Dynamic extensions of peroxisomes, or peroxules, are associated with ROS production under stress and require expression of a key peroxin protein.
Abstract
Peroxisomes are highly dynamic and metabolically active organelles that play an important role in cellular functions, including reactive oxygen species (ROS) metabolism. Peroxisomal dynamics, such as the proliferation, movement, and production of dynamic extensions called peroxules, have been associated with ROS in plant cells. However, the function and regulation of peroxules are largely unknown. Using confocal microscopy, we have shown that treatment of Arabidopsis leaves with the heavy metal cadmium produces time course-dependent changes in peroxisomal dynamics, starting with peroxule formation, followed by peroxisome proliferation, and finally returning to the normal morphology and number. These changes during Cd treatment were regulated by NADPH oxidase (C and F)-related ROS production. Peroxule formation is a general response to stimuli such as arsenic and is regulated by peroxin 11a (PEX11a), as Arabidopsis pex11a RNAi lines are unable to produce peroxules under stress conditions. The pex11a line showed higher levels of lipid peroxidation content and lower expression of genes involved in antioxidative defenses and signaling, suggesting that these extensions are involved in regulating ROS accumulation and ROS-dependent gene expression in response to stress. Our results demonstrate that PEX11a and peroxule formation play a key role in regulating stress perception and fast cell responses to environmental cues.
Peroxisomes are highly versatile organelles that adapt to changes in their cellular environment through morphological and metabolic adjustments (Hu et al., 2012; Sandalio et al., 2013). Plant peroxisomes perform essential functions such as photorespiration and fatty acid β-oxidation, ureide, and phytohormone (auxin and jasmonic acid) metabolisms, and also act as a source of signal molecules such as reactive oxygen and nitrogen species (ROS and RNS; Sandalio and Romero-Puertas, 2015). Peroxisomes contain a large battery of antioxidants to control ROS and RNS accumulation (Sandalio and Romero-Puertas, 2015). These organelles proliferate in response to environmental cues through a complex process involving elongation, constriction, and fission (Hu et al., 2012; Baker and Paudyal, 2014). Deciphering the signaling pathways governing the regulation of peroxisome proliferation under different environmental and metabolic conditions presents a major challenge in this field of research. Before peroxisomal division, organelle elongation occurs in a process regulated by peroxins 11 (PEX11), with the final division requiring dynamin-like or dynamin-related proteins and fission proteins (Hu et al., 2012, Schrader et al., 2012). There is evidence to suggest that ROS are involved in regulating peroxisome proliferation, as some peroxisomal biogenesis genes (PEX) are transcriptionally regulated by H2O2 in both plant and animal cells (López-Huertas et al., 2000). The formation of peroxisomal extensions, known as peroxules, has also been observed to be regulated by exogenous applications of H2O2 (Sinclair et al., 2009; Barton et al., 2013). The term “peroxule” was coined by Scott et al. (2007) to refer to dynamic extensions, which are similar to those observed in chloroplasts (stromules). It has been suggested that peroxules are involved in the elongation of peroxisomes (Sinclair et al., 2009), although little is known about the function and regulation of peroxules. Furthermore, the speed of peroxisome movement can change under stress conditions imposed by Cd and herbicide 2,4-d and is also regulated by ROS (Rodríguez-Serrano et al., 2009, 2014). Specific ROS sensors in plant tissues have not yet been identified. We postulated that if peroxisomal dynamics are regulated by ROS, peroxisomes may act as sensors of ROS/redox changes. Therefore, the aim of this study was to establish the role of ROS induced by Cd in peroxisomal dynamics and specifically to characterize the molecular regulation of peroxule formation and functionality under abiotic stress conditions imposed by heavy metals. We have identified PEX11a as a potential ROS/redox sensor in response to stress and propose a model in which peroxules are controlled by PEX11a in response to ROS produced by RBOHs outside peroxisomes. We therefore suggest that peroxules play a key role in regulating fast cell responses to environmental cues.
RESULTS
Cadmium Treatment Induces Changes in the Dynamics of Peroxisomes
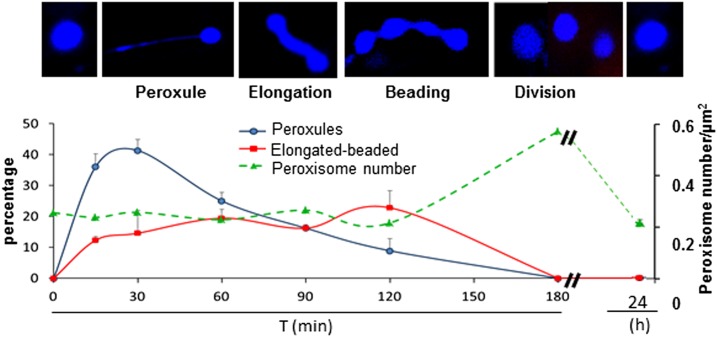
Cadmium (Cd) is a well-known inducer of ROS accumulation and oxidative stress in different plant species (Sandalio et al., 2012; Gallego et al., 2012). To determine the effect of Cd (100 µm) on peroxisomal dynamics, we first analyzed the time course-dependent changes in the organelle using the Arabidopsis px-ck line (Nelson et al., 2007), which contains the peroxisome fluorescent marker protein, CFP-PTS1 (peroxisomal targeting signal type 1, which is composed of SKL). Cd induced peroxule formation after 15 min of treatment (Fig. 1), and the highest frequency of peroxule formation (40% of peroxisomes) occurred after 30 min of treatment. Higly dynamic peroxules were transiently produced and could extend into the cytosol and retract in very short periods of time (Supplemental Video S1). These structures were closely associated with the chloroplast (Supplemental Video S1). Following peroxule production in response to Cd, peroxisomes underwent elongation, constriction, and beading, followed by fragmentation into smaller peroxisomes (Fig. 1). Peroxisomal fragmentation gave rise to proliferation, with the largest number of peroxisomes (3-fold increase) observed after 3 h of treatment with Cd (Fig. 1; Supplemental Fig. S1). After 24 h of exposure to Cd, peroxisomal populations declined to the levels observed in untreated plants (Fig. 1; Supplemental Fig. S1).
Figure 1.
Time course analyses of cadmium (100 μm) effect on peroxisomal dynamics. Effect of Cd on peroxule formation and peroxisomal elongation, constriction, and proliferation.
Peroxule Formation and Peroxisomal Movement Are Regulated by ROS Produced by NADPH Oxidases in Response to Cd
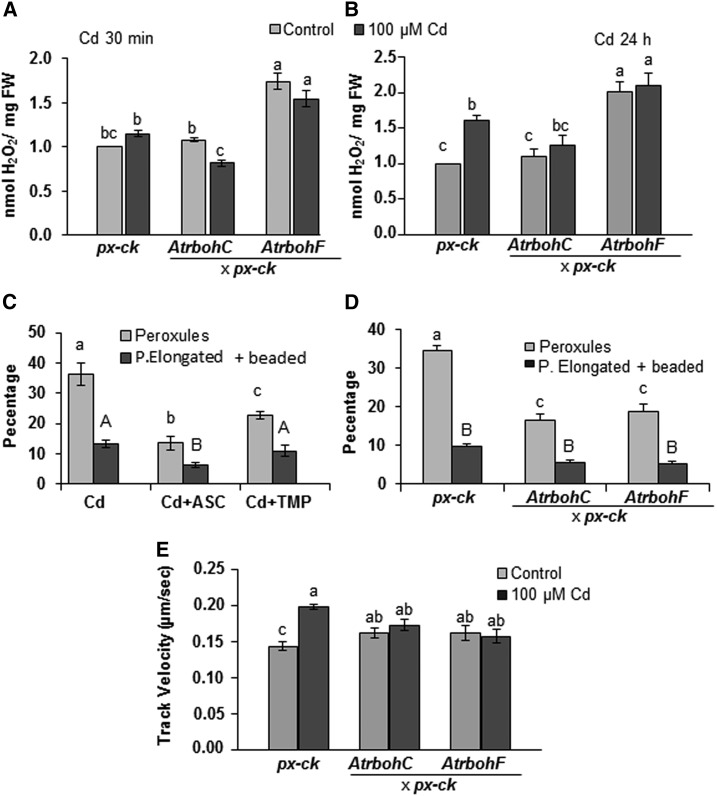
The formation of peroxules has been observed to be induced by exogenous applications of H2O2, a major ROS species (Sinclair et al., 2009). To verify the involvement of ROS in the morphological changes of peroxisomes in response to Cd, we analyzed the production of H2O2 at 30 min and 24 h after Cd treatment in px-ck plants. After 30 min of Cd treatment, a small, although not statistically significant, increase in H2O2 was observed in px-ck plants (Fig. 2a). However, 24 h of treatment with Cd gave rise to an increase in H2O2 (Fig. 2b). To further demonstrate whether ROS are involved in peroxule formation, we treated Arabidopsis plants with ASC (ascorbic acid) and TMP (tetramethylpiperidinooxy; Rodríguez-Serrano et al., 2009). Both of these molecules have previously been shown to reduce H2O2 and O2.− accumulation, respectively, in response to Cd (Rodríguez-Serrano et al., 2009). ASC and TMP caused a 3- and 1.7-fold reduction, respectively, in the number of peroxisomes producing peroxules in response to Cd, while peroxisomal elongation and beading were slightly reduced by ASC (Fig. 2c). These results suggest that peroxule formation is a component of the cell’s fast response to the metal and may be regulated by ROS.
Figure 2.
Hydrogen peroxide production induced by Cd treatment and regulation of peroxisomal dynamics by ROS. a and b, H2O2 production was analyzed using fluorimetry after 30 min (a) and 24 h (b) of Cd treatment. c, Regulation of peroxule formation, peroxisomal elongation, and constriction after 30 min of Cd treatment by the ROS scavengers ASC (H2O2 scavenger) and TMP (O2.− scavenger). d, Role of NADPH oxidases C and F in peroxule formation and peroxisomal elongation and constriction induced by 30-min Cd treatment. e, Effect of Cd on the speed of peroxisomal movement after 24 h of treatment and regulation by NADPH oxidases C and F. Data represent the mean ± se (error bars) of at least three independent experiments, each with five replicates. The asterisk denotes statistically significant differences at P < 0.01 according to Tukey’s multiple range test.
The plasma membrane-associated NADPH oxidase is one of the main sources of ROS produced under Cd toxic conditions (Romero-Puertas et al., 2004; Smeets et al., 2009). To determine whether this enzyme is a major source of ROS involved in this process, we analyzed H2O2 production and morphological changes in peroxisomes in Arabidopsis lines AtrbohCxpx-ck and AtrbohFxpx-ck, both of which express the peroxisomal marker CFP-PTS1 and are deficient in NADPH oxidases (NADPH oxidase C for AtrbohCxpx-ck and F for AtrbohFxpx-ck). The frequency of peroxule formation showed a 2.3- and 1.9-fold decrease, respectively, after 30 min of Cd treatment in AtrbohCxpx-ck and AtrbohFxpx-ck (Fig. 2d). However, the rate of the elongated and beaded peroxisomes did not show any statistically significant changes in Atrboh mutants (Fig. 2d), suggesting that peroxule formation and peroxisomal elongation are not necessarily linked. The reduction of Cd-dependent peroxule formation in Atrboh lines was in line with that of H2O2 production observed in both lines after 30 min of treatment, as compared to px-ck plants (Fig. 2a). This suggests that peroxule formation is regulated by redox signals produced outside the peroxisomes, in this case, by plasma membrane-derived ROS production.
During peroxule production in response to Cd, peroxisome movement ceased (Supplemental Videos S1 and S2), then resumed after peroxisomal proliferation and even showed a 1.4-fold acceleration after 24 h of Cd treatment (Fig. 2e). We thus analyzed the speed of peroxisomes in AtrbohC and AtrbohF after 24 h of Cd treatment. The increase in peroxisomal movement observed in px-ck plants was abolished in Atrboh lines (Fig. 2e), possibly as a result of the reduction of H2O2 production in Atrboh lines as compared to the increase observed in px-ck plants (Fig. 2a). These data demonstrate that ROS produced by RBOHs may also regulate peroxisomal movement after long periods of Cd treatment.
Peroxule Formation Is Regulated by PEX11a
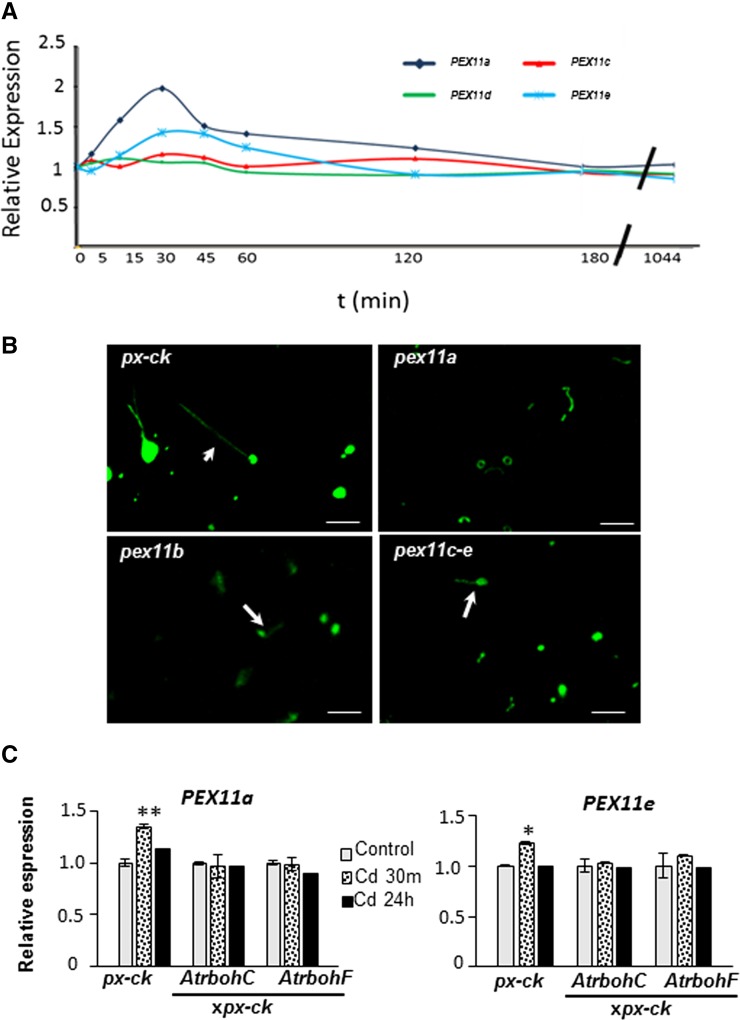
The first stage in peroxisome proliferation is triggered by PEX11 proteins, which are associated with peroxisome membranes and promote peroxisome elongation and division (Lingard and Trelease, 2006; Orth et al., 2007). To characterize the molecular mechanism underlying the regulation of peroxule formation, we determined whether the PEX11 protein family is involved in peroxule formation. Time-course analysis of the steady-state levels of PEX11 gene transcripts in seedlings showed that PEX11 genes were not expressed in untreated plants, while PEX11a and PEX11e expression was up-regulated after 30 min of Cd treatment, with the largest increase (2-fold) being observed for PEX11a (Fig. 3a). PEX11b was not expressed during the treatment period. PEX11 gene expression returned to the level observed in untreated plants after 3 h of exposure to Cd (Fig. 3a).
Figure 3.
Role of PEX11 in peroxule formation. a, Time-course analyses of PEX11a-e expression in response to Cd treatment. b, Confocal microscopy showing peroxule formation in Arabidopsis RNAi lines of PEX11a, PEX11b, and PEX11c-e after 30 min of Cd treatment. c, PEX11a and PEX11e expression in AtrbohCxpx-ck and AtrbohFxpx-ck lines after 30 min and 24 h of Cd treatment. Data represent the mean ± se of at least three independent experiments, each with five replicates. The asterisk denotes statistically significant differences at P < 0.01 according to Tukey’s multiple range test.
Given the increase in PEX11a and PEX11e transcripts in response to Cd, we were interested in determining whether these two peroxins were involved in peroxule formation. We therefore analyzed peroxule formation in the RNAi lines of PEX11a (pex11a), PEX11b (pex11b), and PEX11c-e (pex11c-e; Orth et al., 2007). After 30 min of Cd treatment, peroxules were detected in the pex11b and pex11c-e lines but not in pex11a (Fig. 3b). It is worth noting that pex11a lines treated with Cd showed elongated or donut-shaped peroxisomes, while the number and size of peroxisomes were considerably smaller than in the px-ck, pex11b, and pex11c-e lines (Fig. 3b), as previously observed by Orth et al. (2007). To determine whether peroxules are produced in response to stimuli other than Cd and, if so, whether PEX11a is involved in this process, we analyzed peroxule formation in response to Arsenic (As), another metal that induces H2O2 accumulation (Gupta et al., 2013). After As treatment, peroxules were observed in px-ck plants but not in pex11a lines (Supplemental Fig. S2a). Up-regulation of PEX11a transcripts was also observed in px-ck plants exposed to As for 30 min (Supplemental Fig. S2a).
To demonstrate ROS-dependent induction of PEX11a and PEX11e, we analyzed the steady-state expression of both genes in px-ck as well as AtrbohCxpx-ck and AtrbohFxpx-ck lines after 30 min of Cd treatment. PEX11a and PEX11e expression was induced in px-ck but not in AtrbohCxpx-ck or AtrbohFxpx-ck lines (Fig. 3c), indicating that Cd-induced NADPH oxidase-dependent ROS control PEX11a and PEX11e expression, thus positioning PEX11 downstream from ROS in ROS-mediated pathways.
Peroxules Regulate ROS Accumulation and ROS-Mediated Signaling Networks
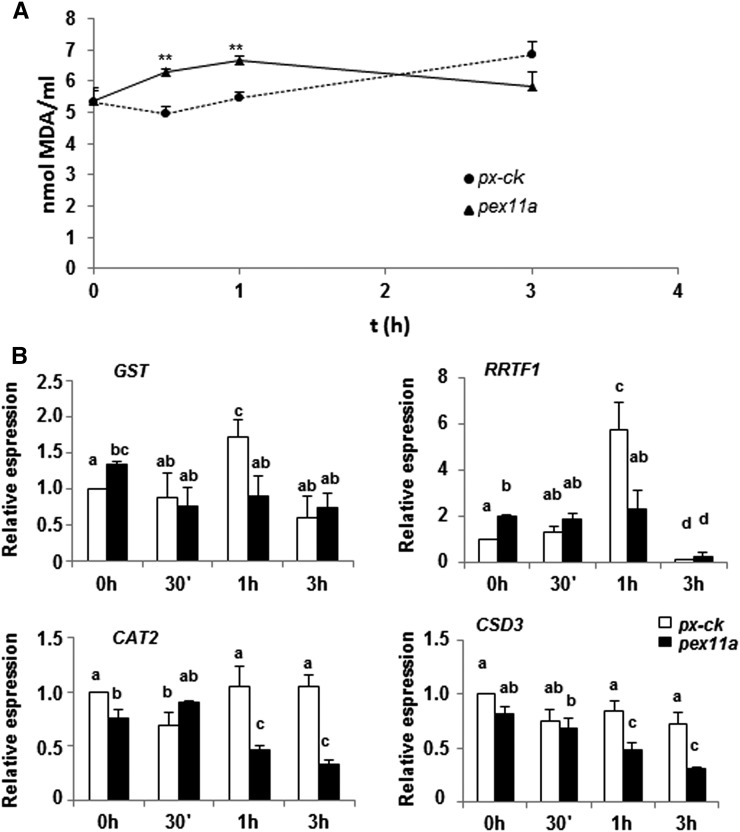
To determine the biological significance of peroxules, we analyzed the level of lipid peroxidation, a widely used oxidative damage marker, in px-ck and pex11a plants in response to Cd. Our results show a higher level of lipid peroxidation in pex11a than in px-ck, indicating a major oxidative stress in the cell immediately after stimulus. ROS-dependent signaling within the plant cell is critical for plant survival under stress and development conditions (Baxter et al., 2014). We then analyzed changes in the expression of Cd-inducible genes involved in cell responses to ROS, namely, glutathione-s-transferase (GST) and redox responsive transcription factor1 (RRTF1), and of the peroxisomal antioxidants catalase 2 (CAT2) and CuZn-superoxide dismutase 3 (CSD3). Compared with px-ck plants, pex11a lines did not show any induction of GST and RRTF1, which are involved in ROS-dependent signaling, after 1 h of Cd treatment (Fig. 4b), suggesting that the fast ROS signaling network is disturbed in pex11a. The absence of PEX11a also promotes a reduction in the expression of antioxidants CAT2 and CSD3, which may favor the accumulation of ROS. These results are consistent with the increase in lipid peroxidation during the same period of treatment and demonstrate that these extensions play a role in antioxidant defenses and ROS-dependent signaling.
Figure 4.
Role of PEX11a and peroxules in ROS metabolism and ROS-derived signaling. a, Lipid peroxidation induced by Cd treatment in px-ck and pex11a lines. b, Expression of GST, RRTF1, CAT2, and CSD3 in px-ck and pex11a in response to Cd treatment. Data represent the mean ± se (error bars) of at least three independent experiments, each with five replicates; error bars represent se. Asterisks denote statistically significant differences at P < 0.01 according to Duncan’s multiple range test. Values indicated by the same letter do not significantly differ (P < 0.05) according to Tukey’s test.
DISCUSSION
Peroxisomes are extremely dynamic organelles. Live imaging of peroxisomes using different fluorescent proteins enabled us to observe environmentally dependent changes in their morphology, speed of movement, and numbers (Sinclair et al., 2009; Rodríguez-Serrano et al., 2009). In this study, with the aid of Arabidopsis lines expressing CFP-SKL in peroxisomes, we identified the pattern of changes induced by Cd toxicity in the dynamics of peroxisomes during periods of exposure to Cd ranging from 5 min to 24 h. Cadmium treatment induced rapid production of peroxules that were visible after 15 min of treatment, reaching a maximum rate after 30 min of treatment. The production of peroxules was a short and transitory response to Cd and then peroxisomes changed from a spherical to a tubular morphology. The elongation of peroxisomes is associated with the addition of new matrix proteins and membrane lipids to peroxisomes as a first step in their further fission and proliferation (Hu et al., 2012). Indeed, beaded peroxisomes can be observed in parallel to the elongated type, with a marked proliferation of peroxisomes being observed after 3 h of Cd treatment. Proliferation of peroxisomes has been associated with stress conditions such as ozone (Oksanen et al., 2003), light (Desai and Hu, 2008), xenobiotics (de Felipe et al., 1988; Palma et al., 1991; Castillo et al., 2008), and salinity (Mitsuya et al., 2010). López-Huertas et al. (2000) have suggested that peroxisomal proliferation is regulated by ROS produced under different stress conditions, probably to cope with oxidative damage induced by various stimuli. Information on the role of peroxules and the mechanisms involved in their formation is currently limited. Peroxules are highly dynamic peroxisomal protrusions that extend and retract very rapidly and are induced in response to Cd and As treatment, as we have observed in this work. In a previous study, Sinclair et al. (2009) observed that peroxules are induced by exogenous applications of H2O2. Cadmium has been demonstrated to induce oxidative stress by accumulating ROS in different cell compartments such as the plasma membrane, mitochondria, and peroxisomes (Olmos et al., 2003; Romero-Puertas et al., 2004; Garnier et al., 2006). By using Arabidopsis lines deficient in RBOHs, it has recently been demonstrated that NADPH oxidases could be a primary source of ROS production in response to Cd treatment (Gupta et al., 2016).
In our study, the following pieces of evidence demonstrate that peroxule production is regulated by ROS induced by cadmium: (1) An increase, although not statistically significant, in H2O2 production after 30 min of Cd treatment was observed. This could be explained by the difficulty in analyzing small changes in H2O2 using fluorimetric techniques or other standard methods, even though the overall tendency is for H2O2 to increase. The increase in H2O2 was in line with the maximum rate of peroxule formation. (2) Preincubation of seedlings with ASC and TMP, acting as antioxidants that remove H2O2 and O2.−, respectively, considerably reduced the number of peroxule-forming peroxisomes. (3) Analysis of Arabidopsis lines defective in RBOHs C and F (Atrboh C and Atrboh F) confirms that the reduction in peroxule formation parallel to the reduction in H2O2 in these lines is in response to Cd. It is worth noting that AtrbohF showed higher basal levels of H2O2 than AtrbohC or px-ck in the absence of treatment. A similar trend has recently been observed in AtrbohF, which, it has been suggested, is caused by differential contributions of each RBOH to ROS production and by differential regulation of antioxidants in each mutant (Gupta et al., 2016). These findings also suggest that peroxule formation is regulated by redox signals produced outside peroxisomes, in this case by plasma membrane-derived ROS. Our findings also suggest that peroxule formation constitutes a strategy of rapid response to punctual stress, while peroxisomal proliferation may be a strategic response to continuous exposure to Cd stress. The possibility that signals different from ROS also contribute to the regulation of peroxule formation cannot be ruled out, although information regarding extremely early responses to Cd in plants is scarce. One hour of Cd treatment induced a 55% to 60% reduction in ion uptake and transpiration (Veselov et al., 2003), and transcriptomic studies after 2 h of Cd treatment have shown that calcium, calmodulin, MAPkinases, sulfur, and ethylene are important components in the signaling events triggered by Cd (Herbette et al., 2006).
Cd not only promoted changes in the morphology of peroxisomes, but also altered their speed of movement. Thus, peroxisomes were mainly stopped when producing peroxules. Similar results have been reported by Sinclair et al. (2009) in Arabidopsis plants exposed to exogenous H2O2. Although the mechanism has not been clearly identified, cytoskeleton has been shown to affect peroxisomal movement (Sinclair et al., 2009), while, more recently, Ruberti et al. (2014) also have observed that peroxule motility is dependent on the cytoskeleton and that microtubules are not involved in peroxule dynamics, whose regulation has been reported to be associated with the endoplasmic reticulum (Barton et al., 2013). However, longer periods of treatment with Cd (24 h) produced the opposite effect, with the speed of peroxisomal movement being observed to increase. The increase of peroxisome movement after 24 h of Cd treatment has demonstrated to be regulated by ROS (Rodríguez-Serrano et al., 2009), although neither the source of ROS nor the functionality of these changes in peroxisomal dynamics have been clearly identified. The reduction in the speed of peroxisomal movement observed in Atrboh lines as compared to the genetic background of wild-type plants after 24 h of Cd treatment suggests that changes in peroxisomal speed are regulated by ROS accumulation outside the peroxisomes. This could constitute an important strategy to regulate ROS accumulation in cells, and thus to protect against oxidative damage or to regulate ROS-dependent signaling to improve cell responses to environmental signals.
Although the mechanism regulating peroxisomal elongation and proliferation has been extensively studied, no information exists on the molecular processes involved in peroxule formation. Arabidopsis peroxin11 (PEX11) have been shown to regulate the elongation and proliferation of peroxisomes. The PEX11 gene family has five members: PEX11a, b, c, d, and e, which were divided into three subfamilies, PEX11a, PEX11b, and PEX11c-e, according to their sequence similarities (Lingard and Trelease, 2006; Orth et al., 2007). Although these isoforms appear to play different roles in peroxisomal proliferation, functional redundancy among various isoforms has been observed in Arabidopsis plants, as deletion of a single isoform does not significantly change the plant’s physiology (Orth et al., 2007). Light-induced peroxisomal proliferation involving the peroxine PEX11b has been well characterized (Desai and Hu, 2008), while salinity-induced peroxisomal proliferation was regulated by PEX11e (Mitsuya et al., 2010). In addition to its role in proliferation, our results demonstrate that PEX11a controls peroxule formation, as the RNAi lines of PEX11a (pex11a) are unable to produce peroxules in response to Cd and As treatment. The important role of PEX11a in peroxules formation is confirmed by the higher expression of PEX11a in line with the formation of peroxules. In addition to PEX11a, PEX11e is also up-regulated by Cd treatment, although its role in regulating peroxules can be ruled out under our experimental conditions, as RNAi lines of pex11c-e showed peroxules as px-ck plants. It is therefore possible to suggest that PEX11a is involved in peroxule formation, while PEX11e controls peroxisomal elongation and proliferation. RNAi pex11a lines exposed to Cd actually showed elongated peroxisomes. Analysis of PEX11a and PEX11e in Atrboh lines demonstrated that both genes are regulated by ROS or RBOH-dependent redox signals. However, the rapid formation of peroxules in response to ROS would suggest that post-translational modifications of PEX11a could be involved in activating peroxin-promoting peroxules. Although no information is available on PEX11 activation in plants, peroxines Pex11p in yeast has been reported to be activated by protein dimerization, which is regulated by redox changes in cysteines (Knoblach and Rachubinski, 2010; Schrader et al., 2012); however, other post-translational modifications of Pex11p such as phosphorylation are a subject of debate (Thomas et al., 2015). Proteomic studies are required to clarify the rapid regulation of peroxule formation and the role played by the post-translational modification of PEX11a.
Concerning the function of peroxules, it has been suggested that they could represent a mechanism to increase the peroxisomal surface area to improve the biochemical functions of these organelles, or may represent a first step in the morphological changes giving rise to elongation and peroxisomal division (Sinclair et al., 2009). After using optical tweezers to investigate peroxule functionality, Gao et al. (2016) have suggested that peroxules play a role in establishing and maintaining connections and communication between organelles in the intracellular environment, particularly with those that have different motilities or are spatially separated. However, we believe that the functionality of these extensions could be also specifically related to the ROS metabolism. Thus, the absence of PEX11a and peroxules promotes an increase in oxidative lipid damage parallel to a reduction in the expression of peroxisomal antioxidants CAT2 and CSD3. These findings indicate that PEX11a is an important component of rapid ROS-mediated stress perception, with peroxules possibly being involved in regulating ROS accumulation to protect cells against oxidative stress. Peroxules have been reported to be associated with chloroplasts (Sinclair et al., 2009; Oikawa et al., 2015; Gao et al., 2016) and mitochondria (Sandalio et al., 2013), suggesting that they may contribute to the regulation of the excess H2O2 produced in these organelles and may also favor interorganelle communication and transport. Peroxisomes contain enzymatic and nonenzymatic defense mechanisms to deal with different types of ROS such as O2.− and H2O2. The enzymes involved in these mechanisms include superoxide dismutases, catalase, and components of the ascorbate-glutathione cycle (ascorbate, glutathione, and ascorbate peroxidase as well as dehydroascorbate, monodehydroascorbate, and glutathione reductase; Jiménez et al., 1997; Sandalio and Romero-Puertas, 2015).
It has been suggested that the peroxisome functions as a sink for H2O2 produced in different parts of the cell (Dat et al., 2000). Peroxules could thus facilitate the removal of ROS accumulated in the cytosol or other organelles by increasing the peroxisomal surface. Nevertheless, peroxisomes also play a key role in regulating redox homeostasis in the cell and in ROS-dependent signaling networks under stress and developmental conditions (Sandalio and Romero-Puertas, 2015; Tiew et al., 2015). Therefore, peroxules may play a role in regulating signaling networks, as the absence of PEX11a and peroxules causes disturbances in the expression of GST and RRTF1 genes, both of which are involved in ROS-dependent signaling networks in response to Cd. To clearly characterize the functionality of these extensions, it is important to determine whether specific proteins or other small molecules are present in peroxules or move from the body of the peroxisome to the peroxule, and also to analyze their role in regulating ROS accumulation.
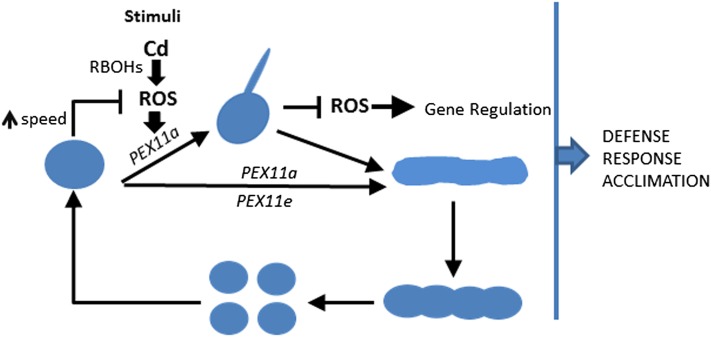
In conclusion, peroxisomal dynamics play an important role in organelle functionality and plant behavior during developmental, physiological, and pathophysiological processes. The molecular mechanisms and biological significance of peroxisomal dynamics remain elusive. We have, however, identified PEX11a as a potential ROS/redox sensor in response to stress, and propose a model consistent with our findings, in which peroxules are produced in response to stress conditions and PEX11a-dependent peroxule formation is regulated by ROS produced outside peroxisomes by RBOHs. We suggest that peroxules are involved in regulating ROS accumulation and signaling to protect cells against oxidative stress. We also demonstrated that peroxule formation is not a necessary step in peroxisome elongation. We show that changes in peroxisome dynamics are a key event in stress response networks and could be critical for cells to identify stress conditions by detecting changes in ROS levels and to ensure survival under adverse environmental conditions (Fig. 5). Further studies of other components involved in peroxule formation and coordination of peroxules with other organelles should enable us to determine how peroxisomes interact with their environment. Another important issue to be resolved is whether redox/ROS signals produced in different organelles trigger specific responses in relation to peroxule formation and transcriptional responses. An interdisciplinary approach, using in vivo imaging and proteomic and specific mutant designs, will be necessary to better understand the function of these protrusions of peroxisomes and other organelles such as mitochondria and chloroplasts.
Figure 5.
A model for changes in peroxisomal dynamics in response to Cd treatment. Cd promotes the generation of ROS by NADPH oxidases (RBOHs), which activate the expression of PEX11a and PEX11e. PEX11a promotes the formation of peroxules, which control ROS accumulation and the expression of ROS-dependent genes. Peroxule formation is followed by peroxisomal elongation, beading, and proliferation. The increase in the speed of peroxisome movement, which is also controlled by NADPH oxidases, contributes to cell responses to Cd after 24 h of treatment. Peroxule formation, peroxisome proliferation, and changes in the speed of peroxisomal movement together contribute to the regulation of cell responses to stress and acclimation.
MATERIALS AND METHODS
Arabidopsis ecotype Columbia-0 (Col-0) constitutes the genetic background for all wild-type plants used in this study. Arabidopsis lines expressing peroxisome-targeted cyan fluorescent protein (CFP) in the AtrbohC and AtrbohF background (AtrbohCxpx-ck and AtrbohFxpx-ck) were obtained by crossing Arabidopsis marker lines px-ck (Nelson et al., 2007), which expresses the CFP-PTS1 peroxisomal marker protein, with AtrbohC and AtrbohF mutants (kindly supplied by Dr. Jonathan Jones at the Sainsbury Laboratory, Norwich, UK). Arabidopsis RNAi lines pex11a, pex11b, and pex11c-e were available from a previous study (Orth et al., 2007). Arabidopsis seeds were surface-sterilized and stratified for 24–48 h at 4°C and then sown on Murashige and Skoog (MS) 0.5× solid medium (Murashige and Skoog, 1962), grown at 22°C in 16 h light and 8 h darkness for 14 d. The plants were then transferred to petri dishes containing 0.5× liquid MS medium and grown for 24 h. The plants were treated with 100-μm CdCl2 for 5 min to 24 h, or with 50 μm Na2HAsO4 for 30 min. To remove reactive oxygen species, 1 mm TMP (2,2,6,6-tetramethylpiperidinooxy; an O2·− scavenger) or 1 mm ascorbate (an H2O2 scavenger) was added to the MS solution 1 d before Cd treatment.
Confocal Microscopic Analyses
All leaves analyzed in the experiments were grown under the same conditions and were the same age and size. The Arabidopsis leaves were sliced using razor blades and mounted between a slide and a coverslip in PBS/glycerol 30:70%. The abaxial sections were examined using a confocal laser scanning microscope model no. TCS SP5 (Leica Microsystems, Wetzlar, Germany).
At least seven confocal images were collected from one leaf of each plant, with at least five plants being used per experiment. Each experiment was repeated five times. At least 125 (n) images were analyzed per treatment, which means that thousands of peroxisomes were examined.
The movement of individual peroxisome stacks was analyzed using the classification and particle-tracking routine of Volocity v. 3.0 (Improvision; Perkin-Elmer, Palo Alto, CA; Rodríguez-Serrano et al., 2009). This software can track the movement of individual fluorescent particles in time-resolved two and three dimensions and automatically generates speed and track length.
For speed analysis, the images were acquired in the x, y, z, and t dimensions. Each video contains 15 z-series, each consisting of 6–9 frames in the z axis (1 μm interval; 512×512 resolution; bidirectional scan mode). The videos, with a resolution of 1024×1024, were generated taking 20 frames in the x, y, and t dimensions. QuickTime (apple.com/quicktime) movies of peroxisome movement were generated from sequential images (five frames per s). Images of peroxule formation were acquired in x, y, and t dimensions, and the number of peroxisomes was analyzed using Leica Lite software (Leica Microsystems).
RT-PCR Analysis of Gene Expression
Total RNA was isolated from seedlings with the aid of the acid guanidine thiocyanate-phenol-chloroform method, as described by Chomczynski and Sacchi (1987), which uses TRIzol reagent (Thermo Fisher, Scientific, Waltham, MA) according to the manufacturer’s instructions. RNA was reverse-transcribed with the aid of an oligo(dT) primer and the Invitrogen SuperScript First-Strand Synthesis System for RT-PCR (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Quantitative real-time PCR was performed on an iCycler iQ5 (Bio-Rad, Hercules, CA). Each 20-μl reaction contained either 1 μl cDNA or a dilution, 200 nm of each primer, and iQ SyBrGreen Supermix (BioRad). Control PCR reactions of the RNA samples not treated with reverse transcriptase were also carried out to confirm the absence of contaminating genomic DNA. The samples were initially denatured by heating at 95°C for 3 min followed by 35-cycle amplification and a quantification program (95°C for 30 s, 55°C for 30 s, and 72°C for 45 s). A melting curve was conducted to ensure amplification of a single product. The amplification efficiency of primers was calculated using the formula E = [10 (1/a) −1] × 100, where a is the slope of the standard curve. The relative expression of each gene was normalized to that of TUB2, and the results were analyzed using the relative expression ratio according to the Pfaffl method (Pfaffl, 2001).
PEX11 expression was analyzed by semiquantitative RT-PCR, as no expression was found in wild-type seedlings under our experimental conditions. cDNAs were amplified by PCR as follows: 1 μl of the cDNA produced was added to 2× PCR Master Mix (Promega, Madison, WI) and 0.2 μmol of each primer in a final volume of 25 μL. Reactions were carried out in a Master Cycler (Eppendorf, Hamburg, Germany). An initial step lasting 2 min at 94°C was followed by 28–32 cycles of 30 s at 94°C, followed by 20–40 s at annealing temperature (55–65°C depending on the gene) and 30 s at 72°C, with a final 10-min extension at 72°C. The amplified PCR products were detected after electrophoresis on 0.8% agarose gels stained with ethidium bromide. The bands were quantified using a Chemidoc system (Bio-Rad) coupled with a high-sensitivity CCD camera. Band intensity was expressed as relative absorbance units. For normalization purposes, each cDNA band density was first divided by the density of the actin II band in the same lane to compensate for variations in cDNA loading onto the gel. The relative increase or decrease in gene expression in the Cd-treated leaves was then calculated by dividing the normalized band density of the gene from the Cd-treated leaves by that of the same gene from the untreated control leaves (Marone et al., 2001). The relative density of the control gene band is therefore 1. All the primers used are described in Supplemental Table S1.
Protein and Lipid Peroxidation Assays
Protein concentration was quantified with the aid of the Bradford Protein Assay kit (Bio-Rad) using BSA (bovine serum albumin) as standard. Lipid peroxidation was estimated by analyzing MDA (malondialdehyde) content obtained using the TBA (thiobarbituric acid) reaction according to the method described by Buege and Aust (1978). The MDA concentration was determined using a calibration curve with commercial MDA.
Determination of Hydrogen Peroxide Levels
The level of hydrogen peroxide (H2O2) was analyzed using fluorimetric techniques as described by Romero-Puertas et al. (2004). Leaf tissues were extracted in 25 mm H2SO4 (1:2 w/v) and centrifuged at 17,000g for 25 min at 4°C. Pigments were removed with activated charcoal. H2O2 content was analyzed using homovanillic acid (excitation of 315 nm; emission of 425 nm) and horseradish peroxidase in 50 mm HEPES pH 7.5, and the H2O2 concentration was estimated with the aid of a standard curve using commercial H2O2.
Statistical Analyses
The mean values in the quantitative experiments described above were obtained from at least three replicates of three independent experiments. Statistical analyses were performed using an ANOVA test. Mean values for the different treatments were compared using Tukey’s multiple comparison tests (P < 0.05).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_564514 (AT1G47750), NP_191666 (AT3G61070).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Time course analyses of the effect of Cd (100 μm) on peroxisomal dynamics.
Supplemental Figure S2. Regulation of peroxule formation by PEX11a in response to Cd and As.
Supplemental Table S1. Primer sequences.
Supplemental Video S1. Peroxule formation after 30 min of Cd treatment (blue). The red chloroplasts are marked in red.
Supplemental Video S2. Peroxisomal movement and peroxule formation after 30 min of Cd treatment.
Supplementary Material
Acknowledgments
The authors thank Juani Muñoz and Katiuska Cárdenas for their technical assistance. The confocal laser fluorescence microscopic analyses were carried out at the Technical Services Department of the University of Granada. The English text was corrected by Michael O’Shea.
Footnotes
Articles can be viewed without a subscription.
References
- Baker A, Paudyal R (2014) The life of the peroxisome: from birth to death. Curr Opin Plant Biol 22: 39–47 [DOI] [PubMed] [Google Scholar]
- Barton K, Mathur N, Mathur J (2013) Simultaneous live-imaging of peroxisomes and the ER in plant cells suggests contiguity but no luminal continuity between the two organelles. Front Physiol 4: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N (2014) ROS as key players in plant stress signalling. J Exp Bot 65: 1229–1240 [DOI] [PubMed] [Google Scholar]
- Buege JA, Aust SD (1978) Microsomal lipid peroxidation. Methods Enzymol 52: 302–310 [DOI] [PubMed] [Google Scholar]
- Castillo MC, Sandalio LM, Del Río LA, León J (2008) Peroxisome proliferation, wound-activated responses and expression of peroxisome-associated genes are cross-regulated but uncoupled in Arabidopsis thaliana. Plant Cell Environ 31: 492–505 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- de Felipe MR, Lucas MM, Pozuelo JM (1988) Cytochemical study of catalase and peroxidase in the mesophyll of Lolium rigidum plants treated with isoproturon. J Plant Physiol 132: 67–73 [Google Scholar]
- Desai M, Hu J (2008) Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 HOMOLOG, and the peroxisomal protein PEROXIN11b. Plant Physiol 146: 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego SM, Pena LB, Barcia RA, Azpilicueta CE, Iannonea MF, Rosalesa EP, Zawoznik MS, Groppa MD, Benavides MP (2012) Unravelling cadmium toxicity and tolerance in plants: insight into regulatory mechanisms. Environ Exp Bot 83: 33–46 [Google Scholar]
- Gao H, Metz J, Teanby NA, Ward AD, Botchway SW, Coles B, Pollard MR, Sparkes I (2016) In vivo quantification of peroxisome tethering to chloroplasts in tobacco epidermal cells using optical tweezers. Plant Physiol 170: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier L, Simon-Plas F, Thuleau P, Agnel JP, Blein JP, Ranjeva R, Montillet JL (2006) Cadmium affects tobacco cells by a series of three waves of reactive oxygen species that contribute to cytotoxicity. Plant Cell Environ 29: 1956–1969 [DOI] [PubMed] [Google Scholar]
- Gupta DK, Inouhe M, Rodríguez-Serrano M, Romero-Puertas MC, Sandalio LM (2013) Oxidative stress and arsenic toxicity: role of NADPH oxidases. Chemosphere 90: 1987–1996 [DOI] [PubMed] [Google Scholar]
- Gupta DK, Pena LB, Romero-Puertas MC, Hernández A, Inouhe M, Sandalio LM (2016) NADPH oxidases differentially regulate ROS metabolism and nutrient uptake under cadmium toxicity. Plant Cell Environ (in press) doi: 10.1111/pce.12711 [DOI] [PubMed] [Google Scholar]
- Herbette S, Taconnat L, Hugouvieux V, Piette L, Magniette M-LM, Cuine S, Auroy P, Richaud P, Forestier C, Bourguignon J, Renou J-P, Vavasseur A, et al. (2006) Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 88: 1751–1765 [DOI] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez A, Hernández JA, Del Río LA, Sevilla F (1997) Evidence for the presence of the ascorbate–glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol 114: 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B, Rachubinski RA (2010) Phosphorylation-dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J Biol Chem 285: 6670–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Trelease RN (2006) Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J Cell Sci 119: 1961–1972 [DOI] [PubMed] [Google Scholar]
- López-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A (2000) Stress induces peroxisome biogenesis genes. EMBO J 19: 6770–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone M, Mozzetti S, De Ritis D, Pierelli L, Scambia G (2001) Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol Proced Online 3: 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuya S, El-Shami M, Sparkes IA, Charlton WL, Lousa CdeM, Johnson B, Baker A (2010) Salt stress causes peroxisome proliferation, but inducing peroxisome proliferation does not improve NaCl tolerance in Arabidopsis thaliana. PLoS One 5: e9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Oikawa K, Matsunaga S, Mano S, Kondo M, Yamada K, Hayashi M, Kagawa T, Kadota A, et al. (2015) Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis, Nature Plants 1: 15035. [DOI] [PubMed] [Google Scholar]
- Oksanen E, Haikio E, Sober J, Karnosky DF (2003) Ozone-induced H2O2 accumulation in field-grown aspen and birch is linked to foliar ultrastructure and peroxisomal activity. New Phytol 161: 791–799 [DOI] [PubMed] [Google Scholar]
- Olmos EO, Martínez-Solano JR, Piqueras A, Hellín E (2003) Early steps in the oxidative burst induced by cadmiumin cultured tobacco cell (BY-2 line). J Exp Bot 54: 291–301 [DOI] [PubMed] [Google Scholar]
- Orth T, Reumann S, Zhang X, Fan J, Wenzel D, Quan S, Hu J (2007) The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma JM, Garrido M, Rodríguez-García MI, del Río LA (1991) Peroxisome proliferation and oxidative stress mediated by activated oxygen species in plant peroxisomes. Arch Biochem Biophys 287: 68–74 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M, Pazmiño DM, Sparkes I, Rochetti A, Hawes C, Romero-Puertas MC, Sandalio LM (2014) 2,4-Dichlorophenoxyacetic acid promotes S-nitrosylation and oxidation of actin affecting cytoskeleton and peroxisomal dynamics. J Exp Bot 65: 4783–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M, Romero-Puertas MC, Sparkes I, Hawes C, del Río LA, Sandalio LM (2009) Peroxisome dynamics in Arabidopsis plants under oxidative stress induced by cadmium. Free Radic Biol Med 47: 1632–1639 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Rodríguez-Serrano M, Corpas FJ, Gómez M, del Río LA, Sandalio LM (2004) Cadmium-induced subcellular accumulation of O2.− and H2O2 in pea leaves. Plant Cell Environ 27: 1122–1134 [Google Scholar]
- Ruberti C, Costa A, Pedrazzini E, Lo Schiavo F, Zottini M (2014) FISSION1A, an Arabidopsis tail-anchored protein, is localized to three subcellular compartments. Mol Plant 7: 1393–1396 [DOI] [PubMed] [Google Scholar]
- Sandalio LM, Rodríguez-Serrano M, Archilla-Ruiz A, Gupta DK, Romero-Puertas M, del Río LA (2012) Reactive oxygen species and nitric oxide under cadmium stress: from toxicity to signalling. In Ahmad P and Prasad MNV, editors, Environmental Adaptation and Stress Tolerance of Plants in the Area of Climate Change, Springer, Berlin, Germany, pp. 199–216. [Google Scholar]
- Sandalio LM, Rodríguez-Serrano M, Romero-Puertas MC, del Río LA (2013) Role of peroxisomes as a source of reactive oxygen species (ROS) signaling molecules. Subcell Biochem 69: 231–255 [DOI] [PubMed] [Google Scholar]
- Sandalio LM, Romero-Puertas MC (2015) Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann Bot (Lond) 116: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, Bonekamp NA, Islinger M (2012) Fission and proliferation of peroxisomes. Biochim Biophys Acta 1822: 1343–1357 [DOI] [PubMed] [Google Scholar]
- Scott I, Sparkes IA, Logan DC (2007) The missing link: inter-organellar connections in mitochondria and peroxisomes? Trends Plant Sci 12: 380–381 [DOI] [PubMed] [Google Scholar]
- Sinclair AM, Trobacher CP, Mathur N, Greenwood JS, Mathur J (2009) Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J 59: 231–242 [DOI] [PubMed] [Google Scholar]
- Smeets K, Opdenakker K, Remans T, van Sanden S, van Belleghem F, Semane B, Horemans N, Guisez Y, Vangronsveld J, Cuypers A (2009) Oxidative stress-related responses at transcriptional and enzymatic levels after exposure to Cd or Cu in a multipollution context. J Plant Physiol 166: 1982–1992 [DOI] [PubMed] [Google Scholar]
- Thomas AS, Krikken AM, van der Klei IJ, Williams CP (2015) Phosphorylation of Pex11p does not regulate peroxisomal fission in the yeast Hansenula polymorpha. Sci Rep 5: 11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiew TW-Y, Sheahan MB, Rose RJ (2015) Peroxisomes contribute to reactive oxygen species homeostasis and cell division induction in Arabidopsis protoplasts. Front Plant Sci 6: 658 10.3389/fpls.2015.00658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselov D, Kudoyarov G, Symonyan M, Veselov S (2003) Effect of cadmium on ion uptake, transpiration and cytokinin content in wheat seedlings. Bulg J Plant Physiol Special Issue: 353–359 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.