Abstract
We examined the influence of the tetratricopeptide repeat factor XAB2 on chromosomal break repair, and found that XAB2 promotes end resection that generates the 3′ ssDNA intermediate for homologous recombination (HR). Namely, XAB2 is important for chromosomal double-strand break (DSB) repair via two pathways of HR that require end resection as an intermediate step, end resection of camptothecin (Cpt)-induced DNA damage, and RAD51 recruitment to ionizing radiation induced foci (IRIF), which requires end resection. Furthermore, XAB2 mediates specific aspects of the DNA damage response associated with end resection proficiency: CtIP hyperphosphorylation induced by Cpt and BRCA1 IRIF. XAB2 also promotes histone acetylation events linked to HR proficiency. From truncation mutation analysis, the capacity for XAB2 to promote HR correlates with its ability to form a complex with ISY1 and PRP19, which show a similar influence as XAB2 on HR. This XAB2 complex localizes to punctate structures consistent with interchromatin granules that show a striking adjacent-localization to the DSB marker γH2AX. In summary, we suggest that the XAB2 complex mediates DNA damage response events important for the end resection step of HR, and speculate that its adjacent-localization relative to DSBs marked by γH2AX is important for this function.
INTRODUCTION
Homologous recombination (HR) repair of chromosomal double-strand breaks (DSBs) is important for tumor suppression and cellular resistance to clastogens. For example, two key mediators of HR, BRCA1 and BRCA2, are essential for breast and ovarian cancer tumor suppression, as well as resistance to several types of DNA damage (1). HR can occur by at least two distinct pathways: homology-directed repair (HDR) and single-strand annealing (SSA) (2). HDR involves strand invasion of a homologous template (usually the sister chromatid) by the RAD51 recombinase to initiate nascent DNA synthesis (2,3). SSA refers to annealing of homologous repeats that flank a DSB to form a bridge for end processing and ligation, which leads to a deletion between the repeats (2,4). Both pathways are likely initiated by DSB end resection to form 3′ single-stranded DNA (ssDNA), which reveals the homology used during repair (5). Namely, factors critical for end resection (e.g. CtIP) are important for both HDR and SSA (6,7). Conversely, suppression of end resection is important for non-homologous end joining (NHEJ) repair, which is likely the preferred pathway when the sister chromatid template is unavailable (8,9). Accordingly, defining the DNA damage response (DDR) factors that influence the end resection step of HR provides insight into the mechanisms that underlie genome maintenance.
XAB2, which is composed of an array of 15 tetratricopeptide repeat (TPR) motifs (10,11), has been implicated in the DDR, but its function in genome maintenance remains poorly understood. Namely, XAB2 has been identified in RNA interference (RNAi) screens for factors important to suppress the accumulation of DNA damage (i.e. γH2AX foci), and for factors that promote resistance to agents that cause replication stress, i.e. cisplatin and an inhibitor of Poly-ADP ribose polymerase (12–14). The study on DDR factors influencing resistance to a Poly-ADP ribose polymerase inhibitor suggested that XAB2 is dispensable for HR, but is likely important for transcription coupled nucleotide excision repair (TC-NER) (12). Consistent with a role during NER, XAB2 was identified in a two-hybrid screen as interacting with the NER factor XP-A (hence XP-A Binding protein 2) (11), and XAB2-depleted cells show hypersensitivity to UV light irradiation (15).
In addition to these DDR studies, XAB2, which is homologous to the S. cerevisiae splicing factor SYF1 (16), has been shown to form a complex with spliceosome-associated factors, including ISY1 and PRP19/PSO4 (15). The composition of this XAB2 complex is conserved, including in S. cerevisiae and D. melanogaster (17,18). While this complex is associated with the spliceosome, PRP19 has also been demonstrated to play a key role in the DDR (19–21). Namely, PRP19 has an evolutionarily conserved role in cellular resistance to DNA damage, promotes the ATR kinase signaling pathway, and is important for HR (22–27).
Thus, we sought to examine the influence of XAB2 on chromosomal break repair. We find that XAB2 is important for both HDR and SSA (using DSB reporter assays), and for the end resection step that is common to these pathways, as measured by induction of chromatin bound Replication Protein A (RPA) by treatment with the topoisomerase I poison camptothecin (Cpt). We also find that XAB2 is important for the dynamics of other HR factors in response to DNA damage: the hyper-phosphorylation of the end resection factor CtIP via Cpt treatment, and recruitment of both BRCA1 and RAD51 to ionizing radiation induced foci (IRIF). Since HR and BRCA1 IRIF are influenced by histone acetylation (28,29), we also examined histone modifications in XAB2-deficient cells, and find that XAB2 is important for histone acetylation levels. We then examined mutant forms of XAB2 (N- and C-terminal truncations) and found that the capacity for XAB2 to promote HR correlates with the ability to form a complex with ISY1 and PRP19, which we find influence HR in a similar manner to XAB2. Finally, through cellular localization studies, we find that XAB2, PRP19, and ISY1 form punctate structures that show a striking adjacent-localization to the DSB marker γH2AX, and we speculate that such localization may be important for the regulation of HR.
MATERIALS AND METHODS
Cell lines, siRNA and plasmids
Establishment and culturing of U2OS reporter cell lines, the pCAGGS-BSKX empty vector, the expression vector for I-SceI (pCBASce), and CtIP siRNA, were each described previously (6,30). The U2OS GFP-CtIP cell line, and GFP-CtIP expression plasmid were generously provided by Dr. Alessandro A. Sartori (7). The pDEST-3xFlag-BRCA1 plasmid (Addgene #52504) was generously provided by Dr Daniel Durocher (31). Sequences of other siRNAs (GE/Dharmacon) are: non-targeting siCTRL (D-001810-01), 5′-ugguuuacaugucgacuaa; siXAB2-4 (D-004914-04), 5′-ccaauucucugucaaaugc; siXAB2-2 (D-004914-02), 5′-acgcagcacucucgaauuu; siPRP19 (D-004668-01), 5′-cagaagagcucagcaaaua; siISY1 (D-013894-04), 5′-gagccgaguuaguggaaaa; siPPIE (J-009466-09), 5′-gccuagaugucuugcggca; siBRCA1 (D-003461-06, 5′gggauaccaugcaacauaa); and si53BP1 (pool of 4 siRNAs: D-003548-01, -02, -04, -05). The colony formation assay is described in Supplementary Figure S1B.
The 3xf-XAB2-WT expression vector was generated from a human XAB2 cDNA in pCMV6-AC (Origene catalog #sc319216, NCBI accession #NM_020196). The N-terminal coding region was replaced with a fragment (gBLOCK, Integrated DNA Technologies) that contains the 3xflag coding sequence (MDYKDHDGDYKDHDIDYKDDDDK) fused to the XAB2 start codon, as well as silent mutations to resist siXAB2-4 (5′-cTaGuuTAGCguGaaGugU, mutations in capital letters). This 3xf-XAB2 fragment was then inserted into the pCAGGS-BSKX expression vector. Truncation mutations were generated by deleting fragments from this plasmid, inserting the relevant stop codons for 3′ truncations, and inserting 3xflag coding sequence fused to the relevant XAB2 coding sequence for the 5′ truncations.
DSB reporter assays
For siRNA transfection, (0.5–1) × 105 U2OS cells were plated in 0.6 ml antibiotic-free media on a 24-well plate with 5 pmol of each siRNA incubated with 1.8 μl RNAiMAX (Invitrogen/Thermofisher), and cultured overnight (20 h). Following the overnight RNAi treatment, cells were transfected with 0.4 μg of the I-SceI expression vector (pCBASce) using 1.8 μl Lipofectamine 2000 (Invitrogen/Thermofisher), in 0.6 ml antibiotic-free media. For the complementation experiments, 0.1 μg of a 3xf-XAB2 expression vector or EV was included. The transfection media was removed after 3 h and replaced with antibiotic media. Three days after the plasmid transfections, GFP+ frequencies were determined by flow cytometery using a CyAn ADP Analyzer (Beckman Coulter, Inc.), as described previously (30). The GFP+ frequency for each transfection was divided by the mean value for the control samples treated in parallel (i.e. siCTRL or siCTRL+EV). Each repair value is the mean of at least three independent transfections, error bars reflect the standard deviation, and statistics were performed with the unpaired t-test. Error bars denote the standard deviation from the mean.
Immunofluoresence analysis
For IRIF analysis, siRNA treatment was performed as described for the reporter assays, and subsequently cells were plated onto chamber slides, which were treated with 10 Gy of IR (Gammacell 3000) and allowed to recover for 6 h prior to fixation. For localization studies of XAB2, PRP19, Pol2-S2P and ISY1, cells were treated with 10 Gy of IR, but allowed to recover for 30 min, and were treated with pre-extraction buffer (20 mM HEPES, 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM sucrose, 0.25% Triton-X 100) just prior to fixation. The same procedure was used for localization studies of 3xfXAB2, using cells treated with siXAB2-4 prior to transfection with 3xfXAB2 expression vectors, with transfection conditions as described for the reporter assays. LASER-induced DNA damage was performed as described (32), and is detailed in Supplementary Figure S5B. Slides were fixed with 4% paraformaldehyde and treated with 0.1 M glycine and 0.5% Triton-X 100 prior to probing with antibodies against BRCA1 (Abcam ab16780), Cyclin A (Abcam ab16726), γH2AX (active motif 39117, or Novus NB100-78356), ISY1 (Santa Cruz Biotech sc398437), RAD51 (Santa Cruz Biotech sc-8349), 53BP1 (Abcam ab36823), PRP19 (Bethyl A300-101A), XAB2 (HCNP, Santa Cruz Biotech sc-271037), Pol2-S2P (Abcam ab5095), ubiquitin conjugates (FK2, Enzo Life Sciences BML-PW8810), Flag (Sigma, F3165), and followed by secondary antibodies (Life Technologies, A-11036 and A-11029), and with DAPI using Vectashield Mounting Medium (Vector Laboratories H1500). IRIF images were acquired using a BX-50 (Olympus) microscope at 40× magnification with Image-Pro software. Confocal microscopy images were acquired at 40× magnification using the Zeiss LSM 700 Confocal Microscope, using the ZEN Black image acquisition software. At least 50 cells from at least three independent treatments per condition were scored for cells with >10 IRIF. Statistics were performed as for the reporter assays.
Immunoblotting analysis and immunoprecipitation
For immunoblotting, cells were lysed with NETN (20 mM Tris pH 8, 100 mM NaCl, 1 mM EDTA, 0.5% IGEPAL, 1.25 mM DTT and Roche Protease Inhibitor), and several freeze/thaw cycles. To examine CtIP phosphorylation and histone modifications, cells were pre-extracted using Triton-buffer (25 mM Hepes pH 7.4, 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM sucrose, 0.5% Triton X-100), scraped into SDS loading buffer (62.5 mM Tris–HCl pH 6.8, 2% SDS, 10% glycerol, 0.01% bromophenol blue, and 140 mM DTT), and then boiled, sonicated (QSonica Q800RS ultrasonic horn), and boiled again. For quantitative analysis of histone acetylation levels, 10 mM sodium butyrate was added to the pre-extraction buffer in several repeat experiments to inhibit deacetylation during extraction.
For co-IP analysis, 106 cells were transfected with 5 μg 3xf-XAB2 expression vectors or EV and 15 μl Lipofectamine 2000 in 3 ml. Subsequently, cells were treated with 1 μM Cpt for 1 h, and lysed in IP buffer: 20 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% IGEPAL, phosSTOP (Roche 04906845001), Protease Inhibitor Cocktail (Roche 11697498001) with 30 units/ml of benzonase (Sigma-Aldrich E1014). Lysates were dounce homogenized, and soluble material was pre-cleared with protein-G Dynabeads (Novex 10003D) prior to incubation with 2 μg of Flag antibody (Sigma catalog F3165), followed by protein-G Dynabeads, which were washed with IP buffer, eluted with 100 mM Glycine pH 2.5, and neutralized with 1 M Tris–HCl pH 10.85. For mass spectrometry analysis of the IPs, the samples were in-gel reduced, alkylated and digested with trypsin as described previously (33). The mass spectrometry analysis is described in Supplementary Figure S4A.
Blots of these extracts or IPs were probed with antibodies described above for the IF analysis, as well as CtIP (Active Motif #61141), CSB (Bethyl Labs A301-345A), Ku70 (Santa Cruz Biotech, sc-1487), FANCD2 (Abcam ab2187), HRP-Flag (Sigma A8592), actin (Sigma A2066), H3K36me3 (Epigentek A4042), H3K56Ac (Abcam ab76307), H4K16Ac (Epigentek A4030), H3K9Ac (Upstate 06-942), H3K9me3 (Upstate 07-442), and HRP-conjugated secondary antibodies (Santa Cruz Biotech, sc-2004, sc-2005, sc-2020). ECL reagent (Amersham Biosciences) were used to develop HRP signals.
Cell cycle and end resection analysis
To examine cell cycle profiles, after siRNA treatment, cells were incubated with 10 μM BrdU for 30 min prior to fixation, and staining with both FITC-labeled anti-BrdU antibody (BD Pharmingen 51-33284X), and propidium iodide. The end resection assay was performed as previously described (26,34). Briefly, cells were treated with 1 μM Cpt in media for 1 h, collected by trypsinization, washed with PBS, treated with 0.2% Triton X-100 in PBS to remove non chromatin-bound RPA, fixed with BD cytofix/cytoperm buffer, stained with RPA34 antibody (antibody 9H8, Abcam ab2175), followed by secondary staining with Alexa Fluor 488 goat anti-mouse (Life Technologies A-11029). Cells were counterstained with DAPI (Sigma, R4642). Staining for both assays was analyzed with a CyAn ADP Analyzer (Beckman Coulter, Inc.) flow cytometer.
RESULTS
XAB2 is important for HR, end resection, and CtIP hyper-phosphorylation
We have sought to define the influence of the TPR motif factor XAB2 on chromosomal break repair, beginning with examining the effect of RNAi depletion of XAB2 on a set of DSB reporter assays. Each reporter contains one or more recognition sites for the rare-cutting endonuclease I-SceI, and is designed such that repair of an I-SceI-induced DSB by a specific repair event restores GFP expression, which can be quantified by flow cytometry. We examined three previously described reporters integrated into the U2OS human osteosarcoma cell line (35): EJ5-GFP to examine end-joining (EJ) repair between two tandem DSBs, DR-GFP for HDR, and SA-GFP for SSA (Supplementary Figure S1A). Regarding SA-GFP, specific HDR events (i.e. long tract HDR resolved by EJ, and/or crossover HDR) have the potential to cause the deletion measured by this reporter, however these HDR events are relatively uncommon (36,37). Furthermore, disruption of the central HDR factor RAD51 causes a substantial increase in the deletion measured by SA-GFP (38), which is inconsistent with HDR making a substantial contribution to this repair product. In contrast, disruption of the end resection factor CtIP causes a reduction in both HDR and SSA (6,39), which we confirmed here using an siRNA targeting CtIP (siCtIP), as compared to a non-targeting siRNA (siCTRL, Figure 1A).
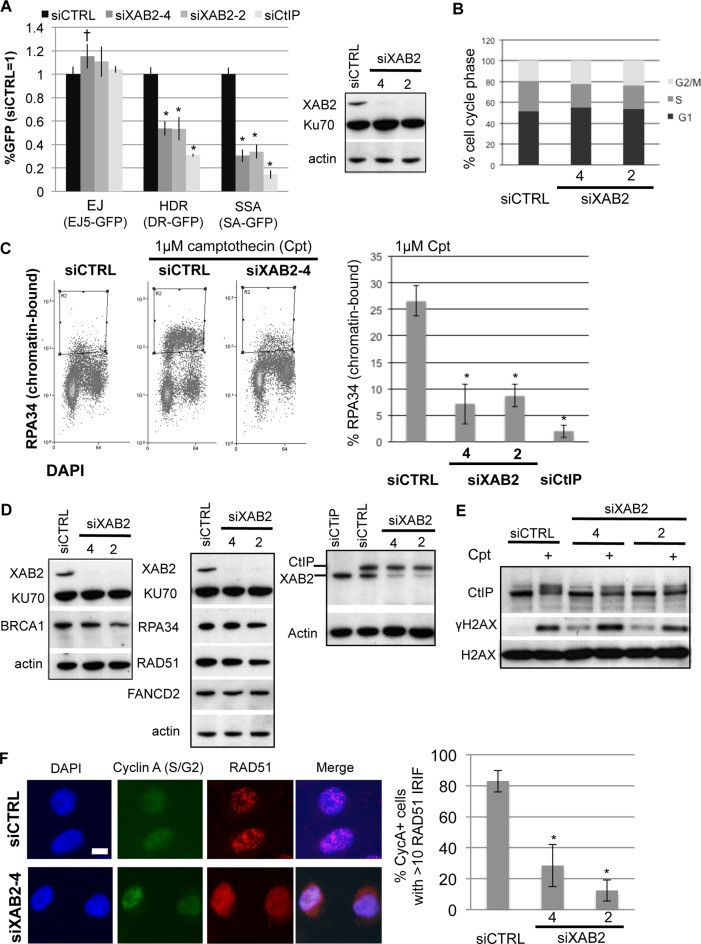
Figure 1.
XAB2 is important for HR DSB repair pathways that require end resection (HDR and SSA), Cpt-induced end resection, hyper-phosphorylation of CtIP, and RAD51 IRIF. (A) RNAi-depletion of XAB2 in U2OS cells causes a reduction in HDR and SSA, but not EJ. Shown are immunoblot signals for XAB2 and two loading controls (Ku70 and actin) after transfection with a non-targeting siRNA (siCTRL), two XAB2 siRNAs (siXAB2-2 and siXAB2-4), and a CtIP siRNA. Shown are the frequencies of GFP+ cells for the EJ5-GFP (EJ), DR-GFP (HDR), and SA-GFP (SSA) reporter lines transfected with an I-SceI expression vector after pre-treatment with the siRNAs shown. GFP+ frequencies are normalized to parallel siCTRL treated samples (= 1). Distinct from siCTRL, *P ≤ 0.0001, †P = 0.013 (N = 6). (B) XAB2 depletion does not obviously affect cell cycle phase distribution. Shown are the percentage of cells in G1, S and G2/M based on propidium iodide/BrdU staining, after treatment with the siRNAs shown, and pulse BrdU labeling (N = 2). (C) XAB2 depletion causes a reduction of Cpt-induced end resection. Following siRNA treatment, cells were treated with Cpt, and mild detergent extracted prior to fixation and staining with RPA34 and DAPI. Shown are representative flow cytometry plots for cells treated with siCTRL and siXAB2-4, as well as the percentage of cells showing detergent resistant (i.e. chromatin bound) RPA34 staining. *Distinct from siCTRL, P < 0.0001, N ≥ 4. (D) XAB2 depletion does not have an obvious effect on the levels of several DDR proteins, except a modest decrease in BRCA1 for only siXAB2-2. Shown are immunoblot signals for several DDR factors for cells treated with the siRNAs shown. (E) XAB2 promotes CtIP hyper-phosphorylation induced by Cpt treatment. Following siRNA treatment, cells were treated with Cpt, and then treated with mild detergent prior to extraction of chromatin-bound proteins. Shown are CtIP, γH2AX and H2AX immunoblotting signals from representative samples. (F) XAB2 depleted cells show reduced RAD51 IRIF. For IRIF analysis, following siRNA treatment, cells were exposed to 10 Gy IR, and allowed to recover for 6 h prior to fixation for immunofluorescence analysis. Shown are representative images of RAD51 and Cyclin-A (S/G2 marker) staining of siCTRL and siXAB2-4 treated cells (scale bar = 10 μm), as well as the percentage of Cyclin-A+ cells showing >10 RAD51 IRIF for each siRNA treatment. *Distinct from siCTRL, P < 0.004 (N = 3).
Similar to siCtIP treatment, depletion of XAB2 by two distinct siRNAs (siXAB2-2; siXAB2-4) caused a significant decrease in HDR (2-fold, P < 0.0001) and SSA (3-fold, P < 0.0001), without causing a substantial decrease in EJ (Figure 1A). In contrast, XAB2 depletion did not show a clear effect on cell cycle phase distribution (Figure 1B), such that the decrease in HR cannot be readily explained by loss of S/G2 cells, which are more dependent on HR for clastogen resistance than G1 cells (9,40). Although, by examining long-term clonogenic survival, we found siXAB2 treatment caused a substantial reduction in colony formation (>30-fold, P ≤ 0.005, Supplementary Figure S1B), which is consistent with the embryonic lethality of both Xab2-/- mice (41), and HR-deficient (e.g. Rad51-/- and Ctip-/-) mice (42,43). These findings indicate that XAB2 is important for the HR pathways HDR and SSA, which share end resection as a common intermediate.
Thus, we tested the influence of XAB2 on a quantitative assay for end resection: recruitment of the ssDNA binding factor RPA to chromatin following treatment with Cpt, which is a topoisomerase I poison that causes replication fork collapse. Specifically, we performed a flow cytometry-based assay that uses a mild-detergent extraction to remove non-chromatin bound proteins, prior to fixation and immunostaining for a subunit of RPA (RPA34) and counterstaining with DAPI for cell cycle phase (34). Such chromatin-bound RPA staining is substantially induced by Cpt treatment in a manner dependent on the end resection factor CtIP (26,34), which we confirmed here (Figure 1C). Using this assay, we found that siXAB2 treatment caused a significant reduction in the frequency of Cpt-treated cells with chromatin-bound RPA staining, compared to siCTRL treatment (3.7-fold for siXAB2-4, and 3-fold for siXAB2-2, Figure 1C, P < 0.0001). Thus, XAB2 appears important for end resection, as measured by Cpt-induction of chromatin-bound RPA.
We next considered that XAB2 could be important for protein levels and/or post-translational modifications of factors that are important for the DDR. First, we examined the levels of several DDR factors in cells treated with siXAB2-2 and siXAB2-4. Apart from a 2-fold decrease in BRCA1 levels with only siXAB2-2 (P = 0.03, N = 3), the levels of other DDR factors were not obviously affected by XAB2 depletion by either siRNA (i.e. levels of Ku70, FANCD2, RAD51, RPA34 and CtIP, Figure 1D). Next, we tested whether XAB2 may affect CtIP hyper-phosphorylation that is induced by Cpt treatment. Such DNA damage induced CtIP hyper-phosphorylation occurs on several S/T residues and causes a gel migration shift that can be detected with immunoblotting (44,45). Furthermore, this CtIP hyper-phosphorylation is mediated by the ATM kinase and cyclin-dependent kinases, and is likely important for controlling CtIP function during HR (44,45). Consistent with these studies, we find that Cpt treatment causes a substantial induction of CtIP hyper-phosphorylation (Figure 1E). However, cells treated with siXAB2-2 and siXAB2-4 showed a significant reduction in CtIP hyper-phosphorylation after Cpt treatment (Figure 1E, 1.8-fold and 1.7-fold, respectively, P ≤ 0.005). Thus, XAB2 is important for hyper-phosphorylation of CtIP, which is likely important for control of end resection, and hence is consistent with a role for XAB2 during the end resection step of HR.
XAB2 is important for IRIF of the HR factors RAD51 and BRCA1, and for histone acetylation that has been linked to HR proficiency
Since XAB2 appears important for HR, we considered that it might also be important for focal accumulation of HR factors to sites of DNA damage. Following end resection, the RAD51 recombinase is recruited to facilitate strand exchange during HDR (2). BRCA1 co-localizes with RAD51 at sites of DNA damage (46), and cells deficient in BRCA1 show reduced HDR and SSA, as well as a partial defect in end resection (39,47,48). Thus, we tested whether XAB2 is important for accumulation of RAD51 and BRCA1 into IRIF. We examined RAD51 accumulation into IRIF in cells that co-stain for the S/G2 phase marker, Cyclin A, and found that cells treated with siXAB2 showed a substantial decrease in Cyclin A+ cells with RAD51 IRIF (6.7-fold for siXAB-2, 2.8-fold for siXAB2-4, P<0.003, Figure 1F). We also found that the frequency of cells with BRCA1 IRIF was markedly reduced by siXAB2 treatment (4-fold for siXAB2-2, 7-fold for siXAB2-4, P < 0.002, Figure 2A and B). In contrast, siXAB2 treatment did not affect IRIF of the DSB marker γH2AX Figure 2A and B).
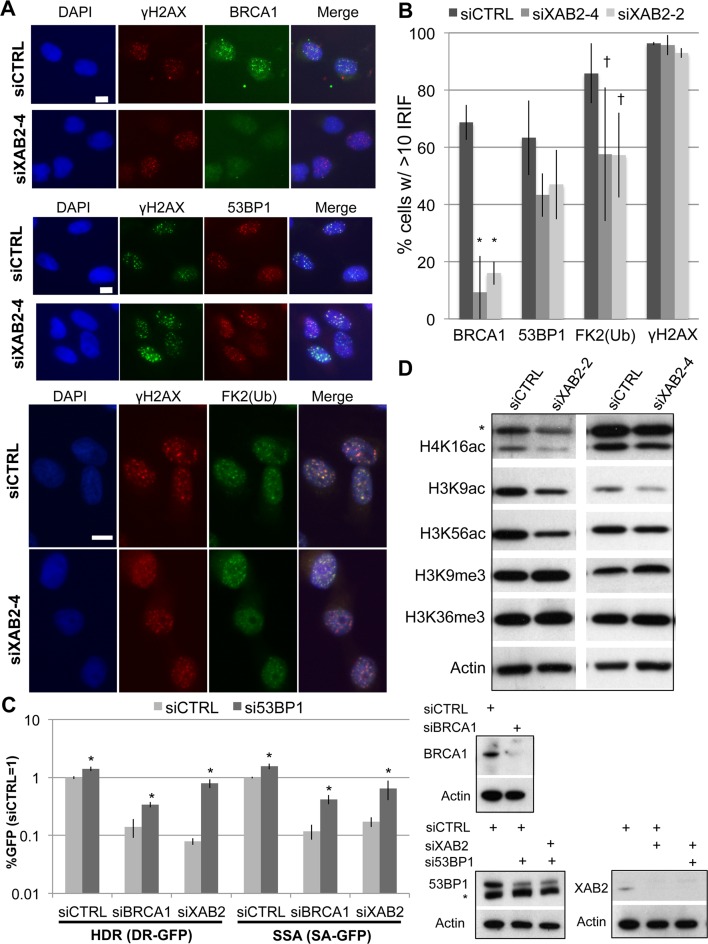
Figure 2.
XAB2 promotes BRCA1 IRIF and histone acetylation events associated with HR proficiency, and HR defects in siXAB2 treated cells can be suppressed by 53BP1 depletion. (A) XAB2 depleted cells show markedly reduced BRCA1 IRIF, a minor reduction in ubiquitin-chain IRIF, and no statistical decrease on 53BP1 or γH2AX IRIF. IRIF experiments were performed as in Figure 1F. Shown are representative images of BRCA1, 53BP1, ubiquitin-chain (FK2), and γH2AX staining of siCTRL and siXAB2-4 treated cells (scale bar = 10 μm). (B) Shown is the percentage cells showing >10 IRIF for each marker shown in (A), and for each siRNA treatment (N ≥ 3). Distinct from siCTRL: *P < 0.002 (N = 3), †P < 0.04 (N = 5). (C) Treating cells with siRNAs targeting 53BP1 can suppress the HR defects of cells depleted of XAB2. DR-GFP and SA-GFP reporter cells were treated with several double siRNA combinations, maintaining the same total siRNA concentration as experiments in Figure 1A, prior to I-SceI expression, and analysis of GFP+ cells (siXAB2; siXAB2-4). The I-SceI transfection was 2 days after siRNA treatment for the DR-GFP experiments to allow for efficient RNAi depletion of 53BP1. Shown are the frequencies of GFP+ cells from these experiments, relative to siCTRL (= 1). *Distinct from the siRNA treatment without si53BP1, P ≤ 0.0007, (HDR N = 5; SSA N = 6). Immunoblots confirming depletion of the target protein by the respective siRNAs are also shown (*non-specific band). (D) XAB2 promotes histone acetylation levels. Following siRNA treatment, cells were treated with mild detergent prior to extraction of chromatin-bound proteins, as in Figure 1E. Shown are immunoblotting signals from representative samples for several histone modifications; *band detected by the H4K16Ac antibody that is non-specific, in that it migrates much slower than histone H4.
Based on these findings that XAB2 is important for BRCA1 IRIF and CtIP hyperphosphorylation, we also examined other aspects of the DDR that relate to BRCA1 and CtIP. For one, we examined recruitment of CtIP to LASER-induced DNA damage, using a previously described U2OS cell line expressing GFP-CtIP (7). From these experiments, we found that siXAB2 treatment did not obviously affect localization of CtIP to LASER-induced DNA damage (Supplementary Figure S2A). Thus, effects of XAB2 on CtIP hyper-phosphorylation are not readily explained by reduced recruitment to damage. Next, we examined the interaction of CtIP with BRCA1, which can be detected by co-IP (49,50). We found that siXAB2 treatment did not obviously disrupt this CtIP–BRCA1 interaction (Supplementary Figure S2B), which is consistent with reports that this interaction appears dispensable for HR (51–53). Finally, since loss of 53BP1 can suppress the HR defects caused by BRCA1-deficiency (54,55), we tested whether 53BP1 depletion could cause a similar suppression in XAB2-deficient cells. Using the DSB reporter assays, we found that 53BP1 depletion caused a significant increase in HDR and SSA in cells depleted of either XAB2 or BRCA1 (Figure 2C). These findings are consistent with XAB2 being important for BRCA1 function during HR.
Since BRCA1 IRIF are promoted by the RNF8-mediated ubiquitin signaling pathway, which is also important for 53BP1 IRIF (56), we also examined the influence of XAB2 on IRIF of ubiquitin conjugates (FK2 antigen) and 53BP1. We found that siXAB2 treatment caused no statistical difference on 53BP1 IRIF, and caused a modest but statistically significant decrease in the frequency of cells with ubiquitin conjugate IRIF (1.5-fold for both siXAB2-2 and -4, P < 0.04, Figure 2A and B). Thus, the substantial defect in BRCA1 IRIF cannot be readily explained by a loss of the RNF8-mediated ubiquitin signaling pathway.
We also tested whether depletion of XAB2 may influence the level of chromatin modifications that are associated with HR proficiency. We examined five different histone modifications: H4K16Ac, which appears important for BRCA1 IRIF (28); H3K9Ac and H3K36me3, which show enriched deposition at sites of enhanced RAD51 recruitment to DSBs (29); H3K56Ac, which is affected by DNA damage (57); and the H3K9me3 mark for heterochromatin, which shows a negative correlation with RAD51 recruitment to DSBs (29), and furthermore appears to have distinct DDR requirements for HR (58). From analyzing the levels of these histone modifications (Figure 2D), we found that siXAB2 treated cells showed a substantial decrease in H3K9Ac (1.8 ± 0.3- and 1.5 ± 0.2-fold for siXAB2-2 and siXAB2-4, respectively, N ≥ 4) and H4K16Ac (2.8 ± 0.85- and 1.6 ± 0.35-fold for siXAB2-2 and siXAB2-4, respectively, N ≥ 3), as well as a modest decrease in H3K56Ac (1.6 ± 0.3- and 1.3 ± 0.25-fold for siXAB2-2 and siXAB2-4, respectively, N ≥ 3). In contrast, siXAB2 treatment did not cause a reduction in either H3K36me3 or H3K9me3 (Figure 2D). Thus, XAB2 appears to promote histone acetylation levels, which have been linked to BRCA1 IRIF and HR proficiency (28,29).
XAB2 regions representing TPR motifs 2–4 and 11–12 are important for HR
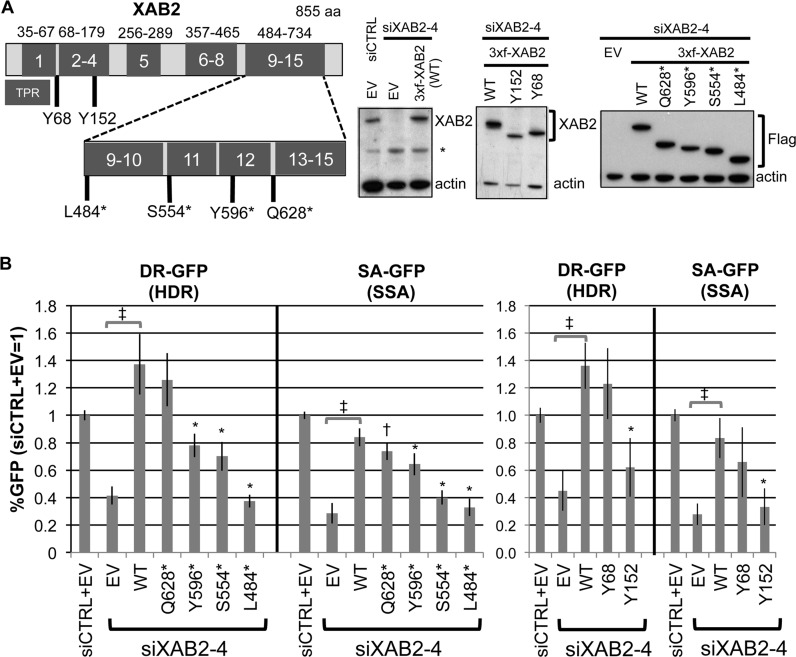
We next sought to define regions of XAB2 important for HR. For this, we performed transient complementation experiments using the HDR and SSA reporter assays. After treatment with siXAB2-4, we co-transfected cells with the I-SceI expression vector and an expression vector for XAB2 that contains an N-terminal 3Xflag tag, as well as silent mutations to evade depletion by siXAB2-4 (3xf-XAB2-WT). We found that 3xf-XAB2-WT is readily expressed in cells treated with siXAB2-4 (Figure 3A), and can substantially rescue the HDR and SSA defects caused by siXAB2-4 treatment (Figure 3B, compared to empty vector, EV, P < 0.0001). We then compared 3xf-XAB2-WT to a series of six truncation mutations in XAB2, which is composed of 15 TPR motifs: Y68 (Δ1-67, ΔTPR1), Y152 (Δ1-151, ΔTPR1-3 and part of TPR4), L484* (Δ484-855, ΔTPR9-15), S554* (Δ554-855, ΔTPR11-15), Y596* (Δ596-855, ΔTPR13-15 and part of TPR12), and Q628* (Δ628-855, ΔTPR13-15). Each of these mutant forms was readily expressed in cells (Figure 3A). Among these mutants, only Y68 and Q628* were highly proficient at promoting HR (Figure 3B, HR not distinct from WT, although SSA for Q628* was statistically lower than WT, P = 0.02). In contrast, Y152 and L484* were not proficient at promoting HR (Figure 3B, not distinct from EV). The mutants S554* and Y596* showed partial proficiency for HR (Figure 3B, distinct from both WT and EV, P≤0.0009, except Y596* versus EV for SSA, P = 0.012). Thus, residues 1–67 (TPR1) and 628–855 (TPR13-15) are dispensable for HR, S554-855 (TPR11-15) are important for full HR function, whereas 1–151 (TPR1-3, part of TPR4) and 484–855 (TPR9-15) are required for HR. These findings indicate that XAB2 regions representing TPR motifs 2–4 and 11–12 are important for HR.
Figure 3.
Defining regions of the XAB2 protein that are important to promote HR using truncation mutant analysis. (A) Expression of XAB2 mutant forms. Shown is a diagram of the XAB2 protein, with TPR motif regions outlined in dark grey, along with the positions of a series of truncation mutations (not to scale). All expression cassettes begin with an N-terminal 3xflag tag (3xf) and silent mutations to resist siXAB2-4. Mutants Y68 and Y152 represent a deletion (Δ) of 1–67 and 1–151, respectively, and the asterisks in the other mutants represent stop codons in place of the residue. Shown are immunoblot signals for XAB2 or Flag from cells transfected with various XAB2 expression vectors or EV following treatment with siXAB2-4 or siCTRL. (B) Distinct XAB2 truncation mutants show a varying capacity to promote HR: Y152 (Δ1–151) and L484* (Δ484-855) are deficient, S554* (Δ554–866) and Y596* (Δ596–855) show partial deficiency, whereas Y68 (Δ1–67) and Q628* (Δ628–855) are proficient. The DR-GFP (HDR) and SA-GFP (SSA) U2OS reporter cell lines were treated with siCTRL or siXAB2-4, and subsequently co-transfected with the I-SceI expression vector combined with the various XAB2 expression vectors shown in (A), or EV. Shown are the frequencies of GFP+ cells from these experiments, relative to parallel siCTRL/EV transfections (= 1). XAB2-WT distinct from EV: ‡P≤0.0001. Mutants distinct from XAB2-WT: *P < 0.001, †P = 0.02.
XAB2 shows overlapping localization with Pol2-S2P, but a striking adjacent-localization relative to the DSB marker γH2AX
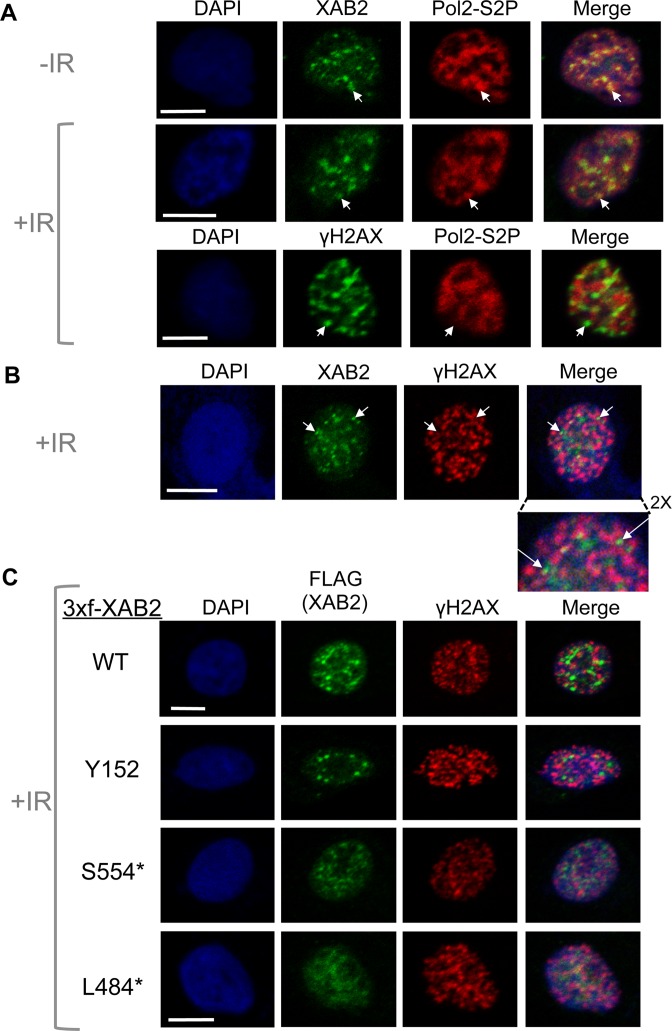
To further define the role of XAB2 in the DDR, we examined the cellular localization of XAB2. In a previous study of cells without DNA damage, XAB2 was shown to form punctate structures that co-localize with phosphorylated forms of RNA polymerase II (15), which have been referred to as interchromatin granules (59,60). We found a similar staining pattern, in that XAB2 localizes to punctate areas of the nucleus, and shows overlap with a phosphorylated form of the large subunit of RNA polymerase II (Pol2-S2P), although Pol2-S2P staining is more diffuse than XAB2 (Figure 4A).
Figure 4.
XAB2 shows punctate nuclear staining, which overlaps with a phosphorylated form of RNA polymerase II (Pol2-S2P), but shows a striking adjacent-localization to the DSB marker γH2AX (i.e. XAB2 and γH2AX often localize to adjacent sites). Cells were either treated with 10 Gy IR (+IR) or mock treated (-IR) and allowed to recover for 30 min prior to mild detergent extraction and fixation for immunofluorescence analysis using confocal microscopy. Scale bars = 10 μm. (A) Shown are immunofluorescence signals for representative cells: -IR treated and stained for Pol2-S2P and XAB2, +IR treated and stained for Pol2-S2P and XAB2, and +IR treated and stained for Pol2-S2P and γH2AX. Arrows in the top panels highlight examples of bright Pol2-S2P signal that co-localizes with punctate XAB2 staining, whereas those in the bottom panel highlight regions of low Pol2-S2P signal with bright γH2AX staining. (B) Shown are immunofluorescence signals for XAB2 and γH2AX of a representative cell, where arrows highlight two examples of punctate XAB2 signals that are adjacent to γH2AX stained regions. Also shown is a 2X magnification of the top of this cell. (C) Exogenously expressed XAB2 shows a similar staining pattern as the endogenous protein, whereas a C-terminal truncation mutant that is deficient in HR (L484*) shows more diffuse nuclear staining. Shown are immunofluorescence signals of Flag and γH2AX for representative cells treated with siXAB2-4, and then transfected with expression vectors for 3xf-XAB2 WT, Y152, S554* and L484*.
We then evaluated the localization of XAB2 in response to DNA damage. We examined XAB2 shortly after treatment with IR (10 Gy, 30 min), and compared its localization to the DSB marker γH2AX. We found that XAB2 remains localized to punctate areas of the nucleus, but shows a striking adjacent-localization pattern relative to γH2AX (Figure 4B). Namely, regions of high punctate XAB2 staining are often adjacent to regions of high γH2AX staining. This staining pattern persists for several hours after IR (30 min to 6 h), and is also found through the depth of the cell, based on examining an orthogonal view of several Z-stacks (Supplementary Figure S3A). Consistent with this localization pattern, we did not observe an obvious IR-induced chromatin enrichment of XAB2 (Supplementary Figure S3B). Pol2-S2P also shows a similar staining pattern as XAB2 relative to γH2AX, but again its localization is more diffuse than XAB2 (Figure 4A). We then performed a similar analysis of IR-treated cells with transiently expressed 3xf-XAB2 using Flag immunostaining, and found that 3xf-XAB2-WT shows a similar pattern as endogenous XAB2 (i.e. punctate staining that is adjacent to γH2AX, Figure 4C). Regarding the HR-deficient mutants described above, Y152 and S554* showed similar staining as WT, albeit with more diffuse staining, whereas L484* lacked clear punctate staining (Figure 4C). These findings indicate that XAB2 localizes in punctate foci that overlap with Pol2-S2P, but are strikingly adjacent to the DSB marker γH2AX, which we speculate could be relevant to the HR function of XAB2 (see Discussion).
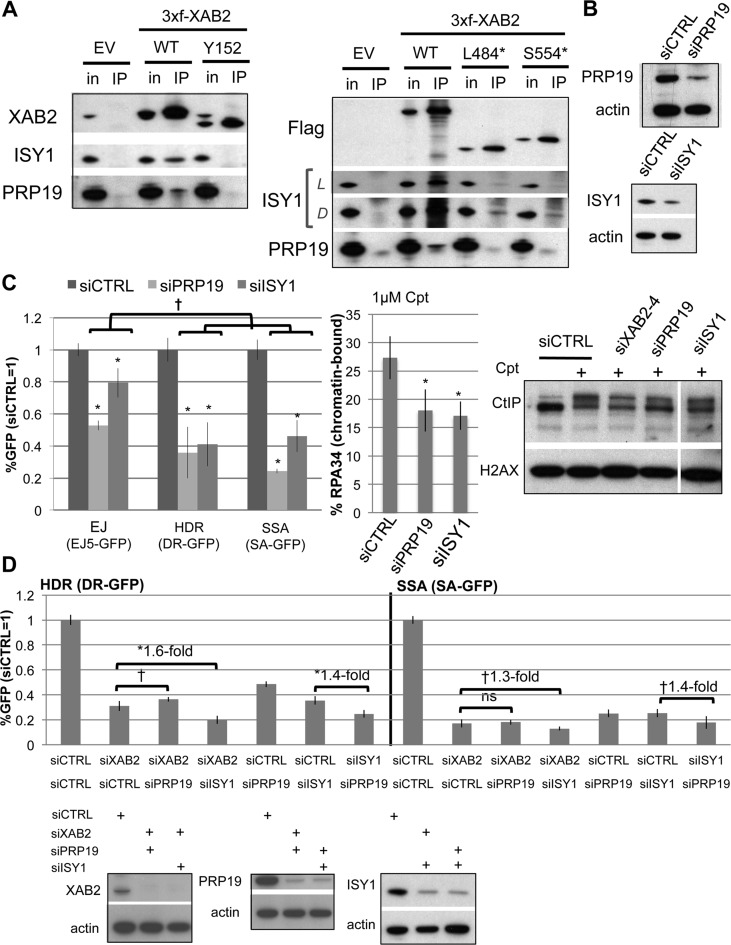
XAB2 interacts with PRP19 and ISY1, which affect HR similarly to XAB2
We next sought to investigate factors that interact with XAB2. While XAB2 was previously shown to form a two-hybrid interaction with XPA, and to co-immunoprecipitation (co-IP) with the NER factor CSB when both are exogenously expressed (11), other studies for components of the XAB2 complex have identified ISY1 and PRP19 (15,17,18). We examined IP samples using anti-Flag antibodies from cells expressing 3xf-XAB2-WT, which were treated with Cpt before harvesting. We confirmed enrichment of XAB2 in the IP, compared to control cells transfected with EV (Figure 5A, Supplementary Figure S4A). From mass spectrometry analysis of these IP samples, we did not identify NER factors, and furthermore immunoblotting analysis of these samples failed to detect CSB (Supplementary Figure S4A, B). Also, we did not detect BRCA1 or CtIP in IPs of XAB2 in either the mass spectrometry results, or from immunoblotting analysis (Supplementary Figure S4B). However, the mass spectrometry analysis identified other factors previously shown to associate with XAB2, which we also confirmed by immunoblotting analysis: ISY1, PRP19 and PPIE (Figure 5A, Supplementary Figure S4A, C).
Figure 5.
XAB2 forms a complex with PRP19 and ISY1, which also promote end resection. (A) XAB2-WT, but not HR-deficient truncation mutants, forms a complex with PRP19 and ISY1. Extracts were prepared from cells with transient expression of 3xf-XAB2 WT, Y152, L484* and S544*, which were treated with Cpt prior to harvesting. A fraction of each extract was used for input analysis (in), and the rest was used for a Flag immunoprecipitation (IP). Shown are immunoblot signals for XAB2, ISY1, PRP19, and Flag (the latter for the C-terminal truncation mutants). The PRP19 immunoblots also show a smaller non-specific band (i.e. also in the EV IP), which is likely due to signal from the Flag IP antibody. L = Light exposure, D = Dark exposure. (B) Depletion of ISY1 and PRP19 with siRNA. Shown are immunoblot signals for cells treated with siRNAs targeting ISY1 (siISY1) and PRP19 (siPRP19). (C) PRP19 and ISY1 affect HR similarly to XAB2. The influence of siISY1 and siPRP19 treatment on DSB reporter assays, end resection of Cpt-induced damage (chromatin-bound RPA34), and CtIP-hyperphosphorylation was performed as described in Figure 1E. Shown are repair values from the reporter assays (N ≥ 5), *distinct from siCTRL P < 0.0001, †P<0.0001 except P = 0.04 for siPRP19 regarding HDR versus EJ. Shown is the percentage of RPA34+ cells after Cpt treatment for each siRNA treatment, *P < 0.0001, N = 9. Also shown are CtIP immmunoblot signals for cells treated with the siRNAs shown, that were subsequently Cpt treated (+), or untreated (–). (D) Double siRNA treatment combinations targeting XAB2 (siXAB2-4), ISY1, and PRP19, each cause a fold-decrease in HR that is less than the addition of the fold-decrease caused by the individual siRNA treatments (i.e. not additive). DSB reporter cell lines were treated with a series of double siRNA combinations, maintaining the same total siRNA concentration as experiments in Figure 1A, prior to I-SceI expression. Shown are the frequencies of GFP+ cells from these experiments, relative to siCTRL (= 1). Also shown is the fold-decrease in HR for double siRNA treatments relative to the respective individual siRNA treatment, which in each case are less than 2-fold. In contrast, each of the individual siRNA treatments cause a decrease in HR that is at least 2-fold. †P = 0.015, *P < 0.0001 (N = 6). Also shown are immunoblots confirming depletion of the target proteins by the respective double siRNA treatments.
Based on these findings, we tested whether PRP19, ISY1, or PPIE influence HR, using siRNAs targeting these factors (siPRP19, siISY1, siPPIE), which we confirmed deplete the target protein (Figure 5B, Supplementary Figure S4C). Beginning with the DSB reporter assays (Figure 5C), we find that siPRP19 and siISY1 treatment each caused a significant decrease in HDR (2.7-fold and 2.4-fold, respectively), as well as SSA (4-fold and 2.2-fold, respectively). Treatment with siPRP19 and siISY1 also caused a reduction in EJ (1.9-fold and 1.3-fold, respectively), but the effects on EJ were significantly less than effects on HDR and SSA (P<0.0001, except P = 0.04 for siPRP19 regarding HDR versus EJ). In contrast, siPPIE treatment did not cause a significant difference among the reporter assays (Supplementary Figure S4C). These findings indicate that ISY1 and PRP19 are important for HDR and SSA to a greater degree than EJ, which is similar to our above findings with XAB2.
Thus, we further examined ISY1 and PRP19. For one, we performed a series of double treatment experiments with all combinations of siXAB2, siISY1, and siPRP19, using the DSB reporter assays. We found that the fold-effect of each double siRNA treatment combination on HR was less than the addition of the fold-effects of the two single siRNA treatments (i.e. not additive, Figure 5D). This finding is consistent with the notion that these factors function in a complex to promote HR. Next, we examined the XAB2 HR-deficient mutants described above (Y152, L484*, S554*) for the capacity to form a complex with these factors. We found that each of these mutants failed to form a detectable complex with PRP19 (Figure 5A). The mutant Y152 failed to form a detectable complex with ISY1, and L484* and S554* showed a markedly reduced capacity to form a complex with ISY1 (Figure 5A). These findings indicate that the capacity for XAB2 to form a complex with ISY1 and PRP19 correlates with the ability to promote HR.
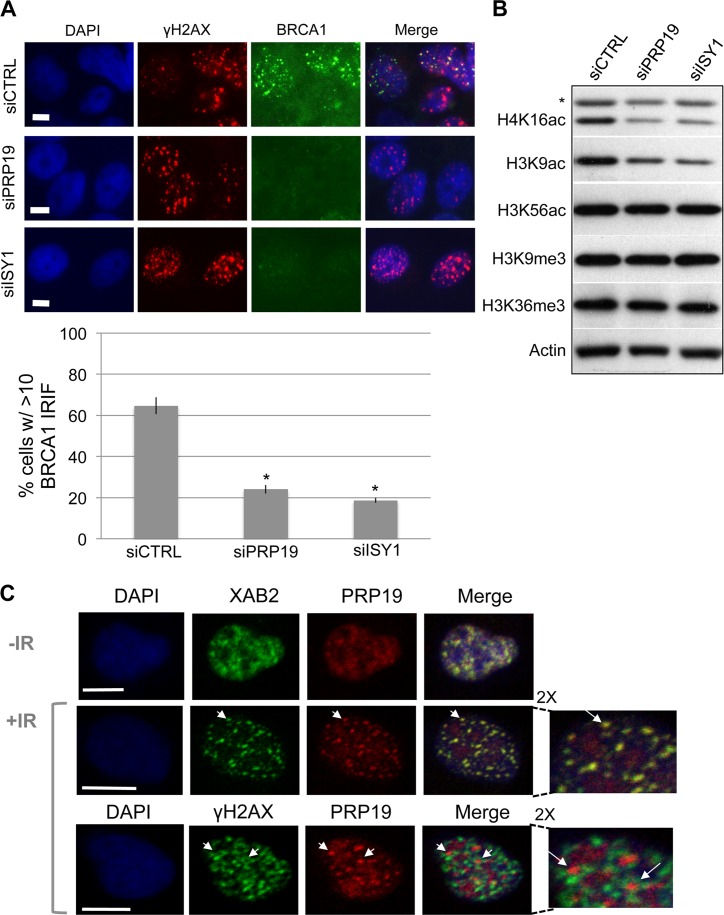
We next examined effects of siPRP19 and siISY1 treatment on other aspects of the DDR that we found were affected by siXAB2 treatment. Beginning with the flow-cytometry based end resection assay, we found that both siPRP19 and siISY1 treatment caused a significant reduction in the frequency of cells with chromatin bound RPA34 (Figure 5C, P < 0.0001, 1.5-fold and 1.6-fold, respectively). Treatment with siPRP19 also caused a significant reduction in CtIP hyper-phosphorylation after Cpt treatment (Figure 5C, 1.5-fold, P = 0.0037), although siISY1 treatment did not obviously cause a significant difference. Next, we examined BRCA1 IRIF and chromatin modifications, and found similar effects as siXAB2 treatment. Namely, we found that siPRP19 and siISY1 caused a significant reduction in BRCA1 IRIF (3-fold, P<0.0001, Figure 6A), as well as a reduction in H4K16Ac (2.7 ± 1.4-fold, 3 ± 2.2-fold, respectively, N = 3) and H3K9Ac levels (1.6 ± 0.1, 2.8 ± 0.6, respectively, N = 3), but not K3K56Ac, H3K9me3 nor H3K36me3 (Figure 6B).
Figure 6.
As with XAB2, ISY1 and PRP19 promote BRCA1 IRIF and histone acetylation. (A) PRP19 and ISY1 promote BRCA1 IRIF. Cells were treated with siRNAs targeting PRP19 and ISY1 as in Figure 5B, and subsequently BRCA1 IRIF were analyzed by immunofluorescence as in Figure 2A. Shown are representative cells stained for γH2AX and BRCA1, as well as the frequency of cells showing BRCA1 IRIF for cells treated with the respective siRNAs. *P < 0.0001 (N = 3). Scale bars = 10 μm. (B) PRP19 and ISY1 are important for H3K9Ac and H4K16Ac levels. Cells were treated with siRNAs targeting PRP19 and ISY1 as in (A), and modified histones were analyzed as in Figure 2D. Shown are representative immunoblot signals for several modified histones and actin from cells treated with the respective siRNAs. (C) PRP19 and XAB2 show substantial co-localization, and PRP19 shows adjacent-localization staining relative to γH2AX. Cells were treated as in Figure 4, and analyzed by immunostaining. Scale bars = 10 μm. Shown are immunofluorescence signals of representative cells for XAB2 and PRP19, and γH2AX and PRP19. Arrows highlight examples of punctate PRP19 signals that co-localize with XAB2 (middle panel), and are adjacent to γH2AX stained regions (bottom panel). Also shown are 2X magnification images from a top segment of the +IR treated cells.
Finally, we examined the cellular localization of PRP19 and ISY1, and found that both of these factors show punctate staining that is often adjacent to γH2AX, similar to XAB2 (Figure 6C, Supplementary Figure S5A). Also, using LASER-induced DNA damage stripes (32), we found that PRP19 fails to enrich at sites of DNA damage (detected by γH2AX), and indeed some cells show instances of reduced localization along the stripe, including cells treated with the transcription inhibitor DRB (Supplementary Figure S5B). We performed the DRB treatment, since some factors that show localization away from a LASER damage stripe have been shown to localize with the stripe in cells pre-treated with DRB (61). We also found that PRP19 and XAB2 show substantial co-localization, as do PRP19 and ISY1 (Figure 6C, Supplementary Figure S5A). Finally, the adjacent-localization pattern of each of these factors relative to γH2AX was not obviously affected by siRNAs targeting the other members of the complex (Supplementary Figure S6). These findings indicate that the HR function of XAB2 correlates with the capacity to form a complex with ISY1 and PRP19, which show a similar influence on HR as XAB2.
DISCUSSION
The TPR motif factor XAB2 has been identified in screens for factors important for the DDR (12–14), but its influence on chromosomal break repair has remained unclear. We have presented evidence that XAB2 mediates the end resection step of HR repair of chromosomal DSBs. We found that XAB2 promotes two distinct HR events that require end resection: HDR and SSA. XAB2 is also critical for Cpt-induced recruitment of RPA34 to chromatin, which is a measure of end resection (34), as well as RAD51 accumulation into IRIF, which requires end resection (5).
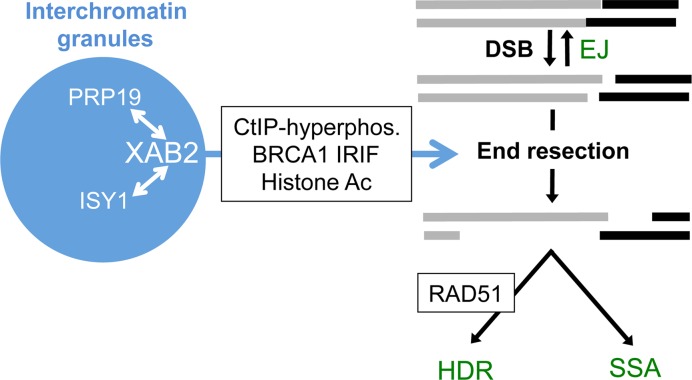
Furthermore, we have found that XAB2 affects regulation of the DDR factors that have been associated with end resection proficiency (Figure 7). Namely, we found that XAB2 is important for hyper-phosphorylation of CtIP induced by Cpt treatment, BRCA1 IRIF, and histone acetylation levels. CtIP hyper-phosphorylation is mediated by the ATM kinase and cyclin-dependent kinases, and several of these phosphorylation sites appear important for CtIP function during end resection (44,45). The cellular pathways that influence the efficiency of CtIP hyper-phosphorylation by these kinases are not well understood, although our findings indicate that XAB2, and associated factor PRP19, promote this process.
Figure 7.
A model for the influence of XAB2 and associated factors on the end resection step of HR. XAB2, along with PRP19 and ISY1, are depicted localizing to interchromatin granules that are apart from DSBs. XAB2 is depicted as promoting a set of DDR events linked to end resection of DSBs, which are important for both HDR (and RAD51 IRIF) and SSA, but are dispensable for EJ.
XAB2 is also critical for IRIF of BRCA1, which is a factor important for proficient end resection, as well as repair by HDR and SSA (39,47,48). We also found that 53BP1 depletion can suppress HR defects caused by siXAB2 treatment, which is similar to the suppression observed with BRCA1-deficient cells (54,55). These findings support the notion that XAB2 is important for BRCA1 function during HR. At least two cellular pathways mediate BRCA1 IRIF: the RNF8-mediated ubiquitin signaling cascade (56), and histone acetylation (28). Our findings are consistent with XAB2 influencing the latter pathway, since cells depleted of the XAB2 show reduced histone acetylation. In contrast, while XAB2 depletion caused a modest decrease in ubiquitin chain (FK2 antibody) IRIF, we found no statistical defect in 53BP1 IRIF, which are dependent on the ubiquitin signaling cascade (56). In addition to the influence of H4K16Ac in promoting BRCA1 IRIF (28), another histone acetylation mark (H3K9Ac) is linked to HR proficiency, based on a correlation with enhanced recruitment of RAD51 to DSBs in chromatin enriched for histone modifications that are linked to actively transcribed regions, which include H3K36me3 and H3K9Ac (29). Accordingly, the role of XAB2 in promoting both H4K16Ac and H3K9Ac could be a significant aspect of the function of this factor in mediating HR.
To further investigate the mechanism of XAB2 function during these aspects of chromosomal break repair, we defined the N- and C-terminal regions of XAB2 that are important for HR, and subsequently found that the capacity to promote HR correlates with the ability for XAB2 to form a complex with PRP19 and ISY1. In contrast, another previously described binding factor of XAB2, the NER protein CSB (11), was not found associated with XAB2 in our analysis. Of course, we cannot eliminate the possibility of a weak interaction with CSB, or loss of the interaction in our experiments, but our findings are also consistent with other reports of XAB2 associated factors (15,17). Furthermore, we found that depletion of PRP19 and ISY1 caused a reduction in HR, end resection, BRCA1 IRIF, and histone acetylation (specifically H4K16Ac and H3K9Ac), which supports a role for the XAB2 complex (i.e. in association with PRP19 and ISY1) during these aspects of the DDR.
Regarding the precise role of this complex to promote end resection via the DDR events described above (Figure 7), we raise several possibilities. Given that XAB2, PRP19, and ISY1 co-purify with the spliceosome (16–18), one model is that these factors are important for efficient mRNA splicing of the transcripts of end resection genes. While plausible, we suggest this mechanism is unlikely for several reasons. For one, such a defect in gene expression would need to be specific for end resection genes without substantially affecting either NHEJ factors or the BRCA2/RAD51-mediated strand invasion step of HDR. Namely, loss of these NHEJ or HDR factors causes an increase in SSA (38,39), whereas we have found that XAB2 depletion causes an even greater decrease in SSA than HDR. Second, we did not identify a substantial or consistent reduction in protein levels for several DDR factors in XAB2 depleted cells. Finally, we found that transient co-expression of siRNA-resistant XAB2 with I-SceI substantially rescues the HR defects in the DSB reporter assays caused by prior siXAB2 treatment. Such simultaneous and substantial complementation of HR function is inconsistent with a model requiring the restoration of an XAB2-dependent gene expression program to facilitate the initiation of HR. While each of these arguments is not necessarily definitive, we suggest that another mechanism may be more likely.
For example, apart from pre-mRNA splicing of specific genes per se, proper regulation of the RNA processing machinery, in particular the control of its sub-nuclear localization, could be important for the end resection step of HR. This model is supported by the sub-nuclear localization of XAB2, along with Pol2-S2P, PRP19, and ISY1, which show a striking adjacent-localization pattern relative to the DSB marker γH2AX in IR-treated cells. Other RNA processing factors implicated in HR (e.g. RBMX and hnRNPUL) have been demonstrated to show a related pattern, namely localization away from LASER-induced DNA damage (61,62). Accordingly, several factors implicated in HR have been shown to localize away from DNA damage marked by γH2AX, to regions of the nucleus referred to as interchromatin granules (59,60).
We suggest that the localization of the XAB2 complex within interchromatin granules may be relevant for its DDR function. Considering one model, factors that inhibit end resection could be negatively regulated within interchromatin granules. For examples, inhibitors of CtIP phosphorylation, such as a phosphatase (63), and/or histone acetylation, such as a deacetylase (57), could be regulated by sequestration and/or degradation within the interchromatin granules. Notably, PRP19 contains a U-box ubiquitin ligase domain (64), which could potentially mediate ubiquitination events important for such regulation. A limitation of this model is that the cellular function of interchromatin granules, particularly as relates to transcription elongation and RNA processing, remains poorly understood (59,60). Accordingly, a more inclusive model is that the functions of interchromatin granules, which may include transcription elongation and/or RNA processing, affect signaling events that are important for end resection, such as histone acetylation, BRCA1 IRIF, and/or CtIP hyperphosphorylation. While these models are speculative, we suggest that the striking degree to which the XAB2 complex shows adjacent-localization to γH2AX, and localization within interchromatin granules, is likely to have functional relevance to the regulation of the DDR, and the end resection step of HR in particular.
Supplementary Material
Acknowledgments
We thank Gabriel Gugiu, Ph.D. for the Mass Spectrometry analysis, and the City of Hope Light Microscopy Core for assistance. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Present address: Diana A. Yanez, Yale University School of Medicine, New Haven, CT 06510, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute of the National Institutes of Health [R01CA120954 and R01CA197506 to J.M.S.]; Ira & Alice Rosenberg Ovarian Cancer Research Fund; City of Hope Analytical Cytometry Core and Mass Spectrometry and Proteomics Core, National Cancer Institute of the National Institutes of Health [P30CA33572]. Funding for open access charge: National Cancer Institute of the National Institutes of Health [R01CA197506 to J.M.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Roy R., Chun J., Powell S.N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kass E.M., Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 4.Lin F.L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol. Cell. Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 6.Bennardo N., Cheng A., Huang N., Stark J.M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonoda E., Hochegger H., Saberi A., Taniguchi Y., Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Zeytuni N., Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Nakatsu Y., Asahina H., Citterio E., Rademakers S., Vermeulen W., Kamiuchi S., Yeo J.P., Khaw M.C., Saijo M., Kodo N., et al. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J. Biol. Chem. 2000;275:34931–34937. doi: 10.1074/jbc.M004936200. [DOI] [PubMed] [Google Scholar]
- 12.Lord C.J., McDonald S., Swift S., Turner N.C., Ashworth A. A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair (Amst.) 2008;7:2010–2019. doi: 10.1016/j.dnarep.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E., et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohni K.N., Thompson P.S., Luzwick J.W., Glick G.G., Pendleton C.S., Lehmann B.D., Pietenpol J.A., Cortez D. A Synthetic Lethal Screen Identifies DNA Repair Pathways that Sensitize Cancer Cells to Combined ATR Inhibition and Cisplatin Treatments. PLoS One. 2015;10:e0125482. doi: 10.1371/journal.pone.0125482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuraoka I., Ito S., Wada T., Hayashida M., Lee L., Saijo M., Nakatsu Y., Matsumoto M., Matsunaga T., Handa H., et al. Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. J. Biol. Chem. 2008;283:940–950. doi: 10.1074/jbc.M706647200. [DOI] [PubMed] [Google Scholar]
- 16.Hegele A., Kamburov A., Grossmann A., Sourlis C., Wowro S., Weimann M., Will C.L., Pena V., Luhrmann R., Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 17.Guilgur L.G., Prudencio P., Sobral D., Liszekova D., Rosa A., Martinho R.G. Requirement for highly efficient pre-mRNA splicing during Drosophila early embryonic development. Elife. 2014;3:e02181. doi: 10.7554/eLife.02181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Almeida R.A., O'Keefe R.T. The NineTeen Complex (NTC) and NTC-associated proteins as targets for spliceosomal ATPase action during pre-mRNA splicing. RNA Biol. 2015;12:109–114. doi: 10.1080/15476286.2015.1008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Legerski R.J. The Pso4 complex splices into the DNA damage response. Cell Cycle. 2009;8:3448–3449. doi: 10.4161/cc.8.21.9760. [DOI] [PubMed] [Google Scholar]
- 20.Mahajan K. hPso4/hPrp19: a critical component of DNA repair and DNA damage checkpoint complexes. Oncogene. 2015 doi: 10.1038/onc.2015.321. doi:10.1038/onc.2015.321. [DOI] [PubMed] [Google Scholar]
- 21.Montecucco A., Biamonti G. Pre-mRNA processing factors meet the DNA damage response. Front Genet. 2013;4:102. doi: 10.3389/fgene.2013.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henriques J.A., Vicente E.J., Leandro da Silva K.V., Schenberg A.C. PSO4: a novel gene involved in error-prone repair in Saccharomyces cerevisiae. Mutat. Res. 1989;218:111–124. doi: 10.1016/0921-8777(89)90017-7. [DOI] [PubMed] [Google Scholar]
- 23.Marechal A., Li J.M., Ji X.Y., Wu C.S., Yazinski S.A., Nguyen H.D., Liu S., Jimenez A.E., Jin J., Zou L. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell. 2014;53:235–246. doi: 10.1016/j.molcel.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan L., Huang J. The PSO4 protein complex associates with replication protein A (RPA) and modulates the activation of ataxia telangiectasia-mutated and Rad3-related (ATR) J. Biol. Chem. 2014;289:6619–6626. doi: 10.1074/jbc.M113.543439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbas M., Shanmugam I., Bsaili M., Hromas R., Shaheen M. The role of the human psoralen 4 (hPso4) protein complex in replication stress and homologous recombination. J. Biol. Chem. 2014;289:14009–14019. doi: 10.1074/jbc.M113.520056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howard S.M., Yanez D.A., Stark J.M. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015;11:e1004943. doi: 10.1371/journal.pgen.1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Z., Kanjanapangka J., Liu N., Liu S., Liu C., Wu Z., Wang Y., Loh T., Kowolik C., Jamsen J., et al. Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol. Cell. 2012;47:444–456. doi: 10.1016/j.molcel.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang J., Cho N.W., Cui G., Manion E.M., Shanbhag N.M., Botuyan M.V., Mer G., Greenberg R.A. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P., et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunn A., Stark J.M. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 2012;920:379–391. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 31.Escribano-Diaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkac J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 34.Forment J.V., Walker R.V., Jackson S.P. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry A. 2012;81:922–928. doi: 10.1002/cyto.a.22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunn A., Bennardo N., Cheng A., Stark J.M. Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J. Biol. Chem. 2011;286:42470–42482. doi: 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stark J.M., Jasin M. Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol. Cell. Biol. 2003;23:733–743. doi: 10.1128/MCB.23.2.733-743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandramouly G., Kwok A., Huang B., Willis N.A., Xie A., Scully R. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat. Commun. 2013;4:2404. doi: 10.1038/ncomms3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stark J.M., Pierce A.J., Oh J., Pastink A., Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munoz M.C., Laulier C., Gunn A., Cheng A., Robbiani D.F., Nussenzweig A., Stark J.M. RING finger nuclear factor RNF168 is important for defects in homologous recombination caused by loss of the breast cancer susceptibility factor BRCA1. J. Biol. Chem. 2012;287:40618–40628. doi: 10.1074/jbc.M112.410951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothkamm K., Kruger I., Thompson L.H., Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yonemasu R., Minami M., Nakatsu Y., Takeuchi M., Kuraoka I., Matsuda Y., Higashi Y., Kondoh H., Tanaka K. Disruption of mouse XAB2 gene involved in pre-mRNA splicing, transcription and transcription-coupled DNA repair results in preimplantation lethality. DNA Repair (Amst.) 2005;4:479–491. doi: 10.1016/j.dnarep.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Lim D.S., Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen P.L., Liu F., Cai S., Lin X., Li A., Chen Y., Gu B., Lee E.Y., Lee W.H. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 2005;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H., Shi L.Z., Wong C.C., Han X., Hwang P.Y., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W., et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013;9:e1003277. doi: 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferretti L.P., Lafranchi L., Sartori A.A. Controlling DNA-end resection: a new task for CDKs. Front. Genet. 2013;4:99. doi: 10.3389/fgene.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D.M. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 47.Schlegel B.P., Jodelka F.M., Nunez R. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006;66:5181–5189. doi: 10.1158/0008-5472.CAN-05-3209. [DOI] [PubMed] [Google Scholar]
- 48.Cruz-Garcia A., Lopez-Saavedra A., Huertas P. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014;9:451–459. doi: 10.1016/j.celrep.2014.08.076. [DOI] [PubMed] [Google Scholar]
- 49.Yu X., Fu S., Lai M., Baer R., Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu X., Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reczek C.R., Szabolcs M., Stark J.M., Ludwig T., Baer R. The interaction between CtIP and BRCA1 is not essential for resection-mediated DNA repair or tumor suppression. J. Cell Biol. 2013;201:693–707. doi: 10.1083/jcb.201302145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polato F., Callen E., Wong N., Faryabi R., Bunting S., Chen H.T., Kozak M., Kruhlak M.J., Reczek C.R., Lee W.H., et al. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Exp. Med. 2014;211:1027–1036. doi: 10.1084/jem.20131939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson S.E., Li Y., Chait B.T., Gottesman M.E., Baer R., Gautier J. Cdk1 uncouples CtIP-dependent resection and Rad51 filament formation during M-phase double-strand break repair. J. Cell Biol. 2011;194:705–720. doi: 10.1083/jcb.201103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bunting S.F., Callen E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Panier S., Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat. Rev. Mol. Cell. Biol. 2013;14:661–672. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- 57.Miller K.M., Tjeertes J.V., Coates J., Legube G., Polo S.E., Britton S., Jackson S.P. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kakarougkas A., Ismail A., Klement K., Goodarzi A.A., Conrad S., Freire R., Shibata A., Lobrich M., Jeggo P.A. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 2013;41:9719–9731. doi: 10.1093/nar/gkt729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bregman D.B., Du L., van der Zee S., Warren S.L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spector D.L., Lamond A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polo S.E., Blackford A.N., Chapman J.R., Baskcomb L., Gravel S., Rusch A., Thomas A., Blundred R., Smith P., Kzhyshkowska J., et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol. Cell. 2012;45:505–516. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., Elledge S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng X.F., Kalev P., Chowdhury D. Emerging role of protein phosphatases changes the landscape of phospho-signaling in DNA damage response. DNA Repair (Amst.) 2015;32:58–65. doi: 10.1016/j.dnarep.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 64.Vander Kooi C.W., Ohi M.D., Rosenberg J.A., Oldham M.L., Newcomer M.E., Gould K.L., Chazin W.J. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry. 2006;45:121–130. doi: 10.1021/bi051787e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.