Abstract
The past 15 years have seen an explosion of discoveries related to the cellular regulation of phenotypes through epigenetic mechanisms. This regulation provides a software that packages DNA, without changing the primary base sequence, to establish heritable patterns of gene expression. In cancer, many aspects of the epigenome, controlled by DNA methylation, chromatin, and nucleosome positioning, are altered as one means by which tumor cells maintain abnormal states of self-renewal at the expense of normal maturation. Epigenetic and genetic abnormalities thus collaborate in cancer initiation and progression, as exemplified by frequent mutations in genes encoding proteins that control the epigenome. There is growing emphasis on using epigenetic therapies to reprogram neoplastic cells toward a normal state. Many agents targeting epigenetic regulation are under development and entering clinical trials. This review highlights the promise that epigenetic therapy, often in combination with other therapies, will become a potent tool for cancer management over the next decade.
Keywords: epigenetics, DNA methyltransferase inhibitor, histone deacetylase inhibitor therapy
INTRODUCTION
Despite our increasing knowledge of the basis of cancer, progress in treatment of the common cancers is still suboptimal (1, 2). Most new therapy approaches focus on genetic abnormalities. Deep sequencing of cancer genomes has allowed targeting of specific driver mutations, which can provide robust initial responses but often has short durability with evolution of resistance (3).
This review considers the promise of therapies based on understanding cancer epigenetic abnormalities. “Epigenetics” refers to heritable changes in gene expression patterns that do not rely on primary DNA sequence changes (4, 5). If DNA is like the hard drive that contains the information to guide every cellular function, then epigenetic modulation acts as software that regulates the packaging of DNA to guide potential for gene expression patterns. Epigenetic control guides cell type fates during embryogenesis and adult cell renewal (4–6).
A key example of epigenetic alteration in cancer is abnormal silencing of nonmutated tumor suppressor genes, which serves as an alternative to mutations for effecting loss of gene function (7–9). A recent exciting indication of the importance of epigenetic changes is the finding, in virtually every cancer type, of mutations in genes that encode many proteins that regulate the epigenome (7, 10–12). Deciphering the ramifications of these genetic changes is a major imperative for cancer research.
The great potential for epigenetic therapies lies in the fact that, unlike genetic abnormalities, epigenetic changes are reversible, allowing recovery of function for affected genes with normal DNA sequences (7, 13). These therapies aim to reprogram cancer cells to a more normal state (4, 7, 13). In normal development, the epigenome is agile, allowing changes in cell phenotypes from an embryonic to a differentiated state. In cancer, abnormal epigenetic states can help lock in cell states that hinder the ability of cells to exit self-renewal and differentiate normally. For example, in colon tumorigenesis, colon crypt cells retain a more primitive, embryonic cell type (14). These reprogramming steps likely evolve over years of cancer initiation and progression (7, 10–12).
Potential for reversing epigenetic alterations began with the discovery in the late 1970s of agents that reverse DNA methylation (15). However, it took until the 1990s for these drugs to gain traction in clinical trials, primarily in treating hematologic malignancies and especially the preleukemic disorder myelodysplasticsyndrome (MDS). Their efficacy emerged as doses of the epigenetic drugs were lowered, improving patient tolerance and perhaps specificity of reprogramming (16, 17). DNA demethylating agents are now approved by the US Food and Drug Administration (FDA) for MDS (18), and signals for efficacy for the lower drug doses in the treatment of solid tumors have been steadily emerging. Rapid advances in our understanding of epigenome regulation are leading to development of many new epigenetic therapeutic drugs.
THE EPIGENOME LANDSCAPE
During the past decade, understanding of epigenetic regulation in both normal and cancer cells has rapidly increased. Technological advancements in genome-wide DNA sequencing, RNA sequencing for coding and noncoding expression patterns, assays of DNA methylation and chromatin, and assessment of all of the above with deep bioinformatics are helping to define the cancer epigenome (4, 7, 11) and enable key insights for developing epigenetic therapies.
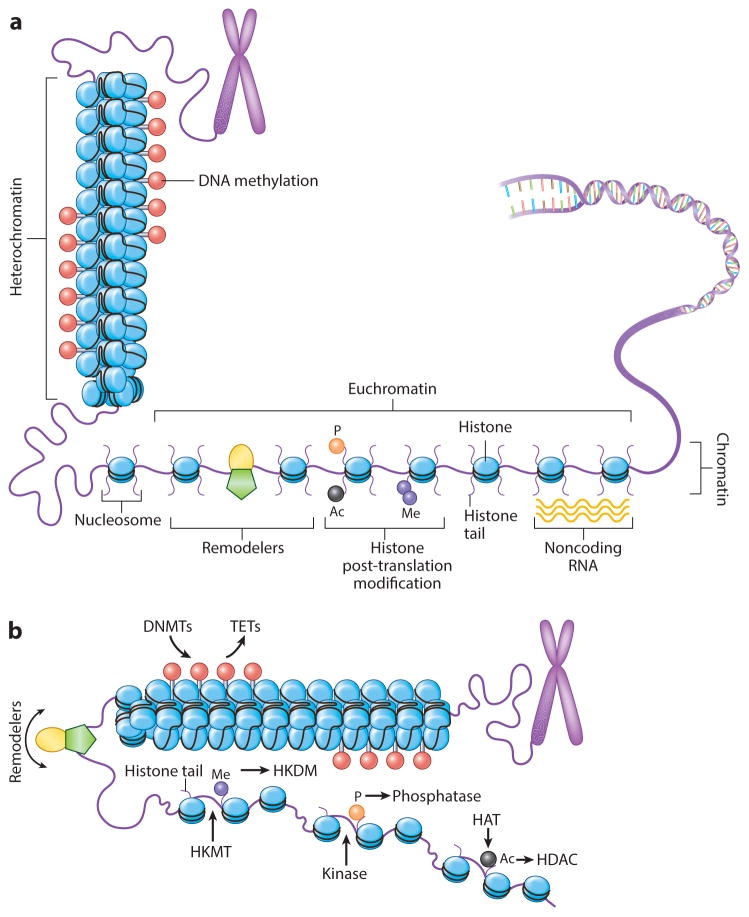
The epigenetic landscape is controlled predominantly by DNA methylation and chromatin, the latter encompassing DNA plus interacting proteins (4, 19). At the heart of this process are nucleosome structures, a core of histone proteins around which ~160 base pairs are wrapped. Nucleosome positioning determines how DNA is packaged to modulate its gene expression (Figure 1), and this is regulated by modifications of the core histones (4, 19, 20). Gene expression is facilitated when transcription start sites are in a nucleosome-free state and is repressed with compacted nucleosome occupancy (Figure 1). This epigenetic control is accomplished by the “four Rs” of epigenetics (Figure 2): the writer, eraser, reader, and remodeler proteins that function within intricate complexes to establish heritable patterns of gene expression (4, 5, 11, 19, 20).
Figure 1.
The epigenome landscape. (a) Chromatin states support either transcriptional activation or silencing of genes, allowing gene regulatory regions to switch these states through positioning of nucleosomes (blue ovals). More open conformations leave the transcription start site nucleosome free. Modifications of nucleosome histone tails ( purple lines extending from ovals) regulate the process, including DNA methylation (red lollipops), serine phosphorylation (orange circle), lysine acetylation (black circle) and lysine methylation ( purple circle), and nucleosome remodeler complexes ( green pentagon with yellow oval). Additionally, noncoding RNAs ( yellow waves) can participate in these regulatory steps through recruitment of chromatin proteins and DNA methylation. (b) Control of histone modifications and of DNA methylation by proteins: writers (DNMTs, HKMTs, HATs, kinases for phosphorylation), readers (shown in subsequent figures for binding to and interpreting each mark for function), erasers (TETs for DNA methylation, HKDMs for lysine methylation, HDACs, phosphatases for removing phosphorylation) and nucleosome remodelers. Red lollipops indicate DNA methylation; green pentagon with yellow oval indicates nucleosome remodeler complexes; purple circle indicates histone lysine methylation; orange circle indicates serine phosphorylation; black circle indicates lysine acetylation. Abbreviations: DNMT, DNA methyltransferase; HAT, histone acetylases; HDAC, histone deacetylases; HKDM, histone lysine demethylase; HKMT, histone lysine methyltransferase; TET, ten-eleven translocation protein.
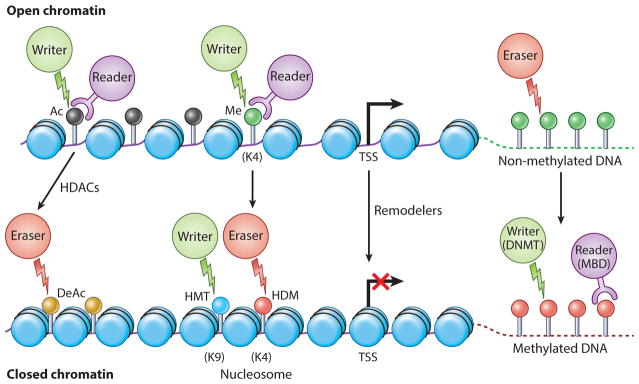
Figure 2.
The four Rs of proteins regulating the epigenome. For open promoter conformation (top), epigenetic signal writers (green circles), readers ( purple circles), and erasers (red circles), and generally no DNA methylation in associated CpG islands ( green lollipops). Nucleosomes (blue ovals) are in an open conformation around the transcription start site (TSS). Writers in the form of histone acetylases (HAT) and histone methyltransferases (HMTs) are enzymes that add acetyl (Ac) and methyl (Me) marks to histone proteins (acetylated lysine, black circles on lollipops; methylated lysine, green circle on lollipop). These modifications to histones cause chromatin conformational changes and gene expression regulation. Readers containing specialized domains bind to these distant marks, which are critical for binding to specific modification states. Erasers such as histone deacetylases (HDACs), lysine demethylases (KDMs), and phosphatases are involved in the removal of epigenetic marks. Transition to an inactive state (bottom) with cancer-specific promoter CpG-island DNA hypermethylation is associated with a more closed nucleosome spacing over the TSS, and HDACs, which erase histone acetylation (gold lollipops), and writers (HMTs) that change active histone methylation marks to repressive ones such as H3K9me3 (blue lollipop) and H3k27me3, as discussed in the text, with HDMs acting as antagonist to HMTs. Another set of writers (DNMT) establish methylation of CpGs at promoter regions (red lollipops), and readers for this methylation are methyl cytosine binding proteins (MBDs). Other abbreviations: DeAc, deacetylation; HDAC, histone deacetylase; HMT, histone methyltransferase; HDM, histone demethylases.
A most exciting development linking genetic and epigenetic abnormalities in cancer suggests that some of the best therapeutic strategies may be to block epigenetic events that result from gene mutations. For virtually all tumor types, there is a very high frequency of mutations in the genes encoding proteins that guide and maintain the normal epigenome, including in the histones themselves (7, 10–12, 21–24). Examples include mutations in an enzyme, DNA (cytosine-5)-methyltransferase 3A (DNMT3A), which can be found in up to 25% of patients with acute myeloid leukemia (AML). Both AML and glioblastoma multiforme are associated with mutations in other genes affecting DNA methylation, including mutations in IDH (encoding isocitrate dehydrogenase) and TET (encoding ten-eleven translocation proteins), as discussed below (11). Mutations in chromatin remodeling proteins occur in a multitude of solid cancers including pancreas, breast, ovarian, gastric, and colon (4, 11, 23, 24). The consequences of many of these genetic changes for the cancer epigenome are still unclear. However, patients harboring such mutations may be particularly suitable for targeted epigenetic therapy.
DNA METHYLATION IN NORMAL AND DISEASE EPIGENOMES
In humans, DNA methylation occurs almost exclusively at cytosines in the sequence CpG. This dinucleotide is dispersed unevenly across the genome; most of our DNA is CpG poor and heavily DNA methylated. In contrast, localized CpG-rich regions, known as CpG islands, remain largely unmethylated, especially in gene promoter regulatory sites (4, 7, 9, 11). DNA methylation is critical for silencing of imprinted gene alleles and transcription from repetitive elements, including retroviral genes (4, 25). Certain diseases can occur as a result of errors associated with imprinting, which generally requires parental allele-specific expression. Disorders in the other allele may result in disorders such as Prader-Willi and Angelman syndrome, and efforts are under way to utilize therapies to reprogram to a normal developmental program (26). DNA methylation and noncoding RNA interference may also play a role in other disease states such as Alzheimer’s disease and frontotemporal dementia (27).
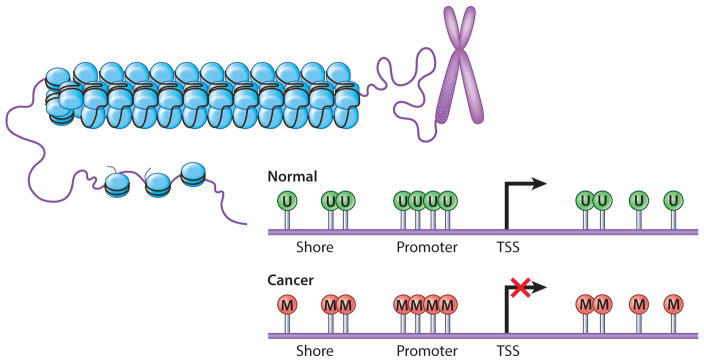
DNA methylation is vital to changes of gene expression during embryogenesis and in normal mature cell renewal (4, 6). In cancer, DNA methylation is severely altered; there are both widespread genomic hypomethylation and focal gains in many normally unmethylated promoter CpG islands, the latter associated with abnormal gene silencing (CpG-island hypermethylation) (4, 7, 9, 11) (Figure 3). Restoring more normal patterns of DNA methylation, especially at the promoter sites, is one cancer therapy goal.
Figure 3.
DNA methylation changes in cancer cells. Promoter CpG islands in normal cells (top) generally lack CpG site DNA methylation ( green U lollipops). In cancer (bottom), many genes gain DNA methylation in promoter-region CpG islands with an accompanying repressive chromatin landscape and abnormal gene silencing (red M lollipops).
DNA methylation patterns are established by three DNA methyltransferases (DNMTs), the “writers” of the modification. Drugs such as 5-azacitidine (5AC or Vidaza®) and 5-aza-2′-deoxycytidine (DAC or Dacogen®) inhibit and degrade these enzymes (4, 7, 8, 28). The most abundant DNMT, DNMT1, maintains existing methylation patterns; DNMT3A and -3B primarily establish new sites of methylation. Proteins that can bind to DNA methylated sites, or methyl cytosine binding proteins (MBDs), can act as “readers” to give functional context to the DNA modification (4, 7, 28). Normal and abnormal removal of DNA methylation, or “erasing,” can occur passively when DNA is replicated and the modification is not re-established. There is much recent excitement over the discovery of an active removal system of this methylation mark, initiated by a set of enzymes termed ten-eleven translocation proteins (TETs) (4, 11, 29, 30).
Abnormalities in DNA methylation are being studied extensively for their role in carcinogenesis (4, 7–9, 11). Distinct patterns associate with various mutations, with much to be determined about causes and effects (11, 12, 21). Hyper- and hypomethylation can be independent of each other (4, 7, 8, 11) but can also simultaneously reside in long-range regions of DNA wherein abnormal focal gains in promoter CpG islands are surrounded by broad zones of demethylation. Both hyper-and hypomethylation lead to alterations in gene expression, and the latter has been associated with genomic instability (4, 7, 11). DNA methylation abnormalities very often manifest early in tumorigenesis, being well set in pre-invasive stages such as nonmalignant colon polyps (4, 7, 8, 13). As mentioned, the focal gains in promoter CpG islands are associated with aberrant silencing of genes, including well-defined tumor suppressor genes, as an alternative to mutations for loss of function (4, 6–9, 11). Hundreds of such affected genes are often present in individual tumors of all types, and a key challenge is to determine the significance of particular genes or groups of genes. These promoter changes affect genes, often multiple ones, in virtually every abnormal signaling pathway driving tumorigenesis, including cell cycle control, apoptosis, DNA damage, immune recognition, and cell self-renewal (4, 7–9, 11, 17).
CHROMATIN ARCHITECTURE IN NORMAL AND CANCER EPIGENOMES
The establishment and function of abnormal DNA methylation are tied to histone modifications regulating chromatin architecture (4, 7–9, 11, 31). As mentioned, these are controlled by proteins known as writers, readers, and erasers (4, 7–9, 11, 31), which regulate histone amino acids for lysine acetylation and methylation, methylation of arginines, and phosphorylation of serines and threonines (Figures 1 and 2). Like DNA methylation, these histone modifications facilitate how cells maintain “memories” for gene expression patterns fundamental to proper embryonic development and balance of cell renewal and differentiation in adult tissues (4, 19, 31).
Histone modifications facilitate active versus repressed states of gene expression and especially lysine acetylation and methylation (4, 19, 31). The writers for acetylation are histone acetylases (HATs); their removal is catalyzed by histone deacetylases (HDACs) (Figure 1). Lysine acetylation, especially at gene start sites, is usually associated with transcriptionally active genes and deacetylation with repressed genes. Key examples of such active marks are H3K9 acetyl and H4K16 acetyl (4, 19, 31, 32). The deacetylated state is especially associated, either on its own or accompanied by DNA methylation, with abnormally silenced genes in cancer (4, 7–9, 11). Reversing such deacetylation with histone deacetylase inhibitors (HDACis) is a much-pursued cancer therapy target discussed in a later section.
A myriad of lysine methylation modifications, written by histone methyltransferases (HTMs) and removed by histone demethylases (HDMs) (4, 19, 31), help determine states of gene expression. Such modifications include dimethylation (H3K4me2) and trimethylation (H3K4me3) of lysine 4 of histone 3 for active promoter sites, while H3K4me1 marks active gene enhancers that can modulate a gene or groups of genes (19). In contrast, methylation of lysine 9 or lysine 27 (H3K9me3, H3K27me3) mark repressed gene promoters (4, 19, 31–35). The latter mark is established and interpreted by the Polycomb complex of proteins (PcG), which are not only important in development but also critical to carcinogenesis (4, 33–35). The PcG enzyme EZH2 is the writer for H3K27me3 (4, 33–35). Its levels are elevated in numerous cancers, and it can be mutated as an apparent oncogenic event (36) that can drive tumorigenesis by increasing stem cell self-renewal (35, 36).
NONCODING RNA
What was long considered “junk” DNA can generate RNA transcripts to play an important role in normal and abnormal epigenetic processes (37). We are still gaining insights into these RNAs and their functions in biological pathways. These transcripts, including small nuclear RNAs (snRNAs), microRNAs (miRNAs), and long noncoding RNAs (lncRNAs), can modulate transcriptional output from the genome. Particularly lncRNAs can help establish chromatin remodeling, transcription, and post-transcriptional processing. These can be generated as anti-sense RNAs transcribed in gene-regulatory regions to repress the canonical transcription from the gene (37). Such transcripts have been associated with attracting normal DNA methylation in association with repressed gene transcription, as for silenced alleles of imprinted genes, as well as establishing abnormal promoter methylation for cancer genes such as p15 (38). It is suggested that such lncRNAs may provide a general mechanism for establishing promoter, CpG-island DNA hypermethylation and gene silencing in cancer (39).
EPIGENETIC THERAPY
Understanding fundamental elements of epigenetic regulation is progressively contributing to concepts of cancer epigenetic therapeutics. Efficacy has been predominantly seen for hematologic malignancies (7, 40, 41). The current epigenetic therapy primarily involves inhibitors of DNA demethylation and histone deacetylation (Table 1). In addition to the FDA approval for use of the former approach for MDS, HDACs are now FDA-approved for T cell cutaneous lymphoma and multiple myeloma (18, 42). These agents are being tried for solid tumors.
Table 1.
Examples of some epigenetic trials in various cancers
| Trial (ClinicalTrials.gov) | Drug | Combination | Target | Study phase | Indication |
|---|---|---|---|---|---|
| NCT01034163 | Panobinostat (LBH589) | None | Pan-HDAC inhibitor | Phase III | Hodgkin’s lymphoma and multiple myeloma |
| NCT01023308 | Bortezomib, dexamethasone | Pan-deacetylase inhibitor | Phase III | Relapsed multiple myeloma | |
| NCT00425555 | None | Phase II/III | Cutaneous T cell lymphoma | ||
| NCT00866333 | Entinostat (SNDX-275) | None | Class I HDAC inhibitor | Phase I/II | Hodgkin’s lymphoma, kidney cancer |
| NCT01038778 | None | Phase II | Clear cell renal cell carcinoma, metastatic renal cell cancer | ||
| NCT01349959 | Azacitidine | Phase II | Advanced breast cancer | ||
| NCT00357032 (Completed) | Belinostat (PXD101) | None | Pan-HDAC inhibitor | Phase II | Relapsed or refractory acute myeloid leukemia or older patients with newly diagnosed acute myeloid leukemia |
| NCT01310244 | Carboplatin, paclitaxel | HDAC inhibitor | Phase I/II | Non–small cell lung cancer | |
| NCT00274651 | None | Phase II | Recurrent or refractory cutaneous and peripheral T cell lymphomas | ||
| NCT00301756 | None | Phase II | Ovarian cancer | ||
| NCT01873703 | Pracinostat (SB939) | Azacitidine | HDAC inhibitor | Phase II | Myelodysplastic syndrome |
| NCT01912274 | Azacitidine | Phase II | Acute myeloid leukemia | ||
| NCT01112384 (Completed) | None | Phase II | Translocation-associated recurrent/metastatic sarcomas | ||
| NCT01761968 | Givinostat | None | HDAC inhibitor | Phase II | Chronic myeloproliferative neoplasms |
| NCT01900730 | Valproic acid | None | HDAC inhibitor | Phase II | Breast cancer |
| NCT00477386 | Carboplatin | Decitabine | Demethylation | Phase I/II | Platinum-resistant ovarian cancer |
| NCT00387465 | Entinostat | Azacitidine | HDAC inhibitor | Phase I/II | Recurrent advanced non–small cell lung cancer |
| NCT01105377 | Entinostat | Azacitidine | HDAC inhibitor | Phase II | Metastatic colorectal cancer |
| NCT02115282 | Exemestane with or without | Entinostat | HDAC inhibitor | Phase III | Recurrent hormone receptor-positive breast cancer that is locally advanced or metastatic |
| NCT00091559 | Vorinostat | None | HDAC inhibitor | Phase II | Advanced cutaneous T cell lymphoma |
| NCT00275080 | Vorinostat | Decitabine | HDAC inhibitor | Phase I | Advanced solid tumors or relapsed or refractory non-Hodgkin’s lymphoma |
| NCT00071799 | Azacitidine | None | Demethylation | Phase III | High-risk myelodysplastic syndromes comparing azacitidine versus conventional care |
| NCT00007345 | Romidepsin | None | HDAC inhibitor | Phase II | Cutaneous T cell lymphoma and peripheral T cell lymphoma |
Abbreviation: HDAC, histone deacetylase.
DNA Methyltransferase Inhibitors
DNA demethylating drugs are modified forms of cytidine and work by incorporating into replicating DNA and covalently binding to the catalytic sites of all three biologically active DNMTs. The DNA methyltransferase inhibitors (DNMTis) irreversibly inhibit the enzymatic activities of DNMTs (42–44) and trigger their proteasomal degradation (13, 43, 44). Three current cytidine analogues are clinically available and/or in clinical trials for various cancer types. 5AC (Vidaza® and DAC (Dacogen® or decitabine) have been in clinical trials for many years (7, 13, 40–45). A newer compound, SGI-110, which combines 5-aza-2′-deoxycytidine with guanosine in a single molecule, essentially functions as a prodrug for DAC, providing a longer half-life and resistance to degradation by cytidine deaminase (46). SGI-110 shows great promise for both MDS and AML and is currently in phase III trials for these diseases (ClinicalTrials.gov NCT02348489). All of the above inhibitors can reactivate, in vitro and in vivo, the expression of silenced cancer genes with promoter, CpG-island hypermethylation (4, 7, 8, 13, 42, 46).
The above DNMTis, synthesized as potential cytotoxic agents in the late 1960s, initially proved so toxic that their therapeutic efficacy could not be assessed (4, 7, 13, 42, 45). Reduced doses in the 1990s proved efficacious for treating MDS, leading to FDA approval for this purpose (18, 47). Our group has stressed that these low doses might behave as true epigenetic therapy by reprogramming cancer cells (17). Low, nanomolar doses of 5AC and DAC can reduce long-term clonogenic or tumorigenic properties of hematopoietic and solid tumor cells with virtually all the key signaling pathways impacted that drive cancer initiation and progression (17).
Histone Deacetylase Inhibitors
The use of histone deacetylase inhibitors (HDACis) in clinical trials has been well reviewed (4, 13, 42, 48), and suberoylanilidehydroxamic acid (SAHA; vorinostat) and depsipeptide (romidepsin) are FDA approved for cutaneous T cell lymphomas (4, 13, 42, 48, 49). HDACis, as noted above, alter the balance between acetylation and deacetylation of histone lysines, and the latter is a key constituent of abnormal gene repression in cancer (4, 13, 42, 49). Relieving this repression is one proposed mechanism for efficacy of HDACis for cancer treatment. However, targeting HDACs is at least as complex as targeting DNMTs.
First, there are multiple families of HDACs, which target not only histones but also other nuclear proteins, such as the tumor suppressor p53 and even non-nuclear proteins (4, 13, 42, 48, 49). There are two general types of HDACis: broad-acting pan-inhibitors and inhibitors that target specific classes of HDACs. For example, vorinostat is a pan-inhibitor whereas romidepsin is a class I–specific drug, but all HDACis lack complete specificity (48, 49). Those with the best activity against the class I HDACs 1–3, those residing in the nucleus and most involved in direct gene repression (48, 49), can inhibit other non-nuclear HDACs less likely involved in epigenetic regulation.
Second, the multiple mechanisms of action of HDACis can be immensely dose dependent and are potentially more cytotoxic than epigenetic. Preclinically, HDACis at high doses induce DNA damage such as double-strand DNA breaks, resulting in early cell cycle arrest and death (4, 7, 48–50) and precluding cellular reprogramming. This could explain why HDACis alone have not generally proven effective clinically, especially for common human cancers, and could account for their toxicities in patients.
CLINICAL TRIALS
Save for the previously outlined FDA-approved uses of DNMTis and HDACis as single agents, their most promising use, especially for solid tumors, may be in combination with each other and/or other drugs. Increasingly these combinations are being derived from ongoing preclinical work aimed at understanding their therapeutically relevant doses and mechanisms of efficacy as outlined below.
Combining DNMTis and HDACis
HDACis given after low doses of 5AC or DAC can augment the latter’s effect of re-expressing genes with cancer-specific DNA hypermethylation of promoter CpG islands, although HDACis alone are not effective in this regard (51–53). Multiple clinical trials are testing this combination, most in hematopoietic settings, with some claiming increased efficacy for MDS and/or AML and others not (54–58). Reasons for these differences are not yet apparent.
A particularly intriguing result is seen for advanced non–small cell lung cancer (NSCLC), the biggest cancer killer across the world. Among 65 patients with treatment-resistant, metastatic NSCLC, low combined doses of 5AC and the HDACi entinostat induced remarkably deep and durable responses lasting 3 to >4 years (59). This number represents only 3% of the group, but if this response could be predicted, many patients with advanced NSCLC would benefit. It is essential to obtain pre- and post-therapy biopsies in all future trials of epigenetic therapy to establish biomarkers for such personalization.
Combining DNMTis and HDACis with Other Therapies
A rapidly emerging theme is to combine epigenetic therapies with other cancer treatments (Table 1). An underlying theme is illustrated by data linking chemoresistance to epigenetic events in cancer stem-like cells (60). The studies implicated an HDM, JARID1, which decreases the active transcription mark H3K4me3, and low doses of HDACis reversed the stem cell–like properties and chemoresistance (60). A close family member, JARID2, may also drive stem-like human melanoma cells (61). Low doses of DNMTis can likewise inhibit cancer stem cell properties, enhance apoptosis, and block entry into the cell cycle (17). These examples illustrate the potential for broad reprogramming of cancer cells with epigenetic therapies to prime for subsequent therapies.
A number of recent clinical trials suggest utility for the above paradigm. The HDACi vorinostat combined with carboplatin and paclitaxel reportedly improves response rates and progression-free and overall survival in patients with metastatic NSCLC (62). Durable responses and disease stability occurred in almost half of the patients in a small phase I/II trial of 5AC plus carboplatin for advanced ovarian cancer (63). DAC and carboplatin may increase survival in a similar patient population, and the combination is being tested in expanded trials. As mentioned, 25% of treated patients with metastatic NSCLC achieved stable disease or responses to several chemotherapy agents subsequent to treatment with 5AC plus the HDACi entinostat (59). Clinical trials are ongoing to test this sensitization paradigm in NSCLC, as well as a study of SGI-110 to restore sensitivity to irinotecan in patients with colorectal cancer who had failed this latter drug. For multiple myeloma, the FDA recently approved the HDACi panobinostat (Farydak®) in combination with a proteasomal inhibitor, bortezomib, and an immunomodulatory drug on the basis of progression-free survival improvement in a phase III clinical trial (64). Phase II clinical trials showed positive efficacy when entinostat was combined with the aromatase inhibitor exemestane for treatment of resistant breast cancer (65), and a larger phase III study is under way.
Combining Epigenetic Therapy with Immunotherapy
An exciting recent development is the possibility that epigenetic therapy may sensitize patients (66, 67) to reversal of immune tolerance (68–70). This tolerance is mediated by chronic interaction between defined ligands on tumor cells and receptors on host immune cells, which renders T cells immunologically inert (69–72). These findings have energized the concept of cancer immunotherapy (68–74). Clinical trials using antibodies to CTLA-4, one key receptor for tolerance on T cells, show extraordinarily durable responses in patients with advanced melanoma (72). Targeting of this molecule is being attempted in clinical trials for lung and prostate cancer (ClinicalTrials.gov NCT00527735, NCT01524991, NCT00323882). Also, targeting the host immune cell receptor, PD-1, and its ligand on tumor cells, PD-L1, is proving immensely promising. Anti-PD-1 therapy is inducing remarkably durable responses in metastatic cancers and especially melanoma (70, 71, 73). Expression of PD-L1 on tumor cells appears to offer a positive biomarker for predicting better responses to anti-PD-1 in patients with NSCLC (70, 74).
Clinical and preclinical data are suggesting how epigenetic therapy might improve even further the efficacy of immune checkpoint therapy (67, 76). In the 65-patient trial for NSCLC introduced above, five patients who progressed after epigenetic therapy were subsequently enrolled in trials of anti-PD-1 and PD-L1 that included patients with advanced NSCLC (70, 71). As compared to 20% of the patients who responded to (or had progression-free survival past 24 weeks on) the latter therapy alone, all five patients who had prior epigenetic therapy passed this time point without disease progression—and three manifested high-grade responses to the immunotherapy as defined by Response Evaluation Criteria In Solid Tumors (RECIST). These responses have persisted for more than two years (66). A much larger clinical trial is now ongoing to see if these results are indeed directly attributable to combining the immune checkpoint therapy with the epigenetic therapy.
Results of preclinical studies concurrent with this clinical trial suggest great promise for understanding the mechanisms involved in combined therapy and for designing biomarker approaches to personalize it for NSCLC and other solid tumor types. It has been known for some time that HDACis and particularly DNMTis can induce expression of individual components of immune attraction of cancer cells (66, 67, 76–78). These effects include upregulation of antigens that are generally expressed only in early embryonic cells and are epigenetically silenced in normal mature cells (67, 76–78). The drugs have also been recognized by others to increase expression of other components of immune attraction generally involving interferon-mediated events (77).
Our group has recognized that low doses of 5AC and DAC induce a remarkably coordinated series of immune responses in NSCLC and other solid tumor type cells (66, 67, 76), not only for the above antigens but also for their processing by major histocompatibility complex class I proteins and for an entire series of interferon-driven immune-related signaling events, which we have termed AZA-induced immune (AIM) genes (76). Upregulation of PD-L1 can accompany these events (66, 76). One key factor for inducing these events is upregulation of a viral defense mechanism triggered by cell cytosolic recognition of double-stranded RNA (67, 79). In part, this may be driven by 5AC and DAC increasing the expression of transcripts emanating from endogenous retroviral genes, harbored in our genomes and epigenetically silenced in mature but not early embryonic cells (67). We hypothesize that these drug-induced responses constitute a potential reversion of tumor cell “immune evasion” and could then provide a mechanism for improving immune checkpoint therapies. In this regard, our viral defense gene signature separates primary samples of breast, colon, NSCLC, ovarian tumors, and melanomas in The Cancer Genome Atlas into low versus high groupings—and the high signature in tumors predicts for beneficial effects of anti-CTLA4 in a small series of patients with advanced melanoma (67). In this setting, the signature correlates well with high tumor mutational burden, which is proving strongly predictive for beneficial effects of immune checkpoint therapy (80, 81).
All of these insights about priming immune checkpoint therapy with epigenetic therapy await verification of efficacy in clinical trials of combinations of these agents. The individual effects of DNMTis and HDACis must be examined not only for tumor cells but for host immune cells. Finally, correlative studies in biopsies are essential to validating the proposed biomarker strategies.
EPIGENETIC THERAPIES WITH NEW AGENTS
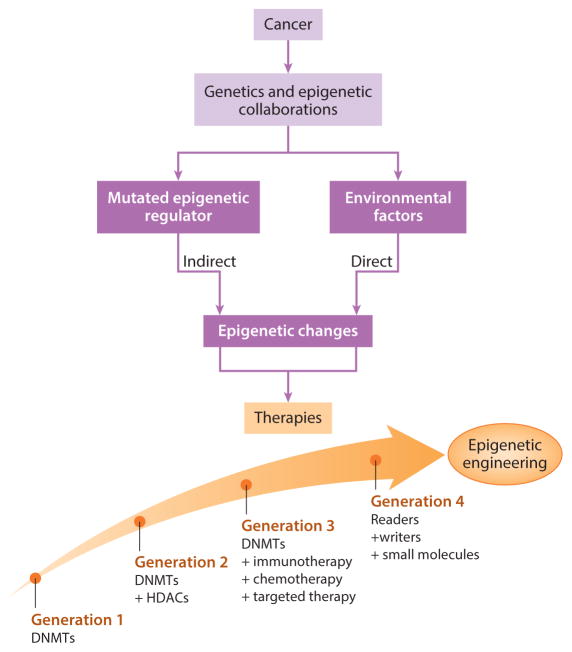
Many new small molecules for epigenetic therapy, some already in clinical trials (82, 83), are becoming available, and these target many of the proteins for control of the epigenome shown in Figures 1 and 2. Especially interesting are those targeting mutated epigenetic regulating genes in cancer. A key example is for isocitrate dehydrogenase genes IDH1 and IDH2, frequently mutated in AML, glioblastomas, and several other tumor types (84–86). These mutations block production of alpha-ketoglutarate, a key cofactor for multiple proteins regulating the epigenome, including the TET proteins and several HDMs (11, 85, 87). One result is a large increase in DNA methylation in the above tumor types in a phenotype known as CIMP (CpG island methylation phenotype) (11, 86, 88). Small molecules targeting these IDH mutations are now in clinical trials with early promise for AML. Another tumor type, carotid body tumor, has mutations in the succinate dehydrogenase enzyme complex, which result in hypermethylation of the promoter for O6-methylguanine-DNA methyltransferase, and epigenetic therapy with DNMTi in this disease is also promising (89). Another mutated regulator is EZH2, for which inhibitors are being introduced in clinical trials, predominantly for hematopoietic tumors (90, 91). Mixed-lineage leukemia translocations in AML, acting through the protein DOT1 (disruptor of telomeric silencing–1), induce inappropriate gene activation, and drugs blocking DOT1 are in clinical trials for AML (92). BRD4 (bromodomain-containing protein–4) is a key reader of lysine acetylation, and when it is overexpressed, abnormal activation occurs for c-Myc oncogene targets (82, 83). Several types of small molecules blocking BRD4 (82, 83) are now being introduced in phase I clinical trials, again predominantly for hematopoietic malignancies. More and more such novel agents are rapidly forthcoming and will enter clinical trials as promising preclinical data emerge (Figure 4).
Figure 4.
Treatment approaches based on juxtaposing mutational events and epigenetic alterations. Cancers can have direct effects on the epigenome, resulting from chronic inflammation, viral infections, or microbiome changes, or indirect effects due to mutations in epigenetic driver genes. Epigenetic therapy regimens are evolving (large arrow with circles) for generations of evolving therapies.
CONCLUSIONS
Only in the past decade have we begun to fully recognize the extent to which cancer cells use epigenetics to abnormally reprogram cells. This new understanding includes the appreciation of the large numbers of mutations in cancer affecting genes that regulate epigenetic control. These coalitions of epigenetic and genetic abnormalities are guiding our thinking about epigenetic therapy strategies for cancer (Figure 4). The advancement of genome sequencing in the clinical arena would enable further advances toward personalized medical care (93). Already there are signs that this form of treatment, using older agents and the novel small molecules that are being developed, could lead to a true change in cancer management. The potential to reverse cancer-associated epigenetic abnormalities to reprogram neoplastic cells is a growing reality, and the next few years may see increasing excitement if signs of clinical efficacy continue to emerge.
Acknowledgments
Portions of N.A.’s work cited in this invited review were supported by K23 CA127141 and R01 CA185357, both from the National Institutes of Health and National Cancer Institute, and by the American College of Surgeons. S.B.B.’s work is supported by grants ES011858 from the National Institutes of Environmental Health Sciences, W81XWH-14-1-0385 Teal from the US Department of Defense, SU2C-AACR-CT0109 from Stand Up 2 Cancer—American Association for Cancer Research, the Miriam & Sheldon Adelson Medical Research Foundation, the Hodson Trust, and the Samuel Waxman Cancer Research Foundation. The authors thank Michael Ruiz and Kathy Bender for manuscript preparation.
Footnotes
DISCLOSURE STATEMENT
N.A. receives grant funding from Astex, Inc., and holds a patent on a methylation marker for pancreas cancer detection. S.B.B. consults for MDxHealth. MSP is licensed to MDxHealth in agreement with Johns Hopkins University (JHU), and S.B.B. and JHU are entitled to royalty shares received from sales.
LITERATURE CITED
- 1.Masters GA, Krilov L, Bailey HH, et al. Clinical cancer advances 2015: annual report on progress against cancer from the American Society of Clinical Oncology. J Clin Oncol. 2015;33:786–809. doi: 10.1200/JCO.2014.59.9746. [DOI] [PubMed] [Google Scholar]
- 2.Sun Y, Ma L. The emerging molecular machinery and therapeutic targets of metastasis. Trends Pharmacol Sci. 2015;36:349–59. doi: 10.1016/j.tips.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mwenifumbo JC, Marra MA. Cancer genome-sequencing study design. Nat Rev Genet. 2013;14:321–32. doi: 10.1038/nrg3445. [DOI] [PubMed] [Google Scholar]
- 4.Allis C, Jenuwein T, Reinberg D, Caparros M. Epigenetics. 2 Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2015. [Google Scholar]
- 5.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–83. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28:812–28. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22:50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahuja N, Easwaran H, Baylin SB. Harnessing the potential of epigenetic therapy to target solid tumors. J Clin Investig. 2014;124:56–63. doi: 10.1172/JCI69736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 16.Silverman LR, Demakos EP, Peterson BL, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–40. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 17.Tsai HC, Li H, Van Neste L, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–46. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminskas E, Farrell AT, Wang YC, et al. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10:176–82. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 19.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2015;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–43. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, McEachron TA, Schwartzentruber J, Wu G. Histone H3 mutations in pediatric brain tumors. Cold Spring Harb Perspect Biol. 2014;6:a018689. doi: 10.1101/cshperspect.a018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones S, Wang TL, Shih IeM, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bestor TH, Edwards JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development. PNAS. 2015;112:6796–99. doi: 10.1073/pnas.1415301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millan MJ. An epigenetic framework for neurodevelopmental disorders: from pathogenesis to potential therapy. Neuropharmacology. 2013;68:2–82. doi: 10.1016/j.neuropharm.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Chendhore SV, Sleiman S, Coppola G. Epigenetics of Alzheimer’s disease and frontotemporal dementia. Neurotherapeutics. 2013;10:709–21. doi: 10.1007/s13311-013-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–55. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 29.Spruijt CG, Gnerlich F, Smits AH, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–59. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–56. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 32.Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–32. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14:853–64. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 35.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 36.Hock H. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev. 2012;26:751–55. doi: 10.1101/gad.191163.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–39. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 38.Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Ruscio A, Ebralidze AK, Benoukraf T, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–76. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Issa JP. DNA methylation in the treatment of hematologic malignancies. Clin Adv Hematol Oncol. 2005;3:684–86. [PubMed] [Google Scholar]
- 41.Silverman LR, Mufti GJ. Methylation inhibitor therapy in the treatment of myelodysplastic syndrome. Nat Clin Pract Oncol. 2005;2(Suppl 1):S12–23. doi: 10.1038/ncponc0347. [DOI] [PubMed] [Google Scholar]
- 42.Azad N, Zahnow CA, Rudin CM, Baylin SB. The future of epigenetic therapy in solid tumours—lessons from the past. Nat Rev Clin Oncol. 2013;10:256–66. doi: 10.1038/nrclinonc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghoshal K, Datta J, Majumder S, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–41. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–78. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silverman LR, Fenaux P, Mufti GJ, et al. Continued azacitidine therapy beyond time of first response improves quality of response in patients with higher-risk myelodysplastic syndromes. Cancer. 2011;117:2697–702. doi: 10.1002/cncr.25774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuang JC, Warner SL, Vollmer D, et al. S110, a 5-Aza-2′-deoxycytidine-containing dinucleotide, is an effective DNA methylation inhibitor in vivo and can reduce tumor growth. Mol Cancer Ther. 2010;9:1443–50. doi: 10.1158/1535-7163.MCT-09-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lubbert M. DNA methylation inhibitors in the treatment of leukemias, myelodysplastic syndromes and hemoglobinopathies: clinical results and possible mechanisms of action. Curr Top Microbiol Immunol. 2000;249:135–64. doi: 10.1007/978-3-642-59696-4_9. [DOI] [PubMed] [Google Scholar]
- 48.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bose P, Dai Y, Grant S. Histone deacetylase inhibitor (HDACI) mechanisms of action: emerging insights. Pharmacol Ther. 2014;143:323–36. doi: 10.1016/j.pharmthera.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robert C, Rassool FV. HDAC inhibitors: roles of DNA damage and repair. Adv Cancer Res. 2012;116:87–129. doi: 10.1016/B978-0-12-394387-3.00003-3. [DOI] [PubMed] [Google Scholar]
- 51.Cai Y, Geutjes EJ, de Lint K, et al. The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene. 2013;33:2157–68. doi: 10.1038/onc.2013.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cameron EE, Bachman KE, Myohanen S, et al. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki H, Watkins DN, Jair KW, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 54.Garcia-Manero G. Demethylating agents in myeloid malignancies. Curr Opin Oncol. 2008;20:705–10. doi: 10.1097/CCO.0b013e328313699c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gore SD, Jiemjit A, Silverman LB, et al. Combined methyltransferase/histone deacetylase inhibition with 5-azacitidine and MS-275 in patients with MDS, CMMoL and AML: clinical response, histone acetylation and DNA damage. Blood (ASH Annu Meet Abstr) 2006;108:517. [Google Scholar]
- 56.Maslak P, Chanel S, Camacho LH, et al. Pilot study of combination transcriptional modulation therapy with sodium phenylbutyrate and 5-azacytidine in patients with acute myeloid leukemia or myelodys-plastic syndrome. Leukemia. 2006;20:212–17. doi: 10.1038/sj.leu.2404050. [DOI] [PubMed] [Google Scholar]
- 57.Voso MT, Santini V, Finelli C, et al. Valproic acid at therapeutic plasma levels may increase 5-azacytidine efficacy in higher risk myelodysplastic syndromes. Clin Cancer Res. 2009;15:5002–7. doi: 10.1158/1078-0432.CCR-09-0494. [DOI] [PubMed] [Google Scholar]
- 58.Prebet T, Sun Z, Figueroa ME, et al. Prolonged administration of azacitidine with or without entinostat for myelodysplastic syndrome and acute myeloid leukemia with myelodysplasia-related changes: results of the US leukemia intergroup trial E1905. J Clin Oncol. 2014;32:1242–48. doi: 10.1200/JCO.2013.50.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roesch A, Fukunaga-Kalabis M, Schmidt EC, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–94. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramalingam SS, Maitland ML, Frankel P, et al. Carboplatin and paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:56–62. doi: 10.1200/JCO.2009.24.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu S, Hu W, Iyer R, et al. Phase 1b-2a study to reverse platinum resistance through use of a hypomethylating agent, azacitidine, in patients with platinum-resistant or platinum-refractory epithelial ovarian cancer. Cancer. 2011;117:1661–69. doi: 10.1002/cncr.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.San-Miguel JF, Hungria VT, Yoon SS, et al. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double-blind phase 3 trial. Lancet Oncol. 2014;15:1195–206. doi: 10.1016/S1470-2045(14)70440-1. [DOI] [PubMed] [Google Scholar]
- 65.Yardley DA, Ismail-Khan RR, Melichar B, et al. Randomized phase II, double-blind, placebo-controlled study of exemestane with or without entinostat in postmenopausal women with locally recurrent or metastatic estrogen receptor-positive breast cancer progressing on treatment with a nonsteroidal aromatase inhibitor. J Clin Oncol. 2013;31:2128–35. doi: 10.1200/JCO.2012.43.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wrangle J, Wang W, Koch A, et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget. 2013;4:2067–79. doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chiappinelli KB, Strissel PL, Desrichard A, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–86. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med. 1995;182:459–65. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hodi FS, O’Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–28. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 75.Deleted in proof
- 76.Li H, Chiappinelli KB, Guzzetta AA, et al. Immune regulation by low doses of the DNA methyl-transferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5:587–98. doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karpf AR, Lasek AW, Ririe TO, et al. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine. Mol Pharmacol. 2004;65:18–27. doi: 10.1124/mol.65.1.18. [DOI] [PubMed] [Google Scholar]
- 78.Khan AN, Tomasi TB. Histone deacetylase regulation of immune gene expression in tumor cells. Immunol Res. 2008;40:164–78. doi: 10.1007/s12026-007-0085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roulois D, Helen Loo Yau RS, Wang Y, et al. Low dose DNA-demethylating agents target colorectal cancer-initiating cells by activation of MDA5/MAVS/IRF7 pathway. Cell. 2015;162:961–73. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–28. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 83.Tanaka M, Roberts JM, Qi J, Bradner JE. Inhibitors of emerging epigenetic targets for cancer therapy: a patent review (2010–2014) Pharm Pat Anal. 2015;4:261–84. doi: 10.4155/ppa.15.16. [DOI] [PubMed] [Google Scholar]
- 84.Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–78. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Venneti S, Felicella MM, Coyne T, et al. Histone 3 lysine 9 trimethylation is differentially associated with isocitrate dehydrogenase mutations in oligodendrogliomas and high-grade astrocytomas. J Neuropathol Exp Neurol. 2013;72:298–306. doi: 10.1097/NEN.0b013e3182898113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yeng FH, Molly NH, Louis JM. Oxygen concentration controls epigenetic effects in models of familial paraganglioma. PLoS ONE. 2015;10:e0127471. doi: 10.1371/journal.pone.0127471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Knutson SK, Wigle TJ, Warholic NM, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–96. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 91.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 92.Daigle SR, Olhava EJ, Therkelsen CA, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caskey CT. Using genetic diagnosis to determine individual therapeutic utility. Annu Rev Med. 2010;61:1–15. doi: 10.1146/annurev-med-011209-132719. [DOI] [PubMed] [Google Scholar]