Abstract
The modular architecture of aureochrome blue light receptors, found in several algal groups including diatoms, is unique by having the LOV-type photoreceptor domain fused to the C-terminus of its putative effector, an N-terminal DNA-binding bZIP module. The structural and functional understanding of aureochromes’ light-dependent signaling mechanism is limited, despite their promise as an optogenetic tool. We show that class I aureochromes 1a and 1c from the diatom Phaeodactylum tricornutum are regulated in a light-independent circadian rhythm. These aureochromes are capable to form functional homo- and heterodimers, which recognize the ACGT core sequence within the canonical ‘aureo box’, TGACGT, in a light-independent manner. The bZIP domain holds a more folded and less flexible but extended conformation in the duplex DNA-bound state. FT-IR spectroscopy in the absence and the presence of DNA shows light-dependent helix unfolding in the LOV domain, which leads to conformational changes in the bZIP region. The solution structure of DNA bound to aureochrome points to a tilted orientation that was further validated by molecular dynamics simulations. We propose that aureochrome signaling relies on an allosteric pathway from LOV to bZIP that results in conformational changes near the bZIP-DNA interface without major effects on the binding affinity.
INTRODUCTION
Light-Oxygen-Voltage (LOV) proteins are one major class of photoreceptors mediating blue/UVA light response in three kingdoms of life. An archetype of LOV domain-dependent photoreceptors is represented by plant phototropins, which mediate phototropism, chloroplast relocation, stomatal opening and photomorphogenesis in plants by regulating the C-terminal Ser/Thr kinase effector domain (1–3). LOV domains are not only found in many eukaryotes, including plants, fungi and algae, but also in the prokaryotic kingdom with >1500 reported orthologs (4). The C-terminus of more than 80% of these LOV domains is fused to histidine kinase, GGDEF/EAL, STAS or helix-turn-helix (HTH) domains as effectors. How the signal is generated and propagated in LOV proteins has been the subject of intense research activities. LOV domains non-covalently bind flavin, mostly Flavin mononucleotide (FMN), sometimes Flavin dinucleotide (FAD) or riboflavin (5,6), as chromophore that absorbs blue light (BL) (λmax ≈ 450 nm). Upon BL illumination, the flavin forms a C4(a) adduct (λmax ≈ 390 nm) with a conserved cysteine residue (7). In phototropin, this event triggers the unfolding of the C-terminal Jα helix in the LOV2 domain (8). Rearrangement of this Jα helix was shown to increase the activity of the kinase fused to the LOV2 domain (9). In contrast, bacterial LOV proteins have been implicated to proliferate signals to the effector by a rotation of LOV protomers within the dimer with an extended arrangement of the Jα helices (10) or without any involvement of Jα at all (11). Moreover, the fungal stand-alone LOV domain vivid (VVD) mediates structural changes in the N-cap region for signal transduction (12).
Unlike other LOV-domain photoreceptors, aureochromes are exceptional because of their inversed effector-sensor topology. The first aureochrome, VfAUREO1, was discovered in the photosynthetic stramenopile Vaucheria frigida (13). Here, an N-terminal bZIP (basic region/leucine zipper) domain of the S/D type precedes the C-terminal LOV domain. Further, aureochromes were reported in other stramenopiles, including the diatom Phaeodactylum tricornutum. This alga comprises, according to phylogenetic analysis, three aureochrome paralogs of class 1 (PtAUREO1a, 1b, 1c), which differ mostly at their N-terminal extensions, and one ortholog of class 2, PtAUREO2 (14). The LOV domain of PtAUREO2 was found to be unable to bind a flavin chromophore because of the steric hindrance by a methionine residue within its binding cavity, which points to a non-sensory function of class II aureochromes (15). Unlike the role of VfAUREO1 in controlling photomorphogenesis in the multinucleate V. frigida (13), PtAUREO1a affects the photoacclimation and cell division of the unicellular P. tricornutum (14,16). Aureochromes have been suggested to act as light-induced transcription factors (13). For VfAUREO1, the cognate DNA-recognition motif was determined to be TGACGT and hereafter referred to as aureo box. A combination of redox-dependent oligomerization and blue light-induced dsDNA binding mode was proposed for VfAUREO1 given the occurrence of two cysteines in its bZIP domain (C162 and C182) (13,17), which are, however, missing in 5 of 11 aureochromes (14). The LOV domain of this aureochrome also found optogenetic applications in synthetically designed receptor tyrosine kinases, for which a spatio-temporal, light-induced dimerization and thus activation could be shown (18,19).
Recently, three independent crystal structures of the LOV domains from VfAUREO1 and PtAUREO1a including their flanking helices were solved (15,20,21). Interestingly, only one of these structures was analyzed to demonstrate a dimeric arrangement that is consistent with the architecture of full-length aureochrome in its dark state (15), whereas the others were presented as monomers or in an inconsistent dimer arrangement. Moreover, Fourier transform infrared spectroscopy (FT-IR) studies of the photosensory module of PtAUREO1a showed that upon illumination both flanking helices of this LOV domain, the N-terminal A′α helix and the C-terminal Jα helix, unfold in an allosterically-dependent manner (22,23). The release of the A′α helix leads to the adoption of an active dimer in the lit state by uncovering the dimerization site (21,23). A lit state crystal structure of the dimeric PtAUREO1a LOV domain confirmed large conformational changes of both helices including a partial unfolding of Jα and revealed changes in the topology of the two protomers (21). For VfAUREO1, a 160 ms change in diffusion coefficient was determined, which implies a change in the shape of the protein and thereby some conformational response of the bZIP/linker region of the-bZIP-LOV module to light (24). Despite these data, our knowledge of the mode of intra-molecular signal transmission within the aureochromes remains limited.
Here, we show that the expression of P. tricornutum aureochromes maintain differential diel rhythms. In addition, class I aureochrome paralogs PtAUREO1a and 1c are expressed in a light-independent circadian rhythm. Given their co-occurrence in the nucleus, we demonstrate that these aureochromes are not only capable to form homo-, but also heterodimers in vitro. The N-terminal, probably unstructured region in PtAUREO1a failed to respond to light illumination, but facilitates further oligomerization. In contrast, light-induced unfolding of the flanking helices of the PtAUREO1a LOV domain occurs also in the context of full-length protein. A light-induced helix unfolding in the bZIP domain was found. However, in the presence of bound duplex DNA, the response of the bZIP region changed instead to a light-induced helix extension. The functional module of PtAUREO1a, bZIP-A′α-LOV-Jα binds not only the aureo box, but it is rather specific to any ACGT containing promoter sequence and binds independent of light conditions but with formation of a super-shift complex in the light. The solution structure of bZIP-LOV module in its lit state, when bound to 20 bp long dsDNA, revealed a stabilization of the bZIP domain flexibility, which was further confirmed by molecular dynamics (MD) simulations including a tilted orientation of bound dsDNA. Overall, we propose that aureochrome, a natural optogenetic module, may not simply act by light-triggered DNA binding, but rather utilizes under in situ conditions a distinct, more complex mechanism, e.g. by building higher oligomeric structures leading to DNA looping.
MATERIALS AND METHODS
Transcript analysis
The expression patterns of four aureochrome paralogs in P. tricornutum were investigated to screen for their light-dependent differential expression using cDNA generated earlier (25). In short, cells had been pre-adapted to 16 h of daily illumination with low light (LL) and had been either kept under the same condition or transferred to continuous darkness for one illumination period. Transcript levels were assayed every 3 h for 33 h under LL or darkness using these pre-adapted cells. HGPRT (hypoxanthine-guanine phosphoribosyltransferase; JGI Protein ID 35566), RPS (ribosomal protein S1; 45451) and TBP (TATA-binding protein; 10199) were chosen as housekeeping genes due to their confirmed stable expression under the studied conditions (26).
Protein purification and determination of oligomeric states using SEC
PtAUREO1a and its variants were expressed in E. coli and purified (Supplementary Figure S1B) as described in the Supplementary Methods. Size exclusion chromatography (SEC) was performed at 4°C using Superdex200 10/300 GL column and 50 mM phosphate buffer, pH 8, 300 mM NaCl. The column was either kept in the dark or continuously illuminated using four 455 nm LEDs. The dsDNA•bZIP-A′α-LOV-Jα complexes were isolated in either 10 mM Tris-HCl pH8, 300 mM NaCl or 0.5X Tris-Borate-EDTA (TBE) (50 mM Tris pH 8, 50 mM boric acid, 1 mM EDTA (Ethylenediaminetetraaceticacid)) as buffer under white light conditions at 4°C. When using TBE buffer the bZIP-A′α-LOV-Jα module, its complex with dsDNA or dsDNA alone eluted differently (see Supplementary Figure S3H) than in Tris-HCl buffer.
Electrophoretic mobility shift assay (EMSA) and microscale thermophoresis
Electrophoretic mobility shift assays (EMSA) were performed using binding buffer containing 10 mM sodium phosphate pH8, 50 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, 1 mM DTT, 20 μg/ml BSA, 6% glycerol and 2 nM IRD700-labeled dsDNA oligonucleotides and increasing protein concentrations (10 nM to 20 μM) in dark and blue light illuminated states (see Supplementary Methods). Thermophoresis binding experiments were performed in a binding buffer containing 10 mM Tris-HCl pH8, 300 mM NaCl and 0.1% (w/v) Tween-20. Dark state experiments were performed under safe light (λmax ≈ 650 nm). A total of 66.7 nM of fluorescently labeled protein was titrated at steady state with increasing concentrations of unlabeled interaction partners (see Supplementary Methods).
UV/Vis, FT-IR and far-UV CD spectroscopy
For UV/Vis spectroscopy, residual free FMN and imidazole were removed in an additional purification step with a PD-10 column. Kinetic traces were recorded in the dark at 447 nm and 20°C after illumination for 500 ms with a 455 nm LED.
Protein samples for FT-IR spectroscopy with or without annealed dsDNA were transferred into 50 mM phosphate buffer, pH 8, 100 mM NaCl and 1–3 μl of the concentrated protein solution were applied onto a BaF2 window and sealed with a second window without any drying. Light-minus-dark difference spectra were recorded before and after illumination for 2 s with a 455 nm LED and scaled to flavin bands at 1727 (+) and 1713 (−) cm−1.
Circular dichroism (CD) spectroscopy was performed in assay buffer comprising either 10 mM Tris-HCl pH8, 300 mM NaCl or 0.5X TBE using the lit state formed by white light illumination (see also Supplementary Methods for details on all spectroscopic experiments).
SAXS data collection, analysis and molecular dynamic simulation
Small angle X-ray scattering (SAXS) data sets in either 10 mM Tris-HCl pH8, 300 mM NaCl or 0.5% TBE buffer were collected at the synchrotron BioSAXS beamline BM29 of the ESRF, Grenoble, France (27). The dark state of the protein was handled under safe light (λmax ≈ 650 nm) and measured in darkness. For the lit state samples were illuminated for at least 10 min using blue light (λmax ≈ 470 nm) and measured under white light. SAXS data sets were analyzed using the ATSAS software package (28). Details of data collection and analysis can be found in Supplementary Methods.
Molecular dynamic simulations were performed with the AMBER14 package, using the amber ff14SB force field (29), TIP3P water model and the corresponding monovalent ion parameters (30). After minimization and temperature and pressure equilibration, self-guided Langevin dynamics were run for 1 ns with moving, rigid body fitted EMAP (31,32) restraints based on an a priori coarse SAXS model, followed by 1 ns of relaxation dynamics. Next, production simulations were run for 145 ns. Details of analysis can be found in Supplementary Methods.
RESULTS
Diatom aureochromes follow a circadian rhythm and form homo- and heterodimers
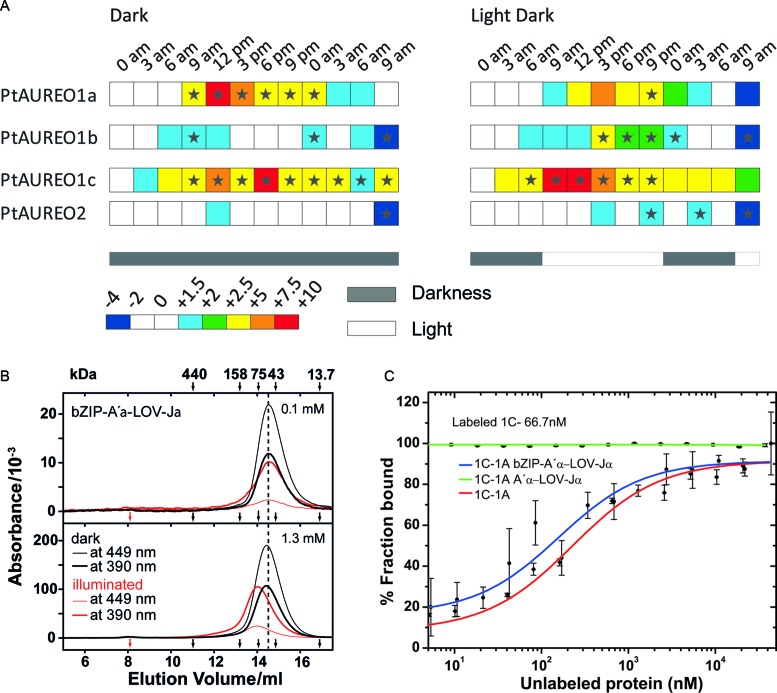
To understand the differential expression of the four aureochrome paralogs from P. tricornutum (PtAUREO1a, 1b, 1c and 2), light- and circadian-dependent transcript analyses were performed. In the resulting transcript patterns, expression levels are shown relative to the first time point of each data set (Figure 1A). The expression of the non-flavin-binding PtAUREO2 appears to occur in a time- and light-independent manner, whereas all other AUREO paralogs showed differential expression patterns throughout the day. PtAUREO1a was upregulated during the day both in day–night cycles as well as in continuous darkness, indicating a light-independent circadian regulation. However, the upregulation during midday was dampened in the illuminated cultures, indicating a residual light-dependent influence on regulation as well. PtAUREO1b transcripts were significantly increased in illuminated cultures compared to the cultures grown in darkness with a peak expression (ca. 5-fold increase relative to midnight) around 3 pm. PtAUREO1c was strongly expressed during the day in both conditions, but illumination shifted the peak of expression from around 6 pm to early day. Overall, we found that the transcript levels of the four PtAUREO paralogs follow different patterns. Whereas PtAUREO2 lacks any significant light/circadian regulation, PtAUREO1b exhibits a moderate light-regulation and PtAUREO1a/c are clearly subjected to circadian regulation. This finding indicates that aureochromes in P. tricornutum may have distinct biological functions. However, the found coexpression of PtAUREO1a/c indicates a synergistic functions as well, e.g. via heterodimerization. Structural bioinformatics based on the sequence of their leucine zipper domains using DrawCoil (33) indeed suggested that PtAUREO1a may be capable to form heterodimers with the other three paralogs. For comparison, the formation of PtAUREO1c heterodimer with the bZIP regions of PtAUREO1b and PtAUREO2 are apparently less stable (Supplementary Figure S1D).
Figure 1.
PtAUREO paralogs harbor a circadian rhythm in the expression and form homo- or hetero-dimers. (A) Transcript patterns of aureochrome genes during a 16 h light and 8 h dark night-day-night cycle (right) or complete darkness (left), relative to the first time point of each data set. Transcripts of PtAUREO1a and PtAUREO1c show a primarily light-independent circadian rhythm, whereas PtAUREO1b shows a primarily light-dependent transcriptional control. Transcription of PtAUREO2 seems to be regulated independently of both light and time. Arrows indicate time points of sampling. Stars indicate significance according to the software REST 2009 (57). (B) Size exclusion chromatography of bZIP-A′α-LOV-Jα at injected concentrations of 0.1 (top) and 1.3 mM (bottom) in the dark and under continuous illumination. Protein with bound flavin was detected at 448 nm and 390 nm. In the dark, bZIP-A′α-LOV-Jα eluted as dimer. Upon illumination, a concentration-dependent shift of the elution peak is observed, from an apparent mass of ∼60 kDa to 78 kDa. The void volume and the elution volumes of protein standards are marked by red and black arrows, respectively. (C) PtAUREO1a interacts with PtAUREO1c using the bZIP region in the lit state. PtAUREO1c interacts with PtAUREO1a and bZIP-A′α-LOV-Jα, but poorly with A′α-LOV-Jα, indicating that the bZIP domain plays a critical role in the interaction of PtAUREO1a and PtAUREO1c.
We produced recombinant PtAUREO1a and 1c as full-length proteins and, in the case of PtAUREO1a, as functional module (bZIP-A′α-LOV-Jα, residues: D145-D378, calculated MM 28.7 kDa) for characterizing their light dependent oligomerization behavior in vitro by SEC. bZIP-A′α-LOV-Jα eluted as dimer in the dark state at 0.1 mM and at 1.3 mM injected concentrations. However, in the lit state a concentration-dependent shift can be observed to higher molecular masses, which indicates light-induced formation of larger oligomeric species (Figure 1B). Interestingly, full-length PtAUREO1a eluted independent of illumination but dependent on the concentration to a higher molecular mass than its theoretical dimer, indicating that the N-terminal extension (M1-S144) causes further oligomerization (Supplementary Figure S1C). In contrast, for VfAUREO1 a redox-dependent dimerization of the bZIP module was evident (17), which depends on a pair of cysteines missing in P. tricornutum aureochromes.
To characterize the ability of PtAUREO1a and 1c for heterodimerization, fluorescence based microscale thermophoresis (MST) was performed. Labeled 1a and 1c aureochromes were kept at nanomolar concentrations assuming a prevalence of their monomeric state. At low concentration, we did not find any major light-triggered effect on homodimerization (Figures 1B, 3B and Supplementary Figure S1C). Hence we performed MST measurements only in the lit state. PtAUREO1c showed a high affinity interaction to PtAUREO1a with a KD of 240 ± 90 nM (Figure 1C). Using labeled protein at a concentration about 5 times higher than the KD, we confirmed that the PtAUREO1a•PtAUREO1c interaction is equimolar (1:1) in vitro (Supplementary Figure S1A). In order to understand which part of the aureochrome affects the heterodimerization ability, we either used the PtAUREO1a bZIP-A′α-LOV-Jα or the A′α-LOV-Jα module with fluorescently labeled PtAUREO1c. The bZIP-A′α-LOV-Jα module interacts with PtAUREO1c with a KD of 450 ± 240 nM, indicating no major role of the N-terminal extension for heterodimerization, whereas the A′α-LOV-Jα domain alone lacks any interaction with full length PtAUREO1c in the lit state (Figure 1C). These data corroborate that the bZIP domain is the dominant factor for the heterodimerization of P. tricornutum aureochromes (Supplementary Figure S1D).
Figure 3.
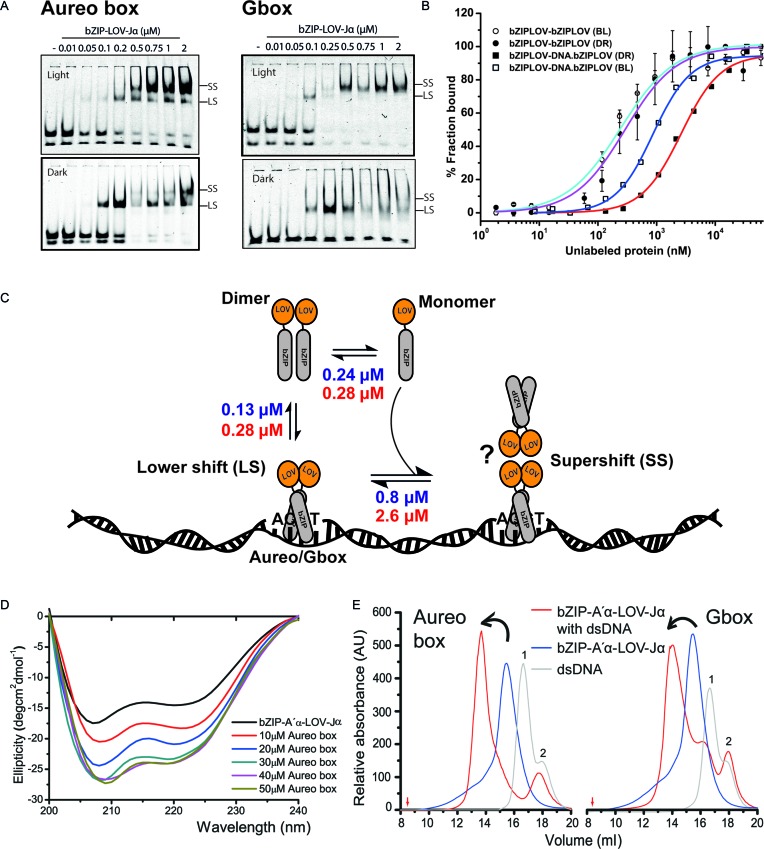
Aureochrome binds ACGT sequence independent of light. (A) Increasing concentration (0.01–2 μM) of bZIP-A′α-LOV-Jα from PtAUREO1a bound to 2 nM aureo box or G-box sequences visualized by an EMSA experiment. Lit state was obtained by illuminating the sample with blue light (λ ≈ 475 nm). LS, lower shift, indicating a 1:1 complex of DNA and bZIP-A′α-LOV-Jα dimer. SS, super shift complex. Free DNA probe can be visualized in two isoforms. The upper isoform bound efficiently to bZIP-A′α-LOV-Jα. The apparent dissociation constants were estimated using the Hill equation and considering bound versus unbound. The bound complex was calculated by summing the intensities of the LS and SS complexes. For the unbound complex, intensities of both dsDNA isoforms were summed up. (B) bZIP-A′α-LOV-Jα forms light-independent homodimer with similar affinity (cyan line for lit state and violet line for dark state). bZIP-A′α-LOV-Jα can build a ternary complex upon DNA binding, as revealed by EMSA (SS complex). The affinity for building a ternary complex is 3-fold lower (blue line) than homodimerization in the lit state and 6-fold lower than in the dark state (red line). (C) Schematic overview of bZIP-A′α-LOV-Jα self-association, dsDNA binding and ternary complex formation in both lit and dark states. A putative structure (question mark) for the SS complex is included. (D) Far UV circular dichroism spectra of bZIP-A′α-LOV-Jα and in presence of increasing concentration of dsDNA. An increment of helicity can be observed, which saturates after formation of a 1:1 complex. (E) Isolation of dsDNA bound bZIP-A′α-LOV-Jα in Tris-HCl buffer in the lit state. Aureo box DNA eluted at 16.6 ml and G-box at 16.7 ml (peak 1). bZIP-A′α-LOV-Jα eluted at 14.9 ml as a dimer with apparent molecular weight of 56.2 kDa. Aureo box-bZIP-A′α-LOV-Jα eluted at 13.8 ml and G-box-bZIP-A′α-LOV-Jα at 14 ml. Red arrows indicate the void volume. The dsDNA preparation contained some fraction of ssDNA (peak 2), which consistent with the EMSA assay did not form any complex with the protein and eluted separately.
LOV and bZIP domains of PtAUREO1a respond to blue light by changes in secondary structure
To further address the role of the N-terminal extension in signaling by aureochromes, we investigated, whether it affects the spectroscopic characteristics of PtAUREO1a. The identical UV/Vis absorption spectra with maxima at 448, 375 and 358 nm for the oxidized flavin state (Supplementary Figure S2A) as well as the similar dark recovery rates with time constants of 1750 s (R2 = 0.9995) and 1560 s (R2 = 0.9996) (Figure 2A) for PtAUREO1a and bZIP-A′α-LOV-Jα, respectively, indicate that the N-terminal extension causes only a limited effect on the sensory module in the dark and the light. In contrast, the C-terminal Jα-helix plays a central role for BL switching in full-length aureochromes, because the truncated AUREO1aΔJα variant exhibited a 5-fold shorter time constant (τ = 360 s, R2 = 0.9995) for dark recovery. Such fast recovery has been associated with the monomeric state of A′α-LOV-Jα and is accordingly also found for the A′α-LOV domain, which fails to form stable lit-state dimers in solution (22). Hence, we concluded that the lit-state dimers of the A′α-LOV-Jα modules alone or in the context of full-length aureochrome are structurally highly similar, with their rearrangement depending on light-triggered changes of the Jα-helix.
Figure 2.
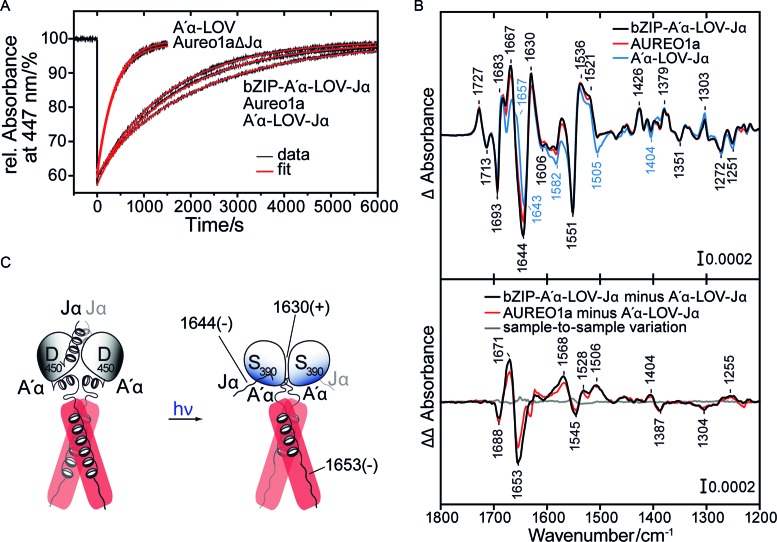
The bZIP domain of aureochrome responds to light in the absence of DNA. (A) Recovery kinetics after illumination of PtAUREO1a, PtAUREO1aΔJα, bZIP-A′α-LOV-Jα, A′α-LOV-Jα and A′α-LOV (latter two taken from Herman et. al, 2013).Traces were analyzed by fitting mono- or biexponential functions. Similar time constants were obtained for full-length PtAUREO1a and for bZIP-A′α-LOV-Jα, close to the major component of the A′α-LOV-Jα recovery assigned to the lit state dimer. In contrast, AUREO1aΔJα exhibited a much faster decay, similar to the minor component of A′α-LOV-Jα recovery and of A′α-LOV, pointing to a monomer-like arrangement of the LOV domains. (B) Top. Light-minus-dark FT-IR difference spectra of the full-length PtAUREO1a, bZIP-A′α-LOV-Jα and A′α-LOV-Jα. Bottom. Segment-resolved double difference spectra obtained by subtraction of the difference spectrum of A′α-LOV-Jα from that of bZIP-A′α-LOV-Jα or from that of the full length. Both spectra show a loss of helical structure and an increase in turn structure in the bZIP-linker region upon illumination. bZIP-A′α-LOV-Jα minus bZIP-A′α-LOV-Jα illustrates the sample-to-sample variation of independent preparations. (C) Resulting model of the photoreaction in the absence of DNA (IR signals indicated in cm−1).
Next, we performed FT-IR spectroscopy on PtAUREO1a and bZIP-A′α-LOV-Jα to investigate BL-induced changes at the molecular level (Figure 2B) and compared the results to previously obtained spectra and assignments for the A′α-LOV-Jα module (22,23). Many difference bands are identical because they originate from structural changes of the flavin and the protein moiety of the A′α-LOV-Jα module. Most relevant are bands in the amide I region between 1615 and 1695 cm–1, in which the different secondary structures show a characteristic absorption (34). Notably, the negative band at 1644 (−) cm−1 in bZIP-A′α-LOV-Jα and full length PtAUREO1a (Figure 2B) is assigned to light-induced unfolding of the Jα helix as for A′α-LOV-Jα (22), in agreement with earlier assignments for other LOV domains (35–37). Likewise, the marker band at 1630 (+) cm−1 is detected in bZIP-A′α-LOV-Jα and PtAUREO1a, which results from changes at the β-sheet surface by formation of the lit-state LOV dimer of A′α-LOV-Jα (23). This assignment was confirmed for bZIP-A′α-LOV-Jα by hydrogen/deuterium (H/D) exchange, whereby contributions from side chains such as the arginines of the bZIP domain were excluded (Supplementary Figure S2B). Moreover, the band at 1630 cm−1 is strongly reduced in intensity in AUREO1aΔJα and in the monomeric A′α-LOV (Supplementary Figure S2C). BL-triggered unfolding of the A′α helix using its marker band at 1656 (−) cm−1 could not be followed for bZIP-A′α-LOV-Jα and full length PtAUREO1a because of overlapping and additional contributions in their spectra. However, it is likely that this conformational change takes place as well, because the dimerization site for formation of the lit-state dimer of the LOV domains was found to be covered by A′α helices.
To address the question if and how the bZIP domain reacts on BL-induced changes in the LOV domain, we calculated a double difference spectrum by subtracting the light-minus-dark difference spectrum of A′α-LOV-Jα from that of the bZIP-A′α-LOV-Jα module (Figure 2B, bottom). Remaining signals are specific for the presence of the bZIP domain and the linker region (I209-D238) between the bZIP and the A′α helix. In the amide I region, bands are detected at 1688 (−), 1671 (+) and 1653 (−) cm−1 and confirmed as secondary structural changes by H/D exchange (Supplementary Figure S2B). The most prominent band at 1653 (−) cm−1 points to a light-induced loss of helicity (34) within the bZIP domain and/or linker region. We confine these changes to the bZIP domain given its general propensity to form helices, in agreement with recent results on the change in accessibility from HDX-MS (21). Further signals at 1688 (−) and 1671 (+) cm−1 can be assigned to light-induced changes in turn structural elements, all the other bands to the corresponding amide II and amide III modes. Only the difference band at 1387 (−)/1404 (+) cm−1 is assigned to the symmetric stretch of an aspartate or glutamate pointing to an altered hydrogen bonding (Supplementary Figure S2B). As before, only very minor contributions are detected from the N-terminal extension, because the difference and double difference spectra of full-length PtAUREO1a were very similar to those of bZIP-A′α-LOV-Jα (Figure 2B). Therefore, the N-terminal truncated bZIP-A′α-LOV-Jα module can be considered as valid model for investigating the interplay of DNA-binding and photoresponse within aureochromes.
bZIP domain of PtAUREO1a binds to ACGT core sequence independent of light
To confirm that the bZIP domain of PtAUREO1a binds the aureo box similar to VfAUREO1, we have performed EMSAs with the 31 bp long aureo box containing DNA using the same conditions as published previously (13). In the lit state, bZIP-A′α-LOV-Jα bound to the IR700-labeled DNA, but interestingly the protein was able to bind DNA even in the dark-state (Figure 3A). The most striking observation was that the bZIP-A′α-LOV-Jα module formed a light-dependent higher order oligomer complex of bound DNA and protein and we named it as supershift (SS) complex. However, the apparent interaction affinity was only 2-fold higher in the lit state. Recently, it was shown that Mg2+ affects formation of a specific complex between bZIP and the aureo box, yielding a 10-fold increase for the lit state affinity to DNA compared to the dark state (21). Electrostatic shielding by Mg2+ ions has been indeed reported to affect specific binding of duplex DNA to CREB (cAMP response element-binding protein), but with non-uniform, conflicting results (38,39). Unlike Heintz et al., we did not rely on Mg2+-based effects by using a non-related sequence, the STAT box (40) as well as competition EMSA with an unlabeled aureo box probe to confirm that also in the absence of Mg2+ in the electrophoresis buffer (21) PtAUREO1a specifically interacts with the aureo box dsDNA (Supplementary Figure S3B and D).
It was previously found that plant bZIP domains regulate gene expression by acting on ACGT sequence-containing promoters (41). To check whether the bZIP-A′α-LOV-Jα recognizes the ACGT core of the aureo box, we analyzed a C→T mutant in the ACGT motif of the aureo box. Indeed, the mutated aureo box was not able to displace the unaltered aureo box from the DNA binding site (Supplementary Figure S3D (ii)). In plants, promoters with G-box cis elements control the expression of various genes in response to light (42,43). Interestingly, the G-box motif (CACGTG) shares the ACGT core of the aureo box (Supplementary Figure S3A). Accordingly, we examined whether PtAUREO1a interacts with the G-box sequence and detected binding to a 32 bp long G-box containing dsDNA, forming a supershift complex in a light-dependent manner. To check for the effect of flanking nucleotides on binding, we performed competition-binding experiments with either non-labeled wild-type or mutated aureo box and G-box motifs. The results demonstrate that the bZIP-LOV module of aureochromes can bind both types of cis elements with no significant difference in affinity (Supplementary Figure S3D and E). Moreover, the calculated apparent affinities for these interactions revealed that bZIP-A′α-LOV-Jα binds to both sequences with an apparent KD of ∼150 nM in the lit state and ∼280 nM in the dark state.
To characterize the supershift complex, the higher oligomer formed by the interaction of bZIP-A′α-LOV-Jα modules in a concentration-dependent manner, we postulated two hypotheses: (i) one dimer binds to the ACGT motif and the other dimer binds to the DNA non-specifically in order to scan for the binding sequence, and (ii) bZIP-A′α-LOV-Jα modules while bound specifically to the DNA interact with each other building a concentration-dependent higher order oligomer. To address the first hypothesis we used 31 bp (3 turn), 21 bp (2 turn) or 14 bp (1.5 turn) dsDNA containing the aureo box. The first hypothesis was found to be inconsistent with the observation that the supershift remained stable, even when using the 1.5 turn dsDNA (Supplementary Figure S3C). To address the second hypothesis, we performed MST and confirmed firstly the homodimerization/oligomerization affinity for bZIP-A′α-LOV-Jα in its lit and dark states (Figure 3B). Homodimerization of the bZIP-A′α-LOV-Jα module clearly resembles the PtAUREO1a•PtAUREO1c heterodimerization and is light independent with affinities of 240–280 nM (Figure 3B). Next, in a similar MST experiment, but with increasing concentrations of unlabeled dsDNA/bZIP-A′α-LOV-Jα complexes, we found that these added protein–DNA complexes form higher order species with the labeled bZIP-A′α-LOV-Jα module (Figure 3B). Interestingly, the affinity was three times higher in the lit state than in the dark (0.8 μM versus 2.6 μM), and thereby resembled the light sensitivity of the EMSA-detected supershift (Figure 3A). Competition EMSA experiments confirm this notion, as the supershift complex disappeared at very low concentrations of competition probe (Supplementary Figure S3D and E). In conclusion, the supershift complex seen in EMSA appears to be a result of the formation of a higher order species of bZIP-A′α-LOV-Jα in the DNA bound state (Figure 3C).
To further characterize the complex of DNA and bZIP-A′α-LOV-Jα, we performed CD spectroscopy and SEC. The experiments were performed in the lit state only, because DNA binding is independent of the light conditions (Figure 3). By CD spectroscopy we found that the calculated helicity of bZIP-A′α-LOV-Jα increased by up to 21% on formation of the dsDNA•bZIP complex (Supplementary Table S1, Figure 3D and Supplementary Figure S3G). A saturation of the helicity increment was observed at a stoichiometry of 1:1.
Recently VfAUREO1 bound to a Cy3-labeled dsDNA complex was isolated using SEC (17). Accordingly, we were also able to purify the dsDNA bound complex of bZIP-A′α-LOV-Jα and a 20 bp long aureo or G-box sequence-containing DNA in Tris-HCl (Figure 3E) and 0.5X TBE buffers (Supplementary Figure S3H). The DNA protein complex eluted at 13.7 ml and 13.9 ml, for the aureo box and G box bZIP-A′α-LOV-Jα complex, respectively (Figure 3E). The theoretical molecular mass of the 1:1 complex is 68.6 kDa but the complex eluted at a higher molecular mass of about 100 kDa. We hypothesize that the higher order complex eluted from size exclusion is a tetrameric bZIP-A′α-LOV-Jα-dsDNA complex (as shown in Figure 3C).
The response of the aureochrome•DNA complex to light
To investigate a possible influence of the bound DNA on the LOV domain kinetics, the recovery of bZIP-A′α-LOV-Jα was recorded in the absence and in the presence of an equimolar concentration of the aureo box at a reduced concentration of 100 mM NaCl (Supplementary Figure S4A). Addition of the aureo box causes a changed kinetics that cannot be fitted by a mono-exponential function anymore. A moderate effect of DNA binding on the LOV domain function is therefore observed in contrast to previous studies on VfAUREO1 (44), but the effect is much weaker than that of the Jα helix truncation (Figure 2A).
To elucidate the effect of DNA binding on the response of the bZIP domain to light, we performed FT-IR spectroscopy in the presence of the aureo box and the G-box in comparison to the STAT box and without DNA (Figure 4A and Supplementary Figure S4B). Additionally, the difference spectrum of full length PtAUREO1a was recorded in the presence of the aureo box and was found to resemble that of the bZIP-A′α-LOV-Jα module (Supplementary Figure S4C). All difference spectra show a good agreement in the bands below 1500 cm–1 (Supplementary Figure S4D) that originate mainly from the chromophore. Furthermore, the marker band of dimerization of the LOV domains at 1630 (+) cm−1 is fully preserved. However, significant differences are induced in the amide I and II regions by the presence of DNA, indicating a strong influence on the light-induced secondary structure changes in the bZIP domain. In the presence of the G-box, the compensating effect of these changes is so strong that two bands at 1657 (−) and 1642 (−) cm−1 are uncovered, which originate from the unfolding of the A′α and the Jα helix in the LOV domain. These differences cannot be simply interpreted as a blockage of light-induced changes in the bZIP domain by the presence of DNA (Supplementary Figure S4E), because the resulting spectra do not match those of A′α-LOV-Jα alone (Figure 2B). The effect by DNA was even detected in the presence of the STAT box as a non-cognate oligonucleotide, although EMSA experiments failed to show any binding. This difference can be explained by the requirement of millimolar concentrations for FT-IR spectroscopy, leading to an unspecific binding of the STAT box to the bZIP domain due to electrostatic interactions with the DNA backbone (45).
Figure 4.
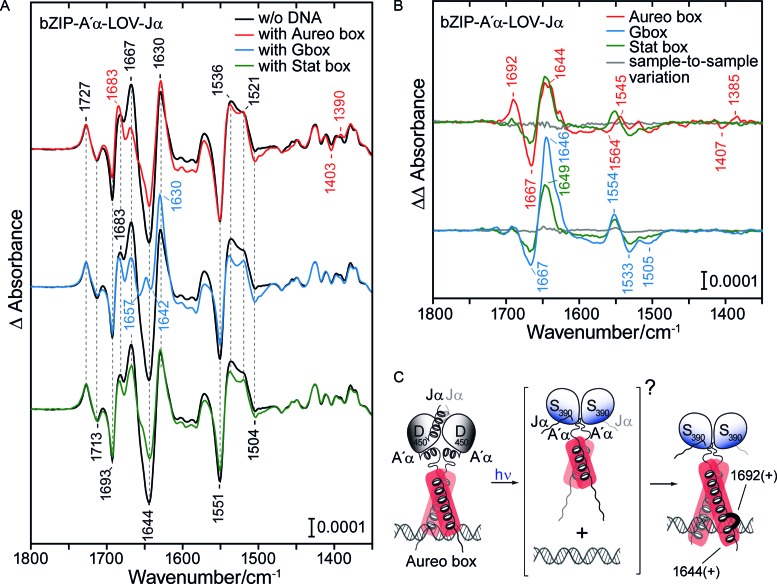
Light induces helix elongation in the bZIP domain in the DNA-bound state. (A) Left. Light-minus-dark FT-IR difference spectrum of bZIP-A′α-LOV-Jα with aureo box, with G-box and with STAT box in comparison to that without DNA. The presence of DNA changes the response in secondary structure, as seen in the amide I and II region (1695–1520 cm−1). (B) Double difference spectra obtained by subtraction of the difference spectrum of bZIP-A′α-LOV-Jα in the absence of DNA from that complexed to the aureo box, G-box and STAT box. Sample-to-sample variations were calculated from recordings on independent preparations of the same sample. (C) Model of the photoreaction of bZIP-A′α-LOV-Jα in the presence of the aureo box. Upon illumination, formation of the lit state dimer by the LOV domains leads to an extension of the helix in the bZIP region (unspecific) and folding of a turn element only observed with the aureo box (IR signals indicated in cm−1). As an intermediate, light-induced unfolding of bZIP helices might take place as it was observed in the absence of DNA.
Double difference spectra of the DNA–protein complex minus protein alone reveal similarities between the effects of the three oligonucleotides (Figure 4B). In the amide I region, a positive band was detected in the spectral region of helices (1644–1649 cm−1) and a negative band in the spectral region of turn structures (1667 cm−1). The shift in the frequencies of these signals from 1653 cm−1 to around 1647 cm−1 in the presence of DNA provides evidence for additional helical folding by light. In general, longer helices absorb at lower frequencies within the spectral range specific for α-helices (46). Therefore, the presence of DNA leads to an additional light-induced folding of the bZIP domain that is countercurrent to the changes in the bZIP domain found in the absence of DNA (Figure 2B). An alternative explanation for the downshift would be a strongly increased bending of the coiled coil (47). In any case, this effect is considered to be unspecific, because it is found irrespective of the sequence but more pronounced in the presence of the G-box than of the STAT box, reflecting that the former binds with much higher affinity (Supplementary Figure S3B).
Sequence-specific effects are found in the double difference spectrum in the presence of the aureo box, it clearly differs from those in the presence of the G- and STAT boxes (Figure 4B). First, a positive band at 1692 cm−1 is detected, which can be assigned to an increase in turn structural element upon illumination (34). Second, bands at 1407 (−) and 1385 (+) cm−1 point to a hydrogen bonding change of a glutamate or aspartate, which only occurs upon illumination without DNA and hence is blocked by the aureo box (Supplementary Figure S4E). It has been shown for the bZIP protein GCN4 that E237 forms a stabilizing interaction to Arg 240, which in turn reinforces the conformation of the central base pairs by hydrogen bonding to the phosphate groups (48). In most bZIP proteins, E237 is replaced by a hydrophobic amino acid, while it is conserved in PtAUREO1a (as E162), PtAUREO1b, PtAUREO1c as well as in VfAUREO1. Interestingly, in the bZIP protein CREB, this amino acid (E295) is found within a turn-like structural element in one of its two strands in the presence of the target DNA. The additional light-induced formation of a turn structure in bZIP-A′α-LOV-Jα and the preservation of hydrogen bonding to an amino acid, possibly E162, might allow for additional sequence-specific recognition of the aureo box. In this context, the response of the bZIP observed in the absence of DNA (Figure 2C) might be regarded as an intermediate, unfolded structure of the basic region on the pathway to the higher affinity complex in the light (Figure 4C).
Solution structure of DNA bound aureochrome
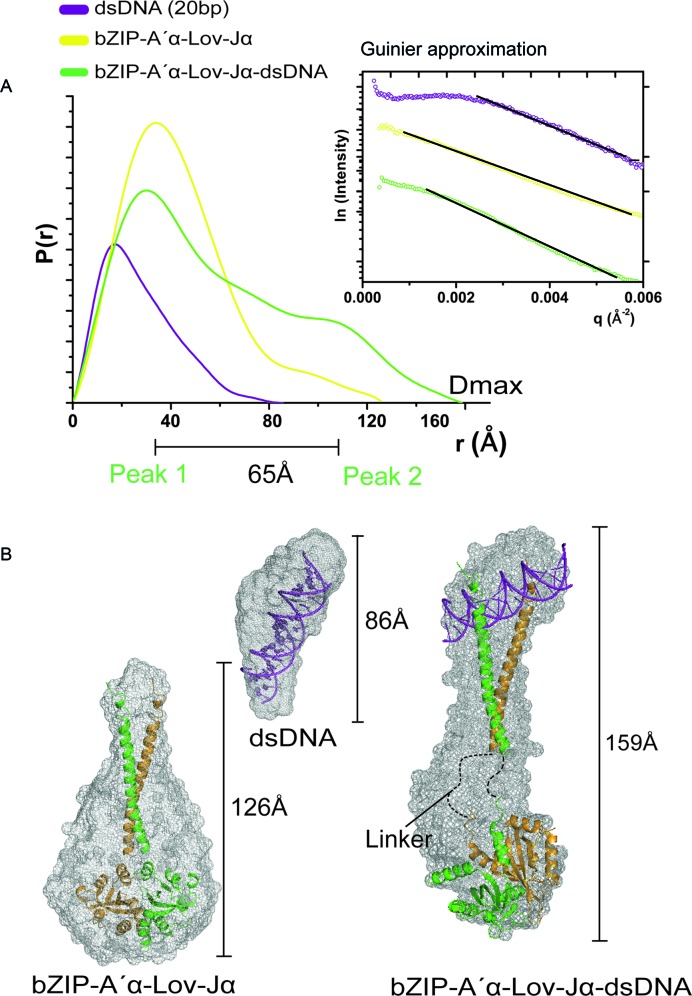
To unravel global conformational differences induced by light, we performed SAXS. In the absence of dsDNA, we were unable to observe large-scale structural rearrangements in the bZIP-A′α-LOV-Jα module in its light and dark-adapted states with an almost identical radius of gyration (Rg) of 38 Å and degree of flexibility (Supplementary Figure S5A and B). As mentioned above, bZIP-A′α-LOV-Jα already attains its dimeric conformation in the dark state, which might restrict large-scale global conformational changes upon illumination. Accordingly, the following measurements were performed only for the lit state. bZIP-A′α-LOV-Jα in Tris-HCl buffer exhibited a concentration dependence, where the Rg increased by 8 Å (Supplementary Table S2), in agreement with the SEC elution profiles (Figure 1B). We obtained the DNA-bound complex not only in Tris-HCl buffer, but also in TBE buffer, and found that the functional module holds less flexibility in TBE (see Supplementary Table S1), presumably due to lower ionic strength. Such lower flexibility is advantageous for a detailed structural analysis. Therefore, we chose a single concentration (1.25 mg.mL−1) in TBE buffer for the representation of SAXS data (Figure 5).
Figure 5.
Solution structure of aureo box-bound bZIP-A′α-LOV-Jα. (A) P(r) distribution profile from small angle X-ray scattering (SAXS) experiments of 20 bp long dsDNA, bZIP-A′α-LOV-Jα and dsDNA-bound bZIP-A′α-LOV-Jα. The Guinier approximation of the respective scattering vectors is represented in the top panel. The dsDNA showed a small extent of inter particle repulsion. The linear region was defined to calculate the Rg and I0. The P(r) distribution of dsDNA bound to bZIP-A′α-LOV-Jα indicates formation of two domains separated by 65 Å. Domain I originate from shorter distances in the globular LOV domain and domain II arises from longer distances in the dsDNA-bound bZIP domain. The inter domain distance is a result of the extended bZIP helix and linker region. (B) The ab initio envelopes of respective scattering vectors and the structural models fitted into the envelope. The maximum particle diameters (Dmax) are shown with the envelopes. The crystal structure of the lit-state LOV domain was used for the dsDNA-bZIP-A′α-LOV-Jα complex.
bZIP-A′α-LOV-Jα revealed a globular shape in the P(r) distribution, and no aggregation was visible as depicted in the Guinier approximation (Figure 5A). However, the ab initio envelope shape appeared to be wider compared to the model of bZIP-A′α-LOV-Jα (χ value of 3) with a Dmax of 126.4 Å (Figure 5B). We assume that the flexibility arises from an unordered bZIP region in the absence of bound dsDNA as indicated before by FT-IR and CD spectroscopy. Nonetheless, the Porod volume and the calculated I0 are close to the expected molecular mass for a dimer of 57.2 kDa (Supplementary Table S2). The 20 bp long aureo box containing dsDNA showed little inter-particle repulsion in the Guinier approximation, and thus appeared to be elongated with a Dmax of 86 Å and Rg of 22 Å. The modeled aureo box dsDNA was fitted into an ab initio envelope with an apparent χ value of 1.9 (Figure 5B).
To obtain structural insight of the dsDNA•bZIP-A′α-LOV-Jα complex, we used different protein:dsDNA ratios (1:1, 2:1 and 1:2) as well as the dsDNA•bZIP-A′α-LOV-Jα complex purified by SEC. As expected, the equimolar mixture of dsDNA and bZIP-A′α-LOV-Jα and the isolated complex gave similar results with identical Rg of ∼48 Å and Dmax ∼160 Å. The P(r) distribution indicated a dumbbell-shaped architecture of the complex, where the equimolar mixture resulted in a more pronounced domain distribution. Accordingly, we chose the latter data for representation of the dsDNA•bZIP-A′α-LOV-Jα complex (Figure 5B). The peaks in the P(r) distribution are separated by ∼65 Å. Peak 1 resulted from shorter intra-domain scattering vectors and peak 2 corresponds to the longer inter-domain scattering vectors. The bZIP dimer was modeled bound to the major groove of the 20 bp long dsDNA, attached to an unstructured linker region because of a lack of any template structure, and combined with the dimeric structure of the PtAUREO1a-A′α-LOV-Jα (lit state, 5DKL (21)). The resulting model was fitted to the scattering data yielding an overall χ value of 2.4 with crysol (49).
The most striking feature was found in the DNA-bound bZIP dimer region where in the best-fitting model consistent with the SAXS data, the dsDNA was not bound in a ‘T’-shaped arrangement, as observed before for dsDNA•bZIP complex crystal structures (50,51). Instead, the DNA appears to be tilted in the bound state. This discrepancy might be related to the fact that our measurements were performed in solution, whereas, to our knowledge, bZIP-DNA complexes other than that of PtAUREO1a were characterized only in the crystalline state.
Accordingly, we performed non-restrained molecular dynamic simulations based on the experimental ‘T-shaped’ conformation of the dsDNA•bZIP complex (PDB ID: 1YSA), which showed strong DNA movement relative to the bZIP domain. We named the DNA movement ‘tilt’ and the bZIP domain flexibility ‘twist’ (Supplementary Figure S5C). The twisting motion of bZIP domains had been thoroughly described previously and is dependent on salt concentration and temperature (52). Further, the flexibility of the bound dsDNA had also been analyzed by MD simulations previously, and revealed dsDNA movement in the bound state (53).
In order to further refine our rigid-body fitted model, as presented above, we performed steered MD simulations. In practice, after 1 ns self-guided Langevin dynamics (Supplementary Figure S5D and E) using the SAXS a priori coarse model as a harmonic restraint, we obtained a stable system, which could be equilibrated and readied for 140 ns MD production trajectories (Figure 6).
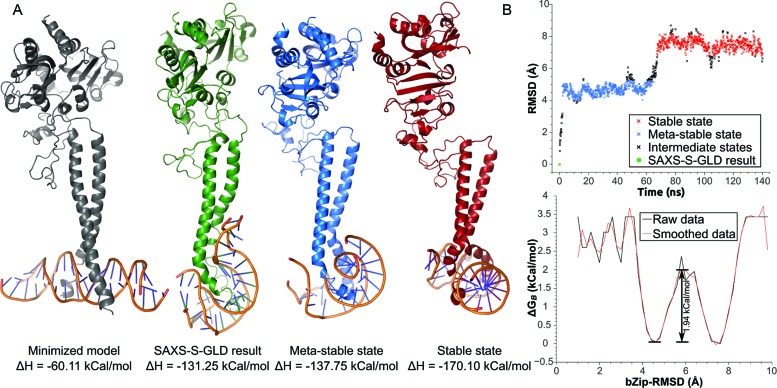
Figure 6.
Molecular dynamic simulation of aureochrome-dsDNA complex. (A) Minimized model (grey), SAXS-Self-guided langevin dynamic restrained model (green) and average meta-stable (blue) and stable (red) poses for the protein–DNA complex. Molecules were aligned by the bZIP domain backbones. Calculated DNA-to-protein binding enthalpies are indicated. (B) bZIP domain RMSD analysis. Top: bZIP backbone RMSD trajectory. Assigned states are color coded corresponding to the poses in A. Black intermediate states do not correspond to the minimized model, but to transitions between states (B). Boltzmann free energies at 300 K based on bZIP-RMSD frequencies. The meta-stable state corresponds to the first major energy minimum, while the stable state to the second. Activation energy of 1.94 kCal/mol separates the two states.
In order to accommodate for the SAXS restraints, both the bound dsDNA and the bZIP domain were distorted, to the point where one side of the DNA double strand attached longitudinally to the bZIP domain, while the other looped and surrounded the tip of the domain (Figure 6, green). When freed from restraints, the simulation converged rapidly (∼2.2 ns, Figure 6B top), and adopted a meta-stable conformation via partial relaxation of the extended dsDNA end (Figure 6 blue). Finally, after 67.3 ns, a stable conformation was attained, in which both DNA ends looped around the tip of the bZIP domain symmetrically (Figure 6 red). Kinetic analysis of the data indicated that a low energy barrier of 1.94 kcal/mol separates these two conformations (Figure 6B bottom). Please refer to the molecular dynamics video (Supplementary Video 1) for simulation traces. Interestingly, MMGBSA energy analysis indicates that the DNA-to-protein binding enthalpy decreased in all steps from −60.1 kcal/mol for the initial conformation (T-shaped, Figure 6, grey) to the stable state (Figure 6, red) with −170.1 kcal/mol.
Overall, our MD simulations indicated that a highly compact binding conformation of dsDNA with the bZIP dimer is stable and energetically favored in solution. Such a binding mode is fully compatible with the SAXS data, further refining the coarse model and provides insight into the variability of the conformational space between crystalline and solution phases.
DISCUSSION
Unlike other light-dependent transcriptional regulators and LOV protein photoreceptors, aureochromes are unique by their inversed effector-sensor topology. To understand their intramolecular signal transmission at the molecular level, three different levels of response to light need to be considered for aureochromes: The BL-triggered change of LOV domain dimerization, subsequent conformational changes transmitted from the LOV domains to the bZIP domain and global changes of the dsDNA•aureochrome complexes. Besides contributing to the elucidation of its natural role as a blue light photoreceptor (13,14,16), such insight is valuable for its perspectives in optogenetic applications: Recently, the C-terminal aureochrome LOV domain was used to develop a synthetic BL-dependent receptor tyrosine kinase (18).
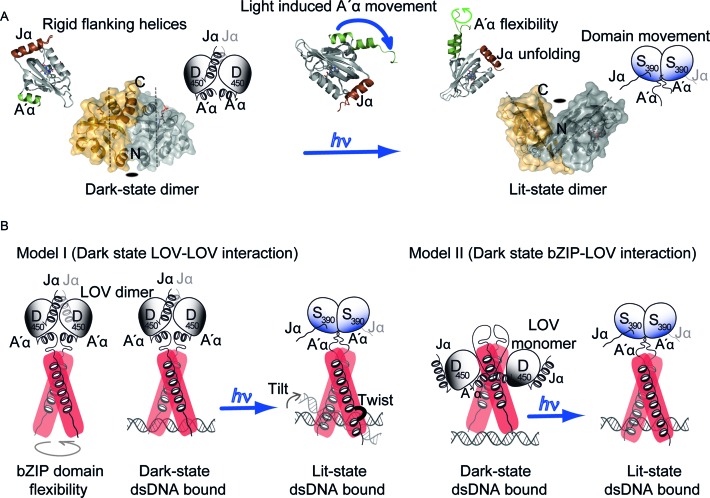
The lit-state structure of the LOV domain has been solved recently (21), which indicates light-induced conformational changes at the A′α helix (Figure 7A) and which accordingly resembles the fungal stand-alone LOV domain VVD (12). This conformational change of the flanking helix was also shown by FT-IR and SAXS measurements using PtAUREO1a A′α-LOV-Jα (15,23). Unlike VVD, intrinsic dark-state dimerization is evident for the PtAUREO1a A′α-LOV-Jα module, as shown by two independent crystal forms and KD values determined to 64 and 14 μM, respectively (15,21). Despite their different orientation in the lit state, these dark state dimers are topologically still consistent with the bZIP dimer arrangement. Light-triggered changes of the quaternary structure of LOV domains, with an overall rotation of 173° for its protomers relative to each other (15,21), clearly exceed the domain movements found in other LOV domains, e.g. YtvA from Bacillus subtilis with a light-triggered change of 4–5° (10). This way, the capability of diatom aureochrome LOV domains to perform light-induced flipping of their domains may be translated to large-scale conformational changes in multi-domain proteins. In addition, unlike in bacterial LOV proteins or VVD, the Jα helix of PtAUREO1a unfolds upon illumination (Figure 2B), at least partially as confirmed by the lit state structure of the LOV domain. Such unfolding more resembles the response of homologous phototropin-LOV from plants (8). The relevance of this unfolding in the context of the full-length protein is underlined by the finding that the Jα-deficient aureochrome does not form the lit-state dimer of LOV anymore (Figure 2A and Supplementary Figure S2C), in agreement with an allosteric regulation between the two flanking helices as suggested before (23). This regulation of light-induced dimerization constitutes the key difference to phototropin-LOV domains.
Figure 7.
Working model for aureochrome function. (A) Aureochrome LOV domains form both lower affinity dark-state and high affinity lit-state dimers. The dimers differ in the orientation of the two protomers and the lit state dimer is tilted along the symmetry axis. Dimerization is a result of light-induced undocking of the A′α and unfolding of Jα, resulting in flexibility of the flanking helices in the lit state. (B) Two different working models can be derived from our present knowledge of aureochrome. The hypothesis I is based on a dark state LOV–LOV interaction and our present data in the manuscript. The bZIP domain holds flexibility in the unbound state. In this conformation, the aureochrome can bind independent of light. Light induces a domain movement similar to bacterial stress response protein YtvA, which in turn rigidifies the bZIP dimer by elongation of the helix and formation of a turn element and hence enhances DNA binding. dsDNA can tilt in the bound state and the bZIP domain can twist while bound to the cognate DNA. Hypothesis II is based on a bZIP-LOV interaction in the dark state (21). In the dark state, the LOV domain dimer interface is hidden in the interaction surface between LOV and bZIP and in the lit state the LOV domain is released from the bZIP domain and forms the high affinity lit-state dimer. This response in turn enhances affinity towards DNA. Position of dsDNA in the bound state is drawn respective to their apparent binding affinity to the basic region of bZIP domain.
Interestingly, FT-IR and UV/Vis measurements revealed only very minor influence of the probably unstructured N-terminal domain upon illumination in aureochromes (Figure 2A and B). Given the low degree of conservation between different aureochromes in this region, it may apparently play a functional role only for the recruitment of other factors affecting transcription. In contrast, the bZIP domain/linker region clearly lost helicity upon BL illumination (Figure 2B) as evidence for conformational changes being transmitted from the LOV domain to the bZIP domain. A recent study (21) using hydrogen-deuterium exchange (HDX) mass spectrometry interpreted analogous changes of the exchange rates in the bZIP domain as an indicator of a potential LOV-bZIP interaction (Figure 7B model II) that would be sufficiently strong to overcome LOV dimerization in the dark state. However, in the present study we observed no significant length-induced changes by SAXS in the global envelope of the bZIP-A′α-LOV-Jα module (Supplementary Figure S5B). Changes were limited to helicity loss within the bZIP domain (Figure 2), which might be triggered by the formation of a tilted lit-state LOV dimer (Figure 7A). The bZIP domain itself was found to be flexible both in the dark and lit states, which was significantly reduced in the presence of bound dsDNA as shown by an increased helicity (Figures 3D and 4). The mostly light-independent dsDNA binding and the lack of significant changes in the scattering envelope of the bZIP-A′α-LOV-Jα module upon light-dark transition, lead us to propose a contrasting model (Figure 7B model I) to model II. Here, aureochromes function at the molecular level, similar to the bacterial stress response protein YtvA, for which stabilization of the STAS dimer was predicted to derive from Jα helix movement and a tilted lit-state LOV dimer (10). In our study, we observed that the affinity for bZIP domain dimerization in the context of full-length protein is in the nanomolar range and hereby independent of the light condition (Figures 1C and 3B). Accordingly, the dark-/light-dependent stability of aureochrome•DNA complexes is only slightly affected by light, in the present study 2-fold and previously 9.6-fold (21), and highly depends on the chosen buffer conditions. Accordingly, one may wonder, whether binding of aureochromes to promoter regions in the nucleus really depends on light or if light instead causes other changes to the aureochrome•DNA complex to regulate transcription. Indeed we found that light induces a further oligomerization of the bZIP-A′α-LOV-Jα dimer leading to a shift under SEC conditions in the absence of DNA (Figure 1B) and to a supershift in EMSA assays (Figure 3). It is unclear, where the additional cross-wise interaction between bZIP-A′α-LOV-Jα dimers is located. It likely involves the LOV domain dimers formed within the framework of the aureochrome scaffold. For example, lit state LOV dimers may associate either with each other, or the remaining portions of the aureochrome•DNA complex. In any case, such cross-wise interactions would promote co-localization of dsDNA-bound aureochromes as a prerequisite for DNA looping. The latter is known as a basic mechanism for transcription enhancer function (54,55).
In this context, it is notable that the bZIP-A′α-LOV-Jα module of PtAUREO1a recognizes not only the aureo box, but also the G-box motif, as both share the ACGT core sequence. Interestingly, light-triggered specific structural rearrangements in turn elements take place only in the presence of the aureo box, but not of the G- and STAT boxes (Figure 4). Nevertheless, these rearrangements do not affect the global affinity to dsDNA, which is similar for both cis elements. One may hence argue that the type of the formed aureochrome•DNA complex also feeds back to the kind of conformational change that is accessible to the bZIP-A′α-LOV-Jα module as a whole.
Finally, P. tricornutum harbors four different aureochrome paralogs, of which we found two as being regulated by a circadian rhythm and capable of forming heterodimer complexes. The latter indicates a synergistic function of PtAUREO1a and 1c. In dsCYC2 and other promoters predicted to be affected by aureochromes in P. tricornutum (16,56) we found more than one aureochrome binding motif (TGACGT/NNACGT) at a fixed distance of 100–150 bp. Hence, we hypothesize that in P. tricornutum, aureochromes might form a loop-structured DNA and in turn regulate transcription in an enhancer-like mechanism. However, further studies are needed to confirm this notion. Our present study speaks in favor of an aureochrome function that includes the formation of a higher order structure and not just light-triggered DNA binding, which further restraints the usage of aureochrome as a natural optogenetic module.
Supplementary Material
Acknowledgments
The authors thank the beamline staff at ESRF BM29, Grenoble, France. The transcript analysis was performed with the help of Vincent Spegg.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG research unit 1261 [ES152/12, KO3580/1-2, KR1661/8-2]; Heisenberg programme [KO35880/4-1]. Funding for open access charge: DFG research unit 1261 [ES152/12].
Conflict of interest statement. None declared.
REFERENCES
- 1.Christie J.M. Phototropin blue-light receptors. Annu. Rev. Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 2.Demarsy E., Fankhauser C. Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 2009;12:69–74. doi: 10.1016/j.pbi.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Herrou J., Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat. Rev. Microbiol. 2011;9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Losi A., Mandalari C., Gärtner W. From plant infectivity to growth patterns: The role of blue-light sensing in the prokaryotic world. Plants. 2014;3:70–94. doi: 10.3390/plants3010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwerdtfeger C., Linden H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 2003;22:4846–4855. doi: 10.1093/emboj/cdg451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green J., Crack J.C., Thomson A.J., LeBrun N.E. Bacterial sensors of oxygen. Curr. Opin. Microbiol. 2009;12:145–151. doi: 10.1016/j.mib.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Salomon M., Christie J.M., Knieb E., Lempert U., Briggs W.R. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 8.Harper S.M., Neil L.C., Gardner K.H. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 9.Harper S.M., Christie J.M., Gardner K.H. Disruption of the LOV-Jα helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 10.Möglich A., Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J. Mol. Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nash A.I., McNulty R., Shillito M.E., Swartz T.E., Bogomolni R.A., Luecke H., Gardner K.H. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9449–9454. doi: 10.1073/pnas.1100262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaidya A.T., Chen C.-H., Dunlap J.C., Loros J.J., Crane B.R. Structure of a light-activated LOV protein dimer that regulates transcription. Sci. Signal. 2011;4:ra50. doi: 10.1126/scisignal.2001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi F., Yamagata D., Ishikawa M., Fukamatsu Y., Ogura Y., Kasahara M., Kiyosue T., Kikuyama M., Wada M., Kataoka H. AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19625–19630. doi: 10.1073/pnas.0707692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schellenberger Costa B., Sachse M., Jungandreas A., Bartulos C.R., Gruber A., Jakob T., Kroth P.G., Wilhelm C. Aureochrome 1a Is Involved in the Photoacclimation of the Diatom Phaeodactylum tricornutum. PLoS One. 2013;8:e74451. doi: 10.1371/journal.pone.0074451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee A., Herman E., Kottke T., Essen L.O. Structure of a native-like Aureochrome 1a LOV domain dimer from phaeodactylum tricornutum. Structure. 2016;24:171–178. doi: 10.1016/j.str.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Huysman M.J.J., Fortunato A.E., Matthijs M., Costa B.S., Vanderhaeghen R., Van den Daele H., Sachse M., Inzé D., Bowler C., Kroth P.G., et al. AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum) Plant Cell. 2013;25:215–228. doi: 10.1105/tpc.112.106377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hisatomi O., Nakatani Y., Takeuchi K., Takahashi F., Kataoka H. Blue light-induced dimerization of monomeric Aureochrome-1 enhances its affinity for the target sequence. J. Biol. Chem. 2014;289:17379–17391. doi: 10.1074/jbc.M114.554618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grusch M., Schelch K., Riedler R., Reichhart E., Differ C., Berger W., Inglés-Prieto A., Janovjak H. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J. 2014;33:1713–1726. doi: 10.15252/embj.201387695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inglés-Prieto Á., Reichhart E., Muellner M.K., Nowak M., Nijman S.M.B., Grusch M., Janovjak H. Light-assisted small-molecule screening against protein kinases. Nat. Chem. Biol. 2015;11:952–954. doi: 10.1038/nchembio.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra D., Yang X., Moffat K. Crystal structures of aureochrome1 LOV suggest new design strategies for optogenetics. Structure. 2012;20:698–706. doi: 10.1016/j.str.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heintz U., Schlichting I. Blue light-induced LOV domain dimerization enhances the affinity of Aureochrome 1a for its target DNA sequence. Elife. 2016;5:e11860. doi: 10.7554/eLife.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herman E., Sachse M., Kroth P.G., Kottke T. Blue-light-induced unfolding of the Jα helix allows for the dimerization of aureochrome-LOV from the diatom Phaeodactylum tricornutum. Biochemistry. 2013;52:3094–3101. doi: 10.1021/bi400197u. [DOI] [PubMed] [Google Scholar]
- 23.Herman E., Kottke T. Allosterically regulated unfolding of the A′α helix exposes the dimerization site of the blue-light-sensing aureochrome-lov domain. Biochemistry. 2015;54:1484–1492. doi: 10.1021/bi501509z. [DOI] [PubMed] [Google Scholar]
- 24.Toyooka T., Hisatomi O., Takahashi F., Kataoka H., Terazima M. Photoreactions of aureochrome-1. Biophys. J. 2011;100:2801–2809. doi: 10.1016/j.bpj.2011.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sturm S., Engelken J., Gruber A., Vugrinec S., Kroth P.G., Adamska I., Lavaud J. A novel type of light-harvesting antenna protein of red algal origin in algae with secondary plastids. BMC Evol. Biol. 2013;13:159. doi: 10.1186/1471-2148-13-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sachse M., Sturm S., Gruber A., Kroth P.G. Identification and evaluation of endogenous reference genes for steady state transcript quantification by qPCR in the diatom Phaeodactlyum tricornutum with constitutive expression independent from time and ligh. J. Endocytobiosis Cell Res. 2013;24:1–7. [Google Scholar]
- 27.Pernot P., Round A., Barrett R., De Maria Antolinos A., Gobbo A., Gordon E., Huet J., Kieffer J., Lentini M., Mattenet M., et al. Upgraded ESRF BM29 beamline for SAXS on macromolecules in solution. J. Synchrotron Radiat. 2013;20:660–664. doi: 10.1107/S0909049513010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D.T., Konarev P.V., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general Amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 30.Li P., Merz K.M. Taking into account the ion-induced dipole interaction in the nonbonded model of ions. J. Chem. Theory Comput. 2014;10:289–297. doi: 10.1021/ct400751u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X., Milne J.L.S., Borgnia M.J., Rostapshov A.V., Subramaniam S., Brooks B.R. A core-weighted fitting method for docking atomic structures into low-resolution maps: Application to cryo-electron microscopy. J. Struct. Biol. 2003;141:63–76. doi: 10.1016/s1047-8477(02)00570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X., Subramaniam S., Case D.A., Wu K.W., Brooks B.R. Targeted conformational search with map-restrained self-guided Langevin dynamics: Application to flexible fitting into electron microscopic density maps. J. Struct. Biol. 2013;183:429–440. doi: 10.1016/j.jsb.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grigoryan G., Keating A.E. Structural specificity in coiled-coil interactions. Curr. Opin. Struct. Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barth A., Zscherp C. What vibrations tell about proteins. Q. Rev. Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 35.Alexandre M.T.A., Van Grondelle R., Hellingwerf K.J., Kennis J.T.M. Conformational heterogeneity and propagation of structural changes in the LOV2/Jα domain from Avena sativa phototropin 1 as recorded by temperature-dependent FTIR spectroscopy. Biophys. J. 2009;97:238–247. doi: 10.1016/j.bpj.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto A., Iwata T., Sato Y., Matsuoka D., Tokutomi S., Kandori H. Light signal transduction pathway from flavin chromophore to the Jα helix of Arabidopsis phototropin1. Biophys. J. 2009;96:2771–2778. doi: 10.1016/j.bpj.2008.12.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeifer A., Mathes T., Lu Y., Hegemann P., Kottke T. Blue light induces global and localized conformational changes in the kinase domain of full-length phototropin. Biochemistry. 2010;49:1024–1032. doi: 10.1021/bi9016044. [DOI] [PubMed] [Google Scholar]
- 38.Craig J.C., Schumacher M.A., Mansoor S.E., Farrens D.L., Brennan R.G., Goodman R.H. Consensus and Variant cAMP-regulated Enhancers Have Distinct CREB-binding Properties. J. Biol. Chem. 2001;276:11719–11728. doi: 10.1074/jbc.M010263200. [DOI] [PubMed] [Google Scholar]
- 39.Moll J.R., Acharya A., Gal J., Mir A.a, Vinson C. Magnesium is required for specific DNA binding of the CREB B-ZIP domain. Nucleic Acids Res. 2002;30:1240–1246. doi: 10.1093/nar/30.5.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu C.L., Meyer D.J., Campbell G.S., Larner a.C., Carter-Su C., Schwartz J., Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 41.Foster R., Izawa T., Chua N.H. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994;8:192–200. doi: 10.1096/fasebj.8.2.8119490. [DOI] [PubMed] [Google Scholar]
- 42.Donald R.G., Cashmore A.R. Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 1990;9:1717–1726. doi: 10.1002/j.1460-2075.1990.tb08295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcotte W.R., Russell S.H., Quatrano R.S. Abscisic acid-responsive sequences from the em gene of wheat. Plant Cell. 1989;1:969–976. doi: 10.1105/tpc.1.10.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerruth S., Ataka K., Frey D., Schlichting I., Heberle J. Aureochrome 1 illuminated: Structural changes of a transcription factor probed by molecular spectroscopy. PLoS One. 2014;9:e103307. doi: 10.1371/journal.pone.0103307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fedorova A.V., Chan I.S., Shin J.A. The GCN4 bZIP can bind to noncognate gene regulatory sequences. Biochim. Biophys. Acta. 2006;1764:1252–1259. doi: 10.1016/j.bbapap.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dousseau F., Pézolet M. Determination of the secondary structure content of proteins in aqueous solutions from their amide I and amide II infrared bands. Comparison between classical and partial least-squares methods. Biochemistry. 1990;18:8771–8779. doi: 10.1021/bi00489a038. [DOI] [PubMed] [Google Scholar]
- 47.Heimburg T., Schünemann J., Weber K., Geisler N. FTIR-spectroscopy of multistranded coiled coil proteins. Biochemistry. 1999;38:12727–12734. doi: 10.1021/bi983079h. [DOI] [PubMed] [Google Scholar]
- 48.Keller W., Konig P., Richmond T.J. Crystal-structure of a bZIP/DNA complex at 2.2 angstrom-determinants of DNA specific recognition. J. Mol. Biol. 1995;254:657–667. doi: 10.1006/jmbi.1995.0645. [DOI] [PubMed] [Google Scholar]
- 49.Svergun D., Barberato C., Koch M.H. CRYSOL - A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 50.Ellenberger T.E., Brandl C.J., Struhl K., Harrison S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: Crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 51.Schumacher M.A., Goodman R.H., Brennan R.G. The structure of a CREB bZIP·somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J. Biol. Chem. 2000;275:35242–35247. doi: 10.1074/jbc.M007293200. [DOI] [PubMed] [Google Scholar]
- 52.Cukier R.I. Simulations of temperature and salt concentration effects on bZIP, a basic region leucine zipper. J. Phys. Chem. B. 2012;116:6071–6086. doi: 10.1021/jp300836t. [DOI] [PubMed] [Google Scholar]
- 53.Derreumaux S., Fermandjian S. Bending and adaptability to proteins of the cAMP DNA-responsive element: molecular dynamics contrasted with NMR. Biophys. J. 2000;79:656–669. doi: 10.1016/S0006-3495(00)76324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrascheck M., Escher D., Mahmoudi T., Verrijzer C.P., Schaffner W., Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 2005;33:3743–3750. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su W., Jackson S., Tjian R., Echols H. DNA looping between sites for transcriptional activation: Self-association of DNA-bound Sp1. Genes Dev. 1991;5:820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 56.Jungandreas A., Costa B.S., Jakob T., Von Bergen M., Baumann S., Wilhelm C. The acclimation of Phaeodactylum tricornutum to blue and red light does not influence the photosynthetic light reaction but strongly disturbs the carbon allocation pattern. PLoS One. 2014;9:e99727. doi: 10.1371/journal.pone.0099727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.