Abstract
Replication protein A (RPA), the major eukaryotic single-stranded DNA (ssDNA) binding protein, is essential for replication, repair and recombination. High-affinity ssDNA-binding by RPA depends on two DNA binding domains in the large subunit of RPA. Mutation of the evolutionarily conserved aromatic residues in these two domains results in a separation-of-function phenotype: aromatic residue mutants support DNA replication but are defective in DNA repair. We used biochemical and single-molecule analyses, and Brownian Dynamics simulations to determine the molecular basis of this phenotype. Our studies demonstrated that RPA binds to ssDNA in at least two modes characterized by different dissociation kinetics. We also showed that the aromatic residues contribute to the formation of the longer-lived state, are required for stable binding to short ssDNA regions and are needed for RPA melting of partially duplex DNA structures. We conclude that stable binding and/or the melting of secondary DNA structures by RPA is required for DNA repair, including RAD51 mediated DNA strand exchange, but is dispensable for DNA replication. It is likely that the binding modes are in equilibrium and reflect dynamics in the RPA–DNA complex. This suggests that dynamic binding of RPA to DNA is necessary for different cellular functions.
INTRODUCTION
Faithful replication of chromosomes and efficient repair of DNA lesions preserve the integrity of the genome (1). The major single-stranded DNA (ssDNA) binding protein in human cells, Replication protein A (RPA) is essential for DNA replication, repair and recombination (2–4). RPA also functions in checkpoint activation and fork restart (5–8). RPA binds to ssDNA intermediates in the cells and interacts with protein partners to coordinate assembly of complexes that synthesize or repair the DNA (3,4,9,10).
RPA is required for both initiation and elongation phases of DNA replication. During initiation, RPA stabilizes the ssDNA regions formed by origin unwinding and helps recruit proteins to the nascent replication fork (11,12). During elongation, RPA promotes DNA polymerase loading and switching on the lagging strand and processing of Okazaki fragments (13–15). RPA is also required for most DNA repair pathways including nucleotide excision repair (NER), recombinational repair and mismatch repair (MMR) (16–18). In NER, RPA (i) interacts with XPA to stabilize the open complex after damage-recognition, (ii) helps position the nuclease for dual incision and (iii) stimulates DNA synthesis after excision (19). During repair of double-strand breaks (DSBs) by homologous recombination (HR), RPA is involved in the DNA end resection and loading of RAD51 recombinase onto the ssDNA to form the RAD51 nucleoprotein filament which mediates DNA strand exchange (20). RPA binding also helps determine which pathway repairs DSBs by down-regulating microhomology-mediated end joining (MMHJ) and competing with Ku protein to inhibit non-homologous end joining (21,22). In MMR, RPA functions to modulate excision and stimulate DNA re-synthesis (17,18,23).
RPA is a stable heterotrimer formed by the three subunits, RPA1, RPA2 and RPA3 (2). It contains a winged helix (Wh) domain and six structurally-related, oligonucleotide binding (OB) folds connected by flexible linkers (24,25). This multi-domain architecture makes the RPA complex very flexible. All six OB fold domains can interact with DNA and are referred to as DNA-binding domains A-F (DBD-A-F) while the winged helix domain primarily interacts with protein partners. RPA1 is composed of four DBDs (F, A, B and C) (26–28). RPA2 is composed of DBD-D and the winged helix domain. RPA3 is composed exclusively of DBD-E. One domain from each subunit interacts to form a trimerization core (DBD-C, -D and -E) (29). The trimerization core and the other domains move independently in the absence of DNA (25). When RPA binds to ssDNA, four DBDs (A-D) form a stable complex with ∼30 nt of ssDNA (30). Each DBD binds directionally, with domains A through D binding from 5′ to 3′ along ssDNA (30–32).
High affinity, stable binding of RPA to ssDNA (Ka∼1010 M−1) involves four DBDs interacting with the DNA: DBD-A, DBD-B, DBD-C and DBD-D (Supplementary Figure S1; (29,30,33–35)). DBD-A and -B in RPA1 have the highest affinity for ssDNA and form the primary binding site in RPA (36–38). Individually DBD-A and DBD-B have Ka for ssDNA of 5 × 105 and 5 × 104 M−1, respectively (37) but when connected by a short linker their affinity is increased ∼100-fold (Ka ∼2 × 107 M−1) (37,38). DBD-C in RPA1 subunit and DBD-D in RPA2 subunit are secondary binding domains with weaker binding affinity (29,38–40). The affinity of RPA for ssDNA differs depending on the length of ssDNA and the number of DBDs engaged with the DNA (33,35,38,41,42).
High affinity, ssDNA binding allows RPA to (i) rapidly coat ssDNA regions, (ii) prevent the formation of secondary structure, (iii) efficiently destabilize duplex DNA and (iv) under some conditions, promote DNA annealing (42–45). These functions are all thought to be important for normal processing of ssDNA in the cell (4,42). In addition to the DBD-A-D, the N-terminal domain of RPA1 (DBD-F) is needed for efficient helix destabilization of duplex DNA (44). DBD-F is located at the end a ∼60 residue flexible linker, and binds both ssDNA (46) and multiple protein partners (e.g. p53, MRE11, ATRIP, RAD9, Dna2) (47–49). Interactions with DFD-F modulate RPA binding to partially duplex DNA structures (28) and regulate DNA repair and checkpoint activation (47–49).
The high affinity domains DBD-A and DBD-B interact with ssDNA through a combination of polar and aromatic residues ((27); Supplementary Figure S1A). Interestingly, while the interacting polar residues are not conserved in evolution, the four interacting aromatic residues, F238 and F269 in DBD A and W361 and F386 in DBD B, are highly conserved (26,50,51). The different types of interactions have different effects on RPA function, and binding affinity does not always correlate with function. Mutation of the polar residues can dramatically reduce affinity (38,51,52) but some low affinity polar residue mutants maintain the full range of cellular RPA functions (52). In contrast, mutation of the aromatic residues had only modest effects on the affinity for ssDNA, but were found to have a separation-of-function phenotype: they were defective in DNA repair but were still able to support replication (51,52). These aromatic residue mutants have no defects in protein interactions, leading us to conclude that their phenotype is caused by altered DNA interactions. This suggests that DNA replication and repair require different RPA–DNA interactions. The aromatic residue mutants have a high affinity for ssDNA but altered interactions with short ssDNA (51). However, the DNA binding defect(s) that disrupts DNA repair remains unknown.
To gain a better understanding of the molecular defects responsible for the loss of activity in DNA repair, we analyzed the DNA interactions of a set DBD-A and DBD-B aromatic residue (Aro) mutants using single molecule total internal reflection fluorescence microscopy (smTIRFM). Our studies show that the interactions of the Aro mutants with linear and partially duplex DNA structures 20 nt or longer are similar to that of the wild-type RPA. However, the Aro mutants were defective in forming complexes with oligonucleotides 15 nt in length. In addition, our kinetic analysis suggests that wild-type RPA has at least two states when bound to ssDNA characterized by a fast and a slow dissociating state. The Aro mutants are defective in forming long-lived complexes and are also not able to efficiently destabilize partially duplex DNA structures. Brownian dynamic simulations suggested that RPA binding to topologically constrained DNA structures, such as single stranded bubbles, promotes DNA unwinding. We conclude that the slow-dissociating state or helix destabilizing activity of RPA is needed for correct processing of ssDNA intermediates in DNA repair and that reduction in these activities cause the separation of function phenotype of the Aro mutants.
RPA binds to DNA dynamically (4,42,53). A recent study by Kemmerich et al., showed that binding of microdomains of RPA contributes to this rapid dynamics and the balance between annealing and unwinding activities of RPA (54). Our studies show that the dynamic modes of RPA binding have a least two kinetic states and that the more stable state is required for repair functions of RPA.
MATERIALS AND METHODS
Protein purification
The construct for expression of the biotinylated RPA3 was constructed by synthesizing a synthetic coding sequence that contained an XbaI site, an N-terminal BirA recognition sequence (BAP:GLNDIFEAQKIEWHW) (55), a six histidine His-Tag and the coding sequence for RPA3 with codon usage optimized for expression in E. coli followed by a BamHI site (GenScript). This sequence was then used to replace the existing RPA3 gene in p11d-tRPA containing wild-type RPA (56) using XbaI and BamHI. The new plasmid, p11d-tRPA•biotinRPA3 directs the expression of RPA1, RPA2 and biotin-RPA3 as a synthetic operon in E. coli. To produce biotinylated Aro mutants, the AroA, AroB and Aro2 coding sequence from pRSF–AroA, –AroB and –Aro2 were each excised with SfiI and AvrII and individually inserted into the wild-type RPA1 subunit in p11d-tRPA•biotinRPA3 (cut at SfiI and NheI sites). Biotinylated proteins were purified as previously described for non-biotinylated RPA (57), with the exception that 100 μM biotin was added to the LB media concomitant with the induction of 0.3 mM IPTG. RAD51 protein was purified as described previously (58).
DNA oligonucleotides
All oligonucleotides were purchased from Integrated DNA Technologies, Coralville, IA. dT35, dT25, dT20, dT15 have a Cy3 fluorophore at the 5′ end. Replication fork like (RFL) branched, Gap, Flap and Bubble DNAs were made by annealing up to four different partially complementary oligonucleotides containing, where indicated, Cy3 and Cy5 dyes. All structures had 20 nt single strand regions adjacent to partially duplex regions (Supplementary Figure S2). The composition and sequences of all the oligonucleotides used are listed in the supplementary methods (see also Supplementary Figure S2).
Reaction conditions for the single-molecule assays
Biotinylated RPA or Aro mutants were immobilized on quartz slides (Finkenbeiner) coated with sparsely biotinylated polyethylene glycol (PEG) to eliminate the non-specific binding. The immobilization was mediated by neutravidin–biotin interaction between biotinylated proteins, neutravidin (Pierce), biotinylated PEG polymer (Laysan Bio, MW5000; mPEG-SVA and biotin-PEG-SVA). Details for preparing the slides and chambers for single molecule experiment were as previously described (59). All single molecule experiments were performed in binding buffer: 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 100 mg/ml BSA. Imaging was carried out in imaging buffer (binding buffer supplemented with 1 mg/ml of trolox (6-hydroxy-2, 5,7,8-tetramethylchroman-2-carboxylic acid; Sigma-Aldrich), 1 mg/ml of glucose oxidase (Sigma-Aldrich), 0.4% (w/v) D-glucose (Sigma-Aldrich) and 0.04 mg/ml of catalase (Calbiochem)). In each experiment, 100 μl of 0.2 mg/ml of Neutravidin dissolved in PBS was flowed into the assembled chamber, incubated for 5 min, followed by 100 μl of 50 pM of Biotinylated protein in binding buffer. After another 5-min incubation, 100 μl of 100 pM of the indicated Cy3- or Cy3- and Cy5-labeled DNA substrate diluted in imaging buffer was injected into the chamber. In some experiments as a final control, the chamber was washed extensively with binding buffer and Cy3-labeled dT35 was added to detect all tethered RPA. The concentrations were optimized to detect 500–1000 individual molecules per experiment (50 pM of biotinylated RPA and 100 pM Cy3-labeled DNA).
Single-molecule smTIRFM
Single molecule experiments were carried out using a prism-type TIRF microscope as previously described (60). Cy3-labeled substrates were excited with the green DPSS laser (532 nm, 75 mW, Coherent). DNA molecules with both Cy3 and Cy5 were also excited at 532 nm and the Cy5 signal observed due to Föster resonance energy transfer (FRET) between Cy3 and Cy5 dyes. The fluorescence signals coming from Cy3 and Cy5 dyes were collected using a water immersion objective 60× (Olympus), separated by a 630-nM dichroic mirror, passed through a 550-nm long-pass filter to block out laser scattering and recorded with an EMCCD camera (Andor).
smTIRFM data analysis
The single molecule trajectories were extracted from the recorded video files. Individual trajectories were visually inspected and picked using MATLAB. The picked individual trajectories were analyzed using QUB software (61,62) using a two-state model, where the presence of a fluorescent signal at the specific location on the surface corresponded to the ‘on’ or bound state and the absence of fluorescence corresponded to the free RPA (‘off’) state. The idealized QUB trajectories yielded the ‘on times’ and ‘off times’. The measured ‘on times’ and ‘off times’ from all RPA molecules were combined and binned to plot histograms, which were fit to single-exponential or double exponential decay using GraphPad Prism 6.0 software to obtain respective rate constants. The extracted ‘off times’ were binned with a bin size of 3 s and ‘on times’ were binned with a bin size of 0.5 s. The dissociation rate constant, koff (s−1), is calculated from fitting the ‘on times’ distribution to a one or two phase exponential decay. The apparent association rate constants kon (M−1 s−1), were calculated by fitting the ‘off times’ to one or two phase exponential decay to obtain the number of events per second (s−1) and divided by the DNA concentration. Ka = kon/koff (M−1). The details of the fitting are described in the supplementary methods.
Gel mobility assay and helix destabilization assays
A gel mobility shift assay (GMSA) was used to determine the binding affinity of RPA and mutants to Replication fork like (RFL) DNA structure, Gap and Bubble DNA, and 6 nM DNA was incubated with increasing amounts of the indicated form of RPA in a total volume of 15 μl binding buffer (50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 100 ng/μl BSA) for 20 min at room temperature. DNA–protein complexes were separated on a non-denaturing 3.5% acrylamide gel (in 1× TAE). The running buffer was 1× TAE (40 mM Tris, 20 mM Acetic acid, 1 mM EDTA). The Cy3 signals from the free and bound DNA were visualized using the Chemidoc MP imaging System (Bio-Rad). In helix destabilization assays, at the end of the 20-min incubation, 2% (w/v) SDS, 1 mg/ml proteinase K (Qiagen) was added to denature protein and a large excess of competitor DNA (5μM) that is complementary to the non-labeled strand of DNA was added to prevent re-annealing. Reactions were separated on a 15% acrylamide gel to distinguish intact DNA structures from melted ssDNA.
Brownian dynamics simulations
Molecular simulations of the association of RPA with Gap and Bubble DNA models were performed using coarse-grained simulation techniques. First, a complete, putative structure of human RPA in complex with ssDNA was constructed with homology-modeling techniques using the following crystal structures: 1EWI (63) for DBD-F of RPA1, 1DPU (64) for the Wh domain of RPA2, and 4GNX (30) for the trimerization core comprising DBDs A, B, C, D and E, and ssDNA. Substitutions of residues to the corresponding human sequences were performed using SwissModel (65) and missing loops and linkers were added using the loop-modeling program Loopy (66). Models of the Gap and Bubble DNAs were constructed from standard B-DNA. Since conducting simulations of RPA:DNA association events using atomically detailed models is not currently feasible due to computational expense, the all-atom representations of RPA and the DNAs were replaced by coarse-grained (CG) simulation models (67). In the case of RPA, a C-alpha-only model was constructed in which one CG ‘bead’ of the model represents one amino acid residue; in the case of DNA, a three-bead per nucleotide model was used, with different CG beads used to represent the base, sugar and phosphate groups of each nucleotide. Standard molecular mechanics terms (bonds, angles, dihedrals) were used to describe interactions of beads involved in bonds (68,69). Interactions of nonbonded beads (e.g. those involved in RPA–DNA interactions) were described using a combination of electrostatic and Lennard-Jones potential functions (details of the Brownian Dynamic model and parameters for the simulations are described in Supplementary Methods). In each simulation, single molecules of RPA and DNA were initially separated from each other by ∼40 Å and simulations of 5 μs duration were performed in order to gauge their tendency to associate.
Throughout each simulation, the stability of both RPA and DNA structures and the degree of their association with each other was monitored by measuring ‘Q’, i.e. the fraction of the expected native contacts that are actually formed (see, e.g. (68)). In the fully formed RPA–DNA complex there are 90 pairs of CG beads that are expected to be in contact with each other. A partially formed RPA–DNA complex with, for example, only 45 of these pairs formed would have QRPA-DNA = 45/90 = 0.50. We use a similar approach to define Q values for RPA-RPA (QRPA-DNA) and DNA-DNA (QDNA-DNA) contacts; in both cases the overall Q value reflects a combination of intramolecular interactions (e.g. intra-subunit/-strand contacts) and intermolecular interactions (e.g. inter-subunit/-strand contacts).
Three-strand DNA exchange assays
RAD51-mediated DNA strand exchange assays were performed essentially as described in (70) using the φX174 virion ssDNA and the φX174 RFI dsDNA as substrates for the reaction. The φX174 RFI dsDNA was linearized using ApaLI to generate 4 nucleotide 5′ overhangs and purified using phenol:chloroform:isoamyl alcohol extraction followed by isopropanol precipitation. DNA strand exchange was initiated by incubating 7.5 μM RAD51 with 30 μM (Nucleotide) φX174 virion ssDNA and 2.5 mM ATP in the Reaction Buffer (20 mM HEPES-NaOH pH 7.5, 10% Glycerol, 1 mM MgCl2 and 1 mM DTT) for 5 min at 37°C. To this, 150 mM of sodium ammonium phosphate (NaNH4PO4) was added along with 2 μM RPA unless otherwise indicated and incubated for another 5 min. This was followed by addition of 15 μM (bp) of ApaLI digested Linear dsDNA and further incubation at 37°C for the indicated time. The samples were then deproteinized by addition of 0.8% SDS and Proteinase K (800 μg/ml) followed by incubation at 37°C for 0.5 h. The samples were then run on a 0.9% agarose gel (UltraPure Agarose, Life Technologies) at 3.5 V/cm at 25°C for 16 h. The gel was then stained for 10 min using SYBR Gold and then destained for 10 min in H2O.
RESULTS
TIRFM measurements with surface-tethered RPA
Mutation of the conserved aromatic residues in the DNA-binding sites of DBD-A and DBD-B causes defects in DNA repair but not in replication. This suggests that replication and repair require different RPA–DNA interactions. While it was known that these aromatic residue mutants alter interactions with short ssDNA regions (51), it was not known how these mutations affect kinetics of binding or the binding to partially duplex DNA intermediates. Therefore, we carried out single-molecule analysis in which biotinylated RPA is immobilized on a slide surface and is allowed to interact with freely diffusing fluorescent-labeled DNA substrates (71). This approach allows direct observation and analysis of binding events in real time (72). We examined three previously characterized mutants that each had two aromatic residues in DBD-A and/or DBD-B mutated to alanine. Two mutant forms, AroA (F238A, F269A) and AroB (W361A, F386A) had both conserved aromatic residues mutated in DBD-A or DBD-B, respectively. The third mutant, Aro2 (F269A, F386A), had one residue in each domain mutated. All three mutant forms function in DNA replication but are defective in DNA repair (51,52).
To make biotinylated RPA and Aro mutants, constructs were made for expressing RPA in which RPA3 contained an N-terminal recognition sequence for BirA biotin ligase (Supplementary Figure S1B) (55,71). The resulting forms of RPA were expressed and purified (Supplementary Figure S1C). The purified biotinylated forms of RPA were active and have apparent association constants similar to the non-biotinylated forms (Supplementary Figure S1D).
Surface-tethered biotinylated RPA or Aro mutants were then incubated with Cy3 and/or Cy5 labeled DNA and the slide was illuminated with the 532 nM green laser in the total internal reflection (TIR) mode. Dye molecules present within the evanescent field generated by the TIR (generally within 100 nM of the slide surface) due to the binding of the fluorescently labeled DNA substrates to the surface-tethered RPAs are directly excited (Figure 1A), while molecules that freely diffuse in solution remain invisible. Thus, fluorescent signal is only observed when a DNA molecule is bound to the immobilized RPA (Figure 1A). (The camera time resolution was set to 100 ms to exclude short events that result from free DNA diffusing into the evanescent field.)
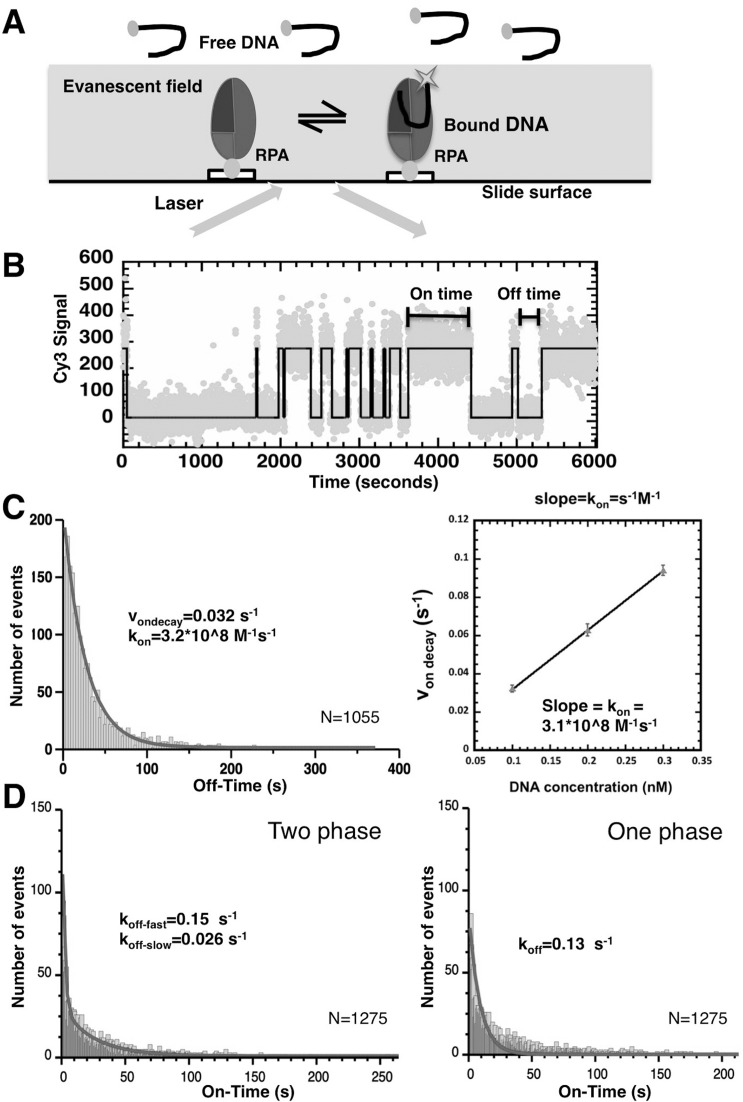
Figure 1.
Surface-tethered RPA DNA-binding activity. (A) Schematic representation of TIRFM-based assay for analysis of DNA binding by tethered RPA. RPA is immobilized on the surface of the microscope flow cell and an evanescent field generated on cell surface by TIR while illuminating with a 530 nM laser. The DNA substrates labeled with Cy3 are only visible when bound to surface-tethered RPA inside the evanescent field. (B) A representative trajectory one RPA molecule monitored in the presence of Cy3-labeled dT35 for 6000 s. Each dot represents a fluorescence reading and black line represents the two-state model determined by QUB. Spikes in the fluorescence intensity correspond to binding and dissociation of different DNA molecules. The length of each binding event is an ‘on time’, and time between binding events is counted as an ‘off time’. In each experiment, 600–1000 trajectories from individual surface-tethered RPA molecules similar to the one depicted here are analyzed. (C) kon and (D) koff determined for RPA binding to dT35 using smTIRF. Distributions of ‘on times’ and ‘off times’ were fit to single or double exponential decay equations to obtain von decay and koff-fast, koff-slow, and koff as shown. The von decay was determined at concentration 0.1, 0.2 and 0.3 nM. The plot shows that von decay had a linear relationship with DNA concentration; kon (s−1M−1) is the slope.
The advantage of this experimental approach is that each surface-tethered RPA molecule is able to bind to multiple ssDNA molecules over time, yielding a fluorescent trajectory (example shown in Figure 1B). The fluorescent signal at the location of a molecule of RPA is characterized by a series of fluorescent pulses in which the duration of each pulse corresponds to an ‘on time’ of a binding event (i.e. the amount of time that a fluorescently labeled DNA molecule spends bound to the RPA). The times between pulses, where the fluorescent signal is minimal, are defined as ‘off times’ and correspond to the free protein (Figure 1B). We consistently observed individual surface-tethered RPA molecules undergoing multiple binding events. No binding events were observed in the absence of biotinylated RPA protein.
Kinetics of the DNA-binding to surface-tethered RPA suggest two distinct nucleoprotein complexes with different stabilities
Initial experiments monitored the interactions of surface-tethered, wild-type RPA and Cy3-labeled dT35. The acquired ‘on times’ and ‘off times’ were binned and plotted as histograms of number of events versus time (Figure 1C & D). The resulting distributions were fit to one or more exponential decay terms, enabling us to determine rate constants and the fraction of each term. The F test-based comparison suggested that for wild-type RPA binding to dT35, the ‘off times’ fit best to a one phase exponential with a von = 0.032 ± 0.001 s−1, which corresponds to kon = von / (100 pM dT35 DNA) = 3.20 ± 0.14 × 108 (M−1s−1). The association rate was also determined for DNA concentrations from 100 to 300 pM (Figure 1C). This confirmed that von linearly depends on the DNA concentration in the analyzed range. The slope of the dependence of von on the DNA concentration yielded a kon of 3.1 × 108 M−1s−1. (Figure 1C). Based on the linear dependence of von, we used 100 pM DNA for all subsequent studies. The observed linear dependence of the von at these low DNA concentrations also indirectly confirms that surface tethering does not adversely affect RPA-ssDNA interactions.
The ‘on times’ of wild-type RPA fit best to a two-phase exponential decay, with koff-fast = 0.15 ± 0.05 s−1 and koff-slow = 0.02 ± 0.01 s−1 and the fast component comprising ∼82% of all events (Figure 1D, Supplementary Table S1A and S1B). Thus the binding of wild-type RPA to dT35 is best described by two equilibrium constants: Ka-fast = 1.47 ± 0.27 × 109 M−1 and Ka-slow = 1.66 ± 0.59 × 1010 M−1. Ka-slow for RPA was similar to the equilibrium binding constants determined from GMSA (Supplementary Figure S1C) and in a previous GMSA analysis of RPA (33,52) which further confirmed that immobilization of RPA did not affect the ability of the protein to bind to ssDNA.
This is the first observation that two distinct types of the RPA-ssDNA complexes are formed, characterized by a fast and a slow dissociation, respectively. Notably, the two complexes could not have been distinguished in the traditional assays such as GMSA.
Aro mutants have reduced affinity for short ssDNA
We then determined the binding parameters for the three aromatic residue mutant with repair defects: AroA, AroB and Aro2. The DNA binding parameters obtained for the Aro mutants with dT35 RPA were similar to those for wild-type RPA; the Aro mutants had on-rates and off-rates for dT35 similar to those obtained for wild-type RPA (Supplementary Table S1A and S1B). As with wild-type RPA, dissociation fit better to a two-phase exponential with the faster off-rate predominating for all three mutants (>78% fast rate, Supplementary Table S1B).
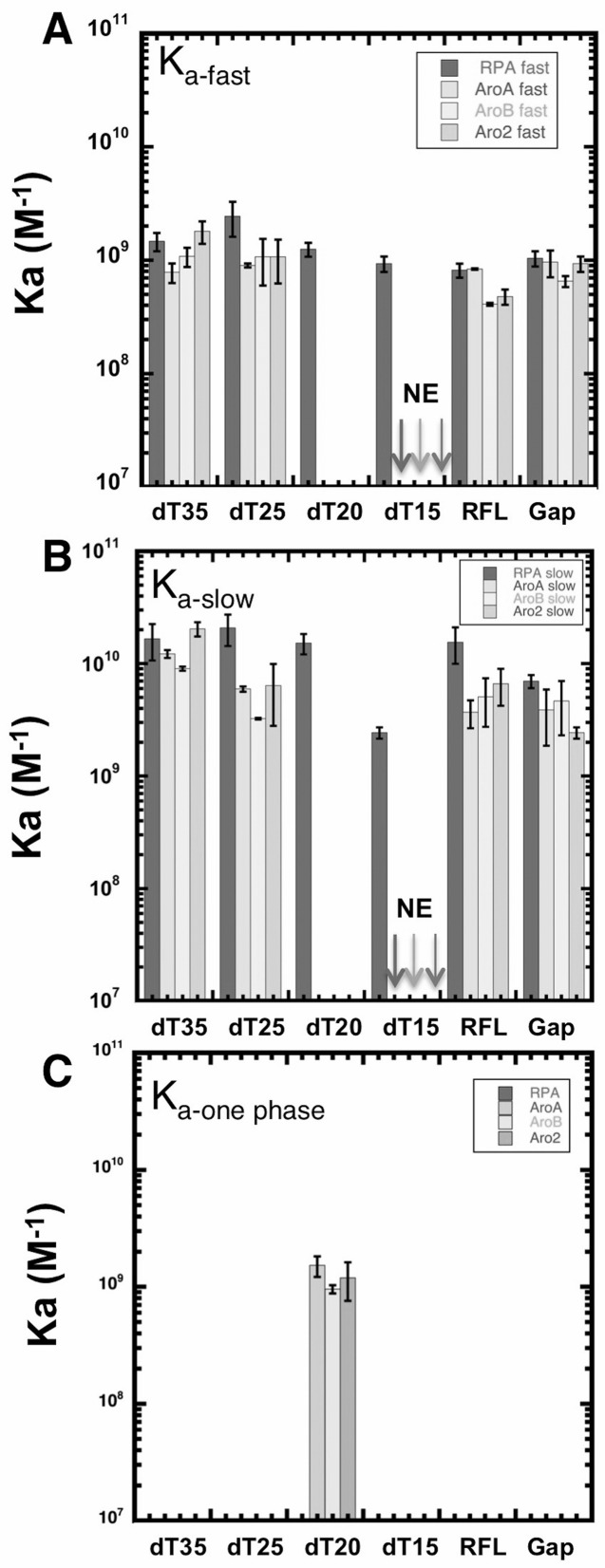
We next examined interactions of the different forms of RPA with oligonucleotides of shorter lengths: dT25, dT20 and dT15. Wild-type RPA, AroA, AroB and Aro2 bound to dT25 and dT20 with an affinity similar to that observed with dT35 (Figure 2). The on-rates (1 × 108 M−1s−1 to 4 × 108 M−1s−1) indicated that association of all forms of RPA occurred at rates close to the diffusion limit. Dissociation of the Aro mutant complexes from dT25 and dT35 was best described by a two-phase exponential. However, with dT20, the dissociation of all three Aro mutants fit best to a single off-rate that was similar to the fast rate observed with dT35 and dT25 (Supplementary Table S1A). This suggests that while having a high affinity for dT20, the Aro mutants interact differently with dT20 and do not form the more stable long-lived complex. With dT15, wild-type RPA bound with an affinity similar to dT35 but no binding events were observed with any of the Aro mutants (Figure 2 and Supplementary Table S1A and S1B). We conclude that like wild-type RPA, the aromatic residue mutants have multiple states with oligonucleotides ≥25 nt but only formed a single short-lived complex with dT20 and were not able to form stable complexes with dT15 at all.
Figure 2.
Length and structure dependence of DNA binding. (A) Binding affinity of RPA and Aro mutants to the indicated DNA substrates was determined by smTIRF. Association constants from two-phase exponential decay, Kafast (A) and Kaslow (B) were calculated based on kon and koff-fast and koff-slow from (Supplementary Table S1B). Data from AroA, AroB and Aro2 binding with dT20 fit best to a one phase exponential decay. So the association constant (Kaone phase) calculated from the kon and koff (Supplementary Table S1A) is shown in (C). No binding events were detected (NE) for AroA, AroB and Aro2 with dT15. Error bars represent standard deviation of three experiments.
Aro mutants are defective in forming ‘long-lived’ complexes
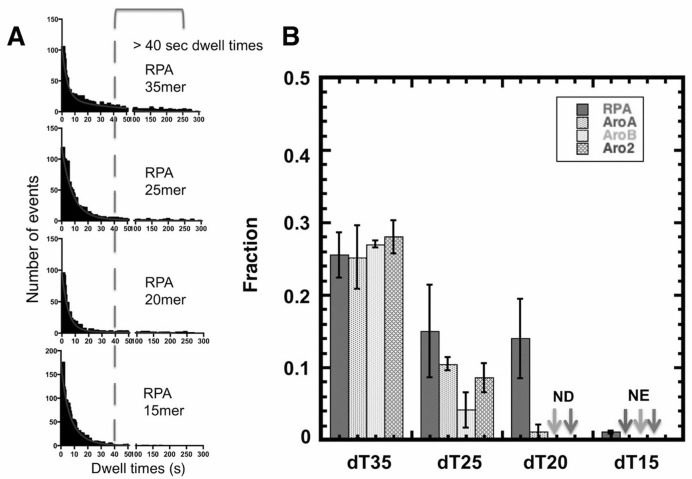
Previously these Aro mutants have been shown in GMSA to have an apparent affinity for dT30 that is approximately one tenth that of wild-type RPA ((51,52); Supplementary Figure S1). However, in our TIRFM studies that directly monitor individual binding events, all three Aro mutants have binding parameters that are the same as wild-type RPA. This suggests that the two assays are measuring different properties. In particular in the GMSA, RPA-DNA complexes must be sufficiently stable during electrophoretic separation to be observed. So a lower apparent affinity in GMSA suggests that the aromatic residue mutations are affecting the stability of the RPA-DNA complexes during electrophoresis. To explore this difference, we quantitated the fraction of ‘long-lived’ RPA-DNA complexes for each form of RPA as a function of DNA length (Figure 3A). We determined the fraction of each population of RPA-DNA complexes with dwell-times longer than 40 s. For wild-type RPA binding to dT35, ∼26% of the binding events were longer that 40 s. This fraction decreased with shorter DNA lengths until only one percent of the wild-type RPA•dT15 complexes had dwell time 40 s or longer (Figure 3B). We conclude that wild-type RPA forms less stable complexes with shorter ssDNA.
Figure 3.
Fraction of long-lived RPA–DNA complexes. (A) The ‘dwell time’ distribution for RPA binding to dT35, dT25, dT20 and dT15 with a 40-s cutoff indicated (dashed line). (B) Quantification of the fraction of events with dwell-times longer than 40 s for RPA, AroA, AroB and Aro2 with each of the oligonucleotides analyzed. Error bars indicate standard errors from three experiments. No long-dwell complex (>40 s) were detected AroB and Aro2 with dT20 (ND) and no binding events were observed with the Aro mutants with dT15 (NE).
The Aro mutants showed a more pronounced length dependence of binding. With dT35, we observed ∼25% of binding events with dwell times longer than 40 s for all three Aro mutants. With dT20, wild-type RPA had still had 14% long events while AroA had 2% and AroB and Aro2 did not have any events longer than 40 s. (Figure 3B). We conclude that the Aro mutants are defective in forming ‘long-lived’ complexes when binding to intermediate and short length ssDNA. The loss of ‘long-lived’ complexes correlated with the disappearance of the slow-off rate phase for the Aro mutants. This suggests that the slow-off phase represents a more stable complex that is not formed when the Aro mutants bind to dT20. Even though the long binding events constitute only a small portion of the total binding events, we speculate that the ability of RPA to have long binding events might be what is being measured in GMSA and that this type of long event is important for RPA function in DNA repair.
Aro mutant binding to partially duplex DNA structures
Intermediates that form during DNA replication and repair have ssDNA regions of different lengths and are either at the end of or adjacent to duplex DNA. To determine whether the structures adjacent to a single stranded region affect Aro mutant binding, we next tested binding to partially duplex DNA structures. DNA structures resembling a stalled replication fork (RFL-replication fork like), a single stranded gap (Gap) and single stranded bubble (Bubble) were examined. Each of these structures was made by annealing appropriate synthetic oligonucleotides and had a 20-nucleotide ssDNA region (Supplementary Figure S2). The oligonucleotides were labeled with Cy3 or Cy5 as indicated so that annealing and subsequent melting of the partially duplex structures could be directly monitored (Supplementary Figure S2).
smTIRFM analysis showed that wild-type and the Aro mutants bound the RFL and Gap DNAs with an affinity similar to that of dT20 (Figure 2) and had similar kinetic parameters (Supplementary Table S1A and S1B). Both Cy3 and Cy5 channels were monitored for all binding events. This showed that under our experimental conditions RPA binding generally does not cause melting of the partial duplex structures that would have manifested in a decrease in the FRET between the two dyes (data not shown and see also below). Rare binding events in which only a single labeled oligonucleotide was present were excluded from the analysis. With both RFL and Gap DNA, all forms of RPA dissociated with two-phase kinetics (Supplementary Table S1A and S1B). For the Aro mutants, this is different than binding to dT20, which was best described by a single off-rate. We speculate that this difference is the result of ‘breathing’ of the duplex adjacent to the ssDNA region, which results in these templates having a slightly longer effective ssDNA region for RPA binding.
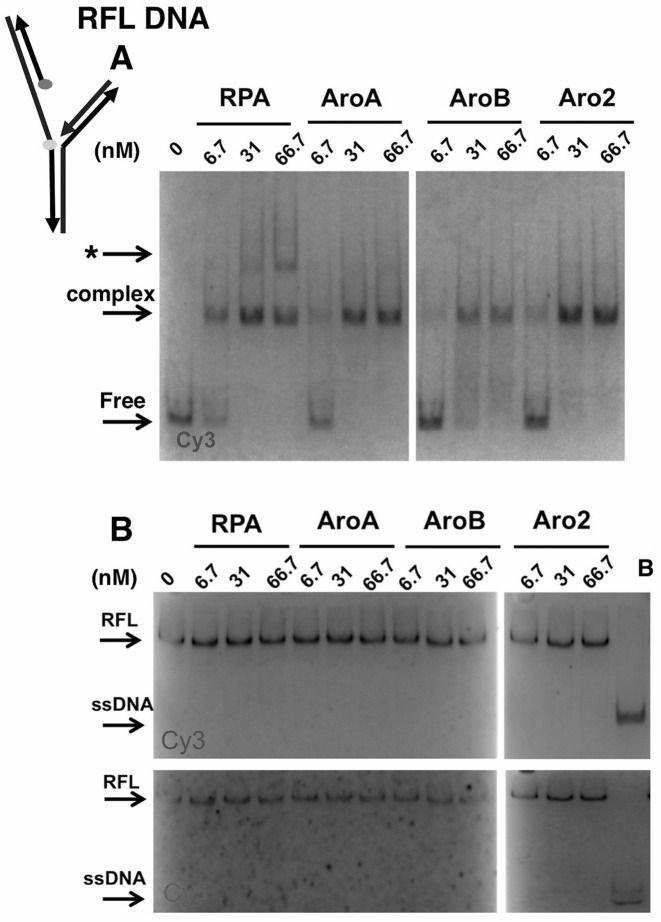
To confirm these findings and explore if there were differences in the DNA complexes formed, we also examined binding in GMSA assays. Wild-type RPA and Aro mutants bound RFL and Gap DNAs with similar apparent affinities (Figure 4A and Supplementary Figure S3A). RPA–DNA complexes formed with similar concentrations of each form of RPA leading to similar apparent association constants (Supplementary Figure S4). In addition, a slower mobility complex was observed with RFL DNA at high concentrations of wild-type RPA (Figure 4A). This complex was not observed with Gap DNA (Supplementary Figure S3A). A very small amount of this complex was also observed at the highest concentrations of AroA and Aro2 but it was never observed with AroB. This suggests that wild-type RPA can form additional complexes with the replication fork-like structure, which are not formed efficiently (or at all) with the Aro mutants.
Figure 4.
DNA-binding to and helix destablization of a replication fork like (RFL) DNA. (A) RFL-binding and (B) helix destabilization activities of RPA and Aro mutants. The indicated amount of RPA or Aro mutant was incubated with 6 nM of RFL at 25° for 25 min. The reactions were separated by electrophoresis under binding (A) or helix destabilization (B) conditions. The positions of the DNA–protein complex, free (RFL) DNA, RFL and ssDNA are indicated. Slow mobility complex indicated with *. The boiled control (B) confirms the position of melted Cy3- and Cy5-labeled ssDNAs. Schematic shows positions of Cy3 (light) and Cy5 (dark) in RFL with arrowheads indicating 3′ ends.
To determine whether RPA and Aro mutants binding to RFL and Gap caused melting of the duplex regions in the gel assays, we carried out helix destabilization assays. In these assays, RFL or Gap DNA was incubated with protein as in GMSA and then complexes were disrupted with SDS and proteinase K in the presence of excess unlabeled ssDNA (to prevent the labeled oligonucleotide from reforming into duplex structures). The DNA was then analyzed on 15% polyacrylamide gels to determine whether any melting had occurred. No melting was observed for RFL and Gap DNAs with either RPA or the Aro mutants (Figure 4B and Supplementary Figure S3B). These experiments suggest that melting was not occurring (and is not required for) RPA binding to the 20 nt gap in these structures.
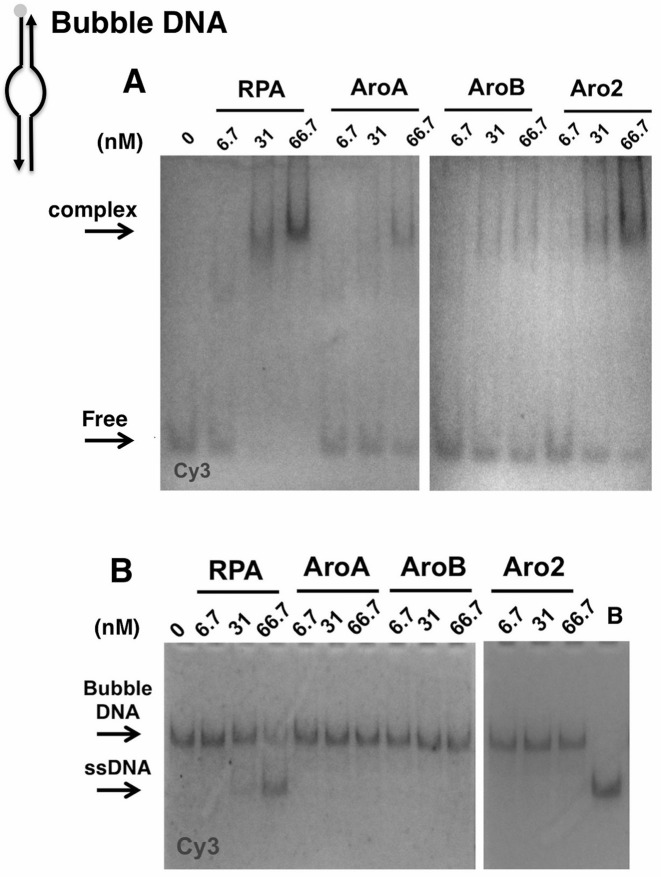
RPA and Aro mutants binding to Bubble DNA
We also examined binding to duplex DNA containing a 20 nt single stranded region surrounded by duplex DNA (Bubble; Supplementary Figure S2). In GMSA assays, wild-type RPA was able to bind the Bubble DNA at high concentrations. Complexes were observed at 5- and 10-fold molar excess of RPA (Figure 5A). In contrast, there was minimal binding by the Aro mutants as the same concentrations (Figure 5A; note the minimal decrease in the amount of free DNA with Aro mutants.) When parallel reactions were analyzed for helix destabilization, wild-type RPA caused melting of the Bubble DNA at the same concentrations as complex formation was observed (Figure 5B). This suggests that the stable RPA–Bubble complex either requires or occurs concurrently with melting. Binding to the Bubble DNA also required higher concentrations than was required for binding to either RFL or Gap DNA (compare Figures 4 and 5) and only occurred when the stoichiometry of RPA to DNA was ≥ 5:1. This suggests that multiple RPA molecules are needed to form a stable complex with and melt Bubble DNA. In contrast to wild-type, the Aro mutants were unable to melt the Bubble DNA. We conclude that the Aro mutants are defective in forming stable complexes with and melting the Bubble DNA.
Figure 5.
DNA-binding to and helix destablization of a 20 nt Bubble DNA. (A) Bubble binding and (B) helix destabilization activities of RPA and Aro mutants. The indicated amount of RPA or Aro mutant was incubated with 6 nM of fluorophore-labeled bubble at 25° for 25 min. The reactions were terminated and separated by electrophoresis under binding (A) or helix destabilization (B) conditions. The positions of the DNA-protein complex, free (Bubble) DNA, Bubble DNA and ssDNA are indicated. The boiled control (B) confirms the position of the position of the Cy3-labeled ssDNA. Schematic shows position of Cy3 in Bubble DNA with arrowheads indicating 3′ ends.
To further examine interactions with Bubble DNA, we examined binding by smTIRFM. Strikingly, no binding events were detected with either wild-type or the Aro mutants. In these smTIRFM experiments, the density of tethered-RPA on the slide surface is low enough that each binding event represents a DNA molecule binding to an individual molecule of RPA. (Note that if multiple RPA molecules were located at a site on a slide, they would have been detected as spots that have binding events with double fluorescence intensity (2 DNA molecules) and this was not observed.) These data confirm that binding of multiple RPA molecules are needed for stable binding to and melting of Bubble DNA.
The binding behavior of the Aro mutants was very different with the Bubble substrate relative to the Gap DNA even though both contain a 20 nt single strand region. To explore this difference we also examined melting of DNAs containing a 20 nt single strand region adjacent to either a 5′ or a 3′ single strand DNA flap (Supplementary Figure S5). No melting was observed with either Flap substrate (Supplementary Figure S5).
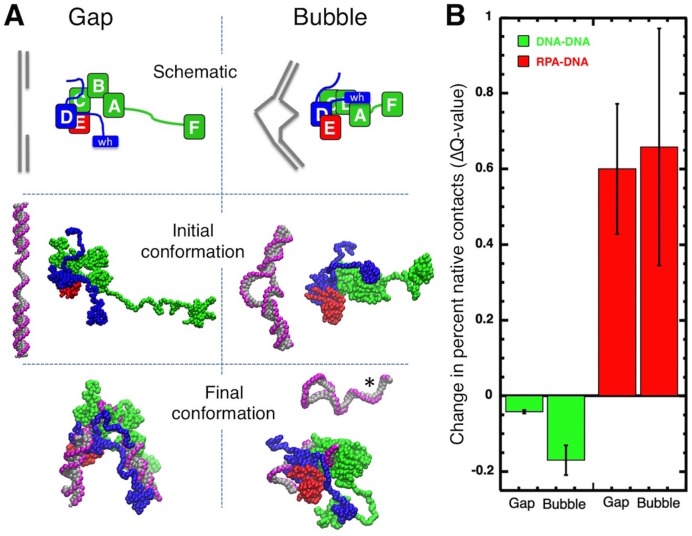
These findings suggest that RPA binding is affected by DNA topology. To explore the effects of RPA binding on DNA with different topologies, Brownian dynamic modeling was used to simulate RPA binding to Gap and Bubble structures. To make the simulations computationally accessible, the models consisted of coarse-grained representations of a single molecule of RPA and the Gap or Bubble DNA. The model of wild-type RPA was complete with all domains and linkers while the models of DNA contained either 3 or 2 oligonucleotides held together by non-covalent interactions. The models were allowed to dynamically change conformations and interact for 5 μs of simulation time. The strengths of RPA–DNA energetic interactions were set sufficiently high to allow formation of stable intermolecular interactions in the time frame of the simulations (see Supplementary Methods).
Our experimental results demonstrated that a single molecule of RPA could bind to the Gap structure but not to the Bubble DNA (Figure 5); multiple RPA molecules were needed to form a stable complex with the Bubble DNA. In our simulations, however, the RPA-DNA binding interactions were deliberately set high enough to allow one molecule to form a stable complex with either DNA structure. This way, we were able to model the dynamics of RPA and DNA, and visualize the consequences of binding of a single molecule of RPA to these two DNA structures.
Figure 6A shows the initial and final configurations of a representative simulation for RPA with either the Gap or Bubble DNA. To quantitate interactions, the fraction of expected native contacts that are actually formed (the Q-value) was quantitated for the RPA with the DNA and for each molecule with itself (RPA with RPA and DNA with DNA; Supplementary Figure S6). In all simulations, RPA ended up bound to the DNA (Figure 6; note the final conformations and the large increase in the RPA-DNA Q-value). The time of the initial interaction varied as did the number of domains of RPA that were engaged with the DNA at the end of the simulation (final RPA-DNA Q-value, Supplementary Figure S6). We also observed that binding of RPA to the Bubble DNA always caused melting of either one or both ends of the duplex (Figure 6A; see also simulation videos in supplementary materials). This strand separation was never observed with the Gap DNA which is less topologically constrained. Melting was characterized by a large decrease in the fraction of native contacts within the DNA during the simulation (Figure 6B). This combination of experimental and simulation data indicate that topological constraints present in the 20 nt Bubble prevent stable binding unless the adjacent duplex regions are melted. We conclude that the Aro mutants are defective in this binding/melting activity.
Figure 6.
Brownian dynamic modeling of RPA interactions with Gap and Bubble DNA. (A) Representative simulation of RPA with Gap or Bubble DNA. Top-schematic of initial conformation, middle-initial conformation and bottom-final conformation after 5 μs. RPA subunits (RPA1, RPA2 and RPA3) shown in green, blue and red, respectively; DNA shown in gray (bases and deoxyribose) and pink (phosphates). *Melted, non-RPA bound strand of DNA. (B) Average change in percent contacts (Q-value) for DNA-DNA (green) or RPA-DNA (red) over five independent 5 μs simulations for Gap and Bubble DNAs. Error bars represent standard deviation.
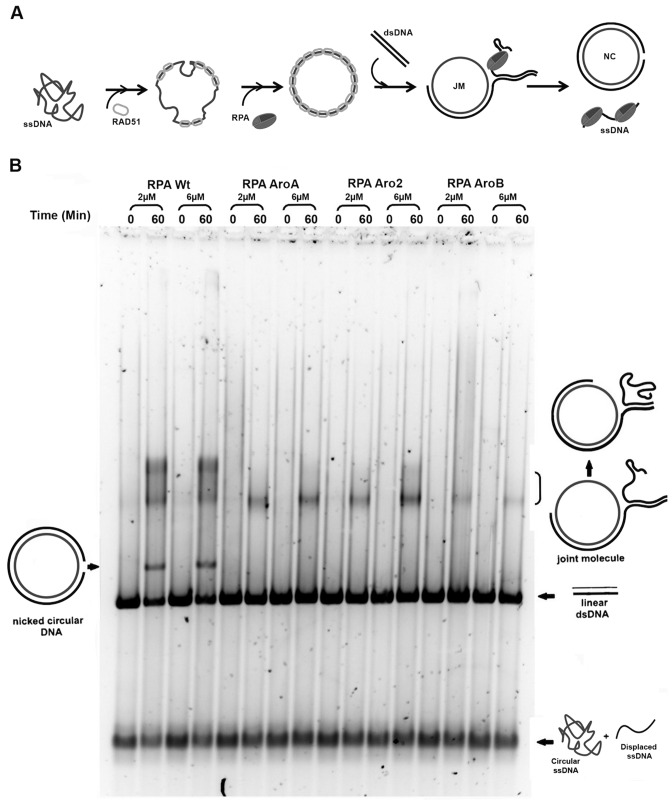
Aro mutants fail to support progression of the RAD51-mediated DNA strand exchange reaction
Our studies described above showed that the Aro mutants have defects in forming stable complexes with ssDNA, melting duplex regions and have reduced binding to short ssDNA regions. It is not clear which of these defects is causing the repair deficiency. While some repair pathways, such as NER and MMR have short ssDNA intermediates, recombinational repair has ssDNA intermediates that are longer than those found at the replication fork. So if the defect in binding to short ssDNA regions is responsible for the repair defect, the Aro mutants should be able to function in recombination. To test this hypothesis, the Aro mutants were tested for their capacity to facilitate RAD51-mediated DNA strand exchange. In these reactions, φX174 closed circular ssDNA is incubated with stoichiometric amounts of RAD51 protein in the presence of ATP and magnesium. RPA is added and allowed to equilibrate and the reaction is initiated by the addition of φX174 RFI linear duplex DNA. RAD51 catalyzes formation of the joint molecule intermediates, which are then converted into nicked circular products. RPA is required for both, the formation of continuous RAD51-ssDNA nucleoprotein filaments and later to ensure the progression of the reaction from the joint molecule intermediates to the final nicked circular products (Figure 7). With wild-type RPA both nicked circle products and a substantial amount of slow-migrating, joint molecules intermediates were observed. In contrast, none of the Aro mutants supported the complete DNA strand exchange; there was no formation of nicked circle products (Figure 7). AroA and Aro2 supported limited initiation of the reaction, as indicated by the formation of a small amount of joint molecules, while AroB reactions had minimal formation of joint molecules. The mobility of the joint molecules forms (73) indicated that the Aro mutants are only forming early intermediates with limited partial exchange. We conclude that the Aro mutants can partially support initiation of RAD51 mediated recombination but fail to support the progression of the strand exchange. This suggests that defects in stable complex formation or DNA melting activity prevent the Aro mutants from forming the necessary ssDNA intermediates needed for the DNA strand exchange.
Figure 7.
Function of Aro mutants in Rad51 mediated strand exchange. (A) Schematic of the RAD51 DNA strand exchange assay. RAD51 coated φX174 circular ssDNA recombines with φX174 RFI linear dsDNA. This leads to formation of joint molecules followed by the nicked circular product, containing the ssDNA circular substrate and the complementary ssDNA strand, and the displaced ssDNA strand. (B) Reactions containing indicated amounts of wild-type or Aro mutant forms of RPA were assembled (0 min) and incubated at 37° for 60 min. Positions of starting DNA (linear duplex), initially formed joint molecule intermediates and completely exchanged nicked circular product are indicated. Some denatured ssDNA (bottom of gel) was present in the starting DNA (0-min points).
DISCUSSION
Dynamic DNA binding by RPA
RPA is a modular protein with multiple DNA-binding domains connected by flexible linkers. The flexible structure of RPA has been suggested to be responsible for multiple modes of DNA binding that differ in length of DNA covered and number of domains that engage DNA (34,38,42,74–76). Formation of stable RPA–DNA complexes requires interactions of the high affinity binding domains, DBD-A and DBD-B, and also the lower affinity DBD-C and DBD-D. This model is supported by mutational analysis (34,38) and structural studies (30,35). Recent studies also suggest that structural flexibility allows RPA to dynamically interact with ssDNA. RPA has been recently shown to diffuse rapidly compared to other ssDNA-binding proteins (diffusion rate of ∼5000 nt2s−1) and to efficiently melt small hairpins and duplex regions as it diffuses (42). RPA–DNA complexes have also been shown to exchange rapidly in the presence of free RPA and other ssDNA-binding proteins (53). Both of these processes are thought to be caused by microscopic dissociation of the individual domains of RPA followed by rebinding to promote diffusion or creation of small ssDNA regions that act as nucleation sites for other ssDNA-binding proteins (4). These findings suggest a model of RPA binding in which the multiple DBDs linked in flexible structure can dynamically interact with ssDNA regions to adopt multiple conformations with different combinations of DBD interacting with ssDNA (4). In spite of these new insights into RPA–DNA interactions, the dynamic interactions of domains needed to support RPA function in different pathways remain poorly understood.
Multiple binding states of RPA
We applied single-molecule total internal reflection fluorescence microscopy (smTIRFM) to analyze DNA binding to surface-tethered wild-type and mutant forms of RPA. This allowed observation of large numbers of individual binding events including multiple binding cycles for individual molecules of RPA. Kinetic analysis of these individual binding events demonstrated, for the first time, that RPA•DNA interaction results in two distinct complexes with different stability. We also found that the more stable state correlates with the observed affinity of different forms of RPA in indirect gel shift assays and that mutants that have reduced ability to form the more stable state also have repair defects. We conclude that the ability to form multiple states when interacting with DNA is critical for RPA function.
What could be the physical basis for the two kinetic states of the RPA-ssDNA complexes? It is known that the high affinity, binding core (DBD-A and –B) of RPA can bind to ssDNA with high affinity (37,38). Also the RPA trimerization core (DBD-C, -D, and –E) is capable of binding to partially duplex ssDNA (29,39). RPA has a binding site size of either 22 or 30 nt at different ionic strengths which is consistent with there being either 3 or 4 DBDs interacting with DNA (42). Also a progressive compaction of RPA has been observed upon binding to different length DNA (35,77). However it is not clear whether the difference in states represents a difference in number or spacing of DBDs engaged with the DNA or different conformations of complex or both (Figure 8).
Figure 8.
Model of possible dynamic RPA binding modes. See Discussion for details.
We hypothesize that when RPA binds to ssDNA, initially one or more DBDs interact with DNA after a diffusion-dependent collision. More DBDs then associate to form a moderately stable complex in which three or four DBDs are generally associating with the DNA. This complex corresponds to the faster dissociating kinetic state and has individual domains microscopically dissociating from and rebinding to the DNA. The complex then undergoes conformational changes which could involve additional DBD association or a conformational change that leads to the more stable state. It seems likely that the two states are in equilibrium and could actually represent a continuum of conformations/complexes. Together these data suggest that RPA dynamically forms at least two kinetically distinct complexes with DNA and that these modes of binding are required for some functions of RPA in cells.
Aro mutant DNA interactions
The Aro mutants function in DNA replication but not in DNA repair. In this study we showed that the Aro mutants have altered DNA binding properties but that these differences were minimal with longer ssDNA regions. We propose that by having multiple DBDs, the affinity of RPA for ssDNA remains high even when a mutation partially disrupts the binding of a single domain. This suggests that reduced binding of mutated single domain can be partially compensated by binding of the other three domains. Further, our results indicate that 20 nt is sufficient to accommodate binding of four DBDs. These results correlate with the previous findings that RPA binds with lower affinity to oligonucleotides shorter than 20 nt and the affinity of RPA for 5′- and 3′–protruding ssDNA is different if the length of the arm is 19 nt or shorter but similar when length of the arm is 23 nt or longer (31,33).
We also identified changes in DNA binding that may be responsible for the defect in DNA repair. Even though the Aro mutants bound oligonucleotides 25 nt and longer with the same affinity as the wild-type protein, we observed changes in binding with short oligonucleotides. The Aro mutants did not form stable complexes with dT15. This supports the model that binding to short DNA is mediated by the high affinity binding domains A and B. This makes binding to dT15 very sensitive to mutations in DBD-A or -B. With intermediate length ssDNA, contribution of domains outside of the high affinity domain is increased, which makes binding of Aro mutants more robust. Aro mutants also showed a decrease in the formation of the more stable binding state with intermediate length DNAs (dT25 and dT20). This causes a reduction in affinity in gel mobility shift assays and reduced helix destabilization activity. We propose that these changes cause the Aro mutants to be defective in DNA repair.
Single strand intermediates in DNA replication and repair differ in size and lifetime. In DNA replication, the ssDNA intermediates are generally short-lived and have lengths usually around 100–200 nt. In contrast the ssDNA intermediates in NER are much shorter (<30 nt), topologically constrained and formation of a stable RPA-containing complex is needed for processing and repairing the DNA damage. In the case of NER, it seems likely that both the reduced affinity for short single strand regions and the less stable complexes will cause the defect in repair.
In homologous recombination, RPA initially competes with RAD51 for ssDNA binding and later assists it by melting ssDNA secondary structures and allowing formation of the continuous RAD51 nucleoprotein filament, which is required for DNA strand exchange. In contrast to the moving replication fork, where the role of RPA is transient, RPA melting of the secondary structures in ssDNA to allow formation of RAD51 nucleoprotein filament, as well as perhaps assisting in progression of the strand exchange reaction, require stable bound RPA–ssDNA complexes capable of duplex destabilization. The Aro mutants were able to support the initiation of the RAD51-mediated three-strand DNA strand exchange reaction, but all failed to support its progression through joint molecule intermediates to the final products. This suggests that more stable binding state is needed for efficient strand exchange.
The formation of the stable state seems to be important for RPA melting of secondary DNA structure (42). Indeed, the Aro mutants were unable to unwind the 20 nt Bubble DNA structure while showing high affinity to RFL, Gap, and Flap containing structures. The Brownian dynamic simulations showed that RPA binding to a 20 nt bubble caused melting of the adjacent duplex. We conclude that a short single strand bubble has topological constraints that make it ‘act’ like a shorter region of DNA. These constraints did not occur with the gapped DNA model. This indicates that binding to topologically constrained DNA structures like Bubble DNA requires either full engagement of DBD-A and DBD-B or formation of the more stable slow-disassociating RPA complex or both.
These results also demonstrate that binding of DBD-A and DBD-B is required for RPA to efficiently melt duplex DNA. It is also known that DBD-F in RPA1 is required for efficient melting of duplex DNA by RPA (28,44). It is interesting that in our Brownian dynamic simulations, we observed DBD-F interacting transiently with the DNA. This raises the possibility that efficient helix destabilization requires interactions of DBD-F with the DNA in addition to formation of the stable state by the rest of the domains of RPA.
The interaction of RPA and DNA are complex (4). Recent work by Kemmerich et al. has shown that step-wise interactions by domains of RPA contribute to rapid dynamics at fork-like structures (54). Our studies show that these dynamics can be characterized kinetically. By studying the four-conserved aromatic resides in high affinity domain of RPA, we have identified that RPA–DNA complexes exist in at least two kinetic states and that formation of the more stable state is required for DNA repair but dispensable for DNA replication. It seems most likely that these states are a consequence of RPA binding dynamically to ssDNA and that full function of DBD-A and –B are required for formation of more stable complexes needed for DNA repair. Additional studies are needed to determine how the domains of RPA contribute to these two states and how these RPA–DNA complexes contribute to processing of different single-stranded DNA intermediates.
Supplementary Material
Acknowledgments
We thank the members of the Wold and Spies lab for discussion and critical suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health Research Grants [GM44721, CA086862 to M.S.W.; GM108617 to M.S.; GM087290 to A.H.E.]. Funding for open access charge: Institutional funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Heyer W.D. Regulation of recombination and genomic maintenance. Cold Spring Harb. Perspect. Biol. 2015;7:195–216. doi: 10.1101/cshperspect.a016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wold M.S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 3.Oakley G.G., Patrick S.M. Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci. 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen R., Wold M.S. Replication protein A: single-stranded DNA's first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays. 2014;36:1156–1161. doi: 10.1002/bies.201400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 6.Lindsey-Boltz L.A., Kemp M.G., Reardon J.T., DeRocco V., Iyer R.R., Modrich P., Sancar A. Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J. Biol. Chem. 2014;289:5074–5082. doi: 10.1074/jbc.M113.542787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashley A.K., Shrivastav M., Nie J., Amerin C., Troksa K., Glanzer J.G., Liu S., Opiyo S.O., Dimitrova D.D., Le P., et al. DNA-PK phosphorylation of RPA32 Ser4/Ser8 regulates replication stress checkpoint activation, fork restart, homologous recombination and mitotic catastrophe. DNA Repair (Amst.) 2014;21:131–139. doi: 10.1016/j.dnarep.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy A.K., Fitzgerald M., Ro T., Kim J.H., Rabinowitsch A.I., Chowdhury D., Schildkraut C.L., Borowiec J.A. Phosphorylated RPA recruits PALB2 to stalled DNA replication forks to facilitate fork recovery. J. Cell Biol. 2014;206:493–507. doi: 10.1083/jcb.201404111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fanning E., Klimovich V., Nager A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugitani N., Chazin W.J. Characteristics and concepts of dynamic hub proteins in DNA processing machinery from studies of RPA. Progress in Biophys. Mol. Biol. 2015;117:206–211. doi: 10.1016/j.pbiomolbio.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melendy T., Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 12.Yeeles J.T., Deegan T.D., Janska A., Early A., Diffley J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuzhakov A., Kelman Z., O'Donnell M. Trading places on DNA-a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 14.Stewart J.A., Miller A.S., Campbell J.L., Bambara R.A. Dynamic removal of replication protein A by Dna2 facilitates primer cleavage during Okazaki fragment processing in Saccharomyces cerevisiae. J. Biol. Chem. 2008;283:31356–31365. doi: 10.1074/jbc.M805965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae S.H., Bae K.H., Kim J.A., Seo Y.S. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 16.Iyama T., Wilson D.M., 3rd DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst.) 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiricny J. Postreplicative mismatch repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spies M., Fishel R. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb. Perspect. Biol. 2015;7:a022657. doi: 10.1101/cshperspect.a022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scharer O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Symington L.S. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol. 2014;6:55–72. doi: 10.1101/cshperspect.a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng S.K., Gibb B., de Almeida M.J., Greene E.C., Symington L.S. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krasner D.S., Daley J.M., Sung P., Niu H. Interplay between Ku and replication protein A in the restriction of Exo1-mediated DNA break end resection. J. Biol. Chem. 2015;290:18806–18816. doi: 10.1074/jbc.M115.660191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G.M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 24.Bochkarev A., Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Brosey C.A., Chagot M.E., Ehrhardt M., Pretto D.I., Weiner B.E., Chazin W.J. NMR analysis of the architecture and functional remodeling of a modular multidomain protein, RPA. J. Am. Chem. Soc. 2009;131:6346–6347. doi: 10.1021/ja9013634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brill S.J., Bastin-Shanower S. Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol. Cell Biol. 1998;18:7225–7234. doi: 10.1128/mcb.18.12.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bochkarev A., Pfuetzner R.A., Edwards A.M., Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 28.Binz S.K., Wold M.S. Regulatory functions of the N-terminal domain of the 70-kDa subunit of replication protein A (RPA) J. Biol. Chem. 2008;283:21559–21570. doi: 10.1074/jbc.M802450200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bochkareva E., Korolev S., Lees-Miller S.P., Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan J., Pavletich N.P. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Laat W.L., Appeldoorn E., Sugasawa K., Weterings E., Jaspers N.G., Hoeijmakers J.H. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998;12:2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iftode C., Borowiec J.A. 5′ –>3′ molecular polarity of human replication protein A (hRPA) binding to pseudo-origin DNA substrates. Biochemistry. 2000;39:11970–11981. doi: 10.1021/bi0005761. [DOI] [PubMed] [Google Scholar]
- 33.Kim C., Paulus B.F., Wold M.S. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 34.Bastin-Shanower S.A., Brill S.J. Functional analysis of the four DNA binding domains of replication protein A - The role of RPA2 in ssDNA binding. J. Biol. Chem. 2001;276:36446–36453. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brosey C.A., Yan C., Tsutakawa S.E., Heller W.T., Rambo R.P., Tainer J.A., Ivanov I., Chazin W.J. A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res. 2013;41:2313–2327. doi: 10.1093/nar/gks1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walther A.P., Gomes X.V., Lao Y., Lee C.G., Wold M.S. Replication protein A interactions with DNA. 1. Functions of the DNA-binding and zinc-finger domains of the 70-kDa subunit. Biochemistry. 1999;38:3963–3973. doi: 10.1021/bi982370u. [DOI] [PubMed] [Google Scholar]
- 37.Arunkumar A.I., Stauffer M.E., Bochkareva E., Bochkarev A., Chazin W.J. Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J. Biol. Chem. 2003;278:41077–41082. doi: 10.1074/jbc.M305871200. [DOI] [PubMed] [Google Scholar]
- 38.Wyka I.M., Dhar K., Binz S.K., Wold M.S. Replication protein A interactions with DNA: differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry. 2003;42:12909–12918. doi: 10.1021/bi034930h. [DOI] [PubMed] [Google Scholar]
- 39.Pestryakov P.E., Khlimankov D.Y., Bochkareva E., Bochkarev A., Lavrik O.I. Human replication protein A (RPA) binds a primer-template junction in the absence of its major ssDNA-binding domains. Nucleic Acids Res. 2004;32:1894–1903. doi: 10.1093/nar/gkh346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bochkarev A., Bochkareva E., Frappier L., Edwards A.M. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blackwell L.J., Borowiec J.A. Human replication protien A binds single-stranded DNA in two distinct complexes. Mol. Cell Biol. 1994;14:3993–4001. doi: 10.1128/mcb.14.6.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen B., Sokoloski J., Galletto R., Elson E.L., Wold M.S., Lohman T.M. Diffusion of human replication protein A along single stranded DNA. J. Mol. Biol. 2014;21:3246–3261. doi: 10.1016/j.jmb.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treuner K., Ramsperger U., Knippers R. Replication protein A induces the unwinding of long double-stranded DNA regions. J. Mol. Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 44.Lao Y., Lee C.G., Wold M.S. Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry. 1999;38:3974–3984. doi: 10.1021/bi982371m. [DOI] [PubMed] [Google Scholar]
- 45.Bartos J.D., Willmott L.J., Binz S.K., Wold M.S., Bambara R.A. Catalysis of strand annealing by replication protein A derives from its strand melting properties. J. Biol. Chem. 2008;283:21758–21768. doi: 10.1074/jbc.M800856200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daughdrill G.W., Ackerman J., Isern N.G., Botuyan M.V., Arrowsmith C., Wold M.S., Lowry D.F. The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res. 2001;29:3270–3276. doi: 10.1093/nar/29.15.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bochkareva E., Kaustov L., Ayed A., Yi G.S., Lu Y., Pineda-Lucena A., Liao J.C., Okorokov A.L., Milner J., Arrowsmith C.H., et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl Acad. Sci. U.S.A. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu X., Vaithiyalingam S., Glick G.G., Mordes D.A., Chazin W.J., Cortez D. The basic cleft of RPA70N binds multiple checkpoint proteins including RAD9 to regulate ATR signaling. Mol. Cell Biol. 2008;28:7345–7353. doi: 10.1128/MCB.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou C., Pourmal S., Pavletich N.P. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. Elife. 2015;4:e09832. doi: 10.7554/eLife.09832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin Y., Lin L.J., Sriratana P., Coleman K., Ha T., Spies M., Cann I.K.O. Engineering of functional replication protein A homologs based on insights into the evolution of oligonucleotide/oligosaccharide-binding folds∇†. J. Bacteriol. 2008;190:5766–5780. doi: 10.1128/JB.01930-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hass C.S., Lam K., Wold M.S. Repair-specific functions of replication protein A. J. Biol. Chem. 2012;287:3908–3918. doi: 10.1074/jbc.M111.287441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haring S.J., Mason A.C., Binz S.K., Wold M.S. Cellular functions of human RPA1. Multiple roles of domains in replication, repair, and checkpoints. J. Biol. Chem. 2008;283:19095–19111. doi: 10.1074/jbc.M800881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibb B., Ye L.F., Gergoudis S.C., Kwon Y., Niu H., Sung P., Greene E.C. Concentration-dependent exchange of replication protein a on single-stranded DNA revealed by single-molecule imaging. PLoS One. 2014;9:e87922. doi: 10.1371/journal.pone.0087922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kemmerich F.E., Daldrop P., Pinto C., Levikova M., Cejka P., Seidel R. Force regulated dynamics of RPA on a DNA fork. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw187. doi:10.1093/nar/gkw187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beckett D., Kovaleva E., Schatz P.J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henricksen L.A., Umbricht C.B., Wold M.S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 57.Binz S.K., Dickson A.M., Haring S.J., Wold M.S. Functional assays for replication protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- 58.Subramanyam S., Jones W.T., Spies M., Spies M.A. Contributions of the RAD51 N-terminal domain to BRCA2-RAD51 interaction. Nucleic Acids Res. 2013;41:9020–9032. doi: 10.1093/nar/gkt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joo C., Ha T. Preparing sample chambers for single-molecule FRET. Cold Spring Harbor Protocols. 2012;2012:1104–1108. doi: 10.1101/pdb.prot071530. [DOI] [PubMed] [Google Scholar]
- 60.Honda M., Park J., Pugh R.A., Ha T., Spies M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Mol. Cell. 2009;35:694–703. doi: 10.1016/j.molcel.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milescu L.S., Nicolai C., Bannen J. 2013. QUB Software.
- 62.Nicolai C., Sachs F. Solving ion channel kinetifs wth the QuB software. Biophys. Rev. Lett. 2013;8:1–21. [Google Scholar]
- 63.Jacobs D.M., Lipton A.S., Isern N.G., Daughdrill G.W., Lowry D.F., Gomes X., Wold M.S. Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker. J. Biomol. NMR. 1999;14:321–331. doi: 10.1023/a:1008373009786. [DOI] [PubMed] [Google Scholar]
- 64.Mer G., Bochkarev A., Gupta R., Bochkareva E., Frappier L., Ingles C.J., Edwards A.M., Chazin W.J. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 65.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 66.Xiang Z., Soto C.S., Honig B. Evaluating conformational free energies: the colony energy and its application to the problem of loop prediction. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7432–7437. doi: 10.1073/pnas.102179699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takada S. Coarse-grained molecular simulations of large biomolecules. Curr. Opin. Struct. Biol. 2012;22:130–137. doi: 10.1016/j.sbi.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 68.Elcock A.H. Molecular simulations of cotranslational protein folding: fragment stabilities, folding cooperativity, and trapping in the ribosome. PLoS Computat. Biol. 2006;2:e98. doi: 10.1371/journal.pcbi.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frembgen-Kesner T., Elcock A.H. Striking effects of hydrodynamic interactions on the simulated diffusion and folding of proteins. J. Chem. Theory Comput. 2009;5:242–256. doi: 10.1021/ct800499p. [DOI] [PubMed] [Google Scholar]
- 70.Amunugama R., He Y., Willcox S., Forties R.A., Shim K.S., Bundschuh R., Luo Y., Griffith J., Fishel R. RAD51 protein ATP cap regulates nucleoprotein filament stability. J. Biol. Chem. 2012;287:8724–8736. doi: 10.1074/jbc.M111.239426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Masuda-Ozawa T., Hoang T., Seo Y.S., Chen L.F., Spies M. Single-molecule sorting reveals how ubiquitylation affects substrate recognition and activities of FBH1 helicase. Nucleic Acids Res. 2013;41:3576–3587. doi: 10.1093/nar/gkt056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Oijen A.M. Single-molecule approaches to characterizing kinetics of biomolecular interactions. Curr. Opin. Biotechnol. 2011;22:75–80. doi: 10.1016/j.copbio.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 73.Tham K.C., Hermans N., Winterwerp H.H., Cox M.M., Wyman C., Kanaar R., Lebbink J.H. Mismatch repair inhibits homeologous recombination via coordinated directional unwinding of trapped DNA structures. Mol. Cell. 2013;51:326–337. doi: 10.1016/j.molcel.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bochkareva E., Belegu V., Korolev S., Bochkarev A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20:612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cai L., Roginskaya M., Qu Y., Yang Z., Xu Y., Zou Y. Structural characterization of human RPA sequential binding to single-stranded DNA using ssDNA as a molecular ruler. Biochemistry. 2007;46:8226–8233. doi: 10.1021/bi7004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumaran S., Kozlov A.G., Lohman T.M. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–11973. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Witosch J., Wolf E., Mizuno N. Architecture and ssDNA interaction of the Timeless-Tipin-RPA complex. Nucleic Acids Res. 2014;42:12912–12927. doi: 10.1093/nar/gku960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.