Abstract
Toll-like receptor 5 (TLR5) expression in the intestinal epithelial cells (IECs) is critical to maintain health, as underscored by multiple intestinal and extra-intestinal diseases in mice genetically engineered for IEC-specific TLR5 knockout. A gradient of expression exists in the colonic epithelial cells from the cecum to the distal colon. Intriguingly, an identical gradient for the dietary metabolite, butyrate also exists in the luminal contents. However, both being critical for intestinal homeostasis and immune response, no studies examined the role of butyrate in the regulation of TLR5 expression. We showed that butyrate transcriptionally upregulates TLR5 in the IECs and augments flagellin-induced immune responses. Both basal and butyrate-induced transcription is regulated by differential binding of Sp-family transcription factors to the GC-box sequences over the TLR5 promoter. Butyrate activates two different protein kinase C isoforms to dephosphorylate/acetylate Sp1 by serine/threonine phosphatases and phosphorylate Sp3 by ERK-MAPK, respectively. This resulted in Sp1 displacement from the promoter and binding of Sp3 to it, leading to p300 recruitment and histone acetylation, activating transcription. This is the first study addressing the mechanisms of physiological TLR5 expression in the intestine. Additionally, a novel insight is gained into Sp1/Sp3-mediated gene regulation that may apply to other genes.
INTRODUCTION
Toll-like receptors (TLRs) are a class of host pattern recognition receptors, which recognize structurally conserved molecules derived from the microbes and activate innate immune responses (1). TLRs expressed by mucosal epithelia are critical for the distinction between resident microbiota and the pathogens (2). While most TLRs are expressed at high levels by the immune cells, TLR5, which recognizes flagellin from Gram-negative and Gram-positive bacteria is also expressed by various epithelial cells (3) and serves pleiotropic functions through the activation of multiple intracellular signaling pathways (2). TLR5 expression in the intestinal epithelium protects against microbial infections and inflammation, tissue injury, radiation colitis, pro-apoptotic stimuli and colon cancers (4). In addition, it helps to maintain tolerance to commensals through the generation of flagellin-specific immunoglobulin A, which suppresses the flagellin gene expression in commensals (5). Functional TLR5 expression is also required to maintain gastrointestinal health, as suggested by the development of intestinal dysbiosis, spontaneous colitis and obesity-associated metabolic disorders in mice with intestinal epithelial cells (IECs)-specific TLR5 knockout (6). Flagellin has been proposed as an adjunct therapy to colon cancers, because it not only boosts the efficacy of radiation and chemotherapy, but also protects the healthy tissues from the adverse effects of such therapies (7). However, despite the central role of TLR5 expression in gut physiology and immune responses, the regulatory mechanisms for its expression in healthy and diseased intestine remain unknown.
Differential expression of TLRs in healthy and diseased gut epithelia has been reported with vital consequences for microbial pathogenesis, immune responses and homeostasis. Aside from TLR expression altering with age, rapid modulation of their expression occurs following the exposure of cells to environmental stress, microbes and microbe-derived factors and host mediators, such as cytokines (8). TLR5 is downregulated in ulcerative colitis patients (9) and in DSS colitis in mice, while the expression of TLR4 and TLR2 is increased and other TLRs remain unchanged (10). Pro-inflammatory cytokines, IFN-γ and GM-CSF upregulate TLR2 and TLR4 expression in monocytes, but suppress TLR5 expression (11). A gradient of TLR expression was reported along the length of the healthy gut with TLR5 maximally expressed in the proximal colon and gradually decreasing toward the distal part. In contrast, TLR4 and TLR2 are maximally expressed in middle and distal colon, respectively, while their expression is lowest in the proximal colon (10). However, the expression gradient is lost during disease states, suggesting its critical importance in maintaining gut physiology and overall health of the host organism.
Butyrate, a short chain fatty acid (SCFA), is naturally produced in the gut by microbial fermentation of dietary fibres. It is established as the most biologically active SCFA, which plays an important role in the maintenance of normal gut physiology and immune homeostasis by regulating cell proliferation and differentiation, tight junction integrity, apoptosis, anti-inflammatory and oxidative stress response, membrane synthesis and sodium absorption (12,13). Depending on the dietary fibre contents, the physiological concentration of butyrate varies between 1 and 20 mM (14). The maximum concentration is found in the proximal colon, which gradually decreases distally (14). This is similar to TLR5 and opposite to TLR4 expression in the colonic epithelium (10). Earlier studies showed that butyrate induces differentiation of HT29 cells, leading to the downregulation of TLR4 expression and suppression of lipopolysaccharide-induced IL-8 expression (15). However, it induces peptidoglycan-mediated chemokines production in Caco-2 cells through the induction of Nod2 expression (16). In addition, a recently published study has reported that butyrate enhances flagellin-activated signals and expression of IL-8 and TNF-α in human bronchial epithelial cells (17). These findings led us to investigate whether butyrate regulates the physiological expression of TLR5 in the intestinal epithelial cells.
We report here that butyrate enhanced flagellin-mediated immune responses in the IECs through the induction of TLR5 expression. It was regulated by differential binding of Sp1 and Sp3 to the TLR5 promoter. We report a novel mechanism regulating Sp1/Sp3 swap at the TLR5 promoter, mediated through simultaneous modifications by dephosphorylation/ acetylation of Sp1 via PKC-dependent activation of serine/threonine (ser/thr) phosphatases and phosphorylation of Sp3 by PKC-δ-mediated ERK activation. These modifications led to enhanced p300 recruitment and histone acetylation at the TLR5 promoter, giving an insight into the specificity of histone acetylation process in gene regulation. This study not only addressed the regulation of physiological expression of TLR5 in the intestine, but also revealed a novel mechanism of Sp1/Sp3-mediated gene regulation.
MATERIALS AND METHODS
Reagents
Sodium butyrate was purchased from Sigma. Pharmacological inhibitors were from Sigma [Actinomycin-D, Cycloheximide, Mithramycin A, H-7, Staurosporin, Rp-CAMP, Sodium orthovanadate (vanadate), Bisindolylmaleimide-I, Go6976, Rottlerin and Okadaic acid], Calbiochem (H-89, U0126, SB203580, JNK-II and MG-132) and Promega (LY294002). Antibodies were purchased from Abcam (TLR5 and Sp1), Santacruz Biotechnology [Sp1 (sc-14027), Sp3 (sc-644 and sc-13018), PKC-δ (sc-213), p300 (sc-584X), p-Ser(4A3) (sc-81516), egr-1 (sc-189), c-myc (sc-789), TLR5 (sc-10742) and tubulin (sc-5286)], Cell Signaling (H3, H4, p-PKC-δ, ERK, p-ERK, p-Thr, p-Tyr, Acetyl-Lys), Sigma (HDAC1) and Upstate (Acetyl-H3 and Acetyl-H4). All others chemicals were purchased from Sigma. Luciferase reporter constructs were bought from Promega. egr-1 overexpression construct and TLR5 promoter deletion reporter constructs were generated in laboratory. pN3-Sp3FL (Addgene plasmid # 24541) and pN3-Sp1FL (Addgene plasmid # 24543) were gifts from Guntram Suske. Primers used for the study are listed in Supplementary Table S1.
Cell culture
HT29, Caco-2, T-84, INT-407, SW480 and HEK293 cell lines were purchased from American type culture collection (ATCC) and maintained as per the provider's instructions. LoVo and COLO 205 cells were kindly provided by Dr P. Tsichlis, Tuft University. These cells were maintained in DMEM or RPMI1640 supplemented with 10% fetal bovine serum (FBS) and 100 U/ml Penicillin/Streptomycin (Invitrogen). Cells were cultured overnight in serum-free medium before subjecting to the experiments.
Mice treatment with butyrate
Mice were maintained according to the National Guidelines for ethical animal treatment, approved by National Institute of Cholera and Enteric diseases (ICMR), Kolkata, India. All the animal experiments were approved by Institutional Animal Ethics Committee. BALB/c mice, 6–8 weeks old, were fed with either phosphate buffered saline (PBS) (vehicle) or butyrate (300 µmol/kg body wt.) twice a day at 12 h intervals for 3 days. On day 4, mice were sacrificed and colon were collected for Immunohistochemistry or for isolation of colonic epithelial cells. Colonic epithelial cells were isolated using low temperature method as described earlier (18).
RNA isolation, reverse transcription PCR and real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer's protocol. One microgram of DNA-free RNA was used for c-DNA synthesis using Superscript II reverse transcriptase (Life technologies) following the manufacturer's protocol. qPCR was performed in StepOnePlus (ABI) using SYBR green master mix (ABI) with annealing/extension temperature at 60°C for 1 min. The specificity of PCR products was verified by melting curve analysis. Relative quantitation was done by the comparative CT method (19). Data were presented after normalization with GAPDH expression.
Immunohistochemistry
Three micrometers thick paraffin sections of colonic tissues were deparaffinized and incubated in 3% hydrogen peroxide. After washing, sections were blocked with TBS containing 10% goat serum and 1% BSA and stained overnight with 1:100 dilution of TLR5 antibody or isotype control antibody. After incubation with HRP-conjugated goat anti-mouse antibody (Pierce) for 30 min, the sections were developed with diaminobenzidine (DAB, Sigma) for 10 min at room temperature. Sections were counterstained with Hematoxylin for 1 min and mounted for microscopic analysis.
Flow cytometry
Cells were fixed in 4% paraformaldehyde and permeabilized with PBS containing 0.1% Triton-X. Washed cells (106) were stained with TLR5 antibody (2 μg, Abcam) or IgG2a isotype (BD biosciences) for 30 min at room temperature. After washing, cells were stained with FITC-labeled anti-mouse IgG antibody (1:200) (Jackson) for 20 min at room temperature and analyzed by flow cytometry.
ELISA
Quantitation of IL-8 and CCL20 in the culture supernatants of the experimental cells were measured using ELISA kits (eBioscience) as per the manufacturer's instructions.
Construction of reporter and expression plasmids
Various lengths of TLR5 putative promoter were amplified by PCR using human genomic DNA as the template. PCR products were cloned into pTZ57R/T vector (Fermentas) and then subcloned into pGL3 basic vector (Promega). egr-1 over-expression construct was generated by cloning the PCR-amplified coding sequence into pCDNA3.1 (Invitrogen). All the recombinant clones were validated by sequencing.
Mutation, transfection and reporter assay
Mutations over the SP-A (M1) and SP-B (M2) sites were introduced using the quick change site directed mutagenesis kit (Stratagene); according to the manufacturer's protocol. Transient transfections were carried out using Lipofectamine-LTX Plus (Invitrogen); according to the manufacturer's protocol. For reporter assays, cells were co-transfected with the reporter construct and the internal control (pRL-TK) vector. Cells were stimulated with butyrate 36 h after transfection and luciferase activity was measured using the Dual Luciferase Reporter Assay system (Promega); according to the manufacturer's protocol. Relative reporter activity was plotted after normalization with renila luciferase activity.
RNA interference and Gene knockdown
Validated siRNAs designed to knockdown the expression of egr-1, c-myc, Sp1, Sp3, PKC-δ or MEK-1 and control siRNA (Santacruz Biotechnology) were transfected into HT29 cells with Lipofectamin-LTX plus reagent (Invitrogen); following the manufacturer's protocol. Efficiency of gene silencing was verified by immunoblotting 48 h post-transfection.
Preparation of cytosolic and nuclear protein
Nuclear and cytoplasmic extracts from variously-treated cells were prepared as described previously (19). Briefly, HT29 cells were harvested by scraping, resuspended in low salt buffer A [10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl] and incubated on ice for 15 min. After the addition of Nonidet P-40 (0.5%, final concentration), the cell suspensions were passed through a 21-gauge needle 5–6 times. Cytoplasmic and nuclear fractions were separated by centrifugation at 14 000 rpm for 10 min at 4°C. Nuclear proteins were extracted by incubating the cell pellet in high salt buffer C [20 mM HEPES (pH 7.9), 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol] for 30 min on ice with vigorous shaking (250 rpm) followed by centrifugation at 14 000 rpm for 10 min at 4°C. All the preparations were made in presence of 1X protease inhibitor (Sigma) and 1X phosphatase inhibitors cocktails (Calbiochem).
Immunoblot
Immunoblots were performed using the standard protocol (19). In brief, post-experimental cells were lysed with Nonidet P-40 lysis buffer [50 mM HEPES (pH 7.4), 100 mM NaCl, 2 mM EDTA, 0.5% Nonidet P-40, 10% glycerol, 50 mM β-glycerophosphate, 1 mM NaF, protease inhibitor cocktail] and proteins in the samples were separated in SDS-PAGE and transferred to a PVDF membrane (Millipore). Blots were incubated overnight at 4°C with primary antibodies followed by HRP-conjugated secondary antibodies for 1 h at room temperature. Blots were developed with ECL plus Western blot detection reagent (Pierce) and the chemiluminescence was transferred to autoradiographs. All the Western blot experiments were repeated at least two times and a representative blot is presented.
Immunoprecipitation (IP)
A total of 400 μg of nuclear proteins from HT29 cells were pre-cleared with Protein A/G plus agarose beads (Santacruz) and the supernatants were incubated with 2 μg rabbit Sp1 or Sp3 antibody or IgG overnight at 4°C with gentle rocking. Immune complexes were precipitated by incubation with 50 μl of Protein A/G plus beads for additional 1 h followed by brief centrifugation. Beads were washed thrice with NP-40 buffer, resuspended in 60 μl of 2X sample loading buffer and boiled for 5 min before SDS-PAGE and immunoblotting.
Electrophoretic mobility shift assay (EMSA)
DNA-binding activity was analyzed by electrophoretic mobility shift assay (EMSA) as described (19). Briefly, nuclear extracts of cells were incubated with 20 fmol of biotin-labeled probes (3′-end biotin labeling was done according to Pierce kit manual) in the binding buffer [10 mM Tris (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 0.5 mM DTT, 10% glycerol, 0.1% NP-40, 0.5 mM EDTA, 0.1 μg/μl poly-dI.dC] for 30 min at room temperature. Supershift was performed by adding respective antibodies 15 min prior to the addition of biotin-labeled probe. Complexes were separated by gel electrophoresis using a 6% non-denaturing polyacrylamide gel, transferred to a Nylon+ membrane and probed with HRP-conjugated streptavidin for 15 min. The substrate was added to the membrane and autoradiogram was taken.
DNA affinity precipitation assay (DAPA)
DNA affinity precipitation assay (DAPA) was performed as described earlier with modifications (20). Briefly, 200 μg of nuclear protein was incubated with 2 μg of biotin-labeled probe in 400 μl of binding buffer [60 mM KCl, 12 mM HEPES (pH 7.9), 4 mM Tris-HCl, 5% glycerol, 0.5 mM EDTA, 1 mM DTT, 1X protease inhibitor mixture] on ice for 45 min. DNA-protein complexes were pelleted down after incubation with 25 μl of streptavidin microbeads (Miltenyi Biotec) for 1 h at 4°C. Proteins were eluted by boiling in 2X sample loading buffer and analyzed by Western blotting.
Chromatin immunoprecipitation (ChIP)
Quantitative chromatin immunoprecipitation (ChIP) assays were performed using EZChIPTM chromatin immunoprecipitation kit (Upstate) following the manufacturer's instructions. Briefly, treated and untreated HT29 cells were crosslinked using formaldehyde and harvested by scraping. The cell pellet was resuspended in the lysis buffer provided with the kit, and sheared chromatin was immunoprecipitated overnight with 2 μg of rabbit antibodies against RNA polymerase II, Sp1, Sp3, Acetyl-H3, Acetyl-H4, p300, HDAC1 or IgG (negative control), followed by incubation with Protein G agarose for additional 1 h. The immunoprecipitated complex were sequentially washed with low salt, high salt, LiCl and Tris-EDTA buffer followed by extraction with elution buffer. The DNA-protein complexes were reversed and DNA was purified by ethanol precipitation. The relative binding of proteins to TLR5 promoter was quantitatively analyzed by qPCR of precipitated DNA and input DNA. Results are presented as percentages of the total input DNA by using percent input method (% input = 100 × 2 ∧ (Adjusted Ct of input − Ct of IP samples).
Statistical analysis
The data are presented as mean±SD of three independent experiments. Statistical significance was calculated by Student's t-test using GraphPad Prism software (La Jolla, CA, USA) and P < 0.05 was considered as significant.
RESULTS
Butyrate enhances flagellin-mediated immune responses in the IECs
We investigated if butyrate enhanced immune responses induced by flagellin in the IECs, as was reported for the airway epithelial cells. Pre-treatment with butyrate significantly stimulated the release of IL-8 and CCL20 in the culture supernatants of HT29 cells treated with rFliC (recombinant flagellin of Salmonella typhimurium) (Figure 1A, left and right). Mouse IL-8 homolog, KC and MIP-1α mRNAs were induced in the colonic epithelial cells isolated from butyrate-treated mice and stimulated ex vivo with rFlic (Figure 1B, left and right). Flagellin-activated immune responses in the IECs are NF-κB- and AP-1-dependent (21). We investigated if butyrate enhanced activation of these pathways by rFliC or activated different signaling mechanisms. Butyrate significantly upregulated rFliC-induced NF-κB and AP-1 consensus binding sequence-driven luciferase reporter activities in HT29 cells (Figure 1C, left and right). To further investigate the role of butyrate in NF-κB and AP-1 activation, we analyzed the activation of signaling pathways by Western blots. Butyrate and rFliC independently phosphorylated ERK and p38 MAPK, while butyrate augmented rFliC-dependent phosphorylation of MAPKs (Figure 1D). In contrast, NF-κB activation as revealed by IκBα (a negative regulator of NF-κB) degradation was induced by rFliC, but not by butyrate. Instead, butyrate enhanced IκBα levels in HT29 cells. However, increased NF-κB activation was found when rFliC-stimulated cells were pre-treated with butyrate. Taken together these results demonstrated that butyrate enhanced flagellin-induced immune responses in the IECs by augmenting flagellin-activated signals.
Figure 1.
Butyrate enhances TLR5 ligand flagellin-mediated responses in IECs. (A) HT29 cells were pre-treated with 0 or 4 mM of butyrate for 24 h prior to treatement with increasing concentration of rFliC for 18 h. IL-8 and CCL20 was measured by ELISA in the culture supernatants. (B) Colonic epithelial cells isolated from mice fed with PBS- (Vehicle) or butyrate (300 μmol/kg body wt.) twice a day at 12 h intervals for 3 days; were treated in vitro with increasing concentration of rFliC for 3 h. qRT-PCR shows expression of KC and MIP-1α mRNA. (C) NF-κB or AP-1 luciferase plasmids transfected HT29 cells were pre-treated with butyrate as above, followed by increasing concentration of rFliC for 6 h and firefly luciferase activities were measured. (D) HT29 cells were pre-treated with butyrate (4 mM) for 24 h, followed by rFliC (1 μg/ml) for 60 min for p-ERK and p-p38 or 30 min for IκBα for immunoblot. ERK, p38 and Tubulin were probed as loading control. *P < 0.05; **P < 0.01; ***P < 0.001 compared with butyrate-untreated samples.
Butyrate transcriptionally induces TLR5 expression that does not require new protein synthesis in the IECs
Butyrate effects on flagellin-induced immune responses in the IECs may be mediated through upregulated TLR5 expression or synergism between butyrate and TLR5-activated signaling pathways. To investigate the modulation of TLR5 expression, we stimulated HT29 cells with physiological concentrations of butyrate for various durations. Butyrate induced TLR5 mRNA expression in a dose- and time-dependent manner with the highest levels of expression noted after treatment with 4 mM of butyrate for 24 h (Figure 2A). This correlated with TLR5 protein expression as studied by Western blots and flow cytometry (Figure 2B). Cell line specificity of the butyrate effects was excluded by enhanced TLR5 expression in other IECs (Supplementary Figure S1A). In vivo induction of TLR5 expression by butyrate was detected by Western blots of the colonic epithelial cells or immunohistochemistry of the colonic tissues of mice orally fed with butyrate (Figure 2C). Significantly enhanced DAB staining corresponding to TLR5 expression was detected in the upper two-thirds of the colonic villi of butyrate-treated mice. These results suggested that TLR5 upregulation by butyrate in the IECs might lead to accelerated immune responses to flagellated intestinal pathogens.
Figure 2.
Butyrate transcriptionally upregulates the TLR5 expression in IEC. (A) qRT-PCR showing TLR5 mRNA expression in HT29 cells treated with physiological concentration of butyrate (1 to 20 mM) for 24 h (left) or with 4 mM of butyrate for indicated times (right). (B) Immunoblot of total lysates from HT29 cells treated with butyrate (4 mM) for indicated times and probed with TLR5 and Tubulin (loading control) antibodies (upper); Histogram showing increment of TLR5 expression in HT29 cells treated with butyrate as above and analyzed by flow cytometry (lower). (C) Immunoblot of TLR5 and Tubulin (loading control) in cell lysates of primary colonic epithelial cells isolated from mice fed with PBS (Vehicle) or butyrate (300 µmol/kg body wt.) twice a day at 12 h intervals for 3 days (left); immunohistochemistry showing TLR5 expression in colonic tissues from mice treated as above (right). (D) qRT-PCR showing TLR5 mRNA expression in HT29 cells treated with actinomycin D (8 μM) and butyrate, either alone or in combination. Actinomycin D was added 1 h before butyrate. (E) ChIP assays showing IgG and RNA polymerase II binding to the TLR5 promoter in HT29 cells. (F) qRT-PCR showing TLR5 mRNAs in HT29 cells treated with cycloheximide (CHX) (50 μg/ml) for 1 h followed by butyrate. *P < 0.05; **P < 0.01 compared with butyrate-untreated samples; ###P < 0.001 as compared with butyrate-treat samples.
To investigate the underlying mechanisms of butyrate-induced TLR5 expression, we analyzed the changes in TLR5 transcript levels after treatment of HT29 cells with butyrate. The biological half life (t1/2) of TLR5 mRNA in both unstimulated and butyrate-stimulated cells was 105 min (Supplementary Figure S1B), excluding any role for increased mRNA stability in the enhanced TLR5 expression. In contrast, inhibition of transcription by actinomycin D (ActD) completely abolished the butyrate effects (Figure 2D), indicating that TLR5 induction was transcriptionally regulated. To further confirm transcriptional regulation, RNA polymerase II recruitment to the TLR5 promoter was examined by ChIP assays. Significant and progressive increase in RNA polymerase II binding to the proximal TLR5 promoter was observed after butyrate treatment (Figure 2E). Interestingly, pre-treatment with cycloheximide (CHX), a global translation inhibitor, failed to suppress butyrate-induced TLR5 mRNA expression (Figure 2F). This suggested that de novo protein synthesis was not required for TLR5 transactivation by butyrate. Together these results illustrated that butyrate transcriptionally upregulated TLR5 expression in the IECs that was independent of de novo synthesis of transcriptional regulators.
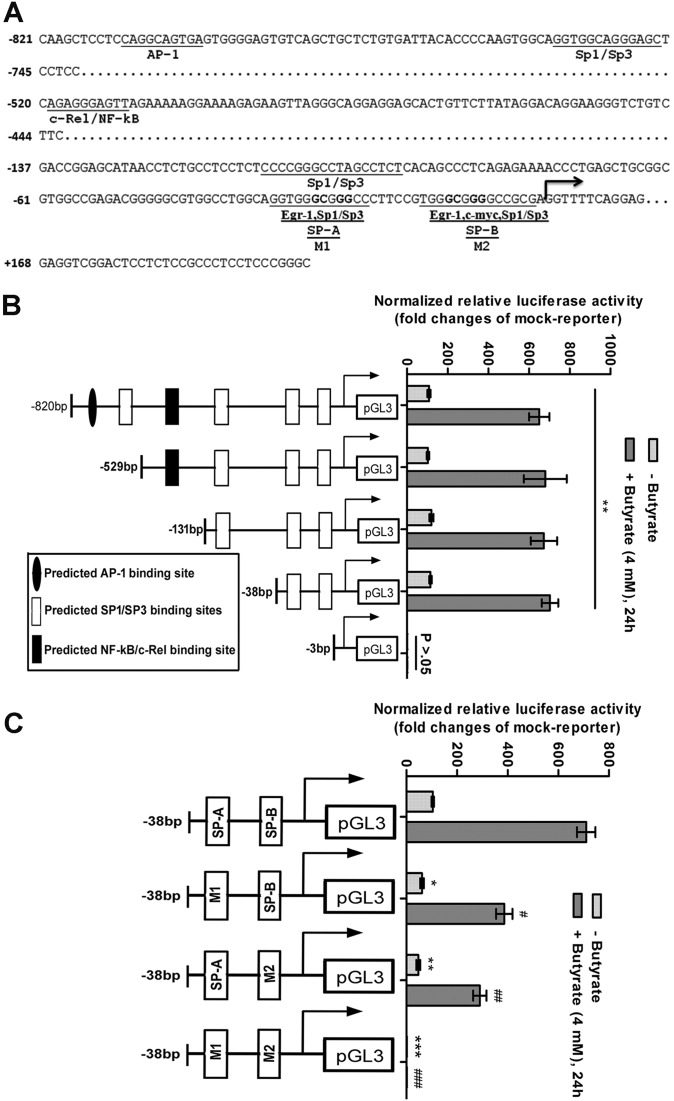
Two GC-box sequences located at the positions -33/-22 and -15/-2 of the TLR5 promoter is required for basal and butyrate-induced transcriptional activities
To study the mechanism of TLR5 transactivation, we analyzed the TLR5 putative promoter for transcription factor binding sites using the software TRANSFAC®Professional (http://www.generegulation.com/pub/programs/alibaba2/index.html) available at the public domain. Consensus binding sequences for several putative regulatory elements, including AP-1, Sp-family of transcription factors (TFs), NF-κB, egr-1 and c-myc were identified within 820 bp upstream of the transcription start site (TSS) (Figure 3A). To experimentally validate promoter activities, luciferase reporter construct of the TLR5 putative promoter (−820 to +200 bp) was transfected into HT29 cells. Basal reporter activities were ∼100-fold higher compared with a promoterless reporter. Promoter activities were further enhanced by 6- to 8-fold following butyrate treatment (Figure 3B). To identify the promoter elements responsible for basal and butyrate-induced TLR5 transcription, luciferase constructs carrying serial deletions of the TLR5 promoter from the 5′ end were transiently transfected into HT29 cells. Reporter assays showed that the minimal promoter containing the cis-regulatory elements for basal as well as butyrate-inducible TLR5 transcription was located within 38 bp upstream of the TSS (Figure 3B). This contained two GC-box sequences (−33 to −22 bp and −15 to −2 bp), designated as SP-A and SP-B, respectively, which constituted the critical cis-regulatory elements for the binding of Sp-family TFs. They were overlapped by egr-1 and c-myc consensus binding sequences (Figure 3A). To further confirm the role of the GC-boxes in TLR5 transcription, reporter constructs with mutated SP-A and SP-B sites were transfected into the cells. Basal as well as butyrate-induced reporter activities were reduced by ∼40% and ∼60% for SP-A and SP-B sites, respectively. However, it was completely abolished when both mutations were present together (Figure 3C). These results indicated that the two GC-rich sequences above, predicted to bind the Sp-family TFs played mandatory and synergistic roles in the transcriptional regulation of TLR5 gene in the IECs.
Figure 3.
Two GC-box near TSS of TLR5 promoter regulate both basal and butyrate-induced promoter activities. (A) Sequence of the human TLR5 gene putative promoter. The predicted transcription factor-binding sites are underlined and labeled. SP-A and SP-B represent Sp1/Sp3 binding sites over the minimal promoter. Arrow indicates the TSS. M1 and M2 denote mutated sequences over SP-A and SP-B, respectively. (B) Firefly reporter assays with HT29 cells transfected with TLR5 promoter deletion constructs. Lengths of the promoters used with predicted TFs binding sites are indicated. **P < 0.01 as compared with pGL3 (empty vector)-transfected samples (mock-reporter). (C) Firefly reporter assays with TLR5 minimal promoter of mutated SP-A and/or SP-B site. *P < 0.05; **P < 0.01; ***P < 0.001 compared with the wild-type promoter; #P < 0.05; ##P < 0.01; ###P < 0.001 as compared with butyrate-treated wild-type promoter.
Sp1 regulates the basal TLR5 transcription, while Sp3 mediates induction by butyrate
To identify the trans-acting factors regulating TLR5 expression, we pre-treated HT29 cells with mithramycin, a pharmacological inhibitor of Sp-family proteins. Increasing doses of mithramycin progressively suppressed TLR5 expression under basal conditions as well as after stimulation with butyrate (Figure 4A). Among the Sp-family proteins, Sp1 and Sp3 are constitutively expressed and involved in the gene regulation in various tissues through binding to GC-rich regions (22). To investigate the role of Sp1 and Sp3 in TLR5 transactivation, we silenced their expression by RNA interference and examined the effects on TLR5 mRNA expression and promoter activities. Knockdown of Sp1 and Sp3 protein expression was confirmed by western blot analysis, which showed significantly reduced levels of Sp1 and all the isoforms of Sp3 proteins in the respective siRNA-transfected cells (Figure 4B). Interestingly, Sp1 knockdown inhibited the basal promoter activities and mRNA expression of TLR5 (Supplementary Figure S2A and Figure 4C), while knockdown of Sp3 suppressed butyrate-induced TLR5 promoter activities and mRNA expression (Supplementary Figure S2B and Figure 4D). However, Sp3 exerted no role in basal TLR5 transcription and Sp1 did not regulate the butyrate-induced effects. In agreement with the above results, ectopic expression of Sp1 and Sp3 dose-dependently enhanced the basal and inducible promoter activities, respectively, driven by the SP-A and SP-B sequences (Figure 4E and F). However, silencing or ectopic expression of egr-1 and c-myc did not regulate TLR5 expression in HT29 cells (Supplementary Figure S2C, S2D). Taken together, these results demonstrated that Sp1 was required to maintain the basal transcriptional machinery for TLR5 expression, while Sp3 was indispensable for butyrate-induced transactivation.
Figure 4.
Sp1 and Sp3 regulate basal and butyrate-induced TLR5 expression, respectively. (A) HT29 cells were pre-treated with increased concentrations of mithramycin for 1 h followed by butyrate; qRT-PCR shows TLR5 mRNA expression. *P < 0.05; **P < 0.01; ***P < 0.001 compared with untreated; #P < 0.05; ##P < 0.01; ###P < 0.001 as compared with butyrate treatment alone. (B) HT29 cells were transfected with control siRNA, Sp1 or Sp3- specific siRNA. After 48 h, Sp1 and Sp3 proteins were detected by immunoblot analysis. (C and D) qRT-PCR showing (C) basal, or (D) butyrate (4 mM, 24 h)-induced expression of TLR5 mRNA in HT29 cells transfected with Sp1 and/or Sp3-specific siRNA. ***P < 0.001 compared with control siRNA transfected; ###P < 0.001 as compared with control siRNA-transfected, butyrate-treated samples. (E and F) TLR5 promoter activities with HT29 cells transfected with increasing concentrations of Sp1 and/or Sp3 expression plasmids and treated with or without butyrate (4 mM, 24 h). *P < 0.05; **P < 0.01; ***P < 0.001 compared with reporter transfection alone; #P < 0.05; ##P < 0.01; ###P < 0.001 compared with reporter-transfected, butyrate-treated samples.
Butyrate triggers decreased Sp1 and increased Sp3 binding to the TLR5 promoter
To further explore the role of Sp-family TFs, we checked their nuclear abundance in the untreated and butyrate-treated HT29 cells. Sp1 and Sp3 were predominantly present in the nuclear extracts of the cells and their relative abundance was unaltered by butyrate treatment (Figure 5A). To investigate the physical binding between the TLR5 promoter and the Sp-family proteins, we performed EMSA assays with double-stranded biotinylated oligonucleotides containing both SP-A and SP-B sequences and the nuclear extracts of HT29 cells. DNA-protein complexes resolved in a non-denaturing PAGE were represented by two distinct bands, a prominent upper band in the un-stimulated cells that had substantially diminished intensity after butyrate treatment and a lower band, which was evident in butyrate-treated cells and increased over time (Figure 5B). Supershift assays using Sp1 and Sp3 antibodies confirmed that the upper band represented Sp1 complex, while DNA-bound Sp3 was responsible for the lower band shift. To further characterize Sp1/Sp3 binding to the GC-box sequences over the TLR5 promoter, we repeated the EMSA with oligonucleotides containing either the SP-A or the SP-B. Binding pattern of Sp1 and Sp3 was similar to that observed with the probe containing the sequences together (Figure 5C). Although weakly-bound complexes were apparent further down the EMSA gel, they did not contain Sp1 and Sp3 as suggested by the supershift assay. To investigate if differential Sp1/Sp3 binding to the TLR5 promoter is unique to HT29 cells, we performed EMSA with the nuclear extracts of the IEC line SW480 and a second epithelial cell line of different origin (HEK293). The results for SW480 cells were similar to HT29 cells (Supplementary Figure S3A); but for HEK293 cells, no Sp3 binding to the TLR5 promoter was observed (Supplementary Figure S3B). These results suggest cell type specific regulation of TLR5 by the Sp-family proteins. TLR5 promoter binding by Sp1 and Sp3 was further studied by DNA affinity precipitation assay (DAPA) coupled with immunoblots using biotin-labeled oligonucleotides containing both SP-A and SP-B sites and the nuclear extracts of HT29 cells. We observed higher amounts of DNA-bound Sp1 at the basal state, which decreased with time after butyrate treatment. In contrast, butyrate induced progressive increase of the long Sp3 isoform binding to the DNA (Figure 5D). Finally, we investigated Sp-family protein binding to the endogenous TLR5 promoter by ChIP assays. A time-dependent decrease of Sp1 and increase of Sp3 occupancy of the TLR5 promoter, but not of the GAPDH promoter after butyrate treatment of the cells was noted (Figure 5E and F). Similar assays with other gene promoters regulated by Sp1 and/or Sp3 revealed that the basal promoters of Sp1 and Sp3 are mostly occupied by Sp1, but not Sp3 protein and the promoter occupancy by the above proteins or RNA pol II remained unaltered after butyrate treatment (Supplementary Figure S3C). In contrast, p21 promoter occupancy shows a similar pattern as the TLR5 promoter, while significantly enhanced Sp3 binding was noted for BAK promoter after butyrate treatment (Supplementary Figure S3D). The above data collectively suggested that butyrate-induced TLR5 expression correlated with a switch in the binding from Sp1 to Sp3 to the TLR5 promoter in HT29 cells. However, this regulation is cell type specific and may be different for other gene promoters.
Figure 5.
Butyrate differentially regulates Sp1 and Sp3 binding to TLR5 promoter. (A) Immunoblots showing Sp1 and Sp3 expression in the cytosolic and nuclear extracts from the untreated- and butyrate-treated (4 mM, 24 h) HT29 cells. Histone H3 and Tubulin were probed as loading controls for the nuclear and cytosolic extracts, respectively. (B and C) EMSA showing retarded bands due to binding of Sp1 and Sp3 present in the nuclear extracts of HT29 cells to the biotin-labeled oligonucleotides containing the SP-A and/or SP-B sequences. Probes used contained both SP-A and SP-B sequences (B). Mutated or 100-fold excess of the unlabeled oligo was used as the competitor. For supershift (ss), reaction was carried out in presence of antibodies (2 μg) to Sp1, Sp3 or both. (D) DAPA. Biotin-labeled TLR5 minimal promoter was incubated with the nuclear extracts of untreated or butyrate-treated (4 mM for indicated times) HT29 cells. DNA-protein complexes were pulled down with streptavidin-coated beads, separated by Western blot and probed with Sp1 and Sp3 antibodies. Total proteins in the nuclear extracts were immunoblotted as loading controls. (E and F) ChIP assays with IgG, Sp1 and Sp3 antibodies in butyrate-treated (4 mM for indicated times) or untreated HT29 cells. qPCR showed IgG, Sp1 and Sp3 binding to endogenous (E) TLR5 promoter or (F) GAPDH promoter. *P < 0.05; **P < 0.01 compared with untreated.
Butyrate-induced TLR5 expression is regulated by ser/thr phosphatases and PKC-δ-mediated ERK activation
Published reports had shown a great diversity in the transcriptional regulatory activities of Sp family transcription factors, which could be determined at least in part by the post-translational modifications, especially phosphorylation/ dephosphorylation (23,24). To investigate the mechanism behind TLR5 expression, we inhibited different kinases and phosphatases using pharmacological inhibitors. Basal expression of TLR5 mRNA was markedly suppressed by pan-Protein Kinase C inhibitors (H7, staurosporin), while butyrate-inducible expression was additionally blocked by the inhibitors of MEK (U0126). In addition, agents that inhibited both conventional and novel isoforms of PKC [bisindolylmaleimide I (BIM 1)] or PKC-δ (rottlerin) effectively countered butyrate effects, while conventional PKC isoforms-specific inhibitor (Go6976) failed to achieve this (Supplementary Figure S4A, Figure 6A). Moreover, butyrate-induced TLR5 expression was dose-dependently suppressed by okadaic acid (oka. acid) and high concentrations (≥100 µM) of vanadate, both of which inhibited ser/thr phosphatases (STPs) (lower doses of vanadate was only effective in suppressing tyrosine phosphatases) (Figure 6B). Role of PKC-δ and MEK-ERK signaling in butyrate-induced TLR5 expression was further confirmed by silencing of the genes in HT29 cells using RNA interference technology. The levels of respective proteins, as detected by western blots were markedly reduced in the siRNA-transfected HT29 cells (Figure 6C, inset), which showed significantly reduced expression of TLR5 upon butyrate treatment (Figure 6C). Accordingly, ectopic expression of PKC-δ dose-dependently enhanced TLR5 promoter activity only when the cells were treated with butyrate (Supplementary Figure S4B). The above findings were further supported by butyrate-induced activation of the kinases and ser/thr phosphatases (STP) (Figure 6D and Supplementary Figure S4C). Interestingly, rottlerin suppressed ERK activation, suggesting a role for PKC-δ (Figure 6E), while phosphatase activation was blocked by okadaic acid and H7 (Figure 6F). The above finding confirmed the activation of ser/thr phosphatases in the pool of butyrate-activated phosphatases and their regulation by PKC signaling. Taken together, these results illustrated the activation of ERK and STP by different PKC isoforms to regulate TLR5 expression in butyrate-treated cells.
Figure 6.
Butyrate induced TLR5 expression depends on activation of PKC-δ/ERK and ser/thr phosphatases. (A and B) qRT-PCR showing TLR5 mRNA expression in HT29 cells, treated with bisindolylmaleimide I (BIM 1, 5 μM), Go6976 (20 μM) or Rottlerin (10 μM) for 1 h followed by butyrate (4 mM) for 24 h (A), or increasing concentrations of vanadate (10, 25, 50 and 100 μM) or okadaic acid (10, 25, 50 and 100 nM) for 1 h before treatment with butyrate (B). (C) qRT-PCR showing basal or butyrate (4 mM, 24 h)-induced expression of TLR5 mRNA in HT29 cells transfected with control siRNA, PKC-δ or MEK-1 siRNA. Immunoblots showing knockdown of PKC-δ and MEK-1 (Inset). ##P < 0.01 as compared with control siRNA transfected but butyrate treated. (D) Immunoblots of untreated and butyrate-treated (4 mM for indicated times) HT29 cell lysates probed with p-PKC-δ and p-ERK. Total PKC-δ and ERK were probed as loading control. (E) Immunoblots showing p-ERK and ERK (loading control) in lysates of untreated or butyrate (4 mM, 6 h)-treated HT29 cells pretreated with Rottlerin (10 μM), U0126 (10 μM) or okadaic acid (100 nM). (F) Specific phosphatase activities (U/mg of protein) measured by calorimetric assay using pNPP as a substrate, in HT29 cells pretreated with vanadate (100 μM), okadaic acid (100 nM), H7 (10 μM), Rottlerin (10 μM) and U0126 (10 μM) for 1 h, followed by butyrate (4 mM, 6 h). **P < 0.01 as compared with untreated; #P < 0.05; ##P < 0.01; ###P < 0.001 compared with butyrate treatment alone.
Butyrate promotes Sp1 and Sp3 modifications through activation of ser/thr phosphatases and PKC-δ/ERK and regulates their binding at TLR5 promoter
Our earlier results showed that TLR5 regulation was independent of de novo protein synthesis, and the expression and nuclear translocation of Sp1 and Sp3 were unaltered by butyrate treatment. To gain insights into the mechanisms underlying butyrate-regulated differential binding of Sp1 and Sp3 to the TLR5 promoter, we investigated Sp1 and Sp3 modifications by the kinases and phosphatases activated by butyrate. Serine phosphorylation of Sp1 was gradually decreased after treatment with butyrate. This was accompanied by progressive serine phosphorylation of Sp3 along with acetylation of both Sp1 and Sp3. However, threonine and tyrosine residue(s) of the above proteins were not differentially phosphorylated (Figure 7A, left and right panel). Although multiple Sp3 isoforms are present in HT29 cells, phosphorylation was increased only for the longer isoform. In contrast, acetylation of both the isoforms was enhanced after butyrate treatment, albeit to a lesser extent for the shorter isoform (Figure 7A, right panel and 7B). Inhibitor treatment of the cells and gene-silencing experiments showed that acetylation of the proteins was regulated by STP, which also controlled Sp1 dephosphorylation. In contrast, phosphorylation of Sp3 was regulated by PKC-δ and ERK (Figure 7B, Supplementary Figure S5A). DAPA and ChIP assays showed that the inhibitors of STP and pan-PKC, but not of PKC-δ and MEK1 reversed butyrate-induced Sp1 de-recruitment from the TLR5 promoter. Sp3 recruitment to the promoter after butyrate treatment, on the other hand, was suppressed by all the kinase and phosphatase inhibitors (Figure 7C and D). The above results were further confirmed by ChIP assays in gene-slienced HT29 cells (Supplementary Figure S5B). Considering that butyrate did not change the nuclear abundance of the proteins, post-translational modifications through phosphorylation/dephosphorylation regulated the binding of Sp-family proteins to the TLR5 promoter. Interestingly, basal Sp1 occupancy of the TLR5 promoter was decreased after pan-PKC and ERK inhibition (Figure 7E), perhaps due to their requirement for basal phosphorylation and promoter recruitment of Sp1 to maintain the basal expression of TLR5 in IECs. Together the above data suggest that butyrate dephosphorylated and acetylated Sp1 by STP, leading to its reduced binding to the TLR5 promoter. Simultaneously, phosphorylation of Sp3 by PKC-δ/ERK enhanced its promoter recruitment. However, STP inhibition led to impaired Sp1 dephosphorylation/ acetylation, resulting in the failure of its replacement by phosphorylated Sp3 (Figure 7B–D).
Figure 7.
Butyrate regulates Sp1 and Sp3 modification and binding dependent on STP and PKC-δ/ERK. (A) Nuclear extracts isolated from untreated and butyrate (4 mM for indicated times)-treated HT29 cells were immunoprecipitated with Sp1 (left), Sp3 (right) or IgG antibody, followed by immunoblotting for phosphorylation and acetylation. Sp1 and Sp3 total proteins were probed as loading controls. (B) Untreated and butyrate (4 mM, 24 h)-treated HT29 cells were pre-treated with or without okadaic acid (100 nM), Rottlerin (10 μM) or U0126 (10 μM). Nuclear extracts from the cells were immunoprecipitated with Sp1, Sp3 or IgG antibody, resolved in SDS-PAGE and immunoblotted as above. (C) DAPA. Biotin-labeled TLR5 minimal promoter was incubated with the nuclear extracts of untreated or butyrate (4 mM, 24 h)-treated HT29 cells, pre-treated with inhibitors. DNA-protein complexes were pulled down as above and probed with Sp1 and Sp3 antibodies. Total proteins in the nuclear extracts were immunoblotted as loading controls. (D and E) ChIP asays. DNA-protein complexes from (D) butyrate-treated or (E) untreated HT29 cells, pre-treated with H7 (10 μM), okadaic acid (100 nM), Rottlerin (10 μM) or U0126 (10 μM) were immunoprecipitated with IgG, Sp1 or Sp3 antibody. DNA corresponding to TLR5 minimal promoter was amplified by qPCR. *P < 0.05, **P < 0.01, ***P < 0.001 as compared with untreated; #P < 0.05, ##P < 0.01 as compared with butyrate-treated but inhibitor untreated samples.
Differential binding of Sp1/Sp3 leads to enhanced p300 recruitment and histone acetylation at the TLR5 promoter and dependent on STP- and PKC-δ/ERK-activation
Sp-group of TFs interact and cooperate with histone deacetylases (HDACs) and histone acetyltransferases (HATs) to form activator/repressor complexes by post-translational modifications of the TFs/co-factors or to induce epigenetic modifications of the gene promoter. We investigated by DAPA followed by immunoblots and ChIP assays for altered p300 and HDAC binding to the TLR5 promoter upon butyrate treatment. The results showed enhanced recruitment of p300, but not HDAC1 to the promoter, although nuclear abundance of the proteins was unaltered after butyrate treatment. Moreover, no acetylated proteins were detected in the regulatory complexes in the untreated and butyrate-treated cells (Figure 8A and B). Furthermore, pre-treatment of the cells with C646, a specific p300/CBP inhibitor significantly suppressed butyrate-induced TLR5 expression (Figure 8C), illustrating its role in TLR5 transactivation. Next, we investigated if p300 recruitment to the promoter resulted in epigenetic modifications due to histone acetylation. Butyrate significantly induced histone H3 and H4 acetylation in HT29 cells (Supplementary Figure S6A). ChIP assays showed that butyrate promoted accumulation of acetylated H3 and H4 at the binding site of Sp1/Sp3 regulatory complexes over the TLR5 promoter (Figure 8D). Since butyrate may exert the above effects through direct HDACi activities without removing HDAC from the promoter, we addressed this issue by pre-treating the cells with STP and PKC-δ/ERK inhibitors. ChIP assays showed that the occupancy of acetyl-H3, acetyl-H4 and p300 to the TLR5 promoter was significantly decreased in the inhibitor-treated cells (Figure 8E). This suggested that histone acetylation at TLR5 promoter was the consequence of enhanced p300 recruitment rather than the direct HDAC inhibitory activities of butyrate. Similar results were found for p21 promoter except that histone acetylation was regulated by STP, but not PKC-δ/ERK, indicating additional acetylation mechanisms. In contrast, histone was not acetylated for BAK promoter despite enhanced recruitment of p300 by butyrate treatment (Supplementary Figure S6B–D). Taken together the above results suggested that butyrate-induced switch between Sp1 and Sp3 in the regulatory complexes was accompanied by p300 recruitment to the TLR5 promoter, leading to TLR5 transactivation through histone acetylation.
Figure 8.
Enhanced p300 recruitment and histone acetylation at TLR5 promoter by butyrate was dependent on STP and PKC-δ/ERK: (A) DAPA. Biotin-labeled TLR5 minimal promoter was incubated with the nuclear extracts of untreated or butyrate (4 mM, 24 h)-treated HT29 cells. DNA-protein complexes were pulled down as above and probed with p300, HDAC1, Sp1, Sp3 or acetyl-Lys antibody. Total proteins in the nuclear extracts were immunoblotted as loading controls. (B) ChIP analysis with antibodies p300, HDAC1 or IgG in untreated or butyrate (4 mM, 24 h)-treated HT29 cells. DNA corresponding to TLR5 minimal promoter was amplified by qPCR. (C) qRT-PCR showing TLR5 mRNA expression in HT29 cells pre-treated with increasing concentrations of p300 inhibitor (10, 25 and 50 μM) for 1 h followed by butyrate (4 mM, 24 h). (D) ChIP analysis with antibodies against acetylated H3, H4 or IgG in untreated or butyrate (4 mM, 24 h)-treated HT29 cells. DNA corresponding to TLR5 minimal promoter was amplified by qPCR. (E) ChIP analysis with antibodies p300, acetyl-H3, acetyl-H4 or IgG in untreated or butyrate (4 mM, 24 h)-treated HT29 cells, pre-treated with different inhibitors (as indicated). DNA corresponding to TLR5 minimal promoter was amplified by qPCR. *P < 0.05; **P < 0.01 as compared with untreated; #P < 0.05; ##P < 0.01; ###P < 0.001 as compared with butyrate-treated alone.
DISCUSSION
TLR5 expression in the gut epithelium is critical for normal gut physiology and immune responses, and the loss of expression has been associated with altered gut microbiota composition, inflammatory bowel diseases, metabolic syndromes and carcinogenesis (4,6,9). However, the expression is restricted to the villi (25), the most differentiated part of the intestinal epithelium and a downward gradient exists from the proximal to the distal colon (10). This underscores the importance of stringent regulation of TLR5 expression in the gut epithelium. However, no study has so far been undertaken to unearth the mechanisms behind the physiological regulation of TLR5 expression. We report here that short chain fatty acid butyrate, one of the most biologically active dietary metabolites in the colon, transcriptionally upregulates TLR5 expression in the IECs and enhances flagellin-mediated immune responses. We identified two GC-box sequences within 38 bp upstream of the TSS, which differentially bind Sp1/Sp3 to regulate both basal and butyrate-induced TLR5 promoter activities. While Sp1 regulates the basal expression of TLR5, Sp3 mediates the butyrate role. Promoter binding of Sp1 was high at the basal state, but diminished after butyrate treatment due to dephosphorylation by ser/thr phosphatases. In contrast, ERK-dependent Sp3 phosphorylation induced by butyrate augmented its binding affinity for the same regulatory regions as Sp1. Two different PKC isoforms were involved in Sp1 and Sp3 modifications and also regulated the recruitment of p300 and acetylayed histones H3 and H4 to the TLR5 promoter, leading to transcriptional activation.
Butyrate induces global changes in gene expression. In the intestine, it is involved in the maintenance of gut physiology and mounting of immune responses (26,27). Recently, flagellin-mediated IL-8 and TNF-α expression in the human bronchial epithelial cells was shown to be enhanced with the treatment of phenylbutyrate (a synthetic derivative of butyrate) (17), although the mechanisms were not studied. We found similar effects with natural butyrate treatment of the IEC line HT29. Butyrate upregulated TLR5 expression and enhanced the flagellin-induced signaling and downstream responses. Functional expression of TLR5 in vivo was also modulated by butyrate. Consistent with the previous reports (25), staining of TLR5 was primarily confined to the vertically more differentiated villus surface rather than the crypt. It is believed that differentiation and migration of intestinal cells in the gut is regulated by butyrate. In addition, maximum TLR5 expression was found in the cecum (10), where butyrate concentration was also reported to be the highest (14). These findings anticipated butyrate as a candidate involved in the differential regulation of the TLR5 expression in the gut lumen.
Species-specific regulation of TLR expression has been reported earlier. Murine TLR2 expression is upregulated by LPS, mycobacterial products and NF-κB activation, whereas TLR2 expression in humans is unaffected by them (28). Moreover, LPS upregulates TLR3 expression in murine macrophages through the induction of autocrine INF-β, but blocks INF-β-induced TLR3 expression in the human myloid dendritic cells (29). However, our study showed similar levels of TLR5 expression in the untreated and butyrate-treated human and murine IECs. TLR5 promoter sequences in the above species showed 53.9% homology with conserved regulatory regions (data not shown), suggesting similar regulatory mechanisms behind TLR5 expression across the species.
Transcriptional regulation by butyrate through GC-rich sequences over the gene promoters was reported for several genes (30–35). These sequences are frequently bound by ubiquitously-expressed Sp-family TFs, leading to augmented or attenuated gene expression in a spatio-temporal manner. Two Sp-family proteins, namely Sp1 and Sp3 working independently or in collaborations, were shown to regulate both constitutive and inducible expression of a large number of genes (23,24). In our study, Sp1 controlled the basal transcriptional machinery of TLR5, while stimulation with butyrate dislodged Sp1 from the promoter and recruited Sp3. Similar observations of differential binding of Sp1/Sp3 were earlier reported for BAK, p21, major vault protein (MVP) and Na+/H+ exchanger (NHE)3 (32,34–36). However, Sp1/Sp3 modifications required for the switch in promoter binding were not well-described. We identified the post-translational modifications that allowed Sp3 binding over Sp1 to the GC-boxes of the TLR5 promoter and transcriptional activation. This demonstrated a novel mechanism of gene regulation by the Sp-family proteins.
Posttranslational modifications of Sp1 and Sp3 may regulate their stability, promoter-binding affinity and transactivation potential (23,24). Lysine acetylation and serine dephosphorylation of Sp1 by butyrate in our study might be responsible for reduced binding to the TLR5 promoter. Sp1 acetylation may increase or decrease the DNA binding affinity, depending on the stimuli and the cell lines tested (24). An acetylated residue (lysine703) in the DNA-binding domain of Sp1 was associated with reduced binding to the promoter and transcriptional downregulation of Neuropilin-1 (NRP-1), a transmembrane receptor for vascular endothelial growth factor (VEGF) (37). In contrast, increased binding of acetylated Sp1 to the tumor suppressor gene PTEN promoter in Hela cells also repressed transcription by recruiting more HDAC1, which resulted in the development and/or aggressiveness of cancers (38). However, no acetyl-Sp1 was detected in the promoter-bound complexes of P21, BAK (32) and TLR5 after butyrate treatment, despite substantial increase in transactivation and the pool of acetylated Sp1 in the nucleus. This suggests that acetylation of Sp1 either suppressed its binding affinity and/or played a minor role in transcriptional activation.
Phosphomodifications of Sp1 protein regulate DNA-binding and transcriptional activity (23,24). We observed more serine phosphorylation of Sp1 and stronger binding to the TLR5 promoter in untreated compared to butyrate-treated HT29 cells. Serine phosphorylation of Sp1 may be regulated by the constitutive cellular signaling pathways regulating the cell growth and cell division. Butyrate-induced dephosphorylation of Sp1 by STP in HT29 cells, leading to reduced binding to the TLR5 promoter was inhibited by okadaic acid, a potent inhibitor of PP1 and PP2A. PP1 was shown to suppress the binding affinity of Sp1 and inhibit the promoter functions of αENaC2 and mouse vesicular glutamate transporter 2 (mVGLUT2) gene in the lung epithelial cells and mouse pancreatic β-cell line, respectively (39,40). Dephosphorylation by other phosphatases, such as protein tyrosine phosphatases and alkaline phosphatase were reported to inhibit recruitment to VEGF and AKR1C1 promoters, respectively, and suppress gene expression (41,42). Since okadaic acid also suppressed Sp1 acetylation, we cannot completely exclude if acetylation contributed to butyrate-mediated de-recruitment of Sp1 from the TLR5 promoter.
Sp3-mediated transcriptional regulation appears to be more complex due to the presence of different isoforms and their post-translational modifications (43). Regulation of gene expression by Sp3 isoforms appears to be gene and promoter specific, as suggested by butyrate-mediated regulation of NHE3 and NHE8 expression in Caco-2 cells. While two Sp3-containing complexes were formed at NHE3 promoter, only one Sp3 complex at NHE8 promoter was detected in the same cell line (35,44). We found expression of multiple Sp3 isoforms in HT29 cells, but butyrate induced phosphorylation/acetylation predominantly of the long isoform. Variations in the expression and post-translational modifications of the isoforms may explain, in part, differential gene regulation by Sp3. Two long isoforms may be transcriptional activators or repressors, whereas small isoforms always function as repressors. All isoforms of Sp3 may be SUMO-modified at lysine551, which represses transcription (43). We found that the relative amounts of different isoforms before and after butyrate treatment in the cytosolic and nuclear extracts remained the same. Waby et al. (2010) concluded that decreased binding of acetylated Sp1 to p21 promoter perhaps allowed access to Sp3, leading to transcriptional upregulation (32). However, knockdown of Sp1 inhibited the basal transcription of TLR5 in HT29 cells. Overexpression of Sp3 didn't influence the basal TLR5 expression, but transactivation was markedly enhanced when the cells were treated with butyrate. Given that nuclear abundance of Sp3 remained unaltered after butyrate treatment, these results suggested that Sp3 required post-translational modifications for DNA binding. Kiela, PR et al. (2007) showed that acetylation of Sp3 and phosphorylation of Sp1 with butyrate might result in enhanced promoter binding of Sp3 and transactivation of NHE3 in Caco-2 cells (35). In contrast, butyrate-mediated Sp3 acetylation suppressed human insulin-like growth factor-binding protein-3 expression in Caco-2 cells (45). We observed enhanced acetylated Sp3 levels in HT29 cells, but not in the TLR5 transactivation complex. Recently, the role of phosphorylation of the longer Sp3 isoform in binding to the promoter and transcriptional activation has been reported (24,46). We found enhanced pool of phosphorylated long isoform of Sp3 in the nuclei of butyrate-treated cells. The kinetics of Sp3 phosphorylation correlated with its recruitment to the TLR5 promoter, suggesting that phosphorylation may potentially underlie enhanced promoter-binding of Sp3. However, Sp1 dephosphorylation continues to be a pre-requisite for its replacement by Sp3 from the promoter, necessitating further investigations into the atomic interactions of Sp-family proteins with the TLR5 promoter and their binding affinity before and after butyrate treatment.
An array of protein kinases (PKA, PKC, MAPK, Casein kinase and Calmodulin kinase) and phosphatases (ser/thr and tyrosine phosphatases) were reported to post-translationally modify Sp1 (23,24). Although PKC phosphorylates a large number of target proteins, its role in dephosphorylation via activation of phosphatases was also reported. Activation of PKC by 4β-phorbol 12-myristate 13-acetate (PMA) enhanced the activity of PP1, a ser/thr phosphatase, in human breast cancer cell line, which dephosphorylated p38 MAPK (47). PKC-δ-activated PP2A reduced the tyrosine hydroxylase activity and dopamine synthesis in PC12 cells (48). In the current study, STPs were activated downstream of PKC in the butyrate-treated cells and regulated not only dephosphorylation, but also acetylation of Sp1. While PKC-δ was not involved, the particular PKC isoform(s) responsible for the above modifications were not identified. In contrast, butyrate-induced phosphorylation of Sp3 and binding to the TLR5 promoter were dependent on PKC-δ induced ERK activation. Limited information exist about Sp3 regulation by different stimuli, although involvement of ERK was reported earlier (46,49). One ERK phosphorylation site in the long isoform of Sp3 (serine73) was predicted as critical for VEGF expression (49). However, Sp3 phosphorylation by PKC-δ-dependent ERK activation is a novel signaling mechanism.
Butyrate modulates the activation of multiple cellular signaling pathways to induce global changes in gene expression. Butyrate activates G-protein coupled receptor (GPCR), GPR43 to regulate the size and function of colonic Treg cell pool in mice and protect from colitis (50). SCFAs including butyrate activate GPR41 and GPR43 in the IECs and mediate protective immunity and tissue inflammation in mice through the activation of ERK1/2 and p38 MAPK signaling pathways (51). We had earlier reported that butyrate induced cathelicidin expression in the intestinal epithelial cells, which was regulated by cAMP-dependent CREB and AP-1 activation (19). While this may be regulated by GPCR activation, GPCR-independent cAMP-PKA-CREB signaling pathway activation by butyrate in Caco-2 cells was also reported (52). Butyrate may augment PKC and MAPKs activity, such as during erythroid proliferation and differentiation (53). Blockade of PKC-δ and p38 MAPK in colorectal adenoma cells abrogated pro-apoptotic effect of butyrate, while Inhibition of ERK diminished Gαi2 and choline acetyltransferase gene activation in K562 and CHP126 cells, respectively (54,55). Moreover, butyrate-activated PKC-δ was also shown to be critical for Epstein–Barr virus (EBV) reactivation or induction of EBV lytic cycle in nasopharyngeal carcinoma (NPC) cell line (56). However, two different isoforms of PKC, regulating differential modification and DNA binding of Sp1 and Sp3 through simultaneous activation of ser/thr phosphatases and kinases is a novel underlying mechanism of butyrate action.
The interaction of TATA-binding protein and associated factors (both activators and/or repressors) with the regulatory transcriptional complexes was proposed as crucial determinant of the specificity of gene regulation (57). Post-translational modifications of TFs enhance their interaction with the regulatory proteins. We observed increased p300 recruitment and histone acetylation of the TLR5 minimal promoter with butyrate treatment. Being a well known HDAC inhibitor, butyrate directly binds to HDACs, leading to enhanced activity of the existing HATs in the complex and increased transcription through histone or non-histone protein acetylation (57). In contrast, enhanced recruitment of p300 and histone acetylation on the TLR5 promoter following butyrate treatment was dependent on the differential regulation of Sp1/Sp3, since STP and PKC-δ/ERK inhibitors reversed the butyrate effects. Suppression of p300 recruitment and histone acetylation with PKC-δ/ERK inhibition suggested that increased amounts of p300 in the TLR5 transactivation complex is likely to be associated with the recruitment of phosphorylated Sp3. This is similar to the regulation of ChM-I gene through the association of YY1 and p300 with Sp1/Sp3. YY1 negatively regulated the expression of ChM-I through deacetylation by Sp1-dependent recruitment of histone deacetylase, whereas transcriptional co-activator p300 binds to core promoter with Sp3 but not Sp1 and activate ChM-I through histone acetylation (58). The differential binding of Sp1/Sp3 and recruitment of p300 was also observed at BAK and p21 promoter. However, histone was not acetylated at the BAK promoter, and p300 recruitment, but not histone acetylation at p21 promoter was regulated by PKC-δ/ERK. Earlier studies suggested involvement of other HATs like GCN5 for p21 regulation (59,60). Progressive accumulation of p300 at the TLR5 transactivation complex through differential regulation of Sp1/Sp3 provides an example of selectivity in the gene regulation by histone modifications with a global regulator like butyrate.
This study presents a novel insight into the transcriptional regulation by Sp1/Sp3 swap that is mediated through differential regulation of Sp1 and Sp3 by PKC signaling pathways. This is the first study that addressed physiological regulation of TLR5 gene expression in the intestinal epithelium, enabling us to further understand the molecular mechanisms of the maintenance of normal gut physiology and homeostasis, which may potentially lead to the development of new therapeutics targeted to TLR5 for the amelioration of intestinal diseases and restoration of normalcy.
Supplementary Material
Acknowledgments
We thank Dr Dhira Rani Saha, from the Division of Electron Microscopy, National Institute of Cholera and Enteric Diseases, Kolkata for her help with microscopy data analysis. We acknowledge the gift of N2 PKC-δ, pcDNA3-cmyc plasmids and LoVo and COLO 205 cell lines by Dr P. Tsichlis, Tufts University, Boston.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology, Govt. of India [extramural project ID: BT/PR-14952/FNS/20/492/2010]; Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) of the Japan Agency for Medical Research and Development (AMED). The open access publication charge for this paper has been waived by Oxford University Press - NAR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gay N.J., Symmons M.F., Gangloff M., Bryant C.E. Assembly and localization of Toll-like receptor signalling complexes. Nat. Rev. Immunol. 2014;14:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho F.A., Aitken J.D., Vijay-Kumar M., Gewirtz A.T. Toll-like receptor-gut microbiota interactions: perturb at your own risk! Annu. Rev. Physiol. 2012;74:177–198. doi: 10.1146/annurev-physiol-020911-153330. [DOI] [PubMed] [Google Scholar]
- 3.Sandor F., Buc M. Toll-like receptors. II. Distribution and pathways involved in TLR signalling. Folia Biol. 2005;51:188–197. doi: 10.14712/fb2005051060188. [DOI] [PubMed] [Google Scholar]
- 4.Vijay-Kumar M., Aitken J.D., Sanders C.J., Frias A., Sloane V.M., Xu J., Neish A.S., Rojas M., Gewirtz A.T. Flagellin treatment protects against chemicals, bacteria, viruses, and radiation. J. Immunol. 2008;180:8280–8285. doi: 10.4049/jimmunol.180.12.8280. [DOI] [PubMed] [Google Scholar]
- 5.Cullender T.C., Chassaing B., Janzon A., Kumar K., Muller C.E., Werner J.J., Angenent L.T., Bell M.E., Hay A.G., Peterson D.A., et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chassaing B., Ley R.E., Gewirtz A.T. Intestinal epithelial cell Toll-like receptor 5 regulates the intestinal microbiota to prevent low-grade inflammation and metabolic syndrome in mice. Gastroenterology. 2014;147:1363–1377. doi: 10.1053/j.gastro.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vijay-Kumar M., Gewirtz A.T. Guardians of the gut: newly appreciated role of epithelial toll-like receptors in protecting the intestine. Gastroenterology. 2008;135:351–354. doi: 10.1053/j.gastro.2008.06.064. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins P.A., Sriskandan S. Mammalian Toll-like receptors: to immunity and beyond. Clin. Exp. Immunol. 2005;140:395–407. doi: 10.1111/j.1365-2249.2005.02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanislawowski M., Wierzbicki P.M., Golab A., Adrych K., Kartanowicz D., Wypych J., Godlewski J., Smoczynski M., Kmiec Z. Decreased Toll-like receptor-5 (TLR-5) expression in the mucosa of ulcerative colitis patients. J. Physiol. Pharmacol. 2009;60(Suppl 4):71–75. [PubMed] [Google Scholar]
- 10.Ortega-Cava C.F., Ishihara S., Rumi M.A., Aziz M.M., Kazumori H., Yuki T., Mishima Y., Moriyama I., Kadota C., Oshima N., et al. Epithelial toll-like receptor 5 is constitutively localized in the mouse cecum and exhibits distinctive down-regulation during experimental colitis. Clin. Vaccine Immunol. 2006;13:132–138. doi: 10.1128/CVI.13.1.132-138.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Mahony D.S., Pham U., Iyer R., Hawn T.R., Liles W.C. Differential constitutive and cytokine-modulated expression of human Toll-like receptors in primary neutrophils, monocytes, and macrophages. Int. J. Med. Sci. 2008;5:1–8. doi: 10.7150/ijms.5.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong J.M., de Souza R., Kendall C.W., Emam A., Jenkins D.J. Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 13.Hamer H.M., Jonkers D., Venema K., Vanhoutvin S., Troost F.J., Brummer R.J. Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 14.Cummings J.H., Pomare E.W., Branch W.J., Naylor C.P., Macfarlane G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S.K., Il Kim T., Kim Y.K., Choi C.H., Yang K.M., Chae B., Kim W.H. Cellular differentiation-induced attenuation of LPS response in HT-29 cells is related to the down-regulation of TLR4 expression. Biochem. Biophys. Res. Commun. 2005;337:457–463. doi: 10.1016/j.bbrc.2005.09.071. [DOI] [PubMed] [Google Scholar]
- 16.Leung C.H., Lam W., Ma D.L., Gullen E.A., Cheng Y.C. Butyrate mediates nucleotide-binding and oligomerisation domain (NOD) 2-dependent mucosal immune responses against peptidoglycan. Eur. J. Immunol. 2009;39:3529–3537. doi: 10.1002/eji.200939454. [DOI] [PubMed] [Google Scholar]
- 17.Kulkarni N.N., Yi Z., Huehnken C., Agerberth B., Gudmundsson G.H. Phenylbutyrate induces cathelicidin expression via the vitamin D receptor: Linkage to inflammatory and growth factor cytokines pathways. Mol. Immunol. 2015;63:530–539. doi: 10.1016/j.molimm.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Kang Y.J., Otsuka M., van den Berg A., Hong L., Huang Z., Wu X., Zhang D.W., Vallance B.A., Tobias P.S., Han J. Epithelial p38alpha controls immune cell recruitment in the colonic mucosa. PLoS Pathog. 2010;6:e1000934. doi: 10.1371/journal.ppat.1000934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakraborty K., Maity P.C., Sil A.K., Takeda Y., Das S. cAMP stringently regulates human cathelicidin antimicrobial peptide expression in the mucosal epithelial cells by activating cAMP-response element-binding protein, AP-1, and inducible cAMP early repressor. J. Biol. Chem. 2009;284:21810–21827. doi: 10.1074/jbc.M109.001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y., Saunders M.A., Yeh H., Deng W.G., Wu K.K. Dynamic regulation of cyclooxygenase-2 promoter activity by isoforms of CCAAT/enhancer-binding proteins. J. Biol. Chem. 2002;277:6923–6928. doi: 10.1074/jbc.M108075200. [DOI] [PubMed] [Google Scholar]
- 21.Rhee S.H., Keates A.C., Moyer M.P., Pothoulakis C. MEK is a key modulator for TLR5-induced interleukin-8 and MIP3alpha gene expression in non-transformed human colonic epithelial cells. J. Biol. Chem. 2004;279:25179–25188. doi: 10.1074/jbc.M400967200. [DOI] [PubMed] [Google Scholar]
- 22.Li L., He S., Sun J.M., Davie J.R. Gene regulation by Sp1 and Sp3. Biochem. Cell Biol. 2004;82:460–471. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]
- 23.Tan N.Y., Khachigian L.M. Sp1 phosphorylation and its regulation of gene transcription. Mol. Cell. Biol. 2009;29:2483–2488. doi: 10.1128/MCB.01828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu S. Transcriptional regulation by post-transcriptional modification–role of phosphorylation in Sp1 transcriptional activity. Gene. 2012;508:1–8. doi: 10.1016/j.gene.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 25.Bambou J.C., Giraud A., Menard S., Begue B., Rakotobe S., Heyman M., Taddei F., Cerf-Bensussan N., Gaboriau-Routhiau V. In vitro and ex vivo activation of the TLR5 signaling pathway in intestinal epithelial cells by a commensal Escherichia coli strain. J. Biol. Chem. 2004;279:42984–42992. doi: 10.1074/jbc.M405410200. [DOI] [PubMed] [Google Scholar]
- 26.Meijer K., de Vos P., Priebe M.G. Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health. Curr. Opin. Clin. Nutr. Metab. Care. 2010;13:715–721. doi: 10.1097/MCO.0b013e32833eebe5. [DOI] [PubMed] [Google Scholar]
- 27.Vinolo M.A., Rodrigues H.G., Nachbar R.T., Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3:858–876. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haehnel V., Schwarzfischer L., Fenton M.J., Rehli M. Transcriptional regulation of the human toll-like receptor 2 gene in monocytes and macrophages. J. Immunol. 2002;168:5629–5637. doi: 10.4049/jimmunol.168.11.5629. [DOI] [PubMed] [Google Scholar]
- 29.Heinz S., Haehnel V., Karaghiosoff M., Schwarzfischer L., Muller M., Krause S.W., Rehli M. Species-specific regulation of Toll-like receptor 3 genes in men and mice. J. Biol. Chem. 2003;278:21502–21509. doi: 10.1074/jbc.M301476200. [DOI] [PubMed] [Google Scholar]
- 30.Zeissig S., Fromm A., Mankertz J., Weiske J., Zeitz M., Fromm M., Schulzke J.D. Butyrate induces intestinal sodium absorption via Sp3-mediated transcriptional up-regulation of epithelial sodium channels. Gastroenterology. 2007;132:236–248. doi: 10.1053/j.gastro.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 31.Jin H., Kanthasamy A., Harischandra D.S., Kondru N., Ghosh A., Panicker N., Anantharam V., Rana A., Kanthasamy A.G. Histone hyperacetylation up-regulates protein kinase Cdelta in dopaminergic neurons to induce cell death: relevance to epigenetic mechanisms of neurodegeneration in Parkinson disease. J. Biol. Chem. 2014;289:34743–34767. doi: 10.1074/jbc.M114.576702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waby J.S., Chirakkal H., Yu C., Griffiths G.J., Benson R.S., Bingle C.D., Corfe B.M. Sp1 acetylation is associated with loss of DNA binding at promoters associated with cell cycle arrest and cell death in a colon cell line. Mol. Cancer. 2010;9:275. doi: 10.1186/1476-4598-9-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amin M.R., Dudeja P.K., Ramaswamy K., Malakooti J. Involvement of Sp1 and Sp3 in differential regulation of human NHE3 promoter activity by sodium butyrate and IFN-gamma/TNF-alpha. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293:G374–G382. doi: 10.1152/ajpgi.00128.2007. [DOI] [PubMed] [Google Scholar]
- 34.Chirakkal H., Leech S.H., Brookes K.E., Prais A.L., Waby J.S., Corfe B.M. Upregulation of BAK by butyrate in the colon is associated with increased Sp3 binding. Oncogene. 2006;25:7192–7200. doi: 10.1038/sj.onc.1209702. [DOI] [PubMed] [Google Scholar]
- 35.Kiela P.R., Kuscuoglu N., Midura A.J., Midura-Kiela M.T., Larmonier C.B., Lipko M., Ghishan F.K. Molecular mechanism of rat NHE3 gene promoter regulation by sodium butyrate. Am. J. Physiol. Cell Physiol. 2007;293:C64–C74. doi: 10.1152/ajpcell.00277.2006. [DOI] [PubMed] [Google Scholar]
- 36.Steiner E., Holzmann K., Pirker C., Elbling L., Micksche M., Berger W. SP-transcription factors are involved in basal MVP promoter activity and its stimulation by HDAC inhibitors. Biochem. Biophys. Res. Commun. 2004;317:235–243. doi: 10.1016/j.bbrc.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 37.Yu D.C., Waby J.S., Chirakkal H., Staton C.A., Corfe B.M. Butyrate suppresses expression of neuropilin I in colorectal cell lines through inhibition of Sp1 transactivation. Mol. Cancer. 2010;9:276. doi: 10.1186/1476-4598-9-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kou X.X., Hao T., Meng Z., Zhou Y.H., Gan Y.H. Acetylated Sp1 inhibits PTEN expression through binding to PTEN core promoter and recruitment of HDAC1 and promotes cancer cell migration and invasion. Carcinogenesis. 2013;34:58–67. doi: 10.1093/carcin/bgs336. [DOI] [PubMed] [Google Scholar]
- 39.Chu S., Cockrell C.A., Ferro T.J. Expression of alpha-ENaC2 is dependent on an upstream Sp1 binding motif and is modulated by protein phosphatase 1 in lung epithelial cells. Biochem. Biophys. Res. Commun. 2003;303:1159–1168. doi: 10.1016/s0006-291x(03)00497-2. [DOI] [PubMed] [Google Scholar]
- 40.Li T., Bai L., Li J., Igarashi S., Ghishan F.K. Sp1 is required for glucose-induced transcriptional regulation of mouse vesicular glutamate transporter 2 gene. Gastroenterology. 2008;134:1994–2003. doi: 10.1053/j.gastro.2008.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prasanna Kumar S., Thippeswamy G., Sheela M.L., Prabhakar B.T., Salimath B.P. Butyrate-induced phosphatase regulates VEGF and angiogenesis via Sp1. Arch. Biochem. Biophys. 2008;478:85–95. doi: 10.1016/j.abb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Selga E., Noe V., Ciudad C.J. Transcriptional regulation of aldo-keto reductase 1C1 in HT29 human colon cancer cells resistant to methotrexate: role in the cell cycle and apoptosis. Biochem. Pharmacol. 2008;75:414–426. doi: 10.1016/j.bcp.2007.08.034. [DOI] [PubMed] [Google Scholar]
- 43.Sapetschnig A., Koch F., Rischitor G., Mennenga T., Suske G. Complexity of translationally controlled transcription factor Sp3 isoform expression. J. Biol. Chem. 2004;279:42095–42105. doi: 10.1074/jbc.M404989200. [DOI] [PubMed] [Google Scholar]
- 44.Xu H., McCoy A., Li J., Zhao Y., Ghishan F.K. Sodium butyrate stimulates NHE8 expression via its role on activating NHE8 basal promoter activity. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;309:G500–G505. doi: 10.1152/ajpgi.00194.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White N.R., Mulligan P., King P.J., Sanderson I.R. Sodium butyrate-mediated Sp3 acetylation represses human insulin-like growth factor binding protein-3 expression in intestinal epithelial cells. J. Pediatr. Gastroenterol. Nutr. 2006;42:134–141. doi: 10.1097/01.mpg.0000189345.31010.89. [DOI] [PubMed] [Google Scholar]
- 46.Jiang Y., Fleet J.C. Phorbol esters enhance 1alpha,25-dihydroxyvitamin D3-regulated 25-hydroxyvitamin D-24-hydroxylase (CYP24A1) gene expression through ERK-mediated phosphorylation of specific protein 3 (Sp3) in Caco-2 cells. Mol. Cell. Endocrinol. 2012;361:31–39. doi: 10.1016/j.mce.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitatani K., Idkowiak-Baldys J., Bielawski J., Taha T.A., Jenkins R.W., Senkal C.E., Ogretmen B., Obeid L.M., Hannun Y.A. Protein kinase C-induced activation of a ceramide/protein phosphatase 1 pathway leading to dephosphorylation of p38 MAPK. J. Biol. Chem. 2006;281:36793–36802. doi: 10.1074/jbc.M608137200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang D., Kanthasamy A., Yang Y., Anantharam V., Kanthasamy A. Protein kinase C delta negatively regulates tyrosine hydroxylase activity and dopamine synthesis by enhancing protein phosphatase-2A activity in dopaminergic neurons. J. Neurosci. 2007;27:5349–5362. doi: 10.1523/JNEUROSCI.4107-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pages G. Sp3-mediated VEGF regulation is dependent on phosphorylation by extra-cellular signals regulated kinases (ERK) J. Cell. Physiol. 2007;213:454–463. doi: 10.1002/jcp.21104. [DOI] [PubMed] [Google Scholar]
- 50.Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly Y.M., Glickman J.N., Garrett W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim M.H., Kang S.G., Park J.H., Yanagisawa M., Kim C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396–406. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 52.Wang A., Si H., Liu D., Jiang H. Butyrate activates the cAMP-protein kinase A-cAMP response element-binding protein signaling pathway in Caco-2 cells. J. Nutr. 2012;142:1–6. doi: 10.3945/jn.111.148155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivero J.A., Adunyah S.E. Sodium butyrate stimulates PKC activation and induces differential expression of certain PKC isoforms during erythroid differentiation. Biochem. Biophys. Res. Commun. 1998;248:664–668. doi: 10.1006/bbrc.1998.9041. [DOI] [PubMed] [Google Scholar]
- 54.McMillan L., Butcher S.K., Pongracz J., Lord J.M. Opposing effects of butyrate and bile acids on apoptosis of human colon adenoma cells: differential activation of PKC and MAP kinases. Br. J. Cancer. 2003;88:748–753. doi: 10.1038/sj.bjc.6600793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espinos E., Weber M.J. Activation of the MAP kinase cascade by histone deacetylase inhibitors is required for the stimulation of choline acetyltransferase gene promoter. Brain Res. Mol. Brain Res. 1998;56:118–124. doi: 10.1016/s0169-328x(98)00036-9. [DOI] [PubMed] [Google Scholar]
- 56.Lee H.H., Chang S.S., Lin S.J., Chua H.H., Tsai T.J., Tsai K., Lo Y.C., Chen H.C., Tsai C.H. Essential role of PKCdelta in histone deacetylase inhibitor-induced Epstein-Barr virus reactivation in nasopharyngeal carcinoma cells. J. Gen. Virol. 2008;89:878–883. doi: 10.1099/vir.0.83533-0. [DOI] [PubMed] [Google Scholar]
- 57.Davie J.R. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 58.Aoyama T., Okamoto T., Fukiage K., Otsuka S., Furu M., Ito K., Jin Y., Ueda M., Nagayama S., Nakayama T., et al. Histone modifiers, YY1 and p300, regulate the expression of cartilage-specific gene, chondromodulin-I, in mesenchymal stem cells. J. Biol. Chem. 2010;285:29842–29850. doi: 10.1074/jbc.M110.116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kobayashi H., Tan E.M., Fleming S.E. Acetylation of histones associated with the p21WAF1/CIP1 gene by butyrate is not sufficient for p21WAF1/CIP1 gene transcription in human colorectal adenocarcinoma cells. Int. J. Cancer. 2004;109:207–213. doi: 10.1002/ijc.11697. [DOI] [PubMed] [Google Scholar]
- 60.Xiao H., Hasegawa T., Isobe K. p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J. Biol. Chem. 2000;275:1371–1376. doi: 10.1074/jbc.275.2.1371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.