Why Study Nasal Microbiota?
Bacterial species that commonly reside on surfaces of the human nasal passages (Fig 1) interact with the host along a continuum from beneficial to harmful, i.e., from mutualist to commensal to pathogen. Likewise, the host responds along a continuum from tolerance to damage [1]. In fact, a small number of bacterial species that are prevalent and often abundant members of the nasal microbiota are important human pathogens, e.g., Staphylococcus aureus and Streptococcus pneumoniae. In the United States alone, S. pneumoniae contributes to ~20,000 deaths [2] and methicillin-resistant strains of S. aureus (MRSA) contribute to ~10,000 deaths annually [3]. Despite this significant mortality, most S. aureus and S. pneumoniae interactions with humans are harmless and do not result in disease, i.e., are commensal. However, benign colonization can be the starting point for disease, host–host transmission, and selection for new microbial traits. This duality of behavior from commensal to pathogen has led to the term pathobiont [4]. The factors that shift the behavior of pathobionts from a commensal to a pathogenic state remain to be identified. However, the interplay of pathobionts with other members of the microbiota might be one such factor. The combination of cultivation and 16S rRNA gene-based approaches has revealed new insights into this possibility and has sparked renewed interest in determining the molecular mechanisms of commensal–pathobiont interactions in the nasal microbiota. One goal of these efforts is to identify potentially beneficial bacteria (mutualists) that might either exclude pathobionts through colonization resistance or shift the behavior of colonizing pathobionts towards commensalism.
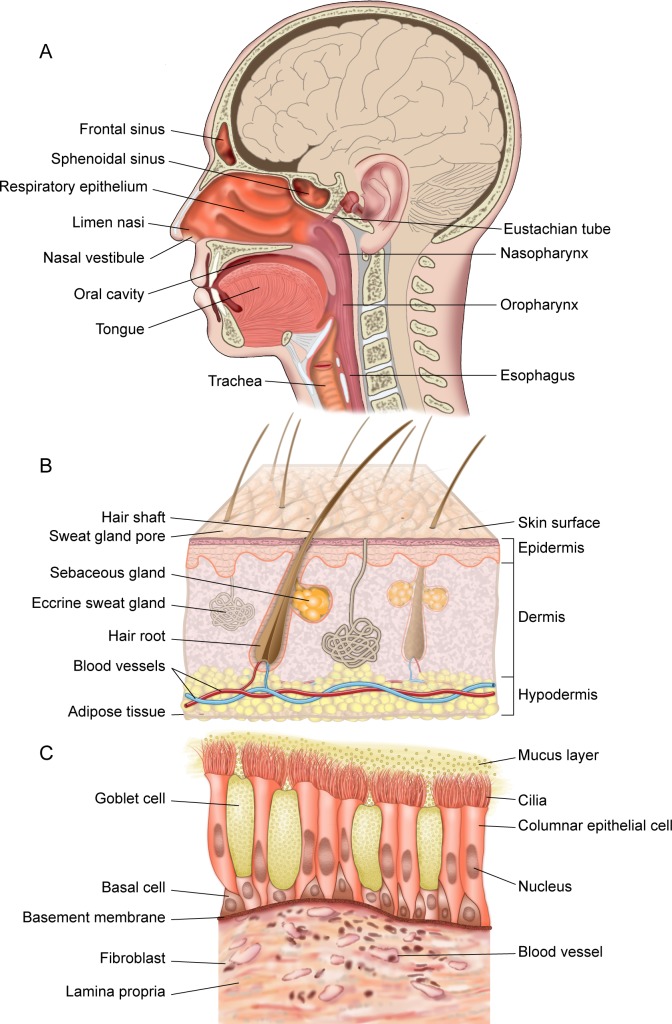
Fig 1. The human nasal passages.
As reviewed in [5], the nostrils (anterior nares) are the entrance into the (A) human nasal passages (sagittal section) and open onto the skin-covered surface of the nasal vestibule, an acidic environment that contains sweat and sebaceous glands. (B) Cross section of the nostril skin. Moving posterior (A), the limen nasi marks the transition from the posterior region of the nasal vestibules to a mucosal surface, which contains mucin-secreting goblet cells and where the pH begins to steadily increase, reaching neutrality before the nasal cavity ends in the nasopharynx, the top of the back of the throat. Respiratory epithelial cells, including cilia that beat towards the esophagus, line the posterior segment of the nasal cavity and the nasopharynx. (C) Cross section of the mucosal surface.
The feasibility of this goal is supported by two studies in adult Danish twins. Data from a large study of 617 twin pairs indicate that host genetics play a limited role in determining S. aureus nostril colonization and suggest a larger role for environmental factors, which could include the microbiota [6]. In a follow-up study of 46 monozygotic and 43 dizygotic twin pairs, the bacterial composition of the nasal microbiota is also predominantly an environmentally derived phenotype with host genetics playing a minor role in composition, but a larger role in determining bacterial density on nasal surfaces [7]. These studies, along with evidence that nasal microbiota composition changes over time, including seasonal variation [8–13], support the hypothesis that nasal microbiota composition could be altered for therapeutic benefit [7,14]. This hypothesis is bolstered by reports of negative correlations in colonization between key pathobionts (e.g., S. pneumoniae and S. aureus) and select benign commensals and of alterations in microbiota composition in disease states, e.g., middle ear infections (otitis media) [7,15–26]. Such studies further highlight the need to understand the role and function of the bacterial species that commonly reside in the human nasal passages and commensal–pathobiont interactions.
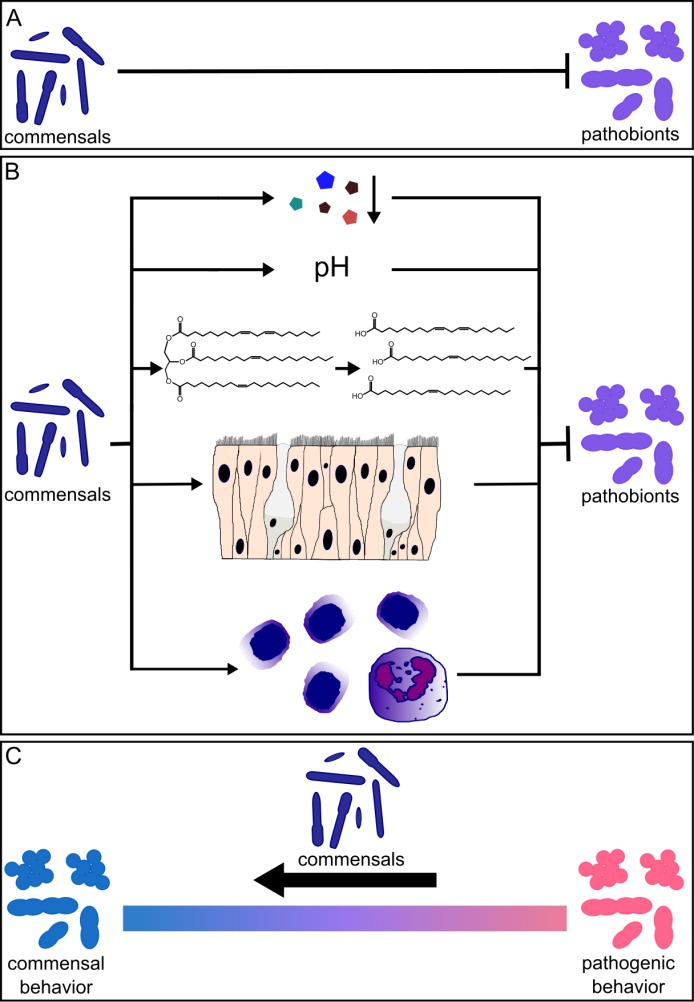
As recently reviewed for gut microbiota [27] and illustrated in Fig 2, beneficial nasal bacteria might impact pathobionts by inhibiting colonization and proliferation (colonization resistance) via (A) direct or (B) indirect mechanisms or by (C) shifting pathogen behavior towards commensalism and away from pathogenesis (Fig 2). Here we review some recent advances in determining the molecular mechanisms of commensal–pathobiont interactions among bacterial members of the microbiota of the nasal passages.
Fig 2. Commensal–pathobiont interactions that are beneficial to the host.
Commensal bacteria can impact pathobionts in a manner beneficial to the host via (A) direct inhibition, e.g., production of antimicrobials; (B) indirect inhibition, e.g., competition for nutrients, modification of the habitat via acidification of environmental pH, alteration of host compounds or secretion of toxic metabolite(s), promotion of host epithelial barrier function, or stimulation of the host immune system; or by (C) behavior modification, e.g., shifting pathogens towards commensalism.
What Is Known about the Composition of the Bacterial Microbiota of the Nasal Passages?
The human nasal passages and nasopharynx (Fig 1) host a distinctive bacterial community that is increasingly well characterized via culture-independent 16S rRNA gene surveys of different age groups in health and disease (e.g., as reviewed in [28]). In prepubertal children, members of the phyla Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes commonly colonize the nostrils and nasopharynx with the latter two exhibiting lower relative abundance on average than the other two [8,9,12,13,17,18,20,23,29,30]. Of the Actinobacteria, Corynebacterium is typically the dominant genus detected [8,12,23,29]. A clear shift in the nostril microbiota occurs during puberty [29] and persists in the majority of healthy adults under age 65 years; it becomes dominated by Actinobacteria, in particular the genera Corynebacterium and Propionibacterium, and Firmicutes, in particular the genus Staphylococcus [7,11,16,19,21,24,25,29,31–35]. In spite of transitions in the type of epithelial surface from skin to respiratory epithelium (Fig 1), a high similarity exists in the bacterial microbiota along the length of the nasal passages [19,33]; however, the nasal cavity is reported to host a more diverse bacterial community [19]. Among the common members of the nasal microbiota in both children and adults are known pathobionts.
What Are the Common Nasal Pathobionts?
The primary middle ear and respiratory bacterial pathogens, S. pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis, commonly colonize the nasal passages of healthy children, with S. aureus usually present less often. In contrast, S. aureus colonization of the nasal passages is much more common in adults, whereas S. pneumoniae and H. influenzae are less common. We favor the term pathobiont for these species because pathobionts are by definition commensal members of the host’s microbiota, whereas opportunistic pathogens may be commensal, environmental, or zoonotic in origin. In addition, these nasal pathobionts can cause infection in healthy hosts, e.g., healthy children and adolescents, whereas opportunistic pathogens generally only infect hosts who are compromised in some manner, e.g., decreased immune function or a significant breakdown in barrier function, such as with traumatic open wounds. (Although of great interest, the research on pathobiont–pathobiont interactions in the upper respiratory tract is beyond the scope of this review.) Recent upper respiratory tract cross-sectional studies that combine culture-based detection of pathobionts with high-throughput-sequencing characterization of microbiota in adults and children reveal inverse correlations between levels of S. aureus or S. pneumoniae and specific commensal bacteria (e.g., [7,12,16,17,19,20,23–26,35]). These correlations provide the impetus for experiments to test for direct antagonism between these specific commensals and pathobionts and, when these hypotheses are verified, to uncover the molecular mechanisms involved, as reviewed below.
What Is Currently Known about Commensal–Pathobiont Interactions in Nasal Microbiota?
Culture-dependent and -independent surveys indicate that non-diphtheriae Corynebacterium spp. commonly colonize the pediatric and adult nasal passages [8,11,12,16,19–26,29–34,36,37]. Yet, the function of Corynebacterium spp. in these habitats is understudied and remains poorly understood. This is likely because most commensal Corynebacterium spp. do not cause disease. Culture-dependent studies indicate that C. accolens, C. tuberculostearicum, C. amycolatum, C. aurimucosum, C. propinquum, and C. pseudodiphtheriticum commonly colonize the adult nose [33,37]. Data on which species colonize the pediatric nose are sparse, but a recent 16S rRNA gene survey of pediatric nasopharyngeal samples detected operational taxonomic units (OTUs) resembling C. accolens and C. pseudodiphtheriticum/C. propinquum [20]. The observed inverse correlation between relative abundances of Corynebacterium spp. and S. pneumoniae in the noses of children under seven years old leads to the hypothesis that antagonism exists between these two groups of bacteria and that Corynebacterium spp. might be protective against pneumococcal colonization [23,26]. We observed that in vitro C. accolens, a lipid-requiring species, releases antipneumococcal free fatty acids from representative human skin surface triacylglycerols; we also identified a primary C. accolens triacylglycerol lipase [26]. This might represent a mechanism by which C. accolens antagonizes S. pneumoniae growth in vivo, thus contributing to colonization resistance against S. pneumoniae.
An inverse correlation between the genus Corynebacterium and S. aureus is reported in some studies of adult nasal microbiota [15,16,19,22,25], a few of which have examined this at the species level for Corynebacterium [16,19]. For example, in a cohort of 40 healthy adults, S. aureus negatively correlated with higher relative abundance of C. accolens and positively correlated with C. pseudodiphtheriticum [16]. In contrast, in a cohort of twelve adults, six with persistent S. aureus nasal colonization, C. accolens positively correlated with S. aureus colonization and in vitro S. aureus enhanced C. accolens growth [19]. In the same study, C. pseudodiphtheriticum negatively correlated with S. aureus and inhibited S. aureus growth during in vitro cocultivation [19]. The variation of results between different studies speaks to the potential complexity of S. aureus–Corynebacterium interactions, including the possibility of strain-level variations, and highlights the need for research to determine the molecular mechanisms involved, which are unlikely to be limited to inhibition.
In considering whether commensal Corynebacterium spp. might have a future role in managing nasal microbiota composition, there are precedents for testing commensal Corynebacterium spp. as probiotics for eradication of S. aureus nasal colonization, albeit in small cohorts [15,38]. For example, Uehara and colleagues report that repeatedly implanting an unidentified Corynebacterium sp. (Co304) eradicated S. aureus colonization in 12 of 17 healthy adult carriers [15].
Additional examples of commensal–pathobiont interactions have been described for cutaneous Propionibacterium spp., which include P. acnes, P. avidum, and P. granulosum. These are common members of the nostril microbiota in late adolescence and adulthood [7,16,19,21,29–34] and are also detected on nasal mucosal surfaces [19,33,36]. Humans appear to be unusual among mammals in hosting Propionibacterium on the skin, and this might relate to the abundance of triacylglycerols in human sebum and skin-surface lipids [39]. P. acnes is the dominant bacterial inhabitant of the pilosebaceous glands (pores) of skin (Fig 1) [40] and is present on the skin of most adults, at least in developed countries [29,31,34]. (A note of caution: the more recent next-generation sequencing (NGS) studies that rely on the V4 region of the 16S rRNA gene fail to detect the same levels of Propionibacterium as prior 454 studies [41], likely because the commonly used 806R primer poorly detects Propionibacterium.) Given the ubiquity of Propionibacterium on adult human skin surfaces, including the nasal vestibules, several studies have examined potential interactions between cutaneous Propionibacterium spp. and S. aureus. For example, coproporphyrin III (CIII), a diffusible small molecule excreted by nostril- and skin-associated Propionibacterium spp., induces S. aureus aggregation and biofilm formation; this activity is dependent on dose, growth phase, and pH [42]. In other work, Huang and colleagues published several studies that describe effects of P. acnes on S. aureus virulence and/or growth in skin wounds [43,44]. Lo et al. showed that P. acnes-secreted Christie–Atkins–Munch-Petersen (CAMP) factor enhances the hemolytic and cytolytic activity of S. aureus-secreted beta-hemolysin (sphingomyelinase C) [44]. In a mouse skin infection model, when compared to monoinfection, coinfection of P. acnes with S. aureus enhanced S. aureus virulence in vivo in a manner dependent on active CAMP factor and beta-hemolysin [44]. Shu et al. demonstrated that, when P. acnes and 13C-labeled glycerol are injected into a mouse ear, P. acnes ferments glycerol, a carbon source available on human skin, to short-chain fatty acids, e.g., propionic acid. These products of P. acnes glycerol fermentation inhibited growth of USA300 CA-MRSA both in vitro and in vivo in a mouse model of skin wounds [43], although it was unclear how much of this was due to low pH alone. It is likely that these interactions observed on skin surfaces outside of the nasal passages will be applicable to commensal–pathobiont interactions on the skin surfaces of the nasal vestibule.
In addition to the Corynebacterium–pathobiont and Propionibacterium–pathobiont interactions described above, interactions between Staphylococcus epidermidis, a commensal, and S. aureus have also been investigated. This is driven largely by the hypothesis that, due to their close phylogenetic relationship, these species might compete for a similar niche within the nasal habitat. Because much of this research has been recently reviewed (e.g., [45,46]), it is not covered here.
What Are Future Research Directions in This Area?
The research discussed here lays the foundation for future testing of harmless nasal bacterial species, e.g., C. accolens, as potential probiotics. These studies also set the stage for identifying commensal-produced small molecules with the potential for use in managing nasal microbiota composition to promote health. For example, recently developed algorithms that can identify biosynthetic gene clusters whose products synthesize small molecules that might be involved in interspecies interactions within human microbiota promise rapid expansion in this area of research [47,48]. In addition, we expect that microbiota composition studies will continue to lead to the recognition of potentially important, yet previously neglected, commensals, e.g., Dolosigranulum pigrum, which is overrepresented in children without S. pneumoniae nasal colonization and which in older adults appears to be an informative predictor for the lack of S. aureus colonization [7,17,26]. This review highlights an exciting early stage in the exploration of the molecular mechanisms of interspecies interactions that sculpt nasal microbiota composition and influence pathobiont colonization. Much of this work will also relate directly to the composition of skin microbiota. Because pathobiont colonization is a prerequisite for infection and transmission, a rational approach to prevent infections is to limit or decrease pathobiont abundance and to shift pathobiont behavior towards commensalism using either commensal-derived compounds or commensals as probiotics. We look forward to an increase in research on commensal–pathobiont interactions within the human microbiome and an ever-increasing understanding of the functional significance of our commensal and mutualist bacterial partners.
Acknowledgments
We thank Matthew Ramsey, Megan Lambert, and Isabel F. Escapa for constructive comments and Virge Kask for the artwork in Fig 1. We apologize to colleagues whose work was not cited due to space limitations, e.g., work on sinus microbiota in health and disease, which has been recently well reviewed.
Funding Statement
This work was supported by the National Institutes of Health through the National Institute of Allergy and Infectious Diseases R01 AI101018 (KPL) and the National Institute of General Medical Sciences R01 GM117174 (KPL), by the McIntosh Foundation (KPL), and by the Swiss National Science Foundation and Swiss Foundation for Grants in Biology and Medicine P3SMP3_155315 (SDB). Funders had no role in the preparation of this manuscript or decision to publish.
References
- 1. Casadevall A, Pirofski LA. What is a host? Incorporating the microbiota into the damage-response framework. Infect Immun. 2015;83(1):2–7. 10.1128/IAI.02627-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang SS, Johnson KM, Ray GT, Wroe P, Lieu TA, Moore MR, et al. Healthcare utilization and cost of pneumococcal disease in the United States. Vaccine. 2011;29(18):3398–412. 10.1016/j.vaccine.2011.02.088 . [DOI] [PubMed] [Google Scholar]
- 3. Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med. 2013;173(21):1970–8. 10.1001/jamainternmed.2013.10423 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–5. Epub 2008/05/30. 10.1038/nature07008 . [DOI] [PubMed] [Google Scholar]
- 5. Wilson M. Microbial inhabitants of humans: their ecology and role in health and disease: Cambridge University Press; 2005. [Google Scholar]
- 6. Andersen PS, Pedersen JK, Fode P, Skov RL, Fowler VG Jr., Stegger M, et al. Influence of host genetics and environment on nasal carriage of Staphylococcus aureus in danish middle-aged and elderly twins. J Infect Dis. 2012;206(8):1178–84. Epub 2012/08/09. jis491 [pii] 10.1093/infdis/jis491 ; PubMed Central PMCID:3448969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu CM, Price LB, Hungate BA, Abraham AG, Larsen LA, Christensen K, et al. Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv. 2015;1(5):e1400216 Epub 2015/11/26. 10.1126/sciadv.1400216 1400216 [pii]. ; PubMed Central PMCID:4640600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS ONE. 2011;6(2):e17035 10.1371/journal.pone.0017035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mika M, Mack I, Korten I, Qi W, Aebi S, Frey U, et al. Dynamics of the nasal microbiota in infancy: a prospective cohort study. J Allergy Clin Immunol. 2015;135(4):905–12 e11. Epub 2015/02/01. S0091-6749(14)03709-9 [pii] 10.1016/j.jaci.2014.12.1909 . [DOI] [PubMed] [Google Scholar]
- 10. Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJ, et al. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci U S A. 2015;112(22):E2930–8. 10.1073/pnas.1423854112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–7. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190(11):1283–92. Epub 2014/10/21. 10.1164/rccm.201407-1240OC . [DOI] [PubMed] [Google Scholar]
- 13. Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17(5):704–15. 10.1016/j.chom.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4(137):137rv5 Epub 2012/06/08. 10.1126/scitranslmed.3004183 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uehara Y, Nakama H, Agematsu K, Uchida M, Kawakami Y, Abdul Fattah AS, et al. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect. 2000;44(2):127–33. Epub 2000/02/09. 10.1053/jhin.1999.0680 . [DOI] [PubMed] [Google Scholar]
- 16. Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, et al. A poke into the diversity and associations within human anterior nare microbial communities. ISME J. 2010;4(7):839–51. Epub 2010/02/26. 10.1038/ismej.2010.15 . [DOI] [PubMed] [Google Scholar]
- 17. Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Applied and Environmental Microbiology. 2012;78(17):6262–70. Epub 2012/07/04. 10.1128/AEM.01051-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hilty M, Qi W, Brugger SD, Frei L, Agyeman P, Frey PM, et al. Nasopharyngeal microbiota in infants with acute otitis media. J Infect Dis. 2012;205(7):1048–55. Epub 2012/02/22. jis024 [pii] 10.1093/infdis/jis024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host & Microbe. 2013;14(6):631–40. Epub 2013/12/18. 10.1016/j.chom.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Biesbroek G, Bosch AATM, Wang X, Keijser BJF, Veenhoven RH, Sanders EAM, et al. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med. 2014;190(3):298–308. 10.1164/rccm.201401-0073OC . [DOI] [PubMed] [Google Scholar]
- 21. Camarinha-Silva A, Wos-Oxley ML, Jauregui R, Becker K, Pieper DH. Validating T-RFLP as a sensitive and high-throughput approach to assess bacterial diversity patterns in human anterior nares. FEMS Microbiol Ecol. 2012;79(1):98–108. Epub 2011/11/10. 10.1111/j.1574-6941.2011.01197.x . [DOI] [PubMed] [Google Scholar]
- 22. Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl Environ Microbiol. 2003;69(1):18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial communities of the upper respiratory tract and otitis media in children. mBio. 2011;2(1):e00245–10. Epub 2011/02/03. 10.1128/mBio.00245-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. The human nasal microbiota and Staphylococcus aureus carriage. PLoS ONE. 2010;5(5):e10598 10.1371/journal.pone.0010598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson RC, Ellis MW, Lanier JB, Schlett CD, Cui T, Merrell DS. Correlation between nasal microbiome composition and remote purulent skin and soft tissue infections. Infect Immun. 2015;83(2):802–11. 10.1128/IAI.02664-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio. 2016; 6:e01725–15(6). 10.1128/mBio.01725-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McKenney PT, Pamer EG. From Hype to Hope: The Gut Microbiota in Enteric Infectious Disease. Cell. 2015;163(6):1326–32. 10.1016/j.cell.2015.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Steenhuijsen Piters WA, Sanders EA, Bogaert D. The role of the local microbial ecosystem in respiratory health and disease. Philos Trans R Soc Lond B Biol Sci. 2015;370(1675). Epub 2015/07/08. rstb.2014.0294 [pii] 10.1098/rstb.2014.0294 ; PubMed Central PMCID:4528492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012;4(10):77 10.1186/gm378 ; PubMed Central PMCID:3580446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stearns JC, Davidson CJ, McKeon S, Whelan FJ, Fontes ME, Schryvers AB, et al. Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. ISME J. 2015;9(5):1246–59. 10.1038/ismej.2014.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–2. Epub 2009/05/30. 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio. 2010;1(3):e00129–10 Epub 2010/08/31. 10.1128/mBio.00129-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaspar U, Kriegeskorte A, Schubert T, Peters G, Rudack C, Pieper DH, et al. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol. 2015. 10.1111/1462-2920.12891 . [DOI] [PubMed] [Google Scholar]
- 34. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–14. 10.1038/nature11234 ; PubMed Central PMCID:3564958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bessesen MT, Kotter CV, Wagner BD, Adams JC, Kingery S, Benoit JB, et al. MRSA colonization and the nasal microbiome in adults at high risk of colonization and infection. J Infect. 2015;71(6):649–57. Epub 2015/09/04. S0163-4453(15)00261-3 [pii] 10.1016/j.jinf.2015.08.008 . [DOI] [PubMed] [Google Scholar]
- 36. Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS ONE. 2010;5(12):e15216 10.1371/journal.pone.0015216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rasmussen TT, Kirkeby LP, Poulsen K, Reinholdt J, Kilian M. Resident aerobic microbiota of the adult human nasal cavity. APMIS. 2000;108(10):663–75. 10.1034/j.1600-0463.2000.d01-13.x . [DOI] [PubMed] [Google Scholar]
- 38. Kiryukhina NV, Melnikov VG, Suvorov AV, Morozova YA, Ilyin VK. Use of Corynebacterium pseudodiphtheriticum for elimination of Staphylococcus aureus from the nasal cavity in volunteers exposed to abnormal microclimate and altered gaseous environment. Probiotics Antimicro Prot. 2013;5:233–8. 10.1007/s12602-013-9147-x [DOI] [PubMed] [Google Scholar]
- 39. Webster GF, Ruggieri MR, McGinley KJ. Correlation of Propionibacterium acnes populations with the presence of triglycerides on nonhuman skin. Appl Environ Microbiol. 1981;41(5):1269–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133(9):2152–60. 10.1038/jid.2013.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nelson MC, Morrison HG, Benjamino J, Grim SL, Graf J. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PLoS ONE. 2014;9(4):e94249 10.1371/journal.pone.0094249 ; PubMed Central PMCID:3983156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wollenberg MS, Claesen J, Escapa IF, Aldridge KL, Fischbach MA, Lemon KP. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. MBio. 2014;5(4):e01286–14. 10.1128/mBio.01286-14 ; PubMed Central PMCID:4120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shu M, Wang Y, Yu J, Kuo S, Coda A, Jiang Y, et al. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus . PLoS ONE. 2013;8(2):e55380 10.1371/journal.pone.0055380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lo CW, Lai YK, Liu YT, Gallo RL, Huang CM. Staphylococcus aureus hijacks a skin commensal to intensify its virulence: immunization targeting beta-hemolysin and CAMP factor. J Invest Dermatol. 2011;131(2):401–9. 10.1038/jid.2010.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krishna S, Miller LS. Host-pathogen interactions between the skin and Staphylococcus aureus . Curr Opin Microbiol. 2012;15(1):28–35. 10.1016/j.mib.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Christensen GJ, Bruggemann H. Bacterial skin commensals and their role as host guardians. Benef Microbes. 2014;5(2):201–15. 10.3920/BM2012.0062 . [DOI] [PubMed] [Google Scholar]
- 47. Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158(2):412–21. 10.1016/j.cell.2014.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158(6):1402–14. 10.1016/j.cell.2014.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]