Abstract
Human immunodeficiency virus–infected people discontinuing therapy experience a rebound in the virus level (hereafter, “rebound virus”) from a persistent reservoir. We examined 10 samples from patients in AIDS Clinical Trials Group study A5068 with rebound virus, using single-genome amplification and Primer ID deep sequencing, to assess env genetic diversity of the virus population. Most rebound-virus populations showed significant diversity. All env examined required high levels of CD4 for entry, consistent with selection of replication in CD4+ T cells. These results indicate that most people discontinuing therapy release a diverse population of virus and that this released virus has entry features of virus selected for replication in CD4+ T cells, rather than in myeloid cells.
Keywords: HIV-1, rebound virus, tropism, genetic diversity, reservoir
Successful antiretroviral therapy (ART) reduces the human immunodeficiency virus type 1 (HIV-1) load in the blood to stable low levels; however, ART does not eliminate virus. Most people discontinuing therapy experience a rebound in the virus level (hereafter, “rebound virus”) from a persistent reservoir [1–4]. The variable timing of rebound, where the virus is coming from, and the complexity of the rebound virus population are poorly understood.
One potential source of virus is latently infected resting CD4+ T cells. There is also a continual production of virus at low levels despite ART, and other cell types could be the source of rebound virus. In a largely quiescent reservoir, rebound virus could be clonal, as the result of virus from a single cell leaving a latent state. However, the number and complexity of rebound virus within a person has recently been predicted to be much higher than predicted on the basis of a largely quiescent reservoir [4].
To this end, we used peripheral blood samples from patients in AIDS Clinical Trials Group study A5068 [5] to investigate the diversity and entry phenotype of rebound virus. Subjects in A5068 discontinued ART as part of an analytic treatment interruption (ATI) and had virologic rebound detectable in the blood 2–4 weeks, on average, after discontinuation of therapy. To study the complexity of rebound virus, we assessed the genetic diversity of viral env and also examined viral entry phenotype. Phylogenetic analysis showed a high degree of diversity in the rebound virus population. In addition, all Env examined required high levels of CD4 for efficient entry, consistent with prior selection for replication in CD4+ T cells. Our analysis suggests that a viral population replicating in myeloid cells is not a significant source of rebound virus in blood early after the discontinuation of ART.
METHODS
A detailed description of the study methods is available in the Supplementary Materials. The parent trial is registered at ClinicalTrials.gov (registration NCT00011011).
Study Population
All samples were collected under institutional review board–approved protocols, and all subjects provided written informed consent. AIDS Clinical Trials Group study A5068 examined whether structured treatment interruptions and/or vaccination with an ALVAC-HIV vector was associated with control of viral replication during a subsequent ATI, compared with no vaccination or structured interruption control (arm A). The study showed a modest effect in reduced peak and set point viral load of rebound virus in the arms that included the structured treatment interruptions [5]. Subjects with the ATI only (arm A) had a median CD4+ T-cell count of 779 copies/mL and viral RNA levels of <50 copies per mL at entry while still receiving therapy. For our study, we used blood plasma samples from 10 of these subjects early after the initiation of the ATI, when viral load was ≥1000 copies/mL. For one subject, a pretherapy sample was also available.
Virus Characterization
Sequences of virion RNA were examined using single-genome amplification (SGA) [6] of full-length env and deep sequencing with Primer ID [7] of the V1–V3 region of env. Phylogenetic analysis of full-length env was used to select a representative sampling of amplicons for cloning to assess viral entry phenotype. env cloned in an expression vector was used in cotransfection with the pNL4–3.LucR-E reporter backbone plasmid into 293 T cells to generate an Env-pseudotyped luciferase reporter virus as described previously [8–10]. By comparing the ability of the pseudotyped virus to infect Affinofile cells expressing high versus low levels of surface CD4, we are able to consistently assess the viral CD4 entry phenotype as largely restricted to CD4+ T cells when high levels of CD4 are required for entry (T-cell tropic), compared with tropism favoring cells with a low density of CD4, such as macrophages (macrophage tropic) [8]. Sensitivity to soluble CD4 (sCD4) was used to confirm the CD4 entry phenotype. Additionally, coreceptor use was analyzed using Maraviroc inhibition of CCR5.
RESULTS
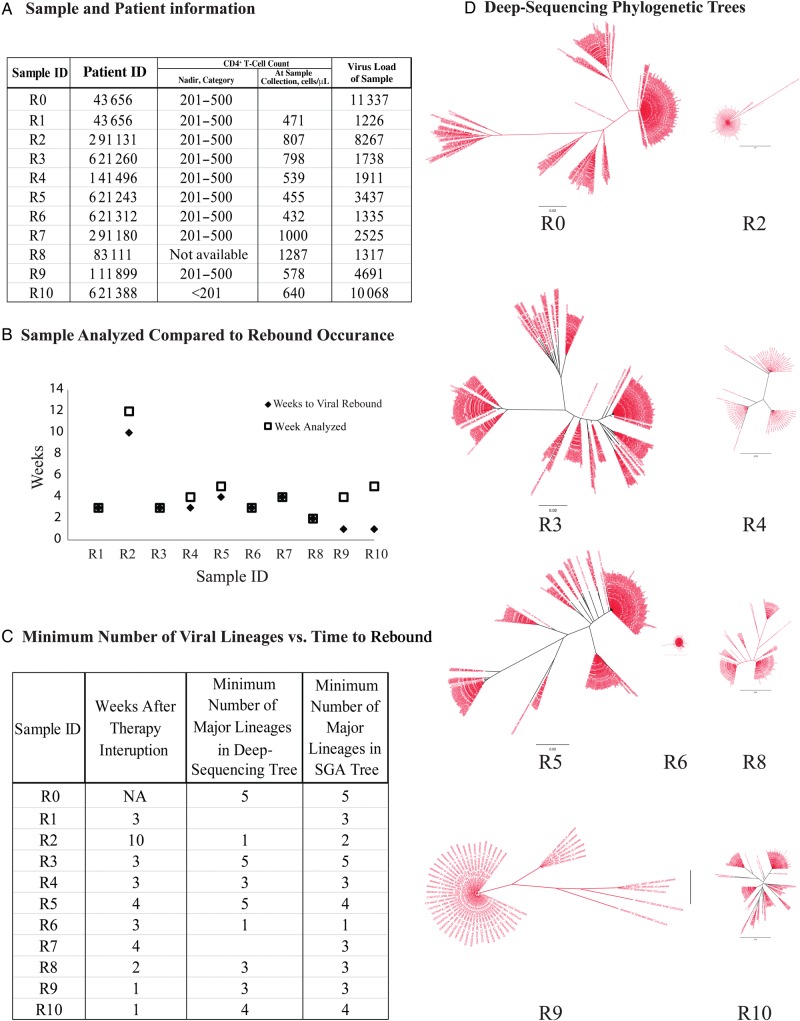
We analyzed 11 plasma samples from 10 subjects (subject 43 656 had a pretherapy [R0] and rebound [R1] sample available; Figure 1A). We used plasma from the first available time point at which the viral load was >1000 copies/mL, to focus on early rebound virus, with most samples yielding a viral load of <10 000 copies/mL (Figure 1B). Eight of ten subjects had nadir CD4+ T-cell counts before therapy of 201–500 cells/mL, while 1 (sample R10) was <201 cells/mL.
Figure 1.
Sample/subject information and phylogenetic analysis of env sequences, using a deep-sequencing approach. A, Sample and subject information includes a key indicating the subjects from whom samples were obtained, as well as nadir CD4+ T-cell counts, by category, over the course of care, CD4+ T-cell counts at the time of sampling, and viral loads. B, Comparison of the time to a rebound in virus level among samples. In the majority of subjects, we were able to analyze the first sample in which virus was detected or a sample very close to rebound detection. C, Comparison of the minimum number of major lineages represented in both deep-sequencing phylogenetic analysis and single-genome amplification (SGA) phylogenetic analysis. D, Phylogenetic trees, created in FigTree, of the sequenced samples were produced using deep sequencing with Primer ID and are presented so all samples are on the same scale. Abbreviation: NA, not applicable.
To determine the viral sequence diversity of the rebound samples, we performed deep-sequencing analysis with Primer ID to sequence the V1–V3 region of env [7] (Figure 1D), in addition to doing SGA of full-length env [6]. Deep-sequencing results were achieved for 8 of 10 rebound samples and the pretherapy sample (however, the rebound sample, R1, from the patient from whom the pretherapy sample was obtained did not yield deep-sequencing results). We estimated the minimum number of rebound viruses as distinct sequence lineages in the phylogenetic tree analysis and highlighter plots (Figures 1D and 2A and Supplementary Figures 1 and 2). We used both distinct lineages and complex clusters of linked sequence differences in forming this estimate while trying to avoid interpreting complexity that could have been the result of postrebound recombination. We consider these values as minimum estimates of rebound virus complexity since the viruses considered as potential recombinants could have preexisted in the reservoir and been independently reactivated. Analysis of the minimum number of lineages versus the time (in weeks) to rebound demonstrated no obvious correlation between the time to rebound and the complexity of the rebound virus population (Figure 1C). For virus that rebounded in the first few weeks, we estimated 1–5 distinct lineages by deep sequencing, with a mean of 3.1 lineages, for these early rebound viruses. R2 took the longest to rebound (10 weeks) and showed a relatively low diversity, with only 2 lineages. SGA of full-length env showed 1–4 distinct lineages, with a mean of 3.1 lineages, similar to the results obtained with deep sequencing (Figure 1C). The mean intrapatient diversity was also determined by pair-wise analysis using full-length env and was found to be 0.2%–3.1%. Additional analyses of 10 subjects with early phase infection (Supplementary Table 2) [11] and 7 with chronic infection who were not receiving therapy (unpublished data; Supplementary Table 3) were also done, revealing mean intrapatient diversity of 0.16%–1.2% (Supplementary Figure 3) and 1.1%–4.1%, respectively. These numbers suggest that rebound virus is more similar to virus in chronically infected subjects, in whom multiple viral lineages form a genetically complex population.
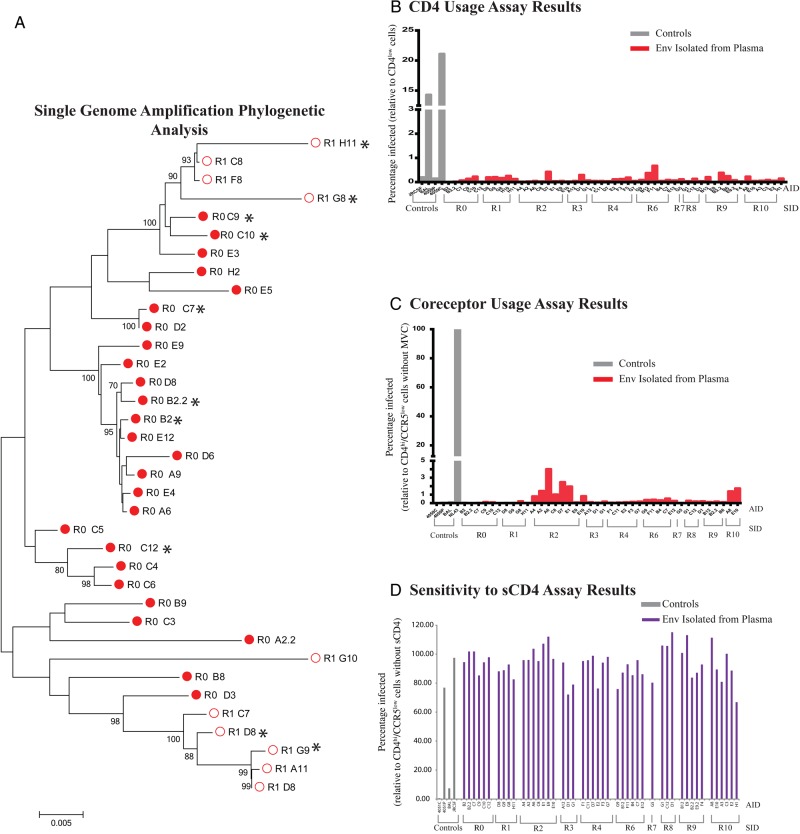
Figure 2.
Phylogenetic analysis of env sequences and tropism determination results. A, Phylogenetic tree, created using MEGA, of the sequences of subject 43 656 for the sample obtained before therapy (R0; closed circles) and after the rebound in virus load (R1; open circles), generated using SGA. Cloned and tropism-analyzed env sequences are marked with an asterisk. B–D, Tissue-culture assay results, using pseudotyped virus to define viral entry tropism phenotypes. The relative infectivity of pseudoviruses (red bars; gray bars denote controls) was determined using Affinofile cells induced to expressing low levels of CD4 as compared to infection at high levels of CD4 (B). The relative infectivity of all pseudotyped virus was <2% and indicates T-cell tropism. The relative infectivity of pseudovirus in the presence of CCR5 antagonist, maraviroc (MVC; red bars; gray bars denote controls), showed nearly complete inhibition of all pseudotyped viruses and indicates use of CCR5 (C). The relative infectivity of TZM-bl cells by pseudotyped virus in the presence of soluble CD4 (sCD4; purple bars; gray bars denote controls) showed resistance to inhibition by all pseudotype viruses, which further confirms a T-cell–tropic phenotype (D).
We subsequently cloned 51 individual env genes (average, 4 amplicons/subject) from the major lineages in each participant to determine the entry phenotype of rebound virus, using the Affinofile cell line (Figure 2A and Supplementary Figure 2). The pretherapy sample (R0) was also included; no clones were obtained for sample R5. Affinofile cells allow controlled expression of CD4 and are currently the most consistent method to assess CD4-dependent entry phenotype [8]. Entry phenotype was evaluated on the basis of the ability of viruses pseudotyped with Env from rebound virus to infect Affinofile cells expressing either high or low levels of CD4. The phenotype was confirmed by sensitivity to inhibition by sCD4 at a concentration of 5.0 µg/mL when infecting TZM-bl cells [12]; this concentration neutralizes macrophage-tropic viruses but not R5 T-cell–tropic viruses. No Env showed any (<2% infection) ability to infect cells expressing low levels of CD4 as compared to their infectivity on cells expressing high levels of CD4 (Figure 2B), and they were also resistant to inhibition by sCD4 (Figure 2D). In addition, all pseudotyped viruses were inhibited by the CCR5 antagonist maraviroc, and therefore all isolated Env used CCR5 as the coreceptor. Taken together, all Env tested from rebound virus from all subjects had a phenotype consistent with that of R5 T-cell–tropic virus.
DISCUSSION
In this report, we describe 2 key features of the initial virus that rebounds from the latent or low-level viremia reservoir after therapy discontinuation. These features are relevant to developing strategies of HIV-1 eradication because they are features of the first virus that must be targeted. We asked what the entry phenotype was of this virus, to determine what cell type it had been predominantly replicating in at the time therapy was initiated and/or it entered the latent reservoir. In addition, we examined the complexity of the rebound population as a measure of the minimum number of viruses that rebound from the reservoir and become detected in the blood. The genetic diversity of the viral reservoir and rebound virus has been debated but it is perceived to be low and to be primarily clonal or multiclonal [13, 14]. Our results are consistent with the observation of Rothenberger et al [4], who proposed a multifocal model of virus reactivation. Counting major lineages with both deep sequencing and SGA, we observed a mean of 3.1 distinct lineages, which suggests a minimum of 3 viruses rebounding within a short period after treatment interruption. While this is significantly less than the 5–25 rebound/founder viruses suggested by Rothenberger et al [4], when we reanalyzed the sequence data from that study for the 3 subjects with SGA of virion RNA by using the same criteria as used in this report, we found that the mean number of distinct lineages was 3.3, similar to what we observed (Supplementary Methods). However, since we did not count potential recombinants in our analysis, this is a minimum estimate.
The number of rebound viruses could be affected by factors that affect viral reservoir size, such as ART duration and time to ART initiation after infection, but such data are not available for these subjects. Reservoir size, as measured by cell-associated viral RNA and DNA, has been shown to be correlated with set point after therapy discontinuation [15]. Sample R2 had one of the lowest sequence diversities (0.59%) and took the longest to have a measurable rebound virus population, but we only analyzed one subject for whom >4 weeks passed before rebound was detected.
The use of viral outgrowth assays from subjects receiving therapy after isolation of resting CD4+ T cells and reactivation in culture has established this cell type as a reservoir for HIV-1 latency [1–3]. We sought to evaluate the rebound virus population by using entry phenotype. Distinct lineages of HIV-1 can evolve the ability to use low levels of CD4 for entry, but these have been found primarily in the cerebral spinal fluid; presumably, this localized evolution is due to selective pressure to enhance replication in cells such as macrophages, which express low levels of CD4. Here we assessed the CD4 entry phenotype in conjunction with sCD4 sensitivity (a feature of low CD4-using Env) to determine that rebound viruses needed high levels of CD4 for efficient entry (Figure 2B). Sensitivity to inhibition by maraviroc of all Env showed that all of the viruses used CCR5 (Figure 2C), making these viruses R5 T-cell tropic. On the basis of this phenotypic characterization, we can conclude that rebound virus was adapted to replicating in CD4+ T cells at the time the reservoir was formed. These results do not define the cell type that harbored the latent virus before rebound (T cells or macrophages), but these results exclude the possibility that rebound virus was replicating extensively in a myeloid cell lineage before latency, since this would have selected for the ability to use low levels of CD4 for entry. It should be noted, however, that viral rebound in isolated compartments of the body, such as the brain, or viruses released later from smaller reservoirs could have phenotypes that are distinct from that of the initial rebound virus that predominates in the blood.
In summary, we have shown that the early rebound virus is more diverse than previously thought. Additionally, we were able to show that all virus isolates from the plasma used CCR5 and required high levels of CD4 for entry and therefore were R5 T-cell tropic. This excludes a population of virus replicating in myeloid cells as a significant contributor to the viral reservoir that repopulates blood early after therapy discontinuation.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank all of the study participants and study team members of AIDS Clinical Trials Group A5068.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, NIH (awards UM1 AI068634, UM1 AI068636, and UM1 AI106701).
Potential conflicts of interest. The University of North Carolina at Chapel Hill is pursuing intellectual property protection for Primer ID, and R. S is listed as a coinventor and has received nominal royalties. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Finzi D, Hermankova M, Pierson T et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278:1295–300. [DOI] [PubMed] [Google Scholar]
- 2.Wong JK, Hezareh M, Gunthard HF et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278:1291–5. [DOI] [PubMed] [Google Scholar]
- 3.Finzi D, Blankson J, Siliciano JD et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999; 5:512–7. [DOI] [PubMed] [Google Scholar]
- 4.Rothenberger MK, Keele BF, Wietgrefe SW et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A 2015; 112:E1126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobson JM, Pat Bucy R, Spritzler J et al. Evidence that intermittent structured treatment interruption, but not immunization with ALVAC-HIV vCP1452, promotes host control of HIV replication: the results of AIDS Clinical Trials Group 5068. J Infect Dis 2006; 194:623–32. [DOI] [PubMed] [Google Scholar]
- 6.Salazar-Gonzalez JF, Bailes E, Pham KT et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol 2008; 82:3952–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou S, Jones C, Mieczkowski P, Swanstrom R. Primer ID validates template sampling depth and greatly reduces the error rate of next-generation sequencing of HIV-1 genomic RNA populations. J Virol 2015; 89:8540–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joseph SB, Arrildt KT, Swanstrom AE et al. Quantification of entry phenotypes of macrophage-tropic HIV-1 across a wide range of CD4 densities. J Virol 2014; 88:1858–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ping LH, Cohen MS, Hoffman I et al. Effects of genital tract inflammation on human immunodeficiency virus type 1 V3 populations in blood and semen. J Virol 2000; 74:8946–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keele BF, Giorgi EE, Salazar-Gonzalez JF et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 2008; 105:7552–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sturdevant CB, Joseph SB, Schnell G, Price RW, Swanstrom R, Spudich S. Compartmentalized replication of R5 T cell-tropic HIV-1 in the central nervous system early in the course of infection. PLoS Pathog 2015; 11:e1004720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol 2005; Chapter 12:Unit 12.11. [DOI] [PubMed] [Google Scholar]
- 13.Kearney MF, Spindler J, Shao W et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog 2014; 10:e1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joos B, Fischer M, Kuster H et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci U S A 2008; 105:16725–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JZ, Heisey A, Ahmed H et al. Relationship of HIV reservoir characteristics with immune status and viral rebound kinetics in an HIV therapeutic vaccine study. AIDS 2014; 28:2649–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.