Abstract
Since the work of Alexander Rich, who solved the first Z-DNA crystal structure, we have known that d(CpG) steps can adopt a particular structure that leads to forming left-handed helices. However, it is still largely unrecognized that other sequences can adopt ‘left-handed’ conformations in DNA and RNA, in double as well as single stranded contexts. These ‘Z-like’ steps involve the coexistence of several rare structural features: a C2’-endo puckering, a syn nucleotide and a lone pair–π stacking between a ribose O4’ atom and a nucleobase. This particular arrangement induces a conformational stress in the RNA backbone, which limits the occurrence of Z-like steps to ≈0.1% of all dinucleotide steps in the PDB. Here, we report over 600 instances of Z-like steps, which are located within r(UNCG) tetraloops but also in small and large RNAs including riboswitches, ribozymes and ribosomes. Given their complexity, Z-like steps are probably associated with slow folding kinetics and once formed could lock a fold through the formation of unique long-range contacts. Proteins involved in immunologic response also specifically recognize/induce these peculiar folds. Thus, characterizing the conformational features of these motifs could be a key to understanding the immune response at a structural level.
INTRODUCTION
Diversity in shape between RNA and DNA is striking. Although DNA can adopt A and B helical forms, RNA double strands are never of the B-form, due to the ribose preference for a C3’-endo over a C2’-endo pucker. Yet, both DNA and RNA can adopt a left-handed Z-form in which C2’-endo and C3’-endo alternate along a CpG sequence (1).
Historically, Z-DNA was crystallized before A-DNA, B-DNA and Z-RNA (2–4). Its structural properties and in particular the repeated 5′-pyrimidine-purine-3′ dinucleotide step along the helix with a purine in syn (Figure 1), were described in detail (1,2,5–9). The most frequently crystallized Z-DNA dinucleotide step is CpG, but other Z-DNA steps have been described (1,10,11). However, because the in vitro formation of both Z-DNA and Z-RNA usually requires a high ionic strength or specific nucleotide modifications, it was assumed for a long time that Z-forms were mere structural artifacts (3,4).
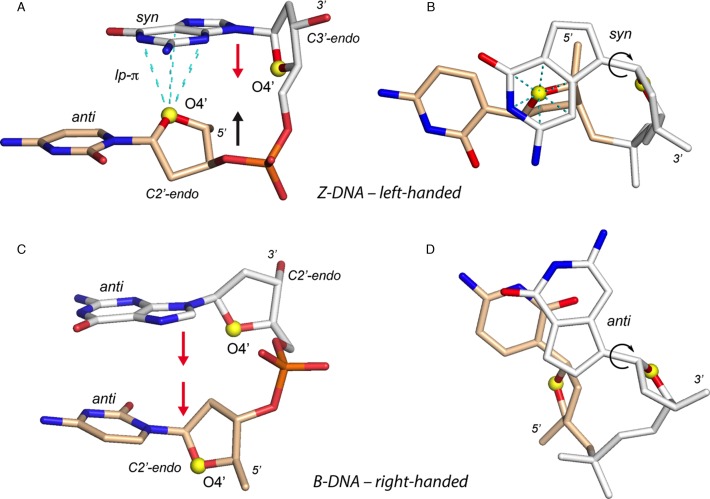
Figure 1.
Structural features of Z-DNA and B-DNA CpG steps (similar rules apply for all d/r(NpN) steps). 3′- and 5′-nucleotides are coloured in white and wheat, respectively; O4’ atoms are shown as yellow spheres. (A) d(CpG) step extracted from a Z-DNA crystal structure (PDB: 3P4J; res: 0.55 Å). Note the characteristic antiparallel orientation of the ribose rings marked by red and black arrows. (B) Orthogonal view of (A) emphasizing the syn orientation of the the 3′-residue. In (A) and (B), the dashed cyan lines correspond to interatomic contact distances (≤3.5 Å) involving the 5′-O4’ atom and the 3′-six-membered ring atoms. These contacts define a ‘capping’ or ‘lp–π’ interaction. (C) d(CpG) step extracted from a B-DNA crystal structure (PDB: 1EN3; res: 0.99 Å). Two red arrows mark the parallel orientation of the ribose rings. (D) Orthogonal view of (C) emphasizing the 3′-residue anti orientation. In (B) and (D), 3′-glycosidic ‘syn/anti’ bonds marked by circular arrows are similarly oriented and all atoms except the N/O atoms on the Watson–Crick edges and the O4’ atoms are in white or wheat colours.
Although both Z-DNA and Z-RNA have been known to be immunogenic since the 1980s, their biological role was questioned (4). We now know of four families of Z-DNA binding proteins that are all involved in the innate immune response such as the interferon induced form of the RNA editing enzyme ADAR1, the innate immune system receptor DLM-1, the fish kinase PKZ and the pox-virus inhibitor of interferon response EL3 (12,13). These proteins were found to recognize Z-DNA in a conformation-specific manner since most of the contacts with the protein involve backbone atoms (13,14). Moreover, evidence that some of these protein domains interact with Z-RNA in vitro have been gathered, which raised issues related to the in vivo role of this RNA form (12,15–17).
Here, we report the unanticipated occurrence of Z-like dinucleotide steps at key locations in single stranded RNA regions following a first identification in CUG-regulator binding proteins (18). We also highlight how r(U/ApA) steps are found more frequently than r(CpG) steps. Since our goal is to better characterize rare conformational features in RNA, we examine in detail the structure of what we refer to as a ‘Z-like’ motif, in particular within the context of r(UNCG) tetraloops where it has never been described although it is an essential component of this fold. We find that the Z-like motif contains a ribose-base or lone pair–π (lp–π) stacking that consists in the close contact of the 5′-ribose O4’ atom with the six-membered 3′-guanine ring as observed in Z-DNA (Figure 1). This ribose-base stacking has been mentioned in the first studies of Z-DNA crystallographic structures (2,6,7) but its implications were only investigated several years later (19–21) and never addressed in RNA systems. More generally, such lp–π interactions are currently subject to strong interest in the chemical field where they are considered as a significant and largely unexplored non-covalent interaction type (22–24). Finally, we describe double and single strand Z-like conformations in RNA/protein systems and, among those, in viral RNA that are specifically recognized in a conformation-dependent manner by specialized proteins from the immune system.
MATERIALS AND METHODS
The Protein Data Bank (PDB) was searched for ‘Z-DNA like’ dinucleotide steps (hereafter named ‘Z-like’ steps) in DNA and RNA crystallographic structures with resolutions ≤3.0 Å. Z-like steps were characterized using the following criteria: (i) the 3′- and 5′-nucleotide adopt a syn and anti orientation, respectively; (ii) the 5′-O4’ ribose atom is at ≤3.5 Å from the 3′-nucleobase plane with its projection on the base plane circumscribed in the polygon defined by the ring atoms (Figure 1A and B); these criteria define a lone pair–π interaction that is associated with an atypical antiparallel orientation of the ribose rings with facing 3′- and 5′-O4’ atoms (6). In order to exclude a few borderline cases, we explicitly imposed antiparallel orientation of the ribose rings in this survey.
Since some Z-like steps have ribose puckers in the north, east or south quadrant, we did not rely for their identification on the classical and more restrictive Z-DNA C2’-endo to C3’-endo ribose pucker sequence. Thus, we avoid issues related to the difficulty to resolve precisely ribose puckers in experimental structures (25,26). The 3DNA/DSSR analysis tool was used to identify ‘lp–π’ capping contacts (Figure 1A and B) as well as to calculate backbone torsion angles, hydrogen bond contacts, ribose puckers and to characterize syn/anti conformations (27).
Z-like steps having atoms with B-factors ≥79 Å2 were excluded from our statistics as well as disordered or terminal steps found at the 3′- or 5′-end of the structures unless otherwise specified. Crystallographic structures with resolutions >3.0 Å as well as some cryo-EM and NMR structures were also inspected, although they were not considered for statistics. As of February 2016, the PDB contains ≈5100 nucleic acid crystal structures including complexes with proteins over a total of ≈96 000 biomolecular structures (resolution ≤ 3.0 Å). Images of 3D structures were generated with PyMOL (Schrödinger, L.L.C.; http://www.pymol.org).
Non-redundant Z-like steps were tagged as follows. If two steps from different structures share identical residue numbers, chain codes and tetranucleotide sequences (including a residue before and after the dinucleotide step) as well as ribose puckers, backbone dihedral angle sequences (following a g+, g-, t categorization) and syn/anti conformations, they are considered as similar and the one with the best resolution is marked as non-redundant. Alike, if in a same structure two Z-like steps share same residue numbers and tetranucleotide sequences (with different chain codes) as well as ribose puckers, backbone dihedral angle sequences and syn/anti conformations, they are considered as similar and the one corresponding to the first biological unit is marked as non-redundant. The former criteria are used to filter similar PDB structures and the latter to filter structures with multiple related biological units. Note that it is impossible to completely eliminate redundancy from a dataset without eliminating at the same time significant data. Here, we provide an upper limit for a truly ‘non-redundant’ set.
RESULTS
Z-like steps are found in both DNA and RNA and are not limited to CpG steps
CpG steps are highly represented in Z-DNA left-handed duplexes (Table 1). Besides, a few Z-like TpG/ApA, CpA and GpG/ApC/TpT steps were identified in quadruplex loops, in a rare DNA tetraloop (28) and in single stranded DNA. Also some telomere end-binding proteins recognize Z-like GpG steps (29).
Table 1. Number of non-redundant Z-like steps found in PDB crystallographic RNA and DNA structures (resolution ≤ 3.0 Å; atomic B-factors ≤ 79 Å2; February 2016 PDB survey) over a total >600 000 nucleotide steps.
| Dinucleotide steps | DNAa | RNAa |
|---|---|---|
| 3′-guanine | ||
| CpG | 124 (187) | 42 (125) |
| (T/U)pG | 9 (9) | 13 (21) |
| ApG | – | 14 (57) |
| GpG | 8 (11) | 1 (6) |
| 3′-adenine | ||
| CpA | 3 (4) | 18 (25) |
| (T/U)pA | 13 (15) | 42 (127) |
| ApA | 1 (1) | 75 (279) |
| GpA | – | 13 (13) |
| 3′-cytosine | ||
| CpCb | – | 1 (1) |
| (T/U)pC | – | 5 (5) |
| ApC | – | 1 (1) |
| GpC | – | – |
| 3′-thymine/uridine | ||
| Cp(T/U) | – | 2 (2) |
| (T/U)p(T/U) | 1 (1) | 1 (1) |
| Ap(T/U) | – | 3 (4) |
| Gp(T/U) | – | – |
| Total: | 159 (228) | 231 (667) |
aThe total number of Z-like steps found in the PDB and with no redundancy considerations is given in parenthesis.
bThe 3′-cytosine of this CpC step displays a 50% syn and anti occupancy (70).
Steps with disordered backbones, usually found in high-resolution Z-DNA structures, were not taken into account. However, modified nucleotides in Z-DNA were considered. Note that these statistics reflect only the step distribution in structures deposited to the PDB and not the in vivo distribution of these steps. The high number of non-redundant CpG steps in Z-DNA is partly related to the incorporation of modified nucleotides in our structural sample and should be considered with caution.
In RNA, over 600 Z-like steps are found in the PDB. Compared with the total number of dinucleotide steps in the database (>600 000), they correspond to rare events, but their presence at key locations in a limited number of RNA families, including ribosomal RNA (see below) makes them noteworthy. Their sequence variety is much greater than in DNA (Table 1). All steps containing a 3′-purine are represented and ApA steps are more frequent than CpG steps contrasting with the dominance of the latter in DNA. Strikingly, in both nucleic acids, 5′-pyrimidine steps remain uncommon. Up to now, as only one crystal structure of a Z-RNA helix has been solved (16), most of the identified Z-like steps are located within non-helical regions. However, since a large diversity of sequences are found in Z-DNA helices, the crystallization of further Z-RNA duplexes might reveal a similar sequence diversity.
Ribose-base stacking or ‘lp–π’ (lone pair–π) interactions define the structure of Z-like steps
A striking characteristic of Z-like steps in DNA relates to the stacking of the O4’ atom of the 5′-deoxyribose ring with the six-membered ring of the 3′-residue, a contact that was to the best of our knowledge never described in NMR or crystallographic structures of RNA systems (Figure 1A and B). This contact is promoted by the large slide of the two bases and by a specific sequence of backbone dihedral angles that leads to an antiparallel arrangement of the ribose rings with facing O4’ atoms (6). This arrangement contrasts with the same strand parallel ribose alignment in B- and A-DNA helical structures (Figure 1C). In addition, this stacking interaction is similar to the stacking of phosphate groups over uridines observed in tRNA anticodon loops (30). Such ribose-base stacking interactions are sometimes called ‘lp–π’ (lone pair–π) stacking (19–22) and where associated with a rare shift of proton signals in Z-DNA NMR spectra (31).
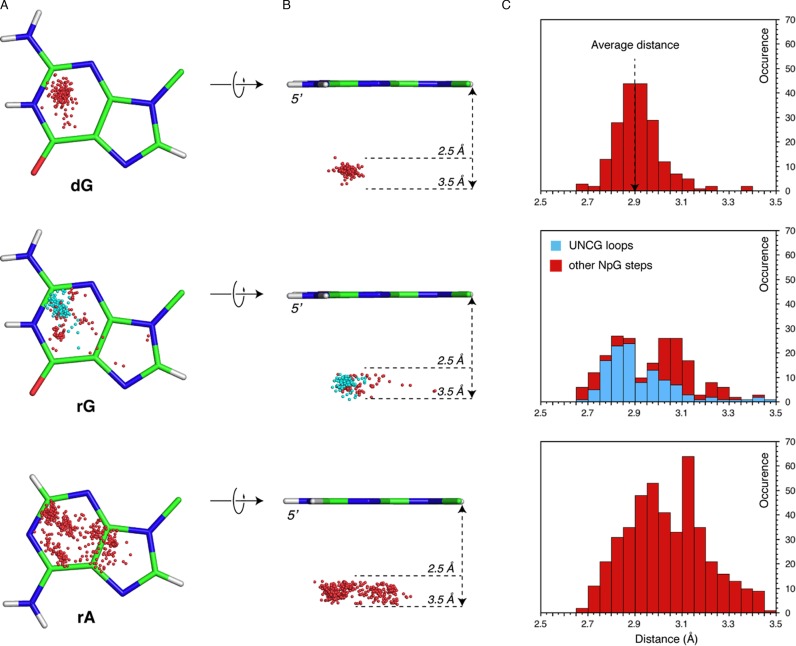
In order to better characterize these stacking interactions, we calculated the O4’ to base plane contact distance (Figure 2). A strong decay towards 3.5 Å in the associated histogram suggests that the O4’ atoms are clustered close to the aromatic nucleobase, and that contacts above this limit should no longer be considered as stacking interactions. The average contact distance for Z-DNA and RNA is 2.9 ± 0.1 and 3.0 ± 0.2 Å, respectively. Interestingly, in these histograms the Z-DNA peak is much sharper than the RNA peak. This might be linked to two factors: (i) Z-DNA structures are generally of a much better resolution (average 1.8 Å for DNA versus 2.6 Å for RNA), and (ii) the structural context of Z-like steps is more diverse in RNA than in DNA, leading to more dispersed positions of the O4’ atoms over the purine rings (Figure 2B and C). Furthermore, the r(NpG) step histogram displays two peaks. The first is associated with UNCG loops (≈2.9 Å) for which the CpG step is very similar to that in Z-DNA (see below). The second is associated with more diverse RNA turns (≈3.1 Å). For r(NpA) steps, the distance distribution is broader as it is associated with a much larger structural diversity in small and large RNAs. The r(NpA) step distance distribution is like that of the second r(NpG) peak, further stressing that both are associated with a broad structural context extending that of Z-helices and UNCG tetraloops.
Figure 2.
Ribose O4’ stacking (‘lp–π’) to dG, rG and rA nucleobases. (A) Top view (5′-side) of the dG, rG and rA nucleobases showing the positions of all O4’ atoms above the nucleobase plane with a contact distance ≤ 3.5 Å. For rG, the O4’ atoms belonging to r(UNCG) loops are in cyan instead of red. (B) 90° rotation of (A); the 2.5-3.5 Å boundaries are marked by arrows and dashed lines. (C) Histogram of the O4’ to nucleobase plane distances drawn from structural sets including all contacts. For rG, O4’ positions and related distances belonging to r(UNCG) loops are in cyan instead of red.
Next to ‘lp–π’ interactions, ribose puckers are very specific in Z-like steps with much stronger constraints on the 5′- than on the 3′-nucleotide (Table 2). The former puckers are mainly in the south quadrant (≈91%) while north dominates for the latter (≈62%) followed by south and east. Puckers in the west quadrant remain exceptional. Overall, the N-S (or C3’-endo-C2’-endo) pucker configuration prevails in both DNA and RNA.
Table 2. Ribose puckers for the 3′- and 5′-nucleotides in non-redundant Z-like steps found in PDB crystallographic RNA and DNA structures (resolution ≤ 3.0 Å; B-factors ≤ 79 Å2).
| Pucker | DNAa | RNAa |
|---|---|---|
| 3′-nucleotide | ||
| North [C3’-endo] | 87 (132) [82 (127)] | 132 (419) [131 (418)] |
| South [C2’-endo] | 27 (34) [21 (27)] | 34 (87) [34 (87)] |
| West | 1 (1) | – |
| East | 44 (61) | 63 (160) |
| 5′-nucleotide | ||
| North [C3’-endo] | – [–] | 12 (13) [8 (9)] |
| South [C2’-endo] | 137 (202) [130 (193)] | 198 (611) [190 (599)] |
| West | 2 (2) | 1 (1) |
| East | 20 (24) | 18 (41) |
aThe total number of ribose puckers in each category with no redundancy considerations is given in parenthesis.
Steps with disordered backbones, usually found in high-resolution Z-DNA structures, were not taken into account. Modified nucleotides were considered.
Z-like steps in r(UNCG) loops are similar to those found in left-handed helices
As stated above, Z-like CpG steps are constitutive of r(UNCG) tetraloops and structurally similar to those found in Z-DNA and Z-RNA (Figure 3) (32). Despite their high thermodynamic stability (33,34), these tetraloops are rare in natural RNA systems where almost all known occurrences are gathered in ribosomal structures (see below). r(UNCG) loops were also artificially grafted to RNA structures to serve as stem capping motifs for stabilization and crystallographic purposes, e.g. the r(UUCG) tetraloops in an RNA-protein complex (32), the group II intron (35) and the RNaseP structure (36). All these structures exhibit tetraloop CpG steps whose structure is consistent with those reported in high-resolution NMR structures (37). The lp–π stacking interaction over more than hundred r(UNCG) instances is associated with an average distance of ≈2.9 ± 0.1 Å that is, as mentioned above, very similar to that calculated for Z-DNA CpG steps (Figure 2A and B). Despite the fact that UNCG loops can have any nucleotide at the second position, we could only identify cUUCGg and cUACGg loops in our PDB set of structures.
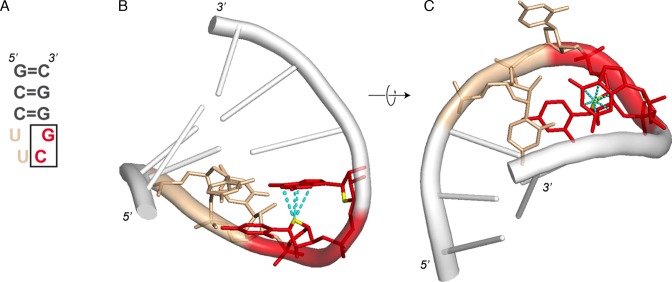
Figure 3.
Z-like step in a r(UUCG) tetraloop. In all panels and subsequent figures, r(CpG) steps are shown in red; the ribose O4’ atoms are shown in yellow; the cyan dashed lines are defined in Figure 1. In this figure, r(UpU) steps are shown in wheat. (A) 2D structure of a r(UUCG) tetraloop. The Z-like r(CpG) step is boxed. (B) 3D structure of a r(UUCG) tetraloop (PDB: 1F7Y; res: 2.8 Å). (C) 90° rotation of (B).
Z-like steps appear at key locations in small RNAs…
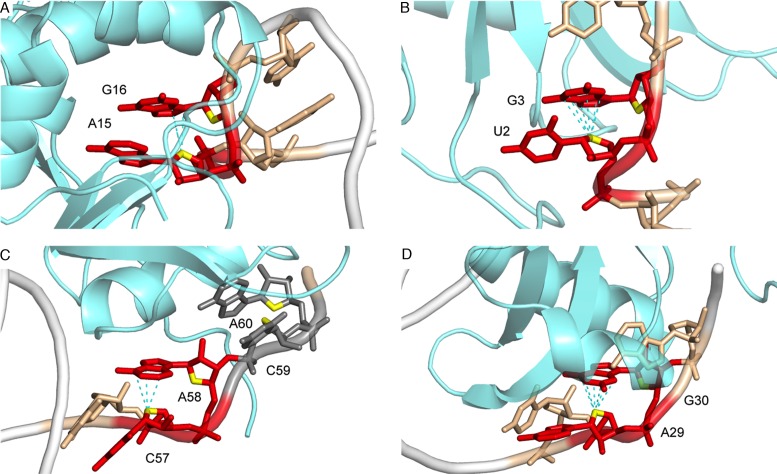
Z-like steps were also identified in a variety of small RNA structures, including: (i) purine riboswitches (38), (ii) classI/II preQ1 riboswitches (39,40), (iii) thiamine pyrophosphate (TPP) riboswitches (41), (iv) lysine riboswitches (42), (v) streptomycin aptamers (43) and (vi) hepatitis delta virus (HDV) ribozymes (44–48) but are absent in other structures like tRNA or group I introns (Figure 4). The diversity of dinucleotide sequences involved in these Z-like steps is unexpected. Contrary to what could be inferred from the dominance of CpG steps in Z-DNA, Z-like steps in RNA form in a large variety of contexts and in the absence of high salt conditions. For instance, >30 purine riboswitches associated with ≈18 different ligands were crystallized, all of them displaying a conserved UpA, ApA or CpA Z-like step (38). In these riboswitches, the 3′-nucleotide stacks with the ligand (Figure 4A), while the conserved 5′-adenine participates in an important base triple. If base triple disrupting mutations of this adenine are detrimental to the structure and activity of the riboswitch, it has been shown that mutations of the 3′-U to C or A are tolerated and preserve the Z-like step structure that is consequently partly sequence independent (49). As such, Z-like steps must shape in a very specific manner the ligand-binding pocket of purine riboswitches.
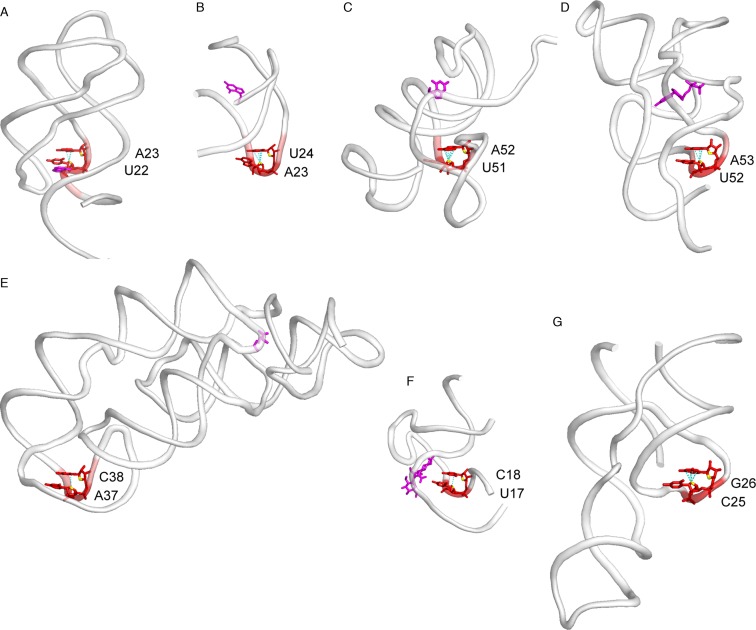
Figure 4.
Z-like steps observed in crystallographic structures of small RNA systems. When present, ligands are shown in magenta. (A) Purine riboswitch (PDB: 4FE5; res: 1.3 Å); (B) class I preQ1 riboswitch (PDB: 3K1V; res: 2.2 Å); (C) class II preQ1 riboswitch (PDB: 4JF2; res: 2.3 Å); (D) TPP riboswitch (PDB: 2GDI; res: 2.1 Å); (E) lysine riboswitch (PDB: 3D0U; res: 2.8 Å); (F) streptomycin aptamer (PDB: 1NTA; res: 2.9 Å); and (G) HDV ribozyme (PDB: 3NKB; res: 1.9 Å).
In other RNA systems, Z-like steps occur in turns similar to those found in UNCG tetraloops (Figure 3B), where the bottom 5′-nucleobase often has a solvent-exposed face. There, Z-like steps are involved in junctions or joining regions where at least one of the nucleotides is pairing with distant residues (e.g. the A53•AZERAZRZ84 base pair in the TPP riboswitch). Together, these observations suggest that Z-like steps occur at key locations where they promote specific turns that are strategic for creating precise and not otherwise possible RNA folds (see Discussion).
… are involved in long-range contacts …
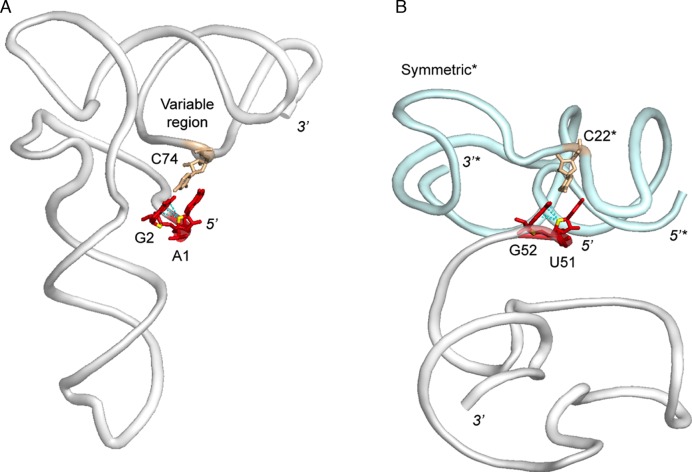
An important long-range interaction involving a Z-like step but no specific turn occurs in a viral tRNA-like structure (TLS; Figure 5A) (50,51). In this RNA, a previously unidentified Z-like ApG step is embedded within a terminal 5′-UUAG sequence, which was historically not recognized as part of the minimal TLS (the two uridines are not visible in the crystallographic structure). However, the presence of this nucleotide sequence has been proven important to stabilize the global tRNA-like TLS fold via a long-range interaction and is required for aminoacylation. The involved base pair is a cis-WC G2=C74 pair with a G in syn. The existence of this single base pair to hold the global fold allows TLS to have a structural and functional flexibility exploited for viral activity, a functional plasticity present in almost all tRNA and tRNA related systems (52). It has been proposed that a loss of this interaction is what enables TLS to more readily unfold to allow viral replication. Indeed, a loss of the TLS structure is observed when the 5′-fragment containing the Z-like step is truncated. A similar Z-like step associated with a cis-WC G=C pair is present in the crystal structure of a fluoride riboswitch (53). There, it involves a symmetry-related molecule, suggesting that such long-range interactions, although uncommon, are modular elements that are of importance in the fold of specialized RNA molecules (Figure 5B).
Figure 5.
Z-like steps establishing long-range contacts in a tRNA like system (TLS) and a fluoride riboswitch (symmetry contact). The nucleotides forming a Watson–Crick pair with the 5′-nucleotide of the Z-like step are shown in wheat. (A) TLS structure (PDB: 4P5J; res: 2.0 Å). (B) Fluoride riboswitch (PDB: 4ENC; res: 2.3 Ǻ). The symmetry related molecule is shown in light blue; asterisks mark annotations for this molecule.
… and are also present in ribosomal structures
In addition to being found in small RNA systems, Z-like steps are also present in all available ribosome structures, an indication that they might occur in ribosomal RNA of all organisms. Several of them are clustered in the conserved core of the ribosomal large subunit (LSU; Figure 6), while others are found in non-conserved peripheral regions of the large and small subunits (SSU). Given the complexity of these ribosomal structures, we report only a few conserved occurrences of Z-like steps in the LSU of E. coli (54), S. cerevisiae (55) and H. sapiens (56), as deduced from their 3D structure and from sequence conservation data based on phylogeny (57). Other Z-like steps found in non-conserved regions including those in the SSU are poorly resolved in available crystallographic and cryo-EM structures and therefore will not be discussed here.
Figure 6.
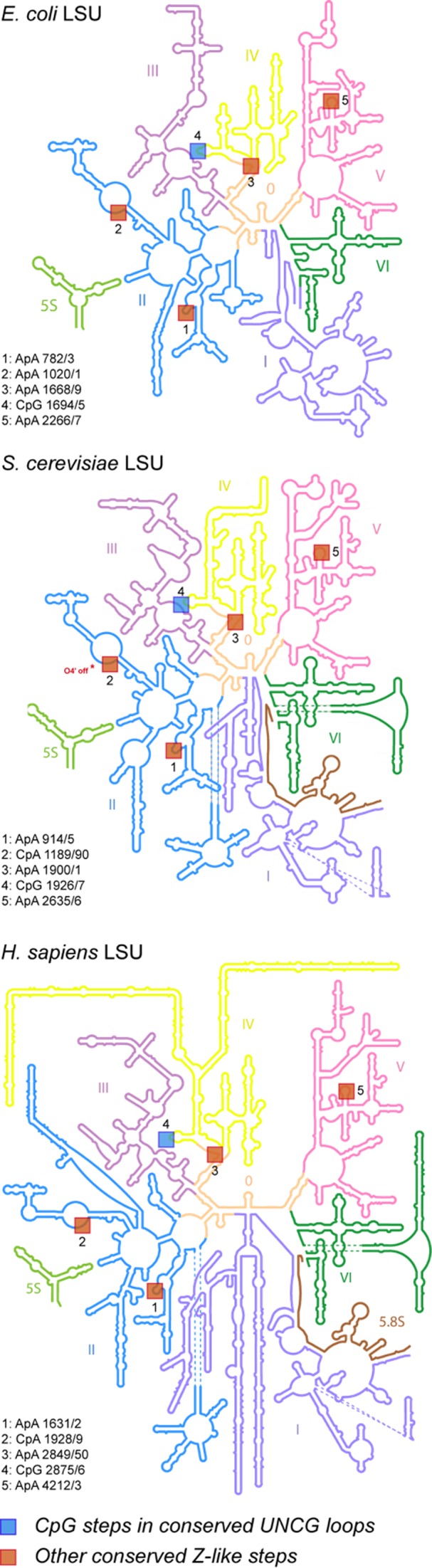
Conserved Z-like steps in 2D structures of three large (LSU) ribosomal subunits from E. coli, S. cerevisiae and H. sapiens. The 2D representations (derived from 3D structures) were adapted from images stored at http://apollo.chemistry.gatech.edu/RibosomeGallery (57). The Z-like steps were inferred from the X-ray E. coli (4YBB; res: 2.1 Ǻ; chain: DA) (54), S. cerevisiae (4U4R, res: 2.8 Ǻ; chain: 1) (55) and cryo-EM H. sapiens (4UG0, res: 3.6 Ǻ; chain: L5) (56) structures. Note that in yeast, for step ‘2’, the O4’ atom is shifted by 0.1 Å and therefore not exactly stacked over the adenine ring, illustrating the difficulties to work with large structures of medium to low resolution that often embed local inaccuracies.
Overall, these ribosomal Z-like steps are similar to those found in small RNAs where they allow for distant pairing between nucleobases. Some of them, like the ones within conserved UUCG tetraloops, are additionally contacting proteins that interact specifically with their Z-like CpG step, pointing out that Z-like steps can be part of RNA-protein recognition schemes.
Specific recognition of Z-like steps by regulatory and RNA modification proteins
Single stranded segments integrate Z-like steps that are directly recognized by specialized RNA binding proteins such as the iron regulatory protein 1 (IRP1) (58,59), CUG-binding proteins (18) and proteins associated with H/ACA box snoRNA (60–62). Hereafter, we will briefly address the variety of recognition patterns in which they are involved (Figure 7).
Figure 7.
Examples of Z-like steps recognized by proteins. (A) IREs mRNA in complex with IRP1 (PDB: 3SNP; res: 2.8 Å). (B) CUG-binding protein in complex with UGU-rich mRNA (PDB: 3NMR; res: 1.9 Å). (C) H/ACA ribonucleoprotein particle (PDB: 3HAX; res: 2.1 Å). This structure displays two consecutive and borderline Z-like steps. The C59pA60 step that displays a lp–π ‘contact’ > 4.0 Å is shown in grey with O4’ atoms in yellow. (D) ToxI RNA-ToxN protein complex (PDB: 2XDB; res: 2.6 Ǻ).
Iron-responsive elements (IREs) are short mRNA stem-loops recognized by the iron regulatory protein 1 (IRP1) at two sites separated by ≈30 Å. One of these sites involves a conserved ApGpU triloop where ApG forms a Z-like step (Figure 7A) (58). These bulged-out A and G nucleotides point toward the protein and are associated with a sharp turn in the RNA backbone. This step contacts five different amino-acids and is specifically sandwiched by two leucine side chains that provide van der Waals contacts to the exposed aromatic surfaces of the A and G residues. Although a subsequent crystal structure with a different IRE element displays the same 2D motif (59), the available NMR structures of this element are not showing a Z-like step. Therefore, this conformation might be protein-induced and/or protein-stabilized (63).
CUG-binding proteins regulate multiple aspects of nuclear and cytosplamic mRNA processing. They are known to preferentially target UGU-rich mRNA elements to accomplish their mRNA processing functions. The UpG step adopts a left-handed Z-RNA conformation (Figure 7B) where the syn guanine is recognized through specific Hoogsteen edge-protein backbone interactions (18). The similarities between UpG steps as found in this complexes and CpG steps in structures of a complex of Z-RNA with the ADAR1 Zα protein were described. Interestingly, the U3-G4 and U8-G9 steps in the GUUGUUUUGUUU sequence in complex with two RNA recognition motifs (bound RRM1 and tandem RRM1/2) share the same Z-like step structural features.
H/ACA ribonucleoprotein particles are a family of pseudouridine synthases that use guide RNAs to specific modification sites (60–62,64). They also participate in eukaryotic ribosomal RNA processing and are a component of vertebrate telomerases. H/ACA RNAs fold into repeats of a consensus hairpin structure that comprise an internal loop and an ACA signature that harbours a 3′-tail. This three single stranded A58pC59pA60 signature forms two consecutive CpA Z-like steps when including the terminal C57 stem nucleotide (Figure 7C). The PUA (PseudoUridine synthase and Archaeosine transglycosylase) domain of Cbf5 recognizes very specifically this Z-like step repeat. Although the two Z-like steps are easily characterized by visual inspection, it appears that they represent also borderline steps regarding their lp–π geometry; the C57 ribose is almost parallel to the A58 nucleobase and the lp–π contact distance of the C59pA60 step exceeds 4.0 Å. Although borderline, these consecutive Z-like steps seem stable since they are recurrently observed in H/ACA box systems.
A further occurrence of Z-like steps in RNA/protein complexes involves a bacteria-to-phage ‘immune response’, i.e. the bacterial phage-resistance system ToxIN involving the protein ToxN that is inhibited in vivo by a specific ToxI antitoxin RNA (65). A crystal structure of the complex shows a Z-like ‘ApG’ step shortly upwards the ToxI 3′-end that binds to the ToxN groove 1 (Figure 7D). The backbone turn associated to the Z-like step allows the two following adenines to directly point towards the protein recognition pocket and establish several specific interactions. There, amino acid side chains interact with the Watson–Crick edges of the Z-like nucleotides like those in the RNA junctions and turns described above.
Z-like steps in immunology-related viral RNAs
The last and most intriguing aspect of Z-like steps, already mentioned for the bacteriophage system, is related to their role in the immune response. Adenosine deaminase (ADAR1) proteins embed a Zα domain able to recognize Z-DNA as well as Z-RNA duplexes (12,16,66,67). The two available crystal structures of a Zα domain in complex with Z-DNA and Z-RNA CpG hexamers display similar characteristics. In these complexes, the central CpG step is essentially recognized through specific amino acid-phosphate group contacts. Additionally, in the Z-DNA complex, a single weak van der Waals contact of the CH…π type between Tyr177 and a guanine is observed. When the RNA replaces the DNA substrate, this contact disappears, suggesting that the Zα domain could recognize any Z-RNA motif (the authors did however not exclude that crystal-packing effects may have slightly altered the structure of this RNA complex). Indeed, three other d(CACGTG)2, d(CGTACG)2 and d(CGGCCG)2, Z-DNA substrates were co-crystalized with the Zα domain, stressing that a Z-like TpA step can be recognized similarly to a CpG step (11). Hence, these protein domains and their analogs (68) could recognize Z-DNA as well as Z-RNA in a non-sequence specific manner, a process that may be associated with the immune response (69).
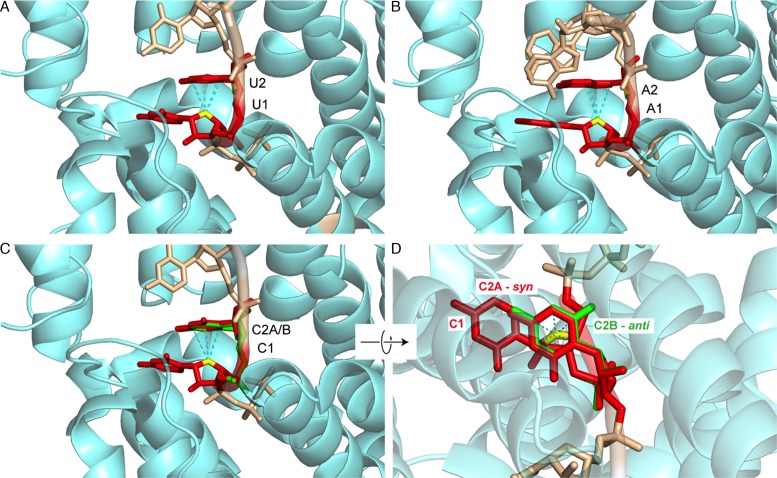
Z-like steps were further identified in single stranded RNA 5′-triphosphate groups (5′-PPP-RNA), a signature of viral RNA, when recognized by interferon-induced proteins with tetratricopeptide repeats of the IFIT5 family. Three crystal structures of the human IFIT5 protein in complex with 5‘-PPP-N1N2N3N4 (N = C, U or A) ligands (Figure 8) (70–72) reveal that the ligand is recognized in a non-sequence- but conformation- and modification-specific manner. Indeed, no nucleobase-to-protein contacts are observed in these structures but only contacts to the ribose-phosphate backbone and the binding pocket of this protein does not seem large enough to accommodate the usual capping modifications found in eukaryotic RNAs.
Figure 8.
Z-like conformations adopted by the N1pN2 step of a 5′-PPP-N1N2N3N4 viral RNA primer in complex with the human interferon induced IFIT5 protein. The 5′-PPP group and the N3pN4 residues are shown in wheat and the protein backbone in aquamarine. (A) 5′-PPP-UUUU (PDB: 4HOS; res: 2.0 Å). (B) 5′-PPP-AAAA (PDB: 4HOT; res: 2.5 Å). Note the rare lp–π stacking involving the five membered ring of A2. (C) 5′-PPP-CCCC (PDB: 4HOR; res: 1.9 Å). (D) 90° rotation of (C). In (C) and (D), the nucleobase C2 adopts a syn (A; red) and an anti (B; green) alternate conformation illustrating the difficulty of unambiguously assigning syn conformations for pyrimidines.
In each of these ligands, the N1pN2 step adopts a Z-like conformation inducing the formation of important contacts between the protein and the RNA backbone. The N1 and N2 bases do not establish specific hydrogen bonds with protein residues, and there is ample space adjacent to the pyrimidine edges, suggesting that the binding pocket can easily accommodate the larger purine nucleobases as seen in the oligo-A complex. Interestingly, the position of the RNA backbone in the oligo-A complex favors the formation of a lp–π interaction involving the five- instead of the more usual six membered ring (Figure 8B). This particular Z-like step arrangement allows the accommodation of all-purine as well as rare all-pyrimidine sequences and eventually combinations of them without the need to adjust backbone conformation. As such, it allows the incorporation of an all–C sequence for which it was difficult to precisely identify the syn/anti nucleobase conformation (Figure 8C and D). Given available structures, it seems very likely that the C2 base is in syn. Yet, these data also imply that it is much more difficult to identify syn pyrimidines than syn purines in crystallographic structures. Consequently, their number might be slightly underestimated in the PDB (26). Indeed, evidence was given very early that pyrimidines could adopt syn conformations in solution and be associated with Z-steps (4,10).
DISCUSSION
In both DNA and RNA, we observe that not only CpG steps but also almost any dinucleotide sequence can adopt similar Z-like structures, with a preference for those with purines on the 3′-side. A difference between Z-like steps in RNA and DNA is that in RNA these steps are usually found in single-stranded regions such as loops and junctions, where they contribute to creating specific backbone kinks and turns. However, we were unable to identify recurring interaction patterns, which point to a great diversity of Z-like step usages. In Z-dinucleotides, we noted that the Watson-Crick sites of the two nucleobases point in the same direction. Hence, in most instances the two nucleobases are alternatively or simultaneously forming hydrogen bonds with distant residues. Significantly, a Z-like step has remained unnoticed in the UNCG tetraloop family, although the similarity between Z-DNA and UNCG backbones was mentioned elsewhere (73). This observation highlights the difficulties of circumventing the complex interaction patterns present in even simple and well-studied RNA motifs.
Dissecting the unusual structure of a Z-like step reveals that it is the result of several uncommon events. Both nucleotides are required to adopt specific conformations (C2’-endo on the 5′-side, syn on the 3′-side) that each occurs at a 2% and 12% frequency, respectively (as estimated from PDB crystal structures). In that respect, the frequency of Z-like steps that combines both conformational features drops to a low ≈0.1%. Moreover, the syn nucleobase is fully flipped over the ribose of the anti nucleobase, which among most but not all Z-like steps results in a specific stacking interaction involving the O4’ of the first ribose where the distance between that O4’ atom and the nucleobase ring is shorter than the average stacking interaction distances between aromatic rings (2.9 Å versus 3.4 Å), due to electrostatic and dispersion effects probably dominated by solvent induced-forces (74). This interaction of the lp–π type has been shown through quantum mechanical calculations to be rather weak and is probably an incidental event rather than a potential folding driver (19,21). If this interaction had been promoting folding, it would have been observed much more frequently. In short, although a Z-like step is made of rare and energetically costly conformations, its structure is induced and stabilized by its surrounding during folding.
We wish to posit that the low frequency and the incidental nature of Z-like steps are compatible with a precise structural function. Since syn and C2’-endo conformations are linked with slow dynamics, they probably need assistance from other elements to overcome the structural stress imposed to the backbone (26,75). We therefore propose that the combination of rare conformations within a Z-like step would create regions that retain their fold once formed. Like other structural elements involved in long-range 3D contacts such as GNRA loops interacting with their receptors (76), Z-like steps could act as conformational locks at strategic locations in RNAs, with the particularity of involving rare nucleotide conformations that need specific structural contexts for their formation. Here, the case of the purine riboswitch is particularly interesting, as the Z-like UpA is part of the ligand-binding site (U22 is directly contacted by the ligand). The Z-like motif is part of the junction J1/2. It stabilizes the final bound structure through long-range interactions with J2/3—J3/1 and is next to the the entry site of the ligand (77,78). Additionally, the preorganized state of the binding pocket that is known to involve J1/2 and precede ligand binding (79) could be in part attributed to the presence of this ‘Z-lock’. We hope that these considerations will encourage studies of the dynamics of formation of Z-like steps, as those may reveal cues to understanding the ligand binding process.
Among all the intramolecular RNA motifs that were described here, the one found in the TLS structure seems the most peculiar since the active fold of the structure requires the formation of a single long-range Watson-Crick base pair involving a Z-like step (50). The study of other folds such as those associated with the purine riboswitch ligand binding pocket or the ion sensitivity reported for the HDV ribozyme (44–48,80–82) and the lysine riboswitch (42) systems may also offer insights about the structural role of these Z-like steps. Exploring Z-like steps may thus suggest how other motifs may turn out to exert their main function through locking 3D folds. It might also be worthwhile to understand how these rare motifs form in the perspective of using them to create specific folds in synthetic biological systems (83).
Regarding interactions with proteins, Z-like steps are found in Z-RNA and Z-DNA duplexes and the viral 5′-PPP-RNA recognition system (70) that are recognized specifically through conformation-dependent interactions. These interactions involve solely backbone atoms, allowing various sequences to be accommodated (11). For example, a family of proteins recognize double stranded Z-DNA and Z-RNA via a common winged helix-turn-helix domain called Zα. In other instances, like the IRE-RNA system (58,59), the CUG binding complexes (18), the H/ACA (62) or the bacteria-to-phage ‘immune response’ system (65), different recognition patterns implicating the nucleobases and the backbone atoms are at play, which suggest sequence-dependent recognition patterns. For these systems however, no unique RNA/protein recognition pattern could be found and it appears certain that proteins are able to induce Z-conformations in single stranded RNA by using various mechanisms.
As noted by Alexander Rich et al., both Z-RNA and Z-DNA are highly antigenic and are stimulated by a particular structural context. The first Z-DNA binding protein was found to be a Z-RNA binding protein called double stranded RNA adenosine deaminase (or ADAR1) (4) and numerous Z-DNA specific antibodies are found in human autoimmune diseases such as systemic lupus erythematosus (3,84). These proteins recognize short double stranded Z-RNA steps by essentially binding in a non-sequence specific manner to the sugar-phosphate backbone. Our exploration of Z-RNA motifs therefore also uncovers strong ties to the immune response. Deeper investigations of the structural characteristics, occurrence, and associated RNA/protein recognition features of Z-like steps could represent a pathway to further our understanding of the immune response.
In particular, elevation of CpG but also UpA frequencies in influenza A viruses used as an RNA genome model system have been recently involved in the attenuation of the viral pathogenicity and in the simultaneous increase in host response to infection (85). We hypothesize here that some of the involved mechanisms could be related to a certain tendency of these pyrimidine-purine sequences to promote Z-conformations.
Acknowledgments
P.A. and Q.V. wish to thank Prof. Eric Westhof for ongoing support and helpful discussions, as well as Prof. Rob Batey and Prof. Richard Giegé for useful comments on the manuscript. This article is dedicated to the memory of Alexander Rich.
CONCLUDING REMARKS
Classical sequence and structure analysis of this new RNA Z-motif is limited due to the low number of RNA crystal structures in the PDB. In that respect, we expect that the current upsurge of medium and high resolution RNA and RNP structures will increase the number and significance of Z-dinucleotide motifs in structural databases, so that a more advanced characterization of their structural and recognition properties will be within reach. With respect to the comments of one referee that wondered about the significance of these rare motifs, we stand by Alexander Rich, who worked hard to convince fellow researchers that Z-DNA was of biological significance and we share his view that Z-DNA and by extension Z-motifs had to be present in cellulo since evolution is opportunistic (4). After all, as demonstrated by this survey, Z-conformations are readily accessible to most dinucleotide sequences, when placed in the appropriate environment. Interestingly, some of these Z-steps are recognized by pattern recognition receptors (PRRs) to distinguish between self and non-self. Thus, the ability of some single or double stranded RNA sequences to be twisted into Z-form steps might be a structural key in some immune system diseases (72). Hence, although Z-steps may currently appear as a ‘black swan’ (86) in the RNA world, we trust that this new motif will find its place in the still incomplete RNA motif library.
FUNDING
French ‘Ministère de la recherche et de l'enseignement’ (to L.D.); Polish Ministry of Higher Education and Science (Mobility Plus programme) [1103/MOB/2013/0 to F.L.]. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ho P.S., Mooers B.H.M. Z-DNA crystallography. Biopolymers. 1997;44:65–90. doi: 10.1002/(SICI)1097-0282(1997)44:1<65::AID-BIP5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 2.Wang A.H.J., Quigley G.J., Kolpak F.J., Crawford J.L., Vanboom J.H., Vandermarel G., Rich A. Molecular-structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 3.Rich A., Zhang S. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 4.Rich A. The excitement of discovery. Annu. Rev. Biochem. 2004;73:1–37. doi: 10.1146/annurev.biochem.73.011303.073945. [DOI] [PubMed] [Google Scholar]
- 5.Drew H., Takano T., Tanaka S., Itakura K., Dickerson R.E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980;286:567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- 6.Sundaralingam M., Westhof E. Structural motifs of the nucleotidyl unit and the handedness of polynucleotide helices. Int. J. Quantum Chem. 1981:287–306. [Google Scholar]
- 7.Wang A.H.J., Quigley G.J., Kolpak F.J., Vandermarel G., Vanboom J.H., Rich A. Left-handed double helical DNA - Variations in the backbone conformation. Science. 1981;211:171–176. doi: 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]
- 8.Gessner R.V., Frederick C.A., Quigley G.J., Rich A., Wang A.H.J. The molecular structure of the left-handed Z-DNA double helix at 1.0 Å atomic resolution. Geometry, conformation, and ionic interactions of d(CGCGCG) J. Biol. Chem. 1989;264:7912–7935. doi: 10.2210/pdb1dcg/pdb. [DOI] [PubMed] [Google Scholar]
- 9.Fuertes M.A., Cepeda V., Alonso C., Perez J.M. Molecular mechanisms for the B-Z transition in the example of poly[d(G-C)•d(G-C)] polymers. A critical review. Chem. Rev. 2006;106:2045–2064. doi: 10.1021/cr050243f. [DOI] [PubMed] [Google Scholar]
- 10.Feigon J., Wang A.H.J., Vandermarel G.A., Vanboom J.H., Rich A. Z-DNA forms without an alternating purine-pyrimidine sequence in solution. Science. 1985;230:82–84. doi: 10.1126/science.4035359. [DOI] [PubMed] [Google Scholar]
- 11.Ha S.C., Choi J., Hwang H.Y., Rich A., Kim Y.G., Kim K.K. The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ(ADAR1) Nucleic Acids Res. 2009;37:629–637. doi: 10.1093/nar/gkn976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savva Y.A., Rieder L.E., Reenan R.A. The ADAR protein family. Genome Biol. 2012;13:252. doi: 10.1186/gb-2012-13-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D., Hur J., Park K., Bae S., Shin D., Ha S.C., Hwang H.Y., Hohng S., Lee J.H., Lee S., et al. Distinct Z-DNA binding mode of a PKR-like protein kinase containing a Z-DNA binding domain (PKZ) Nucleic Acids Res. 2014;42:5937–5948. doi: 10.1093/nar/gku189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D., Lee Y.H., Hwang H.Y., Kim K.K., Park H.J. Z-DNA binding proteins as targets for structure-based virtual screening. Curr. Drug Targets. 2010;11:335–344. doi: 10.2174/138945010790711905. [DOI] [PubMed] [Google Scholar]
- 15.Koeris M., Funke L., Shrestha J., Rich A., Maas S. Modulation of ADAR1 editing activity by Z-RNA in vitro. Nucleic Acids Res. 2005;33:5362–5370. doi: 10.1093/nar/gki849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Placido D., Brown B.A., Lowenhaupt K., Rich A., Athanasiadis A. A left-handed RNA double helix bound by the Zα domain of the RNA-editing enzyme ADAR1. Structure. 2007;15:395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Rosa M., Zacarias S., Athanasiadis A. Structural basis for Z-DNA binding and stabilization by the zebrafish Z-DNA dependent protein kinase PKZ. Nucleic Acids Res. 2013;41:9924–9933. doi: 10.1093/nar/gkt743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teplova M., Song J., Gaw H.Y., Teplov A., Patel D.J. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18:1364–1377. doi: 10.1016/j.str.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egli M., Gessner R.V. Stereoelectronic effects of deoxyribose O4′ on DNA conformation. Proc. Natl. Acad. Sci. U.S.A. 1995;92:180–184. doi: 10.1073/pnas.92.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sponer J., Gabb H.A., Leszczynski J., Hobza P. Base-base and deoxyribose-base stacking interactions in B-DNA and Z-DNA: a quantum-chemical study. Biophys. J. 1997;73:76–87. doi: 10.1016/S0006-3495(97)78049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egli M., Sarkhel S. Lone pair-aromatic interactions: to stabilize or not to stabilize. Acc. Chem. Res. 2007;40:197–205. doi: 10.1021/ar068174u. [DOI] [PubMed] [Google Scholar]
- 22.Mooibroek T.J., Gamez P., Reedijk J. Lone pair–π interactions: a new supramolecular bond? CrystEngComm. 2008;10:1501–1515. [Google Scholar]
- 23.Gadre R.S., Kumar A. In: Noncovalent Forces. Scheiner S, editor. Springer; 2015. pp. 391–418. [Google Scholar]
- 24.Salonen L.M., Ellermann M., Diederich F. Aromatic rings in chemical and biological recognition: energetics and structures. Angew. Chem. Int. Ed. Engl. 2011;50:4808–4842. doi: 10.1002/anie.201007560. [DOI] [PubMed] [Google Scholar]
- 25.Keating K.S., Pyle A.M. Semiautomated model building for RNA crystallography using a directed rotameric approach. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8177–8182. doi: 10.1073/pnas.0911888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokoloski J.E., Godfrey S.A., Dombrowski S.E., Bevilacqua P.C. Prevalence of syn nucleobases in the active sites of functional RNAs. RNA. 2011;17:1775–1787. doi: 10.1261/rna.2759911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu X.J., Bussemaker H.J., Olson W.K. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015;43:e142. doi: 10.1093/nar/gkv716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hickman A.B., James J.A., Barabas O., Pasternak C., Ton-Hoang B., Chandler M., Sommer S., Dyda F. DNA recognition and the precleavage state during single-stranded DNA transposition in D.radiodurans. EMBO J. 2010;29:3840–3852. doi: 10.1038/emboj.2010.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theobald D.L., Schultz S.C. Nucleotide shuffling and ssDNA recognition in Oxytrichanova telomere end-binding protein complexes. EMBO J. 2003;22:4314–4324. doi: 10.1093/emboj/cdg415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auffinger P., Westhof E. An extended structural signature for the tRNA anticodon loop. RNA. 2001;7:334–341. doi: 10.1017/s1355838201002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis P.W., Adamiak R.W., Tinoco I. Z-RNA: The solution NMR structure of r(CGCCCG) Biopolymers. 1990;29:109–122. doi: 10.1002/bip.360290116. [DOI] [PubMed] [Google Scholar]
- 32.Ennifar E., Nikulin A., Tishchenko S., Serganov A., Nevskaya N., Garber M., Ehresmann B., Ehresmann C., Nikonov S., Dumas P. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000;304:35–42. doi: 10.1006/jmbi.2000.4204. [DOI] [PubMed] [Google Scholar]
- 33.Antao V.P., Tinoco I. Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheehy J.P., Davis A.R., Znosko B.M. Thermodynamic characterization of naturally occurring RNA tetraloops. RNA. 2010;16:417–429. doi: 10.1261/rna.1773110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcia M., Pyle A.M. Visualizing group II intron catalysis through the stages of splicing. Cell. 2012;151:497–507. doi: 10.1016/j.cell.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krasilnikov A.S., Yang X., Pan T., Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 37.Nozinovic S., Furtig B., Jonker H.R., Richter C., Schwalbe H. High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res. 2010;38:683–694. doi: 10.1093/nar/gkp956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter E.B., Marcano-Velazquez J.G., Batey R.T. The purine riboswitch as a model system for exploring RNA biology and chemistry. Biochim. Biophys. Acta. 2014;1839:919–930. doi: 10.1016/j.bbagrm.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein D.J., Edwards T.E., Ferre-D'Amare A.R. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat. Struct. Mol. Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liberman J.A., Salim M., Krucinska J., Wedekind J.E. Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold. Nat. Chem. Biol. 2013;9:353–355. doi: 10.1038/nchembio.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serganov A., Polonskaia A., Phan A.T., Breaker R.R., Patel D.J. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garst A.D., Heroux A., Rambo R.P., Batey R.T. Crystal structure of the lysine riboswitch regulatory mRNA element. J. Biol. Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tereshko V., Skripkin E., Patel D.J. Encapsulating streptomycin within a small 40-mer RNA. Chem. Biol. 2003;10:175–187. doi: 10.1016/s1074-5521(03)00024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J.H., Yajima R., Chadalavada D.M., Chase E., Bevilacqua P.C., Golden B.L. A 1.9 Å crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage. Biochemistry. 2010;49:6508–6518. doi: 10.1021/bi100670p. [DOI] [PubMed] [Google Scholar]
- 45.Veeraraghavan N., Ganguly A., Chen J.H., Bevilacqua P.C., Hammes-Schiffer S., Golden B.L. Metal binding motif in the active site of the HDV ribozyme binds divalent and monovalent ions. Biochemistry. 2011;50:2672–2682. doi: 10.1021/bi2000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J., Ganguly A., Miswan Z., Hammes-Schiffer S., Bevilacqua P.C., Golden B.L. Identification of the catalytic Mg2+ ion in the hepatitis delta virus ribozyme. Biochemistry. 2013;52:557–567. doi: 10.1021/bi3013092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golden B.L., Hammes-Schiffer S., Carey P.R., Bevilacqua P.C. In: Biophysics of RNA Folding. Russel R, editor. NY: Springer; 2013. pp. 135–167. [Google Scholar]
- 48.Kapral G.J., Jain S., Noeske J., Doudna J.A., Richardson D.C., Richardson J.S. New tools provide a second look at HDV ribozyme structure, dynamics and cleavage. Nucleic Acids Res. 2014;42:12833–12846. doi: 10.1093/nar/gku992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert S.D., Love C.E., Edwards A.L., Batey R.T. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colussi T.M., Costantino D.A., Hammond J.A., Ruehle G.M., Nix J.C., Kieft J.S. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature. 2014;511:366–369. doi: 10.1038/nature13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akiyama B.M., Eiler D., Kieft J.S. Structured RNAs that evade or confound exonucleases: function follows form. Curr. Opin. Struct. Biol. 2016;36:40–47. doi: 10.1016/j.sbi.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giegé R., Juhling F., Putz J., Stadler P., Sauter C., Florentz C. Structure of transfer RNAs: similarity and variability. WIREs RNA. 2012;3:37–61. doi: 10.1002/wrna.103. [DOI] [PubMed] [Google Scholar]
- 53.Ren A., Rajashankar K.R., Patel D.J. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486:85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., Cate J.H. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015;22:336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garreau de Loubresse N., Prokhorova I., Holtkamp W., Rodnina M.V., Yusupova G., Yusupov M. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014;513:517–522. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- 56.Khatter H., Myasnikov A.G., Natchiar S.K., Klaholz B.P. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- 57.Petrov A.S., Bernier C.R., Hershkovits E., Xue Y., Waterbury C.C., Hsiao C., Stepanov V.G., Gaucher E.A., Grover M.A., Harvey S.C., et al. Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Res. 2013;41:7522–7535. doi: 10.1093/nar/gkt513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walden W.E., Selezneva A.I., Dupuy J., Volbeda A., Fontecilla-Camps J.C., Theil E.C., Volz K. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314:1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- 59.Walden W.E., Selezneva A., Volz K. Accommodating variety in iron-responsive elements: crystal structure of transferrin receptor 1 B IRE bound to iron regulatory protein 1. FEBS Lett. 2012;586:32–35. doi: 10.1016/j.febslet.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 60.Li L., Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 61.Liang B., Xue S., Terns R.M., Terns M.P., Li H. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat. Struct. Mol. Biol. 2007;14:1189–1195. doi: 10.1038/nsmb1336. [DOI] [PubMed] [Google Scholar]
- 62.Duan J., Li L., Lu J., Wang W., Ye K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol. Cell. 2009;34:427–439. doi: 10.1016/j.molcel.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 63.McCallum S.A., Pardi A. Refined solution structure of the iron-responsive element RNA using residual dipolar couplings. J. Mol. Biol. 2003;326:1037–1050. doi: 10.1016/s0022-2836(02)01431-6. [DOI] [PubMed] [Google Scholar]
- 64.Zhou J., Liang B., Li H. Structural and functional evidence of high specificity of Cbf5 for ACA trinucleotide. RNA. 2011;17:244–250. doi: 10.1261/rna.2415811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blower T.R., Pei X.Y., Short F.L., Fineran P.C., Humphreys D.P., Luisi B.F., Salmond G.P.C. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat. Struct. Mol. Biol. 2011;18:185–246. doi: 10.1038/nsmb.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herbert A., Alfken J., Kim Y.G., Mian I.S., Nishikura K., Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwartz T., Rould M.A., Lowenhaupt K., Herbert A., Rich A. Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 68.Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 69.Brown B.A. 2nd, Lowenhaupt K., Wilbert C.M., Hanlon E.B., Rich A. The Zα domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13532–13536. doi: 10.1073/pnas.240464097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abbas Y.M., Pichlmair A., Gorna M.W., Superti-Furga G., Nagar B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vladimer G.I., Gorna M.W., Superti-Furga G. IFITs: emerging roles as key anti-viral proteins. Front. Immunol. 2014;5:94. doi: 10.3389/fimmu.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hull C.M., Anmangandla A., Bevilacqua P.C. Bacterial riboswitches and ribozymes potently activate the human innate immune sensor PKR. ACS Chem. Biol. 2016;11:1118–1127. doi: 10.1021/acschembio.6b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Richardson J.S., Schneider B., Murray L.W., Kapral G.J., Immormino R.M., Headd J.J., Richardson D.C., Ham D., Hershkovits E., Williams L.D., et al. RNA backbone: consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang L.X., Adam C., Nichol G.S., Cockroft S.L. How much do van der Waals dispersion forces contribute to molecular recognition in solution? Nat. Chem. 2013;5:1006–1010. doi: 10.1038/nchem.1779. [DOI] [PubMed] [Google Scholar]
- 75.Mortimer S.A., Weeks K.M. C2 '-endo nucleotides as molecular timers suggested by the folding of an RNA domain. Proc. Natl. Acad. Sci. U.S.A. 2009;106:15622–15627. doi: 10.1073/pnas.0901319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fiore J.L., Nesbitt D.J. An RNA folding motif: GNRA tetraloop-receptor interactions. Q. Rev. Biophys. 2013;46:223–264. doi: 10.1017/S0033583513000048. [DOI] [PubMed] [Google Scholar]
- 77.Gilbert S.D., Stoddard C.D., Wise S.J., Batey R.T. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 78.Buck J., Furtig B., Noeske J., Wohnert J., Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greenleaf W.J., Frieda K.L., Foster D.A., Woodside M.T., Block S.M. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Webb C.H., Riccitelli N.J., Ruminski D.J., Luptak A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Veeraraghavan N., Ganguly A., Golden B.L., Bevilacqua P.C., Hammes-Schiffer S. Mechanistic strategies in the HDV ribozyme: chelated and diffuse metal ion interactions and active site protonation. J. Phys. Chem. B. 2011;115:8346–8357. doi: 10.1021/jp203202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grabow W., Jaeger L. RNA modularity for synthetic biology. F1000Prime Rep. 2013;5:46. doi: 10.12703/P5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lenert P. Nucleic acid sensing receptors in systemic lupus erythematosus: development of novel DNA- and/or RNA-like analogues for treating lupus. Clin. Exp. Immunol. 2010;161:208–222. doi: 10.1111/j.1365-2249.2010.04176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gaunt E., Wise H.M., Zhang H., Lee L.N., Atkinson N.J., Nicol M.Q., Highton A.J., Klenerman P., Beard P.M., Dutia B.M., et al. Elevation of CpG frequencies in influenza A genome attenuates pathogenicity but enhances host response to infection. Elife. 2016;5:e12735. doi: 10.7554/eLife.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taleb N.N. The Black Swan: The Impact of the Highly Improbable. NY: The Random House Publishing Group; 2010. [Google Scholar]