Abstract
Structural studies of amyloidogenic segments by X-ray crystallography have revealed a novel packing motif, consisting of out-of-register β sheets, that may constitute one of the toxic species in aggregation related diseases. Here we sought to determine the presence of such a motif in Islet amyloid polypeptide (IAPP), whose amyloidogenic properties are associated with Type 2 Diabetes. We determined four new crystal structures of segments within IAPP all forming steric zippers. Most interestingly, one of the segments in the fibril core of IAPP forms an out-of-register steric zipper. Analysis of this structure reveals several commonalities with previously solved out-of-register fibrils. Our results provide additional evidence of out-of-register β sheets as a common structural motif in amyloid aggregates.
Introduction
Protein aggregation and its associated cytotoxicity are implicated in a wide range of diseases that affect the nervous system as well as other organs; recently protein aggregates have also been associated with certain forms of cancer1–4. Altogether these conditions account for the majority of diseases with few to no treatment options. One step towards understanding the disease etiology is to identify the molecular structures of the aggregated states of proteins that cause cellular dysfunctions. While the atomic structures of the spine of amyloid fibrils have been shown to be made up of β sheets with inter-digitating side chains termed steric zippers5,6, scientists remain confounded about the structures of intermediates that are formed as amyloid proteins transition from monomeric states to insoluble aggregates. An additional complication is that aggregation of proteins yields a heterogeneous population of species that are difficult to separate and characterize. To date, researchers have identified multiple aggregated species, often termed polymorphs that vary in size, secondary structure and cytotoxicity, but there is as yet no consensus about the molecular structures of the toxic species in amyloid-related diseases7,8.
Recently, structural studies have revealed a novel packing motif, the anti-parallel out-of-register β sheet, that may be associated with cytotoxicity in vitro. In one study, the crystal structure of an 11-residue segment from the amyloid protein αB crystallin (ABC) was deciphered9. The structure, termed cylindrin, is a six-stranded β barrel made up of out-of-register anti-parallel β strands. Cylindrin displayed a novel arrangement of β strands different from the steric zippers seen in amyloid fibrils. In most steric zippers, the strands in each β sheet are stacked directly above one another, an arrangement termed in-register; cylindrin instead has out-of-register strands that shear relative to strands below. The out-of-register strands of cylindrin form hexameric oligomers in solution, which were mildly cytotoxic to cells in vitro9.
In other studies, atomic structures of amyloid β-sheet mimics (BAMs) and a hexameric segment from β2-microglobulin (β2m) were determined showing the cylindrin-like feature of out-of-register β strands10–12. The short segment of β2m with the amino acid sequence KDWSFY formed an unusual out-of-register steric zipper. The segment was mildly cytotoxic to cultured cells in vitro, and it was suggested that the toxicity of out-of-register fibrils might derive from forming cylindrin-like oligomers. In view of these out-of-register structures, we set out to investigate if such a motif can be formed by segments of Islet Amyloid Polypeptide (IAPP), the protein associated with Type 2 Diabetes.
IAPP is a 37-residue peptide secreted by the β-cells of the pancreas13,14. It is the main component of extracellular aggregates that display classic amyloid characteristics and are found in the majority of patients suffering from Type-2 diabetes15,16. The segment from residues 20-29 has been suggested to form the core of IAPP fibrils, as mutating residues within this region blocks fibril formation17. Furthermore, mouse IAPP, which has several different residues in this region, does not aggregate and mice do not get diabetes. Another important aspect of IAPP aggregation is that the protein can adopt different conformations in its fibrillar state, a phenomenon referred to as polymorphism. Depending upon the conditions, IAPP has been found to form different fibrillar structures varying in their width, pitch length and ultrastructure18,19. Our previous work has proposed the molecular basis of extreme polymorphism seen in IAPP-derived fibrils. We have found multiple pathways that can lead to variant fibril assemblies. In IAPP, we find that the same segment can adopt different steric zippers, a phenomenon that we have previously termed “packing polymorphism”. Various segments can also nucleate into different steric zippers, a phenomenon termed “segmental polymorphism” 20,21.
Here, we provide additional atomic resolution structures of segments from the fibril core of IAPP identified previously21, one of which forms an out-of-register steric zipper.
Materials and Methods
Sample preparation and crystallization
Peptides were synthesized at greater than 97% purity from CS. Bio (Menlo Park, CA) and Celtek Bioscience (Nashville, TN). All peptide solutions were filtered through a 0.1 μm Ultrafree-MC centrifugal filter device (Amicon, Bedford, MA) prior to crystallization experiments at 18 °C via hanging-drop vapor diffusion. Hanging drop vapor diffusion was carried out in 6-well plates with 1 ml reservoir solution and 1-1.5 ul peptide/reservoir drop sizes.
Crystallization conditions
IAPP13-18 13-ANFLVH-18
The segment was dissolved at 20 mg/ml in water and mixed with 10% (w/v) PEG-8000, 0.1 M Na/K phosphate pH 6.2, and 0.2 M NaCl at a 1:1 ratio by volume. Needle-like crystals appeared within 24 hours.
IAPP16-21 16-LVHSSN-21
This segment was dissolved at 20 mg/ml in water and mixed with 0.09 M HEPES pH 7.5, 1.26M tri-sodium citrate, and 10% glycerol at a 1:1 ratio by volume. Needle-like crystals appeared in 2-3 days.
IAPP22-28 22-NFGAILS-28
This segment was dissolved at 7 mg/ml in water and mixed with 10% ethanol and 1.5M NaCl at a 1:1 ratio by volume. Needle like crystals appeared in one week.
IAPP23-29 23-FGAILSS-29
The segment was dissolved at 6.4mg/ml in 20mM Lithium hydroxide and mixed with 0.1M HEPES pH 6.5 and 0.5M Sodium Formate at a 2:1 ratio by volume. Short micro crystals appeared in a month.
Data collection and structure refinement
Crystals of IAPP segments (ANFLVH, LVHSSN and NFGAILS) were mounted on 20-50 μm Mitegen LD (Ithaca, NY) loops in the presence of 20% glycerol and flash cooled in liquid nitrogen. Crystals of FGAILLS were mounted on glass capillaries without any cryoprotectant. Data was collected at 100 K using a microfocus beam (5×5 μm2) at beamline 24-ID-E of the Advanced Photon Source (APS) at Argonne National Laboratory. Data indexing, integration and scaling were performed using XDS/XSCALE and DENZO/SCALEPACK22. The merged scaled data was imported into the CCP4 format with programs from the CCP4 program suite organized under the “CCP4i” interface23. Molecular replacement solutions for the segments were obtained using the program PHASER24, using a polyalanine β strand as the search model. Crystallographic refinements were performed with REFMAC5, and PHENIX25. Model building was performed with COOT26 and illustrated with PYMOL27.
Results
In register steric zipper structures from IAPP
Previously we have shown that full-length IAPP is capable of forming at least two different fibril morphologies that originate from distinct regions within the sequence 20,21. In addition, we determined crystal structures of six segments within residues 14-37 from IAPP, showing the large variety of steric-zipper spines that can form from the full-length sequence. Here, we expand on the previous work, elucidating the atomic details of four more IAPP segments that were identified by ZipperDB28 to have high fibrillation propensity (Fig. 1a, Table 2), bringing the total number of molecular structures of IAPP amyloidogenic segments to nine. Data collection and refinement statistics can be found in Table 1, and steric zipper statistics in Table 2. Three segments, located in the central region of the IAPP, crystallize as in-register steric zippers.
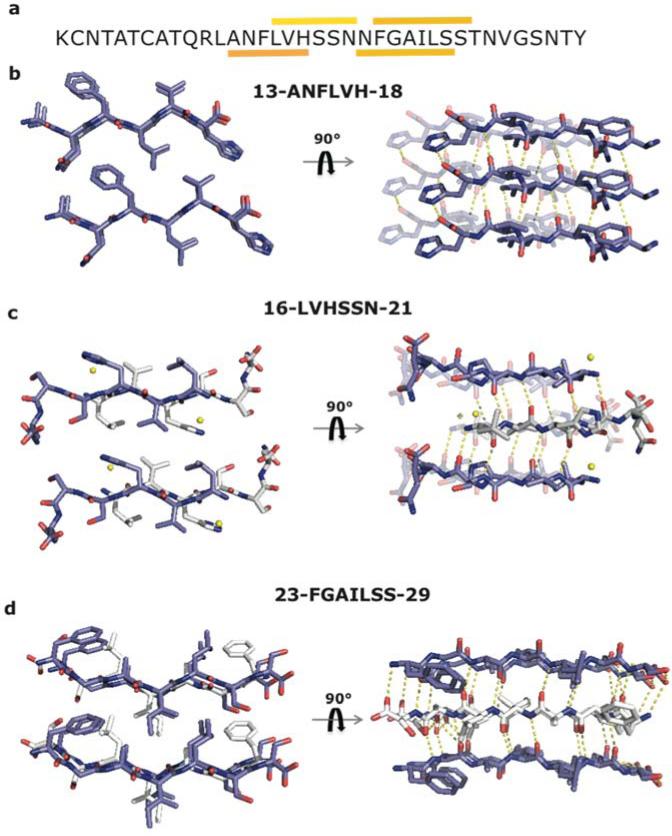
Figure 1.
IAPP segments 13-ANFLVH-18, 16-LVHSSN-21 and 23-FGAILSS-29 form in-register steric zippers. (a) Sequence of human IAPP. Segments characterized here are highlighted by a colored bar over the segment. (b) Crystal structure of segment ANFLVH. View looking down the fibril axis reveals the steric zipper interface involving phenylalanine, leucine, and valine residues. ANFLVH forms a parallel, Class 2 steric zipper6, in which the sheets are related by a pure translation. View perpendicular to the fibril axis reveals hydrogen bond network and stacking of aromatic residues Phe and His, which adds to the stability of the zipper. (c) View down the fibril axis showing the steric zipper interface of segment LVHSSN. This peptide forms a Class 7 steric zipper, in which the strands stack with anti-parallel orientations, while the sheets pack parallel to each other. View perpendicular to the fibril axis, showing the hydrogen bond network of LVHSSN. There is little inter-digitation of side chains. (d) Crystal structure of segment FGAILSS. View down the fibril axis reveals the steric zipper interface. The segment forms a Class 6 steric zipper with the strands in one sheet stacked anti-parallel to each other and the two sheets arranged parallel to each other. Water molecules are shown as yellow spheres.
Table 2.
Structural characteristics of the four steric zippers determined in this work.
| Segment | ZipperDBa (kcal/mol) | Strand Orientation | Steric Zipper Class | Area buried (Å2)b | Shape Complementarityc |
|---|---|---|---|---|---|
| 13-ANFLVH-18 | −22.900 | Parallel | Face-to- back In register β-sheets Symmetry Class 2 | 258 | 0.80 |
| 16-LVHSSN-21 | −22.000 | Anti-parallel | Face-to-back Staggered β-sheets Symmetry Class 7 | 160 | 0.50 |
| 22-NFGAILS-28 | −22.300 | Anti-parallel | Face-to-back Out of register β-sheets Symmetry Class 7 | 293 | 0.83 |
| 23-FGAILSS-29 | −22.500 | Anti-parallel | Face-to-back In register Symmetry Class 6 | 217 | 0.77 |
Estimated energies of steric zippers formed by six-residue segments (starting at the listed residue) of IAPP. Segments having energies of –23 kcal mol–1 or lower are predicted to form fibrils28.
Area buried was calculated using AREAIMOL43 with a probe radius of 1.4Å. The summation of the difference between the accessible surface areas of a) one β-strand alone and in contact with the opposite β-sheet, and of b) the β-sheet alone and in contact with the opposite β-strand, constitutes the reported area buried. In structures with anti-parallel β-strand orientation, as well as in parallel β-strand orientations with different conformations, the average area buried per β-strand is reported.
Lawrence and Colman's shape complementarity index44.
Table 1.
Statistics of structure determination of four segments of IAPP which form steric zippers.
| 13-ANFLVH-18 | 16-LVHSSN-21 | 22-NFGAILS-28 | 23-FGAILSS-29 | |
|---|---|---|---|---|
| Crystal Parameters | ||||
| Space Group | P21 | P21 | P1 | P1 |
| Cell Dimensions | ||||
| a, b, c (Å) | 4.83, 39.73, 9.87 | 9.64, 9.61, 19.03 | 8.7, 11.6, 21.6 | 8.77, 9.5, 24.74 |
| α, β, γ (°) | 90, 103.69, 90 | 90, 101.22, 90 | 86.4, 82.2, 76.4 | 88.22, 80.00, 70.34 |
| Molecules in Asymmetric Unit | 1 | 1 | 2 | 2 |
| Data collection | ||||
| Synchrotron beamline | APS (24-ID-E) | APS (24-ID-E) | APS (24-ID-E) | APS (24-ID-E) |
| Wavelength (Å) | 0.9792 | 0.9792 | 0.9792 | 0.9792 |
| Resolution (Å) | 1.61 | 1.66 | 1.24 | 1.78 |
| Unique Reflections | 433 | 391 | 2227 | 647 |
| Overall Redundancy | 3.1 (3.2) | 3.0(3.0) | 2.9(2.6) | 5.2(4.0) |
| Completeness (%) | 93.8(87.0) | 90.6(97.1) | 97.8(96.9) | 93.4(72.3) |
| Rmerge (%)b | 14.7(13.3) | 7.6(14.9) | 16.6(55.8) | 24.1(69.6) |
| <I/σI> | 6.6(9.7) | 14.1(9.1) | 6.5(1.6) | 4.3(1.4) |
| Refinement | ||||
| Resolution (Å) | 19.86-1.61 | 19.53-1.66 | 21.34-1.24 | 24.35-1.78 |
| Rwork(%)c | 11.2 | 16.7 | 17.3 | 16.7 |
| Rfree(%)d | 16.1 | 19.8 | 20.6 | 21.8 |
| No. atoms | ||||
| Protein | 50 | 46 | 102 | 98 |
| Ligand/ion | 0 | 0 | 0 | 0 |
| Water | 0 | 1 | 7 | 0 |
| Overall B-factors | 7.8 | 4.6 (4.4e) | 2.3(1.9e) | 23.7 |
| R.m.s. deviation | ||||
| Bond length (Å) | 0.003 | 0.004 | 0.008 | 0.016 |
| Bond angle (°) | 0.70 | 1.0 | 1.0 | 2.0 |
a. Values in parentheses correspond to the highest resolution shell.
Rmerge=Σ|I–<I>|/ΣI.
Rwork=Σ|Fo –Fc |/ΣFo.
Rfree= Σ | Fo – Fc | / Σ Fo, calculated using a random set containing 10% reflections that were not included throughout structure refinement.
without water
The segment ANFLVH (residues 13-18) forms β strands that are arranged as parallel, in-register β sheets, with a dry steric zipper interface displaying a face-to-back orientation of the pair of sheets (Fig 1b). This is a Class 2 steric zipper6. The zipper core consists of hydrophobic interactions involving Phe15 and Val17 of one sheet interdigitating with Leu16 of the adjacent sheet. Both the strands and the sheets pack in a parallel orientation, with Phe and His residues stacking on one another along the sheets, adding to the stability of the fibril (Fig. 1b).
The hexameric segment LVHSSN (residues 16-21) forms a staggered in-register steric zipper (Fig 1c) in which the strands stack in an anti-parallel orientation, while the sheets are oriented parallel. Thus, the segment forms a Class 7 zipper. This staggered arrangement of β strands has been seen previously and can be termed “locally out-of-register”29,30. The structure is not “globally out-of-register” because there is no continuous shearing of strands along the sheet. Rather, each pair lies directly above the pair below. This steric zipper lacks the tight inter-digitation seen in ANFLVH. It contains water molecules between mating sheets, hydrogen bonded to serine and histidine residues.
The crystal structure of the segment FGAILSS (residues 23-29) reveals a Class 6 steric zipper with β strands arranged anti-parallel in a β sheet and the two mating sheets running parallel to each other (Fig 1d). The crystal structure is completely devoid of water molecules and the inter-digitation between mating sheets is made up of Ala25 and Leu27 from one sheet and Ile26 and Ser 28 from the opposing sheet.
Crystal structure of IAPP22-28 NFGAILS reveals an out-of-register steric zipper
We identified a fourth segment NFGAILS from IAPP, located in the very amyloidogenic C-terminal region that, interestingly, forms an out-of-register steric zipper (Fig. 2a). The segment forms anti-parallel β strands arranged into parallel sheets, forming a Class 7 steric zipper. The glycine and alanine residues in the center of the segment allow space for the larger phenylalanine, leucine, and isoleucine residues forming the dry, highly complementary stericzipper interface (Figure 2b). Each strand within each sheet of NFGAILS is sheared out of register by two residues (Fig. 2c), as in the previously determined steric zipper structure from β2m, KDWSFY (residues 58-63) forming alternate weak and strong hydrogen bonded interfaces (Fig. 3a, 3c)11. Similar to the KDWSFY structure, the β-strands in the NFGAILS structure are not perpendicular to the fibril axis as in in-register steric zippers11; instead, each strand forms an angle of 40° from the perpendicular (Fig. 3b, 3d). Additionally, similar to the previously determined out-of-register structures, NFGAILS also displays alternating weak and strong hydrogen bonding interfaces. In contrast to β2m structure, the β sheets in NFGAILS have no crossing angle with each other and instead run parallel to each other (Fig. 3b, 3d). This is the first out-of-register structure determined in which the strands completely eclipse each other with a zero crossing angle.
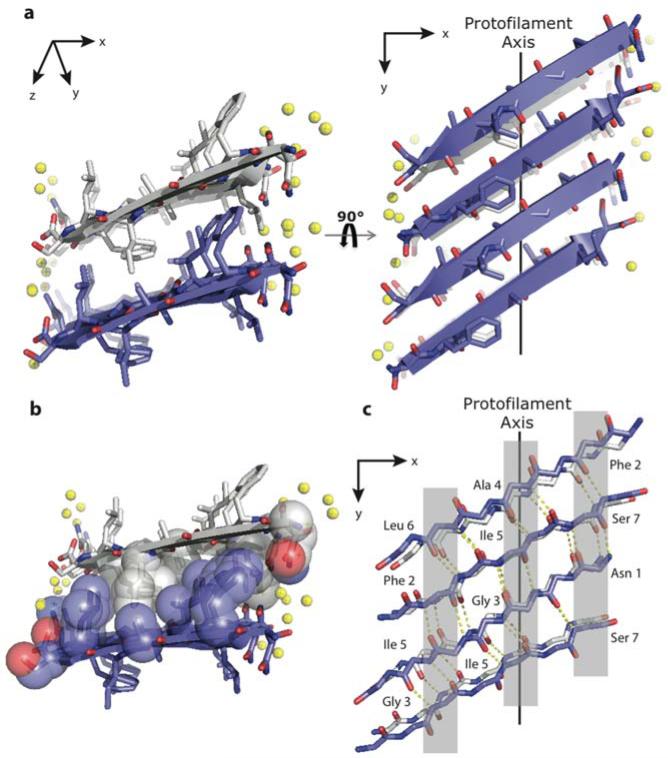
Figure 2.
Segment 22-NFGAILS-28 from IAPP forms an out-of-register steric zipper. (a) View looking down the fibril axis shows mating sheets with side chain inter-digitation. Right panel shows the view rotated 90° to the fibril axis. Notice that the sheets form an acute angle with the protofilament axis as opposed to being exactly perpendicular seen in in-register steric zippers. Water molecules are shown as yellow spheres. (b) View down the fibril axis with side chains in space filling representation shows the dry interface with mating side chains of Ile and Phe (c) Intra-sheet main chain hydrogen bonding along one β sheet. Notice that the strands are sheared such that different amino acids line up over each other (highlighted in gray) in contrast to in register zippers where the strands align such that the same amino acid residues lie over each other.
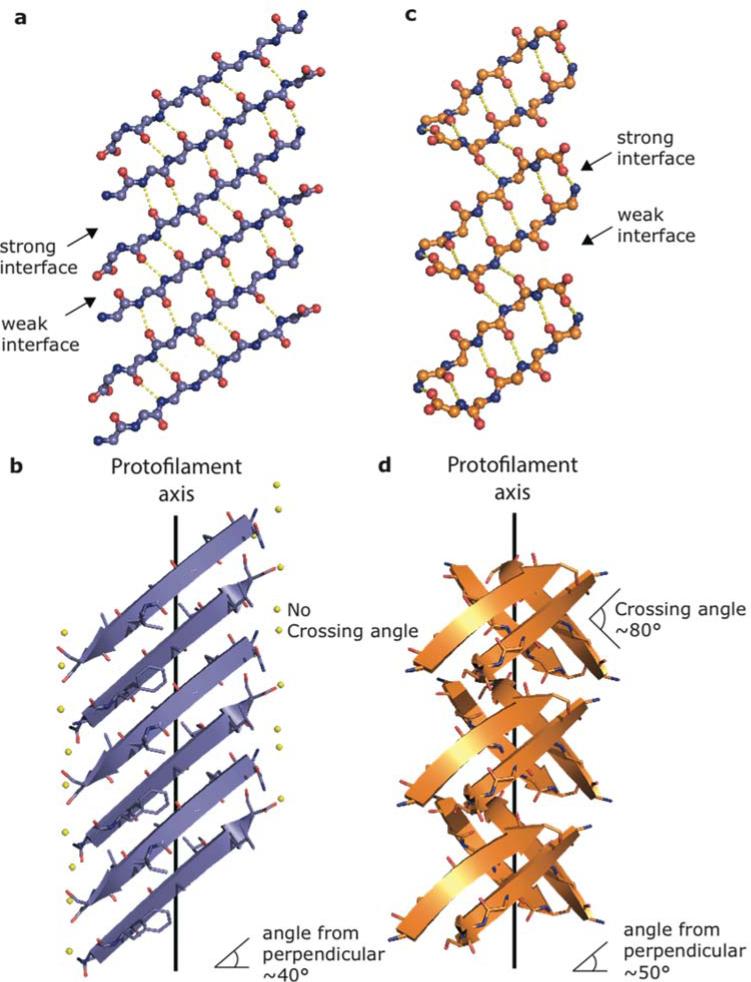
Figure 3.
Structural comparison of 22-NFGAILS-28 of IAPP (left) with the previously determined out-of-register steric zipper from β2-microglobulin (right). View of the hydrogen bond network between strands along a single sheet for NFGAILS (a) and KDWSFY (c) (residues 58-63 of β2-microglobulin, PDB 4E0K). The structure of NFGAILS reveals alternating weak and strong interfaces that run along the sheet, in which the weak interface contains 5 inter-strand hydrogen bonds and the strong interface contains 6 main chain hydrogen bonds similar to the structure of KDWSFY that has a weak interface containing 2 hydrogen bonds and a strong interface containing 6 hydrogen bonds. View perpendicular to the fibril axis shows the β sheets of NFGAILS (b) forming an acute angle with the fibril axis similar to KDWSFY (d). However, the sheets completely eclipse each other in NFGAILS whereas they form an acute angle in KDWSFY.
Discussion
Conformational polymorphism has been hypothesized to be the molecular basis of prion strains. Replication of strains upon addition of new monomers was first reported for the PrP protein, and there is increasing evidence that other amyloid-forming proteins share characteristics of strains, replication and transmission31–36. In our previous work, we showed the atomic basis of polymorphism in IAPP by determining the crystal structures of five amyloidogenic segments that formed different steric zippers20,21. The high degree of segmental polymorphism in IAPP is further highlighted in our current work as the different segments characterized here, even when shifted by only one residue from a previously studied segment, form a different class of steric zipper.
The atomic structures of ANFLVH and LVHSSN determined here further support the role of histidine 18 in promoting fibril formation. Mouse IAPP, which does not aggregate, forms abundant fibrils when a single point replacement R18H is introduced21. From the ANFLVH structure, we deduce that the pi stacking of His18 side chains along each sheet contributes to the stability of the fibril that may be one factor that contributes to mouse IAPP R18H mutant fibrillizing.
The crystal structures of FGAILSS and NFGAILS located in the amyloidogenic core of IAPP reveal anti-parallel zippers. In our previous work, the atomic structure of AILSST, a segment in which four residues overlap with NFGAILS, was also shown to be an anti-parallel zipper21. Together the three structures suggest a propensity of the fibril core of IAPP to form anti-parallel β sheets, consistent with some models proposed for the packing of the amyloidogenic core of IAPP fibrils19,37,38. Interestingly, structural studies of another amyloid protein, Abeta, suggest that some fibril polymorphs and sequence variants are also composed of anti-parallel β sheets 39–41. It has been proposed that because anti-parallel fibrils are typically less stable than parallel fibrils, conversion to potentially toxic, transient morphologies is possible42. However, the physiological consequence of these various fibril architectures is still unclear.
Models of toxic amyloid oligomers and fibrils have emerged from structural studies of cylindrin, amyloid β-sheet mimics (BAMs), and a hexameric segment from β2-microglobulin (β2m)10–12. A notable characteristic shared in all of these structures is that they are composed of out-of-register β strands. This is in contrast to the classic in-register β-sheet packing seen in most amyloid fibrils10–12. The structure of NFGAILS presented here suggests IAPP may also be capable of forming out-of-register fibrils and oligomers.
The structure of NFGAILS reveals several conserved features with previously determined out-of-register structures. First, in all out-of-register structures determined so far including NFGAILS, the β sheets form an acute angle with the protofilament axis. Second, the β sheets are composed of two interfaces of inter-strand hydrogen bond networks: (i) a strong interface in which the hydrogen bond donors and acceptors within the peptide backbone are satisfied; and (ii) a weak interface that leaves unsatisfied donors and acceptors. This is notable in the crystal structure of KDWSFY from β2-microglobulin, the only other solved out-of-register fibril structure to date (Fig. 3c). The structure of NFGAILS also reveals a moderately weak interface, differing by one hydrogen bond (Fig. 3a). One difference between NFGAILS and previously determined out of register zippers is the crossing angle of the mating β sheets. The β sheets in NFGAILS are parallel and do not cross with each other (Fig. 3b). In KDWSFY, the sheets form an 80° crossing angle (Fig. 3c). While it remains to be determined how these characteristics affect the biological properties of these proteins, nevertheless it highlights the variety of different conformations amyloid segments adopt.
In summary, we have further characterized the segments of the fibril core of IAPP, a protein associated with Type 2 diabetes, by crystallizing several of its overlapping segments. Our work provides additional evidence that the fibril cores of IAPP are derived from two distinct regions: one involving Histidine18 and the other, more amyloidogenic region, involving residues 20-29. Furthermore, we show that a segment within residues 20-29 can form an anti-parallel out-of-register zipper, suggesting that out-of-register zippers may be a common motif in amyloid proteins.
Acknowledgements
We thank Michael Collazo and beam line staff at the Advanced Photon Source (APS) Northeastern Collaborative Access Team beamline 24-ID-E for help with experiments. The beamline is funded by the National Institute of General Medical Sciences (P41 GM103403) and U.S. Department of Energy (DOE) Office of Science under Contract No. DE-AC02-06CH11357. We thank HHMI, DOE, and NIH AG 029430 for funding. A.B.S. was supported by UCLA Dissertation year award and S.S. was supported by Whitcome pre-doctoral fellowship.Atomic coordinates and structure factors have been deposited in the Protein Data Bank as 5E5V for NFGAILS, 5E5Z for LVHSSN, 5E61 for FGAILSS, 5E5X for ANFLVH. We thank Pascal Krotee for valuable comments on the manuscript.
References
- 1.Eisenberg D, Jucker M. The Amyloid State of Proteins in Human Diseases. Cell. 2012;148(6):1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006;75(1):333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, Cornelis A, Rozenski J, Zwolinska A, Marine J-C, et al. Gain of Function of Mutant p53 by Coaggregation with Multiple Tumor Suppressors. Nat. Chem. Biol. 2011;7(5):285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- 4.Makin OS, Serpell LC. Structures for Amyloid Fibrils: Structures for Amyloid Fibrils. FEBS J. 2005;272(23):5950–5961. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- 5.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Structure of the Cross-Β Spine of Amyloid-like Fibrils. Nature. 2005;435(7043):773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, et al. Atomic Structures of Amyloid Cross-Β Spines Reveal Varied Steric Zippers. Nature. 2007;447(7143):453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 7.Glabe CG. Structural Classification of Toxic Amyloid Oligomers. J. Biol. Chem. 2008;283(44):29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haass C, Selkoe DJ. Soluble Protein Oligomers in Neurodegeneration: Lessons from the Alzheimer's Amyloid Β-Peptide. Nat. Rev. Mol. Cell Biol. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 9.Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, et al. Atomic View of a Toxic Amyloid Small Oligomer. Science. 2012;335(6073):1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, Sawaya MR, Cheng P-N, Zheng J, Nowick JS, Eisenberg D. Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic Β-Sheet Mimics. J. Am. Chem. Soc. 2011;133(17):6736–6744. doi: 10.1021/ja200222n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Zhao M, Jiang L, Cheng P-N, Park J, Sawaya MR, Pensalfini A, Gou D, Berk AJ, Glabe CG, et al. Out-of-Register -Sheets Suggest a Pathway to Toxic Amyloid Aggregates. Proc. Natl. Acad. Sci. 2012;109(51):20913–20918. doi: 10.1073/pnas.1218792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng P-N, Liu C, Zhao M, Eisenberg D, Nowick JS. Amyloid Β-Sheet Mimics That Antagonize Protein Aggregation and Reduce Amyloid Toxicity. Nat. Chem. 2012;4(11):927–933. doi: 10.1038/nchem.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper GJS, Willis AC, Clark A, Turner RC, Sim RB, Reid KBM. Purification and Characterization of a Peptide from Amyloid-Rich Pancreases of Type 2 Diabetic Patients. Proc. Natl. Acad. Sci. 1987;84:8628–8632. doi: 10.1073/pnas.84.23.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westermark P, Wernstedt C, Wilander E, Sletten K. A Novel Peptide in the Calcitonin Gene Related Peptide Family as an Amyloid Fibril Protein in the Endocrine Pancreas. Biochem. Biophys. Res. Commun. 1986;140(3):827–831. doi: 10.1016/0006-291x(86)90708-4. [DOI] [PubMed] [Google Scholar]
- 15.Kahn SE, Andrikopoulos S, Verchere CB. Islet Amyloid: A Long- Recognized but Underappreciated Pathological Feature of Type 2 Diabetes. Diabetes. 1999;48(2):241–253. doi: 10.2337/diabetes.48.2.241. [DOI] [PubMed] [Google Scholar]
- 16.Cao P, Marek P, Noor H, Patsalo V, Tu L-H, Wang H, Abedini A, Raleigh DP. Islet Amyloid: From Fundamental Biophysics to Mechanisms of Cytotoxicity. FEBS Lett. 2013;587(8):1106–1118. doi: 10.1016/j.febslet.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tenidis K, Waldner M, Bernhagen J, Fischle W, Bergmann M, Weber M, Merkle M-L, Voelter W, Brunner H, Kapurniotu A. Identification of a Penta- and Hexapeptide of Islet Amyloid Polypeptide (IAPP) with Amyloidogenic and Cytotoxic Properties. J. Mol. Biol. 2000;295(4):1055–1071. doi: 10.1006/jmbi.1999.3422. [DOI] [PubMed] [Google Scholar]
- 18.Goldsbury CS, Cooper GJS, Goldie KN, Muller S, Kistler J. Polymorphic Fibrillar Assembly of Human Amylin. Journal of Structural Biology. 1997;119:17–27. doi: 10.1006/jsbi.1997.3858. [DOI] [PubMed] [Google Scholar]
- 19.Madine J, Jack E, Stockley PG, Radford SE, Serpell LC, Middleton DA. Structural Insights into the Polymorphism of Amyloid-Like Fibrils Formed by Region 20−29 of Amylin Revealed by Solid-State NMR and XRay Fiber Diffraction. J. Am. Chem. Soc. 2008;130(45):14990–15001. doi: 10.1021/ja802483d. [DOI] [PubMed] [Google Scholar]
- 20.Wiltzius JJW, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, Eisenberg D. Atomic Structure of the Cross-Β Spine of Islet Amyloid Polypeptide (amylin). Protein Sci. 2008;17(9):1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiltzius JJW, Landau M, Nelson R, Sawaya MR, Apostol MI, Goldschmidt L, Soriaga AB, Cascio D, Rajashankar K, Eisenberg D. Molecular Mechanisms for Protein-Encoded Inheritance. Nat. Struct. 38 Mol. Biol. 2009;16(9):973–978. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. J. Appl. Crystallogr. 1993;26(6):795–800. [Google Scholar]
- 23.The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D Biol. Crystallogr. No. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Read RJ. Pushing the Boundaries of Molecular Replacement with Maximum Likelihood. Acta Crystallogr. D Biol. Crystallogr. 2001;57(10):1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 25.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX : A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66(2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot : Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60(12):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC [Google Scholar]
- 28.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the Amylome, Proteins Capable of Forming Amyloid-like Fibrils. Proc. Natl. Acad. Sci. 2010;107(8):3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta AK, Lu K, Childers WS, Liang Y, Dublin SN, Dong J, Snyder JP, Pingali SV, Thiyagarajan P, Lynn DG. Facial Symmetry in Protein Self-Assembly. J. Am. Chem. Soc. 2008;130(30):9829–9835. doi: 10.1021/ja801511n. [DOI] [PubMed] [Google Scholar]
- 30.Liang Y, Pingali SV, Jogalekar AS, Snyder JP, Thiyagarajan P, Lynn DG. Cross-Strand Pairing and Amyloid Assembly. Biochemistry (Mosc.) 2008;47(38):10018–10026. doi: 10.1021/bi801081c. [DOI] [PubMed] [Google Scholar]
- 31.Prusiner SB. A Unifying Role for Prions in Neurodegenerative Diseases. Science. 2012;336(6088):1511–1513. doi: 10.1126/science.1222951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brundin P, Melki R, Kopito R. Prion-like Transmission of Protein Aggregates in Neurodegenerative Diseases. Nat. Rev. Mol. Cell Biol. 2010;11(4):301–307. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J-X, Qiang W, Yau W-M, Schwieters CD, Meredith SC, Tycko R. Molecular Structure of Β-Amyloid Fibrils in Alzheimer's Disease Brain Tissue. Cell. 2013;154(6):1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda-Suzukake M, Nonaka T, Hosokawa M, Oikawa T, Arai T, Akiyama H, Mann DMA, Hasegawa M. Prion-like Spreading of Pathological -Synuclein in Brain. Brain. 2013;136(4):1128–1138. doi: 10.1093/brain/awt037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, Serpell LC, et al. Distinct Tau Prion Strains Propagate in Cells and Mice and Define Different Tauopathies. Neuron. 2014;82(6):1271–1288. doi: 10.1016/j.neuron.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall KE, Serpell LC. Fibres, Crystals and Polymorphism: The Structural Promiscuity of Amyloidogenic Peptides. Soft Matter. 2010;6(10):2110. [Google Scholar]
- 37.Ashburn TT, Auger M, Lansbury PT. The Structural Basis of Pancreatic Amyloid Formation: Isotope-Edited Spectroscopy in the Solid State. J. Am. Chem. Soc. 1992;114(2):790–791. [Google Scholar]
- 38.Griffiths JM, Ashburn TT, Auger M, Costa PR, Griffin RG, Lansbury PT. Rotational Resonance Solid-State NMR Elucidates a Structural Model of Pancreatic Amyloid. J. Am. Chem. Soc. 1995;117(12):3539–3546. [Google Scholar]
- 39.Lansbury PT, Costa PR, Griffiths JM, Simon EJ, Auger M, Halverson KJ, Kocisko DA, Hendsch ZS, Ashburn TT, Spencer RGS, et al. Structural Model for the Β-Amyloid Fibril Based on Interstrand Alignment of an Antiparallel-Sheet Comprising a C-Terminal Peptide. Nat. Struct. Biol. 1995;2(11):990–998. doi: 10.1038/nsb1195-990. [DOI] [PubMed] [Google Scholar]
- 40.Colletier J-P, Laganowsky A, Landau M, Zhao M, Soriaga AB, Goldschmidt L, Flot D, Cascio D, Sawaya MR, Eisenberg D. Molecular Basis for Amyloid-Β Polymorphism. Proc. Natl. Acad. Sci. 2011;108(41):16938–16943. doi: 10.1073/pnas.1112600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiang W, Yau W-M, Luo Y, Mattson MP, Tycko R. Antiparallel -Sheet Architecture in Iowa-Mutant -Amyloid Fibrils. Proc. Natl. Acad. Sci. 2012;109(12):4443–4448. doi: 10.1073/pnas.1111305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berthelot K, Ta HP, Géan J, Lecomte S, Cullin C. In Vivo and In Vitro Analyses of Toxic Mutants of HET-S: FTIR Antiparallel Signature Correlates with Amyloid Toxicity. J. Mol. Biol. 2011;412(1):137–152. doi: 10.1016/j.jmb.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Lee B, Richards FM. The Interpretation of Protein Structures: Estimation of Static Accessibility. J. Mol. Biol. 1971;55(3):379–IN4. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 44.Lawrence MC, Colman PM. Shape Complementarity at Protein/Protein Interfaces. J. Mol. Biol. 1993;234(4):946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]