Abstract
We describe an as yet unreported neocentric small supernumerary marker chromosome (sSMC) derived from chromosome 1p21.3p21.2. It was present in 80% of the lymphocytes in a male patient with intellectual disability, severe speech deficit, mild dysmorphic features, and hyperactivity with elements of autism spectrum disorder (ASD).
Several important neurodevelopmental genes are affected by the 3.56 Mb copy number gain of 1p21.3p21.2, which may be considered reciprocal in gene content to the recently recognized 1p21.3 microdeletion syndrome. Both 1p21.3 deletions and the presented duplication display overlapping symptoms, fitting the same disorder category. Contribution of coding and non-coding genes to the phenotype is discussed in the light of cellular and intercellular homeostasis disequilibrium. In line with this the presented 1p21.3p21.2 copy number gain correlated to 1p21.3 microdeletion syndrome verifies the hypothesis of a cumulative effect of the number of deregulated genes - homeostasis disequilibrium leading to overlapping phenotypes between microdeletion and microduplication syndromes.
Although miR-137 appears to be the major player in the 1p21.3p21.2 region, deregulation of the DPYD (dihydropyrimidine dehydrogenase) gene may potentially affect neighboring genes underlying the overlapping symptoms present in both the copy number loss and copy number gain of 1p21. Namely, the all-in approach revealed that DPYD is a complex gene whose expression is epigenetically regulated by long non-coding RNAs (lncRNAs) within the locus. Furthermore, the long interspersed nuclear element-1 (LINE-1) L1MC1 transposon inserted in DPYD intronic transcript 1 (DPYD-IT1) lncRNA with its parasites, TcMAR-Tigger5b and pair of Alu repeats appears to be the “weakest link” within the DPYD gene liable to break. Identification of the precise mechanism through which DPYD is epigenetically regulated, and underlying reasons why exactly the break (FRA1E) happens, will consequently pave the way toward preventing severe toxicity to the antineoplastic drug 5-fluorouracil (5-FU) and development of the causative therapy for the dihydropyrimidine dehydrogenase deficiency.
Keywords: Neuronal homeostasis, Neurodevelopmental genes, Overlapping phenotypes, Common fragile site FRA1E, Epigenetics, Non-coding RNAs, Transposons, Tc1/mariner family of transposable elements, Human brain transcriptome, Bones and dental anomalies
1. Introduction
We report a case of a neocentric small supernumerary marker chromosome (sSMC) derived from the 1p21.3p21.2 chromosome in order to provide insight into the molecular processes influencing the phenotype. Most of the genes comprised in sSMC(1) are enriched in the developing human brain (PTBP2, DPYD, miR-137, SNX7, LPPR5, LOC100129620, LPPR4), as revealed by Yale’s genome-wide exon-level transcriptome data base (www.humanbraintranscriptome.org) [1], implicating their role in the processes leading to proper brain organization and functioning.
To date, the genotype-phenotype correlations in genetic disorders have been mostly viewed through coding gene mutations and copy number losses/gains. However, recent studies implicate the important contribution of non-coding RNAs to the phenotype, and a need for an all-in approach in investigating the molecular processes underlying the genetic disorders, necessary for a comprehensive understanding of human disease. Therefore, an all-in approach has been applied in analyzing the genotype-phenotype correlation of 1p21.3p21.2 copy number gain.
2. Material and methods
2.1. Clinical description
The patient was a boy identified by the multiplex ligation-dependent probe amplification (MLPA) screening in individuals with unexplained intellectual disability (ID). He is the fifth child of parents who both function at a borderline intellectual level or mild ID. At the age of 10, the boy and his siblings were placed in foster care homes, as the parents had not been able to take care of them.
According to scant data on his early development, he was born at term after an uneventful pregnancy, and the child’s early motor development was unremarkable. His language development, in contrast, was severely delayed. On two occasions in his 2nd and 3rd year of life he was hospitalized for febrile convulsions. At the age of 2, behavioral problems and hyperactivity were recorded, which became more remarkable from the age of 6. At the age of 8 he was diagnosed with a moderate to severe ID, pervasive developmental disorder with elements of autism spectrum disorder (ASD), undeveloped speech, febrile convulsions and nocturnal enuresis.
Physical examination at the age of 11 years and 10 months, revealed a height of 142 cm (17th percentile), a weight of 42 kg (50th percentile) and a head circumference of 52 cm (50th percentile). Presently at the age of 19, his height is 172 cm (25th percentile), weight 60 kg (20th percentile), and head circumference 55 cm (25th percentile). The patient’s clinical presentation includes the following features:
Face
The face displays mild dysmorphism with deep-set eyes and hooded eyelids, philtrum with upturned upper lip and an open mouth appearance, narrow high arched palate and maxillary prognathism. In each jaw there are 12 permanent teeth: 4 incisors, 2 canines, 2 premolars (without panoramic tomography it was not possible to determine which premolars are missing, teeth no. 4. or no. 5) and 4 molars (3rd molars/“wisdom teeth” are missing). The first maxillary left incisor is mechanically broken, but both maxillary lateral incisors are undergrown and peg-shaped.
Stature
The shoulders are sloping and dropped. A mild thoracic kyphosis (with a hump resembling a buffalo hump) extends to lumbar lordosis.
Hands and feet
Fingers are long and tapering with bilateral clinodactyly of the 5th finger. Instep is very high and rigid. The first toe is long and widely spaced from the 2nd toe, with partial cutaneous syndactyly of 2nd and 3rd toe. Toes appear pointed. His peculiar gait with a tendency toward toe-walking (most evident when running) resembles cock-walk with equinus gait, during which he leans the body forward.
Genitals exhibit one sided cryptorchidism.
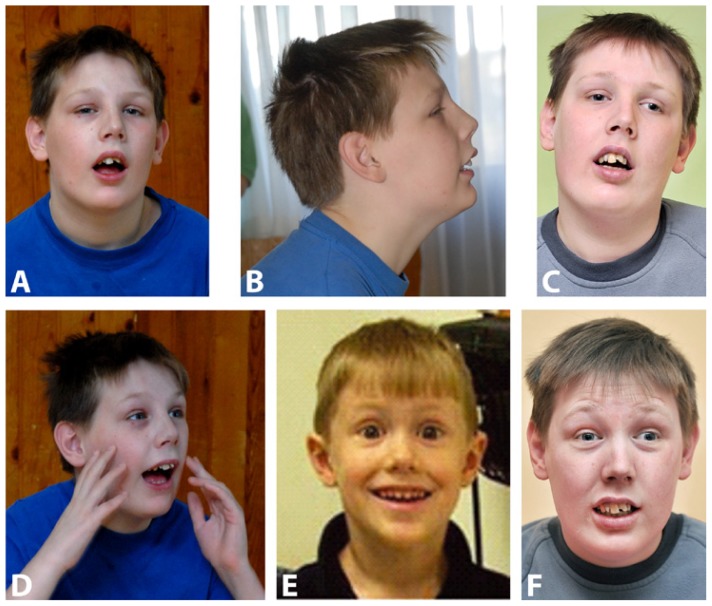
Neurological examination: Psychomotor restlessness and hyperactivity made assessments and evaluations extremely difficult even with multiple examinations. The patient is disoriented, socially unadapted, with very poor concentration and attention. He frequently has an empty gaze, as if he is living in his own world with sudden expressions of a laughing grimace (Fig. 1D and Fig. 1F). Speech is sparse and incomprehensible (at the word level) with echolalia and neologisms. Psychomotor restlessness and hyperactivity, as well as aggressive outbursts, are controlled with neuroleptics. The patient is depicted in Fig. 1A–D at the age of 13 years 10 months, and 19, respectively.
Figure 1.
Patient’s facial appearance. A–C) The facial appearance (front and lateral view) of the patient at 13/10 years and 19 years, respectively. Note the way the boy is holding his head. D) The facial expression in a sudden laughing/smiling grimace at 13/10 years (F) and at 19 years. Informed consent is obtained from the guardian. E) Note the resemblance of the face expression between the present case and the patient 4 with 10 kb DPYD deletion reported by Carter et al. [15]. The reuse of the Figure 2f from the paper by Carter at al. [15] is kindly provided by © 2010 John Wiley & Sons A/S (license number 3435250114502).
2.2. Cytogenetics and molecular cytogenetics
Banding cytogenetics from peripheral blood lymphocytes was done according to standard procedure. The sSMC was microdissected (glass needle based), the obtained DNA was amplified in vitro and labeled by degenerated oligonucleotide-primed polymerase chain reaction (DOP-PCR) as previously reported [2], and applied in standard reverse fluorescence in situ hybridization (FISH) [3].
2.3. Multiplex ligation-dependent probe amplification (MLPA)
MLPA probe sets (SALSA MLPA Kits P036-Human Telomere, P070-Human Telomere, P245-Microdeletion Syndromes-1, P297-Microdeletion Syndromes-2, P343-B1 Autism and ME028-PWS/AS) were purchased from MRC-Holland (Amsterdam, The Netherlands) and used according to the manufacturer’s protocols. We have used commercially available software, Gene Marker from SoftGenetics (State College, PA, USA) to analyze our data.
2.4. Array-comparative genomic hybridization (arrayCGH)
A genome-targeted copy number profile of the patient’s DNA was obtained by subjecting it to microarray analysis using our own custom-designed 8×60 K oligo (60-mer) array platform (Custom CGH Zagreb, ID:061743) manufactured by Agilent Technologies Inc. (Santa Clara, CA, USA), containing most known genes and regions associated with neurodevelopmental disorders (publication in preparation). The array was processed following the manufacturer’s recommended protocol, and a sex-matched non-disease control sample was used as reference.
3. Results
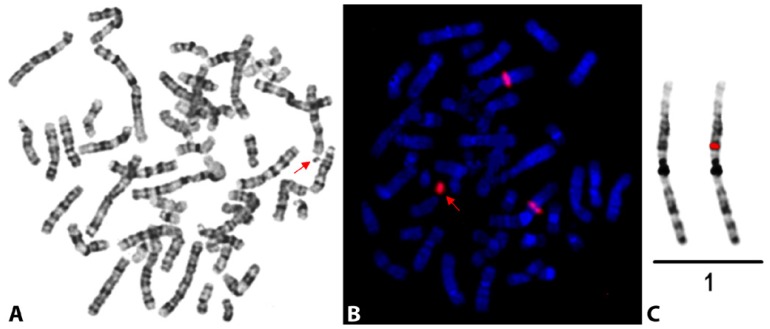
The present case was singled out during MLPA screening in individuals with unexplained ID, which started with subtelomeres screening (P036 and P070) followed by screening for microdeletion syndromes (P245 and P297) and autism (P343-B1), which all showed normal results. Finally, a ME028-PWS/AS probe set was used to exclude possible Angelman syndrome due to uniparental disomy, and the chromosome culture was set up. The methylation pattern was normal. However, the copy number report in repeated experiments revealed a deletion of SNRPN exon1B-b (copy number variation in this region has been described in healthy individuals too), and duplication of the reference 1p21 probe 05330-L04717 (results not shown). Banding cytogenetics disclosed a ring shaped sSMC found in 80% of the lymphocytes (Fig. 2A). Subsequent parallel reverse FISH and arrayCGH, both confirmed what MLPA result already suggested: the sSMC was derived from 1p21 (Fig. 2B and Fig. 2C).
Figure 2.
GTG-banded metaphase spread and reverse FISH displaying sSMC(1). A) GTG-banding revealed a karyoytpe 47,XY,+r[80%]/46,XY[20%]; sSMC is labeled by an arrowhead. B) After the microdissection and reverse FISH the red labeled DNA-probe (midi) stained the sSMC itself and a region in the short arm of both chromosomes 1 (arrowhead). C) Inverted DAPI banding shows the mapping of the microdissection derived DNA probe (red; midi) to 1p21.3~21.2.
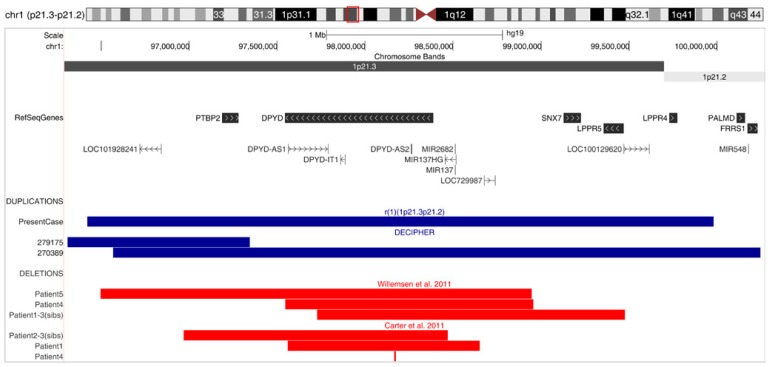
ArrayCGH identified a 3.56 Mb copy number gain on chromosome 1 short arm: arr[hg19] 1p21.3p21.2(96,420,239-99,981,342) x3, containing five annotated RefSeq (http://www.ncbi.nlm.nih.gov/refseq/) coding genes: PTBP2, DPYD, SNX7, LPPR5 and LPPR4, and seven annotated Refseq non-coding RNAs (ncRNAs): antisense long non-coding RNAs (lncRNAs) DPYD-AS1 and DPYD-AS2, microRNAs miR-137 and miR-2682, and uncharacterized long intergenic non-coding RNAs (lincRNAs) LOC10192824, LOC729987 and LOC100129620 (Fig. 3).
Figure 3.
Schematic representations of the deleted and duplicated segments in 1p21. Copy number losses and gains are outlined with the aid of UCSC Genome Browser - Genome Graphs software. Coordinates of the deletions reported by Carter et al. [15] and Willemsen et al. [16] are converted to hg19. The transcription streams and positions of the coding and non-coding genes along 1p21.3 and adjacent distal 1p21.2 are indicated. The deletions studied by Kuilenburg et al. [146] are not included in the figure: patients 1–4 originate from highly consanguineous families; for the patient 5 (~14 Mb, del1p21.3p13.3), no coordinates or most proximal-most distal genes are quoted.
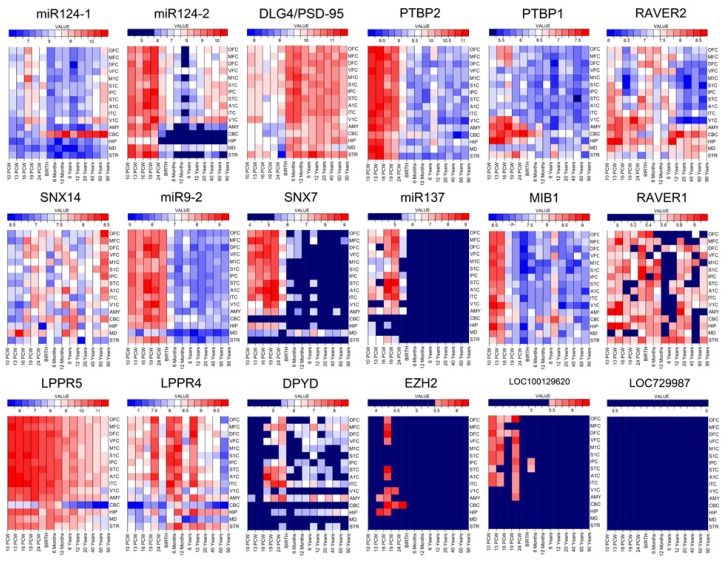
Spatio-temporal expression profiles of the genes involved in 1p21.3p21.2 from the genome-wide exon-level transcriptome data [1] (Fig. 4) revealed their enrichment in human brain during neurogenesis, as well as importance in adult brain functioning (Table 1). In addition, microarray expression data of the transcripts possibly involved in common pathway with the genes comprised in sSMC(1), or commented in the discussion, are shown in Fig. 4.
Figure 4.
Expression patterns of selected transcripts in the human brain. The microarray expression data were analyzed for affected transcripts in 1p21.3p21.2 and those discussed in the paper in 16 brain regions and 13 developmental periods. Samples colored dark blue are considered unexpressed (cutoff value <5.5). Note that the range of expression intensity is displayed with each transcript profile. Spatiotemporal expression profiles of brain regions and neocortical areas (NCX) were obtained from the Yale’s genome-wide exon-level transcriptome data base (www.humanbraintranscriptome.org). MiR124-1 microarray expression data shows overall moderate expression level in the brain, except in cerebral cortex during early mid-fetal period (16–19 PCW). However, from neonatal period until adulthood miR124-1 expression level is consistently high in cerebellum. MiR124-2 expression level is high prenatally during major neurodevelopmental processes in all examined cerebral cortical areas, thalamus, basal ganglia and cerebellum until late mid-fetal period when its expression decreases, especially in cerebellum, hippocampus and striatum. PTBP2 shows similar expression pattern like miR124-2, which is consistent with the finding that miR-124 downregulates PTBP1, leading to upregulation of PTBP2. PTBP2 expression level is consistently high in all brain regions until late mid-fetal period, a crucial neurodevelopmental period for major histogenetic events and formation of neocortical circuits. After 24PCW its expression is downregulated. In contrast, DLG4 (PSP-95) shows moderate expression level in cerebrum and cerebellum until neonatal time, when expression is upregulated in all examined regions (consistent with alternative splicing regulation of PSD-95; PSD-95 is post-transcriptionally repressed by PTBP2, which thus temporarily inhibits the expression of “adult” protein isoforms until neurons have matured) and remain high throughout adulthood. The PTBP1 expression level is consistently low in all neocortical areas, except visual. However, its expression is high prenatally in amygdala, cerebellum and hippocampus until neonatal period when is downregulated. The RAVER1 expression pattern is relatively low throughout lifetime and it shows lowest level during childhood. RAVER2 displays spatiotemporal differential expression pattern in different cortical areas; namely: in prefrontal cortex is highest during perinatal time, while in some cortical areas (like visual) is highly expressed from early fetal until early infancy. RAVER2 expression level is lower from late childhood throughout adulthood in all cortical areas. On a contrary, expression level remains more stable in other brain regions, such as thalamus and basal ganglia. MiR-137 expression is highly expressed prenatally in neocortex and amygdala, but its expression is decreased perinatally and remains low postnatally. According to microarray data, it is not expressed in cerebellum throughout life span. MIB1 shows highest expression during early fetal time in all examined regions of the brain and gradually decreases from early mid-fetal onward in all examined regions, except in cerebellum. SNX7 shows highest expression in neocortical areas, amygdale and hippocampus from early fetal to perinatal period, during the time of major neurodevelopmental processes. Its expression level decreases postnatally. Cerebellum and striatum do not show expression throughout whole life span. MiR9-2 expression level is high from early fetal until neonatal period in all examined brain regions except thalamus. LPPR5 expression is high in all examined brain regions throughout whole life span, except in the cerebellum where it remains lower, except during perinatal period. LPPR4 shows highest expression in neocortical areas, especially during late mid-fetal, neonatal, infancy and childhood during intense synaptogenesis and dendritic differentiation. Its expression is lowest in cerebellum and thalamus throughout life span. DPYD shows substantial spatiotemporal variation in expression pattern throughout lifespan. It is not expressed during early fetal time in the brain and it shows the highest expression level in temporal lobe and amygdale during early and late mid-fetal period. Cerebellum and thalamus do not express it throughout all examined periods. Relatively low LOC100129620 expression can be observed in neocortex from early to late mid-fetal period. LOC729987 is not expressed in any analyzed region or time point. EZH2 is not expressed in the brain throughout lifespan, except in several samples prenatally. SNX14 starts to be expressed at early mid-fetal period and remains to be moderately expressed throughout lifespan. Nomenclature of analyzed brain regions and NCX areas; for the details on ontology see Kang et al. [1]: OFC: Orbital prefrontal cortex, DFC: Dorsolateral prefrontal cortex, VFC: Ventrolateral prefrontal cortex, MFC: Medial prefrontal cortex, M1C: Primary motor (M1) cortex, S1C: Primary somatosensory (S1) cortex, IPC: Posterior inferior parietal cortex, A1C: Primary auditory (A1) cortex, STC: Superior temporal cortex, ITC: Inferior temporal cortex, ITC: Inferior temporal cortex, V1C: Primary visual (V1) cortex, HIP: Hippocampus, AMY: Amygdala, STR: Striatum, MD: Mediodorsal nucleus of the thalamus, CBC: Cerebellar cortex.
Table 1.
Summary table of the genes involved in 1p21.3p21.2 copy number gain* *For the details and references see the discussion on each gene and Fig. 4.
| GENE | PTBP2 | DPYD | miR-137 | SNX7 | LPR5/PRG5 | LPR4/PRG1 |
|---|---|---|---|---|---|---|
| Description | Polypyrimidine Tract Binding Protein 2 | Dihydropyrimidine Dehydrogenase | MicroRNA 137 | Sortin Nexin 7 | Lipid Phosphate Phosphatase-Related Protein Type 5 | Lipid Phosphate Phosphatase-Related Protein Type 4 |
| Multiple transcripts | Multiple transcripts | Highly conserved small noncoding RNA | Multiple transcripts | Multiple transcripts | Multiple transcripts | |
| Tissue specificity | Brain specific (isoform 1 & 2) | Found in most tissues | Neuron-enriched miRNA | Enriched in the brain | Brain & Spinal cord specific | Brain specific |
| Protein | RNA binding protein | Pyrimidine catabolic enzyme | Non-coding; Binds to multiple target mRNAs | Protein binding | Closely related to LPPR1/PRG3; Mediate LPA activity in vitro | Mediate LPA activity in neurons (Hydrolyzes LPA) |
| Function | Mediates negative regulation of exons splicing | Initial and rate-limiting factor in the pathway of uracil and thymidine catabolism; 5-FU degradation | Translational repression or mRNA degradation; Brain: Silences Mib1 important for neurogenesis | ? Exact function (May be involved in several stages of intracellular trafficking) | Involved in neuronal plasticity; Induces filopodia sprouting; Promotes neurite growth; Drives axon elongation | Facilitates axonal outgrowth in the hippocampus; Proper synaptic transmission; Regulator of neuronal plasticity |
| Expression pattern Human brain transcriptome | Consistent with cross-regulatory network PTBP1-miR124-PTBP2-PSD95 | Spatiotemporal variation in expression pattern throughout lifespan | High prenatal expression in neocortex and amygdala | Highest expression in neocortex, amygdale and hippocampus during the time of major neurodevelopmental processes | High in all brain regions throughout whole life span (lower in the cerebellum) | Highest expression in neocortical areas during intense synaptogenesis and dendritic differentiation |
| Knock out/ Knock down/ Null allele/ Homozygous mutation | Lethal shortly after birth (mice); Neurons in culture-fail to develop | DPD deficiency caused by homozygous or compound heterozygous mutation; Large phenotypic variability; Severe toxicity to 5-FU | Aberrant, enriched dendritic tree of fetal and adult hippocampal neurons | n.a. | Inhibits filopodia formation & neurite growth; Attenuates neurite formation and growth | Juvenile epileptic seizures; Pathological increase of synaptic transmission – Hyperexcitability in CA1 pyramidal neurons |
| Copy number loss/ Deletion/ Heterozygous mutation/ Haploinsuficiency | PTBP2 at levels half that of WT animals; Target proteins show half reduced-normal expression | Large phenotypic variability; Severe toxicity to 5-FU | Overexpression of validated target proteins | n.a. | n.a. | LPPR4 at levels half that of WT animals; Intermediate increase of excitatory synaptic transmission |
| Copy number gain/ Duplication/ Overexpression | Repress synaptic activity, spine morphogenesis & reduce PSD-95 transcript in vitro | ? Exact phenotype effect on brain | Reduces the complexity of dendrites and spine density; ‘Deletion effect’ of dosage sensitive targets involved in neuronal differentiation | n.a. | Dramatic morphological changes in neuronal cells & non-neuronal cells in vitro | Cognitive deficits observed in mice and men |
4. Discussion
Small supernumerary marker chromosomes (sSMCs) are found in the general population (0.044%), in infertile (0.125%) and in patients with ID (0.288%) [4]. sSMCs can be present in different shapes, sizes, mosaic states and be derived from different chromosomal regions, predominantly the pericentric ones [5]. Neocentric sSMCs have a centromeric constriction but no detectable alpha-satellite DNA; they “carry newly derived centromeres (or “neocentromeres”) that are apparently formed within interstitial chromosomal sites that have not previously been known to express centromere function” [6]. Neocentric sSMCs [7] can be derived from each region of a chromosome. If they come from more distal, i.e. telomeric parts, they often form inverted duplicated shaped sSMCs; in case they are derived from more proximal parts of chromosome arms they are reported as ring shaped sSMCs [8]. Some of them are also formed by a so called McClintock mechanism [9].
We report the eighth case of a neocentric sSMC(1) in clinical practice; additionally, an inverted duplication-shaped sSMC was seen in leukemia as an acquired aberration. Among 8 clinical cases with a neocentric sSMC(1), all except one were ring-shaped like the present case [10]. Three previously reported neocentric sSMC(1) were formed by the McClintock mechanism [9], and one was even derived from a similar region as the present case. However, it was reported as a balanced cytogenetic aberration and no clinical data was available for that case [10, case McCl-01-N-p21/1-1].
Interestingly, the present case is the second one inducing a gain of copy numbers in the short arm of chromosome 1. In the presented case, sSMC(1) is identified in 80% of lymphocytes, suggesting a possibility of presence in a non-mosaic form and its loss during cell culturing. In addition, it is well known that mosaic cases often display a lower percentage of aberrant cells in lymphocytes than in fibroblasts, the extreme example being Pallister-Killian syndrome [11]. Both fibroblasts and neurons are derived from the same embryonic origin (ectoderm); therefore, it is likely that there are more than 80% of aberrant cells in the brain, even though there is no rule for the distribution of sSMC in different body cells [12]. The expected phenotype with such a high percentage of aberrant cells should not differ much from the non-mosaic form, although the true state of the brain cells remains uncertain. Although it would have been interesting to employ functional magnetic resonance imaging (fMRI) to uncover if any and which cortical areas are affected, due to the risk of performing the procedure under anesthesia, this was not done.
Segmental duplications of 1p are rarely reported. They vary considerably in size and position on 1p and no distinct phenotype has been defined to date for any duplicated segments on 1p. In addition, most previous cases did not have molecular characterization of duplicated segments, so it was not possible to estimate a region of overlap, or genotype-phenotype correlations [reviewed in 13].
4.1. 1p21.3 copy number loss vs. 1p21.3p21.2 copy number gain
Our present case of 1p21.3p21.2 copy number gain is the smallest reported duplication of the proximal short arm of chromosome 1 (Fig. 3). The closest in size are two pathological copy number gains listed in Decipher (http://www.sanger.ac.uk): patient 279175 with 1.04 Mb duplication encompassing lincRNA LOC101928241 and PTBP2 displaying ASD, muscular hypotonia and strabismus, and patient 270389 with 3.68 Mb duplication which, in addition to all genes comprised in sSMC(1), extends to distal 1p21.2 covering flanking PALMD, FRRS1 and miR-548 genes, for whom besides the ID no symptoms are commented (Fig. 3). The only case showing phenotypic similarity (primary teeth anomalies, high arched palate, fingers and toes anomalies, peculiar gait, hyperactivity, no speech at 3 years of age; the image of the patient’s face also being suggestive of an open mouth appearance and maxillary prognathism) is the case of Utkus et al. [14], although the latter duplication appears to be larger.
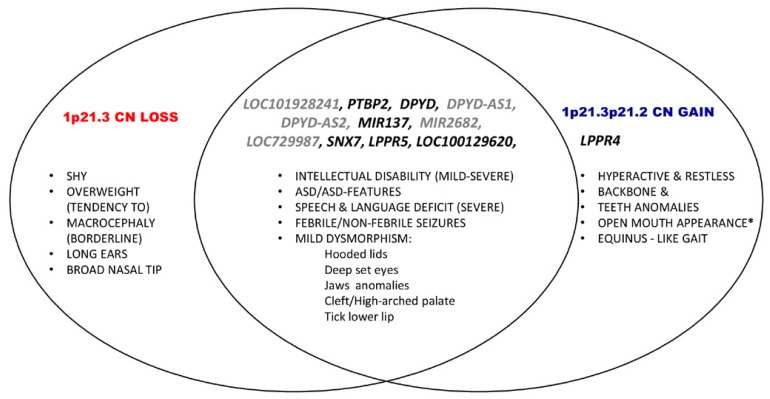
Unexpectedly, the overlap not only of the neurological and behavioral phenotypes (ID, ASD/ASD-like features and severe to profound speech deficit, febrile/non-febrile seizures), but also of the sum of dysmorphic features seen in differently sized 1p21.3 deletions (deep set eyes, tick lower lip, cleft/high-arched palate, hooded lids) [15, 16], becomes evident when compared to the 1p21.3p21.2 duplication phenotype (Fig. 5).
Figure 5.
Neurologic and behavioral phenotypes involving physical features in reciprocal 1p21 CN loss/CN gain. Phenotypes unique to CN loss are shown on the left; phenotypes unique to CN gain are shown on the right; phenotypes common to both CN loss and CN gain are shown in the overlapping part of the two ovals. Note that 1p21.3p21.2 copy number gain in addition to overlapping genes with 1p21.3 CN loss encompasses LPPR4, the first flanking gene in 1p21.2. *Case 5 from Kuilenburg et al. [146] with the deletion involving 1p13.3p21.3, besides profound ID displays macrocephaly, long prominent/upturned filtrum, open mouth appearance, tick lower lip, full nasal tip, high arched palate and large lobules. Eruption of his dentition was delayed, nails were short and thin (in italics are denoted overlapping features with presented 1p21.3p21.2 CN gain; macrocephaly, large lobules, full nasal tip, tick lower lip and high arched palate are the features seen in 1p21.3 CN loss). CN stands for copy number; gray colored are the genes that are either not in the Yale’s transcriptome data base (LOC101928241, DPYD-AS1, DPYD-AS2, miR-2682), or are not enriched in human brain (LOC729987).
The 1p21.3p21.2 copy number gain may be considered reciprocal in gene content to the recently recognized 1p21.3 microdeletion syndrome (Fig. 3) [15, 16, www.orpha.net], characterized by severe speech and language deficit, borderline to moderate and severe ID, ASD features, and minor dysmorphic facial features. Affected individuals have normal gross motor development without major abnormalities. They are often very shy and friendly with a tendency to be overweight.
What could be the rationale behind the fact that both the deletion and duplication of certain genes are capable of producing a similar or overlapping neurological or psychiatric phenotype? According to the hypothesis proposed by Ramocki and Zoghbi [17], a similar or overlapping set of neurological symptoms in reciprocal neurodevelopmental microdeletion and microduplication syndromes can be explained by imbalance of neuronal homeostasis. Briefly, either loss or gain of a certain gene function, which changes synaptic output and neuronal excitability, affects the integrity of the network as a whole, and activates compensation that eventually exhausts homeostatic capacity of the neuronal network and leads to defects of neuronal phenotype and synaptic plasticity. This phenomenon has been described for several genes, such as MECP2 and SHANK3, where loss or gain of function results in overlapping neurological disorders [17].
Therefore, the presented 1p21.3p21.2 copy number gain correlated to 1p21.3 microdeletion syndrome verifies the hypothesis of a cumulative effect of the number of dysregulated genes - homeostasis disequilibrium leading to overlapping phenotypes between microdeletion and microduplication syndromes, since the same conclusion has been drawn from two different points of view.
4.2. Long non-coding RNAs (lncRNAs): LOC10192824, LOC729987 and LOC100129620
The major classes of non-protein coding RNAs (ncRNAs) that are important for the regulation of gene expression include microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), extracellular RNAs (exRNAs), piwi-interacting RNAs (piRNAs), and long-non-coding RNAs (lncRNAs).
Comprehensive analysis of the human transcriptome has revealed that lncRNAs with length >200 nucleotides account for a large fraction of cellular transcripts. The systematization of lncRNAs is still incomplete, as they differ according to genomic localization, size and putative function. Similar to protein coding mRNAs, lncRNA transcripts are capped and polyadenylated, contain multiple exons with large introns and are subject to alternative splicing. Identification of intragenic lncRNAs is hampered due to overlap with protein-coding transcripts or DNA-regulatory elements, and has been originally described as transcription noise [reviewed in 18]. Therefore, they came into the spotlight only recently and their function as key regulators of cellular processes is emerging [19].
Transcription of lncRNAs is cell-type specific and developmentally regulated in the central nervous system (CNS) where they are involved in various roles, such as cell identity, homeostasis, stress responses and synaptic plasticity [20]. Strict temporal and spatial expression of lncRNAs is important for mediating CNS development and function, even though their precise expression pattern and its role are not yet fully known. Many lncRNAs are modulators of gene expression via chromatin modification, and may contain domains for binding other complementary RNAs, protein- and DNA-binding domains that induce conformational changes to other structures in the lncRNA [19].
Hence, it comes as no surprise that some of them are implicated in psychiatric, neurological and neurodegenerative disorders [21]. One of the best studied examples is an antisense long ncRNA BDNF-AS [22] that acts as a regulator of expression of brain derived neurotrophic factor (BDNF), important for neuronal growth, maturation and maintenance, whose expression level is reduced in some psychiatric and neurodegenerative disorders, such as Huntington disease [23]. Interestingly, Sauvageau et al. [24] reported developmental problems and defects in the cerebral cortex in some of the intergenic lncRNA (lincRNA) knockout mice, thus providing strong evidence of lncRNAs role in brain development.
LOC10192824, LOC100129620 and LOC729987, the intergenic lncRNAs that have copy number gain in the present case are yet uncharacterized and their function remains to be elucidated. According to the expression pattern of LOC100129620 in prenatal cerebral cortex (Fig. 4) we can suggest its role in the regulation of epigenetic dynamics in neurodevelopment.
4.3. MicroRNA miR-137
MicroRNAs constitute a class of small, non-coding RNAs that are involved in a subset of biological processes such as developmental programing, cell proliferation, apoptosis, metabolism, cell differentiation, and morphogenesis [25]. The discovery of microRNAs has led to deeper insights into the regulatory mechanisms of gene expression and their complexity. They function as posttranscriptional regulators of gene expression, primarily through gene silencing by binding to their target mRNAs [26], mediating translational repression or mRNA transcript degradation [27].
A single miRNA typically has multiple, up to several hundred mRNA targets, while a gene can have several target sites for different miRNAs [28–32]. Therefore, miRNAs can control the expression of a number of genes, affecting entire signaling pathways at once leading to a stance that modulation of protein levels by miRNAs represents a key epigenetic regulatory mechanism of gene expression [32–34].
Recent studies suggest that expression of miRNAs and their targets are dynamically regulated, both spatially and temporally, contributing to the diversity and plasticity of our brain [35]. It has been shown that many miRNAs also act locally, at the growth cone or at synapses, modulating synaptic plasticity and neuronal connectivity, thereby contributing to the dynamic spatial organization of axonal and dendritic structures and their function [36, 37].
A neuron-enriched miRNA, miR-137 plays an important role in the regulation of cell proliferation and differentiation [38–41]. Micro RNA miR-137 is enriched at the synaptic compartment [36, 37, 39, 42] and regulates neuronal maturation influencing dendritic patterning and spine morphogenesis [43–45] through silencing of Mind bomb one (Mib1), an ubiquitin ligase known to be important for neurogenesis [39, 46–48]. There are a large number of CpG islands in the upstream 2.5 kb promoter region of miR-137 gene [49], suggesting that its expression is epigenetically regulated.
Overexpression of miR-137 results in aberrant morphological maturation of neurons by reducing the complexity of dendrites and spine density, both in brain and cultured primary neurons [39]. In contrast, the knock-down of miR-137 had opposite effects on the dendrite morphogenesis (increases the dendrite length, branch and end points, and number of spines in mouse fetal and adult hippocampal neurons), suggesting that proper expression of miR-137 is required for the normal morphological differentiation and development of dendrites [39]. Furthermore, all histogenetic processes, including neuronal differentiation, need to be precisely spatially and temporally coordinated in order to establish appropriate synaptic contacts and subsequent proper functioning of cortical neural networks. Hence, resulting either reduced or enriched dendritic tree could lead to misrouted axons and misplaced synaptic contacts that eventually lead to similar or identical abnormal cortical functioning.
A number of developmental and adult brain disorders are associated with abnormal changes in synaptic connectivity and plasticity, [29, 50, 51]. Moreover, a connection has been established between abnormalities in miRNA expression and miRNA-mediated gene regulation and cognitive dysfunction [reviewed in 29 and 35]. In this context, large-scale genome-wide association studies (GWAS) have identified miR-137 as one of the leading schizophrenia susceptibility genes [52–54].
Consequently, miRNA gene copy number changes due to genomic deletions and duplications are likely to be involved in neurological disorders as well. While studying the clinical effects of chromosome 1p21.3 microdeletions involving DPYD and miR-137, Willemsen et al. [16] found an association with ID and ASD-like behavior. Furthermore, lymphoblastoid cell lines from these patients were found to have reduced levels of miR-137. The authors also confirmed that miR-137 is highly expressed in the hippocampus, occipital cortex, and frontal cortex in human postmortem tissue, as well as in the synaptosomal fractions of mouse brain preparations, providing further evidence that miR-137 plays a role in synapse formation during brain development and functioning.
Using the lists of putative and experimentally verified targets, we find no gene in sSMC(1) as the verified target of either miR-137 or miR-2682 (http://www.mirbase.org, http://www.targetscan.org, http://mirdb.org). Interestingly, RAVER2, ribonucleoprotein PTB-binding 2, whose expression in adult mice is essentially confined to the brain [55, 56] is the highly ranked miR-137 putative target in miRDB and TargetScan (Agregate PCT 0.96; total context score −0.55). RAVER1 and RAVER2 are co-repressors of PTBP1/PTBP2 (PTB/nPTB) homologs, with a modulating function in PTB-mediated RNA processing [57, 58].
Regulation of RAVER2 by miR-137 would imply its modulatory role of PTBP2 expression through downstream mechanism. However, little is still known about the role of RAVER2 in neurodevelopment due to its restrictive expression pattern and lack of expression in neuronal cell lines [55]. Considering that aberrant expression of miRNAs, leading to either down-regulation or up-regulation of downstream targets, has been implicated in a number of neurodegenerative, neurodevelopmental, as well as psychiatric disorders [59–63], the downstream effect of miR-137 on the alternative splicing pathway seems probable.
4.4. Polypyrimidine Tract Binding Protein 2 (PTBP2)
PTBP2 protein, encoded by PTBP2 (nPTB) gene is a multifunctional RNA binding protein (shuttling between nucleus and cytoplasm) involved in post-transcriptional regulation of gene expression. PTBP2 shows high tissue-specific expression and shares about 74% amino acid homology [64] with PTBP1 (PTB, HnRNP I), a global repressor of alternative splicing in non-neuronal cells. PTBP1 and PTBP2 display specific non-overlapping expression patterns in the brain; PTBP2 is broadly expressed in the developing mouse brain, including neuronal precursors [65–68], while PTBP1 expression is confined to neuronal precursor cells, glia and other non-neuronal cells [66, 69, 70]. The best known function of the two PTB proteins is the regulation of alternative pre-mRNA splicing patterns, which greatly increase the variety of transcripts indispensable for normal brain development and functioning.
PTBP1 and PTBP2 regulate the synapse formation and maintenance through cross-regulatory network and auto-regulation of expressions [67, 69, 71, 72]. This new genetic regulatory program [65, 73], encompasses three sequential changes in alternative splicing regulation, during which postsynaptic density protein 95 (PSD-95; encoded by DLG4, disc large homolog 4 (Drosophila)), essential for synaptic maturation and plasticity, is post-transcriptionally repressed prenatally [67, 68, 71, 72]. The correct switch from general to neuron specific alternative splicing patterns during neuronal differentiation is mediated by neuron specific miR-124 through down-regulation of PTBP1 mRNA, which causes a decreased level of PTBP1 protein, and a dramatic increase in PTBP2 protein leading to production of neuron specific protein isoforms [70] (Fig. 4).
Through cross-regulatory network, PTBP2 temporarily inhibits the expression of “adult” protein isoforms until neurons have matured [65, 73, 74], demonstrating an essential role in controlling the brain’s early development. These proteins all affect neurite outgrowth, axon guidance, synaptic assembly, and synaptic function; their untimely expression would lead to aberrant neuronal network development [73]. The expression of PTBP2 continues after differentiation, and is present in the brain at moderate levels through adulthood [73] (Fig. 4), but its role in differentiating neurons is not fully understood.
The loss of PTBP2, as demonstrated in PTBP2 null generated mice (Ptbp2−/−), does not greatly affect developmental patterning of CNS. However, post-mitotic neuronal maturation and survival are severely impaired, as a result of misexpression of many protein isoforms affecting neurite growth, synapse formation and synaptic transmission [73]. Similarly, when neurons lacking PTPB2 are grown in culture, they fail to develop correctly and die. Overexpression of PTBP1 and PTBP2 in cultured neurons was shown to repress synaptic activity, spine formation/morphogenesis, and reduce PSD-95 transcript [68, 73]. The study of Li et al. [73] showed that PTBP2 is critical to both embryonic and postnatal brain development.
Constitutional gain of PTPB2 gene in the presented case and the continuous excess of PTBP2 during neuronal development and throughout the life, certainly to some extent deregulate the cross-regulatory network and specific pattern of expression of both PTBP1 and PTBP2, affecting also the alternative splicing of a number of pre-mRNAs.
Therefore, in the case of PTBP1 and PTPB2, one would expect that loss of one homologue and its protein leads to overexpression of the other homologue resulting in its protein abundance, and vice versa. However, Li et al. [73] found no changes in the expression of PTBP1 in neuronal progenitor cells, astrocytes, ependymal cells, or other non-neuronal cell types in the brain of PTBP2 null mice (Ptbp2−/−). In spite of this finding, it is still possible that even a relatively mild, but long lasting disproportion of two PTB proteins may impair cognitive function as a result of the cumulative effect of multiple disordered gene expression patterns during development.
In fact, the response of each of the multiple targets to constitutional copy number gain/loss of the sequence(s) which regulate their functioning might depend on the sensitivity of the target to the environmental disequilibrium, and the ability of regulatory mechanisms to overcome disordered homeostasis in the cellular and intercellular milieu. This assumption is in line with the finding that mice with PTBP2 (Ptbp2−/+) deletion/copy number loss express PTBP2 at levels half that of the wild type animals (Ptbp2+/+) [73]. The effect of this heterozygous loss of synaptic protein expression and on target transcript splicing was variable. For some targets, protein levels in the heterozygous brains were intermediate between the wild type and homozygous knockout (as one would expect in one gene-one protein relationship). In other cases, the heterozygotes appeared similar to the wild-type mice, expressing close to normal protein levels.
The same holds true for miR-137, as both PTBP2 and miR-137 affect the expression and function of multiple target sequences in the genome. It has been shown that miR-137 deletion results in an up-regulation/overexpression of its validated targets [16]. On the other side, bioinformatically predicted miR-137 targets showed a small but significant down-regulation/lower level of expression of the genes involved in neuronal differentiation [75] following miR-137 overexpression.
Therefore, the imbalance of sequences with direct and/or downstream influence on multiple genes might, through the cumulative effect of deregulated distant targets (some of which being up-regulated, other being down-regulated, and some being dosage insensitive), be at least partially responsible for the overlapping behavioral and neurologic phenotypes in a number of reciprocal microdeletions and microduplications.
4.5. Bones and dental anomalies: miR-137
Bone organogenesis is a complex process involving the differentiation and crosstalk of multiple cell types, in which the subset of miRNAs has emerged as an important regulator of bone formation and postnatal functions, contributing to every step of osteogenesis [76–78]. The same holds true for tooth development, as the phenotypes associated with mutations in different genes indicate that integrated networks of signaling pathways are the key regulators of tooth morphogenesis [79, 80]. However, the exact mechanism of the regulatory network governed by miRNAs is still poorly understood.
How do the genes within sSMC(1) fit into teeth and osseous abnormalities seen in the present case, and to a lower extent in 1p21.3 deletions? To the best of our knowledge, no known direct connection between the affected genes and the tooth and bone development exists. However, miR-137 again seems to be a player that affects downstream genes.
Micro RNA miR-137 is one of the miRNAs that regulate EZH2 (enhancer of zeste homolog 2), a catalytic component of Polycomb repressive complex 2 (PRC2), which epigenetically regulates chromatin structure to silence gene expressions [81, 82]. An increasing body of evidence suggests that EZH2 plays a critical role in stem cell maintenance and differentiation into specific cell lineages, including neurogenesis, adipogenesis and osteogenesis [81, 83, 84]. Recent studies report that craniofacial skeleton formation in higher vertebrates is crucially dependent on epigenetic regulation [85], and that the switch between adipogenesis and osteogenesis can be epigenetically regulated by phosphorylation of EZH2, which suppresses PCR2 catalytic activity [81, 86, 87]. In addition, recent exome-sequencing studies identified missense mutations and in-frame deletions of EZH2 in patients with Weaver’s syndrome, an autosomal dominant disease characterized by learning disabilities, dysmorphic facial features and general overgrowth, which can include tall stature, obesity and macrocephaly [88]. Mutations of EZH2 are also reported in a cohort of patients with a nonspecific overgrowth syndrome [89]. Interestingly, the patients with 1p21.3 deletions display (borderline) macrocephaly and a tendency to be overweight [15, 16].
Furthermore, among putative miR-137 targets is the transcription factor Twist-related protein 1 (Twist-1) (TargetScan), which together with Twist-2, regulates bone formation through transient suppression of Runx2 gene essential for osteoblastic differentiation and skeletal morphogenesis in mice [90, 91]. Twist-1 or Twist-2 deficiency leads to premature osteoblast differentiation [90].
Micro RNA miR-137 can also influence tooth development. One of the putative targets of miR-137 is AXIN1, which is the key component of canonical Wnt pathway [92]. Vertebrates have two AXIN homologous genes (AXIN1 and AXIN2) [93] which appear to be functionally equivalent and interchangeable in Wnt pathway [94]. Loss of Axin2 function is linked to carcinogenesis as well as abnormal bone and tooth development, including hypodontia [94, 95]. In addition to AXIN1, three other genes are among putative miR-137 targets involved in dental development according to TargetScan: BCOR (BCL6 Corepressor; transcriptional regulator), BCORL1 (BCL6 Corepressor-Like 1) [96, 97] and PVRL1 (NECTIN1), respectively [98].
Thus, gain or loss of miR-137 function could affect both osteogenesis and dentition. However, it appears that osteogenesis and dentition are more affected in 1p21.3/miR-137 copy number gain, though the influence of other genes cannot be ruled out.
Micro RNA miR-137 is an important player in coordinate and complex regulatory events involving a number of genes. In the context of variable sensitivity of target genes/mRNAs/proteins discussed earlier, it is possible to speculate that constitutional gain of miR-137 function, mimics haploinsufficiency/deletion/copy number loss of dosage sensitive targets leading to disordered homeostasis. Consistent with this is finding of a small but significant down-regulation of miR-137 targets following miR-137 over-expression [75]. On the contrary, a copy number loss of miR-137 can lead to upregulation of target genes [16], mimicking their overexpression/duplication/copy number gain. More severely affected jaw/bones and teeth in the present case might therefore be due to the “deletion effect” of dosage sensitive target genes, as a result of constitutional gain of miR-137.
4.6. Plasticity-related genes LPPR4/PRG1 and LPPR5/PRG5
Lipid phosphate phosphatase-related proteins (LPPRs, LPPR1-5), also referred to as plasticity-related genes (PRGs, PRG1-5), are a novel class of integral membrane proteins differentially expressed in the developing brain and reexpressed in regenerating axons [99–101], which belong to the lipid phosphate phosphatase (LPP) family. LPPs interfere with lipid phosphate signaling through mediating the extracellular concentration and signal transduction of lipid phosphate esters, lysophosphatidic acid (LPA) and spingosine-1 phosphate (S1P). LPPRs are predominantly expressed in the brain and may act by modifying bioactive lipids and their signaling pathways [101]. However, the exact functional role of LPPRs is still not fully elucidated.
Among several genes that are duplicated in our case report, there are two members of plasticity-related gene family LPPR4 and LPPR5 (Fig. 4) important for neuronal outgrowth and synaptic transmission, respectively.
The protein encoded by human lipid phosphate phosphatase-related protein type 4 gene (LPPR4), also known as plasticity-related gene 1 (PRG1) protein, is specifically expressed in pyramidal neurons, in the membranes of outgrowing axons and dendrites, where it hydrolyzes lysophosphatidic acid / lysophosphatidate (LPA). LPPR4 localizes in hippocampal neurons exclusively at excitatory postsynaptic endings of dendritic spines in rats [102] and mice [103]. During development and regenerative sprouting, LPPR4 attenuates phospholipid-induced axon collapse in outgrowing axons, thereby facilitating axonal outgrowth in the hippocampus. LPPR4 is considered a putative regulator of neuronal plasticity.
It has been shown that lack of LPPR4 leads to juvenile epileptic seizures in mice [103], suggesting LPPR4 dose-dependent pathological increase of synaptic transmission (hyperexcitability) in CA1 pyramidal neurons [103]. Mice with LPPR4 deletion/copy number loss (Lppr4+/−) shows LPPR4 expression approximately half that of WT (Lppr4+/+) animals, and exhibits intermediate increase of excitatory synaptic transmission (halfway between Lppr4−/− and Lppr4+/+ mice). Trimbuch et al. [103] concluded that the increase in neuronal excitability is due to the lack of LPPR4 at the postsynaptic side. However, seizures can up-regulate LPPR4 gene and increase LPPR4 protein level in hippocampus and cortex by themselves, suggesting LPPR4 might be detrimental after seizures, contributing to post-seizure cognitive deficits observed in mice and men [100, 104–107].
The importance of LPPR4 for proper synaptic transmission was recently demonstrated by analysis of global gene expression in large groups of patients with refractory mesial temporal lobe epilepsy (RMTLE) [108]. In this study LPPR4 was identified as one of the hub genes, interacting with the number of genes, in both subgroups of patients (with and without the history of childhood febrile seizures) indicating not only importance for the etiogenesis of the seizures but also for the clinical outcome.
LPPR5/PRG5, novel LPPR with a high homology with PRG3/LPPR1 is exclusively expressed in nervous system (Fig. 4). As shown by Broggini et al. [109], LPPR5 induces filopodia formation and axon elongation in primary cortical neurons in vitro. Overexpression of LPPR5 induced morphological changes in both non-neuronal cells and neurons. It has been suggested that LPPR5 is involved in axonal fine-tuning and in the final development of neuronal circuitry.
In summary, even though the exact molecular role of these genes is not fully elucidated, we can speculate, in line with the hypothesis of Ramocki and Zoghbi [17], that the imbalance in their expression level disturbs the fine balance which is necessary for axon fine tuning of neuronal circuits and neural connectivity, thus leading to abnormal neural transmission that could contribute to described neurological phenotype.
4.7. Sortin nexin 7 (SNX7)
Sorting nexin 7 (SNX7) belongs to a large family of proteins involved in intracellular trafficking. Its exact function is unknown and apart from a single study on zebrafish [110], no report on SNX7 in rodents/humans is cited in PubMed. The mammalian sorting nexin subgroup of 12 genes coding for SNX–BAR proteins (SNX1, SNX2, SNX4–SNX9, SNX18, SNX32 and SNX33) is characterized by two membrane-binding domains: a phosphoinositide-binding Phox homology (PX) domain and a membrane curvature sensing BAR (for Bin–Amphiphysin–Rvs) domain [111–113]. Several SNX–BAR proteins can elicit vesicle-to-tubule transitions in vitro and in vivo, implicating SNX–BAR proteins as key regulators of tubular-based endosomal sorting [111, 114, 115], an essential process for maintaining cellular homeostasis, with deregulated sorting underlying a variety of pathologies [116, 117].
The contribution of SNX family members to neuronal functioning or disease is poorly understood. However, there are indications that SNXs are disrupted in patients with microcephaly [118], ID [118], and Down syndrome [119]; a link to bipolar disorder [120] and 6q14 microdeletion syndrome [121] has been suggested as well. A very recent finding that sorting nexin14 (Snx14) is imprinted in postnatal mouse visual cortical neurons shed new light on imprinting [122]. Imprinted genes can be regulated in specific cell types and developmental stages [123] which make their identification and validation difficult. To overcome the limitation in identifying new neuron-specific imprinted genes, Huang et al. [122] successfully modified previously employed approaches [124, 125]. SNX14 protein levels increase during mouse brain development exhibiting predominant expression during brain development and maturation; starting in the early mid-fetal period, SNX14 is expressed moderately throughout lifetime in the human brain (Fig. 4). SNX14 localizes to the cytoplasm and dendrites of dissociated mouse cortical neurons where it regulates neuronal intrinsic excitability and promotes synaptic transmission [122].
SNX7 is a putative target of a single broadly conserved microRNA, miR-9 (TargetScan), one of the most highly expressed microRNAs in the developing and adult vertebrate brain [reviewed in 126]. Functional analyses have revealed miR-9 as a versatile regulator of neurogenesis, which together with miR-124, appears to be the core genetic circuit regulating mitotic exit of neural progenitors and the onset of neuronal differentiation [127]. Recent studies link miR-9 with a number of neurodegenerative disorders [128]. The spatiotemporal expression pattern of SNX7 is similar to that of miR-9-2 throughout the life span; the exception is perinatal downregulation of SNX7 in cerebellum and striatum suggesting the possibility of being silenced by miR-9 (Fig. 4).
4.8. Dihydropyrimidine dehydrogenase (DPYD) gene
4.8.1. DPYD: the clinics
The DPYD (dihydropyrimidine dehydrogenase) gene encodes an enzyme (DPD), the initial and rate-limiting factor in the pathway of pyrimidine catabolism, also a key enzyme in the degradation of chemotherapeutic drug 5-fluorouracil (5-FU). Mutations in this gene result in a pharmacogenetic disorder, namely dihydropyrimidine dehydrogenase deficiency, showing large phenotypic variability and ranging from no symptoms to a convulsive disorder with motor and mental retardation in homozygous patients. These individuals also have an increased risk to develop potential life-threatening toxicity to 5-fluorouracil (5-FU) [129, reviewed in 130]. However, it still remains unclear how the excess of uracil and thymine relates to the specific neurological problems that affect some of the people with dihydropyrimidine dehydrogenase deficiency.
Although more than 50 mutations have been characterized in DPYD gene, the majority of them represent variants with unknown biological and clinical significance [131–133]. A splice-site mutation in intron 14 (c.1905+1G>A, IVS14+1G>A, DPYD*2A, rs3918290) as the most prevalent [129], together with two nonsynonymous coding variants [130, 133–136], is the only known functional variant significantly associated with 5-FU-related high-grade (III/IV) toxicity, as shown by case-control studies [137, 138]. Recent comprehensive sequencing of the DPYD, as well as the haplotype-based analyses, revealed deep intronic variants of DPYD gene in patients with severe adverse effects [139, 140]. However, in a significant number of patients with reduced DPD activity, no mutations could be identified in the coding part of DPYD [141, 142]. On the other side, the finding that a DPYD haplotype free of any mutations was associated with 5-FU toxicity, suggested the presence of additional genetic variations in the noncoding region of DPYD [139] and a different underlying mechanism of toxicity. In addition, only 50% of heterozygous carriers of deleterious risk DPYD variants develop 5-FU toxicity. Since the reported genetic variants do not account for most DPD deficiency cases, the epigenetic regulation of DPYD promoter has been suggested as a potential important mechanism in 5-FU toxicity [143]. However, no firm evidence for DPYD promoter hypermethylation has been found so far to corroborate such a premise [144, 145].
Recently, 1p21.3 microdeletion syndrome has been recognized [15, 16], pinpointing miRNA-137 and/or DPYD as underlying causes for the neurological and behavioral phenotype in the affected patients. Here we would like to accentuate the molecular organization of the DPYD gene, and indicate the way the gene is regulated.
Is there a way to explain the obvious similarity in the face expression between the patient with 10 kb deletion of DPYD gene [15], and our patient with 3.56 Mb duplication of 1p21.3p21.2 (Fig. 1D–F)? There is also a partial overlap of phenotypic features seen in 1p21.3 copy number loss and present 1p21.3p21.2 copy number gain, with phenotypic features seen in the severely affected patient having about 14 Mb deletion [146] (Fig. 5).
One explanation could be that it is just a coincidence. If not, does miR-137 or DPYD deregulation affect the chromatin conformation thus influencing the expression of neighboring genes? Is miR-137 or DPYD responsible for the phenotypic resemblance? The regulation of DPYD by miR-137 seems unlikely, since DPYD-001 (NM_000110), the protein coding transcript encompassing full DPYD genomic sequence, is not a putative target of any of the broadly conservative miRNAs, and displays no site with higher probability of preferential conservation, thus implicating its functioning is regulated otherwise. In addition, miRNAs preferentially act through distant downstream targets. However, DPYD-002 (NM_001160301) could be the target of miR-137, as it has an overlapping site with lower probability of preferential conservation, for miR-137/137ab and miR-25/32/92abc/363/363-3p/367, respectively (TargetScan).
4.8.2. DPYD: molecular organization
In accordance with the GRCh38/hg38 annotation, the main known components of DPYD are (Figs. 6–7 and Supplementary Fig. 1):
Figure 6.
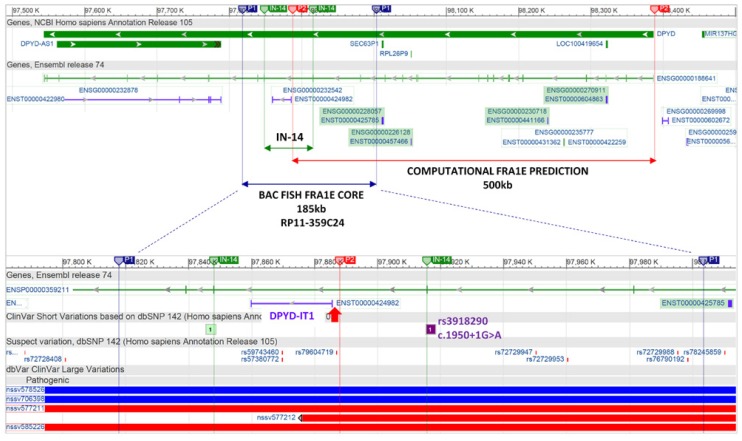
Computational and by BAC FISH predicted coordinates of FRA1E fragile CFS site. FISH predicted FRA1E 185 kb core [195] compared with a computational predicted FRA1E [196]. Note the overlaps between 5′-end of DPYD-IT1 and telomeric border of computational FRA1E prediction marked by red arrow, and between 5′-end of intron 14 and pathogenic splice variant rs391890/c.1905+1G>A, respectively. Coordinates are in GRCh37/hg19 annotation (The NCBI38 has no more capacity to outline the Ensembl annotated genes). P1 stands for BAC FISH predicted core of FRA1E; P2 stands for computational prediction of FRA1E.
Figure 7.
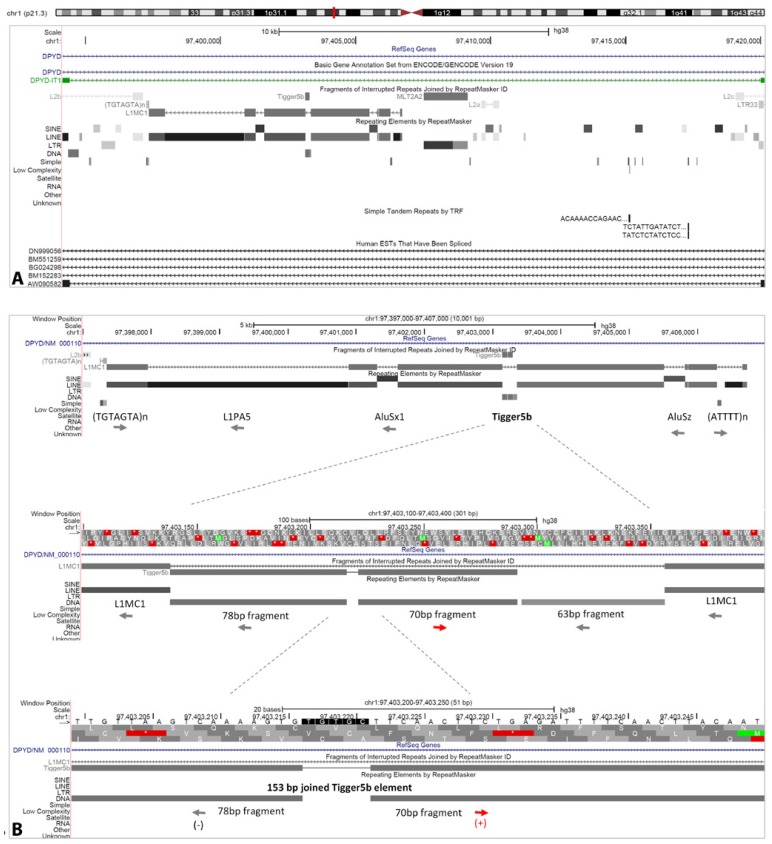
DPYD-IT1 sense intronic lncRNA gene. Overview of repetitive elements comprised in DPYD-IT1 gene (A). The predicted “weakest link”/center of FRA1E: L1MC1 transposon (chr1: 97,397,345–97,406,722 [hg38]). Note Tigger5b DNA element fragments inserted on different genomic strands (B). *GRCh38/hg38 is applied to present the repetitive elements within DPYD-IT1 since it differs from the older version, GRCh37/hg19.
Protein coding sequence which span over entire reverse (−) 843,317 bp long strand of DPYD gene, comprised of 23 exons and processed in four transcripts (protein coding DPYD-001 −DPYD-003 and retained intron DPYD-004).
Two known long non-coding natural antisense RNA genes (NATs), DPYD-AS1 (227 kb, processed in one transcript DPYD-AS1-001 with 5 exons), and DPYD-AS2 (1.15 kb, processed in two transcripts DPYD-AS2-001 with 2 exons, and DPYD-AS-002 with 3 exons); both NATs lay on the forward (+) strand of the DPYD gene.
The fourth gene is a novel sense intronic lncRNA DPYD-IT1 gene (DPYD intronic transcript1; Gene Symbol: RP11-359C24.1; manually annotated by Havana project - Vega 39 Annotations OTTHUMT00000095693, ENSG00000232878). DPYD-IT1 span over 26 kb (chr1: 97,394,154-97,420,141 [hg38]/chr1: 97,859,710-97,885,697 [hg19]) within intron 14 (chr1: 97,450,058 97,382,462 [hg38]/chr1: 97,915,614-97,848,018 [hg19]) of reverse (−) DPYD strand. DPYD-IT1 is comprised of two exons and one long intron, and processed in a 401bp long transcript product (DPYD-IT1-001).
In addition, in NCBI Annotation Release 106, annotated is a new XR_426733.1/LOC102723700/NC_018912.2, 7.7 kb long NAT to the forward strand of DPYD gene, comprised from 3 exons. In GRCh38 Ensembl genebuild, 9 novel EST protein coding transcripts have been annotated; 6 to the forward strand (ENSESTT00000033931–ENSESTT00000033936), and 3 to the reverse strand (ENSESTT00000033938–ENSESTT00000033940). Interestingly, the 5′-end of 352 kb long ENSESTT00000033940 transcript overlaps with 5′-end of intron 14, and 3′-end overlaps more or less with both, 3′-end of DPYD-001 transcript and 5′-end of DPYD-AS1.
4.8.3. Natural antisense RNA transcripts (NATs) and intronic lncRNA involved in the regulation of DPYD
Non-coding RNAs involved in the molecular organization of DPYD locus undoubtedly indicate a complex and multi-layered regulation of the DPYD gene. In addition to the two known NATs, DPYD-AS1 and DPYD-AS2, novel DPYD-IT1 intronic lncRNA gene and NAT XR_426733.1/LOC102723700/ NC_018912.2, are annotated within the DPYD locus.
Characterization of complex mechanisms that regulate DPYD expression is a valuable effort, since the gene expressions regulated by antisense lncRNAs open the possibilities to reverse the process [147–149], thus offering a completely new approach in treating the disease. Understanding the mechanism by which lncRNAs regulate DPYD functioning, will be a step forward in understanding the biological significance of mutations within the gene, which will consequently lead to finding the way to cure the dihydropyrimidine dehydrogenase deficiency and preventing the cytotoxicity of 5-FU.
The following lines are concise notes from the recent work that has been done on NATs and intronic lncRNAs in the regulation of gene functioning.
Natural antisense RNA transcripts (NATs) are lncRNAs which are transcribed from the opposite strand of protein-coding genes [NATs and other lncRNAs are reviewed in 150–153]. It is predicted that up to 70% of protein coding genes in humans are bidirectionally transcribed [154–157]. The primary antisense transcript mRNAs share complementary exons with the related sense transcript, but the degree of complementarity of NATs with corresponding sense transcripts varies greatly [158–160]. Recent studies have shown that antisense RNAs usually regulate complementary sense mRNA by modulating chromatin structure in cis, thereby acting as epigenetic regulators of gene expressions and chromatin remodeling.
Many NATs display opposite/reverse expression patterns with their sense transcript counterparts, implying that they carry the potential to induce allele-specific gene silencing [161, 162]. Actually, the occurrence of NATs correlates with genes that show monoallelic expression [163].
Recent studies have also shown that NATs work in association with chromatin modifiers, mediating their function through transcriptional and epigenetic regulation, RNA-DNA and RNA-RNA interactions, respectively [150, 151]. The extent of the spread of epigenetic silencing may be related to the CTCF binding factor [164], a multifunctional protein that enables and facilitates higher-order chromatin interactions [165].
An interesting example of transcriptional repression by NATs is INK4b/ARF/INK4a locus regulated by ANRIL NAT, where the NAT ANRIL participates in the silencing of two very important tumor suppressor genes via two distinct mechanisms. The alteration of these regulatory circuits has been found in different types of cancers [166–168].
NATs are located within many imprinted loci [169–174] and may be directly involved in modulating gene expression within the imprinted cluster. The classic example is the Angelman syndrome gene, UBE3A, which is subject to genomic imprinting but not by differential DNA methylation at the promoter region [175]. Instead, UBE3A is regulated by its antisense NAT UBE3A-ATS in cis, which is expressed from the paternally inherited chromosome in the brain and is also imprinted [147, 176].
Although up to 80% of protein coding genes have transcriptionally active introns containing intronic lncRNA genes, little is known about their function [177–179]. Intronic ncRNAs are predominantly associated with the sense strand of the unprocessed mRNA, which is also the case with the DPYD-IT1. However, intronic lncRNAs often show expression patterns which are opposite to the processed mRNA [156, 180–182]. This suggests a complex regulatory relationship in which intronic lncRNA transcription is independent from the transcription of protein coding pre-mRNA [177–179]. Intronic lncRNAs may be transcribed from either the sense or antisense strand of the protein-coding gene in which they are encoded [183–185]. Recent work also indicates that many intron-derived RNAs, like many other lncRNAs, function through recruitment of the Polycomb repressive complex 2 (PRC2), leading to subsequent transcriptional repression [186, 187]. Interestingly, DPYD-IT1 is located within the intron 14, which is the major locus responsible for dihydropyrimidine dehydrogenase deficiency. Namely, the most prominent mutation of the DPYD gene that results in severe DPD deficiency is the G to A mutation in the GT 5′-splice recognition site of intron 14 (exon 14-skipping mutation leading to exon 14 deletion). The corresponding mRNA exhibits 165 bp deletion and the enzymatic activity of the translated DPD protein is virtually absent [188].
Another class of lncRNAs, long intergenic ncRNAs (lincRNAs), carries out its regulatory role in trans, affecting chromatin conformation and gene expression at distant loci. Transcription from such an upstream promoter can negatively or positively affect the expression of a downstream gene. For example, HOTAIR is a lincRNA transcribed from the HOXC locus that recruits the chromatin remodeling complex, PRC2, to the HOXD locus where it creates a repressive chromatin conformation across 40 kb of the locus [189]. Therefore, the epigenetic deregulation of DPYD gene may potentially affect neighboring genes underlying the overlapping symptoms present in both the copy number loss and copy number gain of 1p21.
In general, the major role of lncRNAs appears to be the modulation of the epigenetic status of proximal and distal protein-coding genes through cis- and trans-acting mechanisms regulating chromatin structure over a single gene promoter, a gene cluster, or an entire chromosome [190–194].
All aforementioned possibilities are open, including the one where the DPYD and miR-137 expressions may be mutually affected by one of lincRNAs (ENST00000602672.1/RP11-272L13.3, ENST00000561881.1/RP11-490G2.2, MIR-137HG) near the 5′-end of DPYD, acting in trans, as all sequences lie on the same, reverse genomic strand and display the same transcriptional direction.
4.8.4. DPYD: common fragile FRA1E site
By partially overlapping bacterial artificial chromosome (BAC) and P1-derived artificial chromosome (PAC) clones, Hormozian et al. [195] determined that the FRA1E common fragile site extends over 370 kb of genomic region of 1p21 laying within DPYD (from intron 8–18). They estimated that the 185 kb region (BAC clone RP11-359C24) of the highest fragility, which accounts for 86% of all observed breaks at FRA1E, encompasses the central part of DPYD, including exons 13–16.
In a genome-wide analysis of common fragile sites (CFS), Fungtammasan et al. [196] computationally predicted the coordinates of 18 CFS including FRA1E, which was found to span over 500 kb of the 1p21.3 genomic region. The computationally defined FRA1E site was among four out of 18 analyzed CFSs whose coordinates did not overlap with cytogenetically defined coordinates [197, 198]. Discordant results may be explained by both cytogenetic banding and fluorescent mapping methods (on which the multiple standard regression model was derived and estimated), which have inherent technical limitations that contribute to variation among coordinates [196].
In order to correlate both FRA1E range predictions, we have converted the computationally revealed coordinates to hg 19 (chr1: 97,887,980–98,387,979) and retrieved the coordinates for the BAC clone RP11-359C24 (chr1: 97,817,963–98,003,396 [hg19]) from UCSC Browser (http://genome.ucsc.edu/). All the data were loaded in Genome Browsers UCSC, Ensembl (www.ensembl.org) and NCBI Variation viewer (www.ncbi.nlm.nih.gov/variation/view/), respectively. Two unexpected findings became apparent, first being the complexity of DPYD molecular structure discussed above, and second, the overlap of 5′-end of intronic lncRNA DPYD-IT1 gene with the telomeric border of computationally predicted FRA1E fragile site, that prompted us to continue with the analysis (Fig. 6).
The computer search involving almost the whole 843 kb DPYD sequence, both NATs, intron 14 and DPYD-IT1 (retrieved from NCBI GeneBank and/or Ensembl Browser), for specific DNA repeats and secondary structures that can inhibit replication fork progression [197, 199–202], led us to a conclusion that no definite deduction may be drawn, particularly while current evidence suggests that CFSs are caused by an interplay of multiple genomic factors [203–205].
Nevertheless, questions may be raised whether the FRA1E fragile site is a CFS, or where the cluster(s) of loci liable to break precisely map. The most intriguing question is: if such site exists, where the “weakest link”/center of the FRA1E maps? It is our belief that the precise characterization of such particular site is as important, as revealing the mechanisms of DPYD gene transcriptional regulation; whether DPYD is entirely or partly imprinted in the tissue specific manner, or only transiently epigenetically regulated, and in which circumstances.
The immediate focus was on a splice-site, exon skipping mutation at 5′-end of intron 14. Subsequently, other variants also proven to be pathological should be taken into consideration. In such cases only 3–5% [206] of the overall population with true DPD deficiency and additional 2%–3% [140] of the population with partial DPD deficiency due to sequence variation would display the “FRA1E” fragile site; these assumptions may be easily verified in case controlled studies. In favor of such assumption speaks the fact that FRA1E belongs to the group of CFSs with lower expression than FRA3B or FRA16D [197], and that many sequence motifs spread throughout the aCFS region may contribute to fragility [204, 207, 208].
Given that the core of the fragile FRA1E site entails a 185 kb genomic region (RP11-395C24) between intron 12 and intron 16, the intron 14 is again the most suspected region since the computationally defined telomeric border of FRA1E region falls only 2.3 kb upstream from the 5′-end of the DPYD-IT1 intronic lncRNA gene. The 5′-end of DPYD-IT1 maps at chr1: 97,885,697 while telomeric border of computationally predicted FRA1E maps at chr1: 97,887,980 [hg19] (Fig. 6).
We believe that the site most liable to display a breakage, sort of the “weakest link”, maps within L1MC1 transposable autonomous long interspersed nuclear element-1 (LINE-1, L1) element, inserted in DPYD intronic transcript 1 (DPYD-IT1) intronic lncRNA gene (Fig. 7A–B). More precisely, the manner in which the Tigger5b DNA element within L1MC1 is incorporated in the host DNA might be the true “weakest link” within the DPYD gene.
LINE-1 transposons
Transposable elements (TEs) are mobile repetitive sequences that make up at least 45% of the human genome [209]. TEs are classified based upon their method of transposition. Class 1 elements transpose via an RNA intermediate through copy-and-paste fashion using reverse transcriptase and include long and short interspersed nuclear elements (LINEs and SINEs), as well as long terminal repeat elements (LTR). Class 2 elements, or DNA transposons, transpose via a DNA intermediate through a cut-and-paste mechanism [210, reviewed in 211].
Although positive contributions of mobile elements to their host genomes are reported, there is growing evidence of the role of TEs in human disease and genetic instability [reviewed in 212, 213]. The expression of the L1 retrotransposon can damage the genome through insertional mutagenesis, rearrangements generated by non-allelic homologous recombination (NAHR), and the generation of DNA double-strand breaks (DSBs) [214–220].
In order to show the importance of the L1MC1 retrotransposon, with its parasitic Tigger5b and pair of Alu elements, in the putative regulation of the DPYD gene, the following lines are based (citations) on the papers by Belancio et al. [212], Kines et al. [220], Kines and Belancio [221], Belancio et al. [222], Belancio et al. [223], and Wallace et al. [224]; for comprehensive information see the original papers.
The most active autonomous non-LTR element is long interspersed nuclear element-1, LINE-1 (L1), which contains a number of highly successful parasitic elements [225, 226]. Transcription of a L1 generates a retrotranspositionally competent, full-length L1 mRNA [214, 227] and a spectrum of processed L1-related RNA products, the majority of which are not capable of retrotransposition [228, 229].
The full-length L1 mRNA is bicistronic and is influenced by the upstream genomic sequence expression [230]. The functional structure of L1 element includes promoters, 5′ and 3′ UTRs, two open reading frames (ORF1 and ORF2) required for L1 retrotransposition [231], and cis-acting signals for mRNA processing, with RNA polymerase II (pol II) promoter (sense promoter) located in the beginning of the 5′-untranslated region (UTR) [231, 232]. The antisense L1 promoter, also present within the 5′UTR, is demonstrated to drive expression of sequences located upstream of the L1 elements [233, 234]. The biological significance of the antisense promoter is not well established. One of the hypotheses is that its role is to interfere with the transcription initiated within upstream sequences to secure transcription from the sense L1 promoter. Alternatively, the L1 antisense promoter is implicated in the production of small interfering RNAs (siRNAs) that inhibit L1 expression [235, 236]. Both of these promoters can modify the normal gene expression. Independent of the orientation of the L1 insert (forward or reverse relative to gene expression) they have the potential for “gene breaking” by generating 5′-truncated genomic transcripts [237].
The L1 promoter activity is regulated by epigenetic modifications [238, 239]. The short-and long-term consequences of L1 integration (particularly the full-length elements) within or in the vicinity of genes on the epigenetic state and chromatin signature of the gene are not known. Some of the hypotheses dealing with potential contribution of TEs to the epigenetic regulation of the mammalian genome were recently reviewed [240]. L1 elements have been proposed to potentially influence the selective expression of monoallelically expressed genes due to the enrichment of evolutionarily more recent LINE-1 elements in the regions surrounding these genes in human and mouse [241]. Furthermore, L1 promoters contain binding sites for various transcription factors and regulatory proteins that can alter the gene expression in response to various stimuli [242–245]. L1 sequences can exert their influence on the host gene expression by altering the promoter specificity or strength [246–250].
Furthermore, both sense and antisense L1 promoters are reported to exhibit tissue-specificity [245, 251]. While no biological significance has been reported to date for the majority of the known L1/host gene chimeric mRNAs, cancer-specific L1-driven hybrid transcripts were identified in breast and colon cancer cell lines [252].
However, the majority of L1 loci in the human genome are truncated and incapable of retrotransposition. Although thousands of full-length L1 loci remain, most are retrotranspositionally-incompetent due to inactivating mutations. However, some of these retrotranspositionally-incompetent L1 loci previously considered to be inactive and harmless, are indeed expressed [253]. The mutations leading to premature stop codons within the L1 ORF2 sequence may yield truncated proteins that retain a functional endonuclease domain with the potential to generate low levels of chronic genomic instability by introducing double strand breaks (DSBs) and mobilizing Alu sequences [220].
L1 causes insertional mutagenesis through either self retrotransposition or through the mobilization of parasitic non-autonomous transposons, such as Alu elements, which rely on the L1-encoded ORF2 protein for their propagation [226, 254]. Both L1-driven transpositions and L1-induced DSBs depend on the endonuclease activity of the L1 ORF2 protein, which initiates the integration process by nicking the host DNA [219]. Although the origin of the second-strand nick required for completion of the retrotransposition process is unknown, it has been established that expression of the L1 ORF2 protein containing a functional endonuclease domain results in the formation of DSBs [219, 222, 223, 255]. Importantly, it is estimated that L1-induced DSBs are much more frequent than successful L1-retrotransposition events [219]. Though the specific consequences of L1-induced DSBs are not yet fully known, high mutagenic potential of DSBs in mammalian cells is well documented, contributing to genomic instability and cancer progression [219, reviewed in 256–258].
Ongoing endogenous low-level L1 activity has been detected in the germ line, as well as in normal human tissues and adult stem cells [259, 260]. Moreover, the L1 expression is significantly elevated in most human cancers when compared to matched normal tissues [259–264], suggesting a role for L1 as an endogenous mutagen in somatic tissues.
L1 elements, particularly full-length L1s inserted into introns in the forward orientation (like L1MC1 in the intron 14 of DPYD), are poorly tolerated [265, 266] and as a result are significantly underrepresented not only within genes, but also in the 5 kb regions flanking human gene boundaries [210, 221, 267].
Insertions of TE within intronic sequences can interfere with normal gene expression through the introduction of functional (i) promoters and their regulatory elements, (ii) polyadenylation (pA) signals, and (iii) splice donor (SD) and acceptor (SA) sites. Besides the effect of TEs on the expression or function of a single gene through direct insertional interference, some TE integration events can also alter gene or cellular pathway function through indirect mechanisms such as regulation of miRNA expression [221].
DNA transposons
The human genome contains about seven major classes of DNA transposons that are virtually all no longer active, present mostly as fragmented elements in the human genome, and therefore regarded as DNA fossils. Information on human DNA transposons is currently very scarce. However, quite a few functional human genes seem to have originated from DNA transposons, such as genes encoding the RAG1 and RAG2 recombinases and the major centromere-binding protein CENPB. DNA transposons make up 3% of our genome [210]. Two super families, hAT and Tc1/mariner, are predominant in the human DNA transposon population [210, 268, 269].
All complete and autonomous class 2 TEs encode the protein transposases, which are required for insertion and excision. DNA transposons always move on their own, inserting or excising themselves from the genome by means of cut-and-paste mechanism. DNA transposons may also be fully or incompletely inserted into other elements. It has been found that a number of DNA element families, that had copies inserted and integrated into primate-specific L1 elements, also comprise copies nested into dimeric Alu elements, all of which are known to be primate specific [270]; the cohabitation of L1MC1, Tigger5b and a pair of Alu elements comprised in the DPYD-IT1 gene is an example to this.
DNA transposons have been frequently implicated in chromosomal rearrangements, including deletions, inversions, duplications, translocations, and chromosome breakage mediated by recombination or aberrant transposition events [271–274]. In this context, the recurrent partial HUWE1 copy number gain which underlies nonsyndromic ID was recently shown to be caused also by NAHR between two adjacent DNA TcMAR-Tigger2 elements demonstrating that the Xp11.22 region is prone to recombination and replication-based rearrangements [275].
The cut-and-paste transposition mechanism of class II DNA transposons is catalyzed by several transposase enzymes, some of which non-specifically bind to any target site in DNA, whereas others bind to specific DNA sequence targets. The transposase cuts out the DNA transposon (which is then ligated into a new target site) by making a staggered cut at the target site, resulting in single-strand 5′ or 3′ DNA and in most cases in palindromic overhangs or sticky ends. Since the overhangs have to be complementary in order for the ligase to work, the two molecules can only join in one orientation. A transposon or a retroposon that inserts itself into a functional gene will most likely disable that gene, and after a DNA transposon leaves a gene, the resulting gap will probably not be repaired correctly [212].
TEs are also a widely used tool for insertional mutagenesis. Namely, the TE can disrupt the gene’s function in a reversible manner, and after the transposase-mediated excision of the DNA transposon, the gene function may be restored. The first synthetic transposon designed for use in vertebrate cells, the Sleeping Beauty transposon system, is a Tc1/mariner-like transposon. It exists in the human genome as an intron and was activated through reconstruction [276]. The Tc1/mariner-class of TEs Sleeping Beauty transposon system, the Molecule of the Year 2009 [277] is an example of a transposon system that can be adapted for human gene therapy [278–281] and has been extensively used for identifying cancer genes [282–284].
The Tigger5b DNA nested in L1MC1 element belongs to Tc1/mariner superfamily and a family of autonomous DNA transposons and is, like L1 and Alu elements, primate specific [reviewed in 211].
DPYD-IT1: L1MC1, Tigger5b and Alu elements
L1MC1 appears to be a fairly long (9.4 kb) full length L1 LINE element, nested into the genomic DNA, and could therefore be among about a hundred L1 transposons still not dormant in the human genome. The status of L1MC1 activity may depend on the epigenetic (de)regulation of the DPYD, but it could also be the part of epigenetic machinery controlling the DPYD function since it resides within an intronic lncRNA gene. Though probably inactivated epigenetically or by acquired mutation, the environmental stress could potentially awake L1MC1 element, since some TEs contain heat-shock like promoters and their rate of transposition increases if the cell is subjected to stress [285].
Moreover, the whole structure of L1MC1 with nested Tigger5b fragments and pair of Alu repeats (Fig. 7A–B), offers more than one scenario with detrimental outcome for the host DNA, leading to (irreparable) breaks in case of additional mutations and/or environmental stress. In addition, the organization of L1MC1 element annotated in GRCh37/hg19 differs from the annotation in GRCh38/hg38, in particular concerning the Tigger5b element, suggesting that the whole structure is still awaiting its definite characterization.
The Repeat Masker Track (UCSC Browser, hg38) displays three small fragments of Tigger5b DNA transposon: centromeric and telomeric inserted in the reverse/sense strand, while the middle one inserted in the forward/antisense strand, respectively. The fragments are joined in the Interrupted Repeats track in a 153 bp long Tigger5b element, composed of two fragments each lying on the different strand (−,+). In between two fragments is a tgtgc string of 5 bases (Fig. 7A–B).
The manner in which the Tigger5b element is partly inserted in the sense intronic RNA gene, and partly in the antisense DNA strand, suggests an unstable cohabitation between the participants; the situation which potentially may cause problems in the replication processing additionally hampered by agents interfering with DNA transcription, leading to break(s) with sticky ends or overhang which postreplication machinery is unable to repair correctly, ending in the damaged DPYD gene. In such circumstances even the whole Trigger5b element may be cut out from the genomic sequence with unforeseeable consequences.
The Tigger5b is inserted within two L1MC1 fragments flanked by a full-length Alu parasitic TE (lacking ORFs), telomeric AluSz (310 bp) and centromeric AluSx1 (307bp), respectively (Fig. 7A–B). Alu elements are the most abundant of all mobile elements in the human genome [210, 286, reviewed in 221] and as such they may act as a template for homologous recombination [287–289]. However, various inherited disorders have been caused by Alu-mediated recombination including several types of cancer [287]. Overall, ~0.3% of all human genetic diseases seems to be the result of an Alu-mediated unequal homologous recombination. There is also evidence that Alu elements inserted into an inverted orientation are more prone to illegitimate recombination [290–293].
Previous discussion on TEs suggests several possibilities how L1MC1 element with its parasites may potentially affect the functioning of DPYD. In addition, the L1MC1 element seems to be DPYD specific. Searching through clinically important aphidicolin-induced CFS associated genes in the RepeatMasker (UCSC Browser), including PARK2 (MIM 602544)), DMD (MIM 300377), FHIT (MIM 601153), WWOX (MIM 605131), GRID2 (MIM 602368), LARGE (MIM 603590), CTNNA3 (MIM 607667), and CNTNAP2 (MIM 604569), no identical L1 TE element could be found.
5. Conclusion
Even a superficial attempt to comment on the functioning of coding and non-coding sequences comprised in sSMC(1) revealed a complex, and incompletely comprehensible interaction between them, as well as an intricate involvement of these sequences in other cellular and intercellular programs.
Mutation analyses in human disorders have so far been mostly restricted to coding genes. The link between ncRNAs deregulation and neurological diseases is just beginning to emerge. The evolving whole-genome sequencing projects complemented by the arrayCGH studies will undoubtedly reveal that the pathology of neurodevelopmental disorders is indeed affected by mutations and ncRNA copy number gains/losses, thus opening a new avenue for studying the genotype-phenotype correlations in genetic disorders, necessary for a comprehensive understanding of human disease.
Supplementary material
(A–B). Molecular organization of DPYD gene according to GRC38/hg38 Genome Annotation. A) NCBI Annotation Release 106 August 2014. Note the new lncRNA XR_426733.1/LOC102723700/ NC_018912.2; B) GRCh38 Ensembl Release August 2014. Note new 9 EST protein coding transcripts colored in purple. Fig. 1A and 2B are not drawn to scale.
Acknowledgments
We would like to thank Goran Šimić for critical assessments and editing the manuscript, and Zdenka Andrijić for the support during the writing process.
Supported in parts by the Croatian Ministry of Science Education and Sport, project No. 108-1081870-1888 (to LB); Business Innovation Croatian Agency – Croatian Institute for Technology BICRO-HIT, project: Targeted aCGH microarrays for diagnostics of neurodevelopmental diseases (to FB); IBRO RHP (to ŽK); and DAAD and Else Kröner Fresenius Stiftung (2011_A42) (to ABH). Conflict of interest statement: The authors of this work declare that they have no competing interests.
References
- 1.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liehr T, Heller A, Starke H, Rubtsov N, Trifonov V, Mrasek K, et al. Microdissection based high resolution multicolor banding for all 24 human chromosomes. Int J Mol Med. 2002;9:335–339. [PubMed] [Google Scholar]
- 3.Melo JB, Backx L, Vermeesch JR, Santos HG, Sousa AC, Kosyakova N, et al. Chromosome 5 derived small supernumerary marker: towards a genotype/phenotype correlation of proximal chromosome 5 imbalances. J App Genet. 2011;52:193–200. doi: 10.1007/s13353-011-0035-3. [DOI] [PubMed] [Google Scholar]
- 4.Liehr T, Weise A. Frequency of small supernumerary marker chromosomes in prenatal, newborn, developmentally retarded and infertility diagnostics. Int J Mol Med. 2007;19:719–731. [PubMed] [Google Scholar]
- 5.Liehr T, Claussen U, Starke H. Small supernumerary marker chromosomes (sSMC) in humans. Cytogenet Genome Res. 2004;107:55–67. doi: 10.1159/000079572. [DOI] [PubMed] [Google Scholar]
- 6.Choo KH. Centromere DNA dynamics: latent centromeres and neocentromere formation. Am J Hum Genet. 1997;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liehr T, Utine GE, Trautmann U, Rauch A, Kuechler A, Pietrzak J, et al. Neocentric small supernumerary marker chromosomes (sSMC) - three more cases and review of the literature. Cytogenet Genome Res. 2007;118:31–37. doi: 10.1159/000106438. [DOI] [PubMed] [Google Scholar]
- 8.Klein E, Rocchi M, Ovens-Raeder A, Kosyakova N, Weise A, Ziegler M, et al. Five novel locations of neocentromeres in human: 18q22.1, Xq27.1~27.2, acro p13, acro p12, and heterochromatin of unknown origin. Cytogenet Genome Res. 2012;136:163–166. doi: 10.1159/000336648. [DOI] [PubMed] [Google Scholar]
- 9.Mantzouratou A, Mania A, Apergi M, Laver S, Serhal P, Delhanty J. Meiotic and mitotic behaviour of a ring/deleted chromosome 22 in human embryos determined by preimplantation genetic diagnosis for a maternal carrier. Mol Cytogenet. 2009;2:3. doi: 10.1186/1755-8166-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liehr T. Small supernumerary marker chromosomes. 2014. [accessed December 7, 2015]. http://ssmc-tl.com/sSMC.html.
- 11.Yeung A, Francis D, Giouzeppos O, Amor DJ. Pallister-Killian syndrome caused by mosaicism for a supernumerary ring chromosome 12p. Am J Med Genet. 2009;149A:505–509. doi: 10.1002/ajmg.a.32664. [DOI] [PubMed] [Google Scholar]
- 12.Fickelscher I, Starke H, Schulze E, Ernst G, Kosyakova N, Mkrtchyan H, et al. A further case with a small supernumerary marker chromosome (sSMC) derived from chromosome 1-evidence for high variability in mosaicism in different tissues of sSMC carriers. Prenatal Diag. 2007;27:783–785. doi: 10.1002/pd.1776. [DOI] [PubMed] [Google Scholar]
- 13.Piccione M, Antona V, Antona R, Gambino G, Pierluigi M, Malacarne M, et al. Array-CGH defined chromosome 1p duplication in a patient with autism spectrum disorder, mild mental deficiency, and minor dysmorphic features. Am J Med Genet A. 2010;152A:486–489. doi: 10.1002/ajmg.a.33212. [DOI] [PubMed] [Google Scholar]
- 14.Utkus A, Sorokina I, Kucinskas V, Röthlisberger B, Balmer D, Brečević L, et al. Duplication of segment 1p21 following paternal insertional translocation, ins(6;1)(q25;p13.3p22.1) J Med Genet. 1999;36:73–76. [PMC free article] [PubMed] [Google Scholar]
- 15.Carter MT, Nikkel SM, Fernandez BA, Marshall CR, Noor A, Lionel AC, et al. Hemizygous deletions on chromosome 1p21.3 involving the DPYD gene in individuals with autism spectrum disorder. Clin Genet. 2011;80:435–443. doi: 10.1111/j.1399-0004.2010.01578.x. [DOI] [PubMed] [Google Scholar]
- 16.Willemsen MH, Vallès A, Kirkels LA, Mastebroek M, Olde Loohuis N, Kos A, et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J Med Genet. 2011;48:810–818. doi: 10.1136/jmedgenet-2011-100294. [DOI] [PubMed] [Google Scholar]
- 17.Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark BS, Blackshaw S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet. 2014;5:164. doi: 10.3389/fgene.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 20.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Gene Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vučićević D, Schrewe H, Orom UA. Molecular mechanisms of long ncRNAs in neurological disorders. Front Genetics. 2014;5:48. doi: 10.3389/fgene.2014.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vashishtha M, Ng CW, Yildirim F, Gipson TA, Kratter IH, Bodai L, et al. Targeting H3K4 trimethylation in Huntington disease. Proc Natl Acad Sci USA. 2013;110:E3027–E3036. doi: 10.1073/pnas.1311323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLIFE. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambros V. The evolution of our thinking about microRNAs. Nat Med. 2008;14:1036–1040. doi: 10.1038/nm1008-1036. [DOI] [PubMed] [Google Scholar]
- 26.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 27.Prosser HM, Koike-Yusa H, Cooper JD, Law FC, Bradley A. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat Biotechnol. 2011;29:840–845. doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Xu B, Hsu P-K, Karayiorgou M, Gogos JA. MicroRNA dysregulation in neuropsychiatric disorders and cognitive dysfunction. Neurobiol Dis. 2012;46:291–301. doi: 10.1016/j.nbd.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo A, Staahl B, Chen L, Crabtree G. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene. 2010;29:2161–2164. doi: 10.1038/onc.2010.59. [DOI] [PubMed] [Google Scholar]
- 32.Martinez NJ, Gregory RI. MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell. 2010;7:31–35. doi: 10.1016/j.stem.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, Zhan R, He X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 34.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 35.Olde Loohuis NFM, Kos A, Martens GJM, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106:650–661. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smalheiser NR, Lugli G. microRNA regulation of synaptic plasticity. Neuromol Med. 2009;11:133–140. doi: 10.1007/s12017-009-8065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smrt RD, Szulwach K, Pfeiffer R, Li X, Guo W, Pathania M, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun AX, Crabtree GR, Yoo AS. MicroRNAs: regulators of neuronal fate. Curr Opin Cell Biol. 2013;25:10. doi: 10.1016/j.ceb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 43.Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106:650–61. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 45.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choe EA, Liao L, Zhou JY, Cheng D, Duong DM, Jin P, et al. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J Neurosci. 2007;27:9503–9512. doi: 10.1523/JNEUROSCI.1408-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 48.Ossipova O, Ezan J, Sokol SY. PAR-1 phosphorylates Mind bomb to promote vertebrate neurogenesis. Dev Cell. 2009;17:222–233. doi: 10.1016/j.devcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunej T, Godnic I, Horvat S, Zorc M, Calin GA. Cross talk between microRNA and coding cancer genes. Cancer J. 2012;18:223–231. doi: 10.1097/PPO.0b013e318258b771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fiala JC, Spacek J, Harris KM. Dendritic spine pathology: Cause or consequence of neurological disorders? Brain Res Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- 51.Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. Autism and abnormal development of brain connectivity. J Neurosci. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–76. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strazisar M, Cammaerts S, van der Ven K, Forero DA, Lenaerts AS, Nordin A, et al. MIR137 variants identified in psychiatric patients affect synaptogenesis and neuronal transmission gene sets. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.53. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 55.Kleinhenz B, Fabienke M, Swiniarski S, Wittenmayer N, Kirsch J, Jockusch BM, et al. Raver2, a new member of the hnRNP family. FEBS Lett. 2005;579:4254–4258. doi: 10.1016/j.febslet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 56.Henneberga B, Swiniarskia S, Beckea S, Illenbergera S. A conserved peptide motif in Raver2 mediates its interaction with the polypyrimidine tract-binding protein. Exp Cell Res. 2010;316:966–979. doi: 10.1016/j.yexcr.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 57.Gromak N, Rideau AJ, Southby AD, Scadden C, Gooding S, Huttelmaier RH, et al. The PTB interacting protein raver1 regulates alpha-tropomyosin alternative splicing. EMBO J. 2003;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romanelli MR, Diani E, Lievens PM-J. New Insights into Functional Roles of the Polypyrimidine Tract-Binding Protein. Int J Mol Sci. 2013;14:22906–22932. doi: 10.3390/ijms141122906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang S, Wen S, Chen D, Jin P. Small regulatory RNAs in neurodevelopmental Disorders. Hum Mol Genet. 2009;18:R18–R26. doi: 10.1093/hmg/ddp072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitchell KJ. The genetics of neurodevelopmental disease. Curr Opin Neurobiol. 2011;21:197–203. doi: 10.1016/j.conb.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 61.Henshall DC. MicroRNA and epilepsy: profiling, functions and potential clinical applications. Curr Opin Neurol. 2014;27:199–205. doi: 10.1097/WCO.0000000000000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lewis S. Neurological disorders: microRNA gets motoring. Nat Rev Neurosci. 2014;15:67. doi: 10.1038/nrn3672. [DOI] [PubMed] [Google Scholar]
- 63.Yin J, Lin J, Luo X, Chen Y, Li Z, Ma G, Li K. miR-137: a new player in schizophrenia. Int J Mol Sci. 2014;15:3262–71. doi: 10.3390/ijms15023262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 65.Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Gene Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Gene Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang ZZ, Sharma S, Zheng S, Chawla G, Nikolic J, Black DL. Regulation of the mutually exclusive exons 8a and 8 in the CaV1.2 calcium channel transcript by polypyrimidine tract-binding protein. J Biol Chem. 2011;286:10007–10016. doi: 10.1074/jbc.M110.208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng S, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15:381–388. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Nat AcadSci USA. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Q, Zheng S, Han A, Lin C-H, Stoilov P, Fu X-D, et al. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLIFE. 2014;3:01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 75.Hill MJ, Donocik JD, Nuamah RA, Mein CA, Sainz-Fuertes R, Bray NJ. Transcriptional consequences of schizophrenia candidate miR-137 manipulation in human neural progenitor cells. Schizophr Res. 2014;53:225–230. doi: 10.1016/j.schres.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dong S, Yang B, Guo H, Kang F. MicroRNAs regulate osteogenesis and chondrogenesis. Biochem Bioph Res Commun. 2012;418:587–591. doi: 10.1016/j.bbrc.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 77.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taipaleenmäki H, Bjerre Hokland L, Chen L, Kauppinen S, Kassem M. Mechanisms in endocrinology: micro-RNAs: targets for enhancing osteoblast differentiation and bone formation. Eur J Endocrinol. 2012;166:359–371. doi: 10.1530/EJE-11-0646. [DOI] [PubMed] [Google Scholar]
- 79.Nieminen P. Genetic basis of tooth agenesis. J Exp Zool Part B. 2009;312B:320–342. doi: 10.1002/jez.b.21277. [DOI] [PubMed] [Google Scholar]
- 80.Cao H, Wang J, Li X, Florez S, Huang Z, Venugopalan SR, et al. MicroRNAs play a critical role in tooth development. J Dent Res. 2010;89:779–784. doi: 10.1177/0022034510369304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chou RH, Yu YL, Hung MC. The roles of EZH2 in cell lineage commitment. Am J Transl Res. 2011;3:243–250. [PMC free article] [PubMed] [Google Scholar]
- 82.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 83.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 85.Schwarz D, Varum S, Zemke M, Schöler A, Baggiolini A, Draganova K, et al. Ezh2 is required for neural crest-derived cartilage and bone formation. Development. 2014;141:867–877. doi: 10.1242/dev.094342. [DOI] [PubMed] [Google Scholar]
- 86.Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang DC, Tsay HJ, Lin SY, Chiou SH, Li M, Chang TJ, Hung SC. cAMP/PKA regulates osteogenesis, adipogenesis and ratio of RANKL/OPG mRNA expression in mesenchymal stem cells by suppressing leptin. PLoS One. 2008;3:e1540. doi: 10.1371/journal.pone.0001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tatton-Brown K, Hanks S, Ruark E, Zachariou A, Duarte Sdel V, Ramsay E, et al. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2:1127–1133. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, et al. Mutations in EZH2 cause Weaver syndrome. Am J Hum Genet. 2012;90:110–118. doi: 10.1016/j.ajhg.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 91.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, et al. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Song X, Wang S, Li L. New insights into the regulation of Axin function in canonical Wnt signaling pathway. Protein Cell. 2014;5:186–193. doi: 10.1007/s13238-014-0019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chia IV, Costantini F. Mouse axin and axin2/conductin proteins are functionally equivalent in vivo. Mol Cell Biol. 2005;25:4371–4376. doi: 10.1128/MCB.25.11.4371-4376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, et al. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 2004;74:1043–1050. doi: 10.1086/386293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ng D, Thakker N, Corcoran CM, Donnai D, Perveen R, Schneider A, et al. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat Genet. 2004;36:411–416. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- 97.Cai J, Kwak S, Lee JM, Kim EJ, Lee MJ, Park GH, Cho SW, Jung HS. Function analysis of mesenchymal Bcor in tooth development by using RNA interference. Cell Tissue Res. 2010;341:251–258. doi: 10.1007/s00441-010-0996-2. [DOI] [PubMed] [Google Scholar]
- 98.Yoshida T, Miyoshi J, Takai Y, Thesleff I. Cooperation of nectin-1 and nectin-3 is required for normal ameloblast function and crown shape development in mouse teeth. Dev Dynam. 2010;239:2558–2569. doi: 10.1002/dvdy.22395. [DOI] [PubMed] [Google Scholar]
- 99.Brauer AU, Savaskan NE, Kuhn H, Prehn S, Ninnemann O, Nitsch R. A new phospholipid phosphatase, PRG-1, is involved in axon growth and regenerative sprouting. Nat Neurosci. 2003;6:572–578. doi: 10.1038/nn1052. [DOI] [PubMed] [Google Scholar]
- 100.Savaskan NE, Brauer AU, Nitsch R. Molecular cloning and expression regulation of PRG-3, a new member of the plasticity-related gene family. Eur J Neurosci. 2004;19:212–220. doi: 10.1046/j.1460-9568.2003.03078.x. [DOI] [PubMed] [Google Scholar]
- 101.Brauer AU, Nitsch R. Plasticity-related genes (PRGs/LRPs): a brain-specific class of lysophospholipid-modifying proteins. Biochim Biophys Acta. 2008;1781:595–600. doi: 10.1016/j.bbalip.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 102.Tokumitsu H, Hatano N, Tsuchiya M, Yurimoto S, Fujimoto T, Ohara N, et al. Identification and characterization of PRG-1 as a neuronal calmodulin-binding protein. Biochem J. 2010;431:81–91. doi: 10.1042/BJ20100637. [DOI] [PubMed] [Google Scholar]
- 103.Trimbuch T, Beed P, Vogt J, Schuchmann S. Synaptic PRG-1 modulates excitatory transmission via lipid phosphate-mediated signaling. Cell. 2009;138:1222–1235. doi: 10.1016/j.cell.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Strauss U, Bräuer AU. Current views on regulation and function of plasticity-related genes (PRGs/LPPRs) in the brain. Biochim Biophys Acta. 2013;1831:133–138. doi: 10.1016/j.bbalip.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 105.Ni H, Jiang YW, Tao LY, Jin MF, Wu XR. ZnT-1, ZnT-3, CaMK II, PRG-1 expressions in hippocampus following neonatal seizure-induced cognitive deficit in rats. Toxicol Lett. 2009;184:145–150. doi: 10.1016/j.toxlet.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 106.Ni H, Jiang YW, Xiao ZJ, Tao LY, Jin MF, Wu XR. Dynamic pattern of gene expression of ZnT-1, ZnT-3 and PRG-1 in rat brain following flurothyl-induced recurrent neonatal seizures. Toxicol Lett. 2010;194:86–93. doi: 10.1016/j.toxlet.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 107.Ni H, Feng X, Xiao ZJ, Tao LY, Jin MF. Dynamic pattern of gene expression of ZnT-4, caspase-3, LC3, and PRG-3 in rat cerebral cortex following flurothyl-induced recurrent neonatal seizures. Biol Trace Elem Res. 2011;143:1607–1615. doi: 10.1007/s12011-011-8982-4. [DOI] [PubMed] [Google Scholar]
- 108.Bando SY, Alegro MC, Amaro EJr, Silva AV, Castro LH, Wen HT, et al. Hippocampal CA3 transcriptome signature correlates with initial precipitating injury in refractory mesial temporal lobe epilepsy. PLoS One. 2011;10:e26268. doi: 10.1371/journal.pone.0026268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Broggini T, Nitsch R, Savaskan NE. Plasticity-related gene 5 (PRG5) induces filopodia and neurite growth and impedes lysophosphatidic acid- and nogo-A-mediated axonal retraction. Mol Biol Cell. 2010;21:521–537. doi: 10.1091/mbc.E09-06-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu L, Yin W, Xia J, Peng M, Li S, Lin S, Pei D, Shu X. An antiapoptotic role of sorting nexin 7 is required for liver development in zebrafish. Hepatology. 2012;55:1985–1993. doi: 10.1002/hep.25560. [DOI] [PubMed] [Google Scholar]
- 111.van Weering JR, Verkade P, Cullen PJ. SNX-BAR proteins in phosphoinositide-mediated, tubular-based endosomal sorting. Semin Cell Dev Biol. 2010;21:371–380. doi: 10.1016/j.semcdb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Teasdale RD, Collins BM. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem J. 2012;441:39–59. doi: 10.1042/BJ20111226. [DOI] [PubMed] [Google Scholar]
- 113.Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- 114.Haberg K, Lundmark R, Carlsson S. SNX18 is an SNX9 paralog that acts as a membrane tubulator in AP-1-positive endosomal trafficking. J Cell Sci. 2008;121:1495–1505. doi: 10.1242/jcs.028530. [DOI] [PubMed] [Google Scholar]
- 115.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 116.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Van Weering JRT, Verkade P, Cullen PJ. SNX–BAR-mediated endosome tubulation is co-ordinated with endosome maturation. Traffic. 2012;13:94–107. doi: 10.1111/j.1600-0854.2011.01297.x. [DOI] [PubMed] [Google Scholar]
- 118.Vervoort VS, Viljoen D, Smart R, Suthers G, DuPont BR, Abbott A, Schwartz CE. Sorting nexin 3 (SNX3) is disrupted in a patient with a translocation t(6;13)(q21;q12) and microcephaly, microphthalmia, ectrodactyly, prognathism (MMEP) phenotype. J Med Genet. 2002;39:893–899. doi: 10.1136/jmg.39.12.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang X, Zhao Y, Zhang X, Badie H, Zhou Y, Mu Y, et al. Loss of sorting nexin 27 contributes to excitatory synaptic dysfunction by modulating glutamate receptor recycling in Down’s syndrome. Nat Med. 2013;19:473–480. doi: 10.1038/nm.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seelan RS, Khalyfa A, Lakshmanan J, Casanova MF, Parthasarathy RN. Deciphering the lithium transcriptome: microarray profiling of lithium-modulated gene expression in human neuronal cells. Neuroscience. 2008;151:1184–1197. doi: 10.1016/j.neuroscience.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 121.Becker K, Di Donato N, Holder-Espinasse M, Andrieux J, Cuisset JM, Vallée L, Plessis G, et al. De novo microdeletions of chromosome 6q14.1-q14.3 and 6q12.1-q14.1 in two patients with intellectual disability - further delineation of the 6q14 microdeletion syndrome and review of the literature. Eur J Med Genet. 2012;55:490–497. doi: 10.1016/j.ejmg.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 122.Huang HS, YoonBecker K, Di Donato N, Holder-Espinasse M, Andrieux J, Cuisset JM, Vallée L, Plessis G, et al. Snx14 regulates neuronal excitability, promotes synaptic transmission, and is imprinted in the brain of mice. PLoS One. 2014;9:e98383. doi: 10.1371/journal.pone.0098383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8:832–843. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 124.Maenaka S, Hikichi T, Imai MA, Minamoto T, Kawahara E. Loss of imprinting in IGF2 in colorectal carcinoma assessed by microdissection. Oncol Rep. 2006;15:791–795. [PubMed] [Google Scholar]
- 125.Kamikihara T, Arima T, Kato K, Matsuda T, Kato H, Douchi T, et al. Epigenetic silencing of the imprinted gene ZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int J Cancer. 2005;115:690–700. doi: 10.1002/ijc.20971. [DOI] [PubMed] [Google Scholar]
- 126.Coolen M, Katz S, Bally-Cuif L. miR-9: a versatile regulator of neurogenesis. Front Cell Neurosci. 2013;7:220. doi: 10.3389/fncel.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Akerblom M, Sachdeva R, Jakobsson J. Functional studies of microRNAs in neural stem cells: problems and perspectives. Front Neurosci. 2012;7:6–14. doi: 10.3389/fnins.2012.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lau P, de Strooper B. Dysregulated microRNAs in neurodegenerative disorders. Semin Cell Dev Biol. 2010;21:768–773. doi: 10.1016/j.semcdb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 129.van Kuilenburg AB, Vreken P, Abeling NG, Bakker HD, Meinsma R, Van Lenthe H, et al. Genotype and phenotype in patients with dihydropyrimidine dehydrogenase deficiency. Hum Genet. 1999;104:1–9. doi: 10.1007/pl00008711. [DOI] [PubMed] [Google Scholar]
- 130.Amstutz U, Froehlich TK, Largiadèr CR. Dihydropyrimidine dehydrogenase gene as a major predictor of severe 5-fluorouracil toxicity. Pharmacogenomics. 2011;12:1321–1336. doi: 10.2217/pgs.11.72. [DOI] [PubMed] [Google Scholar]
- 131.Collie-Duguid ES, Etienne MC, Milano G, McLeod HL. Known variant DPYD alleles do not explain DPD deficiency in cancer patients. Pharmacogenetics. 2000;10:217–223. doi: 10.1097/00008571-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 132.Maekawa K, Saeki M, Saito Y, Ozawa S, Kurose K, Kaniwa N, et al. Genetic variations and haplotype structures of the DPYD gene encoding dihydropyrimidine dehydrogenase in Japanese and their ethnic differences. J Hum Genet. 2007;52:804–819. doi: 10.1007/s10038-007-0186-6. [DOI] [PubMed] [Google Scholar]
- 133.Froehlich TK, Amstutz U, Aebi S, Joerger M, Largiadèr CR. Clinical importance of risk variants in the dihydropyrimidine dehydrogenase gene for the prediction of early-onset fluoropyrimidine toxicity. Int J Cancer. 2015;136:730–739. doi: 10.1002/ijc.29025. [DOI] [PubMed] [Google Scholar]
- 134.van Kuilenburg ABP, Dobritzsch D, Meinsma R, Haasjes J, Waterham HR, et al. Novel disease-causing mutations in the dihydropyrimidine dehydrogenase gene interpreted by analysis of the three-dimensional protein structure. Biochemical J. 2002;364:157–163. doi: 10.1042/bj3640157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Loganayagam A, Arenas Hernandez M, Corrigan A, Fairbanks L, Lewis CM, Harper P, et al. Pharmacogenetic variants in the DPYD, TYMS, CDA and MTHFR genes are clinically significant predictors of fluoropyrimidine toxicity. Brit J Cancer. 2013;108:2505–2515. doi: 10.1038/bjc.2013.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Terrazzino S, Cargnin S, Del Re M, Danesi R, Canonico PL, Genazzani AA. DPYD IVS14+1G>A and 2846A>T genotyping for the prediction of severe fluoropyrimidine-related toxicity: a metaanalysis. Pharmacogenomics. 2013;14:1255–1272. doi: 10.2217/pgs.13.116. [DOI] [PubMed] [Google Scholar]
- 137.Morel A, Boisdron-Celle M, Fey L, Soulie P, Craipeau MC, Traore S, Gamelin E. Clinical relevance of different dihydropyrimidine dehydrogenase gene single nucleotide polymorphisms on 5-fluorouracil tolerance. Mol Cancer Ther. 2006;11:2895–2904. doi: 10.1158/1535-7163.MCT-06-0327. [DOI] [PubMed] [Google Scholar]
- 138.Schwab M, Zanger UM, Marx C, Schaeffeler E, Klein K, Dippon J, et al. German 5-FU Toxicity Study Group. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J Clin Oncol. 2008;26:2131–2138. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- 139.Amstutz U, Farese S, Aebi S, Largiadèr CR. Dihydropyrimidine dehydrogenase gene variation and severe 5-fluorouracil toxicity: a haplotype assessment. Pharmacogenomics. 2009;10:931–944. doi: 10.2217/pgs.09.28. [DOI] [PubMed] [Google Scholar]
- 140.van Kuilenburg AB, Meijer J, Mul AN, Meinsma R, Schmid V, Dobritzsch D, et al. Intragenic deletions and a deep intronic mutation affecting pre-mRNA splicing in the dihydropyrimidine dehydrogenase gene as novel mechanisms causing 5-fluorouracil toxicity. Hum Genet. 2010;128:529–538. doi: 10.1007/s00439-010-0879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Collie-Duguid ES, Etienne MC, Milano G, McLeod HL. Known variant DPYD alleles do not explain DPD deficiency in cancer patients. Pharmacogenetics. 2000;10:217–223. doi: 10.1097/00008571-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 142.van Kuilenburg AB, van Lenthe H, Tromp A, Veltman PC, van Gennip AH. Pitfalls in the diagnosis of patients with a partial dihydropyrimidine dehydrogenase deficiency. Clin Chem. 2000;46:9–17. [PubMed] [Google Scholar]
- 143.Noguchi T, Tanimoto K, Shimokuni T, Ukon K, Tsujimoto H, Fukushima M, et al. Aberrant methylation of DPYD promoter, DPYD expression, and cellular sensitivity to 5-fluorouracil in cancer cells. Clin Cancer Res. 2004;10:7100–7107. doi: 10.1158/1078-0432.CCR-04-0337. [DOI] [PubMed] [Google Scholar]
- 144.Amstutz U, Farese S, Aebi S, Largiadèr C. Hypermethylation of the DPYD promoter region is not a major predictor of severe toxicity in 5-fluorouracil based chemotherapy. J Exp Clin Canc Res. 2008;27:54. doi: 10.1186/1756-9966-27-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Savva-Bordalo J, Ramalho-Carvalho J, Pinheiro M, Costa VL, Rodrigues A, Dias PC, et al. Promoter methylation and large intragenic rearrangements of DPYD are not implicated in severe toxicity to 5-fluorouracil-based chemotherapy in gastrointestinal cancer patients. BMC Cancer. 2010;10:470. doi: 10.1186/1471-2407-10-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.van Kuilenburg AB, Meijer J, Mul AN, Hennekam RC, Hoovers JM, de Die-Smulders CE, et al. Analysis of severely affected patients with dihydropyrimidine dehydrogenase deficiency reveals large intragenic rearrangements of DPYD and a de novo interstitial deletion del(1)(p13.3p21.3) Hum Genet. 2009;125:581–590. doi: 10.1007/s00439-009-0653-6. [DOI] [PubMed] [Google Scholar]
- 147.Meng L, Person RE, Beaudet AL. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum Mol Genet. 2012;21:3001–3012. doi: 10.1093/hmg/dds130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meng L, Person RE, Huang W, Zhu PJ, Costa-Mattioli M, Beaudet AL. Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS Genet. 2013;9:e1004039. doi: 10.1371/journal.pgen.1004039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Powell WT, Coulsona RL, Gonzalesa ML, Crarya FK, Wonga SS, Adamsa S, et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc Nat Acad Sci USA. 110:13938–13943. doi: 10.1073/pnas.1305426110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Magistri M, Faghihi MA, St Laurent G, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Clark B, Blackshaw S. Long noncoding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet. 2014;5:164. doi: 10.3389/fgene.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Werner A, Cockell S, Falconer J, Carlile M, Alnumeir S, Robinson J. Contribution of natural antisense transcription to an endogenous siRNA signature in human cells. BMC Genomics. 2014;15:1–12. doi: 10.1186/1471-2164-15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 156.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 157.Galante PA, Vidal DO, De Souza JE, Camargo AA, De Souza SJ. Sense antisense pairs in mammals: functional and evolutionary considerations. Genome Biol. 2007;8:R40. doi: 10.1186/gb-2007-8-3-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sun M, Hurst LD, Carmichael GG, Chen J. Evidence for a preferential 1580 targeting of 3′-UTRs by cis-encoded natural antisense transcripts. Nucleic Acids Res. 2005;33:5533–5543. doi: 10.1093/nar/gki852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Seila AC, Calabrese JM, Levine S, Yeo GW, Rahl PB, Flynn RA, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vanhee-Brossollet C, Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/s0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- 162.Alfano G, Vitiello C, Caccioppoli C, Caramico T, Carola A, Szego MJ, et al. Natural antisense transcripts associated with genes involved in eye development. Hum Mol Genet. 2005;14:913–923. doi: 10.1093/hmg/ddi084. [DOI] [PubMed] [Google Scholar]
- 163.Carlile M, Swan D, Jackson K, Preston-Fayers K, Ballester B, Flicek P, et al. Strand selective generation of endo-siRNAs from the Na/phosphate transporter gene Slc34a1 in murine tissues. Nucleic Acids Res. 2009;37:2274–2282. doi: 10.1093/nar/gkp088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 166.Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 167.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Popov N, Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics. 2010;5:685–690. doi: 10.4161/epi.5.8.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Verona RI, Mann MR, Bartolomei MS. Genomic imprinting: intricacies of epigenetic regulation in clusters. Ann Rev Cell Dev Bi. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- 170.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 171.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 172.Terranova R, Yokobayashi S, Stadler MB, Otte AP, Van Lohuizen M, Orkin SH, et al. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 173.Wan LB, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- 174.Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long noncoding RNAs. Epigenetics. 2009;4:277–286. [PubMed] [Google Scholar]
- 175.Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- 176.Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 177.Dermitzakis ET, Reymond A, Antonarakis SE. Conserved nongenic sequences - an unexpected feature of mammalian genomes. Nat Rev Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- 178.Louro R, El-Jundi T, Nakaya HI, Reis EM, Verjovski-Almeida S. Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci. Genomics. 2008;92:18–25. doi: 10.1016/j.ygeno.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 179.St Laurent G, Shtokalo D, Tackett MR, Yang Z, Eremina T, Wahlestedt C, et al. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC Genomics. 2012;13:504. doi: 10.1186/1471-2164-13-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, et al. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8:R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Nat Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, Hartman S, et al. The transcriptional activity of human Chromosome 22. Gene Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 185.Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Guil S, Soler M, Portela A, Carrere J, Fonalleras E, Gomez A, et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19:664–670. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- 187.Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. 2012;19:1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 188.Saif MW, Ezzeldin H, Vance K, Sellers S, Diasio RB. DPYD*2A mutation: the most common mutation associated with DPD deficiency. Cancer Chemoth Pharm. 2007;60:503–507. doi: 10.1007/s00280-006-0392-5. [DOI] [PubMed] [Google Scholar]
- 189.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ng K, Pullirsch D, Leeb M, Wutz A. Xist and the order of silencing. EMBO Rep. 2007;8:34–39. doi: 10.1038/sj.embor.7400871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 194.Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, et al. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development. 2009;136:525–530. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hormozian F, Schmitt JG, Sagulenko E, Schwab M, Savelyeva L. FRA1E common fragile site breaks map within a 370 kilobase pair region and disrupt the dihydropyrimidine dehydrogenase gene (DPYD) Cancer Lett. 2007;246:82–91. doi: 10.1016/j.canlet.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 196.Fungtammasan A, Walsh E, Chiaromonte F, Eckert KA, Makova KD. A genome-wide analysis of common fragile sites: what features determine chromosomal instability in the human genome? Genome Res. 2012;22:993–1005. doi: 10.1101/gr.134395.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lukusa T, Fryns JP. Human chromosome fragility. Biochim Biophys Acta. 2008;1779:3–16. doi: 10.1016/j.bbagrm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 198.Mrasek K, Schoder C, Teichmann A-C, Behr K, Franze B, Wilhelm K, et al. Global screening and extended nomenclature for 230 aphidicolin-inducible fragile sites, including 61 yet unreported ones. Int J Oncol. 2010;36:929–940. doi: 10.3892/ijo_00000572. [DOI] [PubMed] [Google Scholar]
- 199.Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, et al. Molecular basis for expression of common and rare fragile sites. Mol Cell Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Travers AA. The structural basis of DNA flexibility. Philos Trans A Math Phys Eng Sci. 2004;362:1423–1438. doi: 10.1098/rsta.2004.1390. [DOI] [PubMed] [Google Scholar]
- 201.Zhang H, Freudenreich CH. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Shah SN, Opresko PL, Meng X, Lee MYWT, Eckert KA. DNA structure and the Werner protein modulate human DNA polymerase d-dependent replication dynamics within the common fragile site FRA16D. Nucleic Acids Res. 2010;38:1149–1162. doi: 10.1093/nar/gkp1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dillon LW, Burrow AA, Wang Y-H. DNA instability at chromosomal fragile sites in cancer. Curr Genomics. 2010;11:326–337. doi: 10.2174/138920210791616699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 205.Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012;28:22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 206.Gross E, Busse B, Riemenschneider M, Neubauer N, Seck K, Klein HG, et al. Strong association of a common dihydropyrimidine dehydrogenase gene polymorphism with fluoropyrimidine-related toxicity in cancer patients. PLoS One. 2008;3:e4003. doi: 10.1371/journal.pone.0004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, et al. Common chromosomal fragile site FRA16D sequence: Identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 208.Ragland RL, Glynn MW, Arlt MF, Glover TW. Stably transfected common fragile site sequences exhibit instability at ectopic sites. Gene Chromosome Canc. 2008;47:860–872. doi: 10.1002/gcc.20591. [DOI] [PubMed] [Google Scholar]
- 209.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 210.Craig NL, Eickbush TH, Voytas DF. Welcome to mobile DNA. Mob DNA. 2010;1:1. doi: 10.1186/1759-8753-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pace JK, Feschotte C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 213.Belancio VP, Deininger PL, Roy-Engel AM. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH. Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 215.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- 216.Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Han K, Lee J, Meyer TJ, Remedios P, Goodwin L, Batzer MA. L1 recombination-associated deletions generate human genomic variation. Proc Natl Acad Sci USA. 2008;105:19366–19371. doi: 10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Robberecht C, Voet T, Zamani Esteki M, Nowakowska BA, Vermeesch JR. Nonallelic homologous recombination between retrotransposable elements is a driver of de novo unbalanced translocations. Genome Res. 2013;23:411–418. doi: 10.1101/gr.145631.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kines KJ, Sokolowski M, de Haro DL, Christian CM, Belancio VP. Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res. 2014;42:10488–10502. doi: 10.1093/nar/gku687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kines KJ, Belancio VP. Expressing genes do not forget their LINEs: transposable elements and gene expression. Front Biosci (Landmark Ed) 2012;17:1329–1344. doi: 10.2741/3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010;38:3909–3922. doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ‘bout retroelements in cancer. Semin Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wallace NA, Belancio VP, Faber Z, Deininger P. Feedback inhibition of L1 and alu retrotransposition through altered double strand break repair kinetics. Mob DNA. 2010;1:22. doi: 10.1186/1759-8753-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kramerov DA, Vassetzky NS. Short retroposons in eukaryotic genomes. Rev Cytol. 2005;247:165–221. doi: 10.1016/S0074-7696(05)47004-7. [DOI] [PubMed] [Google Scholar]
- 226.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Skowronski J, Singer MF. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc Natl Acad Sci USA. 1985;82:6050–6054. doi: 10.1073/pnas.82.18.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 229.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:51512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lavie L, Maldener E, Brouha B, Meese EU, Mayer J. The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res. 2004;14:2253–2260. doi: 10.1101/gr.2745804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Swergold GD. Identification, characterization, and cell specificity of a human LINE- 1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Severynse DM, Hutchison CA, Edgell MH. Identification of transcriptional regulatory activity within the 5′ A-type monomer sequence of the mouse LINE-1 retroposon. Mamm Genome. 1992;2:41–50. doi: 10.1007/BF00570439. [DOI] [PubMed] [Google Scholar]
- 233.Nigumann P, Redik K, Matlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79:628–634. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- 234.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kazazian HH, Jr, Goodier JL. LINE drive retrotransposition and genome instability. Cell. 2002;110:277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- 236.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 237.Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–1078. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–234. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- 239.Montoya-Durango DE, Liu Y, Teneng I, Kalbfleisch T, Lacy ME, Steffen MC, Ramos KS. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat Res. 2009;665:20–28. doi: 10.1016/j.mrfmmm.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Huda A, Jordan IK. Epigenetic regulation of Mammalian genomes by transposable elements. Ann NY Acad Sci. 2009;1178:276–284. doi: 10.1111/j.1749-6632.2009.05007.x. [DOI] [PubMed] [Google Scholar]
- 241.Allen E, Horvath S, Tong F, Kraft P, Spiteri E, Riggs AD, Marahrens Y. High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc Natl Acad Sci USA. 2003;100:9940–9945. doi: 10.1073/pnas.1737401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Harris CR, Dewan A, Zupnick A, Normart R, Gabriel A, Prives C, Levine AJ, Hoh J. p53 responsive elements in human retrotransposons. Oncogene. 2009;28:3857–3865. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Morales JF, Snow ET, Murnane JP. Environmental factors affecting transcription of the human L1 retrotransposon. I. Steroid hormone-like agents. Mutagenesis. 2002;17:193–200. doi: 10.1093/mutage/17.3.193. [DOI] [PubMed] [Google Scholar]
- 244.Morales JF, Snow ET, Murnane JP. Environmental factors affecting transcription of the human L1 retrotransposon. II. Stressors. Mutagenesis. 2003;18:151–158. doi: 10.1093/mutage/18.2.151. [DOI] [PubMed] [Google Scholar]
- 245.Yang N, Zhang L, Zhang Y, Kazazian HH. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Beier F, Lammi MJ, Bertling W, von der Mark K. Transcriptional regulation of the human type X collagen gene expression. Ann NY Acad Sci. 1996;785:209–211. doi: 10.1111/j.1749-6632.1996.tb56263.x. [DOI] [PubMed] [Google Scholar]
- 247.Landry JR, Medstrand P, Mager DL. Repetitive elements in the 5′ untranslated region of a human zinc-finger gene modulate transcription and translation efficiency. Genomics. 2001;76:110–116. doi: 10.1006/geno.2001.6604. [DOI] [PubMed] [Google Scholar]
- 248.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 249.Shephard EA, Chandan P, Stevanovic-Walker M, Edwards M, Phillips IR. Alternative promoters and repetitive DNA elements define the species-dependent tissue-specific expression of the FMO1 genes of human and mouse. Biochem J. 2007;406:491–499. doi: 10.1042/BJ20070523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Steel G, Lutz EM. Characterisation of the mouse vasoactive intestinal peptide receptor type 2 gene, Vipr2, and identification of a polymorphic LINE-1-like sequence that confers altered promoter activity. J Neuroendocrinol. 2007;19:14–25. doi: 10.1111/j.1365-2826.2006.01498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Matlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol. 2006;2006:1–16. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Cruickshanks HA, Tufarelli C. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics. 2009;94:397–406. doi: 10.1016/j.ygeno.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 253.Dunn CA, van de Lagemaat LN, Baillie GJ, Mager DL. Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate beta3GAL-T5. Gene. 2005;364:2–12. doi: 10.1016/j.gene.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 254.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 255.Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13–25. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11:S52–S59. doi: 10.1016/s0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- 257.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 258.Longhese MP, Mantiero D, Clerici M. The cellular response to chromosome breakage. Mol Microbiol. 2006;60:1099–1108. doi: 10.1111/j.1365-2958.2006.05186.x. [DOI] [PubMed] [Google Scholar]
- 259.Ergün S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 260.Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O’Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 261.Asch HL, Eliacin E, Fanning TG, Connolly JL, Bratthauer G, Asch BB. Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncol Res. 1996;8:239–247. [PubMed] [Google Scholar]
- 262.Nangia-Makker P, Sarvis R, Visscher DW, Bailey-Penrod J, Raz A, Sarkar FH. Galectin-3 and L1 retrotransposons in human breast carcinomas. Breast Cancer Res Treat. 1998;49:171–183. doi: 10.1023/a:1005913810250. [DOI] [PubMed] [Google Scholar]
- 263.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, Burns MB, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22:2328–2338. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, et al. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Smit AF, Riggs AD. Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci USA. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 270.Kapitonov V, Jurka J. The age of Alu subfamilies. J Mol Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- 271.Lim JK, Simmons MJ. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. Bioessays. 1994;16:269–275. doi: 10.1002/bies.950160410. [DOI] [PubMed] [Google Scholar]
- 272.Caceres M, Ranz JM, Barbadilla A, Long M, Ruiz A. Generation of a widespread Drosophila inversion by a transposable element. Science. 1999;285:415–418. doi: 10.1126/science.285.5426.415. [DOI] [PubMed] [Google Scholar]
- 273.Gray YH. It takes two transposons to tango: Transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468. doi: 10.1016/s0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- 274.Zhang J, Peterson T. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics. 2004;167:1929–1937. doi: 10.1534/genetics.103.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Froyen G, Belet S, Martinez F, Santos-Rebouças CB, Declercq M, Verbeeck J, et al. Copy-number gains of HUWE1 due to replication-and recombination-based rearrangements. Am J Hum Genet. 2012;91:252–64. doi: 10.1016/j.ajhg.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 277.Luft FC. Sleeping Beauty jumps to new heights. Mol Med. 2010;88:641–643. doi: 10.1007/s00109-010-0626-1. [DOI] [PubMed] [Google Scholar]
- 278.Ivics Z, Izsvak Z. Transposons for gene therapy! Current Gene Ther. 2006;6:593–607. doi: 10.2174/156652306778520647. [DOI] [PubMed] [Google Scholar]
- 279.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 280.Hackett PB, Largaespada DA, Cooper LJ. A transposon and transposase system for human application. Mol Ther. 2010;18:674–683. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Singh H, Figliola MJ, Dawson MJ, Olivares S, Zhang L, Yang G, et al. Manufacture of clinical-grade CD19-specific T cells stably expressing chimeric antigen receptor using Sleeping Beauty system and artificial antigen presenting cells. PLoS One. 2013;8:e64138. doi: 10.1371/journal.pone.0064138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Carlson CM, Largaespada DA. Insertional mutagenesis in mice: new perspectives and tools. Nat Rev Genet. 2006:568–580. doi: 10.1038/nrg1638. [DOI] [PubMed] [Google Scholar]
- 283.Song G, Cui Z. Mob Genet Elements, Novel strategies for gene trapping and insertional mutagenesis mediated by Sleeping Beauty transposon. Mobile Genetic Elements. 2013;3:e26499. doi: 10.4161/mge.26499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Ranzani M, Annunziato S, Adams DJ, Montini E. Cancer gene discovery: exploiting insertional mutagenesis. Mol Cancer Res. 2013;11:1141–1158. doi: 10.1158/1541-7786.MCR-13-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Strand DJ, McDonald JF. Copia is transcriptionally responsive to environmental stress. Nucleic Acids Res. 1985;13:4401–4410. doi: 10.1093/nar/13.12.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 287.Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 288.Konkel MK, Batzer MA. A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Semin Cancer Biol. 2010;20:211–221. doi: 10.1016/j.semcancer.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Bose P, Hermetz KE, Conneely KN, Rudd MK. Tandem repeats and G-rich sequences are enriched at human CNV breakpoints. PLoS One. 2014;9:e101607. doi: 10.1371/journal.pone.0101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Gebow D, Miselis N, Liber HL. Homologous and nonhomologous recombination resulting in deletion: effects of p53 status, microhomology, and repetitive DNA length and orientation. Mol Cell Biol. 2000;20:4028–4035. doi: 10.1128/mcb.20.11.4028-4035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Stenger JE, Lobachev KS, Gordenin D, Darden TA, Jurka J, Resnick MA. Biased distribution of inverted and direct Alus in the human genome: implications for insertion, exclusion, and genome stability. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- 292.Ade C, Roy-Engel AM, Deininger PL. Alu elements: an intrinsic source of human genome instability. Curr Opin Virol. 2013;3:639–45. doi: 10.1016/j.coviro.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Boone PM, Yuan B, Campbell IM, Scull JC, Withers MA, Baggett BC, et al. The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am J Hum Genet. 2014;95:143–161. doi: 10.1016/j.ajhg.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–B). Molecular organization of DPYD gene according to GRC38/hg38 Genome Annotation. A) NCBI Annotation Release 106 August 2014. Note the new lncRNA XR_426733.1/LOC102723700/ NC_018912.2; B) GRCh38 Ensembl Release August 2014. Note new 9 EST protein coding transcripts colored in purple. Fig. 1A and 2B are not drawn to scale.