Abstract
It has previously been shown that the use of racemic mixtures of naturally chiral macromolecules such as protein and DNA can significantly aid the crystallogenesis process, thereby addressing one of the major bottlenecks to structure determination by X-ray crystallographic methods—that of crystal growth. Although previous studies have provided convincing evidence of the applicability of the racemic crystallization technique to DNA through the study of well-characterized DNA structures, we sought to apply this method to a historically challenging DNA sequence. For this purpose we chose a non-self-complementary DNA duplex containing the biologically-relevant Pribnow box consensus sequence ‘TATAAT’. Four racemic crystal structures of this previously un-crystallizable DNA target are reported (with resolutions in the range of 1.65–2.3 Å), with further crystallographic studies and structural analysis providing insight into the racemic crystallization process as well as structural details of this highly pertinent DNA sequence.
INTRODUCTION
It has been demonstrated that racemic mixtures of macromolecules such as proteins and nucleic acids—that is, mixtures containing equimolar amounts of d and l enantiomers—can aid the crystallization process by giving these chiral molecules access to space groups involving symmetry operators that invert chirality, such as mirror planes or centres of inversion. Wukovitz and Yeates pointed out that the space group preference for protein crystals, for example the common occurrence of P212121, was to be assigned to statistical issues related to protein-protein contacts (1). Symmetry and connectivity constraints associated to a given space group allow a certain number of degrees of freedom for a macromolecule to pack intro a three dimensional network of connections. Space groups that allow a greater number of degrees of freedom such as P212121 are statistically favoured. Several centro-symmetrical space groups allow many degrees of freedom and were thus predicted to facilitate crystallization, in particular P1(bar), P21/c and C2/c (2). The practical use of this prediction has become possible since methods to synthesize non-natural enantiomers have become available. For example, the production costs of l-DNA have decreased and are no longer significantly higher than those of d-DNA. The amenability of a racemic protein mixture to crystallize as racemic crystals was first demonstrated for the iron-binding protein, Rubredoxin (3). Since this seminal work, the racemic crystallization method has been applied to a number of challenging protein targets (2), highlighting the considerable utility of this technique to structural biology. It has also been shown that the potential benefits of racemic crystallography can be applied to DNA, with a recent report describing racemic crystal structures of a variety of DNA motifs, including duplexes, quadruplexes and a four-way junction that had been previously described as single d-enantiomers and were shown to preferentially crystallize as racemates (4).
Following this proof-of-concept study, we have now sought to apply the racemic crystallization method to a previously unreported and historically hard-to-crystallize DNA sequence—that of a DNA duplex containing the Pribnow box consensus sequence. The Pribnow box consensus sequence is a ubiquitous prokaryotic promoter sequence and core element located at the −10 position (consequently also known as the ‘−10 box’) upstream of the bacterial transcription start site. This sequence plays a vital role in the regulation of bacterial transcription (5–7), and is known to be highly consensual, with 5′-TATAAT-3′ as the consensus sequence of the template strand. The degree of homology of a promoter sequence to the consensus determines the promoter strength—that is, the strength with which the transcription-initiating sigma factor binds (8)—yet the underlying structural basis by which the sigma factor recognizes and binds Pribnow box sequences is poorly understood. Indeed, there are no reports of crystal structures of this DNA sequence in a ‘folded’ (i.e. double stranded) state. One possible explanation for the lack of crystal structure of a duplex containing the Pribnow-box sequence may be that crystallization of non-self-complementary duplex sequences is typically challenging, often due to issues of crystallogenesis or static disorder. Indeed the majority of double-stranded DNA structures reported in the Nucleic Acid Database and the Protein Data Bank are composed of self-complementary or palindromic sequences (9–11). These are typically model sequences—such as the seminal Dickerson–Drew dodecamer (12–16) or the decamer d(CCAACGTTGG)2 (17)—and have served the scientific community immeasurably, yet there is a possibility that the unnatural symmetry of self-complementary duplexes masks potentially significant local structural features that may be present in the wild-type DNA sequences (which are rarely self-complementary). In addition, considering the extensive use of the Dickerson–Drew dodecamer and other model duplexes as a structural basis for designing and studying DNA-binding therapeutic compounds (such as anti-cancer drugs) at high-resolution (18–21), the ability to characterize and account for abnormalities arising from unnatural symmetry is of high relevance. The interest in studying the non-self-complementary Pribnow box sequence therefore falls into two categories: (i) understanding the role of this sequence on the initiation of bacterial transcription—how does the sigma factor specifically recognize this sequence? and (ii) the design and study of minor groove binding drugs and small molecules.
We report here the results of our efforts to crystallize a non-self-complementary DNA duplex containing the Pribnow box consensus sequence using a racemic crystallization method. Racemic mixtures of duplexes composed of the sequences, 5′−CGCTATAATGCG−3′ (template strand) and 5′−CGCATTATAGCG−3′ (complementary strand) were used for crystallization trials. Using primarily commercially available screens, these investigations yielded well-ordered racemic crystals encompassing a total of four distinct achiral space groups. Structure determination and subsequent refinement resulted in four high quality models, with resolutions in the range of 1.65–2.3 Å. Comparative non-racemic crystallization trials were also undertaken, yielding poorly-ordered crystals and consequently low-resolution structures. The rapidity and ease with which racemic Pribnow box duplex crystals were obtained, coupled to the high quality of the resulting atomic models, provides strong evidence in support of the potentially considerable utility of the racemic crystallization method in the field of DNA structural biology. Structural analysis of the resulting Pribnow box duplexes and comparison to model duplexes is also provided, revealing differences in certain helical parameters which may be sequence-dependent, and consequently, of potential significance to both bacterial transcription and minor groove-directed drug-design.
MATERIALS AND METHODS
Crystallization and data collection
l- and d-enantiomeric forms of the two strands 5′−CGCTATAATGCG−3′ and 5′−CGCATTATAGCG−3′ were synthesized by ChemGenes Corporation (USA). A 4 mM stock solution of each strand was prepared using ultra-pure water. An annealing solution of the l form was prepared using 2 mM of each of the two complementary strands plus 50 mM sodium cacodylate buffer (pH 7.0) and 50 mM sodium chloride. The solution was heated at 353 K for 20 min and gradually cooled overnight to 293 K to ensure duplex formation. The same protocol was followed to anneal the d-enantiomer sequences. A racemic mixture was prepared following the annealing process by mixing the two enantiopure solutions in an equimolar ratio. Crystallization experiments using commercially available screens (Natrix 2—Hampton Research (22,23)) were performed at 293 K, using the hanging drop vapour diffusion method. Crystals were obtained in four distinct conditions within one week (Supplementary Table S1 and Figure 1), and were used for X-ray diffraction measurements without further optimization. For low temperature measurements (at 100 K), single crystals were flash-frozen directly in liquid nitrogen. Diffraction data were measured on beam line PROXIMA 1 at SOLEIL synchrotron, or using an in-house rotating anode X-ray diffractometer (Rigaku FRX micro-focus rotating anode, generating Cu Kα radiation). The diffraction data were processed using XDS (24), with data statistics described in Supplementary Table S2.
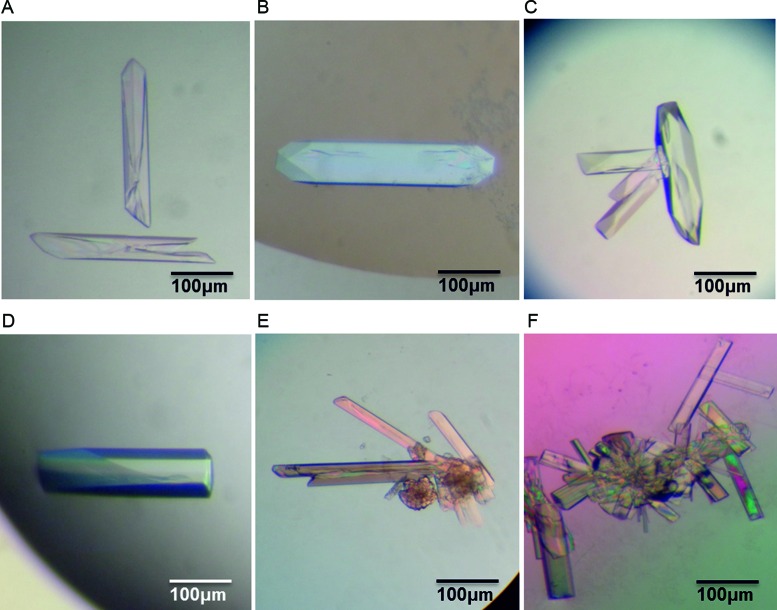
Figure 1.
Crystals of a DNA duplex containing the Pribnow box consensus promoter sequence. Crystals were derived from: (A–D) racemic DNA mixtures using four unique crystallization reagents, and; (E–F) D-enantiopure DNA solutions.
Structure determination and refinement
The structures reported in this work were solved by molecular replacement using the program PHASER (25). The structures were refined using REFMAC5 (26) from the CCP4 software package (27) and phenix.refine (28) from the PHENIX suite (29). The refinement protocol includes simulated annealing, Translation Libration Screw-motion refinement, positional refinement, restrained isotropic B-factor refinement and bulk solvent correction. Full details of the structure determination and refinement process are provided in the Supplementary Data. After each refinement step, visual inspection of the model and the electron-density maps were carried out using Coot (30), using both 2Fo-Fc and Fo-Fc difference maps. Ions and water molecules were added throughout different stages of refinement as indicated by electron density in the appropriate Fo-Fc difference maps. The difference Fourier maps were contoured at 5 σ and 3 σ levels in order to place the ions and water molecules, respectively. Figures were prepared using the program PyMOL (31). The coordinates and structure factors have been deposited in the Protein Data Bank (11) (accession codes are provided in Table 1). Refinement statistics are provided in Supplementary Table S2.
Table 1. Summary of crystal structures of the Pribnow box consensus promoter sequence.
| Crystal structure | DNA solution | Crystal screen (solution no.) | Max. Res. (Å) | Space group | Asymmetric unit | PDB code |
|---|---|---|---|---|---|---|
| 1 | racemic | Natrix 2-39 | 1.83 | P21/n | 1 duplex | 5ET9 |
| 2 | racemic | Natrix 2-36 | 1.69 | P21/c | 2 duplexes (A,B) | 5EWB |
| 3 | racemic | Natrix 2-38 | 2.30 | Pnna | 1 single strand | 5EYQ |
| 4 | racemic | Natrix 2-47 | 1.65 | Pbca | 1 duplex | 5EZF |
| 5 | D-DNA | Natrix 2-39 | 2.90 | P63 | 1 duplex | 5F26 |
| 6 | D-DNA | Custom condition | 2.81 | P32 | 3 duplexes (A,B,C) | 5J0E |
Full crystallization, data collection and refinement details are provided in Supplementary Tables S1 and S2.
RESULTS AND DISCUSSION
Racemic crystallization and structure determination of a Pribnow box duplex
In order to test the ability of the racemic crystallization method to crystallize a DNA sequence which had previously proven challenging to crystallize, we attempted to crystallize a non-self-complementary DNA duplex containing the Pribnow box consensus sequence. The much-studied Dickerson–Drew dodecamer self-complementary sequence—d(CGCGAATTCGCG)—served as a template for the incorporation of the Pribnow box consensus hexamer sequence 5′-TATAAT-3′ within the centre, thereby replacing 5′-GAATTC-3′. This incorporation breaks the sequence symmetry, and gives rise to a duplex composed of two non-self-complementary sequences: d(CGCTATAATGCG) (template strand) and d(CGCATTATAGCG) (complementary strand). The terminal regions are functionally irrelevant, but are needed for duplex stability. Racemic mixtures of preformed duplexes composed of the sequences defined above were then used for crystallization trials. Standard commercially available sparse matrix screens were used, yielding visually well-formed crystals in four distinct crystallization conditions within one week (Figure 1A–D). X-ray diffraction analyses of all four crystal forms indicated the crystals belonged to achiral space groups, implying these crystals to be racemic, in line with previous findings highlighting the tendency of racemic DNA mixtures to form racemic crystals rather than conglomerates (4). Structure determination by molecular replacement following full data collection of all four crystal forms confirmed the achiral space groups, with structures 1–4 belonging to space groups P21/n, P21/c, Pnna and Pbca, respectively (see Table 1, and the Supplementary Data for a discussion on the similarity between structures 1 and 3). All structures were easily refined, resulting in high quality electron density maps in which all residues of the DNA are stable and accurately modelled (Supplementary Figure S1). One may note relatively high R factors (typically around 30%) considering the resolution of the structures. This is inherent to refinement in centrosymmetrical space groups and reflects small differences in the structures of the two enantiomers. The case has been discussed for racemic protein crystals for which it was shown that refining the d-form + l-form in a non-centrosymmetric lattice does not necessarily improve R-factors (32).
In all four crystal forms (structures 1–4), d- and l- DNA enantiomers are related by chirality-inverting symmetry operations (i.e. inversions and glide planes) and chirality-preserving symmetry operations (i.e. rotations, translations and screw axes) relate different molecules of each enantiomer. It has been pointed out by Wukovitz and Yeates (1,2) that the high level of dimensionality of the P1(bar) space group makes it a common and highly favourable packing arrangement for macromolecular racemic/achiral systems (the packing rules are different for small molecules). Surprisingly, we did not observe the P1(bar) space group for the Pribnow box duplex structures reported here. The predicted next best packing arrangements for racemic macromolecules are P21/c and C2/c. Only P21/c and the related P21/n were found in this work. Our previous reports of racemic DNA crystallography (4) and also earlier work on racemic Z-DNA (33) and racemic RNA (34) revealed the apparent prevalence of space group P1(bar) for the DNA sequences studied, particularly for the self-complementary duplex crystal structures reported. Thus, it is possible that the use of non-self-complementary DNA sequences is responsible for the absence of a P1(bar) packing arrangement for the Pribnow box duplexes reported here.
In addition to the considerable number of crystal structures produced by the racemic crystallization screen of the Pribnow box duplex reported above, the ease of crystallization (crystals were obtained within a short space of time using standard commercial crystallization screens, with no optimization of crystal growth conditions needed, see Table 1) and high quality of the crystals (in some cases permitting mid-to-high resolution diffraction data to be collected on home X-ray sources), strongly highlights the potentially considerable utility of adopting a racemic crystallization approach for the structure determination of challenging DNA molecules.
Non-racemic crystal structures of the Pribnow box duplex
In order to measure the true benefit of using the racemic crystallization method and to investigate the principles governing crystal growth, we attempted to determine whether an enantiopure solution of the Pribnow box duplex would crystallize when exposed to the crystallization conditions found to crystallize equivalent racemic mixtures. There is no reason for this to occur a priori since racemic and non-racemic crystals would have distinct packing arrangements. Yet we had observed the occurrence of the reverse: racemates often (but not always) preferentially crystallize under conditions in which the d-enantiomer alone also crystallizes (4). Enantiopure DNA samples were exposed to the same conditions as those used for the racemic crystallization trials, with crystals obtained in just one out of the four racemic crystal conditions. X-ray diffraction analysis and data collection on our home source revealed a crystal structure belonging to space group P63 (crystal structure 5, Table 1, Supplementary Tables S1 and S2) with the data being of lower resolution (2.9 Å) and poorer quality than that obtained for the equivalent racemic mixture. The quality of diffraction data was reflected in the final model, which contains several disordered residues, in contrast to the racemic Pribnow box structures (further details are provided in the Supplementary Data). No attempt was made to optimize crystal growth conditions or to carry out synchrotron measurements.
Thus, it seems that, in the example reported here at least, although it is possible for an enantiopure mixture to crystallize in a condition suitable for crystal growth of a racemic mixture, the conditions most suitable for crystal growth cannot be directly exchanged between racemic and non-racemic samples. In addition, crystals of an enantiopure solution of the Pribnow box duplex were also obtained in a set of crystallization conditions different to those found to crystallize equivalent racemic mixtures—and in which the racemic Pribnow box duplex mixture could not be subsequently crystallized (crystal structure 6, Table 1, Supplementary Tables S1 and S2). This complements earlier results which showed that, in most cases, racemic DNA crystals form under conditions optimized for enantiopure DNA (4). It appears that conditions suitable for crystal growth (even of poorly diffracting crystals) of enantiopure d-DNA samples cannot always be applied to racemic mixtures. As the reverse is also true (as described above), the take home message is clear: if structure determination of a DNA molecule is desired, independent crystallization experiments should be performed using both racemic and enantiopure DNA samples. In doing so, the exploration of all possible space groups is realized.
Overall topology, helical analysis and potential biological significance of racemic Pribnow box crystal structures
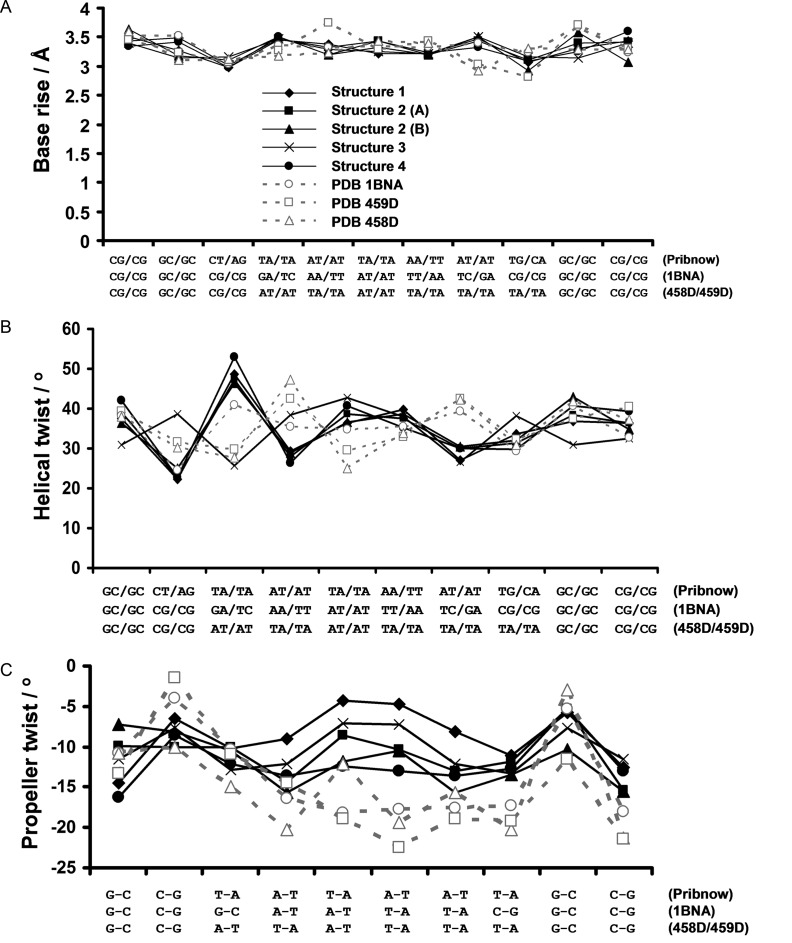
All four crystal forms of the racemic non-self-complementary Pribnow box duplex reported here reveal DNA structures adopting standard B-type DNA conformations (Figure 2, S5). Despite the differences in space group and crystal packing, the four racemic crystal structures share the common feature of their central TATAAT regions taking part in very few crystal packing contacts (a packing feature which is also observed for standard non-racemic duplex crystal structures such as the Dickerson–Drew dodecamer (12–13,35–38)). This therefore reduces the likelihood of crystal packing factors influencing (and potentially perturbing) local structural features of the racemic Pribnow box duplex structures reported here. Helical parameters were calculated using 3DNA (39). In general, the four racemic Pribnow box duplex crystal structures all display helical parameters in line with those expected for standard B-type DNA (Figure 3A–C), however, there are differences in certain helical features that may indicate sequence-specific structural effects. For example, the degree of base propeller-twist (defined as the rotation of one base with respect to the other in the same base pair) (40,41) of the racemic structures is significantly reduced compared to the Dickerson–Drew dodecamer (35) as well as one of the few examples of non-self-complementary duplexes available (42) (Figure 3C). Due to poor model quality, helical parameters for enantiopure Pribnow box structures are unreliable and therefore cannot be used to determine whether the reduced base propeller-twist of the racemic Pribnow box structures is a result of the specific sequence, rather than the (a)chirality of the sample. However, that reduced base propeller-twist values are observed in four distinct packing environments suggests that these observations are meaningful. It is assumed that the Pribnow box sequence possesses a mechanism by which the sigma factor can specifically recognize this key motif amongst the bacterial genome—indeed, local structural variations unique to this specific sequence may well be responsible for such specificity. Although we do not propose the observed reduction in propeller-twist of the Pribnow box structures reported here to be the basis of the mechanism by which the sigma factor recognizes this fundamentally important DNA sequence, the fact that there exists structural differences (albeit subtle) between certain duplexes containing biologically relevant wild-type DNA sequences versus much-studied model duplexes (such as the Dickerson–Drew dodecamer) is noteworthy. Whether such structural differences are indeed of biological significance or not will require further work, however, the findings reported here suggest that the racemic crystallographic method may be able to aid such an endeavour.
Figure 2.
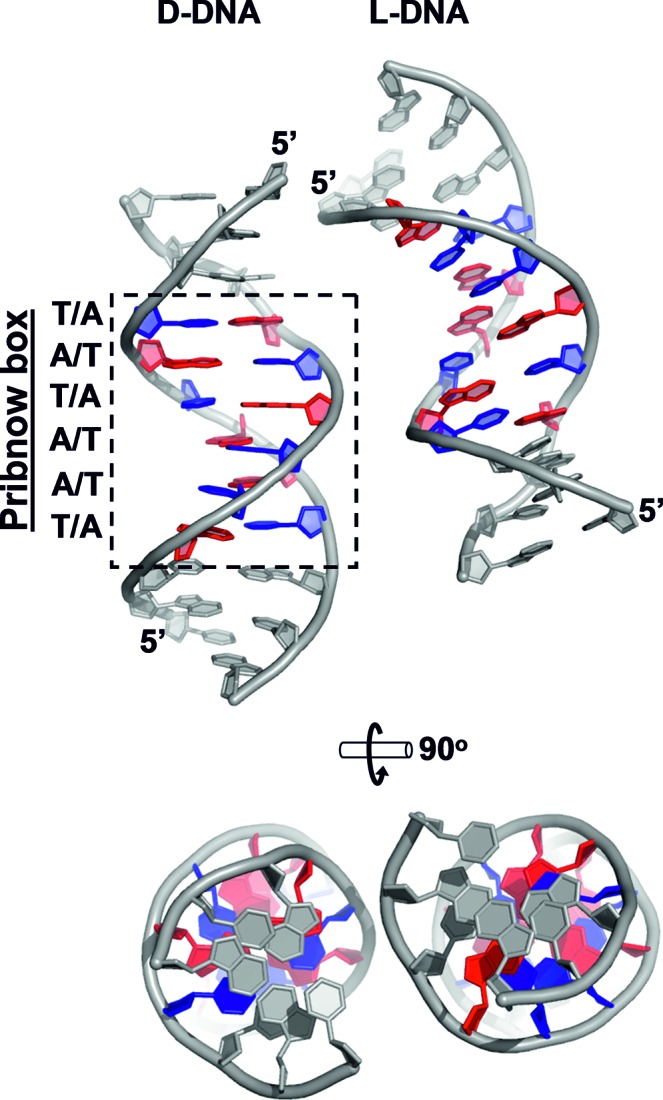
Crystal structure of a DNA duplex containing the Pribnow box consensus promoter sequence (crystal structure 4, space group Pbca, resolution 1.65 Å). Left: asymmetric unit, containing one complete D-DNA duplex. Right: L-DNA symmetry mate. Pribnow box residues are coloured blue (thymine) and red (adenine). All other residues (i.e. all non-Pribnow box residues) are coloured grey.
Figure 3.
Helical parameters of four racemic Pribnow box duplex crystal structures reported here (structures 1–4) and three non-racemic DNA duplex crystal structures (PDB structure 1BNA—a high-resolution Dickerson–Drew dodecamer formed from the sequence d[CGCGAATTCGCG] (35), and PDB structures 458D and 459D—native and ligand-bound dodecamer duplexes, respectively, formed from the non-self-complementary sequences d[CGCATATTTGCG] and d[CGCAAATATGCG]) (42). DNA sequences are indicated along the x axes. Parameters calculated using 3DNA (39). (A) Base-pair rise; (B) local helical-twist; (C) propeller-twist. Overall, when comparing Pribnow box duplexes (structures 1–4) with non-Pribnow box duplexes (PDB structures 1BNA, 458D and 459D) the majority of helical parameters are in agreement, however, the Pribnow box structures show consist variations in propeller twist values of the central TA-rich region (panel C) compared to non-Pribnow box structures. Solid lines with filled markers = racemic Pribnow box structures; dashed lines with empty markers = non-Pribnow box (chiral) structures. Note, the asymmetric unit of structure 2 contains two distinct duplexes which have been analysed separately (referred to as ‘structure 2 (A)’ and ‘structure 2 (B)’).
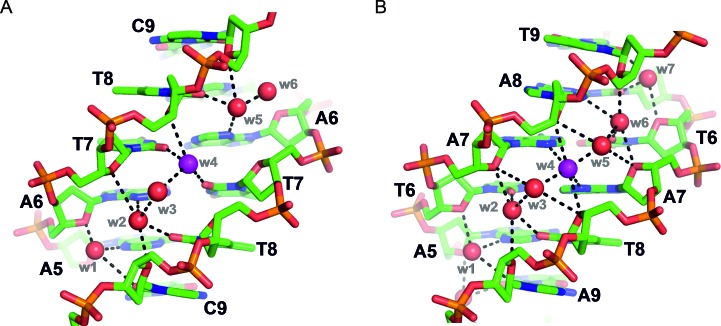
Another aspect worthy of comparison are the minor grooves of these structures. The minor groove of duplex DNA is of particular interest as many anti-cancer (and anti-infective) small-molecule drugs have been designed to recognize and bind specifically to these regions (18–21,43–48). Since crystals structures used to guide rational drug design of such therapeutic agents are almost invariably based on model sequences (18–21,43–48), any differences, even subtle, between model duplexes and those containing wild-type motifs could be significant with respect to DNA recognition by small-molecules. The minor groove widths of the racemic Pribnow box duplexes reported here are comparable to those of model (and other) DNA duplexes of the same size (i.e. same oligomer length) (see Supplementary Table S3). There is also a certain level of conservation of minor groove water structure between Pribnow box and non-Pribnow box duplexes (Figure 4). In particular, some water molecules are found at similar positions despite the difference in sequence, and thus despite the difference in the bases to which water molecules are hydrogen bonded. For example, a water molecule (w4, highlighted in magenta in Figure 4) which is bound tightly to two thymine O2 carbonyl groups within the high-resolution Dickerson–Drew dodecamer crystal structure (PDB ID: 1BNA) (35), occupies an equivalent position within the minor groove of the racemic Pribnow-box structure in the P21/c space group, yet in the latter case, the water molecule is bound instead by the N3 nitrogen atoms of two adenine bases (Figure 4). A small molecule or metal complex minor groove binder may thus also bind at similar positions and its sequence selectivity will be determined only by its inherent affinity for oxygen versus nitrogen acceptors.
Figure 4.
Comparison of minor groove water structure and hydrogen bonding between: (A) a non-racemic model DNA duplex (Dickerson–Drew dodecamer, in space group P212121, PDB ID: 1BNA (35)) and (B) a racemic Pribnow box duplex (in space group P21/c [structure 2 (A) in Table 1]). The water structure of the minor groove is well-conserved overall between these two duplexes—one extra water molecule appears in the Pribnow box duplex—despite the difference in sequence. In the parts shown, minor groove hydrogen bond acceptors may be adenine N3 atoms, or thymine or cytosine O2 carbonyl groups. See for example w4 shown in magenta, hydrogen bonded to thymine-7 bases in (A) and to adenine-7 bases in (B).
CONCLUSIONS
We have reported here the use of a racemic crystallographic approach for the crystallization and subsequent structure determination of a historically intractable, biologically relevant DNA motif—that of the Pribnow box consensus sequence 5’-TATAAT-3’. Four high-quality crystal structures were determined with resolutions in the range of 1.65–2.3 Å, providing an atomic-level view of a DNA motif involved in a biological process of inarguable importance—that of bacterial replication. Although observed differences in certain structural features between the racemic Pribnow box structures reported here and well-studied model sequences cannot, at this point, be correlated to biological processes, our results nevertheless indicate that, by allowing structure determination of non-self-complementary DNA structures, the racemic method may be a suitable means to obtain high-resolution structural data of DNA duplexes formed from biologically meaningful sequences. This may have consequences for those designing and characterizing DNA-interacting therapeutic agents, such as anti-cancer compounds (18,43–45) and anti-infective agents (46–48). In addition, we have described here further investigations into the racemic crystallization process—involving the determination of two additional Pribnow box duplexes—suggesting that crystal growth conditions may not be directly interchangeable between racemic and non-racemic systems.
The racemic crystallization technique used and described here provides a potentially facile method for studying native DNA sequences beyond the classical model sequences, and thus may be of interest to those investigating structural aspects of fundamental biological processes as well as those engaged in structure-based DNA-directed drug discovery programmes.
ACCESSION NUMBERS
PDB IDs: 5ET9, 5EWB, 5EYQ, 5EZF,5F26 and 5J0E.
Supplementary Material
Acknowledgments
We thank SOLEIL synchrotron for providing access to data collection facilities, and are grateful to the beam line staff of PROXIMA 1 for assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union's Seventh Framework Program through the European Research Council [ERC-2012-AdG-320892]; European Union's Seventh Framework Program through the European Research Council Postdoctoral Fellowship (to P.K.M.). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wukovitz S.W., Yeates T.O. Why protein crystals favour some space-groups over others. Nat. Struct. Biol. 1995;2:1062–1067. doi: 10.1038/nsb1295-1062. [DOI] [PubMed] [Google Scholar]
- 2.Yeates T.O., Kent S.B.H. Racemic protein crystallography. Annu. Rev. Biophys. 2011;41:41–61. doi: 10.1146/annurev-biophys-050511-102333. [DOI] [PubMed] [Google Scholar]
- 3.Zawadzke L.E., Berg J.M. The structure of a centrosymmetric protein crystal. Proteins Struct. Funct. Genet. 1993;16:301–305. doi: 10.1002/prot.340160308. [DOI] [PubMed] [Google Scholar]
- 4.Mandal P.K., Collie G.W., Kauffmann B., Huc I. Racemic DNA crystallography. Angew. Chem. Int. Ed. 2014;53:14424–14427. doi: 10.1002/anie.201409014. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu. Rev. Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 6.Siebenlist U., Simpson R.B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 7.Hawley D.K., McClure W.R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Youderian P., Vershon A., Bouvier S., Sauer R.T., Susskind M.M. Changing the DNA-binding specificity of a repressor. Cell. 1983;35:777–783. doi: 10.1016/0092-8674(83)90110-1. [DOI] [PubMed] [Google Scholar]
- 9.Berman H.M., Olson W.K., Beveridge D.L., Westbrook J., Gelbin A., Demeny T., Hsieh S.H., Srinivasan A.R., Schneider B. The Nucleic Acid Database: a comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman H.M., Gelbin A., Westbrook J. Nucleic acid crystallography: a view from the nucleic acid database. Prog. Biophys. Molec. Biol. 1996;66:255–288. doi: 10.1016/s0079-6107(97)00019-9. [DOI] [PubMed] [Google Scholar]
- 11.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wing R.M., Drew H.R., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R.E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 13.Dickerson R.E., Drew H.R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J. Mol. Biol. 1981;149:761–768. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- 14.Coll M., Frederick C.A., Wang A.H.-J., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCGAAATTTGCG) and its complex with distamycin. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon C., Prive G.G., Goodsell D.S., Dickerson R.E. Structure of an alternating B-DNA helix and its relation to A-tract DNA. Proc. Natl. Acad. Sci. U.S.A. 1987;85:6332–6336. doi: 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balendiran K., Rao S.T., Sekharudu C.Y., Zon G., Sundaralingam M. X-ray structures of the B-DNA dodecamer d(CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex. Acta Crystallogr. D Biol. Crystallogr. 1995;51:190–198. doi: 10.1107/S0907444994010759. [DOI] [PubMed] [Google Scholar]
- 17.Prive G.G., Yanagi K., Dickerson R.E. Structure of the B-DNA decamer CCAACGTTGG and comparison with isomorphous decamers CCAAGATTGG and CCAGGCCTGG. J. Mol. Biol. 1991;217:177–199. doi: 10.1016/0022-2836(91)90619-h. [DOI] [PubMed] [Google Scholar]
- 18.Wing R.M., Pjura P., Drew H.R., Dickerson R.E. The primary mode of binding of cisplatin to a B-DNA dodecamer CGCGAATTCGCG. EMBO J. 1984;3:1201–1206. doi: 10.1002/j.1460-2075.1984.tb01951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neidle S. DNA minor-groove recognition by small molecules. Nat. Prod. Rep. 2001;18:291–309. doi: 10.1039/a705982e. [DOI] [PubMed] [Google Scholar]
- 20.Campbell N.H., Evans D.A., Lee M.P.H., Parkinson G.N., Neidle S. Targeting the DNA minor groove with fused ring dicationic compounds: comparison of in silico screening and a high resolution crystal structure. Bioorg. Med. Chem. Lett. 2006;16:15–19. doi: 10.1016/j.bmcl.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 21.Wei D., Wilson W.D., Neidle S. Small-molecule binding to the DNA minor groove is mediated by a conserved water cluster. J. Am. Chem. Soc. 2013;135:1369–1377. doi: 10.1021/ja308952y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott W.G., Finch J.T., Grenfell R., Fog J., Smith T., Gait M.J., Klug A. Rapid crystallization of chemically synthesized Hammerhead RNAs using double screening procedure. J. Mol. Biol. 1995;250:327–332. doi: 10.1006/jmbi.1995.0380. [DOI] [PubMed] [Google Scholar]
- 23.Berger I., Kang C.H., Sinha N., Wolters M., Rich A. A highly efficient 24-condition matrix for the crystallization of nucleic acid fragments. Acta Crystallogr. D Biol. Crystallogr. 1996;52:465–468. doi: 10.1107/s0907444995013564. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storini L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLano W.L. The PyMOL molecular graphics system. Schrödinger LLC; 2002. Version 1.7.4. [Google Scholar]
- 32.Mandal K, Pentelute B.L., Tereshko V., Thammavongsa V., Schneewind O., Kossiakoff A.A., Kent S.B.H. Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods. Protein Sci. 2009;18:1146–1154. doi: 10.1002/pro.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doi M., Inoue M., Tomoo K., Ishida T., Ueda Y., Akagi M., Urata H. Structural characteristics of enantiomorphic DNA: crystal analysis of racemates of the d(CGCGCG) duplex. J. Am. Chem. Soc. 1993;115:10432–10433. [Google Scholar]
- 34.Rypniewski W., Vallazza M., Perbandt M., Klussmann S., DeLucas L.J., Betzel S., Erdmann V.A. The first crystal structure of an RNA racemate. Acta Crystallogr. D. Biol. Crystallogr. 2006;62:659–664. doi: 10.1107/S090744490601359X. [DOI] [PubMed] [Google Scholar]
- 35.Drew H.R., Wing R.M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R.E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. U.S.A. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drew H.R., Samson S., Dickerson R.E. Structure of a B-DNA dodecamer at 16 K. Proc. Natl. Acad. Sci. U.S.A. 1982;79:4040–4044. doi: 10.1073/pnas.79.13.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fratini A.V., Kopka M.L., Drew H.R., Dickerson R.E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J. Biol. Chem. 1982;257:14686–14707. [PubMed] [Google Scholar]
- 38.Shui X., McFail-Isom L., Hu G.G., Williams LD. The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry. 1998;37:8341–8355. doi: 10.1021/bi973073c. [DOI] [PubMed] [Google Scholar]
- 39.El Hassan M.A., Calladine C.R. Two distinct modes of protein-induced bending in DNA. J. Mol. Biol. 1998;282:331–343. doi: 10.1006/jmbi.1998.1994. [DOI] [PubMed] [Google Scholar]
- 40.Dickerson R.E., Bansal M., Calladine C.R., Diekmann S., Hunter W.N., Kennard O., von Kitzing E., Lavery R., Nelson H.C.M., Olson W.K., et al. Definitions and nomenclature of nucleic acid structure components. Nucleic Acids Res. 1989;17:1797–1803. doi: 10.1093/nar/17.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson W.K., Bansal M., Burley S.K., Dickerson R.E., Gerstein M., Harvey S.C., Heinemann U., Lu X.J., Neidle S., Shakked Z., et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 42.Aymami J., Nunn C.M., Neidle S. DNA minor groove recognition of a non-self-complementary AT-rich sequence by a tris-benzimidazole ligand. Nucleic Acids Res. 1999;27:2691–2698. doi: 10.1093/nar/27.13.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kopka M.L., Yoon C., Goodsell D, Pjura P., Dickerson R.E. Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J. Mol. Biol. 1985;183:553–563. doi: 10.1016/0022-2836(85)90171-8. [DOI] [PubMed] [Google Scholar]
- 44.Pjura P.E., Grzeskowiak K., Dickerson R.E. Binding of Hoechst 33258 to the minor groove of B-DNA. J. Mol. Biol. 1987;197:257–271. doi: 10.1016/0022-2836(87)90123-9. [DOI] [PubMed] [Google Scholar]
- 45.Coll M., Frederick C.A., Wang A.H.J., Rich A. A bifurcated hydrogen-bonded conformation in the d(AT) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc. Natl Acad. Sci. U.S.A. 1987;84:8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown D.G., Sanderson M.R., Skelly J.V., Jenkins T.C., Brown T., Garman E., Stuart D.I., Neidle S. Crystal structure of a berenil-dodecanucleotide complex: the role of water in sequence-specific ligand binding. EMBO J. 1990;9:1329–1334. doi: 10.1002/j.1460-2075.1990.tb08242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards K.J., Jenkins T.C., Neidle S. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry. 1992;31:7104–7109. doi: 10.1021/bi00146a011. [DOI] [PubMed] [Google Scholar]
- 48.Nunn C.M., Jenkins T.C., Neidle S. Crystal structure of d(CGCGAATTCGCG) complexed with propamidine, a short-chain homologue of the drug pentamidine. Biochemistry. 1993;32:13838–13843. doi: 10.1021/bi00213a012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.