The analysis of mutants impaired in the formation of K63 polyubiquitin chains indicated that such a linkage is important for responses to iron deficiency, DNA damage, and auxin signaling (Yin et al., 2007; Wen et al., 2008, 2014; Li and Schmidt, 2010; Pan and Schmidt, 2014). However, the nature of the proteins actually modified in these pathways is unknown.
The analysis of isolated proteins using traditional biochemical techniques only provides a glimpse of ubiquitin-mediated processes. Large-scale identification of ubiquitinated proteins, or the ubiquitinome, offers a unique opportunity to obtain a systems view of ubiquitin-dependent processes. Although early proteomic approaches identified only a limited number of proteins, the recent development of proteomic analyses and tools dedicated to study ubiquitinomes has yielded increasingly detailed catalogs of ubiquitinated proteins in a wide variety of organisms, including plants. Affinity purification using ubiquitin-binding domains or antibodies recognizing ubiquitin was instrumental in the characterization of ubiquitinomes containing over 100 proteins (Maor et al., 2007; Manzano et al., 2008; Igawa et al., 2009). However, these strategies are based on nonstringent purification conditions to maintain the interaction between these domains and antibodies, which have low affinity for their ubiquitinated targets and, therefore, are likely to copurify contaminants. To obtain a more accurate profile of proteins carrying ubiquitin moieties, two opposing strategies were developed. The first intended to further purify ubiquitinated proteins by tandem affinity purification using both ubiquitin-binding domains and His-tagged versions of ubiquitin, thus allowing a second nickel-nitrilotriacetic acid agarose (Ni-NTA) purification step of ubiquitin-attached proteins under fully denaturing conditions (Saracco et al., 2009; Kim et al., 2013). These combined approaches identified close to 1,000 Arabidopsis (Arabidopsis thaliana) ubiquitinated proteins. The second strategy was based on combined fractional diagonal chromatography (COFRADIC). In COFRADIC experiments, free primary amine groups are acetylated while ubiquitinated Lys residues are protected. The deubiquitination of ubiquitinated Lys residues is then used to attach a tag that allows the specific isolation of ubiquitinated peptides during subsequent COFRADIC runs. This approach led to the identification of over 3,000 ubiquitination sites in 1,600 proteins (Walton et al., 2016).
Even though our ability to detect ubiquitinated proteins has improved dramatically in recent years (Kim et al., 2013; Walton et al., 2016), a major hurdle in our understanding of ubiquitin biology remains due to the existence of several forms of ubiquitination, associated with many different cellular outputs. The lack of resolution of specific ubiquitin type detection in published ubiquitinomes has prevented us from making biological sense of these large data sets and has greatly hampered our ability to tackle with precision the roles of subtypes of ubiquitin. However, several sensors engineered from TAB2, RAP80, and VPS27, which recognize K63 polyubiquitination events, were characterized recently (Sims et al., 2012; van Wijk et al., 2012). To overcome the limitations associated with current ubiquitinomes, we developed a sensor-based proteomic approach taking advantage of the Vx3K0 K63 polyubiquitin-specific sensor (Sims et al., 2012). Vx3K0 is based on three repetitions of ubiquitin interaction motifs (UIMs) from the baker's yeast (Saccharomyces cerevisiae) VPS27 subunit of the Endosomal Sorting Complex Required for Transport (ESCRT complex), joined by a helical linker that spaces the UIMs for selective binding to extended K63-linked polyubiquitin chains. This sensor exhibits high avidity to K63-linked ubiquitin chains with a Kd of 4 nm for K63 polyubiquitin chains, and a strong selectivity in vitro with a 130-fold preference over K48 polyubiquitin chains. Expressing such a sensor in vivo allowed us not only to visualize cellular compartments enriched in K63 polyubiquitinated proteins but also to identify the K63-specific ubiquitinome. These innovative approaches dramatically increase our resolution of the cellular and biological roles associated with this poorly characterized posttranslational modification.
RESULTS
Generation of Transgenic Plants to Investigate K63 Polyubiquitination in Arabidopsis
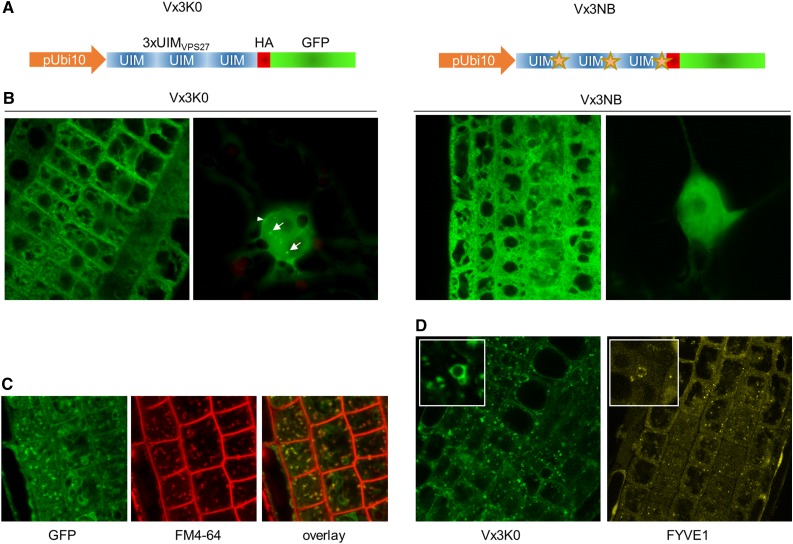
To characterize the role of K63 polyubiquitination in plants, we set out to isolate the proteome modified with K63 polyubiquitin chains. We made use of the K63 polyubiquitin-specific sensor Vx3K0 (Sims et al., 2012) and its nonbinding counterpart Vx3NB as a control for later experiments (Fig. 1A). Both sensor versions are coupled to a hemagglutinin (HA) epitope tag and a GFP for purification and visualization in the cell, respectively.
Figure 1.
Characterization of plants expressing the Vx3K0 and Vx3NB K63 polyubiquitin sensor versions. A, Design of the K63 polyubiquitin sensors expressed in transgenic Arabidopsis plants. The stars represent mutations impairing the ability to recognize K63 polyubiquitin chains in Vx3NB. B, Subcellular localization of Vx3K0 and Vx3NB sensors in the primary root tip (left) or in leaf nuclei (right). White arrows point to nuclear foci. C, Colocalization of Vx3K0 and the endocytic tracer FM4-64. D, Sensitivity of endosome-localized Vx3K0 to wortmannin. Insets show swelling of late endosomes resulting from the inhibition of PI3 kinase by wortmannin.
To evaluate the ability of Vx3K0 to bind to K63 polyubiquitinated proteins in vivo, we generated stable transgenic Arabidopsis plants constitutively expressing the binding and nonbinding sensor versions. Because the sensor strongly binds to K63 polyubiquitin chains in vitro, we anticipated that expressing Vx3K0 in plant cells would likely titrate endogenous K63 polyubiquitinated proteins and interfere with cellular functions dependent on such proteins. In order to limit potential drawbacks associated with the presence of the sensor, Vx3K0 was driven by the promoter of the UBIQUITIN10 (UBI10) gene, which allows a mild expression and greatly limits the silencing and inconsistent expression often observed with the viral 35S promoter (Geldner et al., 2009). Single-insertion homozygous transgenic Ubi10::Vx3K0-HA-GFP and Ubi10::Vx3NB-HA-GFP plants were isolated. None of the selected transgenic lines showed any visible macroscopic phenotypes compared with wild-type plants, indicating that mild constitutive expression does not extensively outcompete endogenous K63 polyubiquitin-binding proteins. For future experiments, care was taken to isolate transgenic lines expressing comparable levels of the Vx3K0-HA-GFP and Vx3NB-HA-GFP sensor versions (Supplemental Fig. S1). The Vx3K0-HA-GFP fusion protein migrated around its expected molecular mass (approximately 39 kD), while its nonbinding counterpart, harboring five amino acid substitutions in each of the three UIMs, showed a slightly slower mobility.
Localization of K63 Polyubiquitinated Proteins
To investigate the cellular processes that involve K63 polyubiquitinated proteins, we first investigated the subcellular localization of Vx3K0-HA-GFP and its nonbinding counterpart in transgenic Arabidopsis plants. Using confocal microscopy, we observed that Vx3NB-HA-GFP accumulated in the cytosol and the nucleus in a pattern reminiscent of free GFP (Fig. 1B). In contrast, in addition to the diffuse cytosolic and nuclear fluorescence distribution, Vx3K0-HA-GFP was found to be enriched in several subcellular compartments, including the plasma membrane, intracellular vesicles, the tonoplast, and nuclear foci (Fig. 1B; Supplemental Fig. S2). This suggests that K63 polyubiquitinated proteins are likely associated with these subcellular compartments or structures, providing insights into where K63 polyubiquitin-dependent processes are found in plant cells.
Since we, and others, recently reported the role of both monoubiquitination and K63 polyubiquitination in the endocytosis of cell surface proteins (Barberon et al., 2011; Kasai et al., 2011; Leitner et al., 2012; Martins et al., 2015), we explored further the possible link between Vx3K0 and endocytosis. We showed that the vast majority of intracellular vesicles positive for Vx3K0 were also positive for the FM4-64 endocytic tracer, indicating that they are of endocytic nature (Fig. 1C). The presence of Vx3K0 in endosomes was further highlighted by its sensitivity to the PI3K inhibitor wortmannin, which creates homotypic fusion and swelling of late-endosome-recruited proteins such as FYVE1 (Barberon et al., 2014; Fig. 1D). Altogether, our observations indicate that K63 polyubiquitinated proteins are widely distributed in the cell and confirm the role of K63 polyubiquitination along the endocytic route.
Sensor-Based Purification of K63 Polyubiquitinated Proteins from Arabidopsis
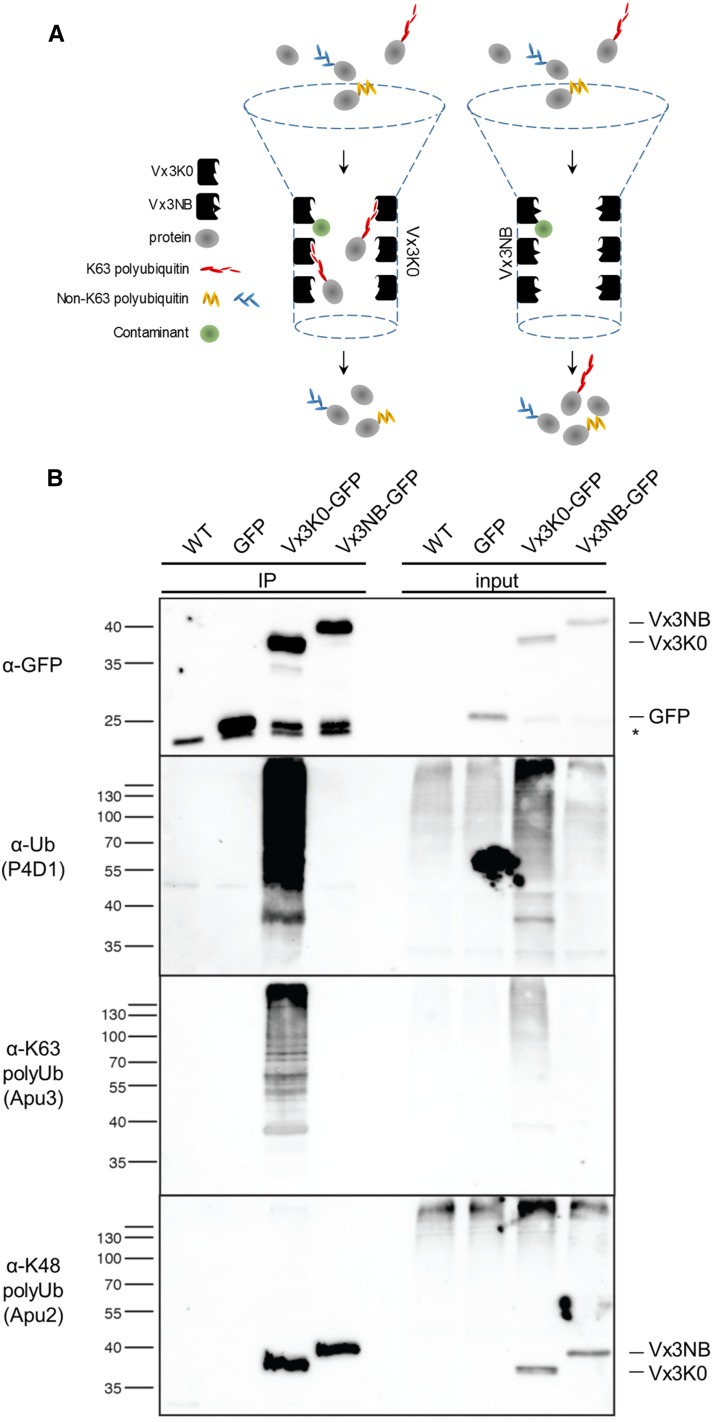
To evaluate the ability of sensor-based approaches to isolate proteins carrying K63 polyubiquitin chains in vivo, crude extracts from wild-type, GFP, Vx3K0-HA-GFP, and Vx3NB-HA-GFP seedlings were prepared with Radioimmunoprecipitation assay (RIPA) buffer amended with protease inhibitors and N-ethylmaleimide to restrict deubiquitinase (DUB) activity. After clarification, the extracts were subjected to immunoprecipitation with GFP antibodycoupled micromagnetic beads to capture K63 polyubiquitinated proteins followed by extensive washes with RIPA buffer and elution (Fig. 2A). The use of the stringent RIPA buffer, which contains 1% Igepal, 0.5% deoxycholate, and 0.1% of the anionic detergent SDS, greatly limits protein-protein interaction and thereby the copurification of interacting proteins. Immunoprecipitates were then probed with different antibodies to characterize pulled-down proteins. To precisely compare the size of the signals obtained, the membranes were stripped between each immunodetection and reprobed in the following order: anti-GFP, anti-K63 polyubiquitin (Apu3), anti-K48 polyubiquitin (Apu2), and the general anti-ubiquitin (P4D1) antibodies. The binding and nonbinding sensors, as well as free GFP, were efficiently immunoprecipitated using our purification procedure, as attested by the immunodetection with anti-GFP antibodies (Fig. 2B). Both sensors showed partial cleavage and release of free GFP during sample preparation. The general P4D1 anti-ubiquitin antibodies that recognize monoubiquitin and various polyubiquitin chain linkage types yielded no signal for the wild type, free GFP, and nonbinding Vx3NB negative controls (Fig. 2B). In contrast, an intense smear ranging across the lane was observed for Vx3K0-expressing plants (Fig. 2B). This indicates not only that ubiquitinated proteins are purified by the K63 polyubiquitin sensor but also that the negative controls including Vx3NB are unable to interact with proteins modified with ubiquitin. The binding of ubiquitinated proteins by Vx3K0 likely protects corresponding proteins from protein degradation or ubiquitin chain cleavage by DUBs, as observed by the slight overaccumulation of ubiquitinated proteins in inputs.
Figure 2.
Biochemical characterization of the Vx3K0 and Vx3NB sensors. A, Scheme representing the principle of K63 polyubiquitinated protein purification using Vx3K0 (left) and the nonbinding Vx3NB as a control (right). B, In vivo characterization of purified proteins using sensor-based immunoprecipitation of proteins carrying K63 polyubiquitin chains. Immunoprecipitation (IP) was performed using anti-GFP antibodies on RIPA buffer-solubilized protein extracts from wild-type (WT) and monoinsertional homozygous plants expressing free GFP, Vx3K0-HA-GFP, and Vx3NB-HA-GFP and subjected to immunoblotting with anti-GFP, anti-ubiquitin P4D1, anti-K63 polyubiquitin Apu3, and anti-K48 polyubiquitin Apu2 antibodies. The membrane was probed with the respective antibodies and stripped in the following order: anti-GFP, anti-K63 polyubiquitin (Apu3), anti-K48 polyubiquitin (Apu2), and anti-ubiquitin (P4D1) antibodies. The sizes of marker proteins in kD are indicated next to the respective blots. The asterisk indicates a nonspecific band recognized by the anti-GFP antibodies. Note that both Vx3K0 and Vx3NB sensors show partial cleavage, as attested by the presence of a signal migrating at the size of free GFP.
To gain further insights into the specificity of the Vx3K0 sensor in vivo, we took advantage of commercially available antibodies that show preferential recognition of different polyubiquitin linkage types. Immunoprecipitates from Vx3K0 showed a strong smear with Apu3 antibodies that favor K63 polyubiquitin chain detection (Fig. 2B), while negative controls failed to show any signals. However, probing the same samples with Apu2 antibodies that are specific for K48 polyubiquitin chains yielded no smear for Vx3K0 and negative controls (Fig. 2B) and only cross-reacted with the sensors themselves. The recognition of both sensors by Apu2 antibodies does not result from signals carried over from anti-GFP immunodetection after stripping, since they also were detected using membranes solely probed with Apu2 (Supplemental Fig. S3). However, these signals observed as a single band are unlikely to represent K48 polyubiquitin chains, since they are not recognized by P4D1 antibodies and may represent nonspecific binding of the antibody to both sensors. Taken together, these observations indicate the Vx3K0 specifically recognizes proteins carrying K63 polyubiquitin chains in vivo. Importantly, the lack of signals observed with antibodies targeting the highly abundant anti-K48 polyubiquitin chains indicates that no K48 polyubiquitinated proteins copurified with K63 polyubiquitin-modified proteins, further demonstrating the stringency of our purification procedure. Similar validation of our purification method was performed with the same four antibodies but using independent membranes for each immunodetection to avoid signals being carried over after stripping and gave the same results (Supplemental Fig. S3). This confirms that the Vx3K0 sensor allows the purification of K63 polyubiquitinated proteins in vivo.
Sensor-Based Proteomic Identification of K63 Polyubiquitinated Proteins
Purified K63 polyubiquitin conjugates from Vx3K0-expressing plants and eluates from the wild-type, free GFP, and nonbinding Vx3NB negative controls were analyzed by tandem mass spectrometry (MS/MS) using the Velos LTQ Orbitrap mass spectrometer, which enables high accuracy and fast sampling times. A likely ubiquitinated conjugate was considered in the K63 polyubiquitin proteome catalog if (1) two or more different matching peptides with protein and peptide thresholds of 95% or more, or (2) a single matching peptide of equivalent stringency with a canonical ubiquitin signature, were identified in both duplicates of Vx3K0 data sets and absent in all three negative controls. The ubiquitin footprints are identified as a missed Lys trypsin cleavage together with an increased mass of 114 D for the modified Lys residue corresponding to the addition of the di-Gly remnant (Peng et al., 2003).
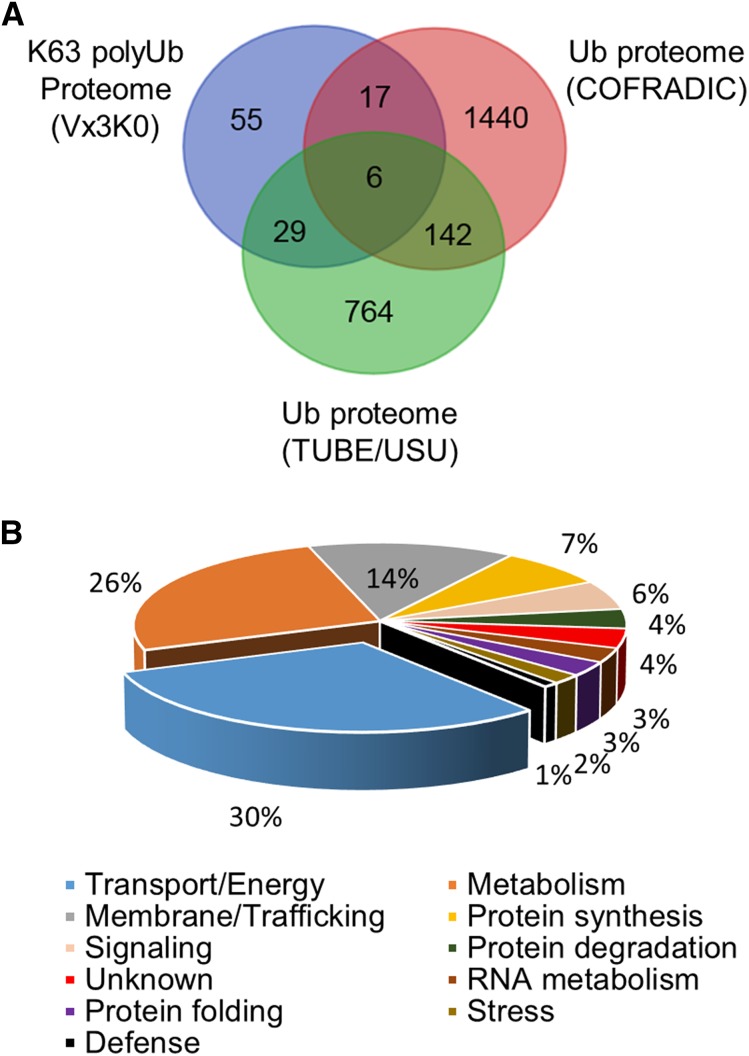
Using these criteria and after eliminating contaminants, we identified 107 Arabidopsis proteins as K63 polyubiquitinated in three replicate experiments (Supplemental Table S1). To evaluate the relevance of the identified proteins, we compared our K63 ubiquitinome with previously published Arabidopsis ubiquitinomes. The recently established TUBE/USU and COFRADIC ubiquitinomes serve as references (Kim et al., 2013; Walton et al., 2016), but these studies monitored ubiquitination with no resolution on linkage types and, therefore, provide no hint of the function of a particular ubiquitination event. Over 32% (35 out of 107 proteins) and 22% (23 out of 107 proteins) of the proteins identified as K63 polyubiquitinated in our analyses also were identified in previous TUBE/USU and COFRADIC proteomes, respectively (Fig. 3A). This not only validates our ability to detect ubiquitinated proteins using Vx3K0-based proteomics but also highlights the existence of proteins carrying different polyubiquitin linkages in existing ubiquitinomes.
Figure 3.
In silico analyses of K63 polyubiquitinated proteins. A, Venn diagram showing the overlap between proteins carrying K63 polyubiquitin chains purified by sensor-based proteomics and the non-linkage-resolutive TUBE/USU and COFRADIC ubiquitinomes (Kim et al., 2013; Walton et al., 2016). B, Functional categorization of K63 polyubiquitinated proteins using Gene Ontogeny (GO) annotations from The Arabidopsis Information Resource (TAIR) and agriGO databases. The percentages of proteins in each bin are shown.
As expected, the dominant peptides identified correspond to ubiquitin itself (Supplemental Table S1). Interestingly, most of the ubiquitin footprints on ubiquitin peptides were found on residue K63, highlighting the specificity of the Vx3K0 sensor in detecting K63 polyubiquitinated proteins. However, some ubiquitin footprints corresponding to K11, and a few targeting K48, also were identified. These non-K63 ubiquitin signatures may result from the ability of Vx3K0 to bind somewhat weakly to other types of polyubiquitin linkages, although in vitro and in vivo evidence indicates that Vx3K0 is highly specific of K63 polyubiquitin chains (Sims et al., 2012; Fig. 2B). Alternatively, the existence of proteins carrying several types of polyubiquitin chains (mixed chains) or branched polyubiquitin chains where a single ubiquitin molecule embedded within a chain is modified at two or more sites likely contributes to K11 and K48 footprints (Nakasone et al., 2013; Meyer and Rape, 2014). The low abundance of such K48 footprints, however, did not allow the detection of proteins carrying mixed or branched chains with K48-linked ubiquitin residues using Apu2 antibodies (Fig. 2B).
Functional Categorization Analyses of K63 Polyubiquitinated Proteins
To obtain a global picture of the molecular functions associated with K63 polyubiquitinated proteins, we binned the 107 targets based on their known or predicted functions using the GO classifications from TAIR and agriGO. To avoid duplicates and obtain more informative ranking, we manually refined the GO annotation listing to assign only the best predicted molecular function to each protein. Interestingly, membrane proteins or subunits of transport complexes involved in the transport of ions and small molecules are highly enriched (false discovery rate [FDR] < 1e-09), representing more than 30% of the K63 ubiquitinome (Fig. 3B; Table I). The fact that transporters are usually of low abundance in the cell and are overrepresented in the crude extract-based isolation of the K63 ubiquitinome suggests that K63 polyubiquitination does play an important role in their function. This is consistent with the recently characterized role of K63 polyubiquitination in plant plasma membrane protein endocytosis (Kasai et al., 2011; Leitner et al., 2012; Martins et al., 2015) and with the preferential localization of Vx3K0 to membranes in vivo (Fig. 1B). Among the few transport proteins known to be ubiquitinated, only the phosphate transporter PHT1;1 and the aquaporin PIP2;1 are found in our K63 ubiquitinome (Table I), further refining the type of ubiquitin linkages targeting these proteins (Lee et al., 2009; Lin et al., 2013). As shown by the K63 ubiquitinome, K63 polyubiquitination likely acts as a major sorting signal for most transporters, thereby controlling ion, water, and small molecule transport in plant cells.
Table I. Examples of molecular functions associated with K63 polyubiquitination targets.
| Locus | Name | Description |
|---|---|---|
| Transport/energy | ||
| AT1G01580 | FRO2 | Ferric reductase |
| AT1G01620 | PIP1;3 | Aquaporin |
| AT1G02520 | PGP11 | P-glycoprotein11 |
| AT1G12110 | NRT1.1 | Nitrate transporter |
| AT1G14870 | PCR2 | Zinc transporter |
| AT1G15690 | AVP1 | Inorganic pyrophosphatase |
| AT1G19910 | AVA-P2 | ATPase, F0/V0 complex, subunit C protein |
| AT1G20260 | ATVAB3 | ATPase, V1 complex, subunit B |
| AT1G30400 | MRP1 | ATP-binding cassette (ABC) transporter |
| AT1G59870 | PEN3 | ABC transporter |
| AT1G78000 | SULTR1;2 | Sulfate transporter |
| AT2G18960 | AHA1 | H+-ATPase |
| AT2G34660 | MRP2 | ABC transporter |
| AT2G36380 | PDR6 | ABC transporter |
| AT2G37280 | PDR5 | ABC transporter |
| AT2G45960 | PIP1;2 | Aquaporin |
| AT2G47000 | MDR4 | ABC transporter |
| AT3G30390 | Transmembrane amino acid transporter | |
| AT3G42050 | VHA-H | Vacuolar ATP synthase subunit H |
| AT3G48740 | SWEET11 | Sucrose efflux transporter |
| AT3G53420 | PIP2;1 | Aquaporin |
| AT3G53480 | PIS1 | ABC transporter |
| AT4G11150 | VHA-E1 | Vacuolar ATP synthase subunit E1 |
| AT4G23700 | CHX17 | Na+/H+ antiporter |
| AT4G23710 | VHA-G2 | Vacuolar ATP synthase subunit G2 |
| AT4G29900 | ACA10 | Ca2+-ATPase |
| AT4G30190 | AHA2 | H+-ATPase |
| AT4G39080 | VHA-A3 | Vacuolar ATPase synthase subunit A3 |
| AT5G09650 | PPa6 | Pyrophosphorylase |
| AT5G43350 | PHT1;1 | Phosphate transporter |
| AT5G60660 | PIP2;4 | Aquaporin |
| ATCG00470 | ATPE | ATP synthase ε-chain |
| Metabolism | ||
| AT1G23190 | PGM3 | Phosphoglucomutase/phosphomannomutase |
| AT1G44170 | Aldehyde dehydrogenase3H1 | |
| AT1G54010 | GDSL-like lipase/acylhydrolase | |
| AT1G70410 | AtBC4 | β-Carbonic anhydrase4 |
| AT1G79550 | PGK | Phosphoglycerate kinase |
| AT2G13360 | AGT1 | Ala:glyoxylate aminotransferase |
| AT2G21170 | TIM | Triosephosphate isomerase |
| AT2G26080 | GLDP2 | Gly decarboxylase P-protein |
| AT2G38230 | PDX1.1 | Gln amidotransferase |
| AT2G42520 | P-loop-containing nucleoside triphosphate | |
| AT3G19820 | DWF1 | Δ24-Sterol reductase |
| AT3G48730 | GSA2 | Glu-1-semialdehyde-2,1-aminomutase |
| AT3G54050 | HCEF1 | Fru-1,6-bisP phosphatase |
| AT3G55440 | TPI | Triosephosphate isomerase |
| AT4G26530 | ATFBA5 | Fru bisphosphate aldolase |
| AT5G01410 | AtPDX1.3 | Gln amidotransferase |
| AT5G61410 | RPE | d-Ribulose-5-phosphate-3-epimerase |
| Trafficking | ||
| AT1G20010 | TUB5 | Tubulin β-chain |
| AT1G22530 | PATL2 | Lipid-binding protein |
| AT1G59610 | DRP2B | Dynamin-like protein |
| AT1G69840 | SPFH/Band7/PHB domain-containing protein | |
| AT1G72150 | PATL1 | Lipid-binding protein |
| AT2G21390 | Coatomer α-subunit | |
| AT2G45820 | Remorin family protein | |
| AT3G01290 | HIR2 | SPFH/Band7/PHB domain-containing protein |
| AT3G08530 | CHC2 | Clathrin H chain |
| AT3G52400 | SYP122 | Syntaxin |
| AT4G31480 | Coatomer, β-subunit | |
| AT4G34660 | SH3 domain-containing protein | |
| AT5G14420 | RGLG2 | RING domain E3 ubiquitin ligase |
| AT5G25250 | FLOT1 | SPFH/Band7/PHB domain-containing protein |
| AT5G42080 | DRP1A | Dynamin-like protein |
| AT5G51570 | SPFH/Band7/PHB domain-containing protein | |
| AT5G62740 | HIR1 | SPFH/Band7/PHB domain-containing protein |
| Protein translation | ||
| AT2G19730 | Ribosomal L28e protein | |
| AT2G34480 | Ribosomal L18ae protein | |
| AT2G41840 | Ribosomal S5 protein | |
| AT2G43030 | Ribosomal L3 protein | |
| AT3G04920 | Ribosomal S24e protein | |
| AT4G11420 | EIF3A | Eukaryotic translation initiation factor 3A |
| AT5G27640 | EIF3B | Eukaryotic translation initiation factor 3B1 |
| AT5G30510 | RPS1 | Ribosomal S1 protein |
Trafficking also emerged as a molecular function overrepresented in our data set (FDR < 2e-09; Table I). The machinery driving endocytosis and vesicular trafficking, such as clathrin, dynamins, syntaxins, coatomer, and microdomain-related proteins, fall into this bin. Such proteins play a role in the dynamics of membrane proteins and transporters, further strengthening the importance of K63 polyubiquitin chains in the regulation of cell surface protein trafficking. By immunoprecipitating SYP122-YFP from transgenic plants and subsequent western blotting using the Apu3 antibodies, we independently confirmed that the syntaxin SYP122 is indeed K63 polyubiquitinated in vivo (Supplemental Fig. S4).
A large number of metabolic enzymes also were found to be enriched among K63 polyubiquitinated proteins (FDR < 5e-05; Table I). These include enzymes localized in the cytosol as well as in various organelles and mostly involved in carbohydrate, amino acid, and vitamin metabolism. Several enzymes were identified in previous global ubiquitinomes, but their K63 ubiquitination was unreported and suggests a role of K63 polyubiquitin chains in their regulation.
The copurification of several ribosomal subunits and elongation factors with Vx3K0 indicates that ribosomes and the protein translation machinery undergo K63 polyubiquitination in plants (Table I). Ribosomal proteins were known to be ubiquitinated from previous ubiquitinomes, but the type of ubiquitination was uncharacterized. The functional relevance of K63 polyubiquitin chain linkage in plant ribosomal subunits is still elusive, but it likely mirrors the role of K63 polyubiquitination in stabilizing the ribosome complex and polysomes recently uncovered in yeast (Silva et al., 2015).
Comparative Analysis of K63 Polyubiquitination Networks in Arabidopsis
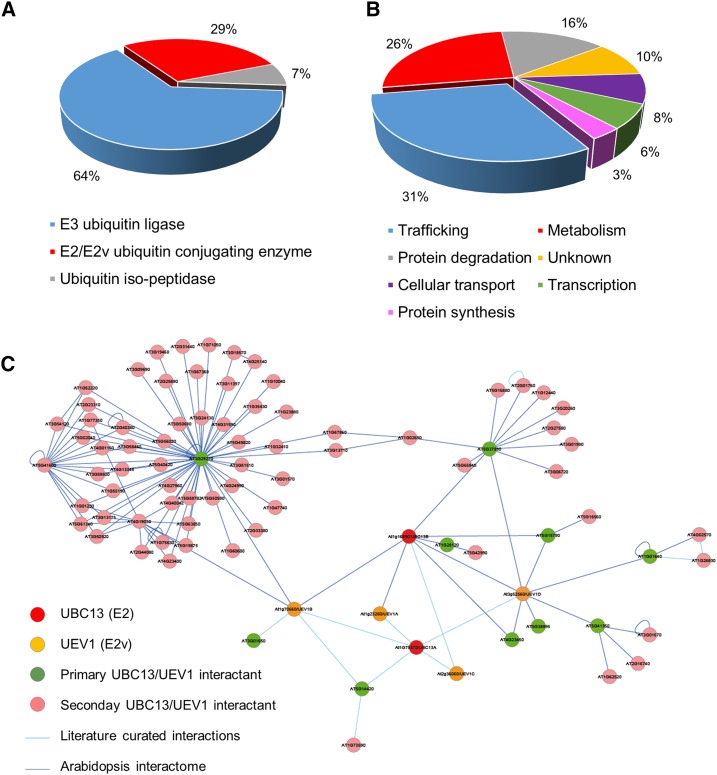
The polyubiquitin chain linkage type is dictated, in most cases, by the E2 ubiquitin-conjugating enzymes involved, while the E3 ubiquitin ligases specify the substrate to be ubiquitinated. E2s from the UBC13 family, with the cooperation of UEV1 E2 variants, catalyze the formation of K63 polyubiquitin chains (Arabidopsis Interactome Mapping Consortium, 2011; Tomanov et al., 2014). The Arabidopsis genome contains two UBC13 genes, UBC13A and UBC13B, and four UEV1 genes, UEV1A to UEV1D. Large-scale interactome analyses by the yeast two-hybrid method using over 6,000 sequenced Arabidopsis open reading frames, including UBC13B and UEV1A, UEV1B, and UEV1D, identified thousands of binary interactions (Arabidopsis Interactome Mapping Consortium, 2011). Protein-protein interactions detected between these four E2/E2v pairs and other Arabidopsis proteins, together with literature curated interactions, identified several primary interactants of these E2/E2v pairs (Supplemental Table S2). Interestingly, all primary interactions were directly related to ubiquitin-dependent processes. Besides interactions between the different UBC13 and UEV1 isoforms, most interactions involved E3 ubiquitin ligases (Fig. 4A; Supplemental Table S2). Based on their ability to interact with UBC13/UEV1, such E3s likely participate in the selection of target proteins to be K63 polyubiquitinated. Among isolated E3s, the RING/U-box proteins are overrepresented, considering that the Arabidopsis genome encodes 64 U-box E3 ligases (Yee and Goring, 2009). More generally, the fact that all UBC13/UEV1-interacting proteins function with E3 ligases or with proteins connected to ubiquitin-related processes demonstrates the robustness of this large-scale interactome and its power in identifying relevant protein-protein interactions.
Figure 4.
Comparative analysis of the K63 ubiquitinome and the Arabidopsis yeast two hybrid-based K63 polyubiquitination networks. A, Functional categorization of proteins interacting with the UBC13/UEV1 E2/E2 variants involved in K63 polyubiquitin chain formation using GO annotations from TAIR and agriGO databases. The percentages of proteins in each bin are shown. B, Functional categorization of secondary partners of the UBC13/UEV1-interacting proteins shown in A. C, Networks of K63 polyubiquitination based on UBC13/UEV1 primary and secondary interacting proteins.
Therefore, the search for proteins pairing with UBC13/UEV1-interacting E3s may reveal additional Arabidopsis proteins undergoing K63 polyubiquitination. Analysis of the Arabidopsis interactome identified 67 targets of UBC13/UEV1-interacting proteins (Supplemental Table S3). Interestingly, with the exception of the U-box family E3 ubiquitin ligase encoded by At3g29270, only a very limited number of targets were identified for each E3. This is consistent with the fact that E3 ligases provide specificity in the ubiquitination reaction. The many interactions identified for At3g29270 likely reflect the stickiness of such proteins in yeast and/or its ability to target several members of the same protein family for ubiquitination, such as five olesins and five Rab GTPase prenylation factors. Interactions were identified between UBC13/UEV1-interacting E3s and membrane traffic-related proteins, metabolic enzymes, and proteins involved in degradative processes dependent on ubiquitin, among others (Fig. 4B; Supplemental Table S3). Altogether, the UBC13/UEV1 primary and secondary interactants define the K63 polyubiquitination yeast two-hybrid interactome (Fig. 4C).
Very little overlap was observed between potential K63 polyubiquitinated proteins coming from the yeast two-hybrid network and our sensor-based K63 ubiquitinome (Supplemental Fig. S5). The only common protein between the two data sets is the RGLG2 E3 ubiquitin ligase. RGLG2 is already known to mediate K63 polyubiquitin chain formation (Yin et al., 2007), and also appears to carry K63 polyubiquitin chains that are likely the result of self-ubiquitination. However, a number of functionally related proteins are found in both lists, including ribosomal subunits, cellulose synthases, and endocytosis- and transport-related proteins. These observations further support the prominent role of K63 polyubiquitin chain formation in the regulation of membrane protein dynamics and protein translation and unravel the identities of some E3 ubiquitin ligases likely involved.
DISCUSSION
Ubiquitin plays a central role in the regulation of a wide variety of biological processes in all eukaryotes. The cataloging of ubiquitinated proteins has expanded rapidly over the past few years. However, our understanding of the biological relevance of these myriad of proteins modified by ubiquitination remains scarce. This is due mostly to the existence of many possible polyubiquitination linkages involving the N-terminal Met and the seven Lys residues from ubiquitin (Komander and Rape, 2012). These different linkages are recognized by different ubiquitin readers in the cell and trigger diverse cellular outputs. Using a K63 polyubiquitin-specific sensor, we established a unique K63 ubiquitinome resource in plants that provides new insights into the roles of K63 polyubiquitin.
The use of the Vx3K0 sensor allowed us to focus on the pool of proteins specifically modified with K63 polyubiquitin chains, greatly refining our resolution compared with previous ubiquitinomes. Compared with strategies using antibodies, genetic sensor-based approaches offer the great advantage of showing high avidity and high specificity for K63 polyubiquitinated proteins (Sims et al., 2012). This allows more stringent purification methods, thus reducing the number of false positives resulting from copurified proteins. We notably employed a stringent purification protocol, using RIPA buffer containing SDS among other detergents, that strips off many secondary interactions. Moreover, expressing Vx3K0 in plant cells stabilizes proteins carrying K63 polyubiquitin chains (Fig. 2C), likely by protecting them from DUB activity, allowing the in vivo detection of rare events for this posttranslational modification. The dissociation constant of Vx3K0 for K63 polyubiquitin trimers and tetramers is comparable, within the nanomolar range. Nevertheless, we cannot rule out that the sensor binds K63 diubiquitin chains, already reported for plant proteins such as the BOR1 boron transporter (Kasai et al., 2011), with a much lower avidity. Therefore, our K63 ubiquitinome may be biased for a certain K63 polyubiquitin chain length.
A first limitation to our approach is the use of nonfully denaturing conditions, which are necessary to completely avoid the copurification of partners interacting with K63 polyubiquitin-modified proteins. Therefore, and despite the many negative controls used in this study, we cannot formally exclude the presence of non-K63 polyubiquitinated proteins within our K63 ubiquitinome. Second, our sensor-based proteomic approach identified over 100 proteins specifically copurifying with Vx3K0, but with very few peptides modified directly with ubiquitin footprints. Although K63 polyubiquitin-modified proteins are greatly enriched in our pipeline, the very large majority of peptides resulting from their tryptic digestion do not carry ubiquitin footprints and render the detection of modified peptides difficult. In addition, nonfully denaturing conditions were previously suggested to limit the detection of ubiquitinated peptides due to residual DUB activity (Matsumoto et al., 2005; Jeon et al., 2007; Danielsen et al., 2011), and may also hamper ubiquitin remnant detection in our approach. However, K63 polyubiquitin chains are detected readily by immunodetection with Apu3 antibodies (Fig. 2B), suggesting that ubiquitin moieties are still attached to target proteins in our specific case.
There are several strategies to improve the cataloging of K63 polyubiquitinated proteins and the detection of ubiquitin remnants. The use of denaturing conditions, which relies on the use of plants overexpressing a His-tagged version of ubiquitin outcompeting endogenous ubiquitin, allows for a second step of affinity purification using Ni-NTA chromatography (Saracco et al., 2009; Kim et al., 2013). A TUBE/Ni-NTA tandem affinity purification demonstrated the power of such an approach to identify ubiquitinated proteins, although without much linkage selectivity (Kim et al., 2013). Combining Vx3K0 purification to Ni-NTA chromatography is, in principle, the most accurate way to isolate a high-quality K63 ubiquitinome. However, the overexpression of a modified form of ubiquitin may lead to several artifacts, including (1) the alteration of the endogenous ubiquitin pool that impacts ubiquitinated proteins, (2) a differential incorporation into specific linkages of polyubiquitin chains, and (3) a poor recognition by K63 polyubiquitin-specific UIMs of Vx3K0. Alternative approaches to enrich for ubiquitinated peptides, using K-ε-GG antibodies recognizing ubiquitin remnants after trypsin digestion, have been used successfully in mammals (Kim et al., 2011; Udeshi et al., 2013) and may be the method of choice to improve the detection of K63 polyubiquitination sites. Recently, a COFRADIC-based ubiquitinome using protein precipitation with a methanol/chloroform mix right after extraction to kill DUB activities also allowed the identification of thousands of ubiquitination sites in Arabidopsis (Walton et al., 2016). Therefore, a lot of effort will be required to optimize linkage-specific ubiquitinomes in the future by adapting these different above-mentioned methods to sensor-based purification. Regardless, the presence in our list of K63 polyubiquitin-modified proteins of about 30% of proteins identified as ubiquitinated in previous studies using denaturing conditions supports the reliability of our approach and provides an unprecedented resolution of plant ubiquitinomes (Fig. 3A).
The vast majority of proteins identified in our plant K63 ubiquitinome are plasma membrane transporters and receptors. This is consistent with the recently characterized role of ubiquitin-mediated endocytosis in regulating plant plasma membrane proteins (Barberon et al., 2011; Kasai et al., 2011; Leitner et al., 2012; Martins et al., 2015). Interestingly, several factors driving protein trafficking also appear to be K63 polyubiquitinated. Of importance are members of the clathrin, syntaxin, coatomer, and dynamin families. In the case of ubiquitin-mediated endocytosis, the concomitant ubiquitination of the cargo and of proteins from the endocytic machinery has already been observed. Several proteins involved in EGFR endocytosis, including CIN85, eps15, Hrs, and epsins, are ubiquitinated after EGF stimulation (Haglund et al., 2002; Polo et al., 2002). This points to the broad role of ubiquitin in the internalization and sorting of cargo proteins. K63 polyubiquitination has been shown to play a role in DNA damage responses (Ciccia and Elledge, 2010). We recovered only a limited amount of proteins found exclusively in the nucleus, likely due to the protein extraction method. Enriching the nuclear fraction from plants subjected to DNA-damaging agents would provide a much better resolution of K63 polyubiquitinated proteins and highlight the DNA repair mechanisms at stake in plants. The ubiquitination of ribosomes was already documented in many organisms, including plants (Kim et al., 2013), but its functional relevance is rather unclear. The identification of several ribosomal subunits as K63 polyubiquitinated or potentially interacting with the K63 polyubiquitin chain formation machinery now refines our vision. K63 polyubiquitination is considered as a proteasome-independent form of ubiquitination, suggesting that it may regulate ribosomal protein activity in a more complex way. In yeast, the attachment of K63 polyubiquitin chains onto ribosomal subunits was shown to stabilize the ribosome complex and polysomes, thereby promoting protein synthesis (Silva et al., 2015). Therefore, an analogous role in plant protein translation is anticipated. Finally, several enzymes involved in carbohydrate, amino acid, and vitamin metabolism were modified by K63 polyubiquitin chains. The attachment of ubiquitin moieties to metabolic enzymes is not novel and was reported already in nonresolutive ubiquitinomes. The finding that these carry K63 polyubiquitin chains raises the question of how K63 polyubiquitination regulates their activity. Surprising is the fact that these proteins show very diverse subcellular localization ranging from the cytosol to different organelles. The connection between ubiquitin and organelles has long been elusive, but recent evidence points to a role of ubiquitin in the control of organelle-localized proteins (Livnat-Levanon and Glickman, 2011; Hua and Vierstra, 2016). Whether metabolic enzymes are modified in the cytosol on their way to organelles or within organelles themselves will have to be clarified in the future.
The analysis of K63 polyubiquitination networks using the Arabidopsis interactome yielded a number of factors potentially involved in or modified by K63 polyubiquitination. We identified several E3 ubiquitin ligases interacting with the UBC13/UEV1 K63 polyubiquitin-specific E2 machinery (Supplemental Table S2). We also identified substrates of such UBC13/UEV1-interacting E3s to depict potential K63 polyubiquitination networks in Arabidopsis (Supplemental Table S3). However, E3 ligases can interact with several E2s, suggesting that only a subset of the identified substrates may undergo K63 polyubiquitination. This explains in part the limited overlap observed between the K63 ubiquitinome and the K63 interactome data sets (Supplemental Fig. S5). More generally, mass spectrometry and yeast two-hybrid approaches are known to generate largely nonoverlapping candidate lists (Gavin et al., 2002), due to inherent differences in the monitored interactions. For example, interactions involving membrane proteins are highly enriched for the K63 ubiquitinome but incompatible with yeast two-hybrid interactome approach. Both approaches, therefore, are complementary in establishing K63 polyubiquitination networks.
Altogether, the K63 ubiquitinome, associated with the K63 interactome, provides a new dimension in our understanding of K63 polyubiquitination in plants.
MATERIALS AND METHODS
Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana) and the various transgenic lines used in this study were grown in sterile conditions on vertical plates containing one-half-strength Linsmaier and Skoog medium at 21°C with a 16-h-light/8-h-dark cycle.
Constructs
The Vx3K0-HA-GFP and Vx3NB-HA-GFP cassettes were a kind gift of Robert Cohen. The two sensor versions and GFP alone were amplified by PCR with primers containing attB1 and attB2 Gateway recombination sites (Supplemental Table S4) and cloned in pDONR-P1P2. Final destination vectors derived from pK7m34GW and carrying Vx3K0-HA-GFP, Vx3NB-HA-GFP, or GFP under the control of the UBI10 promoter were obtained using three-fragment recombination Gateway recombination (Life Technologies). The resulting constructs were transformed into wild-type plants. For each construct, several independent monoinsertional homozygous transgenic lines were selected.
Protein Extraction and Western-Blot Analyses
Total proteins were extracted from 12-d-old plants. Proteins were separated on a 10% Bis-Tris NuPage gel (Life Technologies) and transferred onto a nitrocellulose membrane. For protein detection, the following antibodies were used: monoclonal anti-GFP horseradish peroxidase-coupled (Miltenyi Biotec 130-091-833; 1:5,000), anti-ubiquitin P4D1 (Cell Signaling; 1:2,500), anti-K48 polyubiquitin Apu2 (Millipore; 1:2,000), and anti-K63 polyubiquitin Apu3 (Millipore; 1:2,000) antibodies. To compare the size of signals detected using these various antibodies, the same membrane was stripped and reprobed using the following order: anti-GFP, anti-K63 polyubiquitin (Apu3), anti-K48 polyubiquitin (Apu2), and anti-ubiquitin (P4D1) antibodies. To ascertain that no signal was carried over from one immunodetection to another, each antibody also was incubated on independent membranes. Experiments were done in triplicate.
Confocal Microscopy
Plant samples were mounted in water and imaged on a Leica TCS SP2 confocal laser scanning microscope. Images were taken at the root tip from 7-d-old plants grown in the light or in the leaves. For imaging GFP, the 488-nm laser line was used. FM4-64 (Life Technologies) was applied at a concentration of 5 μm. Wortmannin (Sigma-Aldrich) was used at a concentration of 33 μm for 1 h in liquid medium. Detection settings were kept constant in individual sets of experiments to allow for a comparison of the expression and localization of reporter proteins.
Immunoprecipitation
Immunoprecipitation experiments were performed using 12-d-old-seedlings from wild-type, GFP, Vx3K0-HA-GFP, and Vx3NB-HA-GFP plants. Care was taken to use transgenic lines harboring similar amounts of transgene-expressed proteins. Tissues were ground in liquid nitrogen and resuspended in ice-cold RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Igepal, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitors and 20 mm N-ethylmaleimide to inhibit DUBs. Samples were solubilized at 4°C for 30 min before centrifugation at 14,000g for 15 min at 4°C to eliminate cell debris. Supernatants were subjected to immunoprecipitation with the µMACS GFP isolation kit (Miltenyi Biotec). Immunoprecipitates were eluted off beads using Laemmli buffer. Samples were boiled for 2 min at 95°C and migrated for 15 min on a 10% Bis-Tris NuPage gel (Life Technologies) before bands were cut off.
Mass Spectrometry
In-gel trypsin digestion was carried out as described previously (Shevchenko et al., 2006). Tryptic peptide digests were analyzed by nano-liquid chromatography-MS/MS using the EASY-nLC II HPLC system (Proxeon; Thermo Scientific) coupled to the nanoelectrospray ion source of a Velos LTQ Orbitrap mass spectrometer (Thermo Scientific). Peptide separation was performed on a reverse-phase C18 nano-HPLC column (100 μm i.d., 5-μm C18 particles, 15-cm length; NTCC-360/100-5) from Nikkyo Technos. The peptides were loaded at a pressure-dependent flow rate corresponding to a maximum pressure of 200 bar and eluted at a flow rate of 300 nL min−1 using a gradient of 5% to 35% acetonitrile in 0.1% formic acid. Nano-liquid chromatography-MS/MS experiments were conducted in the data-dependent acquisition mode. The mass of the precursors was measured with a high resolution (60,000 full weight at half maximum) in the Orbitrap mass spectrometer. The 20 most intense ions, above an intensity threshold of 2,000 counts, were selected for collision-induced dissociation fragmentation and analysis in the LTQ Orbitrap mass spectrometer.
Peptide spectra were searched against the Arabidopsis ecotype Columbia-0 protein database containing 35,628 entries (TAIR; www.arabidopsis.org) with Proteome Discoverer 1.3 using the Mascot algorithm version 2.4.1 (Matrix Science) and analyzed using Scaffold 4.3.4 (Proteome Software).
Mass Spectrometry Data Analyses
The GO annotations for the 107 target proteins from the K63 ubiquitinome were determined by hand analysis using TAIR and agriGO databases (Du et al., 2010). The predicting subcellular localization of proteins was performed using SUBA3 (Tanz et al., 2013).
K63 Polyubiquitin Network Analyses
Primary and secondary protein-protein interaction data for UBC13/UEV1 were extracted from the Arabidopsis interactome (Arabidopsis Interactome Mapping Consortium, 2011). Networks were built using Cytoscape.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of transgenic lines expressing similar levels of Vx3K0 and Vx3NB proteins.
Supplemental Figure S2. Confocal images showing the enrichment of Vx3K0 in intracellular vesicles, at the plasma membrane, and at the tonoplast.
Supplemental Figure S3. In vivo characterization of purified proteins using sensor-based immunoprecipitation of proteins carrying K63 polyubiquitin chains.
Supplemental Figure S4. Example of a protein identified as K63 polyubiquitinated in the Vx3K0-derived K63 polyubiquitinome.
Supplemental Figure S5. Venn diagram showing the overlap between proteins from the K63 ubiquitinome and the Arabidopsis K63 polyubiquitin yeast two hybrid networks.
Supplemental Table S1. Complete list of Arabidopsis K63 polyubiquitinated conjugates identified by mass spectrometry.
Supplemental Table S2. Complete list of proteins interacting with UBC13/UEV1 in the yeast two hybrid Arabidopsis interactome.
Supplemental Table S3. Complete list of proteins interacting with UBC13/UEV1 primary interactants in the Arabidopsis yeast two hybrid interactome.
Supplemental Table S4. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank David Cornu from the Institute for Integrative Biology of the Cell SICaPS proteomic facility for mass spectrometry analyses, and Robert Cohen (Colorado State University) and Hans Thordal-Christensen (University of Copenhagen) for sharing the K63 sensor constructs and SYP122-YFP seeds, respectively.
Glossary
- Ni-NTA
nickel-nitrilotriacetic acid agarose
- COFRADIC
combined fractional diagonal chromatography
- UIMs
ubiquitin interaction motifs
- MS/MS
tandem mass spectrometry
- GO
Gene Ontogeny
- TAIR
The Arabidopsis Information Resource
- FDR
false discovery rate
References
- Adhikari A, Chen ZJ (2009) Diversity of polyubiquitin chains. Dev Cell 16: 485–486 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Dubeaux G, Kolb C, Isono E, Zelazny E, Vert G (2014) Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc Natl Acad Sci USA 111: 8293–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G (2011) Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, Jensen LJ, Mailand N, Nielsen ML (2011) Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics 10: M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Shimokawa N, Szymkiewicz I, Dikic I (2002) Cbl-directed monoubiquitination of CIN85 is involved in regulation of ligand-induced degradation of EGF receptors. Proc Natl Acad Sci USA 99: 12191–12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD (2016) Ubiquitin goes green. Trends Cell Biol 26: 3–5 [DOI] [PubMed] [Google Scholar]
- Igawa T, Fujiwara M, Takahashi H, Sawasaki T, Endo Y, Seki M, Shinozaki K, Fukao Y, Yanagawa Y (2009) Isolation and identification of ubiquitin-related proteins from Arabidopsis seedlings. J Exp Bot 60: 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon HB, Choi ES, Yoon JH, Hwang JH, Chang JW, Lee EK, Choi HW, Park ZY, Yoo YJ (2007) A proteomics approach to identify the ubiquitinated proteins in mouse heart. Biochem Biophys Res Commun 357: 731–736 [DOI] [PubMed] [Google Scholar]
- Kasai K, Takano J, Miwa K, Toyoda A, Fujiwara T (2011) High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J Biol Chem 286: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD (2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25: 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT (2009) Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J, Petrášek J, Tomanov K, Retzer K, Pařezová M, Korbei B, Bachmair A, Zažímalová E, Luschnig C (2012) Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc Natl Acad Sci USA 109: 8322–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Schmidt W (2010) A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J 62: 330–343 [DOI] [PubMed] [Google Scholar]
- Lin WY, Huang TK, Chiou TJ (2013) Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livnat-Levanon N, Glickman MH (2011) Ubiquitin-proteasome system and mitochondria: reciprocity. Biochim Biophys Acta 1809: 80–87 [DOI] [PubMed] [Google Scholar]
- Manzano C, Abraham Z, López-Torrejón G, Del Pozo JC (2008) Identification of ubiquitinated proteins in Arabidopsis. Plant Mol Biol 68: 145–158 [DOI] [PubMed] [Google Scholar]
- Maor R, Jones A, Nühse TS, Studholme DJ, Peck SC, Shirasu K (2007) Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol Cell Proteomics 6: 601–610 [DOI] [PubMed] [Google Scholar]
- Martins S, Dohmann EM, Cayrel A, Johnson A, Fischer W, Pojer F, Satiat-Jeunemaître B, Jaillais Y, Chory J, Geldner N, et al. (2015) Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat Commun 6: 6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI (2005) Large-scale analysis of the human ubiquitin-related proteome. Proteomics 5: 4145–4151 [DOI] [PubMed] [Google Scholar]
- Meyer HJ, Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157: 910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D (2013) Mixed-linkage ubiquitin chains send mixed messages. Structure 21: 727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan IC, Schmidt W (2014) Functional implications of K63-linked ubiquitination in the iron deficiency response of Arabidopsis roots. Front Plant Sci 4: 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D (2004) Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8: 610–616 [DOI] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416: 451–455 [DOI] [PubMed] [Google Scholar]
- Saracco SA, Hansson M, Scalf M, Walker JM, Smith LM, Vierstra RD (2009) Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J 59: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Silva GM, Finley D, Vogel C (2015) K63 polyubiquitination is a new modulator of the oxidative stress response. Nat Struct Mol Biol 22: 116–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Scavone F, Cooper EM, Kane LA, Youle RJ, Boeke JD, Cohen RE (2012) Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat Methods 9: 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA (2013) SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanov K, Luschnig C, Bachmair A (2014) Ubiquitin Lys 63 chains: second-most abundant, but poorly understood in plants. Front Plant Sci 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani DR, Qiao JW, Carr SA (2013) Refined preparation and use of anti-diglycine remnant (K-ε-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol Cell Proteomics 12: 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk SJ, Fiskin E, Putyrski M, Pampaloni F, Hou J, Wild P, Kensche T, Grecco HE, Bastiaens P, Dikic I (2012) Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol Cell 47: 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton A, Stes E, Cybulski N, Van Bel M, Iñigo S, Durand AN, Timmerman E, Heyman J, Pauwels L, De Veylder L, et al. (2016) It’s time for some “site”-seeing: novel tools to monitor the ubiquitin landscape in Arabidopsis thaliana. Plant Cell 28: 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen R, Torres-Acosta JA, Pastushok L, Lai X, Pelzer L, Wang H, Xiao W (2008) Arabidopsis UEV1D promotes lysine-63-linked polyubiquitination and is involved in DNA damage response. Plant Cell 20: 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen R, Wang S, Xiang D, Venglat P, Shi X, Zang Y, Datla R, Xiao W, Wang H (2014) UBC13, an E2 enzyme for Lys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. Plant J 80: 424–436 [DOI] [PubMed] [Google Scholar]
- Woelk T, Sigismund S, Penengo L, Polo S (2007) The ubiquitination code: a signalling problem. Cell Div 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D, Goring DR (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Yin XJ, Volk S, Ljung K, Mehlmer N, Dolezal K, Ditengou F, Hanano S, Davis SJ, Schmelzer E, Sandberg G, et al. (2007) Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.