Abstract
A set of 12 fluorogenic glycine riboswitches with different thermodynamic and kinetic response properties was engineered. For the design of functional riboswitches, a three-part RNA approach was applied based on the idea of linking a RNA sensor, transmitter and actuator part together. For the RNA sensor and actuator part, we used the tandem glycine aptamer structure from Bacillus subtillis, and fluorogenic aptamer Spinach, respectively. To achieve optimal signal transduction from the sensor to the actuator, a riboswitch library with variable transmitter was screened with a microfluidic large-scale integration chip. This allowed us to establish the complete thermodynamic binding profiles of the riboswitch library. Glycine dissociation constants of the 12 strong fluorescence response riboswitches varied between 99.7 and 570 μM. Furthermore, the kinetic glycine binding (kon), and dissociation (koff) rates, and corresponding energy barriers of the 10 strongest fluorescence response riboswitches were determined with the same chip platform. kon and koff were in the order of 10−3s−1 and 10−2s−1, respectively. Conclusively, we demonstrate that systematic screening of synthetic and natural linked RNA parts with microfluidic chip technology is an effective approach to rapidly generate fluorogenic metabolite riboswitches with a broad range of biophysical response properties.
INTRODUCTION
Natural riboswitches are structural RNA motifs controlling gene expression by binding metabolites without the need for protein factors (1–7). These functional RNA sequences are primarily found in the 5′ untranslated region of bacterial messenger RNAs (3,5). Since their discovery, riboswitches were attractive targets for engineering sensor systems to detect their corresponding metabolites (8,9). While natural riboswitches have evolved with high specificity and functional selectivity, it remains difficult to engineer synthetic counterparts.
With the recent development of the fluorogenic RNA aptamer Spinach (10), riboswitch designs gained in popularity due to the possibility to engineer fast responding in vivo fluorescence sensors rather than slower protein expression-based readout systems. Upon binding to the fluorogen 3,4-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), the Spinach RNA resembles the green fluorescence protein in fluorescence properties (10). By functional integration of Spinach into natural riboswitches, fluorogenic metabolic sensors have been successfully constructed for sensing thiamine pyrophosphate (TPP), S-Adenosyl methionine (SAM) and guanine (11). Within these three engineered fluorogenic metabolic riboswitches the ligand induced conformational change of the riboswitch is exploited to control the Spinach fluorescence properties. Thereby, the affinity and selectively of the parental riboswitch is retained. The general strategy to link RNA sequence-structure with functional relationships (12) has evolved from early allosteric metabolic sensors with Spinach (13,14). Within an allosteric metabolic sensor only parts rather than whole riboswitch functions including switching state mechanism were used to generate a fluorogenic read out signal with Spinach.
In general, the sequences of metabolic riboswitches can be divided into three functional parts; i.e. a sensor, an actuator and a transmitter part (15). In natural riboswitches, the sensor is mostly an aptamer, which recognizes the metabolite upon direct binding. The actuator is a functional RNA sequence controlling a process or event. A common actuator in prokaryotic organisms is, for example, a gene-expression actuator in the form of a transcriptional terminator sequence (16). The transmitter is a structural RNA element sending or converting signals between the sensor and actuator. The transmitter and actuator are also referred to as the expression platform of a riboswitch.
RNA secondary structural information for shorter actuator parts can be robustly calculated based on thermodynamic folding probabilities (17), whereas the functions of the RNA parts are either taken from natural, characterized riboswitches or designed with targeted mutational approaches in conjugation with high throughput screening technologies (15,18). For longer RNA sequences, secondary structure calculations lose their accuracy. Furthermore, tertiary structure contributions to the RNA function are mostly inaccessible by computational calculations. Thus, for the assembling of synthetic riboswitches, iterative screening rounds for RNA function are still required. Moreover, the transmitter of a riboswitch is shared between the sensor and actuator, and thus they cannot be designed as isolated modules. Design rules for the transmitter can reduce the screening space for functional fluorogenic riboswitches; however, experimental verification remains a necessity. In the first systematic study, the construction of the fluorogenic riboswitch with specificity for TPP demonstrated that the transmitter controls the sensor signal intensity and response kinetics, which are two of the most important sensor parameters (11). Design of the transmitter is thus a central engineering challenge.
Herein, a fluorogenic glycine sensor was engineered by linking the natural glycine sensor from the glycine riboswitch out of Bacillus subtillis (5) with the green fluorogenic Spinach (19). With this we aimed to exploit the riboswitch mechanism to control the fluorescence state of Spinach by maintaining the function of the sensor part. A synthetic DNA library encoding for 94 glycine riboswitches with different transmitter was constructed rapidly with patch oligonucleotide synthesis (POS) (20). All glycine sensor constructs were synthesized and functionally, thermodynamically and kinetically characterized in parallel on a microfluidic screening chip platform. The functional transmitter landscape of the sensor library was recorded using the microfluidic large-scale integration chip platform (21), which is based on polydimethylsiloxane (PDMS). This resulted in 12 fluorogenic glycine riboswitches that could be identified with high fluorescence response signals, but variable dissociation constants for glycine. Furthermore, the systematic kinetic characterization of 10 out of the 12 top selected fluorogenic riboswitches revealed design rules for RNA transmitter with faster fluorescence metabolite responses.
MATERIALS AND METHODS
Construction of the screening library
The fluorogenic glycine riboswitches were constructed from four oligonucleotides O1–O4 (Sigma Aldrich, Germany). O1 contained a T7 promoter and a DNA-pull down anchor sequence. O2 and O3 encoded the natural glycine sensor aptamer 1 and 2 from B. subtilis, respectively. O4 carried the variable transmitter and a previous functionally optimized Spinach actuator sequence. All oligonucleotides in Supplementary Table S1 were ordered from Sigma Aldrich (Taufkirchen, Germany). For the linkage of the complete riboswitch sequences, O1-O4 were combined by POS. In a POS reaction (see Supplementary Figure S1), O1–O4 were mixed with connectors C1–C3 at an equal concentration of 1 μM in Taq DNA Ligase Buffer (New England Biolabs, USA). Only O4 and C3 were specific for each riboswitch. Upon addition of 40 U Taq DNA Ligase per 25 μl reaction vessel, the reverse strand of the fluorogenic glycine riboswitch were ligated together. Constructs of the negative control for all 94 riboswitches were produced in a single POS reaction by omitting the fragments O2 and O3. Purified polymerase chain reaction samples were dissolved in 1x SSC buffer and 1% w/v BSA at a DNA concentration of ∼90 ng/ml and spotted onto an epoxy coated microscope slide (Cel-1, USA) with a microarray contact printer (OmniGrid).
Control experiments outside the microfluidic chip platform
Fluorogenic glycine riboswitches and negative control samples were in vitro transcribed with HiScribe T7 Qiuck High Yield RNA Synthesis Kit (New England Biolabs, USA). RNA products were aliquoted and dissolved in detection buffer (see below). The fluorescent signals of the riboswitches were measured with a Nanodrop 3300 (Thermo Fisher Scientific, USA). The relative fluorescence units (RFU) of all riboswitches were normalized to the RFU in absence of glycine.
Microfluidic chip
The production of microfluidic chips and the experimental workflow for in vitro expression and miniaturized pull-down of RNAs to local detection areas on chip is described in detail elsewhere (19,22) and in the Supplementary Data. In short, the on-chip miniaturized pull-down assay is built upon the epoxy coated glass surface by sequential flush operations. First, NeutrAvidin was deposited locally within a circular region of each micro chamber with a diameter of 80 μm with the help of an integrated PDMS membrane valve. Next, a biotin-labeled capture oligonucleotide was bound to the NeutrAvidin activated surface. After in vitro transcription of the spotted DNA, the fluorogenic glycine riboswitches were captured free of cross contamination from the neighboring unit cells on the pull-down areas. For each positive and negative riboswitch, four repeats on one chip were synthesized and used to calculate standard errors. The functional fluorogenic riboswitch array was then used to perform thermodynamic and kinetic glycine binding studies.
Determination of the apparent glycine dissociation constants on chip
For obtaining the apparent glycine dissociation constants, and absolute fluorescence change of the fluorogenic riboswitch, 15 different concentrations of glycine (0M; 100 nM–100 mM) dissolved in detection buffer were sequentially introduced into the chip. In all experiments, DFHBI-1T ((Z)-4-(3,5-difluoro-4-hydroxybenzylidene)-2- methyl-1-(2,2,2-trifluoroethyl)-1H-imidazol-5(4H)-one)) was used at a concentration of 20 μM. DFHBI-1T is the successor fluorogen of DFHBI with increased fluorescence brightness when bound to Spinach (23). The detection buffer contained 20 μM DFHBI-1T, 24 μM DMSO, 125 mM KCL, 20 mM MgCl2, 50 mM Tris-NaOH pH 8.31. The temperature of the chip was controlled during the whole experiments including the imaging steps with an indium tin oxide (ITO) heated glass slide from below (Tokai Hit, Japan) and with on-stage microscope cell incubator. Between each titration step, the chip was incubated for 10 min at 50°C for refolding. The Spinach fluorescence signals of all samples (excitation 470/20 nm, emission 515/10 nm) were recorded with 10 × Plan-Apochromat on a Zeiss-Axio Observer (Zeiss, Germany) microscope. After the last titration step, the amount of RNA on the pull-down areas was measured with a detection probe against the constant region of the riboswitch 3′ ends. The detection probe was labeled with an ROX fluorophore (excitation 555/20 nm, emission 590/20 nm) and dissolved in 1× phosphate buffered saline.
Determination of the apparent glycine reaction rates on chip
In order to obtain the apparent reaction rates for glycine binding and dissociation from the fluorogenic riboswitches at different temperatures, we measured the real time fluorescence changes of Spinach/DFHBI-1T on chip. The same microfluidic chip set up for the thermodynamic measurements was used at an image frame rate between 1 and 2.5 per miniature. In one real time experiment, a subsection of 26 pull-down areas of the chip were monitored in parallel. Complete buffer exchange for 26 in serial arranged pull-down areas on chip required 5 s at a pressure of 2 psi and were thus more than one order of magnitude faster than the image acquisition rate. Imaging of the forward reaction was started upon introduction of 10 mM glycine dissolved in detection buffer into the chip. Subsequently, the reverse reaction was measured on the same pull-down areas by exchanging the glycine solution with detection buffer without glycine. The amount of RNA on the pull-down areas was determined at the end of the experiment as described above.
Image analysis
The acquired fluorescence images were processed with MATLAB (Mathworks, USA). First, the foreground pixels of the pull-down areas were segmented using an additionally acquired bright field image at each time or titration step. Within the bright field image, the edges of the pneumatic membranes valves used to build up the surface chemistry of the pull-down areas (Supplementary Figure S2). For correction of the local background the pixel positions of an annulus surrounding the pull-down areas were determined. Therefore, an annulus with an inner and outer radius of 80 and 150 μm, respectively, was defined. The foreground (circle with 40 μm radius) and background pixel positions were then used to extract the median fluorescence signal of Spinach/DFHBI-1T, Si, the fluorescence background, Sb and the ROX probe signal Sp of all pull-down areas. The Spinach/DFHBI-1T signals of the glycine riboswitches, SR, were background corrected and normalized to the total amount of RNA on the pull-down areas as SR = (Si-Si,b)/(Sp-Sp,b).
Thermodynamic model
For obtaining the apparent glycine dissociation constants from the glycine riboswitch the following two-state binding model was used,
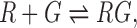 |
(1) |
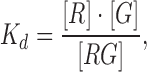 |
(2) |
where R, RG, G denote the concentration of the riboswitch in the unbound glycine state, the riboswitch in the bound glycine state and glycine, respectively. The fluorescence signal obtained from the pull-down areas on the microfluidic chip correspond directly to the concentration of R and thus the titration curves can be fitted following the equation,
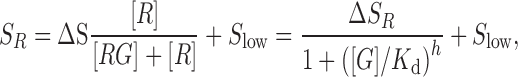 |
(3) |
where SR is the fluorescence signal, ΔS is the fluorescence signal change, Slow the fluorescence signal remaining at the end of the titration at 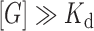 , and h the hill coefficient.
, and h the hill coefficient.
Kinetic model
For obtaining the apparent rate constants of glycine binding and dissociation to the fluorogenic glycine riboswitch, a first order reaction model was used. Thereby, the apparent rate constant for glycine binding 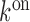 is defined by,
is defined by,
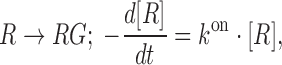 |
(4) |
and the apparent rate constant for glycine dissociation, 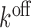 by,
by,
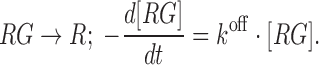 |
(5) |
For fitting the fluorescence time traces we used the solution for the first order differential Equations (4) and (5), which are 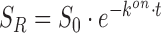 and
and 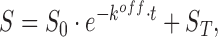 where the concentration of RG was replaced in Equation (5) with the relation
where the concentration of RG was replaced in Equation (5) with the relation 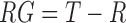 . T is the total immobilized RNA on the pull-down area obtained with a probe molecule against the 3′ end of the riboswitch. The apparent activation energies, Ea, for the glycine binding and dissociation reaction were derived from the Arrhenius equation.
. T is the total immobilized RNA on the pull-down area obtained with a probe molecule against the 3′ end of the riboswitch. The apparent activation energies, Ea, for the glycine binding and dissociation reaction were derived from the Arrhenius equation.
RESULTS
Design of the fluorogenic glycine riboswitch
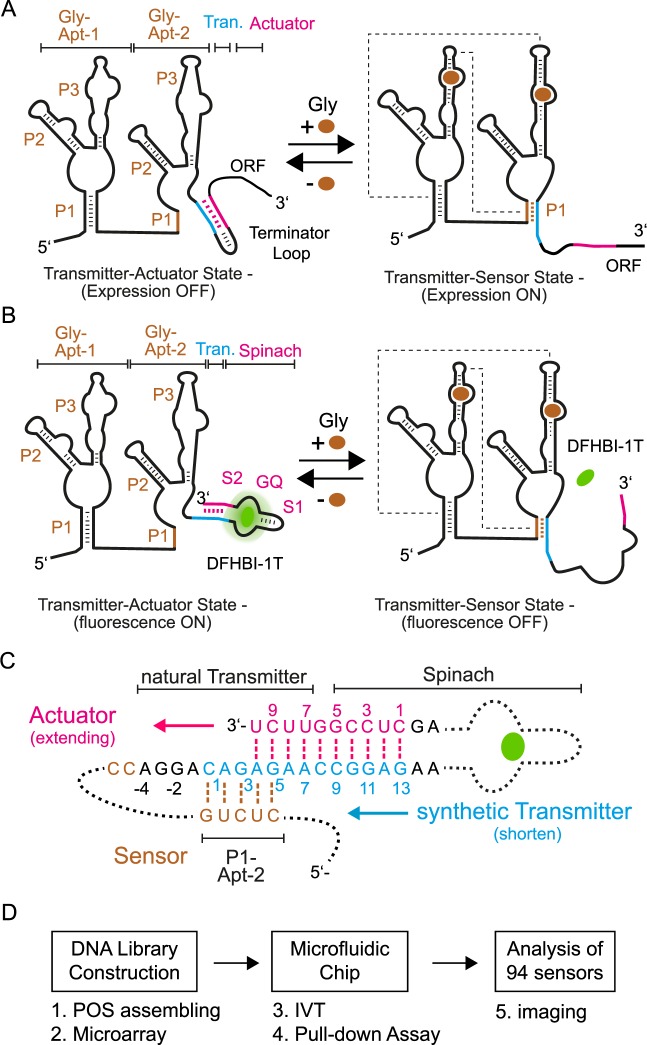
The conserved natural glycine riboswitch of B. subtillis contains two glycine aptamers (Gly-Apt-1 and 2) with a total length of 193 nucleotides (nt), followed by a short transmitter sequence of 11 nt and an RNA stem-loop acting as an actuator of transcriptional inhibition at the 3′ end (37 nt) (5). The two sensor aptamers each bind a separate molecule of glycine with reported dissociation constants between 2 and 30 μM (5,24–26). Each single glycine aptamer exhibited three RNA stems (P1–P3) (Figure 1A). While it is evident that two asymmetrical sensor aptamers dimerize upon glycine binding, the cooperativity of the glycine binding remains controversial (25,27). The transmitter RNA strand is a shared element of the P1 stem of Gly-Apt-2 and the stem-loop of the actuator. In the unbound glycine state, the transmitter participates in the 11 nt long stem-loop of the actuator; whereas in the bound glycine state the transmitter sequence participates in the formation of the 5 nt long P1 stem of Gly-Apt-2 (5). It is thought that the 5-nt long P1 stem is thermodynamically favored over the 11-nt stem-loop due to tertiary folding energy released by dimerization of the two glycine aptamers. The crystal structure of the glycine aptamer dimers revealed an extensive network of inter-aptamer interactions between the P1 stem nucleotides of one aptamer and the P3 of the other (28,29).
Figure 1.
Fluorogenic glycine riboswitch design and engineering approach. (A) Two state mechanism of the natural glycine riboswitch. In absence of glycine the transmitter (blue) hybridizes with the actuator (pink) and forms a transcription terminator loop, which decreases the expression of the following open reading frame (ORF). Upon binding of glycine to the sensor (brown), the two glycine aptamers dimerize. Dotted lines schematically indicate tertiary interactions between the two glycine aptamers. Consequently, the transmitter shifts its binding preference from the actuator strand toward the P1 stem of the Gly-Apt-2 and the transcription terminator loop is released. (B) Transfer of the natural two state folding mechanism of the glycine riboswitch to generate a fluorogenic readout signal. Here, glycine binding to the riboswitch shifts the transmitter binding preference from the Spinach actuator toward the P1 stem of the Gly-Apt-2. Thereby, the fluorogenic compound of Spinach (DFHBI-1T, green) is released. Stems of the two glycine aptamers (Gly-Apt-1 and 2) are labeled with P1-P3, whereas the stem-loop and G-quadruplex elements of the actuator are S1 and GQ, respectively. (C) The riboswitch library contained constructs with variable transmitter and counteracting actuator strands. The natural transmitter and stem sequences (S2) of Spinach are indicated. The nucleotide (nt) numbering starts for the transmitter sequence with the first nt overlap to the P1 stem of the sensor, whereas for the actuator strand it begins with the first complementary nt to the transmitter. (D) Workflow for functional screening of fluorogenic riboswitches with microfluidics (see also Supplementary Data).
To engineer a fluorogenic glycine riboswitch, the actuator part of the natural riboswitch was replaced with the Spinach sequence. The aim was to exploit the riboswitch mechanism to control the fluorescence state of Spinach by maintaining the function of the sensor part. Fluorogenic riboswitches can be designed either in a turn-on or -off configuration (30). In a turn-off configuration, the Spinach actuator shows a fluorescence signal in the unbound glycine state of the riboswitch. This relates thermodynamically to the requirement that the hybridization energy (ΔGH) between the transmitter and actuator strand has to be larger than the interactions between the transmitter and sensor strand. Upon increasing the glycine concentration, the riboswitch turns into the bound glycine state and loses its fluorescence due to destabilization of the Spinach RNA structure and lowered binding affinity for its fluorogenic ligand DFHBI-1T (Figure 1A). In the turn-on configuration, the inverted response is generated. Both fluorogenic riboswitch types are defined by the transmitter. In this work, the turn-off type was developed. For the Spinach aptamer, we used the minimal (24–2 min) (10) functional, and in vitro signal improved sequence (19), which included the central G-quadruplex (GQ), a stem (S2) and stem-loop (S1) element. All sequences are given in the Supplementary Data.
The hybridization region for the transmitter to the Spinach aptamer was designed to complement nucleotides of the S2 stem element. From the complete thermodynamic binding and stability landscape of Spinach, it can be deduced that a short length and single nucleotide point mutations within the S2 stem strongly modulate the binding equilibrium of DFHBI-1T and Spinach (19). On the other end, the hybridization region of the transmitter to the sensor of the natural Glycine riboswitch sequence was maintained. In the starting design, the synthetic transmitter sequence had a length of 13 nt, where 8 nt belonged to the natural transmitter and 5 nt to the minimal Spinach actuator. The transmitter design is shown in Figure 1B. To screen for optimal thermodynamic binding equilibrium between the transmitter-sensor state and the transmitter-actuator state, a fluorogenic glycine riboswitch sequence library was generated. Within the library the length of the transmitter and complementary actuator strand were systematically changed. The transmitter was shortened from 13 to 1 nt. In order to maintain a steady decrease of the hybridization energy between a transmitter/actuator pair with the same length (∼2 kcal/mol), the sequence of the transmitter was changed within the length series. Notably, the shortest transmitter still shared one Watson–Crick base pair with the stem of the sensor. For each transmitter length, the complementary actuator strand was extended to increase hybridization energy of the transmitter-actuator, thereby stabilizing the binding site of Spinach aptamer.
Library construction and microfluidic screening
The riboswitch library contained 94 constructs, where the corresponding linear cDNAs were assembled through POS (20). In a POS reaction, four similarly long oligonucleotides were ligated to a single DNA strand by bridging with oligonucleotide connectors. The first oligonucleotide from the 5′ end contained a T7 promoter and a capture sequence for pulling down the riboswitch after in vitro transcription, whereas the second and third fragment contained Gly-Apt-1 and Gly-Apt-2, respectively. The fourth oligonucleotide encoded the transmitter and the actuator sequence and was the only varying fragment. In the following all fluorogenic glycine riboswitches are named according to their transmitter and actuator length, e.g. the fluorogenic glycine riboswitch with a Transmitter and Actuator length of 8 and 7, respectively is named T8A7. Sanger sequencing positively validated 10% of all linked and amplified POS constructs. Additionally, a negative control was generated for each riboswitch, in which the tandem glycine aptamers were omitted (see Supplementary Figure S2). Combinatorial synthesis of the DNA library reduced the cost and error rates associated with long oligonucleotide synthesis. The entire library was spotted as a DNA microarray onto a microscope slide and bound to a microfluidic chip for screening of functional RNAs.
The chip combined the parallel functional steps of: (i) in vitro transcription of RNAs and (ii) a miniaturized pull-down assay for measuring the interaction between RNAs and small molecules in 640 separated micro chambers. The layout of the PDMS chip and the fluidic control operations has been described before (22,31) and is given in detail in the Supplementary Data.
Functional transmitter-actuator landscape
For the functional screening, we measured the glycine binding curves of all fluorogenic riboswitches in parallel on chip. Curves were generated by the stepwise increase in the glycine concentration over the pull-down areas from 0.1 μM to 100 mM at 37°C. After each titration step, the Spinach/DFHBI-1T fluorescence from each pull-down area was recorded. Signals of the riboswitches were normalized to the total amount of RNA on the pull-down areas. Subsequently, the normalized signal was corrected by the negative control riboswitch lacking a sensor (‘Materials and Methods’ section). The concentration of the DFHBI-1T was left constant at 20 μM, which is higher than the saturating concentration for wild-type Spinach.
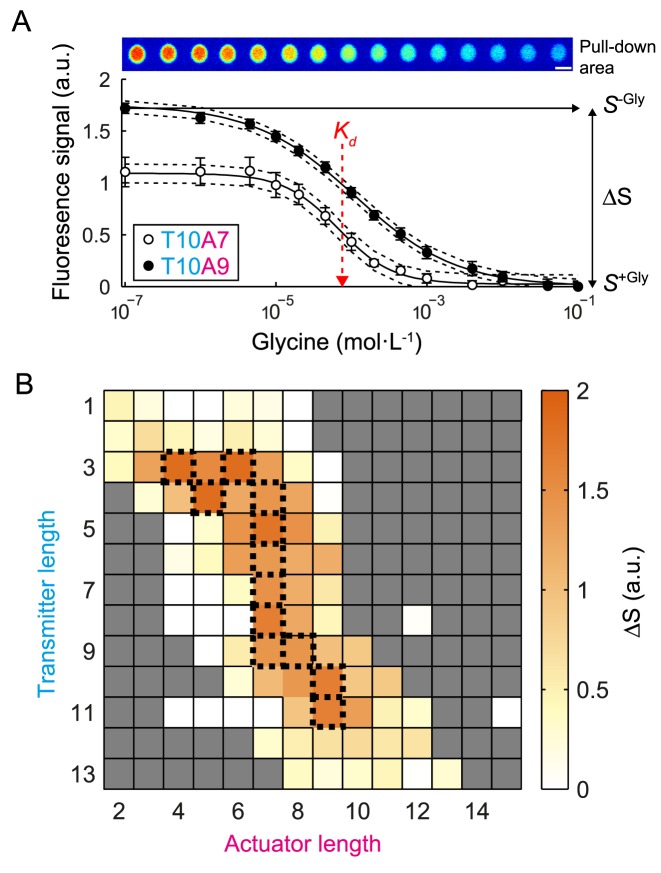
Figure 2A shows two representative glycine titration curves for the fluorogenic glycine riboswitches with a transmitter (T) and actuator (A) length of T10A7 and T10A9. A two-state binding model was fitted to the titration curves to obtain the apparent glycine dissociation constant (KD) and fluorescence signal change (ΔS) for each riboswitch. In Figure 2B, all obtained ΔS values were plotted against the transmitter and actuator length of the glycine riboswitches in order to derive the optimal balance for the length of both strands. The plot reveals that riboswitches with a transmitter length below and above 3 and 11 nt, respectively, showed only a small level of fluorescence in response to glycine concentration. For all other fluorogenic riboswitches, optimal transmitter/actuator pairs were found, which together form a small ridge of 12 riboswitches within the functional landscape.
Figure 2.
Thermodynamic characterization of the fluorogenic glycine riboswitch library on chip. (A) Top: fluorescence images of the riboswitch (T10A9) pulled-down to one out of 640 miniaturized detection areas on chip. From left to right the glycine concentration increased from 0 to 100 mM in sub sequential titration steps over the pull-down area. Bottom: fluorescence signals of two representative fluorogenic glycine riboswitches in dependence of the glycine concentration. Fluorescence signals were normalized to the absolute RNA amount on the pull-down areas and corrected by negative controls (see ‘Materials and Methods’ section). Dotted lines show the 95% confidence interval of the fit of the binding model to the experimental data. The standard errors were calculated from four experimental repeats on one chip. From the fluorescence curves the signal change (ΔS) and the apparent binding constant (Kd) were obtained. (B) Signal response landscape of the fluorogenic glycine riboswitch library. Fluorescence responses are plotted in dependence of the transmitter-actuator chain lengths. The top 12 fluorogenic riboswitches with the highest ΔS are indicated by dotted lines. Grayed out squares were not measured.
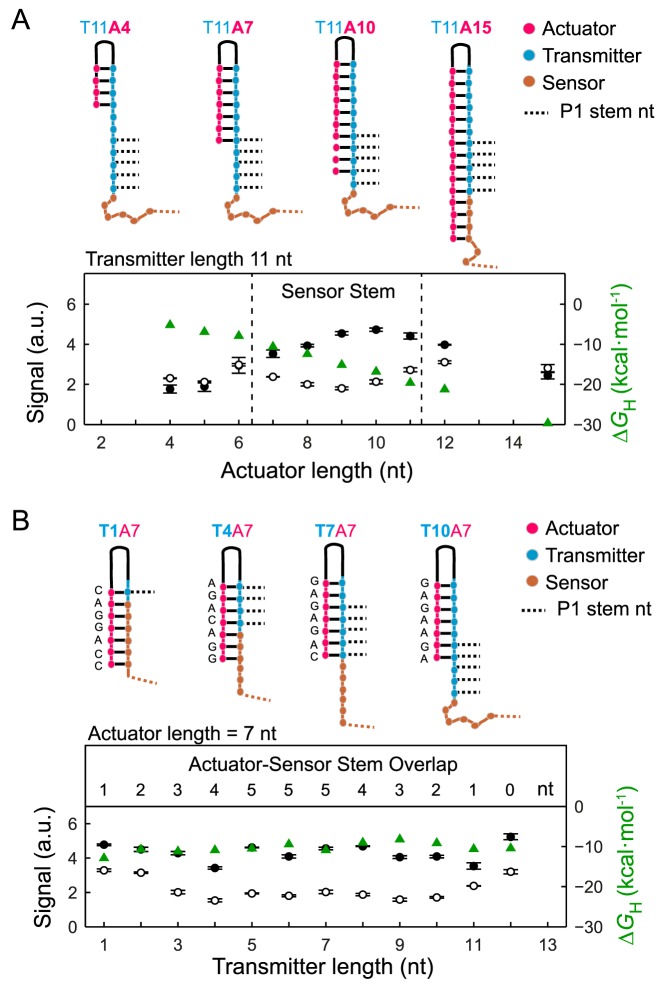
To understand mechanistically how the sensor-transmitter-actuator hybridization equilibrium is altered, representative normalized fluorescence signals of riboswitches were plotted in the absence of (S−Gly) and in saturated glycine concentration (S+Gly) along the vertical and horizontal direction of the functional landscape plot of Figure 2B. The fluorescence signal of the glycine riboswitch relative to the actuator length at a constant transmitter length (nt = 11) is shown in Figure 3A. Upon increasing the actuator chain length, S−Gly steadily increased up to a transmitter length of 10 nt. Expectedly, a longer transmitter-actuator stem stabilized the Spinach structure and enhanced the fluorescence of the Spinach/DFHBI-1T complex. For quantification, the hybridization energies between the transmitter and actuator strand (ΔGH) were calculated with RNAcofold and added to the plot. The S−Gly values of the negative controls and the fluorogenic glycine riboswitches correlated strongly (R = 0.87), which demonstrated that the Spinach/DFHBI-1T fluorescence in the absence of glycine is primarily dependent on the transmitter/actuator stem length. The S−Gly values of the fluorogenic glycine riboswitches and their negative controls were converging into a maximal possible fluorescence signal for transmitter/actuator stems with ΔGH ≲ −10 kcal/mol (see Supplementary Figure S3). Notably, for fluorogenic glycine riboswitches with a longer actuator than transmitter strands, S−Gly was slightly decreased again. This can be explained by the fact that the extension of the actuator beyond the transmitter length leads to sequestering of nucleotides (−1 to −4 nt in Figure 1B), which have been previously shown to form stack interactions to the P3 and P2 stem of the Gly-Apt-2.(28) At saturating glycine concentration, only fluorogenic glycine riboswitches with actuators, which contained an overlap to the 5 nt long P1 stem sequence of the sensor, showed a reduced S+Gly compared to the negative controls. This was found for actuators with a length between 7 and 11 nt, as shown in Figure 3A.
Figure 3.
Dependence of the riboswitch fluorescence signal on the transmitter and actuator length in absence and saturating glycine concentration. (A) Top: RNA secondary structure plots of four representative fluorogenic glycine riboswitches with constant transmitter and changing actuator length. The plot shows the fluorescence signal of the riboswitches with constant transmitter length of 11 nt and variable actuator length in absence (•) and with 0.1 M glycine (∘). Dashed lines in the plot denote for the range where the actuator exhibits a sequence overlap with the P1 sensor/transmitter stem loop. (B) Top: RNA secondary structure plots of four representative fluorogenic glycine riboswitches with constant actuator and changing transmitter length. Fluorescence signal of the riboswitches with constant actuator length of 7 nt and variable transmitter length in absence (•) and with 0.1 M glycine (∘). Additionally, in both plots the hybridization energies (ΔGH) between the transmitter and actuator are added ( ). Further, the competing number of nucleotides of the actuator with the sensor stem P1 for the transmitter is indicated.
). Further, the competing number of nucleotides of the actuator with the sensor stem P1 for the transmitter is indicated.
The fluorescence signal of the glycine riboswitch depended on the transmitter length at a constant actuator length of 7 nt as shown in Figure 3B. Here, the hybridization energy between the actuator and transmitter varied maximal by 3.5 kcal/mol due to a changing actuator sequence. Accordingly, only small changes of S−Gly were observed. In contrast, S+Gly was changed in cases where the riboswitch contained a sequence overlap between its actuator and P1 stem sequence. For the actuator with a length of 7 nt, an overlap of 3 nt was sufficient to destabilize the Spinach/DFHBI-1T binding equilibrium and reduce the fluorescence signal by 60% compared to S−Gly.
Glycine binding thermodynamics
In addition to the normalized and relative fluorescence changes, the dissociation constants of the glycine/riboswitch complex were obtained from the microfluidic titration experiments. Notably, the fluorescence signal changes of Spinach/DFHBI-1T reported the structural transmitter state rather than directly on the glycine binding state of the sensor of the riboswitch. Therefore, the measured dissociation constants, KD, were apparent and expressed the glycine concentration needed to reduce by half the fluorescence signal of Spinach/DFHBI-1T. The KD values for the fully screened fluorogenic glycine riboswitch library varied between 5.3 and 2100 μM (see Supplementary Figure S4). The apparent dissociation constant of the fluorogenic riboswitches from the top 12 glycine signal responders and a transmitter chain length between 5 and 10 was about 127 ± 20 μM. KD values of fluorogenic riboswitches of the top 12 glycine signal responders and a transmitter length below 5 nt and above 10 nt were significantly larger. At the lower transmitter lengths, the apparent KD values decreased due to destabilization of the P1 stem of the Gly-Apt-2, whereas the longer transmitter lengths had inhibition of the Gly-Apt-2 folding due to the relatively high ΔGH.
K D values obtained with the miniaturized pull-down on chip system were higher than those obtained for the same fluorogenic glycine riboswitch in solution off chip (Supplementary Figure S5A). The KD value measured for the fluorogenic glycine riboswitch with the transmitter T8A7 was 71 ± 4 μM in solution and 126 ± 13 μM on chip pull-down assay. The corresponding Hill coefficients for the apparent glycine dissociation reactions were obtained from fitted glycine titration curves. The globally fitted Hill coefficient for the glycine dissociation reaction of the 12 riboswitches with the highest ΔS was 0.7 ± 0.1. A Hill coefficient ≤1 was found in both solution and pull-down binding assays. Therefore, the transmitter hybridization switch induced by glycine was non-cooperative. However, an additional control experiment revealed that the anchor sequence at the 5′ end induced the non-cooperative glycine binding behavior. Indeed, the fluorogenic glycine riboswitch with a T8A7 transmitter without a pull-down anchor sequence showed an increased Hill coefficient of 1.5 ± 0.2. Concomitantly, the KD of the T8A7 riboswitch increased from 71 ± 4 μM to 231 ± 19 μM within a solution binding assay. Nevertheless, the relative thermodynamic binding parameters for fluorogenic glycine riboswitches without anchor sequence measured in solution correlating to the obtained values for the same fluorogenic glycine riboswitches with anchor sequence (see Supplementary Table S4).
Despite a low Hill coefficient, the fluorogenic glycine riboswitches with an anchor sequence required both glycine sensor aptamers for their fluorogenic function. This was demonstrated by synthesizing a glycine riboswitch without the Gly-Apt-1 sequence. The fluorescence signal of the truncated T8A7 construct was not responsive to glycine concentration changes (see Supplementary Figure S5). Consequently, structural changes and dimerization energies of Gly-Apt-1 and 2 were required for changing the equilibrium state of the transmitter.
The binding specificity of the fluorogenic glycine aptamers was also determined. Structural analogs of glycine were tested to assess induction of a fluorescence change in the 12 fluorogenic glycine riboswitches with the highest ΔS. Among the seven glycine analogs tested, only glycine methyl ester and glycine tert-butyl ester were able to reduce the fluorescence signal of the synthetic riboswitches, albeit to a lower extend than glycine (Supplementary Figure S6). This followed the previous trends reveled by single-round transcription assays with the natural glycine riboswitch from B. subtillis (5).
Glycine binding kinetics
For recording the apparent glycine binding (kon) and dissociating (koff) rate constants of fluorogenic glycine riboswitches, we used the same microfluidic chip platform as described above. Here, the fluorescence signal changes of the fluorogenic glycine riboswitches upon changing glycine concentration were recorded in real time.
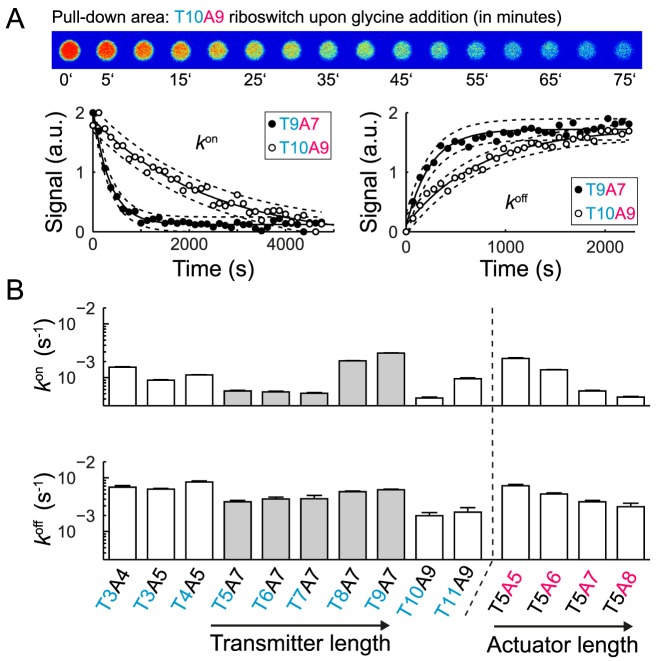
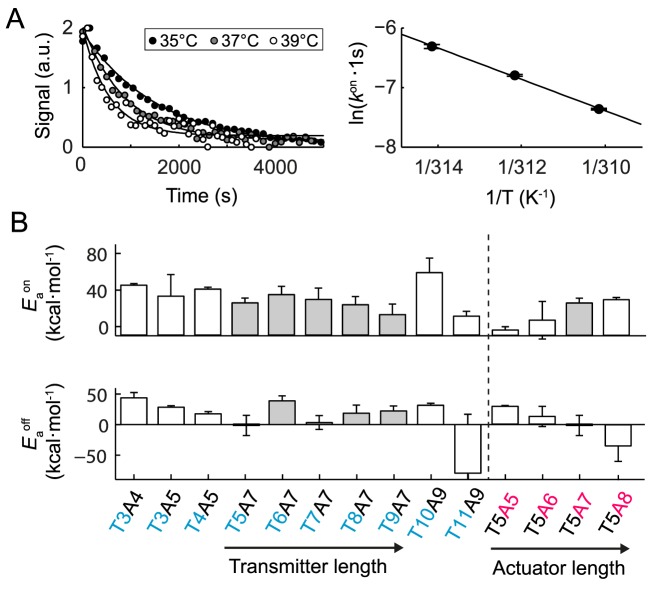
Figure 4A shows representative fluorescence time traces of the pulled-down fluorogenic glycine riboswitches with a transmitter/actuator length of T10A9 and T9A7 upon addition and depletion of glycine at 37°C. The apparent kon and koff rate constants were obtained by fitting the fluorescence traces with a unimolecular reaction model. This was justified based on a finding that both rate constants were independent at glycine concentration above 400 μM (see Supplementary Figure S7). The apparent kon and koff rates for 13 riboswitches were determined (Figure 4B), where 10 of them belong to the top 12 riboswitches with the largest ΔS values. The additional three riboswitches were chosen to complement either a transmitter or actuator length series. Apparent glycine kon rates were in the order of 10−3 s−1 and up to one order of magnitude smaller than the apparent koff rates with values of 10−2 s−1. Again, it was noted that the apparent rate constants were measured since the fluorescence signal changes were indicative of the loosening of the Spinach/DFHBI-1T complex due to the structural rearrangement of the transmitter. The relatively slow kon rates for the fluorogenic glycine riboswitches and the unimolecular reaction behavior supports a model in which the structural rearrangement of the transmitter is the rate limiting step within a more complex binding and folding reaction. While a longer actuator chain length led to a decrease of kon and koff, a longer transmitter chain length led to an increase in kon.
Figure 4.
Kinetic characterization of selected glycine fluorescence riboswitches on chip. (A) Top: fluorescence images representing the response time of the riboswitch (T10A9). For this the glycine concentration was rapid exchanged over the pulled-down areas on the microfluidic chip and the fluorescence signal change recorded. Bottom: representative fluorescence time traces of two riboswitches upon addition of 0.01 M glycine (left) and complete depletion of glycine (right). B) Apparent kon and koff rates for 12 fluorogenic glycine riboswitches with different transmitter-actuator lengths. The rates were obtained by fitting a single exponential function to the fluorescence time traces.
Additionally, the temperature dependencies of the kon and koff rate constants were determined. Figure 5A shows representative fluorescence time traces of the fluorogenic glycine riboswitch with a transmitter/actuator length of T9A7 upon addition of glycine at three different temperatures. By means of an Arrhenius plot, the energy barriers for the formation of the transmitter-actuator and transmitter-sensor states of the riboswitches were determined (Figure 5B). This revealed that an extension of the actuator length increased the energy barrier for the formation of the transmitter-actuator state (EAOn) by 12 kcal/mol per nucleotide and decreased the energy barrier for the formation of the transmitter-sensor state (EAOff) by 21 kcal/mol per nucleotide. Consistent with the thermodynamic results, an extension of the transmitter length resulted in relatively little change to the kinetic energy barriers between the transmitter-sensor and transmitter-actuator states of the fluorogenic glycine riboswitches. All biophysical acquired parameters for the synthetic fluorogenic glycine riboswitch are summarized in Supplementary Table S3.
Figure 5.
Temperature dependence of the apparent kon and koff rates of glycine fluorescence riboswitches with different transmitter-actuator lengths. (A) Representative fluorescence time traces of the riboswitch T10A9 upon addition of 0.01 M glycine (left) at three different temperatures. The corresponding apparent kon rates are plotted against the temperature to obtain the apparent energy barrier for the glycine binding reaction. (B) Apparent energy barriers for glycine binding (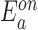 ) and dissociation (
) and dissociation (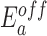 ) to the fluorogenic glycine riboswitches with different transmitter-actuator lengths.
) to the fluorogenic glycine riboswitches with different transmitter-actuator lengths.
DISCUSSION
Here, a set of fluorogenic glycine riboswitches with different thermodynamic and kinetic properties was engineered. Synthetic RNA synthesis and microfluidic large-scale integration technology were combined to screen and characterize functional fluorogenic metabolite biosensors. The generic function of the PDMS-based chip platform has been previously demonstrated for determining complete thermodynamic RNA binding and stability landscapes (19,29). The application of the chip technology was extended by this study, providing the workflow for determining binding and dissociation kinetics of a small molecule to RNA. A systematic and rational engineering approach based on biophysical parameterization was developed for RNA small molecule interactions in vitro. In contrast to evolutional approaches for engineering functional RNAs, including SELEX-based methods, the presented microfluidic technique can only screen over a small library size of hundreds rather than millions of RNA sequences; however, the gained biophysical information of the library is deeper.
Two approaches currently exist for designing fluorogenic metabolite biosensors with the Spinach aptamer as signal actuator. The first approach links Spinach directly to a sensor aptamer with high affinity for a metabolite (14,32). In this RNA two-part system, structural changes of the sensor aptamer induced by metabolite binding are exploited to alter the binding equilibrium of Spinach/DFHBI-1T. The second approach links Spinach via a transmitter sequence to the sensor aptamer (11). This three RNA part design was inspired by the natural riboswitch design and was successfully applied to engineering fluorogenic biosensors, for example for TPP. In both cases, only fluorogenic metabolite biosensors could be engineered with a turn-on configuration.
The three RNA part design approach was used to rapidly identify 12 out of 96 turn-off fluorogenic riboswitches with a high glycine signal response. The top selected riboswitches exhibited a maximum fluorescence signal change compared to the negative control constructs within the miniaturized pull-down assays on chip. For a comparison to existing fluorogenic metabolite sensors, the ΔS value of the strongest fluorogenic glycine riboswitch with transmitter sequence T8A7 was measured in solution at 28°C. Here, ΔS was −59% when normalized to the fluorescence of a wild-type Spinach version (see Supplementary Figure S8). The engineered fluorogenic glycine riboswitches exhibiting fluorescence signal changed within the same range as previously developed Spinach-based RNA sensors with affinities to TPP, guanine, adenine, SAM and cyclic AMP, where ΔS changed by 41, 67, 40, 23–31 and 64%, respectively (11,13). The absolute fluorescence fold change of the turn-off T8A7 glycine riboswitch in solution was 5.2, which is lower than fold changes observed for Spinach based turn-on sensors. This is mainly due to the larger remaining fluorescence signal of the T8A7 in presence of high metabolite concentration. Lower signal-to-noise values are a known drawback of turn-off sensors.
The on chip binding assay determined that the apparent KD values for the 12 fluorogenic glycine riboswitches were in the micromolar range (99.7–570 μM). Reported KD values of the conserved natural glycine riboswitch range from 2 to 311 μM depending on organismic origin of the riboswitch, its sequence length, magnesium concentration, and detection technology (5,24–26,28,33–35). KD values for the natural glycine riboswitch from B. subtillis, however, were consistently reported to be lower than 30 μM (5,24). Higher KD values of riboswitches lacking a pull-down anchor were only partly caused by reduced motional freedom within the pull-down binding assay on chip. However, importantly, the specificity of the synthetic glycine riboswitches was preserved.
The natural glycine riboswitch has drawn attention due to the conserved tandem aptamer structure (1). Initial studies revealed strong cooperative binding (Hill coefficient above 1.4) for glycine to the riboswitch from B. subtillis (5). A similar Hill coefficient for glycine binding to the fluorogenic glycine riboswitches was only obtained upon omitting the pull-down anchor sequence at the 5′ end. For all constructs, which contained the pull-down anchor, a non-cooperative glycine binding reaction with Hill coefficient ≤ 1 was observed. This finding was independent of the detection platform. Non-cooperative glycine binding was also found for the Vibrio cholerae glycine riboswitch containing a small leader sequence at the 5′-end (24,25). However, in Vibrio cholerae, the leader sequence forms a stem loop with the linking sequence of the Gly-Apt-1 and 2. It is unknown if the anchor sequence induces the same structural effect as the leader sequence of the glycine riboswitch from Vibrio cholera but it is obvious that small sequence length alterations at the 5′ end have a strong influence on the glycine binding curves.
Clearly, the tandem glycine aptamers configuration was required to shift the transmitter binding equilibrium from the actuator to sensor at high glycine concentration. In a truncated Gly-Apt-1 construct, no fluorescence signal change could be detected. This suggests that glycine binding to one sensor aptamer alone is not sufficient to overcome the transmitter-actuator hybridization energy barrier, and that further energy from the dimerization process of the Gly-Aptamers is required.
While the glycine binding affinity varied among the top 12 responsive fluorogenic glycine riboswitches by a factor of five, the apparent glycine binding and dissociation rate constants varied by factor of 10. On-rates were an order of magnitude slower than the off-rates. For comparison, fluorescence spectroscopy studies on the majority of metabolite riboswitch classes revealed on- and off-rates on the order of 104–105 M−1s−1 and 10−2-10−3 s−1, respectively (36–40). Thus, the apparent kon and koff values are in good agreement with the previously found microscopic reaction rates for metabolite binding and dissociation from riboswitches at micromolar metabolite concentrations.
Thermodynamic and kinetic data allowed identification of the two most important parameters for the transmitter design of the fluorogenic glycine riboswitches. First, a minimum actuator length of 7 nt is required to stabilize the Spinach fluorescence in absence of glycine. Longer actuator strands were acceptable, but only under the compromise of decreased switching reaction rates of the riboswitch. Second, a sequence overlap of at least 3 nt between the transmitter and the P1 stem region of the Gly-Apt-2 has to be incorporated in the synthetic riboswitch. This overlap is required to couple the energy transfer of the sensor to the actuator in the presence of glycine. Optimal performance relative to both parameters was dependent on the transmitter length and reflected in the functional landscape of the fluorogenic glycine riboswitch (Figure 2B).
Glycine binding to the natural glycine riboswitch is dependent on the magnesium concentration (32). The divalent magnesium ions are needed to neutralize the negative charge of the carboxylate moiety of glycine in the binding pocket of the sensor aptamers. This dependence was also seen for the fluorescence glycine riboswitch (Supplementary Figure S9). At 200 μM glycine concentration the magnesium binding constant of the T8A7 riboswitch was 8.3 ± 0.4 mM. Conversely, lowering the magnesium concentration led to a decrease of the dissociation constant KD between glycine and the riboswitch T8A7. Both natural and synthetic glycine riboswitches have KD values in the micromolar range, whereas the physiological glycine concentration is in the millimolar range (41). Upon changing the glycine concentration to an in vivo relevant value of 5 mM, the synthetic glycine riboswitch exhibits a magnesium KD value of 4.2 ± 0.2 mM. This magnesium dependence of the natural and synthetic riboswitch could explain the mismatch between previously determined in vitro measured glycine KD values and physiological relevant glycine and magnesium concentrations (42,43). The in vivo applications of the fluorogenic glycine riboswitch remain to be discovered; however, the synthetic fluorogenic glycine riboswitches offer a variety of technical possibilities ranging from microfluidic chip integrated sensor systems for analytics of glycine within body fluids to a tool for mutational screening to generate glycine sensor aptamers with variable binding properties.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Excellence Initiative of the German Federal and State Governments [EXC-294, GSC-4]; German Research Foundation (Emmy-Noether Grant) [ME3823/1-1].
Conflict of interest statement. None declared.
REFERENCES
- 1.Breaker R.R. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serganov A., Patel D.J. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu. Rev. Biophys. 2012;41:343–370. doi: 10.1146/annurev-biophys-101211-113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mellin J.R., Cossart P. Unexpected versatility in bacterial riboswitches. Trends Genet. 2015;31:150–156. doi: 10.1016/j.tig.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 5.Mandal M., Lee M., Barrick J.E., Weinberg Z., Emilsson G.M., Ruzzo W.L., Breaker R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 6.Serganov A., Patel D.J. Amino acid recognition and gene regulation by riboswitches. Biochim. Biophys. Acta. 2009;1789:592–611. doi: 10.1016/j.bbagrm.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peselis A., Serganov A. Themes and variations in riboswitch structure and function. Biochim. Biophys. Acta. 2014;1839:908–918. doi: 10.1016/j.bbagrm.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groher F., Suess B. Synthetic riboswitches - a tool comes of age. Biochim. Biophys. Acta. 2014;1839:964–973. doi: 10.1016/j.bbagrm.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Baird N.J., Inglese J., Ferré-D'Amaré A.R. Rapid RNA-ligand interaction analysis through high-information content conformational and stability landscapes. Nat. Commun. 2015;6:8898. doi: 10.1038/ncomms9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paige J.S., Wu K.Y., Jaffrey S.R. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You M., Litke J.L., Jaffrey S.R. Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E2756–E2765. doi: 10.1073/pnas.1504354112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espah Borujeni A., Mishler D.M., Wang J., Huso W., Salis H.M. Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Res. 2016;44:1–13. doi: 10.1093/nar/gkv1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paige J.S., Nguyen-Duc T., Song W., Jaffrey S.R. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellenberger C.A., Chen C., Whiteley A.T., Portnoy D.A., Hammond M.C. RNA-based fluorescent biosensors for live cell imaging of second messenger cyclic di-AMP. J. Am. Chem. Soc. 2015;137:6432–6435. doi: 10.1021/jacs.5b00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang J.C., Chang A.L., Kennedy A.B., Smolke C.D. A high-throughput, quantitative cell-based screen for efficient tailoring of RNA device activity. Nucleic Acids Res. 2012;40:e154. doi: 10.1093/nar/gks636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandal M., Breaker R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 17.Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausländer S., Stücheli P., Rehm C., Ausländer D., Hartig J.S., Fussenegger M. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat. Methods. 2014;11:1154–1160. doi: 10.1038/nmeth.3136. [DOI] [PubMed] [Google Scholar]
- 19.Ketterer S., Fuchs D., Weber W., Meier M. Systematic reconstruction of binding and stability landscapes of the fluorogenic aptamer spinach. Nucleic Acids Res. 2015;43:9564–9572. doi: 10.1093/nar/gkv944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang G., Wang S., Wei H., Ping J., Liu J., Xu L., Zhang W. Patch oligodeoxynucleotide synthesis (POS): a novel method for synthesis of long DNA sequences and full-length genes. Biotechnol. Lett. 2012;34:721–728. doi: 10.1007/s10529-011-0832-0. [DOI] [PubMed] [Google Scholar]
- 21.Melin J., Quake S.R. Microfluidic large-scale integration: the evolution of design rules for biological automation. Annu. Rev. Biophys. Biomol. Struct. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 22.Maerkl S.J., Quake S.R. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 23.Song W., Strack R.L., Svensen N., Jaffrey S.R. Plug-and-play fluorophores extend the spectral properties of Spinach. J. Am. Chem. Soc. 2014;136:1198–1201. doi: 10.1021/ja410819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherman E.M., Esquiaqui J., Elsayed G., Ye J.-D. An energetically beneficial leader-linker interaction abolishes ligand-binding cooperativity in glycine riboswitches. RNA. 2012;18:496–507. doi: 10.1261/rna.031286.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruff K.M., Strobel S.A. Ligand binding by the tandem glycine riboswitch depends on aptamer dimerization but not double ligand occupancy. RNA. 2014;20:1775–1788. doi: 10.1261/rna.047266.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon M., Strobel S.A. Chemical basis of glycine riboswitch cooperativity. RNA. 2008;14:25–34. doi: 10.1261/rna.771608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peselis A., Gao A., Serganov A. Cooperativity, allostery and synergism in ligand binding to riboswitches. Biochimie. 2015;117:100–109. doi: 10.1016/j.biochi.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang L., Serganov A., Patel D.J. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol. Cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler E.B., Xiong Y., Wang J., Strobel S.A. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem. Biol. 2011;18:293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakayama S., Luo Y., Zhou J., Dayie T.K., Sintim H.O. Nanomolar fluorescent detection of c-di-GMP using a modular aptamer strategy. Chem. Commun. 2012;48:9059–9061. doi: 10.1039/c2cc34379g. [DOI] [PubMed] [Google Scholar]
- 31.Martin L., Meier M., Lyons S.M., Sit R.V., Marzluff W.F., Quake S.R., Chang H.Y. Systematic reconstruction of RNA functional motifs with high-throughput microfluidics. Nat. Methods. 2012;9:1192–1194. doi: 10.1038/nmeth.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kellenberger C.A., Wilson S.C., Sales-Lee J., Hammond M.C. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 2013;135:4906–4909. doi: 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipfert J., Das R., Chu V.B., Kudaravalli M., Boyd N., Herschlag D., Doniach S. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J. Mol. Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherman E.M., Elsayed G., Esquiaqui J.M., Elsayed M., Brinda B., Ye J.-D. DNA-rescuable allosteric inhibition of aptamer II ligand affinity by aptamer I element in the shortened Vibrio cholerae glycine riboswitch. J. Biochem. 2014;156:323–331. doi: 10.1093/jb/mvu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erion T.V., Strobel S.A. Identification of a tertiary interaction important for cooperative ligand binding by the glycine riboswitch. RNA. 2011;17:74–84. doi: 10.1261/rna.2271511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert S.D., Stoddard C.D., Wise S.J., Batey R.T. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Wickiser J.K., Cheah M.T., Breaker R.R., Crothers D.M. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 38.Wickiser J.K., Winkler W.C., Breaker R.R., Crothers D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J., Lau M.W., Ferré-D'Amaré A.R. Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry. 2010;49:9123–9131. doi: 10.1021/bi1012645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang K., Rieder R., Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tempest D.W., Meers J.L., Brown C.M. Influence of environment on the content and composition of microbial free amino acid pools. J. Gen. Microbiol. 1970;64:171–185. doi: 10.1099/00221287-64-2-171. [DOI] [PubMed] [Google Scholar]
- 42.Alatossava T., Jütte H., Kuhn A., Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 1985;162:413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipfert J., Sim A.Y.L., Herschlag D., Doniach S. Dissecting electrostatic screening, specific ion binding, and ligand binding in an energetic model for glycine riboswitch folding. RNA. 2010;16:708–719. doi: 10.1261/rna.1985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.