Abstract
The Breast cancer 1, early onset gene (BRCA1) is known to be significantly associated with human familial breast cancer and is identified to play an important role in canine mammary tumors. Here, genetic variations in the coding region and DNA methylation in the 5′ flanking region of BRCA1 in canine mammary tumor samples, 15 each of benign and malignant against 10 normal canine mammary tissue samples, were analyzed using the direct sequencing method. The results indicated two point mutations each in the coding region of canine BRCA1 in one benign mammary tumor sample (4702G >T and 4765G >T) and in one malignant canine mammary tumor sample (3619A >G and 4006G >A). No mutations were detected in the normal canine mammary tissue samples. The 4702G >T mutation was found to terminate further translation. The physical effect of the 4765G >T mutation was found to be the repalacement of the glutamate residue with glutamine. The physical effect of the 3619A >G mutation was found to be the replacement of the threonine residue with alanine, and that of mutation 4006G >A was the replacement of the valine residue with isoleucine in the BRCA1 protein. Bisulfite sequencing detected methylated CpG sites in one canine malignant mammary tumor sample. In conclusion, the present study elucidated the mutational status of the BRCA1 coding region and methylation status of the 5′ flanking region of BRCA1 in canine mammary tumors.
Keywords: Breast cancer 1, canine mammary tumors, DNA methylation, early onset (BRCA1), point mutation
Breast cancer 1, early onset (BRCA1) is one of the most important genes in human familial breast and ovarian cancers. Human BRCA1 was first identified in 1994 by positional cloning methods [17]. The role of the BRCA1 protein is implicated in vital cellular functions. As a tumor suppressor, BRCA1 in association with RAD51 takes part in the repair of DNA double-strand breaks (DSBs) [19, 23]. BRCA1 is involved in maintaining heterochromatin integrity by regulating the ubiquitylation of histone H2A [35]. BRCA1 is known to induce apoptosis in human breast cancer cell lines [24]. Induction of BRCA1 triggers apoptosis through activation of c-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) [9]. The human BRCA1 C-terminal (BRCT) region (amino acids 1560-1863) is recognized as a transcriptional activator in mammalian cells [18]. The integrity of the BRCT domains of BRCA1 is crucial for its role in activating transcription [10]. The importance of BRCA1 in the regulation of translation is also recognized. Inhibition of endogenous BRCA1 by using a small interfering RNA-based approach decreased protein synthesis and overexpression of BRCA1 activated translation, both of which suggested that BRCA1 modulated protein synthesis [4]. BRCA1 regulates the key effectors that control the G2/M checkpoint and is therefore believed to be involved in regulating the onset of mitosis [30, 31].
The well-known function of BRCA1 is its role in mammary tumorigenesis. By age 70, BRCA1-mutation carrying individuals demonstrated 87% cumulative risk for the occurrence of breast cancer [8]. In a study, the prevalence rate of BRCA1 mutation in the patients under 55 was 0.7% [2]. Besides germ line mutation, decreased expression of BRCA1 is an important cause of tumorigenesis in human sporadic cases [28]. DNA methylation of this gene is one of the principle mechanisms for silencing BRCA1 [6].
In 1996, the coding sequence of canine BRCA1 mRNA was determined [26]. Similar to humans, genetic variations in BRCA1 were significantly associated with canine mammary tumors [21]. Extensive studies have been reported to identify the function and mutational status of BRCA1 in human breast cancer. However, only limited data are available on the mutational status in the coding region and the DNA methylation of BRCA1 in canine mammary tumors. The objectives of the present study are to determine the mutational status of the canine BRCA1 coding region and to detect the methylation status of the 5′ flanking region of canine BRCA1 in canine mammary tumors.
MATERIALS AND METHODS
Sample collection
All the canine tissue samples obtained from the Animal Hospital of China Agriculture University were divided into three groups: normal canine mammary tissue group (n=10), benign canine mammary tumor group (n=15) and malignant canine mammary tumor group (n=15). All samples were verified and histopathologically diagnosed by pathologists of the Animal Hospital of China Agriculture University. The study was approved by the Animal Welfare Committee of the Department of Clinical Veterinary Medicine of China Agricultural University.
DNA extraction
Genomic DNA was extracted from mammary tissue samples using Tiangen DNA extracting Kit (Tiangen Co., Beijing, China). All DNA samples were quantified using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Beijing, China). DNA samples with OD 260/280 within 1.8–2.0 were used for the study.
Sequencing the coding region of BRCA1
In order to amplify the entire coding sequence of canine BRCA1, the primers flanking each of the exons were designed (GenBank Accession No. NC_006591 and NM_001013416). The primers used for amplification and sequencing are listed in Table 1. All 22 coding exons were separated into 19 fragments and then amplified. All of the forward and reverse primers used in PCR were used in sequencing. In addition, the sequencing primers 1 and 2 were added to sequence fragments 8 and 9, respectively. For PCR, the initial denaturation was at 95°C for 4 min, followed by 35 cycles at 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec to 2 min 30 sec, were followed by a final elongation step at 72°C for 10 min. For each PCR reaction, 100 ng genomic DNA was used as template. The total volume of each reaction mixture was 25 µl. The extension time of each primer pair is listed in Table 1. The amplified DNA fragments were then separated by agarose gel electrophoresis and purified using the Agarose Gel DNA Extraction Kit (Tiangen Co.) as per the manufacturer’s protocol. The purified PCR products were sent to Beijing Genome Institution (Beijing, China) for sequencing, and the Sanger sequencing methods were used. All the experiments were performed in triplicates.
Table 1. Primer sequences used for PCR and sequencing.
| Primer number | Primers sequence | Sizes | Coding exons | Extension time |
|---|---|---|---|---|
| 1 | F:AAGTGATGTGTGTGTAGGCAT R:TATCCTGGATTTCCAAAGCTC |
331 bp | 1 | 30 sec |
| 2 | F: GTTTGTTGTCTTCATGGAGTGT R: GAGGCAGAATTACAGAGTTAG |
564 bp | 2 | 40 sec |
| 3 | F: GCATGTAAGCTCAGTTTTCATAG R: CTAATACCTTCATAAGACAGACT |
501 bp | 3 | 40 sec |
| 4 | F: GAGCCTACAAGAAAGTACAAGA R: CAAGGTAGGGTTTTCAGGTTCA |
1,022 bp | 4, 5 | 1 min 10 sec |
| 5 | F: CCTGGAGTTTCTCTTCAAG R: CCAATTCAATGTAGACAGAC |
182 bp | 6 | 30 sec |
| 6 | F: GCTCTGATTCAAACTTAGGA R: ATACTACTGTGCTTGAGGAAC |
520 bp | 7 | 40 sec |
| 7 | F: TCCTGTTGCCCTTCCATAAAG R: TCAGCCTTGTGTCATTGAAAC |
511 bp | 8 | 40 sec |
| 8 | F: GGGAGTATGAGTCTGAGGATAGAAC R: ATATGATAAGGGTTTTGGGAAATTC |
2,598 bp | Part of exon 9 | 2 min 30 sec |
| 9 | F: AAGCTAAGAAGCCAGGCGATTTTGC R: CTCTGTAGGCTCCAATTTGGTCCCA |
2,176 bp | Part of exon 9 and exon 10 | 2 min 30 sec |
| 10 | F: CAGAGAGATACCATGCAAGATAAC R: CTCTTTCTGATGCGTTTTGTTCCG |
172 bp | 11 | 30 sec |
| 11 | F: AGCCTCGTCTCAGCCAAGTAT R: TCTCATCTCATAGCTCATGCC |
588 bp | 12 | 40 sec |
| 12 | F: TGAAAAATATCCCACCTCAGC R: AGCACACAAGACTCTCCCAT |
654 bp | 13 | 50 sec |
| 13 | F: GGTCATAAAACAGTTGGAGAAC R: CTTTAGCTTTATCTAGGTCCCA |
697 bp | 14 | 50 sec |
| 14 | F: GTGAGCACAGACTATTTTAG R: CACTACTCTCCAAATCTTAACC |
479 bp | 15 | 30 sec |
| 15 | F: ATGCTGAGTTTGTGTGCGAACGA R: GAGTACCTACCTCATCTAGTATC |
660 bp | 16, 17 | 50 sec |
| 16 | F: AGTCTCAATGAGGTGAAAAG R: TTGGGCTTGGTCTCTCAAAT |
549 bp | 18 | 40 sec |
| 17 | F: CGATGTTCTGATATTAGACTC R: GCACAGGGCTGTTTTTTTGAT |
423 bp | 19 | 30 sec |
| 18 | F: TAGAGGGTCCAGGTCAAGTG R: ACAGGTACACCATCTACTCC |
683 bp | 20, 21 | 50 sec |
| 19 | F: CGTGCTCCTGGTGACTTTTC R: AGTAACTGGGACTGTGGAAG |
502 bp | 22 | 40 sec |
| Sequencing primers | Sequencing primer 1: CTGAAATCAGACATGGAGAG | |||
| Sequencing primer 2: AAGGCATCAACAGTTAGCTC | ||||
| Bisulfite primers | F: TTTAGGGAAAGAATTGATGATTAAT R: TACCCTCACTCAAAAAAACCTTAAC |
217 bp | 30 sec |
Predicting the effects of mutations and the altered amino acids
In order to predict the effects of mutations and the corresponding substituted amino acids, the server PolyPhen was used [1, 3] which was available at http://genetics.bwh.harvard.edu/pph2/index.shtml [1]. The predicted effect of a substitution was reported as unknown, benign, possibly damaging and probably damaging.
Exploring the CpG islands in the 5′ flanking region of BRCA1 and design of methylation primers
The 1,907 bp DNA sequence in the 5′ flanking region present upstream of the BRCA1 coding region was used to search for CpG islands. The sequence was from 19959003 to 19960909 on canine chromosome 9. CpG islands of the region were analyzed using the website “CpG Island Searcher” [27] available at http://cpgislands.usc.edu/. For “CpG Island Searcher”, the following criteria were used: GC% >55%; ObsCpG/ExpCpG >0.65; length >500 bp; and gap between adjacent islands >100 bp. Through the “CpG Island Searcher”, one CpG island was found (Fig. 1A and 1B). This CpG island started from 467 bp, ended at 1091 bp of the input sequence and was used for designing primers.
Fig. 1.
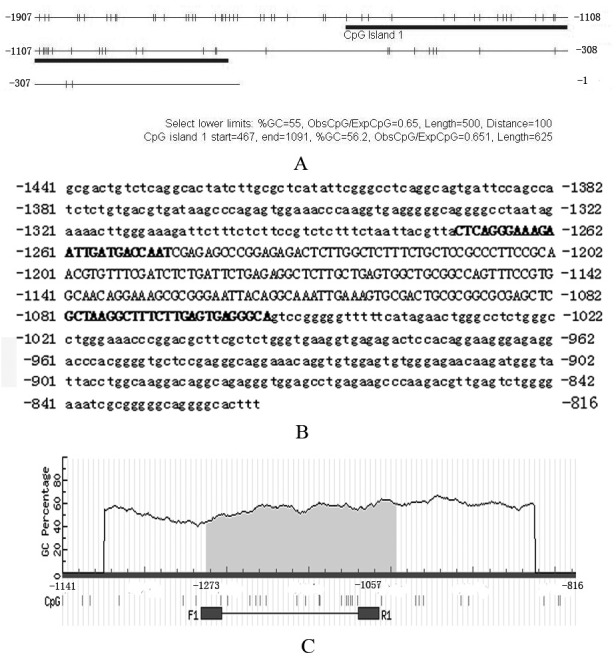
CpG islands in the 5′ flanking region of canine BRCA1 and bisulfite primers used for methylation analysis. Panel A: results of “CpG Island Searcher”. The bold line shows the CpG island in the 5′ flanking region of canine BRCA1. Numbers in panel A indicate the position of corresponding nucleotides relative to translation start (ATG). Panel B: CpG island sequence and bisulfite primer position. The complete sequence in panel B shows the CpG island sequence; position of every nucleotide relative to translation start (ATG) is shown. Capital letters indicate the region amplified by bisulfite PCR. Bold capital letters indicate the position of bisulfite primers. Panel C: partial results of “MethPrimer”. The primer pair “F1” and “R1” was used in the present study. Numbers in panel C indicate the position of corresponding nucleotides relative to translation start (ATG).
For designing bisulfite primers, the website “MethPrimer” [15] available at http://www.urogene.org/methprimer/ was used. The search results are illustrated in Fig. 1B and 1C Bisulfite primers are listed in Table 1.
Methylation analysis of the CpG island
Methylation status of the CpG island was analyzed using a modified DNA bisulfite protocol [7, 34]. DNA samples were denatured in 0.2 M NaOH at 37°C for 10 min. Sodium bisulfite was added to a final concentration of 3.0 M and hydroquinone to a final concentration of 0.5 mM, followed by incubation at 55°C for 16 hr. DNA samples were then desalted by the DNA Clean-Up System (Promega, Madison, WI, U.S.A.) followed by desulfonation in 0.3 M NaOH and ethanol-precipitation.
For PCR, the initial denaturation was at 95°C for 4 min, followed by 40 cycles at 94°C for 30 sec, 53°C for 30 sec, 72°C for 30 sec, was followed by a final elongation at 72°C for 10 min. After PCR, the amplified DNA fragments were separated by agarose gel electrophoresis and purified using gel recovery kit. The purified PCR products were then ligated with pMD-18T vectors (Takara Co., Dalian, China). After amplification in Escherichia coli, the plasmids were isolated and sequenced by Beijing Genome Institution. Three clones from each sample were selected for sequencing.
RESULTS
Point mutations detected in canine BRCA1 tumor samples
All of the 19 fragments of the BRCA1 coding region were amplified (Fig. 2). Sequencing of these PCR products yielded the sequence of each fragment of each sample through assemble program of Vector NTI Suite 7. In alignment with the reference sequence (GenBank Accession No. NC_006591 and NM_001013416), varied mutation sites were found in one benign and in one malignant canine mammary tumor sample. In the benign mammary tumor sample, two point mutations were found (4702G >T and 4765G >T). The 4702G >T mutation resulted in the termination of translation that truncated the BRCA1 protein. The physical effect of the 4765G >T mutation was found to be the repalacement of the glutamate residue with glutamine. In the malignant mammary tumor sample, two point mutations were found (3619A >G and 4006G >A). The 3619A >G mutation inserted an alanine residue (T1207A) instead of a threonine, and the 4006G >A mutation inserted an isoleucine residue (V1336I) instead of valine in the BRCA1 polypeptide.
Fig. 2.
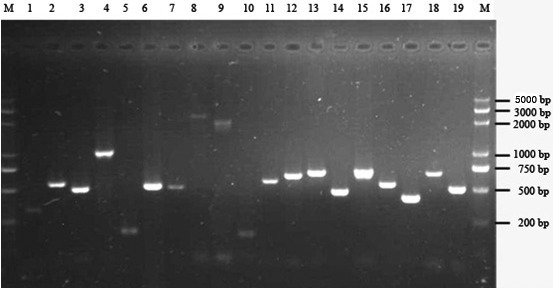
PCR results of different BRCA1 exons. M: DNA molecular weight marker. Lanes 1–19 indicate fragments amplified by primer pairs 1–19. Sizes of amplified fragments were as expected.
Effects of amino acids substitutions on the function of the BRCA1 protein
The effects of the three nonsynonymous point mutations on the function of the BRCA1 protein were predicted by PolyPhen. The results are listed in Table 2. The prediction results showed that the 4765 G >C that leads to the E1589Q substitution was probably damaging. The 4006 G >A mutation that leads to V1336I substitution was probably damaging. The 3619 A >G that leads to T1207A substitution was benign.
Table 2. Prediction of functional effects of the three nonsynonymous point mutations.
| Type of mammary tumors | Positions of mutation | Changes of amino acids | PolyPhen prediction |
|---|---|---|---|
| Benign | 4765 G >C | E1589 Q | Probably damaging |
| Malignant | 3619 A >G | T1207 A | Benign |
| Malignant | 4006 G >A | V1336 I | Probably damaging |
Methylated CpG sites detected in malignant canine mammary tumor
DNA methylation status of the 5′ flanking region of BRCA1 was analyzed by bisulfite sequencing. PCR successfully amplified the target fragments (Fig. 3A). The sequencing results of different clones are shown in Fig. 3B and 3C. No methylated CpG site was found in clones representing benign and normal canine mammary tissues (Fig. 3B), whereas, one clone representing a malignant mammary tumor sample was detected with four methylated CpG sites, representing a methylation rate of 1/15 (6.7%) (Fig. 3C).
Fig. 3.

Methylation status of the 5′ flanking region of BRCA1. Panel A: bisulfite PCR results; M: DNA molecular weight marker. Using bisulfite-modified DNA (bDNA) as the template, the target fragment was successfully amplified. Using unmodified DNA (nDNA) as the template, the target fragment was not amplified, which indicated the specificity of the bisulfite primers. Panel B: the methylation status of normal mammary tissues. Panel C: the methylation status of the malignant tumor sample with methylated CpG sites. In panels B and C, each row of circles represents the methylation status of CpG sites obtained from one plasmid clone. The position of each CpG site relative to translation start (ATG) is shown. Open circles indicate the unmethylated CpG sites. Filled circles indicate the methylated CpG sites. Three clones of each sample were sequenced. As shown in panel B, all CpG sites were unmethylated in normal mammary tissues. As shown in panel C, the first four CpG sites were detected with methylation in one malignant mammary tumor sample.
DISCUSSION
After the canine BRCA1 gene was cloned, only limited data exist indicating its relationship with canine breeds and canine mammary tumors. In the study conducted by Tsuchida et al., microsatellite polymorphism was demonstrated in intron 14 of the canine BRCA1 gene [29]. In several recent reports, single nucleotide polymorphisms (SNPs) of canine BRCA1 gene were found to correlate with canine mammary tumors. Borge et al. identified 64 SNPs in 11 genes including BRCA1 in canine mammary tumors [3]. In their report, they found one SNP in the coding region of BRCA1, which was a synonymous SNP [3] that generally does not alter the structure of the proteins. However, some synonymous SNPs alter RNA folding and may therefore have an impact on protein translation [14]. In the report by Borge et al., the dogs used for the study were from eight different breeds (4 dogs of each breed) without a history of mammary tumors. In the present study, dogs with mammary tumor were used in the experiment, and normal dogs were used as a control. In the normal dog mammary tissue, no nucleotide variations were detected, indicating that genetic variations might be associated with mammary tumorigenesis.
Enginler et al. identified 12 SNPs of BRCA1 and BRCA2 in canine mammary tumors [5]. In their report, two SNPs were identified in intron 8 of BRCA1, and two other SNPs were identified in the coding region of BRCA1 in canine mammary tumors [5]. These two SNPs in the coding region of BRCA1 were also synonymous [5]. In addition, their study indicates a variation in the 5′ UTR, intron 8 and exon 9 of BRCA1, but no data are available on the other coding exons. The present study detected point mutations leading to alteration of the amino acids in BRCA1 corresponding to altered protein function. In addition, all of the coding exons were sequenced to find the genetic variations. Similar to the present study, dogs with mammary tumor were used by Enginler et al. and Sun et al. to identify three SNPs in the 5′- and 3′-UTRs of BRCA1 in canine mammary tumors [25]. In the report by Sun et al., one non-synonymous SNP was found in the coding region of BRCA1 in canine mammary tumors, which leads to a substitution of Ala1851Thr [25]. The present study contributes four additional point mutations in the coding region of BRCA1 with a promising role in canine tumor biology. The above process utilized the direct sequencing technique with the limitation that only the dominant mutation sites could be detected.
The BRCA1 C-terminal (BRCT) is highly conserved among human, mouse and canine [26]. In human, there were two BRCT repeat domains in the C-terminal region [32]. The BRCT repeat domain (amino acids 1560-1863) is an important domain that functions to activate transcription [18]. The 4702G >T mutation detected in the present study, leads to the loss of the BRCT repeat domain. In humans, loss of BRCT function was an important mechanism that promoted tumorigenesis [26]. Considering the high homology of BRCT domains between humans and canine, the truncated BRCA1 might probably lose its function without the C-terminal [10].
The 3619A >G mutation altered the amino acid residue in BRCA1 from threonine to alanine residue (T1207A). Threonine is a hydrophilic amino acid, while alanine is a hydrophobic amino acid. The substitution might therefore affect the structure of the canine BRCA1 protein. The 4006G >A mutation substituted the valine residue with isoleucine residue (V1336I) in the BRCA1 polypeptide. Both valine and isoleucine are hydrophobic amino acid. The molecular weight of valine is similar to the molecular weight of isoleucine. However, there is another trans-activation domain (AD1) of BRCA1 (amino acids 1293-1558), which functions in concert with BRCT domain [11, 12]. The (V1336I) substitution might affect the trans-activation function of AD1 domain.
In order to further predict the effects of the three point mutations with amino acids substitutions, we used the online analysis software “PolyPhen”. The results showed that the 4006 G >A and 4765 G >C mutations were probably damaging. The 3619 A >G mutation was benign. The prediction results implied that two of the three point mutations might cause loss of function of the BRCA1 protein and thereby play a major role in canine mammary tumorigenesis.
In humans, DNA methylation in the promoter region is the most important mechanism responsible for the down regulation of BRCA1 in breast cancer. In human sporadic breast tumor, the methylation rate of BRCA1 promoter ranged from 14.3% to 29.8% [16, 22, 33]. Previous studies indicate that the expression of BRCA1 was altered in canine mammary tumors as compared with normal mammary tissues [13, 20]. The underling mechanism of such a change is not yet elucidated. In this study, one out of fifteen (about 6.7%) malignant canine mammary tumors was found with four methylated CpG sites in the 5′ flanking region of BRCA1 gene. In humans, two CpG sites with methylation were found to affect the expression of BRCA1 [22]. Though the methylation rate was lower in the present study, the methylation mechanism might work in canine BRCA1 regulation.
In conclusion, four novel point mutations were found in the coding sequence along with a methylated CpG island in the 5′ flanking region of BRCA1 in canine mammary tumors, which might alter the function of BRCA1 as a tumor suppressor. The data provide newer insights in understanding the mechanism of BRCA1 mediated regulation of canine mammary tumorigenesis.
REFERENCES
- 1.Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., Kondrashov A. S., Sunyaev S. R.2010. A method and server for predicting damaging missense mutations. Nat. Methods 7: 248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anglian Breast Cancer Study Group2000. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br. J. Cancer 83: 1301–1308. doi: 10.1054/bjoc.2000.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borge K. S., Børresen-Dale A. L., Lingaas F.2011. Identification of genetic variation in 11 candidate genes of canine mammary tumour. Vet. Comp. Oncol. 9: 241–250. doi: 10.1111/j.1476-5829.2010.00250.x [DOI] [PubMed] [Google Scholar]
- 4.Dizin E., Gressier C., Magnard C., Ray H., Décimo D., Ohlmann T., Dalla Venezia N.2006. BRCA1 interacts with poly(A)-binding protein: implication of BRCA1 in translation regulation. J. Biol. Chem. 281: 24236–24246. doi: 10.1074/jbc.M602176200 [DOI] [PubMed] [Google Scholar]
- 5.Enginler S. O., Akış I., Toydemir T. S., Oztabak K., Haktanir D., Gündüz M. C., Kırşan I., Fırat I.2014. Genetic variations of BRCA1 and BRCA2 genes in dogs with mammary tumours. Vet. Res. Commun. 38: 21–27. doi: 10.1007/s11259-013-9577-7 [DOI] [PubMed] [Google Scholar]
- 6.Esteller M., Silva J. M., Dominguez G., Bonilla F., Matias-Guiu X., Lerma E., Bussaglia E., Prat J., Harkes I. C., Repasky E. A., Gabrielson E., Schutte M., Baylin S. B., Herman J. G.2000. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl. Cancer Inst. 92: 564–569. doi: 10.1093/jnci/92.7.564 [DOI] [PubMed] [Google Scholar]
- 7.Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., Paul C. L.1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 89: 1827–1831. doi: 10.1073/pnas.89.5.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford D., Easton D. F., Bishop D. T., Narod S. A., Goldgar D. E., Breast Cancer Linkage Consortium1994. Risks of cancer in BRCA1-mutation carriers. Lancet 343: 692–695. doi: 10.1016/S0140-6736(94)91578-4 [DOI] [PubMed] [Google Scholar]
- 9.Harkin D. P., Bean J. M., Miklos D., Song Y. H., Truong V. B., Englert C., Christians F. C., Ellisen L. W., Maheswaran S., Oliner J. D., Haber D. A.1999. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell 97: 575–586. doi: 10.1016/S0092-8674(00)80769-2 [DOI] [PubMed] [Google Scholar]
- 10.Hayes F., Cayanan C., Barillà D., Monteiro A. N.2000. Functional assay for BRCA1: mutagenesis of the COOH-terminal region reveals critical residues for transcription activation. Cancer Res. 60: 2411–2418. [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Y. F., Li R.2002. JunB potentiates function of BRCA1 activation domain 1 (AD1) through a coiled-coil-mediated interaction. Genes Dev. 16: 1509–1517. doi: 10.1101/gad.995502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y. F., Miyake T., Ye Q., Li R.2000. Characterization of a novel trans-activation domain of BRCA1 that functions in concert with the BRCA1 C-terminal (BRCT) domain. J. Biol. Chem. 275: 40910–40915. doi: 10.1074/jbc.C000607200 [DOI] [PubMed] [Google Scholar]
- 13.Im K. S., Kim I. H., Kim N. H., Lim H. Y., Kim J. H., Sur J. H.2013. Breed-related differences in altered BRCA1 expression, phenotype and subtype in malignant canine mammary tumors. Vet. J. 195: 366–372. doi: 10.1016/j.tvjl.2012.07.014 [DOI] [PubMed] [Google Scholar]
- 14.Gillett A., Bergman P., Parsa R., Bremges A., Giegerich R., Jagodic M.2013. A silent exonic SNP in kdm3a affects nucleic acids structure but does not regulate experimental autoimmune encephalomyelitis. PLoS ONE 8: e81912. doi: 10.1371/journal.pone.0081912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L. C., Dahiya R.2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18: 1427–1431. doi: 10.1093/bioinformatics/18.11.1427 [DOI] [PubMed] [Google Scholar]
- 16.Matros E., Wang Z. C., Lodeiro G., Miron A., Iglehart J. D., Richardson A. L.2005. BRCA1 promoter methylation in sporadic breast tumors: relationship to gene expression profiles. Breast Cancer Res. Treat. 91: 179–186. doi: 10.1007/s10549-004-7603-8 [DOI] [PubMed] [Google Scholar]
- 17.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P. A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L. M., Ding W., Bell R., Rosenthal J., Hussey C., Tran T., McClure M., Frye C., Hattier T., Phelps R., Haugen-Strano A., Katcher H., Yakumo K., Gholami Z., Shaffer D., Stone S., Bayer S., Wray C., Bogden R., Dayananth P., Ward J., Tonin P., Narod S., Bristow P. K., Norris F. H., Helvering L., Morrison P., Rosteck P., Lai M., Barrett J. C., Lewis C., Neuhausen S., Cannon-Albright L., Goldgar D., Wiseman R., Kamb A., Skolnick M. H.1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–71. doi: 10.1126/science.7545954 [DOI] [PubMed] [Google Scholar]
- 18.Monteiro A. N., August A., Hanafusa H.1996. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc. Natl. Acad. Sci. U.S.A. 93: 13595–13599. doi: 10.1073/pnas.93.24.13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moynahan M. E., Chiu J. W., Koller B. H., Jasin M.1999. Brca1 controls homology-directed DNA repair. Mol. Cell 4: 511–518. doi: 10.1016/S1097-2765(00)80202-6 [DOI] [PubMed] [Google Scholar]
- 20.Qiu H. B., Sun W. D., Yang X., Jiang Q. Y., Chen S., Lin D. G.2015. Promoter mutation and reduced expression of BRCA1 in canine mammary tumors. Res. Vet. Sci. 103: 143–148. doi: 10.1016/j.rvsc.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 21.Rivera P., Melin M., Biagi T., Fall T., Häggström J., Lindblad-Toh K., von Euler H.2009. Mammary tumor development in dogs is associated with BRCA1 and BRCA2. Cancer Res. 69: 8770–8774. doi: 10.1158/0008-5472.CAN-09-1725 [DOI] [PubMed] [Google Scholar]
- 22.Rice J. C., Ozcelik H., Maxeiner P., Andrulis I., Futscher B. W.2000. Methylation of the BRCA1 promoter is associated with decreased BRCA1 mRNA levels in clinical breast cancer specimens. Carcinogenesis 21: 1761–1765. doi: 10.1093/carcin/21.9.1761 [DOI] [PubMed] [Google Scholar]
- 23.Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D. M.1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88: 265–275. doi: 10.1016/S0092-8674(00)81847-4 [DOI] [PubMed] [Google Scholar]
- 24.Shao N., Chai Y. L., Shyam E., Reddy P., Rao V. N.1996. Induction of apoptosis by the tumor suppressor protein BRCA1. Oncogene 13: 1–7. [PubMed] [Google Scholar]
- 25.Sun W., Yang X., Qiu H., Zhang D., Wang H., Huang J., Lin D.2015. Relationship between three novel SNPs of BRCA1 and canine mammary tumors. J. Vet. Med. Sci. 77: 1541–1543. doi: 10.1292/jvms.15-0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szabo C. I., Wagner L. A., Francisco L. V., Roach J. C., Argonza R., King M. C., Ostrander E. A.1996. Human, canine and murine BRCA1 genes: sequence comparison among species. Hum. Mol. Genet. 5: 1289–1298. doi: 10.1093/hmg/5.9.1289 [DOI] [PubMed] [Google Scholar]
- 27.Takai D., Jones P. A.2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. U.S.A. 99: 3740–3745. doi: 10.1073/pnas.052410099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson M. E., Jensen R. A., Obermiller P. S., Page D. L., Holt J. T.1995. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat. Genet. 9: 444–450. doi: 10.1038/ng0495-444 [DOI] [PubMed] [Google Scholar]
- 29.Tsuchida S., Ikemoto S., Tagawa M.2001. Microsatellite polymorphism in intron 14 of the canine BRCA1 gene. J. Vet. Med. Sci. 63: 479–481. doi: 10.1292/jvms.63.479 [DOI] [PubMed] [Google Scholar]
- 30.Yamane K., Chen J., Kinsella T. J.2003. Both DNA topoisomerase II-binding protein 1 and BRCA1 regulate the G2-M cell cycle checkpoint. Cancer Res. 63: 3049–3053. [PubMed] [Google Scholar]
- 31.Yarden R. I., Pardo-Reoyo S., Sgagias M., Cowan K. H., Brody L. C.2002. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 30: 285–289. doi: 10.1038/ng837 [DOI] [PubMed] [Google Scholar]
- 32.Vallon-Christersson J., Cayanan C., Haraldsson K., Loman N., Bergthorsson J. T., Brøndum-Nielsen K., Gerdes A. M., Møller P., Kristoffersson U., Olsson H., Borg A., Monteiro A. N.2001. Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum. Mol. Genet. 10: 353–360. doi: 10.1093/hmg/10.4.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei M., Grushko T. A., Dignam J., Hagos F., Nanda R., Sveen L., Xu J., Fackenthal J., Tretiakova M., Das S., Olopade O. I.2005. BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res. 65: 10692–10699. doi: 10.1158/0008-5472.CAN-05-1277 [DOI] [PubMed] [Google Scholar]
- 34.Xiong Z., Laird P. W.1997. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 25: 2532–2534. doi: 10.1093/nar/25.12.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Q., Pao G. M., Huynh A. M., Suh H., Tonnu N., Nederlof P. M., Gage F. H., Verma I. M.2011. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477: 179–184. doi: 10.1038/nature10371 [DOI] [PMC free article] [PubMed] [Google Scholar]


