Abstract
Nearly half of ribosomal proteins are composed of a domain on the ribosome surface and a loop or extension that penetrates into the organelle's RNA core. Our previous work showed that ribosomes lacking the loops of ribosomal proteins uL4 or uL22 are still capable of entering polysomes. However, in those experiments we could not address the formation of mutant ribosomes, because we used strains that also expressed wild-type uL4 and uL22. Here, we have focused on ribosome assembly and function in strains in which loop deletion mutant genes are the only sources of uL4 or uL22 protein. The uL4 and uL22 loop deletions have different effects, but both mutations result in accumulation of immature particles that do not accumulate in detectable amounts in wild-type strains. Thus, our results suggest that deleting the loops creates kinetic barriers in the normal assembly pathway, possibly resulting in assembly via alternate pathway(s). Furthermore, deletion of the uL4 loop results in cold-sensitive ribosome assembly and function. Finally, ribosomes carrying either of the loop-deleted proteins responded normally to the secM translation pausing peptide, but the uL4 mutant responded very inefficiently to the cmlAcrb pause peptide.
INTRODUCTION
Cytoplasmic ribosomes consist of two unequal subunits, referred to as the small subunit (SSU) and the large subunit (LSU), that jointly contain several rRNA molecules and, depending on the organism, 55–80 ribosomal proteins (1,2). Despite differences in the protein composition and the length of the rRNAs, the ribosome structure is largely conserved in prokaryotes, eukaryotes and archaea (3–9). Ribosomal RNA performs key catalytic functions, but genetic evidence accumulating since the 1960's demonstrates that the ribosomal proteins are nevertheless important for ribosome biogenesis and function (10–15).
Ribosome biogenesis in Escherichia coli begins with the transcription of a precursor RNA containing segments that will become the 16S rRNA of the SSU as well as the 23S rRNA and 5S rRNA of the LSU. Interstitial RNA sequences, called spacers, and tRNA genes separate the 16S, 23S and 5S rRNA elements in the primary transcript (16). Formation of functional ribosomes requires the coordination of multiple intertwined processes (17) including the folding, chemical modification, enzymatic cleavage and trimming of rRNA precursor (‘rRNA processing’) which result in extraction of the final mature rRNAs and tRNAs and degradation of the spacer RNA elements (18). The rRNA processing is coordinated with the binding of ribosomal proteins in a hierarchical manner; sequential waves of ribosomal proteins bind to the rRNA and modify the structure of the nascent ribosome to create binding sites for later binding proteins (19,20). The in vivo folding of rRNA into the optimal functional structure requires not only the cleavage of precursor rRNA and interaction between ribosomal proteins (21,22) and rRNA, but also assembly factors that work morphopoetically by binding intermittently to the precursor ribosomes and/or that catalyze the release of energy from nucleoside triphosphates or that modify rRNA and proteins (23–25). Notwithstanding many fundamental studies of protein-induced changes in the rRNA structure, much is still to be learned about the role of ribosomal proteins in the formation and function of ribosomes.
Nearly half of ribosomal proteins are composed of a domain, typically globular, located on the surface of the ribosome and a loop or extension that penetrates into the RNA core of the ribosome and co-folds with the rRNA (3,4,26–28). These loops and extensions represent <20% of the protein mass in the ribosome, but account for more than 40% of the total protein–RNA interactions within the ribosome, suggesting that these extensions may play an important role in stabilizing rRNA tertiary structure. Moreover, the extensions have evolved a higher mol percent of glycine, arginine and lysine than the globular domains residing on the ribosome surface (29,30). The abundance of positively charged basic residues allows the extensions to neutralize the highly negative charge of the rRNA backbone, while the high glycine content confers the flexibility to avoid steric clashes in the very dense rRNA landscape (30).
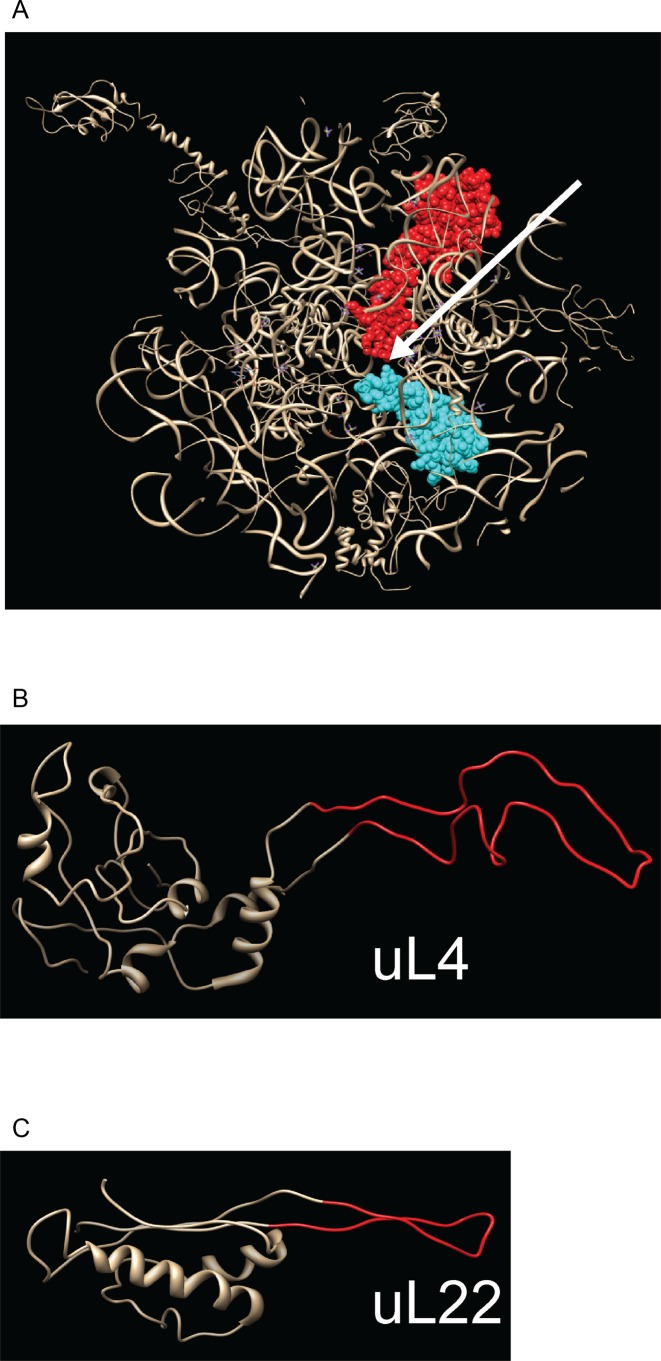
The extended loops of ribosomal proteins uL4 and uL22 (L4 and L22 according to classic E. coli nomenclature, see ref. 1) reach from the globular surface domain all the way into the peptide exit tunnel (Figure 1A), the narrow conduit through which nascent peptides travel from the peptidyl transfer center to the exterior of the ribosome (3,4). Portions of the uL4 and uL22 extended loops are exposed in the interior lumen of the tunnel, which is otherwise comprised of rRNA. Moreover, the tips of the two loops form a constriction that is the narrowest point in the tunnel. Located close to this constriction is the binding pocket for erythromycin and other macrolides and ketolides (31,32). Alterations in the uL4 and uL22 extended loops confer changes to the ribosome sensitivity to these antibiotics, even though the proteins do not directly contact the binding pocket (5,33–35).
Figure 1.
Ribosomal proteins uL4 and uL22. (A) Slab view of the ribosome subunit structure visualizing the exit tunnel from the ribosome solvent side. uL4 is shown in red, uL22 in cyan, other ribosomal components in gray. The arrow points to the ribosome exit tunnel. (B and C) Structure of uL4 (B) and uL22 (C) extracted from the crystal structure of the 50S subunit. Note that two glycine residues replaced the loop in the uL22 mutant. Furthermore, codon 101 was changed from serine to glycine. Structures are rendered from pdb file 2AW4 (54) (superseded by 4V4Q) using Chimera (http://www.cgl.ucsf.edu/chimera).
We previously showed that, despite the extensive interactions of the uL4 and uL22 loops with the rRNA, loop-deleted uL4 or uL22 are incorporated into ribosomes and such ribosomes are associated with polysomes (36). However, these studies focused on mature ribosomes. To gain further insight into the roles of the uL4 and uL22 extensions in ribosomal assembly and translation we have examined mutant strains encoding only loop-less uL4 or uL22 genes.
MATERIALS AND METHODS
Strains and media
Strain uL4-Δloop4, henceforth referred to as uL4-Δloop (Figure 1B), is a spontaneous mutant derived from E. coli A19 (37) (referred to as ‘wild-type’) by selecting for growth on LB containing 2 mg/ml troleandomycin (Oleandomycin phosphate salt dehydrate, Sigma). Polymerase chain reaction (PCR) amplification of the uL4 gene and sequencing showed that the mutant lacks codons 42–90 of the uL4 gene, i.e. the extended loop was completely deleted while leaving flanking sequences intact. The uL22-Δloop3 mutant, referred to as uL22-Δloop (Figure 1C), is also a derivative of E. coli A19. Construction of the L22-ΔLoop3 mutant in the background of DY380 was described previously (38). The L22-Δloop mutation was transferred from DY380 to A19 by the following steps. (i) The pDK46 plasmid carrying genes encoding recombineering enzymes (39) was transformed into E. coli A19. (ii) The L22-ΔLoop3 mutation was amplified from DY380 DNA by PCR. (iii) The PCR fragment was transformed into E. coli A19/pDK46. (iv) The resulting colonies were screened for the presence of the L22-Δloop mutation by PCR and sequencing. (v) The pDK46 was eliminated by growth at high temperature (39). (vi) To minimize the possibility of off-site mutations resulting from recombineering the uL22 mutation was finally P1 transduced into a fresh copy of E. coli A19 according to standard P1 transduction protocols. Sequencing of the resulting strain showed that uL22 codons 82–98, i.e. the entire extended loop, were replaced by two glycine codons. In addition, the uL22 gene harbored an unintended mutation changing codon 101 from encoding serine to encoding glycine. For measurements of translation pausing, pause signals were integrated by recombineering into the lacZ gene of A19 and the loop deletion strains. Cultures were grown with vigorous shaking in LB medium at the indicated temperatures.
Antisera
Rabbit antisera used for experiments in Figure 2 were a gift from Dr Harris D. Bernstein, National Institutes of Health, USA. Antigens for these were synthetic peptides corresponding to the first 20 amino acids of E. coli uL4 and uL22, respectively. Antisera from rabbits used for the rest of the experiments were prepared for our lab by Covance Research Products, Denver, CO, USA using synthetic peptides corresponding to the N-terminal 21 (uL4) or 23 (uL22) amino acids of the respective E. coli proteins. The use of a synthetic peptide as the antigen ensured that the epitope does not overlap with the deleted region of the proteins.
Figure 2.
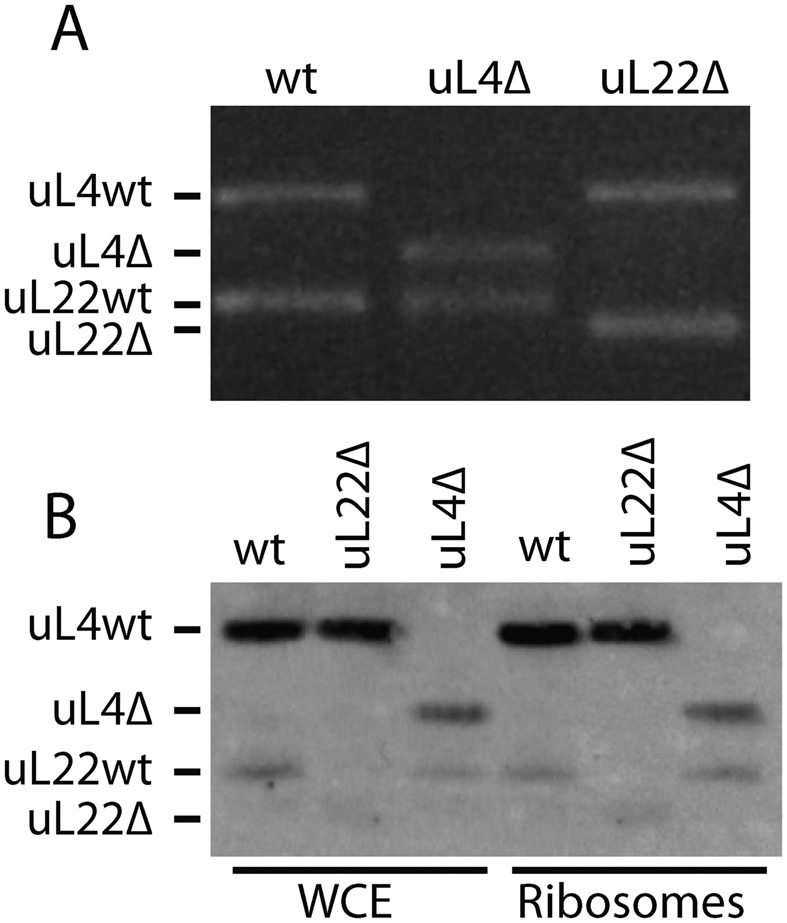
Deletions in and expression of the uL4-Δloop and uL22-Δloop genes. (A) Products from PCR reactions on DNA from parent and mutants colonies. (B) Western analysis of protein from salt washed ribosomes and whole cell extracts (WCE). The membrane was probed with a combination of anti-uL4 and anti-uL22 sera.
Ribosomes preparation
Cultures were grown to OD450 = 1.0–2.0 (Hitachi U-1100 spectrophotometer, 10 mm cuvette path length; corresponding to about 2–4 × 108 cells per ml). Cultures were typically harvested by pouring over ice (‘fast-cool’); where noted, the cultures were allowed to cool to 4°C in an ice bath before harvesting (‘slow-cool’). Cells were collected by centrifugation at 8000 rpm for 10 min in a Beckman JLA 10.5 rotor, resuspended in Buffer A1 (see below) and lysed using a French press at 16 000 psi. Lysates were clarified by spinning at 22 000 rpm for 30 min in a Beckman MLA-80 rotor followed by pelleting the ribosomes at 50 000 rpm for 4 h in the Beckman MLA-80 rotor. The surface of the ribosome pellet was rinsed with Buffer A1 and resuspended overnight at 4°C in Buffer A1. Ribosomes were salt-washed by mixing one part crude ribosome suspension with nine parts salt-wash buffer (20 mM HEPES-KOH pH 7.5, 6 mM MgCl2, 1 M NH4Cl, 6 mM β-mercaptoethanol), incubating on ice for 1 h, and centrifuging at 50 000 rpm for 4 h. Salt-washed ribosomes were resuspended as described above.
Sucrose gradient analysis
Ten A260 units of ribosomes or crude lysates were centrifuged through 10–50% sucrose gradients 4°C for 4–6 h at 40 000 rpm in a Beckman SW40Ti rotor. To maintain 70S and polysomes (associating conditions), we used Buffer A1 (20 mM HEPES-KOH pH 7.5, 6 mM MgCl2, 30 mM NH4Cl, 6 mM β-mercaptoethanol); to separate 70S and polysomes into subunits (dissociating conditions) we used Buffer A2 (same as Buffer A1, except that the MgCl2 concentration was 0.3 mM), or Buffer A3 (20 mM HEPES-KOH pH 7.5, 1 mM MgCl2, 200 mM NH4Cl, 6 mM β-mercaptoethanol), as noted.
Western analysis
Lysates (0.2 A260 units), ribosomes (0.1 A260 units), or portions of sucrose gradient fractions were analyzed following standard protocols. Ribosomal proteins were probed using rabbit antisera against synthetic peptides corresponding to the N-termini of uL4 and uL22 [diluted 1:10 000]. Goat anti-rabbit IgG (Bio-Rad, diluted 1:3000) alkaline phosphatase conjugate was used as the secondary antibody. Probed membranes were incubated in 400 μl of ECF Western Blotting reagent (GE Healthcare) for 5 min. Finally, membranes were visualized using a STORM PhosphorImager (Molecular Dynamics).
Northern analysis
A total of 20 μl of gradient fraction was mixed with 30 μl of formamide (final concentration 60%) and 5 μl of 10×load dye (50 mM Tris–HCl, pH 7.6, 0.25% bromophenol blue, 60% glycerol, 1% sodium dodecyl sulphate (SDS)). Samples were incubated at 65°C for 5 min, then immediately chilled on ice for 5 min before fractionation in 1% agarose gels in 0.5× sodium borate buffer at 150V. Following electrophoresis gels were transferred to Hi-Bond N membranes (Amersham) by the vacuum method. Membrane-bound RNAs were probed by hybridizing 32P 5′-end labeled oligonucleotides at 37°C in ULTRAhyb®-Oligo buffer (Ambion) according to manufacturers’ specifications, then visualized using a STORM phosphoImager (Molecular Dynamics).
Primer Extension were performed as described in (34).
Peptide-induced pausing and β-galactosidase assays were performed as described previously (38).
RESULTS
The extended loops of E. coli ribosomal proteins uL4 and uL22 are not essential for growth
The uL4-Δloop strain, isolated as a spontaneous troleandomycin-resistant derivative of E. coli A19, contains a uL4 gene lacking codons 42–90. This deletion removes the entire tentacle, but leaves the globular domain intact (Figure 1B). The uL22-Δloop strain carries a uL22 gene in which codons 82–98 were replaced by two glycine codons (Figure 1C).
Gel electrophoresis of PCR-amplified copies of the genes confirmed that the mutant strains contained only deletion alleles of the uL4 or uL22 genes (Figure 2A). Moreover, Western blots of whole cell lysates and salt-washed ribosomes from the mutant and parent strains showed that uL4 and uL22 in the respective mutants migrated faster on an SDS gel than the corresponding protein bands from the parent (Figure 2B). These results confirm that the mutant strains express only the deletion versions of uL4 or uL22, and that these loop-less proteins are incorporated into ribosomes. The mere isolation of strains expressing only uL4 or uL22 mutant proteins shows that the extended loops of these proteins are not required for the generation of ribosomes with sufficient function to sustain life.
Since the uL4-Δloop mutant was isolated as a spontaneous mutant, we considered the possibility that the phenotype observed with the mutation (see below) might be affected by secondary (suppressor) mutations elsewhere in the genome that were unknowingly selected for. To determine if the uL4-Δloop deletion alone could generate the observed phenotypes, we used bacteriophage P1 to transduce erythromycin resistance from the original mutant to the A19 parent strain. The transductant recapitulated the phenotype of the original mutant, confirming that the tentacle deletion alone was responsible for the phenotypes.
Lack of the uL4 and uL22 extended loops affects growth
Even though ribosomes containing uL4 or uL22 loop deletion proteins supported growth, both mutant strains grew significantly slower than wild-type on both LB agar plates (not shown) and in liquid LB medium (Figure 3A). The uL22-Δloop mutant grows about half as fast as wild-type at all temperatures (Figure 3B). Interestingly, the uL4-Δloop mutant is cold-sensitive: it grows at almost 50% the wild-type rate at 42°, but growth rates decrease compared to the parent as the temperature is reduced (Figure 3B). At 22°, the uL4 mutant did not grow.
Figure 3.
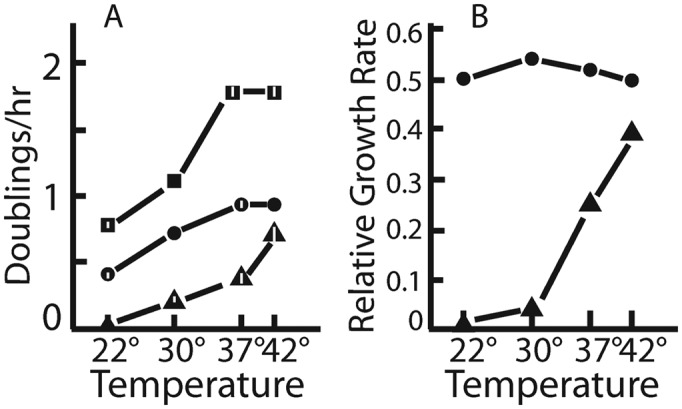
Growth rates of mutants and parent at different temperatures. (A) Actual growth rates. (B) Growth rates of each mutant strain normalized to the growth rate of the parent growth rate at the same temperature. Squares: wild-type; triangles: uL4-Δloop; circles: uL22-Δloop. Verical bars indicate standard error of the mean. Where none is indicated, the standard error of the mean was too close to 0 to show up in the figure. Number of biological replicates: N = 3 for all growth rates measurements, except for uL4-Δloop at 22° (N = 2) and uL4Δloop at 30° (N = 1).
Deletions of the uL4 and uL22 loops disturb ribosomal assembly
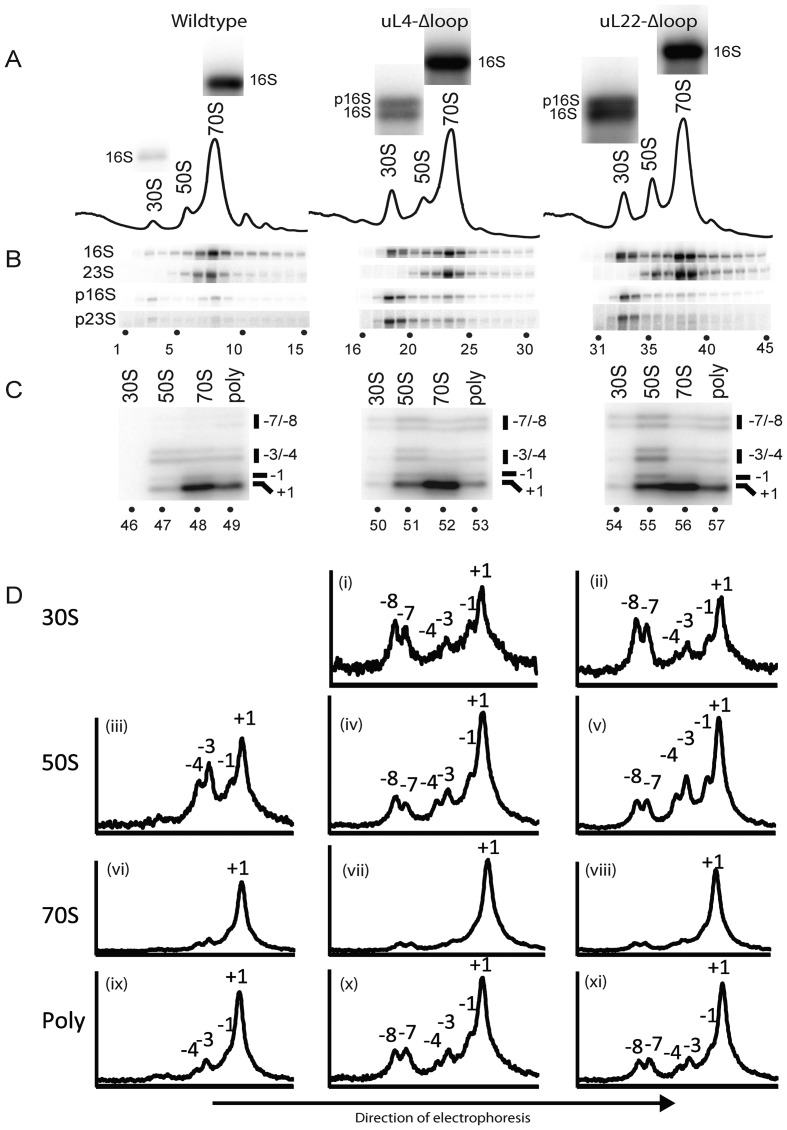
To determine if ribosomal assembly is affected by the uL4-Δloop and uL22-Δloop mutations, we analyzed ribosomes from cells grown at 37° on sucrose gradients in buffer A1, which maintains polysomes and 70S ribosomes (‘associating conditions’). Both mutants showed changes in both the 30S and 50S peaks relative to the wild-type (Figure 4A). In the uL4-Δloop mutant, the 30S was enlarged, while the 50S was asymmetric with an edge trailing toward the 30S peak. In the uL22-Δloop mutant, the 30S and 50S peaks were high relative to the 70S peak when compared to the wild-type cells.
Figure 4.
Analysis of mutant and parent ribosomes. Cultures of the indicated strains were grown at 37°. (A) Sucrose gradient profiles. Ribosomes were fractionated on sucrose gradients in Buffer A1 (associating conditions). The bands shown above each peak are explained in the legend for part of the figure (B). (B) Northern analysis of gradient fractions containing ribosomal peaks. RNA was prepared from each sucrose gradient fraction and used for a northern blot, which was probed with oligonucleotides specific for precursor 16S (p16S), precursor 23S (p23S) and mature 16S and 23S rRNA molecules (see text, Supplementary Table S1 and Supplementary Figure S1 for more information about the probes). The bands from the hybridization with the 16S probe to RNA from the 30S and 70S peak fractions were enlarged and shown above each peak in part A. Note that RNA from the 30S peak from the two mutants, but not from the wild-type, contains two types of rRNA hybridizing to the 16S probe. The top bands are precursor 16S (also called 17S) and the bottom bands are mature 16S rRNA. (C) 5′ end mapping of 23S-type rRNA in sucrose gradient peaks. A second sucrose gradient was run on the ribosome preparations from each of the three strains and 84 fractions were collected from each gradient. Fractions from the 30S, 50S, 70S and polysome regions were then pooled and RNA from each pool was used for primer extension using a primer complementary to nucleotides 72–89 of mature 23S rRNA. The products were fractionated on gels and imaged on a phosphoimager. (D) Quantification of primer extension results shown in C. The pixel density along each lane of the gel image in C was digitized using ImageJ and plotted. The ordinates show the relative pixel density at different points of the gel. Note that the ordinates for different plots (corresponding to different lanes) are not the same. The plots serve only to show the distribution within a lane. The peaks of 23S rRNA with mature and 5′ extensions of 1, 3, 4, 7 and 8 nucleotides are indicated. Panel numbers are referred to in the text.
RNA from each sucrose gradient fraction in the region of ribosomal peaks was fractionated on an agarose gel and transferred to a nylon membrane. The membrane was hybridized successively with probes for precursor rRNAs (p16S and p23S) and mature 23S and 16S rRNAs (Figure 4B). The probe for precursor 23S (p23S) was complementary to the sequence ranging from seven bases upstream of the mature 5′ end to nine bases downstream of the mature end of 23S rRNA (Supplementary Table S1 and Supplementary Figure S1). Under our hybridization conditions, this probe hybridizes to incompletely processed 23S carrying a 7 to 8 nucleotide 5′ extension, but does not hybridize to mature 23S rRNA or to precursor RNA with shorter 5′ end extensions. The p23S probe hybridized to RNA in the 30S peak in the wild-type (Figure 4B, lane 3), as would be expected since one class of precursor 50S ribosomes in wild-type cells cosediments with the 30S subunit under associating buffer conditions (40). However, hybridization of the 30S gradient fractions from both mutants was significantly stronger than in the wild-type (Figure 4B, compare lanes 18–19 and 33–34 with lane 3). We interpret these results as showing that deleting the uL4 or uL22 loop results in elevated levels of precursor 50S particles sedimenting at ∼30S. Unexpectedly, we also observed an increased signal for p16S (17S) in the 30S peak when probing with an oligonucleotide complementary to RNA 95–114 base pairs upstream of the 16S 5′ end (Figure 4B; compare lane 3 with lanes 18–19 and 33–34). Furthermore, the 16S probe generated a double band in the northern analysis of RNA from the 30S peaks of the mutants, but not in the northerns of RNA the 30S peak of the wild-type or the 70S peak from any of the strains, confirming the presence of both p16S rRNA (top band) and mature 16SrRNA (bottom band) in the 30S peaks from the two mutant strains (Figure 3A; see the enlarged northern images of the 16S probing above each peak fraction in the sucrose gradients and the legend to Figure 3B). This suggests that the processing of the p16S rRNA is affected by the changes in the 50S assembly.
To further analyze ribosomal assembly in the mutants, we wanted to characterize the mixture of 5′ ends of 23S rRNA molecules found in the 30S, 50S, 70Sand polysomes. To this end, we ran a second sucrose gradient on the ribosome preparations from each of the three strains and collected 84 fractions from each gradient (rather than 26 collected from the gradients in Figure 4A) to obtain a better separation of the different peaks in the fractions collected. The fractions in the 30S, 50S and 70S peaks were pooled separately; RNA from each pool was purified and used for primer extension experiments using a 32P-labeled primer complementary to a region 72–89 nucleotides downstream of the 5′ end of mature 23S rRNA. The products of the primer extensions were fractionated on an acrylamide gel and visualized on a phosphoimager (Figure 4C). The 30S peak from the parent strain did not contain sufficient RNA to produce a primer extension profile (Figure 4C, lane 46). The results showed that all other peaks contained 23S rRNA with the 5′ ends of mature 23S rRNA (Figure 4C ‘+1’ bands) as well as immature 23S rRNA with ends 1, 3, 4, 7 and 8 nucleotides upstream from the mature 5′ end (Figure 4C, lanes 47–57), although relative intensity of these bands varied between lanes. We refer to these extended molecules as −1, −3, -4, −7 and −8, respectively. To obtain quantitative data, the gel images in Figure 4C were digitized using ImageJ (41) and plotted (Figure 4D). The peaks corresponding to the bands in the images of Figure 4C are indicated in Figure 4D. The results showed that the 30S peaks from both mutants contained significant amounts of immature 23S rRNA with 5′ ends at positions −7 and −8 as well as smaller amounts of molecules with −1, −3 and −4 ends (Figure 4D i and ii). The 50S particles from both mutants also contained immature 23S rRNA, but the balance between −7/−8 and −1/−3/−4 types of 23S rRNA was shifted toward shorter extensions compared with the distribution in the 30S peaks (compare Figure 4D iv and v with panels i and ii), compatible with the notion that the immature rRNA in the 30S peaks is an intermediate on the path to the 23S rRNA in the 50S peak. The higher ratio between longer 5′ extensions (−7 and −8) and shorter 5′ extensions (−3 and −4) in the 30S relative to the 50S peak also shows that the finding of immature 23S rRNA in the 30S peak is not simply due to spillover from the 50S peak to the 30S peak. A significant fraction of the −7 and −8 molecules in the 30S peak must originate from immature particles co-sedimenting with the 30S subunit. The 50S peak from the wild-type contained essentially no −7/−8 5′ ends, but did contain detectable amounts of −4, −3 and −1 ends (Figure 4D iii). Furthermore, it should be noted that very little of the 23S rRNA in the 70S peak has extended 5′ ends (Figure 4D vi–viii)), but significant amounts of 23S rRNA with 5′ extensions was found in the polysomes of the mutant strains (Figure 4Dx and xi).
Deletion of the uL4 extended loop makes ribosome assembly cold-sensitive
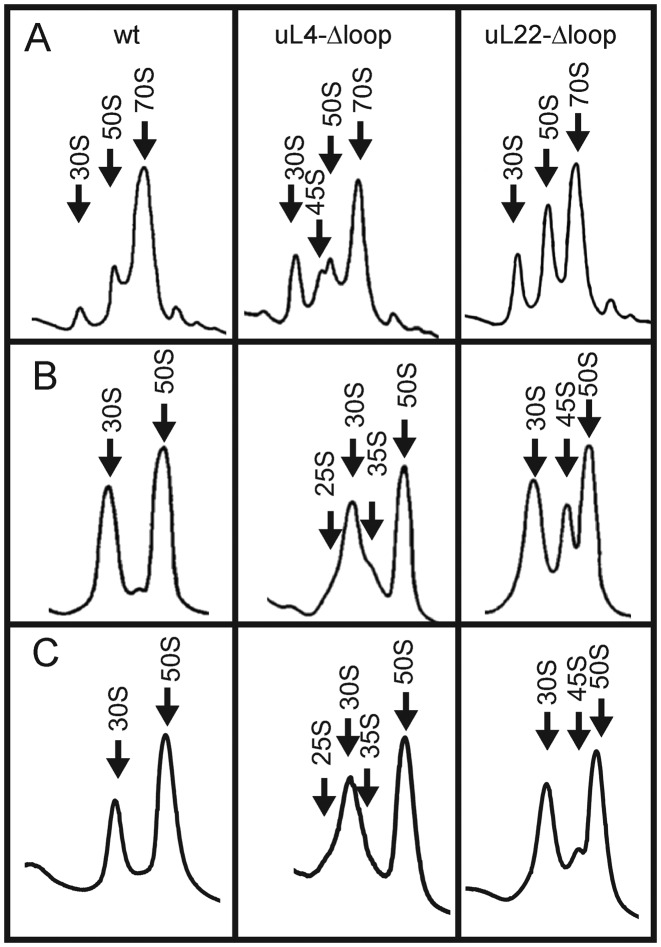
The inability of the uL4-Δloop mutant to grow at 22° prompted us to determine the effect of temperature on ribosome assembly. The ribosome sedimentation profile in the mutant, analyzed on sucrose gradients run for a longer time than the experiment shown in Figure 4A, included an additional peak sedimenting slightly slower than the 50S subunits (Figure 5). This peak, referred to as ‘45S’, was increased in the lysate from the 30°-culture relative to the 42°-culture. Furthermore, shifting the uL4-Δloop culture from 42° to 25° for 6 h revealed ‘chaos’ in the 30S–50S region, with multiple overlapping peaks. We conclude that assembly of the 50S subunit in the uL4-Δloop mutant is indeed cold-sensitive (Figure 5).
Figure 5.
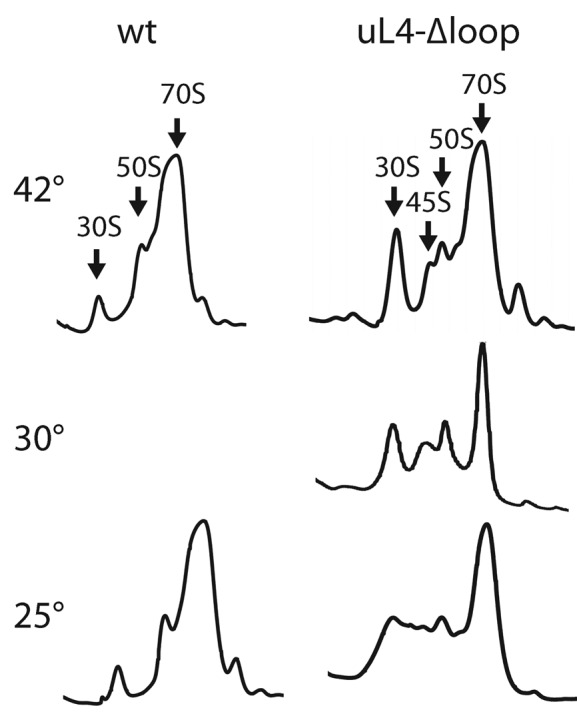
Sucrose gradient analysis of ribosomes from the uL4-Δloop strain grown at different temperatures. Top two rows of gradient traces: cultures of the uL4Δloop and wild-type mutant were harvested after steady state growth at the indicated temperatures. Bottom row: cultures were grown in steady state at 42°, then shifted to 25° for 6 h before harvest. Cultures were diluted as necessary to keep the cell density below ∼108 cells per ml. Whole cell lysates were centrifuged through sucrose gradients in buffer A1.
Effect of buffer conditions on sedimentation of immature mutant ribosomal particles
To investigate the plasticity of the immature particles, we compared the sedimentation profiles of ribosomes under associating and dissociating conditions. The content of immature particles was accentuated by letting the culture cool slowly to 4° before harvest (compare Figure 6A with Figure 4A). Ribosomes were pelleted in buffer A1, then resuspended and analyzed on sucrose gradients in associating or dissociating buffer (Figure 6A and B, respectively). The results show that the 45S peak in the uL4-Δloop mutant in the associating gradient was replaced in the dissociating gradient by two shoulders flanking either side of the 30S peak labeled ‘25S’ and ‘35S’, respectively (compare Figure 6B with Figure 6A), suggesting that the immature ‘45S’ particles change shape and/or lose protein mass under dissociating conditions. The gradient with uL22-Δloop ribosomes revealed a prominent 45S peak under the dissociating conditions, which was not seen under associating conditions (compare Figure 6B with 6A). Apparently some of the 50S particles in this mutant also change shape and/or are somewhat unstable. This is consistent with the finding of substantial amounts of p23S rRNA in the 50S peak under associating conditions (Figure 4B–D), showing that a significant fraction of the material running as 50S under associating conditions is not fully matured 50S subunits. To test whether the properties of the immature particles were modified by the slow-cool harvesting procedure, we also analyzed ribosomes prepared after harvest of cells over ice on dissociating sucrose gradients (Figure 6C). The position of the peaks was the same in ribosomes material from ‘slow-cool’ (Figure 6B) and ‘fast-cool’ (Figure 6C) cells, showing that the sedimentation velocity is determined by the dissociating buffer conditions rather than the protocol for cell harvest.
Figure 6.
Comparison of sucrose gradient profiles after different conditions of harvest and buffers. Cultures were grown at 37° and cooled by submerging the growth vessel into an ice-bath slurry before harvest (A and B; ‘slow cool’) or harvested over ice (C; ‘fast cool’). Ribosomes were extracted and pelleted before being resuspended in Buffer A1, then diluted into Buffer A1 (A), Buffer A3 (B) or Buffer A2 (C) and centrifuged through sucrose gradients in the same buffers.
Competition between proteins with and without the extended loop
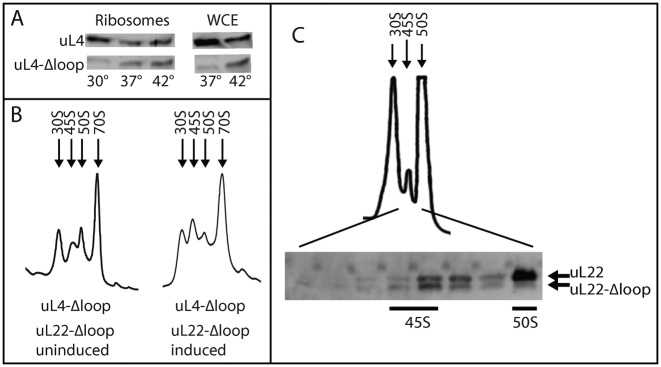
The accumulation of immature particles containing precursor rRNA suggests that ribosome assembly is hampered when the uL4 or uL22 protein lacks the extended loop. To test this model we compared incorporation of uL4 wild-type and loop deletion proteins into ribosomes in cells harboring both uL4 genes. A wild-type strain harboring a plasmid with the uL4-Δloop gene expressed from the araBAD promoter was induced with arabinose for 1.5 doublings at 30°, 37° and 42°. Western analysis of purified ribosomes showed a decreasing ratio between the mutant and parent proteins with decreasing temperature (Figure 7A). Since neither the parental uL4 promoter nor the ara promoter are temperature regulated, it is unlikely that this difference is due to temperature effects on expression of the two genes. Rather, the results support our conclusion that assembly of ribosomes containing the mutant protein is less efficient than assembly of ribosomes containing the wild-type protein. Western analysis of whole cell extracts also showed decreased uL4-Δloop protein at 37° compared to 42° (Figure 7A), suggesting that the uL4-Δloop protein becomes unstable when the temperature is reduced. Apparently, r-proteins not stably incorporated in ribosome particles are targeted by proteases and eliminated.
Figure 7.
Competition between parent and Δloop proteins for incorporation into ribosomal particles. (A) The wild-type was transformed with a plasmid expressing uL4-Δloop from the araBAD promoter and grown in LB arabinose at 30°, 37° and 42°. Proteins from WCE and purified ribosomes were analyzed on a western blot probed with uL4 antiserum. (B) The uL4-Δloop mutant was transformed with a plasmid expressing uL22-Δloop from the araBAD promoter. An aliquot was harvested from a culture in LB medium (left). Another aliquot was harvested after the culture had been then supplemented with 0.2% arabinose to induce synthesis of uL22-Δloop for 1.5 doublings (right). Purified ribosomes were fractionated on a sucrose gradient in Buffer A1. (C) Ribosomes purified from another culture of the uL4-Δloop, uL22 wt/uL22-Δloop strain grown in LB and induced with arabinose for 1.5 h was fractionated on a sucrose gradient in dissociating buffer. Samples from the indicated fractions were analyzed on a western blot probed with uL22 antiserum.
To learn if ribosomes could be assembled with both the uL4 and uL22 loop deletion proteins, we transformed the uL4-Δloop mutant with a plasmid harboring the uL22-Δloop gene expressed from the araBAD promoter, and induced with arabinose for 1.5 doublings at 37°. Sucrose gradient analysis (Figure 7B) shows that the 45S peak increased in size relative to the 50S peak after induction, suggesting that synthesis of the loop-less uL22 protein exacerbates the assembly problems of 50S subunits already caused by the uL4-Δloop protein. To further investigate if 50S subunits can be formed with both loop deletion proteins, ribosomes were prepared from the double mutant strain that had been induced with arabinose for 1.5 doublings and fractionated on a dissociating sucrose gradient. A 45S peak appeared between the 30S and 50S peaks (Figure 7C), as expected from the gradients in Figure 6B. Fractions from the 45S to 50S region of the gradient were analyzed by western blot probed with antibody to uL22. The results show that the 45S peak contains uL22-Δloop and uL22 wild-type protein in approximately the same amount. In contrast, the 50S peak contains only the wt uL22 mutant, suggesting that 45S particles containing both the uL4 and uL22 deletion proteins are formed, but the 45S particles fail to mature into 50S subunits.
Function of ribosomes containing loop-less uL4 or uL22 proteins
To analyze the function of ribosomes containing either loop deletion protein, we induced the lacZ gene with isopropyl thiogalactoside (IPTG) and measured β-galactosidase activity as a function of time after induction (Figure 8). The lag time before the appearance of enzyme activity above the basal level is inversely correlated with the peptide elongation rate (42). Induction kinetics at 42°, 37° and 30° showed that the induction delay in the uL22-Δloop mutant was not significantly longer than in the parent at any temperature. Thus, the deletion of the uL22 loop had little effect on the ribosome translation rate. In contrast, the loss of the uL4 loop significantly impairs the translation speed of the ribosomes in a temperature dependent manner. At 42°, the induction delay in the uL4-Δloop strain was slightly longer than in the wild-type (Figure 8). At 37° the induction time for uL4-Δloop was about 25% longer than for the wild-type, but at 30° the uL4-Δloop ribosomes took almost twice as long as the wild-type to produce enzyme, showing that they translate at only approximately half the speed of wild-type ribosomes. Note that the induction delay is determined by the fastest ribosomes in the cell and is thus not affected by the kinetics of ribosome assembly or heterogeneity of the 70S ribosomes.
Figure 8.
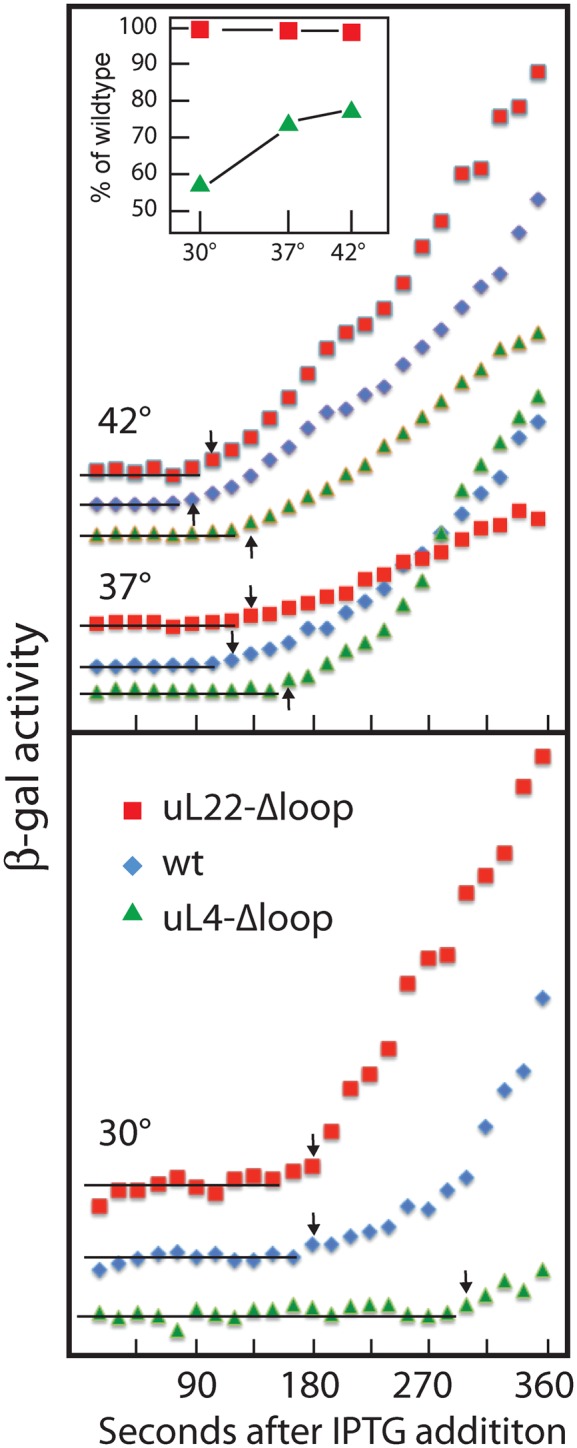
Kinetics of β-galactosidase induction at different temperatures. Cultures were grown at 30°, 37° and 42° and induced with IPTG. Aliquots were withdrawn at the indicated times and analyzed for β-galactosidase. The enzymes activities were graphed as a function of time after induction. Data for 37° and 42° are shown in the top panel and 30° in the bottom panel. The horizontal lines indicate basal levels and the arrows indicate the first point of a monotonous increase of enzyme activity. The time of the last point of the basal level, i.e. the point immediately before the arrow, is taken to be the induction delay. Symbols are defined on the figure. Inset is a plot of the relative translation rates of mutants compared to wild-type at the same temperature.
Previous studies showed that insertion or missense mutations in the uL4 loop strongly reduce ribosome affinity for macrolide antibiotics, while mutations in the uL22 loop have little effect (34,43). We measured erythromycin binding to ribosomes from each of the loop deletion mutants. The loop deletion mutants recapitulated the results from other uL4 and uL22 loop mutants: the uL4-Δloop ribosome bound essentially no drug, while binding to the uL22-Δloop mutant ribosomes was reduced by only 35% (data not shown). The removal of the loops thus generates an erythromycin binding phenotype which is very similar to that observed for the missense and insertion mutants.
Finally, we assessed the ability of mutant ribosomes to respond to pausing signals in nascent peptides. These signals are derived from short amino acid sequences, ‘pausing peptides’, that arrest or pause translation, either spontaneously or in the presence of specific cofactors such as antibiotics.
To measure crb pausing, we inserted a segment containing the gene for the leader peptide (CrbCmlA), the intercistronic region encoding a hairpin between the Crb leader and cmlA, and first 15 codons of cmlA into the chromosomal lacZ gene present on the chromosome (38) (Figure 9A). In the presence of low concentrations of chloramphenicol, ribosomes pause at the end of the crb message and sterically prevent the formation of a hairpin, which, if intact, sequesters the ribosome binding site for the cmlA::lacZ fusion gene (Figure 9A) (38). That is, in this system pausing increases β-galactosidase translation. As found with other uL22 loop mutants (38), the uL22 loop deletion had a modest effect (30% reduction) on crb pausing. In contrast to missense and insertion mutants in the uL4 loop, the uL4-Δloop mutation severely debilitated crb pausing, reducing β-galactosidase synthesis by about 90% (Figure 9B). This is a much stronger effect than previously found for other uL4 loop mutations, which only reduced crb pausing by 10–45% (38).
Figure 9.
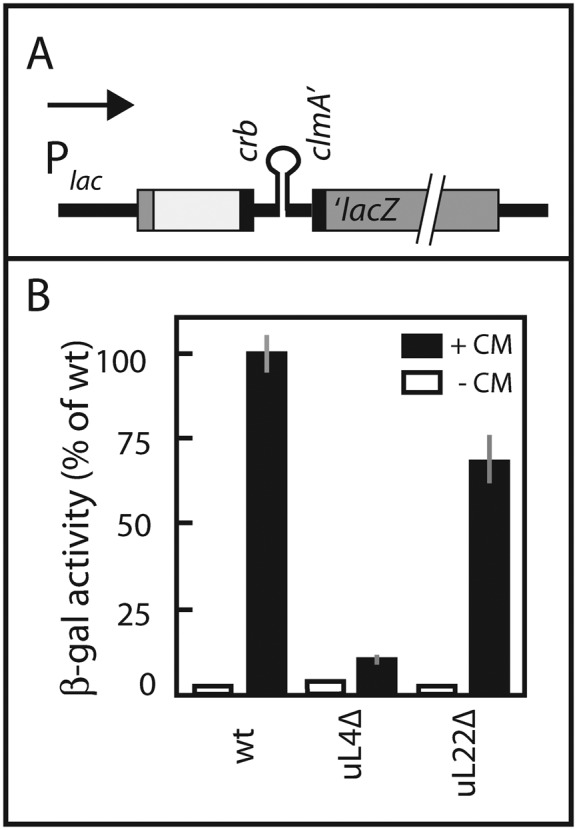
CrbCmlA-mediated translational pausing. (A) Map of construct for quantifying pausing. The construct consists of (from left to right) the lac promoter, N-terminal end of the lacZ gene (gray), a piece of pGEM5 (white box), the sequence for the crb pausing peptide, intergenic hairpin and the N-terminus of cmlA (all black) and finally the remaining part of the lacZ gene (gray). For more details, see ‘Materials and Methods’ section. (B) β-galactosidase enzyme activity. Enzyme activity in the absence (white bars) or presence of 0.8 μg/ml chloramphenicol (CM) (black bars) are shown. The averages and standard errors of the mean (gray vertical lines) of biological triplicates are shown. Three enzyme assays were done for each replicate.
To measure secM pausing, which does not require a cofactor, the DNA sequence for the pausing peptide was inserted into the chromosomal lacZ gene (Supplementary Figure S1A). As previously reported, wild-type ribosomes are efficiently arrested on the secM::lacZ mRNA, resulting in an almost 1000-fold decreased accumulation of β-galactosidase activity (Supplementary Figure S1B) (38). Ribosomes carrying the uL4-Δloop mutation respond almost as well: the secM pausing peptide reduces β-galactosidase production more than 500-fold. The uL22-Δloop deletion strain shows a reduced response to the SecM peptide, with an almost 10-fold increase in β-galactosidase activity relative to the parent. However, this is still only about 1% of the enzyme level in the control strain without the secM pause signal (Supplementary Figure S1B). In conclusion, deletion of the uL4 and uL22 loops has only a marginal effect on the SecM pause peptide, a result similar to what we have reported for other uL4 and uL22 loop mutations (38).
DISCUSSION
Role of the extended loops of uL4 and uL22 in ribosome biogenesis
Our experiments show that the extended loops in uL4 and uL22 are not essential for E. coli viability and growth. This confirms our previous conclusion that 50S ribosomes capable of entering polysomes can be formed with uL4 or uL22 lacking part or all of the extended loops (36). However, in our previous analysis the deletion proteins were expressed temporarily from inducible genes and ‘chased’ into mature 50S ribosomes, preventing detailed analysis of the ribosome assembly process. In the current report, ribosome assembly and function were analyzed in strains without wild-type uL4 or uL22 genes, i.e. containing only the loop deletion alleles. Under that regime, we could show that without the uL4 or uL22 extended loops, cells grow slowly and accumulate immature ribosomal particles, demonstrating that ribosomal assembly is hampered. We conclude that the loops facilitate ribosome formation, although cells can still make functional ribosomes in the absence of the uL4 and uL22 extended loops. Interestingly, while the uL4 and uL22 genes are essential in E. coli (44), the bacterial species Deinococcus radiodurans survives with ribosomes entirely lacking uL4, even though this mutant strain grows at only one-third the rate of wild-type cells and also accumulates immature LSUs (J.M. Zengel and L. Lindahl, manuscript in preparation). On the other hand, the extended loop in yeast ribosomal protein uL4 (RPuL4) is essential for growth and ribosome assembly ((45) and our unpublished results). The difference between bacteria and yeast may relate to the fact that the assembly system in eukaryotes is much more complex than the bacterial system; eukaryotes may have exploited the uL4 loop to perform assembly function(s) that do not exist in bacteria. Indeed, it was recently proposed that part of the uL4 extended loop may be used as a nuclear localization signal in eukaryotes (46).
The uL4 and uL22 mutants show different sucrose gradient profiles of immature particles containing p23S rRNA (Figure 4). Moreover, the sedimentation profiles depend on the buffer conditions. Specifically, analysis of ribosomes from the uL4 mutant revealed immature ribosome peaks at 30S and 45S under ‘associating conditions’, but at 25S and 35S under dissociating conditions, while analysis of uL22 mutant ribosomes revealed 30S and 50S immature particles under associating conditions, but 30S and 45S particles under dissociating conditions (Figure 6). Together these experiments suggest that the immature particles change conformation and/or lose protein material depending on the ionic conditions. They also suggest that the extended loops of uL4 and uL22 are critical for separate steps of ribosome assembly.
Implications of incomplete trimming of rRNA in mutant ribosomes
The incorporation of loop-less uL4 and uL22 proteins into 50S subunits affects rRNA processing; we found incompletely processed 5′ ends in the 23S from both mutants. Interestingly, both mutants accumulated more unprocessed 5′ ends of 23S rRNA in polysomes than in 70S ribosomes. This observation parallels the finding of precursor 16S rRNA in polysomes (47) and supports the notion that the final steps in rRNA processing involve binding to mRNA (48). Possibly, almost-mature precursors particles are tested for translation competence on polysomes, before final maturation and stabilization of the 50S subunits, in a process similar to the translation-like quality control checkpoint in yeast (49).
Contribution of the uL4 extended loop to ribosome assembly
What is the role of the uL4 loop in ribosome assembly? Considering that (i) the uL4 extended loop is intrinsically disordered in the free protein (50), but has a defined structure in the mature ribosome (3,4); (ii) the uL4 loop interacts with several domains in 23S rRNA in the mature ribosome (51); (iii) purified uL4 protein binds specifically to Domain I of 23S rRNA (52,53) and (iv) the globular domain of uL4 is bound to Domain I of 23S rRNA in the mature 50S subunit (54), it appears that the globular uL4 domain is responsible for the initial binding of uL4 to rRNA. This process may be cooperative since L24 binds close to uL4 in Domain I (53). Furthermore, it is important to note that deletion of the gene encoding the RNA helicase SrmB, involved in 50S assembly, generates phenotypes similar to those of the L4 extended loop deletion mutant, including severe cold sensitivity, accumulation of 50S precursor(s), 23S processing defect, and, especially interesting, accumulation of 17S SSU RNA precursor (55). While the effect on 16S rRNA processing may be indirect, it should be noted that this effect is common to the L4 loop and the srmB deletions, but is absent with other mutations that hamper 50S assembly such as deletion of the csdA/deaD gene (56). Moreover, the SrmB helicase co-immunoprecitates with uL4 and uL24, suggesting that SrmB forms a complex with uL4 and uL24 during ribosome assembly (57). We therefore propose that, subsequent to formation of the initial uL4-uL24-pre23S rRNA complex, the uL4 loop and parts of p23S rRNA fold cooperatively, likely with the help of the SrmB RNA helicase, Accordingly, deletion of the uL4 loop would preclude the formation, or perhaps action, of the uL4-L24-SrmB complex, thereby changing the assembly to a slower alternate, cold sensitive path that results in functionally cold sensitive ribosomes. In contrast to uL4, the uL22 loop structure is essentially the same in crystals of both the free protein (58) and the mature ribosome (3,4), suggesting that uL22 modifies the rRNA structure, but is not itself significantly changed in structure.
Loops and extensions of other ribosomal proteins may play roles similar to the uL4 and uL22 loops (59). For example, both the flexible loop of uL11 protein and rRNA are re-organized when the protein binds to rRNA (60). In addition, the extensions of S12 and L20 play roles in the assembly of their respective subunits (61,62).
Why ribosome assembly mutants contain 70S ribosomes
Given the extensive interactions of the amino acids in the uL4 and uL22 extensions with the rRNA core of the 50S subunit (3,4), one might have assumed that their deletion would abort ribosome assembly, preventing cell survival. Yet, cells are viable despite these deletions (albeit with reduced growth rates). Two things may explain this apparent discrepancy. First, the deletion of the loops may slow a particular step in the 23S rRNA normal/major folding pathway without completely preventing it. This would lead to a buildup of normal assembly intermediates, which in the wild-type are below the detection level. Second, it is important to note that alternate ribosome assembly pathways have been observed, i.e. ribosomal assembly occurs in a set of branched pathways (63,64), not a single linear pathway as the process traditionally is represented. Thus, if the folding step facilitated by one of the uL4 or uL22 protein loops has a major effect on 23S rRNA folding, or completely block the normal/major folding pathway, the formation of 70S ribosomes may switch to an alternate pathway in the energy landscape, leading to accumulation of assembly intermediates characteristic of the alternate pathway. We cannot distinguish between these possibilities and it is indeed possible that they both contribute simultaneously to formation of 70S ribosomes in the mutants.
When we induced expression of L22-Δloop in cells carrying a wild-type L22 gene and the uL4-Δloop gene, we observed 45S particles containing both loop deletion proteins. However, the mature 50S subunits contained only the uL4-Δloop protein and not the uL22-Δloop protein (Figure 7). We interpret this to mean that ribosomal particles containing both deletion proteins cannot be converted into 50S subunits suggesting that the uL4 and uL22 extended loops cooperate in some way, which lands the double mutant in a kinetic trap. The uL4-Δloop uL22-Δloop 45S particles appear to be unstable, paralleling the observation of ribosome turnover of ribosomal particles in the presence of sub-lethal neomycin concentrations (65).
Translation by mutant ribosomes
Cold-sensitivity is a frequent feature of ribosome assembly mutants; see, e.g. (66–74). Interestingly, both assembly and function of ribosomes assembled with the uL4-Δloop mutant are cold-sensitive. In contrast, the uL22-Δloop ribosome translates at almost the same rate as the parent ribosome at all temperatures (Figure 8), even though ribosome assembly is also abnormal in this mutant (Figure 4). These results suggest that the extension of uL4 is required for both efficient assembly and optimal translation, particularly at low temperature, while uL22's extended loop plays predominantly a role in assembly. On the other hand, it is interesting to note that insertions into the uL22 loop significantly reduce the ribosome translation rate (34), suggesting that, for this protein, adding extra mass inside the ribosome may be more detrimental to ribosome function than simply removing the loop.
Interdependence of assembly of the two ribosomal subunits
We were surprised that the deletion of the uL4 and uL22 extended loops also affects the processing of 16S rRNA, as indicated by the increased accumulation of p16S rRNA in the 30S peaks of the mutant strains. Previous studies showed that the assembly of the 30S subunits affects the formation of the 50S, but not vice versa (75). Our observations suggest that the prevailing view that the rRNA processing and assembly of the two subunits are largely independent of each other deserves further scrutiny.
Translation pausing in ribosomes lacking the uL4 and uL22 extended loops
Gabashvilli et al. (76) suggested that the constriction formed by the tips of the uL4 and uL22 loops functions as a gate that regulates the migration of the nascent peptide through the tunnel. However, the effect of the loop deletions on pausing is, for the most part, modest, suggesting that passage through the uL4-uL22 constriction is not a necessary part of the pausing mechanism, at least not for the two pausing systems, secM and cmlA, tested here. In fact, recent experiments demonstrated that interaction between the erythromycin pausing peptide and the ribosome changes the structure of the peptidyl transferase center and reduces its activity (77), suggesting that pausing peptides set off cascades of structural modulations that ultimately target the peptidyl transferase center rather than a gate in the exit tunnel.
Comparison of deletion and insertion mutants
We have previously reported that point mutations as well as insertions and short deletions in the uL4 and uL22 loops also affect ribosome assembly and rRNA processing (34). Based on the phenotypes of those mutants, we could not determine whether the phenotype is caused by loss of contact between the uL4 or uL22 loop and 23S rRNA, or by generation of new but inappropriate contacts between rRNA and the mutated loops. In general, there is a strong similarity between the particles accumulating in the mutants with modified loops described previously (34) and the loop-less mutants reported here, suggesting that the loss of the contacts that exist in the parent are the most important for normal assembly. The uL4 and uL22 loop deletion mutants also respond with high efficiency to the secM pause peptide, as do the insertion and deletion mutants (38). However, the uL4 and uL22 loop deletion mutants differ in their response to the crbcmlA pause signal. While the response in the uL22-Δloop cells is similar to the response of uL22 insertion and missense mutations, the uL4-Δloop mutant responds very differently compared to the uL4 loop-insertion and missense mutants. The latter mutations have no major impact on the reading of this pause signal. In contrast, the uL4 loop deletion mutant fails almost entirely to pause in response to the crbcmlA pausing peptide. Thus, the ability to pause during translation of the crbcmlA appears to depend on maintaining contacts that are preserved in the insertions and missense mutants, but removed in the deletion mutant.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Footnotes
Present addresses:
Marlon G. Lawrence, Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC 27599, USA.
Janice M. Zengel, Department of Biological Sciences, Carnegie Mellon University, Pittsburgh, PA 15213, USA.
FUNDING
National Science Foundation Grant [MCB0920578]. Funding for open access charge: Department of Biological Sciences, UMBC, Baltimore account number [1113-021-10423-CV388856-CNV].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ban N., Beckmann R., Cate J.H., Dinman J.D., Dragon F., Ellis S.R., Lafontaine D.L., Lindahl L., Liljas A., Lipton J.M., et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melnikov S., Ben-Shem A., Garreau de Loubresse N., Jenner L., Yusupova G., Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 3.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution [see comments] Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 4.Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 5.Dunkle J.A., Xiong L., Mankin A.S., Cate J.H. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 8.Klinge S., Voigts-Hoffmann F., Leibundgut M., Arpagaus S., Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 9.Khatter H., Myasnikov A.G., Natchiar S.K., Klaholz B.P. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- 10.Gorini L., Jacoby G.A., Breckenridge L. Ribosomal ambiguity. Cold Spring Harb. Symp. Quant. Biol. 1966;31:657–664. doi: 10.1101/sqb.1966.031.01.084. [DOI] [PubMed] [Google Scholar]
- 11.Bjorkman J., Samuelsson P., Andersson D.I., Hughes D. Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol. Microbiol. 1999;31:53–58. doi: 10.1046/j.1365-2958.1999.01142.x. [DOI] [PubMed] [Google Scholar]
- 12.Ozaki M., Mizushima S., Nomura M. Identification and functional characterization of the protein controlled by the streptomycin-resistant locus in E. coli. Nature. 1969;222:333–339. doi: 10.1038/222333a0. [DOI] [PubMed] [Google Scholar]
- 13.Birge E.A., Kurland C.G. Altered ribosomal protein in streptomycin-dependent Escherichia coli. Science. 1969;166:1282–1284. doi: 10.1126/science.166.3910.1282. [DOI] [PubMed] [Google Scholar]
- 14.Rosset R., Gorini L. A ribosomal ambiguity mutation. J. Mol. Biol. 1969;39:95–112. doi: 10.1016/0022-2836(69)90336-2. [DOI] [PubMed] [Google Scholar]
- 15.Guthrie C., Nashimoto H., Nomura M. Studies on the assembly of ribosomes in vivo. Cold Spring Harb. Symp. Quant. Biol. 1969;34:69–75. doi: 10.1101/sqb.1969.034.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Lund E., Dahlberg J.E., Lindahl L., Jaskunas S.R., Dennis P.P., Nomura M. Transfer RNA genes between 16S and 23S rRNA genes in rRNA transcription units of E. coli. Cell. 1976;7:165–177. doi: 10.1016/0092-8674(76)90016-7. [DOI] [PubMed] [Google Scholar]
- 17.Nomura M. Assembly of bacterial ribosomes. Science. 1973;179:864–873. doi: 10.1126/science.179.4076.864. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava A.K., Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Ann. Rev. Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- 19.Held W.A., Ballou B., Mizushima S., Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 20.Röhl R., Nierhaus K.H. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha T., Zhuang X., Kim H.D., Orr J.W., Williamson J.R., Chu S. Ligand-induced conformational changes observed in single RNA molecules. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9077–9082. doi: 10.1073/pnas.96.16.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H., Abeysirigunawarden S.C., Chen K., Mayerle M., Ragunathan K., Luthey-Schulten Z., Ha T., Woodson S.A. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014;506:334–338. doi: 10.1038/nature13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang M., Sullivan S.M., Wout P.K., Maddock J.R. G-protein control of the ribosome-associated stress response protein SpoT. J. Bacteriol. 2007;189:6140–6147. doi: 10.1128/JB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ofengand J., Del Campo M. Modified nucleosides of Escherichia coli Ribosomal RNA. EcoSal Plus. 2004;1 doi: 10.1128/ecosalplus.4.6.1. doi:10.1128/ecosalplus.4.6.1. [DOI] [PubMed] [Google Scholar]
- 25.Sharma S., Lafontaine D.L. ‘View From A Bridge': a new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015;40:560–575. doi: 10.1016/j.tibs.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Selmer M., Dunham C.M., Murphy F.V.t., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 27.Klinge S., Voigts-Hoffmann F., Leibundgut M., Ban N. Atomic structures of the eukaryotic ribosome. Trends Biochem. Sci. 2012;37:189–198. doi: 10.1016/j.tibs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Yusupova G., Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu. Rev. Biochem. 2014;83:467–486. doi: 10.1146/annurev-biochem-060713-035445. [DOI] [PubMed] [Google Scholar]
- 29.Brodersen D.E., Clemons W.M., Jr, Carter A.P., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 30.Klein D.J., Moore P.B., Steitz T.A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 31.Hansen J.L., Ippolito J.A., Ban N., Nissen P., Moore P.B., Steitz T.A. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 32.Tu D., Blaha G., Moore P.B., Steitz T.A. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Chittum H.S., Champney W.S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J. Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaman S., Fitzpatrick M., Lindahl L., Zengel J. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol. Microbiol. 2007;66:1039–1050. doi: 10.1111/j.1365-2958.2007.05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diner E.J., Hayes C.S. Recombineering reveals a diverse collection of ribosomal proteins L4 and L22 that confer resistance to macrolide antibiotics. J. Mol. Biol. 2009;386:300–315. doi: 10.1016/j.jmb.2008.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zengel J.M., Jerauld A., Walker A., Wahl M.C., Lindahl L. The extended loops of ribosomal proteins L4 and L22 are not required for ribosome assembly or L4-mediated autogenous control. RNA. 2003;9:1188–1197. doi: 10.1261/rna.5400703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gesteland R.F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J. Mol. Biol. 1966;16:67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- 38.Lawrence M.G., Lindahl L., Zengel J.M. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J. Bacteriol. 2008;190:5862–5869. doi: 10.1128/JB.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindahl L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J. Mol. Biol. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- 41.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coffman R.L., Norris T.E., Koch A.L. Chain elongation rate of messenger and polypeptides in slowly growing Escherichia coli. J. Mol. Biol. 1971;60:1–19. doi: 10.1016/0022-2836(71)90442-6. [DOI] [PubMed] [Google Scholar]
- 43.Chittum H.S., Champney W.S. Erythromycin inhibits the assembly of the large ribosomal subunit in growing Escherichia coli cells. Curr. Microbiol. 1995;30:273–279. doi: 10.1007/BF00295501. [DOI] [PubMed] [Google Scholar]
- 44.Shoji S., Dambacher C.M., Shajani Z., Williamson J.R., Schultz P.G. Systematic chromosomal deletion of bacterial ribosomal protein genes. J. Mol. Biol. 2011;413:751–761. doi: 10.1016/j.jmb.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gamalinda M., Woolford J.L., Jr Deletion of L4 domains reveals insights into the importance of ribosomal protein extensions in eukaryotic ribosome assembly. RNA. 2014;20:1725–1731. doi: 10.1261/rna.046649.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melnikov S., Ben-Shem A., Yusupova G., Yusupov M. Insights into the origin of the nuclear localization signals in conserved ribosomal proteins. Nat. Commun. 2015;6:7382. doi: 10.1038/ncomms8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mangiarotti G., Schlessinger D. Polyribosome metabolism in Escherichia coli. I. Extraction of polyribosomes and ribosomal subunits from fragile, growing Escherichia coli. J. Mol. Biol. 1966;20:123–143. doi: 10.1016/0022-2836(66)90122-7. [DOI] [PubMed] [Google Scholar]
- 48.Britton R.A., Wen T., Schaefer L., Pellegrini O., Uicker W.C., Mathy N., Tobin C., Daou R., Szyk J., Condon C. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 2007;63:127–138. doi: 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- 49.Strunk B.S., Novak M.N., Young C.L., Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worbs M., Huber R., Wahl M.C. Crystal structure of ribosomal protein L4 shows RNA-binding sites for ribosome incorporations and feedback control of the S10 operon. EMBO J. 2000;19:807–818. doi: 10.1093/emboj/19.5.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Worbs M., Wahl M.C., Lindahl L., Zengel J.M. Comparative anatomy of a regulatory ribosomal protein. Biochimie. 2002;84:731–743. doi: 10.1016/s0300-9084(02)01410-4. [DOI] [PubMed] [Google Scholar]
- 52.Zengel J.M., Lindahl L. Domain I of 23S rRNA competes with a paused transcription complex for ribosomal protein L4 of Escherichia coli. Nucl. Acids Res. 1993;21:2429–2435. doi: 10.1093/nar/21.10.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stelzl U., Nierhaus K.H. A short fragment of 23S rRNA containing the binding sites for two ribosomal proteins, L24 and L4, is a key element for rRNA folding during early assembly. RNA. 2001;7:598–609. doi: 10.1017/s1355838201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 55.Charollais J., Pflieger D., Vinh J., Dreyfus M., Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 56.Charollais J., Dreyfus M., Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trubetskoy D., Proux F., Allemand F., Dreyfus M., Iost I. SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo. Nucleic Acids Res. 2009;37:6540–6549. doi: 10.1093/nar/gkp685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davydova N.L., Gryaznova O.I., Mashchenko O.V., Vysotskaya V.S., Jonsson B.-H., Al-Karadaghi S., Liljas A., Garber M.B. Ribosomal protein L22 from Thermus thermophilus: Sequencing, overexpression and crystallisation. FEBS Lett. 1995;369:229–232. doi: 10.1016/0014-5793(95)00755-x. [DOI] [PubMed] [Google Scholar]
- 59.Timsit Y., Acosta Z., Allemand F., Chiaruttini C., Springer M. The role of disordered ribosomal protein extensions in the early steps of eubacterial 50 S ribosomal subunit assembly. Int. J. Mol. Sci. 2009;10:817–834. doi: 10.3390/ijms10030817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee D., Walsh J.D., Yu P., Markus M.A., Choli-Papadopoulou T., Schwieters C.D., Krueger S., Draper D.E., Wang Y.X. The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes. J. Mol. Biol. 2007;367:1007–1022. doi: 10.1016/j.jmb.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guillier M., Allemand F., Graffe M., Raibaud S., Dardel F., Springer M., Chiaruttini C. The N-terminal extension of Escherichia coli ribosomal protein L20 is important for ribosome assembly, but dispensable for translational feedback control. RNA. 2005;11:728–738. doi: 10.1261/rna.7134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calidas D., Lyon H., Culver G.M. The N-terminal extension of S12 influences small ribosomal subunit assembly in Escherichia coli. RNA. 2014;20:321–330. doi: 10.1261/rna.042432.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen S.S., Williamson J.R. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J. Mol. Biol. 2013;425:767–779. doi: 10.1016/j.jmb.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta N., Culver G.M. Multiple in vivo pathways for Escherichia coli small ribosomal subunit assembly occur on one pre-rRNA. Nat. Struct. Mol. Biol. 2014;21:937–943. doi: 10.1038/nsmb.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sykes M.T., Shajani Z., Sperling E., Beck A.H., Williamson J.R. Quantitative proteomic analysis of ribosome assembly and turnover in vivo. J. Mol. Biol. 2010;403:331–345. doi: 10.1016/j.jmb.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guthrie C., Nashimoto H., Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc. Natl. Acad. Sci. U.S.A. 1969;63:384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nashimoto H., Held W., Kaltschmidt E., Nomura M. Structure and function of bacterial ribosomes. XII. Accumulation of 21 s particles by some cold-sensitive mutants of Escherichia coli. J. Mol. Biol. 1971;62:121–138. doi: 10.1016/0022-2836(71)90135-5. [DOI] [PubMed] [Google Scholar]
- 68.Bryant R.E., Sypherd P.S. Genetic analysis of cold-sensitive ribosome maturation mutants of Escherichia coli. J. Bacteriol. 1974;117:1082–1092. doi: 10.1128/jb.117.3.1082-1092.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ripmaster T.L., Vaughn G.P., Woolford J.L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl. Acad. Sci. U.S.A. 1992;89:11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dammel C.S., Noller H.F. A cold-sensitive mtuation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- 71.Sharpe Elles L.M., Sykes M.T., Williamson J.R., Uhlenbeck O.C. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 2009;37:6503–6514. doi: 10.1093/nar/gkp711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roy-Chaudhuri B., Kirthi N., Culver G.M. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4567–4572. doi: 10.1073/pnas.0912305107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clatterbuck Soper S.F., Dator R.P., Limbach P.A., Woodson S.A. In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol. Cell. 2013;52:506–516. doi: 10.1016/j.molcel.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alix J.H., Nierhaus K.H. DnaK-facilitated ribosome assembly in Escherichia coli revisited. RNA. 2003;9:787–793. doi: 10.1261/rna.5360203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dodd J., Kolb J.M., Nomura M. Lack of complete cooperativity of ribosome assembly in vitro and its possible relevance to in vivo ribosome assembly and the regulation of ribosomal gene expression. Biochimie. 1991;73:757–767. doi: 10.1016/0300-9084(91)90055-6. [DOI] [PubMed] [Google Scholar]
- 76.Gabashvili I.S., Gregory S.T., Valle M., Grassucci R., Worbs M., Wahl M.C., Dahlberg A.E., Frank J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- 77.Ramu H., Vazquez-Laslop N., Klepacki D., Dai Q., Piccirilli J., Micura R., Mankin A.S. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol. Cell. 2011;41:321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.