ABSTRACT
Although it is becoming clear that many microbial primary producers can also play a role as organic consumers, we know very little about the metabolic regulation of photoautotroph organic matter consumption. Cyanobacteria in phototrophic biofilms can reuse extracellular organic carbon, but the metabolic drivers of extracellular processes are surprisingly complex. We investigated the metabolic foundations of organic matter reuse by comparing exoproteome composition and incorporation of 13C-labeled and 15N-labeled cyanobacterial extracellular organic matter (EOM) in a unicyanobacterial biofilm incubated using different light regimes. In the light and the dark, cyanobacterial direct organic C assimilation accounted for 32% and 43%, respectively, of all organic C assimilation in the community. Under photosynthesis conditions, we measured increased excretion of extracellular polymeric substances (EPS) and proteins involved in micronutrient transport, suggesting that requirements for micronutrients may drive EOM assimilation during daylight hours. This interpretation was supported by photosynthesis inhibition experiments, in which cyanobacteria incorporated N-rich EOM-derived material. In contrast, under dark, C-starved conditions, cyanobacteria incorporated C-rich EOM-derived organic matter, decreased excretion of EPS, and showed an increased abundance of degradative exoproteins, demonstrating the use of the extracellular domain for C storage. Sequence-structure modeling of one of these exoproteins predicted a specific hydrolytic activity that was subsequently detected, confirming increased EOM degradation in the dark. Associated heterotrophic bacteria increased in abundance and upregulated transport proteins under dark relative to light conditions. Taken together, our results indicate that biofilm cyanobacteria are successful competitors for organic C and N and that cyanobacterial nutrient and energy requirements control the use of EOM.
IMPORTANCE
Cyanobacteria are globally distributed primary producers, and the fate of their fixed C influences microbial biogeochemical cycling. This fate is complicated by cyanobacterial degradation and assimilation of organic matter, but because cyanobacteria are assumed to be poor competitors for organic matter consumption, regulation of this process is not well tested. In mats and biofilms, this is especially relevant because cyanobacteria produce an extensive organic extracellular matrix, providing the community with a rich source of nutrients. Light is a well-known regulator of cyanobacterial metabolism, so we characterized the effects of light availability on the incorporation of organic matter. Using stable isotope tracing at the single-cell level, we quantified photoautotroph assimilation under different metabolic conditions and integrated the results with proteomics to elucidate metabolic status. We found that cyanobacteria effectively compete for organic matter in the light and the dark and that nutrient requirements and community interactions contribute to cycling of extracellular organic matter.
INTRODUCTION
The extracellular milieu is the interface at which cyanobacteria interact both with their environment and with one another (1), and yet there is much that we do not understand about how these primary producers use this space. Because extracellular composition and activities dictate community resource partitioning and ultimately link to microbial community function, it is important to investigate this extracellular pool of resources (2).
The metabolic foundations behind extracellular processes such as excretion, degradation, and reincorporation are surprisingly complex, particularly for cyanobacteria. Relatively few physiological mechanisms controlling excretions by “leaky” primary producers have been described, although they are thought to account for 25% of primary production in some marine systems (3). For example, excretion of osmolytes (4) (as a response to stress) and excretion of fermentation by-products (5) are well established, but the functional roles of other exopolymers are less clear (6). Cyanobacteria such as Crocosphaera and those dwelling in microbial mats may use exopolymers for oxygen or UV protection (7, 8); others appear to produce exopolysaccharides to remedy carbon-nitrogen imbalances (9). Microbial interactions also likely play a role in cyanobacterial excretions; for example, marine Synechococcus spp. secrete allelopathic chemicals to mediate competition (10).
Cyanobacterial extracellular degradation of organic compounds also plays a role in nutrient cycling. For example, in response to phosphate stress, some marine cyanobacteria secrete alkaline phosphatases to acquire phosphate from organic sources (11, 12). Extracellular nucleases are also common in heterocystous cyanobacterial strains (13). However, extracellular degradation may be the least understood of these processes; genomics-enabled research has revealed a large number of extracellular proteins (exoproteins) of unknown function, implying that a number of extracellular activities remain uncharacterized (14).
Although primarily photoautotrophs, many cyanobacteria also incorporate organic C and N, ranging from phosphonates (15) and amino acids (16, 17) to glucose (18, 19). Light availability is likely to play a role in this incorporation and has been shown to influence the uptake of an important organic sulfur and carbon compound, dimethylsulfoniopropionate, by marine picocyanobacteria and heterotrophs (20). Interestingly, leucine uptake in these coastal picocyanobacterial assemblages is not light-driven (20), but picocyanobacterial amino acid uptake in oligotrophic environments is stimulated by light (21), and the nature of the metabolic regulation behind this light stimulation is unclear (22).
Cyanobacterial biofilms and phototrophic mats provide a relevant and tractable system to study metabolic drivers of photoautotroph organic matter incorporation in the presence of heterotrophs. These systems often contain copious amounts of extracellular polymeric substances (EPS), mainly polysaccharides, proteins, and nucleic acids (23), to which metabolites and inorganic micronutrients may be adsorbed, creating an extracellular pool of resources that is large relative to the size of the active biomass. Members of a single group of organisms, the cyanobacteria, are often the dominant primary producers in phototrophic mats (e.g., 24, and the turnover of organic matter can be rapid (25), suggesting active processes that can be measured to identify contributors to turnover under various conditions.
In a previous study, we characterized extracellular organic matter (referred to here as “EOM”) from a natural microbial mat collected at Elkhorn Slough in Monterey Bay, CA, and a unicyanobacterial biofilm (“cultured biofilm”) (26). The biofilm is comprised of a marine filamentous cyanobacterium, strain ESFC-1, and associated microbes, isolated from the Elkhorn Slough microbial mat into culture. ESFC-1 belongs to a recently identified lineage of active diazotrophs in these mats and can comprise up to 10% of the natural mat cyanobacterial community (27). The biofilms produce exopolysaccharides predominantly composed of either α- or β(1,4)-linked glucans, as does the intact mat (26). Similarly, a number of homologous exoproteins, such as Zn-dependent peptidases and redox-active proteins, are produced by the both the biofilm and the intact mat (26). Accordingly, these biofilms represent a suitable simplified system to study cyanobacterial extracellular processes. Furthermore, in both intact mat and cultured biofilms, enzymes are produced by cyanobacteria that likely function in the degradation of EOM, and ESFC-1 incorporates its own extracellular organic C (26). This led us to hypothesize that the cyanobacteria use the extracellular matrix as a storage reservoir for C and regulate both excretion and assimilation processes.
Here, we used this simplified biofilm system to examine the metabolic and molecular underpinnings of cyanobacterial EOM reuse in cultured biofilms. To test whether the extracellular matrix is used as a storage reservoir, we compared EOM incorporation, composition, and activity using four light regimes: normal diel light-dark treatment, continuous light (C-replete; dissolved inorganic C [DIC] achieved through photosynthesis) treatment, continuous dark (C-starved) treatment, and diel DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea] (oxygenic photosynthesis inhibition) treatment. We also investigated N incorporation from EOM, since little is known about the dynamics of organic C/N incorporation by photoautotrophs in aquatic ecosystems. Applying stable isotope tracing at the single-cell level, using the high-resolution imaging secondary ion mass spectrometry (NanoSIMS) method, we integrated these results with proteomics data to provide a more comprehensive picture of the flow of essential nutrients to different community members.
RESULTS
Uptake of EOM under various light regimes.
Organic matter additions were examined under three distinct illumination regimes: continuous light, continuous dark, and diel (12-h light/12-h dark) treatments over 24 h. 13C-labeled and 15N-labeled cyanobacterial EOM was added to unicyanobacterial biofilms at the start of the light phase. The continuous light regime was sampled at the 24-h time point, as data from earlier time points were identical to those in the diel regime. High-resolution imaging secondary ion mass spectrometry (NanoSIMS) was used to quantify 13C and 15N assimilation in individual cyanobacterial trichomes and in associated microbes (referred to here as “heterotrophs”) (Fig. 1; see also Table S1 in the supplemental material). By 24 h, cyanobacterial C incorporation was higher in the continuous light treatment than in the continuous dark treatment (Fig. 1). To quantify these differences, we calculated mean 13C and 15N enrichment values for each treatment over time (Fig. 2). On the basis of bulk isotope ratio mass spectrometry (IRMS) analysis, the C/N ratio of the added labeled EOM was 4.8, representing an isotopic enrichment of 67.6 13C atom percent excess (APE) and 85.4 15N APE. The EOM (13C APE)/(15N APE) ratio was used to establish the line indicating the proportions of C and N uptake from the added EOM (Fig. 2, dashed line) to calculate the C/N incorporated by individual cells.
FIG 1 .
Representative NanoSIMS images of cyanobacterium ESFC-1 trichomes and heterotrophs 24 h after addition of 13C-labeled and 15N-labeled EOM. Trichomes and associated microbes were identified morphologically by scanning electron microscopy (SEM) prior to NanoSIMS analysis. Left, images (panels A and C and panel E) are paired images from a light-treated biofilm; right, images (panels B and D and panel F) are paired images from a dark-treated biofilm. (A and B) Scanning electron microscope images taken prior to NanoSIMS analysis. White outlines in panels C to F correspond to areas analyzed (ROIs), from which values for individual cells are derived. (C and D) 13C enrichment images. (E and F) 15N enrichment images. APE, atom percent excess.
FIG 2 .
NanoSIMS-derived average levels of 13C and 15N APE (atom percent excess) of cyanobacterial trichomes for four treatments over time following incubation of unialgal biofilms with 13C-labeled and 15N-labeled EOM. Error bars represent 1 standard error of the mean. Each point represents an average of at least 14 cells analyzed via NanoSIMS. The dashed line indicates the 13C/15N ratio of the labeled substrate, as determined by IRMS. *, significant difference between mean 13C APE values (P < 0.05). **, significant difference between mean 13C APE values (P < 0.05) and mean 15N APE values (P < 0.05).
Under all experimental regimes, cyanobacterial cells incorporated EOM-derived C and N, as shown in Fig. 2. Mean 13C and 15N APE values were significantly higher than the killed control mean values (P < 0.05), except for the 2-h and 24-h dark treatments. When photosynthetic electron transfer was inhibited by DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea], 13C and 15N incorporation at 6-h was still significantly higher than that seen with killed controls (P < 0.0001). In the continuous dark treatment, mean cyanobacterial 13C incorporation increased, with the maximum measured at 6 h (0.54 13C APE; 0.46 15N APE), and then decreased by ~50% at 24 h (Fig. 2). In the biofilms incubated with a diel light cycle, 13C incorporation increased significantly (0.5 to 0.9 APE) between 2 hours and 12 hours of light and then decreased to 0.7 APE following the 12-h dark period, although this was not statistically significant. In the continuous light treatment, mean 13C incorporation increased between 12 h and 24 h of light (P = 0.0027).
To assess preferences for C or N assimilation from the substrate, we calculated C/N relative assimilation levels for individual cells, based on the C/N ratio of the organism and the substrate (28), as shown in Fig. 3. A C/N relative assimilation value of 1.0 indicates that C and N were incorporated in equivalent amounts from the substrate. For cyanobacteria, we compared C/N relative assimilation values at the 6-h time point so that we could include the DCMU treatment as well as the continuous dark and diel regimes. Mean C/N relative assimilation values were 1.25 ± 0.1 and 1.29 ± 0.12 at 6 h in the diel and dark regimes, respectively, indicating a significant C preference in assimilation from the substrate. Photosynthesis inhibition with DCMU at 6 h resulted in a cyanobacterial C/N relative assimilation value of 0.57 ± 0.04, indicating that under normal conditions, a fraction of the EOM-derived C was taken up as DIC. This also indicates that chemolithoautotrophy among the noncyanobacterial microbes (referred to as “heterotrophs”) was not occurring, supporting our presumption that the associated microbes in the biofilm were functioning primarily as heterotrophs.
FIG 3 .
C/N relative assimilation from the substrate, accounting for C/N of both substrate and biomass, based on mass balance calculations. Each point represents calculated C/N substrate usage for an individual cell. (Left panel) Cyanobacteria following 6-h incubations using dark (n = 19), diel (n = 9), or DCMU (n = 24) treatments. (Right panel) Heterotrophs following 12-h dark (n = 32) and diel (n = 17) treatments and 6-h DCMU (n = 70) treatment. Error bars indicate 1 standard error of the mean. All means were significantly different from 1.0 (P < 0.05).
We used parallel incubations with 13C bicarbonate to verify the expected utilization patterns of inorganic C and to determine a bulk rate of DIC incorporation. On the basis of bulk IRMS measurements, as expected, the biofilm culture did not incorporate a detectable amount of dissolved inorganic C (DIC) in the dark (13C APE, 0.009 ± 0.003) but incorporated high levels in the light (13C APE, 4.56 ± 0.27). This result supports our interpretation that a portion of the EOM-derived C assimilation is from mineralized EOM in the light.
While cyanobacteria and heterotrophs both incorporated EOM, net levels of C/N assimilation differed between these two groups (see Fig. S1 in the supplemental material). For heterotrophs, we compared C/N relative assimilation levels between diel-, continuous dark-, and DCMU-treated cells. Due to an insufficient number of cells analyzed at the 6-h time point for the dark and diel regimes (see Table S1), we also analyzed the 12-h time point and the DCMU treatment at 6 h. Average heterotroph C/N relative assimilation values were 0.72 ± 0.05 and 0.74 ± 0.05 following 12 h in continuous dark and diel, respectively, indicating a significant N preference, regardless of light conditions (Fig. 3). This was significantly different from the C preference displayed by cyanobacteria (Fig. 3).
Contributions to C and N fluxes in the extracellular reservoir.
To evaluate the effect of light regime on cyanobacterial versus heterotroph C and N uptake, we calculated the average levels of C and N incorporated from the added EOM after 6 h of light, dark, or DCMU treatment conditions and compared the results to those seen with DIC incorporation, measured via bulk IRMS, after 6 h in the light (Table 1). Associated heterotroph cells represented on average 4.2% (±1.36%) of the biomass C, with cyanobacteria accounting for the remaining 95.8%. Since the biovolumes of the replicates were not significantly different over time, we extrapolated the data to calculate the average amounts of C and N (in micrograms) incorporated per well area (in cubic micrometers) (see Materials and Methods). The net fixation of DIC (percent C incorporated relative to the initial DIC pool) was 5.6 µg C incorporated per biofilm. In the light treatment, cyanobacteria incorporated approximately 1.3 µg of C derived from EOM (Table 1). Inorganic nitrogen uptake was not measured, but on the basis of our measured cyanobacterial C/N ratio of 4.0, we estimate that uptake of an additional 1.5 µg N from nitrate (or ammonium) would be necessary to maintain a constant C/N ratio. Given these assumptions, uptake of EOM-derived C and N may account for up to 18% and 10.5% of total cyanobacterial C and N uptake in the light treatment, respectively. In the dark, applying the same assumptions, organic substrate-derived N could account for 59% of N uptake.
TABLE 1 .
Incorporation of C and N by taxonomic groups based on NanoSIMS-derived population averages at 6 h for the cyanobacteria and 12 h for the heterotrophsa
| Treatment | µg C incorporated/well (± SE) |
µg N incorporated/well (± SE) |
||
|---|---|---|---|---|
| Cyanobacteria | Heterotroph | Cyanobacteria | Heterotroph | |
| Dark, extracellular | 0.93 (0.19) | 1.2 (0.23) | 0.13 (0.03) | 0.55 (0.08) |
| Diel, extracellular | 1.27 (0.30) | 1.3 (0.22) | 0.18 (0.04) | 0.65 (0.16) |
| DCMU, extracellular | 0.60 (0.10) | 1.3 (0.23) | 0.18 (0.02) | 0.89 (0.22) |
| Light, bicarbonate | 5.66 (0.75) | |||
| Dark, bicarbonate | NSb | |||
| Nitrate, light (estimated) | 1.5 | |||
| Nitrate, dark (estimated) | 0.09 | |||
The number of cells analyzed (n) was between 9 and 31.
NS, not significantly different from result seen with killed control.
Despite low biovolume relative to the cyanobacteria, heterotrophs accounted for approximately 68% of EOM-derived C uptake in the DCMU treatment, with cyanobacteria incorporating the remaining 32%. This difference was more pronounced for N, where heterotrophs accounted for 80% of the EOM N uptake in the DCMU treatment (Table 1).
The effect of light regime on biofilm composition and enzyme activity.
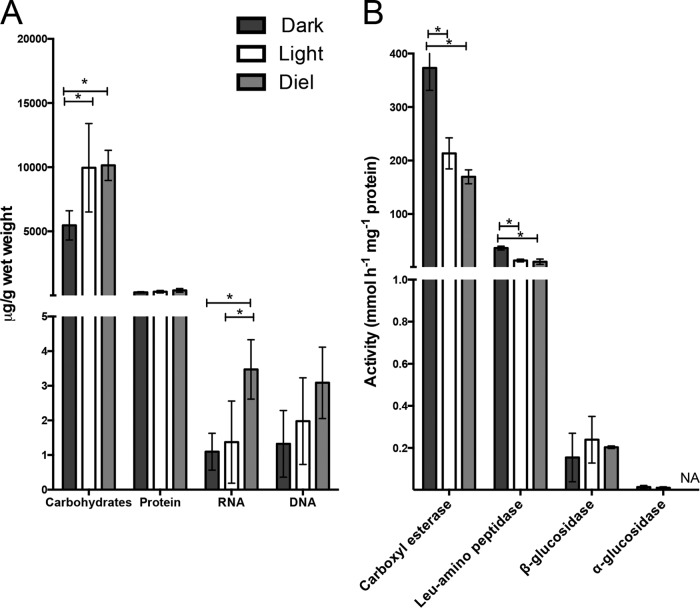
After 3 days of treatment using continuous darkness, we measured extracellular composition and enzyme activity in the biofilms and found evidence for both decreased EPS levels and increased degradation activity relative to the continuous light and diel treatments. Measured levels of extracellular carbohydrate and RNA content were significantly lower in the dark treatment (Fig. 4A). Extracellular enzyme assays revealed that protease and carboxyl esterase activities increased significantly in the dark-treated biofilms (Fig. 4B). Additionally, in biofilms kept in continuous darkness, the signal intensity from the exopolysaccharide fluorescent stain, Congo red, was significantly reduced (see Fig. S2 in the supplemental material), corroborating carbohydrate measurements indicating that EPS levels decreased in the dark.
FIG 4 .
Bulk composition of major EPS polymers and selected enzymatic activities of cyanobacterial biofilm extracellular matrix after 3 days under different light treatments (Dark, 3 days in the dark; Diel, 12-h/12-h light/dark cycle; Light, 3 days in the light. (A) Carbohydrate, protein, and nucleic acids in the extracellular fraction are all normalized to grams of wet weight. Error bars represent 1 standard deviation for results from five biological replicates. *, significant difference between treatments (P < 0.05). (B) Four different degradative enzymes were assayed in the extracellular fraction, and for each enzyme, the specific activity was calculated by the rate of degradation of a fluorescent substrate over 3 h. Error bars represent 1 standard error for results from three biological replicates.
Oxygen levels in the media at the surface of the biofilm were measured after 3 days of incubation and were significantly lower in the dark treatment and higher in the light treatment than in the diel treatment at the 6-h light phase (see Table S2 in the supplemental material). We also measured pH in spent media and found that pH was significantly higher in the light-treated cultures and lower in the dark-treated cultures.
Cyanobacterial exoprotein expression under light and dark conditions.
Using shotgun metaproteomics, we examined protein composition in two operationally defined fractions of the unicyanobacterial biofilms: the extracellular “EOM” fraction, which included all soluble extracellular proteins in the biofilm, and the “total” fraction containing all intracellular and extracellular proteins. We examined five replicate biofilms after 3 days of exposure of the biofilms to three light treatments. Using the cyanobacterial genome (strain ESFC-1 [29]) as a search database, a total of 1,668 unique proteins were identified with high confidence in at least 3 of 5 replicate samples (see Table S3 in the supplemental material). Among these detected proteins, we searched for those significantly overrepresented (P < 0.05, >0.8 log2 fold) in the EOM fraction relative to the total fraction. We identified 168 proteins overrepresented in the EOM fraction (exoproteins) in the continuous light treatment, 147 in the continuous dark treatment, and 248 in the diel light treatment. Of these, 87 exoproteins were overrepresented in the dark treatment relative to the diel and/or light treatments and 107 exoproteins were overrepresented in the light treatment (Fig. 5; see also Table S3).
FIG 5 .
Comparison of the relative abundances determined by shotgun proteomics for the ESFC-1 exoproteome in dark versus light treatments after 3 days. Points represent exoprotein average relative abundances in comparisons between results from 3 and 5 biological replicates and 1 technical replicate (4 to 10 replicates). Relative abundance data were determined from the counts of normalized spectral abundance factors (NSAF). Exoproteins are proteins significantly enriched in the extracellular fractions relative to the total fraction (t test, P < 0.05, log2 fold change > 0.8). Colored points represent proteins with significant differences in abundance between the two treatments (t test, P < 0.05). CMBase, carboxymethylenebutenolidase; Glu-DHase, glucose/sorbosone dehydrogenase; RRM, 4 different RRM domain proteins; ATase, aminotransferase; β-PGM, β-phosoglucomutase; Fe Trans, Fe-regulated ABC transporter ATPase unit; DNA binding, bacterial nucleoid DNA-binding protein; FC, fasciclin domain-containing protein; Ccm, carbon dioxide-concentrating-mechanism protein.
Exoproteins overrepresented in the continuous dark treatment had predicted functions ranging from RNA metabolism to degradation of organic compounds to stress-related activities (see Fig. S3A in the supplemental material). Two carbon breakdown-associated exoenzymes, including a putative carboxymethylenebutenolidase (CMBase), which may function in chloroaromatic degradation, and a glucose/sorbosone dehydrogenase with a carbohydrate binding domain (CBM2), were significantly overrepresented in the dark treatment (Fig. 5; see also Table S3). Three exoproteins containing the RNA recognition motif (RRM) domain were the most abundant in the dark-treated samples (Fig. 5; see also Table S3). These RRM proteins may play a role in RNA degradation, as the biofilm extracellular RNA level was significantly lower in the continuous dark treatment (Fig. 4A).
The putative CMBase had low (≥25%) sequence identity to enzymes with validated activity, so we performed sequence-to-structure modeling to link the enzyme to its potential activity. A homology-based structural model of the ESFC-1_(A2994) gene product was constructed using the crystal structure of putative dienelactone hydrolase (gene name, ysgA) from Klebsiella pneumoniae (PDB chain 3f67_A) as the structural template. It was experimentally solved at the resolution of 1.74 Å and was identified as the closest PDB template, with 36% sequence identity to A2994. Analysis of the constructed model revealed strong conformational similarities (above 85% as measured by the LGA_S score [30]) to the cluster of CMBases (EC 3.1.1.45; see Fig. S4 in the supplemental material). Additionally, the comparison of our constructed model for the ESFC-1 gene product (A2994) with the crystal structure of a dienelactone hydrolase (PDB 4u2b) from Pseudomonas knackmussii revealed high conservation in residues that are critical for enzyme activity (31) (see Fig. S4B in the supplemental material). Enzymes in this cluster have general carboxyl esterase activity (32), which we detected in biofilm EOM extracts, and we measured significantly higher activity in the dark treatment than in the other light regimes (Fig. 4B). Homologs of this protein exist in more than 200 cyanobacterial genomes, and many have multiple paralogs (IMG portal, http://img.jgi.doe.gov).
The continuous-light-treatment exoprotein pool included proteins with predicted functions in transport, CO2 concentration, and breakdown of carbohydrate, nucleic acid, and protein (see Fig. S3B in the supplemental material). This includes an Fe3+ ABC transporter, as well as an abundant fasciclin domain protein that may be involved in high-affinity CO2 uptake (Fig. 5; see also Table S3). Enzymes that may be involved in polysaccharide modification such as beta-phosphoglucomutase and 1,4-alpha-glucan branching were also overrepresented (see Table S3).
To examine the more complex “total” fraction samples, a second (liquid chromatography-tandem mass spectrometry [LC-MS/MS]) analysis was conducted (“total” analysis). For this analysis, we used iTraq isobaric tags to improve relative quantitation (33). We identified 1,838 proteins with high confidence that matched cyanobacterium ESFC-1 in at least 2 of 5 replicate samples. In the dark treatment, there was notable upregulation of a glycolysis protein, a multidrug transporter, and two stress response proteins (see Fig. S3C and S5 in the supplemental material). In the light treatment, the cyanobacterium upregulated transport of Fe3+, phosphate/phosphonate, and amino acids (see Fig. S3D and Fig. S5).
Heterotroph diversity and protein expression under light, dark, and diel conditions.
As the identities of associated cells (referred to as heterotrophs) in this unicyanobacterial biofilm were unknown, we sequenced the 16S V4 region rRNA from the culture to identify the noncyanobacterial community and to compare levels of microbial diversity between biofilms in light and dark treatments. Four cultures were sequenced, two from a 3-day continuous light treatment and two from a 3-day continuous dark treatment. The communities consisted of 18 operational taxonomic units (OTUs) with abundance above 1% (representing the percentage of total rarefied counts), representing 4 phyla and 6 classes (see Table S4 in the supplemental material). The average relative abundance seen with members of two classes, Flavobacteriia and Gammaproteobacteria, was higher in the 3-day dark treatment than in the light treatment. We determined that the cyanobacterial OTUs in samples treated in the dark were lower in average relative abundance than those in samples treated in the light and that the members of the three other classes, Alphaproteobacteria, Phycisphaerae, and Cytophagia, maintained similar relative abundances between the two treatments (Fig. 6).
FIG 6 .
Community composition at the class level of unicyanobacterial ESFC-1 biofilms after 3 days in either continuous dark or continuous light. Points represent combined OTU percent counts of total rarefied counts: 3 OTUs for Flavobacteriia, 2 OTUs for gammaproteobacteria, 10 OTUs for alphaproteobacteria, and 1 OTU for each ESFC-1, Cytophagia, and Phycisphaerae. Error bars represent 1 standard deviation for comparisons between results from the two replicates.
To identify proteins from the heterotroph community, we used a constructed database with 7 genomes that was based on close hits to abundant OTUs. Since our initial shotgun proteomics analysis did not yield a significant number of exoproteins from 1 of the 6 heterotroph genomes, we examined the “total” analysis. Of the 2,267 proteins identified, 429 matched one of the heterotroph genomes, while the rest were from the cyanobacterial genome (Fig. 7; see also Table S3 in the supplemental material). We refer to these 429 noncyanobacterial proteins as “heterotroph proteins.” Using a 2-sigma significance test to compare relative protein abundance levels between treatments, we identified 81 heterotroph proteins that were enriched in the dark relative to either the light treatment or the diel treatment and 5 heterotroph proteins that were overrepresented in the light treatment.
FIG 7 .
Heterotroph total (intracellular and extracellular) proteome; comparison between light and dark treatments after 3 days. Total proteome relative abundances were determined via the use of iTraq isobaric tags; points represent log2 normalized median-centered averages of results from five biological replicates. (Top panel) Proteins identified using Marinobacter sp. strain ES.048 genome. (Bottom panel) Proteins identified using H. phototrophica genome. Points with black diamond border indicate significant differences in average abundances (>2 standard deviations from mean difference). Colored points indicate assigned functional categories for each protein. The “Other” category includes photosynthesis, stress, motility, cell envelope, replication/recombination, and inorganic ion metabolism. aapJ, amino acid ABC transporter substrate-binding protein; livK, an ABC-type branched-chain amino acid transporter; TRAP_CA, TRAP-type mannitol/chloroaromatic compound transport system; ICL, isocitrate lyase; glcB, malate synthase; nrtA, ABC-type nitrate-nitrite transport system protein; GH74, putative endoglucanase; glpV, ABC-type glycerol transport system; TRAP_C4, TRAP-type C4-dicarboxylate transport system; pufM, photosynthetic reaction center M subunit protein; ggtB, ABC-type alpha-glucoside transport system; ABC.SS, ABC-type simple sugar transport system.
Transport and central C metabolism proteins were the most abundant heterotroph proteins detected, with many of these upregulated in the dark (see Fig. S3E and F and Table S3 in the supplemental material). We detected the highest number of heterotroph proteins (208 of the 429 heterotroph proteins; see Table S3) from one particular group (closest genome match to Marinobacter sp. strain ES.048). This one group represented 7% to 18% of all OTUs. Amino acid transporters were the most abundant Marinobacter proteins and were upregulated in the dark treatment (Fig. 7, top panel). Dicarboxylate transport and metabolism proteins, including formate dehydrogenase, anaerobic selenocysteine dehydrogenase, and glyoxylate cycle proteins, were also abundant, as was a dark treatment-upregulated chloroaromatic transporter (Fig. 7, top panel; see also Table S3).
Other heterotroph proteins detected were associated with alphaproteobacterium Hoeflea phototrophica and with Flavobacterium muricauda ES.050 (Muricauda) (111 and 63 proteins, respectively). H. phototrophica expressed amino acid and dicarboxylate transporters, as well as abundant carbohydrate and sugar transporters and chloroaromatic transporters (Fig. 7, bottom panel). H. phototrophica also expressed an anoxygenic photosynthesis protein, PufM, at similar abundances in the light and dark treatments and upregulated an Fe3+ transport protein in the light treatment relative to the diel treatment (see Table S3 in the supplemental material).
DISCUSSION
Cyanobacteria are globally important primary producers, but the fate of their fixed carbon is complicated by their excretion, degradation, and incorporation of extracellular organic matter (EOM), the rates and pathways of which are not well constrained. In previous work, we found that a filamentous cyanobacterium (ESFC-1) in a biofilm degrades and reincorporates its own extracellular organic carbon (26). This led us to hypothesize that the cyanobacterial biofilm extracellular matrix was, among other things, a storage reservoir for C. To test this, here we compared EOM incorporation, composition, and enzymatic activity in a unicyanobacterial biofilm under continuous light (C replete; DIC through photosynthesis), continuous dark (C starved), and DCMU (oxygenic photosynthesis inhibition) treatment conditions. Our study demonstrated distinct light-dependent cyanobacterial EOM management patterns, with storage of C in the extracellular reservoir, metabolic usage at night, and micronutrient demand driving daytime incorporation (Fig. 8). Our results also suggest that community interactions play a role in EOM cycling.
FIG 8 .
Schematic cycling of EOM in ESFC-1 biofilms following different light regimes. The widths of the arrows represent approximate levels of C or N uptake in micrograms. Orange arrows represent C flux, and blue arrows represent N flux. Dashed arrows indicate processes that were not measured or estimated.
Cyanobacterial nighttime reuse of EOM C stores.
On the basis of previous work, we hypothesized that cyanobacteria would use EOM as an extracellular reserve of C for nighttime fermentation. The results of this study support this hypothesis. From flux estimates, we saw assimilation of C-rich EOM in the dark (Fig. 8), and in continuous darkness, with no other source of C, cyanobacteria showed assimilation of EOM that decreased after 6 h, which we interpret as reflecting progressive consumption of readily available EOM. This is consistent with metabolic shifts observed in natural mats, where nighttime cyanobacterial fermentation leads to sharp increases in levels of organic acids, by-products of fermentation that are thought to be excreted by cyanobacteria and utilized by other community members (34, 35). Marine picocyanobacteria in mixed assemblages also incorporate carbon from their own exudates in the dark (36), confirming that this applies to cyanobacteria in other systems. From our previous work, we know that EOM in natural mats and ESFC-1 biofilms is predominantly composed of alpha- and beta-linked glucose oligosaccharides, with smaller amounts of protein and nucleic acids (26). Our C/N relative assimilation analysis demonstrated that, in the dark, organic C was preferentially incorporated from EOM. This could indicate assimilation of oligosaccharides or assimilation of a specific compound with a higher C/N ratio than was seen with the bulk EOM.
Bulk EPS concentrations and degradation activity also supported the hypothesis that the extracellular space is used for cyanobacterial C storage. Our results indicate that EPS accumulation was light dependent in these biofilms, with high levels in the daytime and low levels at night. This is consistent with what has been observed in a benthic diatom, Cylindrotheca closterium (37), where EPS levels decreased in the dark. The increase of EPS levels in the light is also suggestive of a C-sink into exopolysaccharides, as occurs in species of Nostoc (9). Notably, extracellular proteolytic activity and carboxyl esterase activity were detectable in the light and the dark but were higher in the dark, representing a potential mechanism for usage of the extracellular stores when necessary through breakdown and uptake of degraded products.
Further evidence for this breakdown mechanism could be seen in the cyanobacterial dark-induced exoproteome. Dark-treatment-upregulated exoproteins included enzymes that may function in organic C breakdown and acquisition, which is consistent with both the C-rich incorporation data and the decreased levels of carbohydrate and exopolysaccharides in the dark treatment. Of particular interest was an abundant carboxymethylenebutenolidase (CMBase), which, according to our structural modeling, could be the main source of the observed increase in carboxyl esterase activity in the dark and may function in chloroaromatic breakdown (38). Cyanobacterial exoproteomes have been characterized for Nostoc punctiforme PCC 73102, Anabaena sp. strain PCC 7120, Synechocystis sp. strain PCC 6803, and eight marine Synechococcus strains, but CMBase was not detected in these exoproteomes (14, 39–41). However, this enzyme was detected in exoproteomes from Elkhorn Slough microbial mats (26), the ecosystem from which our cyanobacterial strain originated. Interestingly, intact mats have the ability to degrade 2,4-dichlorophenoxyacetic acid and respond to increased levels of this substrate with increased degradation (42). In our cyanobacterial biofilms, it may have a more general function in restructuring or breaking down EOM by-products.
Daytime cyanobacterial EOM reuse for DIC and micronutrient acquisition.
In these experiments, the cyanobacteria assimilated higher and steadily increasing levels of isotope from the EOM during light periods in comparison to dark periods (Fig. 2). These results are in part explained by the fact that daytime data reflect both DIC and organic C uptake. We can disentangle these two processes with the results of our DCMU photosynthesis inhibition experiment. On the basis of those results, we estimate that at 6 h, approximately 50% of cyanobacterial 13C incorporation was from organic C and 50% was from DIC (Fig. 8). Using the rate of carbon fixation from our 13C-DIC experiments, we estimate that approximately 9% of C in the cyanobacteria was from remineralized EOM incorporation at 6 h (Fig. 8).
Past 12 h, the relative uptake of C from the EOM increased with continuous light in comparison to the N results (Fig. 2). We interpret this result as reflecting an increasing proportion of fixed DIC coming from the added EOM following prolonged light exposure, but since the continuation of DCMU experiments past 6 h is not feasible because of cell death, we cannot confirm this. Prolonged light conditions did increase the pH in the spent media (see Table S2 in the supplemental material) and led to an increase of levels of CO2-concentrating exoproteins, which may indicate lower availability of DIC. This suggests that cyanobacteria may preferentially acquire locally remineralized EOM in response to prolonged light exposure.
We estimate that the use of EOM in the other 50% of cyanobacteria under daytime conditions takes place by direct uptake of organic matter (Fig. 8). We hypothesize that the reason that the cyanobacteria take up this organic matter when they are fixing C is that it provides other required nutrients, such as N, P, and Fe, and that the uptake of C, for example, by organic siderophores, is incidental (43). On the basis of the photosynthesis inhibition experiment, this organic matter is N rich, contrasting with the C-rich nighttime incorporation (Fig. 8), which supports this hypothesis. We estimate that EOM was a substantial source of N for cyanobacteria in the light treatment, representing 10% of N assimilation. Uptake and assimilation of nitrate, which was provided in excess, have additional metabolic costs (44, 45), so alternative nitrogen sources (ammonium, amino acids) are often preferred. This hypothesis is further supported by evidence of increased capacity for nutrient uptake in the light. For example, ferric iron transport, phosphate/phosphonate transport, and amino acid transport were all upregulated in the light treatment, suggesting that there may have been increased demands for multiple nutrients, many of which could be counteracted through reuse of EOM. The higher abundance of exoenzymes involved in breakdown of nucleic acids and proteins, perhaps related to acquisition of phosphate (46) and N (47), supports the hypothesis that light-induced organic C incorporation is driven by a need for other micronutrients.
Resource partitioning and interactions with heterotrophs.
Our data provide evidence for both competition and cooperation between heterotrophs and cyanobacteria. Although they constituted only 4% of the biomass, heterotrophs accounted for more than half the C and N uptake from the EOM (Fig. 8). Although we did not determine the compounds used by the two groups, it is clear from our flux estimates that they are competitors for the cyanobacteria-derived EOM (Fig. 8). In accordance with the analysis described in the previous section, the two groups were incorporating EOM with similar C/N ratios, indicating that they both target the nutrients and also express amino acid transporters at high abundances. A form of cooperation is indicated by the EOM-derived DIC used by the cyanobacteria, which likely originates from respiration by the heterotrophs, as suggested for heterotrophs associated with Anabaena (see, for example, reference 48). This highlights the multifactorial nature of these interactions.
We also observed changes in the heterotroph community and in the abundances of cyanobacteria relative to heterotrophs with 3 days of darkness compared to continuous light. This change in the heterotroph community was primarily due to the members of two dominant classes, Gammaproteobacteria and Flavobacteria (Fig. 6). There is evidence in diatoms of C substrate exchange for Fe from associated Marinobacter species (49), as well as in targeted feeding of sulfonate compounds to a marine Roseobacter in exchange for vitamin B12 (50), both secretion mechanisms by which the primary producers can control heterotroph populations. Due to their close spatial relationships with other community members, mat cyanobacteria may be particularly suited to controlling the availability of extracellular compounds to heterotrophs, through uptake as well as secretion. Recent work has shown excretion of a much wider range of metabolites by a mat cyanobacterium, Microcoleus vaginatus, than by an aquatic cyanobacterium, Synechocystis sp. strain PCC 6803, and reuptake by Microcoleus of a large number of those metabolites (51), suggesting that mat organisms may tailor their immediate extracellular environment. In addition to the hypothesis that cyanobacteria outcompete the heterotrophs for certain compounds, they could also be excreting allelopathic compounds, an activity which would be discontinued in the dark due to C limitation.
Heterotroph metaproteome expression.
By comparing proteomes from several groups of heterotrophs, we found that the heterotrophs may have distinct growth strategies for using cyanobacterial products that allow them to circumvent heterotroph-heterotroph competition and to respond to the changing light regimes. The proteomic response from our representative gammaproteobacterium (Marinobacter sp. strain ES.048) suggested a specialist strategy, utilizing amino acids and simple carbon compounds through the glyoxylate pathway. Experimental evidence and genomic predictions support these specializations for this group (47, 52). Marinobacter also upregulated sugar alcohol and chloroaromatic transport proteins in the dark, suggesting that this group may take advantage of cyanobacterial dark extracellular degradation, given the upregulation of extracellular cyanobacterial CMBase in the dark treatment. In contrast, the proteomic response from our representative Rhizobiales genome, H. phototrophica, indicates a more generalist strategy that includes incorporation of a wide range of compounds, supplemented by anoxygenic photosynthesis. For example, H. phototrophica expressed transporter proteins responsible for uptake of carbohydrates, sugars, osmolytes, organic acids, and citrate, which were not detected in Marinobacter, as well as nitrate, amino acid, dicarboxylate transport, and chloroaromatics, which were found in both classes. The H. phototrophica genome also has a full set of genes for anoxygenic photosynthesis (53), and at least one of these photosynthesis proteins was expressed. This may explain why this group did not increase its relative abundance in the dark treatment, since some members may rely on energy from light. Work on other unicyanobacterial consortia with similar levels of heterotroph taxonomic diversity shows successional species composition over biofilm development (54), and this may be related to the availability of extracellular resources. Our work builds upon those findings by describing proteomic responses of some of these groups to changing resources, which we controlled by extended incubations in treatments using continuous light and continuous darkness. The differences in responses between groups provide insight into the partitioning of extracellular resources in this community.
Conclusions.
In summary, cyanobacteria not only produce EOM but also use it directly both in the light and in the dark (Fig. 8). It is likely that cyanobacteria also benefit from the remineralization of EOM by heterotrophic bacteria during periods of photosynthesis. Our biofilm cyanobacterium accounted for 43% of the total EOM C assimilation in the dark and appeared to metabolize extracellular resources under C-starved conditions. On the basis of the proteome and the C/N content of the EOM incorporated by cyanobacteria in the light treatment, it seems likely that light-driven incorporation occurs primarily in acquisition of micronutrients. Furthermore, cyanobacteria produce degradation exoenzymes and the heterotrophs respond with upregulation of transporters, suggesting the presence of a dynamic extracellular pool requiring specialized transport. While primary producer excretions are acknowledged to influence microbial organic matter cycling in many systems, the redirection of complex organic carbon back to the primary producer is not nearly as well documented. This redirection has the potential to substantially influence EOM composition and remineralization rates, and our data suggest that determining primary producer metabolic status is critical for a complete understanding of organic matter turnover in these systems.
MATERIALS AND METHODS
Cell culture and extracellular separation.
Unicyanobacterial cultures of the cyanobacterium ESFC-1 were grown as described by Stuart et al. (26) in artificial seawater media (ASN) (27, 55) at 23°C with 20 µmol m−2 s−1 (4.16 W m−2) light on a 12-h/12-h light/dark cycle. Extraction of EOM from the biofilms was carried out as described by Stuart et al. (26). Briefly, biofilms were weighed and homogenized on ice in sterile 10% NaCl (Wheaton Dounce homogenizer; pestle clearance, 0.114 ± 0.025 mm). Homogenate was incubated at 40°C for 15 min and centrifuged at 15,000 × g at 4°C for 20 min and supernatant filtered through a 0.2-µm-pore-size polycarbonate filter, to separate out cells, forming the extracellular (or “EOM”) fraction.
13C-labeled and 15N-labeled extracellular organic fraction.
13C-labeled and 15N-labeled EOM was generated as described by Stuart et al. (26), excepting that 1.76 mM K15NO3 (Sigma-Aldrich Co., St. Louis, MO, USA) (98 atom percent 15N) was substituted in ASN, along with 2 mM 13C sodium bicarbonate (Cambridge Isotopes, Tewksbury, MA, USA) (13C; 99%). Bulk isotope ratios for 13C/12C and 15N/14N of a sample of 13C-labeled and 15N-labeled substrate were determined by automated nitrogen and carbon analysis-IRMS (ANCA-IRMS) (PDZE Europa Limited, Crewe, England) at the University of California, Berkeley.
Reuptake of EOM.
To quantify differences in uptake of extracellular material in the cyanobacterium and heterotrophs under various light conditions, replicate unicyanobacterial biofilms were grown as a biofilm for 1 month in 13 6-well plates as described by Stuart et al. (26). After 20 days of growth, a total of six treatments were applied: “killed,” “no label,” “dark,” “diel,” “light,” and “DCMU.” “Killed” controls were generated by addition of 4% formaldehyde to wells and incubation overnight before the experiment start. At the start of the light phase of the 12:12 diel incubation cycle, medium was then removed from all cultures and 3 ml of 13C-labeled and 15N-labeled EOM (described above) diluted 1:30 in fresh sterile medium was added to each well. For “no-label” control, only fresh sterile medium was added. For “dark” treatments, 4 plates were wrapped in 2 layers of tin foil and incubated with the 4 “diel” plates under conditions of 12-h/12-h light/dark. The cultures receiving the “light” treatment were moved to a constant-light incubator from the 12-h time period to the 24-h time period following the addition. For the “DCMU” treatment, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) was added at 20 µM along with the labeled substrate addition and the plate was incubated in the light. At 2, 6, and 12 and 24 h, three replicate cultures from each of the dark and diel treatments were washed with sterile unlabeled ASN, incubated with 4% formaldehyde for 1 to 2 h, rinsed with 1× phosphate-buffered saline (PBS), and frozen at −20°C in 50% ethanol. At the 6-h time point, killed, no-label, and DCMU control cultures were also fixed as described above. The cultures that received light treatment were also fixed at the 24-h time point as described above. To prepare these samples for NanoSIMS analysis, subsamples were removed, homogenized gently to separate trichomes, and rinsed with sterile MilliQ water three times to remove unincorporated label. A 0.5-µl volume was then spotted onto a silica wafer, air dried, and stored in an argon dry box.
Parallel 13C bicarbonate additions (2 mM) were run for 6 h on 9 replicate biofilms, using 3 replicates each for a dark incubation, a lighted incubation, and a killed control. Biofilms were fixed as described above and bulk isotope ratios determined by IRMS as described above.
NanoSIMS isotope imaging and scanning electron microscopy (SEM).
Samples were prepared as described by Stuart et al. (26). Briefly, wafers were coated with ~5 nm of gold, and secondary ion mass spectrometry (SIMS) imaging was performed with a Cameca high-resolution imaging SIMS (NanoSIMS) 50 microprobe at Lawrence Livermore National Laboratory. A focused primary ion beam (2 pA, approximately 150 nm, 16 keV 133Cs+) was used in a raster pattern with 15-by-15 µm2 to 25-by-25 µm2 analysis areas of 256-by-256 pixels and a dwell time of 1 ms/pixel for 19 to 30 scans (cycles). Before analysis, samples were presputtered with 90 pA of Cs+ current (equivalent to approximately 50 nm) to reach sputtering equilibrium and to make sure that the isotope analysis targeted intracellular material rather than the surface of the cells. Serial quantitative secondary ion images (maps) were simultaneously collected for 12C2−, 13C12C−, 12C14N−, and 12C15N− using electron multipliers in pulse counting mode. Previous work indicated that the C dimer (e.g., 12C12C) provides yield superior to that of the 12C monomer (56). Secondary electrons were also simultaneously collected as previously described (56).
NanoSIMS ion image data were processed as described by Woebken et al. (27). For each raster, quantitative ion ratio images were generated from the summed ion images to generate 13C and 15N enrichment images, presented in values corresponding to atom percent excess (APE) (56, 57). Regions of interest (ROIs) for quantification of isotopic ratios were selected on the basis of secondary electron images, 13C enrichment images, and 12C14N− ion images, which allowed cells to be specifically selected and hot spots of residual labeled substrate to be excluded. Isotopic ratios were extracted per cycle and averaged.
Biovolume, net fixation, and C/N substrate usage calculations.
Bacterial counts and biovolume measurements were done for all biofilm samples analyzed with NanoSIMS, as described by Stuart et al. (26). Briefly, biofilms were imaged, images were analyzed to calculate biovolume, and C content was estimated using biovolume-to-carbon conversion factors from the literature (2.2 × 10−13 g C µm−3 for other bacteria and 1.8 × 10−13 g C µm−3 for filamentous diazotrophic cyanobacteria) (58, 59). Cyanobacterial biovolumes within the biofilms were not significantly different between treatments (5.57 × 108 µm−3/well ± 1.55 × 108 µm−3/well, 5.61 × 108 µm−3/well ± 1.55 × 108 µm−3/well, and 5.95 × 108 µm−3/well ± 3.29 × 108 µm−3/well for 24-h dark treatment, light treatment, and killed control, respectively). N content was estimated based on C/N ratios determined via elemental analysis of biomass coupled with IRMS analysis and determined from the literature for marine bacteria (C/N of 4.0 for cyanobacteria and 4.4 for heterotrophs) (60). C and N content was extrapolated to micrograms of C or N per well for the 6-well plates used. 13C and 15N APE values were compared between treatments and time points by use of one-way analysis of variance (ANOVA) and Dunnett’s post hoc test for comparison with killed controls and Tukey’s post hoc test for comparison between time points within a given treatment. Net fixation of 13C and 15N (percentage of C or N incorporated relative to initial C or N content) (28) was calculated for both heterotrophs and cyanobacteria and extrapolated to calculate micrograms of C or N fixation per well. C/N relative use efficiency data were calculated as described by Mayali et al. (28), and mean use efficiencies were compared to a theoretical mean of 1.0 using a t test.
Bulk composition and enzyme activity assays and oxygen measurements.
Batch cultures were inoculated into 300 ml of ASN in acid-washed 1-liter glass flasks and grown as described by Stuart et al. (26). At 4 weeks, at the onset of the light phase of the diel cycle, 3 treatments (dark, light, and diel treatments) were initiated in 3 replicate flasks (9 total). Dark-treated biofilms were incubated in the dark for 3 days, light-treated biofilms were incubated under constant illumination for 3 days, and diel-treated biofilms were incubated using a 12-h/12-h light/dark cycle. After 3 days of treatment, all biofilms were harvested and the extracellular fraction was separated as described above. Carbohydrate concentration was determined using the traditional phenol sulfuric acid method (61). Nucleic acid concentrations were measured using PicoGreen and RiboGreen dyes for DNA and RNA, respectively, according to instructions from the manufacturer (Life Technologies, Carlsbad, CA). Protein concentrations were determined using a Bradford assay (Bio-Rad, Hercules, CA) and absorbance at 280 nm. All concentrations were normalized to biomass wet weight.
Enzyme activities were assayed in the extracellular fraction following the protocol established by Bell et al. (62) and described by Stuart et al. (26). Briefly, diluted extracellular fraction samples were combined with a fluorescent substrate. l-Leucine-7-amido-4-methylcoumarin hydrochloride, 4-methylumbelliferyl α-d-glucopyranoside, 4-methylumbelliferyl β-d-glucopyranoside (Sigma Aldrich, St. Louis, MO, USA), and 4-methylumbelliferyl caprylate (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were the substrates for peptidase, α-glucosidase, β-glucosidase, and carboxyl esterase activity, respectively. Controls included each sample with water and sterile 10% NaCl (substrate control) instead of sample. Plates were read every 5 min for 3 h at 380/440 nm.
Congo red staining was performed on biofilms grown for 5 days on glass chamber slides with 3 ml of ASN. Triplicate biofilms were then incubated under dark, diel, or full-light conditions for 3 days and fixed with 4% formaldehyde at 4°C. Biofilms were rinsed with sterile water, stained with DAPI (4′,6-diamidino-2-phenylindole) for 10 min at 23°C (10 µg/ml), rinsed again, and then stained with Congo red (1 mg/ml) for 15 min at 23°C. Biofilms were then rinsed with 1 M NaCl (added to remove excess stain). A total of 10 fields of view for each biofilm were imaged in 9-µm z-stacks in both the Cy3 and DAPI channels at 100% power using a 150-ms exposure. Images were then combined into a maximum projection image, and cyanobacterial filaments were manually outlined. Fluorescence intensity was then quantified, excluding the filaments. Mean intensities were compared using one-way ANOVA and Tukey post hoc tests.
Oxygen measurements were performed on biofilms grown in 6-well plates as described above with triplicate wells for three treatments: light, dark, and diel. After 3 days of treatment, oxygen was measured at the surface of each biofilm using an OXR50 optical oxygen sensor coupled to a PyroScience FireSting optical oxygen meter (Pyro Science GmbH, Aachen, Germany).
16S iTag sequencing.
Illumina MiSeq sequencing was done at Laragen (Culver City, CA) using primers for amplification of the 16S V4 region (obtained from Caporaso et al. [63]). Analysis details are described in Text S1 in the supplemental material.
LC-MS/MS metaproteomics.
Batch cultures were grown and the extracellular “EOM” fraction was extracted as described above, with 5 replicates per treatment (total, 15 flasks). Additionally, following homogenization, a 1/10 vol of the homogenate from each biofilm was separated, sonicated to lyse cells (Misonix, Farmingdale, NY; 50% intensity, 6 cycles of 30 s on and 2 min off on ice), and centrifuged at 12,000 × g at 4°C for 10 min. The supernatant constituted the “total” fraction and included both intracellular and extracellular proteins. This resulted in 30 total samples for proteomic analyses. Run details are described in Text S1 in the supplemental material.
Metaproteomic analyses.
The MSGF+ search algorithm was used to match the spectra to peptide sequences (64) derived from the ESFC-1 genome (61) or a constructed database as described below, each amended with common contaminant sequences (keratins, trypsin, serum albumins, etc.); the search parameters included partially tryptic cleavages, dynamic modification of methionine oxidation, and a ± 20 ppm parent ion mass tolerance.
To identify proteins expressed by the heterotrophic population, we identified genomes with high BLASTn identity scores to abundant OTUs from our iTag data and constructed a search database to query the spectra of the iTraq isobaric tags. The gammaproteobacterial OTU with the highest abundance matched an Elkhorn Slough microbial mat isolate, Marinobacter sp. strain ES.048. For the 10 OTUs in the alphaproteobacterial class, we chose two genomes that had the highest identity (100% and 99% at the 16S rRNA gene level) to 2 alphaproteobacterial OTUs, from the Rhizobiales (H. phototrophica DFL-43) and Rhodobacter (Tropicibacter multivorans DSM 26470) orders, respectively. The Flavobacteriia class was represented by 3 OTUs, and the OTU with the highest abundance did not match any database genome 16S sequence above 90% identity, so we used the next-most-abundant OTU, which matched a bacterium isolated from Elkhorn Slough, Muricauda sp. strain ES.050, with 100% identity. The Phycisphaerae and Cytophagia classes were represented by 1 OTU each, so we selected the most closely related genomes, Tepidiphilus thermophilus JCM 19170 and Algoriphagus terrigena DSM 22685, which had 81% identity and 94% identity to their respective OTUs. We constructed a database using these 6 genomes and the ESFC-1 genome and used it for comparisons to our proteomics samples.
Reporter ion abundance values were extracted using MASIC (65), and their respective sequences were aggregated using an in-house processing pipeline, including the APE software package (http://omics.pnl.gov/software/ape-mdart) with a 1% false discovery rate (FDR) cutoff (MSGF + Q-value ≤ 0.01). Peptide data were combined to protein levels using summed abundances, which were subsequently log2 normalized and zero centered. The significance of data corresponding to protein abundances across treatments was determined based on a 2 sigma deviation (P value, ≤0.05). Cutoffs of at least 2 unique peptides and presence in at least three biological replicates in a given treatment were applied.
Spectral counts were used to generate normalized spectral abundance factors (NSAF) (66), generating relative percent abundances, normalized to protein length for each protein identified. For proteins that were not detected in some of the replicates, we substituted a value of one-quarter of the lowest NSAF calculated (67). We then generated average abundances for proteins found in at least 3 biological replicates and at least 4 of the 10 total replicates (5 biological and 2 technical for each treatment). We compared abundances of proteins between treatments using a Student’s t test. Localization predictions were made using MetaLocGramN (68) and SecretomeP (69) for nonclassical secretion prediction. CAZy enzymes were predicted using dbCAN (70).
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
13C and 15N enrichment of cells analyzed via NanoSIMS. Each point represents 13C and 15N atom percent excess (APE) for a single trichome (solid) or bacterial cell (outlined). The dotted line indicates the ratio of the levels of 13C-labeled and 15N-labeled EOM-substrate, as determined by IRMS. Download
Exopolysaccharide stain Congo red in biofilms grown under different light regimes for 3 days. (A and B) Representative epifluorescent microscopy images of Congo red-stained continuous-light-treated biofilm (A) and continuous-dark-treated biofilm (B). The white scale bar represents 10 µm. The blue coloring is from DAPI DNA staining, and the red coloring is from both autofluorescence (inside filaments) and Congo red stain (extracellular). Yellow outlines indicate lines manually drawn to exclude cyanobacterial cells from analysis of Congo red intensities. (C) Average extracellular Congo red intensities under different light regimes for 3 days. Error bars represent 1 standard deviation of results of comparisons between 3 biological replicates (10 fields of view per replicate analyzed). *, significant difference in means (P < 0.05). Download
Functional predictions for proteins significantly overrepresented in either continuous dark (panels A and C and panel E) or continuous light (panels B and D and panel F) treatments. (A and B) The top panels show functional categories for ESFC-1 exoproteins overrepresented in either continuous dark (A) or continuous light (B) treatments. (C and D) Middle panels show functional categories for ESFC-1 total proteome proteins overrepresented in either continuous dark (C) or continuous light (D) treatments. (E and F) The bottom panels show functional categories for total proteome heterotroph proteins overrepresented in the continuous dark treatment from representative gammaproteobacterium Marinobacter sp. strain ES.048 (E) or representative alphaproteobacterium H. phototrophica (F). Significantly overrepresented, the abundance of the proteins was higher that seen with either the diel treatment or the light or dark treatment (P < 0.05). Download
Sequence-structure-based analyses of the ESFC-1 CMBase. (A) StralCP dendrogram and clusters for proteins identified in PDB with predicted structural similarity to A2994. Proteins with known enzyme activity have the EC number listed together with corresponding PDB chain identifier (ID) labels. (B) List of residue-residue correspondences between the structural model of A2994 and ClcD from Pseudomonas knackmussii (PDB chain 4u2b_A; resolution, 1.70 Å; sequence identity, 26%) in critical functional positions. Catalytic triad residues are colored in red. Mutation positions that improve activity toward substrates are in green. In blue are colored positions where mutations increase substrate accessibility during catalysis by increasing flexibility of the loop containing catalytic triad histidine. The distances between corresponding Cα atoms from the calculated structure superposition are displayed in the last column. Positions marked by an asterisk (*) denote perfect agreement between residues from A2994 and native or mutated residues from 4u2bA. (C) Cartoon representation of the structural superposition of A2994 model (gray) with the crystal structure of ClcD dienelactone hydrolase (yellow). Side-chain conformations of corresponding residues identified as functionally critical are presented in “ball and stick” form using the same coloring scheme as in the table in panel B and numbering from A2994. Download
Cyanobacteria total (intracellular and extracellular) proteome differences between light and dark treatments after 3 days. Total proteome relative abundances were determined via the use of iTraq isobaric tags, and points represent log2 normalized median-centered averages of results from five biological replicates. Colored points represent proteins with significant differences in average abundance (>2 standard deviations from mean difference). pfkA, 6-phosphofructokinase; ABC.2A, ABC-type multidrug transport system; glnB, nitrogen regulatory protein P-II; AfuA, ABC-type Fe3+ transport system; CphX, CO2 hydration protein; phnD, ABC-type phosphate/phosphonate transport system; petJ, cytochrome c6; ABC.PA, ABC-type polar amino acid transport system. Download
Number of cells analyzed via NanoSIMS.
O2 and pH measurements.
Proteomics results. Data represent exoproteins and total proteins identified in cyanobacteria and heterotrophs under all experimental conditions.
Relative abundances of bacterial taxa based on 16S iTag sequencing in biofilms treated in the light and the dark.
ACKNOWLEDGMENTS
We thank Heather Dang (UC Berkeley) for IRMS analysis, Leslie Prufert-Bebout (NASA Ames Research Center) for isolating and providing the ESFC-1 strain, and Tijana Glavina del Rio and the DOE Joint Genome Institute (JGI) staff for sequencing and bioinformatics support (as part of JGI Community Sequencing Project 701).
Funding Statement
Funding was provided by the DOE Genomic Science Program under contract SCW1039. Work at Lawrence Livermore National Laboratory was performed under the auspices of DOE contract DE-AC52-07NA27344. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Stuart RK, Mayali X, Boaro AA, Zemla A, Everroad RC, Nilson D, Weber PK, Lipton M, Bebout BM, Pett-Ridge J, Thelen MP. 2016. Light regimes shape utilization of extracellular organic C and N in a cyanobacterial biofilm. mBio 7(3):e00650-16. doi:10.1128/mBio.00650-16.
REFERENCES
- 1.Helm RF, Potts M. 2012. Extracellular matrix (ECM), p 461–480. In Ecology of cyanobacteria II. Springer Verlag, Berlin, Germany. [Google Scholar]
- 2.Pomeroy L, Williams PJ, Azam F, Hobbie J. 2007. The microbial loop. Oceanography 20:28–33. doi: 10.5670/oceanog.2007.45. [DOI] [Google Scholar]
- 3.Fogg GE. 1983. The ecological significance of extracellular products of phytoplankton photosynthesis. Bot Marina 26:3–14. doi: 10.1515/botm.1983.26.1.3. [DOI] [Google Scholar]
- 4.Hagemann M. 2011. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 35:87–123. doi: 10.1111/j.1574-6976.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- 5.Stal L, Moezelaar R. 1997. Fermentation in cyanobacteria. FEMS Microbiol Rev 21:179–211. doi: 10.1016/S0168-6445(97)00056-9. [DOI] [Google Scholar]
- 6.Pereira S, Zille A, Micheletti E, Moradas-Ferreira P, De Philippis R, Tamagnini P. 2009. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol Rev 33:917–941. doi: 10.1111/j.1574-6976.2009.00183.x. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Pichel F, Castenholz RW. 1991. Characterization and biological implications of scytonemin, a cyanobacterial sheath pigment. J Phycol 27:395–409. doi: 10.1111/j.0022-3646.1991.00395.x. [DOI] [Google Scholar]
- 8.Foster RA, Sztejrenszus S, Kuypers MM, Raven J. 2013. Measuring carbon and N2 fixation in field populations of colonial and free-living unicellular cyanobacteria using nanometer-scale secondary ion mass spectrometry. J Phycol 49:502–516. doi: 10.1111/jpy.12057. [DOI] [PubMed] [Google Scholar]
- 9.Otero A, Vincenzini M. 2004. Nostoc (Cyanophyceae) goes nude: extracellular polysaccharides serve as a sink for reducing power under unbalanced C/N metabolism. J Phycol 40:74–81. doi: 10.1111/j.0022-3646.2003.03-067.x. [DOI] [Google Scholar]
- 10.Paz-Yepes J, Brahamsha B, Palenik B. 2013. Role of a microcin-C-like biosynthetic gene cluster in allelopathic interactions in marine Synechococcus. Proc Natl Acad Sci U S A 110:12030–12035. doi: 10.1073/pnas.1306260110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Block MA, Grossman AR. 1988. Identification and purification of a derepressible alkaline phosphatase from Anacystis nidulans R2. Plant Physiol 86:1179–1184. doi: 10.1104/pp.86.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stihl A, Sommer U, Post AF. 2001. Alkaline phosphatase activities among populations of the colony-forming diazotrophic cyanobacterium Trichodesmium spp. (Cyanobacteria) in the Red Sea. J Phycol 37:310–317. doi: 10.1046/j.1529-8817.2001.037002310.x. [DOI] [Google Scholar]
- 13.Wolk CP, Kraus J. 1982. Two approaches to obtaining low, extracellular deoxyribonuclease activity in cultures of heterocyst-forming cyanobacteria. Arch Microbiol 131:302–307. doi: 10.1007/BF00411176. [DOI] [Google Scholar]
- 14.Christie-Oleza JA, Armengaud J, Guerin P, Scanlan DJ. 2015. Functional distinctness in the exoproteomes of marine Synechococcus. Environ Microbiol 17:3781–3794. doi: 10.1111/1462-2920.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyhrman ST, Chappell PD, Haley ST, Moffett JW, Orchard ED, Waterbury JB, Webb EA. 2006. Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439:68–71. doi: 10.1038/nature04203. [DOI] [PubMed] [Google Scholar]
- 16.Church M, Ducklow H, Letelier R, Karl D. 2006. Temporal and vertical dynamics in picoplankton photoheterotrophic production in the subtropical North Pacific Ocean. Aquat Microb Ecol 45:41–53. doi: 10.3354/ame045041. [DOI] [Google Scholar]
- 17.Mary I, Garczarek L, Tarran GA, Kolowrat C, Terry MJ, Scanlan DJ, Burkill PH, Zubkov MV. 2008. Diel rhythmicity in amino acid uptake by Prochlorococcus. Environ Microbiol 10:2124–2131. doi: 10.1111/j.1462-2920.2008.01633.x. [DOI] [PubMed] [Google Scholar]
- 18.Paerl HW, Bebout BM, Joye SB, Des Marais DJ. 1993. Microscale characterization of dissolved organic-matter production and uptake in marine microbial mat communities. Limnol Oceanogr 38:1150–1161. doi: 10.4319/lo.1993.38.6.1150. [DOI] [PubMed] [Google Scholar]
- 19.Gómez-Baena G, López-Lozano A, Gil-Martínez J, Lucena JM, Diez J, Candau P, García-Fernández JM. 2008. Glucose uptake and its effect on gene expression in Prochlorococcus. PLoS One 3:e3416. doi: 10.1371/journal.pone.0003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz-González C, Simó R, Vila-Costa M, Sommaruga R, Gasol JM. 2012. Sunlight modulates the relative importance of heterotrophic bacteria and picophytoplankton in DMSP-sulphur uptake. ISME J 6:650–659. doi: 10.1038/ismej.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paerl HW. 1991. Ecophysiological and trophic implications of light-stimulated amino acid utilization in marine picoplankton. Appl Environ Microbiol 57:473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zubkov MV. 2009. Photoheterotrophy in marine prokaryotes. J Plankton Res 31:933–938. doi: 10.1093/plankt/fbp043. [DOI] [Google Scholar]
- 23.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 24.Revsbech NP, Ward DM. 1984. Microelectrode studies of interstitial water chemistry and photosynthetic activity in a hot spring microbial mat. Appl Environ Microbiol 48:270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decho AW, Visscher PT, Reid RP. 2005. Production and cycling of natural microbial exopolymers (EPS) within a marine stromatolite. Palaeogeogr Palaeoclimatol Palaeoecol 219:71–86. doi: 10.1016/j.palaeo.2004.10.015. [DOI] [Google Scholar]
- 26.Stuart RK, Mayali X, Lee JZ, Craig Everroad R, Hwang M, Bebout BM, Weber PK, Pett-Ridge J, Thelen MP. 2016. Cyanobacterial reuse of extracellular organic carbon in microbial mats. ISME J 10:1240–1251. doi: 10.1038/ismej.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woebken D, Burow LC, Prufert-Bebout L, Bebout BM, Hoehler TM, Pett-Ridge J, Spormann AM, Weber PK, Singer SW. 2012. Identification of a novel cyanobacterial group as active diazotrophs in a coastal microbial mat using NanoSIMS analysis. ISME J 6:1427–1439. doi: 10.1038/ismej.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayali X, Weber PK, Pett-Ridge J. 2013. Taxon-specific C/N relative use efficiency for amino acids in an estuarine community. FEMS Microbiol Ecol 83:402–412. doi: 10.1111/j.1574-6941.12000.x. [DOI] [PubMed] [Google Scholar]
- 29.Everroad RC, Woebken D, Singer SW, Burow LC, Kyrpides N, Woyke T, Goodwin L, Detweiler A, Prufert-Bebout L, Pett-Ridge J. 2013. Draft genome sequence of an oscillatorian cyanobacterium, strain ESFC-1. Genome Announc 1:e00527. doi: 10.1128/genomeA.00527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zemla A, Geisbrecht B, Smith J, Lam M, Kirkpatrick B, Wagner M, Slezak T, Zhou CE. 2007. STRALCP—structure alignment-based clustering of proteins. Nucleic Acids Res 35:e150. doi: 10.1093/nar/gkm1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porter JL, Boon PL, Murray TP, Huber T, Collyer CA, Ollis DL. 2015. Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search. ACS Chem Biol 10:611–621. doi: 10.1021/cb500809f. [DOI] [PubMed] [Google Scholar]
- 32.Park YJ, Yoon SJ, Lee HB. 2010. A novel dienelactone hydrolase from the thermoacidophilic archaeon Sulfolobus solfataricus P1: purification, characterization, and expression. Biochim Biophys Acta 1800:1164–1172. doi: 10.1016/j.bbagen.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Zieske LR. 2006. A perspective on the use of iTRAQ™ reagent technology for protein complex and profiling studies. J Exp Bot 57:1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 34.Burow LC, Woebken D, Marshall IP, Lindquist EA, Bebout BM, Prufert-Bebout L, Hoehler TM, Tringe SG, Pett-Ridge J, Weber PK, Spormann AM, Singer SW. 2013. Anoxic carbon flux in photosynthetic microbial mats as revealed by metatranscriptomics. ISME J 7:817–829. doi: 10.1038/ismej.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JZ, Burow LC, Woebken D, Everroad RC, Kubo MD, Spormann AM, Weber PK, Pett-Ridge J, Bebout BM, Hoehler TM. 2014. Fermentation couples Chloroflexi and sulfate-reducing bacteria to Cyanobacteria in hypersaline microbial mats. Front Microbiol 5:61. doi: 10.3389/fmicb.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson CE, Carlson CA. 2012. Tracking differential incorporation of dissolved organic carbon types among diverse lineages of Sargasso Sea bacterioplankton. Environ Microbiol 14:1500–1516. doi: 10.1111/j.1462-2920.2012.02738.x. [DOI] [PubMed] [Google Scholar]
- 37.De Brouwer J, Wolfstein K, Stal L. 2002. Physical characterization and diel dynamics of different fractions of extracellular polysaccharides in an axenic culture of a benthic diatom. Eur J Phycol 37:37–44. doi: 10.1017/S0967026201003419. [DOI] [Google Scholar]
- 38.Schmidt E, Knackmuss HJ. 1980. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem J 192:339–347. doi: 10.1042/bj1920339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira P, Martins NM, Santos M, Couto NA, Wright PC, Tamagnini P. 2015. The Anabaena sp. PCC 7120 exoproteome: taking a peek outside the box. Life (Basel) 5:130–163. doi: 10.3390/life5010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vilhauer L, Jervis J, Ray WK, Helm RF. 2014. The exo-proteome and exo-metabolome of Nostoc punctiforme (Cyanobacteria) in the presence and absence of nitrate. Arch Microbiol 196:357–367. doi: 10.1007/s00203-014-0974-2. [DOI] [PubMed] [Google Scholar]
- 41.Gao L, Huang X, Ge H, Zhang Y, Kang Y. 2014. Profiling and compositional analysis of the exoproteome of Synechocystis sp. PCC 6803. J Metabolomics Syst Biol 1:8. [Google Scholar]
- 42.Hogan ME, Ward BB. 1998. Response of a marine sediment microbial community exposed to 2,4-dichlorophenoxyacetic acid. Microb Ecol 35:72–82. [DOI] [PubMed] [Google Scholar]
- 43.Hutchins DA, Witter AE, Butler A, Luther GW. 1999. Competition among marine phytoplankton for different chelated iron species. Nature 400:858–861. doi: 10.1038/23680. [DOI] [Google Scholar]
- 44.Collier JL, Lovindeer R, Xi Y, Radway JC, Armstrong RA. 2012. Differences in growth and physiology of marine Synechococcus (Cyanobacteria) on nitrate versus ammonium are not determined solely by nitrogen source redox state. J Phycol 48:106–116. doi: 10.1111/j.1529-8817.2011.01100.x. [DOI] [PubMed] [Google Scholar]
- 45.Berube PM, Biller SJ, Kent AG, Berta-Thompson JW, Roggensack SE, Roache-Johnson KH, Ackerman M, Moore LR, Meisel JD, Sher D, Thompson LR, Campbell L, Martiny AC, Chisholm SW. 2015. Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J 9:1195–1207. doi: 10.1038/ismej.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ammerman JW, Azam F. 1985. Bacterial 5-nucleotidase in aquatic ecosystems: a novel mechanism of phosphorus regeneration. Science 227:1338–1340. doi: 10.1126/science.227.4692.1338. [DOI] [PubMed] [Google Scholar]
- 47.Mayali X, Weber PK, Brodie EL, Mabery S, Hoeprich PD, Pett-Ridge J. 2012. High-throughput isotopic analysis of RNA microarrays to quantify microbial resource use. ISME J 6:1210–1221. doi: 10.1038/ismej.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiefer GE, Caldwell DE. 1982. Synergistic interaction between Anabaena and Zoogloea spp. in carbon dioxide-limited continuous cultures. Appl Environ Microbiol 44:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amin SA, Green DH, Hart MC, Küpper FC, Sunda WG, Carrano CJ. 2009. Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc Natl Acad Sci U S A 106:17071–17076. doi: 10.1073/pnas.0905512106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, Dearth SP, Van Mooy BA, Campagna SR, Kujawinski EB, Armbrust EV, Moran MA. 2015. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci U S A 112:453–457. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baran R, Brodie EL, Mayberry-Lewis J, Hummel E, Da Rocha UN, Chakraborty R, Bowen BP, Karaoz U, Cadillo-Quiroz H, Garcia-Pichel F, Northen TR. 2015. Exometabolite niche partitioning among sympatric soil bacteria. Nature J Commun 6:8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowman J, McMeekin T. 2005. Alteromonadales ord. nov., p 443–491. In Brenner D, Krieg N, Staley J, Garrity G, Boone D, De Vos P, Goodfellow M, Rainey F, Schleifer K-H (ed), Bergey’s Manual of Systematic Bacteriology. Springer, New York, NY. [Google Scholar]
- 53.Fiebig A, Pradella S, Petersen J, Michael V, Päuker O, Rohde M, Göker M, Klenk HP, Wagner-Döbler I. 2013. Genome of the marine alphaproteobacterium Hoeflea phototrophica type strain (DFL-43(T)). Stand Genomic Sci 7:440–448. doi: 10.4056/sigs.3486982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole JK, Hutchison JR, Renslow RS, Kim YM, Chrisler WB, Engelmann HE, Dohnalkova AC, Hu D, Metz TO, Fredrickson JK, Lindemann SR. 2014. Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: model systems for the study of autotroph-heterotroph interactions. Front Microbiol 5:109. doi: 10.3389/fmicb.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rippka R. 1988. Isolation and purification of cyanobacteria. Methods Enzymol 167:3–27. [DOI] [PubMed] [Google Scholar]
- 56.Pett-Ridge J, Weber PK. 2012. NanoSIP: NanoSIMS applications for microbial biology. Microb Syst Biol 881:375–408. [DOI] [PubMed] [Google Scholar]
- 57.Popa R, Weber PK, Pett-Ridge J, Finzi JA, Fallon SJ, Hutcheon ID, Nealson KH, Capone DG. 2007. Carbon and nitrogen fixation and metabolite exchange in and between individual cells of Anabaena oscillarioides. ISME J 1:354–360. doi: 10.1038/ismej.2007.44. [DOI] [PubMed] [Google Scholar]
- 58.Bratbak G. 1985. Bacterial biovolume and biomass estimations. Appl Environ Microbiol 49:1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goebel NL, Edwards CA, Carter BJ, Achilles KM, Zehr JP. 2008. Growth and carbon content of three different-sized diazotrophic cyanobacteria observed in the subtropical North Pacific. J Phycol 44:1212–1220. doi: 10.1111/j.1529-8817.2008.00581.x. [DOI] [PubMed] [Google Scholar]
- 60.Fukuda R, Ogawa H, Nagata T, Koike I. 1998. Direct determination of carbon and nitrogen contents of natural bacterial assemblages in marine environments. Appl Environ Microbiol 64:3352–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. 1951. A colorimetric method for the determination of sugars. Nature 168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 62.Bell CW, Fricks BE, Rocca JD, Steinweg JM, McMahon SK, Wallenstein MD. 2013. High-throughput fluorometric measurement of potential soil extracellular enzyme activities. J Vis Exp 2013:e50961. doi: 10.3791/50961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S, Gupta N, Pevzner PA. 2008. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res 7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monroe ME, Shaw JL, Daly DS, Adkins JN, Smith RD. 2008. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput Biol Chem 32:215–217. doi: 10.1016/j.compbiolchem.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res 5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 67.Adkins JN, Monroe ME, Auberry KJ, Shen Y, Jacobs JM, Camp DG II, Vitzthum F, Rodland KD, Zangar RC, Smith RD, Pounds JG. 2005. A proteomic study of the HUPO Plasma Proteome Project’s pilot samples using an accurate mass and time tag strategy. Proteomics 5:3454–3466. doi: 10.1002/pmic.200401333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Magnus M, Pawlowski M, Bujnicki JM. 2012. MetaLocGramN: a meta-predictor of protein subcellular localization for gram-negative bacteria. Biochim Biophys Acta 1824:1425–1433. doi: 10.1016/j.bbapap.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 69.Bendtsen JD, Kiemer L, Fausbøll A, Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol 5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
13C and 15N enrichment of cells analyzed via NanoSIMS. Each point represents 13C and 15N atom percent excess (APE) for a single trichome (solid) or bacterial cell (outlined). The dotted line indicates the ratio of the levels of 13C-labeled and 15N-labeled EOM-substrate, as determined by IRMS. Download
Exopolysaccharide stain Congo red in biofilms grown under different light regimes for 3 days. (A and B) Representative epifluorescent microscopy images of Congo red-stained continuous-light-treated biofilm (A) and continuous-dark-treated biofilm (B). The white scale bar represents 10 µm. The blue coloring is from DAPI DNA staining, and the red coloring is from both autofluorescence (inside filaments) and Congo red stain (extracellular). Yellow outlines indicate lines manually drawn to exclude cyanobacterial cells from analysis of Congo red intensities. (C) Average extracellular Congo red intensities under different light regimes for 3 days. Error bars represent 1 standard deviation of results of comparisons between 3 biological replicates (10 fields of view per replicate analyzed). *, significant difference in means (P < 0.05). Download
Functional predictions for proteins significantly overrepresented in either continuous dark (panels A and C and panel E) or continuous light (panels B and D and panel F) treatments. (A and B) The top panels show functional categories for ESFC-1 exoproteins overrepresented in either continuous dark (A) or continuous light (B) treatments. (C and D) Middle panels show functional categories for ESFC-1 total proteome proteins overrepresented in either continuous dark (C) or continuous light (D) treatments. (E and F) The bottom panels show functional categories for total proteome heterotroph proteins overrepresented in the continuous dark treatment from representative gammaproteobacterium Marinobacter sp. strain ES.048 (E) or representative alphaproteobacterium H. phototrophica (F). Significantly overrepresented, the abundance of the proteins was higher that seen with either the diel treatment or the light or dark treatment (P < 0.05). Download
Sequence-structure-based analyses of the ESFC-1 CMBase. (A) StralCP dendrogram and clusters for proteins identified in PDB with predicted structural similarity to A2994. Proteins with known enzyme activity have the EC number listed together with corresponding PDB chain identifier (ID) labels. (B) List of residue-residue correspondences between the structural model of A2994 and ClcD from Pseudomonas knackmussii (PDB chain 4u2b_A; resolution, 1.70 Å; sequence identity, 26%) in critical functional positions. Catalytic triad residues are colored in red. Mutation positions that improve activity toward substrates are in green. In blue are colored positions where mutations increase substrate accessibility during catalysis by increasing flexibility of the loop containing catalytic triad histidine. The distances between corresponding Cα atoms from the calculated structure superposition are displayed in the last column. Positions marked by an asterisk (*) denote perfect agreement between residues from A2994 and native or mutated residues from 4u2bA. (C) Cartoon representation of the structural superposition of A2994 model (gray) with the crystal structure of ClcD dienelactone hydrolase (yellow). Side-chain conformations of corresponding residues identified as functionally critical are presented in “ball and stick” form using the same coloring scheme as in the table in panel B and numbering from A2994. Download
Cyanobacteria total (intracellular and extracellular) proteome differences between light and dark treatments after 3 days. Total proteome relative abundances were determined via the use of iTraq isobaric tags, and points represent log2 normalized median-centered averages of results from five biological replicates. Colored points represent proteins with significant differences in average abundance (>2 standard deviations from mean difference). pfkA, 6-phosphofructokinase; ABC.2A, ABC-type multidrug transport system; glnB, nitrogen regulatory protein P-II; AfuA, ABC-type Fe3+ transport system; CphX, CO2 hydration protein; phnD, ABC-type phosphate/phosphonate transport system; petJ, cytochrome c6; ABC.PA, ABC-type polar amino acid transport system. Download
Number of cells analyzed via NanoSIMS.
O2 and pH measurements.
Proteomics results. Data represent exoproteins and total proteins identified in cyanobacteria and heterotrophs under all experimental conditions.
Relative abundances of bacterial taxa based on 16S iTag sequencing in biofilms treated in the light and the dark.