Abstract
Mitochondria have their own translation machinery that produces key subunits of the OXPHOS complexes. This machinery relies on the coordinated action of nuclear-encoded factors of bacterial origin that are well conserved between humans and yeast. In humans, mutations in these factors can cause diseases; in yeast, mutations abolishing mitochondrial translation destabilize the mitochondrial DNA. We show that when the mitochondrial genome contains no introns, the loss of the yeast factors Mif3 and Rrf1 involved in ribosome recycling neither blocks translation nor destabilizes mitochondrial DNA. Rather, the absence of these factors increases the synthesis of the mitochondrially-encoded subunits Cox1, Cytb and Atp9, while strongly impairing the assembly of OXPHOS complexes IV and V. We further show that in the absence of Rrf1, the COX1 specific translation activator Mss51 accumulates in low molecular weight forms, thought to be the source of the translationally-active form, explaining the increased synthesis of Cox1. We propose that Rrf1 takes part in the coordination between translation and OXPHOS assembly in yeast mitochondria. These interactions between general and specific translation factors might reveal an evolutionary adaptation of the bacterial translation machinery to the set of integral membrane proteins that are translated within mitochondria.
INTRODUCTION
Mitochondria possess their own translational machinery required for the expression of the few genes encoded by the mitochondrial DNA (mtDNA). This machinery is associated with the mitochondrial inner membrane allowing the co-translational insertion of mtDNA-encoded membrane subunits of the three respiratory complexes I, III and IV as well as the ATP synthase (oxidative phosphorylation (OXPHOS) complexes). The components of the mitochondrial protein synthesis apparatus are distinct from their cytoplasmic counterparts and in general share homologies with their bacterial equivalents.
The different steps of the translation process involve a limited number of auxiliary protein factors that ensure the speed and accuracy of each step. While the essential role of these auxiliary factors has been extensively analyzed in bacteria, two major difficulties have complicated the study of the corresponding mitochondrial factors. First, no in vitro translation system able to carry out the complete translation of a mitochondrial mRNA (mt-mRNA) has been established despite a number of years of effort with human and yeast mitochondria. Second, the in vivo analysis of the phenotype of Saccharomyces cerevisiae mutants of these mitochondrial factors has not been very informative since defects in translation components generally lead to mtDNA instability (1). To circumvent these difficulties, the auxiliary factors in human mitochondria have been studied in vitro using purified factors and mitochondrial or bacterial ribosomes (2).
Few studies have focused on the mitochondrial ribosome-recycling step that should a priori differ from the bacterial step due to the binding of the mitochondrial ribosome to the inner membrane. In bacteria, there have been many debates about the mechanism of ribosomal recycling and the role of the three factors involved: ribosome recycling factor (RRF), elongation factor G (EF-G) and initiation factor 3 (IF3) (3,4). To summarize, the ribosome-recycling factor RRF interacts with the large ribosomal subunit and dissociates the ribosome through interaction with the GTPase EF-G, that also acts at the elongation step. Subsequently, the initiation factor IF3 binds to the small ribosomal subunit and prevents the re-association of free ribosomal subunits. In human and yeast mitochondria, the homologs of RRF and IF3 have been identified (5–8) and there are two homologs of EF-G, one (hEFG1/Mef1) acting at the elongation step and the other (hEFG2/Mef2) at the recycling step (9,10). Purified human mitochondrial mtRRF and hEFG2 disassemble ribosomes into the small and large subunits and both factors remain on the large subunit until GTP is hydrolyzed. Depletion of either mtRRF or hEFG2 causes profound mitochondrial dysfunctions (5,11). In addition, purified human mitochondrial mtIF3 cross-links with proteins of the small ribosomal subunit, assists the dissociation of mitochondrial ribosomes and promotes the formation of the initiation complex in the presence of the mitochondrial initiation factor 2 (2). Thus, mtIF3 is active at the interface between recycling and initiation. In S. cerevisiae, deletion of Rrf1, Mef2 or Mif3/Aim23 (the RRF, EF-G2 and IF3 homologs, respectively) was reported to lead to mtDNA loss in a yeast strain with an intron-containing mtDNA, although recently it has been shown that a mtDNA with a reduced intron content is more stable in a Δmif3 background. Interestingly, some mutants like S. pombe Δrrf1 or S. cerevisiae Δmif3 can be complemented by the human homologs (5,6,8,11–13).
Moreover, specific translational activator proteins absent in bacterial translation are also required at the initiation step of mitochondrial translation. They have been extensively studied in S. cerevisiae mitochondria (14–16). They are specific for each mitochondrial mRNA, are often associated with the ribosomes and tether mRNA to the inner membrane. For example, the translational activator Mss51 controls the synthesis of Cox1, a catalytic subunit of complex IV through its interaction with Pet309 and the COX1 mRNA. In addition, Mss51 is recruited to early assembly intermediates containing the newly synthesized Cox1 polypeptide. In a wild type strain, Mss51 is predominantly found in assembly competent complexes that also contain assembly factors such as Cox14 or Coa3 (Cox25) that control the first step of Cox1 assembly (17–21). This form is known as Mss51A. In the absence of these assembly factors, Mss51 accumulates in a low molecular weight complex Mss51T that was proposed to be the source of the translationally-active form. Thus, Mss51 is a key player in a translational feedback control loop that coordinates the synthesis of Cox1 and its assembly with the other subunits of complex IV (22,23).
In human cells, an assembly intermediate, called MITRAC for "mitochondrial translation regulation assembly intermediate of cytochrome c oxidase" plays a similar role in the coupling between COX1 synthesis and assembly and its dysfunction results in severe human mitochondrial disorders (24–27). MITRAC contains homologs of assembly factors that are known to interact with Cox1 and Mss51 in yeast, such as C12ORF62 and MITRAC12 (Cox14 and Coa3 in yeast). A homolog of Mss51 has been also recently shown to be localized in the mitochondria of skeletal muscle and to modulate cellular metabolism (28,29); however, to date there is no evidence to suggest that it is part of the human MITRAC assembly intermediate.
Generally, mutations in yeast specific translation factors do not destabilize mtDNA while defects in general translation lead to mtDNA loss. Surprisingly, we previously reported that in an S. cerevisiae strain carrying an intron-less mtDNA, the absence of the ribosome-recycling factor Rrf1 is respiratory competent and does not lead to mtDNA instability (Supplementary Material S2 from (5)), thus allowing us to study in vivo defects in the ribosome recycling step and make comparisons with other steps of mitochondrial translation. In this paper, we have analyzed the phenotype of yeast strains devoid of mitochondrial introns and carrying mutations in, or deletions of, genes coding for mitochondrial auxiliary translation factors. We find that the loss of factors acting at the ribosome recycling step (Rrf1) or at the interface between recycling and initiation (Mif3) increases the synthesis of several OXPHOS subunits while compromising the assembly of complex IV and adenosine triphosphate (ATP) synthase (complex V). We further show a functional interaction between Rrf1 and the specific translational activator Mss51. Taken together, our results demonstrate that Rrf1 and Mif3 despite their role in the ribosome recycling step are not essential for mitochondrial translation but actively participate in coupling the synthesis and the assembly of OXPHOS subunits.
MATERIALS AND METHODS
Media and strain construction
Media were prepared as in (30). The respiratory medium contains 2% glycerol; fermentable media contain either 2% glucose or 2% galactose and 0.1% glucose. Galactose is a fermentable substrate that does not cause the repression of genes encoding mitochondrial proteins while glucose does.
All the strains are listed in Supplementary Table S1. Each mutant was constructed in the strain CW252 that has the W303-1B nuclear background (MAT alpha ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100) and an intron-less mitochondrial genome (31). Each mutation was introduced at the chromosomal locus by integrative transformation. Only the Δmef2 and Δnam2 mutants lost their mtDNA (rho°). Thus, the point mutants mef2-K54A and nam2-W238C were constructed. In both cases, the wild-type genes were mutated on centromeric plasmids and introduced into the corresponding deletion mutants, which were then re-populated with the intron-less mitochondrial genome by cytoduction. Cytoduction experiments were performed by crossing the mutants with JC8/252 (MAT a kar1-1 leu1, rho+) with an intron-less mtDNA; kar1-1 mutation delays nuclear fusion in the zygotes and allows the budding of mutant cells with an intron-less mtDNA. The percentage of mtDNA deletion mutants (rho− mutants) accumulating in glucose medium at 28°C after about 15 generations was estimated by spreading haploids cells on YPD plates at a dilution giving single colonies after 2 days of incubation at 30°C. Colonies were crossed with a lawn of rho° haploid strain (KL14-4A MAT a his1, trp2, rho°) and incubated overnight at 30°C. The resulting diploid cells were tested for their respiratory growth on non-fermentable medium. Rho− mutants crossed with the rho° tester strain give respiratory deficient diploids, whereas diploids resulting from crossing rho+ with the rho° tester strain are respiratory competent.
Rrf1 and Mss51 were tagged at their C-termini with three copies of the HA, or 13 copies of the c-Myc epitope respectively using the Schizosaccharomyces pombe his5 gene as selection marker (Sphis5 complements the S. cerevisiae mutation his3) as described in (32). In each case, the correct integration of the mutation or the tag was confirmed by polymerase chain reaction (PCR) amplification and sequencing. We verified that the addition of the tags did not modify the respiratory phenotype.
In vivo labeling of mitochondrial translation products
The S. cerevisiae cells were grown to early exponential phase in galactose medium (0.8 OD600 units) and for each strain precisely 1.6 OD600 units were collected and incubated at 30°C for 2.5 to 10 min with 35S methionine and cysteine in the presence of cycloheximide that blocks cytoplasmic translation. When a chase was performed the cells were labeled for 10 min and then washed in galactose medium containing an excess of cold methionine and cysteine before further incubation in the same medium supplemented with cycloheximide. Total proteins were extracted according to (33) and two identical gels were subjected to SDS-PAGE (17% acrylamide 0.5% bis-acrylamide), each with half of the samples. One gel was stained with Coomassie blue, dried and the labeled proteins were revealed by autoradiography; the other gel was transferred onto a membrane, analyzed by Western blotting and also revealed by autoradiography.
Northern blot analyses
Total RNAs were purified from galactose grown cells using the ‘hot phenol’ technique (34). The RNAs were separated on 1.2% agarose formaldehyde gels and were transferred onto Hybond-N membrane (Amersham). The blots were successively hybridized with different probes at 42°C in 50% formamide in the presence of Denhardt's solution (see Supplementary Table S2 for a description of the probes). For detection of the mitochondrial RNAs, PCR-amplified fragments internal to each mitochondrial gene were generated and used to produce radiolabeled probes by random priming (Invitrogen).
Cytochrome absorption spectra
Cytochrome absorption spectra of whole cells grown in galactose medium were recorded at liquid nitrogen temperature after reduction by dithionite using a Cary 400 spectrophotometer (Varian, San Fernando, CA, USA). Cytochromes c1 and b are part of complex III while cytochromes a + a3 are part of complex IV. Absorption maxima for the alpha-bands of cytochromes c, c1, b and a + a3 are expected at 546, 552, 558 and 602 nm, respectively.
SDS-PAGE, antibodies
Mitochondria were isolated from mid-phase galactose grown cells by differential centrifugations after digestion of cell walls with Zymoliase-100T (35). Mitochondrial proteins were resolved on SDS-PAGE followed by immunoblotting with various antibodies.
Polyclonal antibodies against Rrf1 were raised against a recombinant protein expressed in E. coli. The RRF1 sequence between the nucleotides 57 and 690 was cloned in the Pet24b bacterial expression vector in order to produce a recombinant protein with a 6-His tag. The recombinant protein was expressed in Escherichia coli after isopropyl beta-D-1-thiogalactopyranoside (IPTG) induction and was purified on Ni-NTA agarose. Polyclonal antibodies against Cyt1 (31) and Cytb (36) had been previously raised in the laboratory against recombinant proteins expressed in E. coli. Other polyclonal antibodies were gifts: Atp2 and Atp9 (37), Mrp20 (38), Ssc1 (39) and Tom40 (40). Monoclonal antibodies against yeast Cox1 (11D8-B7), Cox2 (4B12-A5) and porin (16G9-E6) were purchased from Molecular Probes (Eugene, USA), anti-human Hsp60 (LK2) and anti-HA (HA-7) were from Sigma-Aldrich and the anti-cMyc (9E10) was from Roche Life Sciences. Bound antibodies were detected using enzyme-linked secondary antibodies and a chemiluminescent substrate (Pierce).
BN-PAGE
Mitochondria were lysed in a buffer containing either 2% digitonin or 2% dodecyl maltoside in presence of phenylmethane sulfonyl fluoride (PMSF) and the complete protease inhibitor cocktail (Roche Life Sciences). Mitochondrial complexes were separated on 3–12% or 4–16% BN-PAGE gels, as described in (41) and modified in (35) or using the precast polyacrylamide gel system from Life Technology. Protein markers from Amersham Biosciences were used to estimate the molecular weights of the complexes. The blue-native gels were transferred to nitrocellulose and immunoblotted with four antibodies to reveal the positions of each complex: anti-Atp2 (complex V), anti-Cyt1 (respiratory complex III), anti-Cox2 (respiratory complex IV), anti-Tom40 (outer-membrane import complex).
Immunoprecipitation
Mitochondria were isolated from the tagged strains and solubilized in 1% digitonin. The suspension was centrifuged for 15 min at 4°C at 100 000 g. HA or c-Myc-antibodies coupled to agarose beads (Sigma-Aldrich) were added to the supernatant. Samples were incubated with gentle shaking for 90 min at 4°C in presence of PMSF and the complete protease inhibitor cocktail (Roche Life Sciences). The beads were washed twice with the lysis buffer, the immunoprecipitates were eluted with 50 μl of sample loading buffer, without ß-mercaptoethanol, and the different fractions were analyzed by SDS-PAGE and Western blotting experiments.
Structural modeling
To obtain a structural model of S. cerevisiae Rrf1 we used the Phyre2 web portal for protein modeling, prediction and analysis (42). A reported structure of Mycobacterium tuberculosis RRF ((43), PDB entry 1wqg) was selected as the highest scoring template to model the Rrf1 input sequence. Rrf1 residues 48–230 (79%) were aligned with bacterial RRF and among those, 176 residues (76% of the whole protein) were modeled on the template with 100% confidence, meaning that the match between the template and the input sequence was a true homology. Molecular graphics and analyses were then performed with the UCSF Chimera package (44). Chimera is developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
RESULTS
Mutants devoid of the ribosome recycling factor Rrf1, or translation initiation factors Ifm1 and Mif3, can maintain an intron-less mitochondrial genome
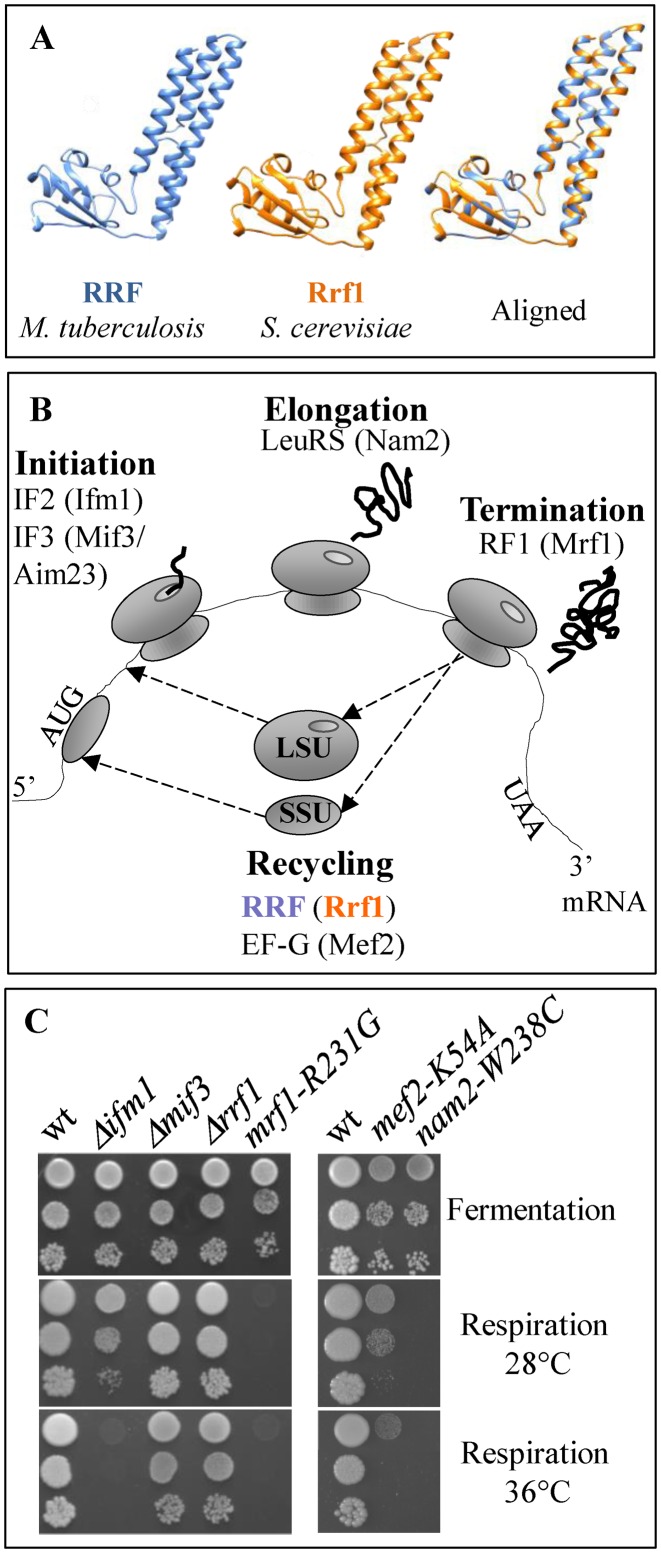
In bacteria, the ribosome recycling factor RRF is essential for translation and viability (45). The structure of RRF is virtually super-imposable on the predicted structure of its yeast homolog, Rrf1, suggesting that Rrf1 plays the same role in mitochondrial ribosome recycling (Figure 1A). However, despite this strong structural homology to bacterial RRF, a deletion mutant of RRF1 can maintain an intron-less mtDNA and is able to grow on respiratory medium (5), showing that Rrf1 is not essential for mitochondrial translation.
Figure 1.
MtDNA stability and respiratory growth in mitochondrial translation mutants. (A) The structure of the M. tuberculosis RRF and the predicted structure of the last 183 residues of S. cerevisiae Rrf1 are shown together with the superposition. (B) The names of bacterial translation factors are indicated on a schematic representation of the main steps of translation; corresponding yeast mitochondrial factors are given in parentheses. LSU: large ribosomal subunit, SSU: small ribosomal subunit. (C) Dilution series of cells from wild type (wt) and a series of mitochondrial translation factor mutants were spotted onto fermentable (glucose) and respiratory (glycerol) media and incubated for four days at 28°C and 36°C.
To further investigate the role of Rrf1 in mitochondrial translation, we decided to construct additional yeast mutants in homologs of bacterial factors known to control the recycling of ribosomes: the recycling GTPase Mef2, and the initiation factor Mif3/Aim23. As a comparison, we have also analyzed mutants in factors acting in other steps of mitochondrial translation: the initiation factor Ifm1 (IF2 in bacteria), the leucine tRNA synthetase Nam2 (LeuRS in bacteria) that provides charged leutRNA essential for elongation, and the release factor Mrf1 (RF1 and RF2 in bacteria), which recognizes the stop codons during termination (Figure 1B and Table 1).
Table 1. Summary of the mitochondrial translation factors studied, including the bacterial homologs and mutant phenotypes.
| Mitochondrial translation factors | Bacterial homologs | % identity S. cerevisiae/E. coli | S. cerevisiae mutants used in this study | % rho- 28°C |
|---|---|---|---|---|
| Ifm1 | IF2 | 33 | Δifm1 | 20 ± 10 |
| Mif3/Aim23 | IF3 | <20 | Δmif3 | <5 |
| Rrf1 | RRF | 20 | Δrrf1 | <5 |
| Mrf1 | RF1 | 35 | Δmrf1 | 100 |
| mrf1 (R231G) | 20 ± 10 | |||
| Mef2 | EF-G | 29 | Δmef2 | 100 |
| mef2 (K54A) | 10 ± 5 | |||
| Nam2 | LeuRS | 32 | Δnam2 | 100 |
| nam2 (W238C) | <5 |
The parental strain for all our constructions carried an intron-less mitochondrial genome (31). As expected, Δrrf1 strains did not lose their mtDNA, but in this background, Δifm1 and Δmif3 were also able to maintain their mtDNA and were capable of respiratory growth (Figure 1C). Thus, Rrf1 is not the only mitochondrial translation factor that is dispensable for the maintenance of the mtDNA. Conversely, deletions of NAM2, MRF1 and MEF2 compromised the stability of mtDNA and were incapable of respiratory growth, as previous described (11,46,47). To investigate this further, we used point mutations in these genes: nam2-W238C (48) and mrf1-R231G (49) that were already available, and we constructed mef2-K54A that changes a lysine in the G motif conserved in all GTPases (50). All three point mutations induce no, or only mild loss of the mtDNA (Table 1); however, only mef2-K54A shows a very weak and temperature sensitive respiratory growth, the others were respiratory deficient under all conditions (Figure 1C).
Deletion of MIF3, or RRF1 does not lead to an overall decrease in mitochondrial protein synthesis
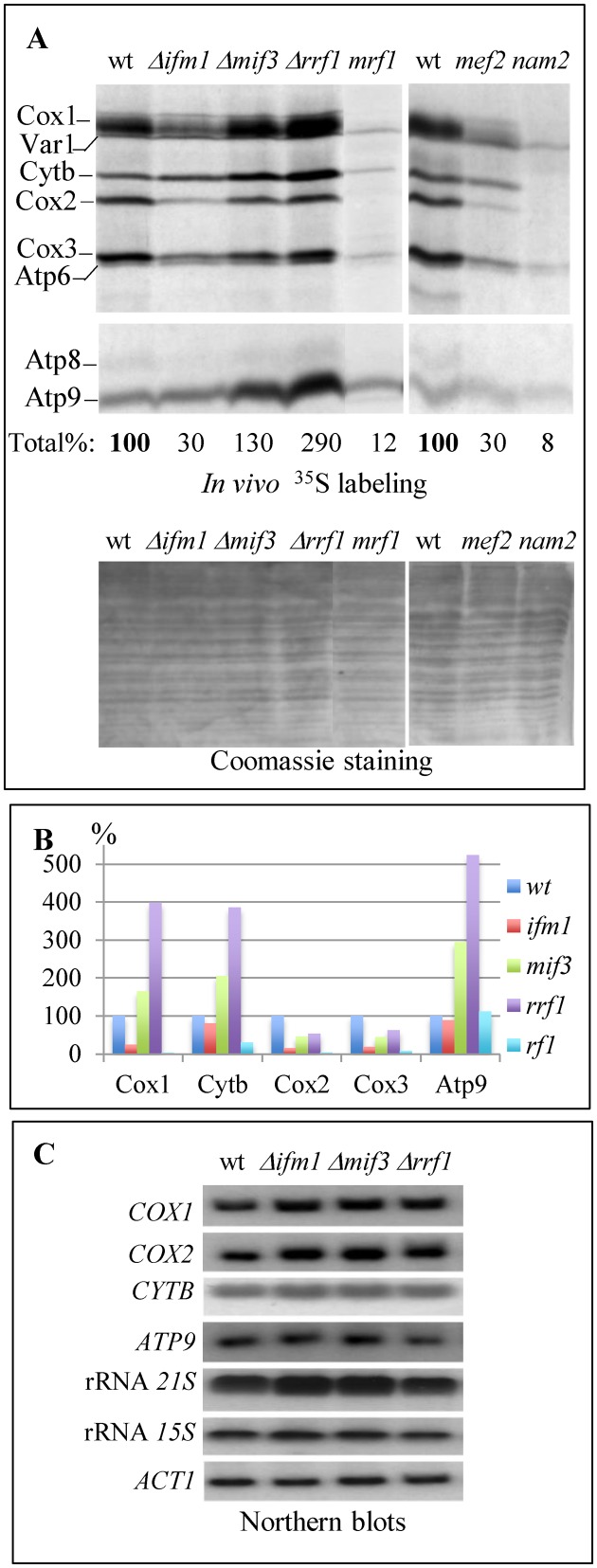
To further characterize all the mutants retaining mtDNA at the molecular level, we blocked cytoplasmic translation with cycloheximide, used a short incubation in the presence of 35S amino acids to label mitochondrial translation products and equal amounts of total proteins were subjected to SDS PAGE (Figure 2A). As expected, the three point mutants that had a compromised respiratory growth showed a lower level of labeling than the wild type. However, for the three deletion mutants, all mitochondrial translation products were detected and the Δmif3 and Δrrf1 strains displayed stronger labeling of Cox1, Cytb and Atp9 when compared to the wild type (Figure 2B). In the Δifm1 strain, Cytb and Atp9 levels were similar to the wild type while labeling of the other mitochondrial proteins was decreased. Thus, in all three deletion mutants, we observed a differential effect on the labeling of specific mitochondrial proteins.
Figure 2.
In vivo mitochondrial translation and mRNA accumulation in mutants devoid of ribosome recycling (Rrf1) or translation initiation factors (Ifm1 or Mif3). (A) Products of mitochondrial translation were labeled in vivo for 10 min with [35S]-methionine and cysteine. Cox1, Cox2, Cox3, Cytb, Atp6, Atp8, Atp9 are the mitochondrially encoded subunits of respiratory complexes IV (Cox), III (Cytb) and V (Atp). Var1 is a mitochondrially-encoded small ribosomal subunit. Upper panel: autoradiography of the gel. Lower panel: Coomassie staining. The total incorporation, obtained by quantifying with the Image J. software and summing up all bands of the autoradiography corresponding to mitochondrial proteins, is given as a percentage of the wild type labeling normalized using the Coomassie staining. (B) The labeling of individual proteins was also quantified with the Image J. software and normalized to the Coomassie staining. (C) Total RNAs (10 μg) were analyzed by Northern blotting with probes specific for the protein-encoding mitochondrial genes COX1, COX2, CYTB and ATP9, and the mitochondrial rRNAs 21S and 15S; actin (ACT1) was used as a loading control.
To determine if these effects could be due to changes in the steady state levels of the corresponding mRNAs, we performed Northern blots on total RNA using the nuclear-encoded actin RNA as a loading control (Figure 2C). Δifm1, Δmif3 and Δrrf1 showed a very small increase in the 21S, COX1 and COX2 RNAs, while the ATP9 and CYTB mRNAs remained largely unchanged. Thus, the differential effects on mitochondrial protein labeling observed in the three deletion mutants are not correlated with changes in RNA abundance, suggesting that they are due to translational or post-translational events. This is also consistent with the fact that the labeling of Cox1 increased while Atp8 remained unchanged although the corresponding RNAs are co-transcribed.
The Δmif3 and Δrrf1 mutants exhibit strong assembly defects in complexes IV and V
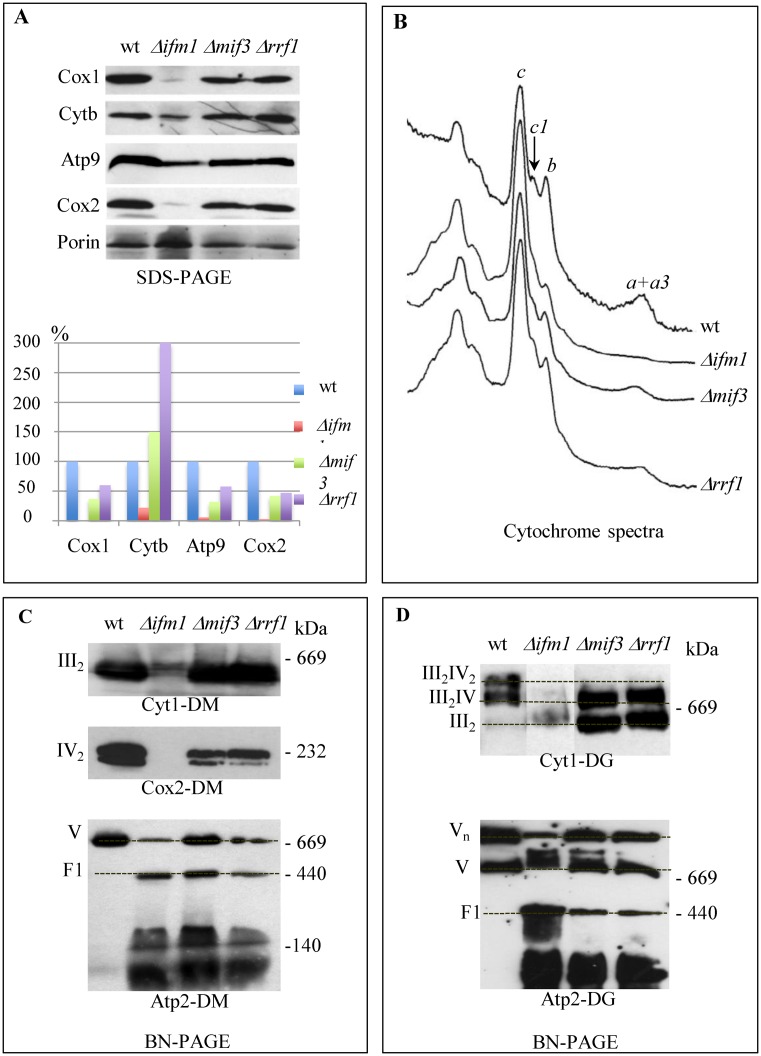
To determine if the stronger labeling of specific mitochondrial proteins observed in the Δmif3 and Δrrf1 deletion mutants resulted in an elevated steady state level of the proteins, a fraction of the same samples as in Figure 2A was analyzed by Western blotting to reveal the Cox1, Cox2, Cytb and Atp9 proteins; porin was used as a loading control (Figure 3A). As expected, low levels of mitochondrial proteins were detected in Δifm1. However, accumulation of Cox1, Atp9 and Cox2 was decreased in the Δmif3 and Δrrf1 mutants while the Cytb level remained higher compared to the wild type strain. Thus, the increase in protein labeling observed in Δmif3 and Δrrf1 for Cox1 and Atp9 is clearly not correlated with an increased steady state level of the proteins, suggesting that only a fraction of the proteins synthesized are assembled into their final OXPHOS complex.
Figure 3.
Assembly of the OXPHOS complexes in mutants devoid of ribosome recycling (Rrf1) or translation initiation factors (Ifm1 or Mif3). (A) Total proteins from [35S]-labeling experiments were analyzed by Western blotting with antibodies against Cox1, Cytb, Atp9, Cox2; Porin was used as a loading control (upper panel). Quantification was done using the Image J software (lower panel). (B) Cytochrome absorption spectra were recorded on whole cells. The position of the absorption maxima of cytochromes a + a3 (complex IV), cytochrome b and c1 (complex III) and c are indicated. (C and D) Mitochondrial proteins were solubilized with either dodecylmaltoside (DM, panel C) or digitonin (DG, panel D), separated on BN-PAGE 4–16% (left) or 3–12% (right) and immunoblotted with antibodies against Cyt1, Cox2 and Atp2. Positions of respiratory complexes III and IV, of supercomplexes III2 + IV2, III2 + IV as well as those of multimers (Vn), monomers (V) and F1-part of complex V are indicated. The protein molecular mass markers are also indicated (kDa).
To test this hypothesis, cytochrome spectra were recorded and clearly showed that the assembly of complex IV (cytochromes a+a3) was strongly compromised in the Δmif3 and Δrrf1 mutants whereas complex III (cytochromes b and c1) seemed less affected (Figure 3B). In addition, blue native gels were run to see individual OXPHOS complexes as well as the supra-molecular organization of the complexes, called super-complexes (super-complexes III/IV, complex V oligomers (Vn)). The results correlated well with those from the cytochrome spectra, showing a decrease in complex IV for Δmif3 and Δrrf1 and a complete absence in Δifm1, whereas the level of complex III was similar to wild type in Δmif3 and Δrrf1 (Figure 3C). Super-complexes III2IV2 were absent in Δmif3 and Δrrf1, instead super-complexes III2IV and complex III dimers were detected (Figure 3D). Complex V was severely affected since oligomers were reduced, and the free F1 sub-complex was clearly detected in the mutants but not the wild type. All these data show that the assembly of complexes IV and V is strongly compromised in the absence of the translation factors Mif3 or Rrf1.
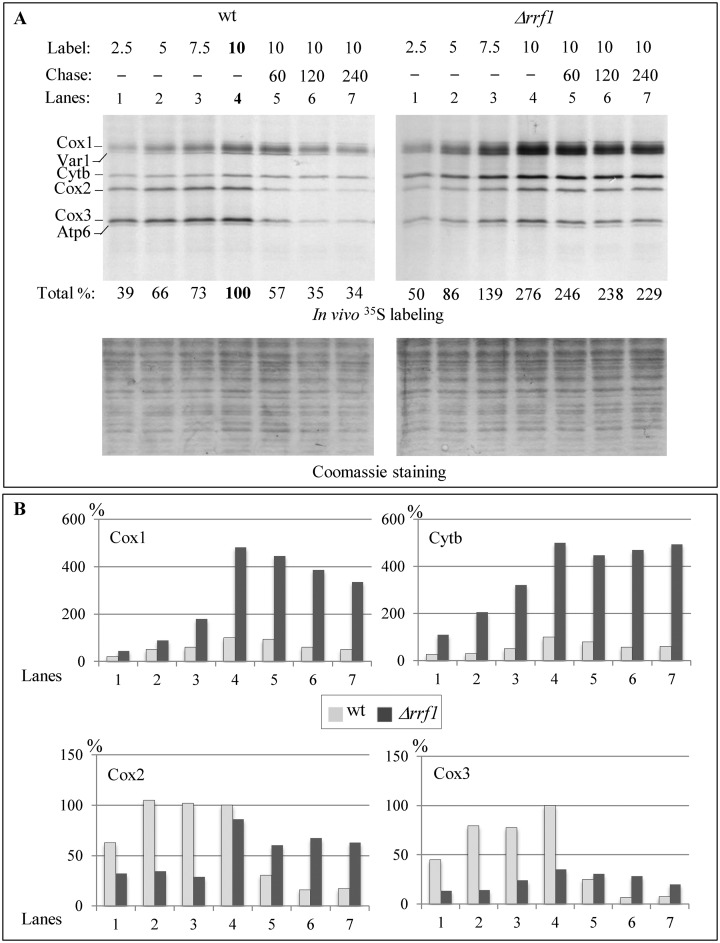
The synthesis rate of Cox1 and Cytb is enhanced in the Δrrf1 mutant
To further understand the paradoxical effects observed on labeling and assembly, we decided to focus on the fate of Cox1 and Cytb in the Δrrf1 mutant, because several factors coordinating Cox1 and Cytb synthesis and assembly are known. We performed a time course of protein synthesis focused on Cox1 and Cytb in the wild type and Δrrf1 mutant; this clearly showed a linear increase of the labeling of both proteins (Figure 4A). The synthesis rate of Cox1 and Cytb was strongly enhanced in the Δrrf1 mutant compared to the wild type (as quantified in Figure 4B). On the contrary Cox2 and Cox3 showed a reduced synthesis rate in the Δrrf1 mutant.
Figure 4.
Pulse-chase analysis of mitochondrial translation in the wile type and Δrrf1 strains. (A) Products of mitochondrial translation were labeled in vivo for 2.5, 5, 7.5 or 10 min with [35S]-methionine and cysteine and then chased for 60, 120 or 240 min in presence of an excess of cold methionine and cysteine. Total incorporation was calculated as described in Figure 2A. (B) Histograms showing the quantification of individual proteins calculated as described in Figure 2B.
Despite this increased synthesis of Cox1 and Cytb, we observed a low steady state level of Cox1 in Δrrf1 cells while the Cytb level remained elevated (Figure 3A). To test whether this low accumulation of Cox1 was due to an enhanced degradation of mitochondrially-encoded proteins, we carried out a pulse-chase experiment of up to 240 min to analyze the degradation of Cox1, Cox2, Cox3 and Cytb in the Δrrf1 mutant compared to the wild type (Figure 4A and B). Cytb was very stable in both the wild type and the mutant, while Cox2 and Cox3 appeared more stable in the Δrrf1 mutant than in the wild type. In absolute terms, the degradation of Cox1 was higher in the Δrrf1 mutant than in the wild type. The fact that the degradation was not sufficient in the Δrrf1 mutant to bring Cox1 and Cytb levels back to the wild type levels after a 240-min chase could be explained by the fact that the mitochondrial proteases (such as Afg3) are not sufficient to degrade the excess proteins. To test this hypothesis, we constructed a Δrrf1 Δafg3 double mutant in a mitochondrial intron-less context and found that it rapidly lost its mtDNA, whereas both single mutants maintained a wild type level of mtDNA. Whereas a cumulative effect on translation cannot be excluded in the double mutant, it is possible that the selection of cells devoid of mtDNA allows Δrrf1 Δafg3 strains to avoid a toxic over-accumulation of Cox1 and Cytb.
Taken together, our data suggest that the Δrrf1 mutation leads to a strong increase in the synthesis of Cox1 and Cytb which cannot be corrected by degradation via the mitochondrial proteases, and so in turn leads to an imbalance in subunit stoichiometry that might particularly affect the assembly of complexes IV and V, which contain several mitochondrially-encoded subunits.
The absence of Rrf1 alters the formation of Mss51 containing complexes involved in the regulation of complex IV assembly
To further understand the mechanisms underlying increased translation and reduced assembly of the OXPHOS subunits, we have investigated in the Δrrf1 mutant the fate of specific translational activators known to coordinate translation and assembly. Cox1 and Cytb are the targets of a feedback synthesis mechanism mediated by Mss51 and Cbp6/Cbp3, respectively (14,22,23,38). Mss51 and Cbp6/Cbp3 play a dual role, they are specific translational activators of the COX1 and CYTB mRNAs, but they also interact with the translated proteins and promote their assembly into their respective complexes. First, double deletions of RRF1 and one of the translational activators were constructed and de novo mitochondrial translation products were labeled and compared with the respective single deletions and the wild type (Figure 5A). Cox1 was essentially absent in the Δmss51 strain and Cytb was severely reduced in the Δcbp6 strain, and these effects were independent of presence or absence of Rrf1. However, the synthesis of Cox1 or Cytb was still increased in the Δrrf1 Δcbp6 or Δrrf1 Δmss51 strain, respectively. Thus, the higher translation levels of the COX1 and CYTB mRNAs in the Δrrf1 strain are not interdependent, and the Δmss51 and Δcbp6 mutants are epistatic to Δrrf1 since they prevent up-regulation of their target messenger.
Figure 5.
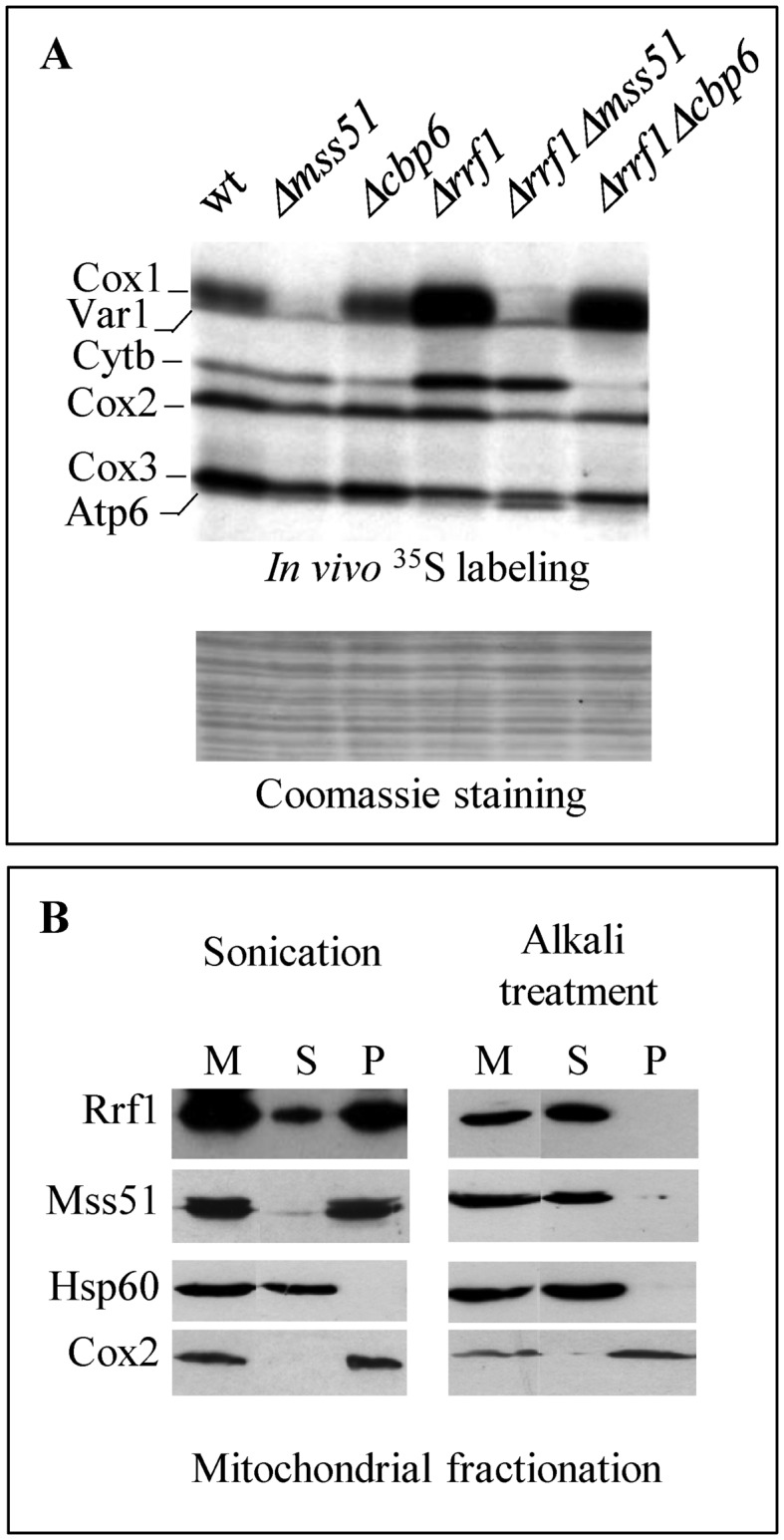
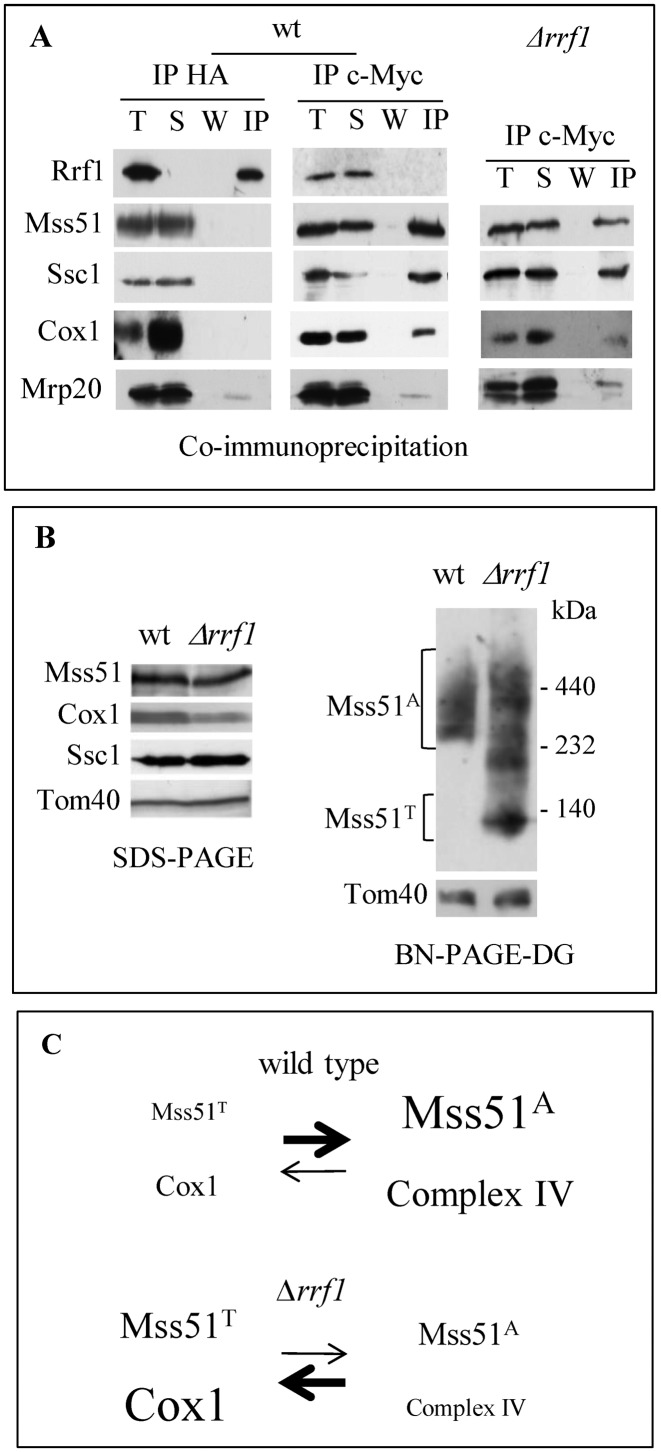
Genetic interactions and localization of Rrf1 and the translational activators. (A) In vivo [35S] incorporation as in Figure 2A. (B) Mitochondria were purified from cells producing Mss51-c-Myc. Mitochondrial proteins (M) were either sonicated (left) or alkali treated (right) and after centrifugation the soluble proteins were recovered in the supernatant (S) whereas the membrane-associated or membrane-spanning proteins remained in the pellet (P). Proteins were analyzed by Western blotting with antibodies recognizing Rrf1 or the epitope c-Myc. The soluble matrix protein Hsp60 and the integral mitochondrial membrane protein Cox2 were used as controls.
Next we decided to focus on the increase in Cox1 production in the Δrrf1 mutant to try understanding how a mutant affected in mitochondrial ribosome recycling could lead to an increase in the translation of a specific mitochondrial protein. Since Mss51 plays a key role in the regulation of Cox1 synthesis, we analyzed the possible interactions between Mss51 and Rrf1 and whether the presence or absence of Rrf1 can modify the functional state of Mss51.
First, we over-expressed the MS551 gene in the wild type and Δrrf1 strains, to test whether we could on the one hand mimic the effects of Rrf1 deletion in the wild type, and on the other, restore complex IV assembly in the Δrrf1 mutant. However, no significant effect was observed on cytochrome spectra or in a BN PAGE analysis of OXPHOS complexes upon over-expression of MSS51, whether in the wild type or Δrrf1 strain (Supplementary Figure S1).
Second, mitochondria were purified from a strain carrying a c-Myc tagged version of Mss51 (MSS51-c-Myc) and the localization of Mss51 and Rrf1 within the mitochondria was determined using anti-c-Myc and polyclonal anti-Rrf1 antibodies (Figure 5B). Both proteins were part of the mitochondrial pellet after sonication, although they became soluble after alkaline carbonate treatment. Thus, Rrf1 and Mss51 are both peripheral membrane proteins, loosely associated with the mitochondrial membrane.
To test whether Rrf1 and Mss51 could interact directly, co-immunoprecipitation experiments were performed using mitochondria from a strain containing an HA-tagged version of Rrf1 and the Mss51-c-Myc strain (Figure 6A). Immunoprecipitation of the HA tag pulled down only Rrf1, whereas the immunoprecipitation of the c-Myc tag retrieved Mss51, the heat shock protein Ssc1 and Cox1, as previously described (19,22) but no co-immunoprecipitation of Rrf1 and Mss51 was detected. Thus, Rrf1 does form a stable interaction with Mss51.
Figure 6.
Functional interaction between Rrf1 and Mss51. (A) Mitochondrial proteins from RRF1-HA or Δrrf1 cells producing Mss51-c-Myc were immunoprecipitated using anti-HA or anti-c-Myc-agarose beads. T: total; S: supernatant; W: wash; IP: immunoprecipitate. Proteins were analyzed by Western blotting with antibodies recognizing Rrf1, Ssc1, Cox1, Mrp20, HA or c-Myc. The ribosomal protein Mrp20 undergoes some degradation during these immunoprecipitation experiments unless a cocktail of protease inhibitors is added. (B) Mitochondrial proteins of wt or Δrrf1 cells producing Mss51-c-Myc were analyzed by SDS-PAGE (left panel) or were lysed in digitonin, separated on BN-PAGE 3–12% (right panel) and immunoblotted with antibodies recognizing c-Myc, Cox1, Ssc1, Tom40. The protein molecular mass markers are indicated. Mss51A: high molecular weight assembly-competent complexes; Mss51T: low molecular weight complex thought to be the source of translationally-active complex. (C) Model of the effect of an absence of Rrf1 on Cox1 synthesis and complex IV assembly. We hypothesize that in the absence of Rrf1, the dissociation of the ribosomal subunits is slowed down, reducing the accessibility of Mss51 to its binding site on the C-terminus of Cox1 and concomitantly preventing the formation of the high molecular weight complex MSS51A. This would displace the equilibrium between Mss51T and Mss51A toward Mss51T, thus reducing the assembly of complex IV.
Next, we asked whether the steady state level of Mss51 could be increased in the Δrrf1 mutant, thereby explaining the elevated synthesis of Cox1. However, the Mss51 level remained unchanged in the Δrrf1 mutant compared to the wild type (Figure 6B, left panel). Last, since the assembly-promoting form of Mss51 (Mss51A) and the low molecular-weight forms previously observed in assembly defective mutants belong to complexes of different sizes, we analyzed the behavior of Mss51 on blue native gels in the Δrrf1 mutant compared to the wild type (Figure 6B, right panel). Only the Mss51A complex was detected in the wild type, confirming that under efficient growth conditions, most Mss51 is found in Cox1 assembly intermediates. However, in the absence of Rrf1, a substantial fraction of Mss51 was detected in low molecular weight complexes, including a complex of approximately 120 kDa (Mss51T) known to correspond to a heterodimer of Mss51 and Ssc1, proposed to be the source of the translationally-active form (19).
DISCUSSION
It is generally accepted that mitochondrial protein synthesis is required for the maintenance of an intact mitochondrial genome in S. cerevisiae (1). Stringent mutations occurring in genes encoding essential components of the mitochondrial translation apparatus such as ribosomal proteins, tRNA synthetases but also elongation or release factors produce a high frequency of rho− deletion mutations that rapidly lead to the loss of the mtDNA. We have shown in this paper that this is also true for some representative mutations constructed in an intron-less mtDNA and affecting different steps of mitochondrial translation. For example, the absence of the recycling GTPase Mef2 leads to mtDNA loss and a point mutant located in a conserved GTPase motif shows a lower level of translation. Binding of Mef2-GTP to the ribosome is sufficient for ribosome splitting, but GTP-hydrolysis is required to remove Mef2 itself (and probably Rrf1) from the large subunit of the human mitochondrial ribosome (10). Therefore, the absence of Mef2 is likely to abolish the dissociation of ribosomal subunits and block all the subsequent steps of the mitochondrial translation cycle.
Strikingly, the absence of the other auxiliary factors controlling the recycling step (Rrf1) or the interface between recycling and initiation (Mif3) does not lead to mtDNA loss in a strain with an intron-less mtDNA while it does in an isonuclear strain with an intron-containing mtDNA. This suggests that the absence of Rrf1 and Mif3 has only a slight effect, if any, on mitochondrial translation. The higher instability of mtDNA observed with an intron-containing mtDNA is probably due to the fact that several introns encode maturases that are needed for the excision of the intron by which they are encoded (51). Thus, the expression of the intron-containing genes requires several rounds of translation and splicing. Therefore, if translation is slowed or less efficient, the expression of these genes could act as a trap for ribosomes and translation factors, leading to a more stringent translation deficiency and mtDNA loss.
More interestingly, in an intron-less mtDNA background that more closely models the mammalian mtDNA which is devoid of introns, the two mutants Δrrf1 and Δmif3 not only maintain their mtDNA, they are also are respiratory sufficient. Moreover, analysis of in vivo mitochondrial translation in these mutants shows that they do not decrease mitochondrial translation but rather lead to an increased synthesis of three OXPHOS subunits (Cytb, Cox1 and Atp9). In a recent paper, analysis of the mif3/aim23 deletion has also revealed an imbalance of mitochondrial protein synthesis with a very high production of Atp9 but no increase in Cox1 and Cytb (12). We believe that this difference is due to the strain used by these authors, which contains eight mitochondrial introns (52), five of which contain maturase ORFs that need to be translated to promote splicing of the COX1 or CYTB mRNAs.
Thus, despite their bacterial ancestry, the S. cerevisiae Rrf1 and Mif3 factors are not essential for mitochondrial translation, while their bacterial homologs are. An important difference between bacterial and mitochondrial translation is that the second occurs in close proximity to the inner membrane in order to synthesize the few hydrophobic proteins encoded by the mtDNA. The mito-ribosomes are indeed associated with the membrane (53,54) and one can easily imagine that this might spatially modify the steps of recycling and re-initiation. In addition, there is no equivalent to the Shine–Dalgarno ribosome-binding site on mitochondrial mRNAs; instead, specific translational activators participate in anchoring of mRNAs the mitochondrial inner-membrane, the recruitment of the ribosomes and the initiation step. As described above Mss51 targets both the COX1 mRNA and the Cox1 protein, thus controlling a feedback loop that coordinates the rates of Cox1 synthesis with the subsequent assembly of the protein into complex IV (14,22,23).
We have further shown that Rrf1, like Mss51, is loosely associated with the inner membrane in accordance with the idea that the recycling step of mitochondrial translation also occurs at proximity of the inner membrane. In the absence of Rrf1 we see an increase in de novo Cox1 synthesis but a decrease in the level of assembled complex IV. One possibility is that Rrf1 could serve to localize mitochondrial translation in membrane sites where complex assembly occurs. Translation in random locations in the Δrrf1 mutant would result in a deficit of complex assembly and a compensatory increase in translation. Although we cannot exclude this hypothesis, Rrf1 is only loosely associated with the inner membrane and no physical interactions between Rrf1 and plausible candidate partners for this localization have been identified (BioGRID3.4, (55)). We favor an alternative hypothesis based on structural data from bacterial models, showing that binding of RRF to the ribosome loosens the interactions between the large and the small subunits through a conformational change, before binding of EF-G (56). Thus, it is tempting to propose that in the absence of Rrf1 the dissociation of the ribosomal subunits is slowed down; as this occurs at the inner membrane, it is possible that this reduces the accessibility of Mss51 to its binding site on the C-terminus of Cox1, preventing the formation of the high molecular weight assembly complex MSS51A, and leading to a reduced assembly of complex IV. If this hypothesis is correct, the effect of deleting RRF1 would be to displace the equilibrium between Mss51T and Mss51A toward Mss51T, thus reducing assembly despite the excess of Cox1 produced (see proposed model in Figure 6C). In addition, the uncoordinated synthesis of mitochondrially-encoded subunits belonging to the same complex might also further perturb complex assembly.
Similar effects on the translational activators specific for Cytb and Atp9 might also explain their increased synthesis. For Cytb, some of the translational activators, like the complex Cbp3/Cbp6, are also required to coordinate synthesis and assembly (38), in an analogous way to Mss51 for Cox1. However, complex III assembly is not severely decreased in the Δrrf1 mutant, possibly because Cytb is remarkably stable (38). For Atp9, two potential translational activators have been identified (57,58) but no feedback control loop has been reported until now, thus the regulation of Atp9 translation remains to be investigated.
In human cells, the depletion of the mitochondrial ribosome recycling factor mtRRF leads to a partial translation defect, a decreased levels of fully assembled OXPHOS complexes, changes in mitochondrial morphology and ultimately cell death (5). While numerous pathogenic mutations have been reported in other mitochondrial translation factors (59), no human mutation has been identified in the mtRRF or mtIF3 proteins to our knowledge. Our work suggests that the main phenotype of such mutations could be a defect in the assembly of OXPHOS complexes IV and V (and maybe of complex I that is absent in yeast) rather than a general decrease of mitochondrial translation. Interestingly, EFG2 (mef2 in yeast) mutations have been recently reported in patients; in skin fibroblasts they lead to a decrease in the activity of complexes III and IV, but have no effect on complex I, although seven subunits of this complex are synthesized within mitochondria (60).
Altogether our results show that in S. cerevisiae, auxiliary factors acting at the recycling (Rrf1) and interface between recycling and initiation (Mif3) steps of mitochondrial translation are not essential for translation in an intron-less strain, rather their absence strongly impairs the assembly of complex IV and the ATP synthase. We also present the first evidence of a functional interaction between a general recycling factor, Rrf1, and a specific translational activator, Mss51. This interaction might actively participate in the coupling between mitochondrial translation and OXPHOS assembly in yeast. In the future, it will be interesting to further explore the nature of the interactions existing between general and specific auxiliary translation factors that might reveal an evolutionary adaptation of the translation machinery to the membranous environment within mitochondria.
Supplementary Material
Acknowledgments
The authors are grateful to C. Meisinger, M. Ott, N. Pfanner and J. Velours for the gifts of antibodies.
Footnotes
Present address: Jelena Ostojić, The Salk Institute, 10010 North Torrey Pines Road, La Jolla, CA 92037, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique. J.O. was a PhD student from the University Evry Val d'Essonne and she was the recipient of a fellowship from the Ministère de la Recherche et de la Technologie. Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
REFERENCES
- 1.Myers A.M., Pape L.K., Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christian B.E., Spremulli L.L. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim. Biophys. Acta. 2012;1819:1035–1054. doi: 10.1016/j.bbagrm.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirokawa G., Demeshkina N., Iwakura N., Kaji H., Kaji A. The ribosome-recycling step: consensus or controversy? Trends Biochem. Sci. 2006;31:143–149. doi: 10.1016/j.tibs.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Seshadri A., Varshney U. Mechanism of recycling of post-termination ribosomal complexes in eubacteria: a new role of initiation factor 3. J. Biosci. 2006;31:281–289. doi: 10.1007/BF02703921. [DOI] [PubMed] [Google Scholar]
- 5.Rorbach J., Richter R., Wessels H.J., Wydro M., Pekalski M., Farhoud M., Kuhl I., Gaisne M., Bonnefoy N., Smeitink J.A., et al. The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res. 2008;36:5787–5799. doi: 10.1093/nar/gkn576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teyssier E., Hirokawa G., Tretiakova A., Jameson B., Kaji A., Kaji H. Temperature-sensitive mutation in yeast mitochondrial ribosome recycling factor (RRF) Nucleic Acids Res. 2003;31:4218–4226. doi: 10.1093/nar/gkg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koc E.C., Spremulli L.L. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J. Biol. Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- 8.Atkinson G.C., Kuzmenko A., Kamenski P., Vysokikh M.Y., Lakunina V., Tankov S., Smirnova E., Soosaar A., Tenson T., Hauryliuk V. Evolutionary and genetic analyses of mitochondrial translation initiation factors identify the missing mitochondrial IF3 in S. cerevisiae. Nucleic Acids Res. 2012;40:6122–6134. doi: 10.1093/nar/gks272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammarsund M., Wilson W., Corcoran M., Merup M., Einhorn S., Grander D., Sangfelt O. Identification and characterization of two novel human mitochondrial elongation factor genes, hEFG2 and hEFG1, phylogenetically conserved through evolution. Hum. Genet. 2001;109:542–550. doi: 10.1007/s00439-001-0610-5. [DOI] [PubMed] [Google Scholar]
- 10.Tsuboi M., Morita H., Nozaki Y., Akama K., Ueda T., Ito K., Nierhaus K.H., Takeuchi N. EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis. Mol. Cell. 2009;35:502–510. doi: 10.1016/j.molcel.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Callegari S., McKinnon R.A., Andrews S., de Barros Lopes M.A. The MEF2 gene is essential for yeast longevity, with a dual role in cell respiration and maintenance of mitochondrial membrane potential. FEBS Lett. 2011;585:1140–1146. doi: 10.1016/j.febslet.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 12.Kuzmenko A., Derbikova K., Salvatori R., Tankov S., Atkinson G.C., Tenson T., Ott M., Kamenski P., Hauryliuk V. Aim-less translation: loss of Saccharomyces cerevisiae mitochondrial translation initiation factor mIF3/Aim23 leads to unbalanced protein synthesis. Sci. Rep. 2016;6:18749. doi: 10.1038/srep18749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuzmenko A., Atkinson G.C., Levitskii S., Zenkin N., Tenson T., Hauryliuk V., Kamenski P. Mitochondrial translation initiation machinery: conservation and diversification. Biochimie. 2013;100:132–140. doi: 10.1016/j.biochi.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mick D.U., Fox T.D., Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontanesi F. Mechanisms of mitochondrial translational regulation. IUBMB Life. 2013;65:397–408. doi: 10.1002/iub.1156. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann J.M., Woellhaf M.W., Bonnefoy N. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim. Biophys. Acta. 2013;1833:286–294. doi: 10.1016/j.bbamcr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Pierrel F., Bestwick M.L., Cobine P.A., Khalimonchuk O., Cricco J.A., Winge D.R. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 2007;26:4335–4346. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mick D.U., Vukotic M., Piechura H., Meyer H.E., Warscheid B., Deckers M., Rehling P. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 2010;191:141–154. doi: 10.1083/jcb.201007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fontanesi F., Soto I.C., Horn D., Barrientos A. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol. Cell. Biol. 2010;30:245–259. doi: 10.1128/MCB.00983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontanesi F., Clemente P., Barrientos A. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 2011;286:555–566. doi: 10.1074/jbc.M110.188805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McStay G.P., Su C.H., Tzagoloff A. Stabilization of Cox1p intermediates by the Cox14p-Coa3p complex. FEBS Lett. 2013;587:943–949. doi: 10.1016/j.febslet.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Martinez X., Broadley S.A., Fox T.D. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrientos A., Zambrano A., Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter-Dennerlein R., Dennerlein S., Rehling P. Integrating mitochondrial translation into the cellular context. Nat. Rev. Mol. Cell Biol. 2015;16:586–592. doi: 10.1038/nrm4051. [DOI] [PubMed] [Google Scholar]
- 25.Mick D.U., Dennerlein S., Wiese H., Reinhold R., Pacheu-Grau D., Lorenzi I., Sasarman F., Weraarpachai W., Shoubridge E.A., Warscheid B., et al. MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell. 2012;151:1528–1541. doi: 10.1016/j.cell.2012.11.053. [DOI] [PubMed] [Google Scholar]
- 26.Weraarpachai W., Sasarman F., Nishimura T., Antonicka H., Aure K., Rotig A., Lombes A., Shoubridge E.A. Mutations in C12orf62, a factor that couples COX I synthesis with cytochrome c oxidase assembly, cause fatal neonatal lactic acidosis. Am. J. Hum. Genet. 2012;90:142–151. doi: 10.1016/j.ajhg.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostergaard E., Weraarpachai W., Ravn K., Born A.P., Jonson L., Duno M., Wibrand F., Shoubridge E.A., Vissing J. Mutations in COA3 cause isolated complex IV deficiency associated with neuropathy, exercise intolerance, obesity, and short stature. J. Med. Genet. 2015;52:203–207. doi: 10.1136/jmedgenet-2014-102914. [DOI] [PubMed] [Google Scholar]
- 28.Szklarczyk R., Wanschers B.F., Cuypers T.D., Esseling J.J., Riemersma M., van den Brand M.A., Gloerich J., Lasonder E., van den Heuvel L.P., Nijtmans L.G., et al. Iterative orthology prediction uncovers new mitochondrial proteins and identifies C12orf62 as the human ortholog of COX14, a protein involved in the assembly of cytochrome c oxidase. Genome Biol. 2012;13:R12. doi: 10.1186/gb-2012-13-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moyer A.L., Wagner K.R. Mammalian Mss51 is a skeletal muscle-specific gene modulating cellular metabolism. J. Neuromuscul. Dis. 2015;2:371–385. doi: 10.3233/JND-150119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dujardin G., Pajot P., Groudinsky O., Slonimski P.P. Long range control circuits within mitochondria and between nucleus and mitochondria. I. Methodology and phenomenology of suppressors. Mol. Gen. Genet. 1980;179:469–482. doi: 10.1007/BF00271736. [DOI] [PubMed] [Google Scholar]
- 31.Saint-Georges Y., Bonnefoy N., di Rago J.P., Chiron S., Dujardin G. A pathogenic cytochrome b mutation reveals new interactions between subunits of the mitochondrial bc1 complex. J. Biol. Chem. 2002;277:49397–49402. doi: 10.1074/jbc.M207219200. [DOI] [PubMed] [Google Scholar]
- 32.Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Yaffe M.P. Analysis of mitochondrial function and assembly. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]
- 34.Racki W.J., Becam A.M., Nasr F., Herbert C.J. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 2000;19:4524–4532. doi: 10.1093/emboj/19.17.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemaire C., Dujardin G. Preparation of respiratory chain complexes from Saccharomyces cerevisiae wild-type and mutant mitochondria : activity measurement and subunit composition analysis. Methods Mol. Biol. 2008;432:65–81. doi: 10.1007/978-1-59745-028-7_5. [DOI] [PubMed] [Google Scholar]
- 36.Mathieu L., Marsy S., Saint-Georges Y., Jacq C., Dujardin G. A transcriptome screen in yeast identifies a novel assembly factor for the mitochondrial complex III. Mitochondrion. 2011;11:391–396. doi: 10.1016/j.mito.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Velours J., Arselin G. The Saccharomyces cerevisiae ATP synthase. J. Bioenerg. Biomembr. 2000;32:383–390. doi: 10.1023/a:1005580020547. [DOI] [PubMed] [Google Scholar]
- 38.Gruschke S., Kehrein K., Rompler K., Grone K., Israel L., Imhof A., Herrmann J.M., Ott M. Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J. Cell Biol. 2011;193:1101–1114. doi: 10.1083/jcb.201103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanjuan Szklarz L.K., Guiard B., Rissler M., Wiedemann N., Kozjak V., van der Laan M., Lohaus C., Marcus K., Meyer H.E., Chacinska A., et al. Inactivation of the mitochondrial heat shock protein Zim17 leads to aggregation of matrix Hsp70s followed by pleiotropic effects on morphology and protein biogenesis. J. Mol. Biol. 2005;351:206–218. doi: 10.1016/j.jmb.2005.05.068. [DOI] [PubMed] [Google Scholar]
- 40.Dekker P.J., Ryan M.T., Brix J., Muller H., Honlinger A., Pfanner N. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol. Cell Biol. 1998;18:6515–6524. doi: 10.1128/mcb.18.11.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schagger H., Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saikrishnan K., Kalapala S.K., Varshney U., Vijayan M. X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications. J. Mol. Biol. 2005;345:29–38. doi: 10.1016/j.jmb.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 44.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Janosi L., Ricker R., Kaji A. Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie. 1996;78:959–969. doi: 10.1016/s0300-9084(97)86718-1. [DOI] [PubMed] [Google Scholar]
- 46.Labouesse M., Dujardin G., Slonimski P.P. The yeast nuclear gene NAM2 is essential for mitochondrial DNA integrity and can cure a mitochondrial RNA-maturase deficiency. Cell. 1985;41:133–143. doi: 10.1016/0092-8674(85)90068-6. [DOI] [PubMed] [Google Scholar]
- 47.Pel H.J., Maat C., Rep M., Grivell L.A. The yeast nuclear gene MRF1 encodes a mitochondrial peptide chain release factor and cures several mitochondrial RNA splicing defects. Nucleic Acids Res. 1992;20:6339–6346. doi: 10.1093/nar/20.23.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li G.Y., Becam A.M., Slonimski P.P., Herbert C.J. In vitro mutagenesis of the mitochondrial leucyl tRNA synthetase of Saccharomyces cerevisiae shows that the suppressor activity of the mutant proteins is related to the splicing function of the wild-type protein. Mol. Gen. Genet. 1996;252:667–675. doi: 10.1007/BF02173972. [DOI] [PubMed] [Google Scholar]
- 49.Towpik J., Chacinska A., Ciesla M., Ginalski K., Boguta M. Mutations in the yeast mrf1 gene encoding mitochondrial release factor inhibit translation on mitochondrial ribosomes. J. Biol. Chem. 2004;279:14096–14103. doi: 10.1074/jbc.M312856200. [DOI] [PubMed] [Google Scholar]
- 50.Verstraeten N., Fauvart M., Versees W., Michiels J. The universally conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 2011;75:507–542. doi: 10.1128/MMBR.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazowska J., Jacq C., Slonimski P.P. Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell. 1980;22:333–348. doi: 10.1016/0092-8674(80)90344-x. [DOI] [PubMed] [Google Scholar]
- 52.Shaw L.C., Lewin A.S. The Cbp2 protein stimulates the splicing of the omega intron of yeast mitochondria. Nucleic Acids Res. 1997;25:1597–1604. doi: 10.1093/nar/25.8.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu M., Spremulli L. Interaction of mammalian mitochondrial ribosomes with the inner membrane. J. Biol. Chem. 2000;275:29400–29406. doi: 10.1074/jbc.M002173200. [DOI] [PubMed] [Google Scholar]
- 54.Pfeffer S., Woellhaf M.W., Herrmann J.M., Forster F. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat. Commun. 2014;6:6019. doi: 10.1038/ncomms7019. [DOI] [PubMed] [Google Scholar]
- 55.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yokoyama T., Shaikh T.R., Iwakura N., Kaji H., Kaji A., Agrawal R.K. Structural insights into initial and intermediate steps of the ribosome-recycling process. EMBO J. 2012;31:1836–1846. doi: 10.1038/emboj.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Payne M.J., Schweizer E., Lukins H.B. Properties of two nuclear pet mutants affecting expression of the mitochondrial oli1 gene of Saccharomyces cerevisiae. Curr. Genet. 1991;19:343–351. doi: 10.1007/BF00309594. [DOI] [PubMed] [Google Scholar]
- 58.Ackerman S.H., Gatti D.L., Gellefors P., Douglas M.G., Tzagoloff A. ATP13, a nuclear gene of Saccharomyces cerevisiae essential for the expression of subunit 9 of the mitochondrial ATPase. FEBS Lett. 1991;278:234–238. doi: 10.1016/0014-5793(91)80124-l. [DOI] [PubMed] [Google Scholar]
- 59.Perez-Martinez X., Funes S., Camacho-Villasana Y., Marjavaara S., Tavares-Carreon F., Shingu-Vazquez M. Protein synthesis and assembly in mitochondrial disorders. Curr. Top Med. Chem. 2008;8:1335–1350. doi: 10.2174/156802608786141124. [DOI] [PubMed] [Google Scholar]
- 60.Fukumura S., Ohba C., Watanabe T., Minagawa K., Shimura M., Murayama K., Ohtake A., Saitsu H., Matsumoto N., Tsutsumi H. Compound heterozygous GFM2 mutations with Leigh syndrome complicated by arthrogryposis multiplex congenita. J. Hum. Genet. 2015;60:509–513. doi: 10.1038/jhg.2015.57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.