Abstract
Motivation: DisGeNET-RDF makes available knowledge on the genetic basis of human diseases in the Semantic Web. Gene-disease associations (GDAs) and their provenance metadata are published as human-readable and machine-processable web resources. The information on GDAs included in DisGeNET-RDF is interlinked to other biomedical databases to support the development of bioinformatics approaches for translational research through evidence-based exploitation of a rich and fully interconnected linked open data.
Availability and implementation: http://rdf.disgenet.org/
Contact: support@disgenet.org
1 Introduction
Advancements in experimental technologies give an unprecedented capacity of description of a patient from a molecular point of view. Translational bioinformatics envisions to push forward biomedical discoveries and to enhance healthcare practice by bridging the gap between these two worlds (Altman, 2012). To create this synergistic translation between molecular data and clinical events, it is crucial a comprehensive understanding of the complex relationships between genotype, phenotype and environment that underlie human diseases. To explore these complex relationships, current biomedical research requires leveraging and linking different types of information such as genetic basis of diseases, disease biomarkers, drug therapeutic applications and side effects, or effects of exposure to environmental factors. But, this integration is challenging as the information is fragmented in resources dispersed and often technology or domain-specific or, importantly, hidden in free text. The emerging Semantic Web is gaining momentum in Life Sciences as it provides standards to set a semantic and syntactic interoperable infrastructure for data integration over the Web. The increasing publication of open biomedical databases structured and interlinked using the W3C Resource Description Framework (RDF) and Web Ontology Language (OWL) technologies through projects such as Bio2RDF (Belleau et al., 2008) and the EBI RDF platform (Jupp et al., 2014) paves the way to answer more complex and sophisticated cross-domain questions. In this paper, we present DisGeNET-RDF a new open resource in the Semantic Web and a new facet of DisGeNET (Piñero et al., 2015). DisGeNET is one of the most comprehensive databases on gene-disease associations (GDAs) for the study of the molecular mechanisms underpinning human diseases. Most of the GDAs in DisGeNET (82% in version 3.0) have been identified by text mining the literature using BeFree (Bravo et al., 2015), and are integrated with curated GDAs from a variety of authoritative sources on human genetics data. Each GDA is explicitly annotated with its supporting evidence, which makes DisGeNET a resource of reference for evidence-based knowledge discovery. With the publication of DisGeNET-RDF, we aim to foster the development of bioinformatic tools to leverage biomedical Big Data, and to facilitate knowledge navigation and discovery to support translational research.
2 Data model
DisGeNET-RDF is a linked dataset that represents GDAs as entities described by different properties such as the annotated gene and disease, supporting article(s), SNP and the DisGeNET score (Piñero et al., 2015). The data model, illustrated in Figure 1, makes extensive reuse of standard identifiers, common vocabularies and ontologies, which include OWL ontologies like the NCI thesaurus (NCIT) for medical vocabulary and SIO for general science. GDAs are semantically harmonized using the DisGeNET association type ontology, which formally defines as concepts the different types of associations between a gene and a disease. The goal of the ontology is to harmonize the different association types provided by the databases integrated in DisGeNET, within a hierarchical structure of a directed acyclic graph. In the current version, the ontology consists of seven classes represented in OWL (see the schema modeling in Fig. 1). The DisGeNET ontology is integrated into the Semanticscience Integrated Ontology (http://sio.semanticscience.org) (SIO) (Dumontier et al., 2014) that provides ontology support for Bio2RDF Linked Data among other projects. Furthermore, SIO and thus the DisGeNET ontology have been integrated in other ontologies such as the Biological Observation Matrix Ontology (https://bioportal.bioontology.org/ontologies/BIOMO) and the Orthology Ontology (https://bioportal.bioontology.org/ontologies/ORTH), which expand DisGeNET semantic interoperability with other data sources. All resources in DisGeNET-RDF are identified by Uniform Resource Identifiers (URIs), a Web-based global identification system used in RDF, that are dereferenceable (it is possible to get a representation about the referenced resource on the Web). These URIs are from authoritative data providers whenever possible, otherwise the Identifiers.org registry of scientific identifiers is used (Juty et al., 2012). Consistently, DisGeNET-RDF makes available GDAs as unique digital objects identified by URIs. We implemented a harmonized URI identification scheme for DisGeNET GDAs based on the http://rdf.disgenet.org/ domain and a unique identifier built on association attributes. These minted URIs are dereferenceable, and support content negotiation for both human HTML and machine-processable RDF views. In addition, our RDF dataset is interlinked to Linked Open Data (LOD) implementations of biomedical databases and several disease terminologies such as MeSH, OMIM, Orphanet or ICD9-CM available through Linked Data projects such as Bio2RDF (see statistics at Datahub (http://datahub.io/ca/dataset/disgenet)). Consequently, DisGeNET appears in the LOD cloud diagram (2014-08-30 version). In the context of Big Data integration and large-scale analysis over the Web, discoverability, reliability and reproducibility are major concerns for linked datasets. To address this issue, DisGeNET-RDF is available with a full provenance dataset description conformant to the W3C HCLS specification (http://www.w3.org/TR/hcls-dataset/) in order to ease its discoverability and data reuse (Fig. 1). The W3C recommended Vocabulary of Interlinked Datasets (VoID) is used for describing the metadata of the DisGeNET-RDF dataset. The detailed description of the RDF schema, ontologies used and the URI scheme for the normalization of GDAs identification are available at the DisGeNET-RDF web site (see http://rdf.disgenet.org/).
Fig. 1.
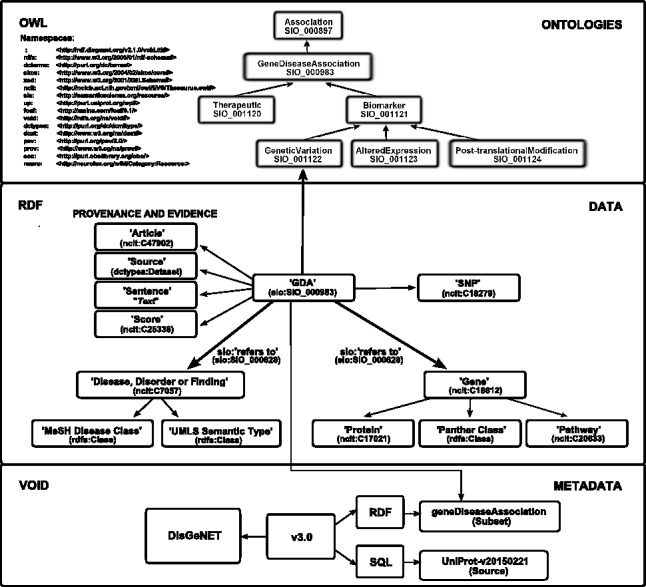
DisGeNET-RDF data model. Simplified version of the DisGeNET-RDF schema modeling. Top: the DisGeNET ontology. Central: the DisGeNET-RDF Schema. Bottom: the provenance metadata model conformant to the W3C HCLS specification. See the full schema in the RDF web site
3 Implementation and availability
DisGeNET-RDF is an open access resource of machine-processable GDAs published on the Web as Linked Data supported by a full provenance dataset description. It is created by the data providers following the Open PHACTS guidelines for exposing data as RDF (http://www.openphacts.org/specs/2013/WD-rdfguide-20131007/). The current implementation of DisGeNET-RDF (v3.0.0) consists of 21,730,060 triples and is accessible as an RDF dump serialized in Turtle syntax for download. This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/) terms. The RDF dump is generated using a production system based on the D2RQ platform (http://d2rq.org/). We have also implemented a Faceted browser and a SPARQL endpoint that makes our RDF available for Linked Data navigation, information retrieval and, importantly, federated interrogation with external resources. These services are supported by Triple Store technology and they are provided by an instance of the open source edition of the OpenLink Virtuoso server (http://virtuoso.openlinksw.com/). Noticeably, we provide a web site for supporting users with documentation, links to access points, SPARQL query examples on how to retrieve integrated data, and contact details. In recognition of the interest in the RDF representation of DisGeNET, we note that from January 1st, 2015 to January 1st, 2016 the dataset web site had more than 10 000 page views according to Google Analytics report. As a proof of concept, DisGeNET-RDF is currently implemented in several applications: (i) it is integrated in the Open PHACTS Discovery Platform (https://dev.openphacts.org) (Gray et al., 2014), which is an application for drug discovery based on Semantic Web technology and RDF linked datasets; (ii) it is used in KNIME workflows to answer sophisticated research questions in drug discovery (http://www.myexperiment.org/workflows/4513.html), and for in silico target validation of cellular phenotypic screening (Digles, 2016); (iii) it is available as one of the databases in Bioqueries, where a user can access and share, edit or publish queries over DisGeNET-RDF (http://bioqueries.uma.es/endpoint/disgenet); and (iv) it is used in the disgenet2r R package (https://bitbucket.org/albags/disgenet2r).
4 Applications
To identify the biological mechanisms responsible for disease aetiology, pharmacological treatment and toxicological events we need to exploit biomedical data integrated in a multifaceted way. The possible applications of DisGeNET-RDF are numerous and diverse. Our SPARQL endpoint allows query federation to interrogate DisGeNET with several LOD resources with a single query. These include data on gene expression, drugs and other chemicals, biological pathways and networks, kinetic models, to just mention some of the information covered. Some examples of sophisticated research questions that can be solved using DisGeNET-RDF and its linkage with other resources are:
What are the pathways associated with Lafora disease?
Which of the proteins associated with Aarskog syndrome are potential drug targets?
What are the other diseases associated with genes differentially expressed in Pancreatic cancer?
A comprehensive list of examples of DisGeNET-RDF use cases, with supporting information on how to formulate the SPARQL queries from our endpoint service are provided on the web site (http://disgenet.org/web/DisGeNET/menu/rdf#sparql). For instance, to solve the previous questions (1), (2) and (3), we can cross DisGeNET-RDF with WikiPathways, ChEMBL and Gene Expression Atlas, respectively. See examples of SPARQL queries for these particular use cases in the web site (queries Q2.1.12, Q2.3.5 and Q2.2.3, respectively). These queries could be used to explore the underlying molecular mechanisms of a disease, to explore repurposing opportunities for drugs, or to identify drug targets associated with adverse effects.
5 Conclusion
DisGeNET-RDF is a new open resource to harness the Semantic Web for new discovery opportunities on the genetic basis of human diseases. The publication of DisGeNET-RDF and the implementation of an SPARQL endpoint offer the possibility to integrate DisGeNET with other LOD resources to answer complex biomedical questions. DisGeNET-RDF web site supplies supporting documentation and query examples to help users getting started. Our aim is to make DisGeNET information more discoverable and to integrate it with the current open life science knowledge in order to support projects on the aetiology of human diseases, drug discovery and toxicological research.
Acknowledgements
The authors thank the Open PHACTS partners, Michel Dumontier and the OpenLink staff for their input, collaboration and help.
Funding
We received support from ISCIII-FEDER (PI13/00082, CP10/00524), from the IMI-JU under grants agreements no. 115002 (eTOX), no. 115191 (Open PHACTS), no. 115372 (EMIF) and no. 115735 (iPiE), resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007–2013) and EFPIA companies’ in kind contribution, and the EU H2020 Programme 2014–2020 under grant agreements no. 634143 (MedBioinformatics) and no. 676559 (Elixir-Excelerate). The Research Programme on Biomedical Informatics (GRIB) is a node of the Spanish National Institute of Bioinformatics (INB).
Conflict of Interest: none declared.
References
- Altman R.B. (2012) Translational bioinformatics: linking the molecular world to the clinical world. Clin. Pharmacol. Ther., 91, 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleau F. et al. (2008) Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J. Biomed. Inf., 41, 706–716. [DOI] [PubMed] [Google Scholar]
- Bravo À. et al. (2015) Extraction of relations between genes and diseases from text and large-scale data analysis: implications for translational research. BMC Bioinformatics, 16, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digles D. et al. (2016) Open PHACTS Computational Protocols for in silico Target Validation of Cellular Phenotypic Screens: Knowing the Knowns, in press. [DOI] [PMC free article] [PubMed]
- Dumontier M. et al. (2014) The Semanticscience Integrated Ontology (SIO) for biomedical research and knowledge discovery. J. Biomed. Seman., 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray A.J.G. et al. (2014) Applying linked data approaches to pharmacology: architectural decisions and implementation. Semant. Web, 5, 101–113. [Google Scholar]
- Jupp S. et al. (2014) The EBI RDF platform: linked open data for the life sciences. Bioinformatics, 30, 1338–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juty N. et al. (2012) Identifiers.org and MIRIAM Registry: community resources to provide persistent identification. Nucleic Acids Res., 40, D580–D586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J. et al. (2015) DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database, 2015, bav028–bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]


