Abstract
Recently, MGME1 was identified as a mitochondrial DNA nuclease with preference for single-stranded DNA (ssDNA) substrates. Loss-of-function mutations in patients lead to mitochondrial disease with DNA depletion, deletions, duplications and rearrangements. Here, we assess the biochemical role of MGME1 in the processing of flap intermediates during mitochondrial DNA replication using reconstituted systems. We show that MGME1 can cleave flaps to enable efficient ligation of newly replicated DNA strands in combination with POLγ. MGME1 generates a pool of imprecisely cut products (short flaps, nicks and gaps) that are converted to ligatable nicks by POLγ through extension or excision of the 3′-end strand. This is dependent on the 3′-5′ exonuclease activity of POLγ which limits strand displacement activity and enables POLγ to back up to the nick by 3′-5′ degradation. We also demonstrate that POLγ-driven strand displacement is sufficient to generate DNA- but not RNA-flap substrates suitable for MGME1 cleavage and ligation during replication. Our findings have implications for RNA primer removal models, the 5′-end processing of nascent DNA at OriH, and DNA repair.
INTRODUCTION
The human mitochondrial genome is a ∼16 kb double stranded circular molecule that encodes several essential subunits of the oxidative phosphorylation system. Each strand of the mitochondrial DNA (mtDNA) contains one origin of replication: the origin of the heavy strand (OriH) from which replication of the leading H-strand initiates, and the origin of the light strand (OriL) from which the lagging L-strand initiates (1). Replication from OriH also produces a preterminated strand, 7S DNA, which remains annealed to the template. The mitochondrial genome is maintained by a specialized replication machinery and mutations affecting the components of this machinery are associated with mtDNA defects and mitochondrial disease (2–5). The replisome includes the polymerase POLγ, the helicase TWINKLE, mitochondrial single-stranded DNA binding protein (mtSSB), and the mitochondrial RNA polymerase (POLRMT) that in addition to transcribing genes is also the mitochondrial primase (6,7). POLγ is a heterotrimeric complex comprising the catalytic A subunit and two accessory B subunits required for processivity (8–10). The POLγA subunit contains the 5′-3′ polymerase domain and the 3′-5′ exonuclease domain required for proofreading. POLγ also associates with nucleases that can cleave flap intermediates, suggesting that flap removal and replication may be coupled (11,12).
Flaps are believed to be essential intermediates for the removal of the RNA primers at the origins of DNA replication (13–15). Analysis of mtDNA from knockout models and recent patient studies strongly indicate that RNase H1 is involved in removing the RNA primers at OriH and OriL (16–19). However, RNase H1 alone is insufficient since this enzyme cannot remove the last two ribonucleotides at the RNA–DNA junction (20). The remaining RNA is believed to be displaced into a short or long flap intermediate which the nucleases DNA2, FEN1 and most recently MGME1, have been implicated in cleaving away (3,13–15,21). Some disagreement surrounds the mitochondrial localization of FEN1 (12,22–24) and the roles of the different nucleases are not yet fully defined. The fact that loss of MGME1 function in humans causes mitochondrial disease but is not lethal indicates that other nucleases can at least partially compensate (11,25). There are several differences in the substrate specificities of the nucleases. For example, unlike DNA2 and MGME1, which only cleave DNA, FEN1 can also cut RNA (26). Also, FEN1 can cleave flaps as short as 1 nucleotide, while DNA2 requires longer flaps (27). MGME1 has been shown to cleave long flaps, but its activity on shorter flaps has not yet been studied (25).
POLγ itself can create short flaps during mtDNA synthesis – instead of terminating when reaching a duplex region, POLγ continues to polymerize a few nucleotides into the duplex, thereby creating short 5′-flaps in the displaced strand (28,29). To avoid uncontrolled formation of flaps, coordination between the polymerase and 3′-5′ exonuclease domains of POLγ carefully regulates strand displacement. Through 3′-5′ excision at the 3′-end of the DNA, POLγ can return to the nick position and the flap can reanneal (28,29). This idling process is important for ligation since inactivation of POLγ's exonuclease activity leads to excessive strand displacement synthesis and the formation of unligatable 5′-flaps (29). In the nucleus, maturation of Okazaki fragments has also been shown to depend on idling by the replicative polymerase (Pol δ) (30,31), and to involve the nucleases RNase H2, FEN1 and DNA2 (15,32).
MGME1 is a mitochondrial DNA nuclease of the RecB family with a strong preference for single-stranded DNA (25,33). MGME1 cleaves DNA with a free end, but there are conflicting data on whether it has a preference for 5′ or 3′ ends (25,33). MGME1 can cut 5′ flaps of substrates that resemble replication/repair intermediates as well as 3′ flaps (11,25). Loss-of-function mutations in the MGME1 gene can cause mitochondrial disease and are associated with mtDNA depletion, deletions, duplications and rearrangements. Interestingly, MGME1-deficient patient cells contain a truncated (11 kb) linear mtDNA fragment spanning the two origins which closely resembles that found in mice and flies which express a 3′-5′ exonuclease deficient version of POLγ (11,34,35). The lack of 3′-5′ exonuclease activity leads to unregulated POLγ strand displacement activity and extended flap formation (29). This effect may cause persistent flaps in the 5′-end of the nascent H-strand that pose a ligation block, leaving an unligatable nick in the mtDNA close to the OriH region. The nick will cause the formation of a double-strand break in the next round of replication, which can explain the formation of the 11-kb linear fragment (29). The presence of a similar, linear fragment in MGME1 patients therefore indicates that the nuclease may be involved in the same process, i.e. the formation of ligatable ends at, or near, OriH during mtDNA replication. The 11 kb linear mtDNA fragment has also been suggested to result from replication arrest near OriL and OriH and subsequent chromosomal breakage at these sites (36).
RNase H1 is involved in primer removal at both origins (17), but the situation at OriH may be more complicated compared to OriL. Primer synthesis for OriH replication is initiated at the light-strand promoter (LSP) (37), and the shift from RNA to DNA, based on the detection of covalently attached RNA to DNA, is located mainly at a conserved sequence element located about 100 bp downstream of the promoter (conserved sequence block 2, CSB2) (38,39). An additional 100 nt of DNA downstream of CSB2 are presumably removed to mature the 5′-end of the nascent H-strand. This conclusion is based on the observation that the free 5′-ends of H-strand DNA map primarily to position 191 (a position classically referred to as OriH), which is located about 100 bp downstream of the RNA to DNA transition at CSB2 (40). The mechanisms that allow the removal of a long stretch of DNA from CSB2 to the OriH region are still not understood. Evidence from MGME1-deficient patients suggests that a flap pathway involving MGME1 is linked to the 5′-end processing of the nascent H-strand (11). In vivo analysis has demonstrated that loss of MGME1 causes the formation of elongated 7S DNA species due to incomplete processing of 5′ ends. Interestingly, the 5′ ends observed upon loss of MGME1 are not only located near CSB2 but are also found in a range of positions, including a very strong signal near CSB1 (11). Therefore, even if MGME1 is required to process DNA flaps in the CSB1 to OriH region, additional factors are most likely involved, since even in the absence of MGME1, the RNA primer initiated at LSP can be removed together with the DNA up to CSB1 at least. At OriH, the main role of MGME1 may therefore be to participate in the processing of the last 20–50 nt, involving a region from CSB1 to OriH.
Here, we set out to define the function of MGME1 in flap removal processes during mtDNA replication. We reconstitute MGME1 activity in an in vitro system for mtDNA replication and characterize its interplay with other mitochondrial replication factors. We find that MGME1 can process flap intermediates during mtDNA replication. Nuclease assays reveal that MGME1 activity improves with increasing flap length with final cleavages targeted around the flap base. Only a fraction of the cleaved products are nicked and directly ligatable. In combination with POLγ however, ligation efficiency is greatly enhanced by the ability of POLγ to remodel the cleaved products.
MATERIALS AND METHODS
Cloning and protein purification
The coding sequence of human MGME1 lacking the predicted MTS (residues 1–20) but carrying a 6xHis tag coding sequence at the C terminus was cloned into the pET17-b expression plasmid. The catalytic mutant K253A was generated by site-directed mutagenesis following the manufacturer's instructions (Agilent Technologies). All sequences were confirmed by sequencing. Expression of recombinant MGME1 was induced overnight at 16°C in E. coli Rosetta pLysS cells with 0.1 mM IPTG. Cells were lysed by mechanical homogenization (ULTRA-TURRAX Ika). MGME1 was purified from lysates over His-Select Nickel Affinity Gel (Sigma-Aldrich), HisTrap FF (GE Healthcare), Hi-Trap Heparin HP (GE Healthcare) and Hi-Trap Q HP columns (GE Healthcare). For a more detailed MGME1 purification protocol, please see Supplementary Materials. All other recombinant proteins were purified as previously described (29). All protein dilutions prior to use were done in 25 mM Tris pH 7.5, 1 mM dithiothreitol (DTT), 0.1 mg/ml bovine serum albumin (BSA), 200 mM NaCl and 10% glycerol.
DNA substrates
All oligonucleotides were purchased from Eurofins MWG Operon and were PAGE purified where necessary. Supplementary Table S1 lists all oligonucleotide sequence details. For assays using linear substrates, different 80-bp long substrates were created by annealing three different oligonucleotides together. For strand displacement assays, the templates contained a 20-nt long single-stranded gap flanked by double stranded DNA. For nuclease and gel shift assays the flap substrates contained a nick with or without a downstream 5′-flap of varying length. Oligonucleotides were labeled as indicated using T4 polynucleotide kinase and [γ-32P] ATP or were 3′-end labeled using Klenow fill-in and [α-32P] dCTP. To form the biotin-streptavidin substrates, streptavidin was added at 28 times molar excess to the amount of biotinylated oligonucleotide and incubated at 37°C for 10 min. For assays using circular templates, different primers were annealed to single-stranded circular pBluescript SK (+). Labeling was achieved during experimental reactions by incorporation of radioactive [α-32P] dCTP.
Strand displacement assay
Strand-displacement reactions were performed on an 80-bp gapped substrate with the upstream oligonucleotide radioactively labeled at the 5′-terminus. Reactions (20 μl) contained 25 mM Tris-HCl, 10 mM MgCl2, 0.1 mg/ml BSA, 1 mM DTT, 0.5 mM ATP, 100 μM each of dNTPs, 25 fmol DNA substrate, 200 fmol mtSSB, 600 fmol POLγB and 150 fmol POLγA. Reactions were incubated for the indicated times at 32°C and stopped with 2x stop buffer (formamide with 10 mM ethylenediaminetetraacetic acid (EDTA), 0.025% bromophenol blue, 0.025% xylene cyanol). Electrophoresis of samples was performed in 7 M urea/10% polyacrylamide gels and signals were visualized by autoradiography or PhosphorImaging.
Nuclease and coupled nuclease-ligation assays
The nuclease assay was adapted from (25). In short, the reactions were typically performed in a volume of 20 μl containing 10 mM Tris-HCl (pH 7.5), 1 mM DTT, 0.1 mg/ml BSA, 10 mM MgCl2, 25 fmol DNA substrate and varying amounts of MGME1. Reactions were incubated at 37°C for 30 min and stopped with 2x stop buffer (formamide with 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol). Samples were run in 7 M urea/10% polyacrylamide gels and visualized with a PhosphorImager or autoradiography. For high resolution separation, sequencing gels were used and dried prior to visualization. When coupled to ligation, reactions also contained 1 mM ATP, 1 unit T4 DNA ligase, 100 μM dNTPs and where indicated, 150 fmol POLγA and 600 fmol POLγB. When stated, the substrate was pre-incubated with the indicated amount of mtSSB for 15 min on ice prior to the nuclease assay. As a control experiment, EMSA was performed on a 40 nt single-stranded oligonucleotide to confirm that mtSSB was still active after prolonged incubation times when MGME1 activity is detected, e.g. after 30 min incubation (data not shown).
EMSA
Electrophoretic mobility shift assays (EMSA) were performed in 25 mM Tris-HCl, 0.1 mg/ml BSA, 1 mM DTT, 1 mM EDTA and 10% glycerol. Ten fmol of a specific DNA substrate (as indicated in the figure legends) was incubated with increasing amounts of MGME1 (0, 1000, 10 000 fmol) or mtSSB (0, 2.5, 10, 40, 2500 fmol) on ice for 15 min. Reactions were separated by 6% PAGE in 0.5x TBE buffer at 4°C. Signals were visualized by autoradiography or PhosphorImaging.
Coupled mtDNA replication-ligation assay
Templates were prepared by annealing circular single-stranded pBluescript SK (+) with different oligonucleotide primers. The primers are indicated in the text and figures, and sequences are listed in Supplementary Table S1. Twenty microliters reactions were performed similar to a previous report (29) with 10 fmol template, 150 fmol POLγA, 600 fmol POLγB and 10 pmol of mtSSB tetramers in 25 mM Tris-HCl (pH 7.8), 1 mM DTT, 10 mM MgCl2, 0.1 mg/ml BSA, 1 mM ATP, 100 μM of each dNTP. Unless stated otherwise, 300 fmol MGME1 and DNA ligase (1 unit T4 DNA ligase or 100 fmol human DNA ligase 3) were added where indicated. Reactions were incubated at 37°C for 30 min and stopped by the addition of 4 μl stop buffer (90 mM EDTA, 6% sodium dodecyl sulphate (SDS), 30% glycerol, 0.25% bromophenol blue and 0.25% xylene cyanol). When indicated, reactions were initiated for 20 min, then MGME1 was added and reactions proceeded for an additional 30 min. Samples were separated on 0.75% agarose gels containing 0.5 μg/ml ethidium bromide (EtBr). Dried gels were visualized by autoradiography or PhosphorImaging.
RESULTS
MGME1 cleaves longer flaps more efficiently
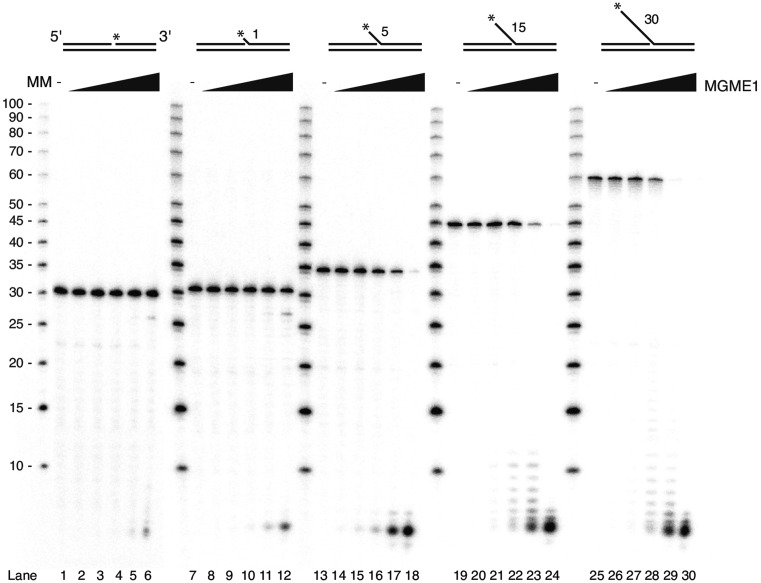
MGME1 DNA cleavage has been shown on long 5′-flap (30 nt) substrates (25). However, the cleavage by MGME1 of substrates containing shorter flap lengths that more closely resemble flap intermediates formed when POLγ encounters a free 5′-end during DNA synthesis (29) has not been investigated. We therefore tested the nuclease activity of MGME1 on substrates with 5′-flap lengths ranging from 1 to 30 nt.
The substrates investigated consisted of an 80 nt oligonucleotide annealed to an upstream 50 nt long oligonucleotide and a downstream 30 nt complementary oligonucleotide with either 0, 1, 5, 15 or 30 nt non-complementary 5′-flaps that had been radioactively labeled at the 5′-terminus. MGME1 was purified in recombinant form by sequential affinity column chromatographies to apparent homogeneity as judged by SDS-PAGE and immunoblotting. Nuclease assays with MGME1 on these templates showed that cleavage efficiency increased with longer flaps (Figure 1). Almost no activity was seen on the 1-nt flap or on the nicked substrate, while cleavage was observed on longer flaps. As expected, the catalytic mutant version of MGME1 (K253A, (25,33)) had no cleavage activity (Supplementary Figure S1).
Figure 1.
MGME1 cleavage activity increases with flap length. MGME1 nuclease assay showing cleavage of flap intermediates with different flap lengths (0, 1, 5, 15 and 30 nucleotides, respectively). A total of 25 fmol of substrate with increasing amounts of MGME1 (0, 5, 20, 50, 100, 200 fmol) were used in the reactions. The asterisks indicate the position of the radiolabel and the numbers indicate the length of the flaps. MM, molecular marker (values indicate the number of nucleotides).
MtSSB is a highly abundant protein in the mitochondrial matrix (41,42) and could in principle bind to longer flaps that are the preferred substrate for MGME1 cleavage. The nuclear counterpart of mtSSB, RPA, has been shown to inhibit FEN1 cleavage but stimulate DNA2 cleavage when bound to 5′-flaps (43). We therefore asked what effect mtSSB has on MGME1 nuclease activity. A 5′ end-labeled 40 nt long sDNA oligonucleotide was pre-incubated with increasing amounts of mtSSB tetramers before the addition of MGME1. Binding of mtSSB was confirmed by EMSA (Supplementary Figure S2A). We found that MGME1 cleavage of the oligonucleotide was inhibited at the earliest time points (2 and 5 min) with increasing amounts of mtSSB but that this inhibition was overcome with longer incubation times (Supplementary Figure S2B). We also repeated the nuclease assay with a 40 nt long flap substrate with similar results (Supplementary Figure S2C). These experiments demonstrated that MGME1 can process flaps even in the presence of mtSSB.
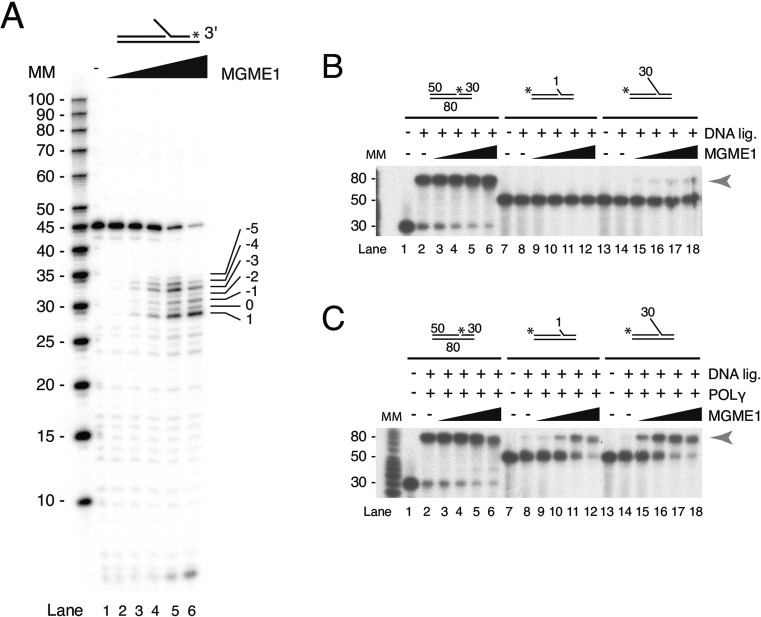
MGME1 cuts imprecisely around the flap base
The binding and cleavage patterns of MGME1 suggest that it enters the flap from the 5′-end for cleavage (Supplementary Figures S3 and S4, and (25,33)) since a block on the 5′-end of the flap structure (biotin–streptavidin or a hairpin structure) inhibits MGME1. To examine the final cleavage products, a substrate with a 15 nt long 5′-flap was made in which the flap-oligonucleotide was radioactively labeled at the 3′-terminus (Figure 2A). The template was incubated with increasing amounts of MGME1. MGME1 cleavage generated a heterogeneous pool of products comprising short (1-6 nt) 5′-flaps, nicks and 1-nt gaps (Figure 2A). The relative abundance of products with a 1 nt gap increased with increasing concentrations of MGME1. However, at all concentrations there was only a small amount of nicked product (annotated with a ‘0’ in Figure 2A) that could potentially be directly ligated without further processing of the template.
Figure 2.
MGME1 cleaves imprecisely around the flap base. (A) Mapping of MGME1 final cleavage sites shows a pool of products with short flaps, nicks and 1 nt gaps. Nuclease assays were performed on 15 nt flap substrates that had been radioactively labeled at the 3′-terminus on the downstream oligonucleotide. The numbered values in the rightmost lane indicate how close to the base MGME1 can cleave (0 = the whole flap is cleaved off and a ligatable nick is created). (B) Substrates (10 fmol) containing no flaps (lanes 1–6), 1 nt flaps (lanes 7–12) and 30 nt flaps (lanes 13–18) were incubated with increasing amounts of MGME1 (0, 12.5, 50, 250, 1250 fmol) in the presence of T4 DNA ligase. Ligation of the upstream and downstream primers results in an 80 nt product (arrowhead). MM, molecular marker. (C) As in (B), except repeated in the presence of POLγ. Note that in both (A) and (B), reaction buffer included dNTPs and ATP.
POLγ promotes ligation of flap substrates in a nuclease-ligation assay
We next directly assessed the ligation potential of MGME1 cleavage products using a coupled nuclease-ligation assay. A no-flap, 1 nt flap or 30 nt 5′-flap substrate was incubated with T4 DNA ligase and increasing amounts of MGME1 as indicated (Figure 2B). Ligation was very efficient with nicked substrates containing no flaps. No ligation was observed with the 1 nt flap substrate, which is in agreement with the nuclease assays where 1 nt flaps are not cleaved by MGME1 (Figure 1). Low levels of ligated product were produced from the 30 nt flap substrate in the presence of high amounts of MGME1. This inefficient ligation reflects the nuclease assay findings where MGME1 cleaves imprecisely around the base, with only a minor fraction of reactions producing precisely nicked products (Figure 2A).
We reasoned that flap cleavage is likely to occur in the context of POLγ idling at the 3′-end of the nascent DNA during mtDNA replication and that this could enable more efficient ligation. To test our prediction, the coupled nuclease-ligation assay was repeated in the presence of POLγ (note that dNTPs are also present). The addition of POLγ caused a strong increase in the amount of 80 nt ligated product formed from both the 1 nt flap and 30 nt flap substrates (Figure 2C). Ligation of the non-complementary 1 nt flap substrate, which is not a substrate for MGME1 cleavage itself (Figure 1), can only have occurred if the flap was extended by POLγ strand displacement synthesis so that a preferred substrate for MGME1 was created. In this way the 5′-3′ polymerase activity of POLγ promotes MGME1 flap cleavage. Together these data suggest that the efficient production of ligatable nicks involves both MGME1 and POLγ activity.
Reconstitution of DNA replication, primer flap removal and ligation
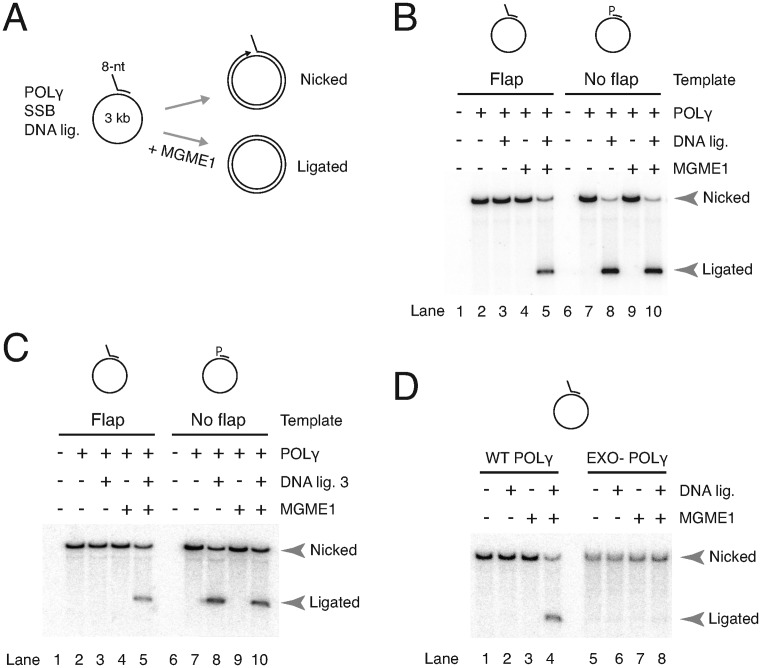
We next reconstituted an mtDNA replication system coupled to flap removal and ligation. A long (∼3 kb) circular single-stranded template primed with a DNA oligonucleotide containing an 8 nt flap at the 5′-end was used. DNA synthesis by POLγ will convert the template into a nicked double stranded circular molecule that can only be ligated to a closed circular molecule if the flap is removed (illustrated in Figure 3A and described in the Materials and Methods section). Mitochondrial SSB, POLγ and different combinations of MGME1 and T4 DNA ligase were added and reactions were separated by ethidium bromide agarose gel electrophoresis to allow topological analysis.
Figure 3.
Reconstitution of flap removal in a coupled replication-ligation assay. (A) Schematic of assay showing the flap-template. Per reaction, 10 fmol of template was incubated with mtSSB, and where indicated, POLγ, T4 DNA ligase and MGME1 (300 fmol). Products were labeled with [α-32P] dCTP during the reaction and run on agarose gels containing EtBr to separate closed circular (ligated) products from nicked products. (B) MGME1 is required for ligation in the presence of a 5′-primer flap. Double stranded nicked products (nicked arrowhead) are formed after DNA synthesis. Supercoiled (ligated arrowhead) DNA is only formed in the presence of MGME1 and T4 DNA ligase. Templates are shown above the gel: 5′-flap primed template (lanes 1–5); control 5′-phosphorylated (P) fully annealed primed template (lanes 6–10). (C) As in (B), but reconstitution of mtDNA replication using only human mitochondrial enzymes, specifically DNA ligase 3 instead of T4 DNA ligase. (D) The 3′-5′ exonuclease activity of POLγ is required for ligation. Essentially same assay as in (B), but using either WT POLγ or EXO- POLγ. The flap template is shown above.
When ligase was added to the flap substrate reactions, ligated closed circular products only formed when MGME1 was also present (Figure 3B, compare lane 3 with lane 5). In contrast, ligation using a 5′-phosphorylated non-flap control template (Figure 3B, lanes 6–10) was completely independent of MGME1 (compare lanes 8 and 10). The amount of ligation using the flap template was slightly less but comparable to the control (compare lanes 5 and 10). With the flap template, over half of the nicked products were ligated after 5 min in the presence of MGME1 (Supplementary Figure S5A). A concentration curve revealed that a small amount of ligated product was formed with a 2-fold molar excess of MGME1 and progressively increased with MGME1 concentration (Supplementary Figure S5B). That the flap is cleaved by MGME1 is supported by the absence of ligation with the catalytically inactive K253A MGME1 mutant (Supplementary Figure S5C). Finally, to fully reconstitute mtDNA replication with human mitochondrial enzymes, T4 DNA ligase was substituted with the mitochondrial DNA ligase (Lig3). Using the flap substrate, the nicked circular products could be ligated by Lig3 only in the presence of MGME1, similar to the results with T4 DNA ligase (Figure 3C). These experiments demonstrated that MGME1 can process preformed flaps to ligatable ends in a reconstituted mtDNA replication system.
Flap removal and ligation are dependent on the 3′-5′ exonuclease activity of POLγ
In the coupled nuclease-ligation assay with MGME1 we found that POLγ enhanced the generation of ligatable nicks. In all likelihood POLγ is able to fine-tune the cleaved products via its 5′-3′ polymerase and 3′-5′ excision activities, similar to Pol δ (30,31,44). To further address this point, we used a POLγA mutant lacking 3′-5′ excision capacity and with associated increased strand displacement activity (29). This ‘EXO-’ POLγ contains a single amino acid substitution in the second exonuclease motif in the POLγA subunit (D274A) (35). We previously showed using a similar circular replication assay that DNA synthesis by EXO- POLγ is unaffected by the mutation, but that ligation is abolished due to the creation of very long 5′-flaps in the downstream primer (29). We therefore now asked whether MGME1 would be able to restore ligation by cleaving these longer flaps into ligatable ends. Our results showed that this is not the case: no ligated products were formed in the EXO- reactions even in the presence of MGME1 (Figure 3D). Failure of MGME1 to compensate is possibly due to the rapid displacement of any cleaved 5′-ends into new flaps by EXO- POLγ. In summary, MGME1 can process flap intermediates and support ligation in a reconstituted coupled replication-ligation assay and this requires the 3′-5′ exonuclease activity of POLγ.
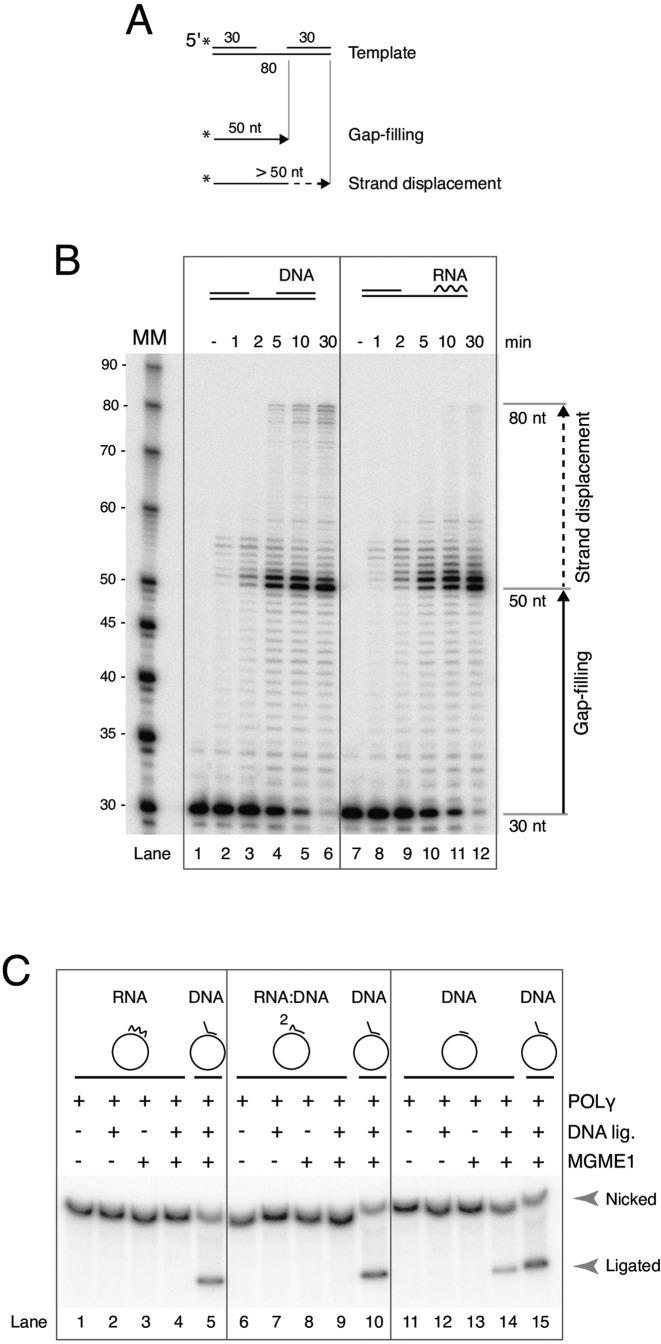
POLγ strand displacement creates short 5′-flaps in downstream DNA and RNA primers
In our assays we have taken advantage of DNA primers with preformed 5′-flaps. In mitochondria, the formation of flaps may be driven by POLγ strand displacement. While POLγ has been shown to have limited strand displacement activity on downstream DNA duplexes (28,29) this has never been tested on RNA:DNA duplexes that mimic annealed RNA primers. To examine this we used a gapped linear substrate in which an 80-nt DNA template was annealed to a 30 nt upstream DNA oligonucleotide that had been radioactively labeled at the 5′-terminus, and a 30 nt downstream DNA or RNA oligonucleotide (Figure 4A). POLγ will extend the primer across the gap until reaching the 5′-end of the downstream oligonucleotide, producing a 50-nt long product. Strand displacement synthesis by POLγ will result in longer products. As with the control downstream DNA template, we found that POLγ could displace several nucleotides of the downstream RNA oligonucleotide, though complete displacement was not as extensive as seen with the DNA control (Figure 4B).
Figure 4.
Strand displacement and flap processing by POLγ and MGME1. (A) Diagram of the linear gapped substrate used in strand-displacement assays, with products shown below. (B) Strand displacement by POLγ over time on a DNA or RNA containing template. Polymerase performs limited strand displacement on both templates. Complete displacement (the uppermost bands) is more prominent with the downstream DNA than RNA. Reactions were started by the addition of POLγ. Time points are indicated above in minutes (− represents reactions where no POLγ was added). MM, molecular marker. (C) Reactions are as described in Figure legend 3. Per reaction, 10 fmol of template was incubated with mtSSB, and where indicated, POLγ, T4 DNA ligase and MGME1 (300 fmol). Products were labeled by [α-32P] dCTP incorporation and run on agarose gels containing EtBr. Templates are shown with annotated primers above the gel. Unphosphorylated 30 nt fully annealed RNA primer (lanes 1–4), a 30 nt fully annealed unphosphorylated RNA(2):DNA(28) chimeric primer (lanes 6–9), a 30 nt fully annealed unphosphorylated DNA primer (lanes 11–14) or, a control template containing an 8 nt flap DNA primer (lane 5, 10 and 15).
Reconstitution of flap formation in coupled replication-ligation assays
We next asked whether the strand displacement activity of POLγ shown above is sufficient to create flaps that can be cleaved by MGME1 and enable ligation. To this end we used primers with unligatable unphosphorylated 5′-ends that fully anneal to the circular template in our reconstituted circular replication-ligation system. With these templates, due to the unphosphorylated 5′-ends, formation of a cleavable flap is a prerequisite for ligation – the ligation block can only be removed if the primer is displaced into a flap by POLγ strand displacement synthesis followed by MGME1 cleavage to expose a ligatable 5′-phosphate end.
We initially used RNA oligonucleotides to test whether MGME1 and POLγ are sufficient to reconstitute RNA primer removal during replication. First, a 30 nt long RNA primer that was fully complementary to the template strand was used (Figure 4C, lane 1–4). As a positive control, a template containing a preformed DNA flap primer was used (Figure 4C, lane 5, 10 or 15). DNA synthesis was initiated by addition of POLγ for 20 min to allow POLγ to reach the 5′-end of the primer, then MGME1 and T4 DNA ligase were added and reactions were incubated for a further 30 min. Compared to the control template, no closed circular products were formed from templates containing the RNA primer (Figure 4C, compare lane 4 with 5). This is consistent with our data showing that POLγ displacement of RNA is mainly limited to <10 nt (Figure 4B) and that MGME1 only cleaves within DNA and not RNA (25,33). Next, we used a chimeric primer of the same sequence as the RNA primer above, but comprised of only 2 ribonucleotides at the 5′-end followed by 28 deoxyribonucleotides (Figure 4C, lane 6–9). This mimics an RNase H1 primer intermediate product where all but the last two RNase H1-resistant ribonucleotides have been degraded (20). Compared to the control template, essentially no ligated band was observed in the presence of MGME1 and DNA ligase (Figure 4C, lane 9; a barely detectable band is visible in Supplementary Figure S5D with longer exposure). This result indicates that POLγ and MGME1 are not sufficient for removal of the remaining 2 ribonucleotides.
Finally, we constructed a template with a fully annealed unphosphorylated DNA primer. Contrary to the templates containing RNA primers, this template could be converted to a ligated product in the presence of MGME1 and DNA ligase during replication (Figure 4C, lane 14). A previous report showed that MGME1 can cut RNA:DNA chimeric flaps 2–4 nt downstream of the RNA:DNA junction (25). MGME1 cleavage of an RNA-containing primer therefore necessitates the formation of a longer flap than if the primer only contains DNA and this is consistent with our finding that ligation was only achieved with DNA primers but not those containing RNA.
DISCUSSION
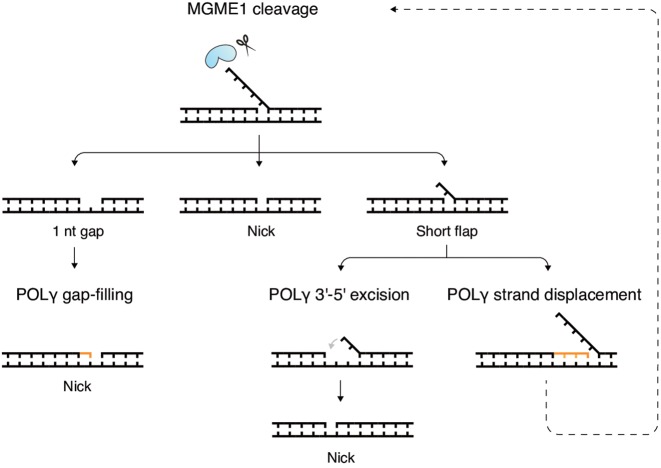
We have characterized the role of MGME1 in flap processing during mtDNA replication by using in vitro reconstituted systems. A previous study showed that MGME1 can cleave DNA or RNA:DNA chimeric flaps and might therefore be involved in processing flap intermediates that can arise during DNA repair or primer removal (25). Here, we directly examined this by coupling MGME1 cleavage activity to DNA synthesis and ligation. We found that while MGME1 alone is poor at generating ligatable ends, it does so very efficiently in combination with POLγ. We show that MGME1 cuts imprecisely around the flap base: only a fraction of products are nicked while others contain short flaps or gaps. Nonetheless, it seems not to be strictly necessary for MGME1 to cut exactly at the flap base in the context of DNA replication (or repair) since we found that POLγ can extend or excise the 3′-end strand and thus edit the imprecise cleavage products (Figure 5 for schematic model). Short gaps are filled in and short flaps are displaced into longer flaps that are optimal for further rounds of MGME1 cleavage. Meanwhile, the 3′-5′ exonuclease activity of POLγ can degrade from the 3′-end of the newly synthesized strand and allow any complementary flaps to reanneal to the template and thereby create a nick instead of a flap.
Figure 5.
Model of flap removal and the creation of a ligatable nick by MGME1 and POLγ. MGME1 is shown together with a flap substrate at the top. The outcome of cleavage is a pool of three types of products containing 1 nt gaps, nicks or short flaps (second row). POLγ can edit the imprecisely cleaved products to create ligatable nicks (bottom rows). Gaps can be filled in (left). Short flaps can be removed by two alternative pathways: through 3′-5′excision of the 3′-end strand a complementary flap can reanneal to the temple (middle), or, through strand displacement synthesis the flap can be extended to a long flap to enable a second round of MGME1 cleavage (right).
Poor ligation coupled to flap cleavage has also been observed with DNA2, which like MGME1, cleaves indistinctly around the flap base (43). Reflecting our MGME1 findings, DNA2 was shown to nonetheless efficiently support flap removal and ligation in a reconstituted budding yeast Polδ nuclear replication system (44). Previous studies had reported only very limited ligation with DNA2 alone, necessitating FEN1 as an additional nuclease to complete flap removal (31,45,46). In a reconstituted human mitochondrial replication system DNA2 was not sufficient for flap removal and ligation, but required the addition FEN1, which was explained by the fact that DNA2 cleavage leaves behind a short flap (12). That MGME1, but not DNA2, can act as the sole nuclease in removing mtDNA flaps may indicate that POLγ coordination with flap cleavage is specific to MGME1.
The apparent coordination between POLγ and MGME1 was dependent on the 3′-5′ exonuclease activity of POLγ. Exonuclease deficient (EXO-) POLγ has excessive strand displacement activity so that even if MGME1 is able to cut flaps, new flaps are presumably rapidly regenerated. Also, it is not possible to generate a ligatable nick by re-annealing of the 5′-flap because EXO- POLγ is unable to perform 3′-5′ excision of the upstream DNA. Our findings are in line with reports showing that DNA2 cannot rescue ligation in nuclear replication systems using exonuclease deficient Polδ (31,44). The requirement for exonuclease activity may only apply to nucleases such as MGME1 and DNA2 that cleave the flaps in an imprecise manner since FEN1 was reported to rescue ligation in reconstituted nuclear replication assays with exonuclease deficient Polδ (31). One explanation is that FEN1 cuts flaps more precisely, leaving nicks that can be rapidly ligated before further strand displacement takes place (30,31). However, this still remains unclear since another study using a similar system found that FEN1 was not able to restore ligation (44).
Our finding that MGME1 cannot rescue ligation with EXO- POLγ is also consistent with aberrant replication intermediates observed in the EXO- mutator mouse and suggests that the presence of flap nucleases may not fully overcome the toxic effects of uncontrolled POLγ strand displacement. Normally, POLγ idles at the nick position when reaching the 5′-end of downstream DNA (28,29). EXO- POLγ however has been shown to synthesize DNA several hundred nucleotides past the 5′-end (47). We speculate that in mutator mice, when EXO- POLγ completes replication from OriH and reaches the mature 5′-end of the nascent H-strand at OriH position 191, the polymerase does not stop, but continues DNA synthesis, thus forming 5′-flaps downstream of OriH. These flaps may continue to be regenerated even after MGME1 cleavage. Consequently, the 5′-end of the nascent H-strand in EXO- mice would be shifted downstream of OriH, which is in agreement with the mapping of 5′-ends in EXO- mice to a ∼600 nt region downstream of OriH (36).
The dependence on a free 5′-end for MGME1 binding and cleavage (Supplementary Figures S3 and S4) may be an important aspect of possible coordination between POLγ and MGME1 activities. FEN1 and DNA2 also require a free 5′-end for flap cleavage (32), however unlike MGME1, they can still bind a 5′-end blocked flap by binding at the flap base (48,49). Both FEN1 and DNA2 flap binding involve contacts with the downstream duplex region near the flap base (48–50). In contrast, MGME1 does not appear to have dsDNA binding activity ((33) and Supplementary Figure S3C, lane 1–3). This potentially means that MGME1 never ‘blocks’ the idling activity of POLγ at the nick. Instead, the activities seem to be coordinated; MGME1 can cut displaced flaps while the polymerase is idling at the 3′-end of the nascent DNA. That their activities may be coordinated while coexisting on the same molecule is further suggested by direct interactions reported between MGME1 and POLγ (11).
During primer removal it is thought that the RNase H1-resistant ribonucleotides (the last 2 ribonucleotides at the RNA–DNA junction) are displaced into a short flap by POLγ strand displacement (short flap pathway). Our data showing that POLγ can synthesize a few nucleotides (<10 nt) into an RNA:DNA duplex support this view. However, MGME1 was unable to cleave these POLγ-generated RNA flaps. Presumably this is because MGME1 cleavage of RNA:DNA chimeric flaps only occurs a few nucleotides downstream of the RNA (11), which means that in the case of RNA-containing primers a longer flap is necessary for MGME1 cleavage. MGME1 may therefore remove RNA primers or remnants thereof via a long flap pathway analogous to the long flap pathway of Okazaki fragment maturation involving DNA2. This model is also more consistent with MGME1's preference for longer flaps. The activities that would generate the longer flaps in mitochondria have yet to be identified. Finally, it cannot be ruled out that some ribonucleotides are not removed, but ligated to DNA. Mitochondrial DNA ligase 3 ligates DNA ends very efficiently in vitro, but it also has some weak ligation activity between upstream DNA and downstream RNA (51). Evidence from mouse embryonic fibroblasts lacking RNase H1 suggests that in a notable proportion of mtDNA molecules the RNA primers are ligated to the DNA (17).
A long flap pathway may also operate in the 5′-end processing of the nascent H-strand. As noted in the introduction, it is thought that ∼100 nt of nascent DNA is removed downstream of the RNA primer based on observations that the RNA:DNA transitions occur near CSB2, at position 301–299 and 292–289 of mtDNA (39), while free 5′-DNA ends map mainly to the downstream OriH sequence (position 191). In MGME1-deficient patients, the 5′-ends are extended upstream of OriH in the direction of CSB2, suggesting that this region is removed via a flap pathway involving MGME1 (11). How the long flaps needed for MGME1 or DNA2 cleavage are generated is not known. In the case of DNA2, this could be solved by its intrinsic helicase activity (52,53). Since MGME1 does not possess helicase activity, additional factor/s need to be involved in order to lengthen the flaps. We here demonstrate that POLγ can assist in creating flaps that are processed by MGME1. Others have suggested that secondary structures formed in DNA near CSB1, may also be important for this effect (11). Clearly, in vivo, there must be additional activities that assist in processing the 5′-ends of 7S DNA, since these can mature even in the absence of full-length H-strand DNA synthesis.
Finally, the role of MGME1 may not be limited to only mtDNA replication. Our finding that MGME1 is able to cut DNA flaps created by POLγ strand displacement synthesis supports the possibility that MGME1 may also be involved in DNA repair processes. The long patch base excision repair (LP-BER) pathway has been identified in mitochondrial extracts and shown to involve FEN1, DNA2 or as yet unidentified nucleases (12,23,24,54). It remains to be seen whether our system also supports the removal of 5′ abasic sites and whether MGME1-deficiency is associated with abnormally high levels of mtDNA damage in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council grants [2013-3621 to M.F., 2012-2583 to C.M.G.]; Swedish Cancer Foundation (to M.F. and C.M.G.); European Research Council [261248 to M.F., 268897 to C.G.]; Wallenbergs foundation (to M.F. and C.M.G.); Wilhelm and Martina Lundgrens foundation (to J.P.U.). Funding for open access charge: ERC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Clayton D.A. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 2.Copeland W.C. Defects in mitochondrial DNA replication and human disease. Crit. Rev. Biochem. Mol. Biol. 2012;47:64–74. doi: 10.3109/10409238.2011.632763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Copeland W.C., Longley M.J. Mitochondrial genome maintenance in health and disease. DNA Repair (Amst) 2014;19:190–198. doi: 10.1016/j.dnarep.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKinney E.A., Oliveira M.T. Replicating animal mitochondrial DNA. Genet. Mol. Biol. 2013;36:308–315. doi: 10.1590/S1415-47572013000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wanrooij S., Falkenberg M. The human mitochondrial replication fork in health and disease. Biochim. et Biophys. Acta. 2010;1797:1378–1388. doi: 10.1016/j.bbabio.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Fuste J.M., Wanrooij S., Jemt E., Granycome C.E., Cluett T.J., Shi Y., Atanassova N., Holt I.J., Gustafsson C.M., Falkenberg M. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 7.Korhonen J.A., Pham X.H., Pellegrini M., Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrodeguas J.A., Kobayashi R., Lim S.E., Copeland W.C., Bogenhagen D.F. The accessory subunit of Xenopus laevis mitochondrial DNA polymerase gamma increases processivity of the catalytic subunit of human DNA polymerase gamma and is related to class II aminoacyl-tRNA synthetases. Mol. Cell. Biol. 1999;19:4039–4046. doi: 10.1128/mcb.19.6.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan L., Sanschagrin P.C., Kaguni L.S., Kuhn L.A. The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: implications for a dual role as a primer recognition factor and processivity clamp. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9527–9532. doi: 10.1073/pnas.96.17.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray H., Wong T.W. Purification and identification of subunit structure of the human mitochondrial DNA polymerase. J. Biol. Chem. 1992;267:5835–5841. [PubMed] [Google Scholar]
- 11.Nicholls T.J., Zsurka G., Peeva V., Scholer S., Szczesny R.J., Cysewski D., Reyes A., Kornblum C., Sciacco M., Moggio M., et al. Linear mtDNA fragments and unusual mtDNA rearrangements associated with pathological deficiency of MGME1 exonuclease. Hum. Mol. Genet. 2014;23:6147–6162. doi: 10.1093/hmg/ddu336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng L., Zhou M., Guo Z., Lu H., Qian L., Dai H., Qiu J., Yakubovskaya E., Bogenhagen D.F., Demple B., et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt I.J. Mitochondrial DNA replication and repair: all a flap. Trends Biochem. Sci. 2009;34:358–365. doi: 10.1016/j.tibs.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Uhler J.P., Falkenberg M. Primer removal during mammalian mitochondrial DNA replication. DNA Repair (Amst) 2015;34:28–38. doi: 10.1016/j.dnarep.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Zheng L., Shen B. Okazaki fragment maturation: nucleases take centre stage. J. Mol. Cell Biol. (Oxford, U.K.) 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerritelli S.M., Frolova E.G., Feng C., Grinberg A., Love P.E., Crouch R.J. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 17.Holmes J.B., Akman G., Wood S.R., Sakhuja K., Cerritelli S.M., Moss C., Bowmaker M.R., Jacobs H.T., Crouch R.J., Holt I.J. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9334–9339. doi: 10.1073/pnas.1503653112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyes A., Melchionda L., Nasca A., Carrara F., Lamantea E., Zanolini A., Lamperti C., Fang M., Zhang J., Ronchi D., et al. RNASEH1 mutations impair mtDNA replication and cause adult-onset mitochondrial encephalomyopathy. Am. J. Hum. Genet. 2015;97:186–193. doi: 10.1016/j.ajhg.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruhanen H., Ushakov K., Yasukawa T. Involvement of DNA ligase III and ribonuclease H1 in mitochondrial DNA replication in cultured human cells. Biochim. et Biophys. Acta. 2011;1813:2000–2007. doi: 10.1016/j.bbamcr.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lima W.F., Rose J.B., Nichols J.G., Wu H., Migawa M.T., Wyrzykiewicz T.K., Siwkowski A.M., Crooke S.T. Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol. Pharmacol. 2007;71:83–91. doi: 10.1124/mol.106.025015. [DOI] [PubMed] [Google Scholar]
- 21.Kang Y.H., Lee C.H., Seo Y.S. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2010;45:71–96. doi: 10.3109/10409230903578593. [DOI] [PubMed] [Google Scholar]
- 22.Kazak L., Reyes A., He J., Wood S.R., Brea-Calvo G., Holen T.T., Holt I.J. A cryptic targeting signal creates a mitochondrial FEN1 isoform with tailed R-Loop binding properties. PloS One. 2013;8:e62340. doi: 10.1371/journal.pone.0062340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu P., Qian L., Sung J.S., de Souza-Pinto N.C., Zheng L., Bogenhagen D.F., Bohr V.A., Wilson D.M., 3rd, Shen B., Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell. Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szczesny B., Tann A.W., Longley M.J., Copeland W.C., Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornblum C., Nicholls T.J., Haack T.B., Scholer S., Peeva V., Danhauser K., Hallmann K., Zsurka G., Rorbach J., Iuso A., et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat. Genet. 2013;45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao H.I., Henricksen L.A., Liu Y., Bambara R.A. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 27.Kao H.I., Veeraraghavan J., Polaczek P., Campbell J.L., Bambara R.A. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 2004;279:15014–15024. doi: 10.1074/jbc.M313216200. [DOI] [PubMed] [Google Scholar]
- 28.He Q., Shumate C.K., White M.A., Molineux I.J., Yin Y.W. Exonuclease of human DNA polymerase gamma disengages its strand displacement function. Mitochondrion. 2013;13:592–601. doi: 10.1016/j.mito.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macao B., Uhler J.P., Siibak T., Zhu X., Shi Y., Sheng W., Olsson M., Stewart J.B., Gustafsson C.M., Falkenberg M. The exonuclease activity of DNA polymerase gamma is required for ligation during mitochondrial DNA replication. Nat. Commun. 2015;6:7303. doi: 10.1038/ncomms8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garg P., Stith C.M., Sabouri N., Johansson E., Burgers P.M. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Y.H., Ayyagari R., Resnick M.A., Gordenin D.A., Burgers P.M. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol delta in the creation of a ligatable nick. J. Biol. Chem. 2003;278:1626–1633. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- 32.Balakrishnan L., Bambara R.A. Okazaki fragment metabolism. Cold. Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a010173. doi:10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szczesny R.J., Hejnowicz M.S., Steczkiewicz K., Muszewska A., Borowski L.S., Ginalski K., Dziembowski A. Identification of a novel human mitochondrial endo-/exonuclease Ddk1/c20orf72 necessary for maintenance of proper 7S DNA levels. Nucleic Acids Res. 2013;41:3144–3161. doi: 10.1093/nar/gkt029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bratic A., Kauppila T.E., Macao B., Gronke S., Siibak T., Stewart J.B., Baggio F., Dols J., Partridge L., Falkenberg M., et al. Complementation between polymerase- and exonuclease-deficient mitochondrial DNA polymerase mutants in genomically engineered flies. Nat. Commun. 2015;6:8808. doi: 10.1038/ncomms9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly Y.M., Gidlof S., Oldfors A., Wibom R., et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 36.Bailey L.J., Cluett T.J., Reyes A., Prolla T.A., Poulton J., Leeuwenburgh C., Holt I.J. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Res. 2009;37:2327–2335. doi: 10.1093/nar/gkp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang D.D., Clayton D.A. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl. Acad. Sci. U.S.A. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang D., Miyako K., Kai Y., Irie T., Takeshige K. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J. Biol. Chem. 1997;272:15275–15279. doi: 10.1074/jbc.272.24.15275. [DOI] [PubMed] [Google Scholar]
- 39.Pham X.H., Farge G., Shi Y., Gaspari M., Gustafsson C.M., Falkenberg M. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J. Biol. Chem. 2006;281:24647–24652. doi: 10.1074/jbc.M602429200. [DOI] [PubMed] [Google Scholar]
- 40.Tapper D.P., Clayton D.A. Mechanism of replication of human mitochondrial DNA. Localization of the 5′ ends of nascent daughter strands. J. Biol. Chem. 1981;256:5109–5115. [PubMed] [Google Scholar]
- 41.Miralles Fuste J., Shi Y., Wanrooij S., Zhu X., Jemt E., Persson O., Sabouri N., Gustafsson C.M., Falkenberg M. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 2014;10:e1004832. doi: 10.1371/journal.pgen.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takamatsu C., Umeda S., Ohsato T., Ohno T., Abe Y., Fukuoh A., Shinagawa H., Hamasaki N., Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bae S.H., Bae K.H., Kim J.A., Seo Y.S. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 44.Levikova M., Cejka P. The Saccharomyces cerevisiae Dna2 can function as a sole nuclease in the processing of Okazaki fragments in DNA replication. Nucleic Acids Res. 2015;43:7888–7897. doi: 10.1093/nar/gkv710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ayyagari R., Gomes X.V., Gordenin D.A., Burgers P.M. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 46.Pike J.E., Burgers P.M., Campbell J.L., Bambara R.A. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J. Biol. Chem. 2009;284:25170–25180. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farge G., Pham X.H., Holmlund T., Khorostov I., Falkenberg M. The accessory subunit B of DNA polymerase gamma is required for mitochondrial replisome function. Nucleic Acids Res. 2007;35:902–911. doi: 10.1093/nar/gkl1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gloor J.W., Balakrishnan L., Campbell J.L., Bambara R.A. Biochemical analyses indicate that binding and cleavage specificities define the ordered processing of human Okazaki fragments by Dna2 and FEN1. Nucleic Acids Res. 2012;40:6774–6786. doi: 10.1093/nar/gks388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart J.A., Campbell J.L., Bambara R.A. Dna2 is a structure-specific nuclease, with affinity for 5′-flap intermediates. Nucleic Acids Res. 2010;38:920–930. doi: 10.1093/nar/gkp1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsutakawa S.E., Classen S., Chapados B.R., Arvai A.S., Finger L.D., Guenther G., Tomlinson C.G., Thompson P., Sarker A.H., Shen B., et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cotner-Gohara E., Kim I.K., Hammel M., Tainer J.A., Tomkinson A.E., Ellenberger T. Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry. 2010;49:6165–6176. doi: 10.1021/bi100503w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J.H., Kim H.D., Ryu G.H., Kim D.H., Hurwitz J., Seo Y.S. Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res. 2006;34:1854–1864. doi: 10.1093/nar/gkl102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masuda-Sasa T., Imamura O., Campbell J.L. Biochemical analysis of human Dna2. Nucleic Acids Res. 2006;34:1865–1875. doi: 10.1093/nar/gkl070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akbari M., Visnes T., Krokan H.E., Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.