Abstract
Floral thermogenesis has been found in dozens of primitive seed plants and the reproductive organs in these plants produce heat during anthesis. Thus, characterization of the molecular mechanisms underlying flowering is required to fully understand the role of thermogenesis, but this aspect of thermogenic plant development is largely unknown. In this study, extensive database searches and cloning experiments suggest that thermogenic skunk cabbage (Symplocarpus renifolius), which is a member of the family Araceae, possesses two genes encoding phosphatidyl ethanolamine-binding proteins (PEBP), FLOWERING LOCUS T (SrFT) and MOTHER OF FT AND TFL1 (SrMFT). Functional analyses of SrFT and SrMFT in Arabidopsis indicate that SrFT promotes flowering, whereas SrMFT does not. In S. renifolius, the stage- and tissue-specific expression of SrFT was more evident than that of SrMFT. SrFT was highly expressed in flowers and leaves and was mainly localized in fibrovascular tissues. In addition, microarray analysis revealed that, within floral tissues, SrFT was co-regulated with the genes associated with cellular respiration and mitochondrial function, including ALTERNATIVE OXIDASE gene proposed to play a major role in floral thermogenesis. Taken together, these data suggest that, among the PEBP genes, SrFT plays a role in flowering and floral development in the thermogenic skunk cabbage.
Floral thermogenesis has been found in dozens of flowering plants including basal angiosperms (Nymphaeaceae and Schisandraceae)1,2,3,4, magnoliids (Magnoliaceae)5,6,7, monocots (Araceae)8,9,10, and eudicots (Nelumbonaceae and Rafflesiaceae)10,11. According to the updated Angiosperm Phylogeny Group Classification (APG III)12, basal angiosperms are the most primitive group of angiosperms, and the magnoliids diverged early. In addition, the Araceae and Nelumbonaceae are early diverging monocots and eudicots, respectively. Heat production is not restricted to angiosperms but also occurs in several families of gymnosperms (i.e., Cycadaceae and Zamiaceae)10,13,14, which are more primitive than angiosperms among seed plants. Thus, it is reasonable to suggest that plant species that are able to produce heat in their reproductive organs belong to relatively primitive plant families. These plants differ from well-studied annual herbaceous plants, in aspects of growth and development, in that they flower after several years of vegetative growth and exhibit thermogenesis when they bloom. Thus, it is of interest to determine the mechanisms by which thermogenic plants initiate flowering, progress through floral development, and begin thermogenesis.
Extensive studies on the roles of floral thermogenesis in physiology, ecology and pollination biology have suggested that thermogenesis may be related to reproductive functions such as attracting pollinators by spreading odor15, providing a warm environment for pollinators16, and protecting the inflorescences from freezing damage17. In addition, extensive studies using arum species have revealed that thermogenesis is positively correlated with oxygen consumption rate10,18. However, the physiological roles of floral thermogenesis and the causal relationship between thermogenesis and respiration have not been completely established due to the lack of molecular genetic studies on thermogenic arum species and techniques to control their flowering.
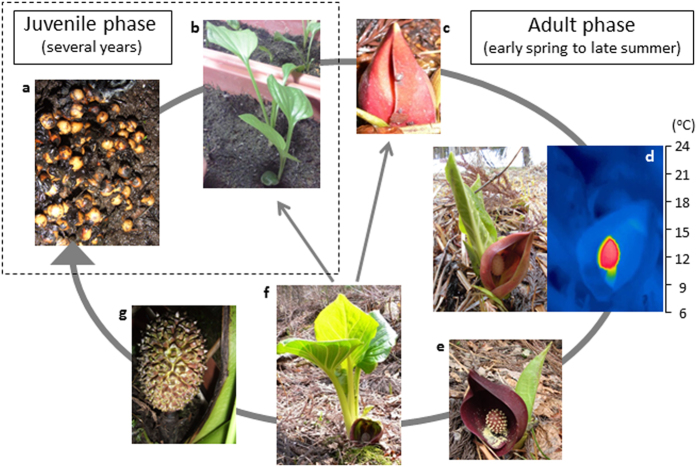
Thermogenic skunk cabbage (Symplocarpus renifolius), which is a member of the family Araceae, is a thermoregulatory plant that can maintain spadix temperatures at approximately 20 °C for 1–2 weeks in early spring, when the ambient temperature falls below freezing19. S. renifolius is a polycarpic plant that flowers after several years of vegetative growth (Fig. 1a,b). The inflorescences begin heat production when they bloom (Fig. 1d), and terminate it when the pollen is released from the anthers (Fig. 1e). Interestingly, floral development precedes leaf development in the adult phase (Fig. 1c–f). In S. renifolius, the developmental features of the flowers and leaves generally contrast those of well-studied plants, in which flowering and floral development occur after leaf development. During the floral development from the thermogenic stage (Fig. 1d) to the post-thermogenic stage (Fig. 1e), intracellular structures within the floral tissues change significantly; the mitochondrial content is reduced, especially in the petals and pistils, whereas the vacuolar volume increased20,21. Consistent with the cellular change, the genes involved in cellular respiration and mitochondrial function are up-regulated in the former thermogenic spadices, whereas the genes involved in stress responses and protein degradation are up-regulated in the latter post-thermogenic spadices21,22. To uncover the molecular mechanisms underlying floral thermogenesis, much effort has been made to characterize an alternative oxidase (AOX), an energy-dissipating mitochondrial protein, and AOX has been proposed to play a pivotal role due to the correlation between heat production and AOX concentration as well as activity23,24,25,26,27.
Figure 1. The life cycle of skunk cabbage (Symplocarpus renifolius).
S. renifolius is a polycarpic plant that flowers after several years of vegetative growth. In early spring, the flowers begin producing heat when they bloom and terminate heat production when the pollen is released from the anthers. At the end of spring, leaf development is initiated at floral senescence, and environmental conditions determine whether or not flowers will bloom in the following year. In summer, if pollination is successful, the flower becomes a fruit, which contains many seeds. (a) Seeds, (b) seedlings, (c) immature-stage flowers (pre-thermogenic stage), (d) female-stage flowers (thermogenic stage), (e) male-stage flowers (post-thermogenic stage), (f) developed leaves, and (g) fruits. The thermal image of the right panel in (d) was taken using an FLIR SC 620 thermal imager (FLIR SYSTEMS).
Plant phosphatidyl ethanolamine-binding proteins (PEBP) family is divided into three main clades, FLOWERING LOCUS T (FT)-like, TERMINAL FLOWER1 (TFL1)-like, and MOTHER OF FT AND TFL1 (MFT) -like28. These members have been extensively studied in Arabidopsis, especially FT and TFL1. Despite an amino acid similarity of over 88%, these two proteins have antagonistic functions; FT promotes flowering by mediating both photoperiod and temperature signals, whereas TFL1 represses flowering29,30,31. The remaining MFT-like clade is generally thought to be the evolutionary ancestor to the other two clades32,33. A first duplication event resulted in two families of plant PEBP genes (MFT-like and FT/TFL1-like), and the second duplication resulted in the FT-like and TFL1-like clades. Thus, the MFT-like clade contains not only angiosperms but also more primitive plants, including mosses, spikemosses and gymnosperms, whereas the FT/TFL1-like clade contains only angiosperms. Given that the evolution of plant PEBP genes seems to coincide with the evolution of seed plants, it is of interest to determine which PEBP genes exist in skunk cabbage since it is presumably one of the earliest diverging monocots among angiosperms.
Recent molecular studies have shown that, in several species, FT and its orthologs that are translated in the leaves act as florigens34,35,36. In Arabidopsis, FT moves to the shoot apical meristem (SAM) via the phloem and, once there, forms a transcriptional complex with a basic leucine zipper (bZIP) transcription factor, FD. Transcription of floral regulator genes, such as APETALA1 (AP1), is then activated, leading to flowering37. Thus, as the major function, FT-like genes have been proposed to control the switch from the vegetative phase to the reproductive phase. However, recent functional studies have demonstrated more divergent roles for FT-like genes. In sugar beet, for example, two orthologous FT genes, BvFT1 and BvFT2, exhibit antagonistic functions; BvFT2 is essential for flowering, whereas BvFT1 represses flowering, in both transgenic sugar beet and Arabidopsis plants38. In potato, two orthologous FT genes, SP3D and SP6A, have different functions, as well; SP3D promotes flowering, but SP6A promotes tuberization39. Furthermore, in Arabidopsis, AtFT is expressed in guard cells and regulates stomatal opening40. Therefore, FT seems to be involved in various other processes, in addition to flowering.
In this study, we characterized the expression and function of two PEBP genes, FT and MFT, designated as SrFT and SrMFT, from thermogenic skunk cabbage. Our study suggests that SrFT may play a principal role in the regulation of flowering in thermogenic skunk cabbage, and the expression of SrFT mRNA in floral tissues may have additional roles in processes other than flowering, through the complicated gene network associated with energy metabolism.
Results
Isolation and identification of SrFT and SrMFT cDNA from S. renifolius
An S. renifolius cDNA fragment library was searched to identify the DNA sequences of orthologous PEBP genes, including FT-like, TFL1-like, and MFT-like. Two PEBP genes, FT and MFT, were found in the cDNA fragment library, but TFL1 homologs and other FT/MFT isoforms could not be found. To isolate SrFT and SrMFT genes from the tissues of S. renifolius, cloning primers were designed based on the SrFT and SrMFT sequences, and RT-PCR was performed. The amplified 600-bp region of SrFT cDNA contained a 525-bp protein coding sequence (LC030437) flanked by a 21-bp 5′-untranslated region and a 54-bp 3′-untranslated region. The predicted SrFT protein consisted of 174 amino acid residues and had a calculated molecular mass of 19.6 kDa. In contrast, the amplified 700-bp region of SrMFT cDNA contained a 639-bp protein coding sequence (LC030438) flanked by a 33-bp 5′-untranslated region and a 28-bp 3′-untranslated region. The predicted SrMFT protein consisted of 212 amino acid residues with a calculated molecular mass of 22.9 kDa. To isolate TFL1-like genes from S. renifolius, RT-PCR was performed using the degenerate primers (Supplementary Figs 1a,c, and 2); however, no fragments were amplified. The amplification of other PEBP genes using the other sets of degenerate primers were also unsuccessful (Supplementary Fig. 1b).
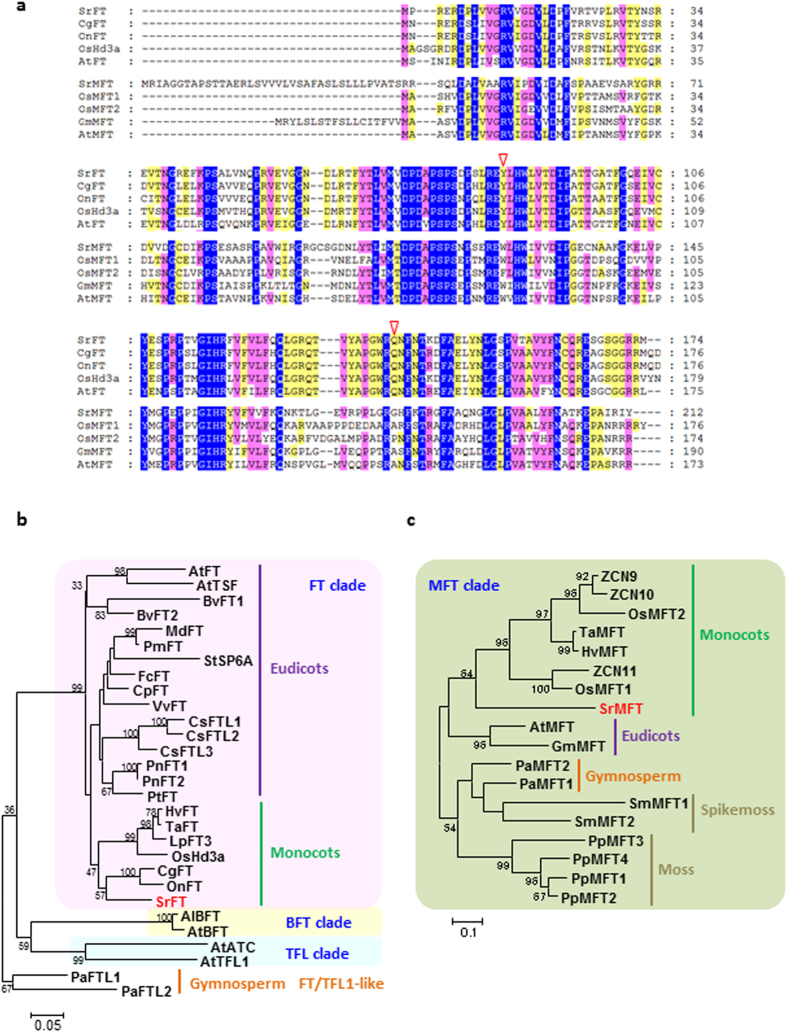
The SrFT showed significant homologies to Cymbidium and Oncidium FT proteins (87% and 85%, respectively; Fig. 2a). SrFT was also highly homologous to rice and Arabidopsis FT proteins (83% and 76%, respectively). In addition, the conserved key amino acid residues Tyr and Gln in FT homologs, which contribute to FT function, were identified at positions 84 and 139 of the SrFT protein, respectively (Fig. 2a). In Arabidopsis, mutational and structural analyses of chimeric proteins revealed that two amino acid residues are possibly the most critical for distinguishing FT and TFL1 activity29,30,31. However, the key amino acids were not conserved in SrMFT, leading to the hypothesis that SrMFT does not play a central role in the regulation of flowering. The SrMFT sequence exhibited 57% homologies to the Glycine max MFT protein (GmMFT) and contained an amino acid extension at its N-terminus that was similar to GmMFT (Fig. 2a). The sequence was also orthologous to rice and Arabidopsis MFT proteins (54% and 55%, respectively).
Figure 2. Sequence analysis of SrFT, SrMFT, and related proteins.
(a) Alignment of the predicted amino acid sequences of SrFT, SrMFT, and related proteins. The predicted SrFT sequence was aligned with CgFT (Cymbidium goeringii), OnFT (Oncidium Gower Ramsey), OsHd3a (rice), and AtFT (Arabidopsis). The SrMFT sequence was aligned with OsMFT1 and 2 (rice), GmMFT (Glycine max), and AtMFT (Arabidopsis) sequences. Amino acids highlighted in blue, pink, or yellow are conserved across all, more than eighty percent, or more than half of the protein sequences in the alignment, respectively. Arrowheads indicate amino acids (Y84 and Q139) that are critical to FT function. (b) A phylogenetic tree of the predicted amino acid sequences of SrFT and related proteins. (c) A phylogenetic tree of the predicted amino acid sequences of SrMFT and related proteins. The tree was created using MEGA 6.06 with the Clustal W and maximum likelihood method. Bootstrap values above 30 are shown near each branch. The consensus phylogenic trees are shown with bootstrap values from 1000 replications. The accession numbers, locus IDs, and species abbreviations are listed in Supplementary Table 2.
A phylogenetic tree for the PEBP genes was constructed using several amino acid sequences of other FT/TFL orthologs (Fig. 2b), and other MFT orthologs (Fig. 2c). The tree was divided into four major clades: FT, TFL, BFT (BROTHER OF FT AND TFL1), and MFT clades. The SrFT protein was located in the monocot group of the FT clade and was also located close to the FT proteins from orchids. Although FT genes have been isolated from numerous plant species, SrFT is the first to be isolated from a primitive flowering plant (i.e., S. renifolius). According to the updated APG III12, the Araceae, which includes S. renifolius, seems to be more ancient than other plant families that are included in the monocot group of FT sequence. The SrMFT protein was also located in the monocot group of the MFT clade but was also located close to the MFT proteins from grasses.
Expression analyses of SrFT and SrMFT mRNA in S. renifolius
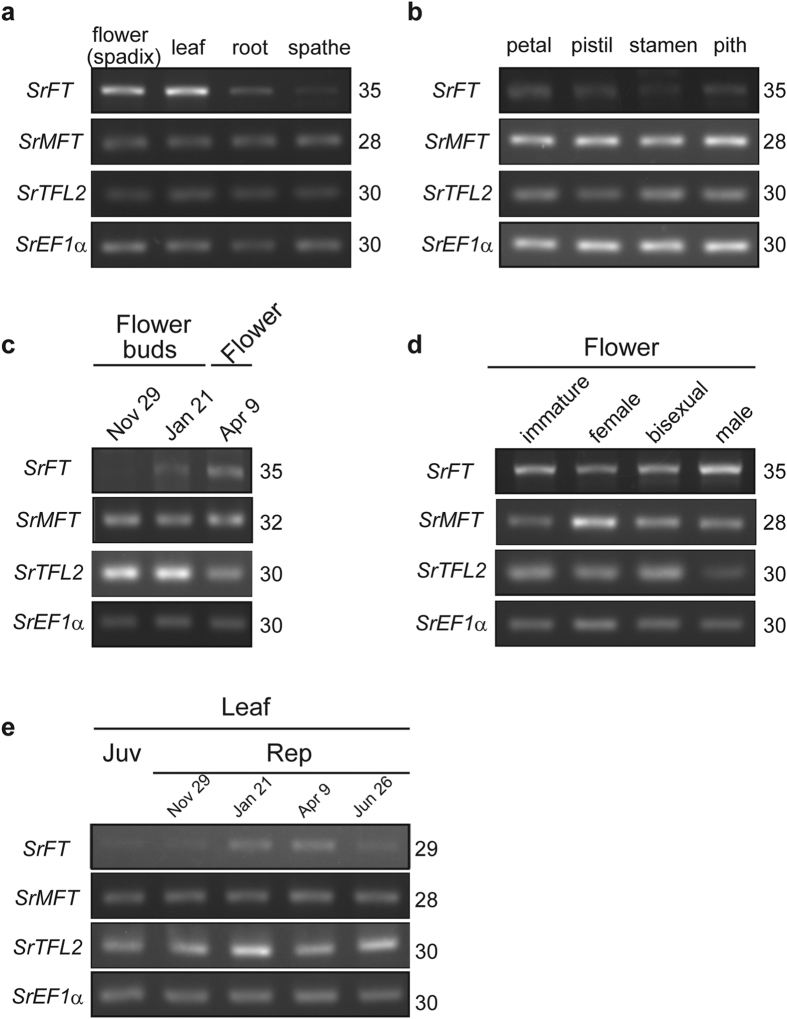
To investigate the distribution of SrFT mRNA, semi-quantitative RT-PCR was performed using various tissues of S. renifolius at the thermogenic female stage. SrFT mRNA was highly expressed in flowers and leaves but not in roots or spathes (Fig. 3a). To further examine the expression profile of SrFT mRNA in the flower, semi-quantitative RT-PCR was also performed using different floral tissues, including petals, pistils, stamens and the pith of female-stage flowers (Fig. 3b). SrFT was expressed in all floral tissues, but the expression was slightly lower in stamens and pith than in petals and pistils.
Figure 3. The tissue- or stage-specific expression of two PEBP genes, SrFT and SrMFT.
(a) Tissue-specific expression in plant tissues. (b) Tissue-specific expression in floral tissues. (c) Stage-specific expression in flowers during the transition from floral buds to mature flowers. (d) Stage-specific expression in flowers at the late stage of floral maturation, including immature (pre-thermogenesis), female (strong thermogenesis), bisexual (weak thermogenesis), and male (post-thermogenesis) stages. (e) Stage-specific expression in leaves of juvenile (Juv)- and reproductive (Rep)-phase plants. To prevent saturation, semi-quantitative RT-PCR was analysed with different PCR cycles (Supplementary Fig. 7) and the appropriate numbers of PCR cycles were shown on the right side of each gel. The signals detected here were not from contaminated genomic DNA (Supplementary Fig. 8). In (c,e), potted plants were used to collect samples (Supplementary Fig. 9).
Next, the stage-specific expression of SrFT mRNA in flowers was examined using the spadices of different developmental stages in S. renifolius (Fig. 3c,d). During the former stage of floral maturation, levels of SrFT mRNA increased and reached a peak in mature flowers (Fig. 3c), and then, during the late stages of floral maturation, SrFT levels remained high (Fig. 3d). We also examined the seasonal changes in SrFT mRNA in leaves, using semi-quantitative RT-PCR (Fig. 3e). We found that SrFT mRNA was expressed in all leaves examined here, and was also more highly expressed during the reproductive stage in winter and spring than in early summer.
In contrast to SrFT, SrMFT transcripts were constitutively expressed in various tissues and developmental stages, except that the level of SrMFT was higher in female-stage spadices than in the other stages investigated. We also investigated the expression of the TFL2-like gene, designated as SrTFL2. SrTFL2 contained highly conserved regions among Arabidopsis and apple TFL2 proteins (Supplementary Fig. 3). Although TFL2 does not belong to the PEBP family, the protein is active in the regulation of flowering in both Arabidopsis41,42 and apple43. SrTFL2 transcripts were also constitutively expressed in various tissues and developmental stages, but interestingly, during the former stage of floral maturation, SrTFL2 transcripts decreased contrary to the increase of SrFT mRNA levels (Fig. 3c). Therefore, instead of TFL1-like genes, SrTFL2 may function in S. renifolius.
In situ localization of SrFT mRNA in S. renifolius
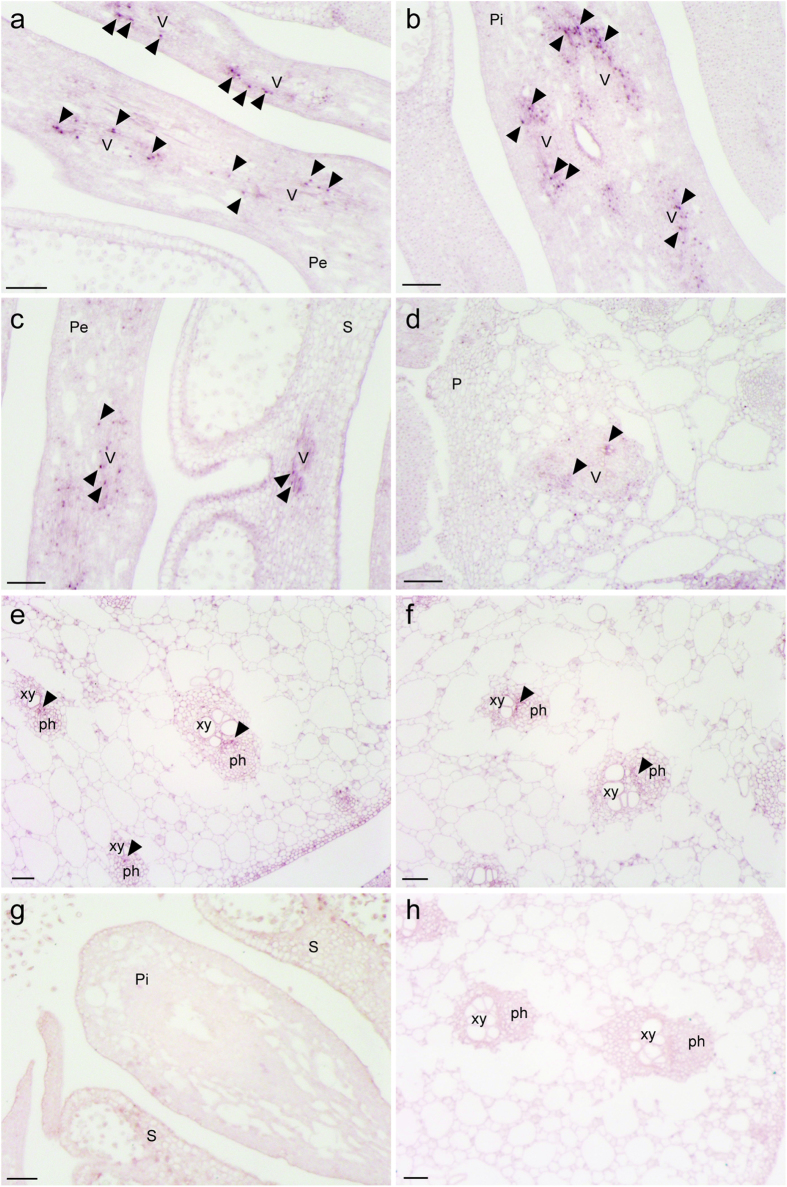
In Arabidopsis and rice, FT mRNA is localized in cotyledons but not in meristems35,44, but there have been very few studies of the in situ localization of FT mRNA in mature floral organs. Thus, we carried out in situ hybridization using an antisense probe to examine the tissue-specific localization of SrFT mRNA in detail. In floral tissues (Fig. 4a–d), the signals derived from SrFT mRNA were mainly observed in the vascular bundles in petals (Fig. 4a), pistils (Fig. 4b), stamens (Fig. 4c), and the outer layer of the pith (Fig. 4d). In leaf tissues (Fig. 4e,f), SrFT mRNA was also mainly localized in vascular bundles. A sense probe used as the control did not produce any signals in flowers (Fig. 4g) and leaves (Fig. 4h). Together, these results indicated that SrFT mRNA was localized mainly in the vascular tissues of flowers and leaves.
Figure 4. Localization of SrFT mRNA in flowers and leaves.
DIG-labeled antisense RNA probes were hybridized on cross-sections of flowers (a–d) and leaves (e,f). As the control, sense RNA probes were hybridized on cross-sections of flowers (g) and leaves (h). Arrowheads indicate the locations of signals. Pe, petal; Pi, pistil; P, pith; S, stamen; V, vascular bundle; xy, xylem; ph, phloem. Bar =100 μm.
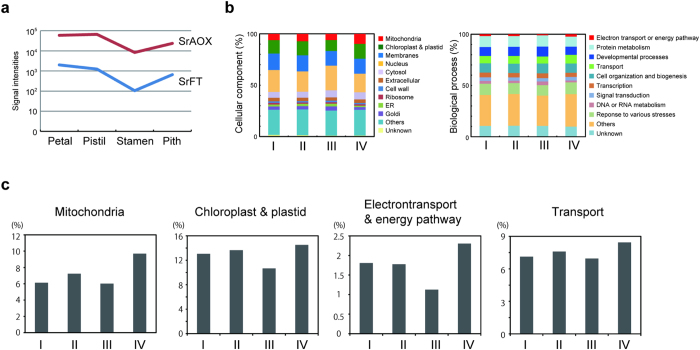
Microarray analysis of SrFT and other co-regulated genes
To examine the expression profiles of SrFT and co-regulated genes in mature floral tissues, microarray analysis was carried out using labeled cRNA that had been prepared from different floral tissues (Fig. 5). Consistent with RT-PCR analyses (Fig. 3b), the expression of SrFT mRNA was found to be higher in petals and pistils than in stamens and pith. Interestingly, we observed that the tissue-specific expression of SrFT was quite similar to that of SrAOX (Fig. 5a). The microarray result of SrAOX mRNA was validated with RT-PCR (Supplementary Fig. 7b at the bottom). Thus, to identify additional co-regulated genes with these two genes, we performed cluster analysis using a Poisson approach. As a result, 6523, 7259, 7867, and 5535 genes were grouped into clusters I, II, III, and IV (Supplementary Fig. 4). Two genes, SrFT and SrAOX, were classified into cluster IV along with the other co-regulated genes (Supplementary Table 4).
Figure 5. Microarray analysis of four different floral tissues.
(a) SrFT and SrAOX, representative genes for flowering and respiration/thermogenesis, respectively, had similar expression profiles among the different floral tissues. (b) 27,184 genes were classified into four clusters (I-IV) according to their expression profiles in the different floral tissues. Of the resulting 6523, 7259, 7867, and 5535 genes in clusters I, II, III and IV, respectively, 2052, 1981, 2438, and 1808 were annotated with AGI codes, respectively. The annotated genes were classified based on the GO annotation that assigns cellular components and biological processes to each sequence. (c) Categories that had the highest percentage representation in cluster IV of all clusters were extracted. Most notably, many genes involved in mitochondrial function and in electron transport and the energy pathway were found in cluster IV.
To further assess the major function of these co-regulated genes in the cellular processes associated with thermogenesis in S. renifolius, 8279 genes that could be annotated with the Arabidopsis Gene Identifier (AGI) codes were analyzed using Gene Ontology analysis (Fig. 5b,c). Interestingly, the percentage of genes encoding mitochondrial proteins in cluster IV was the highest among the four clusters, and the percentage of genes associated with electron transport or the energy pathway in cluster IV was also the highest among the four clusters (Fig. 5c and Supplementary Tables 5 and 6). Among the four clusters, transcripts that encoded chloroplast and plastid proteins as well as transporters were also overrepresented in cluster IV (Fig. 5c). These data suggest that the expression of SrFT is co-regulated with other genes associated with energy metabolism.
Analysis of Arabidopsis transformants expressing SrFT or SrMFT
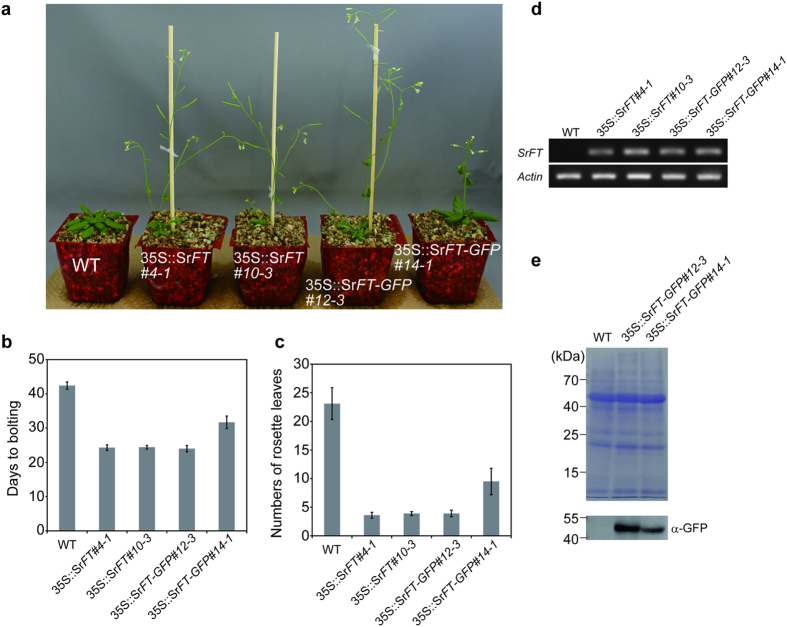
FT genes isolated from many plants exhibit an FT-like function in Arabidopsis; however, Picea abies, which is a primitive seed plant, has PEBP genes that occupy an intermediate phylogenetic position between the FT-like and TFL1-like (FT/TFL1-like) genes and exhibit a TFL1-like function32. Thus, it is of interest to determine whether the FT genes of skunk cabbage, an ancestral flowering plant, encode an activity that promotes flowering. To examine SrFT function, SrFT and SrFT-GFP cDNA constructs driven by the CaMV 35S promoter were individually transformed into Arabidopsis plants. In the transgenic lines, the 35S::SrFT (#4-1 and #10-3) and 35S::SrFT-GFP (#12-3) constructs caused very early flowering and the 35S::SrFT-GFP (#14-1) constructs caused modestly early flowering, compared with the empty vector (Fig. 6a).
Figure 6. Analysis of transgenic Arabidopsis plants that ectopically expressed SrFT or SrFT-GFP.
(a) Phenotypes of 33-day-old wild type (empty vector), 35S::SrFT, and 35S::SrFT-GFP transgenic plants grown in soil under LD conditions. (b,c) Flowering response of wild type, 35S::SrFT, and 35S::SrFT-GFP transgenic plants in soil under LD conditions. Data are means ± standard deviation (SD; N = 10). (d) SrFT mRNA expressed in rosette leaves of 33-day-old wild type, 35S::SrFT, and 35S::SrFT-GFP transgenic plants were analyzed using RT-PCR. Actin is shown as a loading control. (e) Rosette leaves of 33-day-old wild type and 35S::SrFT-GFP transgenic plants were analyzed using SDS-PAGE and then visualized using CBB-staining or immunoblotting with α-GFP antibodies.
To assess the flowering response quantitatively, the number of days required for the main shoot to attain a length of 1 cm (days to bolting) and the number of rosette leaves observed on that day were measured for ten plants from each transgenic line (Fig. 6bc, and Table 1). In agreement with Fig. 6a, three transgenic lines (35S::SrFT #4-1, and #10-3, and SrFT-GFP #12-3) exhibited a very early flowering phenotype, and another line (35S::SrFT-GFP #14-1) exhibited a modestly early flowering phenotype, compared to that of the wild type.
Table 1. Measurements of flowering time in transgenic Arabidopsis plants overexpressing SrFT, SrFT-GFP, SrMFT, and SrMFT-GFP.
| Genotype | Nos of plantsa | Days to boltingb | Numbers of rosette leavesc |
|---|---|---|---|
| WT 1-6 | 10 | 42.40 ± 1.08 | 23.10 ± 2.77 |
| SrFT 4-1 | 10 | 24.30 ± 0.82 | 3.60 ± 0.52 |
| SrFT 10-3 | 10 | 24.40 ± 0.52 | 3.90 ± 0.32 |
| SrFT-GFP 12-3 | 10 | 24.00 ± 0.94 | 3.90 ± 0.57 |
| SrFT-GFP 14-1 | 10 | 31.70 ± 1.83 | 9.50 ± 2.32 |
| SrMFT 6-8 | 10 | 45.10 ± 2.69 | 23.5 ± 2.87 |
| SrMFT 14-3 | 10 | 40.50 ± 1.58 | 20.2 ± 2.78 |
| SrMFT-GFP 4-6 | 10 | 40.30 ± 2.60 | 17.3 ± 1.41 |
| SrMFT-GFP 16-10 | 10 | 43.30 ± 2.54 | 21.4 ± 2.55 |
aPlants were grown on the potted soil at 22 °C under the day length conditions at 16 h of light/8 h of darkness (long-day conditions). Data are means ± standard deviation (SD; N = 10).
bThe number of days required for the main shoot to attain a length of 1 cm.
cThe number of rosette leaves observed on the day when plants start bolting.
To explore the correlation of the early flowering phenotype with SrFT expression in the 35S::SrFT and SrFT-GFP transgenic lines, RT-PCR was performed. As shown in Fig. 6d, the expression of SrFT mRNA was observed in the transgenic lines, but not in wild type. To analyze the expression of SrFT protein, SrFT-GFP proteins expressed in rosette leaves of two SrFT-GFP transgenic lines (#12-3 and #14-1) were detected using immunoblotting with anti-GFP antibodies (Fig. 6e). The levels of SrFT-GFP protein were higher in #12-3 than #14-1. This result suggests that the very early flowering phenotype observed in 35S::SrFT-GFP (#12-3) is due to higher SrFT protein expression.
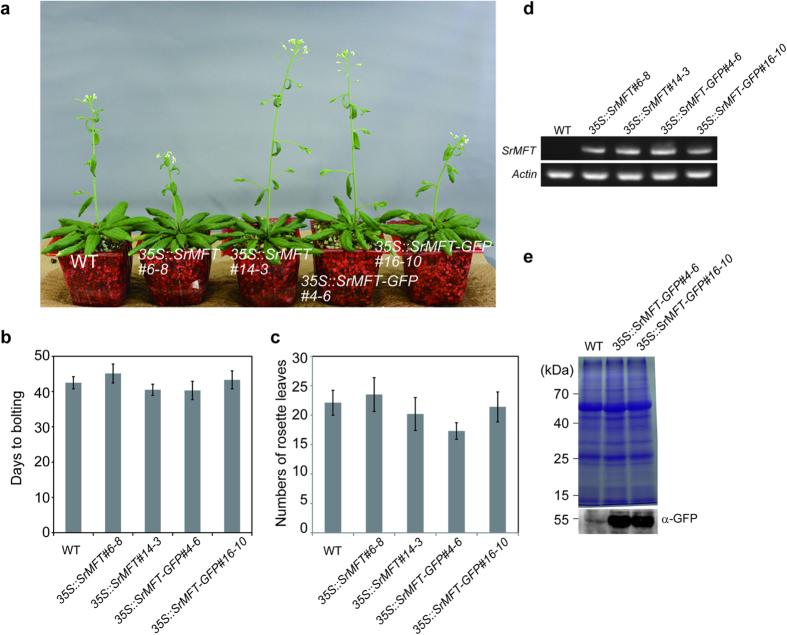
We also measured the days to bolting and the numbers of rosette leaves observed on that day using 35S::SrMFT and 35S::SrMFT-GFP lines (Fig. 7 and Table 1). The results showed that SrMFT did not promote flowering in Arabidopsis.
Figure 7. Analysis of transgenic Arabidopsis plants that ectopically expressed SrMFT or SrMFT-GFP.
(a) Phenotypes of 46-day-old wild type (empty vector), 35S::SrMFT, and 35S::SrMFT-GFP transgenic plants grown in soil under LD conditions. (b,c) Flowering response of wild type, 35S::SrMFT, and 35S::SrMFT-GFP transgenic plants in soil under LD conditions. Data are means ± standard deviation (SD; N = 10). (d) SrMFT mRNA expressed in rosette leaves of 29-day-old wild type, 35S::SrMFT, and 35S::SrMFT-GFP transgenic plants were analyzed using RT-PCR. Actin is shown as a loading control. (e) Rosette leaves of 29-day-old wild type and 35S::SrMFT-GFP transgenic plants were analyzed using SDS-PAGE and then visualized using CBB-staining or immunoblotting with α-GFP antibodies.
Taken together, these data clearly show that SrFT proteins promote flowering and that elevated levels of SrFT protein cause earlier flowering in Arabidopsis.
Discussion
In this study, we isolated two PEBP genes, SrFT and SrMFT, from thermogenic skunk cabbage (S. renifolius). Although skunk cabbage is a primitive seed plant, the primary structure of the proteins encoded by the two genes was similar to those found in other plants. SrFT mRNA was highly expressed in both flowers and leaves, and its level in flowers significantly increased during floral maturation. In contrast, SrMFT mRNA was constitutively expressed in various tissues and stages. Interestingly, within floral tissues, SrFT and several genes related to mitochondrial function and cellular respiration, including SrAOX, had similar expression profiles. The expression of SrFT in Arabidopsis resulted in an early flowering phenotype, indicating that SrFT promotes flowering, whereas the expression of SrMFT in Arabidopsis failed to promote flowering. Taken together, these data suggest that, among PEBP genes, SrFT plays a role in the regulation of flowering and floral development in the thermogenic skunk cabbage, through the complicated gene network associated with energy metabolism.
S. renifolius is a polycarpic plant that flowers after several years of vegetative growth (Fig. 1). Once the plant flowers, it can bloom almost every year under optimal environmental conditions, and one of the most interesting features of S. renifolius is that floral development precedes leaf development in the adult phase (Fig. 1c–f). In S. renifolius, the flower blooms in early spring, whereas leaf development begins at the end of spring. The developmental features in the flowers and leaves contrast those of well-studied plants, in which flowering and floral development occurs after leaf development. In the present study, the expression of SrFT was high in both flowers and leaves; therefore, we hypothesize that SrFT expressed in each organ has distinct functions. In leaves, the SrFT protein is likely to function as a florigen, and we speculate that SrFT is transported to the SAM, where it stimulates the formation of flower buds that will mature in the following season. However, in flowers, SrFT may regulate floral or fruit maturation. This proposal is consistent with the observation that the level of SrFT mRNA increased during the maturation of floral buds (Fig. 3c). Highly expressed FT genes in floral tissues were observed not only in skunk cabbage (Figs 3 and 4) but also in Arabidopsis (Supplementary Fig. 6), and the levels of FT mRNA increased significantly from flower buds to mature flowers in both plants. Thus, highly expressed FT genes in these tissues may have additional roles in floral development in various plants. In addition, although it would be of interest to study the levels and the localization of SrFT mRNA during the transition from the vegetative stage to the reproductive stage, we did not undertake such a study because of the difficulty in culturing plants in laboratory conditions during this transition.
The plant PEBP gene family is divided into three main clades: FT-like, TFL1-like, and MFT-like. Among PEBP genes, we identified SrFT and SrMFT as the major skunk cabbage genes of the FT-like and MFT-like subfamilies, respectively. However, there were no fragments of TFL1-like genes in our skunk cabbage cDNA database, and we could not amplify any TFL1-like fragments using RT-PCR and degenerate primers (Supplementary Fig. 1). Interestingly, gymnosperms possess only two types of PEBP genes, MFT-like and a group that occupies an intermediate phylogenetic position between the FT-like and TFL1-like (FT/TFL1-like)32. It is, therefore, of great interest to determine whether there are two or three types of PEBP genes in skunk cabbage. However, the complete sequence of the skunk cabbage genome will be needed to determine whether TFL1-like genes exist or not. Interestingly, in the skunk cabbage inflorescences, the levels of SrFT mRNA increased during floral maturation, whereas the levels of SrTFL2 mRNA decreased (Fig. 3c). Since TFL2, which does not belong to the PEBP family, functions in the repression of flowering in Arabidopsis41,42 and apple43, TFL2, instead of TFL1, may regulate flowering and floral development together with SrFT in thermogenic skunk cabbage.
In Arabidopsis and rice, FT/Hd3a expression is much higher in leaves than in the shoot apex, and is observed in phloem tissues of both cotyledons and leaves35,44,45. Based on the expression studies and analyses of transgenic Arabidopsis/rice expressing GFP-tagged FT/Hd3a, it became clear that FT/Hd3a proteins are produced in the leaves and subsequently move to the shoot apex, where they promote flowering34,35. Interestingly, the number of studies describing FT transcripts in tissues other than leaves has increased recently. For example, in grapes, VvFT is highly expressed in inflorescences and developing fruit and is especially prominent in developing seeds46, and in poplar trees, PnFT1 and PnFT2 are highly expressed in capsules47. In addition, the expression levels of OnFT and CgFT in orchids are significantly higher in young flower buds and decrease during floral maturation48,49. Although OnFT and CgFT exhibited the highest homologies to SrFT, their expression patterns were in marked contrast to those of skunk cabbage. SrFT expression in skunk cabbage increased during floral maturation (Fig. 3c), whereas FT expression in orchids decreased. In addition, two potato FT genes, SP3D and SP6A, also exhibit different functions; SP3D promotes flowering, whereas SP6A promotes tuberisation39, and accumulates to high levels in stolons and leaves of short-day induced plants. These observations suggest that FT functions may be more diverse than previously thought and that other functions may be found with future investigation.
Extensive research on the expression and activity of AOX has shown that the protein plays a pivotal role in floral thermogenesis23,24,25,26,27. In contrast, several studies have suggested that uncoupling protein (UCP), which plays a major role in mammalian thermogenesis, likely plays a minor role in plant thermogenesis23,25. Interestingly, we found that SrAOX transcripts were co-regulated with SrFT transcripts within floral tissues (Fig. 5a). Classification analysis based on gene expression profiles also revealed that SrAOX and SrFT were classified into cluster IV (Supplementary Table 4). In addition, SrMFT was also co-regulated with SrAOX and SrFT and classified into cluster IV; however, SrUCPA was not co-regulated with these genes and was classified into cluster II (Supplementary Table 4 and Supplementary Fig. 5). In previous studies, genes involved in cellular respiration and mitochondrial function were significantly enhanced during the female stage of skunk cabbage floral development21,22. Of these genes, several were classified into cluster IV in this study, and were co-regulated with SrFT and SrAOX (Supplementary Fig. 5). Thus, defining the mechanisms that underlie the co-regulation of the genes involved in flowering and respiration in floral tissues will be a future research objective.
Photoperiodism in plants from the Araceae family, including skunk cabbage, remains largely unknown. Only a few plants, such as Spirodera polyrhiza (duckweed), are known to be day-neutral plants with respect to turion formation50. However, since skunk cabbage has a long juvenile phase and is buried under heavy snow during winter, it is difficult to examine the effects of environmental factors, including day length, on the timing of flowering. It is also difficult to control the flowering of skunk cabbage by changing environmental conditions. Thus, altering endogenous conditions by overexpressing SrFT is probably the best strategy to promote skunk cabbage flowering. To achieve this goal, recent advances in transformation techniques and tissue culture will be useful; especially since the number of reports using these techniques in the Araceae family has been gradually increasing51,52,53. In addition, since floral thermogenesis is always observed in reproductive organs, such as flowers and inflorescences, technical advances in the control of flowering will elucidate the molecular mechanisms that underlie floral thermogenesis and will likely reveal many unanswered questions.
Methods
Plant materials
Skunk cabbages, Symplocarpus renifolius, tissues were sampled from plants growing in the marshlands of Iwate and Nagano, Japan with the exception of a few potted plants that were transferred from outdoors at the end of autumn, in order to study gene expression in floral buds.
cDNA sequencing
A cDNA fragment library was prepared from S. renifolius by using a GS FLX Titanium Rapid Library Preparation Kit (Roche Diagnostics), and over 450,000 cDNA fragments were sequenced with an FLX+ genome sequencer (Roche Diagnostics). Sequences that were highly homologous to the FT and MFT of several plant species were screened from ~35,000 contigs using BLAST-X searches, and cloning primers were designed to isolate the genes. TFL1 homologs and other PEBP genes were not identified in these datasets. However, partial sequences of TFL2 homologs were identified (Supplementary Fig. 3), and primers were designed to analyze their gene expression.
Cloning of FT and MFT genes from S. renifolius
Total RNA was extracted from leaves of S. renifolius using the RNeasy Plant Mini Kit (QIAGEN), and cDNA was synthesized from 1 μg total RNA using a PrimeScriptTM RT reagent kit (Takara Bio Inc.). The resulting cDNA was then used as the template in subsequent RT-PCR experiments with KOD-Plus DNA polymerase (TOYOBO). For the cloning of SrFT genes, nested-PCR was carried out using two sets of primers: SrFT-F4 and SrFT-R4 for 1st PCR, and SrFT-F1 and SrFT-R1 for 2nd PCR. To clone the SrMFT genes, 5′-RACE was conducted using a FirstChoice RLM-RACE Kit (Life Technologies) and the gene-specific outer and inner primers SrMFT-R3 and SrMFT-R4, respectively, which was designed from partial SrMFT sequences on our cDNA fragment library. The full-length clone of SrMFT was isolated using RT-PCR and the primers SrMFT-F5 and SrMFT-R1. After being cloned into the pZErO-2 vector, the full-length SrFT and SrMFT genes were sequenced using the primers M13F and M13R. The nucleotide sequences of SrFT and SrMFT have been submitted to DNA Data Bank of Japan (DDBJ) with accession number LC030437 and LC030438, respectively. Although considerable effort was made to isolate TFL1 homologs and other PEBP genes (Supplementary Figs 1 and 2), these genes were not isolated. All primers used in this study are summarized in Supplementary Table 1.
Construction of plant transformation vectors
To generate the 35S::SrFT construct, the PCR products that were amplified using the primers SrFT-F1 and SrFT-R1 were cloned into the NcoI-NheI site of the pCAMBIA 1301 vector (pCAM1301) under the control of the cauliflower mosaic virus (CaMV) 35S promoter using an In-Fusion HD Cloning Kit w/Cloning Enhancer (Clontech Laboratories, Inc). The pCAM1301 vector was partially digested because it contains two NheI digestion sites. To generate the 35S::SrFT-GFP construct in a similar way, PCR was carried out using the primers SrFT-F1 and SrFT-R2. The GFP gene was amplified from pUC35S::GFP using PCR with KOD-Plus DNA polymerase (TOYOBO) and the primers GFP-F1 and GFP-R1. The two amplified PCR products were mixed and cloned into the NcoI-NheI site of pCAM1301 using an In-Fusion HD Cloning Kit w/Cloning Enhancer (Clontech Laboratories, Inc), and the resulting DNA inserts were sequenced with the primers pCAM1301-F1 and pCAM1301-R1. To generate 35S::SrMFT and 35S::SrMFT-GFP constructs, the pZerO-2 vector that contained the SrMFT gene was used as a template. The SrMFT gene for 35S::SrMFT was amplified using the primers SrMFT-IF-F1 and SrMFT-IF-R1, and the SrMFT gene for 35S::SrMFT-GFP was amplified using the primers SrMFT-IF-F1 and SrMFT-IF-R2.
Arabidopsis transformation
All pCAMBIA constructs were introduced into Arabidopsis (ecotype Columbia) via Agrobacterium tumefaciens-mediated transformation, using the floral dip method54. The flowering time of the T3 line was evaluated.
Molecular phylogenetic and evolutionary analyses
The predicted amino acid sequences of FT/MFT and other related proteins from angiosperms were collected by searching the National Center for Biotechnology Information (NCBI) databases. In addition, the protein sequences of MFT orthologous genes from gymnosperms (P. abies), spikemosses (S. moellendorffii), and mosses (P. patens) were identified and obtained from the HMMER web server (http://hmmer.janelia.org/)55 by using the SrMFT gene sequence as the query and selecting sequences with the lowest E values. A phylogenetic tree of the full-length sequences of these proteins was constructed based on the alignment from CLUSTAL W using the maximum likelihood (ML) method in MEGA 6.0656. The consensus phylogenic trees are shown with bootstrap values from 1000 replications.
Semi-quantitative RT-PCR of SrFT, SrMFT, and SrTFL2 transcripts
Total RNA was extracted from the spadices, leaves, roots, and spathes of S. renifolius using an RNeasy Plant Mini Kit (QIAGEN), and cDNA was synthesized from 1 μg total RNA using a PrimeScriptTM RT reagent kit (Takara Bio Inc.). the resulting cDNA was then used as the template in subsequent RT-PCR experiments using ExTaq DNA polymerase (Takara Bio Inc.) with primers SrFT-F4 and SrFT-R4 for amplifying a 600-bp region containing the SrFT gene, SrMFT-F3 and SrMFT-R3 for amplifying a 177-bp region within SrMFT, and SrTFL2-F3 and SrTFL2-R3 for amplifying a 152-bp region within SrTFL2. As a control, the primers qPCR60-F and qPCR60-R were used to amplify a 160-bp region containing the SrEF1α gene. The primers SrFT-F3 and SrFT-R3 were used to amplify a 190-bp region within SrFT when examining the mRNA levels of cultured S. renifolius leaves. The primers SrFT-F1-rt and SrFT-R1-rt were used to amplify a 142-bp region within SrFT when examining the mRNA levels of Arabidopsis plants transformed with SrFT.
In situ hybridization of SrFT transcripts
To generate an antisense probe construct, 600-bp target regions containing the SrFT coding sequence (525 bp) and the 5′ and 3′ flanking sequences (21 bp and 54 bp, respectively) were amplified from S. renifolius leaf cDNA using KOD-Plus DNA polymerase (TOYOBO) and the primers revSrFT-f1 and revSrFT-r1. The antisense probe construct was then used as a template to generate a sense probe construct, and 600-bp regions were amplified using the orSrFT-f1 and orSrFT-r1 primers. Amplified DNA fragments were inserted into the PstI-XhoI site of the pBlueScript SK(+) vector and sequenced. In situ hybridization was subsequently conducted as previously described using a Hybrimaster HS-300 (ALOKA)57,58. DIG-labeled antisense and sense probes were then generated using a T3/T7 digoxigenin RNA labeling kit (Roche Diagnostics) and were hybridized to cross-sections of spadices and leaves from female-stage S. renifolius. Tissue sections were observed with a light microscope (SteREO Discovery V20; Carl Zeiss).
SDS-PAGE and immunoblotting
SDS-PAGE was carried out as described previously59. After electrophoretic separation using SDS-PAGE, proteins were electrotransferred to a PVDF membrane (Millipore) using a semi-dry electrophoretic transfer system (ATTO) and blotting buffer (50 mM TRIS-HCl, 1.44% glycine, 20% methanol). Rabbit polyclonal antibodies against GFP were produced using His-tagged GFP as the antigen. The anti-GFP IgG was affinity-purified using a GFP antigen column and the purified IgG was used as the primary antibody to detect SrFT-GFP and SrMFT-GFP. Horseradish peroxidase-conjugated secondary antibodies were detected using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific).
Microarray analysis
Total RNA was isolated from individual floral tissues including petals, pistils, stamens, and the pith using an RNeasy Plant Mini Kit (QIAGEN). The RNA samples were quantified using an ND-1000 spectrophotometer (NanoDrop Technologies) and the quality was confirmed with an Experion System (Bio-Rad Laboratories). The cRNA was then amplified, labeled, and hybridized to a 4 × 44k Agilent 60-mer custom oligomicroarray according to the manufacturer’s instructions. The probes on this custom array were designed based on our in-house cDNA database, which was prepared from female-stage spadices of S. renifolius. All hybridized microarray slides were scanned using a G2505B Microarray Scanner (Agilent). Relative hybridization intensities and background hybridization values were calculated using Agilent Feature Extraction Software (9.5.1.1), and the microarray data have been deposited in the NCBI Gene Expression Omnibus repository with the accession number GSE68011.
Cluster analysis
To identify genes that were co-regulated with SrFT and SrAOX, cluster analysis was performed60, using 27,184 genes from floral tissues including petals, pistils, stamens, and the pith, and the “Correlation” clustering algorithm. According to their expression profiles, 6523, 7259, 7867, and 5535 genes were classified into four clusters I, II, III and IV, respectively.
Gene ontology analysis
The Arabidopsis dataset for BLAST analysis was obtained from the Arabidopsis information Resource (TAIR) website (http://www.arabidopsis.org/index.jsp), and the S. renifolius dataset prepared in this study was analyzed using the Blast2.2.27+ program from NCBI. A homolog search was performed by using tBLASTx to search the Arabidopsis dataset for sequences that were similar to those in the S. renifolius dataset (DB: Arabidopsis thaliana, query: S. renifolius), and vice versa (DB: S. renifolius, query: A. thaliana). The genes that were among the top hits of both homolog searches and that had E values less than e−10 were selected, and AGI codes for the selected Arabidopsis orthologs were obtained from the TAIR database. Of 27,184 genes, a total of 8279 were annotated with AGI codes and analyzed using the Gene Ontology tool on the TAIR website, and 2052, 1981, 2438, and 1808 of the genes were classified in clusters I, II, III, and IV, respectively.
Additional Information
How to cite this article: Ito-Inaba, Y. et al. Characterization of two PEBP genes, SrFT and SrMFT, in thermogenic skunk cabbage (Symplocarpus renifolius). Sci. Rep. 6, 29440; doi: 10.1038/srep29440 (2016).
Supplementary Material
Acknowledgments
We thank Ms. Yoko Katayama (University of Miyazaki) for her technical support. This work was supported by The Naito Foundation to YII, The Inamori Foundation to YII, Grants-in-Aid for Young Scientists (B, Nos 26850073 to TI and 26850065 to YII), Grant-in-Aid for Scientific Research (C, No. 15K07843 to TI), Grants-in-Aid for Scientific Research on Innovative Areas (Nos 23113001 and 23113006 to MW), Grant-in-Aid for Challenging Exploratory Research (No. 16K15085 to MW), Grant-in-Aid for Scientific Research (B, 16H04854 to MW), and the Program to Disseminate Tenure Tracking System from the Japanese Ministry of Education, Culture, Sports, Science and Technology to YII, and by a grant for Scientific Research on Priority Areas from the University of Miyazaki to YII.
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.I.-I. contributed to the experimental design and wrote the manuscript. Y.I.-I., H.M.-S. and H.M. conducted the experiments. Y.I.-I., M.W. and T.I. analyzed the data. All authors read and approved the manuscript.
References
- Thien L. B. et al. Pollination biology of basal angiosperms (ANITA grade). Am J Bot 96, 166–182, doi: 10.3732/ajb.0800016 (2009). [DOI] [PubMed] [Google Scholar]
- Yuan L.-C. et al. Pollination of Kadsura longipedunculata (Schisandraceae), a monoecious basal angiosperm, by female, pollen-eating Megommata sp. (Cecidomyiidae: Diptera) in China. Biol J Linn Soc 93, 523–536, doi: 10.1111/j.1095-8312.2007.00897.x (2008). [DOI] [Google Scholar]
- Ervik F. & Knudsen J. T. Water lilies and scarabs: faithful partners for 100 million years? Biol J Linn Soc 80, 539–543, doi: 10.1046/j.1095-8312.2003.00258.x (2003). [DOI] [Google Scholar]
- Seymour R. S. & Matthews P. G. The role of thermogenesis in the pollination biology of the Amazon waterlily Victoria amazonica. Ann Bot 98, 1129–1135, doi: 10.1093/aob/mcl201 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour R. S., Silberbauer-Gottsberger I. & Gottsberger G. Respiration and temperature patterns in thermogenic flowers of Magnolia ovata under natural conditions in Brazil. Funct Plant Biol 37, 870–878, 10.1071/FP10039 (2010). [DOI] [Google Scholar]
- Dieringer G., Cabrera R. L., Lara M., Loya L. & Reyes-Castillo P. Beetle pollination and floral thermogenicity in Magnolia tamaulipana (Magnoliaceae). Int J Plant Sci 160, 64–71 (1999). [Google Scholar]
- Wang R. et al. Thermogenesis, flowering and the association with variation in floral odour attractants in Magnolia sprengeri (Magnoliaceae). PLoS One 9, e99356, doi: 10.1371/journal.pone.0099356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibernau M., Barabe D., Moisson M. & Trombe A. Physical constraints on temperature difference in some thermogenic aroid inflorescences. Ann Bot (Lond) 96, 117–125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubatz H., Nelson T. A., Dong A. M., Meeuse B. J. D. & Bendich A. J. Infrared thermography of Arum lily inflorescences. Planta 182, 432–436 (1990). [DOI] [PubMed] [Google Scholar]
- Seymour R. S. Scaling of heat production by thermogenic flowers: limits to floral size and maximum rate of respiration. Plant Cell Environ 33, 1474–1485, doi: 10.1111/j.1365-3040.2010.02190.x (2010). [DOI] [PubMed] [Google Scholar]
- Patiño S., Grace J. & Bänziger H. Endothermy by flowers of Rhizanthes lowii (Rafflesiaceae). Oecologia 124, 149–155, doi: 10.1007/s004420050001 (2000). [DOI] [PubMed] [Google Scholar]
- The Angiosperm Phylogeny, G. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161, 105–121, doi: 10.1111/j.1095-8339.2009.00996.x (2009). [DOI] [Google Scholar]
- Tang W. Heat production in cycad cones. Bot Gaz 148, 165–174 (1987). [Google Scholar]
- Skubatz H., Tang W. & Meeuse B. J. D. Oscillatory heat-production in the male cones of cycads. J Exp Bot 44, 489–492, doi: 10.1093/jxb/44.2.489 (1993). [DOI] [Google Scholar]
- Meeuse B. J. D. & Raskin I. Sexual reproduction in the arum lily family, with emphasis on thermogenicity. Sex Plant Reprod 1, 3–15 (1988). [Google Scholar]
- Seymour R. S., White C. R. & Gibernau M. Environmental biology: heat reward for insect pollinators. Nature 426, 243–244 (2003). [DOI] [PubMed] [Google Scholar]
- Seymour R. S. & Blaylock A. J. Switching off the heater: influence of ambient temperature on thermoregulation by eastern skunk cabbage Symplocarpus foetidus. J Exp Bot 50, 1525–1532 (1999). [Google Scholar]
- Seymour R. S. Biophysics and physiology of temperature regulation in thermogenic flowers. Biosci Rep 21, 223–236 (2001). [DOI] [PubMed] [Google Scholar]
- Knutson R. M. Heat production and temperature regulation in eastern skunk cabbage. Science 186, 746–747 (1974). [DOI] [PubMed] [Google Scholar]
- Ito-Inaba Y. et al. Developmental changes and organelle biogenesis in the reproductive organs of thermogenic skunk cabbage (Symplocarpus renifolius). J Exp Bot 60, 3909–3922 (2009a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Inaba Y. Thermogenesis in skunk cabbage (Symplocarpus renifolius): New insights from the ultrastructure and gene expression profiles. Adv Hortic Sci 28, 73–78 (2014). [Google Scholar]
- Ito-Inaba Y. et al. The gene expression landscape of thermogenic skunk cabbage suggests critical roles for mitochondrial and vacuolar metabolic pathways in the regulation of thermogenesis. Plant Cell Environ 35, 554–566, doi: 10.1111/j.1365-3040.2011.02435.x (2012). [DOI] [PubMed] [Google Scholar]
- Grant N. M., Miller R. E., Watling J. R. & Robinson S. A. Synchronicity of thermogenic activity, alternative pathway respiratory flux, AOX protein content, and carbohydrates in receptacle tissues of sacred lotus during floral development. J Exp Bot 59, 705–714, doi: 10.1093/jxb/erm333 (2008). [DOI] [PubMed] [Google Scholar]
- Ito-Inaba Y., Hida Y. & Inaba T. What is critical for plant thermogenesis? Differences in mitochondrial activity and protein expression between thermogenic and non-thermogenic skunk cabbages. Planta 231, 121–130 (2009b). [DOI] [PubMed] [Google Scholar]
- Miller R. E. et al. In the heat of the night–alternative pathway respiration drives thermogenesis in Philodendron bipinnatifidum. New Phytol 189, 1013–1026, doi: 10.1111/j.1469-8137.2010.03547.x (2011). [DOI] [PubMed] [Google Scholar]
- Watling J. R., Robinson S. A. & Seymour R. S. Contribution of the alternative pathway to respiration during thermogenesis in flowers of the sacred lotus. Plant Physiol 140, 1367–1373, doi: 10.1104/pp.105.075523 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. M., Krab K., Wagner M. J. & Moore A. L. Regulation of thermogenesis in flowering Araceae: the role of the alternative oxidase. Biochim Biophys Acta 1777, 993–1000, doi: 10.1016/j.bbabio.2008.04.001 (2008). [DOI] [PubMed] [Google Scholar]
- Chardon F. & Damerval C. Phylogenomic analysis of the PEBP gene family in cereals. J Mol Evol 61, 579–590, doi: 10.1007/s00239-004-0179-4 (2005). [DOI] [PubMed] [Google Scholar]
- Ahn J. H. et al. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J 25, 605–614, doi: 10.1038/sj.emboj.7600950 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y., Money T. & Bradley D. A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102, 7748–7753, doi: 10.1073/pnas.0500932102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W. W. & Weigel D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26, 552–564, doi: 10.1105/tpc.113.115220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlgren A. et al. Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol 156, 1967–1977, doi: 10.1104/pp.111.176206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman H., Källman T. & Lagercrantz U. Early evolution of the MFT-like gene family in plants. Plant Mol Biol 70, 359–369, doi: 10.1007/s11103-009-9478-x (2009). [DOI] [PubMed] [Google Scholar]
- Corbesier L. et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033, doi: 10.1126/science.1141752 (2007). [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H. L., Yokoi S. & Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036, doi: 10.1126/science.1141753 (2007). [DOI] [PubMed] [Google Scholar]
- Notaguchi M. et al. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol 49, 1645–1658, doi: 10.1093/pcp/pcn154 (2008). [DOI] [PubMed] [Google Scholar]
- Zeevaart J. A. Leaf-produced floral signals. Curr Opin Plant Biol 11, 541–547, doi: 10.1016/j.pbi.2008.06.009 (2008). [DOI] [PubMed] [Google Scholar]
- Pin P. A. et al. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400, doi: 10.1126/science.1197004 (2010). [DOI] [PubMed] [Google Scholar]
- Navarro C. et al. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122, doi: 10.1038/nature10431 (2011). [DOI] [PubMed] [Google Scholar]
- Kinoshita T. et al. FLOWERING LOCUS T regulates stomatal opening. Curr Biol 21, 1232–1238, doi: 10.1016/j.cub.2011.06.025 (2011). [DOI] [PubMed] [Google Scholar]
- Kotake T., Takada S., Nakahigashi K., Ohto M. & Goto K. Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol 44, 555–564 (2003). [DOI] [PubMed] [Google Scholar]
- Larsson A. S., Landberg K. & Meeks-Wagner D. R. The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149, 597–605 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimida N., Kidou S. & Kotoda N. Constitutive expression of two apple (Malus x domestica Borkh.) homolog genes of LIKE HETEROCHROMATIN PROTEIN1 affects flowering time and whole-plant growth in transgenic Arabidopsis. Mol Genet Genomics 278, 295–305, doi: 10.1007/s00438-007-0250-0 (2007). [DOI] [PubMed] [Google Scholar]
- Takada S. & Goto K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856–2865, doi: 10.1105/tpc.016345 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P. A. et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059, doi: 10.1126/science.1114358 (2005). [DOI] [PubMed] [Google Scholar]
- Carmona M. J., Calonje M. & Martinez-Zapater J. M. The FT/TFL1 gene family in grapevine. Plant Mol Biol 63, 637–650, doi: 10.1007/s11103-006-9113-z (2007). [DOI] [PubMed] [Google Scholar]
- Igasaki T., Watanabe Y., Nishiguchi M. & Kotoda N. The FLOWERING LOCUS T/TERMINAL FLOWER 1 family in Lombardy poplar. Plant Cell Physiol 49, 291–300, doi: 10.1093/pcp/pcn010 (2008). [DOI] [PubMed] [Google Scholar]
- Hou C. J. & Yang C. H. Functional analysis of FT and TFL1 orthologs from orchid (Oncidium Gower Ramsey) that regSulate the vegetative to reproductive transition. Plant Cell Physiol 50, 1544–1557, doi: 10.1093/pcp/pcp099 (2009). [DOI] [PubMed] [Google Scholar]
- Xiang L. et al. Functional analysis of FLOWERING LOCUS T orthologs from spring orchid (Cymbidium goeringii Rchb. f.) that regulates the vegetative to reproductive transition. Plant Physiol Biochem 58, 98–105, doi: 10.1016/j.plaphy.2012.06.011 (2012). [DOI] [PubMed] [Google Scholar]
- Appenroth K.-J., Hertel W. & Augsten H. Phytochrome Control of Turion Formation in Spirodela polyrhiza L. Schleiden. Ann Bot 66, 163–168 (1990). [Google Scholar]
- Matsumoto T. et al. Screening promoters for Anthurium transformation using transient expression. Plant Cell Reports 32, 443–451, doi: 10.1007/s00299-012-1376-z (2013). [DOI] [PubMed] [Google Scholar]
- He X., Miyasaka S. C., Fitch M. M., Moore P. H. & Zhu Y. J. Agrobacterium tumefaciens-mediated transformation of taro (Colocasia esculenta (L.) Schott) with a rice chitinase gene for improved tolerance to a fungal pathogen Sclerotium rolfsii. Plant Cell Rep 27, 903–909, doi: 10.1007/s00299-008-0519-8 (2008). [DOI] [PubMed] [Google Scholar]
- Ban H. et al. Transgenic Amorphophallus konjac expressing synthesized acyl-homoserine lactonase (aiiA) gene exhibit enhanced resistance to soft rot disease. Plant Cell Rep 28, 1847–1855, doi: 10.1007/s00299-009-0788-x (2009). [DOI] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–743, doi: 10.1046/j.1365-313x.1998.00343.x (1998). [DOI] [PubMed] [Google Scholar]
- Finn R. D., Clements J. & Eddy S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39, W29–W37, doi: 10.1093/nar/gkr367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 30, 2725–2729, doi: 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwabe K. et al. Separated transcriptomes of male gametophyte and tapetum in rice: validity of a laser microdissection (LM) microarray. Plant Cell Physiol 49, 1407–1416, doi: 10.1093/pcp/pcn124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Inaba Y., Masuko H., Watanabe M. & Inaba T. Isolation and gene expression analysis of a papain-type cysteine protease in thermogenic skunk cabbage (Symplocarpus renifolius). Biosci Biotechnol Biochem 76, 1990–1992, doi: 10.1271/bbb.120434 (2012). [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970). [DOI] [PubMed] [Google Scholar]
- Huang H., Cai L. & Wong W. H. Clustering analysis of SAGE transcription profiles using a Poisson approach. Methods Mol Biol 387, 185–198 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.