Abstract
Shewanella putrefaciens CN-32 contains a single Type I-Fv CRISPR-Cas system which confers adaptive immunity against bacteriophage infection. Three Cas proteins (Cas6f, Cas7fv, Cas5fv) and mature CRISPR RNAs were shown to be required for the assembly of an interference complex termed Cascade. The Cas protein-CRISPR RNA interaction sites within this complex were identified via mass spectrometry. Additional Cas proteins, commonly described as large and small subunits, that are present in all other investigated Cascade structures, were not detected. We introduced this minimal Type I system in Escherichia coli and show that it provides heterologous protection against lambda phage. The absence of a large subunit suggests that the length of the crRNA might not be fixed and recombinant Cascade complexes with drastically shortened and elongated crRNAs were engineered. Size-exclusion chromatography and small-angle X-ray scattering analyses revealed that the number of Cas7fv backbone subunits is adjusted in these shortened and extended Cascade variants. Larger Cascade complexes can still confer immunity against lambda phage infection in E. coli. Minimized Type I CRISPR-Cas systems expand our understanding of the evolution of Cascade assembly and diversity. Their adjustable crRNA length opens the possibility for customizing target DNA specificity.
INTRODUCTION
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas (CRISPR-associated) modules are adaptive immune systems found in archaea and bacteria. CRISPR arrays consist of short repeat sequences that flank unique spacer sequences, which can be derived from viral genomes and conjugative plasmids (1–3). These arrays are transcribed into long precursor molecules that are further processed into multiple small crRNAs (CRISPR RNAs). Mature crRNAs contain a single spacer sequence and parts of the repeat sequences at their termini (4–8). The acquisition of new genomic spacers requires cleavage of the foreign DNA, which is mediated by the nearly universal Cas proteins Cas1 and Cas2 (9–11). Recognition of the source DNA, i.e. the protospacer, depends on the presence of a short conserved 2–5 base pairs (bp) sequence, termed protospacer adjacent motif (PAM). The crRNAs are bound by Cas proteins to form a CRISPR ribonucleoprotein (crRNP) surveillance complex. These complexes utilize crRNA spacers as guides to recognize foreign genetic material, e.g. during a recurring viral infection (6,12–16). Target DNA recognition and subsequent degradation requires the presence of PAM sequences and Watson–Crick base pairing between the crRNA and the unwound target strand (17–19).
CRISPR-Cas systems are diverse and were divided into three main types, which are defined by their conserved signature proteins Cas3 (Type I), Cas9 (Type II) and Cas10 (Type III) (20,21). DNA interference in Type I CRISPR-Cas systems is mediated by a multi-protein crRNP complex termed Cascade (CRISPR associated complex for antiviral defense) (13,22,23), while Type II systems utilize the single nuclease Cas9 and a trans-activating RNA for target recognition and degradation (22,24,25). Both Types employ PAM-dependent target recognition mechanisms (19,26,27). Type III systems generate crRNP complexes (termed Csm or Cmr) which are able to target ssRNA in a PAM-independent manner (12,28). Recently, three additional CRISPR-Cas Types have been classified (29). Type IV systems lack Cas1 and Cas2 and the Type V and Type VI signature proteins function as standalone DNA nucleases (30).
Type I CRISPR-Cas systems are most widespread in nature and the Cas protein composition of the employed Cascade interference complexes differs between seven subtypes (A-F,U) (29). Most insights were obtained for the Type I-E Cascade complex from Escherichia coli and several crystal structures are available (17,18,31). The Type I-E Cascade crRNP consists of the five Cas proteins Cas5, Cas6, Cas7, Cse1 and Cse2 and a 61 nucleotide (nt) mature crRNA (17,18,32). The crescent-shaped structure of this complex has a molecular mass of 405 kDa and displays an uneven stoichiometry: (Cse1)1-(Cse2)2-(Cas5)1-(Cas7)6-(Cas6e)1 (6). The 61 nt mature crRNA contains a 32 nt spacer sequence that is flanked by a 8 nt long handle-region at the 5′-end and a 21 nt long repeat sequence with a terminal hairpin at 3′-end. The crRNA is generated by the RNA endonuclease Cas6. After processing, Cas6 stays tightly associated with the 3′-end, while Cas5 caps the crRNA's 5′-terminal repeat tag (6,33). The helical backbone of Type I-E Cascade is formed by six copies of Cas7, which generates a groove to bind and protect the crRNA. The large subunit, Cse1, is responsible for PAM recognition and is involved in recruitment of the target nuclease Cas3 (6,31,32,34). A dimer of two small subunits, Cse2, stabilizes the formation of an R-loop structure and binds the displaced DNA strand (6,17,18). The general architecture of Cascade crRNPs appears to be conserved in Type I systems, even though their Cas protein composition can differ (35).
One example of a strikingly altered minimalistic Cascade architecture was recently described as a Type I-F variant (Type I-Fv). This subtype was initially identified in several beta- and gamma-proteobacteria and suggested to rely on a minimal set of five Cas proteins (20). Type I-Fv systems contain Cas1, the integrase that mediates spacer acquisition (11,36), Cas3, the target nuclease and Cas6f, the crRNA endonuclease (7). The two additional Cas proteins showed no apparent sequence similarity to known Cas protein families and small and large Cascade subunits were not detected (20). Our first characterization of this Type I-Fv CRISPR-Cas system revealed that the minimal Cascade can target conjugative plasmids in Shewanella putrefaciens CN-32 (37). Recombinant Cascade was found to contain the Cas6f endonuclease, the backbone-forming protein Sputcn32_1821 and the protein of unknown function Sputcn32_1822 (37). With support of our current knowledge on the roles of these two proteins and in analogy to the E.coli Cascade nomenclature, Sputcn32_1821 was renamed as Cas7fv (Cas7 backbone protein of the Type I-F variant CRISPR-Cas) and Sputcn32_1822 was renamed as Cas5fv (Cas5 protein of the Type I-F variant CRISPR-Cas). However, it remained unclear whether additional host proteins substitute for the missing small and large subunits. In the present study, we addressed this question and transferred the S. putrefaciens Cascade into E. coli. Heterologous interference activity against lambda phage infections was observed. The minimal architecture of this active Cascade system allowed for the modulation of the crRNA's spacer length and synthetic Cascade assemblies with elongated and shortened backbones were created. In addition, requirements for proper Cascade assembly were screened. Surprisingly, some of the synthetic Cascade complexes can retain DNA interference activity. These results shed light on the evolution of minimalist Cascade complexes without small and large subunits. The observed possibility of altering the spacer length highlights a potential for the modulation of the DNA target specificity.
MATERIALS AND METHODS
Bacterial strains and growth conditions
E. coli BL21 (DE3) cells (Novagen) were used for the production of recombinant proteins and cultures were grown in either LB or NZ-amine (1% NZ-amine, 0.5% yeast extract and 1% NaCl) media with respective antibiotics until an OD600 of ∼0.6 was reached. Gene expression was induced by addition of 1 mM IPTG and cultures were grown overnight at 18°C.
E. coli BL21-AI strains (F- ompT hsdSB (rB-mB-) gal dcm araB::T7RNAPtetA, Invitrogen) were used for phage assays and grown in 2YTL media (16 g/l tryptone, 10 g/l yeast extract, 5 g/l NaCl, 10 mM MgSO4, 0.2% maltose) supplemented with appropriate antibiotics at 37°C. A virulent variant of lambda phage (NCCB 3467) was obtained from Centraalbureau voor Schimmelcultures (Utrecht, the Netherlands).
Production and purification of recombinant proteins
The gene cassette cas7fv, cas5fv, cas6f was cloned into pRSFDuet-1 which allows for the simultaneous production of all three proteins with Cas7fv having an N-terminal His-tag fusion. The cas gene containing plasmids (pCas) were recloned to obtain variants without Cas5fv (pCas3) or with a His-tag fusion at Cas6f (pCas2). The pCas plasmid variants were co-transformed into E. coli with a second pUC19 vector containing the repeat-spacer4-repeat sequence of the single S. putrefaciens CN-32 CRISPR array downstream of a T7 RNA polymerase promoter. The CRISPR sequences were generated by cloning annealed oligonucleotides into the pUC19 vector using BamHI and HindIII restriction sites. Plasmids harboring extended (+18/+15) or shortened (-18/-15) spacer sequences contained either additional random nucleotides at the 5′-end of the spacer sequence or deleted nucleotides at the 3′-end of the spacer sequence. All sequences are listed in Supplementary Table S1. Cas protein production and the purification of recombinant Cascade variants via Ni-NTA chromatography and size exclusion chromatography (Superdex 200) in a buffer containing 50 mM HEPES-NaOH pH 7.0, 150 mM NaCl, 1 mM DTT and 1 mM EDTA was performed as described (37).
Identification of protein–RNA interactions via mass-spectrometry analysis
Protein–RNA crosslinking was performed using UV irradiation at 254 nm followed by the enrichment of cross-linked peptide–RNA heteroconjugates as described in (38,39). Around 1 nmol of the Type I-Fv Cascade complex was resuspended in 100 μl of 50 mM HEPES (pH 7.0), 300 mM NaCl, 1 mM EDTA and 1 mM DTT. The complex was incubated at 4°C for 30 min. The samples were then transferred to black polypropylene microplates (Greiner Bio-One) and irradiated at 254 nm for 10 min on ice as described in (38). The samples were ethanol precipitated and the pellet was dissolved in 4M Urea and 50 mM Tris-HCl pH 7.9. The final concentration of urea was then adjusted to 1 M with 50 mM Tris-HCl pH 7.9 and the RNA was hydrolyzed using 1 μg RNase A and T1 (Ambion, Applied Biosystems) for 2 h at 52°C followed by digestion with Benzonase at 37°C for 1 h. The proteolysis was carried out using trypsin (Promega) at 37°C, overnight. The sample was desalted to remove non-cross-linked RNA fragments using an in-house prepared C18 (Dr. Maisch GmbH) column and the cross-linked peptides were enriched on an in-house prepared TiO2 (GL Sciences) column using the protocol described in (38). The samples were then dried and resuspended in 5% v/v ACN, 1% v/v FA for mass spectrometry analysis. The sample was injected onto a nano-liquid chromatography system (Dionex, Ultimate 3000, Thermo Fisher Scientific) coupled with Q-Exactive HF instrument (Thermo Fisher Scientific) (40). Online ESI-MS was performed in data-dependent mode using a TOP20 HCD method. All precursor ions as well as fragment ions were scanned in the Orbitrap, and the resulting spectra were measured with high accuracy (<5 ppm) both in the MS and MS/MS level. Data analysis was done using a dedicated database search tool (39).
Small-angle X-ray scattering
Recombinant Cascade complexes for SAXS analysis were produced and purified as described above, snap frozen in liquid nitrogen and stored at −80°C. SEC buffer aliquots of the individual purifications were kept and treated equally for sample dilution and solvent blanking. SAXS data were collected at the European Synchrotron Radiation Facility (Grenoble, France) on beamline BM29. Detailed SAXS statistics are listed in Supplementary Table S2. Dilution series of the complexes were measured to assess sample quality in respect to inter-particle interference and aggregation. Measurements exhibiting radiation damage were discarded. Buffer subtracted SAXS curves of 10 mg ml−1 for the short variant, 20 mg ml−1 for the wild type (WT) complex and 25 mg ml−1 for the elongated version were subjected to data analysis and ab initio shape restoration bead modeling using the Primus implemented program Dammif (41). A total of 20 independent shape restorations were calculated in slow mode to generate average and filtered models. SAXS data, one individual ab initio shape restoration model for each construct and the averaged and filtered models were deposited at the curated repository SASBDB under accession SASDBK4 (short Cascade), SASDBL4 (wild-type Cascade) and SASDBM4 (long Cascade complex). The volume of the Cas7fv protein was estimated using the peptide property calculator tool (Northwestern University) based on (42).
Construction of E. coli strains with heterologous Type I-Fv CRISPR-Cas systems
A minimal CRISPR was designed with a single spacer targeting the positions 6463–6494 (GA PAM) or 6630–6662 (GG PAM) of the lambda phage gene E, flanked by two repeat sequences and a T7 RNA polymerase promoter (pCRISPRλGA and pCRISPRλ, Supplementary Table S1). The CRISPR sequence was generated by cloning annealed oligonucleotides into pCDFDuet-1 vector using BamHI and HindIII restriction sites. In addition, a second CRISPR with a spacer corresponding to a random 32 bp sequence of the S. putrefaciens genome was generated as a negative control (pCRISPR NT, Supplementary Table S1). CRISPR variants were generated by adding 1, 6, 9, 12 or 18 nt (complementary to the phage genome) to the targeting spacer; or by removing 6 or 18 nt (pCRISPRλ variants). The spacer sequences have no significant complementarity to the E. coli genome. A mutation in the HD nuclease motif of Cas3 (H156A/D157A) was introduced into pCas6 by QuikChange site-directed mutagenesis according to the manufacturer's protocol (pCas7). E. coli BL21-AI cells were transformed with pCDFDuet-1 carrying one variant of the CRISPR array and pCas6 or pCas7. After transformation, single colonies were picked for each strain, grown as a lawn for 12 h in 2YTL agar and colony-forming cells were isolated for phage assays. In parallel, a strain with empty pRSFDuet-1 and pCDFDuet-1 vectors was generated as a control.
Lambda phage-assays for DNA interference analyses
Phage assays were essentially performed as described (13). Briefly, a 1/100 dilution of an E. coli BL21-AI overnight culture was grown until OD600 nm = 0.3 and induced with 0.2% arabinose (Sigma-Aldrich) and 0.1 mM IPTG for 30 min. Then, cells were pelleted and resuspended in 10 mM MgSO4. A 1:1 mix of cells and serial phage dilutions was incubated for 20 min at 37°C and added to 2YTL top-agar containing the mentioned inducers, plated over 2YTL agar plates with antibiotics and incubated overnight at 37°C. Plates were then stained with 0.2% crystal violet and plaque number was determined. Efficiency of plaquing (EOP) was defined as the ratio between the plaque count of the strain of interest and the plaque count of the strain carrying empty plasmids. Phage assays were performed in triplicate and error bars were calculated as standard error of the mean (SEM).
Electrophoretic mobility shift assays (EMSA)
The recombinant Cascade complex was tested for its ability to bind target DNA in electrophoretic mobility shift assays (EMSAs). Utilized target oligonucleotides are detailed in Supplementary Table S1. A total of 100 pmol of each substrate was 5′-labeled with [γ-32P]-ATP (5000Ci/mmol, Hartmann Analytic) and T4 PNK (Ambion) for 1 h at 37°C. The reaction was stopped by addition of formamide loading buffer and substrates were separated by denaturing-PAGE (10% polyacrylamide, 8 M Urea). After autoradiographic exposure, RNA bands were cut from the gel, eluted and EtOH precipitated. A total of 20 nM (∼20 000 cpm) of labeled substrate was incubated with varying concentrations of Cascade (0–60 nM) for 30 min at 30°C in 50 mM HEPES-NaOH pH 7.0, 150 mM NaCl, 1 mM DTT and 1 mM EDTA. Samples were separated via non-denaturing TBE-PAGE (6% polyacrylamide, 1x TBE).
RESULTS
Recombinant production of the minimal S. putrefaciens CN-32 Type I-Fv Cascade
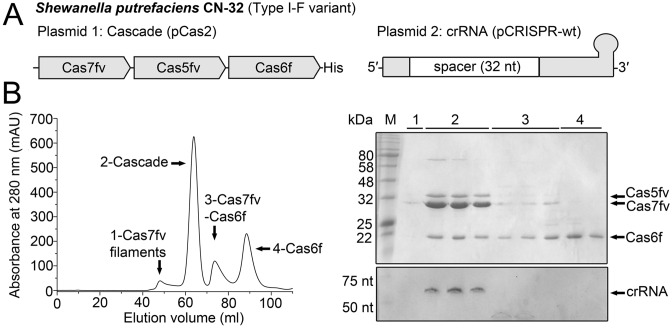
In our earlier study, we demonstrated that recombinant Type I-Fv Cascade can be produced from His-tagged Cas7fv, Cas5fv, Cas6f and crRNA in E.coli (37). We improved this purification procedure and also shifted the location of the His-tag from the N-terminus of Cas7fv to the C-terminus of Cas6f to be able to evaluate the Cas5fv:Cas7fv protein ratio (Figure 1A). Affinity chromatography of Cas6f was successfully used to co-purify mature 60 nt long crRNA and the untagged Cas5fv and Cas7fv proteins. The eluted complex was subjected to size-exclusion chromatography, which revealed four distinct peaks (Figure 1B) that were investigated via SDS-PAGE (Figure 1B). A small first peak contains Cas7fv filaments that were shown to form due to the unspecific binding of RNA molecules. The largest second peak contains the fully assembled Cascade ribonucleoprotein complex. This verifies that the pre-crRNA was produced, cleaved by Cas6f and incorporated into Cascade. An increased abundance of the backbone protein Cas7fv was evident. Two smaller peaks contained either Cas6f-Cas7fv complexes or Cas6f alone. Mature crRNA was not detected in the corresponding fractions. EMSAs verified that the recombinant Cascade complex specifically binds complementary DNA target-strands (Supplementary Figure S1).
Figure 1.
Purification of recombinant Type I-Fv Cascade interacting with His-Cas6f. (A) Schematic overview of the investigated recombinant Cas proteins and crRNA. (B) His-tagged Cas6f co-eluted with the Cascade components Cas7fv, Cas5fv and mature crRNA during size-exclusion chromatography (peak 2). Cas7fv filaments were observed in the void volume (peak 1) and Cas7fv-Cas6f complexes (peak 3) and Cas6f (peak 4) were separated. SDS-PAGE (top) and 8 M urea PAGE with toluidine blue staining (bottom) was used to analyze the protein and RNA content of the fractions corresponding to the peaks indicated in the gel-elution chromatogram (left).
The presence of Cas5fv and crRNA is required for proper Cascade assembly
Next, we asked which components are required for proper Type I-Fv Cascade assembly and investigated if a Cas-protein complex forms in the absence of crRNA. The three recombinant proteins Cas6f, Cas5fv and Cas7fv with an N-terminal His-tag were produced in E. coli and subjected to affinity- and subsequent size-exclusion chromatography. Two distinct peaks were obtained which were analyzed via SDS-PAGE. The first peak fractions contain the previously observed Cas7fv filaments and the largest second peak corresponds to Cas5fv-Cas7fv dimers (Figure 2A). Multimeric Cascade-like structures were not observed, which indicates that the Cas5fv-Cas7fv might be Cascade building blocks that require mature crRNAs as assembly signals. It is evident that Cas6f is necessary to process the pre-crRNA transcripts and Cas7fv is required to form the filamentous Cascade backbone (37). However, the role of Cas5fv during Cascade assembly is not known. Thus, we constructed a plasmid lacking the gene encoding for Cas5fv and attempted to produce recombinant Cascade structures. It was shown that Cascade cannot be reconstituted in the absence of Cas5fv (Supplementary Figure S2). Precipitation of Cas7fv was observed which suggests that it requires the interaction partner Cas5fv for stability. The small and indistinct peaks revealed Cas7fv-Cas6f interactions. Finally, we asked if the proteins Cas1 and Cas2-3 are stable subunits of the minimal Cascade. A larger plasmid was constructed that contains genes encoding all five Type I-Fv Cas proteins. The construct contained only one His-tag at the N-terminus of Cas1. Affinity chromatography revealed that Cas1 was co-purified with Cas2-3 and Cas6f (Supplementary Figure S3). However, crRNA was not present in the elution fractions and Cas5fv-Cas7fv-Cas6f Cascade-complexes were eluted in the flow-through. Taken together, these results suggest that the minimal DNA surveillance complexes are formed by three Cas proteins interacting with a second Cas1-Cas2-3 complex during target DNA degradation.
Figure 2.

Size-exclusion chromatography analysis of Cas proteins in the absence of crRNA. The Cas proteins were produced and purified via size-exclusion chromatography (see Figures 1 and 4) in the absence of crRNA. Cas7fv filaments (peak 1) and Cas5fv-Cas7fv dimers (peak 3) were observed but the characteristic middle peak (peak 2, elution volume 60–70 ml) was absent indicating the lack of Cascade backbone formation.
Analysis of Cascade protein–RNA interactions via mass-spectrometry
In order to pinpoint specific amino acids and nucleotides that interact within the minimal Type I-Fv Cascade complex, we utilized UV-induced cross-linking and mass spectrometry. Upon UV irradiation at 254 nm a covalent bond forms between the amino acid side chain and nucleic acid bases when both components are in close spatial proximity. The cross-linked peptide–RNA heteroconjugates can then be identified via LC-MS/MS analysis and a dedicated database search (38,39). This approach allowed for the identification of 13 peptides with different cross-links between amino acids and RNA fragment, which provides first insights into RNA recognition sites of the Cas proteins (Supplementary Figures S4-S8). The analyses confirmed extensive interactions of all three Cascade subunits with the crRNA (Figure 3). Most interactions were identified for Cas7fv, which is in agreement with the unspecific RNA binding function of this Cascade backbone protein. The crRNA was also found to interact with a distinct C-terminal region of Cas6f and an N-terminal region and the C-terminus of Cas5fv. The shortness of the identified cross-linked RNA moieties does not permit their unambiguous assignment to crRNA nucleotides. However, it is plausible that all identified Cas5fv interactions are with nucleotides (i.e. CU, UU, AAU) that are also found at the 5′-tag of the crRNA. Structural data are not available for Cas5fv and Cas7fv and only the structure of Cas6f can be modeled due its similarity to available Cas6 structures. Here, the observed interaction of the C-terminal domain of Cas6f with the crRNA is in agreement with the Cas6f-crRNA co-crystal structure from Pseudomonas aeruginosa (43). The identified cross-linked nucleotides (i.e. U, GU) are found at the very beginning of the 3′-terminal repeat tag of the crRNA.
Figure 3.
Mass-spectrometry analysis of Cas protein–crRNA interactions in Type I-Fv Cascade. Identified Cas protein peptides (red) cross-linked to the indicated nucleotides were subjected to tandem mass spectrometry fragmentation to identify which amino acid is cross-linked (blue). Potential interaction sites of Cas5fv (5.1–5.4) and Cas6fv (6.1 and 6.2) have been projected onto the crRNA sequence. Repeat tags flanking the spacer are indicated in green.
Biochemical analysis of synthetic Cascade variants with shortened and elongated crRNAs
Type I-E Cascade complexes form crescent-shaped structures with small subunits located along its belly. These small subunits were shown to interact with Cas7 and Cas6e (18). The absence of the small and large subunits suggests that the minimal Type I-Fv Cascade might facilitate the presence of a more variable backbone. To test if the Cascade backbone can be altered, we created pre-crRNA constructs with shortened and extended spacer sequences. The investigated crRNA contained a 32 nt spacer which was engineered into a drastically shortened crRNA with a 14 nt spacer (−18 nt) and a long 50 nt crRNA with an extended spacer (+18 nt). We verified that all three transcripts were produced in E. coli and processed by Cas6f into mature crRNA species (Supplementary Figure S9). Recombinant Cascade complexes were produced in the presence of the three crRNA variants. Surprisingly, all three variants resulted in stable ribonucleoprotein complex formation. The complexes were purified and subjected to size-exclusion chromatography. Each complex purification procedure resulted in three distinct peaks that were analyzed via SDS-PAGE (Figure 4). The first peak always corresponded to long Cas7fv filaments and the third peak corresponded to the previously observed Cas5fv-Cas7fv dimers. The constant distance between these two peaks demonstrated that the size-exclusion chromatography runs were performed under identical conditions to allow for their comparison. The highest absorbance value was always identified as a distinct middle peak. The corresponding fractions revealed complexes consisting of Cas6f, Cas5fv, multiple subunits of Cas7fv and the mature crRNA. The peak was shifted in accordance to the crRNA size. The gel-filtration column was calibrated to evaluate the approximate sizes of the eluted Cascade complexes (Supplementary Figure S9). The WT complex has a calculated size of 215 kDa, the short crRNA (−18 nt) is incorporated in a synthetic Cascade complex of 125 kDa and the long crRNA (+18 nt) is incorporated in a complex of 280 kDa. SDS-PAGE analysis demonstrated that the intensity of the Cas7fv bands varied. Thus, we conclude that the alteration of the crRNA spacer length can be compensated in synthetic Cascade complexes. Shortened crRNAs can result in the omission of Cas7fv subunits and elongated crRNAs can result in additional Cas7fv subunits. The S. putrefaciens Cas7fv has a molecular mass of 36 kDa which suggests that at least two Cas7fv subunits were added in the long Cascade complex (crRNA +18 nt) and three Cas7fv subunits were omitted in the short Cascade complex (crRNA −18 nt). To investigate the importance of the 6 nt periodicity that was observed for the E. coli Cascade, we repeated these experiments with crRNAs whose length was altered by 15 nt. Again, stable Cascade complexes were formed with short crRNAs (−15 nt) and long crRNAs (+15 nt) and the size-exclusion peaks correspond to Cascade assemblies with varied numbers of Cas7fv subunits (Supplementary Figure S10).
Figure 4.
Purification of recombinant synthetic Cascade variants with shortened and elongated spacers. Variants of crRNAs with wild type (WT) spacer (32 nt), short spacer length (14 nt) and long spacer length (50 nt) were designed. Production and maturation of the crRNA variants in E. coli was verified. Recombinant Cascade complexes were produced and purified via size-exclusion chromatography. Cas7fv filaments (peak 1) and Cas5fv-Cas7fv dimers (peak 3) were observed and the middle peak corresponded to fully assembled Cascade ribonucleoproteins (peak 2). The relative shift of this peak during identical size-exclusion chromatography runs and SDS-PAGE revealed that additional spacer nucleotides result in additional Cas7fv subunits.
Structural analyses of synthetic Cascade variants
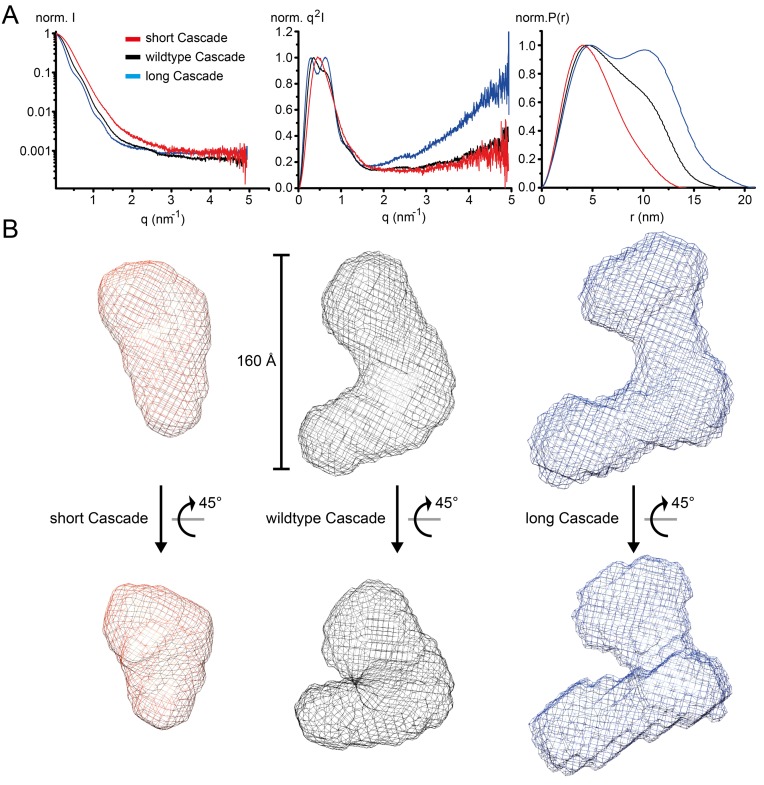
SAXS analysis was performed to assess the synthetic assemblies on a structural level in comparison to WT Cascade (Figure 5A). Ab initio shape restoration bead modeling revealed that distinct Cascade assemblies are formed, depending on the crRNA length (Figure 5B). WT Cascade forms a crescent assembly (Figure 5B, middle), reminiscent in size and shape of the sea-horse-shaped E. coli Cascade. Truncation of the scaffold crRNA by 18 nt resulted in a contracted structure (Figure 5B, left) and elongation of the RNA by 18 nt consequently manifested in an elongated assembly (Figure 5B, right). Kratky analysis of the scattering indicates that the long Cascade might be more flexible and structurally less folded, as judged from the increased q2I in the high q-range, compared to short and WT Cascade (Figure 5A, middle). Porod volume estimation revealed that WT Cascade features a volume of ∼360.28 nm3, whereas the short version is characterized by a volume of ∼207.82 nm3 and the long version by a volume of ∼519.09 nm3 (Supplementary Table S2). The volume difference of 152.46 nm3 (WT-short) and 158.81 nm3 (long-WT) suggests that an equal amount of His6-Cas7fv subunits was removed or added in respect to the crRNA length variation. To address how many subunit less or more were accommodated, we estimated the volume of His6-Cas7fv to be 44.972 nm3 ((1.21x MW)A3/molecule), arguing for the volume difference to correspond to three subunits.
Figure 5.
Small-angle X-ray scattering analyses of synthetic Cascade assemblies. (A) SAXS of the short (red), WT (black) and long (blue) Cascade constructs. Left: scattering curve, normalized to max I; middle: Kratky plot, normalized to max q2I, illustrating the ‘random coil likeliness’ differences in the high q-range; right: P(r) distance distribution curve, normalized to max P(r), highlighting the different domain organization. (B) Surface grid representations of averaged and filtered ab initio bead models calculated by Dammif (41).
A minimal Type I-Fv CRISPR-Cas system provides heterologous protection against lambda phage infections
The Type I-Fv CRISPR-Cas system was shown to interfere with plasmid conjugation in S. putrefaciens CN-32 (37). However, it remained possible that S. putrefaciens contained unidentified Cas proteins that could substitute the roles of the missing small and large subunits. We aimed to transfer the minimal system into E. coli to investigate this possibility and to screen if the system is able to target invading viruses. Two plasmids were transformed into E. coli BL21-AI, which does not contain endogenous cas genes. One plasmid (pCas6) harbored a cassette for the production of Cas1, Cas6f, Cas5fv, Cas7fv and the DNA nuclease Cas3. The second plasmid (pCRISPRλ) was designed to produce precursor-crRNA with a single spacer targeting gene E lambda phage (Supplementary Table S1, Figure 6A). Cell sensitivity to lambda phage was tested using standard plaque assays. The presence of all Type I-Fv Cascade compounds and a crRNA with a target ‘GG’ PAM reduced the EOP by 77% in comparison to the strain carrying empty plasmids (Figure 6). A crRNA with full spacer complementarity and a target ‘GA’ PAM resulted in an EOP of 0.94, demonstrating PAM recognition. In addition, a crRNA with a random spacer sequence showed an EOP of 0.98, indicating sequence-specific targeting of the lambda phage genome (Figure 6B). As a second control, a plasmid (pCas7) was constructed that allows the production of a nuclease-deficient Cas3 mutant (Cas3 H156A/D157A). This inactive Cas3 mutant did not confer immunity against lambda phage (EOP = 0.94) (Figure 6B). Taken together, these results show that the minimal Type I-Fv CRISPR-Cas subtype can be transferred into E. coli where it provides heterologous protection against lambda phage infections. The interference reaction is sequence- and PAM-specific, depends on Cas3 nuclease activity and does not require the presence of small and large subunits.
Figure 6.
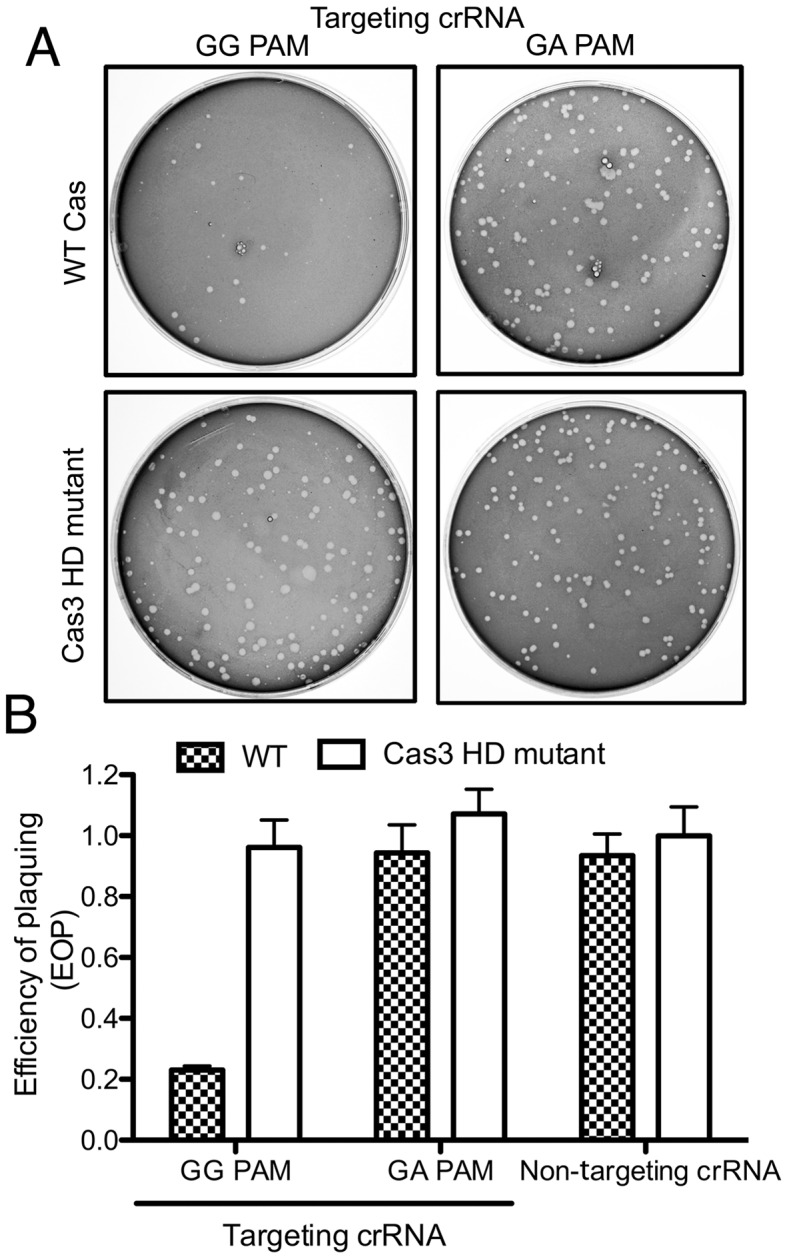
The S. putrefaciens Type I-Fv CRISPR-Cas system provides heterologous protection against lambda phage infection in E. coli. (A) Plaque formation by lambda phage was observed in E. coli BL21-AI strains carrying two plasmids: a first plasmid encoding a crRNA with a spacer of 32 nt complementarity to the lambda phage genome flanked either by a ‘GG’ or ‘GA’ PAM (targeting crRNA) and a second plasmid encoding all Cas proteins (pCas6) or pCas7 (Cas3 HD mutant). (B) Quantification of plaque formation was performed in triplicate (represented as efficiency of plaquing, EOP) of strains carrying the WT or the Cas3 HD mutant plasmid, in addition to a second plasmid producing either the targeting crRNA (in the presence of a target ‘GG’ PAM or a target ‘GA’ PAM) or a crRNA with a 32 nt non-targeting random spacer without complementarity to the phage genome. EOP is defined as the ratio between the plaque count of the strain of interest and the strain carrying empty plasmids. Bars represent mean ± SEM.
Cascade assemblies with modulated Cas7 backbones can retain functionality
Finally, we investigated the functionality of the synthetic Cascade variants by lambda phage interference assays. We changed the spacer length of the crRNA provided by the pCRISPRλ plasmid, while retaining all five cas genes on the pCas6 plasmid. Added nucleotides were designed to extend base complementarity with the target sequence. The plaque assay results showed that crRNAs with spacers elongated by 1 nt (EOP = 0.11), 6 nt (EOP = 0.23) or 9 nt (EOP = 0.09) retained activity, which was comparable to WT spacer activity (EOP = 0.18). Further elongation of the spacer reduced interference activity (Figure 7). Therefore, spacer length can be increased to a certain degree and crRNAs with a spacer length that is not found in native Type I-Fv CRISPRs can be functional. The reduction of the crRNA spacer length by either 6 or 18 nt resulted in Cascade variants that did not confer immunity against lambda phage (Figure 7).
Figure 7.
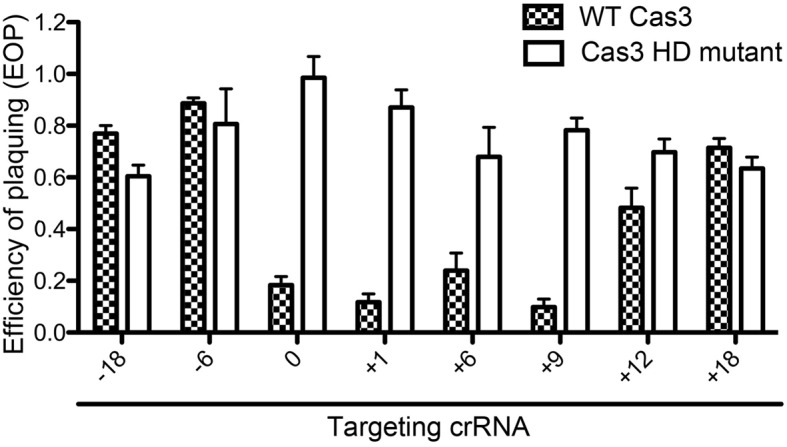
Cascade-mediated interference activity with extended and shortened crRNAs. Shown is the efficiency of plaquing of Type I-Fv CRISPR-Cas system variants expressed in E. coli. The 32 nt spacer sequence (0) was modulated by removal or addition of the indicated number of nucleotides (−18 to +18) while maintaining complementarity to the lambda phage sequence. These arrays were co-expressed in E. coli BL21-AI with pCas6 (in checkers) or pCas7 (Cas3 HD mutant, white bars), which was then subjected to plaque assays with lambda phage. Bars represent mean ± SEM of three independent experiments.
DISCUSSION
The three proteins Cas6f, Cas5fv and Cas7fv are sufficient to form a Cascade structure that can interact with Cas3 to target viral DNA in E. coli. Heterologous immunity was previously shown to be provided by Type II CRISPR-Cas, e.g. derived from Streptococcus thermophilus (16), and CRISPR-Cas systems usually exist as compact modules that have the potential to be transferred between species. However, all investigated Type I systems contain additional subunits, termed large subunits and the best studied system, the Type I-E CRISPR-Cas from E. coli, contains both, a large subunit, Cse1 and two small subunits, Cse2. Our results show that a functional Type I Cascade system can be transferred from S. putrefaciens into E. coli and can provide a DNA targeting mechanism in the absence of these additional subunits. Cse2 is described to be necessary for efficient binding of Cascade to dsDNA and Cse1 was found to be involved in PAM recognition and target DNA duplex destabilization (31,44). A crystal structure of the E. coli Cascade bound to a foreign double-stranded DNA (dsDNA) target revealed that distinct structural features of the large subunit Cse1 recognize the PAM duplex sequence (31). However, these structures also highlight that Cas5e is located in close vicinity of the PAM. Thus, we hypothesize that the highly divergent Cas5fv protein might play a critical role in DNA target binding and its presence might abrogate the need for a large subunit. Modeling attempts of the Cas5fv structure did not succeed, underlining its dissimilarity to other Cas5 proteins. The minimal Type I-Fv Cascade requires Cas3 HD nuclease activity to function and interactions between Cas1 and Cas3 were observed, in agreement with previous studies of the Type I-F system of Pectobacterium atrosepticum (36) and Type I-A systems (45). It is suggested that PAM-dependent foreign DNA processing involves the Cascade-mediated recruitment of Cas3, which creates a nick in the R-loop (46,47). Our results indicate that the Type I-Fv Cascade is sufficient for this Cas3 recruitment activity and other S. putrefaciens host proteins are not required. This minimal Cascade architecture might represent its most basic assembly model as the involved Cas5, Cas6 and Cas7 proteins were all proposed to be derived from a single ancestral RNA-binding protein (20). It is possible that cellular processes that generate ssDNA are coupled to the Type I-Fv Cascade, which could also explain its moderate activity. These minimal Cascade complexes are only observed in mesophilic bacteria, which might not require the most active defense measures as evidenced by down-regulated or absent CRISPR-Cas systems. Instead, they might benefit from CRISPR-Cas variants that effectively balance foreign DNA targeting activity and their fitness cost, e.g. by reducing the self-targeting potential.
E. coli Type I-E Cascade was shown to be produced in the absence of crRNAs (48). In this system, the Cas proteins Cas5, Cas7 and Cas6e form a stable complex with small and large subunits which could be loaded in vitro with synthetic crRNAs to generate functional DNA interference complexes. Similarly, Type I-A Cascade complexes can be purified without bound crRNAs (15). These observations are in stark contrast to the results obtained for the assembly of the minimal Type I-Fv Cascade, as this complex was shown to require crRNA for proper assembly. Furthermore, the length of the crRNA was identified to define the length of the Cas7fv backbone. It is possible that Type I-E Cascade can assemble without a crRNA scaffold if non-crRNAs provide a substituted scaffold. Thus, the defined length of the Cas7e backbone might be a result of protein–protein interaction between the capping enzymes Cas5e and Cas6e and the large and small subunits along the Cascade belly. These interactions are absent in the minimal Type I-Fv Cascade, which supports the notion that crRNA guidance is necessary for the Cascade assembly process. In addition, these minimal complexes are likely maintained as stable ribonucleoprotein units that do not permit the cycling of crRNAs. Our purification procedures of Cascade complexes without crRNAs yielded Cas5fv-Cas7fv dimers and the isolated Cas proteins are not stable. This suggests that these dimers could function as Cascade building blocks, following the Cas6f-mediated release of the crRNA's 5′-tag. Cas6f remains bound to the 3′-terminal repeat hairpin and would serve as a roadblock to crRNA-guided Cas7fv filament formation. This model entails that the alteration of the crRNA's spacer length results in modulated Cas7fv filaments. In agreement, drastically shortened crRNAs resulted in Cascade complexes with fewer Cas7fv subunits and elongated crRNAs resulted in added Cas7fv subunits. Flexibility of the Cas7 backbone was not yet observed for Type I Cascade systems, but is well documented for Type III systems. In these systems, Csm3 or Cmr4 act as ruler proteins that span the crRNA spacer in 6 nt increments (49,50). Variant crRNAs whose length differs by 6 nt are found in nature, which indicates that both forms can function in active Type III complexes. Thus, crRNA-guided backbone filament formation is a shared feature of multi-Cas protein complexes. In contrast, Type II, V and VI CRISPR-Cas systems each contain a single large Cas protein that spans the crRNA and filaments are absent. Nevertheless, the length of the crRNA in these systems can be altered without adjustment of the Cas protein content. It was shown that the length of the guide RNA–DNA interaction in Streptococcus pyogenes Cas9 can be reduced by 3 nt without affecting activity, while a reduction of 5 nt was not tolerated (51). Interestingly, the truncated guide RNAs improved the specificity of the Cas9 nuclease and the recognition of off-target sites was decreased. Our results demonstrate that the length of a Type I Cascade can be modulated and that only some alterations affect activity. Thus, the degree of modulating the Cascade backbone is likely limited by its requirement for productive Cas3 docking. Nevertheless, Cascade complexes with added spacer nucleotides will influence DNA targeting specificity. It has been shown that Cascade can be used as a gene silencing tool (52), but its large size might be a disadvantage for its transfer into heterologous hosts. However, endogenous Type I and Type III CRISPR-Cas systems can be efficiently utilized for genome editing purposes (53,54). In these cases, a minimal Cascade system without small and large subunits would provide the most compact CRISPR-Cas system with an adjustable Cas protein filament backbone. The variability of the spacer length of recombinant Type I-Fv Cascade opens the possibility of influencing DNA targeting specificity.
Supplementary Material
Acknowledgments
The authors thank Andre Plagens for helpful discussions. The authors also thank the European Synchrotron Radiation Facility (ESRF), Grenoble, France for beamline support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Max Planck Society and the Deutsche Forschungsgemeinschaft [DFG FOR 1680 to L.R. and H.U.]; LOEWE programme of the state of Hessen (to G.B. and L.R); P.P. is a fellow of the International Max Planck Research School for Environmental, Cellular and Molecular Microbiology (IMPRS-MIC). Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pourcel C., Salvignol G., Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 2.Mojica F.J., García-Martínez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 3.Grissa I., Vergnaud G., Pourcel C. Clustered regularly interspaced short palindromic repeats (CRISPRs) for the genotyping of bacterial pathogens. In: Caugant DA, editor. Molecular Epidemiology of Microorganisms. NY: Springer; 2009. pp. 105–116. [DOI] [PubMed] [Google Scholar]
- 4.Gesner E.M., Schellenberg M.J., Garside E.L., George M.M., MacMillan A.M. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Mol. Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 5.Richter H., Zoephel J., Schermuly J., Maticzka D., Backofen R., Randau L. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012;40:9887–9896. doi: 10.1093/nar/gks737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., Wiedenheft B., Pul Ü., Wurm R., Wagner R. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 7.Haurwitz R.E., Jinek M., Wiedenheft B., Zhou K., Doudna J.A. Sequence-and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carte J., Pfister N.T., Compton M.M., Terns R.M., Terns M.P. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nuñez J.K., Kranzusch P.J., Noeske J., Wright A.V., Davies C.W., Doudna J.A. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter C., Dy R.L., McKenzie R.E., Watson B.N., Taylor C., Chang J.T., McNeil M.B., Staals R.H., Fineran P.C. Priming in the Type IF CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuñez J.K., Lee A.S., Engelman A., Doudna J.A. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rouillon C., Zhou M., Zhang J., Politis A., Beilsten-Edmands V., Cannone G., Graham S., Robinson C.V., Spagnolo L., White M.F. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., Van Der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plagens A., Tripp V., Daume M., Sharma K., Klingl A., Hrle A., Conti E., Urlaub H., Randau L. In vitro assembly and activity of an archaeal CRISPR-Cas type IA Cascade interference complex. Nucleic Acids Res. 2014;42:5125–5138. doi: 10.1093/nar/gku120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulepati S., Héroux A., Bailey S. Crystal structure of a CRISPR RNA–guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson R.N., Golden S.M., van Erp P.B., Carter J., Westra E.R., Brouns S.J., van der Oost J., Terwilliger T.C., Read R.J., Wiedenheft B. Crystal structure of the CRISPR RNA–guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maier L.-K., Lange S.J., Stoll B., Haas K.A., Fischer S.M., Fischer E., Duchardt-Ferner E., Wöhnert J., Backofen R., Marchfelder A. Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system IB. RNA Biol. 2013;10:865–874. doi: 10.4161/rna.24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova K.S., Aravind L., Wolf Y.I., Koonin E.V. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garneau J.E., Dupuis M.-È., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 24.Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S., et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chylinski K., Le Rhun A., Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westra E.R., Semenova E., Datsenko K.A., Jackson R.N., Wiedenheft B., Severinov K., Brouns S.J. Type IE CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders C., Niewoehner O., Duerst A., Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staples J., Qiao D., Cho M.H., Silverman E.K., Nickerson D.A., Below J.E. PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet. 2014;95:553–564. doi: 10.1016/j.ajhg.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H., et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayes R.P., Xiao Y., Ding F., van Erp P.B., Rajashankar K., Bailey S., Wiedenheft B., Ke A. Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature. 2016;530:499–503. doi: 10.1038/nature16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiedenheft B., van Duijn E., Bultema J.B., Waghmare S.P., Zhou K., Barendregt A., Westphal W., Heck A.J., Boekema E.J., Dickman M.J., et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niewoehner O., Jinek M., Doudna J.A. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014;42:1341–1353. doi: 10.1093/nar/gkt922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sashital D.G., Wiedenheft B., Doudna J.A. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Judith R., James H.N., Malcolm F.W. CRISPR interference: a structural perspective. Biochem. J. 2013;453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter C., Gristwood T., Clulow J.S., Fineran P.C. In vivo protein interactions and complex formation in the Pectobacterium atrosepticum subtype I-F CRISPR/Cas System. PLoS One. 2012;7:e49549. doi: 10.1371/journal.pone.0049549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dwarakanath S., Brenzinger S., Gleditzsch D., Plagens A., Klingl A., Thormann K., Randau L. Interference activity of a minimal Type I CRISPR-Cas system from Shewanella putrefaciens. Nucleic Acids Res. 2015;43:8913–8923. doi: 10.1093/nar/gkv882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma K., Hrle A., Kramer K., Sachsenberg T., Staals R.H., Randau L., Marchfelder A., van der Oost J., Kohlbacher O., Conti E., et al. Analysis of protein-RNA interactions in CRISPR proteins and effector complexes by UV-induced cross-linking and mass spectrometry. Methods. 2015;89:138–148. doi: 10.1016/j.ymeth.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Kramer K., Sachsenberg T., Beckmann B.M., Qamar S., Boon K.L., Hentze M.W., Kohlbacher O., Urlaub H. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods. 2014;11:1064–1070. doi: 10.1038/nmeth.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirli K., Karaca S., Dehne H.J., Samwer M., Pan K.T., Lenz C., Urlaub H., Gorlich D. A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. eLife. 2015;4:e11466. doi: 10.7554/eLife.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 42.Harpaz Y., Gerstein M., Chothia C. Volume changes on protein folding. Structure. 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 43.Haurwitz R.E., Sternberg S.H., Doudna J.A. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012;31:2824–2832. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Erp P.B., Jackson R.N., Carter J., Golden S.M., Bailey S., Wiedenheft B. Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res. 2015;43:8381–8391. doi: 10.1093/nar/gkv793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plagens A., Tjaden B., Hagemann A., Randau L., Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinkunas T., Gasiunas G., Waghmare S.P., Dickman M.J., Barrangou R., Horvath P., Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulepati S., Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J. Biol. Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beloglazova N., Kuznedelov K., Flick R., Datsenko K.A., Brown G., Popovic A., Lemak S., Semenova E., Severinov K., Yakunin A.F. CRISPR RNA binding and DNA target recognition by purified Cascade complexes from Escherichia coli. Nucleic Acids Res. 2015;43:530–543. doi: 10.1093/nar/gku1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hatoum-Aslan A., Samai P., Maniv I., Jiang W., Marraffini L.A. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J. Biol. Chem. 2013;288:27888–27897. doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rath D., Amlinger L., Hoekzema M., Devulapally P.R., Lundgren M. Efficient programmable gene silencing by Cascade. Nucleic Acids Res. 2015;43:237–246. doi: 10.1093/nar/gku1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y., Pan S., Zhang Y., Ren M., Feng M., Peng N., Chen L., Liang Y.X., She Q. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2015;44:e34. doi: 10.1093/nar/gkv1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo M.L., Mullis A.S., Leenay R.T., Beisel C.L. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 2015;43:674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.