Abstract
Non-canonical transforming growth factor β (TGFβ) signaling through protein kinase B (Akt2) induces phosphorylation of heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1) at serine-43 (p-hnRNP E1). This post-translational modification (PTM) of hnRNP E1 promotes its dissociation from a 3′ untranslated region (UTR) nucleic acid regulatory motif, driving epithelial to mesenchymal transition (EMT) and metastasis. We have identified an hnRNP E1 consensus-binding motif and genomically resolved a subset of genes in which it is contained. This study characterizes the binding kinetics of the consensus-binding motif and hnRNP E1, its various K-homology (KH) domains and p-hnRNP E1. Levels of p-hnRNP E1 are highly upregulated in metastatic cancer cells and low in normal epithelial tissue. We show a correlation between this PTM and levels of Akt2 and its activated form, phosphorylated serine-474 (p-Akt2). Using cellular progression models of metastasis, we observed a signature high level of Akt2, p-Akt2 and p-hnRNP E1 protein expression, coupled to a significantly reduced level of total hnRNP E1 in metastatic cells. Genes that are translationally silenced by hnRNP E1 and expressed by its dissociation are highly implicated in the progression of EMT and metastasis. This study provides insight into a non-canonical TGFβ signaling cascade that is responsible for inducing EMT by aberrant expression of hnRNP E1 silenced targets. The relevance of this system in metastatic progression is clearly shown in cellular models by the high abundance of p-hnRNP E1 and low levels of hnRNP E1. New insights provided by the resolution of this molecular mechanism provide targets for therapeutic intervention and give further insight into the role of the TGFβ microenvironment.
INTRODUCTION
Epithelial to mesenchymal transition (EMT) is a process in which an epithelial cell reverts to a mesenchymal state, typically through cytokine stimulation (1–3). EMT is highly associated with promoting tumor formation, tumor metastasis and the overall progression of cancer through the loss of the cell's epithelial characteristics, and the induction of mesenchymal properties (3). EMT is promoted by non-canonical transforming growth factor β (TGFβ) signaling and downstream activation of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/protein kinase B (Akt) pathway (4–6). This non-canonical pathway of TGFβ drives processes such as EMT, tumor formation and progression and most importantly, metastasis (7,8). We have previously identified a mechanism of translational repression by heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1) binding to a conserved 3′ untranslated region (UTR) nucleic acid motif in the mRNA of EMT-inducing genes. This interaction is responsible for stalling peptide synthesis at the elongation stage, and can be disrupted by the phosphorylation of serine-43 on hnRNP E1 (p-hnRNP E1) causing dissociation of the translational repression complex (9–11).
Akt is a serine/threonine kinase with three different isoforms, Akt1, Akt2 and Akt3 (12,13). This enzyme plays a central role in cellular physiology and its various isoforms have significantly different function when compared to one another. Tumor formation and metastatic progression are highly correlated to increased expression of Akt, as well as an increase in activity, measured by serine-474 phosphorylation (p-Akt2). When compared to normal epithelial tissue, tumor samples show an increased expression of Akt2 and p-Akt2 (14,15). Akt2 is a downstream activation target of non-canonical TGFβ signaling, and is implicated in the activation of multiple pathways associated with cellular growth and proliferation (16). We have previously implicated Akt2 as the only isoform of this enzyme that is capable of inducing p-hnRNP E1, and demonstrated the ability to halt TGFβ-induced EMT by inhibiting p-Akt2 kinase activity with a small molecule inhibitor (LY294005) (9,11).
RNA binding proteins (RBPs) are regulatory proteins that bind RNA, typically in its 3′ or 5′-UTR, and most frequently modulate protein expression (17,18). There have been multiple advances in high-throughput genomic sequencing allowing for the resolution of RBP target binding sites that typically conform to a structural motif. When these motifs are analyzed in an evolutionary context across divergent species orthologous genes, they show a high degree of motif conservation in a non-conserved UTR (18–20). Current genomic approaches to resolve RBP motifs involve the use of exonuclease digestion coupled to high-throughput sequencing (19,21,22). The region of RNA that is covered by RBP interaction is protected from exonuclease digestion through steric interference. Computational analysis of these results makes possible the resolution of a structural binding motif for a given RBP. hnRNP E1 is a highly expressed RBP that also has the ability to bind DNA, it has multiple sites of post-translational modification (PTM) that effect its binding characteristics differently. We have implicated a single hnRNP E1 PTM at serine-43 to cause a loss of affinity for a subset of EMT-inducing genes while not disturbing other interactions of hnRNP E1 targets (11). To this end, we have developed a novel in vitro exonuclease digestion assay that is capable of resolving a nucleic acid binding motif with specificity for a single RBP PTM. Pathway enrichment analysis of genes displaying increased translation upon dissociation of hnRNP E1 from their 3′-UTR regulatory motif correlates with the activation of multiple pathways implicated in metastatic progression. The correlation between high Akt2 and p-hnRNP E1 provides a functional basis for the overstimulation of non-canonical TGFβ signaling during metastatic progression in human breast and colon cancer cell lines.
MATERIALS AND METHODS
Cell culture
NMuMG, 67NR, 4TO7 and 4T1 cells were maintained and grown in Dulbecco's modified Eagle's medium (DMEM) media supplemented with 10% fetal bovine serum. MDA231, MDA435, MDA453, MDA468, SW480, SW620, HCT8, HCT116, HT29 and CaCO2 cells were maintained and grown in DMEM media supplemented with 5% fetal bovine serum.
Cloning of 3′-UTRs for select mRNA
Sequences of primers used for polymerase chain reaction (PCR) amplification of 3′ UTRs are shown in Table 1. For cloning of 3′-UTRs cDNAs were synthesized from mRNA in the total RNA pool extracted from NMuMG cells by reverse transcriptase (RT)-PCR according to manufacturer's instructions. DNA samples were further amplified using Vent DNA polymerase (New England Biolabs). The PCR products were cloned at the SmaI site of pUC18. All the clones contained the 3UTRs sequences under a T7 promoter.
Table 1. Primers used in the work.
| Gene | Direction | Sequence |
|---|---|---|
| Dab2 | Forward | AGATCTCGATCCCGCGAAATTAATACGACTCACTATAGGGGACCTCTGTCCTCGCTCCAGCTTTGACG |
| Dab2 | Reverse | GTAGGGCATAATGTCATCAGGGTCAAACAGCTGC |
| Egfr | Forward | GCGAAATTAATACGACTCACTATAGGGACCAGAAGCCATCTCTGACTCCCC |
| Egfr | Reverse | ATCAATAGTAAGAATTTATCAGAAACAAAATGATGAGAG |
| Fam3c | Forward | AGATCTCGATCCCGCGAAATTAATACGACTCACTATAGGGGACATGCTGCCCTTTGTGGGATGAGGCC |
| Fam3c | Reverse | CCCCACTGGGCCACAGCAGTTACAGAACC |
| Jak2 | Forward | GCGAAATTAATACGACTCACTATAGGGAAGAGATGGCCTTCACTCAGAGACCAAGC |
| Jak2 | Reverse | GACATGACATTGTCTAAGAGGGAGCAGCAC |
Transcription of synthetic mRNA
pUC18 based 3′-UTR clones under T7 promoter were transcribed by T7 RNA polymerase(New England Biolabs) according to manufacturer's instructions with or without [α-32P]-UTP and [α-32P]-CTP. Transcribed RNA samples were treated with RNase-free DNaseI, extracted with trizol solution, precipitated with ethanol and finally dissolved in TE.
Expression of recombinant proteins
Escherichia coli DH5α cells containing an Isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible GST-tagged protein expression plasmid were grown to an absorbance at 600 nm (A600) of 0.6, and induced with 0.4 μM IPTG for 4 h at 37°C with rotation at 270 rpm. After induction, bacterial cells were harvested and lysed with a bacterial lysis buffer (50 mM Tris–HCl pH 8, 300 mM NaCl, 20 mM Imidazole, 0.05% β-mercaptoethanol, 0.5% triton X-100, 1 mM PMSF, 0.5 μg/ml and 10 μg/ml aprotinin) for 30 min at 4°C. To clear lysed bacterial debris, samples were centrifuged at 100 000 × g for 30 min at 4°C. Cleared lysate was incubated with glutathione-agarose beads for a minimum of 1 h at 4°C with shaking. Purified protein was obtained by either elution with a reduced glutathione buffer (50 mM Tris–HCl pH 7.5, 10 mM reduced-glutathione), a minimum of 10-volumes of elution buffer was incubated for 5 min at 4°C, eluted protein was concentrated in a 3 kDa molecular weight cutoff centrifugation filter to a desired volume. For protein that was to be purified without its GST tag, washed protein bound glutathione-agarose beads were incubated for 18 h with PreScission protease (GE Healthcare), expressed protein was recovered by separating the supernatant of the reaction and concentration was performed on an as-needed basis.
RNA–protein binding assays
GST-hnRNP E1 protein purified from E. coli DH5α cells were immobilized on glutathione agarose beads. Radiolabeled or unlabeled RNA samples were bound to the immobilized beads and were incubated at 0°C for 10 min. Beads were washed with RNA–protein binding buffer (40 mM Tris–HCl pH 7.5, 30 mM KCl, 1 mM MgCl2, 0.01% NP40, 1 mM dithiothreitol) and finally eluted with 5 mM reduced glutathione in the RNA-protein binding buffer. The samples were extracted with Trizol and analyzed by 7M urea-10% polyacrylamide gel electrophoresis.
Recombinant protein exonuclease digestion protection assays
Samples of RNA and GST-tagged recombinant protein were incubated in an RNA–protein binding buffer (50 mM Tris–HCl pH 7.5, 100 mM KCl, 4% glycerol, 1 mM ethylenediaminetetraacetic acid (EDTA)) for 40 min at 4°C. Glutathione-agarose beads were added to the reaction and allowed to bind for 1 h at 4°C, with gentle shaking. A series of 50, 100, 150, 200 mM NaCl washes were performed to remove unbound RNA. RNase A was added at manufacturer recommended concentration, and allowed to digest samples for 10 min at 15°C. Samples were washed with the RNA–protein binding buffer and subjected to further analysis.
Preparation of digested mRNA for high-throughput sequencing
Fragments of RNA from recombinant protein exonuclease digestion protection assays were removed from bound protein by a combination of proteinase-K digestion, followed by trizol extraction and resolved by 7M urea-10% polyacrylamide gel electrophoresis (PAGE). To append a primer to the 3′ end of recovered, digested RNA, a modified (5′-/5rApp/ TTT AAC CGC GAA TTC CAG/3SpC3/-3′) primer was appended to RNA by ligation with RNA-ligase-2 (Rnl2) for 18 h at 4°C. Ligated oligonucleotides were resolved using PAGE. 5′ end ligation was achieved by incubation with a modified (5′-ACG GAA TTC CTC ACT rArArA-3′) primer appended to the hybrid DNA/RNA-oligonucleotide by incubation with T4-ligase for 18 h at 4°C. Ligated oligonucleotides were resolved using PAGE and subjected to further analysis.
Computational analysis of high-throughput sequencing results
Transformation from a .bam to .fastq was performed utilizing a custom conversion script implemented in Python (included in supplemental). A reference nucleic acid sequence database was constructed utilizing custom Python implementations (included in Supplementary Data), this database was subsequently used for Bowtie2 analysis (23) to determine gene of origin for respective reads. Evolutionary conservation and finite mapping of sequence was obtained through utilization of custom Python script implementations (included in Supplementary Data). Analysis of read data generated from scripts were further analyzed for statistical conservation, a minimal descriptor sequence describing a unique nucleic acid sequence responsible for the TGFβ-induced reversible binding with hnRNP E1.
In vitro translation assays
Luciferase open-reading frame (ORF) was obtained through PCR amplification of luciferase from pGL3-basic, with HindIII and BamHI cleavage sites (3′ and 5′ respectively) placed on ends of ORF. The ORF was amplified, and cloned into a pcDNA 3.1+ plasmid vector with the multiple-cloning site oriented at the 3′-UTR of Luciferase-ORF. Small, ∼50 nucleotide regulatory oligonucleotides were ligated to the cleaved pcDNA 3.1+-Luciferase plasmid to allow for transcription of RNA using T7 RNA polymerase. RNA was transcribed containing a luciferase promoter, ORF and 3′-UTR regulatory element. Transcribed RNA was incubated with recombinant hnRNP E1/p-hnRNP E1 and recombinant eEF1A1 protein. Incubation was performed for 5 min at room temperature and the RNA subsequently translated using Promega rabbit reticulocyte lysate protein translation system per manufacturers specifications.
RNA electromobility shift assays
Synthetic RNA and recombinant proteins were prepared as previously described (Transcription of synthetic mRNA) and allowed to incubate for 10 min at 4°C in RNA–protein binding buffer (40 mM Tris–HCl pH = 7.5, 30 mM KCl, 1 mM MgCl2, 0.01% NP40, 1 mM dithiothreitol). After binding, a loading buffer composed of 50% glycerol and bromophenol blue/xylene cyanol was added to samples. Samples were loaded into varying percentages of non-denaturing polyacrylamide gel, electrophoresed and autoradiographed.
RNA pull-down assays
Synthetically transcribed RNA was prepared as previously described (Transcription of synthetic mRNA). RNA was fixed to activated CNBr-agarose beads (GE Healthcare) according to manufacturer recommended procedures. RNA-bead mixture was washed with a wash buffer (50 mM Tris–HCl pH = 7.5, 4% glycerol, 1 mM EDTA) and added to mammalian cell lysate. Beads and lysate were incubated for 1 h at 4°C, and subsequently washed with wash buffer. Protein was eluted from beads using PAGE loading buffer, containing β-mercaptoethanol. Samples were subsequently transferred to membrane and analyzed for protein interaction using western blot analysis.
Phosphorylation of recombinant hnRNP E1
Recombinant substrate proteins were prepared as previously described (Expression of recombinant proteins), and incubated with recombinant Akt2 enzyme (Signal Chem) in kinase buffer (25 mM Tris–HCl pH = 7.5, 5 mM β-glycerophosphate, 10 μM ATP, 2 mM dithiothreitol, 0.1 mM Na3VO4, 10 mM MgCl2) and incubated for 15 min at 30°C. Recombinant substrates were removed by extraction with glutathione-agarose, washing and cleavage from GST-tag by PreScission (GE Healthcare) protease. Phosphorylated recombinant proteins were used in various analyses.
Plasmid construction and protein expression
pSilencer puro-shRNA-mouse hnRNP E1-3′-UTR (shRNA against 3′ UTR of hnRNP E1) clone was constructed by annealing shRNA template oligonucleotides (target selected through engine at Ambion and cloning into pSilencer neo vector as described (9). sh-RNA hnRNP E1 was transfected into NMuMG cells using Lipofectamine (Thermo), following manufacturer specifications. Silencing plasmid was selected using puromycin resistance and a stable mutant cell line was generated. This line is designated NMuMG-/-hnRNP E1. S43A (phosphomutant) and S43E (phosphomimetic) forms of hnRNP E1 were generated using Quick Change Site Directed Mutagenesis Kit, Stratagene, as described (9), and wild-type (WT) and mutant hnRNP E1 forms were cloned into pEGFP-C1 plasmid (neomycin selection) as Flag-tagged fusions. These constructs were transfected into hnRNP E1-deficient NMuMG-/-hnRNP E1 cells using Lipofectamine (Thermo), following manufacturer specifications and stable clones were selected in neomycin and puromycin selection media. The design of the hnRNP E1 sh-RNA against a portion of the 3′-UTR was with the intent of subsequent re-expression of exogenous hnRNP E1 forms that were resistant to the effects of the sh-RNA.
RESULTS
TGFβ regulated EMT-inducing genes are translationally regulated
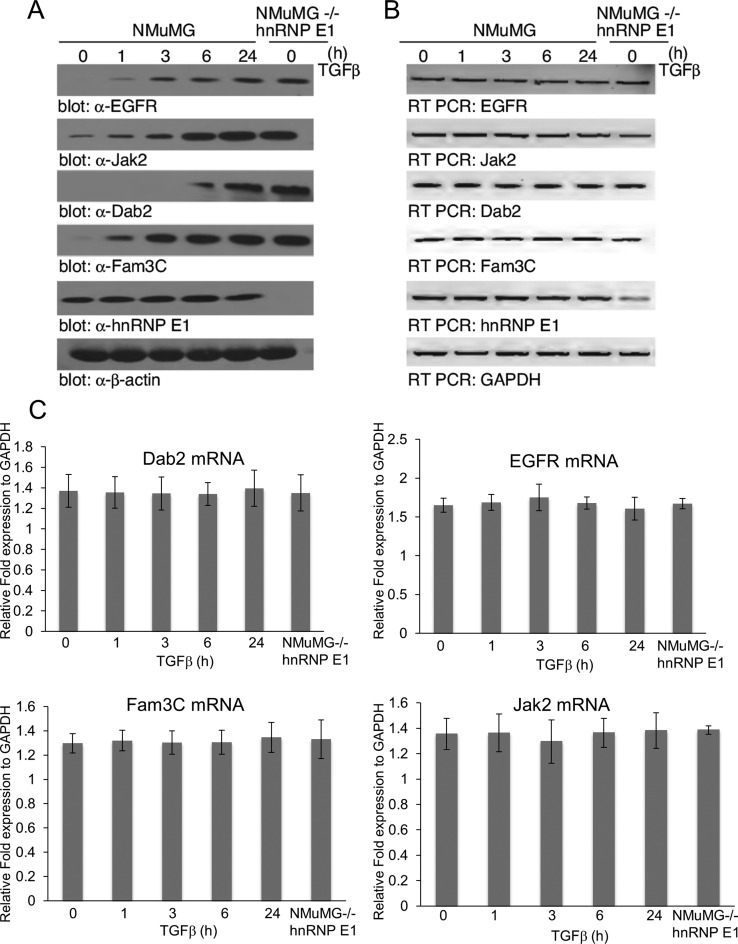
Previous work in our laboratory has identified a subset of EMT-inducing genes that show a signature non-canonical TGFβ translational upregulation through the dissolution of a regulatory complex containing hnRNP E1 (11). From these identified genes, we utilized a biased approach of choosing four candidates that showed a signature TGFβ/hnRNP E1 interaction. To validate these candidates, we first analyzed mRNA and protein levels with respect to TGFβ treatment and hnRNP E1 knockdown (-/- hnRNP E1) (Figure 1A). As shown, at 24 h of TGFβ treatment we observed a significant accumulation of Egfr, Dab2, Fam3c and Jak2 protein levels. In contrast, mRNA levels of these genes do not change in response to the silencing of hnRNP E1 or TGFβ treatment (Figure 1B and C). The discrepancy associated with an increase in protein translation and lack of increase in mRNA shows a translational regulation mechanism rather than transcriptional increase.
Figure 1.
Translational regulation of EMT-inducing genes is upregulated by TGFβ and knockout of hnRNP E1. (A) Immunoblot analysis of candidate motif containing genes where mRNA was previously identified as showing an inherent interaction with hnRNP E1 and whose polyribosomal induction was mediated by TGFβ stimulation. NMuMG cells and a mutant hnRNP E1 knockout version, NMuMG-/-hnRNP E1 were treated with 5 ng/ml TGFβ up to 24 h. (B) Semi-quantitative PCR analysis of mRNA levels from lysates used in (A). (C) Quantitative real-time PCR analysis of mRNA candidate genes, normalized to expression of GAPDH mRNA levels. Results are depicted as means +/− s.e.m. of three independent biological experiments.
TGFβ regulated EMT-inducing genes interact with hnRNP E1 in their 3′-UTR
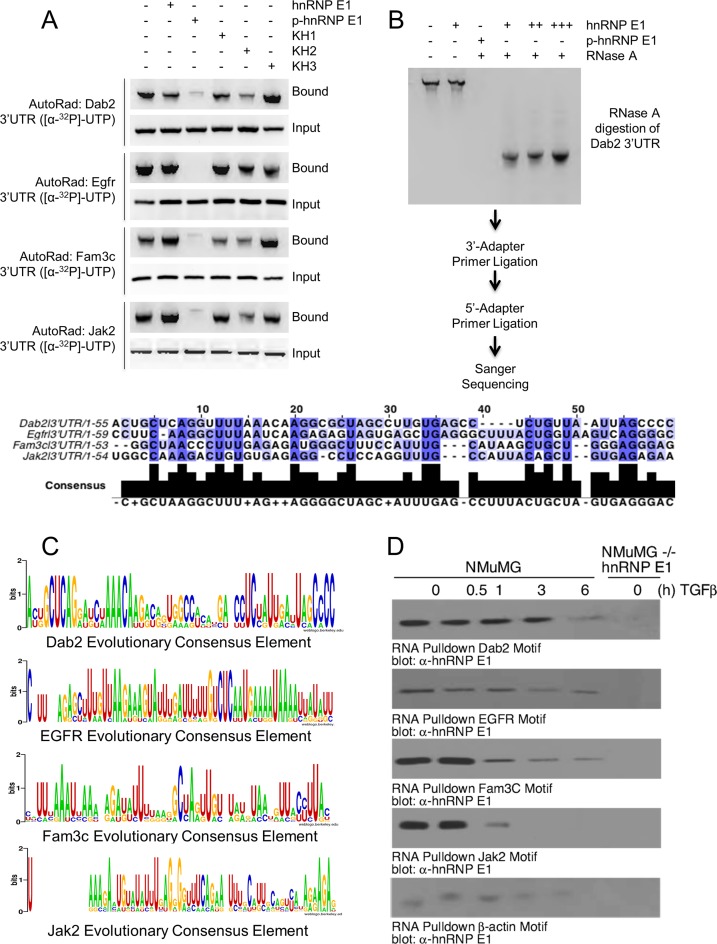
To assess the specificity of binding for 3′-UTRs of candidate genes, we developed an in vitro assay to determine binding affinity for hnRNPE1, its various KH domains (KH1, KH2, KH3) and p-hnRNP E1. 3′-UTRs of candidate genes were transcribed from plasmids containing a T7 RNA polymerase promoter and radioactively labeled. In this assay, GST-tagged proteins were attached to reduced glutathione-agarose beads and incubated with radioactively labeled RNA. Lanes labeled ‘input’ were an aliquot of the reaction prior to precipitation and washing of the beads to show equal loading of RNA. Lanes labeled ‘bound’ were samples of precipitated beads that were washed with a high salt buffer, and subsequently eluted with reduced glutathione buffer. If strong protein–RNA binding occurred, these complexes would stay together until the elution step, where the total volume was run on a gel. We observed (Figure 2A) a high affinity between full length 3′-UTR RNAs and hnRNP E1 as well as high affinity between 3′-UTRs and the three KH domains; however, p-hnRNP E1 samples had a nearly complete lack of affinity for binding 3′-UTR RNA.
Figure 2.
EMT-inducing genes bind hnRNP E1 through their 3′-UTR and are protected from exonuclease degradation. (A) Autoradiograph analysis of RNA bound to hnRNP E1. Rows marked with ‘Bound’ contain sample that was obtained through elution of recombinant protein by reduced glutathione elution buffer. Rows marked ‘Input’ contain sample that was obtained by aliquoting a portion of the reaction prior to elution to ensure equal loading of radioactive RNA. (B) Autoradiograph of exonuclease treated samples that were bound with either hnRNP E1 or p-hnRNP E1, titration is indicated by additional ‘+’ characters. Sanger sequencing of digested fragments protected by hnRNP E1 are shown in alignment below autoradiograph. (C) Evolutionary conservation analysis of sequences obtained by Sanger. Homologene (NCBI) database was used to obtain orthologous gene sequences for evolutionarily divergent organisms. These sequences were aligned by Clustal-omega using the fragment sequence as an alignment template. Height of nucleic acid base in consensus logo indicates its degree of conservation. (D) Immunoblot analysis of hnRNP E1 pulled down by synthetic RNA for each candidate gene from TGFβ (up to 24 h) treated lysate. The data presented are typical of three independent experiments demonstrating similar loss and gain of interactions.
To determine the region of 3′-UTR that was in contact with hnRNP E1, we utilized a modified exonuclease protection assay with homologous samples of synthetic RNA and recombinant hnRNP E1 / p-hnRNP E1 (Figure 2B). As hnRNP E1 protein was titrated into samples containing 3′-UTR RNA, we observed a smaller molecular weight RNA band appear and increase in intensity concomitant with hnRNP E1 titration (Figure 2B). When p-hnRNP E1 was used to protect RNA from digestion, the smaller band observed in hnRNP E1 samples disappeared, supporting our previous findings for a lack of affinity w/ p-hnRNP E1 (Figure 2B). After excising hnRNP E1 protected bands, we ligated 3′ and 5′ primers to these digested RNA fragments and performed Sanger sequencing. When sequencing results of these 4 candidate UTR digestion products were aligned with Clustal-omega, we observed a high degree of conservation, particularly with pyrimidine residues (Figure 2B).
A hallmark of previously resolved regulatory motifs (for other RBPs) is the conservation of the motif sequence throughout evolutionarily divergent species orthologous genes (24–26). Utilizing this observation, we developed a custom computational algorithm implemented in Python to analyze the conservation of our sequenced element in comparison to orthologous genes across evolutionary divergent species. We show a graphical presentation generated using WebLogo (27,28) for the degree of conservation of each base we obtained from our exonuclease protection assay compared to orthologous genes of evolutionarily divergent species (Figure 2C). Alignments revealed further conservation of pyrimidine bases throughout evolution, supporting the data obtained from aligning our four candidate genes sequenced fragments. To provide in vivo evidence of TGFβ induced loss-of-affinity between a putative regulatory motif and hnRNP E1, we utilized an RNA pull down assay where RNA transcripts were synthesized according to our candidate gene sequencing results and attached to CNBr beads. When these RNA motif-coupled beads were incubated with total cellular lysate from NMuMG cells treated with TGFβ, we observed a significant loss of interaction with hnRNP E1 (Figure 2D) as a function of TGFβ stimulation. These data provide further evidence for a loss of affinity between this 3′-UTR RNA motif and p-hnRNP E1.
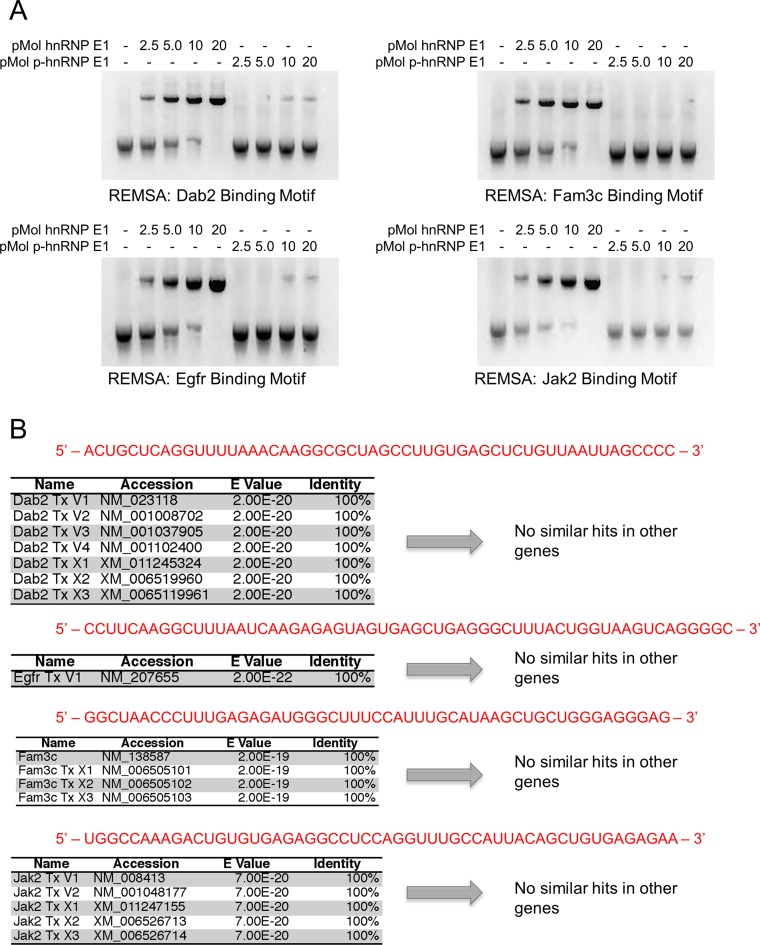
Affinity characterization of putative motifs and hnRNP E1
To further characterize the interaction between putative RNA motif sequences and hnRNP E1, we performed RNA electromobility shift assays (REMSA). We observed a strong interaction for putative motifs and hnRNP E1 (Figure 3A), and a near total loss of interaction with p-hnRNP E1. After generalized characterization of binding kinetics and interactions between newly resolved consensus elements, we set out to determine the uniqueness of each resolved element by using the basic local alignment search tool (BLAST) (available at https://blast.ncbi.nlm.nih.gov/) against genomic mRNA databases. We observed no significant hits outside the candidate gene isoforms of mRNA when the sequenced motif element was used as a search parameter (Figure 3B). There are similar features found in each of the four candidate motifs; however, their sequence homology to one another is low. In order to determine a global consensus motif, we needed to alter our biased approach of resolving individual 3′-UTRs to an unbiased approach using genomic mRNA from cells.
Figure 3.
hnRNP E1 binding nucleic acid motifs show a total loss of affinity for hnRNP E1 when in its phosphorylated state. (A) RNA electromobility shift assay (REMSA) analysis of synthetic regulatory motif RNA. hnRNP E1 and p-hnRNP E1 were titrated (0–20 pMol) with 1 pMol of motif RNA. RNA was labeled with [α-32P]-UTP during transcription, samples were run on non-denaturing polyacrylamide gel. The REMSA data presented are typical of three independent experiments demonstrating similar binding trends (data not shown). (B) BLASTn analysis of conserved motif results to determine uniqueness of these regulatory motifs in a genomic context. Sanger sequencing results were used as a query sequence (highlighted in red), stringency parameters were lowered to allow for the identification of patterns, rather than specific bases. Queries were performed against a mouse genomic database, containing genomic mRNA sequences.
Genomic binding motif identification of TGFβ/hnRNP E1-regulated genes
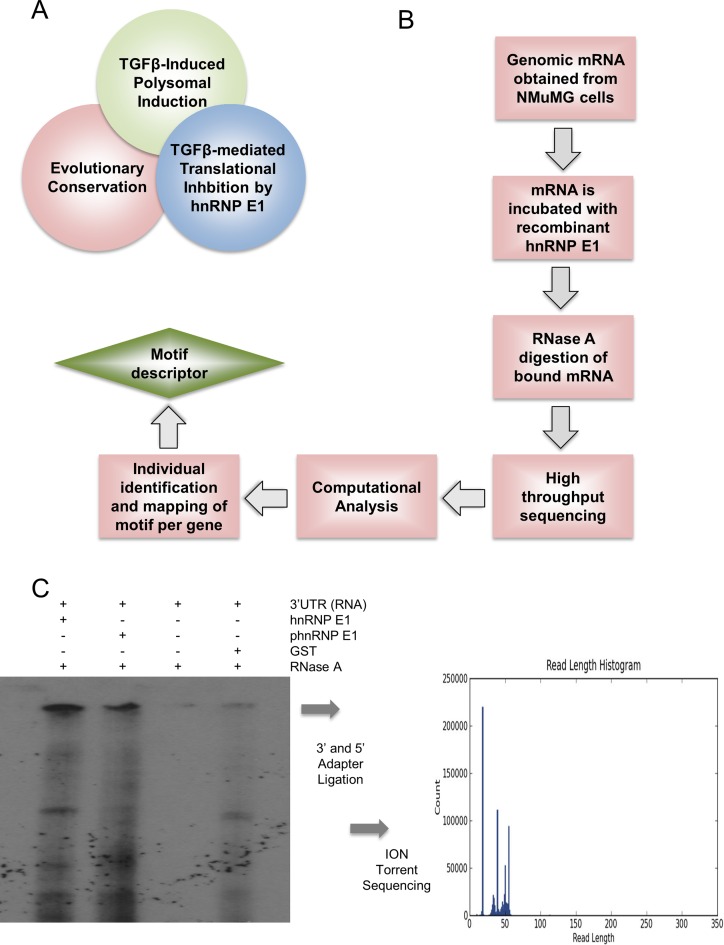
Figure 4A is a Venn diagram depicting the distinguishing features of p-hnRNP E1-regulated mRNAs: (i) exhibit translational silencing by hnRNP E1; (ii) display polyribosomal induction (high protein translation) when the hnRNP E1 regulatory complex is dissociated; and (iii) have evolutionary conserved binding motifs that are responsible for interaction with hnRNP E1. We set out to identify the conserved nucleic acid regions that interact with hnRNP E1 on a genomic scale similar to our approach using synthetic RNA. We replaced synthetic RNA with genomic mRNA obtained from NMuMG cells and incubated with hnRNP E1 or p-hnRNP E1 in an effort to cancel out non Ser43 domain protected fragments. To better illustrate the methods that were utilized to obtain a genomic profile of motif sequences and structure, we have included a flow chart showing the procedure and analysis techniques (Figure 4B). After exonuclease digestion, we observed multiple protected fragments of RNA in both hnRNP E1 and p-hnRNP E1 samples; however, a unique band of protected RNA was only present in hnRNP E1 incubated samples (Figure 4C). This nearly 50-nucleotide length band was excised and ligated with terminal 5′ and 3′ sequencing primers to undergo Ion Torrent sequencing. We obtained approximately 800 000 reads of sufficient quality that mapped to our genomic mRNA database. These reads ranged in size from ∼22 to 54 nucleotides in length and showed similar characteristics amongst them when aligned using simple Clustal-omega multiple sequence alignment (data not shown). Combining these newly resolved motif sequences with previous data from our laboratory demonstrating that p-hnRNP E1-regulated mRNAs are found to be enriched in polyribosomal pools when stimulated with TGFβ has confirmed the three distinct features that these mRNAs uniquely share (Figure 4A).
Figure 4.
Unbiased exonuclease digestion of hnRNP E1 bound target mRNA allows for high-throughput analysis of protected RNA fragment sequences. (A) Venn diagram depiction of genomic characteristics displayed by p-hnRNP E1 responsive genes. (B) Flow diagram of unbiased, genomic approach for resolving 3′-UTR consensus motif. This modification to common exonuclease assays utilizes homologous recombinant proteins for the sake of lowered background and increased specificity of sequencing results. (C) Exonuclease digestion assay for genomic RNA obtained from NMuMG cells (using trizol extraction) and bound to either hnRNP E1 or p-hnRNP E1. p-hnRNP E1 serves as a control to account for the promiscuous nature of hnRNP E1 for binding other RNAs that are not specific to its serine-43 reversible binding region. A unique band found only in samples protected by hnRNP E1 was ligated with 3′ and 5′ sequencing primers and sequenced using ION-torrent sequencing.
Analysis of high-throughput sequencing results
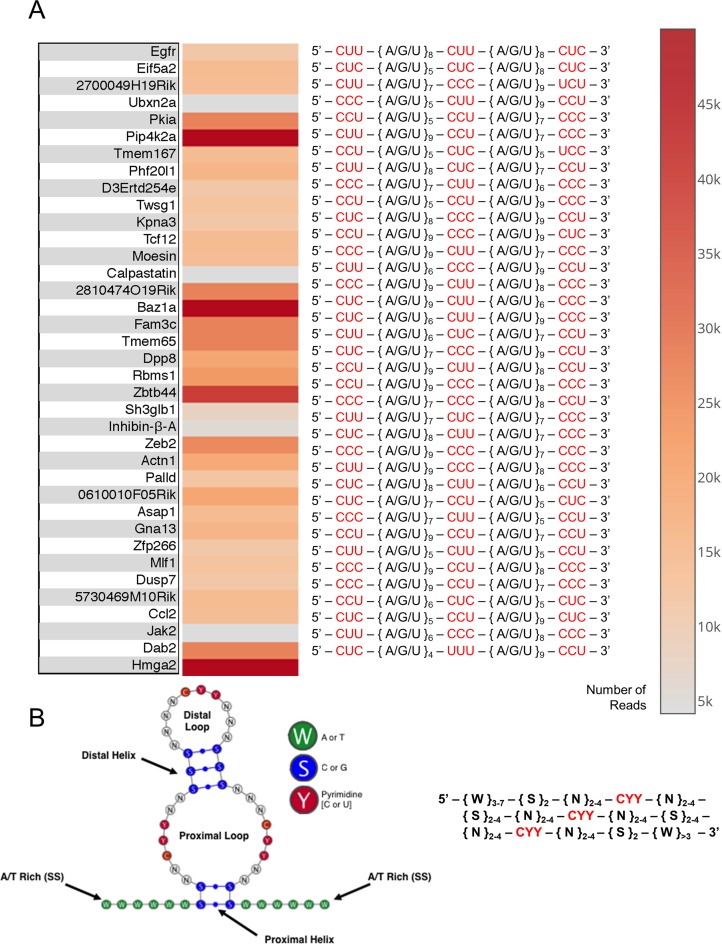
We further analyzed Ion-torrent sequencing results by developing a computational algorithm that utilizes Bowtie2 and Bioconductor packages to align sequence reads with their parent genes and derive positional information of the read sequence in the context of parent gene mRNA (23,29). After stringent analysis composed of mapping reads to their parent genes, analyzing for evolutionary conservation and resolving motif position, we obtained positions for each of our EMT-inducing genes (Figure 5A) putative binding element. We have created a heat map to correlate the number of reads from ION sequencing for individual genes validated to contain this motif and listed the nucleic acid sequence of the motif, with its highly conserved residues highlighted in red (Figure 5A). We observed three-triple pyrimidine repeats that are spaced by 5–9 variable nucleotides, supporting our hypothesis that the motif responsible for binding hnRNP E1 (serine-43 region) is highly variable in sequence, yet rich in pyrimidine bases.
Figure 5.
Genomic exonuclease ION torrent analysis reveals 37 consensus motifs with conserved pyrimidine residues interspaced by variable structural regions. (A) Genes that were identified to be bound to hnRNP E1 and protected from exonuclease digestion. Heat map indicates number of times a sequenced read was mapped to this parent mRNA. Annotated nucleic acid sequences show conserved pyrimidine residues highlighted in red, interspaced regions annotated to better show alignment. Parent genes were determined for each sequenced read by a custom Python script and custom 3′-UTR database containing mouse genomic 3′-UTR sequences from (UTRdb). (B) Structural depiction of consensus motif predicted by custom Python structural prediction algorithm, generalized hnRNP E1 specific binding motif descriptor. Descriptor sequence was inferred by a statistical comparison of all sequenced reads, followed by evolutionary conservation analysis, performed by a custom Python script.
To further annotate these newly resolved binding motifs, we developed a computational algorithm that analyzed each resolved motif and determined a minimal consensus motif descriptor sequence (Figure 5B). When this minimal descriptor sequence was analyzed for secondary structure, we observed the formation of a dual single strand loop formation, with two G-C helices of variable length (Figure 5B). This newly determined descriptor motif was analyzed with BLASTn and found to contain an approximately 92.7% overlap with BLASTn predicted genes and genes contained in Figure 5A.
hnRNP E1 binding characterization of consensus motif
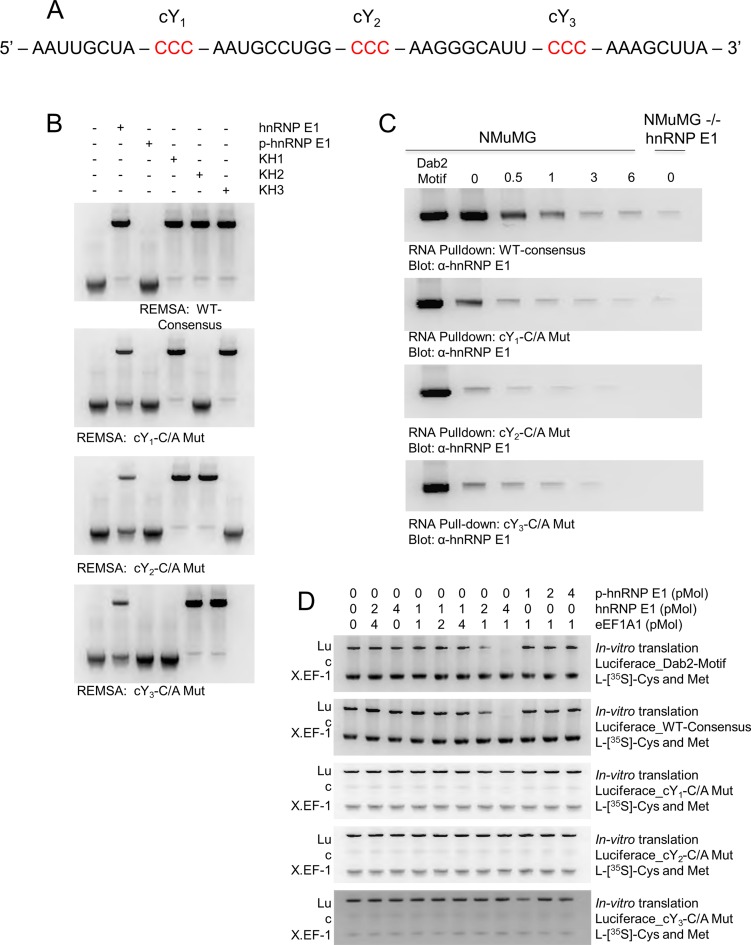
We developed a computational script implemented in python to generate a randomized nucleic acid sequence following the constraints of our descriptor motif. We tested six randomized consensus elements and found similar binding characteristics with no significant changes in affinity for hnRNP E1, KH1, KH2, KH3 or p-hnRNP E1, and carried out the remainder of experiments utilizing a randomized consensus element with the sequence described in Figure 6A. We hypothesized the highly conserved bases (highlighted in red, Figure 6A) were the main residues in the motif that were responsible for binding to hnRNP E1 and set out to test this hypothesis using REMSA analysis. Previous data in our laboratory suggested the point mutation of a residue contained in a putative Dab2/hnRNP E1 binding motif (determined by systematically breaking apart and shortening the 3′-UTR of Dab2) could significantly lower the affinity for hnRNP E1. Using this principle, we mutated the first conserved C residue in each of the 3-pyrimidine rich regions (termed cY1, cY2, and cY3, 5′ to 3′, respectively) to determine the overall effect of binding affinity for hnRNP E1, and to determine domain specificity for the various KH domains (Figure 6B). Our WT consensus motif showed a high affinity for hnRNP E1, KH1, KH2 and KH3, and near total loss of affinity for p-hnRNP E1. Upon mutation of cY1, we observed a significantly lower affinity for full-length hnRNP E1, and a complete loss of affinity for KH2, suggesting an interaction between cY1 and KH2. Systematic mutation of a conserved base in cY2 and cY3 reveals interaction between cY2 and KH3, and cY3 and KH1 (Figure 6B).
Figure 6.
Conservation of descriptor motif features in randomized synthetic RNA maintains regulatory motif properties in protein translation. (A) A randomized hnRNP E1-specific consensus motif generated by a custom Python script capable of randomizing oligonucleotides based on descriptor constraints. This sequence was used as the ‘WT-Consensus’ RNA for experiments contained in this figure. (B) RNA electromobility shift assay (REMSA) for WT-Consensus and subsequent mutations at conserved pyrimidine residues. Recombinant K-homology (KH) domains of hnRNP E1 were used to determine interactions with conserved motif regions. The REMSA data presented are typical of three independent experiments demonstrating similar binding trends (data not shown). (C) RNA pull-down analysis of WT-consensus motif compared to a bonafide Dab2 Motif element. Cellular lysate from NMuMG cells were treated with 5 ng/ml TGFβ for up to 6 h, hnRNP E1 interactions were measured through pull-down by various synthetic RNA constructs. Pull-down assays were performed a minimum of three times, similar binding trends were noted in each replicate (data not shown). D)In vitro translation assay of luciferase mRNA coupled to various 3′-UTR regulatory elements. Luciferase_Dab2-Motif mRNA was used as a control to compare strength of translation inhibition amongst WT-consensus and its various mutations. These assays were performed a minimum of three times, measurements were taken using luminescent units and similar trends were noted among replicates. For visual interpretation, we present radiolabeled samples analyzed by polyacrylamide gel electrophoresis.
To characterize this randomized consensus motif in the context of in vivo protein binding, we attached a WT and subsequent cY mutations to CNBr beads to assess their interactions with cellular hnRNP E1 from lysates treated with TGFβ. We observed a high-affinity interaction of hnRNP E1 and our WT motif, along with a weakening of affinity for each of the mutations induced in cY regions (Figure 6C). Mutations to conserved regions in motif elements cause a high loss of affinity for hnRNP E1 and could potentially mimic the effect of p-hnRNP E1.
In vitro translation effects of conserved motif mutations and p-hnRNP E1
We have developed a reporter assay for protein translation utilizing luciferase mRNA with a consensus regulatory element appended to its 3′ end. This system mimics the translational repression mechanism and allows for a better contextual analysis of translational inhibition induced through hnRNP E1 and the nucleic acid motif. Translation of luciferase was completely inhibited with the addition of 1 pMol luciferase-motif RNA, 4 pMol of hnRNP E1 and 1 pMol eEF1A1 (Figure 6D). When a similar ratio of p-hnRNP E1 to eEF1A1 was incubated with luciferase-motif RNA, no inhibition of protein translation occurred, further strengthening our observation of this motif's lack of binding affinity to p-hnRNP E1. Subsequent mutations to conserved pyrimidine residues caused a slight decrease in the translation of luciferase; however, they were incapable of complete inhibition (Figure 6D).
Differential expression of p-hnRNP E1 and p-Akt in cells
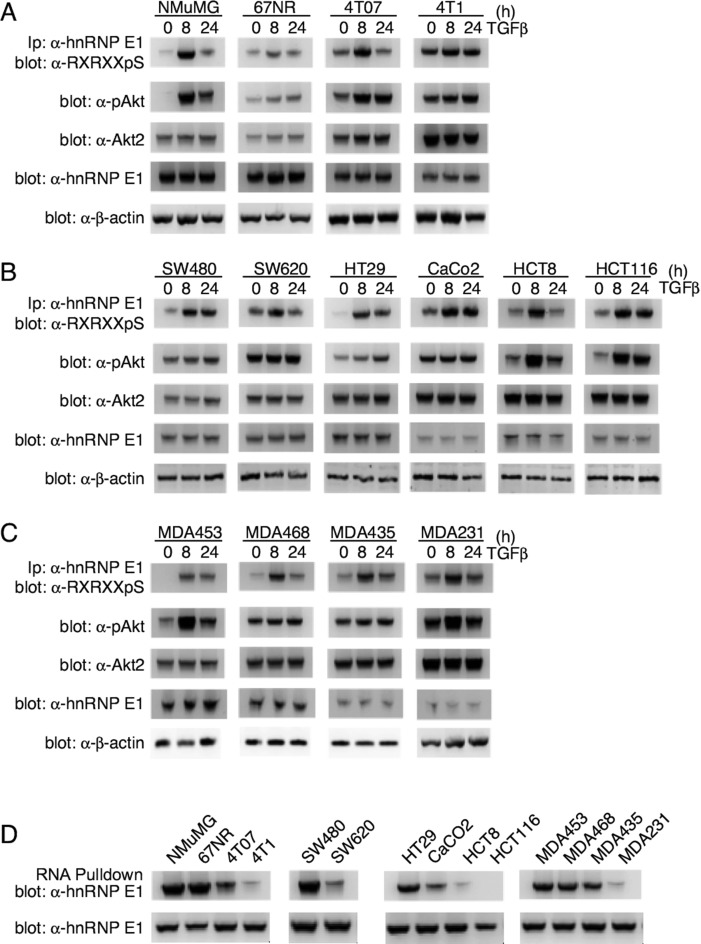
We have characterized and identified nucleic acid motifs that are responsible for the reversible translational stalling through binding to hnRNP E1. To determine the physiological significance of this mechanism, we set out to analyze key members of the non-canonical TGFβ pathway, which drive the phosphorylation of hnRNP E1Ser43. We utilized the 4T1 breast cancer progression model which includes the 67NR that are tumorigenic and not metastatic, the 4TO7 which are metastatic, but the metastases do not form secondary tumors, and the 4T1 which are metastatic and form secondary tumors (30). This progression model was used to compare relative levels of p-hnRNP E1, p-Akt, Akt2 and hnRNP E1, to a normal murine mammary gland (NMuMG) cell line as a means of assessing the role of these proteins and their PTMs in cancer progression.
We observed a low expression of p-hnRNP E1 in NMuMG lines that increased across the progression series (Figure 7A). These p-hnRNP E1 levels are greatly increased upon TGFβ treatment in NMuMG and 4T07 lines, and not significantly altered in 67NR and 4T1 cells (Figure 7A). To assess the activity of Akt2, we measured levels of phosphorylated Ser474 residue (p-Akt2). NMuMG cells show virtually no p-Akt2 levels without TGFβ treatment, and when stimulated, exhibit a significant increase in its activity (Figure 7A). While responsive to TGFβ, the progression cell lines do not exhibit such a large shift in the activity of Akt2; it remains constitutively active in the progression series (Figure 7A). Total Akt2 levels were not affected by TGFβ treatment but basal Akt2 levels did appear higher in the more aggressive 4T07 and 4T1 cells. Similarly, total hnRNP E1 levels were unaffected by TGFβ but did appear to be lower in the aggressive 4T07 and 4T1 cells compared to NMuMG and 67 NR cells (Figure 7A).
Figure 7.
pAkt2 and p-hnRNP E1 are significantly upregulated in metastatic cancer lines. (A) Immunoblot analysis of p-hnRNP E1, p-Akt, Akt2 and hnRNP E1 in NMuMG cells and the 4T01 metastatic progression series. 67NR cells are tumorigenic and not metastatic, 4TO7 are metastatic, yet the metastases do not form secondary tumors and 4T1 cells are metastatic and form secondary tumors. Akt2 activation is measured by assaying phosphorylated Ser474 (p-Akt2), and p-hnRNP E1 levels are analyzed through a combinatorial α-hnRNP E1 immunoprecipitation followed by α-Akt2 phospho-substrate immunoblotting. The immunoblot data presented are typical of three independent experiments demonstrating similar levels of protein expression levels. (B and C) Human colon (B) and breast (C) cancer cell lines of varying metastatic potential were analyzed as in (A). We utilized the progression series analysis as a means of correlating metastatic potential to human tumor cell lines. The immunoblot data presented are typical of three independent experiments demonstrating similar levels of protein expression levels. (D) To demonstrate the effect that p-hnRNP E1 : hnRNP E1 ratios play on interaction with a consensus motif element, we performed an RNA pulldown assay followed by α-hnRNP E1 immunoblot analysis. When compared with total hnRNP E1 levels, a significant loss of interaction is noted in highly aggressive cell lines.
We next validated this Akt2/hnRNP E1 mechanism in the context of well-characterized human cancer lines (Figure 7B and C). We utilized multiple colon (Figure 7B) and breast cancer (Figure 7C) cell lines of varying metastatic capacity and, similarly to the 4T1 progression model, observed a correlation of high p-hnRNP E1, p-Akt2 and Akt2, along with low hnRNP E1 expression, with the metastatic and aggressiveness of the colon (Figure 7B) and breast cancer cell lines (Figure 7C). These observations support our previous demonstration that TGFβ-mediated Akt2 activation (p-Akt2) leads to the phosphorylation of hnRNP E1 (9,10), and is suggestive of a correlation between the high constitutive levels of p-Akt2 and high p-hnRNP E1/hnRNP E1 ratio observed in metastatic cells compared to normal and non-metastatic epithelium.
We next used our identified consensus RNA element to access the affinity of hnRNP E1 binding in cellular lysates prepared from the various normal and cancer lines, reasoning that hnRNP E1 binding affinity should negatively correlate with the p-Akt2/Akt level and metastatic potential of the cells. Using the identified RNA consensus element CNBr-coupled to agarose beads to pull down hnRNP E1 from cell lysate, the data of Figure 7D demonstrate the affinity of hnRNP E1 binding decreases with the metastatic potential of the cells. In normal and less aggressive cell lines (NMuMG, 67NR, SW480), hnRNP E1 can be precipitated by the RNA consensus whereas in aggressive and metastatic cells (HCT8, HCT116, MDA231) less hnRNP E1 is pulled down. These relative binding affinities correlate with the relative level of p-hnRNP E1 observed in each cell line (Figure 7A–C) and suggest that low interaction between this RNA element and hnRNP E1 plays an important role in the metastatic process.
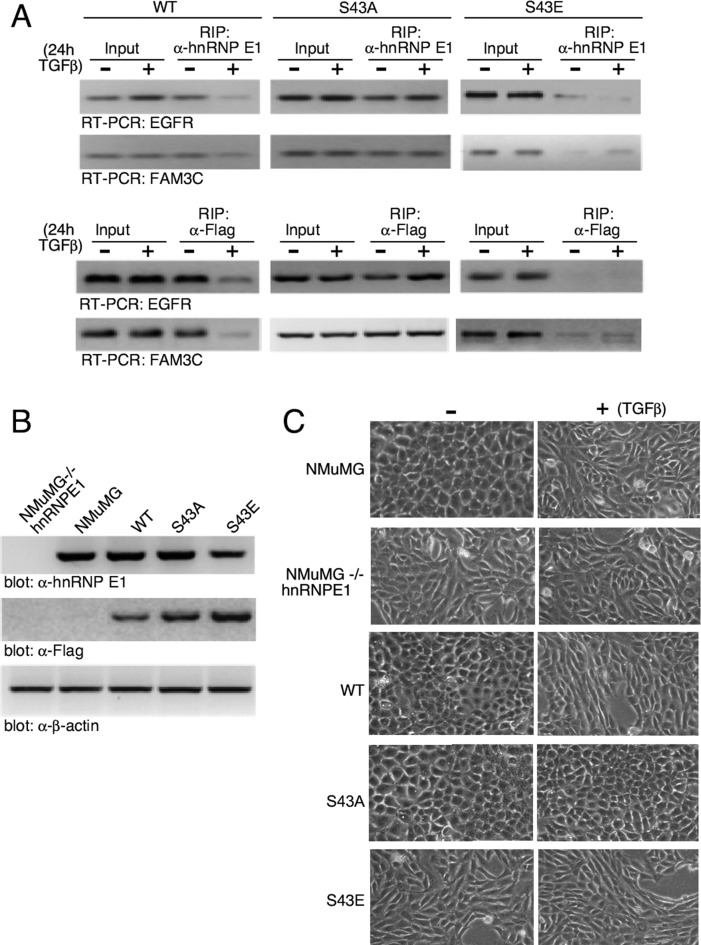
To confirm the relevance of our findings in cells we performed RNA immunoprecipitations (RIPs) in NMuMG cells modulated for hnRNP E1. We generated a stable NMuMG cell line in which hnRNP E1 was attenuated by sh-RNA-mediated silencing (-/-hnRNP E1) and stable clones re-expressing WT, a Ser to Glu (S43E), or a Ser to Ala mutant at position 43 (S43A). The sh-RNA was designed to target the 3′-UTR of hnRNP E1 such that re-expression of exogenous hnRNP E1 constructs, lacking the 3-UTR, were resistant to the effects of the sh-RNA. The expression of hnRNP E1 and of the Flag-tagged constructs in the various cell lines is depicted in Figure 8B. For RIP analyses, WT, S43A and S43E cells were treated −/+ TGFβ for 24 h and cellular lysates were prepared, immunoprecipitated with α-hnRNP E1 (Figure 8A; upper panel) or α-Flag (Figure 8A; lower panel), and EGFR and FAM3C mRNA levels were analyzed by RT-PCR analysis. We observed that in cells expressing WT hnRNP E1 (WT), TGFβ treatment led to a decrease in the abundance of EGFR and FAM3C transcripts immunoprecipitated compared to control untreated cells. In the S43A hnRNP E1 reconstituted cells, we observed no change in the binding of these transcripts in the presence or absence of TGFβ. Cells reconstituted with S43E saw a near total loss of interaction between transcripts that was independent of TGFβ treatment. Similar effects were observed whether the immunoprecipitations were performed with α-hnRNP E1 or α-Flag antibodies suggesting that the observed effects were being mediated through exogenously expressed hnRNP E1 forms. Further demonstration that this identified motif, and more specifically its Ser43 site, is of physiological significance is provided in Figure 8C where we demonstrate the morphological effects of modulating Ser43 phosphorylation levels. As shown, NMuMG cells have an epithelial, cobblestone monolayer appearance which transitions to a more spindle-like fibroblastic morphology upon TGFβ treatment. This represents the classic TGFβ-mediated EMT. Stable knockout of hnRNP E1 (NMuMG-/-hnRNP E1) renders the mesenchymal phenotype to cells irrespective of TGFβ treatment. Exogenous expression of WT hnRNP E1 into hnRNP E1 knockdown cells rescues the epithelial phenotype and TGFβ treatment is able to induce EMT in these rescued clones. Re-expression of the S43A phosphomutant, rescues the epithelial phenotype in the hnRNP E1 attenuated cells; however, TGFβ is unable to induce the transition to the mesenchymal state in this clone. On the other hand, mere re-expression of the S43E phosphomimetic mutant does not rescue the epithelial phenotype to the hnRNP E1 cells and TGFβ is without effect when added to these cells. These data suggest that TGFβ-induced phosphorylation of Ser43 of hnRNP E1 results in loss of hnRNP E1 binding to the identified consensus motif in these transcripts, resulting in loss of translational repression that is required for TGFβ-mediated EMT.
Figure 8.
hnRNP E1 silenced cells reconstituted with WT, phosphomutant S43A or phosphomimetic S43E forms of hnRNP E1. (A) hnRNP E1-deficient NMuMG cells (NMuMG-/-hnRNP E1) were stably reconstituted with WT, S43A, S43E forms of hnRNP E1 and stable clones were treated −/+ TGFβ for 24 h. Cellular lysates were prepared and immunoprecipitated with α-hnRNP E1 (upper panel) or α-Flag (lower panel) to precipitate protein–mRNA complexes and RT/PCR was used to amplify mRNAs for EGFR and FAM3C. The data presented are typical of three independent experiments demonstrating similar binding trends (data not shown). (B) Immunoblot analysis confirming sh-RNA-mediated knockdown of hnRNP E1 in NMuMG-/-hnRNP E1 cells and reconstituted expression of wild-type (WT) and S43A (phosphomutant) and S43E (phosphomimetic) forms of Flag-tagged hnRNP E1. Blots depict levels of endogenous or overexpressed forms of hnRNP E1 using either α-hnRNP E1 or α-Flag immunoblotting. (C) Phase contrast images of unstimulated and TGFβ-treated (24 h) cells examining morphological changes after stimulation with TGFβ.
DISCUSSION
This study has identified a nucleic acid descriptor sequence capable of identifying nucleic acid sequences that specifically binds hnRNP E1 in a phospho-serine 43-dependent manner. We have further identified, on a genomic scale, all genes that contain this unique motif and precisely mapped its location within their respective 3′-UTRs. We have identified three conserved regions of triple-pyrimidine repeats that are separated by a highly variable region and mapped their interaction with the various KH domains of hnRNP E1. The mutation of a conserved pyrimidine residue to a purine causes a significant loss of affinity for hnRNP E1, opening the possibility for mutations that are incorporated into the genome to cause a loss of translational repression for EMT-inducing genes. Since hnRNP E1, also known as polyC binding protein 1 (PCBP1), has previously been shown to be involved in mRNA stability and splicing (31,32), it will be of interest to examine whether our identified binding consensus mediates these effects through non -3′-UTR motifs.
The observation of a single nucleotide mutation to conserved residues in our regulatory motif, coupled to the genomic identification of all genes in which it is contained allows for the prospective of analyzing single nucleotide polymorphisms (SNPs) in a clinical context. The growing number of publicly available SNP arrays in databases such as The Cancer Genome Atlas (TCGA, http://www.cancergenome.nih.gov/) provide a source of data to be correlated to these newly identified and mapped regulatory elements. As shown by in vitro translation assays, a single pyrimidine to purine mutation in CY1, CY2 or CY3 causes a complete loss of translational inhibition, we hypothesize a similar mutation in vivo could be responsible for inducing tumor formation and metastasis. Currently, we are developing a computational algorithm to address this hypothesis and provide further clinical evidence of the misregulation of these motif-containing genes’ role in the development of cancer.
We provide further insight into the kinetics of binding for hnRNP E1 and its target binding motifs. We have identified two aspects of this repression system that can induce its dissociation and cause a loss of translational repression: (i) mutations to conserved pyrimidine bases in target 3′-UTR motifs; and (ii) increased p-hnRNP E1 levels. As demonstrated by our group and others, the interaction between Akt2 and hnRNP E1 is of profound importance in driving EMT and cancer metastasis (2,4,6). We have provided compelling evidence that Akt2 levels shift TGFβ signaling to induce the phosphorylation of serine-43 (hnRNP E1), facilitating its release from a transnational silencing complex, and have shown a clear correlation between the metastatic potential of human cancer cells and their levels of Akt2 and hnRNP E1.
Levels of p-Akt2 and p-hnRNP E1 are constitutively low in normal epithelial tissue, yet treatment with TGFβ increases their expression to levels observed in tumorigenic and metastatic cell lines. There are currently no protein expression profiles in any of the larger cancer databases that assay all of the EMT-inducing genes identified by this study. A future goal of this group is to develop a high-throughput assay capable of detecting Akt2, p-Akt2, hnRNP E1 and p-hnRNP E1 in patient tumor samples to be employed as a prognostic indicator. This assay would couple to a validation screen that assesses the protein levels of EMT-inducing genes with an over-abundance of p-hnRNP E1. A current hurdle to the development of such a high-throughput assay is the detection of p-hnRNP E1. Currently the only method to detect this modification involves utilizing a time-consuming method of immunoprecipitation coupled to immunoblot analysis with an Akt2 substrate recognition antibody. There is no commercially available antibody capable of detecting p-hnRNP E1 and use of the current two-pronged detection method would not be suitable in a high throughput screen.
The development of diagnostic clinical screens and implication of p-hnRNP E1 in cancer will prove useful for identifying a new target for therapeutic treatment. Current drugs on the market capable of lowering p-hnRNP E1 levels target this pathway at early activation points by inhibiting Akt2 or PI3K. Unfortunately, these drugs have many adverse side effects due to the pleiotropic effects of Akt2 and PI3K in cells and postulate that a small molecule capable of specifically binding hnRNP E1 and masking and inhibiting its phosphorylation at serine-43 might prove be more efficacious and specific as an anti-metastatic therapy.
We hypothesize the protein expression of p-hnRNP E1 regulated genes is necessary to the development of an organism, yet catastrophic at later stages of life. As an alternative to a drug-based therapy, we propose a synthetic gene replacement technique whereby a mutated hnRNP E1 (serine-43 to alanine) would tightly bind and silence protein expression disallowing for TGFβ induced phosphorylation. Our laboratory has in vitro evidence of this hnRNP E1 mutation's ability to irreversibly bind targets and inhibit the accumulation of p-hnRNP E1 (data not shown), and we are currently developing this system to be introduced to an in vivo model.
Supplementary Material
Acknowledgments
We thank Drs George Hussey, Breege Howley and other members of our laboratory for helpful comments and critical insights.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute [CA55536, CA154664 to P.H.H ]. Funding for open access charge: Medical University of South Carolina, Department of Biochemistry and Molecular Biology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Huber M.A., Kraut N., Beug H. Molecular requirements for epithelial–mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Thiery J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 3.Xu J., Lamouille S., Derynck R. TGF-β-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hills C.E., Squires P.E. TGF-beta 1-induced epithelial-to-mesenchymal transition and therapeutic intervention in diabetic nephropathy. Am. J. Nephrol. 2010;31:68–74. doi: 10.1159/000256659. [DOI] [PubMed] [Google Scholar]
- 5.Lee J.M., Dedhar S., Kalluri R., Thompson E.W. The epithelial–mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miettinen P.J., Ebner R., Lopez A.R., Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massagué J. How cells read TGF-β signals. Nat. Rev. Mol. Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 8.Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhury A., Hussey G.S., Ray P.S., Jin G., Fox P.L., Howe P.H. TGF-β-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussey G.S., Chaudhury A., Dawson A.E., Lindner D.J., Knudsen C.R., Wilce M.C., Merrick W.C., Howe P.H. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol. Cell. 2011;41:419–431. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussey G.S., Link L.A., Brown A.S., Howley B.V., Chaudhury A., Howe P.H. Establishment of a TGFβ-induced post-transcriptional EMT gene signature. PLoS One. 2012;7:e52624. doi: 10.1371/journal.pone.0052624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agarwal E., Brattain M.G., Chowdhury S. Cell survival and metastasis regulation by Akt signaling in colorectal cancer. Cell. Signal. 2013;25:1711–1719. doi: 10.1016/j.cellsig.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin Y.M.R., Balk S., Toker A. Akt2-specific signaling in prostate cancer maintenance. Cancer Res. 2013;73:4090. [Google Scholar]
- 14.Mulholland D.J., Kobayashi N., Ruscetti M., Zhi A., Tran L.M., Huang J., Gleave M., Wu H. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72:1878–1889. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue G., Restuccia D.F., Lan Q., Hynx D., Dirnhofer S., Hess D., Rüegg C., Hemmings B.A. Akt/PKB-mediated phosphorylation of Twist1 promotes tumor metastasis via mediating cross-talk between PI3K/Akt and TGF-β signaling axes. Cancer Discov. 2012;2:248–259. doi: 10.1158/2159-8290.CD-11-0270. [DOI] [PubMed] [Google Scholar]
- 16.Chen X.-F., Zhang H.-J., Wang H.-B., Zhu J., Zhou W.-Y., Zhang H., Zhao M.-C., Su J.-M., Gao W., Zhang L. Transforming growth factor-β1 induces epithelial-to-mesenchymal transition in human lung cancer cells via PI3K/Akt and MEK/Erk1/2 signaling pathways. Mol. Biol. Rep. 2012;39:3549–3556. doi: 10.1007/s11033-011-1128-0. [DOI] [PubMed] [Google Scholar]
- 17.Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994:615–615. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 18.Glisovic T., Bachorik J.L., Yong J., Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jungkamp A.-C., Munschauer M. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho J., Marsden P.A. Competition and collaboration between RNA‐binding proteins and microRNAs. Wiley Interdiscip. Rev. 2014;5:69–86. doi: 10.1002/wrna.1197. [DOI] [PubMed] [Google Scholar]
- 21.Corcoran D.L., Georgiev S., Mukherjee N., Gottwein E., Skalsky R.L., Keene J.D., Ohler U. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jungkamp A.-C., Munschauer M. PAR-CliP-a method to identify transcriptome-wide the binding sites of RNA binding proteins. J. Vis. Exp. 2010;41 doi: 10.3791/2034. doi:10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grange T., de Sa C.M., Oddos J., Pictet R. Human mRNA polyadenylate binding protein: evolutionary conservation of a nucleic acid binding motif. Nucleic Acids Res. 1987;15:4771–4787. doi: 10.1093/nar/15.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vardhanabhuti S., Wang J., Hannenhalli S. Position and distance specificity are important determinants of cis-regulatory motifs in addition to evolutionary conservation. Nucleic Acids Res. 2007;35:3203–3213. doi: 10.1093/nar/gkm201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie X., Lu J., Kulbokas E., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aslakson C.J., Miller F.R. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 31.Zhang T., Huang XH., Dong L., Hu D., Ge C., Zhan YQ., Xu WX., Yu M., Li W., Wang X., et al. PCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Mol. Cancer. 2010;9:72. doi: 10.1186/1476-4598-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho J.J., Robb G.B., Tai S.C., Turgeon P.J., Mawji I.A., Man H.S., Marsden P.A. Active stabilization of human endothelial nitric oxide synthase mRNA by hnRNP E1 protects against RNA and microRNAs. Mol. Cell. Biol. 2013;33:2029–2046. doi: 10.1128/MCB.01257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.