Abstract
Bioinformatic analysis often produces large sets of genomic ranges that can be difficult to interpret in the absence of genomic context. Goldmine annotates genomic ranges from any source with gene model and feature contexts to facilitate global descriptions and candidate loci discovery. We demonstrate the value of genomic context by using Goldmine to elucidate context dynamics in transcription factor binding and to reveal differentially methylated regions (DMRs) with context-specific functional correlations. The open source R package and documentation for Goldmine are available at http://jeffbhasin.github.io/goldmine.
INTRODUCTION
Many bioinformatics workflows, especially those that process genomic and epigenomic next generation sequencing (NGS) data, produce expansive data sets in the form of genomic ranges defined by chromosome, start position and end position and can represent phenomena such as somatic mutations, copy number variations, DNA– or RNA–protein binding sites and epigenetic state changes. The objective of Goldmine is to provide biologically-relevant annotation to genomic range sets and is motivated by two characteristics of such data. First, the range sets can be very large in size and require automated processing. For example, the number of peaks from a ChIP-seq experiment can range from the 100s to the 100 000s (1) and differentially methylated regions (DMRs) among human tissues can number in the 700 000s, even after stringent filtering criteria (2). Second, genomic ranges are not limited to gene bodies and can overlap with non-gene regulatory elements distal to genes, such as those established by large scale reference sequencing efforts (1,3,4). Analyzing how query genomic ranges from new studies relate to both known gene models and genomic features present in reference data can greatly facilitate hypothesis generation (Supplementary Figure S1A). Goldmine addresses the need to add interpretability, summarization and filtering to large sets of genomic ranges by annotating user-supplied genomic ranges with respect to known gene models and putative functional elements (Supplementary Figure S1B).
Existing tools for the analysis of genomic ranges fall into three categories. Goldmine belongs to a class of tools that provide detailed annotation of a query set of ranges to reference sets of ranges and gene models. Two existing tools with a similar concept are ChIPpeakAnno (5) and HOMER's annotatePeaks.pl (6). While these tools are ChIP-seq centric and provide nearest gene annotations, Goldmine is designed to accept genomic ranges from any source and also provides detailed feature annotation (Supplementary Table S1). A second category links genomic ranges to genes for the purposes of performing gene ontology enrichment and includes tools such as GREAT (7) and ChIP-ENRICH (8). Goldmine complements these tools by providing additional annotation of non-gene elements from reference data and can be used as a pre-filter to create query range sets to be provided to these other tools. For example, Goldmine could be used to stratify all query ranges that fall into known exons, and only these ranges are provided to GREAT for gene ontology analysis. The third category performs statistical enrichment calculations globally between a query set of ranges and a reference set of ranges to determine if range overlaps occur more than expected by chance. These tools include LOLA (9), GenometriCorr (10) and regioneR (11). Goldmine can work together with these existing tools for global enrichment calculations by providing a companion annotation that details the exact overlap for each individual range with combinations of reference set ranges, enabling the next level of candidate filtering and prioritization after an enrichment has been observed. Additionally, the range set enrichment tools can establish statistical significance if a frequent overlap is observed from manual inspection of a Goldmine annotation table. In summary, Goldmine provides detailed annotations and accountings of overlaps with both gene models and features, and automates complex tasks that would otherwise require manual download of data tables and custom programming.
MATERIALS AND METHODS
Obtaining and caching reference genomic and epigenomic data
In addition to user-supplied genomic ranges, Goldmine supports the direct loading into R of data from UCSC Genome Browser tables (12,13). These tables (viewable at https://genome.ucsc.edu/cgi-bin/hgTables) contain the bulk of extant annotation available for most species with assembled genomes. In the case of the human genome, the hg19 assembly contains data for multiple gene databases (including RefSeq, UCSC knownGene and ENSEMBL), non-coding RNA databases, the GWAS catalog, dbSNP, all data from the ENCODE project and various other feature sets including repeat elements, CpG islands and conserved elements. All tables can be loaded directly into R using the Goldmine function getUCSCTable(). The fread() function from the data.table package (http://CRAN.R-project.org/package = data.table) is employed for memory-efficient storage of large tables. To conserve bandwidth, tables can be cached and stored in a local repository, and only re-downloaded if an updated version is available. Each version downloaded by Goldmine is named with a date stamp corresponding to the last modified date on UCSC's FTP server, and specific tables can be loaded by date stamp or synchronization can be disabled to ensure reproducibility of results. Otherwise, the latest available versions of tables are always obtained. Goldmine is not limited to using feature sets from UCSC, and any desired range set from other sources can be utilized. The input features list to Goldmine can be a user-generated list of GenomicRanges objects from any source. GenomicRanges objects can be created from BED files using the included makeGRanges() function.
Annotating sets of genomic ranges
Automated annotation of a set of genomic ranges is performed using the goldmine() function (Supplementary Figure S1B). Internally, functions and data structures from the GenomicRanges package are employed for fast overlap operations (14). Goldmine reports quantitative overlap results, enabling the user to filter the extent of overlap as desired for downstream analysis. For the analyses presented here, we have defined overlap as any overlap between a query range and a range from a reference set (1 bp or more). The functions getGenes() and getFeatures() can be used to customize the gene and feature sets used by goldmine() and allow simplified loading of commonly used tables. The goldmine() function reports two types of annotation tables. The ‘wide’ format has the same number of rows as the query set. The much more detailed ‘long’ format reports each pair of overlapping query range and gene/feature range as a row (analogous to an inner join in a relational database, keyed by range overlap). The ‘wide’ format provides an easy to view summary of contexts, where each query range is annotated with the percent overlap with each gene model component or feature set. These percentages can be used to divide query ranges into categories based on genomic and feature context. A simple category call is made based on the gene models automatically, and multiple overlaps are resolved using the priority order promoter > gene 3′ end > exon > intron > intergenic. By default, promoters are defined as −1000 bp to +500 bp of a transcription start site and gene 3′ ends are defined as 1000 bp flanks both upstream and downstream of a transcription end site. Both definitions are user-adjustable. The distance to nearest gene, genes directly overlapped by the range and the genes that generated the context call are also reported. The ‘long’ format is useful for viewing individual overlaps with certain features or gene isoforms in full detail, as it captures all the complexity of the overlaps that produced the percentages reported in the ‘wide’ format.
Annotation of ENCODE ChIP-seq peaks using goldmine
The Goldmine function getFeatures() was used to obtain the ‘wgEncodeRegTfbsClusteredV3’ supertrack table. A copy of this data was generated and split into a list with one range set per factor using split(). The list of all sites was given as the query to goldmine(), and the split list was given as the features list. The context calls were aggregated to fractions of binding sites called by Goldmine in each context using data.table and plotted using ggplot2. Because this run annotated each binding site with the fraction of overlap with binding sites from each other factor, this output was also used to analyze co-occurrence biases on a per-context level. The feature annotation fraction columns were extracted and made into a matrix. The matrix was made Boolean by considering any overlap (fraction > 0) as TRUE, and FALSE otherwise. The fractions of these overlaps were then aggregated within each context for pairwise combinations of each factor with each other factor. Fractions were computed as the number of sites where factor A sites overlap with factor B sites divided by the total number of factor A sites (1). Experiment-specific peak lists used to ascertain context changes for the same factor across individual cells and conditions were created by parsing the supertrack based on the index available in the ‘wgEncodeRegTfbsClusteredInputsV3’ table at the UCSC genome browser. Goldmine was then applied separately for the list of peaks from each cell line. Results were aggregated by cell line, factor and context using data.table and plotted using ggplot2.
DNA methylation sequencing data processing
Methylated DNA immunoprecipitation sequencing (MeDIP-seq) read alignments in BED format were obtained from the Roadmap Epigenomics Projects under GEO accessions GSM543025, GSM613913, GSM669607, GSM543027, GSM613917 and GSM669609. The samples used are pairs of ‘CD4, Naïve Primary Cells’ and ‘CD8, Naïve Primary Cells’ from three human donors. The BED format alignments were converted to BAM format using the ‘bamtobed’ function of bedtools (15) and were sorted and indexed using samtools (16). The read depths for both cell types from one donor (TC009) were nearly twice as large as the other two donors, and these samples were downsampled to match the mean of the depth (57 349 008 reads) from the other samples using samtools.
Detection of differentially methylated regions (DMRs)
DMRs were detected using MethylAction (17). A window size of 50 bp and a fragment size of 266 bp were selected based on the protocol referenced in the GEO records. A subject-level effect was added to the testing model to account for the paired nature of the samples. Chromosome X and Y were excluded from the analysis. The methylaction() function was run with all other options as default, and the resulting DMR list was filtered to retain only those with ANODEV.padj < 0.01 and |log2(fold change)| > log2(1.5).
Annotation of DMRs
The getGenes() function was used to obtain the ENSEMBL genes for hg19 for annotation. The getFeatures() function was used to obtain the feature sets for annotation from the tables: ‘wgEncodeRegDnaseClusteredV3’, ‘wgEncodeRegTfbsClusteredV3’ and ‘gwasCatalog’. These features were concatenated with the output from getCpgFeatures() resulting in one feature list with the genomic ranges each for ENCODE DNaseI hypersensitive sites, ENCODE ChIP-seq peaks, GWAS catalog SNPs and CpG islands/shores/shelves. The drawGenomePool() option was used to draw a length-matched genomic null set of regions to the DMRs, sampling 100 times more regions than the query. These ranges were concatenated with the filtered MethylAction DMR set. The goldmine() function was run using these gene and feature lists and the GenomicRanges object containing both the DMRs and null regions as the query ranges. Resulting annotated data was saved for viewing using the gmWrite() function. The frequencies of each DMR pattern and the null set for overlapping with each gene model or feature context were aggregated and plotted using the R packages data.table and ggplot2.
Curation of DMRs with known and potential functions
The ‘call_genes’ column in the ‘context.csv’ file saved by Goldmine's gmWrite() function was used to search for DMRs in the promoters of known lineage factors for the CD4+ versus CD8+ fate decision. The ‘genes.csv’ file was filtered for rows with promoter fraction > 0, and a list of all unique ENSEMBL gene IDs (ENSG numbers) was saved. This list was provided to GeneMANIA (18) with the setting of zero related genes and attributes. The gene ontology (GO) term enrichment table was saved and plotted using ggplot2 for all terms with FDR < 15%. Plots of DMR regions were generated using ggbio (19) and the UCSC genome browser (13). ChromHMM and H3K27ac data for CD4+ and CD8+ T cells were obtained from the ‘Roadmap Epigenomics Data Complete Collection at Wash U VizHub’ track hub available from the UCSC browser.
Enrichment of consensus ChIP-seq peaks in DMRs
Enrichment was computed as the odds ratio between observed and expected frequencies of per-bp overlap rates between DMRs and all-methylated regions using a standard equation (20). The all-methylated regions were defined as the set of 50 bp windows with 4 or more reads in all 6 samples. These windows were overlapped with the genome-wide context GenomicRanges produced by the getGeneModels() function from Goldmine and categorized using the same priority order as Goldmine (promoter > gene 3′ end > exon > intron > intergenic). The ‘wgEncodeRegTfbsClusteredV3’ was obtained using getFeatures() and was converted into a list with one range set per factor using the split() function. For each DMR pattern and for each ChIP-seq factor, the fraction of bp in the DMR set that overlap with the given factor was compared to the same fraction in the all-methylated region set, and the odds ratio was calculated. Therefore, the background set used for each enrichment calculation is the set of all-methylated regions that fall in the same genomic context. This comparison is justified because promoters are compared to promoters, introns to introns and so forth. In other words, each enrichment is above that expected for any non-differentially methylated region in the same genomic context. Enrichments were considered significant if the lower bound of the 95% confidence interval (CI) of the odds ratio was >1 and more than 5% of base pairs in the DMR set were covered by the factor's ChIP-seq binding site ranges. The lower bound of the 95% CI was plotted on the heatmap. Non-significant enrichments were excluded from the heatmap (white squares).
RESULTS
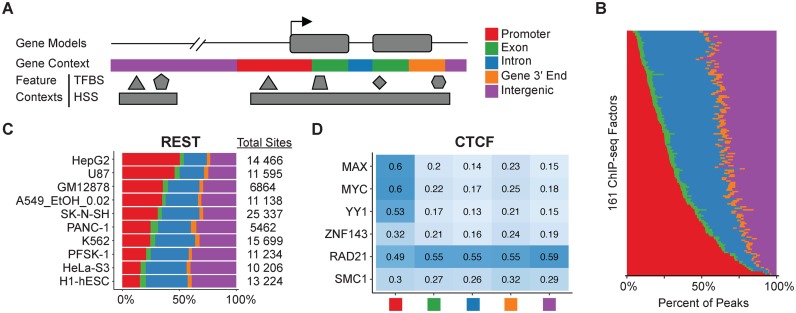
Using a reference gene database, Goldmine classifies genomic ranges as one of promoter, exon, intron, gene 3′ end, or intergenic (Figure 1A). To illustrate how this classification can capture biological information, we annotated a supertrack of cross-cell line ChIP-seq peaks from the ENCODE project. This analysis was enabled by Goldmine's capability to annotate any set of ranges with the percent overlap with any set of query ranges. The annotation revealed a spectrum of context-biased binding profiles (Figure 1B and Supplementary Table S2). We also found numerous examples where genomic contexts shift across cell lines and cell treatments for a given transcription factor (Supplementary Figure S2), and these suggest that the DNA-binding properties of such factors can be dynamic across biological conditions. For example, the binding sites of RE1-silencing transcription factor (REST) show a range of context biases from 15.5% promoter in H1-ESC cells to 50.5% promoter in HEPG2 cells, without a substantial change in total binding site number (Figure 1C). Because REST has known developmental roles (21), these shifts in context may reveal a balance between distal regulatory versus promoter regulatory functions throughout development (22).
Figure 1.
Goldmine automates the annotation of gene model and feature contexts for any set of genomic ranges. (A) Schematic of Goldmine's annotation approach. For gene context annotation, promoter and gene 3′ end regions are user-specified flanks surrounding annotated transcription start and end sites from gene databases that the tool can automatically download and synchronize. In cases of overlapping contexts, regions are classified using the priority order of promoter > 3′ end > exon > intron > intergenic. For feature contexts, Goldmine can take as input any number of user-specified feature sets of ranges or automatically download any table from the UCSC genome browser, including the ENCODE supertracks and GWAS catalog, and reports the percent overlap with these sets. (B) Proportion of ENCODE supertrack ChIP-seq peaks that annotate into the Goldmine gene contexts defined in (A). Each row is a proportional bar graph for an individual factor. (C) The proportion of REST ChIP-seq peaks across the named cell lines within each Goldmine gene model context. The total number of peaks for the factor in a cell line is given in the column next to the graph. (D) Each heatmap square is valued with the fraction of binding sites for CTCF that overlap with each co-binding partner given on the heatmap rows. Fractional overlaps are computed between the unions of all peaks across all available cell lines in ENCODE for each factor. Each column stratifies this relationship across the Goldmine genomic contexts.
DNA binding factors often function in complexes that can be revealed by ChIP-seq peak co-occurrence modules (1), and we identified multiple examples of co-binding preferences that are specific to certain genomic contexts using Goldmine (Supplementary Figure S3). A single command produced a co-occurrence matrix among all peaks by requiring a simple filter of >1 bp overlap. Because Goldmine provides detailed raw annotation, users can select custom thresholds for filtering and analyzing overlaps. One example of a factor with context-specific co-binding preferences is CCCTC-Binding Factor (CTCF) (Figure 1D). The genomic context and binding partners of given CTCF sites can delineate among the multiple transcriptional and structural functions of the protein (23). Using Goldmine, we identified that co-binding occurs with transcription factors MAX, MYC and YY1 highly at promoters but at lower levels in all other genomic contexts, suggesting promoters may be co-regulated by these factors and CTCF acting as a transcription factor (24). CTCF can also function as an insulator element at the boundaries of topological domains (25), and in contrast to the aforementioned transcription factors, Goldmine detected a co-occurrence module with chromatin interaction regulators ZNF143 (26), RAD21 and SMC1 (27) relatively evenly across all genomic contexts. This demonstrates how Goldmine can help stratify a set of genomic ranges based on co-occurrence with factors associated with distinct functions. By providing a unified and simple tool for annotating any set of genomic ranges with respect to gene model contexts, Goldmine enables global insights into the dynamics of phenomena mappable using NGS.
To further demonstrate how Goldmine's annotation can facilitate biological and functional interpretation of a genomic range set, we derived DMRs between CD4+ and CD8+ T cells from Roadmap Epigenomic Project MeDIP-seq data using MethylAction (17) and annotated the results using Goldmine (Supplementary Table S3). As the CD4+ versus CD8+ lineage decision is a model for bivalent differentiation patterns (28), an analysis of this methylome-wide data can reveal how epigenetics interacts with both known and novel drivers of this developmental process. MethylAction provided as output the genomic ranges for 910 CD4+ hypermethylation DMRs and 1005 CD8+ hypermethylation DMRs (Figure 2A), and Goldmine annotation showed that the DMRs distributed in all genomic contexts (Figure 2B). While DNA methylation is commonly studied in a promoter-centric manner, the context analysis reveals widespread DNA methylation changes outside of the promoter. Such revelation can aid in context-specific hypothesis generation. For example, gene body DMRs may be associated with gene activation (29) or alternative splicing (30). Intergenic DMRs may target distal regulatory elements, such as enhancers and repressors, that could regulate many genes and be dynamic throughout developmental processes (31). Goldmine can also annotate input genomic ranges with reference feature ranges. Using this functionality on the T cell DMR data, a bias against CpG islands is evident (Figure 2C), and the DMRs are also enriched for overlap with ENCODE ChIP-seq and DNaseI-seq data (Figure 2D). Such feature annotation can be particularly useful for generating functional hypotheses for intergenic genomic ranges, as feature sets that capture regulatory elements and variation can be employed.
Figure 2.
Goldmine gene annotation links genomic ranges to known gene models. (A) DMRs were detected between CD4+ and CD8+ T cells. Each heatmap row represents a DMR, each column a donor, and each value the fold change between the two cell types for paired samples from a given donor. (B) Percent of DMRs that fall in gene model contexts as compared to a length-matched random genomic null region set. (C) Proportion of DMRs between CD4+ and CD8+ T cells that overlap with CpG-island centric features by Goldmine. CpG islands are annotated in the ‘cpgIslandExt’ table of the UCSC genome browser, shores are ±2 kb from these islands, and shelves are ±2 kb from shores. (D) Proportion of DMRs between CD4+ and CD8+ T cells that overlap with ENCODE ChIP-seq peaks (‘TFBS’) or DNaseI hypersensitive sites (‘DNaseI’) as reported by Goldmine. (E) Regional perspective of a promoter DMR for key lineage factor gene CD4 that was identified using Goldmine's annotation. (F) GO term enrichment for promoter DMR genes. ENSEMBL gene IDs were directly copied from Goldmine's gene-level table and pasted into GeneMANIA (http://www.genemania.org/). (G) An intergenic CD4+ hypermethylation DMR (chr8:2,162,901-2,163,500) with hypothesized function based on Goldmine annotation. This DMR correlates with the activity of an enhancer as predicted by ChromHMM segmentation and the presence of H3K27ac (data from the Roadmap Epigenomics Project). A cluster of ENCODE ChIP-seq peaks (‘TFBS’) and a DNaseI hypersensitive site (‘DNaseI’) that overlap with the DMR as reported by Goldmine are shown. (H) Variable enrichment of ENCODE supertrack ChIP-seq peaks in CD4+ hypermethylation DMRs across the contexts as compared to when the DMR set is not stratified by context (‘All’). The background set used for the enrichment calculation is the set of all-methylated regions genome-wide that also fall in the given genomic context. Significance was assigned when >5% of base pairs in a DMR overlapped with peaks of a given factor, and the lower bound of the 95% confidence interval (CI) of the odds ratio between the DMRs and all non-DMR methylated regions was above 1. Non-significant comparisons are plotted as white, and significant comparisons are colored by the value of the lower bound of the 95% CI of the odds ratio.
By sectioning gene models into components (Figure 2B), Goldmine reveals detailed information about the overlap of the DMRs with transcription units that could be missed by simply overlapping with gene bodies as single units. The annotation immediately revealed the presence of promoter hypermethylation at key lineage genes CD4 (32) (Figure 2E) and CD8A, which corresponds to the expected expression patterns of these genes in the two T cell lineages. The promoter-overlapping gene list saved directly from Goldmine is enriched for gene ontology terms related to T cell receptor and immunity (Figure 2F). Additionally, Goldmine's ‘long format’ annotation provides a detailed accounting of the complex relationships between query regions and gene models by describing all isoforms, introns, exons, nested and overlapping genes. This demonstrates how Goldmine's gene model annotation streamlines and automates the process of deriving biologically relevant loci from a large set of anonymous genomic ranges.
In addition to summary annotations, Goldmine provides detailed descriptions for each pair of overlaps between query ranges and genomic feature sets. This is important because intergenic regulatory elements often involve multiple DNA binding factors with side-by-side binding sites. By filtering the DMR list to those annotated as intergenic and overlapping with both ChIP-seq and DNaseI-seq sites, we identified an intergenic DMR that correlates with a putative enhancer (Figure 2G). Goldmine's gene model and feature annotations can also function in tandem to provide information about regions that might otherwise be overlooked. We computed enrichment of CD4+ hypermethylation DMRs for consensus ChIP-seq peaks from all of ENCODE and found variable enrichment levels across gene model contexts (Figure 2H). When performing the analysis on all DMRs together, only 3 factors achieve statistical significance. However, when the analysis is stratified by context annotations, enrichments unique to each gene context are discovered. Of note, binding sites for RUNX3, a known master regulator in the CD4+ versus CD8+ fate decision (33,34) were found to be enriched in promoters and introns. Additionally, RUNX3 expression is known to be repressed in CD4+ cells, and Goldmine annotation identified RUNX3 promoter to be hypermethylated in CD4+ cells (Supplementary Table S3). Taken together, these observations, made possible by Goldmine, suggest that RUNX3 expression may be directly regulated by promoter methylation and that its transcriptional function may also be modulated by DNA methylation in CD8+ cells. The application of Goldmine to the DMR's between CD4+ and CD8+ T cells illustrates the usefulness of annotation to divide relevant subsets of genomic ranges into those of biological interest and to work in tandem with existing tools for motif and gene set enrichment analysis.
DISCUSSION
Compared with other tools for genomic range annotation, a key distinction of Goldmine is that it enables real-time synchronization with the latest annotation tables, so gene models can be used from the latest builds of reference databases. While this automation applies to any tables available from the UCSC Genome Browser, the user has complete flexibility to use any set of genomic ranges as a reference. Any ranges that can be input to R and stored as GenomicRanges (14) can be used, enabling Goldmine to annotate with respect to any reference range sets of interest that can be derived from existing Bioconductor (35) annotation packages or generated from external file formats such as BED files. In Figure 2G, we used the ChromHMM (3) calls derived from a BED file available from the Roadmap Epigenomics Project (4). Another unique feature is that Goldmine provides annotation on the level of transcripts in the format of a detailed table with a list of the specific introns and exons overlapped by a genomic range. To our knowledge, no existing tool provides this level of detail. Transcript-level annotation can be valuable, such as in the case of linking epigenetic phenomena to co-transcriptional RNA processing (30,36). Additionally, RNA-centric techniques such as HITS-CLIP can map RNA–protein binding, and Goldmine can be a valuable tool to examine the diversity of transcripts produced by each peak event. While we note the limitation of this analysis is that the results are correlative, candidate selection is a requisite step before embarking on detailed experimental studies of novel mechanisms at specific loci. Goldmine should be considered as a tool to establish and prioritize these candidate sets. Moreover, Goldmine's detailed annotation complements existing genomic range analysis tools focused on global gene set and region set enrichment.
As demonstrated in Figure 2G, Goldmine is also distinguished by the fact that it is not limited to gene-centric annotation. In this example, Goldmine was used to identify an epigenetic change that directly overlaps with an annotated enhancer and correlates with the activity of this enhancer as derived from reference histone modification data in the relevant cell types. Not only can Goldmine detect individual loci with such specific overlaps of interest to the biologist, it can comprehensively catalog sites with desired overlap characteristics, whether they involve gene models or not. Thus, Goldmine is an information integration tool that can narrow large sets of genomic ranges into those that match specific configurations in relationship to reference data, and can be used to derive comprehensive lists of candidate loci for further experimental testing.
In summary, because Goldmine is designed to work for any set of genomic ranges, regardless of source or type, it is widely applicable to genomic ranges produced from genetic mutation data, RNA-focused assays such as Ribo-seq and HITS-CLIP, and many epigenome-wide sequencing data including DMRs, ChIP-seq peaks, differential histone modification/positioning, and DNaseI hypersensitive sites. Goldmine also simultaneously provides feature-level annotations that can be used to leverage recent epigenome-wide data sets, which are of particular utility in describing intergenic genomic ranges that may co-occur with functional elements. As a result, Goldmine reduces the complexity of the extant genomic and epigenomic annotation to aid in the prioritization of candidate loci for experimental testing.
Supplementary Material
Acknowledgments
The authors acknowledge the Roadmap Epigenomics Consortium (http://nihroadmap.nih.gov/epigenomics/) and the ENCODE Consortium (https://www.encodeproject.org/) for generating data used in this report.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute of the National Institutes of Health [R01CA154356 to A.T., F31CA195887 to J.M.B]; Scott Hamilton CARES Initiative; J.M.B. is a predoctoral student in the Molecular Medicine PhD Program of the Cleveland Clinic Lerner College of Medicine of Case Western Reserve University, funded in part by the ‘Med into Grad’ initiative of the Howard Hughes Medical Institute (HHMI). Funding for open access charge: National Cancer Institute [R01CA154356].
Conflict of interest statement. None declared.
REFERENCES
- 1.Griffon A., Barbier Q., Dalino J., van Helden J., Spicuglia S., Ballester B. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res. 2015;43:e27. doi: 10.1093/nar/gku1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziller M.J., Gu H., Müller F., Donaghey J., Tsai L.T.-Y., Kohlbacher O., De Jager P.L., Rosen E.D., Bennett D.A., Bernstein B.E., et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ernst J., Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S., Green M.R. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch R.P., Lee C., Imbriano P.M., Patil S., Weymouth T.E., Smith R.A., Scott L.J., Sartor M.A. ChIP-Enrich: gene set enrichment testing for ChIP-seq data. Nucleic Acids Res. 2014;42:e105. doi: 10.1093/nar/gku463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheffield N.C., Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Favorov A., Mularoni L., Cope L.M., Medvedeva Y., Mironov A.A., Makeev V.J., Wheelan S.J. Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput. Biol. 2012;8:e1002529. doi: 10.1371/journal.pcbi.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gel B., Díez-Villanueva A., Serra E., Buschbeck M., Peinado M.A., Malinverni R. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics. 2016;32:289–291. doi: 10.1093/bioinformatics/btv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhasin J.M., Hu B., Ting A.H. MethylAction: detecting differentially methylated regions that distinguish biological subtypes. Nucleic Acids Res. 2016;44:106–116. doi: 10.1093/nar/gkv1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin T., Cook D., Lawrence M. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 2012;13:R77. doi: 10.1186/gb-2012-13-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao L., Shen H., Laird P.W., Farnham P.J., Berman B.P. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol. 2015;16:105. doi: 10.1186/s13059-015-0668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thakore-Shah K., Koleilat T., Jan M., John A., Pyle A.D. REST/NRSF knockdown alters survival, lineage differentiation and signaling in human embryonic stem cells. PloS One. 2015;10:e0145280. doi: 10.1371/journal.pone.0145280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldmann A., Ivanek R., Murr R., Gaidatzis D., Burger L., Schübeler D. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet. 2013;9:e1003994. doi: 10.1371/journal.pgen.1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holwerda S.J.B., de Laat W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013;368:20120369. doi: 10.1098/rstb.2012.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubois-Chevalier J., Oger F., Dehondt H., Firmin F.F., Gheeraert C., Staels B., Lefebvre P., Eeckhoute J. A dynamic CTCF chromatin binding landscape promotes DNA hydroxymethylation and transcriptional induction of adipocyte differentiation. Nucleic Acids Res. 2014;42:10943–10959. doi: 10.1093/nar/gku780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey S.D., Zhang X., Desai K., Aid M., Corradin O., Cowper-Sal Lari R., Akhtar-Zaidi B., Scacheri P.C., Haibe-Kains B., Lupien M. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat. Commun. 2015;2:6186. doi: 10.1038/ncomms7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sofueva S., Yaffe E., Chan W.-C., Georgopoulou D., Vietri Rudan M., Mira-Bontenbal H., Pollard S.M., Schroth G.P., Tanay A., Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Germain R.N. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2002;2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 29.Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H.J., Lowdon R.F., Maricque B., Zhang B., Stevens M., Li D., Johnson S.L., Wang T. Developmental enhancers revealed by extensive DNA methylome maps of zebrafish early embryos. Nat. Commun. 2015;6:6315. doi: 10.1038/ncomms7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sellars M., Huh J.R., Day K., Issuree P.D., Galan C., Gobeil S., Absher D., Green M.R., Littman D.R. Regulation of DNA methylation dictates Cd4 expression during the development of helper and cytotoxic T cell lineages. Nat. Immunol. 2015;16:746–754. doi: 10.1038/ni.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohu K., Sato T., Ohno S.-I., Hayashi K., Uchino R., Abe N., Nakazato M., Yoshida N., Kikuchi T., Iwakura Y., et al. Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single-positive lineage. J. Immunol. 2005;174:2627–2636. doi: 10.4049/jimmunol.174.5.2627. [DOI] [PubMed] [Google Scholar]
- 34.Woolf E., Xiao C., Fainaru O., Lotem J., Rosen D., Negreanu V., Bernstein Y., Goldenberg D., Brenner O., Berke G., et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y.C., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yearim A., Gelfman S., Shayevitch R., Melcer S., Glaich O., Mallm J.-P., Nissim-Rafinia M., Cohen A.-H.S., Rippe K., Meshorer E., et al. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015;10:1122–1134. doi: 10.1016/j.celrep.2015.01.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.