Abstract
RecBCD is a multifunctional enzyme that possesses both helicase and nuclease activities. To gain insight into the mechanism of its helicase function, RecBCD unwinding at low adenosine triphosphate (ATP) (2–4 μM) was measured using an optical-trapping assay featuring 1 base-pair (bp) precision. Instead of uniformly sized steps, we observed forward motion convolved with rapid, large-scale (∼4 bp) variations in DNA length. We interpret this motion as conformational dynamics of the RecBCD–DNA complex in an unwinding-competent state, arising, in part, by an enzyme-induced, back-and-forth motion relative to the dsDNA that opens and closes the duplex. Five observations support this interpretation. First, these dynamics were present in the absence of ATP. Second, the onset of the dynamics was coupled to RecBCD entering into an unwinding-competent state that required a sufficiently long 5′ strand to engage the RecD helicase. Third, the dynamics were modulated by the GC-content of the dsDNA. Fourth, the dynamics were suppressed by an engineered interstrand cross-link in the dsDNA that prevented unwinding. Finally, these dynamics were suppressed by binding of a specific non-hydrolyzable ATP analog. Collectively, these observations show that during unwinding, RecBCD binds to DNA in a dynamic mode that is modulated by the nucleotide state of the ATP-binding pocket.
INTRODUCTION
RecBCD is a multifunctional enzyme that helps carry out a number of critical processes in the DNA metabolism of Escherichia coli. RecBCD executes the first step in homologous recombination for double-stranded break repair and degrades foreign double-stranded DNA (dsDNA). To initiate homologous recombination, RecBCD binds to a dsDNA break and unwinds the DNA with high processivity [>36 000 base pairs (bp)] (1) and high speed (1700 bp/s) (2). Concurrent with unwinding, RecBCD's nuclease activity nicks both of the resulting single-stranded DNA (ssDNA) strands. Upon encountering a specific sequence—the χ-site (3), which distinguishes host from foreign DNA—RecBCD promotes the loading of RecA onto the 3′ strand (4) while degrading the 5′ strand. This multi-functional enzyme's key role in repairing dsDNA breaks has spawned over two decades of research (1,4–17). This present work focuses on RecBCD's helicase activity, which is central to its biological function.
Elucidating the unwinding mechanism of RecBCD (18–20), and more generally, of helicases (18), is a long standing goal that has benefited from a multifaceted approach that includes biochemical, X-ray crystallographic, and, more recently, single-molecule studies. For instance, ensemble measurements probe RecBCD's kinetic rate over a wide range of adenosine triphosphate (ATP) concentrations (14). In addition, ensemble studies have also found that, on average, 2–3 ATPs were hydrolyzed per bp unwound (21). This elevated ATP usage arises because both RecB and RecD are SF1 helicases that can simultaneously translocate along the ssDNA. RecB moves 3′→5′, while RecD moves with the opposite polarity (5′→3′) (11,22). Crystallographic studies showed RecBCD bound to dsDNA in a zipper-like structure (12). Specifically, the RecC subunit was found to bridge both the RecB and RecD subunits, and contain a ‘pin’ structure at the ssDNA–dsDNA junction (12). Unwinding is thought to occur via a forward translocation by one of the two helicase domains along their respective ssDNA strands pulling the dsDNA against the pin and thereby promoting strand separation (12). Finally, single-molecule studies have shown that the translocation rates of RecB and RecD are different (23). Hence, the two helicases do not move in lock-step along the DNA. RecD is the lead helicase before χ-site recognition, while RecB is the lead helicase after χ-site recognition. As a result, the entire RecBCD complex moves at a slower rate after χ-site recognition (23). This structural and enzymatic complexity facilitates RecBCD's functional diversity, but potentially complicates elucidating its unwinding mechanism.
The advent of high-precision optical-trapping studies with 1-bp resolution offers a powerful tool to probe the mechanism of enzymes translocating along DNA (24–27). Researchers using such state-of-the-art instruments have identified 1-bp steps of RNA polymerase (RNAP) along DNA (28), 2.5-bp steps of the bacteriophage Φ29 DNA packaging motor (29), and 1-bp steps of NS3 (30) and XPD (31) helicases. The detection of such steps and characterization of the underlying enzymatic kinetics provide crucial information on the mechano-chemical cycle of these DNA-based molecular motors. In the simplest scheme for RecBCD, crystallographic studies suggest a uniform series of 1-bp steps where the protein–DNA complex goes through a set of well-ordered states that lead to 1 bp of unwinding per ATP consumed (20). Indeed, crystallographic studies of the RecD-like monomeric helicase RecD2 (32) and the structurally similar AddAB helicase (33) complexed with DNA in different nucleotide states also suggest a similar mechanism consisting of a well ordered set of states. More complicated models also exist. For instance, in one proposed model, RecBCD is thought to repetitively melt ∼4-6 bp of dsDNA before translocating along the resulting ssDNA (16,34). We note that high-resolution optical-trapping studies measure what is known as the mechanical step size as opposed to what is now referred to as a kinetic step size based on ensemble studies (35). Kinetic step sizes are altered by static heterogeneity in enzyme kinetics (36). More generally, the measured mechanical motion of the enzyme can convolve discrete mechanical steps with internal enzymatic dynamics that depend on the location at which the enzyme is anchored to the surface (see below).
To elucidate RecBCD's mechanism, we studied it with a surface-coupled optical-trapping assay featuring 1-bp resolution. Typically, in high-resolution studies of DNA-based molecular motors, a number of techniques are used to facilitate detection of angstrom-scale displacements (24–27). First, a substantial force (10–25 pN) is applied, improving the spatiotemporal resolution by increasing both the stiffness of the system and roll-off frequency of the trap. Second, low ATP concentrations are also used to increase the time between steps, allowing the substantial Brownian motion of the trapped bead to be averaged. In other words, higher spatial precision is achieved at the cost of temporal resolution. Both of these strategies were employed in a pioneering study by Abbondanzieri et al. that successfully resolved RNAP's 1-bp steps (28). In that study, RNAP moved against loads of >20 pN and occasionally persisted in a single state for >10 s. Finally, an additional technique to improve detection of helicase motion is a hairpin-unwinding assay, which leads to a ∼3-fold mechanical amplification of the detected signal (37) and facilitated the detection of 1-bp steps of the NS3 helicase (30).
Unfortunately, high-resolution studies of RecBCD could not benefit from application of these techniques. In previous work, we showed that the RecBCD–DNA complex can support only ∼6 pN of applied load before it backslides long distances (150–2700 bp) along its DNA substrate (9). Additionally, in the present study, we found that an actively unwinding RecBCD at low ATP does not persist in a single state between successive ATP arrivals. Rather, there were multiple types of dynamics in the protein–DNA complex, including a kinetic competition between RecBCD moving forward and backward, similar to prior optical-trapping studies of the monomeric XPD (31) and single-molecule FRET studies of the hexameric T7 helicase (38). Finally, RecBCD's nuclease cleaves the resulting ssDNA strands, making RecBCD incompatible with a hairpin-unwinding assay. As a result, there is a narrow window of experimental conditions to perform high-resolution studies on RecBCD.
Despite these challenges, the present high-resolution study of RecBCD yielded insight into its mechanism of motion along DNA. We observed forward unwinding by the enzyme, including discrete changes in DNA length, convolved with rapid, large-scale fluctuations in DNA length (∼4 bp at 10 Hz). We interpret these fluctuations as conformational dynamics of the RecBCD–DNA complex when in an unwinding-competent state arising, in part, from enzyme-induced, back-and-forth motion relative to the dsDNA that opens and closes the duplex. Five observations support this interpretation. First, these dynamics were present in the absence of ATP. Second, the onset of the dynamics was coincident with the previously determined initiation of unwinding activity, which required a sufficiently long 5′ strand to engage the RecD helicase (39). Third, the dynamics were modulated by the GC content of the DNA immediately upstream of the ss-dsDNA junction. Fourth, these dynamics were suppressed by an interstrand cross-link (40) that was placed 2 bp ahead of the junction. Finally, the dynamics were also suppressed in the presence of a specific non-hydrolyzable ATP analog, adenosine diphosphate beryllium fluoride (ADP-BeFx). Collectively, these observations are not consistent with models of RecBCD unwinding the DNA in a series of 1-bp steps via a set of discrete, well-ordered protein–DNA states. Rather, our data show a much more dynamic RecBCD–DNA complex in which RecBCD binds to DNA in both a dynamic and a more static mode that is modulated by the nucleotide state of the ATP-binding pocket.
MATERIALS AND METHODS
DNA preparation
For measuring RecBCD motion along the DNA, we generated a 7249-bp (2450 nm) blunt-ended DNA construct via polymerase chain reaction (PCR), similar to prior work (9). One of the primers contained a 5′- biotin to anchor the DNA to a streptavidin-coated polystyrene sphere (Supplementary Figure S1, see also DNA constructs in Supplementary Data). After PCR, the DNA was gel purified and the concentration was determined by using a spectrophotometer (Genova, Jenway). To determine the stability and precision of the instrument, we directly linked a 2979-bp (1007 nm) DNA molecule to the streptavidin-coated polystyrene sphere via a biotin and to the coverslip via a digoxigenin:anti-digoxigenin bond following established protocols (41).
To investigate the difference in RecBCD–DNA dynamics arising from binding to blunt-ended DNA versus RecBCD binding to DNA in a catalytically active unwinding state, we made a series of tailed DNA constructs based on prior pre-steady-state unwinding results (39). These tailed constructs contained a 3′-ssDNA tail of fixed length [6 thymidine (dT)6] and a 5′-ssDNA tail of varying length [(dT)6, (dT)8, (dT)10 or (dT)20]. Initially, blunt-ended DNA with a length of 2938 bp (993 nm) was prepared, as described above. To make tailed constructs, we first cut the 2938-bp blunt-ended DNA with BsmBI restriction endonuclease (New England BioLabs). We next annealed two oligonucleotides together to form the desired tailed geometry at one end and a BsmBI overhang at the other end. The two annealed oligonucleotides were then ligated to the 2667-bp (901-nm) long DNA using T4 DNA ligase (Invitrogen, Life Technologies) to create a tailed construct that was 2686 bp (908 nm). The resulting DNA was gel purified, and concentrations were determined using a spectrophotometer (see DNA Constructs in Supplementary Data).
We investigated the sequence specificity of the observed RecBCD–DNA dynamics by varying the GC content of the dsDNA immediately upstream of the ssDNA–dsDNA junction. Specifically, the 5 bp immediately ahead of the junction in the CG-rich or AT-rich construct were comprised of either all GC base pairs or AT base pairs, respectively.
We also prepared a tailed DNA construct with an engineered interstrand cross-link (Supplementary Figure S1) positioned in the duplex region 2 bp upstream of the ssDNA–dsDNA junction. The cross-link was made by bridging two directly opposed thymine bases, one on each tailed oligonucleotide. Synthesis required first incorporating a cross-linked phosphoramidite unit at the 5′-end of the oligonucleotide and selectively removing protecting groups to allow construction of the cross-linked duplex via conventional solid-phase oligonucleotide synthesis (42,43). This cross-linked oligonucleotide was ligated onto the longer DNA construct following the protocol outlined above.
Protein expression and purification
A biotinylated version of RecBCD with the biotin tag on RecD, the lead helicase, was prepared using established protocols (44). The unwinding rates for this modified enzyme agreed with the wild-type unwinding rates in both bulk (14) and single-molecule experiments (8,9).
Single-molecule RecBCD assay
To prepare the assay, we first constructed a flow chamber with ∼15 μl volume from coverslips patterned with an array of fiducial marks (45,46), double-sided sticky tape, 5-min epoxy and a microscope slide, similar to prior studies (9,47). Next, we incubated biotinylated bovine serum albumin [BSA, (Vector Laboratories)] at 125 μg/ml in 100 mM sodium phosphate buffer (pH = 7) within the flow chamber for 20 min at room temperature. After incubation, the flow chamber was rinsed by flowing through 400 μl of wash buffer (WB), which consisted of 25 mM Tris–HCl (pH = 7.5), 1 mM magnesium acetate, 1 mM sodium chloride, 1 mM dithiothreitol, 0.4% Tween-20 (Bio-Rad) and 3 mg/ml bovine serum albumin (concentration cited is that before filtration through a 0.2-μm filter). All subsequent buffers are based on WB, and all incubations are at room temperature, unless noted. After waiting 10 min, another 400 μl of WB was flowed through. This rinse, wait, rinse procedure with a total of 800 μl of WB is referred to as washing. We next incubated 50 μl of 10 μg/ml streptavidin (Molecular Probes, Life Technologies) for 20 min, followed by another wash. RecBCD-bio was coupled to this streptavidin-coated surface by flowing through 20 μl of 15 nM RecBCD and letting it incubate for 2 h before washing. Concurrent with the RecBCD incubation, we prepared a bead-DNA mixture by incubating streptavidin-coated beads (320-nm-diameter, Interfacial Dynamics, Life Technologies) for 1 h with biotin-labeled DNA at a 1:9 ratio of DNA to beads (450 pM beads). We then added the bead-DNA mixture to the sample chamber and let it incubate for 1 h. The unbound bead–DNA complexes were removed by a final wash.
Enzymatic reactions under the optical-trapping microscope were initiated by flowing in 50 μl of WB supplemented with 2 (or 4) μM ATP, 1.1 μM E. coli single-stranded DNA binding protein (SSB) (Promega), and an oxygen-scavenging system [6 mg/ml glucose (MP Biomedicals), 0.2 mg/ml glucose oxidase (Roche), 30 units/ml catalase]. Using this assay, the percentage of RecBCD molecules that move varied from 25–75% (see Tethered bead assay in Supplementary Data) depending on the RecBCD protein preparation. Data acquired on RecBCD bound to tailed DNA constructs were taken in WB supplemented with the oxygen-scavenging system but without SSB. To investigate the effect of various nucleotide analogs on the RecBCD dynamics, we used 2 mM ADP, 2 mM adenylyl-imidodiphosphate (AMP-PNP), 2 mM ADP-BeFx, or 2 mM ADP with 2 mM inorganic phosphate (Pi) in place of the ATP.
Optical-trapping instrument and data analysis
Our high-resolution studies of RecBCD utilized an optical-trapping instrument featuring 1-bp precision and stability. The details and calibration of this instrument have previously been published (41). Briefly, the instrument used three intensity-stabilized lasers: one high-powered trapping laser (Nd:YVO4; λ = 1064 nm, ∼150 mW at the focus) and a pair of lower-powered detector lasers (∼0.5 mW at the focus). The first detector laser (λ = 785 nm) measured the position of the bead in the optical trap (xbd, ybd, zbd) (48,49). We scattered the second laser (λ = 850 nm) off a reference mark (or fiducial mark) to measure the position of the surface (xfid, yfid, zfid) and thereby actively stabilized its position to 1 Å in three dimensions over 100 s using a three-axis piezo-electric (PZT) stage (46). The coverslip was fabricated with an array of such fiducial marks using e-beam lithography and a low-index glass [hydrogen silsesquioxane (HSQ, FOx-16, Dow Corning)] (46). The stability of this assay relied upon the excellent differential pointing stability between these lasers (26,50).
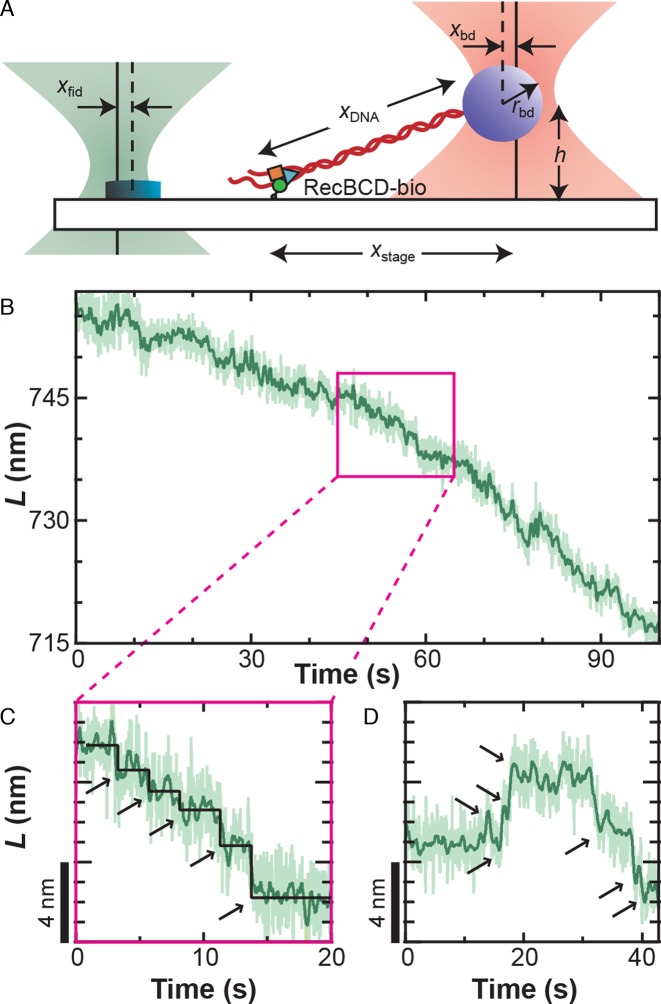
We established a well-defined geometry to study a single RecBCD molecule (Figure 1A) using previously developed techniques (9,41,47,51). Specifically, we first determined the vertical location of the surface by monitoring zbd as an optically trapped, tethered bead was brought into contact with the surface. The stage was then lowered 300 nm. We found the lateral tether-point position by performing a two-dimensional (2D) elasticity-centering procedure, which also returned the persistence length (9,47). Bead–DNA complexes anchored by multiple DNA molecules (determined by a low persistence length) were not studied. The 850-nm laser was next centered on a nearby fiducial mark using a computer-controlled PZT mirror. Finally, we stretched the DNA along the y-axis using the PZT stage to a specified force (F) (with a corresponding move of the stage-tracking 850-nm laser). For unwinding assays, we used either a force clamp (52) or a ‘pseudo-force clamp’ in which the bead position was allowed to vary from its set position (70 nm) by 10 nm in either direction, clamping the force to within 15% of the initial value. The motivation for this algorithm was to reduce rapid stage movements and avoid convolving additional noise into the measurement. In assays in the absence of ATP, the sample was stabilized using a software-based feedback loop with a 100-Hz update rate (41). We note that in both cases xfid and xbd were simultaneously measured and used to determine the contour length (L) of the DNA based on an algorithm that accounts for the 2D optical-trapping geometry (53), since the trapped bead was displaced both laterally and vertically. Finally, as discussed below, this computed L assumes that all measured changes represent a change in the dsDNA length. However, variations in this deduced L can arise from any structural dynamics that occur between the trapped bead and the anchor point of the single-molecule assay to the cover slip, including variations in the RecBCD–DNA conformation (see Figure 2E).
Figure 1.
RecBCD motion shows variably sized discrete translocations. (A) Precision optical-trapping assay measures RecBCD motion. A DNA molecule is stretched between the RecBCD helicase and an optically trapped bead. One detection laser (pink) measures the bead position (xbd), which is used to calculate DNA length. The other detection laser (green) measures the position of a fiducial mark (xfid, yfid, zfid), which is used to stabilize the surface in three dimensions. (B) DNA contour length (L) decreases as RecBCD moves forward. Traces taken at 6 pN of force, 2 μM ATP and smoothed to 10 Hz (light green) and 1 Hz (dark green). (C) Magnification of the record shown in (B) (pink box) shows forward motion fit by a step-finding algorithm (55) (black line, arrows) that finds variably sized discrete translocations. (D) Occasionally, sets of discrete backward steps (∼2–3) were observed followed by subsequent forward motion.
Figure 2.
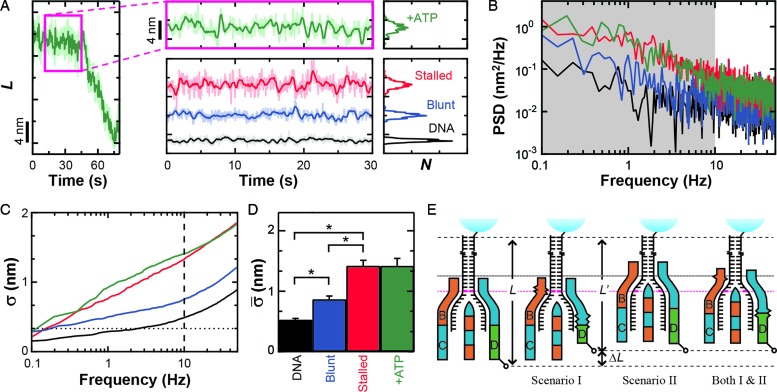
Conformational dynamics in the RecBCD–DNA complex. (A) Measurement of L for RecBCD unwinding DNA in the presence of ATP (green) shows large variations in DNA length between fitted steps. We compare this motion to RecBCD stalled on partially unwound DNA due to removal of ATP (red), RecBCD bound to blunt-ended DNA without ATP (blue) and DNA attached directly to the surface (black). Pink box is a zoom-in. Traces smoothed to 10 Hz (light) and 1 Hz (dark). Histograms of the 1 Hz data are shown at the right. Color scheme is the same in (B–D). (B) Power spectral density (PSD) of the data in (A), with the gray region highlighting the frequency range 0.1–10 Hz. (C) Integral of the PSD, which according to Parseval's Theorem is the equivalent of the standard deviation (σ). Dashed line at 10 Hz and dotted line at 1 bp. (D) Bar graph displays the mean standard deviation (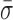 , bandwidth 0.1–10 Hz, N ≥ 5) for the conditions in (A). Error bars are the standard error in the mean. An asterisk represents a statistical significance of P < 0.05. (E) Conformational dynamics (ΔL) could be due to conformational changes within RecBCD (Scenario I), motion of RecBCD relative to the dsDNA (Scenario II) or both (Scenario I & II).
, bandwidth 0.1–10 Hz, N ≥ 5) for the conditions in (A). Error bars are the standard error in the mean. An asterisk represents a statistical significance of P < 0.05. (E) Conformational dynamics (ΔL) could be due to conformational changes within RecBCD (Scenario I), motion of RecBCD relative to the dsDNA (Scenario II) or both (Scenario I & II).
Quantifying RecBCD conformational dynamics
We developed a simple metric to quantify the conformational dynamics of RecBCD and thereby compare the dynamics under different experimental conditions. To do so, we first measured the contour length of at least five molecules (except where noted in Supplementary Table S1) at 6 pN and selected a 30-s portion of the record that exhibited the lowest long-term drift (<0.5 nm). Power spectral density (PSD) were computed from the raw data (4 or 10 kHz) and integrated from 0.1 Hz to 10 Hz. According to Parseval's theorem (54), this analysis corresponds to the standard deviation (σ) for the position trace over the same bandwidth. We then calculated the average standard deviation (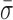 ) from multiple traces. To compute the uncertainty in this measurement, standard error of the mean (SEM) of this average standard deviation was calculated. Traces were excluded if they were <30 s (20.5 s for the unwinding traces) or if they were the incorrect length (<900 nm for the tailed-DNA substrates). Also, a small percentage (∼15%) of the records in the absence of nucleotide but in an unwinding-competent state [e.g., RecBCD bound to 5′-(dT)10] showed significantly elevated conformational dynamics (σ > 3 nm); the origin of this heterogeneity is not known, and these traces were excluded from this analysis.
) from multiple traces. To compute the uncertainty in this measurement, standard error of the mean (SEM) of this average standard deviation was calculated. Traces were excluded if they were <30 s (20.5 s for the unwinding traces) or if they were the incorrect length (<900 nm for the tailed-DNA substrates). Also, a small percentage (∼15%) of the records in the absence of nucleotide but in an unwinding-competent state [e.g., RecBCD bound to 5′-(dT)10] showed significantly elevated conformational dynamics (σ > 3 nm); the origin of this heterogeneity is not known, and these traces were excluded from this analysis.
RESULTS
High-resolution records of RecBCD motion at low ATP
To gain insight into the mechanism of RecBCD's helicase activity, RecBCD's motion was studied using a single-molecule optical-trapping assay (Figure 1A) featuring 1-bp precision and stability (41). We expected such high-resolution records to reveal a series of well-ordered steps. Contrary to this expectation, the resulting records did not show uniform steps. Rather, direct measurements of RecBCD motion at this high-spatiotemporal resolution showed forward motion of the enzyme with several interesting features (Figure 1B–D and Supplementary Figure S2). We observed discrete forward translocations (Figure 1C and D), backward steps interspersed with such forward motion (Figure 1D), and, most notably, rapid nm-scale motion significantly above the instrumental noise floor (41). We note that the average rate of forward translocation agreed with prior single-molecule assays (8,9) and ensemble results (14) at low ATP, suggesting no loss in activity due to surface-anchoring of the enzyme.
Nanometer-scale dynamics obscure high-resolution analysis of stepping
As discussed in the ‘Introduction’ section, our high-resolution studies of RecBCD were limited to an applied load of 6 pN to prevent backsliding (9). At these conditions, the instrument achieved a 1-bp (0.34 nm) precision over a relatively broad bandwidth (Δf = 0.03–2 Hz) when measuring a 556-nm long DNA molecule (41). In other words, steps longer than 0.5 s would have a signal-to-noise ratio greater than 1. To analyze steps with an average duration >0.5 s, we significantly slowed down RecBCD by lowering the ATP concentration to 2–4 μM ATP, similar to prior work (9). At ATP concentrations <2 μM, net forward progress of the enzyme was significantly slowed, presumably due to a kinetic competition between forward and backward steps (Figure 1D). Hence, our studies of RecBCD motion were limited to a narrow window of assay conditions (F ≤ 6 pN & [ATP] > 2 μM), and these limitations were enzymatic in origin. Notwithstanding this narrow window, if RecBCD existed in a static state between steps, such steps would be detectable.
Instead, the observed nm-scale variations in L masked determining a unitary step size from the data. To illustrate the effect of conformational dynamics on resolving steps, we computationally introduced discrete 1-bp steps and an even larger 4-bp step (Supplementary Data and Supplementary Figure S3A) into a 30-s record of stalled RecBCD (Figure 2A, Stalled). The resulting simulated-stepping data was analyzed with a widely used step-finding algorithm (55). As expected for steps in the presence of such large-scale dynamics, the histograms of the simulated data sets of 1 and 4 bp were indistinguishable with an average step size of 5.5 ± 2.8 bp (mean ± standard deviation) and 5.5 ± 2.3 bp, respectively (Supplementary Figure S3B). In addition, analysis of records with 1- and 4-bp steps yielded some backward steps, though not multiple consecutive backward steps exhibited by RecBCD (Figure 1D).
We applied the same stepping analysis to 15 segments of RecBCD unwinding from 8 molecules at 2 μM ATP ranging in duration from 30 to 1870 s for a total of 4286 s of data. The same analysis was also applied to seven segments of unwinding of five molecules at 4 μM ATP ranging in duration from 10 to 125 s long for a total of 430 s. The resulting step-size histograms were similar to the simulated steps in the presence of our observed nm-scale dynamics. Specifically, the peak and the width of the distributions were 5.3 ± 2.3 and 5.4 ± 2.3 bp for 2 and 4 μM ATP, respectively (Supplementary Figure S2B). The notable differences between the two sets of RecBCD data were the expected 2-fold higher velocity and a reduction in the fraction of backward steps from 36% at 2 μM ATP to 30% at 4 μM ATP. Summarizing, the measured nm-scale dynamics prevented a firm conclusion about a putative fundamental mechanical (or physical) step size. However, any mechanism for RecBCD's helicase activity would need to account for these interesting and unexpected large-scale dynamics if such dynamics arose from the RecBCD–DNA complex. Hence, we sought to determine the origin of these dynamics and, if associated with the RecBCD–DNA complex, to characterize them.
Nanometer-scale length changes arise from conformational dynamics of the RecBCD–DNA complex
The observed variations in L could arise from a number of sources beyond the unwinding of duplex DNA, including instrumental noise, conformation changes within RecBCD or changes in RecBCD's anchoring to the coverslip. We first demonstrated that the nm-scale length changes were not instrumental in origin by comparing traces of RecBCD unwinding containing particularly long pauses (>20 s) (Figure 2A, green) to the instrumental noise floor (Figure 2A, black). The long pauses used in this analysis showed similar nm-scale variations in L to those seen during the shorter pauses (<10 s) shown in Figure 1B–D. The instrumental noise floor was established by stretching a DNA molecule at 6 pN that was anchored to the coverslip through a digoxigenin:anti-digoxigenin bond, rather than through RecBCD. Control records of such taut DNA showed variations in L that were significantly smaller than those observed during pauses in enzymatic motion. To quantify the data, we computed the PSD of each trace (Figure 2B) and integrated the PSD over a useful bandwidth (0.1–10 Hz) (Figure 2B, gray shading). This bandwidth encompassed the majority of the region over which these records differed. According to Parseval's theorem (54), the integrated PSD noise is equal to the standard deviation (σ) of the record over the same bandwidth (or smoothing). For conciseness in presentation, we therefore refer to the integrated noise over the specified bandwidth as σ. As expected, Figure 2C shows that there is a significant difference in σ between records containing pauses in RecBCD motion and the control record up to 10 Hz. Over 10 Hz, σ for both records increases at approximately the same rate. We next computed the mean standard deviation (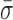 ) over the interval 0.1–10 Hz and the standard error in that mean under each condition (Figure 2D). This quantification shows that pauses in RecBCD unwinding records exhibited fluctuations in L which were almost 3-fold larger than the instrumental noise floor (1.42 ± 0.12 versus 0.53 ± 0.02 nm, respectively). Hence, the observed nm-scale fluctuations were not instrumental in origin.
) over the interval 0.1–10 Hz and the standard error in that mean under each condition (Figure 2D). This quantification shows that pauses in RecBCD unwinding records exhibited fluctuations in L which were almost 3-fold larger than the instrumental noise floor (1.42 ± 0.12 versus 0.53 ± 0.02 nm, respectively). Hence, the observed nm-scale fluctuations were not instrumental in origin.
Enzymes, in general, and molecular motors, in particular, undergo conformational changes during catalysis. Hence, a reasonable expectation for these nm-scale length changes is that they depend on ATP hydrolysis. More explicitly, there may be several rounds of futile ATP hydrolysis per successful mechanical step. To test this hypothesis, we stalled the motion of actively unwinding RecBCD molecules by removing the ATP after they had unwound a few kbp (∼1 μm) of DNA. Visual inspection of such records (Figure 2A, red; labeled as ‘Stalled’) showed no significant change in dynamics from actively unwinding records (Figure 2A, green). Quantitatively, these records of stalled complexes showed fluctuations in L of 1.42 ± 0.09 nm, identical to records in the presence of ATP (1.42 ± 0.12 nm) (Figure 2D). Hence, the observed fluctuations were not driven by ATP-hydrolysis-induced conformational changes in the RecBCD–DNA complex.
Finally, these nm-scale length changes could be associated with conformational fluctuations arising from the surface anchoring of biotinylated RecBCD to streptavidin which, in turn, is bound to biotinylated BSA adhered to the coverslip. To test for this possibility, we measured RecBCD bound to blunt-ended DNA molecules in the absence of ATP. The resulting records (Figure 2A, blue; labeled as ‘Blunt’) showed variations in L (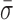 = 0.87 ± 0.05 nm) that were 64% higher than the instrumental noise floor, but 63% lower than the average standard deviation that occurs during unwinding or when the complex is stalled. Both differences were statistically distinct (P < 0.05). As a result, we conclude that the majority of the rapid, nm-scale length changes arise from conformational dynamics of the RecBCD–DNA complex, but only after the complex has unwound some DNA. These conformational dynamics could be due to a conformational change within RecBCD (Figure 2E, Scenario I), a variation in the length of the dsDNA between the enzyme and the bead (Figure 2E, Scenario II), or both. Moreover, the data show that such dynamics were not driven by ATP-hydrolysis-induced conformational changes.
= 0.87 ± 0.05 nm) that were 64% higher than the instrumental noise floor, but 63% lower than the average standard deviation that occurs during unwinding or when the complex is stalled. Both differences were statistically distinct (P < 0.05). As a result, we conclude that the majority of the rapid, nm-scale length changes arise from conformational dynamics of the RecBCD–DNA complex, but only after the complex has unwound some DNA. These conformational dynamics could be due to a conformational change within RecBCD (Figure 2E, Scenario I), a variation in the length of the dsDNA between the enzyme and the bead (Figure 2E, Scenario II), or both. Moreover, the data show that such dynamics were not driven by ATP-hydrolysis-induced conformational changes.
Onset of conformational dynamics is coupled to entering an unwinding-competent state
Prior work by Wu and Lohman has shown that RecBCD bound to blunt-ended DNA is not yet in an unwinding-competent state (39). Rather, the RecBCD–DNA complex undergoes an initiation process to enter into its unwinding-competent state (Figure 3A) (39), with an initial melting of 5–6 terminal bp as seen in earlier work (12,56). More generally, their work establishes a general kinetic scheme where RecBCD's first step upon binding to the end of a blunt-ended DNA molecule is to ‘melt out’ ∼6 bp of DNA in an ATP-independent fashion (kNP). RecBCD then undergoes 2–3 initiation steps with a rate constant kc to change into its unwinding-competent state. After initiation, RecBCD unwinds DNA in a repetitive mechano-chemical cycle with an unwinding rate constant ku such that the length of the DNA ahead of the helicase changes from position L to L - N × m where N is the number of steps and m is the step size for unwinding. Given this kinetic scheme and the aforementioned single-molecule data showing that RecBCD bound to blunt-ended DNA (Figure 2A, blue) shows less conformational dynamics than RecBCD after it had unwound DNA (Figure 2A, red and green), we hypothesized that the nm-scale conformational dynamics in the RecBCD–DNA complex only occurred after entering into an unwinding-competent state.
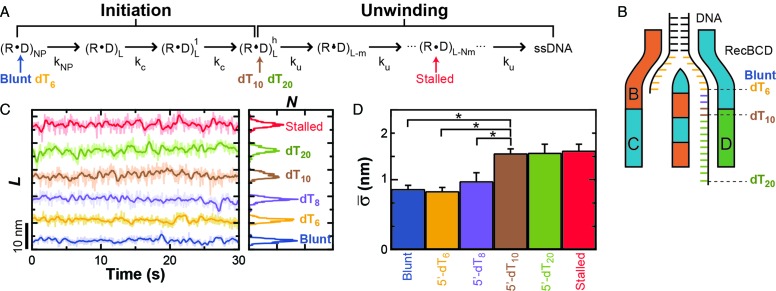
Figure 3.
Onset of conformational dynamics is coincident with entering an unwinding-competent state. (A) Kinetic diagram for RecBCD unwinding adapted from Wu and Lohman (25). Labels are defined as: RecBCD (R), DNA (D), non-productive state (NP), DNA length (L), size of step (m), number of steps (N), rate of slow isomerization (kNP), rate of initiation (kc) and rate of unwinding (ku). The unwinding-competent state is reached once the RecBCD–DNA complex is initiated. Initiation requires either ATP-dependent movement of RecBCD or a DNA substrate with a pre-existing single-stranded 5′ DNA tail of 10 thymidines, denoted 5′-(dT)10. Different tailed-DNA constructs (Blunt, dT6, dT8, dT10, dT20) positioned the RecBCD–DNA complex at different states within this kinetic diagram in the absence of added ATP (Indicated by arrows colored to indicate the length of the dT-tail). A stalled complex has unwound DNA and is thus positioned on the diagram to indicate such unwinding. (B) Cartoon of RecBCD bound to the tailed-DNA ends. (C) DNA length measurement for RecBCD bound to the following DNA substrates: blunt-ended (blue), 5′-(dT)6 (orange), 5′-(dT)8 (purple), 5′-(dT)10 (brown), 5′-(dT)20 (light green) and partially unwound (red). (D) Bar graph of the mean standard deviation for the conditions in (C).
Wu and Lohman also demonstrate that DNA substrates containing different length 5′ tails mimic different kinetic states in this pathway, including bypassing the initiation phase (39). In their experiments, they investigated constructs containing a 3′-ssDNA tail of fixed length [6 thymidine (dT)6] and 5′-ssDNA tail of varying length [(dT)6, and (dT)10]. Their work shows that RecBCD bound to a 5′-(dT)6 substrate exhibits the same initiation kinetics as RecBCD bound to blunt-ended DNA. In contrast, RecBCD bound to a 5′-(dT)10 bypasses the initiation process and is in an unwinding-competent state, presumably because the longer 5′ tail engages the RecD helicase. Importantly, these different constructs provide us a controlled method to put the RecBCD–DNA complex into the initiation or unwinding-competent state in the absence of ATP.
We next prepared an expanded set of constructs and used them to show that the onset of the nm-scale conformational dynamics is coincident with entering an unwinding-competent state. The constructs contained a 3′-(dT)6 tail and a 5′ tail of varying length [(dT)6, (dT)8, (dT)10 or (dT)20 (Figure 3B)]. We bound surface-anchored RecBCD to these constructs in the absence of ATP and measured them at 6 pN of applied load. As shown in Figure 3C, the position-versus-time records show that dynamics of the 5′-(dT)6 were similar to RecBCD bound to blunt-ended DNA (Figure 2A, Blunt) and the 5′-(dT)10 traces were similar to RecBCD stalled midway along a substrate (Figure 2A, Stalled). More quantitatively, the mean standard deviation (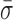 ) in L, which we will refer to as the conformational dynamics, of RecBCD bound to a 5′-(dT)6 tail was the same as RecBCD bound to blunt-ended DNA (0.84 ± 0.05 versus 0.87 ± 0.05 nm). The two longer 5′-tailed substrates [(dT)10 and (dT)20] had average conformational dynamics (1.38 ± 0.06 and 1.39 ± 0.12 nm, respectively) indistinguishable from those of the stalled RecBCD–DNA complex (1.42 ± 0.09 nm, Stalled). We also note that the unwinding assays (Figure 1) were done in the presence of SSB whereas the studies on forked substrates were not (Figure 3). Hence, the observed quantitative agreement in conformational dynamics between these different conditions was not associated with SSB-mediated dynamics. This independence of the nm-scale dynamics on the presence or absence of SSB is recapitulated in a control experiment on a stalled complex (Supplementary Figure S4). Finally, the difference in the conformational dynamics between complexes in states before and after initiation was statistically significant (P < 0.05). As a control, we also measured a 5′-(dT)10 tail directly linked to the surface (Supplementary Figure S5). This construct, as expected, had an average standard deviation (0.57 ± 0.03 nm) indistinguishable from the control record where blunt-ended DNA was directly linked to the surface (Figure 2, black). In summary, the data show that the onset of these large conformational dynamics of the RecBCD–DNA complex is coupled to RecBCD entering into an unwinding-competent state, which argues that the observed dynamics require contact between the 5′ strand and the RecD helicase.
) in L, which we will refer to as the conformational dynamics, of RecBCD bound to a 5′-(dT)6 tail was the same as RecBCD bound to blunt-ended DNA (0.84 ± 0.05 versus 0.87 ± 0.05 nm). The two longer 5′-tailed substrates [(dT)10 and (dT)20] had average conformational dynamics (1.38 ± 0.06 and 1.39 ± 0.12 nm, respectively) indistinguishable from those of the stalled RecBCD–DNA complex (1.42 ± 0.09 nm, Stalled). We also note that the unwinding assays (Figure 1) were done in the presence of SSB whereas the studies on forked substrates were not (Figure 3). Hence, the observed quantitative agreement in conformational dynamics between these different conditions was not associated with SSB-mediated dynamics. This independence of the nm-scale dynamics on the presence or absence of SSB is recapitulated in a control experiment on a stalled complex (Supplementary Figure S4). Finally, the difference in the conformational dynamics between complexes in states before and after initiation was statistically significant (P < 0.05). As a control, we also measured a 5′-(dT)10 tail directly linked to the surface (Supplementary Figure S5). This construct, as expected, had an average standard deviation (0.57 ± 0.03 nm) indistinguishable from the control record where blunt-ended DNA was directly linked to the surface (Figure 2, black). In summary, the data show that the onset of these large conformational dynamics of the RecBCD–DNA complex is coupled to RecBCD entering into an unwinding-competent state, which argues that the observed dynamics require contact between the 5′ strand and the RecD helicase.
Interestingly, the DNA substrate with a 5′-(dT)8 tail showed some quiet segments and some segments with larger-scale dynamics (Figure 3C, purple). The resulting analysis showed an intermediate level of conformational dynamics (0.98 ± 0.12 nm) as compared to RecBCD complexed with 5′-(dT)6 and 5′-(dT)10 constructs. We speculate that with a 5′-(dT)8 substrate, there may be transient contacts between the 5′-(dT)8 tail and the RecD helicase that account for this behavior.
Conformational dynamics depend on GC content of dsDNA
As discussed above, two general schemes for the observed conformational dynamics of the RecBCD–DNA complex are (i) a conformational change in the protein–DNA complex between the ssDNA–dsDNA junction and the anchor point to the coverslip (Figure 2E, Scenario I) and (ii) motion of RecBCD relative to the upstream dsDNA, resulting in unwinding of the duplex DNA (Figure 2E, Scenario II). In Scenario I, RecBCD in an unwinding competent state is not in a single static conformation but rather in a manifold of thermally accessible conformations. Indeed, a recent single-molecule study of RecBCD has argued for such conformations in the RecBCD–DNA complex in the absence of ATP (2). In that work, Liu et al. showed that removing ATP allowed the RecBCD–DNA complex to sample various kinetic states that were otherwise inaccessible at high ATP concentrations and that reintroducing ATP locked the enzyme into a new state with a different, but constant, kinetic-unwinding rate. Hence, it is possible that our observed conformational dynamics arise from the RecBCD–DNA complex sampling various conformational sub-states due to the absence of ATP. These conformational changes would lead to variations in the extension of the RecBCD between the biotinylation site on the N-terminal of RecD and the ssDNA–dsDNA junction. Such variations in protein conformation would manifest themselves as the observed variations in L, but not correspond to a change in the length of the dsDNA between the enzyme and the bead. Thus, a prediction of Scenario I is that the observed conformational dynamics should be independent of the dsDNA sequence immediately upstream of the ssDNA–dsDNA junction, since the enzyme would not be unwinding and rewinding the dsDNA.
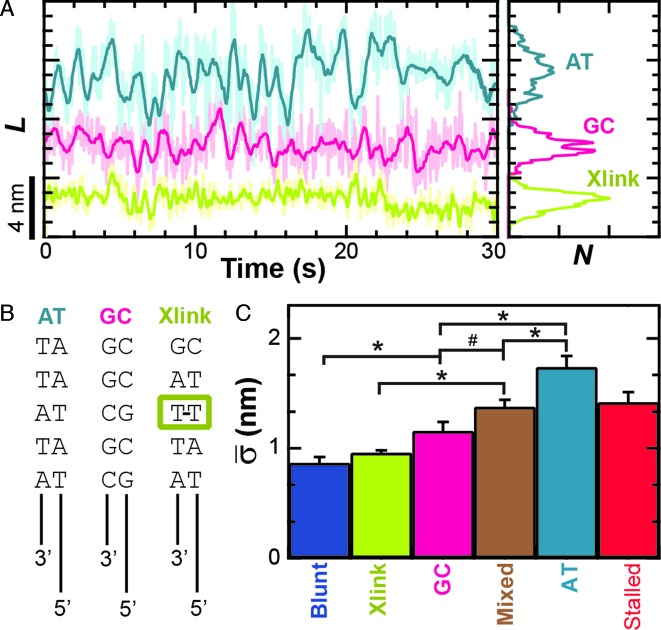
In contrast to this expectation, varying the GC content of the dsDNA sequence immediately ahead of the ssDNA–dsDNA junction affected the conformational dynamics (Figure 4A). In this experiment, we prepared the enzyme in an unwinding-competent state by using constructs that contained a 3′-(dT)6 and a 5′-(dT)10 tail. These tailed DNA constructs had either five GC or five AT base pairs immediately in front of the ssDNA–dsDNA junction. As before, we bound surface-anchored RecBCD to these constructs in the absence of ATP and measured them at 6 pN of applied load. Quantification of the average conformational dynamics (Figure 4C) showed that the AT-rich construct exhibited a 50% larger conformational dynamics (1.74 ± 0.10 nm) than the GC-rich substrate (1.16 ± 0.08 nm) and this difference was statistically significant (P < 0.05). Our earlier DNA construct with a 3′-(dT)6 and a 5′-(dT)10 tail, used in Figure 3, had alternating GC and AT base pairs for the first 12 bp ahead of the ssDNA junction and therefore represented a ‘mixed’ 50% GC sequence. The average conformational dynamics of those mixed substrates (1.38 ± 0.06 nm) was intermediate in magnitude to those on AT- and GC-rich substrates. Additionally, the sequence-dependent dynamics were maintained over a range of applied loads (F = 2, 4, 6 and 10 pN, Supplementary Figure S6). Overall, this set of experiments demonstrates that the nm-scale conformational dynamics of the RecBCD–DNA complex are dependent on the dsDNA sequence in front of the ssDNA–dsDNA junction. Hence, the conformational dynamics cannot arise from Scenario I alone, since that scenario predicts the dynamics should be sequence independent. More generally, the data clearly show that the RecBCD–DNA complex in an unwinding-competent state is dynamic with the enzyme opening and closing multiple base pairs of dsDNA in the absence of ATP, an unexpected result.
Figure 4.
Conformational dynamics are sequence dependent. (A) Measurement of L for RecBCD bound to 5′-(dT)10 tailed DNA ends show increased dynamics when the first five bp ahead of the ssDNA–dsDNA junction are all AT bp (light blue), whereas the dynamics are suppressed when those base pairs are all GC (pink). Conformational dynamics were further suppressed by an interstrand cross-link 2 bp ahead of the ssDNA–dsDNA junction (light green). (B) Schematic of constructs used in (A). (C) Bar graph of the mean standard deviation for the conditions in (A). Also shown are the cases for RecBCD bound to: blunt-ended DNA (blue), stalled DNA after partial unwinding (red) and 5′-(dT)10 DNA with a mixed upstream double-stranded DNA sequence (brown). Conditions with asterisks have P < 0.05 while the number symbol (#) corresponds to P = 0.08.
Conformational dynamics are suppressed by an interstrand DNA cross-link
To further verify this result, we tested another prediction of Scenario II. According to Scenario II, the conformational dynamics exhibited by RecBCD should be reduced if RecBCD was prevented from unwinding the duplex DNA ahead of the ssDNA–dsDNA junction. To test this prediction, we introduced a site-specific interstrand cross-link 2 bp ahead of the ssDNA–dsDNA junction in our DNA construct containing a single-stranded 3′-(dT)6 and 5′-(dT)10 tail. A bulky interstrand cross-link could, in principle, lead to steric hindrance at the point where dsDNA enters into RecBCD's zipper-like structure. Hence, we used an N3T-butylene-N3T interstrand cross-link that exhibits very little deviation in the duplex DNA structure relative to a non-cross-linked DNA duplex in circular dichroism spectroscopy measurements (42). Moreover, molecular dynamics calculations indicated a widening of the major groove of the B-DNA stem without disruption of Watson–Crick base pairing compared to a non-cross-linked base-paired thymine (40,57). We also note that RecBCD can translocate along two non-complementary ssDNA strands simultaneously (16) after initiating on dsDNA, so the entrance to RecBCD's zipper-like structure can accommodate significant variation away from ideal B-form DNA. Collectively, this set of prior work argues that our interstrand cross-link does not create a bulge in the duplex that would prevent the duplex DNA from entering the leading edge of RecBCD.
We expected the interstrand cross-link to prevent the motion of the pin in RecC relative to the upstream dsDNA. Binding efficiency to this DNA substrate in a tethered particle assay was similar to binding to the 3′-(dT)6 and 5′-(dT)10 tailed construct, but the percent of individual RecBCD molecules showing unwinding activity was reduced from 67 to 4%. We note that the remaining activity most likely arises from incomplete ligation of the cross-link to the long dsDNA rather than unwinding through the cross-link (see Supplementary Data). As shown in Figure 4, high-resolution measurements of L for RecBCD bound to the DNA substrate with the interstrand cross-link show a 44% reduction in dynamics (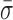 = 0.96 ± 0.02 nm) compared to the mixed DNA sequence (
= 0.96 ± 0.02 nm) compared to the mixed DNA sequence (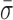 = 1.38 ± 0.06 nm). However, the level of conformational dynamics measured with the cross-link was slightly higher than RecBCD bound to blunt-ended DNA (
= 1.38 ± 0.06 nm). However, the level of conformational dynamics measured with the cross-link was slightly higher than RecBCD bound to blunt-ended DNA (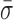 = 0.87 ± 0.05 nm) perhaps, in part, from the repetitive opening and closing of the first two base-pairs before the cross-link. Overall, these data verify the initial observation that the conformational dynamics primarily arise from enzyme-induced back-and-forth motion of RecBCD along the dsDNA in the absence of ATP hydrolysis. As a result, the dsDNA ahead of the ssDNA–dsDNA junction is being unwound and rewound by this motion. Thus, a mechanistic consequence of RecBCD's conformational dynamics is that the RecBCD–DNA complex—at least at low ATP—does not go through a repetitive, well-ordered set of states within the protein–DNA complex.
= 0.87 ± 0.05 nm) perhaps, in part, from the repetitive opening and closing of the first two base-pairs before the cross-link. Overall, these data verify the initial observation that the conformational dynamics primarily arise from enzyme-induced back-and-forth motion of RecBCD along the dsDNA in the absence of ATP hydrolysis. As a result, the dsDNA ahead of the ssDNA–dsDNA junction is being unwound and rewound by this motion. Thus, a mechanistic consequence of RecBCD's conformational dynamics is that the RecBCD–DNA complex—at least at low ATP—does not go through a repetitive, well-ordered set of states within the protein–DNA complex.
Conformational dynamics are suppressed by a specific nucleotide analog
Since the conformational dynamics are coincident with entering into an unwinding-competent state, an intriguing possibility is that they are related to RecBCD's helicase mechanism. A formally possible mechanism for helicase motion that includes substantial Brownian motion is a thermal ratchet model that has been demonstrated in model physical systems (58) (Supplementary Figure S7). Although such a model is most likely too simplistic to capture RecBCD's mechanistic complexity, it motivated us to look for a modulation of the conformational dynamics of RecBCD as a function of the chemical state of the nucleotide-binding pocket.
To do so, RecBCD's conformational dynamics were measured over a range of nucleotide conditions and analogs (Figure 5). We chose conditions that led to no net motion so we could precisely quantify the change in the conformational dynamics. As before, surface-anchored RecBCD was bound to a DNA substrate with a 3′-(dT)6 and a 5′-(dT)10 ssDNA tail and a mixed upstream sequence. We then washed through buffer with one of the following conditions: 2 mM ADP, 2 mM ADP supplemented with 2 mM Pi, 2 mM AMP-PNP or 2 mM ADP-BeFx (Supplementary Figure S8). Binding of different ATP analogs has led to different structural states when complexed with helicases (59). Hence, in the present study, testing the different ATP analogs AMP-PNP and ADP-BeFx allows access to potentially different states in the mechano-chemical cycle; the precise interpretation of the exact nature of such state requires future structural studies since AMP-PNP and ADP-BeFx can mimic either more ADP- or ATP-like states in different helicases (59). More generally, in studies of the classic molecular motor myosin, inclusion of high levels of Pi led to observed changes in force production due to population of presumed ADP + Pi bound state (60), hence we also tested ADP supplemented with Pi. The resulting records showed reduced conformational dynamics for ADP (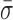 = 1.24 ± 0.14 nm), ADP and Pi (
= 1.24 ± 0.14 nm), ADP and Pi (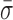 = 1.22 ± 0.07 nm) and AMP-PNP (
= 1.22 ± 0.07 nm) and AMP-PNP (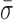 = 1.22 ± 0.19 nm) (Figure 5) when compared to stalled traces (1.42 ± 0.09 nm), but not a statistically significant one (P < 0.05). However, when ADP-BeFx was present, RecBCD's conformational dynamics did exhibit a statistically significant decrease (
= 1.22 ± 0.19 nm) (Figure 5) when compared to stalled traces (1.42 ± 0.09 nm), but not a statistically significant one (P < 0.05). However, when ADP-BeFx was present, RecBCD's conformational dynamics did exhibit a statistically significant decrease (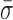 = 0.92 ± 0.05 nm). Hence, of the analogs tested, only a particular nucleotide state, ADP-BeFx, suppresses the conformational dynamics in a statistically significant manner. The interpretation of this observation within the broader mechano-chemical cycle of RecBCD requires additional studies, in particular a structure of RecBCD bound to DNA in an unwinding competent state. Finally, we note that no variation in these dynamics were observed over a range of divalent salt concentrations (Supplementary Figure S9).
= 0.92 ± 0.05 nm). Hence, of the analogs tested, only a particular nucleotide state, ADP-BeFx, suppresses the conformational dynamics in a statistically significant manner. The interpretation of this observation within the broader mechano-chemical cycle of RecBCD requires additional studies, in particular a structure of RecBCD bound to DNA in an unwinding competent state. Finally, we note that no variation in these dynamics were observed over a range of divalent salt concentrations (Supplementary Figure S9).
Figure 5.
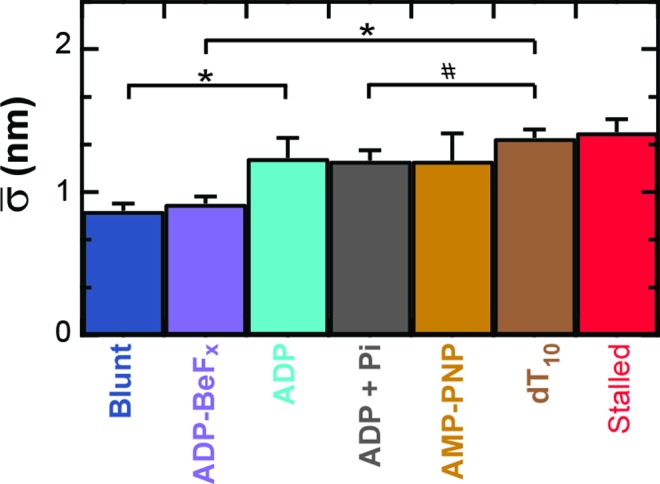
Conformational dynamics are modulated by the chemical state of the ATP-binding pocket. The conformational dynamics are significantly reduced upon introducing 2 mM ADP-BeFx (light purple), a non-hydrolyzable ATP analog, in comparison to other nucleotides: 2 mM ADP (light blue), 2 mM ADP and Pi (gray), 2 mM AMP-PNP (light brown) and in the absence of ATP (brown). For comparison, we also show RecBCD bound to blunt-ended DNA (blue) and RecBCD stalled in the middle of the DNA (red). Conditions with asterisks have P < 0.05. Number symbol (#) represents P = 0.13.
DISCUSSION
High-resolution studies of RecBCD moving along DNA yielded a striking and unexpected result. Rather than elucidating a fundamental step size for RecBCD, the data revealed nm-scale motion of the enzyme relative to the unwound DNA. Hence, at low ATP, RecBCD motion along the DNA, as monitored by a tag on RecD, is not a simple series of discrete steps. Instead, the RecBCD–DNA complex is much more dynamic, with 0.5–3 nm peak-to-peak motion occurring on 1-s time scales. This dynamic motion is due to conformational changes in the RecBCD–DNA complex that requires RecBCD to be in an unwinding-competent state, presumably via DNA engagement of the RecD helicase. Such dynamics, which occurred in the absence of ATP hydrolysis, must have been suppressed in X-ray structures, since the complex needs to be static to crystalize. More generally, our observation of suppressed RecBCD's dynamics when bound to ADP-BeFx suggests a potential path toward structural studies of RecBCD in an unwinding-competent state. If successful, such studies could then in turn provide guidance on the particular state in mechanochemical cycle captured by RecBCD bound to ADP-BeFx.
One plausible explanation for the RecBCD–DNA complex's conformational dynamics is that they arise from conformational fluctuations that occur exclusively within the protein–DNA complex without motion along the dsDNA (Figure 2E, Scenario I). To review, such variations in protein conformation would manifest themselves as the observed variations in L, because the experiment measures the length from the anchor point on RecD to the trapped bead. Indeed, such an explanation would build upon recent work by Liu et al., which postulated structural fluctuations in the RecBCD–DNA complex in the absence of ATP (2). If Scenario I alone were correct, then RecBCD's conformational dynamics should not depend on the sequence of the DNA immediately ahead of the ss-dsDNA junction (Figure 4). However, experiments yielded the opposite result: the AT-rich construct exhibited 50% larger conformational dynamics (1.74 ± 0.10 nm) than the GC-rich substrate (1.16 ± 0.08 nm), a result that is both visually distinctive (Figure 4A) and statistically significant (P < 0.05). This data, in conjunction with the suppression of such dynamics by introduction of a cross-link that is 2 bp ahead of the ssDNA–dsDNA junction conclusively demonstrates that the enzyme is rapidly opening and closing dsDNA ahead of the junction in the absence of ATP hydrolysis. Hence, RecBCD's nm-scale dynamics are inconsistent with an explanation relying exclusively upon internal conformational dynamics of the RecBCD–DNA complex (Scenario I) without motion of the complex relative to the duplex DNA (Scenario II).
An intriguing paradox in our results is that RecBCD's entering into an unwinding-competent state promotes the observed back-and-forth dynamics along the dsDNA, even while RecBCD maintains a well-defined average position along the DNA at substantial applied load in the absence of ATP. Specifically, records of RecBCD stalled in the middle of the DNA (Figure 2A, Stalled) did not show net backward motion at 6 pN. Yet, it is energetically favorable both to reform the duplex DNA and to move backward with the applied load. Additionally, from our prior work (9), we know that rapid long-range backward motion is induced if the force is raised to 8–10 pN. Hence, this lack of backward motion at 6 pN in the present study suggests that a portion of RecBCD must be tightly bound to the DNA in the absence of ATP.
The simplest explanation for RecBCD to rapidly unwind and rewind dsDNA in the absence of ATP hydrolysis is that the enzyme employs a so-called ‘active’ mechanism to open the DNA. In such an active mechanism, RecBCD does not passively wait for a thermal fluctuation to open the duplex, but promotes the opening of dsDNA by creating energetically favorable protein–DNA contacts. Such a helicase mechanism is termed ‘active’ as opposed to ‘passive’ (18,61–64).
One way RecBCD could maintain its average position along the DNA while exhibiting these nm-scale dynamics would be to use an inchworm-like mechanism. In such a mechanism, a ‘toehold’ would enable RecBCD to maintain an average position along the dsDNA, even in the presence of a substantial (6 pN) load. We speculate that such a toehold occurs via RecB's interaction with the dsDNA more than 12 bp ahead of the ssDNA–dsDNA junction (12). The observed conformational dynamics would then arise as the ssDNA–dsDNA junction (or pin structure) moves relative to such a toehold. This mechanism could accommodate both Scenario I and II (Figure 2E) and lead to the observed nm-scale conformational dynamics. Future experiments could test this hypothesis by developing and validating a RecBCD mutant with a biotin near this putative toehold. In such an assay, our hypothesis stipulates that the observed conformational dynamics and their sequence dependence would be suppressed.
However, we note that a simple version of a spring-loaded inchworm model does not describe our data. Specifically, in such a mechanism, the toehold would move forward in larger steps, followed by smaller ATP-driven steps of the trailing portion of the enzyme, akin to the model proposed for NS3 helicase (65). Based on RecBCD's structure, we would expect that RecD, the surface-anchor point to the coverslip in this assay, to be in the trailing portion of such an inchworm. The observed motion would then represent the dynamics between RecD and the toehold. Yet, our unwinding records do not show a repetitive progressive reduction of conformational dynamics followed by a discrete increase that would be expected in an inchworm-based model where RecD moves closer to the toehold and then releases.
In summary, high-resolution studies of RecBCD have revealed an unusually dynamic helicase at low ATP. Future studies will be needed to gain a complete mechanistic understanding of unwinding. One interesting set of experiments would be to measure RecBCD unwinding on an engineered DNA substrate with a repetitive sequence, similar to prior work on RNAP (66). Such experiments would enable correlating velocity with the variation in the underlying GC content (31). Finally, experiments that combine optical trapping with FRET (67,68) would enable simultaneous measurement of conformational dynamics and unwinding.
Supplementary Material
Acknowledgments
The authors thank Dr Anne Noronha for purifying the DNA containing the interstrand cross-link, Marileen Dogterom for the step-finding algorithm and Wayne Halsey for DNA preparation. Mention of commercial products is for information only; it does not imply National Institute of Standard and Technology (NIST) recommendation or endorsement. T.T.P. is a staff member of NIST's Quantum Physics Division.
Footnotes
Present address: Maasa H. Seaberg, Mayo Clinic, Rochester, MN 55905, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Burroughs Wellcome Fund Career Award in the Biomedical Sciences (to T.T.P.); National Science Foundation [Phy-0404286, Phys-1125844, IGERT to A.R.C., M.H.S.]; National Physical Science Consortium Fellowship (to A.R.C.); W.M. Keck Initiative in the RNA Sciences, National Institute of Standard and Technology, Amherst College, Ministry of Science and Technology of Taiwan [NSC 102-2114-M-002-007 to H.F.F., H.W.L.]; Natural Sciences and Engineering Research Council of Canada and Canada Research Chair Program (299384-2011 and 950-213807 to C.J.W.). Funding for open access charge: NSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bianco P.R., Brewer L.R., Corzett M., Balhorn R., Yeh Y., Kowalczykowski S.C., Baskin R.J. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 2.Liu B., Baskin R.J., Kowalczykowski S.C. DNA unwinding heterogeneity by RecBCD results from static molecules able to equilibrate. Nature. 2013;500:482–485. doi: 10.1038/nature12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixon D.A., Kowalczykowski S.C. The recombination hotspot chi is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 1993;73:87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- 4.Anderson D.G., Kowalczykowski S.C. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 5.Amundsen S.K., Neiman A.M., Thibodeaux S.M., Smith G.R. Genetic dissection of the biochemical activities of RecBCD enzyme. Genetics. 1990;126:25–40. doi: 10.1093/genetics/126.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowalczykowski S.C., Dixon D.A., Eggleston A.K., Lauder S.D., Rehrauer W.M. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu M., Souaya J., Julin D.A. The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1998;95:981–986. doi: 10.1073/pnas.95.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dohoney K.M., Gelles J. Chi-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature. 2001;409:370–374. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 9.Perkins T.T., Li H.W., Dalal R.V., Gelles J., Block S.M. Forward and reverse motion of single RecBCD molecules on DNA. Biophys. J. 2004;86:1640–1648. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianco P.R., Kowalczykowski S.C. Translocation step size and mechanism of the RecBC DNA helicase. Nature. 2000;405:368–372. doi: 10.1038/35012652. [DOI] [PubMed] [Google Scholar]
- 11.Dillingham M.S., Spies M., Kowalczykowski S.C. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 12.Singleton M.R., Dillingham M.S., Gaudier M., Kowalczykowski S.C., Wigley D.B. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 13.Lucius A.L., Vindigni A., Gregorian R., Ali J.A., Taylor A.F., Smith G.R., Lohman T.M. DNA Unwinding Step-size of E.coli RecBCD Helicase Determined from Single Turnover Chemical Quenched-flow Kinetic Studies. J. Mol. Biol. 2002;324:409–428. doi: 10.1016/s0022-2836(02)01067-7. [DOI] [PubMed] [Google Scholar]
- 14.Lucius A.L., Lohman T.M. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E.coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol. 2004;339:751–771. doi: 10.1016/j.jmb.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Spies M., Bianco P.R., Dillingham M.S., Handa N., Baskin R.J., Kowalczykowski S.C. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 16.Xie F., Wu C.G., Weiland E., Lohman T.M. Asymmetric regulation of bipolar single-stranded DNA translocation by the two motors within Escherichia coli RecBCD helicase. J. Biol. Chem. 2013;288:1055–1064. doi: 10.1074/jbc.M112.423384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung C., Li H.W. Direct observation of RecBCD helicase as single-stranded DNA translocases. J. Am. Chem. Soc. 2013;135:8920–8925. doi: 10.1021/ja401626t. [DOI] [PubMed] [Google Scholar]
- 18.Lohman T.M., Tomko E.J., Wu C.G. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 19.Dillingham M.S., Kowalczykowski S.C. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wigley D.B. Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 2013;11:9–13. doi: 10.1038/nrmicro2917. [DOI] [PubMed] [Google Scholar]
- 21.Roman L.J., Kowalczykowski S.C. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry. 1989;28:2863–2873. doi: 10.1021/bi00433a018. [DOI] [PubMed] [Google Scholar]
- 22.Taylor A.F., Smith G.R. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 23.Spies M., Amitani I., Baskin R.J., Kowalczykowski S.C. RecBCD enzyme switches lead motor subunits in response to chi recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffitt J.R., Chemla Y.R., Smith S.B., Bustamante C. Recent advances in optical tweezers. Annu. Rev. Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 25.Chemla Y.R. Revealing the base pair stepping dynamics of nucleic acid motor proteins with optical traps. Phys. Chem. Chem. Phys. 2010;12:3080–3095. doi: 10.1039/b920234j. [DOI] [PubMed] [Google Scholar]
- 26.Perkins T.T. Angstrom-precision optical traps and applications. Annu. Rev. Biophys. 2014;43:279–302. doi: 10.1146/annurev-biophys-042910-155223. [DOI] [PubMed] [Google Scholar]
- 27.Greenleaf W.J., Woodside M.T., Block S.M. High-resolution, single-molecule measurements of biomolecular motion. Annu. Rev. Biophys. Biomol. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbondanzieri E.A., Greenleaf W.J., Shaevitz J.W., Landick R., Block S.M. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffitt J.R., Chemla Y.R., Aathavan K., Grimes S., Jardine P.J., Anderson D.L., Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng W., Arunajadai S.G., Moffitt J.R., Tinoco I.J., Bustamante C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011;333:1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Z., Pugh R.A., Spies M., Chemla Y.R. Sequence-dependent base pair stepping dynamics in XPD helicase unwinding. Elife. 2013;2:e00334. doi: 10.7554/eLife.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saikrishnan K., Powell B., Cook N.J., Webb M.R., Wigley D.B. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 33.Krajewski W.W., Fu X., Wilkinson M., Cronin N.B., Dillingham M.S., Wigley D.B. Structural basis for translocation by AddAB helicase-nuclease and its arrest at chi sites. Nature. 2014;508:416–419. doi: 10.1038/nature13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu C.G., Xie F., Lohman T.M. The primary and secondary translocase activities within E. coli RecBC helicase are tightly coupled to ATP hydrolysis by the RecB motor. J. Mol. Biol. 2012;423:303–314. doi: 10.1016/j.jmb.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ha T., Kozlov A.G., Lohman T.M. Single-molecule views of protein movement on single-stranded DNA. Annu. Rev. Biophys. 2012;41:295–319. doi: 10.1146/annurev-biophys-042910-155351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J., Myong S., Niedziela-Majka A., Lee K.S., Yu J., Lohman T.M., Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dumont S., Cheng W., Serebrov V., Beran R.K., Tinoco I.J., Pyle A.M., Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Syed S., Pandey M., Patel S.S., Ha T. Single-molecule fluorescence reveals the unwinding stepping mechanism of replicative helicase. Cell Rep. 2014;6:1037–1045. doi: 10.1016/j.celrep.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C.G., Lohman T.M. Influence of DNA end structure on the mechanism of initiation of DNA unwinding by the Escherichia coli RecBCD and RecBC helicases. J. Mol. Biol. 2008;382:312–326. doi: 10.1016/j.jmb.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.da Silva M.W., Bierbryer R.G., Wilds C.J., Noronha A.M., Colvin O.M., Miller P.S., Gamcsik M.P. Intrastrand base-stacking buttresses widening of major groove in interstrand cross-linked B-DNA. Bioorg. Med. Chem. 2005;13:4580–4587. doi: 10.1016/j.bmc.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 41.Carter A.R., Seol Y., Perkins T.T. Precision surface-coupled optical-trapping assays with 1 base-pair resolution. Biophys. J. 2009;96:2926–2934. doi: 10.1016/j.bpj.2008.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilds C.J., Noronha A.M., Robidoux S., Miller P.S. Mispair-aligned N3T-alkyl-N3T interstrand cross-linked DNA: synthesis and characterization of duplexes with interstrand cross-links of variable lengths. J. Am. Chem. Soc. 2004;126:9257–9265. doi: 10.1021/ja0498540. [DOI] [PubMed] [Google Scholar]
- 43.Sun G., Noronha A., Wilds C. Preparation of N3-thymidine-butylene-N3-thymidine interstrand cross-linked DNA via an orthogonal deprotection strategy. Tetrahedron. 2012;68:7787–7793. [Google Scholar]
- 44.Fan H.F., Li H.W. Studying RecBCD helicase translocation along Chi-DNA using tethered particle motion with a stretching force. Biophys. J. 2009;96:1875–1883. doi: 10.1016/j.bpj.2008.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter A.R., King G.M., Perkins T.T. Back-scattered detection provides atomic-scale localization precision, stability, and registration in 3D. Opt. Express. 2007;15:13434–13445. doi: 10.1364/oe.15.013434. [DOI] [PubMed] [Google Scholar]
- 46.Carter A.R., King G.M., Ulrich T.A., Halsey W., Alchenberger D., Perkins T.T. Stabilization of an optical microscope to 0.1 nm in three dimensions. Appl. Opt. 2007;46:421–427. doi: 10.1364/ao.46.000421. [DOI] [PubMed] [Google Scholar]
- 47.Perkins T.T., Dalal R.V., Mitsis P.G., Block S.M. Sequence-dependent pausing of single lambda exonuclease molecules. Science. 2003;301:1914–1918. doi: 10.1126/science.1088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Visscher K., Gross S.P., Block S.M. Construction of multiple-beam optical traps with nanometer-resolution position sensing. IEEE J. Sel. Top. Quant. Electron. 1996;2:1066–1076. [Google Scholar]
- 49.Pralle A., Prummer M., Florin E.-L., Stelzer E.H.K., Horber J.K.H. Three-dimensional high-resolution particle tracking for optical tweezers by forward scattered light. Microsc. Res. Tech. 1999;44:378–386. doi: 10.1002/(SICI)1097-0029(19990301)44:5<378::AID-JEMT10>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 50.Nugent-Glandorf L., Perkins T.T. Measuring 0.1-nm motion in 1 ms in an optical microscope with differential back-focal-plane detection. Opt. Lett. 2004;29:2611–2613. doi: 10.1364/ol.29.002611. [DOI] [PubMed] [Google Scholar]
- 51.Perkins T.T., Li H.W. Single-molecule studies of RecBCD. Methods Mol. Biol. 2010;587:155–172. doi: 10.1007/978-1-60327-355-8_11. [DOI] [PubMed] [Google Scholar]
- 52.Visscher K., Schnitzer M.J., Block S.M. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 53.Wang M.D., Yin H., Landick R., Gelles J., Block S.M. Stretching DNA with optical tweezers. Biophys. J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laws E.A. Mathematical Methods for Oceanographers: an Introduction. NY: John Wiley & Sons; 1997. [Google Scholar]
- 55.Kerssemakers J.W., Munteanu E.L., Laan L., Noetzel T.L., Janson M.E., Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 56.Farah J.A., Smith G.R. The RecBCD enzyme initiation complex for DNA unwinding: enzyme positioning and DNA opening. J. Mol. Biol. 1997;272:699–715. doi: 10.1006/jmbi.1997.1259. [DOI] [PubMed] [Google Scholar]
- 57.da Silva M.W., Wilds C.J., Noronha A.M., Colvin O.M., Miller P.S., Gamcsik M.P. Accommodation of mispair aligned N3T-ethyl-N3T DNA interstrand cross link. Biochemistry. 2004;43:12549–12554. doi: 10.1021/bi0486435. [DOI] [PubMed] [Google Scholar]
- 58.Faucheux L.P., Bourdieu L.S., Kaplan P.D., Libchaber A.J. Optical thermal ratchet. Phys. Rev. Lett. 1995;74:1504–1507. doi: 10.1103/PhysRevLett.74.1504. [DOI] [PubMed] [Google Scholar]
- 59.Thomsen N.D., Berger J.M. Crystallization and X-ray structure determination of an RNA-dependent hexameric helicase. Methods Enzymol. 2012;511:171–190. doi: 10.1016/B978-0-12-396546-2.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Debold E.P., Walcott S., Woodward M., Turner M.A. Direct observation of phosphate inhibiting the force-generating capacity of a miniensemble of myosin molecules. Biophys. J. 2013;105:2374–2384. doi: 10.1016/j.bpj.2013.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lohman T.M. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol. Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 62.Amaratunga M., Lohman T.M. Escherichia coli rep helicase unwinds DNA by an active mechanism. Biochemistry. 1993;32:6815–6820. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- 63.Betterton M.D., Julicher F. Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys. Rev. E. 2005;71:011904. doi: 10.1103/PhysRevE.71.011904. [DOI] [PubMed] [Google Scholar]
- 64.Manosas M., Xi X.G., Bensimon D., Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38:5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Myong S., Bruno M.M., Pyle A.M., Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herbert K.M., La Porta A., Wong B.J., Mooney R.A., Neuman K.C., Landick R., Block S.M. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Comstock M.J., Ha T., Chemla Y.R. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat. Methods. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Comstock M.J., Whitley K.D., Jia H., Sokoloski J., Lohman T.M., Ha T., Chemla Y.R. Protein structure. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science. 2015;348:352–354. doi: 10.1126/science.aaa0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.