Abstract
Understanding how basic structural units influence function is identified as a foundational/core concept for undergraduate biological and biochemical literacy. It is essential for students to understand this concept at all size scales, but it is often more difficult for students to understand structure–function relationships at the molecular level, which they cannot as effectively visualize. Students need to develop accurate, 3‐dimensional mental models of biomolecules to understand how biomolecular structure affects cellular functions at the molecular level, yet most traditional curricular tools such as textbooks include only 2‐dimensional representations. We used a controlled, backward design approach to investigate how hand‐held physical molecular model use affected students' ability to logically predict structure–function relationships. Brief (one class period) physical model use increased quiz score for females, whereas there was no significant increase in score for males using physical models. Females also self‐reported higher learning gains in their understanding of context‐specific protein function. Gender differences in spatial visualization may explain the gender‐specific benefits of physical model use observed. © 2016 The Authors Biochemistry and Molecular Biology Education published by Wiley Periodicals, Inc. on behalf of International Union of Biochemistry and Molecular Biology, 44(4):326–335, 2016.
Keywords: physical models, gender differences, protein structure–function, external representations, assessment
Introduction
Undergraduate biology and biochemistry instructors have the challenge of helping students understand structure–function relationships, which has been identified as a foundational/core concept for undergraduate biological and biochemical literacy 1, 2. It is essential for students to understand this concept at all size scales; yet understanding cellular processes has been identified as potentially “troublesome” content knowledge because it requires an expert level of understanding of spatial scale and dynamic interactions 3, 4, 5. An expert level of understanding of these kinds of “threshold” concepts is achieved when learning is “transformative, irreversible, and integrative” 6, 7. For example, biochemists have identified the physical basis of molecular interactions as a key threshold concept because it requires an expert level of understanding of how molecular electrostatic properties ultimately determine interactions between biomolecules 8. Understanding difficult threshold concepts requires accommodation. This means that the information cannot be simply assimilated into existing knowledge, but instead students must reorganize their knowledge and transform their thinking, allowing them to cross conceptual thresholds 5, 9. Therefore, it is important for instructors to provide students with effective curricular tools and learning environments as they grapple with these difficult concepts.
Biology and biochemistry instructors use a variety of external representations, such as pictures, animations, and physical biomolecular models of different complexities and size scales, in order to represent biomolecular structures, and the representation used impacts student learning 10, 11. The type of representation chosen by an instructor should depend on desired student learning outcomes. Typical 2‐dimensional (2D) imagery often depicts complex biomolecules as schematic blobs, which may be adequate if the goal is for students to understand a chronological sequence of molecular events, such as cell signaling pathways. However, for many learning outcomes, these schematic blobs may not suffice. These blobs may lead to “deceptive clarity”—images that allow for only a superficial understanding, which can lead students to believe they understand something that they actually do not understand 12. Schematics often obscure basic principles that experts know and therefore infer from the image, and instructors may incorrectly assume that students are able to infer this information as well 13, 14, 15. Complex biomolecular representations may offer students visual “desirable difficulties”, which help overcome “deceptive clarity” by giving students an opportunity to engage with visual representations 12. For example, more complex representations of receptor–ligand binding events which depict random, nondirectional ligand movement instead of directed movement leads to a deeper understanding of these dynamic molecular events 16. Another study found that students who studied with 2D molecular representations combining schematic blob images with realistic images based on Protein Data Bank (PDB) structure entries more accurately labeled novel PDB images than students who studied only with schematic blob images 17. Additionally, realistic representations with many details do not negatively affect student performance on other assessments 17, 18.
Scientific experts tend to have high spatial visualization ability, which is the ability to understand the spatial relationship between objects and to mentally manipulate images 19, 20. Translating a 2D external representation into an accurate 3D mental model is identified as a key cognitive skill possessed by biochemistry experts, yet this is typically a difficult skill for undergraduate students to develop 11. This is particularly true for females, who typically have lower spatial visualization ability than males 21, 22, 23. It is important to support students' development of skills such as visualizing orders of magnitude and relative size and scale 10, 11, as these visual literacy skills are fundamental for students to accurately interpret and learn from a variety of external representations. As such, it is important to identify curricular tools and/or activities that help students to think like an expert about biomolecules, presumably by helping students hone their spatial visualization skills of the molecular level. Specifically, what curricular tools enhance students' understanding of biomolecules as dynamic structural entities whose interactions determine key cellular functions?
Hand‐held physical models are common in the chemistry classroom and multiple studies have documented their effectiveness, particularly for higher‐order chemistry questions 24, 25, 26. “3D‐like” computer imagery, which allows students to manipulate and rotate an image on a 2D screen, has also been shown to improve student understanding of chemical representations 27. The use of physical models and/or 3D‐like imagery in biology and biochemistry classrooms is less well studied; however, a combination of physical molecular model use with computer 3D‐like molecular imagery over several weeks increased student performance on high‐order interview questions about molecular structure–function relationships 28. Additionally, after a semester‐long undergraduate biochemistry class, students rated physical models as the most helpful of seven learning tools for understanding molecular structure–function relationships 29. However, little is known about the optimal learning environment for using 3D physical models and/or 3D‐like imagery in a biology or biochemistry classroom, particularly details such as the impact of each tool alone, the number of exposures students should have to the tools, and the optimal time spent with the tools.
We are interested in identifying instructional approaches that help students to deeply understand molecular structure–function relationships (i.e. acquire an expert level of understanding). Specifically, we want to use student performance as evidence to identify instructional tools that help students develop a sophisticated understanding of molecular structure–function. In the present study, students used either hand‐held physical models, a 3D‐like molecular imaging tutorial (Jmol), both tools, or neither tool in a single discussion class meeting to supplement lecture material about the structure and function of a membrane‐binding protein. We hypothesized that students using both physical models and 3D‐like imagery would exhibit superior understanding of molecular structure–function relationships, as compared to students who used either tool alone, and that students using either tool alone would perform better than those using neither tool. Furthermore, because of the gender differences in spatial visualization skills 21, 22, 23, we examined gender differences in the effectiveness of these tools.
Methodology
Curriculum and Study Sample
Curricular materials in this study were developed as part of the NSF‐CCLI funded Connecting Researchers, Educators, and Students (CREST) project. The University of Wisconsin‐Madison is one of several institutional partners participating in the CREST project. UW‐Madison Biology Core Curriculum (Biocore) honors undergraduate students worked with faculty researchers and educators to develop classroom materials, including hand‐held physical models and a 3D‐like molecular imaging tutorial. Biocore is a four semester undergraduate honors biology program that emphasizes group learning and the process of science 30. Cell and molecular biology (Biocore 383) is the second lecture course in the program sequence. Students apply to the program as freshman and begin as sophomores, thus the majority of students in the cell and molecular biology course are sophomores. Students attend three 50‐min lectures and one required 50‐min discussion section per week. Lectures were taught by three faculty members to the entire group of 111 students and discussion sections were led by three graduate Teaching Assistants (TAs) for groups of 15–16 students. However, during the discussion section when our treatment was applied, the faculty member in charge of this courses segment, E.W.D., was present and led all seven discussion sections, with the section TAs assisting. Our research protocol was approved by the UW‐Madison Education Research Institutional Review Board (protocol 2012‐1060).
Conceptual Goals
Prior to the start of the spring 2014 semester, we used a backward‐design research framework to align three conceptual goals (i.e. what do we want students to understand?) with learning outcomes (i.e. what do we want students to be able to know or do?), instructional activities, and assessments (Supporting Information Appendix A). E.W.D., M.A.H., and three senior undergraduate Biocore students who had previously taken Biocore 383 designed materials used to supplement E.W.D.'s lecture material on a membrane‐binding protein, Cdc42‐interacting protein 4 (CIP4). CIP4's role in regulating cell membrane shape was used as a model system to help students achieve key molecular structure–function learning outcomes. CIP4 is an intracellular protein involved in regulating plasma membrane structure, primarily during endocytosis, but also in protrusions in developing neurons 31. Our three conceptual goals were for students to understand: (1) how biomolecular structure affects function, (2) that endocytosis requires several different proteins, and (3) that “one protein ≠ one function” but depends on where it is located in the cell and on what other proteins/lipids/small molecules are present in that part of the cell.
Instructional Activities
The following instructional activities were designed based on our three conceptual goals: a worksheet with information on CIP4, PDB images of CIP4, and questions for students to answer as they used the tools (Supporting Information Appendix B), an online clickable endocytosis landscape (http://cbm.msoe.edu/teachingResources/jmol/coatedPit/), CIP4 hand‐held physical models, and a Jmol molecular imaging tutorial (http://cbm.msoe.edu/teachingResources/jmol/fbar/). The clickable endocytosis landscape incorporates an original watercolor by David S. Goodsell of an endocytosing vesicle which was created as part of the CREST Project (Fig. 1 A). Students were able to electronically interact with 13 proteins in the landscape, thus providing a 3D‐like view of each, both during and outside of discussion section. Four CIP4 physical models were designed to highlight different features of the protein and were built using 3D printing technologies (Fig. 2). All of the content contained in the physical models and Jmol tutorial was present in the worksheet and/or clickable landscape, including PDB images of CIP4, but the physical models and Jmol tutorial allowed for a 3D or 3D‐like visualization, respectively, of the protein. E.W.D. began all discussion sections by providing a brief overview of CIP4's role in forming an elongated vesicle, using bananas to demonstrate how the CIP4 dimers orient (multimerize) around the vesicle (Fig. 1 B). In sections using the physical models, he also used the physical CIP4 models in this overview. Students were then instructed to work their way through the worksheet questions, using all of the tools available. For sections using the physical models, each of the four CIP4 models was rotated between groups of 2–4 students so that each group had an approximately equivalent amount of time to examine each model. Students were encouraged to discuss the material with their peers. Students given access to the Jmol tutorial were instructed not to share the website address for the tutorial with peers outside of their discussion section.
Figure 1.
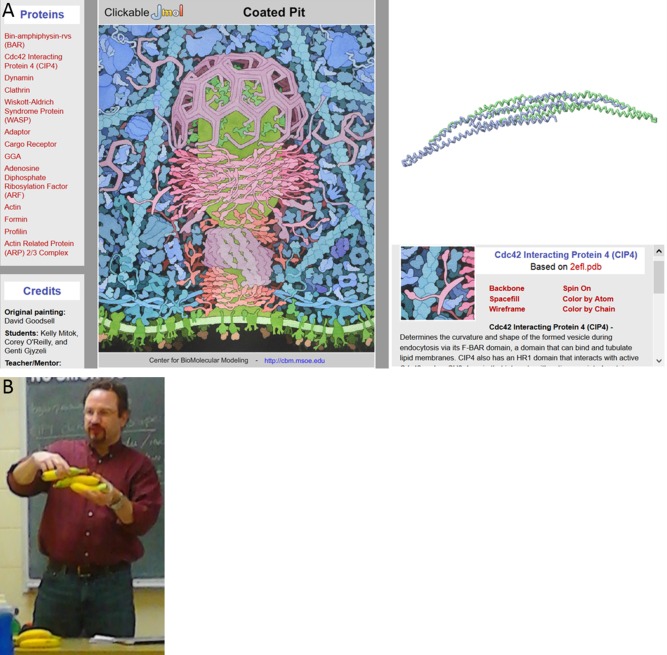
Instructional materials available to all students. (A) Clickable landscape based on David Goodsell's painting. (B) E.W.D demonstrating how multiple CIP4 dimers orient around an endocytosing vesicle using bananas. Each banana represents a CIP4 dimer.
Figure 2.
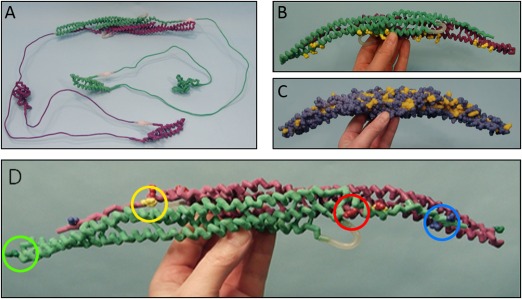
Physical protein models. The four hand‐held physical models used in 4 of the 7 discussion sections (“physical model” treatment groups). Purple and light green backbones represent separate identical monomers connected by magnets to create the CIP4 dimer. (A) Entire CIP4 dimer including FBAR (alpha helix), HR1 (alpha helix) and SH3 domains (beta sheet), made of nylon. Thin metal connections represent uncrystallized regions of the protein. Their length accurately represents the number of amino acids known to span these regions. (B ) F‐BAR dimer of CIP4, made of nylon. The positively charged amino acid side chains (lysine and arginine) important for binding to negatively charged phospholipid head groups in the membrane are displayed on the F‐BAR domain in yellow. (C ) F‐BAR dimer of CIP4 in spacefill, made of plaster. Yellow amino acid residues are hydrophobic and blue amino acid residues are hydrophilic. (D ) F‐BAR dimer of CIP4, made of nylon. The yellow amino acid visible on the purple F‐BAR monomer (the other yellow amino acid is not visible) is the hydrophobic phenylalanine (F276) residue involved in lateral interactions of CIP4 dimers. Red amino acids are the positively charged lysine residues (K273 and K66) involved in ionic interactions with the negatively charged blue amino acids glutamate (E285) and aspartate (D286) of an adjacent CIP4 dimer. The dark green amino acids at the end of each monomer are the positively charged lysine (K166) residues involved in end‐to‐end interactions of CIP4 dimers.
Treatment Groups
Students were randomly assigned by discussion section to one of four treatment groups: physical models only, Jmol tutorial only, physical models and Jmol tutorial, and neither physical models nor Jmol tutorial. Each treatment was assigned to two different discussion sections, except the Jmol tutorial only treatment, which was only assigned to one section (Table 1). Again, all four treatment groups had access to the worksheet with PDB images (Supporting Information Appendix B) and the online clickable endocytosis landscape (Fig. 1 A), and saw E.W.D. stack bananas that represented the multimerized CIP4 dimers (Fig. 1 B).
Table 1.
Treatment groups
| Treatment | TA | Number of students participating |
|---|---|---|
| Physical models | A | 13 (10F, 3M) |
| Physical models + Jmol tutorial | A | 13 (3F, 10M) |
| No physical models or Jmol tutorial | A | 14 (5F, 9M) |
| No physical models or Jmol tutorial | B | 15 (10F, 5M) |
| Jmol tutorial | C | 13 (7F, 6M) |
| Physical models | C | 13 (8F, 5M) |
| Physical models + Jmol tutorial | B | 14 (5F, 9M) |
Discussion sections, listed by treatment and in order of time throughout the day. Teaching Assistants (TAs) are coded A, B, and C. Each section had 15–16 students enrolled and last column states the number included in the study (those who consented and were present for both the intervention and assessment). F = female, M = male. Total N for each treatment group is physical models, N = 26 (18F, 8M); physical models + Jmol tutorial, N = 27 (8F, 19M); Jmol tutorial, N = 13 (7F, 6M); no physical models or Jmol tutorial N = 29 (15F, 14M). Gray rows represent groups with physical models, N = 53 (26F, 27M); and white rows represent groups with no physical models, N = 42 (22F, 20M).
Early in our analysis, we decided to remove Jmol as a treatment factor, leaving two levels of treatment: physical models and no physical models. The three primary reasons for the removal are as follows: (1) Based on classroom observations, we noted that, when Jmol was available, students spent little to no time using it during discussion section. This could be due, in part, to the fact that in those sections for which the Jmol tutorial was available, E.W.D. did not spend time introducing Jmol to the sections. (2) The online clickable landscape provided students in all four treatment groups an opportunity to manipulate 3D‐like imagery to a limited extent, so we felt that we were not able to evaluate the effects of using 3D‐like interactive computer tools alone. (3) Due to the imbalance in our design (there were 7 sections with four treatment combinations), the treatment of Jmol with no physical models could only be assigned to a single section. Thus, there was a strong possibility of a confounding between TA/discussion section and treatment effect. Therefore, our final statistical model do not include Jmol.
Treatment group and sample size information are summarized in Table 1. Informed consent was obtained from ∼95% of enrolled Biocore 383 students. Students who did not consent or were not present for either the CIP4 instructional activities or the assessment were not included in the study.
Performance Assessment
Baseline performance was assessed by the averages of exams 1 and 2, which were both taken before the CIP4 instructional materials were used. A quiz was designed to assess student understanding of how biomolecular structure affects function (Supporting Information Appendix C) and students took this quiz one week following treatment. The quiz was graded independently by R.F.L. and M.A.H., who were blinded to treatment group, using a detailed rubric with a maximum of 27 points (Supporting Information Appendix C). This 27 point rubric was designed to deeply assess a maximal level of student understanding; therefore a high score was not obtained by most students. There was a high degree of interrater reliability (Pearson's correlation = 0.94), so the average of the two scores was used.
Student Assessment of Learning Gains (SALG)
Students rated how well instructional tools helped their understanding of molecular structure/function relationships, as well as several other self‐reported measures, using a 5‐point Likert scale. Several open ended questions were also included. These were not scored but relevant comments are included below. The survey was administered using Qualtrics Online Survey Software during the last week of class. Five students were not present for the SALG survey.
Statistical Analysis
Data were analyzed using the software environment R (version 3.1.3) 32, 33. The two main performance variables measured were quiz score and SALG score. In the study, the two treatments were “applied” to discussion sections. The indicator of TA is also associated with section. All student covariates are unique to individual students. The proper analysis of a nested design of this type requires use of a mixed model with an error term for section and another error term for the individual student.
Initial analyses (initial model described below) indicated that the section error term had negligible magnitude and was insignificant. Thus, for the remainder of our analyses, we dropped the section error term and used standard multiple regression for analysis.
In our primary multiple regression analysis, we considered a number of covariates in addition to the treatment effect. These were TA, student gender, average of exams 1 and 2, GPA, and enrollment in a related lab course. We also considered an interaction between treatment and gender. Backwards elimination was used to remove nonsignificant terms (p > 0.10, for instance GPA was removed). R‐squared values are reported for completeness. In order to directly determine the effect of physical models on each gender, the model was fit two ways: one with females as baseline and the other with males as baseline.
Results
Assessment of Our Initial Model
Our initial mixed model analysis of quiz score indicated that the error term for section had zero variance. A formal likelihood ratio test (not shown) confirmed that the null hypothesis of zero section variance could not be rejected. In addition, in examining the residuals of the model with section excluded, we grouped the residuals by section and found no patterns that could suggest the importance of a section effect. Thus, as noted above, our primary analysis relied on multiple regression with no nested effects.
Assessment of Our Design
Although we decided to remove the Jmol treatment factor due to design issues discussed above, we carried out a preliminary multiple regression analysis with this variable included in order to fully explore the data and assess this decision. There was a Jmol treatment effect of −7.3 points and an interaction between the Jmol and physical model use, accounting for an increase in 5.9 points, after accounting for significant covariates (Table 2). This indicates that students who did use the Jmol tutorial, but did not use physical models, had lower quiz scores. However, this group is composed of a single discussion section, and because of the possible confounding between the treatment group and TA/discussion, we do not have confidence in these results. This supports our decision (presented above) to exclude Jmol as a treatment factor based on design.
Table 2.
Results from multiple regression analysis, with section removed but including four treatments as separate fixed effects
| Fixed effect coeff. | Effect size | Standard error | t score | p value |
|---|---|---|---|---|
| Intercept | −12.55 | 6.31 | −1.87 | 0.06 |
| Jmol tutorial | −7.31 | 3.18 | −2.30 | 0.02 |
| Physical models | 0.94 | 2.14 | 0.44 | 0.66 |
| Jmol + models | 5.93 | 2.17 | 2.73 | 0.008 |
| Gender (male) | 2.27 | 1.92 | 1.19 | 0.24 |
| Exam average | 0.26 | 0.08 | 3.35 | 0.00 |
| TA “A” | −1.08 | 1.36 | −0.80 | 0.48 |
| TA “C” | 4.65 | 2.42 | 1.93 | 0.06 |
| Male: Jmol | 0.19 | 3.40 | 0.06 | 0.96 |
| Male: models | −2.87 | 2.83 | −1.01 | 0.31 |
| Male: both | −5.80 | 2.77 | −2.10 | 0.04 |
Residual standard error was 4.91 on 84 degrees of freedom. F(10,84) = 3.03, p = 0.003. Adjusted R‐square was 0.18. The baseline was set to female with no physical model. This analysis was done to assess our decision to remove the Jmol treatment group in the final model.
Assessment of Our Final Model
The primary multiple regression analysis indicated that there was a treatment effect, a gender effect, and an interaction between the two (p < 0.10) after accounting for significant covariates (Table 3; Fig. 3). One of the covariates, the average of exams 1 and 2, was positively related to quiz score (p = 0.004), and the TA “A” corresponded to lower quiz scores than the other two TAs (p = 0.06). There were no gender differences in performance on exams 1 and 2, using TA as a covariate (data not shown).
Table 3.
Linear regression coefficients and other important statistical information where quiz score is the response variable
| Coefficient | Effect size | Standard error | t score | p value |
|---|---|---|---|---|
| Intercept | −9.91 | 6.58 | −1.51 | 0.14 |
| Physical models | 5.03 | 1.51 | 3.34 | 0.001 |
| Gender (male) | 2.72 | 1.59 | 1.72 | 0.09 |
| TA “A” | −2.37 | 1.27 | −1.87 | 0.06 |
| TA “C” | −0.66 | 1.38 | −0.48 | 0.63 |
| Exam Average | 0.22 | 0.08 | 2.97 | 0.004 |
| Gender:model | −4.01 | 2.12 | −1.89 | 0.06 |
Residual standard error was 5.04 on 88 degrees of freedom. F(6,88) = 3.38, p = 0.005. Adjusted R‐squared was 0.13. The baseline gender was set to female with no physical model use.
Figure 3.
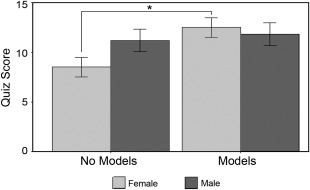
Quiz performance by gender. The minimum value of the quiz was 0 and the maximum was 27. Physical model use increased quiz scores in females by 5 points (p = 0.001), while the effect of physical models in males was not significant (p = 0.60). Error bars represent ±1 SEM.
The students who did not have access to physical models had a mean quiz score of 9.1 points (out of 27 possible points), and the group with physical models had a mean of 12.1 points For students who did not use physical models, males without physical models scored approximately 2.5 points higher than females. For females, physical models significantly increased quiz scores by about 5 points (p = 0.001), while the increase in male scores was only about 1 point and was not significant (p = 0.60, Fig. 3).
Student Assessment of Learning Gains (SALG)
We used the final model described above to analyze SALG data. The multiple regression analysis indicated that there was a treatment effect, a gender effect, and an interaction between the two (p < 0.10) after accounting for significant covariates (Table 4; Fig. 4) on self‐reported gain in the understanding in response to the statement “My understanding that certain proteins may have context‐specific functions in different cell types”. Similar to quiz performance, this effect was specific to females (Fig. 4, p = 0.006) while for males it was not significant (p = 0.98). There were no significant effects of physical model use on any other survey questions.
Table 4.
Linear regression coefficients and other important statistical information where SALG score is the response variable
| Coefficient | Effect size | Standard error | t score | p value |
|---|---|---|---|---|
| Intercept | 3.85 | 1.04 | 3.71 | 0.0004 |
| Physical models | 0.67 | 0.24 | 2.81 | 0.006 |
| Gender (male) | 0.46 | 0.25 | 1.84 | 0.07 |
| TA “A” | 0.01 | 0.20 | 0.06 | 0.95 |
| TA “C” | 0.06 | 0.22 | 0.28 | 0.78 |
| Exam average | −0.003 | 0.01 | −0.27 | 0.79 |
| Gender:model | −0.68 | 0.34 | −2.03 | 0.05 |
Residual standard error was 0.77 on 83 degrees of freedom. F(6,83) = 1.49, p = 0.19. Adjusted R‐squared was 0.03. The baseline gender was set to female with no physical model use.
Figure 4.
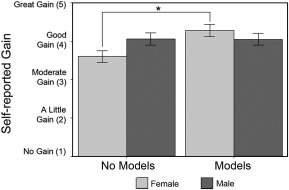
Student assessment of learning gains. Females who used physical models rated larger gains in their understanding in response to the statement “My understanding that certain proteins may have context‐specific functions in different cell types” compared to females who did not use physical models (p = 0.006), while for males there was no difference with physical model use (p = 0.98). Error bars represent ± 1 SEM.
Discussion
We found that short term use of hand‐held physical models improved female students' ability to predict how specific changes in biomolecular structure affect function, as measured by quiz score. For students who did not use physical models, there was a trend for males to perform better than females, while physical models increased quiz scores in females such that they performed equivalently to their male peers. Interestingly, females also reported larger gains in their understanding of the context‐specific function that proteins can have in different types of cells. For students who did not use physical models, there was a trend for males to rate their gains as higher than females, while females who used the physical models reported learning gains that were equivalent to those of their male peers.
In addition to the physical models, students had numerous other tools to support their achievement of the three learning outcomes. All students had access to the clickable endocytosis landscape, which was interactive in that it allowed students to electronically manipulate 3D‐like protein images. Students' comments about these other curricular materials on the open ended questions on the SALG survey were generally positive. For example, one student wrote that, “The cellular landscape was extremely helpful in visualizing what was going on.” Some students, however, pointed out that the static nature of the physical models was a key limitation in their understanding of dynamic inter‐molecular interactions: “The handheld molecules were not helpful because you could not see how it interacted with the other proteins, since it was just the F‐BAR molecule”. Indeed, while students with higher spatial visualization ability learn better from static images than students with low spatial visualization ability, dynamic animations have been shown to help students with lower spatial visualization skills compensate 34. In the current study, the worksheet with PDB images and the 3D‐like clickable landscape were sufficient tools only for males, suggesting that there is something specific about the physical models that enhanced understanding in females.
Models could provide a mental scaffold for students to better visualize molecular structures (although only significantly for females in our study), and could also actually improve spatial visualization ability. The superior gain in understanding of molecular structure–function relationships documented in this study for females who used physical models may be due to the models helping females better visualize the 3D structures represented in the 2D images of molecular structures. Males tend to have higher spatial visualization skills than females 21, 22, 23 and the physical models may help females overcome lower spatial visualization ability by providing them with the 3D structure, thus providing a mental scaffold to learn the content related to this structure. This idea is supported by the correlation between spatial visualization and students' learning, particularly in chemistry 35, 36. Wu and Shah (36) propose that “manipulating these models could help students understand the underlying concepts of visual representations” (p. 478).
In addition to providing a 3D representation for low spatial ability, physical models may also directly improve spatial visualization ability in general. The use of physical chemistry models improved spatial visualization ability and performance on high Bloom's taxonomy questions 24. Furthermore, students who have opportunities to hold and manipulate hand held chemistry models perform better on chemistry concept assessments, even on assessments that do not directly relate to visualizations 36. These findings emphasize the importance of identifying and providing students with tools that allow them to improve their spatial visualization skills; however, it is not known whether the one‐time use of physical models in the present study improved spatial visualization skills in female students.
Interestingly, the difference between males and females in their self‐reported learning gains to the SALG question “My understanding that certain proteins may have context‐specific functions in different cell types” mirrored the differences found in quiz performance when grouped by exposure to physical models. Perhaps even one‐time exposure to physical models provides students, particularly females, with a memorable mental scaffold on which to organize their conceptual understanding at the molecular level. Student reflections from the SALG survey provide some evidence for this assumption; one student wrote, “The physical models don't necessarily explain it better but they help to recall the information.” This suggests that physical models help students to create lasting mental representations at the molecular level.
When instructors use learning tools such as physical models, they should be mindful of assumptions they make about students' comprehension of external representations of molecular structure. Students need basic prior knowledge and visualization abilities when interpreting external representations, particularly complex representations based on PDB images 35, 37, 38: (36, p. 485). It is important to note that the students in this study were honors biology and biochemistry students, so even if there was a gender difference in spatial visualization ability, most students presumably had high spatial visualization ability compared to a typical undergraduate biology population. Our findings suggest that in these honors students, the physical models were accessible to both male and female students.
Cognitive load theory (CLT) may also provide insights into the benefits of physical models to student learning. An understanding of cellular processes has been identified as potentially “troublesome” content knowledge because it requires an expert level of understanding of spatial scale and dynamic interactions 3, 4, 5. According to CLT, experts use complex, long‐term memories to greatly expand the capacity of their working memory 39. Physical models may facilitate students', particularly female students', assimilation of complex biomolecular structures into organized, long term memory units or “schemata”, thus reducing cognitive load when learning about new functions for familiar biomolecular structures.
There were several limitations of the present study that should be noted and improved upon in future studies if possible. First, because of the problem with the imbalance of discussion section, lack of time for students to use Jmol in discussion section, and other study design flaws, we were not able to examine the effects of the 3D‐like imagery provided by the Jmol tutorial. Additionally, the approach of assigning treatment to discussion section is limiting from a design perspective. However, due to the nature of the instructional tools and importance of group work in the Biocore program, this could not be avoided. E.W.D. attempted to teach the same concepts in the same manner in each discussion section, but because of student questions, he provided unique clarifications and examples for some discussion sections. Despite these challenges, we still saw a significant effect of physical model use on quiz score and self‐reported understanding of context‐specific protein functions in females. It should be noted, however, that the generalizability of these findings is limited, as they represent data from only one semester of an honors undergraduate class. We are carrying out follow up studies to investigate the relationship between spatial visualization ability, gender, and use of physical models on student learning. Additionally, future studies should investigate whether these findings are reproducible and perhaps more pronounced, in non‐honors and non‐majors courses.
Future studies are needed to further inform our understanding of curricular tools that help students understand molecular structure–function relationships. Undergraduate biology and biochemistry instructors would benefit from investigation of the following questions: Would multiple exposures to a variety of physical models and/or interactive 3D‐like images improve students' ability to translate 2D molecular images into accurate 3D mental models? Would this improved spatial visualization ability help students understand molecular structure–function relationships at a deeper level, so that they achieve a more expert‐like appreciation for the dynamic molecular interactions implied in typical 2D external representations? Are the benefits of using interactive 3D‐like computer imagery equivalent to the benefits of physical models, or is a combination of these two tools optimal? Researchers should keep in mind that evidence addressing these and other related questions may very well be gender‐dependent.
In conclusion, we found that a single exposure to hand‐held physical models improved the understanding of protein structure–function relationships in female students, increasing quiz performance and self‐reported understanding to levels equivalent to those of their male peers. These findings add to an invaluable and growing body of literature on identifying and closing gender gaps in STEM.
Acknowledgements and Disclaimer
We would like to thank David Goodsell for the clickable landscape and Mark Hoelzer for his help with the online tutorials and graphics. This material is based upon work supported by the National Science Foundation under award numbers DUE‐1022793 and DUE‐1323414. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the National Science Foundation. E.W.D. is funded by NIH grant NS080928.
References
- 1. Tansey, J. T. Baird, T. Cox, M. M. Fox, K. M. Knight, J. Sears, D. and Bell, E. (2013) Foundational concepts and underlying theories for majors in “biochemistry and molecular biology.” Biochem. Mol. Biol. Educ. 41, 289–296. [DOI] [PubMed] [Google Scholar]
- 2. American Association for the Advancement of Science (2011) Vision and Change in Undergraduate Biology Education: A Call to Action, Washington, DC. [Google Scholar]
- 3. Ross P.M., Taylor C. E., Hughes C., Kofod M., Whitaker N., Lutze‐Mann L. and Tzioumis V., in Meyer J. H. F., Land R. and Baillie C., Eds. (2010) Threshold Concepts and Transformational Learning, Sense Publishers, Rotterdam, Boston, pp. 165–178. [Google Scholar]
- 4. Taylor C., in Meyer J. H. F. and Land R., Eds. (2006) Overcoming Barriers to Student Understanding: Threshold Concepts and Troublesome Knowledge, Routledge, New York, NY, pp. 87–99. [Google Scholar]
- 5. Ross, P. M. , Taylor, C. E. , Hughes, C. , Whitaker, N. , Lutze‐Mann, L. , Kofod, M. and Tzioumis, V. (2010) Threshold concepts in learning biology and evolution. Biol. Int. 47, 47–54. [Google Scholar]
- 6. Land R., Cousin G., Meyer J. H. F., Davies P., in Rust C., Ed. (2005) Improving Student Learning Diversity and Inclusivity, Oxford Centre for Staff and Learning Development, Oxford. [Google Scholar]
- 7. Meyer J. H. F. and Land R., in Rust C., Ed. (2003) Improving Student Learning—Ten Years On, OCSLD, Oxford. [Google Scholar]
- 8. Loertscher, J. , Green, D. , Lewis, J. E. , Lin, S. and Minderhout, V. (2014) Identification of threshold concepts for biochemistry. Cell Biol. Educ. 13, 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Posner, G. , Strike, K. , Hewson, P. and Gertzog, W. (1982) Accommodation of a scientific conception: Toward a theory of conceptual change. Sci. Educ. 66, 211–227. [Google Scholar]
- 10. Schonborn, K. J. and Anderson, T. R. (2006) The importance of visual literacy in the education of biochemists. Biochem. Mol. Biol. Educ. 34, 94–102. [DOI] [PubMed] [Google Scholar]
- 11. Schönborn, K. J. and Anderson, T. R. (2010) Bridging the educational research–teaching practice gap. Biochem. Mol. Biol. Educ. 38, 347–354. [DOI] [PubMed] [Google Scholar]
- 12. Linn M., Chang H., Chiu H., Zhang H. and McElhaney K., in Benjamin A., Ed. (2010) Successful Remembering and Successful Forgetting: A Festschrift in Honor of Robert A. Bjork, Routledge, New York, NY, pp. 239–258. [Google Scholar]
- 13. Schonborn, K. J. Anderson, T. R. and Grayson, D. J. (2002) Student difficulties with the interpretation of a textbook diagram of immunoglobulin G (IgG). Biochem. Mol. Biol. Educ. 30, 93–97. [Google Scholar]
- 14. Kozma, R. , Chin, E. , Russell, J. and Marx, N. (2000) The roles of representations and tools in the chemistry laboratory and their implications for chemistry learning. J. Learn. Sci. 9, 105–143. [Google Scholar]
- 15. Kozma, R. B. and Russell, J. (1997) Multimedia and understanding: Expert and novice responses to different representations of chemical phenomena. J. Res. Sci. Teach. 34, 949–968. [Google Scholar]
- 16. Jenkinson, J. and McGill, G. (2012) Visualizing protein interactions and dynamics: Evolving a visual language for molecular animation. CBE Life Sci. Educ. 11, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kramer, I. M. , Dahmani, H.‐R. , Delouche, P. , Bidabe, M. and Schneeberger, P. (2012) Education catching up with science: Preparing students for three‐dimensional literacy in cell biology. Cell Biol. Educ. 11, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dahmani, H.‐R. Schneeberger, P. and Kramer, I. M. (2009) Analysis of students' aptitude to provide meaning to images that represent cellular components at the molecular level. Cell Biol. Educ. 8, 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siemankowski, F. T. and MacKnight, F. C. (1971) Spatial cognition, a success prognosticator in college science courses. J. Coll. Sci. Teach. 1, 56–59. [Google Scholar]
- 20. Wai, J. Lubinski, D. and Benbow, C. P. (2009) Spatial ability for STEM domains: Aligning over 50 years of cumulative psychological knowledge solidifies its importance. J. Educ. Psychol. 101, 817–835. [Google Scholar]
- 21. Linn, M. C. and Petersen, A. C. (1985) Emergence and characterization of sex differences in spatial ability: A meta‐analysis. Child Dev 56, 1479–1498. [PubMed] [Google Scholar]
- 22. Voyer, D. , Voyer, S. and Bryden, M. P. (1995) Magnitude of sex differences in spatial abilities: A meta‐analysis and consideration of critical variables. Psychol. Bull 117, 250–270. [DOI] [PubMed] [Google Scholar]
- 23. Maeda, Y. and Yoon, S. Y. (2013) A meta‐analysis on gender differences in mental rotation ability measured by the Purdue spatial visualization tests: Visualization of rotations (PSVT:R). Educ. Psychol. Rev. 25, 69–94. [Google Scholar]
- 24. Talley, L. H. (1973) The use of three‐dimensional visualization as a moderator in the higher cognitive learning of concepts in college level chemistry. J. Res. Sci. Teach. 10, 263–269. [Google Scholar]
- 25. Gabel, D. and Sherwood, R. (1980) The effect of student manipulation of molecular models on chemistry achievement according to Piagetian level. J. Res. Sci. Teach. 17, 75–81. [Google Scholar]
- 26. Copolo, C. E. and Hounshell, P. B. (1995) Using three‐dimensional models to teach molecular structures in high school chemistry. J. Sci. Educ. Technol. 4, 295–305. [Google Scholar]
- 27. Wu, H.‐K. , Krajcik, J. S. and Soloway, E. (2001) Promoting understanding of chemical representations: Students' use of a visualization tool in the classroom. J. Res. Sci. Teach 38, 821–842. [Google Scholar]
- 28. Harris, M. A. , Peck, R. F. , Colton, S. , Morris, J. , Chaibub Neto, E. and Kallio, J. (2009) A combination of hand‐held models and computer imaging programs helps students answer oral questions about molecular structure and function: A controlled investigation of student learning. Cell Biol. Educ. 8, 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roberts, J. R. , Hagedorn, E. , Dillenburg, P. , Patrick, M. and Herman, T. (2005) Physical models enhance molecular three‐dimensional literacy in an introductory biochemistry course. Biochem. Mol. Biol. Educ. 33, 105–110. [DOI] [PubMed] [Google Scholar]
- 30. Batzli, J. M. (2005) Points of view: A survey of survey courses: Are they effective? A unique approach? Four semesters of biology core curriculum. Cell Biol. Educ. 4, 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saengsawang, W. , Taylor, K. L. , Lumbard, D. C. , Mitok, K. , Price, A. , Pietila, L. , Gomez, T. M. and Dent, E. W. (2013) CIP4 coordinates with phospholipids and actin‐associated proteins to localize to the protruding edge and produce actin ribs and veils. J. Cell Sci. 126, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. The R Team (2014) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 33. Bates, D. , Maechler, M. , Bolker, B. and Walker, S. (2014) _lme4: Linear mixed‐effects models using Eigen and S4_., R package version 1.1‐7.
- 34. Höffler, T. N. (2010) Spatial ability: Its influence on learning with visualizations—A meta‐analytic review. Educ. Psychol. Rev. 22, 245–269. [Google Scholar]
- 35. Harle, M. and Towns, M. (2011) A review of spatial ability literature, its connection to chemistry, and implications for instruction. J. Chem. Educ. 88, 351–360. [Google Scholar]
- 36. Wu, H. ‐K. and Shah, P. (2004) Exploring visuospatial thinking in chemistry learning. Sci. Educ. 88, 465–492. [Google Scholar]
- 37. Harle, M. and Towns, M. H. (2012) Students' understanding of external representations of the potassium ion channel protein, part I: Affordances and limitations of ribbon diagrams, vines, and hydrophobic/polar representations. Biochem. Mol. Biol. Educ. 40, 349–356. [DOI] [PubMed] [Google Scholar]
- 38. Harle, M. and Towns, M. H. (2012) Students' understanding of external representations of the potassium ion channel protein part II: Structure–function relationships and fragmented knowledge. Biochem. Mol. Biol. Educ. 40, 357–363. [DOI] [PubMed] [Google Scholar]
- 39. Van Merriënboer, J. J. G. and Sweller, J. (2005) Cognitive load theory and complex learning: Recent developments and future directions. Educ. Psychol. Rev. 17, 147–177. [Google Scholar]


