Abstract
A simple and cost effective method to independently and stereo-specifically incorporate [1H,13C]-methyls in Leu and Val in proteins is presented. Recombinant proteins for NMR studies are produced using a tailored set of auxotrophic E. coli strains. NMR active isotopes are routed to either Leu or Val methyl groups from the commercially available and scrambling-free precursors α-ketoisovalerate and acetolactate. The engineered strains produce deuterated proteins with stereospecific [1H,13C]-methyl labeling separately at Leu or Val amino acids. This is the first method that achieves Leu-specific stereospecific [1H,13C]-methyl labeling of proteins and scramble-free Val-specific labeling. Use of auxotrophs drastically decreases the amount of labeled precursor required for expression without impacting the yield. The concept is extended to Thr methyl labeling by means of a Thr-specific auxotroph that provides enhanced efficiency for use with the costly L-[4-13C,2,3-2H2,15N]-Thr reagent. The Thr-specific strain allows for the production of Thr-[13CH3]γ2 labeled protein with an optimal isotope incorporation using up to 50% less labeled Thr than the traditional E. coli strain without the need for 2H-glycine to prevent scrambling.
Keywords: Methyl labeling, Large proteins, Auxotrophic strains, NMR
Introduction
Deuteration, selective [1H,13C]-methyl labeling and methyl TROSY effect have expanded the scope and applicability of liquid state NMR to protein systems up to several hundreds kilo Daltons (kDa). This technology, pioneered by the group of Lewis Kay and co-workers (Gardner and Kay, 1997; Goto et al., 1999; Tugarinov et al., 2006) yielded new insights into the function of large proteins (Audin et al., 2013; Gelis et al., 2007; Karagoz et al., 2011; Kato et al., 2011; Kerfah et al., 2015b; Religa et al., 2010; Rosenzweig and Kay, 2014; Rosenzweig et al., 2013; Saio et al., 2014; Sprangers and Kay, 2007; Tzeng and Kalodimos, 2012). Alpha-keto acid precursors that allow for highly specific and scramble-free labeling of Ile, Leu and Val methyls are commercially available (Tugarinov and Kay, 2005). Additional methods for selective [1H,13C]-methyl labeling of proteins on the Ala (Ayala et al., 2009; Isaacson et al., 2007; Popovych et al., 2009), Met (Gelis et al., 2007), Ile-γ2 (Ruschak et al., 2010) and most recently Thr residues (Saio et al., 2014; Sinha et al., 2011; Velyvis et al., 2012) have been reported to date.
The six methyl-bearing amino acids - Ala, Ile, Leu, Met, Thr and Val - are highly abundant, accounting for 35–45% of the primary structure and they are typically well dispersed throughout the protein structure. Full incorporation of [1H,13C]-methyl-labeled reagents and complete deuteration of the remaining protons are essential for optimal sensitivity and resolution in large systems (Kay, 2005). Reducing proton density also prevents 1H-1H spin diffusion and enables the detection of NOEs between more distal atoms (Sounier et al., 2007). Ala, Ile, Met, and Thr rely on distinct reagent precursors and can be independently incorporated while Leu and Val rely on the same α-keto acid for biosynthesis (Fig. 1). Leu and Val methyl groups typically account for more than 50% of all methyl probes in a protein and their correct identification is critical for the success of any NMR study. Owing to their relatively small chemical shift dispersion and overlapping chemical shift ranges, simultaneous labeling of Leu and Val can often result in crowded and difficult to analyze spectra, especially as protein size increases. Simultaneous stereospecific labeling of pro-S or pro-R methyl groups in both Leu and Val via synthesis of specifically labeled acetolactate precursors was shown to alleviate spectral crowding and provides significant sensitivity enhancement in large proteins (Gans et al., 2010). However, there are currently no methods available to separately and stereospecifically label Leu or Val. Doing so would improve the accuracy of the divide-and-conquer strategy (Gelis et al., 2007; Pickford and Campbell, 2004; Sprangers and Kay, 2007), simplify assignment by point mutations (Amero et al., 2011), and empower the structure- and NOESY-based automatic assignment of methyl groups using recently reported methods (Chao et al., 2012; Pickford and Campbell, 2004; Xu and Matthews, 2013).
Figure 1.
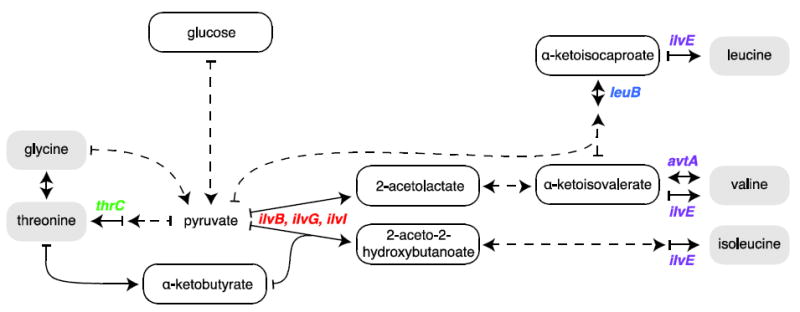
Simplified pathways for Ile, Leu, Val and Thr biosynthesis is outlined starting from glucose, as the main carbon source, according to the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/). The commercially available isotope-labeled amino acid precursors (α-ketobutyrate, α-ketoisovalerate, α-ketoisocaproate, 2-aceto-2-hydroxybutanoate and 2-acetolactate) are also encircled. Each solid arrow is indicative of one enzymatic reaction. Dashed arrows indicate multi-step reactions and double arrows represent reversible enzymatic reaction. The deleted genes are shown for the AcL- (red), leucine- (blue), valine- (purple) and Thr- (green) strain phenotypes.
Specific labeling is generally achieved using the proper precursor to route the isotope-containing reagent into the amino acid of interest. Val is synthesized via transamination of α-ketoisovalerate and thus it is not possible to isolate a biosynthesis-specific precursor (Fig. 1). Conversely, the branched metabolism responsible for Leu synthesis includes five more steps from α-ketoisovalerate to Leu production. (Lichtenecker et al., 2013) reported the synthesis of one labeled intermediate, the α-ketoisocaproate, which can be used for Leu labeling. Deuterated [1H,13C]-monomethyl and [1H,13C]-dimethyl α-ketoisocaproate can also be synthesized. However the [1H,13C]-monomethyl compound is produced as a racemic mixture and therefore cannot be used as a stereo-array isotope labeled precursor. A simple method to achieve Val-only methyl labeling, with minimal decrease in overall incorporation, is to inhibit Leu biosynthesis by supplementing the minimal media with either 10% rich labeled media (e.g. Bioexpress or Isogrow) (Tzeng et al., 2012) or 20 mg/L of deuterated Leu (Mas et al., 2013). Unfortunately, Leu-specific labeling cannot be obtained by analogous means since adding deuterated Val would result in its reversible conversion to α-ketoisovalerate and negatively impact the [1H,13C]-methyl incorporation on Leu. Finally, the strategy of direct addition of labeled amino acids to the culture medium (Metzler et al., 1996) was further demonstrated recently with the synthesis of stereo-specifically labeled Leu and Val, and their subsequent use to achieve stereospecific residue-specific methyl labeling of proteins (Miyanoiri et al., 2013). However, the cost and the availability of these labeled amino acids are still a major hurdle.
Thr methyl labeling has proved to be somewhat challenging (Velyvis et al., 2012). For instance, isotope scrambling into Ile-δ 1 through 13Cγ2-Thr conversion to 13C-α-ketobutyrate by the Thr dehydratases (ilvA and tdcB) was reported. In addition, its isotope incorporation was found to be suboptimal, likely because the 13Cγ2-Thr bulk was isotopically diluted by its conversion to Gly by Thr aldolase (ltaE) or by its endogen production by Thr synthase (thrC) (Fig. 1). Velyvis et al. (2012) reported the simultaneous addition of 50 mg/L of 13Cγ2-Thr with 50 mg/L of 13C-α-ketobutyrate and 100 mg/L of d2-Gly to obtain around 88% isotope incorporation into Thr-γ2 methyl along with close to full labeling of Ile-δ1 methyl. However, it is often beneficial to only label single methyl-bearing amino acids of interest. Last but not least, 13Cγ2-Thr is costly and therefore optimal use of this reagent is important.
We present a method for residue- and stereo-specific labeling of Leu and Val as well as scramble-free Thr methyl labeling with minimal reagent usage. Specialized strains were designed that route the isotope incorporation pathways to either Leu, Val or Thr methyl groups from commercially available precursors (α-ketoisovalerate, acetolactate and Thr). We show successful stereospecific and residue-specific Leu and Val methyl labeling using four different proteins ranging from 10 to 80 kDa: the peptidyl-prolyl isomerase domain (PPD) of Trigger Factor, the calmodulin, the catabolite activator protein (CAP) and the malate synthetase G (MSG). The group of auxothrophs engineered yield deuterated proteins with [1H,13C]-methyl labeling on any of Val-γ1/γ2, Leu-δ1/δ2, Val-γ1/Leu-δ1, Val-γ2/Leu-δ2, Val-γ1, Val-γ2, Leu-δ1 or Leu-δ2. In this method, the amount of precursor is drastically decreased without affecting the yield. In addition, our strains are compatible with Thr, Ala and Met [1H,13C]-methyl labeling protocols, enabling useful combinations of methyl probes that can facilitate residue type identification, and simplify assignment by computational or mutagenic approaches. In the case of Thr, the use of an auxotrophic strain improves the incorporation efficiency and selectivity. The resulting engineered strain can produce fully Thr-γ2 methyl-labeled protein using 50% of the 13Cγ2-Thr reagent as compared to the previous method, and it does so without the need for 2H-Gly reagent to prevent scrambling.
Materials and Methods
Isotopes
The following reagents were used in this study. Names are given in IUPAC and common name in parentheses is used throughout the text and in Supp. Table S1 for simplicity. The 2-hydroxy-2-[13C]methyl-3-oxo-[4,4,4-2H3]butanoate (13C-proS-acetolactate), 2-hydroxy-2-[2H3]methyl-3-oxo-[2,4-13C2]butanoate (13C-proR-acetolactate) and 2-[2H3]aceto-2-hydroxy-[4-13C,3,3-2H2]butanoate (13C-aceto-hydroxybutanoate) were obtained from NMR-bio. The 2-oxo-3-[2H]-3-[2H3]methyl-[4-13C]butanoate (13C-monomethyl α-ketoisovalerate), 2-oxo-[4-13C,3,3-2H2]butanoate (13C-α-ketobutyrate), [2H10]-L-leucine (d10-Leu), [2H10]-L-isoleucine (d10-Ile) and [2H8]-L-valine (d8-Val) were obtained from Cambridge Isotope Laboratory (CIL). The ε-[13C]methyl-L-methionine (13Cε-Met) were obtained from Sigma Aldrich. The L-[4-13C,2,3-2H2,15N]threonine (13Cγ2-Thr) was synthesized in our laboratory following the protocol of (Velyvis et al., 2012).
Engineering of E. coli strains for specific methyl labeling
The gene deletion mutants [JW5605(ΔilvD); JW5606(ΔilvE); JW5652(ΔavtA); JW5807(ΔleuB); JW2004(ΔhisB); JW1254(ΔtrpC); JW0003(ΔthrC); JW0076(ΔilvI); JW3704(ΔilvG) and JW3646(ΔilvB)] were obtained from the Keio collection (University of Keio, Japan). To perform the gene deletion mutagenesis (P1 transduction), BL21(DE3) were grown in 10 ml of LB medium supplemented with 5 mM CaCl2, and when the cell density (OD600) reached 0.5 to 0.6, 100 μL of P1 phage lysate were added. After 30 min incubation at 30°C, 100 mM sodium citrate (pH5.5) was added and the culture was incubated another 30 min at 37 °C. After pelleting by centrifugation, the cells were suspended into LB supplemented with 200 mM sodium citrate (pH5.5) and the cells were plated onto the LB-kanamycin (Km)(50 μg/ml) plates. The colonies were tested for the gene deletion by polymerase chain reactions (PCR). As a final step, the Km cassette was removed from the genome. The deletion mutants were grown in LB medium supplemented with Km (50 μg/ml). The cells were then transformed with pCP20 by electroporation and plated on LB-ampicillin (Amp) (100 μg/ml) plates, which were incubated at 30°C for plasmid selection. The resulting colonies were re-streaked on LB plates, which were incubated at 42°C overnight. The loss of the Km cassette and pCP20 was confirmed using three different plates: LB, LB with Km and LB with Amp.
In this study, the following E. coli strains were used and their naming is given according to their phenotype (within parenthesis) and genotype (in italic): BL21(Val-) for ΔilvE, ΔavtA; BL21(Leu-) for ΔleuB; BL21(AcL-) for ΔhisB, ΔtrpC, ΔilvB, ΔilvG, ΔilvI; BL21(AcL-/Val-) for ΔhisB, ΔtrpC, ΔilvB, ΔilvG, ΔilvI, ΔilvE, ΔavtA; BL21(AcL-/Leu-) for ΔhisB, ΔtrpC, ΔilvB, ΔilvG, ΔilvI, ΔleuB; and BL21(Thr-) for ΔthrC.
Production of Leu/Val methyl-labeled protein samples using engineered strains
Either BL21(AcL-) or BL21(AcL-/Leu-) strains were used to specifically label Val methyl groups. Leu methyl labeling was achieved using the BL21(AcL-/Val-) strain. The engineered BL21 strain cells, transformed with the PPD- or MSG-containing plasmid, were grown at 37°C in 50 mL (or 250 mL for MSG) of M9-D2O medium supplemented with ampicillin (100 mg/L), glucose (2 g/L), Leu (30 mg/L), Ile (30 mg/L), Val (30 mg/L), Trp (30 mg/L), and His (30 mg/L). When the OD600 reached 0.3 to 0.4, the cells were pelleted and washed by M9-D2O medium twice and re-suspended in 50 ml (or 250 mL for MSG) of M9-D2O medium containing ampicillin (100 mg/L), glucose (2 g/L), Trp (30 mg/L), His (30 mg/L) and 13Cε-Met at 80 mg/L. In addition, the medium was supplemented with Leu (30 mg/L) or Val (30 mg/L), and 13C-proS-acetolactate or 13C-monomethyl α-ketoisovalerate, accordingly to the desired specific labeling. After incubation for 1.5 hours at 37 °C, protein overexpression was induced with IPTG to a final concentration of 1 mM and the cells were harvested 2.5 hours later for PPD (or after overnight incubation for MSG). For MSG production, 20 mg/L of 13C-proS-acetolactate was added after cell re-suspension and together with IPTG induction (40 mg/L in total). For Leu labeling, 0.1 g/L of [2H]-celtone base powder (CIL) was added during the growth step. For PPD sample, deuteration is not necessary, thus we used only protonated nutriments to supplement the growth media. Conversely, for MSG sample production, deuterated Ile, Leu, Val and glucose were used during both growth and induction step. As a cost saving measure, protonated His and Trp were used in place of deuterated versions. PPD (Saio et al., 2014) and MSG (Kerfah et al., 2015a) were purified as previously described.
Each strain employs different amino acids and [1H,13C]-methyl-containing precursors in order to obtain the desired labeling. To determine how the reagent concentrations impact the yield and the isotope incorporation, we tested different medium compositions. After growing and centrifuging the cells, we split the pellet into a set of induction media containing different amount of single metabolites. We used BL21(Acl-) to produce U-[12C,1H], Leu/Val-[13CH3]pro-S PPD samples using 10, 15, 20, 25, 30 or 60 mg/L of 13C-proS-acetolactate (with 30 mg/L of Ile); to produce unlabeled PPD samples using 10, 20, 30, 60 and 120 mg/L of Ile (with 30 mg/L of Leu and Val); We used BL21(Acl-/Val-) to produce U-[12C,1H], Leu-[13CH3/12CD3] PPD samples using 17, 25, 50, or 120 mg/L of 13C-monomethyl α-ketoisovalerate (with 30 mg/L of Ile and Val). Protonated glucose and protonated amino acids were used in both growth and induction steps. The concentration of recombinant proteins (UV absorption at 280nm) and the volume of the protein sample obtained at the end of the purification were measured to assess the protein yield. 13Cε-Met at 80 mg/L was used as internal standard for all labeled proteins unless specified otherwise. The relevant methyl signals were characterized by 1H,13C-HMQC.
Production of Thr methyl-labeled protein samples using engineered strains
Thr [1H,13C]-methyl labeling was achieved using the BL21(Thr-) strain. The engineered BL21 strain cells, transformed with either PPD- or Trigger Factor-containing plasmids, were grown at 37°C in 200 mL of M9-D2O medium supplemented with ampicillin (100 mg/L), 13Cγ2-Thr (25 mg/L) and deuterated glucose (2 g/L). When either the signal suppression or the labeling of Ile-δ1 methyls was targeted, either protonated Ile (20 mg/L) or 13C-α-ketobutyrate (25 mg/L), respectively, was added at OD600 = 0.3, and cells were kept growing for one hour at 37 °C. The production was induced by the addition of IPTG to a final concentration of 0.5 mM at 18 °C and the induced cells were harvested 16 hours later. Incorporation was estimated based on signal intensity to the corresponding PPD sample of equal concentration labeled with Ile, Val and Leu methyls.
NMR spectroscopy
Spectra were recorded at 305 K on Bruker Avance III 700 MHz or 600 MHz spectrometers equipped with triple resonance cryo and ambient temperature probes, respectively. Topspin 3.2 (Bruker BioSpin) was used for data collection and NMRPipe (Delaglio et al., 1995) for spectra processing followed by analysis with Sparky 3.115 (T. D. Goddard and D. G. Kneller, University of California, San Francisco, CA).
RESULTS
Engineering of E. coli strains for specific Leu/Val/Thr methyl labeling
In order to achieve residue- and stereo-specific labeling of either Leu or Val using commercially available isotope precursors, and also to decrease the cost of stereospecific labeling of Leu/Val amino acids, we designed auxotrophic strains to route isotopes to either Leu or Val. In the biosynthetic pathway, the Val amino acid is derived directly from α-ketoisovalerate by two transaminases (Fig. 1), coded by the genes ilvE and avtA. We deleted these two genes to block Val biosynthesis and thus exclusively label Leu. The biosynthesis of Leu from α-ketoisovalerate (Fig. 1) involves five additional reaction steps. The gene leuB codes for one of the involved enzymes and was deleted to block Leu biosynthesis and thus exclusively label Val residues. We validated gene deletions and viability of the strains by confirming that the ΔilvE/ΔavtA and the ΔleuB strains only grow in the presence of Val and Leu amino acids, respectively. The transaminase coded by ilvE gene is reported by the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) to be involved in the biosynthesis of both Leu and Ile (Fig. 1). As anticipated, the ΔilvE/ΔavtA strain failed to grow in the absence of Leu and Ile. Nonetheless, cells growth occurred in the absence of Leu when the medium was supplemented with either α-ketoisovalerate or acetolactate, albeit at a much lower growth rate. It appears that transamination of Leu-precursor is carried out by alternative endogenous transaminases, which are not as efficient as IlvE. Thus, using the ΔilvE/ΔavtA strain the isotope incorporation into Leu methyls was very low (data not shown) with either 13C-monomethyl α-ketoisovalerate or 13C-proS-acetolactate as [1H,13C]-methyl source. It is likely that very slow Leu biosynthesis allows for the precursor to recycle into other metabolic pathways thereby triggering isotope dilution by 12C-glucose. On the other hand, no isotope incorporation issues were noted for Val labeling using the ΔleuB strain fed with 120 mg/L of 13C-monomethyl α-ketoisovalerate. In order to avoid precursor recycling and to decrease the overall α-ketoisovalerate amount needed, we engineered strains that are auxotroph to acetolactate and α-ketoisovalerate. The only enzyme that could be removed from the Leu/Val biosynthesis pathway without affecting pyruvate metabolism was acetolactate synthetase (ALS), an enzyme that is closely involved in Ile synthesis as well as in Leu and Val synthesis. Therefore, altering ALS genes was expected to disrupt Ile along with Leu/Val biosynthesis. The ALS enzyme consists of large and small subunits, coded by six genes in the E. coli genome. To obtain an auxotrophic strain, we attempted to delete all three genes that code for the large subunit of ALS (ilvI, ilvB, ilvG). After preparing the deletion mutant strain (ΔilvI, ΔilvB, ΔilvG), the cells were unable to grow in the absence of Ile, Leu and Val in the medium; however, the growth was recovered in the presence of acetolactate and Ile. To achieve Val specific methyl labeling, we blocked the conversion from acetolactate to Leu by the gene deletion ΔleuB. After this gene deletion, the cells were unable to grow in the presence of the acetolactate and Ile; however, the growth resumed when Leu was supplied.
In order to achieve Leu specific methyl labeling, additional gene deletion mutations (ΔilvE, ΔavtA) were also included to block the conversion of acetolactate to Val. After the additional gene deletions, the cells were unable to grow in the presence of acetolactate and Ile; however, growth resumed when Val was added to the medium. We constructed the strains based on the His and Trp auxotroph strain; so the final genotype for these two strains are ΔhisB, ΔtrpC, ΔilvI, ΔilvB, ΔilvG, ΔleuB for Val labeling and ΔhisB, ΔtrpC, ΔilvI, ΔilvB, ΔilvG, ΔilvE, ΔavtA for Leu labeling.
For Thr methyl labeling, we designed an auxotroph strain by deleting the thrC gene that acts in the final step of Thr biosynthesis. The goal was to decrease both isotopic scrambling and the minimal amount of 13Cγ2-Thr to reach full isotope incorporation. The resulting engineered BL21 (DE3) ΔthrC cells were unable to grow without Thr supplement and the growth resumed when Thr was added, confirming the strain is auxotrophic for Thr.
Val/Leu-specific methyl-labeling using auxotroph strains
Each labeling scheme requires both the specific auxotroph and an appropriately supplemented medium for cell growth and protein overexpression. Because the strains are auxotrophs for both Leu/Val precursors, the cells are grown in two different M9-D2O-based media, one for the growth step and another for the induction step. Both media are supplemented with Ile, Val and Leu. However, for the induction medium, the amino acids to be labeled need to be substituted by the appropriate precursors. An optimization round was conducted to determine the optimal amount of reagent to insure proper yield and high [1H,13C]-methyl incorporation. All experiments performed to optimize the protocols and reagent dosages have been conducted using the catalytic N-terminal PPI domain from E. coli Trigger Factor (PPD). To check isotope incorporation, several methyl-labeled PPD samples were produced with increasing amounts of isotope precursors. The methyl signal intensities were measured by recording 13C-HMQC spectra. Intensities of each methyl type (pro-R or pro-S for Leu and Val) were normalized against the mean signal intensity of the Met ε-methyls used as an internal incorporation standard. The relative intensities with respect to the methionine signals were then plotted against the amount of precursors utilized (Fig. 2). We produced U-[1H,12C], Val/Leu-[13CH3]proS PPD samples using the BL21(AcL-) strain with different amounts of 13C-proS-acetolactate (Fig. 2A). Maximum isotope incorporation was reached when just ~30 mg/L of precursors were added, an order of magnitude less than the 300 mg/L reported in the original paper (Gans et al., 2010). We tested isotope incorporation into Leu using the strain BL21(AcL-/Val-) with different concentrations of 13C-monomethyl α-ketoisovalerate and, in contrast to the poor isotope incorporation observed for the ΔilvE/ ΔavtA strain, we reached maximum incorporation using only 25 mg/L of 13C-monomethyl α-ketoisovalerate (Fig. 2B).
Figure 2.
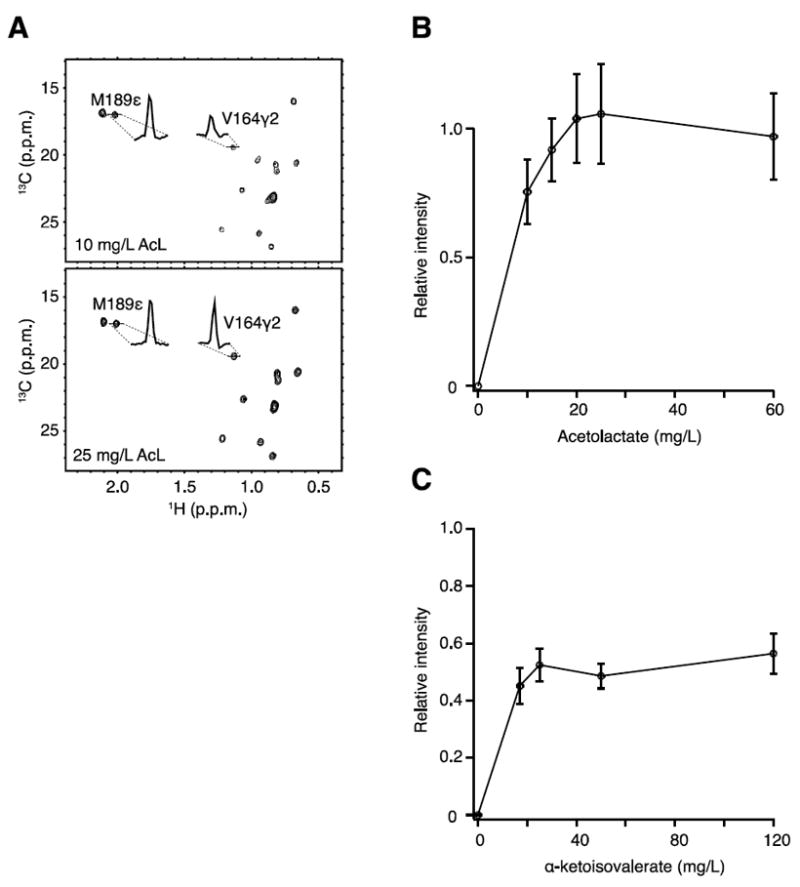
Peptidyl prolyl isomerase domain (PPD) of Trigger Factor is expressed by (A, B) BL21(AcL-) or (C) BL21(AcL-,Val-) using increasing concentrations of either (A, B) 13C-proS-acetolactate or (C) α-13C-ketoisovalerate. (A) Two spectra of U-[1H, 12C,], Leu/Val-[13CH3]-proS PPD are shown with the proton cross-sections depicted for Met189 ε-methyl and Val164 γ2-methyl. The signal of Met ε-methyls is used as internal standard to assess the level of isotope incorporation for the methyls of interest. The intensities of Val/Leu peaks in the 13C-HMQC (B, 9 Leu/Val-proS peaks; C, 6 Leu-proS/proR peaks) have been normalized using the average intensities of Met ε-methyls, and the mean ± sd is plotted against the concentration of precursor. Relative intensity above ‘1’ should be taken as 100% incorporation and is within the variation in intensity of the methyl peaks examined as indicated by error bars.
Expression yields using different conditions in this work were compared to the standard protocol consisting of protein overexpression in BL21 strain with 120 mg/L of 13C-monomethyl α-ketoisovalerate to assess whether or not our method impacts the expression yield. Protein expression using BL21(AcL-) increased with increasing amounts of 13C-proS-acetolactate. The yield of U-[1H, 12C], Leu/Val-[13CH3]proS PPD at 40 mg/L 13C-proS-acetolactate matched the yield of the standard protocol. Interestingly, when using 60 mg/L of 13C-proS-acetolactate, twice as much protein was produced compared to the standard protocol. In this case, the contributing cost of acetolactate to the overall protein production cost was decreased by an order of magnitude. Protein yield dependence on Ile amount added was tested and no significant improvement was observed in the range from 10 mg/L to 120 mg/L. Regarding the high price of d10-Ile, 10 mg/L is a cost-effective use of this reagent. Concerning the BL21(AcL-/Val-) strain, 50 mg/L of 13C-monomethyl α-ketoisovalerate yields an equivalent amount of protein compared to the standard protocol. However, 40 mg/L of 13C-proS-acetolactate used for stereospecific and Leu-specific labeling of MSG yielded about 40% of the standard protocol production.
Using the optimized conditions described above, we produced different methyl-labeled samples of the 82 kDa malate synthetase G (MSG), which is one of the largest monomeric proteins with available methyl assignments(Gans et al., 2010; Tugarinov and Kay, 2003). The MSG sequence contains 70 Leu and 46 Val residues. The Val-proS labeled MSG samples were produced using BL21(Acl-/Leu-) along with 40 mg/L of 13C-proS-acetolactate. The Leu-proS labeled MSG was expressed using the BL21(AcL-/Val-) strain along with 40 mg/L of 13C-proS-acetolactate. The Leu/Val methyl-labeled MSG sample was produced using regular BL21(DE3) along with 120 mg/L of 13C-monomethyl α-ketoisovalerate. Figure 3 shows the 13C-HMQC spectra of Leu/Val methyl-labeled sample, Val-proS labeled sample and Leu-proS labeled sample. The intensity ratio of either Val γ2-methyl or Leu δ2-methyl vs. the Met ε-methyl internal standard was used to quantify isotope incorporation. For both Leu and Val specific labeled samples, the isotope incorporation was near 100%. When compared, the Leu-proS and Val-proS MSG spectra recorded here matched the spectra in the literature (Tugarinov and Kay, 2003).
Figure 3.
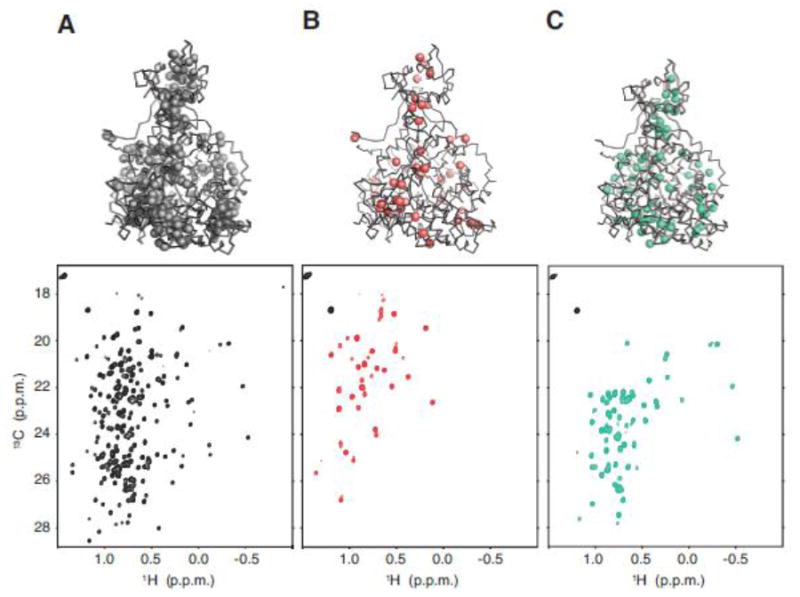
Methyl 13C-HMQC spectra recorded on samples of MSG expressed with (A) BL21 and 120 mg/L of α-13C-ketoisovalerate, containing Leu-δ1/δ2 and Val-γ1/γ2 crosspeaks, (B) BL21(AcL-/Leu-) with 40 mg/L of 13C-acetolactate, containing only Val-γ2 crosspeaks, or (C) BL21(AcL-/Val-) with 40 mg/L of 13C-acetolactate, containing only Leu-δ2 crosspeaks. The MSG crystal structures still show the highly spatially dispersed positions in the Leu-δ2 (green) and Val-γ2 (red) methyls in spite of the large reduction in the absolute number of NMR active probes. This indicates that spatial coverage is maintained while the sub-spectra are greatly simplified compared to the crowded regular spectrum (grey). The crosspeaks that appear in black on (B) and (C) panels are from Met ε-methyl used as internal standard.
During the course of optimization, we frequently noticed that using the BL21(AcL-) strain along with 30 mg/L of d10-Leu for Val-specific methyl labeling results in up to 6% of residual Leu methyl signals (in the case of MSG, Supp. Fig S1.A). We decided to test different concentrations of Leu, from 20 mg/L to 100 mg/L but the amount of residual Leu signals remained unaffected. This is in line with the 98% Leu signal suppression previously published (Mas et al., 2013). Although this signal contamination is low, it could yield heterogeneous peak shape that would complicate data analysis, lead to erroneous conclusions or wrong NOE crosspeak assignment. On the contrary, using the strain BL21(AcL-/Leu-) designed for Val-specific labeling, we can produce spectra without any residual Leu signal (Supp. Fig S1B).
The main limitation of all BL21(AcL-)-related strains is their inability to use α-ketobutyrate for Ile-δ1 labeling. This moiety is a very useful probe in most NMR studies; it gives intense signals and has a well-defined spectral window that does not overlap with methyl resonances from other residues. This shortcoming can be addressed, in part, using 13C-aceto-hydroxybutanoate, a recently developed Ile precursor that is a product of ALS in vivo (Fig. 1). It was developed to prevent isotope scrambling from 13C-Ala to Ile-γ2 methyl when producing Ile-[13CH3]δ1, Ala-[13CH3]β samples (Kerfah et al., 2015a). Here, it could in principle be used to introduce Ile in conjunction with Leu and Val. However, the Ile precursor and acetolactate are both substrates of the keto-l-acid reductoisomerase (KARI), the downstream enzyme in the branched-chain amino acid biosynthesis pathways. As a result, simultaneous addition of Ile precursor and acetolactate leads to a decrease in Leu/Val isotope incorporation. To overcome this issue, Kerfah et al. (2015) delayed the addition of Ile precursor by ~1 hour to enable the cells to metabolize acetolactate first. Since the new strains cannot grow without either 13C-aceto-hydroxybutanoate or Ile, this method is not suitable here. However, considering that the kcat/Km is 5-8 times higher for 13C-aceto-hydroxybutanoate than for 13C-proS-acetolactate (Dumas et al., 2001), it should be in principle possible to force the enzyme to equally process both precursors by adding more acetolactate than 13C-aceto-hydroxybutanoate. Regrettably, the strain for Leu-specific labeling (including ilvE deletion) did not grow in the presence of Ile-precursor. The ilvE gene codes for the transaminase that finalizes both Leu and Ile synthesis. In the case of Leu, we have shown that the biosynthesis from precursors still occurs without this enzyme, likely because other transaminases compensate for the lack of ilvE; however, this compensation is not sufficient in the case of Ile. In the Val-specific strain, the ilvE gene is not deleted and Ile can still be synthesized from 13C-aceto-hydroxybutanoate and so for example an Ile-[13CH3]δ1, Val-[13CH3]proS sample could be obtained. Based on the optimized amount of deuterated Ile in terms of production yield (see above), we expected to only need ~10 mg/L of 13C-aceto-hydroxybutanoate to reach full isotope labeling. Testing for Val-proS and Ile-δ1 isotope incorporation was conducted using 40:40, 10:80, 20:100, 20:130 or 20:160 mg/L of 13C-aceto-hydroxybutanoate:13C-proS-acetolactate. The Val-proS isotope incorporation was only 15% in the case of simultaneous addition of 40 mg/mL of both Ile-precursor and 13C-proS-acetolactate. This rate is in agreement with the 8-times higher substrate specificity of KARI for 13C-aceto-hydroxybutanoate. Increasing the concentration of 13C-proS-acetolactate made the enrichment rate reach 38%. This rate was not dependent on the 13C-proS-acetolactate concentration used beyond 100 mg/mL (test cases 20:100, 20:130 or 20:160 mg/L). In the case of 10:80 mg/L of 13C-aceto-hydroxybutanoate:13C-proS-acetolactate, the Val-proS isotope incorporation was closed to 100% but that of Ile-δ1 dramatically dropped down to 50%. Although, we were unsuccessful in producing samples with full labeling of both Val-proS (or proR) and Ile-δ1 using BL21(Acl-,Leu-). Although the experiment was not attempted, it is worth noting that the strain which is not auxotroph for acetolactate but only for Leu, namely BL21(Leu-), could be used along with 300 mg/L of 13C-proS-acetolactate and 80 mg/L of 13C-ketobutanoate to achieve this particular labeling scheme following the method of Kerfah and cowoker since the biosynthetic pathways for Ile and Val remain undisturbed. Even if no benefit in term of cost is realized, this strain still makes full Val-specific stereospecific labeling along with Ile methyl labeling of deuterated proteins possible without any scrambling to Leu methyls.
Using the proper combinations of auxotroph strains and precursors four different proteins ranging from 10 kDa to 82 kDa were produced with either Val-γ1/γ2, Leu-δ1/δ2, Val-γ1/Leu-δ1, Val-γ2/Leu-δ2, Val-γ1, Val-γ2, Leu-δ1 or Leu-δ2 methyl-labeled. Results are shown for CAP (Supp. Fig S4), PPD (Supp. Fig S5), calmodulin (Supp. Fig S6) and MSG (Fig 3). A summary of all auxotrophic strains and labeling combinations possible is listed in Supp. Table S1.
Thr-specific methyl-labeling using auxotroph strain
First, the amount of Thr necessary to obtain the same growth rate as the standard BL21 (DE3) strain was determined for BL21(Thr-). We tested 10, 20, 30, 40, 50 and 100 mg of Thr per liter of BL21(Thr-) culture and full growth was reached using at least 50 mg/L. Consequently the protein yield would be the same regardless of the Thr auxotrophy, and thereby the engineered strain would not provide any cost advantage. However, when the isotope incorporation was tested by producing different MSG samples using either 10, 25 or 50 mg per liter of 13Cγ2-Thr, and without any 2H-Gly supplement, the full isotope incorporation was reached with 25 mg/L (Supp. Fig S2). The reagent amount may need to be optimized on a protein by protein basis but, compared to the protocol of Velyvis et al. (2012), it is possible to use two-fold less 13Cγ2-Thr and obtain full Thr incorporation without significant isotope scrambling except for Ile-δ1 (Supp. Fig. S3). Moreover, Ile-methyl labeling can be achieved or suppressed by either 25 mg/L of 13C-α-ketobutyrate or 20 mg/L of deuterated Ile, respectively, which is 2-times or 4-times less, respectively, than previously reported(Sinha et al., 2011; Velyvis et al., 2012). The Thr specific information is also listed in Supp. Table S1.
CONCLUSION
We present an efficient and cost effective strategy that achieves residue- and/or stereospecific [1H,13C]-methyl labeling of Leu and Val (either Val-γ1/γ2, Leu-δ1/δ2, Val-γ1/Leu-δ1, Val-γ2/Leu-δ2, Val-γ1, Val-γ2, Leu-δ1 or Leu-δ2) based on new auxotroph E. coli strains and available Val/Leu precursors. The BL21(AcL-) auxotroph strain improved protein yield while requiring only one fifth the amount of acetolactate reagent compared to current methods, leading to significant cost reduction. This is currently the only method available to obtain stereospecific Leu-only methyl-labeled proteins. Analogously to Met shown in this work, Ala and Thr reagents do not interfere with pathways altered in our engineered strains. Although we have not tested it, we anticipate no issues with the incorporation of Ala and Thr along with Leu and Val using the new strains presented herein. Simultaneous complete Ile and Val methyl labeling is also possible but only with the Leu-only auxotroph strain, BL21(Leu-), along with the protocol reported by Kerfah et al. (2015), in order to take advantage of the robust total suppression of Leu signals. Unfortunately, Ile methyl labeling along with stereospecific [1H,13C]-methyl labeling of Leu is not possible using this protocol.
In addition, [1H,13C]-methyl-labeling of Thr is improved by the tailored auxothroph reported here. The isotope dilution of supplemented 13Cγ2-Thr is significantly decreased compared to the traditional strain and that allows for full Thr methyl labeling using only half the labeled reagent and no need for labeled glycine supplementation. The new strain gives near 100% isotope incorporation of Thr methyl without significant scrambling, except into Ile-δ1. This strain can be used to label Thr methyl in combination with any other methyl labeling of methyl-containing residues (Saio et al., 2014) and provide similar expression yield than the regular E. coli BL21 strain when using 50 mg/L of labeled Thr.
Supplementary Material
Footnotes
Statement
Auxotroph strains produced in this work are freely available upon request.
References
- Amero C, Asuncion Dura M, Noirclerc-Savoye M, Perollier A, Gallet B, Plevin MJ, Vernet T, Franzetti B, Boisbouvier J. A systematic mutagenesis-driven strategy for site-resolved NMR studies of supramolecular assemblies. J Biomol NMR. 2011;50:229–236. doi: 10.1007/s10858-011-9513-5. [DOI] [PubMed] [Google Scholar]
- Audin MJ, Dorn G, Fromm SA, Reiss K, Schutz S, Vorlander MK, Sprangers R. The archaeal exosome: identification and quantification of site-specific motions that correlate with cap and RNA binding. Angew Chem Int Ed Engl. 2013;52:8312–8316. doi: 10.1002/anie.201302811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala I, Sounier R, Use N, Gans P, Boisbouvier J. An efficient protocol for the complete incorporation of methyl-protonated alanine in perdeuterated protein. J Biomol NMR. 2009;43:111–119. doi: 10.1007/s10858-008-9294-7. [DOI] [PubMed] [Google Scholar]
- Chao FA, Shi L, Masterson LR, Veglia G. FLAMEnGO: a fuzzy logic approach for methyl group assignment using NOESY and paramagnetic relaxation enhancement data. J Magn Reson. 2012;214:103–110. doi: 10.1016/j.jmr.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dumas R, Biou V, Halgand F, Douce R, Duggleby RG. Enzymology, Structure, and Dynamics of Acetohydroxy Acid Isomeroreductase. Accounts of Chemical Research. 2001;34:399–408. doi: 10.1021/ar000082w. [DOI] [PubMed] [Google Scholar]
- Gans P, Hamelin O, Sounier R, Ayala I, Dura MA, Amero CD, Noirclerc-Savoye M, Franzetti B, Plevin MJ, Boisbouvier J. Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew Chem Int Ed Engl. 2010;49:1958–1962. doi: 10.1002/anie.200905660. [DOI] [PubMed] [Google Scholar]
- Gardner KH, Kay LE. Production and incorporation of N-15, C-13, H-2 (H-1-delta 1 methyl) isoleucine into proteins for multidimensional NMR studies. J Am Chem Soc. 1997;119:7599–7600. [Google Scholar]
- Gelis I, Bonvin AM, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated N-15-, C-13-, H-2-labeled proteins. Journal of Biomolecular Nmr. 1999;13:369–374. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- Isaacson RL, Simpson PJ, Liu M, Cota E, Zhang X, Freemont P, Matthews S. A New Labeling Method for Methyl Transverse Relaxation-Optimized Spectroscopy NMR Spectra of Alanine Residues. J Am Chem Soc. 2007;129:15428–15429. doi: 10.1021/ja0761784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagoz GE, Duarte AM, Ippel H, Uetrecht C, Sinnige T, van Rosmalen M, Hausmann J, Heck AJ, Boelens R, Rudiger SG. N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc Natl Acad Sci U S A. 2011;108:580–585. doi: 10.1073/pnas.1011867108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, van Ingen H, Zhou BR, Feng H, Bustin M, Kay LE, Bai Y. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc Natl Acad Sci U S A. 2011;108:12283–12288. doi: 10.1073/pnas.1105848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay LE. NMR studies of protein structure and dynamics. J Magn Reson. 2005;173:193–207. doi: 10.1016/j.jmr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Kerfah R, Plevin MJ, Pessey O, Hamelin O, Gans P, Boisbouvier J. Scrambling free combinatorial labeling of alanine-beta, isoleucine-delta1, leucine-proS and valine-proS methyl groups for the detection of long range NOEs. J Biomol NMR. 2015a;61:73–82. doi: 10.1007/s10858-014-9887-2. [DOI] [PubMed] [Google Scholar]
- Kerfah R, Plevin MJ, Sounier R, Gans P, Boisbouvier J. Methyl-specific isotopic labeling: a molecular tool box for solution NMR studies of large proteins. Curr Opin Struct Biol. 2015b;32:113–122. doi: 10.1016/j.sbi.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Lichtenecker RJ, Coudevylle N, Konrat R, Schmid W. Selective isotope labelling of leucine residues by using alpha-ketoacid precursor compounds. Chembiochem. 2013;14:818–821. doi: 10.1002/cbic.201200737. [DOI] [PubMed] [Google Scholar]
- Mas G, Crublet E, Hamelin O, Gans P, Boisbouvier J. Specific labeling and assignment strategies of valine methyl groups for NMR studies of high molecular weight proteins. J Biomol NMR. 2013;57:251–262. doi: 10.1007/s10858-013-9785-z. [DOI] [PubMed] [Google Scholar]
- Metzler WJ, Wittekind M, Goldfarb V, Mueller L, Farmer BT. Incorporation of 1H/13C/15N-{Ile, Leu, Val} into a Perdeuterated, 15N-Labeled Protein: Potential in Structure Determination of Large Proteins by NMR. J Am Chem Soc. 1996;118:6800–6801. [Google Scholar]
- Miyanoiri Y, Takeda M, Okuma K, Ono AM, Terauchi T, Kainosho M. Differential isotope-labeling for Leu and Val residues in a protein by E. coli cellular expression using stereo-specifically methyl labeled amino acids. J Biomol NMR. 2013;57:237–249. doi: 10.1007/s10858-013-9784-0. [DOI] [PubMed] [Google Scholar]
- Pickford AR, Campbell ID. NMR studies of modular protein structures and their interactions. Chem Rev. 2004;104:3557–3565. doi: 10.1021/cr0304018. [DOI] [PubMed] [Google Scholar]
- Popovych N, Tzeng SR, Tonelli M, Ebright RH, Kalodimos CG. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. P Natl Acad Sci USA. 2009;106:6927–6932. doi: 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Religa TL, Sprangers R, Kay LE. Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science. 2010;328:98–102. doi: 10.1126/science.1184991. [DOI] [PubMed] [Google Scholar]
- Rosenzweig R, Kay LE. Bringing dynamic molecular machines into focus by methyl-TROSY NMR. Annu Rev Biochem. 2014;83:291–315. doi: 10.1146/annurev-biochem-060713-035829. [DOI] [PubMed] [Google Scholar]
- Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science. 2013;339:1080–1083. doi: 10.1126/science.1233066. [DOI] [PubMed] [Google Scholar]
- Ruschak AM, Velyvis A, Kay LE. A simple strategy for (1)(3)C, (1)H labeling at the Ile-gamma2 methyl position in highly deuterated proteins. J Biomol NMR. 2010;48:129–135. doi: 10.1007/s10858-010-9449-1. [DOI] [PubMed] [Google Scholar]
- Saio T, Guan X, Rossi P, Economou A, Kalodimos CG. Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science. 2014;344:1250494. doi: 10.1126/science.1250494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha K, Jen-Jacobson L, Rule GS. Specific labeling of threonine methyl groups for NMR studies of protein-nucleic acid complexes. Biochemistry. 2011;50:10189–10191. doi: 10.1021/bi201496d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sounier R, Blanchard L, Wu ZR, Boisbouvier J. High-accuracy distance measurement between remote methyls in specifically protonated proteins. J Am Chem Soc. 2007;129:472–473. doi: 10.1021/ja067260m. [DOI] [PubMed] [Google Scholar]
- Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kay LE. Ile, Leu, and Val Methyl Assignments of the 723-Residue Malate Synthase G Using a New Labeling Strategy and Novel NMR Methods. J Am Chem Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kay LE. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. Chembiochem. 2005;6:1567–1577. doi: 10.1002/cbic.200500110. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Pai MT, Kalodimos CG. NMR studies of large protein systems. Methods Mol Biol. 2012;831:133–140. doi: 10.1007/978-1-61779-480-3_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velyvis A, Ruschak AM, Kay LE. An economical method for production of (2)H, (13)CH3-threonine for solution NMR studies of large protein complexes: application to the 670 kDa proteasome. PLoS One. 2012;7:e43725. doi: 10.1371/journal.pone.0043725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Matthews S. MAP-XSII: an improved program for the automatic assignment of methyl resonances in large proteins. J Biomol NMR. 2013;55:179–187. doi: 10.1007/s10858-012-9700-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


