Abstract
HER2 gene-protein assay (GPA) is a new method for the simultaneous evaluation of HER2 immunohistochemistry (IHC) and HER2 dual in situ hybridization (DISH) on single tissue sections of breast cancer. We investigated the presence of HER2 gene and protein discrepancy and HER2-heterogeneity using HER2-GPA. HER2 status was analyzed for the correlation between the presence of HER2-heterogeneity and patient prognosis. Consecutive 280 invasive breast cancer were examined. Statuses of HER2 protein and gene were evaluated in whole tumor sections of HER2 GPA slides. HER2 protein and gene combination patterns were classified to six phenotypic and genotypic types for each case, as well as at individual cell levels: (A) IHC and DISH positive; (B) IHC positive and DISH negative; (C) IHC equivocal and DISH positive; (D) IHC equivocal and DISH negative; (E) IHC negative and DISH positive; and (F) IHC and DISH negative. The presence of HER2-heterogeneity was determined by the existence of at least two of six types within one tumor. HER2-IHC positive patients had significantly worse survival than IHC negative patients and HER2-DISH positive patients had significantly worse survival than DISH negative patients. HER2 IHC negative and DISH positive patients had significantly worse recurrence-free survival than IHC and DISH negative patients. In the HER2 IHC and DISH negative group, the HER2 heterogeneous group had significantly worse survival than the nonheterogeneous group. Notably, among triple negative breast cancer (TNBC), the HER2 heterogeneous group had significantly worse survival than the nonheterogeneous group. Our study suggests that the presence of HER2-heterogeneity might be a prognostic factor in HER2 negative breast cancer patients, especially in TNBC.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-016-3856-2) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, HER2, Intratumoral heterogeneity, Trastuzumab, Gene-protein assay
Introduction
Breast cancer is the most commonly diagnosed cancer in women and the second leading cause of cancer-related death in women [1]. Breast cancer is a diverse disease with different histological tumor subtypes that can be further characterized on the basis of specific markers. The most commonly examined immunohistochemical markers are estrogen receptor (ER), progesterone receptor (PgR), and human epidermal group factor receptor 2 (HER2) [2]. HER2 is a tyrosine kinase member of the epidermal growth factor receptor (EGFR) family and HER2 gene is located on the long arm of chromosome 17 (17q12–21.32). HER2 protein is activated upon receptor dimerization that causes autophosphorylation and subsequent downstream signaling [3, 4]. Patients with HER2 positive disease typically have a worse prognosis with characteristics of aggressive tumor progress and shorter patient survival.
HER2 positive status of breast cancer patients is assessed by the HER2 protein overexpression or HER2 gene amplification. HER2 protein expression is examined using bright filed HER2 immunohistochemistry (IHC) with 3,3′-diaminobenzidine (DAB) detection and HER2 gene level is evaluated by HER2 in situ hybridization (ISH) methods, utilizing various detection approaches such as dual color fluorescent ISH (FISH), chromogenic ISH (CISH), silver ISH (SISH), and dual color ISH (DISH) [5, 6]. Recently, the gene-protein assay (GPA) has been introduced as a new method for simultaneous evaluation of U.S. Food and Drug Administration (FDA)-approved HER2 IHC and HER2 DISH assays in a single tissue section, allowing pathologists to examine both HER2 protein and HER2 gene statuses simultaneously at the single cell level [7]. HER2 intratumoral heterogeneity in breast cancer can be detected effectively by the concurrent observations of HER2 IHC and DISH results in breast cancer [7].
Trastuzumab, a humanized monoclonal antibody to HER2 protein, binds to the extracellular domain of HER2 in the cell membrane of carcinoma cells for the suppressions of HER2 signaling and the inhibition of cell proliferation by arresting the cell cycle during the G1 phase [8, 9]. In addition, antibody binding to the HER2 extracellular domain leads to antibody-dependent cell-mediated cytotoxicity (ADCC) triggering the carcinoma cell death by immune cells [10, 11]. The strategy of breast cancer treatment changed drastically after trastuzumab was accepted by FDA in 1998 for the treatment of HER2 overexpressing breast cancer. Moreover, additional HER2-targeted therapy agents such as lapatinib [12], pertuzumab [13], and ado-trastuzumab emtansine, known as T-DM1 [14], have been approved for the treatment of HER2 overexpressing breast cancers. On the other hand, National Surgical Adjuvant Breast and Bowel Project (NSABP) trial B-31 that compared a standard chemotherapy and a trastuzumab adjuvant therapy with chemotherapy suggested some HER2 negative patients may benefit moderately from adjuvant trastuzumab therapy [15, 16]. A randomized phase III trial of adjuvant trastuzumab therapy (NSABP B-47) with 3260 patients who are HER2 IHC 1+ or 2+ scores, but HER2 FISH negative will reveal how the HER2 targeted therapy is effective to treat HER2 negative breast cancer patients [17].
HER2 tumor heterogeneity is a major challenge for accurate evaluation of HER2 status in breast cancer and the heterogeneity can result in discordant between HER2 IHC and ISH assays [18]. The HER2 tumor heterogeneity might affect the management of early and advanced breast cancer patients [19]. However, theories of HER2 tumor heterogeneity evaluations are mainly based on expert opinions, but not based on clinical outcome of breast cancer patients. Though, recent studies reported that breast cancer patients with the HER2 intratumoral heterogeneity had reduced disease-free survival in HER2 positive invasive breast cancers [20] and influenced the effectiveness of trastuzumab therapy in metastatic HER2 positive breast cancer [21]. HER2 tumor heterogeneity is more often observed with HER2 IHC 2+/equivocal cases, mainly in HER2 gene negative status assessed cases containing individual amplified HER2 gene tumor cells [22]. Furthermore, breast cancer with HER2 heterogeneity presented an aggressive phenotype [23]. However, the significance of HER2 heterogeneity evaluation has not been established for a long-term breast cancer patient outcome after the surgical procedure yet. In the current study, we investigated the relationship between HER2 gene and protein status of invasive breast cancer cases using the HER2 GPA method and analyzed the correlation between the presence of HER2 heterogeneity and the prognosis of these breast cancer patients after surgery.
Methods
Patient backgrounds and eligibility
Two hundred eighty (280) consecutive breast cancer patients with invasive lesions larger than 5 mm diagnosed at Saitama Cancer Center from January 2000 to December 2001 were evaluated in this study. However, male and bilateral breast cancer patients were excluded. All patients underwent breast-conserving surgery or modified radical mastectomy. Clinicopathological parameters including tumor size and nodal status were retrieved from medical reports and follow-up data were obtained from the database of the breast cancer clinic division. Because adjuvant administration had not been approved in Japan before 2008, no HER2 positive patient received adjuvant trastuzumab therapy. However, 76 % of HER2 positive breast cancer patients who presented with the distant metastasis during a follow-up duration received trastuzumab therapy combined with chemotherapy.
This study was conducted in accordance with the Declaration of Helsinki and the study protocol was approved by the Institutional Review Board of Saitama Cancer Center. All patients included in this study provided written informed consents for a comprehensive scientific examination of clinical samples. This retrospective translational study was performed as a collaborative research project between Saitama Prefectural Government, Japan and Ventana Medical Systems, Inc., the United States after execution of a formal contract. In order to keep the integrity of results and analyses of the current study, study activities were performed separately by three groups: (1) the pathological examination group; (2) the GPA examination group; and (3) the clinical analysis group. No clinical information was provided to the GPA analysis group and the GPA analysis data were completely closed before data were transferred to the clinical analysis group.
Histopathological and immunohistochemical analyses
Histopathological analyses and immunohistochemical evaluation of ER and PgR were performed by the pathological examination group. Tumor specimens obtained by surgery were stored in the refrigerated (4 °C) physiological saline solution for approximately 30 to 90 min and then they were fixed in 20 % buffered formalin solution for 3 to 4 days. Formalin-fixed, paraffin-embedded (FFPE) tissue Sects. (4 µm) were prepared for hematoxylin and eosin (H&E) and hormone receptor IHC stains. Pathological diagnosis and evaluation of histological grade were performed using H&E stained sections. ER and PgR IHC stains were performed by an automated instrument (Autostainer, Dako, Denmark) using primary antibodies for ER (clone 1D5, Dako) and PgR (clone PgR636, Dako). ER and PgR status was determined according to the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guideline.
HER2 GPA procedure
HER2 GPA performance and analyses were performed by the HER2 GPA exanimation group. HER2 GPA for the simultaneous visualization of HER2 protein, HER2 gene, and chromosome 17 centromere (CEN17) was performed as described before using FFPE tumor tissue Sects. (4 µm) using FDA-approved HER2 IHC and DISH reagents (Ventana Medical Systems, Inc., USA) on an automated staining platform (BenchMark XT, Ventana) [7]. Briefly, deparaffinized tissue sections were subjected to heat pretreatment and endogenous biotin blocking treatment for HER2 IHC detection. Tissue sections were incubated with anti-HER2 antibody (clone 4B5, Ventana) and the antibody binding site was visualized using a 3,3′ diaminobenzidine IHC kit. After the completion of HER2 IHC staining, HER2 DISH was performed for the detection of HER2 gene and CEN17 targets. HER2 IHC tissue sections were subjected to a second heat pretreatment followed by a protease digestion. A cocktail of 2,4-dinitrophenol (DNP)-labeled HER2 probe and digoxigenin (DIG)-labeled CEN17 probe was hybridized on HER2 IHC stained tissue section. After three stringency wash steps, HER2 and CEN17 probe hybridization sites were visualized using the SISH DNP detection kit and the red ISH DIG detection Kit, respectively. After counterstained with hematoxylin, the slides were cover-slipped for light microscopical observations.
Interpretation of HER2 GPA results
Immunohistochemical expression of HER2 protein and status of HER2 gene were simultaneously analyzed in each section according to the package insert of FDA-approved HER2 IHC and DISH assays by a well-experienced pathologist (MP) with both HER2 tests. HER2 IHC staining was scored as followings: score 0 (negative)—no membrane staining is observed; score 1+ (negative)—faint, partial staining of the membrane in any proportion of the cancer cells; score 2+ (equivocal)—weak complete staining of the membrane, greater than 10 % of cancer cells; and score 3+ (positive)—intense complete staining of the membrane, greater than 10 % of cancer cells. HER2 DISH was analyzed by a HER2 to CEN17 signal ratio as followings: HER2/CEN17 <2.0 (negative) and HER2/CEN17 ≥2.0 (positive). In addition, CEN17 polysomy was defined as an average CEN17 copy number ≥3 per cell.
Evaluation of HER2 discordance and heterogeneity
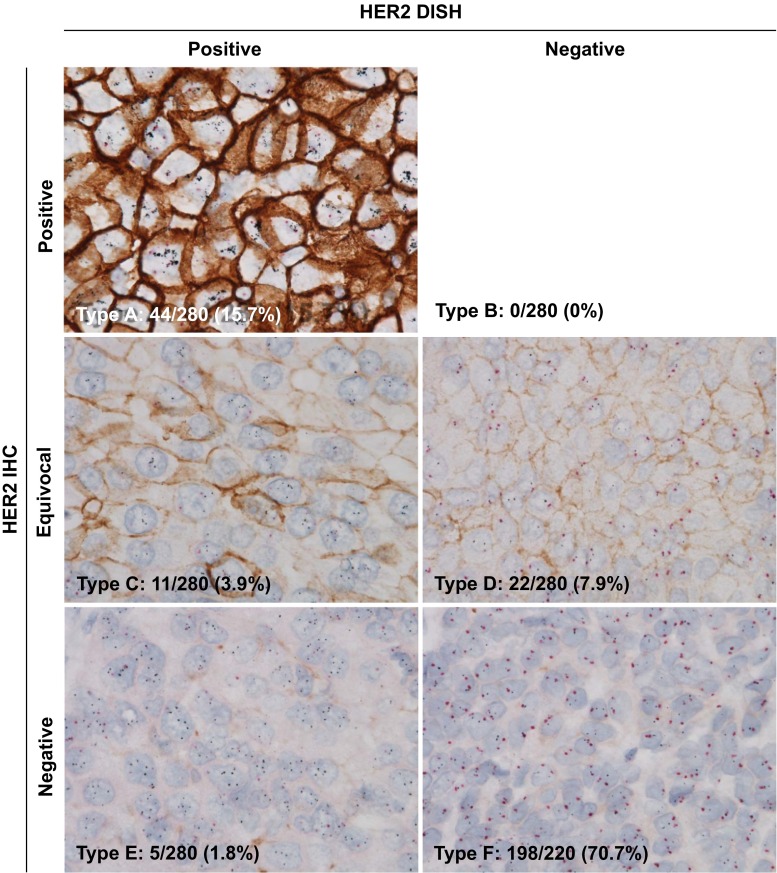
Patterns of the HER2 protein and gene status analyzed with the GPA approach were divided into the following six types among individual cases (see Fig. 1): Type A) IHC & DISH positive; Type B) IHC positive & DISH negative; Type C) IHC equivocal & DISH positive; Type D) IHC equivocal & DISH negative; Type E) IHC negative & DISH positive; and Type F) IHC & DISH negative. Status of HER2 discordance was defined as following two patterns: Type B) IHC positive & DISH negative and E) IHC negative & DISH positive. Cases were categorized HER2 nonheterogeneous when only one type of HER2 protein and gene status combination was observed within one tumor tissue section. On the other hand, cases were categorized HER2 heterogeneous when at least two of the six types of HER2 gene and protein status combinations were observed with individual tumor cells in a tumor tissue section. For example, when a Type F HER2 negative breast case contained individual tumor cells which are not Type F cells, the case is a HER2 heterogeneous case. Before starting this study, the evaluation method of the heterogeneity of HER2 protein expression and HER2 gene status by GPA were confirmed by investigators and the GPA slides of the present study were primarily evaluated by one pathologist (MP) who is one of HER2 GPA induction members.
Fig. 1.
Phenotypic and genotypic human epidermal growth factor receptor 2 (HER2) pattern analyses of breast cancer by the HER2 gene-protein assay method (n = 280). Type A: HER2 immunohistochemistry (IHC) positive and dual in situ hybridization (DISH) positive (n = 44); Type B: HER2 IHC positive and DISH negative (n = 0); Type C: HER2 IHC equivocal and DISH positive (n = 11); Type D: HER2 IHC equivocal and DISH negative (n = 22); Type E: HER2 IHC negative and DISH positive (n = 5); and Type F: HER2 IHC and DISH negative (n = 198). Brown color indicates HER2 protein expression, black dots in the nuclei are HER2 gene, and red dots in the nuclei are chromosome 17 centromere
Clinical outcome investigation
Clinical correlation analyses were conducted by the clinical examination group. Recurrence-free survival (RFS) and cancer-specific survival (CSS) were analyzed with HER2 IHC scores among IHC positive, equivocal, and negative populations and HER2 DISH results between DISH positive and negative populations. RFS and CSS were also analyzed between HER2 heterogeneous and nonheterogeneous populations. Because of the size of samples for statistical analyses, patient outcome data were compared between HER2 nonheterogeneous and heterogeneous groups only in Type F (HER2 IHC & DISH negative) patient population.
Statistical analyses
Statistical analyses were performed using SPSS v22.0 (IBM Corp., USA) software. The Kaplan–Meier survival curves and log-rank test were used to estimate RFS and CSS. Chi square and Fisher’s exact tests were used to analyze the associations between clinicopathological characteristics and HER2 heterogeneity statues. These factors were included in the univariate and multivariate survival analysis using Cox proportional hazards regression model, and these 95 % confidence interval were assessed for each factor. P value <0.05 were defined statistically significant.
Results
Clinicopathological characteristics
The clinicopathological characteristics of the 280 cases are summarized in Supplementary File 1. The median age of the patients was 55 years (range, 25–87 years) and the median follow-up duration was 130 months (range, 4 to 149 months) at the time of diagnosis. One hundred nineteen patients (42.5 %) were in pre-menopausal status and 197 (70.4 %) patients had ER positive tumors.
HER2 phenotypic and genotypic analyses
HER2 IHC staining of HER2 GPA showed that 91 cases (32.5 %) were scored 0 (negative), 112 cases (40.0 %) were 1+ (negative), 33 cases (11.8 %) were 2+ (equivocal) and 44 cases (15.7 %) were 3+ (positive). Sixty (21.4 %) tumor specimens were HER2 DISH positive and 220 (78.6 %) were negative (Table 1). Six subtypes of HER2 GPA results were identified among 280 cases as follows (Fig. 1): (1) Type A (IHC & DISH positive) comprised 44 cases (15.7 %); (2) Type B (IHC positive & DISH negative) comprised no cases (0.0 %); (3) Type C (IHC equivocal & DISH positive) comprised 11 cases (3.9 %); (4) Type D (IHC equivocal & DISH negative) comprised 22 cases (7.9 %); (5) Type E (IHC negative & DISH positive) comprised 5 cases (1.8 %); and 6) Type F (IHC & DISH negative) included 198 cases (70.7 %). Incident of HER2 phenotypic and genotypic discordant was low (0 Type B and 5 Type E cases among 280 cases, 1.8 %) within our cohort (Fig. 2).
Table 1.
Scores of HER2 IHC and HER2 DISH separately 1 analyzed by GPA
| HER2 status | No. of patients | % |
|---|---|---|
| (n = 280) | ||
| HER2 IHC score | ||
| 0 | 91 | 32.5 |
| 1+ | 112 | 40.0 |
| 2+ | 33 | 11.8 |
| 3+ | 44 | 15.7 |
| HER2 DISH score | ||
| Negative | 220 | 78.6 |
| Positive | 60 | 21.4 |
HER2 human epidermal growth factor receptor 2, IHC immunohistochemistry, DISH dual in situ hybridization, GPA gene-protein assay
Fig. 2.
Human epidermal growth factor receptor 2 (HER2) intratumoral heterogeneity of HER2 negative breast cancer revealed by the HER2 gene-protein assay. a Breast cancer cells showing no HER2 protein expression and nonheterogeneous HER2 gene status. b Breast cancer cells showing no HER2 protein expression and heterogeneous HER2 gene status. Yellow dotted circles show tumor cells with amplified HER2 gene. Black dots and red dots in the nuclei are HER2 gene and chromosome 17 centromere, respectively
Survival rate stratified by HER2 protein and gene status
Patients with HER2 IHC 0,1+ (negative) tumors had significantly better survival than those with IHC 3+ (positive) tumors (RFS: HR, = 8.31; P = .0039; CSS: HR, 4.39; P = .036) (Fig. 3a, b). RFS of the HER2 IHC 0,1+ group was better than that of the HER2 IHC 2+ (equivocal) group (RFS: HR, 2.93; P = .087; CSS: HR, .19; P = .66), but survival was not significantly different between the HER2 IHC 2+ (equivocal) group and the HER2 IHC 3+ (positive) group (RFS: HR, .64; P = .43; CSS: HR, 1.15; P = .28). In addition, patients with HER2 DISH positive tumors had significantly worse survival than those with negative tumors (RFS: HR, 7.67; P = .0056; CSS: HR, 3.85; P = .050) (Fig. 3c, d).
Fig. 3.
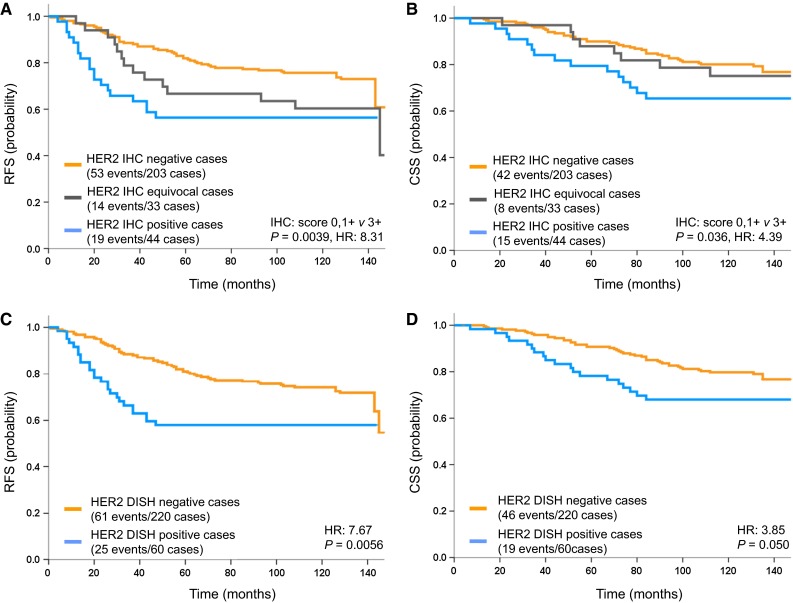
Survival curves of breast cancer patients (n = 280) stratified by the status of human epidermal growth factor receptor 2 (HER2) protein and HER2 gene separately evaluated by the HER2 gene-protein assay. Association of HER2 immunohistochemistry (IHC) scores with a recurrence-free survival (RFS) and b cancer-specific survival (CSS). Association of HER2 dual in situ hybridization (DISH) status with c RFS and d CSS
Survival curves stratified by HER2 phenotypic and genotypic types A–F
The comparison of survival between each group was shown in Table 2 and Supplementary File 2. The HER2 IHC & DISH positive Type A had significantly worse survival than the HER2 IHC & DISH negative Type F (RFS: HR, 4.39; P = .0025; CSS: HR, 4.77; P = .029). The HER2 IHC negative & DISH positive Type E had significantly worse survival than the HER2 IHC & DISH negative Type F (RFS: HR, 5.34; P = .021; CSS: HR, 1.48; P = .18). Finally, the HER2 IHC equivocal & DISH negative Type D had significantly worse RFS than the HER2 IHC & DISH negative Type F (RFS: HR, 4.65; P = .031; CSS: HR, 0.52; P = .47).
Table 2.
Comparison of survival among A–F groups 1 valuated by GPA
| Group | Recurrence-free survival | Cancer-specific survival | ||||||
|---|---|---|---|---|---|---|---|---|
| A | C | D | E | A | C | D | E | |
| C | HR: 1.18 | HR: 1.06 | ||||||
| P = 0.28 | P = 0.30 | |||||||
| D | HR: 0.10 | HR: 0.64 | HR: 0.51 | HR: 0.22 | ||||
| P = 0.75 | P = 0.43 | P = 0.47 | P = 0.64 | |||||
| E | HR: 0.25 | HR: 1.40 | HR: 0.77 | HR: 0.08 | HR: 1.26 | HR: 0.56 | ||
| P = 0.61 | P = 0.24 | P = 0.38 | P = 0.78 | P = 0.26 | P = 0.46 | |||
| F | HR: 4.39 | HR: 0.05 | HR: 4.65 | HR: 5.34 | HR: 4.77 | HR: 0.01 | HR: 0.52 | HR: 1.83 |
| P = 0.0025 | P = 0.82 | P = 0.031 | P = 0.021 | P = 0.029 | P = 0.31 | P = 0.47 | P = 0.18 | |
HR hazard ratio, GPA gene-protein assay
Association of HER2 heterogeneity with the tumor clinicopathological characteristics
HER2 heterogeneity was recognized in 34 (17.2 %) of 198 HER2 IHC & HER2 DISH negative cases (Type F). As shown in Table 3, the HER2 heterogeneous group contained significantly higher HER2 IHC score 1+ cases (P < .001) than HER2 IHC 0 cases and a higher HER2/CEN17 ratio (P = .023) compared to HER2 nonheterogeneous group. Moreover, the presence of HER2 heterogeneity significantly correlated with the presence of chromosome 17 polysomy (P = .0011). Other clinicopathologic factors (histological grade, tumor stage, node stage, ER IHC status, and PR IHC status) were not associated with the presence of HER2 heterogeneity.
Table 3.
Relationship between HER2 heterogeneity and clinicopathological characteristics in HER2 IHC 0, 1+ and HER2 DISH negative breast cancer
| Characteristic | HER2 heterogeneous | HER2 | P |
|---|---|---|---|
| (n = 34) | Nonheterogeneous | ||
| (n = 164) | |||
| Age | 56 (25–81) | 55 (26–86) | .80 |
| Tumor stage | |||
| T1-2 | 32 | 144 | .29 |
| N1-3 | 2 | 20 | |
| Node stage | |||
| N0 | 16 | 90 | .41 |
| N1-3 | 18 | 74 | |
| Histological grade | |||
| 1–2 | 16 | 90 | .41 |
| 3 | 18 | 74 | |
| HER2 IHC score | |||
| 1+ | 28 | 79 | <.001 |
| 0 | 6 | 85 | |
| HER2/CEN17 ratio | 1.339 (0.798–1.972) | 1.151 (0.455–1.810) | .023 |
| CEN17 polysomy | |||
| No | 30 | 162 | .0011 |
| Yes | 4 | 2 | |
| ER IHC score | |||
| Negative | 26 | 127 | .90 |
| Positive | 8 | 37 | |
| PgR IHC score | |||
| Negative | 24 | 111 | .74 |
| Positive | 10 | 53 | |
| TNBC | |||
| No | 27 | 132 | .89 |
| Yes | 7 | 32 |
HER2 human epidermal growth factor 2, IHC immunohistochemistry, DISH dual in situ hybridization, CEN17 chromosome 17 centromere, ER estrogen receptor, PgR progesterone receptor, TNBC triple negative breast cancer
Survival of HER2 IHC & DISH negative patients stratified by presence of HER2 heterogeneity
The HER2 heterogeneous group had significantly worse survival than HER2 nonheterogeneous group (RFS: HR, 7.83; P = .0051; CSS: HR, 4.92; P = .027; Fig. 4). Multivariate analyses showed that HER2 heterogeneity is one of independent prognostic factors (RFS: P = .0076, CSS: P = .041, Table 4). Particularly in patients with triple negative breast cancer (TNBC: ER negative, PgR negative and HER2 negative), the HER2 heterogeneous group had significantly worse survival than HER2 nonheterogeneous group (RFS: HR, 13.44; P = .00025; CSS: HR, 13.72; P = .00021; Fig. 4).
Fig. 4.

Survival curves of breast cancer patients (n = 280) stratified by the presence of human epidermal growth factor receptor 2 (HER2) intratumoral heterogeneity based on the HER2 gene-protein assay analyses. Association of the status of HER2 intratumoral heterogeneity with a recurrent-free survival (RFS) and b cancer-specific survival (CSS) in Type F (HER2 protein and gene negative) breast cancer (BC) patients. Association of the status of HER2 intratumoral heterogeneity with c RFS and d CSS in triple negative breast cancer (TNBC) patients
Table 4.
Results of univariate and multivariate survival-analysis of clinicopathological variable influences including presence of HER2 heterogeneity in HER2 IHC 0, 1+ and HER2 DISH negative breast cancer
| Characteristic | Univariate analysis | Multivariable analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Recurrence-free survival | Cancer-specific survival | Recurrence-free survival | Cancer-specific survival | |||||||||
| HR | P | 95 % CI | HR | P | 95 % CI | HR | P | 95 % CI | HR | P | 95 % CI | |
| Age | 1.41 | .37 | 0.66–3.01 | 1.59 | .26 | 0.71–.3.60 | 1.43 | .38 | 0.64–3.21 | 1.91 | .14 | 0.81–4.51 |
| 40> versus 40≤ | ||||||||||||
| Tumor stage | 2.41 | .013 | 1.20–4.83 | 2.4 | .027 | 1.11–5.21 | 2.29 | .038 | 1.05–5.02 | 2.31 | .061 | 0.96–5.55 |
| T1-2 versus T3-4 | ||||||||||||
| Node stage | 2.75 | .00085 | 1.52–4.99 | 2.31 | .012 | 1.21–4.42 | 2.46 | .0081 | 1.26–4.77 | 1.95 | .074 | 0.94–4.05 |
| N0 versus N1-3 | ||||||||||||
| Histological grade | 1.18 | .56 | 0.68–2.06 | 1.47 | .22 | 0.79–2.75 | 0.84 | .59 | 0.44–1.61 | 0.85 | .68 | 0.41–1.79 |
| 1–2 versus 3 | ||||||||||||
| CEN17 polysomy | 2.67 | .099 | 0.83–8.61 | 3.26 | .05 | 1.00–10.62 | 2.31 | .24 | 0.57–9.39 | 3.48 | .077 | 0.87–13.89 |
| No versus yes | ||||||||||||
| ER IHC score | 1.26 | .48 | 0.67–2.36 | 1.73 | .11 | 0.89–3.36 | 2.56 | .21 | 0.58–11.19 | 3.60 | .096 | 0.80–16.34 |
| Positive versus negative | ||||||||||||
| PgR IHC score | 1.86 | .03 | 1.06–3.25 | 2.35 | .0071 | 1.26–4.36 | 3.37 | .0015 | 1.59–7.14 | 3.69 | .0023 | 1.59–8.54 |
| Positive versus negative | ||||||||||||
| TNBC | 1.2 | .59 | 0.62–2.35 | 1.65 | .16 | 0.82–3.30 | 0.20 | .062 | 0.036–1.09 | 0.20 | .072 | 0.04–1.15 |
| No versus yes | ||||||||||||
| HER2 heterogeneity | 2.36 | .0067 | 1.27–4.39 | 2.16 | .031 | 1.07–4.33 | 2.65 | .0076 | 1.30–5.40 | 2.24 | .041 | 1.03–4.85 |
| No versus yes | ||||||||||||
HR hazard ratio, 95% CI 95% confidence interval, HER2 human epidermal growth factor 2, IHC immunohistochemistry, DISH dual in situ hybridization, CEN17 chromosome 17 centromere, ER estrogen receptor, PgR progesterone receptor, TNBC triple negative breast cancer
Discussion
It is well established that 15–20 % of breast cancer patients are HER2 positive and it is associated with poor prognosis of breast cancer patients [24]. After the discovery of HER2 gene, the significance of HER2 gene role was demonstrated by the correlation of overall survival and time to relapse with the HER2 gene amplification in breast cancer [24]. Additionally, the association of the HER2 gene amplification with the HER2 protein overexpression was proved by immunohistochemical and Western blotting analyses [25]. In our current research using the HER2 GPA method for detecting both HER2 gene and protein targets, 21.4 % of breast cancer patients (n = 280) were classified as HER2 positive by HER2 gene status and HER2 DISH positive patients had significantly worse survival than HER2 gene negative patients. Also, 15.7 % of breast cancer patients were HER2 IHC positive and HER2 IHC positive patients had significantly worse survival than HER2 IHC negative patients. Consequently, our HER2 GPA study confirmed that HER2 positive breast cancer by either HER2 gene amplification or HER2 protein overexpression demonstrated a poor prognosis. Our cohort consisted with 11.8 % of HER2 IHC 2+ (equivocal) cases that require a reflex test if a traditional single HER2 IHC assay was conducted in a real world. Accurate and prompt HER2 status assessment of HER2 equivocal cases is the major challenge of HER2 testing for improving breast cancer patient care. The ASCO/CAP HER2 testing guideline states that HER2 equivocal breast cancer cases must be reexamined for the HER2 status with a different tissue block, if available, with the same HER2 testing method or by an alternative HER2 testing approach with the same tissue block [26]. Li et al. [27] reported that the HER2 GPA approach accurately divided breast cancer cases of a HER2 equivocal enriched cohort to HER2 positive, equivocal, and negative statuses without additional testing. Thus, HER2 GPA method can be utilized effectively for both HER2 positive and equivocal breast cancer cases.
Because HER2 IHC 0, 1+ (negative) cases would not be retested for the HER2 gene status with an ISH assay, if IHC assay was performed first, 1.8 % of patients who were IHC negative & DISH positive with the current cohort would not be eligible for a HER2-targeted therapy. Our statistical analyses showed the HER2 IHC negative & DISH positive group (Type E) had significantly worse survival than the HER2 IHC & DISH negative group (Type F). There are no solid data that if the patient group with HER2 protein negative and gene positive would respond to a HER2-targeted therapy. However, several studies suggested that a subgroup of HER2 negative patients benefit from HER2-targeted therapy [15, 16, 28]. In the near future, the results of the ongoing NSABP-B47 trial for adjuvant trastuzumab in low HER2 expression breast cancer will be reported and the efficacy of HER2-targeted therapy in low HER2 expression patients will be clarified [17].
The HER2 gene is localized on q21 of chromosome 17 [29]. Chromosome 17 polysomy sometimes exists in breast cancer cells [30] while the majority of CEN17 copy number gains are due to focal pericentromeric gains [18]. Copy number gain of CEN17 is thought to be a leading reason for discrepancy between HER2 IHC and HER2 ISH status [31, 32]. It has been suggested that the copy number of CEN17 is important for the accurate assessment of HER2 status [31, 33]. Even though the definition of CEN17 copy number gain has not been well-defined yet, in general, a mean of >3 CEN17 signals per nucleus is commonly adopted threshold [18]. The prevalence rates of CEN17 copy number gain in breast cancer were reported between 3 % and 46 % [18, 34–49]. In our study, approximately 3 % of HER2 IHC & HER2 DISH negative tumors showed CEN17 copy number gain and the presence of CEN17 copy number gain was significantly associated with the presence of HER2 intratumoral heterogeneity. It is suggested that the HER2 intratumoral heterogeneity is caused by the chromosomal instability [20] and our study also demonstrated that CEN17 copy number gain was significantly correlated with the HER2 heterogeneity. The importance of HER2 gene copy numbers is stated for determining for HER2 status in addition to the ratio of HER2 gene/CEN17 in 2013 ASCO/CAP HER2 [26]. However, the CEN17 ISH signal copy numbers may still be significant for subtyping breast cancer cases with the HER2 heterogeneity.
Even though the presence of HER2 genetic heterogeneity has been described [18, 50–52], there are no published data on simultaneous phenotypic and genotypic heterogeneity analyses in breast cancer. Hanna et al. [18] illustrated that HER2 genotypic heterogeneity is categorized into two types: 1) clustered HER2 genotypic heterogeneity and 2) intermixed HER2 genotypic heterogeneity. The clustered heterogeneity type is defined as a cell population of HER2 amplified tumor cells bordered by a HER2 nonamplified tumor cell population. The intermixed genotypic heterogeneity type is defined as comingled HER2 amplified and nonamplified tumor cells within a tumor. In an earlier study, Nitta et al. also reported the findings of two HER2 genotypic heterogeneity types using HER2 DISH assay [6]. The ASCO/CAP 2013 guidelines state that breast cancer containing between 5 % and 50 % of total cells with HER2/CEN17 ratio >2.0 or more than 6 HER2 signals per cell should be reported as the HER2 gene amplification heterogeneity [50]. Allison et al. [52] reported that HER2 heterogeneity defined by the ASCO/CAP guidelines was present in 23 % of breast cancer. Seol et al. [20] suggested that HER2 heterogeneity cases are mainly observed among low grade or equivocal HER2 status breast cancer cases. Furthermore, they reported that HER2 intratumoral heterogeneity is associated with short disease-free survival of the patients [52]. However, our study showed HER2 negative breast cancer cases also showed the HER2 heterogeneity (17 %), although we used a different approach for evaluating the heterogeneity to the ASCO/CAP guidelines. In the present study, initially we evaluated the presence of HER2 heterogeneity only in patients group determined as IHC 0, 1+/DISH negative using FDA criteria as to the routine examination. And then we examined the presence of heterogeneity revealing at least 2 of 6 patterns of HER2-GPA expression such as, (A) IHC 3+/DISH positive; (B) IHC 3+/DISH negative; (C) IHC 2+/DISH positive; (D) IHC 2+/DISH negative; (E) IHC 0, 1+/DISH positive; and (F) IHC 0, 1+/DISH negative, in focal regions less than 10 % area within one tumor. However according to the experts’ opinion, HER2 genetic heterogeneity is defined as “more than 5 % but less than 50 % of infiltrating tumor cells with HER2/CEN17 ratio higher than 2.2″ based on HER2 ISH assay results with dual probes [51]. Because HER2 GPA method allows investigating 6 different HER2 gene and protein combinations at individual cell levels, we anticipate that we will evolve the HER2 tumor heterogeneity definition to another dimension with simultaneous observations of HER2 protein and gene heterogeneity, such as the coexistence of HER2 DISH positive & IHC negative cells with HER2 DISH & IHC positive tumor cells (data not shown). Significance of such the HER2 heterogeneity at the gene and protein levels in breast cancer needs to be investigated for evaluating the efficacy of HER2 targeted therapy with further studies as the therapy target is the protein, not the gene.
Our HER2 GPA data analyses showed that the HER2 heterogeneous group had significantly worse survival than the HER2 nonheterogeneous group among the HER2 IHC & DISH negative breast cancer population. Particularly, TNBC patients with the HER2 intratumoral heterogeneity had significantly worse survival than TNBC patients without the HER2 heterogeneity. TNBC is characterized by ER and PgR IHC staining (<1 % of positive tumor cells) and HER2 IHC (0 and 1+ scores) or ISH (no gene amplification). TNBC is an aggressive metastatic breast cancer factor like targetable HER2 positive breast cancer, but there are no targeted therapies available for TNBC patients. A systematic review study reported that the 5 year relative survivals of TNBC patients and non-TNBC patients were 77 and 93 %, respectively, among 6370 breast cancer patients [53]. Unexpectedly, our current HER2 GPA study analyses showed that the HER2 heterogeneity is a significant factor for short patient survival of TNBC and the majority of HER2 heterogeneous TNBC patients died within 5 years. Therefore, we speculate that the HER2 heterogeneity has a significant role for tumor progression in TNBC. In our study, the HER2 heterogeneity of TNBC was defined by having individual tumor cells of non-HER2 IHC & DISH negative in HER2 IHC & DISH negative cell populations. Sweeney et al. reported that 20.3 % TNBC patients were subtyped as HER2-enriched type by PAM50 test for 50 breast cancer classifier gene expression analyses using RT-qPCR [54]. Intriguingly, our HER2 GPA data analyses showed that 17.9 % of TNBC cases exhibited the HER2 intratumoral heterogeneity. Because breast cancer cases that showed incomplete membrane staining within >10 % of tumor cells are considered as HER2 negative according to the ASCO/CAP HER2 testing guideline and some HER2 negative breast cancer cases have HER2 IHC staining, it is not a surprising observation to detect the expression of HER2 mRNA with TNBC. Thus, it might be possible that amplified HER2 gene tumor cells without obvious HER2 protein expression start overexpressing the HER2 protein later in the cancer progression of TNBC. A combination of HER2 mRNA and DNA ISH assays on the same tissue sections allows further analyses of HER2 gene expression at individual cell levels in TNBC. To our knowledge we are the first team to report that the HER2 intratumoral heterogeneity might be a poor prognosis factor in TNBC patients. If this is a true breast cancer biological phenomenon, we might be able to develop a new therapy strategy for TNBC patients comparing between HER2 heterogeneous and nonheterogeneous patient groups. It might be a reasonable approach to treat HER2 heterogeneous TNBC patients with HER2-targeted therapy. Further studies are commanded to confirm our observations with TNBC patients and investigate opportunity to develop a new therapy approach for TNBC with the heterogeneity.
Conclusions
We demonstrated that the HER GPA approach, which allows the simultaneous evaluation of HER 2 gene and protein at individual cell levels, is useful for more accurate HER2 status assessment, particularly with HER2 IHC negative & DISH positive cases (HER2 IHC & DISH discordant cases) and the HER2 intratumoral heterogeneity evaluation. Our study suggested that the HER2 intratumoral heterogeneity detected with the GPA is a poor prognostic factor among HER2 negative breast cancer cases, especially with TNBC. Our study also advocates that an improvement of HER2 testing methods are required for comprehensive analyses of the HER2 gene and protein discrepancy and the HER2 intratumoral heterogeneity comparing to clinical outcome data for the refinement of breast cancer patient selections for targeted therapies. However, since the examination criteria for the HER2 discrepancy and intratumoral heterogeneity have not been standardized yet, further studies are required for the establishment of HER2 GPA scoring algorithm.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was presented in part at the 37th San Antonio Breast Cancer Symposium, San Antonio, TX, December, 9–13, 2014. We would like to thank Peter Banks, MD (Medical Affairs, Ventana Medical Systems, Inc.) and Bryan Jones, JD (Legal Affairs, Ventana Medical Systems, Inc.) for their critical reviews of the manuscript.
Glossary
Abbreviations
- ADCC
Antibody-dependent cell-medicated cytotoxicity
- ASCO/CAP
The American Society of Clinical Oncology and College of American Pathologists
- CISH
Chromogenic in situ hybridization
- CEN17
Chromosome 17 centromere
- CSS
Cancer-specific survival
- DAB
3,3′-Diamnobenzidine
- DIG
Digoxigenin
- DISH
Dual in situ hybridization
- DNP
2,4′-Dinitrophenol
- EGFR
Epidermal growth factor
- ER
Estrogen receptor
- FDA
U.S. Food and Drug Administration
- FFPE
Formalin-fixed, paraffin-embedded
- FISH
Fluorescent in situ hybridization
- GPA
Gene-protein assay
- H&E
Hematoxylin and eosin
- HER2
Human epidermal growth factor receptor 2
- HR
Hazard ratio
- IHC
Immunohistochemistry
- NSABP
National Surgical Adjuvant Breast and Bowel Project
- PAM50
Prosigna Breast Cancer Prognostic Gene Signature Assay
- PR
Progesterone receptor
- RFS
Recurrence-free survival
- ISH
In situ hybridization
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- SISH
Silver in situ hybridization
- T-DM1
Ado-trastuzumab emtansine
- TNBC
Triple negative breast cancer
Compliance with ethical standards
Conflict of interests
MP, JRM, DCA, ED, and HN are employees of Ventana Medical Systems, Inc. HM, KI, JH, IT, MK received research funding and/or remuneration from Chugai Pharmaceutical Co, Ltd. There were no competing interests for all other authors.
Contributor Information
Sasagu Kurozumi, Email: chachagot-a-mail@hotmail.co.jp.
Mary Padilla, Email: mary.padilla@roche.com.
Masafumi Kurosumi, Email: mkurosumi@cancer-c.pref.saitama.jp.
Hiroshi Matsumoto, Email: hiromatsu@cancer-c.pref.saitama.jp.
Kenichi Inoue, Email: ino@cancer-c.pref.saitama.jp.
Jun Horiguchi, Email: junhorig@gunma-u.ac.jp.
Izumi Takeyoshi, Email: takeyoshi@gunma-u.ac.jp.
Tetsunari Oyama, Email: tetsunari.oyama@gmail.com.
Jim Ranger-Moore, Email: jim.ranger-moore@roche.com.
D. Craig Allred, Email: craig.allred@contractors.roche.com.
Eslie Dennis, Email: eslie.dennis@roche.com.
Hiroaki Nitta, Email: hiro.nitta@roche.com.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Bertos NR, Park M. Breast cancer—one term, many entities? J Clin Invest. 2011;121:3789–3796. doi: 10.1172/JCI57100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 4.Graus-Porta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francis GD, Jones MA, Beadle GF, Stein SR. Bright-field in situ hybridization for HER2 gene amplification in breast cancer using tissue microarrays: correlation between chromogenic (CISH) and automated silver-enhanced (SISH) methods with patient outcome. Diagn Mol Pathol. 2009;18:88–95. doi: 10.1097/PDM.0b013e31816f6374. [DOI] [PubMed] [Google Scholar]
- 6.Nitta H, Hauss-Wegrzyniak B, Lehrkamp M, Murillo AE, Gaire F, Farrell M, et al. Development of automated brightfield double in situ hybridization (BDISH) application for HER2 gene and chromosome 17 centromere (CEN 17) for breast carcinomas and an assay performance comparison to manual dual color HER2 fluorescence in situ hybridization (FISH) Diagn Pathol. 2008;3:41. doi: 10.1186/1746-1596-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nitta H, Kelly BD, Padilla M, Wick N, Brunhoeber P, Bai I, et al. A gene-protein assay for human epidermal growth factor receptor 2 (HER2): brightfield tricolor visualization of HER2 protein, the HER2 gene, and chromosome 17 centromere (CEN17) in formalin-fixed, paraffin-embedded breast cancer tissue sections. Diagn Pathol. 2012;7:60. doi: 10.1186/1746-1596-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaczek A, Brandt B, Bielawski KP. The diverse signaling network of EGFR, HER2, HER3 and HER4 tyrosine kinase receptors and the consequences for therapeutic approaches. Histol Histopathol. 2005;20:1005–1015. doi: 10.14670/HH-20.1005. [DOI] [PubMed] [Google Scholar]
- 9.Lane HA, Beuvink I, Motoyama AB, Daly JM, Neve RM, Hynes NE. ErbB2 potentiates breast tumor proliferation through modulation of p27(Kip1)–Cdk2 complex formation: receptor overexpression does not determine growth dependency. Mol Cell Biol. 2000;20:3210–3223. doi: 10.1128/MCB.20.9.3210-3223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurvitz SA, Betting DJ, Stern HM, Quinaux E, Stinson J, Seshagiri S, et al. Analysis of Fcγ receptor IIIa and IIa polymorphisms: lack of correlation with outcome in trastuzumab-treated breast cancer patients. Clin Cancer Res. 2012;18:3478–3486. doi: 10.1158/1078-0432.CCR-11-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sliwkowski MX, Mellman I. Antibody therapeutics in cancer. Science. 2013;341:1192–1198. doi: 10.1126/science.1241145. [DOI] [PubMed] [Google Scholar]
- 12.Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 13.Baselga J, Cortés J, Kim SB, Im SA, Hegg R, Im YH, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366:109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verma S, Miles D, Gianni L, Krop IE, Welslau M, Baselga J, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367:1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paik S, Kim C, Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358:1409–1411. doi: 10.1056/NEJMc0801440. [DOI] [PubMed] [Google Scholar]
- 16.Pogue-Geile KL, Kim C, Jeong JH, Tanaka N, Bandos H, Gavin PG, et al. Predicting degree of benefit from adjuvant trastuzumab in NSABP trial B-31. J Natl Cancer Inst. 2013;105:1782–1788. doi: 10.1093/jnci/djt321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehrenbacher L, Jeong J-H, Rastogi P, Geyer CE, Paik S, Costantino JP et al (2011) NSABP B-47: a randomized phase III trial of adjuvant therapy comparing chemotherapy alone (six cycles of docetaxel plus cyclophosphamide or four cycles of doxorubicin plus cyclophosphamide followed by weekly paclitaxel) to chemotherapy plus trastuzumab in women with node-positive or high-risk node-negative HER2-low invasive breast cancer. Cancer Res 71:OT1-02-07
- 18.Hanna WM, Rüschoff J, Bilous M, Coudry RA, Dowsett M, Osamura RY, et al. HER2 in situ hybridization in breast cancer: clinical implications of polysomy 17 and genetic heterogeneity. Mod Pathol. 2014;27:4–18. doi: 10.1038/modpathol.2013.103. [DOI] [PubMed] [Google Scholar]
- 19.Cottu PH, Asselah J, Lae M, Pierga JY, Diéras V, Mignot L, Sigal-Zafrani B, Vincent-Salomon A. Intratumoral heterogeneity of HER2/neu expression and its consequences for the management of advanced breast cancer. Ann Oncol. 2008;19:595–597. doi: 10.1093/annonc/mdn021. [DOI] [PubMed] [Google Scholar]
- 20.Seol H, Lee HJ, Choi Y, Lee HE, Kim YJ, Kim JH, Kang E, Kim SW, Park SY. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Mod Pathol. 2012;25:938–948. doi: 10.1038/modpathol.2012.36. [DOI] [PubMed] [Google Scholar]
- 21.Lee HJ, Seo AN, Kim EJ, Jang MH, Suh KJ, Ryu HS, et al. HER2 heterogeneity affects trastuzumab responses and survival in patients with HER2-positive metastatic breast cancer. Am J Clin Pathol. 2014;142:755–766. doi: 10.1309/AJCPIRL4GUVGK3YX. [DOI] [PubMed] [Google Scholar]
- 22.Ohlschlegel C, Zahel K, Kradolfer D, Hell M, Jochum W. HER2 genetic heterogeneity in breast carcinoma. J Clin Pathol. 2011;64:1112–1116. doi: 10.1136/jclinpath-2011-200265. [DOI] [PubMed] [Google Scholar]
- 23.Shafi H, Astvatsaturyan K, Chung F, Mirocha J, Schmidt M, Bose S. Clinicopathological significance of HER2/neu genetic heterogeneity in HER2/neu non-amplified invasive breast carcinomas and its concurrent axillary metastasis. J Clin Pathol. 2013;66:649–654. doi: 10.1136/jclinpath-2012-201403. [DOI] [PubMed] [Google Scholar]
- 24.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 25.Venter DJ, Tuzi NL, Kumar S, Gullick WJ. Overexpression of the c-erbB-2 oncoprotein in human breast carcinomas: immunohistological assessment correlates with gene amplification. Lancet. 1987;2:69–72. doi: 10.1016/S0140-6736(87)92736-X. [DOI] [PubMed] [Google Scholar]
- 26.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH et al (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer. American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31:3997–4013 [DOI] [PubMed]
- 27.Li Z, Dabbs DJ, Cooper KL, Bhargava R. Dual HER2 gene protein assay: focused study of breast cancers with 2+ immunohistochemical expression. Am J Clin Pathol. 2015;143:451–458. doi: 10.1309/AJCPKIVVW4OBPX6I. [DOI] [PubMed] [Google Scholar]
- 28.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 29.Negri T, Tarantino E, Orsenigo M, Reid JF, Gariboldi M, Zambetti M, Pierotti MA, Pilotti S. Chromosome band 17q21 in breast cancer: significant association between beclin 1 loss and HER2/NEU amplification. Genes Chromosomes Cancer. 2010;49:901–909. doi: 10.1002/gcc.20798. [DOI] [PubMed] [Google Scholar]
- 30.Reinholz MM, Bruzek AK, Visscher DW, Lingle WL, Schroeder MJ, Perez EA, et al. Breast cancer and aneusomy 17: implications for carcinogenesis and therapeutic response. Lancet Oncol. 2009;10:267–277. doi: 10.1016/S1470-2045(09)70063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dal Lago L, Durbecq V, Desmedt C, Salgado R, Verjat T, Lespagnard L et al (2006) Correction for chromosome-17 is critical for the determination of true Her-2/neu gene amplification status in breast cancer. Mol Cancer Ther 5:2572–2579 [DOI] [PubMed]
- 32.Vranic S, Teruya B, Repertinger S, Ulmer P, Hagenkord J, Gatalica Z. Assessment of HER2 gene status in breast carcinomas with polysomy of chromosome 17. Cancer. 2011;117:48–53. doi: 10.1002/cncr.25580. [DOI] [PubMed] [Google Scholar]
- 33.Sauter G, Lee J, Bartlett JM, Slamon DJ, Press MF. Guidelines for human epidermal growth factor receptor 2 testing: biologic and methodologic considerations. J Clin Oncol. 2009;27:1323–1333. doi: 10.1200/JCO.2007.14.8197. [DOI] [PubMed] [Google Scholar]
- 34.Bose S, Mohammed M, Shintaku P, Rao PN. Her-2/neu gene amplification in low to moderately expressing breast cancers: possible role of chromosome 17/Her-2/neu polysomy. Breast J. 2001;5:337–344. doi: 10.1046/j.1524-4741.2001.21018.x. [DOI] [PubMed] [Google Scholar]
- 35.Cuadros M, Cano C, López FJ, Talavera P, García-Peréz I, Blanco A, et al. HER2 status in breast cancer: experience of a Spanish National Reference Centre. Clin Transl Oncol. 2011;13:335–340. doi: 10.1007/s12094-011-0663-4. [DOI] [PubMed] [Google Scholar]
- 36.Downs-Kelly E, Yoder BJ, Stoler M, Tubbs RR, Skacel M, Grogan T, Roche P, Hicks DG. The influence of polysomy 17 on HER2 gene and protein expression in adenocarcinoma of the breast: a fluorescent in situ hybridization, immunohistochemical, and isotopic mRNA in situ hybridization study. Am J Surg Pathol. 2005;29:1221–1227. doi: 10.1097/01.pas.0000165528.78945.95. [DOI] [PubMed] [Google Scholar]
- 37.Kokate P, Sawaimoon S, Bhatia S, Mandava S. Evaluation of genetic status of HER-2/neu and aneusomy 17 by fluorescence in situ hybridization and comparison with immunohistochemistry assay from Indian breast cancer patients. Genet Test Mol Biomarkers. 2012;16:239–245. doi: 10.1089/gtmb.2011.0125. [DOI] [PubMed] [Google Scholar]
- 38.Hofmann M, Stoss O, Gaiser T, Kneitz H, Heinmöller P, Gutjahr T, et al. Central HER2 IHC and FISH analysis in a trastuzumab (Herceptin) phase II monotherapy study: assessment of test sensitivity and impact of chromosome 17 polysomy. J Clin Pathol. 2008;61:89–94. doi: 10.1136/jcp.2006.043562. [DOI] [PubMed] [Google Scholar]
- 39.Krishnamurti U, Hammers JL, Atem FD, Storto PD, Silverman JF. Poor prognostic significance of unamplified chromosome 17 polysomy in invasive breast carcinoma. Mod Pathol. 2009;22:1044–1048. doi: 10.1038/modpathol.2009.61. [DOI] [PubMed] [Google Scholar]
- 40.Merola R, Mottolese M, Orlandi G, Vico E, Cognetti F, Sperduti I, Fabi A, Vitelli G, Cianciulli AM. Analysis of aneusomy level and HER-2 gene copy number and their effect on amplification rate in breast cancer specimens read as 2+ in immunohistochemical analysis. Eur J Cancer. 2006;42:1501–1506. doi: 10.1016/j.ejca.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Salido M, Tusquets I, Corominas JM, Suarez M, Espinet B, Corzo C, et al. Polysomy of chromosome 17 in breast cancer tumors showing an overexpression of ERBB2: a study of 175 cases using fluorescence in situ hybridization and immunohistochemistry. Breast Cancer Res. 2005;7:R267–R273. doi: 10.1186/bcr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takehisa M, Sasa M, Bando Y, Hirose T, Morimoto T, Nagao T, et al. Chromosomal aneusomy (chr 1, 11, 17) detected by fluorescence in situ hybridization may be a prognostic factor in breast cancer. Anticancer Res. 2007;27:1073–1078. [PubMed] [Google Scholar]
- 43.Torrisi R, Rotmensz N, Bagnardi V, Viale G, Curto BD, Dell’orto P, et al. HER2 status in early breast cancer: relevance of cell staining patterns, gene amplification and polysomy 17. Eur J Cancer. 2007;43:2339–2344. doi: 10.1016/j.ejca.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 44.Tsukamoto F, Miyoshi Y, Egawa C, Kasugai T, Takami S, Inazawa J, et al. Clinicopathologic analysis of breast carcinoma with chromosomal aneusomy detected by fluorescence in situ hybridization. Cancer. 2001;93:165–170. doi: 10.1002/cncr.9024. [DOI] [PubMed] [Google Scholar]
- 45.Tubbs RR, Hicks DG, Cook J, Downs-Kelly E, Pettay J, Hartke MB, et al. Fluorescence in situ hybridization (FISH) as primary methodology for the assessment of HER2 Status in adenocarcinoma of the breast: a single institution experience. Diagn Mol Pathol. 2007;16:207–210. doi: 10.1097/PDM.0b013e318064c72a. [DOI] [PubMed] [Google Scholar]
- 46.Vanden Bempt I, Van Loo P, Drijkoningen M, Neven P, Smeets A, Christiaens MR et al (2008) Polysomy 17 in breast cancer: clinicopathologic significance and impact on HER-2 testing. J Clin Oncol 26:4869–4874 [DOI] [PubMed]
- 47.Varshney D, Zhou YY, Geller SA, Alsabeh R. Determination of HER-2 status and chromosome 17 polysomy in breast carcinomas comparing HercepTest and PathVysion FISH assay. Am J Clin Pathol. 2004;121:70–77. doi: 10.1309/FUQH92B039025LHG. [DOI] [PubMed] [Google Scholar]
- 48.Zaczek A, Markiewicz A, Supernat A, Bednarz-Knoll N, Brandt B, Seroczyńska B, et al. Prognostic value of TOP2A gene amplification and chromosome 17 polysomy in early breast cancer. Pathol Oncol Res. 2012;18:885–894. doi: 10.1007/s12253-012-9518-8. [DOI] [PubMed] [Google Scholar]
- 49.Zhu X, Lu Y, Lu H, Yang W, Tu X, Cai X, et al. Genetic alterations and protein expression of HER2 and chromosome 17 polysomy in breast cancer. Hum Pathol. 2011;42:1499–1504. doi: 10.1016/j.humpath.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 50.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4014. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 51.Vance GH, Barry TS, Bloom KJ, Fitzgibbons PL, Hicks DG, Jenkins RB, et al. Genetic heterogeneity in HER2 testing in breast cancer: panel summary and guidelines. Arch Pathol Lab Med. 2009;133:611–612. doi: 10.5858/133.4.611. [DOI] [PubMed] [Google Scholar]
- 52.Allison KH, Dintzis SM, Schmidt RA. Frequency of HER2 heterogeneity by fluorescence in situ hybridization according to CAP expert panel recommendations: time for a new look at how to report heterogeneity. Am J Clin Pathol. 2011;136:864–871. doi: 10.1309/AJCPXTZSKBRIP07W. [DOI] [PubMed] [Google Scholar]
- 53.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 54.Sweeney C, Bernard PS, Factor RE, Kwan ML, Habel LA, Quesenberry CP, Jr, et al. Intrinsic subtypes from PAM50 gene expression assay in a population-based breast cancer cohort: differences by age, race, and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2014;23:714–724. doi: 10.1158/1055-9965.EPI-13-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.