Abstract
Summary: We introduce SharpViSu, an interactive open-source software with a graphical user interface, which allows performing processing steps for localization data in an integrated manner. This includes common features and new tools such as correction of chromatic aberrations, drift correction based on iterative cross-correlation calculations, selection of localization events, reconstruction of 2D and 3D datasets in different representations, estimation of resolution by Fourier ring correlation, clustering analysis based on Voronoi diagrams and Ripley’s functions. SharpViSu is optimized to work with eventlist tables exported from most popular localization software. We show applications of these on single and double-labelled super-resolution data.
Availability and implementation: SharpViSu is available as open source code and as compiled stand-alone application under https://github.com/andronovl/SharpViSu.
Contact: klaholz@igbmc.fr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
The first step in processing of stochastic super-resolution microscopy data, the single-molecule localization, recently became a routine operation (Small and Stahlheber, 2014) and is often implemented by the manufacturer with the software operating the microscope. However, the further processing workflow of single-molecule localization microscopy (SMLM) data is much less standardized. Coordinates of individual molecules in space and time with their localization precisions are contained in a table of events obtained after fitting the Gaussian-like spots in the first step of the processing. Most available software for processing of single-molecule data such as rapidSTORM (Wolter et al., 2012), QuickPALM (Henriques et al., 2010), the Localization Microscopy plugin for µManager (Edelstein et al., 2014), RainSTORM (Rees et al., 2013) and ThunderSTORM (Ovesný et al., 2014) are dedicated to fitting of camera images (Sage et al., 2015), while few software, such as PALMsiever (Pengo et al., 2015) and ViSP (El Beheiry and Dahan, 2013) are designed for processing of localization tables. The best way to analyze stochastic microscopy data is to work directly with eventlists (Deschout et al., 2014) for which the development of new specialized and integrated tools is required.
2 Results and discussion
We have developed the SharpViSu software that combines the most important steps from our experience that are required for the treatment of localization data, namely: (i) multi-step correction of sample drift by cross-correlation with or without fiducial markers (Mlodzianoski et al., 2011); (ii) sieving of event lists by merging consecutive events and removal of imprecise localizations; (iii) reconstruction of 2D super-resolution images in different modes (histogram, Gaussian (Huang et al., 2008), quad-tree (Baddeley et al., 2010), local density and hue-coded time) with selectable sampling; (iv) estimation of resolution by Fourier ring correlation (FRC; Banterle et al., 2013; Nieuwenhuizen et al., 2013); (v) correction of chromatic aberrations for multi-color experiments (Erdelyi et al., 2013); and (vi) reconstruction of 3D datasets with astigmatism (Huang et al., 2008). The software also allows for calibration of localization data with chromatic aberrations and astigmatism. The output of SharpViSu can be saved in widespread formats such as .tiff (pictures), .png (graphs), .ascii or ViSP’s .3dlp (El Beheiry and Dahan, 2013) (tables) allowing further analysis or preparation of publications.
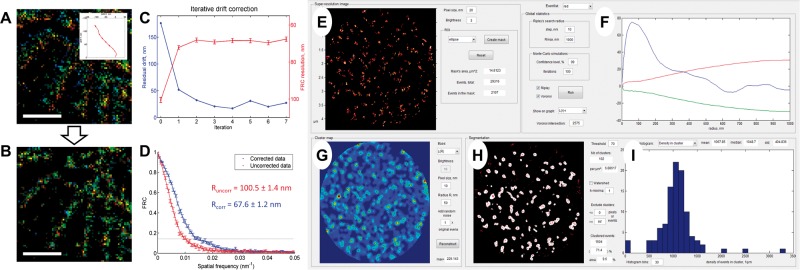
SharpViSu provides a user-friendly integrated software package for corrections, analysis and visualization of super-resolution microscopy data (Supplementary Table S1). It uses localization tables as input which results in very small data sizes compared to raw time-lapse acquisitions, and a high precision of the contained information as compared to uncorrected, preliminarily reconstructed super-resolution images. For example, it handles iterative cross-correlation-based drift correction (without requiring fiducial markers) which shows progressive reduction of the estimated residual drift (Fig. 1A–D). The super-resolution image reconstructed from the corrected data looks much sharper, shows no detectable residual drift in the color-coded time representation (Fig. 1A, B) and demonstrates a significant improvement in resolution as quantified by FRC (Fig. 1D). SharpViSu allows correction of chromatic aberrations (Supplementary Fig. S2) and determination of Z-positions of fluorophores based on fitted data (Supplementary Fig. S4). Additionally, we introduced a novel local density visualization method based on Voronoi diagrams (Andronov et al., 2016) that effectively improves the appearance of data and does not require any user-adjustable parameters that may be non-obvious to determine (Baddeley et al., 2010). Finally, SharpViSu includes direct quantitative resolution evaluation with FRC.
Fig. 1.
Features of SharpViSu. (A, B) 1.5 µm × 1.5 µm fragment of a super-resolution image of β-tubulin in a HeLa cell reconstructed in the color-coded time mode before (A) and after 7 iterations of drift correction (B). The drift trace obtained by SharpViSu is shown in the inset. Scale bars: 500 nm. (C) Reduction of the estimated residual drift (blue) and corresponding improvement of FRC-resolution (red) by iterative drift correction. The curves converge after 2–4 iterations. (D) FRCs of the initial and the corrected datasets show statistically significant improvement in resolution. (E–I) Interface of ClusterViSu, a plugin for comprehensive segmentation of SMLM data. (E) Selected region of interest. (F) Statistics on localizations with Ripley’s L(r)-r functions for the experimental data (blue) and 99% confidence interval for randomly distributed data (red and green) demonstrating statistically significant clustering. (G) Cluster density map calculated on the basis of Ripley’s L(R = 50 nm) function. (H) Cluster map, binarized at the threshold L = 70. (I) Histogram representing distribution of density of localizations in clusters. Data: nucleopore protein TPR, detected with Alexa-647-conjugated secondary antibodies (Lemaître et al., 2014)
The functionality of SharpViSu is extendable via plugins, such as ClusterViSu for comprehensive cluster analysis of SMLM data (Fig. 1E–I). It includes tools such as calculations of Voronoi and Ripley statistics (Owen et al., 2010) with Monte-Carlo simulations, different modes of reconstruction (e.g. based on Gaussian blur or Ripley’s functions) and segmentation of density maps, retrieval of geometrical properties of detected clusters, segmentation based on Voronoi tessellation (Andronov et al., 2016; Levet et al., 2015). SharpViSu is routinely used at the CBI/IGBMC for correction of super-resolution data and visualization of chromatin complexes (Lemaître et al., 2014) and is largely applicable. SharpViSu is a timely contribution for the analysis of data from super-resolution microscopy, a research field in biology which is providing unprecedented insights into cellular and molecular function.
Supplementary Material
Funding
This work was supported by CNRS, the European Research Council (ERC Starting Grant 243296), Association pour la Recherche sur le Cancer (ARC), Université de Strasbourg and Investissement d’Avenir (IDEX), Institut National du Cancer (INCa), the French Infrastructure for Integrated Structural Biology (FRISBI) [ANR-10-INSB-05-01], Instruct as part of the European Strategy Forum on Research Infrastructures (ESFRI) and IGBMC facilities.
Conflict of Interest: none declared.
References
- Andronov L. et al. (2016) ClusterViSu, a method for clustering of protein complexes by Voronoi tessellation in super-resolution microscopy. Sci. Rep., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddeley D. et al. (2010) Visualization of Localization Microscopy Data. Microsc. Microanal., 16, 64–72. [DOI] [PubMed] [Google Scholar]
- Banterle N. et al. (2013) Fourier ring correlation as a resolution criterion for super-resolution microscopy. J. Struct. Biol., 183, 363–367. [DOI] [PubMed] [Google Scholar]
- Deschout H. et al. (2014) Precisely and accurately localizing single emitters in fluorescence microscopy. Nat. Methods, 11, 253–266. [DOI] [PubMed] [Google Scholar]
- Edelstein A.D. et al. (2014) Advanced methods of microscope control using μManager software. J. Biol. Methods, 1, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Beheiry M., Dahan M. (2013) ViSP: representing single-particle localizations in three dimensions. Nat. Methods, 10, 689–690. [DOI] [PubMed] [Google Scholar]
- Erdelyi M. et al. (2013) Correcting chromatic offset in multicolor superresolution localization microscopy. Opt. Express, 21, 10978–10988. [DOI] [PubMed] [Google Scholar]
- Henriques R. et al. (2010) QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods, 7, 339–340. [DOI] [PubMed] [Google Scholar]
- Huang B. et al. (2008) Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science, 319, 810–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C. et al. (2014) Nuclear position dictates DNA repair pathway choice. Genes Dev., 28, 2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levet F. et al. (2015) SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat. Methods, 12, 1065–1071. [DOI] [PubMed] [Google Scholar]
- Mlodzianoski M.J. et al. (2011) Sample drift correction in 3D fluorescence photoactivation localization microscopy. Opt. Express, 19, 15009–15019. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuizen R.P.J. et al. (2013) Measuring image resolution in optical nanoscopy. Nat. Methods, 10, 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesný M. et al. (2014) ThunderSTORM: a comprehensive ImageJ Plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics, 30, 2389–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.M. et al. (2010) PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J. Biophoton., 3, 446–454. [DOI] [PubMed] [Google Scholar]
- Pengo T. et al. (2015) PALMsiever: a tool to turn raw data into results for single-molecule localization microscopy. Bioinformatics, 31, 797–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E.J. et al. (2013) Elements of image processing in localization microscopy. J. Opt., 15, 094012. [Google Scholar]
- Sage D. et al. (2015) Quantitative evaluation of software packages for single-molecule localization microscopy. Nat. Methods, 12, 717–724. [DOI] [PubMed] [Google Scholar]
- Small A., Stahlheber S. (2014) Fluorophore localization algorithms for super-resolution microscopy. Nat. Methods, 11, 267–279. [DOI] [PubMed] [Google Scholar]
- Wolter S. et al. (2012) rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods, 9, 1040–1041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.